Abstract
Carbapenems are β‐lactam antibiotics used in healthcare settings as last resort drugs to treat infections caused by antibiotic‐resistant bacteria. Carbapenem‐resistant bacteria are increasingly being isolated from healthcare facilities; however, little is known about their distribution or prevalence in the environment, especially in the United States, where their distribution in water environments from the West Coast has not been studied before. The aim of this study was to determine the prevalence of carbapenem‐resistant bacteria and carbapenemase genes in water bodies from the Los Angeles area (California, USA). All samples that were analyzed contained carbapenem‐resistant bacteria with a frequency of between 0.1 and 324 carbapenem‐resistant cfu per 100 mls of water. We identified 76 carbapenem‐resistant or ‐intermediate isolates, most of which were also resistant to noncarbapenem antibiotics, as different strains of Enterobacter asburiae, Aeromonas veronii, Cupriavidus gilardii, Pseudomonas, and Stenotrophomonas species. Of them, 52 isolates were carbapenemase‐producers. Furthermore, PCR and sequence analysis to identify the carbapenemase gene of these carbapenemase‐producing isolates revealed that all Enterobacter asburiae isolates had a bla IMI ‐2 gene 100% identical to the reference sequence, and all Stenotrophomonas maltophlia isolates had a bla L1 gene 83%–99% identical to the reference bla L1. Our findings indicate that water environments in Southern California are an important reservoir of bacteria‐resistant to carbapenems and other antibiotics, including bacteria carrying intrinsic and acquired carbapenemase genes.
Keywords: Aeromonas, carbapenem, carbapenemase, carbapenem‐resistant, Cupriavidus, Enterobacter, Pseudomonas, Stenotrophomonas
1. INTRODUCTION
Carbapenems (ertapenem, imipenem, meropenem and doripenem) are broad‐spectrum β‐lactam antibiotics. Unlike other β‐lactams such as penicillins and cephalosporins, carbapenems are resistant to hydrolysis by β‐lactamases and extended spectrum β‐lactamases (Martin & Kaye, 2004; Papp‐Wallace, Endimiani, Taracila, & Bonomo, 2011; Vardakas, Tansarli, Rafailidis, & Falagas, 2012). The use of carbapenems is generally restricted to hospitals and other healthcare settings, where they are used as last resort drugs to treat serious infections caused by antibiotic‐resistant bacteria (Bradley et al., 1999; Nordmann, Dortet, & Poirel, 2012; Papp‐Wallace et al., 2011; Paterson, 2000, 2002; Paterson & Bonomo, 2005; Torres, Villegas, & Quinn, 2007).
Carbapenem‐resistant bacteria represent a major challenge to public health. These bacteria are primarily considered as nosocomial pathogens (Bratu, Landman, et al., 2005; Bratu, Mooty, et al., 2005; Centers for Disease Control and Prevention, 2013a), and their isolation in healthcare settings is on the rise (Centers for Disease Control and Prevention, 2018; Correa et al., 2012; Cuzon et al., 2011; Guh et al., 2015; Gupta, Limbago, Patel, & Kallen, 2011; Kallen, Hidron, Patel, & Srinivasan, 2010; Khuntayaporn, Montakantikul, Mootsikapun, Thamlikitkul, & Chomnawang, 2012; Prabaker & Weinstein, 2011; Queenan et al., 2012; Rhomberg & Jones, 2009; Rizek et al., 2014; Rodríguez‐Martínez, Poirel, & Nordmann, 2009; van Duijn, Dautzenberg, & Oostdijk, 2011; Viehman, Nguyen, & Doi, 2014). For example, carbapenem‐resistant Enterobacteriaceae have been designated as an urgent threat by the CDC (Centers for Disease Control and Prevention, 2013a, 2013b) and are associated with very high mortality rates (Papp‐Wallace et al., 2011; Paterson, 2000; van Duin, Kaye, Neuner, & Bonomo, 2013). Likewise, Acinetobacter baumannii and Pseudomonas aeruginosa strains that are resistant to multiple antibiotics, including carbapenems, have been designated by the CDC as serious threats, and are often untreatable (Centers for Disease Control and Prevention, 2013a, 2013b). Stenotrophomonas maltophilia, another hard‐to‐treat emerging pathogen that causes pneumonia and blood infections among other diseases (Brooke, 2012), is usually resistant to most antibiotics, including carbapenems (Brooke, 2012; Yang et al., 2014).
A variety of mechanisms can contribute to carbapenem resistance. These include decreased outer membrane permeability (Livermore, Mushtaq, & Warner, 2005; Rizek et al., 2014; Shin et al., 2012; Sho et al., 2013; Warner et al., 2013), overexpression of efflux pumps or chromosomal β‐lactamases (Papp‐Wallace et al., 2011; Rodríguez‐Martínez et al., 2009; Warner et al., 2013), and production of carbapenemases, which are enzymes that degrade carbapenems and other β‐lactams (Marsik & Nambiar, 2011; Queenan & Bush, 2007). Carbapenemase production is especially worrisome because of the strong activity of these enzymes against carbapenems, and the fact that carbapenemase genes are frequently found in genetic mobile elements such as plasmids, which favors their spread (Mathers et al., 2011; Walsh, 2010).
Despite their public health importance and increased incidence in healthcare facilities (Guh et al., 2015; Gupta et al., 2011; Papp‐Wallace et al., 2011; Prabaker & Weinstein, 2011; Rhomberg & Jones, 2009; van Duijn et al., 2011), knowledge about carbapenem‐resistant bacteria and genes in the environment is very limited, especially in the United States. Most efforts to detect these bacteria have focused on healthcare (Conlan et al., 2014; Doi & Paterson, 2015; Guh et al., 2015; Gupta et al., 2011) or immediately related settings such as hospital wastewater (Chagas, Seki, da Silva, & Asensi, 2011; Nasri et al., 2017; White et al., 2016). However, recent findings in Europe, Africa and Asia have revealed carbapenem‐resistant bacteria and genes in freshwater and other environmental samples (Di, Jang, Unno, & Hur, 2017; Girlich, Poirel, & Nordmann, 2010; Henriques et al., 2012; Isozumi et al., 2012; Poirel et al., 2012; Potron, Poirel, Bussy, & Nordmann, 2011; Tacão, Correia, & Henriques, 2015; Zurfluh, Hachler, Nuesch‐Inderbinen, & Stephan, 2013).
In the United States, carbapenem‐resistant bacteria were isolated from 7 out of 16 rivers from the Midwest sampled between 1999 and 2001 (Ash, Mauck, & Morgan, 2002; Aubron, Poirel, Ash, & Nordmann, 2005). To this date, this study remains the only specific analysis about the distribution and characteristics of carbapenem‐resistant bacteria in water environments not directly related to healthcare facilities in the United States. Given the importance of carbapenem‐resistant bacteria, further studies on other areas of the United States and on different types of aquatic environments, are needed to gain a better understanding of the environmental distribution and molecular mechanisms of carbapenem‐resistant bacteria in the United States. To contribute to address this gap in knowledge, we report here the first study about the distribution and characteristics of carbapenem‐resistant bacteria and carbapenemase genes in aquatic environments on the West Coast of the United States, as well as the first study about these bacteria and genes in ponds and lakes in the United States. All samples analyzed contained carbapenem‐resistant bacteria — most of which were also resistant to other antibiotics — which we identified as Enterobacter, Aeromonas, Cupriavidus, Pseudomonas or Stenotrophomonas species. Many of the carbapenem‐resistant isolates further characterized carried a carbapenemase gene. These findings suggest that carbapenem‐resistant bacteria and carbapenemase genes are widely distributed on diverse water environments on the West Coast of the United States.
2. MATERIALS AND METHODS
2.1. Sample collection and isolation of carbapenem‐resistant bacteria
We collected 10 different water samples from ponds and lakes in the Los Angeles (California) area between June of 2016 and March of 2017. The location (Figure 1) and characteristics of the sampling sites are summarized in Table 1. Four liters of surface‐level water were collected in sterile bottles and immediately transported to the laboratory. The total count of gram‐negative bacteria was determined by direct plating 100 μl of water (and also by spot plating 10 μl of a 100 to 10–4 dilution bank of each sample in sterile water) on MacConkey agar (Fisher Scientific, Hampton, NH) plates, followed by incubation in aerobic conditions for 24 hr at 37°C, and colony counting. The count of carbapenem‐resistant gram‐negatives was determined by the same procedure except for using MacConkey agar plates supplemented with 4 μg/ml of meropenem (Ark Pharm, Inc., Arlington Heights, IL), which is the meropenem minimum inhibitory concentration (MIC) clinical breakpoint for Enterobacteriaceae according to the Clinical and Laboratory Standards Institute (CLSI) (Clinical and Laboratory Standards Institute, 2017). Meropenem was the second carbapenem approved for medical use in the United States, and has stronger activity than imipenem against most gram‐negatives — the main target in our studies — such as Enterobacteriaceae (Papp‐Wallace et al., 2011). In addition, we concentrated the bacteria present in 2 L of water sample by filtration, using 0.45 μm filters (Merck Millipore, Billerica, MA), and placed the filters onto MacConkey‐meropenem plates for incubation as described above.
Figure 1.
