Abstract
Despite their ubiquity, relatively few bacteriophages have been characterized. Here, we set out to explore Caulobacter bacteriophages (caulophages) in the rhizosphere and characterized Kronos, the first caulophage isolated from the rhizosphere. Kronos is a member of the Siphoviridae family since it has a long flexible tail. In addition, an analysis of the Kronos genome indicated that many of the predicted proteins were distantly-related to those of bacteriophages in the lambdoid family. Consistent with this observation, we were able to demonstrate the presence of cos sites that are similar to those found at ends of lambdoid phage genomes. Moreover, Kronos displayed a relatively rare head and tail morphology compared to other caulophages but was similar to that of the lambdoid phages. Taken together, these data indicate that Kronos is distantly related to lambdoid phages and may represent a new Siphoviridae genus.
Introduction
Bacteriophages (phages) are the most diverse and abundant organisms within the entire biosphere. However, only a small percentage of phages have been isolated and characterized. As of February 2019, there are approximately 2200 completely sequenced phage genomes in the NCBI database, which amounts to about 1% of the completely sequenced bacterial genomes. Given that there are about 10 phages for every bacterium [13] and that phage-mediated horizontal gene transfer contributes to bacterial function and evolution [8], it is clear that the study of phages needs to be increased.
Understanding how phages interact with their hosts is innately linked to the way in which phages package their genomes, since phage genome packaging is dependent on host machinery. Although some phages have RNA or single-stranded DNA genomes, most phages have a double-stranded DNA genome. Three different mechanisms are commonly used by phages to package their DNA: headful packaging, cos site packaging, and T7-like packaging. Phages such as P1, T4, SPP1, P22 and the caulophage Cr30 employ a headful packaging system [10, 21], where packaging is initiated by a terminase at a pac site on the substrate genomic DNA concatemer, and a genome that is longer than unit length is packaged and completed by sequence-independent cleavage [5]. Upon infection the genome is circularized by recombination between the duplicated segments at the ends of the packaged DNA. The second mode, termed cos site packaging, is also initiated on concatemeric genomic DNA by terminase cleavage; however, packaging is completed at a sequence-specific cos site, generating a unit length monomer with the cohesive termini used for circularization [24]. In contrast to both headful and cos site packaging, T7-like packaging results in well-defined terminally redundant sequences that can be thousands of base pairs in length [14]. This form of DNA packaging is considered to be more complex because, unlike headful and cos site packaging, phages employing this form of replication reduce their reliance on host replication machinery by encoding most of their own replication proteins [25]. Recent studies have indicated that most phages that infect Caulobacter strains use the T7-like packaging method [14, 19].
Members of the genus Caulobacter were originally isolated from freshwater and were thought to be restricted to aquatic environments [22]. However, several recent studies [18, 23, 29] have demonstrated that caulobacters are present in the rhizosphere as well, and that some species are considered to be plant growth-promoting bacteria (PGPB). PGPB can increase plant growth in a variety of ways including: increased IAA production, increased ACC deaminase production, siderophore production, nitrogen fixation, and phosphate solubilization [12].
Despite the recent interest in rhizosphere caulobacters, there have been no reports of caulophages that have been isolated from the rhizosphere. This lack of rhizosphere phage research neglects potentially insightful information about the plant growth-promoting abilities of bacteria such as caulobacters since PGPB research is being conducted to thwart the imminent world-wide shortage of biofertilizers [34] and provide food for the rising world population. Currently, all of the known caulophages have been isolated from aquatic sources and most belong to the phiCbk Siphoviridae family, the Myoviridae family, or the Podoviridae family and include only phages that utilize either T7-like or headful DNA packaging mechanisms. Since phages are thought to be present in all environments, we have initiated a study of phages that infect rhizosphere caulobacters. We hypothesized that some rhizosphere caulophages would be significantly divergent from the currently characterized caulophages, since the typical rhizosphere environment drastically differs from most aquatic environments. Here we report the characterization of a novel rhizosphere caulophage that has cos sites at the termini of its genome and appears to be distantly related to the lambdoid family of phages.
Methods
Bacterial and caulophage isolation and growth
Dichotomous plant roots were extracted from riparian land on the banks of the Congaree River in Columbia, SC and brushed clean to remove remnant dirt. The roots were then soaked for 24 h in room-temperature sterilized tap water to allow the release of root-associated bacteria. In order to effectively isolate caulophages from the rhizosphere, we first isolated a rhizosphere Caulobacter wild-type strain, since phage-host interactions are generally highly specific. Small amounts (20 μl) of water from the soaked root were pipetted onto PYE plates containing 2 μM of ampicillin, since Caulobacters are generally ampicillin resistant. The plates were incubated at 30°C for 24 h, and colonies with characteristics of Caulobacter strains were selected and suspended in 3 ml of PYE broth [11]. After overnight incubation at 30°C, the bacterial culture was visualized with a compound light microscope to observe cellular shape and motility. Presumptive Caulobacter strains were subjected to repeated single colony isolation to produce a pure culture. One strain, designated CBR1, was confirmed to be a species of Caulobacter by analyzing a portion of its 16S rRNA gene. CBR1 DNA was isolated using a Qiagen DNA isolation kit according to the manufacturer’s instructions. The CBR1 16S rRNA gene was amplified by PCR using 5’-GGTTACCTTGTTACGACTT-3’ and 5’-GTGCCAGCMGCCGCGGTAA-3’ as forward and reverse primers, respectively. The nucleotide sequence of the amplified ~ 900 bp product was determined by Sanger sequencing, and the resulting 16S rDNA sequence was compared to other bacterial 16S rDNA sequences using BLASTn to verify that CBR1 was a Caulobacter isolate (Fig. S1).
