Abstract
The use of small-molecule fluorophores to label proteins with minimal perturbation in response to an external stimulus is a powerful tool to probe chemical and biochemical environments. Herein we describe first the use of a coumarin modified triazabutadiene that can deliver aryl diazonium ions to fluorescently label proteins via tyrosine selective modification. The labeling can be triggered by low pH induced liberation of the diazonium species, making the fluorophore specifically useful in labeling biochemical surroundings such as those found within the late endosome. Additionally, we show that a variety of coumarin triazabutadienes may also be prone to releasing their diazonium cargo via irradiation with UV light.
Keywords: Fluorescent probes, Protein modifications, Electrophilic substitution, Protecting groups, Substituent effects
Graphical Abstract
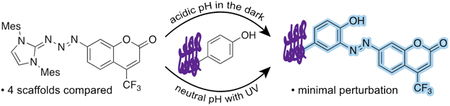
Releasable pro-fluorophores lighten the area while not stepping on toes.
Fluorescence imaging is a crucial tool for measuring complex systems in biology, from surfaces, to cells, to whole organisms. While fluorescent proteins are boon to this area, the ability to covalently modify proteins with small molecules remains essential for many modern imaging experiments.[1] At the outset, a goal of this work was to develop a fluorophore that would have a minimal impact on both the size and structure of the target protein. The attachment of the fluorophore using an aryl diazonium ion to tyrosine (Scheme 1a) was seen as potentially useful because: 1) the amino acid side chain would become part of the fluorophore, and, 2) the resulting modification would be charge neutral. In assessing these criteria, coumarin-based fluorophores garnered attention due to their small size, and proven utility for imaging.
Scheme 1.
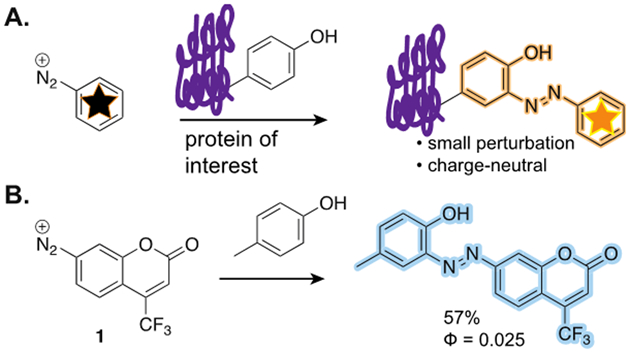
A. Aryl diazonium ions can react with proteins and generate a fluorescent product that is minimally perturbing. B. Coumarin diazonium 1 reacted with p-cresol to form a fluorescent azobenzene.
The modification of tyrosine with aryl diazonium ions via electrophilic aromatic substitution is one of the oldest selective bioconjugation strategies, and one that is still used in modern applications.[2, 3] An early concern was that azobenzenes are often found in fluorescent quencher motifs,[4] but upon investigation there are examples of azobenzene-coumarin compounds that have fluorescent properties.[5, 6] One example in particular that offered encouragement was a reported coumarin-derived aryl diazonium ion that acted as a covalent inhibitor of acetyl choline esterase which became fluorescent upon modification of tyrosine in the enzymatic pocket.[5] Moreover, there are marked difference observed in fluorescence between solution-phase and solid immobilized azo dyes.[7] While promising for the desired application, challenges of storage and delivery exist with aryl diazonium ions, often requiring in situ generation. Our recent work masking aryl diazonium ions as triazabutadienes led us to investigate their usage as a convenient source of this fluorophore-generating moiety.[8] Herein, we report the synthesis and utility of several coumarin triazabutadienes that can be environmentally, or photochemically triggered to release aryl diazonium ions to fluorescently label proteins.
Our previous work with triazabutadienes, a functional group that can release aryl diazonium ions in protic environments, led us to hypothesize that a fluorophorogenic diazonium ion could be protected and liberated as desired. Prior to the synthesis of the triazabutadiene coumarin compound, we sought to test the reactivity of the diazonium ion that would be released. Diazonium salt, 1 (Scheme 1b), was synthetized from its respective amino coumarin precursor following standard diazotization conditions. We confirmed fluorescence of the adduct by making the p-cresol derivative (shown Scheme 1b). While the quantum yield was low (Φ = 0.025), we moved forward to evaluate if there was sufficient fluorescence to be of use in labeling proteins. Using bovine serum albumin (BSA) as a model protein, we examined the pH dependence of the reaction between freshly made 1 and the protein (Scheme 1a). Consistent with previous reports on diazonium ion chemistry on proteins, we observed increased labeling as pH increased.[3] Optimal labeling occurred at pH 8, but even at pH 5, substantial labeling was present (Figure S1 and S2).