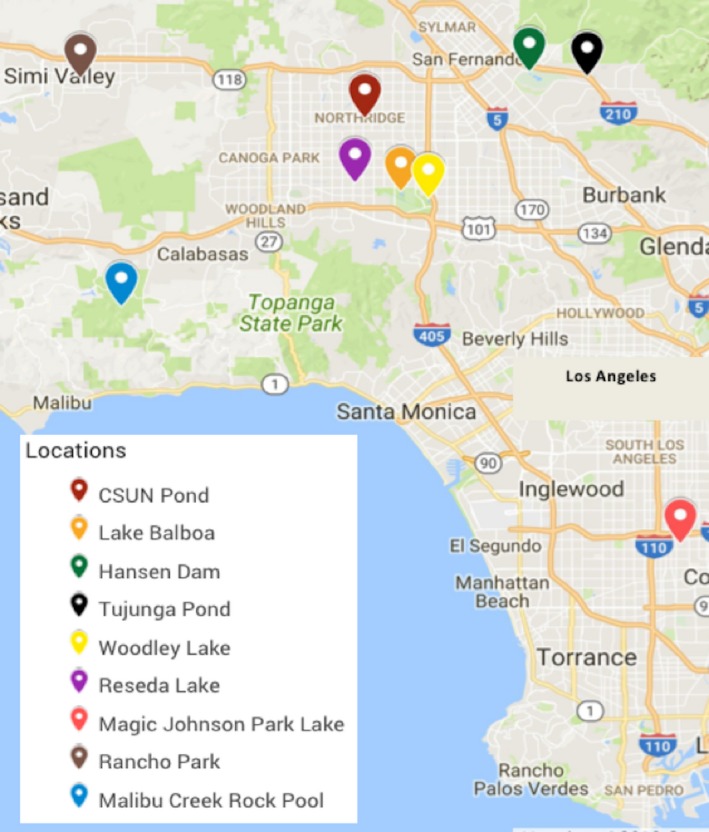
Map of the location of the ponds and lakes from the Los Angeles‐Southern California area sampled in this study
Table 1.
Summary of the origin, total gram‐negative and carbapenem‐resistant gram‐negative bacterial counts obtained in this study
| Sample | Date | Location (Type) | GPS Location | Total bacteria (cfu/100 ml) | Carbapenem‐resistant Bacteria (cfu/100 ml) |
|---|---|---|---|---|---|
| W1 | 6/7/2016 | CSUN Duck Pond (artificial ponda) | 34.2367024, ‐118.5261293 | 4.2 × 105 | 150.0 |
| W2 | 8/2/2016 | CSUN Duck Pond (artificial ponda) | 34.2367024, ‐118.5261293 | 1.2 × 105 | 10.0 |
| W3 | 8/17/2016 | Lake Balboa (reclaimed water from DCTWRPb) | 34.182312, ‐118.495627 | 2.4 × 104 | 22.5 |
| W4 | 9/29/2016 | Hansen Dam (flood control reservoir) | 34.271505, ‐118.388383 | 8.3 × 104 | 42.4 |
| W5 | 10/5/2016 | Tujunga Ponds Wildlife Sanctuary (spring waterc) | 34.268050, ‐118.340026 | 8.8 × 105 | 16.0 |
| W6 | 10/7/2016 | Woodley Wildlife Lake (reclaimed water from DCTWRPb) | 34.177256, ‐118.472841 | 4.0 × 104 | 324.0 |
| W7 | 10/11/2016 | Reseda Park Lake (artificial lake with potable waterd) | 34.188714, ‐118.534383 | 1.0 × 104 | 0.1 |
| W8 | 1/29/2017 | Magic Johnson Park lake (Potable waterd) | 33.919458, ‐118.261776 | 1.2 × 105 | 11.0 |
| W9 | 1/17/2017 | Rancho Simi Community Park Duck Pond (Potable waterd) | 34.266453,‐118.764119 | 3.9 × 104 | 36.8 |
| W10 | 3/1/2017 | Malibu Creek Rock Pool (natural poole) | 34.096555, ‐118.729879 | 4.9 × 104 | 9.2 |
Note.We obtained two water samples from this artificial pond, one before (June of 2016) and one after (August 2016) it was cleaned and the water–pumping system fixed. This artificial pond uses circulation of potable water.
Lake Balboa and Woodley Wildlife Lake are filled with reclaimed water from the Tillman Water Reclamation Plant (DCTWRP). Lake balboa is a recreational lake, and Woodley Wildlife lake is a wild wetland habitat with many species of birds.
The water from the Tujunga Ponds Wildlife Sanctuary is spring water from the Tujunga Canyon delivered to the pond via a small stream.
Reseda Park Lake, Magic Johnson Park lake, and Rancho Simi Community Park Duck Pond use circulation of potable water. Reseda park lake is an asphalt‐lined urban lake.
Malibu Creek Rock Pool is a natural pool filled with rain run‐off.
Up to 50 meropenem‐resistant colonies per sample, choosing different colony morphologies whenever possible, were patched the next day in Mueller‐Hinton (Fisher Scientific) agar plates supplemented with meropenem at 4 μg/ml, which is the CLSI meropenem MIC clinical breakpoint for Enterobacteriaceae, and 16 μg/ml, which is the CLSI meropenem MIC clinical breakpoint for other non‐Enterobacteriaceae gram‐negatives (Clinical and Laboratory Standards Institute, 2017), to confirm their resistance to meropenem. Over 90% of the colonies patched were confirmed as meropenem‐resistant and grew at both 4 and 16 μg/ml of meropenem. For each water sample, eight to twelve different carbapenem‐resistant isolates were restreaked on Mueller–Hinton‐meropenem‐16 μg/ml plates and incubated as described above to obtain isolated colonies. One colony per isolate was used to inoculate Mueller–Hinton broth supplemented with meropenem‐16 μg/ml. These cultures were incubated in aerobic conditions with 200 rpm agitation for 18–24 hr at 37°C. A portion of each overnight culture was saved with 20% (v/v) glycerol at −80°C for long‐term storage, and the remainder was washed and used for PCR analysis. Cells were washed twice by centrifugation for 1 min at 13,000 rpm (16,200g), supernatant removal, and resuspension in DNA grade water (Fisher Scientific). Washed cells were then stored at −20°C until their use as PCR template for amplification of 16S rDNA or carbapenemase genes.
2.2. Identification of carbapenem‐resistant bacteria by 16S rDNA sequencing and oxidase test
The 16S rDNA genes from the 76 selected isolates were amplified using reagents and Dreamtaq polymerase purchased from Thermo Fisher Scientific (Canoga Park, CA), and using the previously described primers 8F and U1492R (Eden, Schmidt, Blakemore, & Pace, 1991), which were purchased from IDT (Coralville, IA). The PCR mixture (50 μl) contained DNA grade water, colorless DreamTaq Buffer, 0.2 mM dNTPs, 1.25 DreamTaq polymerase units, 0.5 μM of each primer, and 5 μl of isolate template (washed cells) prepared as described above. Washed E. coli BW25113 cells were used as template positive control, and DNA grade water was used as the nontemplate control. The amplification reaction was performed in a Simpliamp thermal cycler (Applied Biosystems/Thermo Fisher Scientific), using the following program: one cycle at 95°C for 2 min, 35 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 90 s, with a final cycle of 72°C for 7.5 min and 4°C for infinite. PCR products were then visualized by DNA electrophoresis before sequencing them at Laragen Inc. (Culver city, CA), and analyzing the resulting sequences by BLAST (Altschul et al., 1997).
Because several species of the genera Pseudomonas and Stenotrophomonas are closely related and are difficult to distinguish based only on their 16S rDNA sequences, isolates in which their 16S rDNA closely matched both genera were further identified using the oxidase test. This test detects the production of the cytochrome C oxidase enzyme, and is positive for Pseudomonas and negative for Stenotrophomonas (Bergey & Holt, 1994). The oxidase test was performed using the Becton Dickinson BBL DrySlide Oxidase reagent (Sparks, MD). Briefly, strains were grown at 37°C, overnight on Mueller–Hinton agar plates. A plastic pipette tip was used to transfer a large clump of cells to the DrySlide. Isolates that turned blue within 10 s were scored as positives for the oxidase test. Lab strains of Pseudomonas stutzeri and E. coli BW25113 were used as our positive and negative controls, respectively.
2.3. Determination of the antibiotic susceptibility profile of carbapenem‐resistant isolates
Determination of the antibiotic susceptibility profile for carbapenems and other antibiotics (Table 2; Table 3; and Figure 2) for each selected isolate was performed using the disk diffusion method as described by CLSI (Clinical and Laboratory Standards Institute, 2017), using cells grown 16–18 hr at 37°C on Mueller‐Hinton agar plates, and using the reference strain E. coli ATCC 25922 as a quality control (Clinical and Laboratory Standards Institute, 2017). All antibiotic disks (meropenem 10 μg, imipenem 10 μg, cefotaxime 30 μg, ciprofloxacin 5 μg, gentamicin 10 μg, and tetracycline 30 μg) were purchased from Becton Dickinson (Franklin Lakes, NJ). We used the CLSI zone diameter breakpoint values (Clinical and Laboratory Standards Institute, 2017) to determine whether our isolates were resistant, intermediate, or sensitive to the different antibiotics tested. For taxa in which the CLSI zone diameter breakpoint values were not available, we used the Enterobacteriaceae values.
Table 2.
Number and characteristics of carbapenem‐resistant isolates identified from water samples described in Table 1
| Species | Sample of Origin | Number of isolates | Number of CPa isolates | Carbapenemase genea | Antibiotic Resistance (number of isolates)b |
|---|---|---|---|---|---|
| Enterobacter asburiae | W6 | 7 | 7 | bla IMI‐2 | MP (7), IM (7) |
| Aeromonas veronii | W7 | 2 | 0 | N/A | MP (1), IM (1), TE (1) |
| Cupriavidus gilardii | W2, W8 | 2 | 0 | N/A | MP (2), GE (2) |
| Pseudomonas alcaligenes | W1, W4, W5 | 5 | 0 | N/A | MP (5), IM (1), CF (4) |
| Pseudomonas cedrina | W9 | 1 | 0 | N/A | MP (1), IM (1), CF (1) |
| Pseudomonas geniculata | W8 | 1 | 0 | N/A | MP (1), GE (1) |
| Pseudomonas otitidis | W3–5 | 9 | 0 | N/A | MP (9), IM (2) |
| Pseudomonas stutzeri | W3 | 1 | 0 | N/A | MP (1), IM (1), CI (1) |
| Stenotrophomonas maltophilia | W1–4, W8–10 | 45 | 45 | bla L1 | MP (45), IM (45), CF (44), GE (25), TE (6) |
| Stenotrophomonas pavanii | W5 | 3 | 0 | N/A | MP (3), IM (3), CF (3), GE (2) |
| Total | 76 | 52 | 52 | MP (75), IM (61), CF (52), CI (1), GE (30), TE (7) |
Note.CP = carbapenemase‐producing as determined by the CarbaNP test. CarbaNP‐positive isolates were further tested by PCR and sequencing to identify their carbapenemase gene, whereas CarbaNP‐negative isolates were not further tested and are shown as N/A in the carbapenemase gene column.
