Abstract
In the mammary gland, it is widely believed that the luminal cells are unipotent after birth, contributing only to the luminal compartment in normal development. Here, by lineage tracing, we uncovered an unexpected potential of luminal cells that can give rise to basal cells during pregnancy. These luminal-derived basal cells (LdBCs) persisted through mammary regression and generated more progeny in successive rounds of pregnancies. LdBCs express basal markers as well as estrogen receptor α (ERα). In ovariectomized (OVX) mice, stimulation with estrogen and progesterone promoted the formation of LdBCs. In serial transplantation assays, LdBCs were able to reconstitute new mammary glands in a hormone-dependent manner. Transcriptome analysis and genetic experiments suggest that Wnt/β-catenin signaling is essential for the formation and maintenance of LdBCs. Our data uncover an unexpected bi-potency of luminal cells in a physiological context. The discovery of ERα+ basal cells, which can respond to hormones and are endowed with stem cell-like regenerative capacity in parous mammary gland, provides new insights into the association of hormones and breast cancer.
Subject terms: Developmental biology, Stem cells
Introduction
The mammary epithelium is a bi-layered structure that comprises the cytokeratin K5+/K14+ basal cell layer and the K8+/K18+/K19+ luminal cell layer. Mammary stem cells (MaSCs) are found in the basal layer.1,2 Upon transplantation, MaSCs are capable of regenerating both basal and luminal cells, and reconstituting the whole mammary gland.3,4 In contrast, luminal cells are unable to reconstitute in transplantation.3,4 The in vivo differentiation potential of basal cells has attracted much attention. By lineage tracing, bi-potent MaSCs that can contribute to both basal and luminal cells have been documented,5,6 while basal lineage-restricted stem/progenitor cells also have been discovered.7–12 In contrast, the hierarchy of luminal cells and their plasticity has not yet been fully understood.
The luminal cell layer comprises a subgroup of hormone-sensing cells.13 In the mammary epithelium, the expression of estrogen receptor alpha (ERα) and progesterone receptor alpha (PRα) are confined to the luminal layer.14–19 Estrogen and progesterone are fundamental to postnatal mammary development and homeostasis.13,20 During early and mid-pregnancy, hormonal stimulation drives extensive changes in mammary gland morphology with increasing complexity of ductal branches.13,20 Sustained exposure to estrogen and progesterone is a well-established risk factor for breast cancer.21–23
The hierarchy within the luminal compartment has drawn considerable interest. While some lineage tracing experiments suggested distinct progenitor populations for hormone receptor (HR)-positive and HR-negative luminal cells,24–28 others supported the existence of a common luminal progenitor population.5,29 More recently, single cell RNAseq studies also supported the existence of a common luminal progenitor population.30,31 Regardless of controversy of the hierarchy within the luminal compartment, luminal cells as a whole have been shown to be luminal lineage-restricted during postnatal development using various transgenic mouse models, including K8-CreER, K8-rTTA, K18-CreER or Elf5-CreER lines.5,7,11
Luminal plasticity, in particular luminal-to-basal conversion, has been exploited in certain conditions. In vitro, luminal cells can be reprogramed to become basal cells by ectopic expression of transcription factors, e.g. Sox9, Slug and Yap.32,33 In vivo, luminal-to-basal conversion is mainly associated with pathological conditions, i.e. oncogenic stress, under which luminal cells can give rise to basal cells.34–38 Although such a luminal-to-basal plasticity has not been reported during normal development,5,7,11,12 we are mindful that a negative result in lineage tracing does not necessarily mean that the cell type in question does not exist.2
In this study, we utilized a different K8-CreER BAC transgenic line,39,40 and through lineage tracing, we discovered an unexpected bi-potency of luminal cells that is endowed during pregnancy by hormones and Wnt/β-catenin signaling. The basal progeny derived from luminal cells, named luminal-derived basal cells (LdBCs), possess stem cell-like characteristics, capable of regenerating a new mammary gland upon transplantation. Most interestingly, these cells expressed ERα and responded to hormonal stimulation during regeneration. Our study reveals events of luminal-to-basal cell lineage conversion in normal development, explores the molecular mechanisms involved, and provides new insights into mammary epithelial cell plasticity.
Results
Pregnancy induces the generation of luminal-derived basal cells (LdBCs)
Keratin 8 (K8) expression is restricted to luminal cells.41 To conduct lineage tracing of luminal cells, a K8-CreERT2;Rosa26-mTmG strain was generated through genetic crosses. The 4th mammary glands were harvested at various time points followed by fluorescence activated cell sorting (FACS) analysis (Supplementary information, Fig. S1a). No GFP expression was detected in un-induced mice (Supplementary information, Fig. S1b). When tamoxifen (TAM) was administered to adult mice (9 weeks old, TAM: 2 mg per 25 g body weight), luminal cells were examined after 2 days. We found that luminal cells were efficiently labelled with GFP expression (61.33 ± 10.17% luminal cells were GFP+), and labelled cells were restricted to luminal layer (Supplementary information, Fig. S1c, d). After long-term tracing (for 8 weeks or 7 months), GFP+ cells were still restricted in the luminal compartment (Supplementary information, Figs. S1c, 1e, 1g). This was validated by immunostaining (Supplementary information, Fig. S1f). Similar results were observed when TAM was induced in pubertal mice (5-week old) (Supplementary information, Fig. S1h–j). These observations are consistent with previous reports showing that luminal cells are indeed luminal-fate restricted during postnatal development in nulliparous mice.5,7
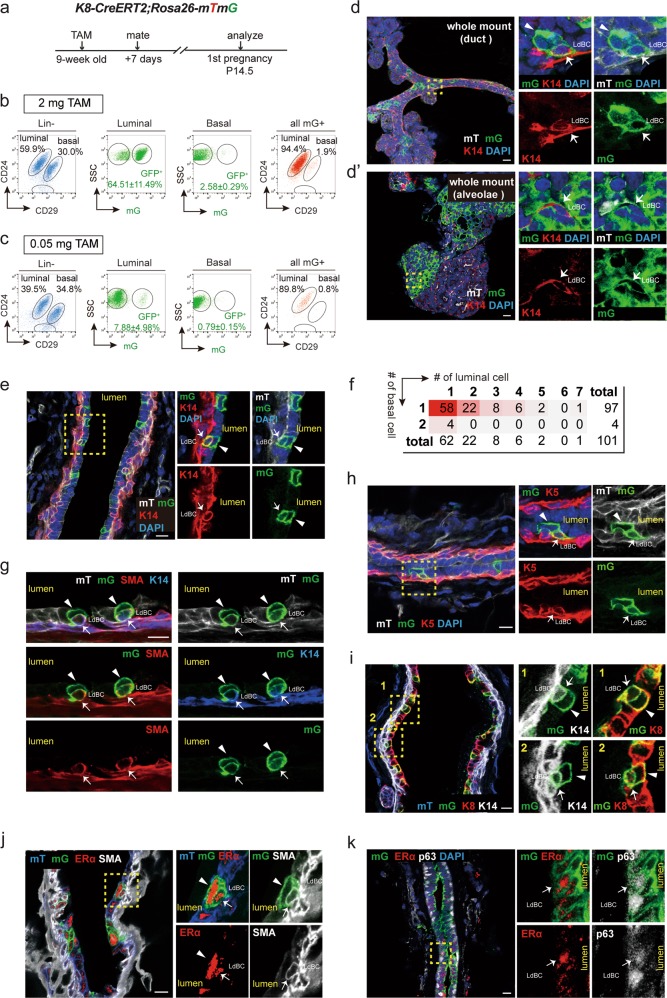
Next, we investigated whether luminal cells remain unipotent during pregnancy. The K8-CreERT2;Rosa26-mTmG mice (9 weeks old) were mated at 7 days post TAM induction. Mammary glands were harvested at pregnancy 14.5 day (P14.5) and underwent wholemount carmine staining. TAM administration had no obvious effect on alveolar development compared with the oil treatment (Supplementary information, Fig. S2a). The distribution of GFP+ cells were analysed (Fig. 1a). FACS analysis indicated that 64.51 ± 11.49% of luminal cells were GFP+ post TAM induction (Fig. 1b). Interestingly, GFP+ cells also appeared in a small portion of basal cells (2.58 ± 0.29%) (Fig. 1b), suggesting that luminal cell may have contributed to basal cell formation during pregnancy. To visualize this potential bi-potent event, a low dose of TAM (0.05 mg/25 g body weight) was administered in order to label luminal cells in clonal density. At this dosage, fewer luminal cells (7.88 ± 4.98%) were labelled, yet GFP+ basal cells (0.79 ± 0.15%) were still present at pregnancy day 14.5 (Fig. 1c). Next, whole mount confocal imaging was performed. In addition to the majority of clones that contain only luminal cells as expected, a few clones consisted of keratin 14 (K14)-expressing basal cells (7.66%, n = 966 clones). These bipotent GFP+ clones resided in both the ductal tree and alveoli (Fig. 1d–d’). Immunostaining on cryosections confirmed the existence of bi-lineage clones (Fig. 1e). Clonal analysis indicated that the majority of the bi-lineage clones contained only one basal cell (96.0%, n = 101 bi-lineage clones) (Fig. 1f). Staining with additional basal markers, i.e. smooth muscle actin (SMA) (Fig. 1g), keratin 5 (K5) (Fig. 1h) and transformation related protein 63 (p63) (Fig. 1k, Supplementary information, S2b–b’) further confirmed the basal cell identity in the bi-lineage clone. We also examined mammary glands at pregnancy day 8.5, and observed a similar luminal-to-basal transition (2% GFP+ basal cells) (Supplementary information, Fig. S2c–e), suggesting that these cells emerge in early pregnancy.
