Abstract
The effectiveness of malaria screening and treatment highly depends on the low-cost access to the highly sensitive and specific malaria test. We report a real-time fluorescence nucleic acid testing device for malaria field detection with automated and scalable sample preparation capability. The device consists a compact analyzer and a disposable microfluidic reagent compact disc. The parasite DNA sample preparation and subsequent real-time LAMP detection were seamlessly integrated on a single microfluidic compact disc, driven by energy efficient non-centrifuge based magnetic field interactions. Each disc contains four parallel testing units which could be configured either as four identical tests or as four species-specific tests. When configured as species-specific tests, it could identify two of the most life-threatening malaria species (P. falciparum and P. vivax). The NAT device is capable of processing four samples simultaneously within 50 min turnaround time. It achieves a detection limit of ~0.5 parasites/μl for whole blood, sufficient for detecting asymptomatic parasite carriers. The combination of the sensitivity, specificity, cost, and scalable sample preparation suggests the real-time fluorescence LAMP device could be particularly useful for malaria screening in the field settings.
Keywords: nucleic acid tests, malaria, point-of-care, blood, centrifuge-free, lab-on-a-disc
1. Introduction
Malaria is a mosquito-borne disease caused by Plasmodium parasites, predominately in resource-limiting areas of low- and middle-income countries. Among 5 parasite species, P. falciparum (Pf) and P. vivax (Pv) pose the greatest threat to the human. Pf is the most prevalent malaria parasite on the African continent. Pv is the dominant malaria parasite in most countries outside of sub-Saharan Africa (WHO, 2017). Increased malaria control efforts have resulted in dramatic reduction in the global malaria incidence over the past decade. The World Health Organization (WHO) thus endorsed the ambitious goal of achieving worldwide malaria elimination and eradication (WHO, 2017). A change in focus from malaria control to elimination requires identification and treatment of both symptomatic and asymptomatic carriers to reduce the parasite reservoir and interrupt the malaria transmission. As a result, the effectiveness of elimination strategies highly depends on the low-cost access to the sensitive and specific malaria screening tests (Slater et al., 2015). Current screening test methods rely almost exclusively on microscopy (thin and thick blood smears) and immunological rapid diagnostic tests (RDTs, detecting antigens in human blood). They perform sufficiently well in high transmission regions for diagnosing people with symptomatic malaria. Nevertheless, both methods could miss a significant portion of asymptomatic parasite carriers in low-transmission areas thanks to the detection limit of ~100 parasites/μl (Moody, 2002; Wongsrichanalai et al., 2007). During the course of malaria elimination, the proportion of low-density and asymptomatic infections increases, thus rapid and highly sensitive point-of-care field test is increasingly needed to identify all infected individuals for treatment.
Lower parasite density can be identified by nucleic acid tests (NATs), often by PCR, which has an excellent detection limit of <1 parasite/μl depending on the assay type (Han et al., 2007; Reddy et al., 2012; Snounou et al., 1993). However, PCR-based assays are poorly suited to perform in field settings as they require specialized equipment for sample preparation and skilled personnel (Britton et al., 2016b). Alternative NATs, such as loop-mediated isothermal amplification (LAMP) assays (Abdul-Ghani, 2014; Goyal et al., 2015; Han, 2013; Hsiang et al., 2014; Morris et al., 2015; Oriero et al., 2015; Patel et al., 2014; Sattabongkot et al., 2014; Singh et al., 2013; Sirichaisinthop et al., 2011; Surabattula et al., 2013), recombinase polymerase amplification (RPA) assays (Cordray and Richards-Kortum, 2015; Kersting et al., 2014), as well as helicase dependent amplification (HAD) assays (Li et al., 2013) have shown high analytical sensitivity and great potential for field deployment by integrating with microfluidic (Liu et al., 2016) and paper based devices (Xu et al., 2016). Among isothermal methods, LAMP is the most studied assay for malaria detection and holds great promise for commercialization. For instance, the illumigene malaria LAMP assays could reliably and sensitively detect Plasmodium by combining a manual blood sample preparation step with an incubator reader (Lucchi et al., 2016). Despite great effort and progress towards field deployment of malaria NATs assays, highly integrated DNA sample preparation from raw peripheral blood for molecular assays remains a bottleneck (Dineva et al., 2007; Kolluri et al., 2018). Current sample preparation usually involves lengthy or error-prone manual processes such as gravity-driven filtration (Lucchi et al., 2016), centrifugation (Sema et al., 2015). Although a few emerging point of care NAT devices, such as Alere q system (Hsiao et al., 2016; Jani et al., 2016), cobas Liat system (Nolte et al., 2016) and Cepheid Xpert Omni platform (Gous et al., 2016), have successfully integrated the sample preparation step, a low-cost LAMP-based malaria NATs is not readily available for screening test. While centrifuge-based lab-on-a-disc (Kim et al., 2014; Kong et al., 2016; Madou et al., 2006; Nolte, 2009) is a promising technology to integrate DNA extraction on the chip, centrifugal force is non-linear in nature and energy-hungry thanks to the rotational frequencies in the range of several thousand revolutions per minute (RPM) (Madou et al., 2006). For the promising LAMP technologies to be used as malaria screening tests in field settings, the DNA extraction method should be simple, rapid, scalable, fully automated, free of cross-contamination and seamlessly integrated with the amplification for immediate analysis.
In this work, we report a real-time fluorescence LAMP device suitable for field detection of Pf and Pv with automated and scalable sample preparation capability. The device uses a non-centrifugal method for solid phase DNA extraction by actuating the DNA-carrying magnetic beads against the stationary reagent droplets. The device consists of a palm-sized analyzer and an enclosed microfluidic reagent compact disc. The reagents were preloaded and separated on the microfluidic reagent disk by teeth-shaped passive valves. The pre-loaded and ready-to-use microfluidic reagent disc contains four parallel testing units. It could be configured either as four identical tests to increase the testing throughput or as four species-specific tests to distinguish Plasmodium genus, Pf and Pv species. Each test unit automatically performs the parasite DNA binding, washing, elution and immediate real-time isothermal amplification and fluorescence detection. This seamless integration from sample to result on a single microfluidic reagent compact disc greatly minimized the manual workload needed for performing the NATs. The device could deliver sensitive (~0.5 parasites/μl) NAT results directly from a small volume of whole blood samples within 50 minutes for a material cost around $1/test.