In parentheses, the number of isolates that were resistant (intermediate isolates are not included) to meropenem (MP), imipenem (IM), cefotaxime (CF), ciprofloxacin (CI), gentamicin (GE), and tetracycline (TE). The detailed antibiotic susceptibility profile, CarbaNP result and carbapenemase gene detected for each individual isolate are provided in Table 3.
Table 3.
Carbapenem‐resistant isolates identified and characterized in this study
| Closest species identified by BLAST using 16S rDNA genea | Isolate# | Inhibition zone (diameter in mm)b | Carba NPc | Carbape nemase gene (%identityd) | |||||
|---|---|---|---|---|---|---|---|---|---|
| MP | IM | CF | CI | GE | TE | ||||
| Aeromonas veronii | W7‐1 | 23 | 20 | 40 | 39 | 27 | 25 | – | N/A |
| Aeromonas veronii | W7‐2 | 5 | 0 | 31 | 34 | 22 | 11 | – | N/A |
| Cupriavidus gilardii | W2‐2 | 1 | 20 | 40 | 36 | 0 | 28 | – | N/A |
| Cupriavidus gilardii | W2‐5 | 2 | 20 | 40 | 36 | 0 | 29 | – | N/A |
| Enterobacter asburiae | W6‐1 | 0 | 0 | 32 | 39 | 27 | 28 | + | bla IMI‐2 (100%) |
| Enterobacter asburiae | W6‐2 | 0 | 0 | 34 | 39 | 27 | 27 | + | bla IMI‐2 (100%) |
| Enterobacter asburiae | W6‐3 | 0 | 0 | 31 | 39 | 25 | 26 | + | bla IMI‐2 (100%) |
| Enterobacter asburiae | W6‐4 | 0 | 0 | 32 | 34 | 25 | 25 | + | bla IMI‐2 (100%) |
| Enterobacter asburiae | W6‐5 | 0 | 0 | 36 | 41 | 26 | 29 | + | bla IMI‐2 (100%) |
| Enterobacter asburiae | W6‐7 | 0 | 0 | 35 | 39 | 27 | 28 | + | bla IMI‐2 (100%) |
| Enterobacter asburiae | W6‐8 | 0 | 0 | 37 | 36 | 28 | 28 | + | bla IMI‐2 (100%) |
| Pseudomonas alcaligenes | W1‐4 | 18 | 26 | 29 | 39 | 24 | 22 | – | N/A |
| Pseudomonas alcaligenes | W4‐5 | 11 | 26 | 15 | 47 | 29 | 27 | – | N/A |
| Pseudomonas alcaligenes | W5‐5 | 8 | 15 | 10 | 38 | 25 | 24 | – | N/A |
| Pseudomonas alcaligenes | W5‐7 | 17 | 24 | 9 | 34 | 25 | 21 | – | N/A |
| Pseudomonas alcaligenes | W5‐8 | 14 | 25 | 12 | 40 | 26 | 20 | – | N/A |
| Pseudomonas cedrina | W9‐8 | 11 | 13 | 16 | 27 | 33 | 33 | – | N/A |
| Pseudomonas geniculata | W8‐10 | 0 | 21 | 43 | 37 | 0 | 31 | – | N/A |
| Pseudomonas otitidis | W3‐5 | 11 | 20 | 26 | 33 | 26 | 25 | – | N/A |
| Pseudomonas otitidis | W4‐1 | 15 | 22 | 27 | 41 | 29 | 23 | – | N/A |
| Pseudomonas otitidis | W4‐2 | 14 | 24 | 29 | 39 | 32 | 21 | – | N/A |
| Pseudomonas otitidis | W4‐3 | 1 | 21 | 27 | 41 | 31 | 25 | – | N/A |
| Pseudomonas otitidis | W4‐6 | 10 | 18 | 24 | 39 | 28 | 22 | – | N/A |
| Pseudomonas otitidis | W4‐7 | 10 | 21 | 27 | 39 | 30 | 23 | – | N/A |
| Pseudomonas otitidis | W4‐8 | 10 | 21 | 25 | 37 | 29 | 21 | – | N/A |
| Pseudomonas otitidis | W5‐3 | 14 | 22 | 25 | 30 | 24 | 20 | – | N/A |
| Pseudomonas otitidis | W5‐4 | 0 | 18 | 21 | 30 | 22 | 15 | – | N/A |
| Pseudomonas stutzeri | W3‐4 | 11 | 19 | 22 | 14 | 23 | 25 | – | N/A |
| Stenotrophomonas maltophilia | W1‐2 | 0 | 0 | 0 | 19 | 0 | 8 | + | bla L1 (99%) |
| Stenotrophomonas maltophilia | W1‐3 | 0 | 0 | 8 | 26 | 21 | 20 | + | bla L1 (84%) |
| Stenotrophomonas maltophilia | W1‐5 | 0 | 0 | 10 | 26 | 24 | 22 | + | bla L1 (83%) |
| Stenotrophomonas maltophilia | W1‐6 | 0 | 0 | 0 | 26 | 22 | 21 | + | bla L1 (90%) |
| Stenotrophomonas maltophilia | W2‐1 | 0 | 0 | 13 | 27 | 12 | 14 | + | bla L1 (92%) |
| Stenotrophomonas maltophilia | W2‐3 | 0 | 0 | 24 | 26 | 19 | 19 | + | bla L1 (84%) |
| Stenotrophomonas maltophilia | W2‐4 | 0 | 0 | 11 | 25 | 11 | 15 | + | bla L1 (89%) |
| Stenotrophomonas maltophilia | W2‐6 | 2 | 0 | 12 | 29 | 11 | 13 | + | bla L1 (89%) |
| Stenotrophomonas maltophilia | W2‐7 | 0 | 0 | 12 | 25 | 11 | 12 | + | bla L1 (89%) |
| Stenotrophomonas maltophilia | W2‐8 | 0 | 0 | 13 | 37 | 12 | 13 | + | bla L1 (89%) |
| Stenotrophomonas maltophilia | W3‐1 | 0 | 0 | 11 | 25 | 11 | 11 | + | bla L1 (89%) |
| Stenotrophomonas maltophilia | W3‐2 | 0 | 0 | 11 | 22 | 5 | 11 | + | bla L1 (92%) |
| Stenotrophomonas maltophilia | W3‐6 | 0 | 0 | 0 | 22 | 12 | 12 | + | bla L1 (99%) |
| Stenotrophomonas maltophilia | W3‐7 | 0 | 0 | 12 | 26 | 11 | 15 | + | bla L1 (89%) |
| Stenotrophomonas maltophilia | W3‐8 | 0 | 0 | 12 | 28 | 14 | 14 | + | bla L1 (89%) |
| Stenotrophomonas maltophilia | W4‐4 | 0 | 0 | 0 | 24 | 16 | 13 | + | bla L1 (93%) |
| Stenotrophomonas maltophilia | W8‐1 | 0 | 0 | 12 | 37 | 21 | 20 | + | bla L1 (89%) |
| Stenotrophomonas maltophilia | W8‐2 | 0 | 0 | 0 | 26 | 7 | 14 | + | bla L1 (94%) |
| Stenotrophomonas maltophilia | W8‐3 | 0 | 0 | 9 | 31 | 23 | 22 | + | bla L1 (84%) |
| Stenotrophomonas maltophilia | W8‐4 | 0 | 0 | 0 | 24 | 19 | 11 | + | bla L1 (94%) |
| Stenotrophomonas maltophilia | W8‐5 | 0 | 0 | 0 | 24 | 17 | 11 | + | bla L1 (94%) |
| Stenotrophomonas maltophilia | W8‐7 | 0 | 0 | 0 | 37 | 15 | 20 | + | bla L1 (94%) |
| Stenotrophomonas maltophilia | W8‐8 | 0 | 0 | 0 | 37 | 18 | 20 | + | bla L1 (92%) |
| Stenotrophomonas maltophilia | W8‐9 | 0 | 0 | 13 | 37 | 25 | 20 | + | bla L1 (88%) |
| Stenotrophomonas maltophilia | W8‐11 | 0 | 0 | 13 | 37 | 20 | 19 | + | bla L1 (89%) |
| Stenotrophomonas maltophilia | W8‐12 | 0 | 0 | 13 | 37 | 27 | 21 | + | bla L1 (89%) |
| Stenotrophomonas maltophilia | W9‐1 | 0 | 0 | 11 | 32 | 11 | 20 | + | bla L1 (84%) |
| Stenotrophomonas maltophilia | W9‐2 | 13 | 0 | 11 | 29 | 12 | 18 | + | bla L1 (83%) |
| Stenotrophomonas maltophilia | W9‐3 | 0 | 0 | 9 | 37 | 25 | 23 | + | bla L1 (89%) |
| Stenotrophomonas maltophilia | W9‐4 | 0 | 0 | 9 | 30 | 0 | 16 | + | bla L1 (88%) |
| Stenotrophomonas maltophilia | W9‐5 | 0 | 0 | 11 | 32 | 27 | 21 | + | bla L1 (84%) |
| Stenotrophomonas maltophilia | W9‐6 | 0 | 0 | 0 | 28 | 11 | 13 | + | bla L1 (94%) |
| Stenotrophomonas maltophilia | W9‐7 | 0 | 0 | 16 | 29 | 11 | 15 | + | bla L1 (92%) |
| Stenotrophomonas maltophilia | W9‐12 | 0 | 0 | 10 | 34 | 26 | 24 | + | bla L1 (83%) |
| Stenotrophomonas maltophilia | W10‐1 | 0 | 0 | 0 | 28 | 10 | 17 | + | bla L1 (88%) |
| Stenotrophomonas maltophilia | W10‐2 | 0 | 0 | 0 | 27 | 8 | 16 | + | bla L1 (89%) |
| Stenotrophomonas maltophilia | W10‐3 | 0 | 0 | 0 | 28 | 11 | 15 | + | bla L1 (89%) |
| Stenotrophomonas maltophilia | W10‐5 | 0 | 0 | 0 | 29 | 6 | 14 | + | bla L1 (89%) |
| Stenotrophomonas maltophilia | W10‐6 | 0 | 0 | 0 | 27 | 10 | 15 | + | bla L1 (89%) |
| Stenotrophomonas maltophilia | W10‐7 | 0 | 0 | 0 | 27 | 9 | 15 | + | bla L1 (89%) |
| Stenotrophomonas maltophilia | W10‐8 | 0 | 0 | 0 | 27 | 16 | 14 | + | bla L1 (88%) |
| Stenotrophomonas maltophilia | W10‐9 | 0 | 0 | 0 | 28 | 4 | 14 | + | bla L1 (89%) |
| Stenotrophomonas maltophilia | W10‐10 | 0 | 0 | 0 | 27 | 15 | 11 | + | bla L1 (85%) |
| Stenotrophomonas maltophilia | W10‐11 | 0 | 0 | 10 | 28 | 0 | 15 | + | bla L1 (84%) |
| Stenotrophomonas maltophilia | W10‐12 | 0 | 0 | 13 | 28 | 0 | 14 | + | bla L1 (92%) |
| Stenotrophomonas pavanii | W5‐1 | 0 | 0 | 12 | 32 | 13 | 18 | – | N/A |
| Stenotrophomonas pavanii | W5‐2 | 0 | 0 | 11 | 27 | 9 | 17 | – | N/A |
| Stenotrophomonas pavanii | W5‐6 | 0 | 0 | 10 | 28 | 0 | 17 | – | N/A |
Note.For each isolate, we obtained their 16S rDNA sequence and used BLAST (Altschul et al., 1997) to determine the closest known strain. In all cases, the DNA identity between our isolate and the top BLAST known strain hit was ≥98% (≥99% for most isolates).
