Summary
The intracellular redox environment of Staphylococcus aureus is mainly buffered by bacillithiol (BSH), a low molecular weight thiol. The identity of enzymes responsible for the recycling of oxidized bacillithiol disulfide (BSSB) to the reduced form (BSH) remains elusive. We examined YpdA, a putative bacillithiol reductase, for its role in maintaining intracellular redox homeostasis. The ypdA mutant showed increased levels of BSSB and a lower bacillithiol redox ratio vs. the isogenic parent, indicating a higher level of oxidative stress within the bacterial cytosol. We showed that YpdA consumed NAD(P)H; and YpdA protein levels were augmented in response to stress. Wild type strains overexpressing YpdA showed increased tolerance to oxidants and electrophilic agents. Importantly, YpdA overexpression in the parental strain caused an increase in BSH levels accompanied by a decrease in BSSB concentration in the presence of stress, resulting in an increase in bacillithiol redox ratio vs. the vector control. Additionally, the ypdA mutant exhibited decreased survival in human neutrophils (PMNs) as compared with the parent, while YpdA overexpression protected the resulting strain from oxidative stress in vitro and from killing by human neutrophils ex vivo. Taken together, these data present a new role for YpdA in Staphylococcus aureus physiology and virulence through the bacillithiol system.
Keywords: Staphylococcus aureus, bacillithiol, bacillithiol disulfide reductase, oxidative stress, virulence
Graphical Abstract
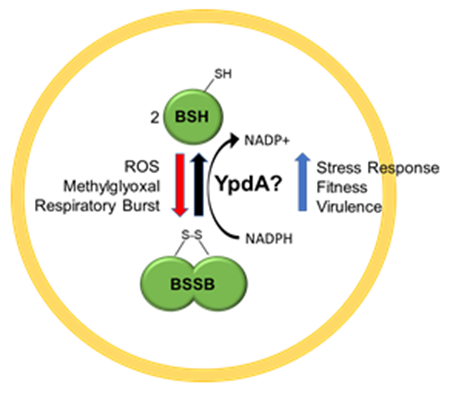
Gram positive bacteria use the low molecular weight thiol bacillithiol to buffer their cytoplasm to a reduced state. While the genes for biosynthesis bacillithiol have been well described, characterization of bacillithiol disulfide reductase responsible for turnover of oxidized bacillithiol disulfide to reduced bacillithiol is still missing. We show that Staphylococcus aureus likely employs YpdA for such role; and the protein has roles in stress response, fitness, and virulence.
Introduction
Staphylococcus aureus (S. aureus) is an opportunistic human pathogen that has become persistent sources of hospital- and community- acquired infections (Chambers and DeLeo, 2009). S. aureus causes a wide range of infections ranging from bacteremia, endocarditis, osteomyelitis, to skin and soft tissue infections (Tong et al., 2015). In addition to acquiring antibiotic resistance, the pathogenic success of S. aureus relies on its diverse arsenal of virulence factors including proteases and toxins, and its ability to adapt to a wide spectrum of environmental and host associated stresses (Archer, 1998; Watkins et al., 2012). During infections, S. aureus initially encounters PMNs capable of generating reactive oxygen species (ROS), including superoxide generated by NADPH oxidases and other ROS products such as hydrogen peroxide (H2O2) produced in secondary reactions (Winterbourn and Kettle, 2013; Spaan et al., 2013). While S. aureus utilizes enzymatic strategies (e.g. superoxide dismutase and catalase) to reduce the damage from host-generated ROS (DeLeo et al., 2009), it also relies on low molecular weight (LMW) thiols to limit the extent of the damage (Pöther et al., 2013; Posada et al., 2014).
Bacterial cells use LMW thiols to maintain cellular redox potential and confer protection against ROS. Glutathione (GSH), the most well studied and wide-spread LMW thiol, is found in eukaryotes and Gram-negative bacteria (Masip et al., 2006). Gram-positive bacteria, however, do not produce GSH and instead utilize alternative LMW thiols for these functions. Actinobacteria produce mycothiol (MSH) (Newton et al., 1996), while Firmicutes such as S. aureus rely on the recently discovered molecule called bacillithiol (BSH) (Newton et al., 2009). Before the discovery of BSH, it was believed that S. aureus used the amino acid cysteine (Cys) and coenzyme-A (CoA-SH) in place of GSH (Chandrangsu et al., 2017). But, BSH has been implicated to be the dominant LMW thiol in Firmicutes because BSH is significantly more reactive under physiological conditions than other LMW thiols (Sharma et al., 2013). BSH, an anomeric glycoside of L-cysteinyl-D-glucosamine with L-malic acid, is produced in a three-step reaction by BshA, BshB, and BshC enzymes in S. aureus (Gaballa et al., 2010). BSH appears to have multiple roles in cellular physiology, and so far has been implicated in Fe-S cluster biogenesis (Rosario-Cruz et al., 2015), the detoxification of reactive oxygen and electrophilic species (Chandrangsu et al., 2014), ameliorating antibiotic damages (Gaballa et al., 2010), and playing a role in virulence (Posada et al., 2014). Additionally, BSH is used in protein S-bacillithiolation as a means of modulating or protecting protein function (e.g. glyceraldehyde-3-phosphate dehydrogenase in metabolism) (Chi et al., 2013; Imber et al., 2018). These modifications are then removed by a class of enzymes called bacilliredoxins (Brx) which act analogously to glutaredoxins (Gaballa et al., 2014).
Under conditions of oxidative stress, BSH is oxidized to form bacillithiol disulfide (BSSB), entailing a disulfide bond between two molecules of BSH. A hallmark of LMW thiol systems is the ability to reduce oxidized molecules, such as BSSB, back to the functional reduced state (Couto et al., 2016). Reduction of LMW thiols is typically carried out by a class of enzymes known as reductases. For GSH, this is accomplished by glutathione reductases (GRs) and by mycothiol reductase (Mtr) for MSH. Organisms that utilize GSH but lack GR display increased susceptibility to oxidative stresses (Carmel-Harel and Storz, 2000). Similarly, overexpression of Mtr has been shown to increase antioxidative capacity (Si et al., 2016). Mutations in GR have been shown to increase susceptibility of pathogenic fungi to macrophage killing and result in decreased virulence in infection models (Tillmann et al., 2015). Despite the clear importance of reductases as part of the LMW thiol cycle, their counterparts for the BSH system in Gram-positive bacteria have not been characterized. Phylogenomic profiling in B. subtilis identified a putative candidate enzyme, YpdA, that has been proposed to be the bacillithiol disulfide reductase (Gaballa et al., 2010). Gene expression data in S. aureus have revealed ypdA transcription to be increased in response to exposure to nitrite (Schlag et al., 2007), nitric oxide (Hochgräfe et al., 2008), diamide (Posada et al., 2014), acid shock (Bore et al., 2007), early adaptation to lung environment (Chaffin et al., 2012), and infection-mimicking conditions (Mäder et al., 2016), suggesting this enzyme may play a role in stress response.
In this work, we characterize the role of YpdA in S. aureus physiology, virulence, and its relationship to the BSH system. We show that YpdA responds to stress and its loss leads to a fitness defect in competition assays. We present the first evidence that YpdA is a putative bacillithiol disulfide reductase that influences intracellular BSSB levels and decreases the BSH/BSSB redox ratio. Additionally, overexpression of YpdA increases BSH level and confers added resistance to oxidative and electrophilic stress. Finally, we demonstrate that YpdA has a role in virulence and survival in ex vivo neutrophil (PMN) experiments.
RESULTS:
ypdA mutant has a fitness defect.
All known S. aureus genomes contain a highly conserved gene, designated as SAOUHSC_01499 in the 8325 background (S. Fig. 1), that encodes a protein with high homology to the predicted bacillithiol disulfide reductase, ypdA, in Bacillus subtilis (Gaballa et al., 2010). However, biochemical and genetic evidence for the reductase function is missing from either organism. Accordingly, we mutated ypdA by replacing the ORF with a kanamycin cassette through homologous recombination in an SH1000 strain corrected for BSH production (Posada et al., 2014), and constructed the complement strain by replacing the kanamycin cassette with the native ypdA gene. We next examined growth phenotypes of the wild type (WT), ypdA mutant, and complement in the presence of stress in Tryptic Soy Broth (TSB) (Figure 1A). In this experiment, we were not able to detect any obvious growth defect of the mutant vs. the parent or the complemented mutant in TSB. Similar observations were also made in media such as TSB, and CDM (chemically defined medium) in the absence of stress (Figure 1B) or under different combinations of thiol and oxidative stresses, using diamide and hydrogen peroxide (data not shown). We reasoned that fitness defects might be masked by other metabolic pathways in rich media that neutralize the effect of stress. Accordingly, the fitness of the ypdA mutant was evaluated by a competition assay which has been shown to be useful for teasing out subtle fitness defects between closely related strains (Le Lam et al., 2017). Despite identical growth kinetics of the mutant vs. the parent in complex media such as TSB and CDM (Figure 1B), overnight competition assays revealed that the WT strain out-competed the ypdA mutant in competitive growth in CDM (Figure 1C) while the competitive index did not change in TSB media. These results suggest that while the ypdA mutation had a limited effect on growth in complex media in vitro, the mutant exhibited a fitness defect vs. the WT when grown competitively in chemically defined medium.
Figure 1: YpdA mutant has a fitness defect in competition assay.
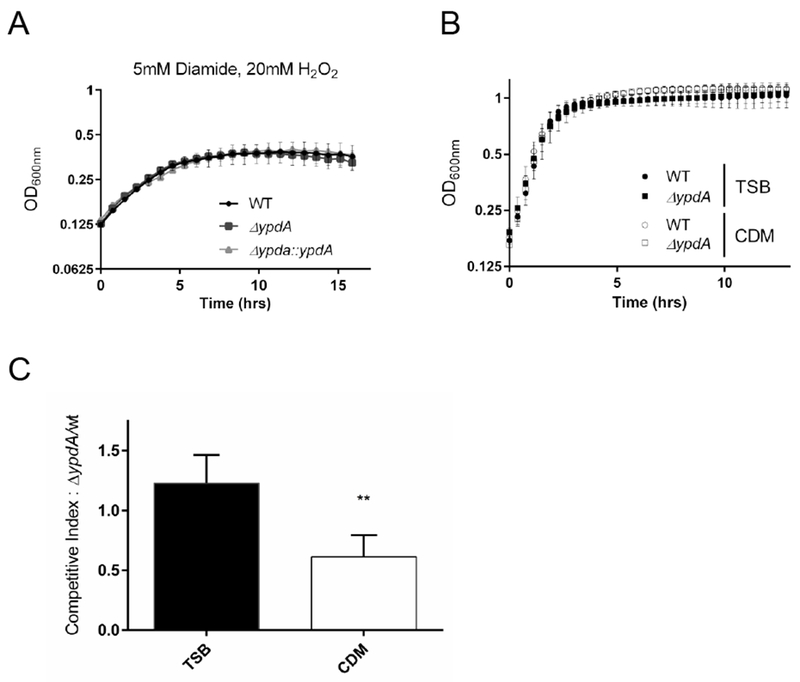
(A) S. aureus growth dynamics in the presence of hydrogen peroxide and diamine. Cell density (OD600) was measured in WT (circle), ypdA mutant (square), and complemented strain (triangle) in the presence of 5mM Diamide, 20mM H2O2 in TSB.