To isolate a phage that infected CBR1, 10 ml of the root-soaked suspension was filtered using a 0.45 μm filter to remove bacteria and other particulate matter. The filtrate was then combined with 50 μl of a CBR1 culture, 2.5 μl of 5X PYE broth and 0.67 mg of ampicillin. The mixture was then incubated at 30°C on an orbital shaker for 24 h. The resulting phage enrichment was poured into a 15 ml sterile tube and centrifuged at 5,000 x g for 10 min. The supernatant was collected and 1 ml of chloroform was added, shaken, and left at room temperature for 30 min. Twenty microliters of the resulting supernatant (lysate) was used to perform a spot test for the presence of phages, and this plate was incubated overnight at 30°C. After observing phage lysis, serial dilutions were prepared and mixed with CBR1 in 3.5 ml of soft agar to obtain single plaques. A single plaque was stabbed with a sterile needle and suspended in 1 ml of PYE broth. To further purify the phage, the resulting phage suspension was diluted 1:100, and 100 μl of the diluted suspension was mixed with 100 μl of CBR1 in 3.5 ml of soft agar. After overnight incubation at 30°C, a single phage plaque was cut out from the soft agar overlay, placed in 1 ml of PYE broth and stored at 4°C overnight to allow the phage to diffuse out of the agar. The resulting phage solution was diluted 1:100 and again plated in a soft agar overlay with CBR1 to produce confluent lysis after overnight incubation at 30°C. The resulting phage particles were eluted with the addition of 7 ml of 1X PYE broth and incubated at room temperature for 24 h. The plates were then decanted, and the lysate was centrifuged at 5,000 x g for 10 min to remove residual bacteria. Approximately 1 ml of chloroform was added to the mixture and then vortexed for 10 s to kill any additional residual bacteria. Some of the lysate was used to make DNA plugs, and the remaining lysate was stored at −72°C.
Host range
The host range for Kronos was determined by mixing 100 μl of a selected Caulobacter species (i.e. CB1, CB2, CB3, CB13, CB15, C. segnis, C. henricii and AP07) with 3.5 ml of soft agar. Once the agar solidified, 20 μl of a 109 plaque forming units (PFU) ml−1 Kronos phage lysate was pipetted onto the surface of the agar plate: this was performed for each Caulobacter species. Plates were incubated overnight at 30°C. After incubation, lysis was determined by a transparent zone of lysis at the site where the lysate was deposited.
To determine which phages infect CBR1, a volume of 100 μl was mixed with 3.5 ml of soft agar and poured onto a PYE agar plate. After the mixture solidified, 20 μl of a 109 PFU ml−1 phage lysate was pipetted onto the surface of the agar. The plates were then incubated overnight at 30°C. The following phiCbk phages were used for the host range experiments: phiCbk, BL-1, and BL-9. Lullwater and Cr30 were used as representatives of the Podoviridae family and Myoviridae family, respectively. Infection was measured by a transparent zone of lysis at the site where the 20 μl of phage lysate was placed.
Genome size determination
The size of the phage genome was determined using pulsed-field gel electrophoresis (PFGE). Agarose plugs containing phage DNA were made using the protocol of Dingwall et al. [9]. Briefly, a 1:1 ratio of phage lysate and 1% agarose in SBA was mixed in a microfuge tube and drawn up into a 1 cc syringe. After solidification, the resulting mixture was cut into 2 mm thick discs (plugs). The resulting plugs were incubated overnight at 50°C in 2 ml of lysis buffer (1.9 ml of 1% Sarkosyl in 0.5 M EDTA and 0.1 ml of proteinase K (20 mg ml−1)). After incubation, the plugs were washed twice with 2 ml of TE buffer and 30 μl of PMSF (17.4 mg ml−1). The plugs were then washed an additional three times with 2 ml of TE buffer. After the completion of the plug washes, one of the plugs was subjected to PFGE at 6 V for 14 h with an initial switch time of 8 s and a final switch time of 20 s and compared to a lambda ladder (BioRad, Hercules, CA) to determine the size of the genome.