MP: meropenem; IM: imipenem; CF: cefotaxime; CI: ciprofloxacin; GE: gentamicin; TE: tetracycline. To determine whether our isolates were Resistant (highlighted in red), Intermediate (highlighted in yellow) or Sensitive (no highlight) to the antibiotics tested, we used the CLSI zone diameter clinical breakpoint values (Clinical and Laboratory Standards Institute, 2017). For taxa in which the CLSI zone diameter breakpoint values were not available, we used the Enterobacteriaceae values.
All CarbaNP‐positive isolates (carbapenemase‐producing isolates) were positive when the test was performed measuring the hydrolysis of both meropenem and imipenem.
Only carbapenemase‐producing isolates (CarbaNP‐positive isolates) were tested by PCR to identify their potential carbapenemases. The rest of isolates were not tested because they were CarbaNP‐negative and are shown as N/A. “%identity” indicates % DNA identity (shown in parenthesis) between the reference bla IMI‐2 or bla L1 gene and the isolate bla IMI‐2 or bla L1 sequence obtained for that isolate.
Figure 2.
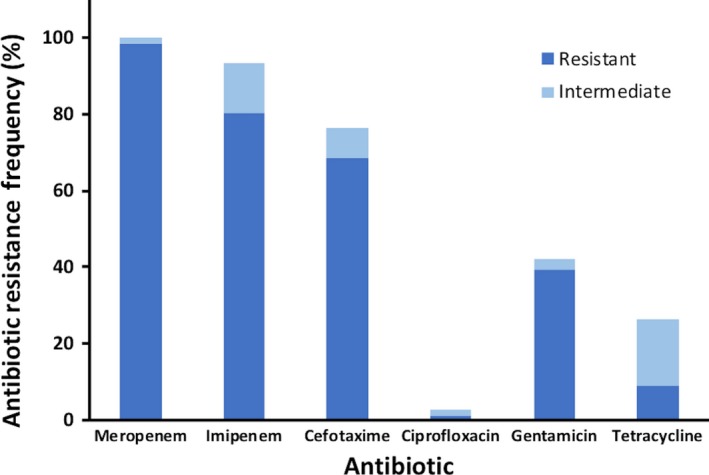
Antibiotic resistance frequency of the water isolates characterized in this study for carbapenem (meropenem and imipenem) and non‐carbapenem (cefotaxime, ciprofloxacin, gentamicin and tetracycline) antibiotics. For each antibiotic tested, the percentage of resistant isolates is shown in dark blue, and the percentage of intermediate isolates is shown in light blue
2.4. Identification of carbapenemase‐producing isolates by the CarbaNP assay
We used the CarbaNP assay (Dortet, Poirel, & Nordmann, 2012a, 2012b; Nordmann, Poirel, & Dortet, 2012) to identify which carbapenem‐resistant isolates produced carbapenemases. The assay was performed as described by CLSI (Clinical and Laboratory Standards Institute, 2017) using 6 mg/ml of either meropenem or imipenem, and using colonies from isolates grown overnight at 37°C on Mueller–Hinton agar without (to detect for noninducible carbapenemases) or with (to detect for carbapenem‐inducible carbapenemases) meropenem at 4 or 16 μg/ml. Isolates that hydrolyzed meropenem and/or imipenem, and thus turned yellow at 37°C within 2 hr, but did not turn yellow in the absence of meropenem or imipenem, were considered positive for carbapenemase production. Carbapenemase production was considered carbapenem‐inducible when the CarbaNP test was positive only when using cells grown in Mueller–Hinton‐meropenem plates.
2.5. PCR, sequencing and phylogenetic analysis of carbapenemases
PCR amplification and sequencing of carbapenemases from CarbaNP‐positive isolates were performed as described in section 2.2, with the following modifications: We used the primers and PCR program described by Henriques et al. (2012) to amplify bla L1. To amplify bla IMI, we designed the primers Imi2‐F1 (5’‐CAA GTA GAA TAG CCA TCT TGT TTA G) and Imi2‐R1 (5’‐AGG TTA TCA ATT GCG ATT CTT G), which amplify 853 out of 870 bp of the bla IMI‐1 (U50278) and bla IMI‐2 (DQ173429) genes, and performed the PCR step using a Tm of 55°C and an extension time of 1 min. For each PCR, washed cells of strains carrying each bla gene were used as positive control, E. coli BW25113 was used as negative control, and DNA grade water was used as nontemplate control. We used Geneious R11 software to perform multiple sequence alignments (MUSCLE alignment tool) of the bla IMI‐2 sequences obtained and the bla IMI‐2 reference sequence (DQ173429), and of the bla L1 sequences obtained and the bla L1 reference sequence (NG_047502), as well as to build a bla L1 phylogenetic tree based on the Jukes–Cantor genetic distance model and the Neighbor‐Joining method.
2.6. Nucleotide accession numbers
All 16S rDNA, bla IMI‐2, and bla L1 sequences obtained in this study have been deposited in GenBank (https://www.ncbi.nlm.nih.gov/genbank/) under the following accession numbers: MG905248–MG905292, MG905294–MG905307, MG905309–MG905321, and MH200608–MH200614 for 16SrDNA sequences, MH203307–MH203307 for bla IMI‐2, and MG882588–MG882609 and MG882611–MG882634 for bla L1 sequences.
3. RESULTS
3.1. Distribution, frequency and identification of carbapenem‐resistant gram‐negative bacteria in water bodies in the Los Angeles area
We analyzed 10 different water samples from ponds and lakes in the Los Angeles area (California, United States) and found that all of them contained carbapenem (meropenem)‐resistant gram‐negative bacteria. The frequency of gram‐negative meropenem‐resistant bacteria was between 0.1 and 324 meropenem‐resistant cfu per 100 mls of water, which represented between 0.002% and 0.8% of the total gram‐negative bacteria found in these water samples (Table 1). The two samples with the highest count of meropenem‐resistant bacteria per 100 ml were W1 and W6. Location W1 (CSUN Duck pond) is an artificial pond with a large number of animals (ducks, geese and turtles) which uses circulated potable water. However, the circulation system was not functioning at the time of sampling (the same location sampled after cleaning the pond and fixing the circulation system had more than a 10‐fold decrease in the number of carbapenem‐resistant bacteria, whereas the total number of gram‐negatives was only reduced by less than 4‐fold). Location W2 (Woodley Wildlife Lake) has an extensive population of animals, particularly birds, and uses reclaimed water from a nearby water treatment facility. The rest of the water samples, which include natural ponds as well as ponds and lakes that use circulation of potable or reclaimed water had comparable total numbers of carbapenem‐resistant bacteria, except for Reseda Park Lake, an asphalt‐lined lake that had a very low number of carbapenem‐resistant bacteria.
We selected a total of 76 meropenem‐resistant/intermediate isolates (about 8 per sample) for further identification and characterization. Using their 16S rDNA sequence (and oxidase test results when necessary), we preliminarily identified them as 7 Enterobacter asburiae, 2 Aeromonas veronii, 2 Cupriavidus gilardii, 5 Pseudomonas alcaligenes, 1 Pseudomonas cedrina, 1 Pseudomonas geniculata, 9 Pseudomonas otitidis, 1 Pseudomonas stutzeri, 45 Stenotrophomonas maltophilia, and 3 Stenotrophomonas pavanii strains (Table 2). Among the isolates selected for identification and characterization, the genera Stenotrophomonas and Pseudomonas were both the most abundant and widespread; that is, 48 selected isolates collected from nine different water samples were Stenotrophomonas, and 17 isolates collected from 6 different water samples were Pseudomonas.