2. Materials and methods
2.1. Pf and Pv-infected blood sample
Complete RPMI 1640 medium with type O+ human red blood cells (RBCs) was used to continuously culture Pf 3D7 (Trager and Jensen, 1976). The parasite synchronization was performed by treating 5% D-sorbitol to remove the mature stages and only maintain the ring-stage parasites. The fresh culture medium was replaced on a daily basis. The ring stage parasites were adjusted to 45% hematocrit to mimic the whole blood sample obtained from patients. Before the nucleic acid testing, parasitemia was assessed by the Giemsa-stained blood smears. Pv-infected blood samples were obtained from BEI Resources Repository (MRA-369 Pv strain Achiote, and MRA-383 Pv strain Chesson). The genomic DNA from these blood samples was purified by phenol-chloroform extraction procedure as described before (Miao et al., 2010).
2.2. LAMP reaction mix
The LAMP reaction mix (25 μl) contains isothermal buffer (20 mM Tris-HCl, 10 mM (NH4)2SO4, 50 mM KCl, 2 mM MgSO4, 0.1% Tween 20, pH 8.8), PCR grade H2O, Betaine (0.4 M), MgSO4 (7 mM), MnCl2 (0.75 mM), calcein (25 μM), deoxyribonucleotide triphosphates (dNTPs, 1.4 mM), Bst 2.0 DNA polymerase, DNA template, and primer sets (0.2 mM F3 and B3c, 1.6 mM FIP and BIP, 0.8 mM LPF and LPB). For pan-Plasmodium detection, we adopted a primer set targeting mitochondrial gene common to all Plasmodium species (Polley et al., 2010). For Pf-specific detection, a primer set to specifically detect the mitochondrial gene of Pf was used (Choi et al., 2016; Polley et al., 2010). For Pv-specific detection, we used a primer set targeting cox1 gene of Pv (Britton et al., 2016a). All primer sets were listed in Supplementary Table S1. It is noteworthy the Pv primers used here could also cross-react with P. knowlesi species (Britton et al., 2016a).
2.3. Instrumentation
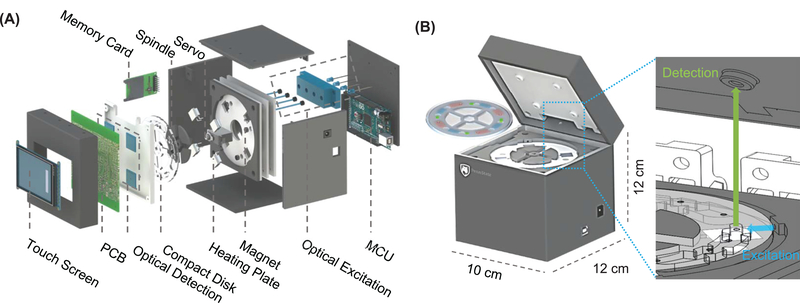
Figure 1A shows an exploded view of the real-time fluorescence LAMP device. The mobile platform was designed in SolidWorks and prototyped by a 3D printer. The device has four parallel testing units and has a small footprint of 10×12×12 cm3 (Figure 1B). The fully integrated device consists of thermal, optical, electromechanical and data subsystems. Thermal subsystem. Four resistive-heating elements were connected in series to maintain the uniform temperature in each testing unit. Each power resistor was bonded to the backside of an aluminum heating plate by thermal paste. A micro-thermistor was embedded in each heating plate for real-time temperature monitoring. Negative thermal feedback was used to maintain the desired temperature during the DNA amplification. Optical subsystem. For real-time tracking of the DNA amplification, we used four LED light sources (λ=488nm). Each of them was directed towards individual reaction chambers through polymer optical fibers. Each LED was connected with a potentiometer to ensure the uniformity of the excitation light. The incidence of the excitation LED lights was perpendicular to the optical sensors to minimize the excitation interference (zoomed illustration in Figure 1B). Electromechanical subsystem. A customized printed circuit board (PCB) with an embedded microcontroller unit (MCU) operates the whole device from sample preparation to result report. A rechargeable 9V portable Lithium-ion battery powers the entire system and could last for >15 hours before recharging. Data subsystem. The fluorescence intensity from four optical sensing units was sampled at a constant interval and displayed on the LCD touchscreen in real-time. The signal was smoothed by a built-in moving average algorithm with background noise removed. The threshold time (Tt) was obtained when the slope of RFU (dRFU/dt) reached the peak (Supplementary Figure S1). Detailed bill of material to replicate our device is listed in Supplementary Table S1.
Figure 1.
Overview of the device. (A) Exploded view of the device, showing the assembly of various components. (B) Schematic of the assembled device and the quadplex microfluidic reagent compact disc. The form factor of the analyzer is palm-sized. The reagent compact disc is secured to the spindle platter. A real-time fluorescence sensing scheme is integrated on the analyzer.