(B) S. aureus growth dynamics in rich and chemically defined media. Cell density (OD600) was measured in WT (circle) and ypdA mutant (square) in TSB (shaded) and CDM (open) media. Each value represents the mean of three biological replicates that were performed in triplicate.
(B) Competition assay shows YpdA fitness defect in chemical defined medium. Overnight cultures were back diluted and allowed to reach OD600=0.6, diluted to 1×105 CFU, and mixed 1:1 volumes. Cultures were grown at 37°C for 18 hrs. and serially diluted to enumerate CFU. Competition index was calculated from number of ypdA mutant divided by number of WT colonies. Each value represents the mean and standard deviation of three biological replicates that were performed in triplicate, statistical significance between media conditions was determined using Student t-test (** P<0.01)
YpdA levels are induced by stress.
Given the fitness defect of the ypdA mutant in CDM, we wondered if expression of ypdA is responsive to challenges with stress. Primer extension assay divulged that the ypdA transcript was initiated 22 bases directly upstream of the ypdA translation start site (Figure 2A). Northern blot data revealed that the ypdA transcript is monocistronic, with increased transcription during the post-exponential phase and upon the addition of thiol stressor diamide (Figure 2A, B). Based on these data, a transcriptional fusion in shuttle plasmid pSK236 containing a 180-bp ypdA promoter fragment fused to venus, which encodes a rapidly folding YFP variant, was created. This plasmid was then introduced into the WT strain and fluorescence was measured upon exposure to various stresses. WT cells exposed to strong oxidants such as H2O2 and hypochlorous acid (HOCl), nitric oxide-generating NOC-12, the toxic electrophilic metabolic byproduct methylglyoxal (MG), and thiol stress-inducing diamide at experimentally determined non-lethal concentrations all showed significant increases in fluorescence attributable to enhanced ypdA promoter activity (Figure 3A). To ensure that these increases in transcriptional activity result in enhanced YpdA protein expression, Western blots were conducted on whole cell lysates probed with polyclonal serum from mice immunized with purified YpdA protein. As expected, the WT strain expressed the YpdA protein at a low level, while no YpdA was detected in the ypdA mutant (Figure 3B). Under identical stress conditions as for the promoter fusion studies, YpdA protein levels were observed to be significantly enhanced upon exposure to these stresses compared to the untreated control (Figure 3B). The YpdA protein levels appear to trend with those of the transcription fusion, with higher protein expression in cells exposed to H2O2 and HOCl (relative densitometric units of 3.26 and 3.04, respectively) as compared to those exposed to methylglyoxal and diamide (relative densitometric units of 1.25 and 2.11, respectively). Taken together, these data suggest that YpdA participates in stress response upon exposure to specific physiologically relevant stresses.
Figure 2: Analysis of ypdA operon.
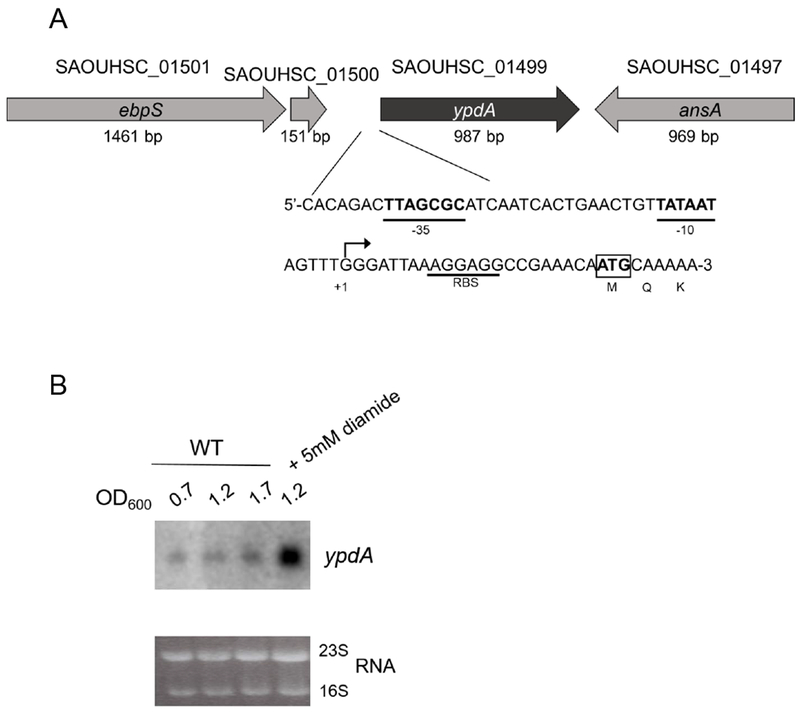
(A) Schematic model of the ypdA genomic region. Loci names and their respective lengths are from the 8325-4 genome annotation. Shown in detail is the promoter region of ypdA with transcriptional start site marked (*), −10 and −35 promoter boxes (bolded and underlined), the RBS (underlined), and ATG translational start codon (boxed).
(B) Northern blot analysis of ypdA at different growth phases with and without diamide stress. Ten μg of total RNA from WT S. aureus was loaded onto each lane and transferred to PVDF membrane. Blots were probed with 295-bp radiolabeled ypdA DNA fragment. The 23S and 16S rRNA bands of the ethidium bromide-stained gel used for loading control are shown below. Experiment was performed three times; a representative experiment is shown.
Figure 3: Oxidative and electrophilic stresses increase YpdA expression.
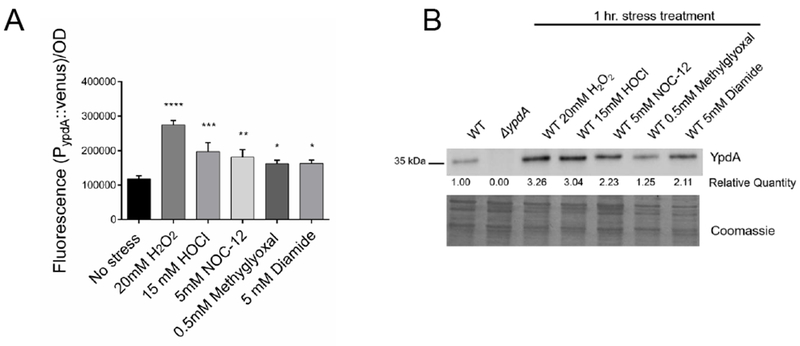
(A) ypdA promoter activity in S. aureus upon exposure to oxidizing and electrophilic agents. Overnight cultures were diluted in fresh TSB and allowed to grow to OD600=0.4, stressor agents were then added. Venus YFP fluorescence (515 nm excitation, 533nm emission) generated from the ypdA promoter activity was measured at 1-hour after the addition of stressor agent. Fluorescence data were normalized by optical densities. Values represent the mean and standard deviation of three biological replicates that were conducted in triplicate. Statistical significance between stress and no stress conditions was calculated using 1-way ANOVA Dunnett’s multiple comparisons test (* P< 0.05; ** P< 0.01; *** P< 0.001; **** P< 0.0001).
(B) YpdA protein levels in S. aureus increase with stress. Overnight cultures were diluted in fresh TSB and allowed to reach OD600=0.4 followed by the addition of stressor agents and cultures were further incubated for 1 hr. Fifteen ug of whole cell lysate were immunoblotted and probed with mouse anti-YpdA serum diluted 1:1000 followed by secondary antibody and substrate. Relative quantification of YpdA were analyzed by ImageJ; data were shown below the blot with WT set at 1.00. A parallel run gel stained with Coomassie is shown in the bottom panel to demonstrate comparable loading. Experiments were performed at least three times; a representative experiment is displayed.
Initial characterization of YpdA.
Based on sequence analysis, YpdA belongs to the pyridine nucleotide-disulfide oxidoreductase family of proteins that use flavin adenine dinucleotide (FAD) to transport reducing equivalents from NAD(P)H to cysteine residues. To purify YpdA, we cloned the ORF into pET14b, and expressed the protein in E. coli BL21. After IPTG induction, the YpdA protein was purified from the cell lysate with a nickel affinity column. The purified YpdA protein had a distinct yellow color, suggesting it likely purified with bound FAD, which is responsible for this distinct color. We measured fluorescence spectra of our purified protein and found the peak to correspond to that of FAD standards (S. Fig 2A). To evaluate whether YpdA functions in a manner consistent with an oxidoreductase, NAD(P)H consumption was monitored. Specifically, NAD(P)H has a distinct absorbance at 340 nm, while the oxidized forms do not (Carlberg and Mannervik, 1985). As shown in Figure 4A and B, purified YpdA can consume both NADPH and NADH. In reactions lacking YpdA, NAD(P)H levels remained constant, while the addition of YpdA resulted in a steady decrease in the level of the NAD(P)H cofactor. To further test our hypothesis that YpdA can consume cofactor, we purified a mutated YpdA protein with a glycine to alanine substitution at position 10. This mutation is predicted to completely abolish cofactor binding activity as glycine 10 is the first glycine residue in a putative Rossman fold domain (GxGxxG), which is characteristic of cofactor binding enzymes and presumed to function in the reduction of BSSB (Bragg et al., 1997). The mutant and WT proteins were analyzed by circular dichroism, which showed very minor changes, ensuring both proteins were similarly folded (S. Fig 2B). When examined in the consumption assay, the YpdA G10A protein was indeed unable to consume NADPH or NADH (Figure 4A, B). Collectively, these data suggest that YpdA, as a reductase, can consume NAD(P)H, consistent with other proteins in the oxidoreductase family.
Figure 4: YpdA mutation influences BSH/BSSB ratio and consumes NADPH and NADH.
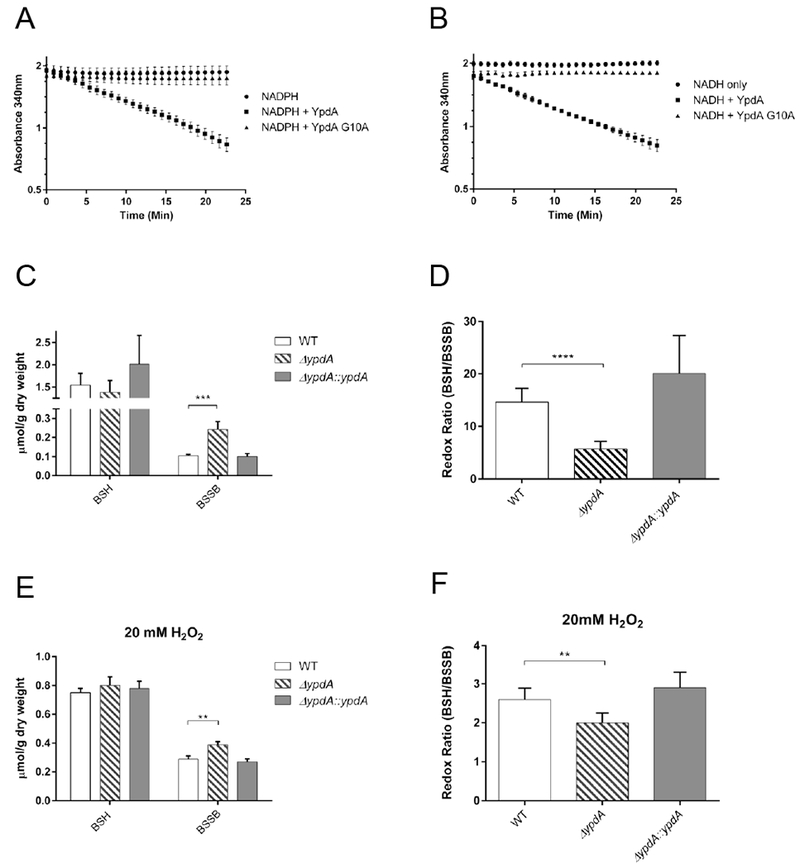
(A, B) In vitro YpdA assay measuring NADPH and NADH consumption. Reactions were set up using 500 μM of NADPH or NADH (●), and 2.5 μM of YpdA protein (■ wild type YpdA; ▲YpdA G10A mutation). Reactions were conducted at room temperature; data are expressed as the mean with standard deviation of three biological replicates conducted in triplicate.