3.2. Characterization of the antibiotic susceptibility profile of carbapenem‐resistant isolates
We characterized the antibiotic susceptibility profile of the 76 identified carbapenem‐resistant or ‐intermediate isolates using disk diffusion experiments with 2 carbapenems (meropenem and imipenem) and 4 non‐carbapenem (cefotaxime, ciprofloxacin, gentamicin, and tetracycline) antibiotics (Table 2; Table 3; and Figure 2). Overall, 99% of our isolates (all except for one intermediate A. veronii) were resistant to meropenem, as expected by our use of meropenem as the selective agent to obtain these isolates. Most isolates (all except for four P. alcaligenes and one P. otitidis) were also resistant (80%) or intermediate (13%) to imipenem. For the non‐carbapenem β‐lactam cefotaxime, most isolates were also resistant (68%) or intermediate (8%). These were all Stenotrophomonas and about two thirds of all Pseudomonas. In contrast, resistance to non‐β‐lactam antibiotics was much lower (Table 2; Table 3; and Figure 2). For ciprofloxacin, 97% of the isolates (all isolates except for one S. maltophilia and one P. stutzeri) were sensitive. For gentamicin, 39% of the isolates were resistant and 3% were intermediate. Gentamicin‐resistant/intermediate isolates were C. gilardii, P. geniculata, and two thirds of all Stenotrophomonas. For tetracycline, 9% and 17% of the isolates were resistant or intermediate, respectively. These were mostly S. maltophilia and one A. veronii isolate (Table 2; Table 3; and Figure 2). Overall, these findings highlight the importance of different aquatic environments in the Los Angeles‐Southern California area as reservoirs of bacteria that are resistant to carbapenems and other antibiotics.
3.3. Identification of carbapenemases from carbapenem‐resistant isolates
We next used the CarbaNP test to identify which carbapenem‐resistant or ‐intermediate isolates produce carbapenemases. We found that 52 out of the 76 isolates studied were positive for carbapenemase production when tested using meropenem and/or imipenem (Table 2; Table 3). CarbaNP‐positive isolates included 7 E. asburiae and all 45 S. maltophilia. In all these isolates, their carbapenemases were inducible.
We then used PCR and sequencing to identify the carbapenemase genes present in the 52 CarbaNP‐positive isolates (Table 2; Table 3; and Figure 3). All seven E. asburiae isolates had a bla IMI‐2 gene 100% identical to the bla IMI‐2 reference sequence (DQ173429). All 45 carbapenemase‐producing S. maltophilia isolates had the bla L1 gene. Analysis of the bla L1 genes identified revealed that environmental L1 carbapenemases are very diverse. All bla L1 DNA sequences obtained for our isolates had between 83% to 99% identity to the reference S. maltophilia bla L1 gene (NG_047502) — a variability similar to that found in S. maltophilia clinical isolates (Avison, Higgins, von Heldreich, Bennett, & Walsh, 2001).
Figure 3.
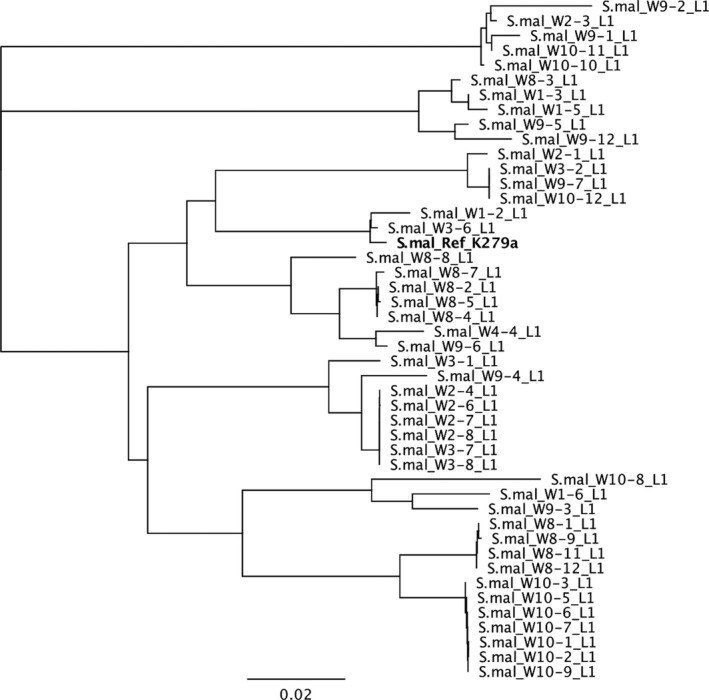
Phylogenetic tree showing relatedness between the reference bla L1 gene sequence and the L1 carbapenemases gene sequences obtained in this study. The tree was constructed using the Neighbor‐Joining method. The scale bar at the bottom represents the number of nucleotide substitutions per site. In bold the S. maltophilia strain K279a bla L1 reference sequence (NG_047502). Abbreviations: S.mal: Stenotrophomonas maltophilia
4. DISCUSSION
Carbapenem resistance is one of the major threats to public health worldwide (Centers for Disease Control and Prevention, 2013a, 2013b; Guh et al., 2015; Gupta et al., 2011; Papp‐Wallace et al., 2011; Prabaker & Weinstein, 2011; Rhomberg & Jones, 2009; van Duijn et al., 2011). Despite the significance of carbapenem‐resistant bacteria, there is little information about these bacteria outside healthcare or immediately related facilities (Chagas et al., 2011; Conlan et al., 2014; Doi & Paterson, 2015; Guh et al., 2015; Gupta et al., 2011; Nasri et al., 2017; White et al., 2016), especially in the United States. To this date, the only study about these bacteria in aquatic environments in the United States focused on rivers from the Midwest sampled between 1999 and 2001 (Ash et al., 2002; Aubron et al., 2005), a time when carbapenem use and the spread of carbapenem‐resistant bacteria in clinical settings were much lower than they are today (Centers for Disease Control and Prevention, 2018; Pakyz, MacDougall, Oinonen, & Polk, 2008).
Our study is the first one in more than a decade to investigate the distribution, frequency, antibiotic susceptibility profile, and carbapenemase genes of carbapenem‐resistant bacteria in aquatic environments in the United States. This study is also the first one to study carbapenem‐resistant bacteria in environmental water bodies on the West Coast of the United States, as well as the first one to study the distribution, frequency and characteristics of carbapenem‐resistant bacteria in non‐riverine water environments such as ponds and lakes in the United States. We found that gram‐negative bacteria resistant to carbapenems and other antibiotics are widespread in water bodies in the Los Angeles‐Southern California area. We could detect and isolate carbapenem‐resistant bacteria from all ponds and lakes tested with a frequency ranging from 0.002% to 0.8% of the total gram‐negative bacteria present in the samples analyzed. Although this frequency cannot be directly compared to the results found for rivers in the midwestern United States sampled between 1999 and 2001 because imipenem‐resistant isolates were identified by screening isolates first identified as ampicillin resistant (Ash et al., 2002; Aubron et al., 2005), the frequency of carbapenem‐resistant bacteria found in our study is similar to that found Tacão et al. (2015) in Portuguese rivers.
Characterization of a total of 76 isolates from these samples showed that carbapenem‐resistant or ‐intermediate bacteria in ponds and lakes from the Los Angeles area are quite diverse, and include different species preliminarily identified as Enterobacter asburiae, Aeromonas veronii, Cupriavidus gilardii, Pseudomonas alcaligenes, Pseudomonas cedrina, Pseudomonas geniculata, Pseudomonas otitidis, Pseudomonas stutzeri, Stenotrophomonas maltophilia, and Stenotrophomonas pavanii. These results have some similarities and differences with previous studies. The most abundant carbapenem‐resistant bacterium among our isolates (found in most of our samples) was S. maltophilia, which is common in aquatic environments and intrinsically resistant to carbapenems (Brooke, 2012). However, we also found S. pavanii in the Tujunga pond (a natural pond filled with spring water). S. pavanii is a bacterium previously found in plants (Ramos et al., 2011) and in bird feces (Kenzaka & Tani, 2018), which to our knowledge has not been found before in aquatic environments.
The second most abundant and widespread (present in most of our samples) group of carbapenem‐resistant bacteria we found was Pseudomonas. Interestingly, carbapenem‐resistant Pseudomonas isolates (P. geniculata and P. otitidis among other Pseudomonas, but not P. cedrina or P. stutzeri) were also found to be abundant in Portuguese rivers (Tacão et al., 2015), but were not found in the midwestern United States rivers (Aubron et al., 2005). In contrast with the results for Pseudomonas, Enterobacter asburiae represented the most abundant and widespread (they were found in 4 different rivers) carbapenem‐resistant isolate found in rivers from the U.S. Midwest (Aubron et al., 2005), but was only found in one sample both in the study of Portuguese rivers (Tacão et al., 2015) as well as in our study (Woodley Wildlife lake).
The other carbapenem‐resistant or ‐intermediate isolates that we found are Aeromonas veronii and Cupriavidus gilardii. Aeromonas, including A. veronii, are common water inhabitants and are often intrinsically resistant to carbapenems (Aubron et al., 2005; Lupo, Coyne, & Berendonk, 2012; Tacão et al., 2015). In contrast, this is the first time that carbapenem‐resistant C. gilardii isolates — which we identified in a location (W2) that uses recirculated potable water — have been reported outside of clinical settings (Karafin et al., 2010; Kobayashi et al., 2016).