Confident that a coumarin-derived fluorophore could be conjugated onto proteins and visualized, we synthesized a bis-mesityl variant of the triazabutadiene scaffold (Figure 1a). We hypothesized that it would have a good combination of synthetic ease, reactivity, and long-term stability. Triazabutadiene 2 was synthesized from the corresponding azido coumarin (3), and N-heterocyclic carbene precursor 4 in good yield (Figure 1a). To confirm that 2 could release 1 and label proteins, BSA was treated with 2 at various pH. We observed substantial labeling only occurring at pH lower than 6, with intensity increasing as pH decreases (Figure 1b). These findings are consistent with the hypothesis that 1 is released only under acidic conditions. Rate data from NMR studies suggested that 2 is more stable to presence of acid than the simple benzene-substituted triazabutadiene (Figure S3). While the resulting electron deficient aryl diazonium ion is not fluorescent, the triazabutadiene does have mild fluorescence, with an observed quantum yield of 0.004. We attribute this fluorescence to the electron-donating nature of the carbene-derived half. These observations are consistent with this class of fluorophores where electron-donation at the 7 position is essential for robust fluorescence. The labeling of proteins with 2 is not fluorogenic, but we were encouraged by the level of labeling and tested general applicability of the probe. As a result of the mild fluorescence of 2, extra caution was taken to ensure that labeling was the result of diazonium ion release, and not simply non-specific binding.
Figure 1.
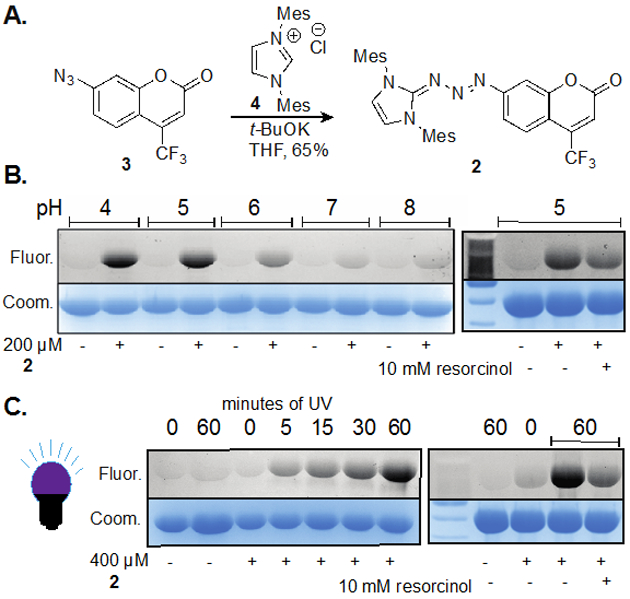
A. Coumarin triazabutadiene 2 was synthesized from the corresponding aryl azide, 3. B. and C. are scans from SDS-PAGE gels. Fluor. = fluorescence scan; Coom. = coomassie blue staining. B. Bovine serum albumin (BSA) labeled with 2 (0.2 mM) in 100 mM PBS buffer with increasing pH (4, 5, 6, 7 and 8) for 12 hours. At far right, resorcinol is added to trap diazonium ion intermediate and reduces fluorescence signal. The samples were then run on a gel and analyzed. C. Samples of BSA in 100 mM PBS (pH 7) are irradiated with 350 nm light for varying times. At far right, resorcinol is added to trap diazonium ion intermediate and reduces fluorescence signal. The samples were then run on a gel and analyzed.
Recognizing that low pH is not appropriate for all protein labeling studies and lacks applicability in typical biological environments, we employed an alternative approach for release of the coumarin aryl diazonium at neutral pH. To accomplish this, we explored the use of photochemistry of the triazabutadiene scaffold to release the diazonium ion. This had previously been reported by both our group and others to facilitate release of aryl diazonium for protein labeling via increasing the basicity of the compound.[9] We were relieved to find that the presence of a fluorophore in conjugation with the triazabutadiene did not prevent the photochemistry. Once more we utilized BSA as a model protein to show labeling could be attained at pH 7 by irradiation at 350 nm (Figure 1c). Labeling was evident following as little as 5 minutes of irradiation. Not surprisingly, we also showed that the presence of excess resorcinol attenuated labeling of BSA. These data are consistent with our understanding that this chemistry is primarily operating through diazonium ion chemistry and not other photochemically generated species (such as nitrenes or aryl radicals). This result is consistent with previous work on using light to release aryl diazonium ions from triazabutadienes.[9]
To ensure fluorescence of the BSA could be attributed to covalent modification several experiments were performed. First, the labeling in the presence or absence of urea showed similar levels of labeling (Figure S4). Additionally, mass spectral data supported covalent modification, and was consistent with the extent of BSA modification reported by others.[3] BSA was treated with 2 and irradiated for 25 min. After prolonged incubation, the sample was analyzed by MALDI mass spectrometry and overall mass shifts indicated that just under two modifications per BSA on average were made in a reagent and light-dependent manner (Figure S5).