Fig. 1.
Pregnancy results in generation of luminal-derived basal cells (LdBCs). a Illustration of lineage tracing strategy using K8-CreERT2;R26-mTmG mice during tracing of 1st pregnancy. b FACS analysis showing that, with normal dose of TAM injection (2 mg per 25 g body weight), 64.51 ± 11.49% of luminal cells are GFP+, and 2.58 ± 0.29% of basal cells are GFP+ at 14.5 day of 1st pregnancy. Another way of analysis was also shown. Total mGFP+ cells were applied to CD24 and CD29 gates, showing that 1.9% of mGFP+ cells are in basal compartment. c FACS analysis showing that, with low dose of TAM administration (0.05 mg per 25 g body weight), 7.88 ± 4.98% of luminal cells are GFP+, and 0.79 ± 0.15% of basal cells are GFP+. Total mGFP+ cells were applied to CD24 and CD29 gates, showing that 0.8% of mGFP+ cells are in basal compartment. d–d’ Whole-mount confocal imaging of mammary gland showing a bi-lineage clone that contains one luminal cell (GFP+ K14−, arrowhead) and one basal cell (GFP+ K14+, arrow). Such bi-lineage clone can be seen in both duct (d) and alveolae (d’). Scale bars, 20 μm. e Section confocal imaging showing that luminal-derived clones contain K14+ basal cells. Within the clone, luminal cell is indicated by arrowhead, basal cell is indicated by arrow. Scale bars, 10 μm. f Clonal analysis showing the number of basal and luminal cells in individual bi-lineage clones. Basal cell numbers are shown along the y-axis, luminal cell numbers are shown along the x-axis. Red shading indicates the relative frequency of certain clone composition, with deeper shading indicating higher frequency. g–h Section confocal imaging showing that the basal cell found in the bi-lineage clone is positive for basal marker SMA (g) and K5 (h). Within the GFP clone, luminal cell is indicated by arrowhead, basal cell is indicated by arrow. Scale bars, 10 μm. i Section confocal imaging showing that the basal cell found in the bi-lineage clone expresses both basal marker K14 and luminal marker K8. 69.4% of these cells are double positive for K14 and K8 (n = 104 GFP+ basal cells). Within the clone, luminal cell is indicated by arrowhead, basal cell is indicated by arrow. Scale bars, 10 μm. j Section confocal imaging showing that the luminal-derived basal cell (LdBC) expresses ERα and basal marker SMA (arrow). Within the clone, luminal cell is indicated by arrowhead, basal cell is indicated by arrow. Scale bars, 10 μm. k Section confocal imaging showing that LdBC expresses ERα and basal marker p63 (arrow). Scale bars, 10 μm. Data represent the mean ± SD, n = 4 mice
The generation of these basal-like cells during pregnancy was further investigated using the multicolour K8-CreERT2;Rosa26-Rainbow reporter mouse, in which luminal cells upon TAM induction can randomly adopt one of three colours (Cherry, BFP or Orange) and pass the colour gene to their progeny (Supplementary information, Fig. S3a). Using the same lineage tracing strategy described above, bi-lineage clones in all three Rainbow colours were observed in pregnant mammary glands, and the clones consisted of both luminal and basal cells (Supplementary information, Fig. S3b–d). These bipotent events occurred in both the mammary duct (Supplementary information, Fig. S3c, d), and the alveolae (Supplementary information, Fig. S3b).
Interestingly, these labelled basal cells displayed hybrid features of typical basal and luminal cells. Using K8-CreERT2;Rosa26-mTmG mice, we found that first, most of the labelled basal cells still retained the expression of luminal maker keratin 8 (K8) (69.4%, n = 104 GFP+ basal cells) (Fig. 1i). Second, the majority of the labelled basal cells adopted a physical location between the basal and the luminal layer (82.25%, n = 144 GFP+ basal cells) (Fig. 1d, e, g–i). Third, the GFP-labelled basal cells did not express the tight junction protein ZO-1, a trait similar to basal cells yet different from luminal cells (Supplementary information, Fig. S2f). Together, these data suggest that during pregnancy, luminal cells exhibit unexpected plasticity and generate basal-like cells. We referred these basal-like cells as luminal-derived basal cells (LdBCs). Intriguingly, some LdBCs appeared to be ERα+, shown by co-expression of ERα with SMA or p63 (Fig. 1j, k). However, it is challenging to quantify ERα-expressing cells at this stage because the overall ERα expression is low during pregnancy due to the inverse correlation between ERα levels and estrogen levels in vivo,42–44 as well as the known negative regulation of ERα by estrogen.45 The ERα expression of LdBCs was later examined in non- pregnancy stages, namely during involution of the mammary gland following pregnancy.