To further characterize the 76 selected carbapenem‐resistant or ‐intermediate isolates, we used disk diffusion antibiotic susceptibility experiments with carbapenem (meropenem and imipenem) and noncarbapenem antibiotics (cefotaxime, ciprofloxacin, gentamicin, and tetracycline). The antibiotics we chose have different cellular targets, entry routes, and resistance mechanisms. Therefore, even if not all these antibiotics are clinically used to treat all of the genera that we identified, they provide very important information about the potential antibiotic resistance mechanisms found in these isolates. For example, strains generally very resistant to most or all antibiotics suggest an important role of general antibiotic resistance mechanism such as decreased outer membrane permeability and/or increased efflux by multidrug efflux pumps, in addition to more specific mechanisms. Strains that are only resistant to one antibiotic or class of antibiotics suggest that such resistance is likely to be predominantly caused by specific mechanisms such as target mutations or antibiotic degrading enzymes such as carbapenemases. In general, resistance to carbapenems, β‐lactams (cefotaxime), aminoglycosides (gentamicin) and tetracyclines (tetracycline) was widespread among our isolates, whereas resistance to fluoroquinolones (ciprofloxacin) was very rare among them.
Resistance to carbapenems can occur by different mechanisms such as production of carbapenemases, overexpression of efflux pumps, and decreased outer membrane permeability (Livermore et al., 2005; Marsik & Nambiar, 2011; Papp‐Wallace et al., 2011; Queenan & Bush, 2007; Rizek et al., 2014; Rodríguez‐Martínez et al., 2009; Shin et al., 2012; Sho et al., 2013; Warner et al., 2013). Production of carbapenemases seems to be a major contributing mechanism for carbapenem‐resistance in most of our isolates because 52 of the 76 isolates characterized were carbapenemase‐producers. The carbapenemase gene of these carbapenemase‐producing isolates was identified by PCR and sequencing as bla IMI‐2 100% identical to the reference sequence for all Enterobacter asburiae isolates and as bla L1 for all Stenotrophomonas maltophilia isolates. The bla L1 carbapenemase genes identified showed varying diversity (83%–99% DNA identity) compared to the bla L1 reference sequence. The presence of the L1 carbapenemase in Stenotrophomonas maltophilia is well documented in both clinical and environmental isolates and is a major contributor to its untreatability (Brooke, 2012; Tacão et al., 2015; Youenou et al., 2015).
Of greater concern is the identification of seven E. asburiae isolates carrying the bla IMI‐2 carbapenemase gene. This gene is an inducible plasmid‐encoded carbapenemase gene that was first identified in carbapenem‐resistant E. asburiae isolates from four different U.S. Midwest rivers (Aubron et al., 2005), and was later found in an Enterobacter cloacae isolate recovered from river sediment in Spain in 2017 (Piedra‐Carrasco et al., 2017). bla IMI‐2 has also recently been found in clinical isolates of E. asburiae (Czech Republic) (Rotova et al., 2017), E. cloacae (China) (Yu, Du, Zhou, Chen, & Li, 2006), Escherichia coli (Spain and China) (Rojo‐Bezares, Martin, Lopez, Torres, & Saenz, 2012; Zhang et al., 2017), and Klebsiella variicola (United Kingdom) (Hopkins, Findlay, Doumith, Mather, & Meunier, 2017). In both environmental and clinical isolates, bla IMI‐2 was usually found in transposable elements located in transferable plasmids (Aubron et al., 2005; Hopkins et al., 2017; Piedra‐Carrasco et al., 2017; Rojo‐Bezares et al., 2012; Rotova et al., 2017; Yu et al., 2006; Zhang et al., 2017); however, one E. asburiae clinical isolate carrying the bla IMI‐2 gene in its chromosome (the transposable element was not characterized) was identified in South Africa in 2015 (Gqunta et al., 2015). Our results show that this acquired carbapenemase gene is also spread outside of clinical and river environments, which may be related to the presence of bla IMI‐2 in transposable elements located in transferable plasmids (Aubron et al., 2005; Hopkins et al., 2017; Piedra‐Carrasco et al., 2017; Rojo‐Bezares et al., 2012; Rotova et al., 2017; Yu et al., 2006; Zhang et al., 2017). Further, surveillance is necessary to better characterize the role of freshwater environments as a source of IMI‐2‐producing E. asburiae, which can be both an opportunistic pathogen, as well as a reservoir of this transferable carbapenemase.
In conclusion, our findings show for the first time that freshwater environments in Los Angeles‐Southern California represent an underappreciated reservoir of bacteria resistant to carbapenems and other antibiotics, many of which carry intrinsic or acquired carbapenemase genes.
ACKNOWLEDGMENTS
This work was supported by the California State University Northridge start‐up funds to C. Ruiz, and was also partially supported by the National Institutes of Health BUILD PODER 5RL5GM118975‐03 and the California State University CSUPERB New Investigator grants to C. Ruiz. We thank William Jackson for his help in the analysis of bla L1 sequences.
CONFLICT OF INTEREST
The authors declare that the research was conducted in the absence of any potential sources of conflict of interest.
DATA ACCESSIBILITY STATEMENT
The authors adhere to all policies on sharing data and materials described in the guidelines for authors.
Harmon DE, Miranda OA, McCarley A, Eshaghian M, Carlson N, Ruiz C. Prevalence and characterization of carbapenem‐resistant bacteria in water bodies in the Los Angeles–Southern California area. MicrobiologyOpen. 2019;8:e692 10.1002/mbo3.692
REFERENCES
- Altschul, S. F. , Madden, T. L. , Schäffer, A. A. , Zhang, J. , Zhang, Z. , Miller, W. , & Lipman, D. J. (1997). Gapped BLAST and PSI‐BLAST: a new generation of protein database search programs. Nucleic Acids Research, 25(17), 3389–3402. 10.1093/nar/25.17.3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ash, R. J. , Mauck, B. , & Morgan, M. (2002). Antibiotic resistance of gram‐negative bacteria in rivers, United States. Emerging Infectious Diseases, 8(7), 713–716. 10.3201/eid0807.010264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubron, C. , Poirel, L. , Ash, R. J. , & Nordmann, P. (2005). Carbapenemase‐producing Enterobacteriaceae, U.S. rivers. Emerging Infectious Diseases, 11(2), 260–264. 10.3201/eid1102.030684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avison, M. B. , Higgins, C. S. , von Heldreich, C. J. , Bennett, P. M. , & Walsh, T. R. (2001). Plasmid location and molecular heterogeneity of the L1 and L2 beta‐lactamase genes of Stenotrophomonas maltophilia. Antimicrobial Agents and Chemotherapy, 45(2), 413–419. 10.1128/AAC.45.2.413-419.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergey, D. H. , & Holt, J. G . (1994). Bergey's manual of determinative bacteriology, 9th Ed . Baltimore, MD: Williams & Wilkins. [Google Scholar]
- Bradley, J. S. , Garau, J. , Lode, H. , Rolston, K. V. , Wilson, S. E. , & Quinn, J. P. (1999). Carbapenems in clinical practice: A guide to their use in serious infection. International Journal of Antimicrobial Agents, 11(2), 93–100. 10.1016/S0924-8579(98)00094-6 [DOI] [PubMed] [Google Scholar]
- Bratu, S. , Landman, D. , Haag, R. , Recco, R. , Eramo, A. , Alam, M. , & Quale, J. (2005). Rapid spread of carbapenem‐resistant Klebsiella pneumoniae in New York City: a new threat to our antibiotic armamentarium. Archives of Internal Medicine, 165(12), 1430–1435. 10.1001/archinte.165.12.1430 [DOI] [PubMed] [Google Scholar]
- Bratu, S. , Mooty, M. , Nichani, S. , Landman, D. , Gullans, C. , Pettinato, B. , … Quale, J. (2005). Emergence of KPC‐possessing Klebsiella pneumoniae in Brooklyn, New York: epidemiology and recommendations for detection. Antimicrobial Agents and Chemotherapy, 49(7), 3018–3020. 10.1128/AAC.49.7.3018-3020.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooke, J. S. (2012). Stenotrophomonas maltophilia: An emerging global opportunistic pathogen. Clinical Microbiology Reviews, 25(1), 2–41. 10.1128/CMR.00019-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention . Vital Signs: Carbapenem‐Resistant Enterobacteriaceae. (2013a). Morbidity and Mortality Weekly Report, 62(9), 165–170. Available: https://www.cdc.gov/mmwr/pdf/wk/mm6209.pdf [PMC free article] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention . (2013b). Antibiotic Resistance Threats in the United States, 2013. Available: http://www.cdc.gov/drugresistance/threat-report-2013/pdf/ar-threats-2013-508.pdf
- Centers for Disease Control and Prevention , 2018. Tracking CRE. Available: https://www.cdc.gov/hai/organisms/cre/TrackingCRE.html
- Chagas, T. P. , Seki, L. M. , da Silva, D. M. , & Asensi, M. D. (2011). Occurrence of KPC‐2‐producing Klebsiella pneumoniae strains in hospital wastewater. Journal of Hospital Infection, 77(3), 281 10.1016/j.jhin.2010.10.008 [DOI] [PubMed] [Google Scholar]
- Clinical and Laboratory Standards Institute . (2017). Performance Standards for Antimicrobial Susceptibility Testing; 27th Ed. CLSI Supplement M100. Wayne, PA.