(C) Bacillithiol measurements in whole cell lysates grown in TSB. BSH and BSSB were measured in WT (open), ypdA mutant (striped), complement (closed) strains. Data are expressed as the mean with standard deviation (n=6). Significance was determined using Student t-test (*** P ≤ 0.001).
(D) ypdA mutant has an altered redox ratio. Ratio was calculated by dividing reduced amount of BSH by oxidized BSSB for WT (open), ypdA mutant (striped), complement (closed) strains. Data are expressed as the mean with standard deviation, significance was determined using Student’s t-test (**** P≤ 0.0001).
(E) Bacillithiol measurements in whole cell lysates grown in TSB after 45 min exposure to hydrogen peroxide. BSH and BSSB were measured in WT (open), ypdA mutant (striped), complement (closed) strains. Data are expressed as the mean with standard deviation (n=3). Significance was determined using Student t-test (** P ≤ 0.01).
(F) Redox ratio after 45min hydrogen peroxide exposure. Redox ratio was calculated by dividing reduced amount of BSH by oxidized BSSB for WT strain (open) ypdA mutant (striped), complement (closed) strains). Data are expressed as the mean with standard deviation, significance was determined using Student t-test (** P ≤ 0.01).
S. aureus ypdA mutant displays altered bacillithiol redox levels.
The role of YpdA in regulating BSH and BSSB levels was further examined by analysis of whole cell thiol content. We postulated that if YpdA functions as a reductase, there would be differences in BSH and BSSB levels. Alternatively, the redox ratio of BSH to BSSB could be diminished in the ypdA mutant vs. parent. Accordingly, oxidized and reduced levels of BSH were measured by HPLC in the WT, ypdA mutant, and complemented mutant. While all three strains had comparable levels of BSH at the level of 1.5-2.0 μmol g−1 dry weight, only the ypdA mutant displayed significantly higher levels of BSSB, the oxidized form of BSH, vs. the parent and complemented mutant (Figure 4C). We surmise that the redox ratio of BSB/BSSB may be a more sensitive indicator of the changing relationship between the BSH and BSSB pool in the ypdA mutant, in a manner similar to GSH/GSSG ratio that has been shown to be a sensitive proxy for the indicator of intracellular oxidative stress levels (Zitka et al., 2012). As shown in Figure 4D, WT and complemented strains showed much higher BSH/BSSB redox ratio than the ypdA mutant, implying that mutant cells are likely under a higher level of intracellular redox stress. Under conditions of oxidative stress, the ypdA mutant displayed a higher level of BSSB (0.4 vs. 0.2 μmol/g dry weight without H2O2 stress), a lower level of BSH (0.8 vs. ~1.5 μmol/g dry weight without H2O2 stress). Additionally, upon H2O2 exposure, the BSH/BSSB ratio decreases from ~6 (no stress) to 2 (oxidative stress) in the ypdA mutant which is significantly lower than the WT ratios of >10 and 2.6 respectively, further highlighting the susceptibility of the mutant to stress conditions (Figure 4E, F). To ensure that the loss of ypdA specifically targeted the BSH system and not all LMW thiols, the intracellular pool of cysteine and cysteine disulfide was measured and found to have no significant differences between the WT, ypdA mutant and complemented mutant (S. Table 1). Similarly, the cysteine redox ratios remained unchanged among all three strains and conditions (S. Table 1). Together, the BSH measurements and cofactor consumption assays suggest that YpdA functions as an oxidoreductase in the BSH system.
YpdA overexpression confers greater resistance to stress.
It was recently shown that overexpression of Mtr, the mycothiol disulfide reductase, increases tolerance of Corynebacterium glutamicum to oxidative stress (Si et al., 2016). We reasoned that if YpdA functions as a bacillithiol disulfide reductase, overexpression of YpdA could conceivably enable us to discern differences in BSH; we speculated that this increased contribution to BSH pool, accompanied by reduced BSSB pool, due to ypdA overexpression would lead to an enhanced redox-protective phenotype. Accordingly, an overexpression plasmid with ypdA under the control of an anhydrotetracycline inducible promoter, designated pALC7926 [pALC7925::ypdA (Table 1)], was constructed. Following validation that YpdA expression was increased under inducing condition with anhydrotetracycline at 160 ng ml−1 (Figure 5A), we verified that this increase in expression in WT strain did not have an impact on bacterial growth as compared with the empty vector control (Figure 5B). To determine if YpdA overexpression allowed for greater tolerance of oxidative and electrophilic stress, cells were exposed to H2O2 (Figure 5C), hypochlorous acid (Figure 5D), methylglyoxal (Figure 5E), and diamide (Figure 5F) at non-lethal concentration that were determined experimentally. The overexpression strain showed improved growth over time vs. the vector control in all tested conditions while there was no change in growth dynamics with nitric oxide-generating compound NOC-12 (data not shown). As a control, overexpression of YpdA did not improve growth of the BSH null strain SH1000 under stress conditions compared to the vector control (S. Fig 3).
Table 1:
Strains and Plasmids used in this study.
| Strains or plasmids | Genotype/description | Source/Reference |
|---|---|---|
| E.coli | ||
| DH5α | E. coli DH5α for cloning | Novagen |
| BL21 | E. coli for protein expression | Novagen |
| ALC8856 | E. coli BL21 with pET14a-6x-His-YpdA | This study |
| ALC8857 | E. coli BL21 with pET14a-6x-His-YpdA-G10A | This study |
| S. aureus | ||
| RN4220 | Restriction minus NCTC8325-4 | (Kreiswirth et al., 1983) |
| ALC7446 | SH1000 corrected for BSH production | (Posada et al., 2014) |
| ALC8254 | ΔypdA::kan, ALC7446 with ypdA replaced by kanamycin cassette | This study |
| ALC8638 | ΔypdA::Δkan::ypdA, ALC8254 chromosomally complemented with ypdA | This study |
| ALC8858 | ALC7446 containing pALC8858 (promoter of ypdA driving venus) | This study |
| ALC8859 | ALC7446 containing pALC7925 (empty vector) | This study |
| ALC8860 | ALC7446 containing pALC7926 | This study |
| Plasmids | ||
| pET14b | E. coli expression vector | Novagen |
| pMAD | E. coli/S. aureus shuttle plasmid with the ori pE194ts; bgaB Ampr Ermr | (Arnaud et al., 2004) |
| pSK236 | Shuttle vector containing pUC19 cloned into the HindIII site of pC194 | (Gaskill and Khan, 1988) |
| pALC7925 | pALC2073 derivative with improved −10 binding site as described by Corrigan and Foster | This study, (Bateman et al., 2001; Corrigan and Foster, 2009) |
| pALC7926 | pALC7925 with ypdA for overexpression (pALC7925::ypdA) | This study |
| pALC8858 | pSK236 with promoter of ypdA driving venus | This study |
Figure 5: Overexpression of YpdA enhances S. aureus fitness.
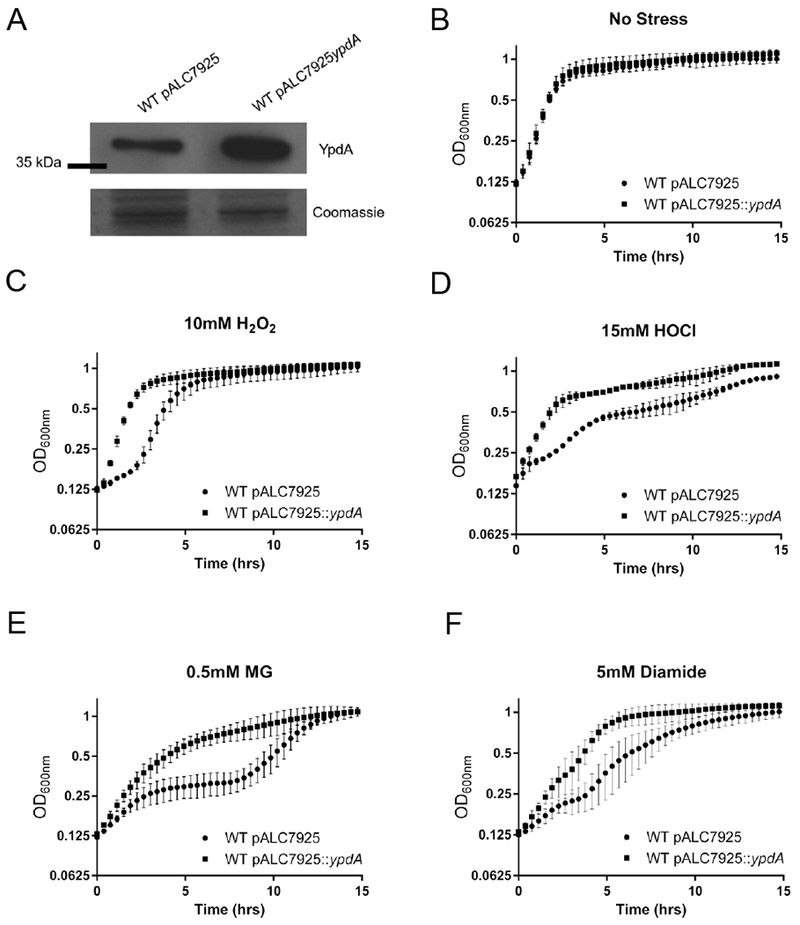
(A) YpdA expression induced with 160 ng/ml of anhydrotetracycline. Cells were harvested at OD600=0.6. Fifteen ug of whole cell lysates were immunoblotted and probed with mouse anti-YpdA serum. A parallel run gel stained with Coomassie is shown below. Experiment was repeated three times, with a representative experiment shown.
(B-F). Growth dynamic of YpdA-overexpression strain in the presence of oxidizing and electrophilic agents. WT overexpressing YpdA or with empty vector were induced with 160 ng/ml of anhydrotetracycline and growth assayed in TSB with conditions of no stress (B), 20 mM hydrogen peroxide, H2O2 (C); 15 mM hypochlorous acid, HOCl (D); 0.5 mM Methylglyoxal, MG (E) and 5 mM Diamide (F). Values represent the mean with standard deviation of three biological replicates conducted in triplicate.