LdBCs persist through involution and expand in next pregnancy
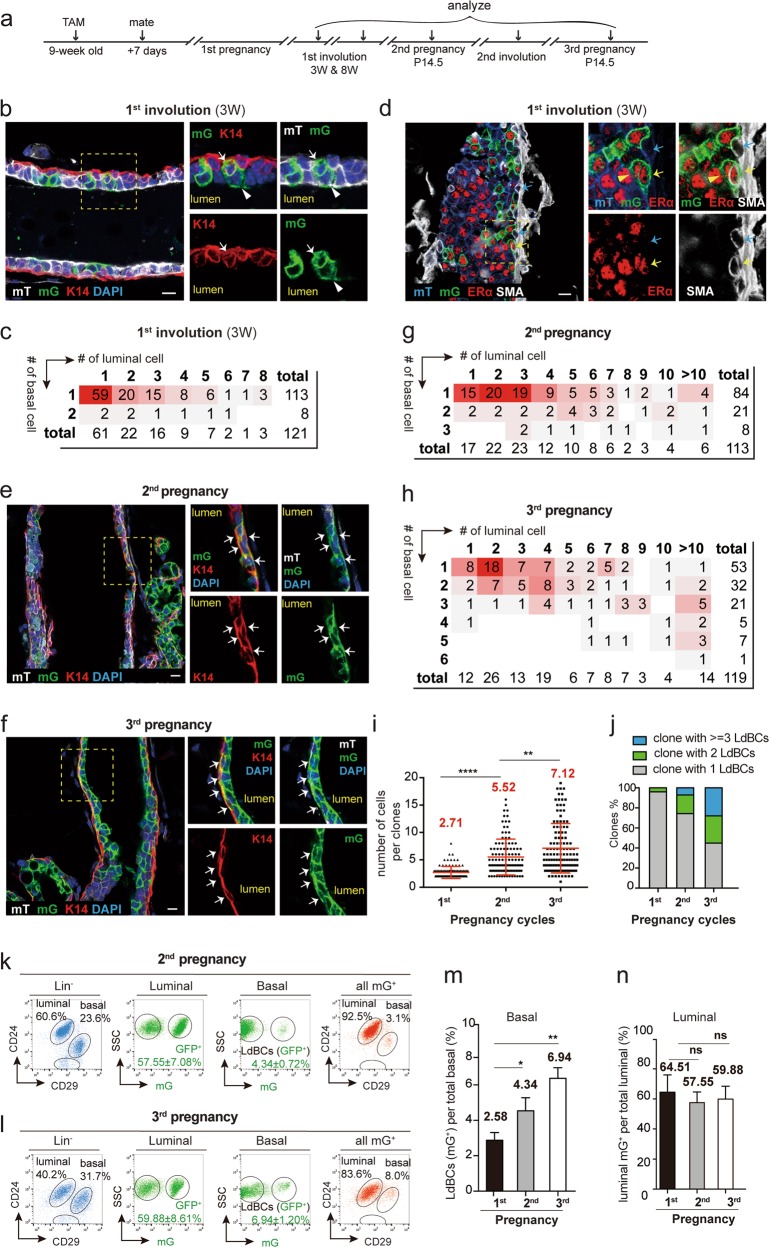
To investigate whether the appearance of LdBCs is a transient event during pregnancy, the behavior of these cells was studied during involution and in multiple rounds of pregnancies using K8-CreERT2;Rosa26-mTmG mice (Fig. 2a). After 3 weeks of involution, GFP+ LdBCs remained in the ducts, as shown by immunostaining (Fig. 2b). Clonal analysis indicated that majority of the bi-lineage clones contained a single LdBC (93.4%, n = 121 clones) (Fig. 2c). We observed that during involution, K8 expression in LdBCs had decreased, only 18.3% LdBCs remained K8+ (n = 131 LdBCs) (Supplementary information, Fig. S4a), compared to 69.4% during pregnancy. Interestingly, ERα expression in LdBCs remained robust, with 63.8% ERα+ cells in the involuted mammary gland (n = 116 LdBCs) (Fig. 2d). FACS analyses revealed that the percentages of GFP+ LdBCs at 3 weeks of involution (1.59 ± 0.23%) are similar to those at 8 weeks of involution (1.66 ± 0.46%) (Supplementary information, Fig. S4b, c), suggesting that these cells persisted through involution, and their percentages remained similar.
Fig. 2.
LdBCs persist through involution and expand in succeeding rounds of pregnancies. a Illustration of lineage tracing strategy using K8-CreERT2;R26-mTmG mice during tracing of 1st and 2nd involution, and 2nd and 3rd pregnancies. b Confocal imaging showing that at 3 weeks of involution, luminal-derived GFP+ clones contain K14+ basal cells (arrow). Luminal cell in the clone is indicated by arrowhead. Scale bars, 10 μm. c Clonal analysis showing the number of basal and luminal cells in individual bi-lineage clones at 3 weeks of involution. Red shading indicates the relative frequency of certain clone composition, with deeper shading indicating higher frequency. d Confocal imaging showing that at 3 weeks of involution, LdBCs express ERα and basal marker SMA (yellow arrow). A normal basal cell is SMA+, ERα− (blue arrow). Within the clone, luminal cell is indicated by a yellow arrowhead. Scale bars, 10 μm. e, f Immunostaining images of mammary tissues upon tracing during 2nd pregnancy (e) and 3rd pregnancy (f). Indicated GFP+ clones contain multiple LdBCs (GFP+, K14+). Scale bars, 10 μm. g–j Clonal analysis showing the number of basal and luminal cells in individual bi-lineage clones in 2nd pregnancy (g) and in 3rd pregnancy (h). Red shading indicates the relative frequency of certain clone composition, with deeper shading indicating higher frequency. Quantification indicates that the average clone sizes increase along the pregnancy cycles (i), and that the percentages of clones with 2 LdBCs and 3 or more LdBCs increase along the pregnancy cycles (j). Bi-potent clones with n = 101, 113 and 119 were analysed in 1st, 2nd and 3rd pregnancy respectively. k–n FACS analysis of 2nd pregnancy (k) and of 3rd pregnancy (l) showing the percentages of GFP+ cells in luminal and basal compartments. Another way of analysis was also shown. Total mGFP+ cells were applied to CD24 and CD29 gates, showing the distribution of mGFP+ cells in basal and luminal compartments. Quantification indicates that the percentages of GFP+ cells in basal compartment (LdBCs) increase along pregnancy cycles (m), while GFP+ cells in luminal compartment remain stable (n). **P < 0.01, * P < 0.05, ns P > 0.1. Data represent the mean ± SD for n = 4 mice (2nd pregnancy) and n = 3 mice (3rd pregnancy)
Next, the progeny clones of LdBCs induced prior to the first pregnancy were examined in subsequent rounds of pregnancies (Fig. 2e, f). In the 2nd pregnancy, clonal analysis revealed that the average size of GFP+ bi-lineage clones is bigger compared to that in the 1st pregnancy (Fig. 2g, i), and the size further increases in the 3rd pregnancy (Fig. 2h, i). Importantly, GFP+ clones containing two or more LdBCs also increased, from 4.0% at the 1st pregnancy to 25.7% at the 2nd pregnancy, and to 55.1% at the 3rd pregnancy (Fig. 2j). FACS analysis validated the increased percentages of GFP+ LdBCs in the 2nd and 3rd pregnancies compared to the 1st pregnancy (Fig. 2k–m). The percentages of GFP+ luminal cells remained stable throughout tracing (Fig. 2k, l, n).
In the 2nd involution, LdBCs remained K14+ and ERα+ (Supplementary information, Fig. S4d, e). LdBC numbers also increased compared to the 1st involution (Supplementary information, Fig. S4f, g). This is likely due to the expansion of LdBCs in the 2nd pregnancy compared to the 1st pregnancy. It is noteworthy that the locations of LdBCs become undistinguishable with typical basal cells after 1st involution and in succeeding pregnancies. Together, these results suggest that LdBCs, once emerge during 1st pregnancy, can persist through involution and give rise to more progeny in successive rounds of pregnancy.