Electron microscopy
A 100 μl aliquot of a 109 PFU ml−1 phage lysate was mixed with 100 μl of a log phase culture of CBR1 for 30 min at 30°C. The mixture was then centrifuged and resuspended in 100 μl of autoclaved, purified water. A total of 20 μl of the resulting mixture was combined with 20 μl of 2% uranyl acetate. A formvar/carbon 300 mesh, copper grid was then floated on the mixture for í30 s. Free and adsorbed phages were then observed in a JEOL 200CX transmission electron microscope.
DNA isolation
Phage genomic DNA was prepared from 3-5 ml of a concentrated, high-titer (> 109 PFU ml−1) phage lysate using a Qiagen DNA isolation kit (Germantown, MD) according to the manufacturer’s instructions.
Genome sequence analysis and assembly
The nucleotide sequence of the phage DNA was determined at the Delaware Bioinformatics Institute using a PacBio RSII single-molecule sequencer. The resulting sequence reads were assembled using HGAP 3 and was polished using Quiver. The consensus sequence was then annotated using the Rapid Annotation using Subsystem Technology (RAST) [4]. The annotated sequence was analyzed in Artemis [28] and trimmed to remove duplicate sequences. The annotated genes were analyzed using BLASTn, BLASTp, and the NCBI CD-search tool. The ends of the genome were determined by digesting a phage plug containing DNA in a mixture that consisted of 5 μl of the restriction endonuclease SacII, 10 μl of NEB Cutsmart buffer, and 55 μl of autoclaved deionized water (diH2O) in a 37°C water bath overnight. The fragments of digested DNA were then separated and visualized by subjecting the plug to PFGE with a pulse time of 1 s for 14 h at 14°C. Cos sites were identified by using Sanger sequencing to determine the nucleotide sequence of the ends of the genome using Kronos genomic DNA as the template and the following primers: 5’-CGAGAAGAACACGGAAGCCAT-3’ and 5’-AGCCGACTTGCCTTGATGGTCTG-3’, to sequence each end of the genome. The NCBI accession number for the Kronos genome is MH884648.
Lysogen detection
Bacterial colonies were selected from phage-bacteria overlay plates containing Kronos plaques. Bacteria from the center of a plaque were streaked onto a PYE plate and incubated at 30°C for 48 h. Isolated colonies were then selected from the streak plate and restreaked onto another PYE plate and were incubated at 30°C for 48 h. The resulting streak plate was then transferred to a velveteen template and then onto a PYE plate containing 3.5 ml of soft agar and 100 μM of CBR1. Lysogens would have been detected by the presence of phage lysis.
Burst size
Phages were added at a final concentration of 107 PFU ml−1 to a10 ml of host bacteria (CBR1) at a concentration of 5×108 colony forming units (CFU) ml−1. After 30 min of incubation at 30°C, the sample was diluted 1:1000 in 3 ml of PYE broth and plated immediately. Every 15 min for 240 min, a 100 μl sample of this dilution was mixed with 100 μl of CBR1 and 3.5 ml of soft agar and plated on a PYE plate. After solidification, the plates were incubated overnight at 30°C. Plaques were counted, and a one-step growth curve was generated by plotting the PFU over time. The assay was repeated three times, and the average was calculated to generate a representative burst size.
Adsorption rate
Phages were added at a final concentration of 105 PFU ml−1 to a 10 ml culture of CBR1 at a concentration of 5×108 CFU ml−1 and was vortexed for 15 s. The mixture was incubated at 30°C, and aliquots of 100 μl were pipetted into a mixture containing 1 ml of chloroform and 9 ml of diH2O at 15 min intervals. The diluted samples were shaken vigorously for 10 s and allowed to rest until the chloroform and diH2O separated. Once the chloroform settled to the bottom of the tube, 100 μl of each sample was mixed with 100 μl of CBR1 and 3.5 ml of soft agar and poured onto a PYE plate and incubated overnight at 30°C. The experiment was performed three times, with the adsorption rate and adsorption rate constant averaged from the three experiments.
Results and Discussion
Phage isolation and characterization
A phage designated Kronos was isolated from plant roots anchored on the banks of the Congaree River in Columbia, SC in August of 2017. To date, Kronos has been shown to infect only the rhizosphere Caulobacter CBR1. Kronos failed to infect C. crescentus isolates including CB1, CB2, CB3, CB13, and CB15 as well as members of other Caulobacter species such as C. segnis and C. henricii. Interestingly, Kronos also failed to infect AP07—a soil derived Caulobacter species isolated from the roots of a poplar tree [30]. The specificity exhibited by Kronos for its host could be the result of a unique receptor present on the cell surface of CBR1, since not only did Kronos fail to infect other C. crescentus strains, but other C. crescentus phages including several phicbkviruses, Podoviridae phage Lullwater, and myophage Cr30 failed to infect CBR1.