To examine if YpdA overexpression was also modulating levels of BSH and BSSB and the redox ratio during challenge with stress, the thiol pools and redox ratio for the YpdA-overexpressing strain and empty vector control were measured in the presence of H2O2. In unstressed cells, normal or overexpression of YpdA in WT did not appear to influence either the levels of BSH or BSSB (Figure 6A) or the resulting redox ratio (Figure 6B). However, following treatment with 20 mM H2O2, the drop in BSH levels was less in the YpdA overexpression strain than the corresponding strain with the empty vector, while levels of BSSB appeared further reduced in the overexpression strain vs. the vector control (Figure 6A). Importantly, the BSH level in the overexpression strain was higher than the vector control under H2O2 stress (Fig. 6A). These alterations led to higher redox ratio during stress in WT cells overexpressing YpdA than the vector control (Figure 6B). Together, these data imply that the overexpression of YpdA increases the redox ratio by keeping BSH levels high and BSSB levels low.
Figure 6: YpdA overexpression enhances cellular redox homeostasis.
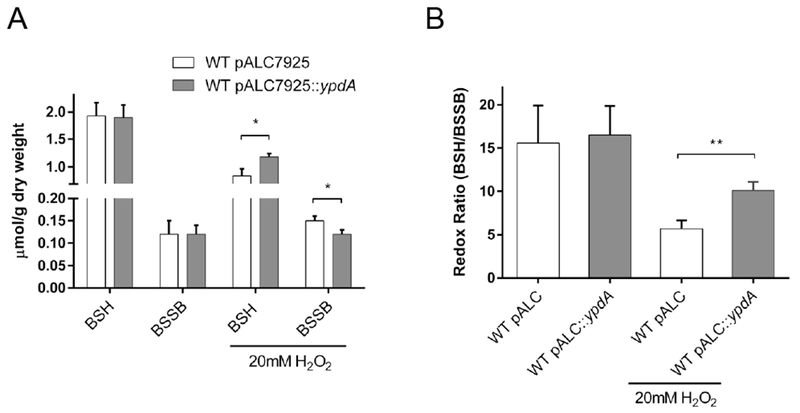
(A) Bacillithiol levels during YpdA overexpression. BSH and BSSB were measured in WT carrying empty vector (open) or vector with overexpression of YpdA (closed), under normal and stress conditions after 45 min exposure. Data are expressed as the mean and standard deviation (n=3). Significance was determined using Student t-test (* P ≤ 0.05).
(B) Redox ratio during YpdA overexpression. Redox ratio was calculated by dividing reduced amount of BSH by oxidized BSSB for WT strain carrying empty vector (open) or carrying overexpression of YpdA (closed). Data are expressed as the mean with standard deviation, significance was determined using Student t-test (** P ≤ 0.01).
Similar measurements were made on intracellular levels of cysteine, another source of LMW thiol (S. Table 1). In the presence of oxidative stress, cysteine concentrations were increased in both empty vector and overexpression strains compared to strains without stress treatment. As with the case of BSH, oxidized cysteine levels were lower in the overexpression strain under H2O2 stress, hinting at potential crosstalk between the two LMW thiol pools. Altogether, our results indicate that oxidative and electrophilic stresses upregulate YpdA expression and the resultant increase in intracellular redox buffer leads to enhanced fitness of S. aureus in the presence of these stresses (Fig. 5).
Decreased fitness of the ypdA mutant results in reduced survival in PMNs.
BSH has previously been shown to contribute to S. aureus virulence (Pöther et al., 2013; Posada et al., 2014). Strains, like SH1000 with a defect in bshC, that do not produce BSH were found to survive less well in human upper airway cells and murine macrophages (Pöther et al., 2013), and are more susceptible to killing in whole blood survival assays (Posada et al., 2014). We wondered if reduced redox ratio and altered BSSB levels due to the ypdA mutation would have similar effects in whole blood killing assays. The ypdA mutant was found to survive less well than the parent and the complemented mutant (Figure 7A). Interestingly, the survival of SH1000 (BSH null strain), akin to the ypdA mutant, was also less than the parent and complemented mutant in this assay (Figure 7A). We reasoned that PMNs were likely to be responsible for the killing phenotype as they represent an important early host response to limit the spread of S. aureus infection. To test this hypothesis, WT, ypdA mutant, and complemented mutant were incubated with purified human PMNs at a MOI of 1. After one hour of co-incubation followed by CFU enumeration, the ypdA mutant was found to be killed more efficiently than WT or complemented strains (Figure 7B). PMNs can generate a number of defenses against bacterial pathogens, including intraluminal oxidative burst, antimicrobial peptides, and induction of NETosis outside apoptotic PMNs (DeLeo et al., 2009). Since YpdA is a putative BSH disulfide reductase that responds and protects against oxidative stress, we hypothesized that oxidative burst would be the likely killing mechanism of S. aureus by PMNs under this scenario. Therefore, we incubated PMNs with diphenyleneiodonium (DPI), an inhibitor of oxidative burst that does not influence phagocytosis capacity (Hampton and Winterbourn, 1995), before co-incubation with S. aureus strains as described above. DPI was able to alleviate the differential killing by human PMNs, with comparable levels of survival among WT, the ypdA mutant and complemented mutant (Figure 7B).
Figure 7: YpdA contributes to virulence in whole blood and neutrophil killing assays.
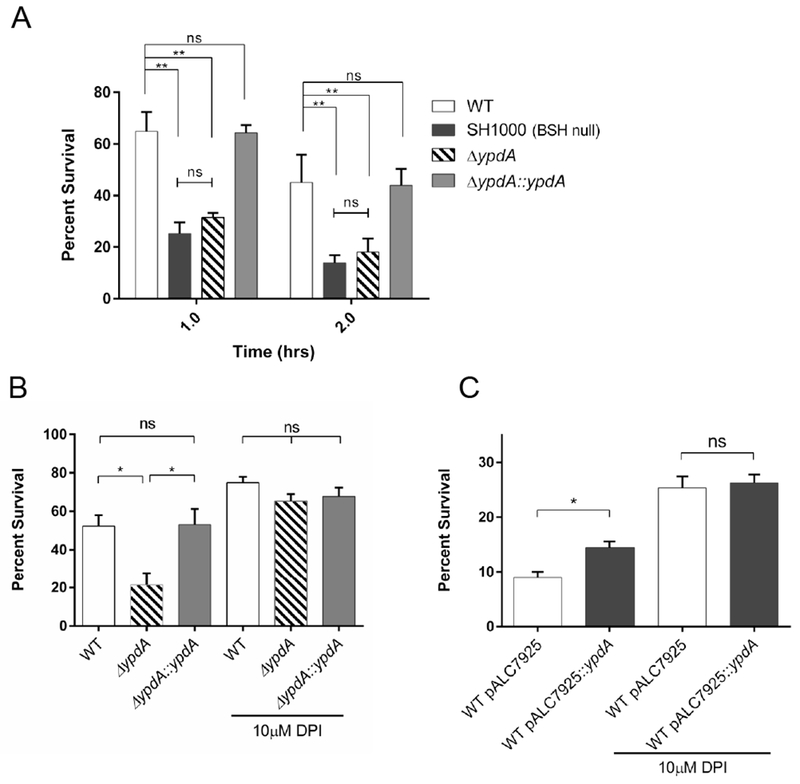
(A) Bacterial survival in whole blood. Whole blood from healthy human volunteers was inoculated with WT (open), SH1000 (black), ypdA mutant (striped), and complement strains (grey) at 1 × 105 CFU/ml. Data represents the mean and standard deviation of three biological replicates. Statistical significance was calculated using 2-way ANOVA with post-hoc Tukey test (** P ≤ 0.01).
(B, C) Bacterial survival in purified human neutrophils. Neutrophils were purified from whole blood from healthy human volunteers using Hypaque-Ficoll gradients. DPI treatment was applied to a portion of the neutrophils for 10 minutes. (B) WT (open), ypdA mutant (striped), and complemented strains (closed) were incubated with neutrophils at a MOI of 1 and surviving CFUs were plated. Data for the 1-hour incubation period are presented. Data represent the mean and standard error of three biological replicates. Statistical significance was calculated for each neutrophil treatment condition using 1-way ANOVA with post hoc Tukey test (* P ≤ 0.05). (C) WT carrying empty vector (open) or vector with YpdA overexpression (closed) was incubated with human neutrophils for 1 hour, at an MOI of 1:1 and surviving CFUs were assessed by plating on TSA. Data represent the mean and standard error of three biological replicates. Statistical significance was calculated for WT with empty vector vs WT with YpdA overexpression as determined by Student t-test (* P ≤ 0.05).
These results led us to hypothesize that overexpression of YpdA in WT cells, conferring a greater fitness upon exposure to oxidative stress, would improve survival in human PMNs. Using WT cells with either overexpression of YpdA or an empty vector for co-incubation with purified human PMNs, it was observed that cells overexpressing YpdA were able to survive better than cells with just the empty vector, and this difference in survival was abolished when oxidative burst of PMNs was blocked by DPI (Figure 7C). Together, these data highlight the important role that YpdA plays in protecting S. aureus cells from the oxidative killing by human PMNs.
RNA-seq suggests metabolic differences.
While YpdA is crucial for BSH-mediated redox balance in S. aureus, we were curious as to its overall impact on S. aureus gene expression as determined by RNA-Seq. Overall, there were 15 up-regulated and 47 down-regulated transcripts in the ypdA mutant vs. the WT (Sup Table 2, 3). Among the 15 up-regulated genes, there were four genes with over 30-fold increase (SAOUHSC_1886, SAOUHSC_1887, SAOUHSC_1888, SAOUHSC_1889) involved in the riboflavin biosynthesis pathway. This was a particularly interesting result since riboflavin levels have long been tied to glutathione reductase activity (Beutler, 1969). Similarly, riboflavin has been proposed to have antioxidant properties in certain bacteria and fungi (Protchenko et al., n.d.; Khan et al., 2014). These results were confirmed independently by qRT-PCR analysis. Another interesting hit from the RNA-Seq experiments was the downregulation of glycerol-3-phosphate transporter (SAOUHSC_00438), glycerol uptake facilitator (SAOUHSC_01275) and glycerol kinase (SAOUHSC_01276) genes. Glycerol metabolism is critical in the production of energy through pyruvate entry into the TCA cycle and glycolysis, and has been implicated as important for L-form formation and persistence in S. aureus (Han et al., 2014). Additionally, glycerol metabolism has been shown to be important in pathogenicity and phospholipid biosynthesis in other organisms (Joseph et al., 2006; Yoshimura et al., 2007).
Using KEGG pathway analysis, we identified several metabolic pathways that appeared to be affected by the ypdA mutation (Table 2). Interestingly, it seemed that carbohydrate metabolism was mostly down regulated in the mutant, potentially illuminating another reason for the reduced fitness of the ypdA mutant in the competitive fitness assays with the WT in chemical defined medium. Similarly, analysis of the protein classes in our dataset suggests that transferases and transporters are most affected in the mutant strain, again emphasizing the effect of YpdA on bacterial metabolism (Sup Figure 4).