Hormonal treatment promotes the formation of LdBCs
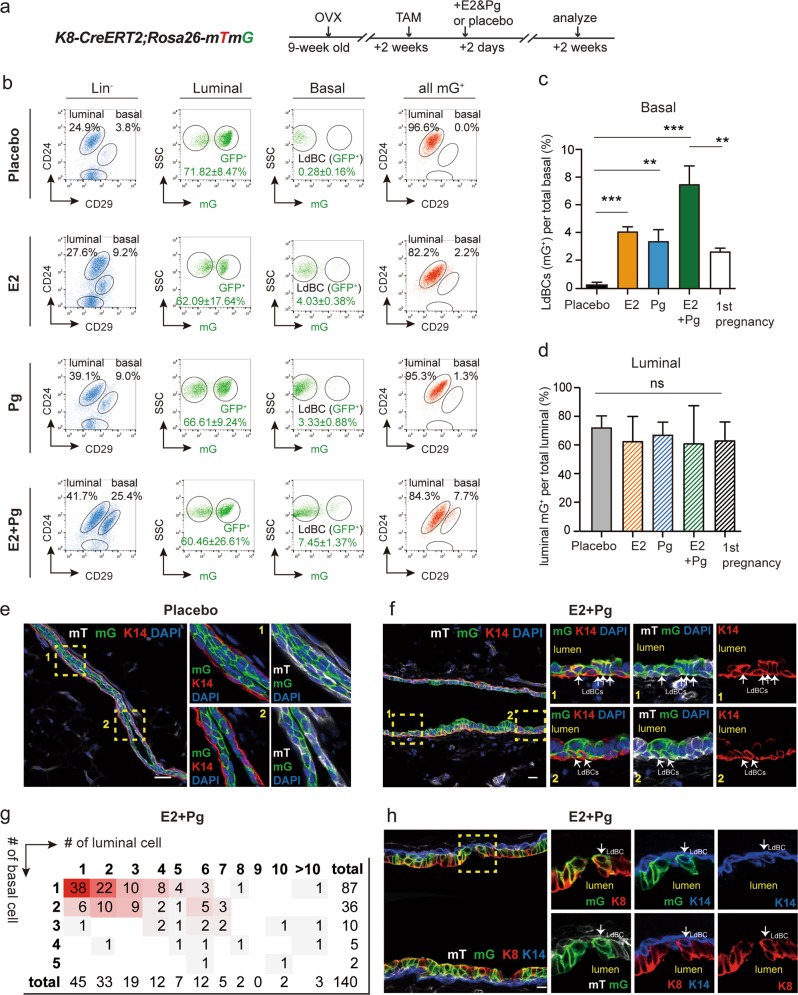
Estrogen (E2) and progesterone (Pg) levels increase drastically during early and mid pregnancy. We next tested whether these ovarian hormones regulate the formation of LdBCs. K8-CreERT2;Rosa26-mTmG adult mice (9 weeks old) were ovariectomized to deplete endogenous ovarian hormones. Luminal cells were labelled by TAM injection at 2 weeks after ovariectomy, and 2 days later, placebo or E2 and/or Pg pellets were implanted and the mammary cells were examined 2 weeks after pellet implantation (Fig. 3a). FACS analyses revealed that 60–72% of luminal cells were GFP+ in each condition, a labelling efficiency that is comparable to previous experiments in normal pregnancy (Fig. 3b, d). In the placebo mice, no GFP+ cells were found in the basal population (Fig. 3b). Either E2 or Pg treatment was sufficient for the generation of LdBCs, inducing 4.03 and 3.33% GFP+ LdBCs of total basal cells respectively (Fig. 3b, c). Combination treatment with E2 and Pg remarkably promoted the efficiency, inducing 7.45% of GFP+ LdBCs (Fig. 3b, c) The E2 + Pg treatment appeared to have higher LdBCs induction efficiency as compared with normal pregnancy (2.58% LdBC) (Fig. 3c), probably due to the sustained stimulation of hormones in high levels for two weeks. Notebaly, the E2 + Pg treatment also resulted in increased percentage of basal cells (Fig. 3b), as previously reported.44 Consistent with FACS results, no LdBC was found in the placebo group by immunostaining (Fig. 3e). In contrast, LdBCs were readily detected in E2 + Pg treated mammary tissues (Fig. 3f). Clonal analysis indicated that the majority of the bi-lineage clones contained one LdBC (62.1%, n = 140 bi-potent clones) (Fig. 3g), an observation similar to normal pregnancy. In addition, 85.7% of LdBCs were K14+ K8+ (n = 112 LdBCs) (Fig. 3h), reminiscent of those found in normal pregnancy. Together, these data suggest that ovarian hormones estrogen and progesterone induce the formation of LdBCs.
Fig. 3.
Hormones promote the formation of LdBCs. a Illustration of lineage tracing strategy upon hormonal stimulation using K8-CreERT2;R26-mTmG mice. Mice were ovariectomized, followed by TAM induction and implantation with E2 or Pg pellet alone or in combination. The mammary glands were harvested at 2 weeks after implantation. b–d FACS analyses indicating that E2 or Pg alone can lead to the generation of LdBCs, while E2 + Pg combination increases the frequency to 7.45 ± 1.37% (b). Another way of analysis was also shown. Total mGFP+ cells were applied to CD24 and CD29 gates, showing the distribution of mGFP+ cells in basal and luminal compartments (b). Quantification indicates that all conditions of hormonal treatment significantly enhance the frequency of LdBCs compared to Placebo (c). E2 + Pg treatment also led to significant higher percentage of LdBCs compared to E2 or Pg treatment alone or normal pregnancy (c). *** P < 0.001, ** P < 0.01. The percentages of luminal GFP+ cells were similar, suggesting that labeling efficiencies were comparable in various treatment conditions (d). One-way ANOVA, ns P > 0.5. e, f Immunostaining showing that no LdBCs (GFP+ K14+) are seen in Placebo (e), while LdBCs (arrows) are readily detected upon E2 + Pg treatment (f). Scale bars, 10 μm. g Clonal analysis showing the number of basal and luminal cells in individual bi-lineage clones upon E2 + Pg treatment. Red shading indicates the relative frequency of certain clone composition, with deeper shading indicating higher frequency. n = 140 bi-lineage clones. h Immunostaining with luminal marker K8 and basal marker K14 showing that the LdBC (arrow) induced by E2 + Pg treatment is K8+ K14+. Scale bars, 10 μm. K8 expressions were detected in 85.7% of LdBCs (n = 112 LdBCs). Data represent the mean ± SD for n = 4 mice (OVX, +E2, +Pg) and n = 6 mice (+E2 + Pg)
LdBCs display repopulating capacity upon transplantation
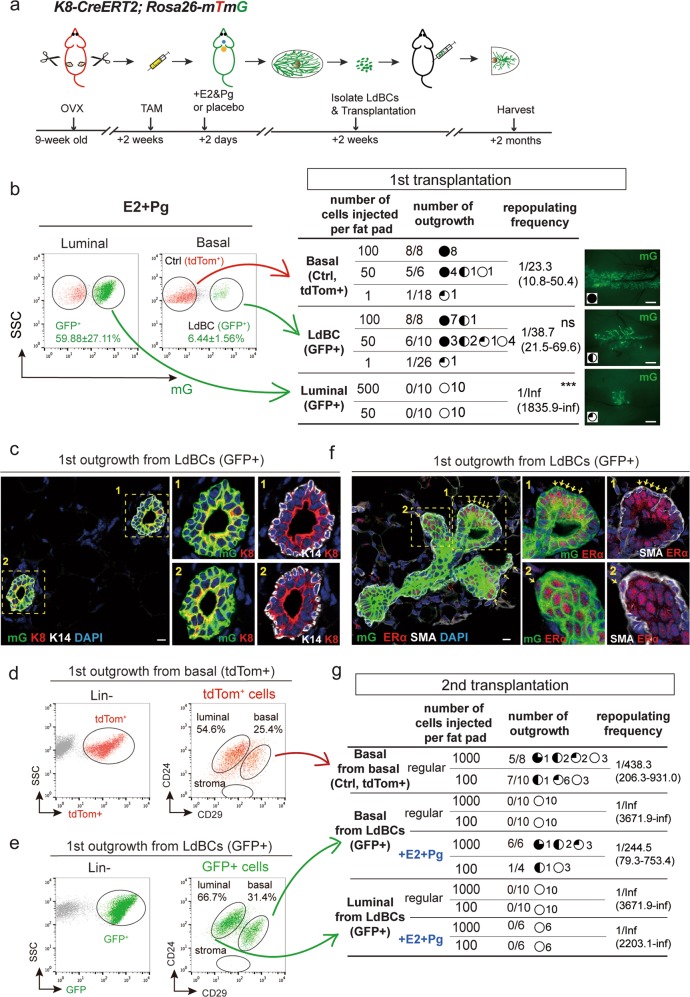
Next, we investigated whether LdBCs possess stem cell-like in vivo repopulation capabilities. K8-CreERT2;Rosa26-mTmG adult mice were ovariectomized, labeled with TAM and treated with hormone, and LdBCs were isolated and transplanted in limiting dilution into cleared fat pads of nude recipients (illustrated in Fig. 4a). Normal basal cells (tdTomato+) and luminal cells (GFP+) were also isolated and transplanted as positive and negative controls respectively (Fig. 4b). The outgrowths were analyzed 2 months post transplantation. We found that both tdTomato+ basal cells and GFP+ LdBCs were able to generate new mammary glands with normal morphology in both nulliparous and pregnancy stages (Fig. 4b, Supplementary information, S5a, b). The GFP+ luminal cells had no reconstitution ability as expected (Fig. 4b). GFP+ LdBCs exhibited a repopulating frequency (1/38.7), which is slightly lower than that of tdTomato+ basal cells (1/23.3, p > 0.1) (Fig. 4b). Immunostaining of cryosections showed that the GFP+ outgrowths have the normal bi-layered structure with normal basal (K14) and luminal (K8) marker expression (Fig. 4c). FACS analysis of outgrowths of GFP+ LdBCs indicated a basal and luminal cell composition, similar to outgrowths from normal basal cells (tdTomato+) (Fig. 4d, e). In outgrowths derived from normal basal cells, ERα expression is restricted to luminal cells (Supplementary information, Fig. S5c). Most intriguingly, in the GFP+ LdBCs-derived mammary gland, ERα expression was readily detected in basal cells (13.8%, n = 752 SMA+ basal cells) (Fig. 4f), reminiscent of ERα expression seen in LdBCs (Figs. 1j, k, 2d). Collectively, these data suggest that LdBCs possess stem cell-like capability that can regenerate a new mammary gland upon transplantation, an ability that luminal cells lack.