Kronos plaques were transparent and uniform in size (< 1 mm). This exceptionally small plaque size produced by Kronos counters the typical inverse relationship of plaque size to genome size among caulophages. For example, giant caulophages that possess larger genomes such as phiCbk (205 kbp) and Colossus (297 kbp) produce small plaques (1 mm) [3, 14]. In contrast, caulophages with small genomes (40-50 kbp) and small capsid sizes (< 60 nm), such as Lullwater [20] produce variable-sized larger plaques (1-3.5 mm). Although not entirely understood, plaque size is most likely associated with a combination of diffusivity, burst size, adsorption efficiency and phage morphology [2].
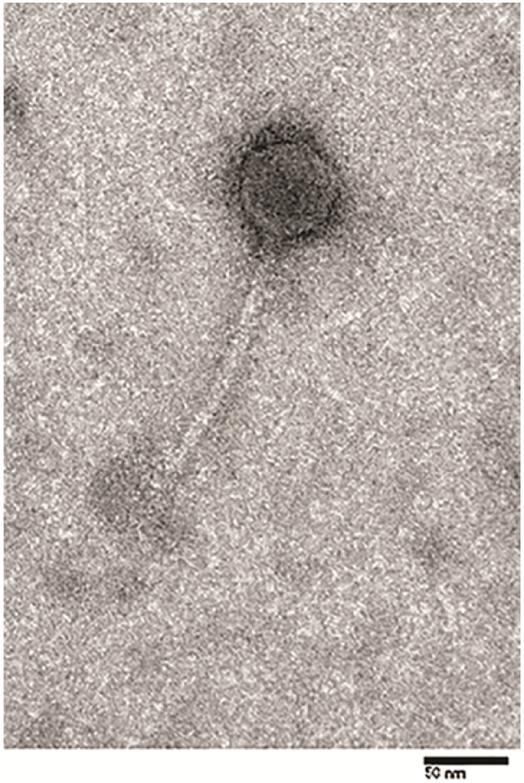
The Kronos phage particle consists of a 50 nm icosahedral head with a 150 nm flexible tail (Fig. 1). This size and morphology is relatively rare among caulophages, but it is similar to that of Ccr1 [15]. However, Ccr1 was lost when a −70°C freezer malfunctioned, and further analyses could not be conducted. Interestingly, Kronos possesses a head and tail morphology strikingly similar to those of the lambdoid phages such as λ phage, HK97, and N15 [6]. Based on the head and tail morphology, we concluded that Kronos is a member of the Siphoviridae family.
Figure 1.
Electron micrograph of Kronos. Bar = 50 nm.
Phage host interactions
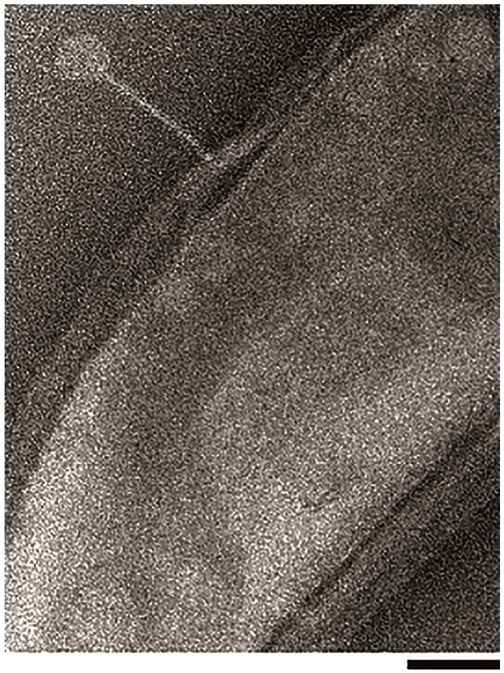
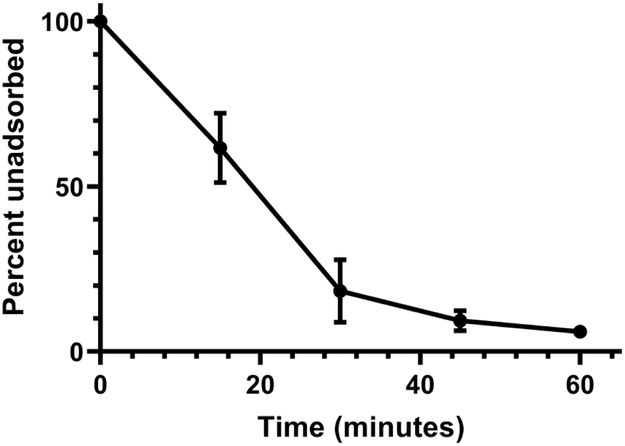
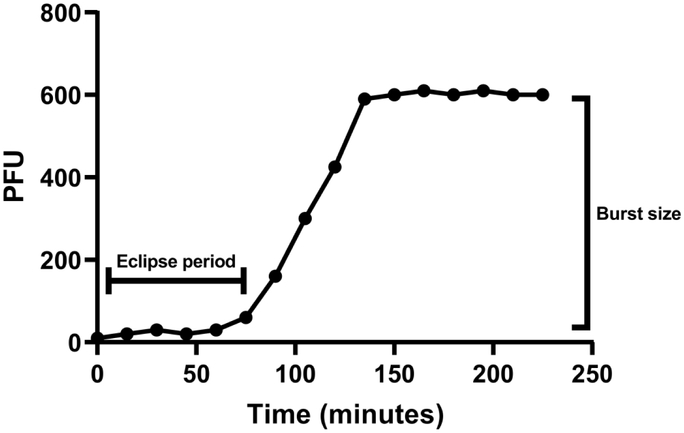
Kronos binds to CBR1 at apparently random locations similar to the binding of Cr22 and Cr30, which adsorb independently of the host flagellum and is in contrast to the polar adsorption method used by the Phicbkviruses (Fig. 2) [15]. The adsorption rate of Kronos was also relatively fast, with 82% of the phages adsorbed after 30 min with an adsorption rate constant of ~ 5.4 × 10−9 ml min−1 (Fig. 3).This adsorption rate was consistent with that of other rhizosphere phages [31], but was in stark contrast to the adsorption rate of the transducing phage Cr30, which was previously shown to adsorb at a rate that was 2.3-fold slower [11]. In addition, Kronos exhibited an eclipse period of 55 min with a burst size of 28 PFU per adsorbed phage (Fig. 4). This eclipse period of Kronos is similar to that of lambdoid phages such as λ phage and HK97 which have an eclipse period of 45 min and 53 min, respectively [35]. In contrast, Cr30 had an extended eclipse period of 90 min [11]. Moreover, despite multiple attempts to detect the ability of Kronos to form a prophage in CBR1, no lysogens were detected. However, some phages have been shown to require hyper-specific, unfavorable physiochemical interactions to induce lysogeny and others have genetic mutations that render lysogenic capabilities ineffective [1].