- Conlan, S. , Thomas, P. J. , Deming, C. , Park, M. , Lau, A. F. , Dekker, J. P. , … Tsai, Y. C. (2014). Single‐molecule sequencing to track plasmid diversity of hospital‐associated carbapenemase‐producing Enterobacteriaceae. Science Translational Medicine, 6(254), p.254ra126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correa, A. , Montealegre, M. C. , Mojica, M. F. , Maya, J. J. , Rojas, L. J. , De La Cadena, E. P. , … Villegas, M. V. (2012). First report of a Pseudomonas aeruginosa isolate coharboring KPC and VIM carbapenemases. Antimicrobial Agents and Chemotherapy, 56(10), 5422–5423. 10.1128/AAC.00695-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuzon, G. , Naas, T. , Villegas, M. V. , Correa, A. , Quinn, J. P. , & Nordmann, P. (2011). Wide dissemination of Pseudomonas aeruginosa producing beta‐lactamase blaKPC‐2 gene in Colombia. Antimicrobial Agents and Chemotherapy, 55(11), 5350–5353. 10.1128/AAC.00297-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di, D. Y. , Jang, J. , Unno, T. , & Hur, H. G. (2017). Emergence of Klebsiella variicola positive for NDM‐9, a variant of New Delhi metallo‐beta‐lactamase, in an urban river in South Korea. Journal of Antimicrobial Chemotherapy, 72(4), 1063–1067. [DOI] [PubMed] [Google Scholar]
- Doi, Y. , & Paterson, D. L . (2015). Carbapenemase‐producing Enterobacteriaceae. Seminars in Respiratory and Critical Care Medicine, 36(1), 74–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dortet, L. , Poirel, L. , & Nordmann, P. (2012a). Rapid identification of carbapenemase types in Enterobacteriaceae and Pseudomonas spp. by using a biochemical test. Antimicrobial Agents and Chemotherapy, 56(12), 6437–6440. 10.1128/AAC.01395-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dortet, L. , Poirel, L. , & Nordmann, P. (2012b). Rapid detection of carbapenemase‐producing Pseudomonas spp. Journal of Clinical Microbiology, 50(11), 3773–3776. 10.1128/JCM.01597-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eden, P. A. , Schmidt, T. M. , Blakemore, R. P. , & Pace, N. R. (1991). Phylogenetic analysis of Aquaspirillum magnetotacticum using polymerase chain reaction‐amplified 16S rRNA‐specific DNA. International Journal of Systematic Bacteriology, 41(2), 324–325. 10.1099/00207713-41-2-324 [DOI] [PubMed] [Google Scholar]
- Girlich, D. , Poirel, L. , & Nordmann, P. (2010). Novel ambler class A carbapenem‐hydrolyzing beta‐lactamase from a Pseudomonas fluorescens isolate from the Seine River, Paris, France. Antimicrobial Agents and Chemotherapy, 54(1), 328–332. 10.1128/AAC.00961-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gqunta, K. , van Wyk, J. , Ekermans, P. , Bamford, C. , Moodley, C. , & Govender, S . 2015. First report of an IMI‐2 carbapenemase‐producing Enterobacter asburiae clinical isolate in South Africa. Southern African Journal of Infectious Diseases, 30, 34–35. 10.1080/23120053.2015.1103963 [DOI] [Google Scholar]
- Guh, A. Y. , Bulens, S. N. , Mu, Y. , Jacob, J. T. , Reno, J. , Scott, J. , … Kallen, A. J. (2015). Epidemiology of Carbapenem‐Resistant Enterobacteriaceae in 7 US Communities, 2012‐2013. JAMA, 314(14), 1479–1487. 10.1001/jama.2015.12480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta, N. , Limbago, B. M. , Patel, J. B. , & Kallen, A. J. (2011). Carbapenem‐resistant Enterobacteriaceae: Epidemiology and prevention. Clinical Infectious Diseases, 53(1), 60–67. 10.1093/cid/cir202 [DOI] [PubMed] [Google Scholar]
- Henriques, I. S. , Araújo, S. , Azevedo, J. S. , Alves, M. S. , Chouchani, C. , Pereira, A. , & Correia, A. (2012). Prevalence and diversity of carbapenem‐resistant bacteria in untreated drinking water in Portugal. Microbial Drug Resistance, 18(5), 531–537. 10.1089/mdr.2012.0029 [DOI] [PubMed] [Google Scholar]
- Hopkins, K. L. , Findlay, J. , Doumith, M. , Mather, B. , & Meunier, D. (2017). D'arcy S, Pike R, Mustafa N, Howe R, Wootton M, Woodford N. IMI‐2 carbapenemase in a clinical Klebsiella variicola isolated in the UK. Journal of Antimicrobial Chemotherapy., 72(7), 2129–2131. 10.1093/jac/dkx103 [DOI] [PubMed] [Google Scholar]
- Isozumi, R. , Yoshimatsu, K. , Yamashiro, T. , Hasebe, F. , Nguyen, B. M. , Ngo, T. C. , … Arikawa, J. (2012). bla(NDM‐1)‐positive Klebsiella pneumoniae from environment, Vietnam. Emerging Infectious Diseases, 18(8), 1383–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallen, A. J. , Hidron, A. I. , Patel, J. , & Srinivasan, A. (2010). Multidrug resistance among gram‐negative pathogens that caused healthcare‐associated infections reported to the National Healthcare Safety Network, 2006‐2008. Infection Control and Hospital Epidemiology, 31(5), 528–531. 10.1086/652152 [DOI] [PubMed] [Google Scholar]
- Karafin, M. , Romagnoli, M. , Fink, D. L. , Howard, T. , Rau, R. , Milstone, A. M. , & Carroll, K. C. (2010). Fatal infection caused by Cupriavidus gilardii in a child with aplastic anemia. Journal of Clinical Microbiology, 48(3), 1005–1007. 10.1128/JCM.01482-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenzaka, T. , & Tani, K. (2018). Draft Genome Sequence of Multidrug‐Resistant Stenotrophomonas pavanii BWK1, Isolated from Mareca penelope Feces. Genome Announcements, 6(12), pii:.e00187‐18 10.1128/genomeA.00187-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khuntayaporn, P. , Montakantikul, P. , Mootsikapun, P. , Thamlikitkul, V. , & Chomnawang, M. T. (2012). Prevalence and genotypic relatedness of carbapenem resistance among multidrug‐resistant P. aeruginosa in tertiary hospitals across Thailand. Annals of Clinical Microbiology and Antimicrobials, 11, 25 10.1186/1476-0711-11-25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi, T. , Nakamura, I. , Fujita, H. , Tsukimori, A. , Sato, A. , Fukushima, S. , … Matsumoto, T. (2016). First case report of infection due to Cupriavidus gilardii in a patient without immunodeficiency: a case report. BMC Infectious Diseases, 16, 493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livermore, D. M. , Mushtaq, S. , & Warner, M. (2005). Selectivity of ertapenem for Pseudomonas aeruginosa mutants cross‐resistant to other carbapenems. Journal of Antimicrobial Chemotherapy, 55(3), 306–311. 10.1093/jac/dki009 [DOI] [PubMed] [Google Scholar]
- Lupo, A. , Coyne, S. , & Berendonk, T. U. (2012). Origin and evolution of antibiotic resistance: The common mechanisms of emergence and spread in water bodies. Frontiers in Microbiology, 3, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsik, F. J. , & Nambiar, S. (2011). Review of carbapenemases and AmpC‐beta lactamases. The Pediatric Infectious Disease Journal, 30(12), 1094–1095. 10.1097/INF.0b013e31823c0e47 [DOI] [PubMed] [Google Scholar]
- Martin, S. I. , & Kaye, K. M. (2004). Beta‐lactam antibiotics: Newer formulations and newer agents. Infectious Disease Clinics of North America, 18(3), 603–619. 10.1016/j.idc.2004.04.006 [DOI] [PubMed] [Google Scholar]
- Mathers, A. J. , Cox, H. L. , Kitchel, B. , Bonatti, H. , Brassinga, A. K. , Carroll, J. , … Sifri, C. D. (2011). Molecular dissection of an outbreak of carbapenem‐resistant enterobacteriaceae reveals Intergenus KPC carbapenemase transmission through a promiscuous plasmid. MBio, 2(6), e00204–e00211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasri, E. , Subirats, J. , Sanchez‐Melsio, A. , Mansour, H. B. , Borrego, C. M. , & Balcazar, J. L. (2017). Abundance of carbapenemase genes (blaKPC, blaNDM and blaOXA‐48) in wastewater effluents from Tunisian hospitals. Environmental Pollution, 229, 371–374. 10.1016/j.envpol.2017.05.095 [DOI] [PubMed] [Google Scholar]
- Nordmann, P. , Dortet, L. , & Poirel, L. (2012). Carbapenem resistance in Enterobacteriaceae: Here is the storm!. Trends in Molecular Medicine, 18(5), 263–272. 10.1016/j.molmed.2012.03.003 [DOI] [PubMed] [Google Scholar]
- Nordmann, P. , Poirel, L. , & Dortet, L. (2012). Rapid detection of carbapenemase‐producing Enterobacteriaceae. Emerging Infectious Diseases, 18(9), 1503–1507. 10.3201/eid1809.120355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pakyz, A. L. , MacDougall, C. , Oinonen, M. , & Polk, R. E. (2008). Trends in antibacterial use in US academic health centers: 2002 to 2006. Archives of Internal Medicine, 168(20), 2254–2260. 10.1001/archinte.168.20.2254 [DOI] [PubMed] [Google Scholar]
- Papp‐Wallace, K. M. , Endimiani, A. , Taracila, M. A. , & Bonomo, R. A. (2011). Carbapenems: Past, present, and future. Antimicrobial Agents and Chemotherapy, 55(11), 4943–4960. 10.1128/AAC.00296-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson, D. L. (2000). Recommendation for treatment of severe infections caused by Enterobacteriaceae producing extended‐spectrum beta‐lactamases (ESBLs). Clinical Microbiology & Infection, 6(9), 460–463. 10.1046/j.1469-0691.2000.00107.x [DOI] [PubMed] [Google Scholar]
- Paterson, D. L. (2002). Serious infections caused by enteric gram‐negative bacilli–mechanisms of antibiotic resistance and implications for therapy of gram‐negative sepsis in the transplanted patient. Seminars in Respiratory Infections, 17(4), 260–264. 10.1053/srin.2002.36446 [DOI] [PubMed] [Google Scholar]