Fig. 4.
LdBCs reconstitute mammary glands upon transplantation. a Illustration of OVX, labeling, hormonal stimulation and transplantation strategies using K8-CreERT2;R26-mTmG mice. b 2 weeks post TAM labeling and hormone stimulation, the mammary cells were FACS isolated. 3 groups, normal basal (Lin−, CD24+, CD29hi, tdTom+), LdBC (Lin−, CD24+, CD29hi, GFP+), and luminal (Lin−, CD24+, CD29low, GFP+) were transplanted to cleared fad pads of Nude recipients in limiting dilution as indicated. The degree of outgrowth was evaluated based on the occupancy of mammary fat pad. The representative images were shown on the right. Repopulating frequency in each group is indicated. Data were pooled from three independent experiments. *** P < 0.01, ns P > 0.1, Scale bars, 1 mm. c The outgrowths derived from LdBC (GFP+) were analyzed by immunostaining, showing normal basal (K14+) and luminal (K8+) composition. Scale bars, 10 μm. d, e FACS analysis of the outgrowths derived from normal basal cells (tdTom+) (d) and the outgrowths derived from LdBC (GFP+) (e) showing LdBC-derived outgrowths have normal basal and luminal composition. f LdBC-derived outgrowths (GFP+) were analyzed by immunostaining, showing ERα-expressing basal cells (ERα+ SMA+) (arrows). Scale bars, 10 μm. g Three groups, basal cells isolated from normal basal-derived outgrowths (Lin−, CD24+, CD29hi, tdTom+), basal cells isolated from LdBC-derived outgrowths (Lin−, CD24+, CD29hi, GFP+), and luminal cells isolated from LdBC-derived outgrowths (Lin−, CD24+, CD29low, GFP+) were transplanted to cleared fad pads of Nude recipients as indicated. Hormone stimulation with E2 + Pg pellets was also added to the latter two groups. The degree of outgrowth was evaluated based on the occupancy of mammary fat pad. Repopulating frequency in each group is indicated. Data were pooled from two independent experiments
The regenerative capacity of LdBCs was further examined in a 2nd transplantation experiment. Basal cells were FACS-isolated from LdBC-derived outgrowths and re-transplanted (Fig. 4e, g). Basal cells from normal basal cell-derived outgrowths (tdTomato+) were also transplanted as a positive control (Fig. 4d, g). We found that LdBCs had exhausted their renewing capacity and failed to generate a 2nd outgrowth, while normal basal cells were still able to reconstitute in a 2nd transplantation (Fig. 4g). Considering the intriguing ERα expression in the LdBC-derived 1st outgrowths, E2 and Pg hormone pellets were supplemented to recipients during the 2nd transplantation. Interestingly, basal cells isolated from LdBC-derived 1st outgrowths were able to generate 2nd outgrowths under these conditions (Fig. 4g). As expected, luminal cells isolated from LdBC-derived 1st outgrowth could not regenerate a mammary gland with or without hormonal stimulation (Fig. 4g). These data further support that LdBCs possess regenerative capacity, and their stem-like capability is dependent on ovarian hormones.
Transcriptome analysis revealed LdBCs as a distinct cell population
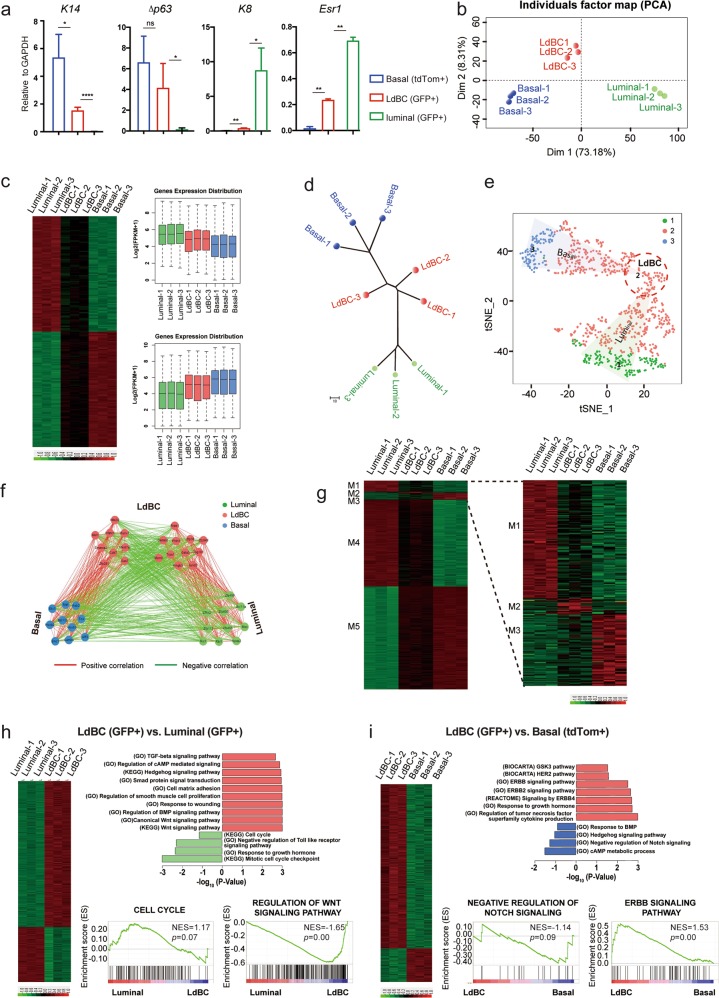
Next we characterized LdBCs at the molecular level using transcriptomic analyses. Three isolated populations, normal basal cells, LdBCs, and normal luminal cells, were isolated from mammary glands undergoing the 1st involution. qPCR analyses indicated that LdBCs expressed basal markers K14 and Δp63, as well as ERα (Esr1), but not luminal marker K8 (Fig. 5a). These are consistent with K14, K8 and ERα staining of LdBCs shown in Fig. 2. To further investigate the property of LdBCs, RNA-sequencing (RNA-seq) analysis was performed using these isolated populations. Principle component analysis (PCA) revealed LdBCs better correlated with normal basal cells, and were relatively more distinct from normal luminal cells along PC1 axis (Fig. 5b). Clustering analysis using the highest PC-loading genes in the PC1 (top 100 PC1-negative and -positive genes) supported the notion that LdBCs isolated in this stage (1st involution) lie in an intermediate state between basal and luminal cells (Fig. 5c). Next, we performed a neighbour-joining (NJ) tree analysis to detect the phylogenetic trajectory of cell populations. By computing the topological distance and the distance matrix specifying different cell populations, we characterized the differentiation branches from basal to luminal cells. LdBCs located between normal basal and luminal cells in the phylogenetic tree (Fig. 5d), slightly closer to basal cells, which further suggests that LdBC is in an intermediate and pro-basal state.