Table 2: Kegg pathway analysis.
Kegg pathway analysis of differentially expressed genes by RNA-seq of ypdA mutant and WT S. aureus cells was performed using KOBAS3.0.
| KEGG Pathway | Upregulated Genes | Downregulated Genes | FDR corrected P-Value |
|---|---|---|---|
| Riboflavin metabolism | 4 | 0 | 1.87E-03 |
| Phosphotransferase system (PTS) | 1 | 4 | 5.03E-03 |
| Starch and sucrose metabolism | 0 | 4 | 6.45E-03 |
| Glycolysis/Gluconeogenesis | 1 | 4 | 2.00E-02 |
| Amino sugar and nucleotide sugar metabolism | 1 | 3 | 3.65E-02 |
| Fructose and mannose metabolism | 1 | 2 | 3.99E-02 |
| Chloroalkane and chloroalkene degradation | 1 | 1 | 4.31E-02 |
Discussion
S. aureus is equipped with a wide range of tools to protect itself from the bactericidal activities of reactive oxygen species, both native and foreign. For instance, superoxide dismutase converts superoxide to hydrogen peroxide and catalase converts hydrogen peroxide to innocuous oxygen and water (Mandell, 1975), while 168 genes have been identified as essential for full nitric oxide resistance (Grosser et al., 2018). In addition, S. aureus also synthesizes carotenoid pigments that function as antioxidants through free-radical scavenging and ability to reduce singlet oxygen (Liu et al., 2005). The thioredoxin system, composed of thioredoxin reductase and thioredoxin, can also maintain the intracellular protein thiol-disulfide balance (Uziel et al., 2004). Despite this wide arsenal of antioxidants and redundancy with the thioredoxin system, S. aureus still possesses a functional LMW thiol pool. In Gram-negative bacteria and eukaryotes, the glutathione system is utilized, whereas in Gram-positive Firmicutes, this system is notably absent. Instead, bacteria such as S. aureus and Bacillus subtilis deploy bacillithiol as the major LMW thiol due to its reactive ability at physiological pH; thus leaving cysteine and coenzyme A, the other LMW thiol pool, to function in metabolism (Leonardi and Jackowski, 2007; Sharma et al., 2013). Although the genes responsible for BSH synthesis have been well characterized, the reduction pathways and enzymes involved remain elusive.
The goal of this study was to examine if YpdA functions as a bacillithiol disulfide reductase as postulated by Gaballa et al. (Gaballa et al., 2010). While the existence of this reductase has been postulated for some time, the direct identification of this enzyme has not been achieved so far. Based on phylogenomic studies in B. subtilis (Gaballa et al., 2010), a YpdA homolog was identified in S. aureus. To characterize YpdA in S. aureus, we constructed a ypdA mutant and its chromosomal complement. While the ypdA mutant did not differ from the parent in growth dynamics in complex media, we were able to tease out a fitness defect in the mutant vs. the parent in growth competition assay in CDM (Figure 1C). It is possible that the ypdA mutant of S. aureus, with a reduced redox ratio, can cope with growth-related stress in complex media due to redundant rescuing mechanisms (e.g. staphyloxanthin, cysteine etc.) but not in CDM. Previously, it was shown that an E. coli glutathione reductase mutant is sensitive to H2O2 only in a catalase minus background (Masip et al., 2006). Additionally, glutathione reductase mutant in E.coli has been shown to keep levels of GSH and GSSG constant in the presence or absence of stress (Smirnova et al., 2005). Thus, it is conceivable that the full impact of bacillithiol disulfide reductase on S. aureus growth in complex media may not be completely understood unless we evaluate a combination of double and triple mutants that target the alternative enzymatic stress response pathways.
As an additional proof that YpdA responds to oxidative and electrophilic stresses, we performed promoter fusion assays and analyzed YpdA protein levels with Western blots (Figure 3). In E. coli, glutathione reductase has been shown to respond to oxidative stress through activation by OxyR, a regulator of antioxidant genes (Storz et al., 1992; Michán et al., 1999). In many Gram-positive bacteria such as S. aureus and B. subtilis, Spx is the main regulator of intracellular redox-related genes (Nakano et al., 2003; Pamp et al., 2006). More specifically, Spx has been shown to control BSH synthesis genes in B. subtilis (Gaballa et al., 2013). Whether Spx in S. aureus would regulate ypdA, akin to increased YpdA expression upon induction of Spx in Bacillus anthracis (Barendt et al., 2013), is not clear. As spx is an essential gene in S. aureus, it would necessitate a conditional mutation to answer this query.
To verify biochemically that YpdA acts as a reductase, we measured intracellular levels of BSH and BSSB in WT and isogenic ypdA mutant. While there was a distinct increase in BSSB levels in the ypdA mutant vs. the parent and under oxidizing conditions, the BSH levels did not differ between the mutant and the parent (Figure 4 C, D, E, F). While there may be several plausible explanations for this finding, we believe the most obvious one is that the basal BSH pool is almost 10 times the size BSSB pool under routine laboratory growth conditions (Figure 4C); consequently, the differential reduction of BSH in the absence of YpdA may not be easily detected within the native BSH pool, while those of the BSSB can rise beyond the low basal level to enable detectable difference in the ypdA mutant vs. the parent and complemented mutant (Figure 4C). Upon challenge with hydrogen peroxide, there was consumption of BSH to yield a lower level of BSH in all strains accompanied by higher level of BSSB. The amount of BSSB remained highest in the ypdA mutant compared to parent or complemented mutant. To overcome the effect of a large basal pool of BSH in S. aureus, we elected to over-express ypdA from an exogenous promoter in the parental strain under stress-inducing condition. Under this condition, we were able to increase BSH production and reduce levels of BSSB, resulting in a higher redox ratio under H2O2 stress in the WT strain overexpressing YpdA (Fig. 6A). This result is consistent with the notion that YpdA is likely a bacillithiol disulfide reductase that only impacts the redox potential of S. aureus cells under stress.
We also considered other explanations for a lack of BSH effect in the ypdA mutant. For instance, it is plausible that there are redundant enzymes for the recycling of BSSB besides YpdA. The Rawat group identified a number of enzymes that disrupted the BSH/BSSB ratio but these enzymes remain to be further characterized to tease out their contributory roles (Rajkarnikar et al., 2013). Additionally, oxidized glutathione has been shown to be reduced by thioredoxin-thioredoxin reductase systems in Saccharomyces cerevisiae (Tan et al., 2010); a similar function is also possible in S. aureus. It is plausible that YpdA de-bacillithiolates other S-bacillithiolated proteins (i.e. a bacilliredoxin) and lacks bacillithiol disulfide reductase activity. However, we believe the data here are more consistent with YpdA’s primary function as a bacillithiol disulfide reductase rather than as a bacilliredoxin based on the following reasons: 1) it has been estimated in B. subtilis that only 3% of total cellular BSH pool was bound to protein during growth (Chi et al., 2013). Bacilliredoxin proteins (e.g. BrxA and BrxB in B. subtilis), responsible for de-bacillithiolating S-bacillithiolated protein, are thus minor contributors to the BSH pool and in the ability to augment global redox buffering capacity (Gaballa et al., 2014). This conjecture holds true for Saccharomyces because deletion of glutaredoxins in this species has no influence on cellular levels of GSH or GSSG (Luikenhuis et al., 1998). The appreciable increase in BSSB levels in the ypdA mutant would be inconsistent with YpdA functioning solely as a bacilliredoxin. 2) Even if S. aureus has a higher BSH pool bound to cellular proteins than that found in B. subtilis, one would predict higher BSSB levels in the WT (i.e. S-bacillithiolated protein +BSH → de-conjugated protein + BSSB) but reduced in the ypdA mutant (Figure 8). Importantly, our data revealed higher BSSB level in our ypdA mutant vs. the parent, in contrast to this prediction. An additional possibility is that NAD(P)H may be donating electrons to reduce S-bacillithiolated protein; in this reaction, one would expect a noticeable decrease in BSH level as a byproduct [S-bacillithiolated protein + 2 NAD(P)H → protein + BSH + NAD(P)] in the ypdA mutant, which we do not observe in our data. Finally, we do not believe that the biosynthesis pathway becomes upregulated in the ypdA mutant to neutralize the reduction in BSH level; we do not favor this explanation because we did not observe noticeable differences in expression of BSH synthesis genes between WT and the ypdA mutant in the RNA-Seq dataset. Additionally, since expression of ypdA is up-regulated in stress, we contend that the moderate increase in BSH levels and redox ratio in the overexpression strain (Figure 6) is what enables greater resistance to oxidative and electrophilic stresses, as seen by faster recovery and growth under stressful conditions. Since the difference in BSH levels between overexpression plasmid and empty vector control was only evident in the presence of stress, this would imply a potential activation mechanism such as oxidation or bacillithiolation of one of the cysteine residues in YpdA. A similar effect has been observed with overexpression of mycothiol reductase in Corynebacterium glutamicum (Si et al., 2016) but an activation mechanism remains elusive. It has previously been argued that S. aureus retains its BSH level in response to oxidative stress by hydrogen peroxide and that this response differs from B. subtilis which displays reduced levels of BSH in its response (Rajkarnikar et al., 2013). Our data here suggest that S. aureus BSH levels are reduced during exposure to hydrogen peroxide, highlighting a discrepancy in the field.
Figure 8: Proposed model for the function of YpdA in the context of bacillithiol cycling.
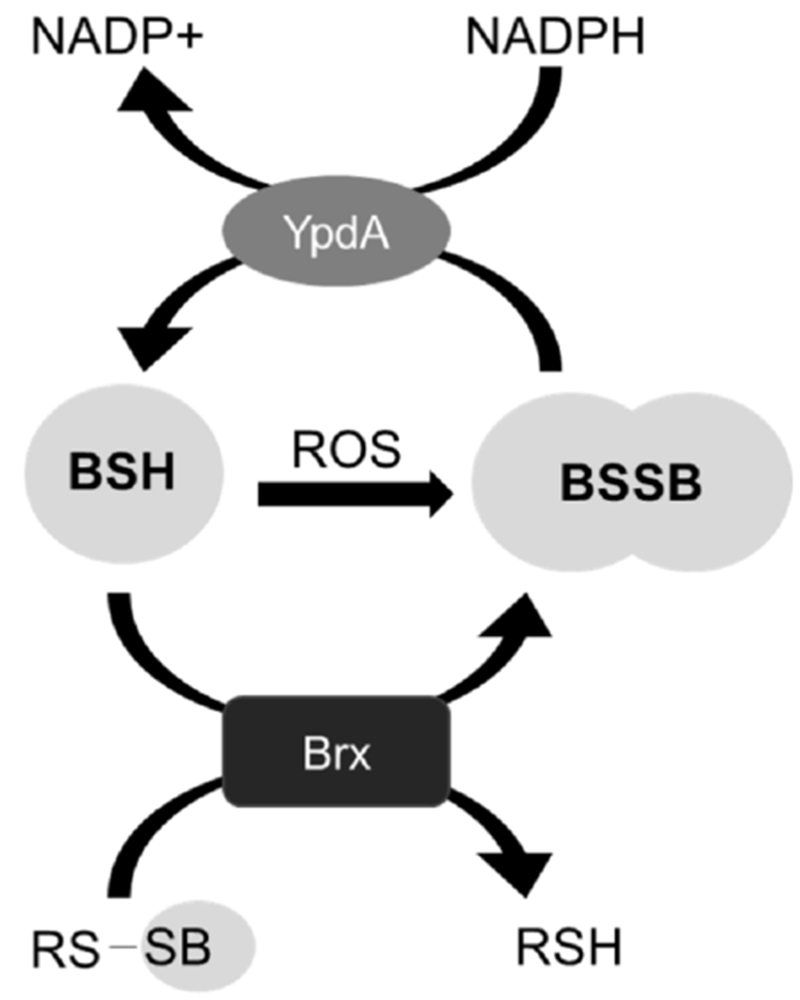
Bacillithiol is oxidized to bacillithiol disulfide through reaction with oxidants or through consumption by enzymes (e.g. Brx enzymes reducing S-bacillithiolated proteins to yield BSSB). YpdA recycles BSSB to maintain the reduced BSH pool.