Figure 2.
Electron micrograph of phage Kronos binding to a CBR1 host cell. Bar = 100 nm.
Figure 3.
Kronos adsorption efficiency. Each point represents the average of three experiments. The error bars represent the range of each data point.
Figure 4.
A representative one-step growth curve of Kronos to determine burst size.
Kronos genome
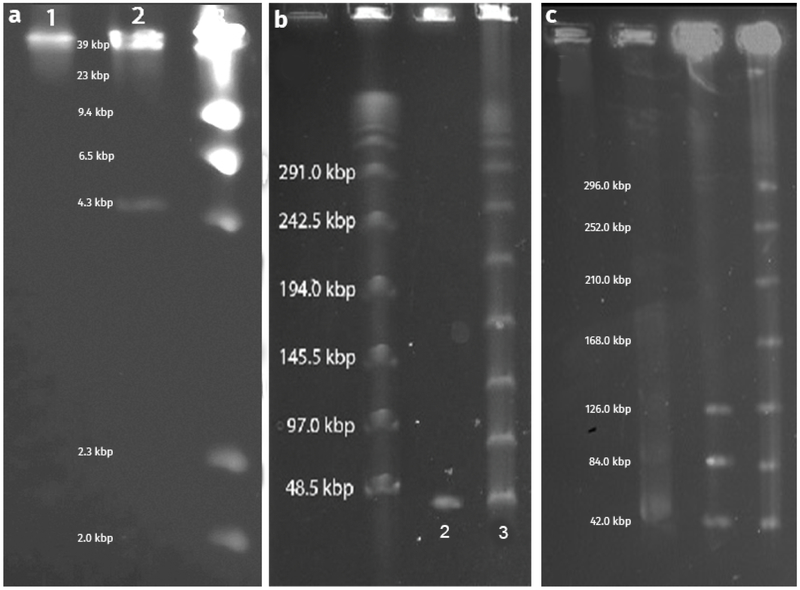
The Kronos genome nucleotide sequence contains 42,424 bp in a linear chromosome. Terminal repeats were not observed when the raw Kronos sequence data was analyzed with the Tablet graphical genome viewer. The N50 read length for the data was ~26 kbp with a read count of ~90 kbp and coverage of ~178X. The absence of double coverage sequence reads in the sequencing data indicated the absence of terminal repeats. In order to verify the sequence and assembly of the Kronos genome, plugs containing Kronos DNA were digested with the restriction endonuclease SacII and were subjected to PFGE. The resulting band pattern did not match the predicted band pattern (4,366 bp and 38,058 bp), since only a single ~42 kbp band was present (Fig. 5a, lane 1). However, undigested plugs containing Kronos DNA produced a ladderlike band pattern similar to that of plugs containing λ phage DNA, implying the presence of cos sites that could anneal to each other (Fig. 5b). Since the presence of cos sites would cause the termini of the ~4 kbp and the ~38 kbp bands to reanneal, we decided to heat the plugs to 70°C for 60 min to denature any annealed cos sites, as demonstrated previously with BstEII digested λ DNA [27]. After heating the plugs and subjecting them to PFGE, the band sizes matched the predicted 4,366 bp and 38,058 bp band sizes (Fig. 5a, lane 2). Additionally, when Kronos DNA was extracted in liquid form, heated to 70°C, and then mixed at a 1:1 ratio with molten 1% agarose, the resulting plugs produced a single ~42 kbp band, which confirmed that the cos sites had been separated (Fig. 5b, lane 2). We also confirmed previous data showing that the formation of the ladder is concentration dependent [36], since an increased DNA concentration increased the ladder appearance (Fig. 5c).