Fig. 5.
Transcriptome analyses of LdBCs in involuted mammary glands. a qPCR analysis indicating that LdBCs isolated from mammary gland undergoing involution express basal marker K14 and Δp63, luminal marker Esr1, but not luminal marker K8. **** P < 0.0001, ** P < 0.01, * P < 0.05, ns P > 0.1. b PCA (Principle Component Analysis) of transcriptome of basal cells (Lin−, CD24+, CD29hi, tdTomato+), luminal cells (Lin−, CD24+, CD29low, GFP+) and LdBCs (Lin−, CD24+, CD29hi, GFP+). c Clustering analysis using the highest PC-loading genes in PC1. d Neighbour-joining (NJ) tree analysis inferring the phylogenetic trajectory of normal basal cells, luminal cells and LdBCs. e t-SNE analysis seperate PC1 transcription factors into three clusters, corresponding to three population of cells. f CSI (Connection Specificity Index) network of PC1 hub TFs. Positive correlations are marked in red, and negative correlations are marked in green. g DEGs (Differentially Expressed Genes) of three populations illustrating the common and distinct features of basal cells, luminal cells and LdBCs. h–i GSEA (Gene Set Enrichment Analysis) analyses reavel distinct signaling pathways when comparing LdBCs with luminal cells (h), or LdBCs with basal cells (i)
Given that intrinsic regulators, especially transcription factors (TFs), play important roles in development, we focused on key TFs which may drive the differentiation process. To identify cell specific TFs, t-SNE (t-distributed stochastic neighbour embedding) analysis on collated development-related TFs was conducted. TFs were classified into 3 clusters, corresponding to the 3 cell populations, with 1 representing luminal cell, 2 representing LdBC, and 3 representing basal cell. The signature TFs of LdBC (Cluster 2) emerged at the continuously changing basal-luminal transition (Fig. 5e). To unravel inter-group connection of TFs in each cluster, we analysed the Connection Specificity Index (CSI) of the three TF groups, and generated a TF co-expression network (CSI > 0.9). Positive correlations between different TFs were marked in red, negative correlations were marked in green. TFs within basal cell group showed positive interactions with each other, indicating that TFs within the group may form a synergistic regulatory circuitry and act in a combinatory manner. Similar synergy was observed in luminal cell group. Interestingly, TFs in LdBC group were further clustered into two sub-groups, and the two sub-groups showed negative correlation, revealing the transition from luminal to basal feature (Fig. 5f). Furthermore, differentially expressed genes were identified for the isolated cell populations, and the signature genes were characterized in five modules to illustrate common and distinct features of LdBCs as compared with the other two populations. The disctinct features were especially zoomed in and shown on the right (Fig. 5g). We further examined the intra-population profiles. GSEA (Gene Set Enrichment Analysis) revealed significant upregulation of Wnt, BMP and TGF-β signaling pathways in LdBCs compared to luminal cells (Fig. 5h), and an increase of ERBB, decrease of negative regulation of Notch signaling signatures in LdBCs relative to basal cells (Fig. 5i).
LdBCs were also isolated from hormonal stimulated mammary gland and compared to basal and luminal populations. Similarly, LdBCs were found in an intermediate stage between basal and luminal cells by PCA, clustering analysis, and CSI of TFs (Supplementary information, Fig. S6b-d). In addition, Wnt and TGF-β signaling pathways were also increased in LdBCs relative to luminal cells (Supplementary information, Fig. S6e), and Notch signaling pathway was upregulated when comparing with basal cells according to KEGG analyses (Supplementary information, Fig. S6f). It should be noted that LdBCs at this stage retained K8 expression (Supplementary information, Fig. S6a), which is consistent with the immunostaining results seen in pregnancy (Fig. 1i) and during hormonal stimulation (Fig. 3h). The transcriptomic analyses, together with the immunostaining and functional study, suggest that LdBC is a novel cell population distinct from the known basal or luminal cells.
Wnt/β-catenin signaling is critical for LdBCs formation
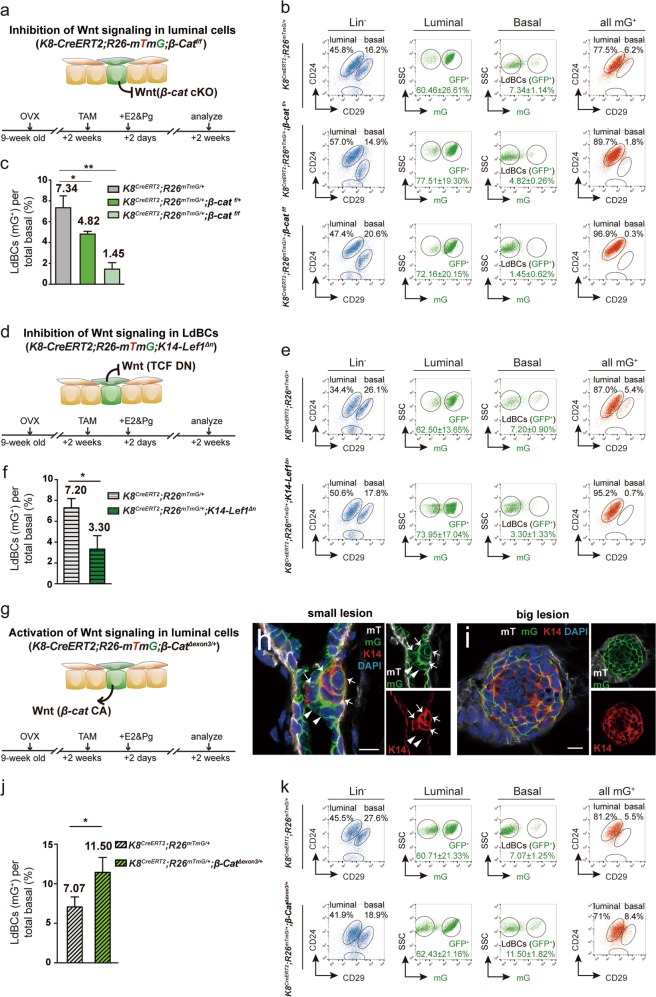
Considering the increased Wnt signaling signature in LdBCs relative to luminal cells, we investigated whether Wnt/β-catenin signaling is involved in the formation of LdBCs. Three different mouse models were utilized to modulate the activity of Wnt/β-catenin signaling in either the luminal or basal compartments. First, to investigate whether Wnt/β-catenin signalling is required for the luminal-to-LdBC conversion, we generated K8-CreERT2;Rosa26-mTmG;β-catf/f mice. Adult mice (9 weeks old) were ovariectomized. After two weeks, TAM was administered to both delete β-catenin in luminal cells and initiate lineage tracing. 2 days later, E2 and Pg pellets were implanted, and the mammary cells were examined at 2 weeks after pellet implantation (illustrated in Fig. 6a). FACS analysis revealed a marked decrease in the percentage of LdBCs compared to the control, suggesting that deletion of β-catenin in luminal cells inhibited the formation of LdBCs (Fig. 6b, c). Deletion of β-catenin displayed a dose-dependent effect as deletion of one copy of β-catenin also reduced the incidence of LdBC formation (Fig. 6b, c). It is worth noting that the whole mount morphology was not obviously changed after β-catenin deletion (Supplementary information, Fig. S7a, b). In addition, neither the absolute cell numbers of basal or luminal population (Supplementary information, Fig. S7d, e), nor the percentage of GFP+ luminal cells (Fig. 6b, Supplementary information, Fig. S7c) were affected by β-catenin deletion. Therefore, the reduced incidence of LdBC formation was not likely due to a broad influence on the epithelium or lower labelling efficiency of luminal cells. This suggests that β-catenin deletion directly regulates LdBC formation.