To move beyond BSA and illustrate general utility we analyzed the labeling of egg yolk proteins. In several experiments, analogous to those run with BSA, egg yolk proteins were labeled by 2 in a pH-dependent manner (Figure S6). Furthermore, we treated proteins from egg yolk extracts with 2 at pH 8 in the presence or absence of light (Figure S7). The fluorescence of several proteins in the irradiated samples confirmed that 2 could label a variety of proteins in a light-dependent manner. The lack of fluorescence in several protein bands suggests that protein labeling by 2 is contained to proteins with accessible tyrosine. This is of consequence for future applications, where the environment of tyrosine will dictate the efficiency and extent of labeling. Also of note is that there is a wide range of non-specific binding observed for 2, from proteins that have no observable background binding, to those that appear equivalent to covalent binding.
Prompted to further broaden applicability, we synthesized, and tested a series of coumarin triazabutadienes (Figure 2a). We had previously established that structures lacking mesityl substituents were more reactive in the presence of low pH than those containing them, and we hypothesized that removal of the trifluoromethyl would also increase release rates.[10] Each of the four triazabutadienes was evaluated for labeling via liberation by both pH and irradiation at 350 nm. Interestingly, treatment of BSA by each triazabutadiene at pH 5 provided similar levels of fluorescence for all four compounds (Figure 2b). While it is possible that we would have seen a difference in labeling at a different pH, a noteworthy trend emerged upon diazonium ion liberation via irradiation. The bis-methyl substituted compounds, 5 and 7, showed nearly zero response to the light, but the bis-mesityl compounds both showed significant amounts of labeling in a light-dependent manner (Figure 2c).[11] The UV/vis absorption peaks for 2 and 6 are broader than those for 5 and 7, but none of the four compounds absorb well at 350 nm. As such, we hypothesize that the mesityl portions of the molecule are potentially forcing 2 and 6 to be ideally positioned to absorb the energy and isomerize. The underlying reason for this phenomenon is unknown at this time, but warrants further investigation. Given the strong increase in fluorescence observed when using 6 under photochemical conditions, we challenged the probe with a complex Escherichia coli (E. coli) lysate.[12] To our delight we observed robust labeling of the sample with minimal background fluorescence (Figure 2d).
Figure 2.
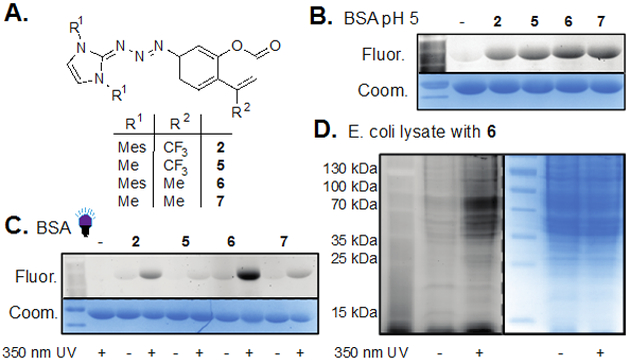
A. A series of triazabutadienes was synthesized and evaluated for their ability to label proteins. B., C. and D. are scans from SDS-PAGE gels. Fluor. = fluorescence scan; Coom. = coomassie blue staining. B. Minimal difference was observed when bovine serum albumin (BSA) was treated with compounds 2, 5-7 at pH 5 for 2 hours. The samples were quenched with resorcinol and then run on a gel and analyzed. C. BSA was incubated with 2, 5-7 in pH 7 PBS buffer and UV irradiation for either 0 or 15 minutes by incubation for an additional 45 minutes in the absence of light. The samples were then run on a gel and analyzed. D. A sample of E. coli lysate was incubated with 6 in Tris-HCl buffer pH 8.8 for 40 minutes in the presence or absence of UV light followed by incubation for an additional 2 hours in the absence of light. The samples were then run on a gel and analyzed.
In conclusion, we have reported the synthesis and general utility of new class of fluorophore-embedded triazabutadienes as delivery tool for aryl diazonium salts to fluorescently label proteins of interest. In the process, we have shown multiple strategies for eliciting environmentally triggered modification of proteins, including the use of low pH and irradiation at 350 nm, which afford these compounds the versatility to be used in a wide range of pH and biologically relevant environments. Taken together, our series of masked coumarin diazonium ions provide bench stable, environmentally triggered fluorophores that pose potential for imaging applications. A current limitation of these molecules is that their absorbance and emission spectra will not allow for imaging of tissue; but more broadly, this strategy should be extendable to other classes of fluorophores.