- Paterson, D. L. , & Bonomo, R. A. (2005). Extended‐spectrum beta‐lactamases: A clinical update. Clinical Microbiology Reviews, 18(4), 657–686. 10.1128/CMR.18.4.657-686.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piedra‐Carrasco, N. , Fàbrega, A. , Calero‐Cáceres, W. , Cornejo‐Sánchez, T. , Brown‐Jaque, M. , Mir‐Cros, A. , … González‐López, J. J. (2017). Carbapenemase‐producing enterobacteriaceae recovered from a Spanish river ecosystem. PLoS ONE, 12(4), e0175246 10.1371/journal.pone.0175246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirel, L. , Barbosa‐Vasconcelos, A. , Simões, R. R. , Da Costa, P. M. , Liu, W. , & Nordmann, P. (2012). Environmental KPC‐producing Escherichia coli isolates in Portugal. Antimicrobial Agents and Chemotherapy, 56(3), 1662–1663. 10.1128/AAC.05850-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potron, A. , Poirel, L. , Bussy, F. , & Nordmann, P. (2011). Occurrence of the carbapenem‐hydrolyzing beta‐lactamase gene blaOXA‐48 in the environment in Morocco. Antimicrobial Agents and Chemotherapy, 55(11), 5413–5414. 10.1128/AAC.05120-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prabaker, K. , & Weinstein, R. A. (2011). Trends in antimicrobial resistance in intensive care units in the United States. Current Opinion in Critical Care, 17(5), 472–479. 10.1097/MCC.0b013e32834a4b03 [DOI] [PubMed] [Google Scholar]
- Queenan, A. M. , & Bush, K . (2007). Carbapenemases: The versatile beta‐lactamases. Clinical Microbiology Reviews, 20(3), 440–458. 10.1128/CMR.00001-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queenan, A. M. , Pillar, C. M. , Deane, J. , Sahm, D. F. , Lynch, A. S. , Flamm, R. K. , … Davies, T. A. (2012). Multidrug resistance among Acinetobacter spp. in the USA and activity profile of key agents: results from CAPITAL Surveillance 2010. Diagnostic Microbiology and Infectious Disease, 73(3), 267–270. 10.1016/j.diagmicrobio.2012.04.002 [DOI] [PubMed] [Google Scholar]
- Ramos, P. L. , Van, Trappen S. , Thompson, F. L. , Rocha, R. C. , Barbosa, H. R. , De, Vos P. , & Moreira‐Filho, C. A. (2011). Screening for endophytic nitrogen‐fixing bacteria in Brazilian sugar cane varieties used in organic farming and description of Stenotrophomonas pavanii sp. nov. International Journal of Systematic and Evolutionary Microbiology, 61(Pt 4), 926–931. 10.1099/ijs.0.019372-0 [DOI] [PubMed] [Google Scholar]
- Rhomberg, P. R. , & Jones, R. N. (2009). Summary trends for the Meropenem Yearly Susceptibility Test Information Collection Program: A 10‐year experience in the United States (1999–2008). Diagnostic Microbiology and Infectious Disease, 65(4), 414–426. 10.1016/j.diagmicrobio.2009.08.020 [DOI] [PubMed] [Google Scholar]
- Rizek, C. , Fu, L. , dos Santos, L. C. , Leite, G. , Ramos, J. , Rossi, F. , … Costa, S. F. (2014). Characterization of carbapenem‐resistant Pseudomonas aeruginosa clinical isolates, carrying multiple genes coding for this antibiotic resistance. Annals of Clinical Microbiology and Antimicrobials., 13, 43 10.1186/s12941-014-0043-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez‐Martínez, J. M. , Poirel, L. , & Nordmann, P. (2009). Molecular epidemiology and mechanisms of carbapenem resistance in Pseudomonas aeruginosa. Antimicrobial Agents and Chemotherapy, 53(11), 4783–4788. 10.1128/AAC.00574-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojo‐Bezares, B. , Martin, C. , Lopez, M. , Torres, C. , & Saenz, Y. (2012). First detection of blaIMI‐2 gene in a clinical Escherichia coli strain. Antimicrobial Agents and Chemotherapy, 56(2), 1146–1147. 10.1128/AAC.05478-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotova, V. , Papagiannitsis, C. C. , Chudejova, K. , Medvecky, M. , Skalova, A. , Adamkova, V. , & Hrabak, J. (2017). First description of the emergence of Enterobacter asburiae producing IMI‐2 carbapenemase in the Czech Republic. Journal of Global Antimicrobial Resistance, 11, 98–99. 10.1016/j.jgar.2017.10.001 [DOI] [PubMed] [Google Scholar]
- Shin, S. Y. , Bae, I. K. , Kim, J. , Jeong, S. H. , Yong, D. , Kim, J. M. , & Lee, K. (2012). Resistance to carbapenems in sequence type 11 Klebsiella pneumoniae is related to DHA‐1 and loss of OmpK35 and/or OmpK36. Journal of Medical Microbiology, 61(Pt 2), 239–245. 10.1099/jmm.0.037036-0 [DOI] [PubMed] [Google Scholar]
- Sho, T. , Muratani, T. , Hamasuna, R. , Yakushiji, H. , Fujimoto, N. , & Matsumoto, T. (2013). The mechanism of high‐level carbapenem resistance in Klebsiella pneumoniae: Underlying Ompk36‐deficient strains represent a threat of emerging high‐level carbapenem‐resistant K. pneumoniae with IMP‐1 β‐lactamase production in Japan. Microbial Drug Resistance, 19(4), 274–281. 10.1089/mdr.2012.0248 [DOI] [PubMed] [Google Scholar]
- Tacão, M. , Correia, A. , & Henriques, I. S. (2015). Low Prevalence of Carbapenem‐Resistant Bacteria in River Water: Resistance Is Mostly Related to Intrinsic Mechanisms. Microbial Drug Resistance, 21(5), 497–506. 10.1089/mdr.2015.0072 [DOI] [PubMed] [Google Scholar]
- Torres, J. A. , Villegas, M. V. , & Quinn, J. P. (2007). Current concepts in antibiotic‐resistant gram‐negative bacteria. Expert Review of Anti‐Infective Therapy, 5(5), 833–843. 10.1586/14787210.5.5.833 [DOI] [PubMed] [Google Scholar]
- van Duijn, P. J. , Dautzenberg, M. J. , & Oostdijk, E. A. (2011). Recent trends in antibiotic resistance in European ICUs. Current Opinion in Critical Care, 17(6), 658–665. 10.1097/MCC.0b013e32834c9d87 [DOI] [PubMed] [Google Scholar]
- van Duin, D. , Kaye, K. S. , Neuner, E. A. , & Bonomo, R. A. (2013). Carbapenem‐resistant Enterobacteriaceae: A review of treatment and outcomes. Diagnostic Microbiology and Infectious Disease, 75(2), 115–120. 10.1016/j.diagmicrobio.2012.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vardakas, K. Z. , Tansarli, G. S. , Rafailidis, P. I. , & Falagas, M. E. (2012). Carbapenems versus alternative antibiotics for the treatment of bacteraemia due to Enterobacteriaceae producing extended‐spectrum β‐lactamases: A systematic review and meta‐analysis. Journal of Antimicrobial Chemotherapy, 67(12), 2793–2803. 10.1093/jac/dks301 [DOI] [PubMed] [Google Scholar]
- Viehman, J. A. , Nguyen, M. H. , & Doi, Y. (2014). Treatment options for carbapenem‐resistant and extensively drug‐resistant Acinetobacter baumannii infections. Drugs, 74(12), 1315–1333. 10.1007/s40265-014-0267-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh, T. R. (2010). Emerging carbapenemases: A global perspective. International Journal of Antimicrobial Agents, 36(Suppl 3), S8–S14. 10.1016/S0924-8579(10)70004-2 [DOI] [PubMed] [Google Scholar]
- Warner, D. M. , Yang, Q. , Duval, V. , Chen, M. , Xu, Y. , & Levy, S. B. (2013). Involvement of MarR and YedS in carbapenem resistance in a clinical isolate of Escherichia coli from China. Antimicrobial Agents and Chemotherapy, 57(4), 1935–1937. 10.1128/AAC.02445-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White, L. , Hopkins, K. L. , Meunier, D. , Perry, C. L. , Pike, R. , Wilkinson, P. , … Woodford, N. (2016). Carbapenemase‐producing Enterobacteriaceae in hospital wastewater: a reservoir that may be unrelated to clinical isolates. Journal of Hospital Infection, 93(2), 145–151. 10.1016/j.jhin.2016.03.007 [DOI] [PubMed] [Google Scholar]
- Yang, Z. , Liu, W. , Cui, Q. , Niu, W. K. , Li, H. , Zhao, X. N. , … Lu, S. J. (2014). Prevalence and detection of Stenotrophomonas maltophilia carrying metallo‐beta‐lactamase blaL1 in Beijing, China. Frontiers in Microbiology, 5, 692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youenou, B. , Favre‐Bonte, S. , Bodilis, J. , Brothier, E. , Dubost, A. , Muller, D. , & Nazaret, S. (2015). Comparative Genomics of Environmental and Clinical Stenotrophomonas maltophilia Strains with Different Antibiotic Resistance Profiles. Genome Biology and Evolution, 7(9), 2484–2505. 10.1093/gbe/evv161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, Y. S. , Du, X. X. , Zhou, Z. H. , Chen, Y. G. , & Li, L. J. (2006). First isolation of blaIMI‐2 in an Enterobacter cloacae clinical isolate from China. Antimicrobial Agents and Chemotherapy, 50(4), 1610–1611. 10.1128/AAC.50.4.1610-1611.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, F. , Wang, X. , Xie, L. , Zheng, Q. , Guo, X. , Han, L. , & Sun, J. (2017). A novel transposon, Tn6306, mediates the spread of blaIMI in Enterobacteriaceae in hospitals. International Journal of Infectious Diseases, 65, 22–26. 10.1016/j.ijid.2017.09.014 [DOI] [PubMed] [Google Scholar]
- Zurfluh, K. , Hachler, H. , Nuesch‐Inderbinen, M. , & Stephan, R. (2013). Characteristics of extended‐spectrum beta‐lactamase‐ and carbapenemase‐producing Enterobacteriaceae Isolates from rivers and lakes in Switzerland. Applied and Environment Microbiology, 79(9), 3021–3026. 10.1128/AEM.00054-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors adhere to all policies on sharing data and materials described in the guidelines for authors.