2.4. Microfluidic reagent compact disc
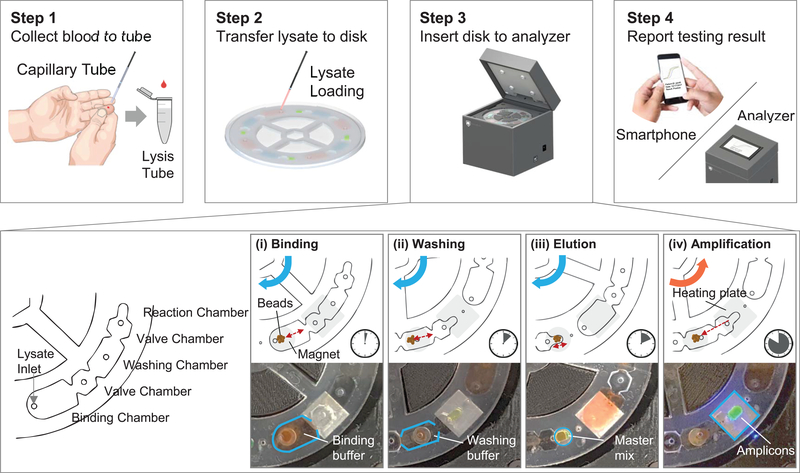
The microfluidic reagent compact disc has a diameter of 9.6 cm and thickness of 3.2 mm. The top, spacer, and bottom polymethyl methacrylate (PMMA) layers were designed using AutoCAD and patterned by a CO2 laser cutting machine (Epilog Helix 24 Laser System). All three layers were aligned using alignment hole and permanently laminated with adhesive solvent. The assembled disc has four testing units and each unit contains three working chambers (Figure 2): binding (210 μl), washing (150 μl), and reaction chambers (25 μl). Liquid phase reagents (beads, washing buffer and master mix) were loaded into the disc immediately prior to use. The reagents could also be pre-loaded into the disc and would last for at least 1 week if stored at 4 °C (Figure 5). Each chamber was isolated by a valve chamber (80 μl) to prevent the reagent mixing. The valving chambers were filled with FC-40 oil or air. The FC-40 oil, which seals the LAMP reaction chamber, helped prevent master mix evaporation during the thermal process. The air-filled valve was surface treated with water-oil repellent to create a barrier for the amphiphilic lysis buffer. After the sample loading, the whole disc was sealed with a PMMA cap to avoid potential cross-contamination.
Figure 2.
Workflow of the device. In step 1, whole blood is collected into the lysis buffer by the capillary tube. In step 2, the lysate is loaded into the binding chamber of the four testing units. In step 3, the disc is sealed and inserted into the analyzer, which then performs automated sample preparation and amplification. This automated process consists of four steps: binding (3 min), washing (4 min), elution (3 min) and amplification (40 min). During the amplification process, the real-time fluorescence signal from each testing unit was recorded and analyzed. Finally, in step 4, the testing results are reported.
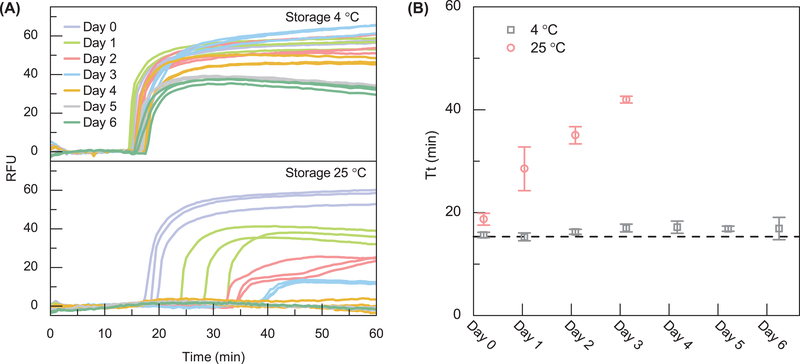
Figure 5.
LAMP reagent thermostability test. (A) The real-time amplification curve for LAMP reagent stored at 4 °C and 25 °C. (B) Extracted amplification threshold time (Tt). When stored at 4 °C, the reagent shows a negligible threshold time drift for a week. When stored at 25 °C, the reagent shows a drift towards higher Tt (decreased activity) over time and no amplification is seen after day 3. The dashed line denotes the freshly prepared LAMP reagent.
2.5. Testing workflow
The workflow of the device consists of four steps (Figure 2 and Supplementary Video S1). The 20 μl of finger-prick blood was collected using a capillary tube and lysed in the collection tube filled with 1000 μl of lysis buffer. 180 μl of blood lysate was transferred into each binding chamber of the testing units on the reagent compact disc. After loading the sample, the disc was sealed with PSA tape and inserted into the mobile analyzer for a streamlined nucleic acid sample preparation and amplification process (enlarged view of step 3). During the amplification, the fluorescence intensity data were recorded on a non-volatile memory card and displayed on the LCD screen in real-time. Users also have an option to receive the results using a smartphone.
3. Results and Discussion
3.1. Non-centrifugal based streamlined sample preparation
One of the significant challenges for NATs at the point of care is related to the front end of the assays - nucleic acid extraction from raw samples (Dineva et al., 2007). For malaria mass screening applications, the ideal sample preparation should be simple, scalable and easy-to-operate. We realized a streamlined process for preparing four samples in parallel on a single enclosed microfluidic disc. In contrast to the conventional lab-on-a-disk devices that rely on energy-hungry centrifugal forces (Kim et al., 2014; Madou et al., 2006; Nolte, 2009) (Kong et al., 2016), our device operates with a non-centrifugal and energy efficient magnetic interaction method. The process for each sample consists of the following three steps: binding, washing, and elution (step 3 of Figure 2 and Supplementary Video S2). The negatively charged parasite DNAs first bind to the pH-sensitive charge-switchable magnetic beads (ChargeSwitch® Forensic DNA Purification Kits, Invitrogen) at pH 5. During the binding process (~3 min), the reagent compact disc was rotated back and forth slowly to ensure thorough mixing of the beads and the lysate. The DNA-binding magnetic beads were then transferred to the washing chamber by magnetic actuation. The washing process lasts for about 4 min, and the magnetic beads with purified DNAs were further transferred to the reaction chamber (LAMP master mix). The LAMP master mix has a pH of 8.8, which switches the surface charge of the magnetic beads towards negative. The negatively charged DNAs were therefore repelled off from the magnetic beads and eluted into the master mix. After that, the residual magnetic beads were removed from the reaction chamber before initiating the LAMP reaction. The entire sample preparation was multiplexed for four samples and could be finished in less than 10 minutes with minimal user intervention. The sample numbers could be easily scaled up if needed in the future.
3.2. Fluorescence sensing
Uniformity.