Figure 5.
PFGE images depicting SacII digested Kronos DNA and undigested DNA. Image (A) represents Kronos DNA digested with SacII. Lane 1 depicts digested DNA that has not been heated, and lane 2 depicts DNA that has been heated (>70°C). Lane 3 is λ DNA digested with HinDIII. Image (B) represents Kronos DNA that has been heated to denature the cos ends (lane 2) and Kronos DNA that has not been heated, leaving the cos ends annealed (lane 3). The λ ladder is appropriately labeled to illustrate relative band sizes. Image (C) illustrates the concentration dependency for concatemer formation of Kronos DNA. DNA concentration is increasing from left to right (~0.05 μg/μl to ~0.40 μg/μl).
Visual inspection of the annotated Kronos genome with Artemis revealed a 13 bp region (5’-GGGGCAGAGCACT-3’) located at the 5’ end of the genome, approximately 1 kbp upstream from the terminase genes. This presumptive cos site is similar to those of phages such as HK97, HK022, and λ phage, since these phages have cos sites that are located at the start of their genomes, are 10-14 bp long and are located approximately 1 kbp upstream of the terminase genes. In addition, they have a GGGGC sequence at positions 1-5 and either CT or CG at the terminal two bases [16, 32, 33]. To verify the presumptive cos site, we sequenced the ends of the genome by Sanger sequencing and observed the sequencing reads ended at the cos site. Based on the similarities to other cos sequences and our end-sequencing results, we concluded that this 13 bp sequence is the Kronos cos site, and the Kronos genome was arranged so that this sequence was at the beginning.
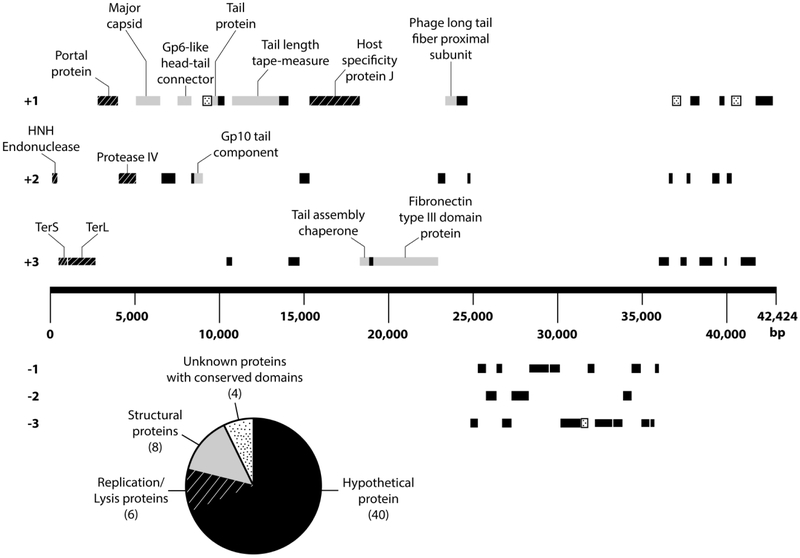
The Kronos genome codes for 58 genes with > 68% of the genes located on the forward strand (Table 1 and Fig. 6). Interestingly, no nucleotide or predicted amino acid sequence matches were identified between any Kronos gene and any previously characterized caulophage gene. In addition the gene arrangement for Kronos was consistent with the gene arrangement of the lambdoid phages with the terminase genes situated within the first couple thousands bases relative to the start of the 5’ plus strand upstream and relatively close to the major capsid genes [7, 16]. This general similarity is consistent with the cos site packaging mechanism and suggests that the Kronos DNA replication mechanism may be similar to that of lambdoid phages; however, this genome arrangement is not entirely exclusive to lambdoid phages, considering other virulent siphophages such as coliphage T1 and Salmonella phage Chi also possess genomic structural modules similar to that of λ phage despite their terminase genes being located ~10,000 bp upstream from the start of 5’ end of the plus strand [17, 26]. Nonetheless, the top BLASTp match for the Kronos small subunit terminase (TerS) was with the TerS gene of the E. coli-infecting phage P27, which is a lambdoid phage that is capable of lysogeny [19]. Similarly, BLASTn analyses identified a ~ 1 kbp region in the Kronos TerL gene that is homologous with the TerL gene of the virulent Halomonas phage QHHSV-1. Halomonas phage QHHSV-1 possesses a phage Cro repressor and a putative integrase that is homologous to λ phage; but due to mutations in its recombination cassette, Elalomonas phage QHHSV-1 has repressed lysogenic activities [1]. Moreover, the Kronos major capsid protein had the most significant BLASTp match with the lambdoid phage HK97. These data combined with the strikingly similar PFGE ladder band pattern between λ phage and Kronos (Fig. 5) strengthens the idea that Kronos uses a cos site DNA packaging mechanism, which is similar to the one used by λ phage.