Fig. 6.
Modulation of Wnt/β-catenin signaling affects the formation of LdBCs. a, d, g Illustration of lineage tracing strategies in various genetic models in which Wnt/β-catenin signaling is modulated in basal or luminal cells. b, c FACS analysis of K8-CreERT2;R26-mTmG;β-catfl/fl mammary tissues showing that inhibition of Wnt signaling in luminal cells suppresses the percentage of LdBCs in a dose-dependent manner. ** P < 0.01, * P < 0.05. e, f FACS analysis of K8-CreERT2;R26-mTmG;K14-Lef1Δn mammary tissues showing that inhibition of Wnt signaling in basal cells results in reduced LdBCs. * P < 0.05. h, i Immunostaining of K8-CreERT2;R26-mTmG;β-CatΔexon3/+ mammary tissues showing that activation of Wnt signaling in luminal cells leads to hyperplasia consisting of many LdBCs (K14+, GFP+). Examples of small lesion (h) and big lesion (i) are shown. Scale bars, 10 μm. j, k FACS analysis of K8-CreERT2;R26-mTmG;β-CatΔexon3/+ mammary tissues showing that activation of Wnt signaling promotes LdBC expansion. * P < 0.05. Data represent the mean ± SD for n = 3 mice (K8-CreERT2;R26-mTmG;β-catfl/fl; K8-CreERT2;R26-mTmG;β-catfl/+; K8-CreERT2;R26-mTmG;β-CatΔexon3/+; K8-CreERT2;R26-mTmG;K14-Lef1Δn), n = 5 mice (K8-CreERT2;R26-mTmG)
Next, we asked whether Wnt signaling is required for the maintenance of LdBC after their formation. We generated K8-CreERT2;Rosa26-mTmG;K14-Lef1Δn mice. TAM induction and hormone treatment followed the same scheme stated above (illustrated in Fig. 6d). Since LdBCs express K14, upon LdBC formation, a dominant negative form of Lef1 (ΔN-Lef1) is expressed in these cells to attenuate Wnt signaling.46 Indeed, significantly fewer LdBCs were observed when blocking Wnt signaling by Lef1Δn in K14-expressing cells (Fig. 6e, f), suggesting that Wnt signaling is important for the maintenance of LdBCs. Although the absolute basal cell number and basal cell percentage beared small decreases in K14-Lef1Δn model, these reductions were not as significant as the reductions of LdBCs (Supplementary information, Fig. S7f, g), suggesting blocking Wnt signalling has more pronounced impact on LdBCs than on total basal cells.
Conversely, we also examined whether upregulation of Wnt signaling in luminal cells could promote the formation of LdBC using a constitutively activated form of β-catenin.47 TAM induction and hormone treatment in K8-CreERT2;Rosa26-mTmG;β-catΔexon3/+ mice were similarly performed as stated above (Fig. 6g). The constitutively activated form of β-catenin was induced in luminal cells and the LdBC fate was followed by lineage tracing. After 5 days of induction, small lesions were formed. We observed small clones that contained both GFP+ K14− luminal cells (arrowheads in Fig. 6h) and GFP+ K14+ LdBCs (arrows in Fig. 6h). After 14 days of induction, we observed bigger lesions that mostly consisted of LdBCs, suggesting an active expansion of LdBCs (Fig. 6i). FACS analysis confirmed the increased percentage of LdBCs in β-catΔexon3/+ mice at day 14 post induction compared to the control mice that had been through the same E2 + Pg treatment (Fig. 6j, k). These results suggest that activation of Wnt signaling in luminal cells promotes LdBC formation. Together, these lineage tracing experiments in various genetic models support the hypothesis that Wnt signaling is critical for LdBC formation and maintenance.
Discussion
It is widely accepted that luminal cells are unipotent (luminal lineage-restricted) during postnatal mammary development, and that ERα expression is confined to the luminal compartment.14–19 In the present study, we investigated luminal cell fate plasticity and revealed that luminal cells can give rise to basal cells during pregnancy or upon estrogen and progesterone stimulation. These LdBCs persisted through involution and generated more progeny in succeeding rounds of pregnancy. Intriguingly, these LdBCs remained ERα+ yet acquired stem cell-like repopulation capability. LdBCs were able to reconstitute new mammary glands upon 1st transplantation. The reconstituted mammary gland retained ERα+ basal cells, which can be serially transplanted when supplemented with estrogen and progesterone. To our best knowledge, this is the first report of bi-potential luminal cells in development, and ERα+ basal cells in physiological conditions. Our data also suggest that the formation of LdBCs introduces an irreversible change in parous mammary glands due to estrogen and progesterone stimulation during early- and mid-pregnancy.
The transcriptome profiling of the LdBCs offers insights into possible molecular mechanisms that account for the luminal-to-basal transition. LdBCs have increased Wnt/β-catenin signaling activities compared to luminal cells, and increased Notch signaling activities compared to basal cells. This is consistent with the previous notion that Wnt/β-catenin signaling is critical for basal fate determination,48–50 and Notch signaling dictates luminal lineage determination.50,51 Indeed, inhibition of Wnt/β-catenin signaling in either luminal or basal cells impeded the formation of LdBCs, whereas activation of Wnt/β-catenin signaling in luminal cells promoted the luminal to basal transition.