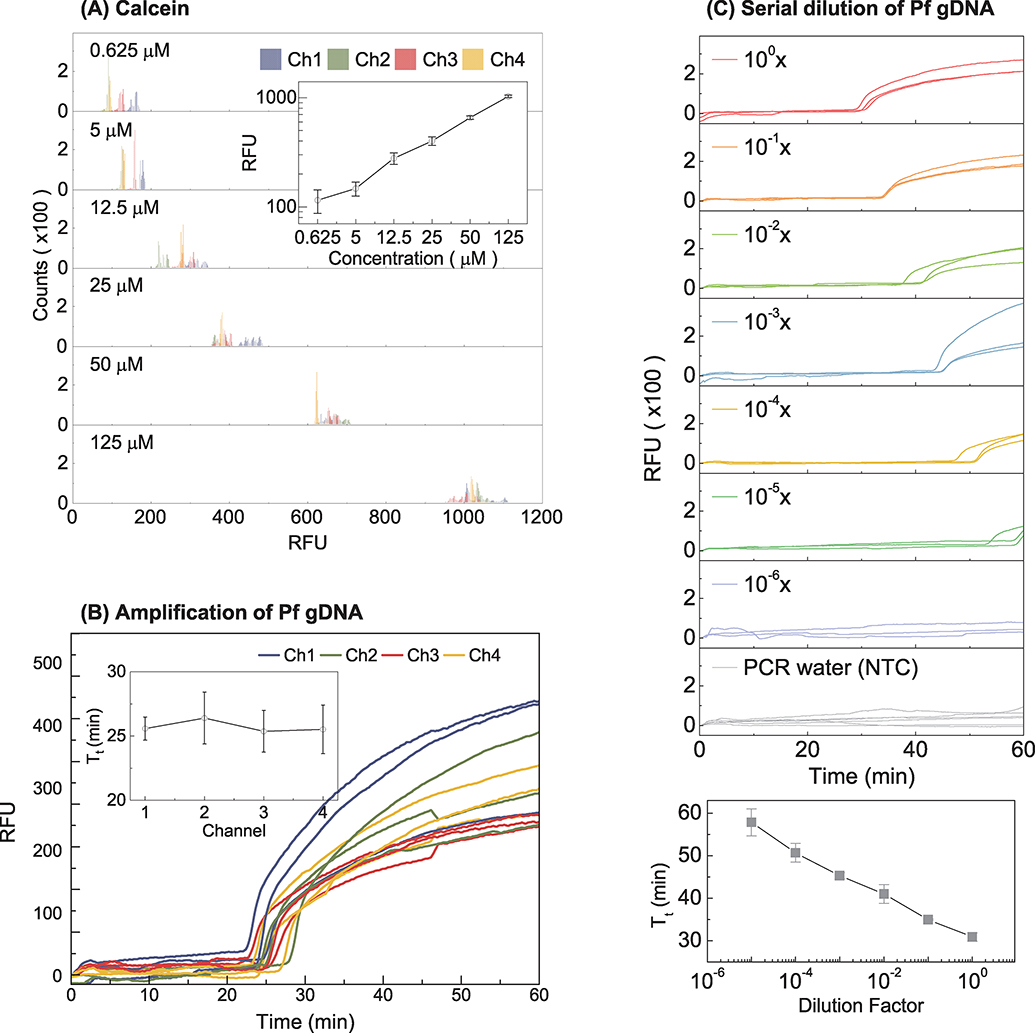
For the quadruplex parallel NAT device, the fluorescence sensing consistency among different channels is essential for quantitative measurement. We tested fluorescent calcein dye at various known concentrations. At each concentration, identical calcein aliquots were loaded into the four reaction chambers for fluorescence intensity measurement. Figure 3A shows the relative fluorescence unit (RFU) distribution from each optical channel at different concentrations. The quantitative uniformity among the four channels is excellent, as seen by the small standard deviation for the RFU values. Moreover, as expected, the mean fluorescence intensity was proportional to the calcein concentration and 2-fold calcein concentration difference could be discriminated (inset in Figure 3A). To further validate the fluorescence sensing uniformity during the real-time LAMP process, four identical 1 μl of Pf genomic DNA was directly loaded into each reaction chamber, and the real-time amplification curve was monitored (Figure 3B). We repeated each test three times. As shown in the inset of Figure 3B, the variation of threshold time (Tt) among different fluorescence sensing channels was ~1.5 minutes. These results validate the fluorescence sensing uniformity among different optical channels.
Figure 3.
Validation of the optical sensing uniformity. (A) With fluorescent calcein dye, the RFU distribution for the four optical channels was evaluated at a series of calcein concentration. A linear dependence of the RFU on the calcein concentration was observed in the range of 0.625–125 μM. The RFU variation from the four channels is small. (B) With Pf genomic DNA at constant concertation, the variations of the amplification threshold time (Tt) obtained from the real-time curve is ~1.5 min. (C) Amplification curves for 10-fold serially diluted Pf genomic DNA samples. The bottom plot shows the calibration curve for the Pf genomic DNA. Standard deviation values are from triplicates.
Quantitative.
A series of 10-fold dilutions of Pf genomic DNA in Tris-EDTA buffer was used to validate the quantitative ability of the device. For each concentration, a set of three identical Pf genomic DNA samples and one internal negative control were loaded into each of the four reaction chambers on the disc. The DNA sample volume is 1 μl, and the LAMP master mix is 24 μl. Figure 3C shows real-time amplification results from various concentrations of Pf genomic DNA. The mean and standard deviation of the amplification threshold time (Tt) was obtained from the triplicates for each concentration. As shown in the bottom subplot of Figure 3C, a clear linear relationship was observed between Tt and the dilution factor, which could be used as a reference curve for quantification. The quantitative ability is important for assessing parasite load in the blood, a useful indicator for determining the proper antimalarial drug dosage (Dormond et al., 2015 ).