We purified WT YpdA and YpdA G10A carrying a mutation known to disrupt cofactor binding in the GxGxxG motif of reductases (Figure 3 C, D). Indeed, WT YpdA readily consumes NAD(P)H while the G10A mutant does not. However, an in vitro reaction using purified YpdA to directly reduce BSSB was not successful, primarily due to technical limitation and the commercial unavailability of reducible BSSB compound. Besides the GxGxxG motif, YpdA also harbors the conserved CxxS motif, located at the N-terminus and close to the cofactor binding domain. CxxS is a derivative of the redox sensitive CxxC motif thought to be commonly associated with glutaredoxins (Fomenko and Gladyshev, 2003). Additionally, C14 of the CxxS motif has been shown to be oxidized under NaOCl stress, suggesting potential increased S-bacillithiolation (Imber et al., 2018). In Saccharomyces cerevisiae, proteins containing this motif can have dual function in reducing oxidized glutathione and various mixed disulfides (Fomenko and Gladyshev, 2002). It is plausible that YpdA could also be a multifunctional protein in reducing different substrates, but further experiments are needed to confirm this possibility.
S. aureus strains that do not produce BSH have been observed to survive less well in whole blood killing assay vs. the parental strain (Figure 7A) (Posada et al., 2014). To date, no data exist as to whether a shifted BSH/BSSB redox ratio would have a similar detrimental effect. Our data suggest that altered BSH redox ratio is indeed linked to reduced survival of S. aureus in human blood and more specifically purified PMNs (Figure 7A, B). PMNs are the dominant cells in the human innate immune response, generating ROS such as H2O2 and HOCl during oxidative burst to kill invading pathogens (Nauseef, 2007b). Interestingly, treatment of PMNs with DPI, an oxidative burst inhibitor, helps abolish the difference in survival between the WT and the ypdA mutant. In addition, overexpression of YpdA also augments S. aureus survival in PMNs. We attribute this increase in S. aureus survival to the enhanced ability of the WT overexpressing YpdA to deal with oxidative stress generated by PMNs. M. tuberculosis mutants with altered MSH/MSSM ratios are also hindered during infection (Nambi et al., 2015). Likewise, Salmonella mutants with altered GSH/GSSG ratios are attenuated in virulence in models of acute salmonellosis (Henard et al., 2010). Taken together, these data emphasize the importance of maintaining redox homeostasis to bacterial survival during host infections.
The rise in antibiotic resistant infections has increased the need to identify new targets for antibiotic development (Spellberg et al., 2008). BSH synthesis pathway has been proposed to be a new target since S. aureus strains lacking BSH production are more susceptible to oxidative stress (Rajkarnikar et al., 2013) and hence survive less well in whole blood killing assays (Posada et al., 2014). Glutathione reductases have long been considered an attractive target for development of antimalarial drugs (Sarma et al., 2003). Our results suggest a related target might be the putative bacillithiol disulfide reductase in S. aureus and possibly in other Gram-positive pathogens.
METHODS AND MATERIALS
Strain construction and growth conditions/assays
Table 1 contains a list of strains used in this study. S. aureus strains were grown in Trypticase Soy Broth (TSB) with shaking at 250 rpm at 37°C unless indicated otherwise. S. aureus strain SH1000 that has been corrected for bacillithiol production, ALC7446, (Posada et al., 2014) was used as the parental strain and is referred to as WT. The temperature sensitive plasmid pMAD was used to achieve homologous recombination in S. aureus; the ypdA (SAOUHSC_01499) gene was replaced by a kanamycin cassette as described previously (Arnaud et al., 2004). Chromosomal deletions were verified by PCR and DNA sequencing. Complemented strain was constructed via pMAD replacement of the kanamycin marker with the WT allele at its native chromosomal locus.
Growth competition assay
Competition experiments were performed as previously described (Wichelhaus et al., 2002). Overnight cultures were diluted 1:500 and allowed to grow to an OD600nm of 0.6 in fresh TSB. Strains were collected by centrifugation, washed in PBS and diluted to 1×105 in media to be used for competition assay. Strains for competition were then mixed 1:1 in a final volume of 5 ml and allowed to grow at 37°C for 18 hrs. Cultures at the beginning and end of experiments were diluted to enumerate CFUs on TSB for both strains or TSB with 30 μg ml−1 kanamycin to select for ypdA mutant. Competition index is calculated as number of mutant colonies divided by WT colonies.
Construction of YpdA overexpressing strains and growth analysis
The ypdA gene was cloned into pALC7925 expression vector, which is a pALC2073 (Bateman et al., 2001) derivative with improved −10 binding site as described by Corrigan and Foster (Corrigan and Foster, 2009), using restriction enzymes KpnI and SacI, transformed into DH5a, and subsequently RN4220, and ALC7446 (WT), selecting for growth on chloramphenicol at 10 μl ml-1. Overnight cultures were diluted 1:500 in fresh TSB and chloramphenicol and induced for expression using 160 ng ml−1 of anhydrotetracycline. Cultures were allowed to reach an optical density of 0.2 at 600nm as measured by Spectronic 20D+ spectrophotometer. Cultures were then divided into 96 well microtiter plates, with oxidative stress and electrophilic stresses added and growth monitored using Tecan Infinite M1000 microplate reader (Tecan Inc.).
Isolation of RNA and Northern blot hybridization
To obtain RNA from cells at different growth phases, overnight S. aureus cultures were diluted 1:1000 in 100 ml of TSB and cell density monitored by absorbance at 600nm. At desired densities, cells were withdrawn and pelleted by centrifugation at 4°C and flash frozen in liquid nitrogen. For stress treatment, cells were stressed with 5mM diamide at OD600 1.2 for 30 minutes, pelleted and frozen. Pellets were resuspended in 1 ml TRIzol (Invitrogen) with 0.25 ml of 0.1 mm diameter glass beads in microcentrifuge tubes and lysed in a Mini-beadbeater 8 (Biospec Products) for two one minutes pulses accompanied by cooling on ice in between. RNA purification and Northern blot analysis were performed as previously described (Cheung et al., 1994).
Primer extension analysis
The 5’ end of the ypdA transcript was mapped by primer extension using anti-sense primer located at position +50 from the ATG start codon. 10 μg of total RNA was extracted from WT cells grown to exponential phase and purified as described above. Primer extension was carried out as previously described (Bayer et al., 1996).
Promoter Fusion
The promoter region comprising 180-bp upstream of the ypdA translation start site was cloned using Gibson assembly (NEB) into a pSK236 plasmid containing a promoterless venus (YFP variant) gene. Recombinant plasmids were verified by restriction digest with EcoRI and XbaI and DNA sequencing, transformed into RN4220, and ALC7446 (WT) and selected on TSB with 10 μg ml−1 chloramphenicol. Overnight cultures were diluted 1:1000 in fresh TSB with chloramphenicol and grown to an OD600nm 0.4, at which point oxidative and electrophilic stresses were added. Fluorescence and growth were monitored simultaneously using Tecan Infinite M1000 microplate reader.
Measuring bacillithiol levels
Levels of reduced and oxidized thiols were measured by HPLC of fluorescent thiol adducts with monobromobimane (mBBr) as previously described (Rajkarnikar et al., 2013). Briefly, overnight cells were diluted 1:1000 in fresh TSB medium, allowed to grow to OD600nm 1.0 and pelleted for analysis. YpdA over-expression and empty vector strains were grown in the presence of 10 μg ml−1 chloramphenicol and 160 ng ml−1 anhydrotetracycline and then exposed to 20 mM H2O2 for 45 min.
Protein purification and antibody generation
The ypdA gene was cloned into pET14b expression vector using NdeI and XhoI cloning sites, transformed into DH5α and subcloned into E. coli BL21. Overnight cultures were diluted 1:50 in LB with 100 μg ml−1 ampicillin. When cells reached an OD600mn of 0.6, protein expression was induced with 0.2 mM IPTG and allowed to grow for 4 more hrs. followed by protein purification. Briefly, cultures were harvested, resuspended in binding buffer [0.5 M NaCl, 20 mM Tris, 5 mM Imidazole, 2.5 mg ml−1 lysozyme and cOmplete mini protease inhibitor cocktail (Roche)] and sonicated with intermittent cooling in an ice bath. Lysate was then loaded on a Ni-NTA column and allowed to flow through. Column was then washed three times with wash buffer (0.5 M NaCl, 20 mM Tris, 20 mM Imidazole), and protein was eluted twice with elution buffer (0.5 M NaCl, 20 mM Tris, 1M Imidazole). Purified protein was dialyzed overnight (40 mM Tris, 200 mM NaCl, 1 mM DTT). Authentication of the protein was rendered by Mass spectrometry. To generate YpdA incapable of binding cofactor (G10A mutation), site directed mutagenesis was performed using Q5 site directed mutagenesis kit (NEB) and purified using the protocol detailed above.
Antibody was raised in BL-6 mice with 3 immunizations of 500 μg each of purified YpdA. Complete Freund’s adjuvant was administered at week 0 whereas incomplete Freund’s adjuvant was given with the protein at 2nd and 4th weeks. On week 6, blood was collected by cardiac puncture after a pre-bleed one week prior to sacrifice demonstrated a titer of >100,000 compared to control sera at week 0 as determined by Western blot with serially diluted sera. Serum fraction from collected blood was then checked for utility by Western blotting against purified YpdA and whole cell lysate.
NAD(P)H consumption assay
To examine enzymatic consumption of cofactor, in vitro reactions were performed with 2.5 μM of purified YpdA, 500 μM of NADPH or NADH in 1× PBS with 1 mM EDTA. Reactions were set up on ice and transferred to Tecan plate reader and monitored at 340 nm for 25 min at 37°C.