Supplementary Material
Acknowledgements
The authors would like to thank Dr. Chad Park, for assistance with instrumentation and helpful conversations, and Dr. Michael Marty for assistance with supplies. Research reported in this publication was supported by the Office of the Director, National Institutes of Health of the National Institutes of Health under award number S10OD013237, and to JCJ by the National Science Foundation under award number CHE-1552568.
Footnotes
Experimental Section
Experimental details and syntheses of all compounds discussed can be found in the supporting information files.
Supporting information for this article is given via a link at the end of the document.
References
- [1].Lavis LD, Biochemistry 2017, 56, 5165–5170. [DOI] [PubMed] [Google Scholar]
- [2] a).Pauly H, Hoppe-Seyler’s Z. Physiol. Chem. 1915, 94, 284–290; [Google Scholar]; b) Higgins HG, Fraser D, Aust. J. Sci. Res., Ser. A 1952, 5, 736–753; [Google Scholar]; c) Lillie RD, Cason JC, Grecohenson JP, J. Histochem. Cytochem. 1961, 9, 11; [DOI] [PubMed] [Google Scholar]; d) Riordan JF, Vallee BL, in Methods in Enzymology: Enzyme Structure, Part B, Vol. 25 (Eds.: Hirs CHW, Timasheff SN), Academic Press, 1972, pp. 521–531; [DOI] [PubMed] [Google Scholar]; e) Pieroni O, Fissi A, Houben JL, Macromol. Chem. Phys. 1975, 176, 3201–3209; [Google Scholar]; f) Hooker JM, Kovacs EW, Francis MB, J. Am. Chem. Soc. 2004, 126, 3718–3719; [DOI] [PubMed] [Google Scholar]; g) Schlick TL, Ding Z, Kovacs EW, Francis MB, J. Am. Chem. Soc. 2005, 127, 3718–3723; [DOI] [PubMed] [Google Scholar]; h) Jones MW, Mantovani G, Blindauer CA, Ryan SM, Wang X, Brayden DJ, Haddleton DM, J. Am. Chem. Soc. 2012, 134, 7406–7413. [DOI] [PubMed] [Google Scholar]
- [3].Gavrilyuk J, Ban H, Nagano M, Hakamata W, Barbas CF, Bioconjugate Chem. 2012, 23, 2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Hangauer MJ, Bertozzi CR, Angew. Chem. Int. Ed. Engl. 2008, 47, 2394–2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Schalk I, Ehret-Sabatier L, Le Feuvre Y, Bon S, Massoulie J, Goeldner M, Mol. Pharmacol. 1995, 48, 1063–1067. [PubMed] [Google Scholar]
- [6] a).Wang MX, Meng XM, Zhu MZ, Guo QX, Chinese Chemical Letters 2007, 18, 1403–1406; [Google Scholar]; b) Karami B, Kiani M, Polycyc. Aromat. Comp. 2017, 37, 257–266. [Google Scholar]
- [7].Puchtler H, Sweat F, Gropp S, J. R. Microsc. Soc. 1967, 87, 307–328. [DOI] [PubMed] [Google Scholar]
- [8] a).Kimani FW, Jewett JC, Angew. Chem. Int. Ed. 2015, 54, 4051–4054; [DOI] [PubMed] [Google Scholar]; b) He J, Kimani FW, Jewett JC, Synlett 2017, 28, 1767–1770. [Google Scholar]
- [9] a).He J, Kimani FW, Jewett JC, J. Am. Chem. Soc. 2015, 137, 9764–9767; [DOI] [PubMed] [Google Scholar]; b) Jensen SM, Kimani FW, Jewett JC, Chembiochem 2016, 17, 2216–2219.; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Addy PS, Erickson SB, Italia JS, Chatterjee A, J. Am. Chem. Soc. 2017, 139, 11670–11673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].NMR studies confirmed that 5 does indeed release much faster than 2 in acidic environments, see Figures S3 and S7
- [11].Unlike with our previous work where the isomerization could be quantified by NMR, these compounds were always observed in the more stable conformation
- [12].Quite remarkably, small molecule azo products of 6 had nearly zero quantum yield, but the protein-bound conjugates were clearly fluorescent on SDS-PAGE gels. To narrow down why this was happening a series of experiments were done in parallel with small molecule azo-coumarins and BSA labeled with 6 (Figure S9). Neither small molecule azobenzene, nor BSA-conjugated fluorophore showed marked fluorescence in solution. However, a large fluorescence increase was observed in SDS-gel for only the BSA-bound azo bonds. This fluorescence increase is consistent with previous reports detailing coumarin derivatives. See, Wagner BD Molecules 2009, 14, 210–237.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