3.3. LAMP assays for Pf, Pv and pan-Plasmodium
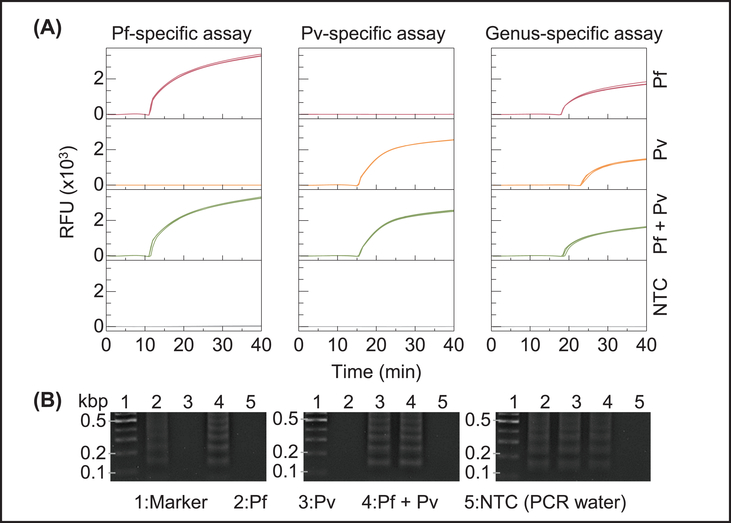
To validate the LAMP primer sets for specifically detecting Pf, Pv and pan-Plasmodium, we performed a cross-reactivity test using extracted Pf and Pv genomic DNA on a bench-top real-time PCR instrument (Bio-Rad CFX96). Each analysis was performed with triplicates. As shown in Figure 4A, Pf- and Pv- specific assays can pick up the corresponding genomic sample specifically without cross-reactivity, while the pan-Plasmodium assays can pick up any Plasmodium species (Pf and Pv). No amplification was observed with the negative control (PCR grade water). We performed gel electrophoresis in 2% agarose gel to further evaluate each amplicon (Figure 4B). A clear ladder-like pattern with multiple bands of different molecular sizes was observed due to the stem-loop DNA structures with several inverted repeats within LAMP amplicons (Notomi et al., 2000). The length of the bands showed confirmative agreement with the length of the target sequence (213-, 127-, and 220-bp bands were expected from Pf, Pv, and pan-Plasmodium products respectively) (Britton et al., 2016a; Polley et al., 2010).
Figure 4.
Validation of the species- and genus-specific LAMP assays using laboratory purified Pf and Pv genomic DNA. (A) Amplification curves obtained from real-time PCR. PCR grade water was used as a no template control (NTC). (B) Gel electrophoresis image (2% agarose gel). Ladder-like bands in the gel image confirm the amplicons from species- and genus-specific LAMP reactions.
3.4. Thermostability of LAMP reagents
Although most of our experiments used liquid phase reagents loaded into the disc immediately prior to use, the reagents could also be pre-loaded before use. We studied the thermostability of LAMP reagents (especially Bst 2.0 DNA polymerase) when stored at 25°C (ambient temperature) and at 4°C (refrigerator temperature) to evaluate their applicability for field use. During a week-long test, LAMP reagents stored at 25°C and 4°C were evaluated every 24 hours to detect the model Pf genomic DNA. These experiments were done in 3 repetitions. As shown in Figure 5, the enzymes and reagents were found to retain sufficient activity to achieve successful DNA amplification when stored at 4°C for a week. There was no significant shift in the average threshold time for reagents stored at 4°C for one week. In contrast, when stored at 25°C, although the enzymes and reagents were active and had the ability to efficiently amplify DNA in less than 50 min for 3 days (no activity afterward), the threshold time (Tt) needed to obtain the positive/negative results were delayed (Figure 5B). These results indicate that a qualitative yes/no amplification could be achievable for reagents storage at room temperature. However, for a consistent real-time quantitative LAMP detection, cold-chain transport and storage of the pre-filled microfluidic reagent disc would be preferred. The stability issue could also be addressed by using reagent lyophilization. It was found the lyophilized reagents could remain stable for 24 months when stored at 4 °C, 28 days at 25 °C, and 2 days at 37 °C (Chen and Ching, 2017).
3.5. Sensitivity estimation with whole blood
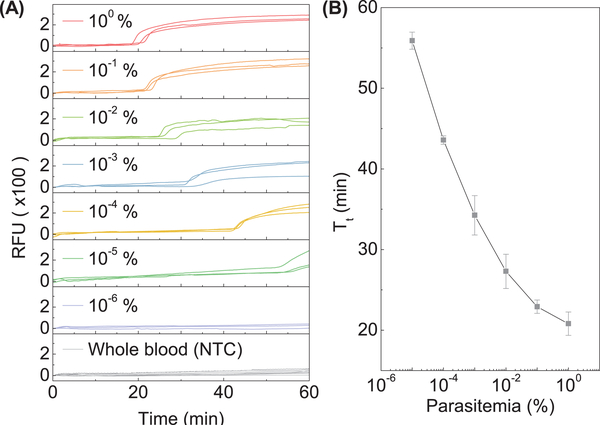
To evaluate the analytical sensitivity of our device in the real world settings, we 10-fold diluted the Pf-infected whole blood with healthy blood to create mock samples with 10−6% to 1% parasitemia. The parasite DNA samples were automatically prepared on the compact disc. Each parasitemia sample was performed in triplicates, together with negative control on a single disc (four reactions per run). As shown in Figure 6A, a whole blood sample with parasitemia higher than 10−5% could be identified. Since parasitemia is the ratio of the parasitized RBCs to the total RBCs, 10−5% parasitemia would correspond to 0.5 parasites/μl (normal RBC count is ~5×106 cells/μl (Moody, 2002)). Although a rigorous report of the limit of detection (i.e., analytical sensitivity) requires a statistical comparison with the analytical blank and should be expressed as a probability with confidence intervals (Long and Winefordner, 1983), a quick eyeball of the data shown in Figure 6A suggests the whole blood sensitivity is around 0.5 parasites/μl. WHO estimates that analytical sensitivity needs to be lower than 2 parasites/μl for identifying low-level infection in a pre-elimination setting(2017). This sensitivity achieved in our device is comparable to other NAT methods (1–5 parasites/μl) (Kolluri et al., 2018) and suitable for detecting low-level asymptomatic carriers (Hopkins et al., 2013; Modak et al., 2016; Vallejo et al., 2015; Wongsrichanalai et al., 2007). No amplification was observed for the healthy whole blood sample, which suggests the background human genomic DNA has negligible interferences. The amplification threshold time (Tt) was extracted for each parasitemia, and the results were shown in Figure 6B. The inversely proportional relationship between Tt and parasitemia confirmed that the quantitative ability is still valid with whole blood samples. It is interesting to note that the calibration curve shown in Figure 6B is not exactly linear which is likely because the DNA extraction efficiency is nonlinear for different cell numbers (Katevatis et al., 2017).