Stress response Western blotting
Overnight cultures were diluted 1:500 and allowed to grow to OD600 of 0.4. Stressing agents were added at indicated concentrations and cultures were further incubated for 1 hr. Cells were collected and whole cell lysates prepared using Bug Buster (Millipore Sigma) supplemented with 25 units gram−1 cell paste of Benzonase nuclease, 2.5 mg ml−1 lysozyme, and cOmplete mini protease inhibitor cocktail (Roche). Protein concentrations were determined by BCA assays (Pierce). Fifteen μg of lysate was run on a 12.5% acrylamide gel and transferred to PVDF membrane using the iBlot2 system (Invitrogen). Membranes were blocked with 5% Milk for 1 hr. at room temperature (RT), incubated with anti-YpdA serum (1:1000 dilution) overnight at 4°C, and probed with anti-mouse HRP (Jackson labs, 1:5000 dilution) for 1 hr. at RT. Blots were developed using Pierce ECL western blotting substrate.
Whole blood survival assay
Blood was collected from healthy human volunteers in accordance with protocol approved by the Institutional Review Board for Human Subjects. All human subjects were given informed consent prior to participation in the study. Whole blood assays were performed as previously described (Liu et al., 2005). Briefly, venous blood was collected into sterile heparin coated tubes and mixed gently. Overnight bacterial cultures were diluted 1:200 in fresh TSB. Cells were grown for several hours, washed with PBS, diluted to an inoculum of 1 × 105 CFU ml−1. 25 μl of bacteria was mixed with 75 μl whole blood and rotated at 37°C for 2 hours. At specific timepoints, 20 μl aliquots of blood and bacteria reactions were serially diluted in PBS, plated on TSB agar and incubated at 37°C overnight. Initial inoculum was also determined by serial dilutions. Colonies were counted the following morning and percent survival from initial inoculum was calculated.
Neutrophil isolation and assay
PMNs were purified according to previously described protocol (Nauseef, 2007a). Briefly, fresh heparinized venous blood was diluted with 1 volume of prewarmed RPMI and overlaid on 10 mL of Hypaque-Ficoll; blood components were separated by 40 min spin at 400 RCF at RT. Supernatant, mononuclear band, and gradient above the PMN-erythrocyte pellet were discarded. Pellet was resuspended in Hank’s balanced salt solution (HBSS) without Mg2+/Ca2+, and 25 ml of 6% dextran added. Tubes were incubated for 20 min at RT and PMN layer was collected and centrifuged. Contaminating erythrocytes were lysed with sterile H2O and tonicity restored with saline solution. PMNs were washed with HBSS and resuspended to 1 ×105 cells ml−1 in RPMI supplemented with 10% fetal bovine serum. Bacterial cells were then opsonized in 10% human serum for 20 min at 37°C prior to infection of PMNs at a multiplicity of infection of 1 as determined by pilot studies and rotated at 37°C for 1 hour. Initial bacterial inoculum was determined by serial dilutions. Aliquots of PMNs and bacteria were serially diluted in H2O at pH 11, plated on TSA with 10 μg ml−1 of chloramphenicol, incubated at 37°C overnight. Colonies were enumerated, and percent survival of the initial inoculum calculated.
RNA-Seq experiment.
Samples for RNA-Seq were prepared from the following strains, the WT, and its isogenic derivative ypdA mutant. Briefly, overnight cultures were diluted 1:200 in fresh TSB (an initial OD600 of 0.02 UA was obtained) and grown at 37ºC under constant agitation (250 rpm) for about 3 h until an OD600 of 0.8 UA. In specific samples, freshly prepared diamide was added at a final concentration of 5 mM for 30 minutes. Two independent cultures for each sample were grown and total cellular RNAs from two biological replicates were performed as described previously (Kim et al., 2014) using the BioSpec reciprocating device in the presence of Trizol. RNA was quantified by using Qubit (Life Technologies) and RNA integrity was assessed with a 2100 Bioanalyzer (Agilent Technologies). One µg of total RNA was ribo-depleted with the bacterial Ribo-Zero kit from Illumina. The truseq total RNA stranded kit from Illumina was used for the library preparation. Library quantity was measured by the Qubit and quality was assessed on a Tapestation on a DNA High sensitivity chip (Agilent Technologies). The libraries were pooled at equimolarity and loaded at 2 nM for clustering. Oriented 50 bases single-read sequencing was performed on the Illumina HiSeq 4000 sequencer yielding a minimum of 4 million mapped reads per sample. Final RNA-Seq analysis and data analysis were carried out using previous described procedures (Robinson et al., 2010; Anders et al., 2013). To detect differentially expressed transcripts between various strains, pairwise comparisons were performed and statistical significance (FDR-adjusted P<.0.05) determined with R v3.2.3 using the edgeR package as described previously (Robinson et al., 2010; Anders et al., 2013).
Supplementary Material
Acknowledgments
We would like to acknowledge Adhar Manna for help with primer extension and Northern blot experiments and Niles Donegan and John Helmann for critical reading of the manuscript. Mass spectrometry was performed at the VGN Proteomics facility which was supported by an Institutional Development Award (IDeA) from the National Institute of General Medical Sciences of the National Institutes of Health under grant number P20GM103449. Research reported in this publication was supported by National Institutes of Health under T32-AI007519 to IVM, R21 AI119579 to ALC and SC3GM-100855-03 to MR.
References
- Anders S, McCarthy DJ, Chen YS, Okoniewski M, Smyth GK, Huber W, and Robinson MD (2013) Count-based differential expression analysis of RNA sequencing data using R and Bioconductor. Nat Protoc 8: 1765–1786. [DOI] [PubMed] [Google Scholar]
- Archer GL (1998) Staphylococcus aureus: A well-armed pathogen. Clin Infect Dis 26: 1179–1181. [DOI] [PubMed] [Google Scholar]
- Arnaud M, Chastanet A, and Débarbouillé M (2004) New vector for efficient allelic replacement in naturally nontransformable, low-GC-content, gram-positive bacteria. Appl Environ Microbiol 70: 6887–6891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barendt S, Lee H, Birch C, Nakano MM, Jones M, and Zuber P (2013) Transcriptomic and phenotypic analysis of paralogous spx gene function in Bacillus anthracis Sterne. Microbiologyopen 2: 695–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman BT, Donegan NP, Jarry TM, Palma M, and Cheung AL (2001) Evaluation of a tetracycline-inducible promoter in Staphylococcus aureus in vitro and in vivo and its application in demonstrating the role of sigB in microcolony formation. Infect Immun 69: 7851–7857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayer MG, Heinrichs JH, and Cheung AL (1996) The molecular architecture of the sar locus in Staphylococcus aureus. J Bacteriol 178: 4563–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beutler E (1969) Effect of flavin compounds on glutathione reductase activity: in vivo and in vitro studies. J Clin Invest 48: 1957–1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bore E, Langsrud S, Langsrud Ø, Rode TM, and Holck A (2007) Acid-shock responses in Staphylococcus aureus investigated by global gene expression analysis. Microbiology 153: 2289–2303. [DOI] [PubMed] [Google Scholar]
- Bragg PD, Glavas NA, and Hou C (1997) Mutation of Conserved Residues in the NADP(H)-Binding Domain of the Proton Translocating Pyridine Nucleotide Transhydrogenase of Escherichia coli. Arch Biochem Biophys 338: 57–66. [DOI] [PubMed] [Google Scholar]
- Carlberg I, and Mannervik B (1985) Glutathione reductase. Methods Enzymol 113: 484–490. [DOI] [PubMed] [Google Scholar]
- Carmel-Harel O, and Storz G (2000) Roles of the glutathione- and thioredoxin-dependent reduction systems in the Escherichia coli and Saccharomyces cerevisiae responses to oxidative stress. Annu Rev Microbiol 54: 439–461. [DOI] [PubMed] [Google Scholar]
- Chaffin DO, Taylor D, Skerrett SJ, and Rubens CE (2012) Changes in the Staphylococcus aureus transcriptome during early adaptation to the lung. PLoS One 7: e41329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers HF, and DeLeo FR (2009) Waves of resistance: Staphylococcus aureus in the antibiotic era. Nat Rev Microbiol 7: 629–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrangsu P, Dusi R, Hamilton CJ, and Helmann JD (2014) Methylglyoxal resistance in Bacillus subtilis: contributions of bacillithiol-dependent and independent pathways. Mol Microbiol 91: 706–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrangsu P, Loi V Van, Antelmann H, and Helmann JD (2017) The Role of Bacillithiol in Gram-Positive Firmicutes. Antioxid Redox Signal 28: 445–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung AL, Eberhardt KJ, and Fischetti VA (1994) A Method to Isolate RNA from Gram-Positive Bacteria and Mycobacteria. Anal Biochem 222: 511–514. [DOI] [PubMed] [Google Scholar]
- Chi BK, Roberts AA, Huyen TTT, Bäsell K, Becher D, Albrecht D, et al. (2013) S-bacillithiolation protects conserved and essential proteins against hypochlorite stress in firmicutes bacteria. Antioxid Redox Signal 18: 1273–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corrigan RM, and Foster TJ (2009) An improved tetracycline-inducible expression vector for Staphylococcus aureus. Plasmid 61: 126–129. [DOI] [PubMed] [Google Scholar]
- Couto N, Wood J, and Barber J (2016) The role of glutathione reductase and related enzymes on cellular redox homeostasis network. Free Radic Biol Med 95: 27–42. [DOI] [PubMed] [Google Scholar]
- DeLeo F, Diep B, and Otto M (2009) Host Defense and Pathogenesis in Staphylococcus aureus Infections. Infect Dis Clin North Am 23: 17–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fomenko DE, and Gladyshev VN (2002) CxxS: fold-independent redox motif revealed by genome-wide searches for thiol/disulfide oxidoreductase function. Protein Sci 11: 2285–2296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fomenko DE, and Gladyshev VN (2003) Identity and functions of CxxC-derived motifs. Biochemistry 42: 11214–11225. [DOI] [PubMed] [Google Scholar]
- Gaballa A, Antelmann H, Hamilton CJ, and Helmann JD (2013) Regulation of Bacillus subtilis bacillithiol biosynthesis operons by Spx. Microbiology 159: 2025–2035. [DOI] [PubMed] [Google Scholar]
- Gaballa A, Chi BK, Roberts AA, Becher D, Hamilton CJ, Antelmann H, and Helmann JD (2014) Redox regulation in Bacillus subtilis: The bacilliredoxins BrxA(YphP) and BrxB(YqiW) function in de-bacillithiolation of S-bacillithiolated OhrR and MetE. Antioxid Redox Signal 21: 357–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaballa A, Newton GL, Antelmann H, Parsonage D, Upton H, Rawat M, et al. (2010) Biosynthesis and functions of bacillithiol, a major low-molecular-weight thiol in Bacilli. Proc Natl Acad Sci 107: 6482–6486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaskill ME, and Khan SA (1988) Regulation of the enterotoxin B gene in Staphylococcus aureus. J Biol Chem 263: 6276–80. [PubMed] [Google Scholar]