Table 1.
Kronos genome summary table.
| Genome size | 42,424 bp |
| GC content percentage | 66.22% |
| Average gene length | 699 bp |
| Coding percentage | 95.6% |
| Number of predicted genes | 58 |
| Number of non-hypothetical proteins | 18 |
| Hypothetical proteins | 40 |
| tRNA genes | 0 |
| Genes on forward strand | 40 |
| Genes on reverse strand | 18 |
| Genes with significant nucleotide homology to genes in the GenBank database | 5 |
| Proteins with significant amino acid homology to proteins in the GenBank database | 26 |
Figure 6.
Genome map of Kronos. All genes that are not labeled code for either hypothetical proteins or proteins with a conserved unknown domain. A comprehensive list of all the genes and positions is available in Table S1.
Since the Kronos genome organization resembles that of lambdoid phage genomes and has significant BLASTp homologies to lambdoid proteins, the ~11 kbp section of genes located on the reverse strand may include a collection of genes functioning in lysis and possibly lysogeny. Recent analyses have demonstrated that lambdoid repressors that induce lysogeny and repress the lytic phase have highly variable amino acid sequences and are often not recognizable as homologous [6]. which may explain some of the amino acid sequence dissimilarities with lambdoid proteins throughout the Kronos genome, particularly on the minus strand. In fact, only 45% of the predicted amino acid sequences in the Kronos genome returned statistically significant (E ≤ 10−5 and query coverage ≥ 40%) BLASTp/BLASTcd matches with phage proteins (Table 1 and Table S1). Since 76% of the genes encoded either hypothetical proteins or proteins with an unknown conserved domain, the overall gene function for most of the genes in the genome remains unknown. Taken together, our data support the idea that Kronos is distantly related to, but genetically distinct from the lambdoid family of phages, and may represent a new genus of Siphoviridae cos phages.
Supplementary Material
Acknowledgments
Funding information: This work was funded in part by National Institutes of Health grant GM076277 to BE.
Contributor Information
Louis Berrios, Department of Biological Sciences, University of South Carolina, Columbia, SC 29208.
Bert Ely, Department of Biological Sciences, University of South Carolina, Columbia, SC 29208.
References
- 1).Aalto AP, Bitto D, Ravantti JJ et al. (2012) Snapshot of virus evolution in hypersaline environments from the characterization of a membrane-containing salisaeta icosahedral phage 1. Proc Natl Acad Sci USA 109:7079–7084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2).Abedon ST, Yin J (2009) Bacteriophage Plaques: Theory and Analysis In: Clokie MR, Kropinski AM (eds) Methods in Molecular Biology™, vol. 501 Humana Press, pp.161–174. [DOI] [PubMed] [Google Scholar]
- 3).Ash KT, Drake KM, Gibbs WS et al. (2017) Genomic diversity of Type B3 bacteriophages of Caulobacter crescentus. Curr Microbiol 74:779–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4).Aziz RK, Bartels D, Best AA et al. (2008) The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9:75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5).Black LW (1989) DNA packaging in dsDNA bacteriophages. Annu Rev Microbiol 43:267–292. [DOI] [PubMed] [Google Scholar]
- 6).Casjens SR, Hendrix RW (2015) Bacteriophage lambda: early pioneer and still relevant. Virology 479:310–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7).Catalano CE (2000) The terminase enzyme from bacteriophage lambda: a DNA-packaging machine. Cell Mol Life Sci 57:128–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8).Chen J, Novick RP (2009) Phage-mediated intergeneric transfer of toxin genes. Science 323:139–141. [DOI] [PubMed] [Google Scholar]
- 9).Dingwall A, Shapiro L, Ely B (1990) Analysis of bacterial genome organization and replication using pulsed-field gel electrophoresis. Methods 1:160–168. [Google Scholar]
- 10).Ely B, Gibbs W, Diez S et al. (2015) The Caulobacter crescentus transducing phage Cr30 is a unique member of the T4-like family of myophages. Curr Microbiol 70:854–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11).Ely B, Johnson RC (1977) Generalized transduction in Caulobacter crescentus. Genetics 87:391–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12).Etesami H, Maheshwari DK (2018) Use of plant growth promoting rhizobacteria (PGPRs) with multiple plant growth promoting traits in stress agriculture: Action mechanisms and future prospects. EcotoxicolEnviron Saf 156:225–246. [DOI] [PubMed] [Google Scholar]