Figure 6.
Sensitivity test with Pf-infected whole blood sample (A) Amplification curves for 10-fold serially diluted Pf-infected whole blood samples. Healthy human blood was used as an NTC. (B) The resulting calibration curve for the whole blood sample. Standard deviation values are from triplicates.
3.6. Whole blood-based Pf, Pv and pan-Plasmodium identification
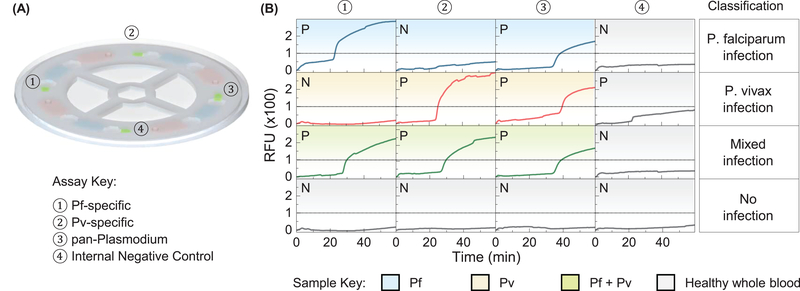
To evaluate the specificity of the device, we prepared whole blood samples spiked randomly with Pf and Pv. Those sample’s species information is recorded but blinded to the tester. The reagent compact disc was configured as species-specific tests as shown in Figure 7A. Each test disc incorporates an internal negative control unit to monitor the test quality. A threshold of 100 RFU is experimentally determined to differentiate positive and negative results. Figure 7B shows the result from a representative set of samples (i.e., Pf, Pv, mixed, and healthy). The species information for a particular infected whole blood sample can be derived from four qualitative results on the single microfluidic disc. For example, the Pf-infected sample (first row of Figure 7B) can be identified by the Pf-specific assay as well as the genus-specific assay, whereas the mixed infection sample can be detected when Pf-, Pv-, and genus-specific assays all show positive. Since Pf and Pv are the two most prevalent species that pose the greatest threat to the human, identification of these major malaria parasite species in the field could provide the malaria transmission profile to the healthcare workers and enable the effective malaria eradication strategy (Kolluri et al., 2018).
Figure 7.
Species- and genus-specific tests using spiked whole blood samples. (A) The reagent compact disc was configured as species-specific tests. The testing unit 1 and 2 contains Pf- and Pv- specific primer sets, respectively. The testing unit 3 has a genus-specific primer set. Testing unit 4 is for internal negative control. (B) The result from a representative set of samples (i.e., Pf, Pv, mixed, and healthy). A threshold of 100 RFU (dashed line) is experimentally determined for positive and negative differentiation. The species information for a particular infected whole blood sample can be derived from four qualitative results on the single microfluidic disc (each row).
4. Conclusion
The palm-sized nucleic acid testing device with quadruplex parallel reactions was developed and validated towards low-cost malaria screening test. The device could perform the integrated and automatic sample preparation for parasite DNA extraction and streamlined real-time amplification on a single microfluidic reagent disc. The quadplex device could be configured either as four identical tests to increase the throughput or as four species-specific tests. The parallelization could be easily scaled up if needed in the future. The quantitative ability of the device could enable parasite load assessment for prescribing correct dose of antimalarial drugs. The analytical sensitivity against Pf-infected whole blood samples is ~0.5 parasites/μl, adequate for detecting asymptomatic parasite carriers. The combination of the sensitivity, specificity, cost, and scalable sample preparation suggests the real-time fluorescence LAMP device could be particularly useful for malaria screening in the field settings. Considering the limited reagent lifetime at room temperature, the challenge of storage and transport of the liquid phase on the microfluidic reagent disc needs to be addressed in our future studies, preferably with reagent lyophilization.
Supplementary Material
5. Acknowledgments
This project was partially supported by a grant from National Institutes of Health, USA (U19AI089672) and a grant from National Science Foundation, USA (ECCS-1710831). Support from Penn State Award ‘Materials Matter at the Human Level’ was also acknowledged. We express our gratitude to Xiaolian Li for providing malaria samples and to Jiho Noh for the 3D printing work. The following reagent was obtained through BEI Resources Repository, NIAID, NIH: Plasmodium vivax, Strain Chesson, MRA-383 and strain Achiote, MRA-369, contributed by W. E. Collins.