- Grosser MR, Paluscio E, Thurlow LR, Dillon MM, Cooper VS, Kawula TH, and Richardson AR (2018) Genetic requirements for Staphylococcus aureus nitric oxide resistance and virulence. PLoS Pathog 14: e1006907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampton MB, and Winterbourn CC (1995) Modification of neutrophil oxidant production with diphenyleneiodonium and its effect on bacterial killing. Free Radic Biol Med 18: 633–639. [DOI] [PubMed] [Google Scholar]
- Han J, He L, Shi W, Xu X, Wang S, Zhang S, and Zhang Y (2014) Glycerol Uptake Is Important for L-Form Formation and Persistence in Staphylococcus aureus. PLoS One 9: e108325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henard CA, Bourret TJ, Song M, and Vázquez-Torres A (2010) Control of redox balance by the stringent response regulatory protein promotes antioxidant defenses of Salmonella. J Biol Chem 285: 36785–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochgräfe F, Wolf C, Fuchs S, Liebeke M, Lalk M, Engelmann S, and Hecker M (2008) Nitric oxide stress induces different responses but mediates comparable protein thiol protection in Bacillus subtilis and Staphylococcus aureus. J Bacteriol 190: 4997–5008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imber M, Huyen NTT, Pietrzyk-Brzezinska AJ, Loi V Van, Hillion M, Bernhardt J, et al. (2018) Protein S-Bacillithiolation Functions in Thiol Protection and Redox Regulation of the Glyceraldehyde-3-Phosphate Dehydrogenase Gap in Staphylococcus aureus Under Hypochlorite Stress. Antioxid Redox Signal 28: 410–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph B, Przybilla K, Stühler C, Schauer K, Slaghuis J, Fuchs TM, and Goebel W (2006) Identification of Listeria monocytogenes genes contributing to intracellular replication by expression profiling and mutant screening. J Bacteriol 188: 556–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan MT, Dijl J.M. van, and Harmsen HJM (2014) Antioxidants Keep the Potentially Probiotic but Highly Oxygen-Sensitive Human Gut Bacterium Faecalibacterium prausnitzii Alive at Ambient Air. PLoS One 9: e96097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Reyes D, Beaume M, Francois P, and Cheung A (2014) Contribution of teg49 small RNA in the 5’ upstream transcriptional region of sarA to virulence in Staphylococcus aureus. Infect Immun 82: 4369–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreiswirth BN, Löfdahl S, Betley MJ, O’Reilly M, Schlievert PM, Bergdoll MS, and Novick RP (1983) The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature 305: 709–12. [DOI] [PubMed] [Google Scholar]
- Lam TN Le, Morvan C, Liu W, Bohn C, and Jaszczyszyn Y (2017) Finding sRNA-associated phenotypes by competition assays: An example with Staphylococcus aureus. Methods 117: 21–27. [DOI] [PubMed] [Google Scholar]
- Leonardi R, and Jackowski S (2007) Biosynthesis of Pantothenic Acid and Coenzyme A. EcoSal Plus 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu GY, Essex A, Buchanan JT, Datta V, Hoffman HM, Bastian JF, et al. (2005) Staphylococcus aureus golden pigment impairs neutrophil killing and promotes virulence through its antioxidant activity. J Exp Med 202: 209–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luikenhuis S, Perrone G, Dawes IW, and Grant CM (1998) The Yeast Saccharomyces cerevisiae Contains Two Glutaredoxin Genes That Are Required for Protection against Reactive Oxygen Species. Mol Biol Cell 9: 1081–1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mäder U, Nicolas P, Depke M, Pané-Farré J, Debarbouille M, Kooi-Pol MM van der, et al. (2016) Staphylococcus aureus Transcriptome Architecture: From Laboratory to Infection-Mimicking Conditions. PLoS Genet 12: e1005962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandell GL (1975) Catalase, superoxide dismutase, and virulence of Staphylococcus aureus. In vitro and in vivo studies with emphasis on staphylococcal--leukocyte interaction. J Clin Invest 55: 561–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masip L, Veeravalli K, and Georgiou G (2006) The Many Faces of Glutathione in Bacteria. Antioxid Redox Signal 8: 753–762. [DOI] [PubMed] [Google Scholar]
- Michán C, Manchado M, Dorado G, and Pueyo C (1999) In vivo transcription of the Escherichia coli oxyR regulon as a function of growth phase and in response to oxidative stress. J Bacteriol 181: 2759–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano S, Küster-Schöck E, Grossman AD, and Zuber P (2003) Spx-dependent global transcriptional control is induced by thiol-specific oxidative stress in Bacillus subtilis. Proc Natl Acad Sci U S A 100: 13603–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nambi S, Long JE, Mishra BB, Baker R, Murphy KC, Olive AJ, et al. (2015) The Oxidative Stress Network of Mycobacterium tuberculosis Reveals Coordination between Radical Detoxification Systems. Cell Host Microbe 17: 829–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nauseef WM (2007a) Isolation of human neutrophils from venous blood. Methods Mol Biol 412: 15–20. [DOI] [PubMed] [Google Scholar]
- Nauseef WM (2007b) How human neutrophils kill and degrade microbes: an integrated view. Immunol Rev 219: 88–102. [DOI] [PubMed] [Google Scholar]
- Newton GL, Arnold K, Price MS, Sherrill C, Delcardayre SB, Aharonowitz Y, et al. (1996) Distribution of thiols in microorganisms: mycothiol is a major thiol in most actinomycetes. J Bacteriol 178: 1990–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton GL, Rawat M, Clair J.J. La Jothivasan VK, Budiarto T, Hamilton CJ, et al. (2009) Bacillithiol is an antioxidant thiol produced in Bacilli. Nat Chem Biol 5: 625–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pamp SJ, Frees D, Engelmann S, Hecker M, and Ingmer H (2006) Spx is a global effector impacting stress tolerance and biofilm formation in Staphylococcus aureus. J Bacteriol 188: 4861–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posada AC, Kolar SL, Dusi RG, Francois P, Roberts AA, Hamilton CJ, et al. (2014) Importance of bacillithiol in the oxidative stress response of Staphylococcus aureus. Infect Immun 82: 316–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pöther DC, Gierok P, Harms M, Mostertz J, Hochgräfe F, Antelmann H, et al. (2013) Distribution and infection-related functions of bacillithiol in Staphylococcus aureus. Int J Med Microbiol 303: 114–123. [DOI] [PubMed] [Google Scholar]
- Protchenko OV, Boretsky YuR, Romanyuk TM, and Fedorovych DV Oversynthesis of riboflavin by yeast Pichia guilliermondii in response to oxidative stress. Ukr biokhimichnyi zhurnal (1999) 72: 19–23. [PubMed] [Google Scholar]
- Rajkarnikar A, Strankman A, Duran S, Vargas D, Roberts AA, Barretto K, et al. (2013) Analysis of mutants disrupted in bacillithiol metabolism in Staphylococcus aureus. Biochem Biophys Res Commun 436: 128–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, and Smyth GK (2010) edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosario-Cruz Z, Chahal HK, Mike LA, Skaar EP, and Boyd JM (2015) Bacillithiol has a role in Fe-S cluster biogenesis in Staphylococcus aureus. Mol Microbiol 98: 218–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarma GN, Savvides SN, Becker K, Schirmer M, Schirmer RH, and Karplus PA (2003) Glutathione Reductase of the Malarial Parasite Plasmodium falciparum: Crystal Structure and Inhibitor Development. J Mol Biol 328: 893–907. [DOI] [PubMed] [Google Scholar]
- Schlag S, Nerz C, Birkenstock TA, Altenberend F, and Götz F (2007) Inhibition of staphylococcal biofilm formation by nitrite. J Bacteriol 189: 7911–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma SV, Arbach M, Roberts AA, Macdonald CJ, Groom M, and Hamilton CJ (2013) Biophysical features of bacillithiol, the glutathione surrogate of Bacillus subtilis and other firmicutes. ChemBioChem 14: 2160–2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Si M, Zhao C, Zhang B, Wei D, Chen K, Yang X, et al. (2016) Overexpression of Mycothiol Disulfide Reductase Enhances Corynebacterium glutamicum Robustness by Modulating Cellular Redox Homeostasis and Antioxidant Proteins under Oxidative Stress. Sci Rep 6: 29491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnova GV, Muzyka NG, and Oktyabrsky ON (2005) Effects of cystine and hydrogen peroxide on glutathione status and expression of antioxidant genes in Escherichia coli. Biochemistry (Mosc) 70: 926–34. [DOI] [PubMed] [Google Scholar]
- Spaan AN, Surewaard BGJ, Nijland R, and Strijp JAG van (2013) Neutrophils Versus Staphylococcus aureus : A Biological Tug of War. Annu Rev Microbiol 67: 629–650. [DOI] [PubMed] [Google Scholar]
- Spellberg B, Guidos R, Gilbert D, Bradley J, Boucher HW, Scheld WM, et al. (2008) The Epidemic of Antibiotic-Resistant Infections: A Call to Action for the Medical Community from the Infectious Diseases Society of America. Clin Infect Dis 46: 155–164. [DOI] [PubMed] [Google Scholar]
- Storz G, Tartaglia LA, G S, and La T (1992) OxyR: A Regulator of Antioxidant Genes. J Nutr 122: 627–630. [DOI] [PubMed] [Google Scholar]
- Tan S-X, Greetham D, Raeth S, Grant CM, Dawes IW, and Perrone GG (2010) The thioredoxin-thioredoxin reductase system can function in vivo as an alternative system to reduce oxidized glutathione in Saccharomyces cerevisiae. J Biol Chem 285: 6118–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tillmann AT, Strijbis K, Cameron G, Radmaneshfar E, Thiel M, Munro CA, et al. (2015) Contribution of Fdh3 and Glr1 to Glutathione Redox State, Stress Adaptation and Virulence in Candida albicans. PLoS One 10: e0126940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong SYC, Davis JS, Eichenberger E, Holland TL, Fowler VG, and Jr (2015) Staphylococcus aureus infections: epidemiology, pathophysiology, clinical manifestations, and management. Clin Microbiol Rev 28: 603–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uziel O, Borovok I, Schreiber R, Cohen G, and Aharonowitz Y (2004) Transcriptional regulation of the Staphylococcus aureus thioredoxin and thioredoxin reductase genes in response to oxygen and disulfide stress. J Bacteriol 186: 326–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins RR, David MZ, and Salata RA (2012) Current concepts on the virulence mechanisms of meticillin-resistant Staphylococcus aureus. J Med Microbiol 61: 1179–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wichelhaus TA, Böddinghaus B, Besier S, Schäfer V, Brade V, and Ludwig A (2002) Biological cost of rifampin resistance from the perspective of Staphylococcus aureus. Antimicrob Agents Chemother 46: 3381–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winterbourn CC, and Kettle AJ (2013) Redox Reactions and Microbial Killing in the Neutrophil Phagosome. Antioxid Redox Signal 18: 642–660. [DOI] [PubMed] [Google Scholar]
- Yoshimura M, Oshima T, and Ogasawara N (2007) Involvement of the YneS/YgiH and PlsX proteins in phospholipid biosynthesis in both Bacillus subtilis and Escherichia coli. BMC Microbiol 7: 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zitka O, Skalickova S, Gumulec J, Masarik M, Adam V, Hubalek J, et al. (2012) Redox status expressed as GSH:GSSG ratio as a marker for oxidative stress in paediatric tumour patients. Oncol Lett 4: 1247–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