- 13).Fuhrman JA, Noble RT (1995) Viruses and protists cause similar bacterial mortality in coastal seawater. Limnol Oceanogr 40:1236–1242. [Google Scholar]
- 14).Gill JJ, Berry JD, Russell WK et al. (2012) The Caulobacter crescentus phage phiCbK: genomics of a canonical phage. BMC Genomics 13:542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15).Johnson RC, Wood NB, Ely B (1977) Isolation and characterization of bacteriophages for Caulobacter crescentus. J Gen Virol 37:323–335. [Google Scholar]
- 16).Juhala RJ, Ford ME, Duda RL et al. (2000) Genomic sequences of bacteriophages HK97 and HK022: pervasive genetic mosaicism in the lambdoid bacteriophages. J Mol Biol 299:27–51. [DOI] [PubMed] [Google Scholar]
- 17).Lee JH, Shin H, Choi Y, Ryu S (2013) Complete genome sequence analysis of bacterial-flagellum-targeting bacteriophage chi. Arch Virol 158:2179–2183. [DOI] [PubMed] [Google Scholar]
- 18).Naveed M, Mitter B, Yousaf S et al. (2014) The endophyte Enterobacter sp. FD17: a maize growth enhancer selected based on rigorous testing of plant beneficial traits and colonization characteristics. Biol Fertil Soils 50:249–262. [Google Scholar]
- 19).Nedialkova LP, Sidstedt M, Koeppel MB et al. (2016) Temperate phages promote colicin-dependent fitness of Salmonella enteric serovar Typhimurium. Enviro Microbiol 18:1591–1603. [DOI] [PubMed] [Google Scholar]
- 20).Nguyen D, Ely B (2018) A genome comparison of T7-like Podoviruses that infect Caulobacter crescentus. Curr Microbiol 75:760–765. [DOI] [PubMed] [Google Scholar]
- 21).Oliveira L, Tavares P, Alonso JC (2013) Headful DNA packaging: bacteriophage SPP1 as a model system. Virus research 173:247–259. [DOI] [PubMed] [Google Scholar]
- 22).Poindexter JS (1964) Biological properties and classification of the Caulobacter group. Bacteriological reviews 28:231–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23).Prischl M, Hackl E, Pastar M et al. (2012) Genetically modified Bt maize lines containing cry3Bb1, cry1A105 or cry1Ab2 do not affect the structure and functioning of root-associated endophyte communities. Appl Soil Ecol 54:39–48. [Google Scholar]
- 24).Rao VB, Feiss M (2008) The bacteriophage DNA packaging motor. Annu Rev Genet 42:647–681. [DOI] [PubMed] [Google Scholar]
- 25).Richardson CC (1983) Bacteriophage T7: minimal requirements for the replication of a duplex DNA molecule. Cell 33:315–317. [DOI] [PubMed] [Google Scholar]
- 26).Roberts MD, Martin NL, Kropinski AM (2004) The genome and proteome of coliphage T1. Virology 318:245–66. [DOI] [PubMed] [Google Scholar]
- 27).Rosenvold EC, Honigman A (1977) Mapping of AvaI and XmaI cleavage sites in bacteriophage DNA including a new technique of DNA digestion in agarose gels. Gene 2:273–288. [Google Scholar]
- 28).Rutherford K, Parkhill J, Crook J et al. (2000) Artemis: sequence visualization and annotation. Bioinformatics 10:944–945. [DOI] [PubMed] [Google Scholar]
- 29).Saraf M, Jha CK, Patel D (2010) The role of ACC deaminase producing PGPR in sustainable agriculture In: Maheshwari DK (ed) Plant growth and health promoting bacteria, vol. 1 Springer-Verlag, pp. 365–385. [Google Scholar]
- 30).Schaefer AL, Lappala CR, Morlen RP et al. (2013) LuxR-and LuxI-type quorum sensing circuits are prevalent in members of the Populus deltoides microbiome. J Appl Environ Microbiol 79:5745–5752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31).Sharaf A, Mercati F, Elmaghraby I et al. (2017) Functional and comparative genome analysis of novel virulent actinophages belonging to Streptomyces flavovirens. BMC Microbiology 17:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32).Takeshi M, Kenichi M (1983) Lambda phage DNA sequences affecting the packaging process. Gene 24:199–206. [DOI] [PubMed] [Google Scholar]
- 33).Vahanian N, Oh CS, Sippy J et al. (2017) Natural history of a viral cohesive end site: cosN of the λ-like phages. Virology 509:140–145. [DOI] [PubMed] [Google Scholar]
- 34).Vance CP (2011) Phosphorus as a Critical Macronutrient In: Barraclough P, Hawkesford MJ (eds) The Molecular and Physiological Basis of Nutrient Use Efficiency in Crops, vol. 1 John Wiley & Sons, Inc., pp. 227–264. [Google Scholar]
- 35).Wang N (2006) Lysis timing and bacteriophage fitness. Genetics 172:17–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36).Waterbury PG, Lane MJ (1987) Generation of lambda phage concatemers for use as pulsed field electrophoresis size markers. Nucleic Acids Res 15:3930. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.