6 Reference
- Abdul-Ghani R (2014). Towards rapid genotyping of resistant malaria parasites: could loop-mediated isothermal amplification be the solution? Malaria journal 13, 237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britton S, Cheng Q, Grigg MJ, Poole CB, Pasay C, William T, Fornace K, Anstey NM, Sutherland CJ, Drakeley C, et al. (2016a). Sensitive Detection of Plasmodium vivax Using a High-Throughput, Colourimetric Loop Mediated Isothermal Amplification (HtLAMP) Platform: A Potential Novel Tool for Malaria Elimination. Plos Neglected Tropical Diseases 10, e0004443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britton S, Cheng Q, and McCarthy JS (2016b). Novel molecular diagnostic tools for malaria elimination: a review of options from the point of view of high-throughput and applicability in resource limited settings. Malaria journal 15, 88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HW, and Ching WM (2017). Evaluation of the stability of lyophilized loop-mediated isothermal amplification reagents for the detection of Coxiella burnetii. Heliyon 3, e00415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi G, Song D, Shrestha S, Miao J, Cui L, and Guan W (2016). A field-deployable mobile molecular diagnostic system for malaria at the point of need. Lab Chip 16, 4341–4349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordray MS, and Richards-Kortum RR (2015). A paper and plastic device for the combined isothermal amplification and lateral flow detection of Plasmodium DNA. Malaria journal 14, 472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dineva MA, MahiLum-Tapay L, and Lee H (2007). Sample preparation: a challenge in the development of point-of-care nucleic acid-based assays for resource-limited settings. The Analyst 132, 1193–1199. [DOI] [PubMed] [Google Scholar]
- Dormond L, Jaton K, de Valliere S, Genton B, and Greub G (2015). Malaria real-time PCR: correlation with clinical presentation. New Microbes New Infect 5, 10–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gous N, Scott L, Berrie L, and Stevens W (2016). Options to Expand HIV Viral Load Testing in South Africa: Evaluation of the GeneXpert(R) HIV-1 Viral Load Assay. Plos One 11, e0168244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goyal K, Kaur H, Sehgal A, and Sehgal R (2015). RealAmp Loop-Mediated Isothermal Amplification as a Point-of-Care Test for Diagnosis of Malaria: Neither Too Close nor Too Far. J Infect Dis 211, 1686. [DOI] [PubMed] [Google Scholar]
- Han ET (2013). Loop-mediated isothermal amplification test for the molecular diagnosis of malaria. Expert Rev Mol Diagn 13, 205–218. [DOI] [PubMed] [Google Scholar]
- Han ET, Watanabe R, Sattabongkot J, Khuntirat B, Sirichaisinthop J, Iriko H, Jin L, Takeo S, and Tsuboi T (2007). Detection of four Plasmodium species by genus- and species-specific loop-mediated isothermal amplification for clinical diagnosis. J Clin Microbiol 45, 2521–2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins H, Gonzalez IJ, Polley SD, Angutoko P, Ategeka J, Asiimwe C, Agaba B, Kyabayinze DJ, Sutherland CJ, Perkins MD, et al. (2013). Highly sensitive detection of malaria parasitemia in a malaria-endemic setting: performance of a new loop-mediated isothermal amplification kit in a remote clinic in Uganda. J Infect Dis 208, 645–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiang MS, Greenhouse B, and Rosenthal PJ (2014). Point of care testing for malaria using LAMP, loop mediated isothermal amplification. J Infect Dis 210, 1167–1169. [DOI] [PubMed] [Google Scholar]
- Hsiao NY, Dunning L, Kroon M, and Myer L (2016). Laboratory Evaluation of the Alere q Point-of-Care System for Early Infant HIV Diagnosis. Plos One 11, e0152672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jani IV, Meggi B, Vubil A, Sitoe NE, Bhatt N, Tobaiwa O, Quevedo JI, Loquiha O, Lehe JD, Vojnov L, et al. (2016). Evaluation of the Whole-Blood Alere Q NAT Point-of-Care RNA Assay for HIV-1 Viral Load Monitoring in a Primary Health Care Setting in Mozambique. J Clin Microbiol 54, 2104–2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katevatis C, Fan A, and Klapperich CM (2017). Low concentration DNA extraction and recovery using a silica solid phase. Plos One 12, e0176848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kersting S, Rausch V, Bier FF, and von Nickisch-Rosenegk M (2014). Rapid detection of Plasmodium falciparum with isothermal recombinase polymerase amplification and lateral flow analysis. Malaria journal 13, 99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TH, Park J, Kim CJ, and Cho YK (2014). Fully Integrated Lab-on-a-Disc for Nucleic Acid Analysis of Food-Borne Pathogens. Analytical chemistry 86, 3841–3848. [DOI] [PubMed] [Google Scholar]
- Kolluri N, Klapperich CM, and Cabodi M (2018). Towards lab-on-a-chip diagnostics for malaria elimination. Lab on a chip 18, 75–94. [DOI] [PubMed] [Google Scholar]
- Kong LX, Perebikovsky A, Moebius J, Kulinsky L, and Madou M (2016). Lab-on-a-CD: A Fully Integrated Molecular Diagnostic System. Jala-J Lab Autom 21, 323–355. [DOI] [PubMed] [Google Scholar]
- Li Y, Kumar N, Gopalakrishnan A, Ginocchio C, Manji R, Bythrow M, Lemieux B, and Kong HM (2013). Detection and Species Identification of Malaria Parasites by Isothermal, tHDA Amplification Directly from Human Blood without Sample Preparation. J Mol Diagn 15, 634–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Nam J, Kim S, Lim CT, Park MK, and Shin Y (2016). Two-stage sample-to-answer system based on nucleic acid amplification approach for detection of malaria parasites. Biosensors & bioelectronics 82, 1–8. [DOI] [PubMed] [Google Scholar]
- Long GL, and Winefordner JD (1983). Limit of detection. A closer look at the IUPAC definition. Anal Chem 55, 712A–724A. [Google Scholar]
- Lucchi NW, Gaye M, Diallo MA, Goldman IF, Ljolje D, Deme AB, Badiane A, Ndiaye YD, Barnwell JW, Udhayakumar V, et al. (2016). Evaluation of the Illumigene Malaria LAMP: A Robust Molecular Diagnostic Tool for Malaria Parasites. Scientific reports 6, 36808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madou M, Zoval J, Jia GY, Kido H, Kim J, and Kim N (2006). Lab on a CD. Annu Rev Biomed Eng 8, 601–628. [DOI] [PubMed] [Google Scholar]
- Miao J, Li JF, Fan Q, Li XL, Li XY, and Cui LW (2010). The Puf-family RNA-binding protein PfPuf2 regulates sexual development and sex differentiation in the malaria parasite Plasmodium falciparum. Journal of Cell Science 123, 1039–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modak SS, Barber CA, Geva E, Abrams WR, Malamud D, and Ongagna YS (2016). Rapid Point-of-Care Isothermal Amplification Assay for the Detection of Malaria without Nucleic Acid Purification. Infect Dis (Auckl) 9, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moody A (2002). Rapid diagnostic tests for malaria parasites. Clin Microbiol Rev 15, 66–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris U, Khamis M, Aydin-Schmidt B, Abass AK, Msellem MI, Nassor MH, Gonzalez IJ, Martensson A, Ali AS, Bjorkman A, et al. (2015). Field deployment of loop-mediated isothermal amplification for centralized mass-screening of asymptomatic malaria in Zanzibar: a pre-elimination setting. Malaria journal 14, 205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolte DD (2009). Invited Review Article: Review of centrifugal microfluidic and bio-optical disks. Rev Sci Instrum 80, 101101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolte FS, Gauld L, and Barrett SB (2016). Direct Comparison of Alere i and cobas Liat Influenza A and B Tests for Rapid Detection of Influenza Virus Infection. J Clin Microbiol 54, 2763–2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notomi T, Okayama H, Masubuchi H, Yonekawa T, Watanabe K, Amino N, and Hase T (2000). Loop-mediated isothermal amplification of DNA. Nucleic Acids Res 28, E63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oriero EC, van Geertruyden JP, Nwakanma DC, D’Alessandro U, and Jacobs J (2015). Novel techniques and future directions in molecular diagnosis of malaria in resource-limited settings. Expert Review of Molecular Diagnostics 15, 1419–1426. [DOI] [PubMed] [Google Scholar]
- Patel JC, Lucchi NW, Srivastava P, Lin JT, Sug-Aram R, Aruncharus S, Bharti PK, Shukla MM, Congpuong K, Satimai W, et al. (2014). Field evaluation of a real-time fluorescence loop-mediated isothermal amplification assay, RealAmp, for the diagnosis of malaria in Thailand and India. J Infect Dis 210, 1180–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polley SD, Mori Y, Watson J, Perkins MD, Gonzalez IJ, Notomi T, Chiodini PL, and Sutherland CJ (2010). Mitochondrial DNA targets increase sensitivity of malaria detection using loop-mediated isothermal amplification. J Clin Microbiol 48, 2866–2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy V, Ravi V, Desai A, Parida M, Powers AM, and Johnson BW (2012). Utility of IgM ELISA, TaqMan real-time PCR, reverse transcription PCR, and RT-LAMP assay for the diagnosis of Chikungunya fever. J Med Virol 84, 1771–1778. [DOI] [PubMed] [Google Scholar]
- Sattabongkot J, Tsuboi T, Han ET, Bantuchai S, and Buates S (2014). Loop-mediated isothermal amplification assay for rapid diagnosis of malaria infections in an area of endemicity in Thailand. J Clin Microbiol 52, 1471–1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sema M, Alemu A, Bayih AG, Getie S, Getnet G, Guelig D, Burton R, LaBarre P, and Pillai DR (2015). Evaluation of non-instrumented nucleic acid amplification by loop-mediated isothermal amplification (NINA-LAMP) for the diagnosis of malaria in Northwest Ethiopia. Malaria journal 14, 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R, Savargaonkar D, Bhatt R, and Valecha N (2013). Rapid detection of Plasmodium vivax in saliva and blood using loop mediated isothermal amplification (LAMP) assay. J Infect 67, 245–247. [DOI] [PubMed] [Google Scholar]
- Sirichaisinthop J, Buates S, Watanabe R, Han ET, Suktawonjaroenpon W, Krasaesub S, Takeo S, Tsuboi T, and Sattabongkot J (2011). Evaluation of loop-mediated isothermal amplification (LAMP) for malaria diagnosis in a field setting. Am J Trop Med Hyg 85, 594–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slater HC, Ross A, Ouedraogo AL, White LJ, Nguon C, Walker PGT, Ngor P, Aguas R, Silal SP, Dondorp AM, et al. (2015). Assessing the impact of next-generation rapid diagnostic tests on Plasmodium falciparum malaria elimination strategies. Nature 528, S94–S101. [DOI] [PubMed] [Google Scholar]
- Snounou G, Viriyakosol S, Zhu XP, Jarra W, Pinheiro L, Dorosario VE, Thaithong S, and Brown KN (1993). High-Sensitivity of Detection of Human Malaria Parasites by the Use of Nested Polymerase Chain-Reaction. Molecular and Biochemical Parasitology 61, 315–320. [DOI] [PubMed] [Google Scholar]
- Surabattula R, Vejandla MP, Mallepaddi PC, Faulstich K, and Polavarapu R (2013). Simple, rapid, inexpensive platform for the diagnosis of malaria by loop mediated isothermal amplification (LAMP). Exp Parasitol 134, 333–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trager W, and Jensen JB (1976). Human Malaria Parasites in Continuous Culture. Science 193, 673–675. [DOI] [PubMed] [Google Scholar]
- Vallejo AF, Martinez NL, Gonzalez IJ, Arevalo-Herrera M, and Herrera S (2015). Evaluation of the loop mediated isothermal DNA amplification (LAMP) kit for malaria diagnosis in P. vivax endemic settings of Colombia. PLoS Negl Trop Dis 9, e3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO (2017). World Malaria Report 2017. 1–196. [Google Scholar]
- Wongsrichanalai C, Barcus MJ, Muth S, Sutamihardja A, and Wernsdorfer WH (2007). A review of malaria diagnostic tools: Microscopy and rapid diagnostic test (RDT). American Journal of Tropical Medicine and Hygiene 77, 119–127. [PubMed] [Google Scholar]
- Xu G, Nolder D, Reboud J, Oguike MC, van Schalkwyk DA, Sutherland CJ, and Cooper JM (2016). Paper-Origami-Based Multiplexed Malaria Diagnostics from Whole Blood. Angewandte Chemie 55, 15250–15253. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.