It is still not clear which subgroup of luminal cells generates LdBCs. Considering the hormonal impact and the resulting ERα+ status of LdBCs, it is plausible that ERα+ luminal cells are responsible for the formation of LdBCs. The rare population of LdBCs appearing particularly during pregnancy may have been missed in previous lineage tracing studies using ERα+ luminal cells.7,26,27 It remains puzzled that previsous lineage tracing studies using K8+ luminal cells also did not reveal these LdBCs. However, it should be noted that only 3.5 kb upstream of ATG of murine K8 gene was cloned in the previous K8-CreER,7 as well as the K8-rTTA transgeneic lines.11 In the current study, we used a different K8-CreER BAC transgenic line that contains the full-length murine K8 gene, plus 60 kb 5’ as well as 100 kb 3’ flanking sequences.39,40 As is known, not only the longer 5’and 3’ flanking sequences, but also the intragenic sequences are essential for sufficient and specific expression of the endogenous K8 gene. We conducted rigorous lineage tracing and immunostaining experiments, and present direct evidence for the existence of luminal-to-basal plasticity during normal development. Most interestingly, the resulting unique ERα+ basal cells have stem-like capabilities, and can be multiplied during succeeding pregnancies or upon transplantation. Unlike MaSCs, the reconstitution ability of these ERα+ basal cells was quickly exhausted in the 2nd transplantation. It is intriguing that hormonal supplements can reinitiate LdBC-dependent reconstitution in 2nd transplantation, providing a beneficial environment. The benefit did not apply to ERα+ luminal cells, as hormones did not promote reconstitution in luminal cell transplantation. These results suggest that ERα+ status is not sufficient for reconstitution in such an environment, and that the stem-like properties of LdBC are critical. To be noted, the advantage of the hormonal environment for reconstitution has been previously reported, in that ERα−, Notch1+ alveolar progenitors gain reconstitution ability when transplanted into pregnant recipients.25
A recent single cell RNA profiling study suggested a rare mixed-lineage cluster amongst basal cells in adult mammary glands, potentially primed for a luminal fate,31 although this population was not seen in another similar study using a different method.30 In the current study, with K8-CreERT2 mediated lineage tracing, we cannot completely exclude the possibility that these “mixed-lineage cells” in the basal compartment were labeled. However several lines of evidence suggest that it is unlikely. First, when K8-CreERT2 lineage tracing was conducted in pubertal mice or nulliparous adults, the labeled progeny were confined to the luminal compartment, and no basal cells were labeled. In addition, LdBCs emerged only when the tracing was extended to pregnancy or upon hormonal supplementation, further suggesting that LdBCs are absent in nulliparous mice and are induced particularly during pregnancy. One may argue that the “mixed-lineage cells” also expand during pregnancy as reported,31 making them easier to detect. However, in our study, TAM was injected in nulliparous adults to initiate the tracing, thus the targeted cells are K8-expressing cells in the nulliparous stage, not K8-expressing cells during pregnancy. If during pregnancy there is any undocumented plasticity, in that basal cells could switch on K8 expression, these basal cells could not be labelled in our labelling strategy. Finally, ERα expression is an intriguing feature of LdBC, while the “mixed-lineage cells” do not appear to express ERα.31 Together, these observations support the idea that LdBCs are derived from bona fide luminal cells, not from “mixed-lineage cells”.
Based on our studies, we believe that LdBCs are distinct from the previously reported temporarily augmented stem cell population during pregnancy.44 The transient stem cell population during pregnancy, demonstrated by enhanced reconstitution ability upon transplantation, contributes to an 11-fold increase in the absolute number of stem cells. They have been hypothesized to drive expansion of alveolar epithelial cells during the proliferative phase of pregnancy. Their capability in regeneration is considered temporary as their reconstitution ability is drastically decreased by 2nd transplantation.44 LdBCs are cells that acquire basal cell fate during the first pregnancy, but do not proliferate actively during the first round of pregnancy and the following involution. LdBCs are capable of reconstituting in serial transplantation in a hormone-dependent manner. LdBCs are not transient, as they persist through involution and contribute to more basal cells in the subsequent rounds of pregnancies.
LdBCs are also distinct from the previously identified parity-induced mammary epithelial cells (PI-MEC).52–54 PI-MEC refers to cells labeled by WAP-Cre during lactation in alveolae, yet these cells survive involution remodeling and remain in parous glands. They contribute to the development of secretory lobules upon successive pregnancies. They reside exclusively within the luminal layer of terminal ducts and developing alveoli. Importantly, they generate ER− cells within the alveolus, but do not contribute to the ER+ cells or the basal lineage.24 Moreover, PI-MEC can be induced by prolactin that rises in the second half of pregnancy (combined with dexamethasone and insulin), but not by estrogen and progesterone that elevate in early pregnancy.53 The LdBCs identified in the current study are different in many aspects. LdBCs are found in early and mid pregnancy, reside in both the duct and alveolae, and express basal markers (K14+, K5+, SMA+, p63+ and CD29hi). Consistent with their early appearance during pregnancy, LdBCs can be induced by estrogen and progesterone. Despite the differences, LdBC and PI-MEC share some similarities. Both populations can expand in multiple pregnancies, and are capable of mammary gland reconstitution upon transplantation.
LdBCs may also be distinct from the rare luminal cells found in nulliparous mice which have certain yet limited reconstitution ability in transplantation assay.19 First, LdBCs are not found in nulliparous mice, and only emerge during pregnancy. Second, LdBCs exhibit characteristics of basal cells. Third, LdBCs are able to reconstitute with an efficiency comparable to normal basal cells. In addition, based on our observation, position and molecular markers of LdBCs change along time. They originate during 1st pregnancy, and locate between basal and luminal layer in the duct, expressing K8 as well as basal markers. However, post involution and in succeeding pregnancies, LdBCs gradually become undistinguishable with normal basal cells in location and basal cell marker expression, further suggesting that LdBCs are generating new basal cells after origination, a property distinct from luminal cells.
Luminal cell plasticity has been implicated in tumor progression both in humans and in mice, demonstrated either by ex vivo luminal cell enrichment and alteration, followed by transplantation,32,55–57 or by imposing oncogenic stress in vivo.34–38 In the current study, we discovered a luminal-to-basal contribution under normal physiological conditions. LdBCs accounted for 2.6% of basal cells in the first pregnancy and 1.6% of basal cells in the involuted mammary gland. Importantly, their contribution increases in successive rounds of pregnancies. Given that luminal cell tracing studies showed Cre efficiency is approximately 65% in the current experiments, these numbers probably represent an under-estimate of the true contribution of LdBCs.
Pregnancy is the most significant modifiable factor known for breast cancer risk in women.58–60 The discovery of LdBCs reveals a new population of basal stem-like cells that emerge upon early pregnancy or hormone stimulation, which express ERα and can repopulate in response to hormonal stimuli. This population could be a potential cellular target of ER+ breast cancer transformation. Our work provides new insights into the association of pregnancy, hormone, Wnt signaling and breast cancer.
Materials and methods
Experimental animals
K8-CreERT2,39 Rosa26-mTmG,61 Rosa26-Rainbow,62 β-catΔexon3/+ 47, βcat flox/+,63 K14- Lef1ΔN 46 and Nude strains were used in this study. For lineage tracing experiments induced in pubertal and mature adult mice, animals received a single intraperitoneal injection of 2 mg or 0.05 mg tamoxifen (TAM; Sigma-Aldrich) diluted in sunflower oil. The Animal Care and Use Committee of Shanghai Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences approved experimental procedures.
Ovariectomy and hormone stimulation
Ovariectomy and hormone stimulation were conducted using conventional protocol. In brief, mature adult mice were bilaterally ovariectomized and allowed to recover for 2 weeks. 21-day 17β-oestradiol (0.05 mg per pellet) and/or progesterone (35 mg per pellet) slow-release pellets (Innovative Research of America) or carrier compound placebos were implanted subcutaneously on the neck in recipient mice. Mammary glands were harvested two weeks later after hormone stimulation.
Antibodies
Antibodies used were: rat anti-K8 (1:250, Developmental Hybridoma Bank, TROMA-I), rabbit anti-K14 (1:1,000, Covance), rabbit anti-K5 (1:1000, Covance), mouse anti-SMA (1:1000, Sigma-Aldrich), rabbit anti-ZO-1 (1:50, Abcam), rabbit anti-ERα (1:100, Millipore), mouse anti-p63 (1:100, Abcam), chicken anti-GFP (1:500, Invitrogen). The secondary antibodies used were goat anti-mouse Alexa Fluor 647, goat anti-rat Alexa Fluor 647, goat anti-rabbit Alexa Fluor 647 (Life Technologies), goat anti-rabbit Alexa Fluor 405 (Invitrogen), donkey anti-rabbit Cy3, donkey anti-mouse Cy3, donkey anti-chicken 488 (Life Technologies).
Primary cell preparation and flow cytometry
Mammary glands from 5-week-old, or 8- to 12-week-old virgin or pregnant female mice were isolated. The minced tissue was placed in culture medium (RPMI 1640 with 25 mM HEPES, 5% fetal bovine serum, 1% penicillin−streptomycin-glutamine (PSQ), 300 U/ml collagenase III (Worthington)) and digested for 2 h at 37 °C. After lysis of the red blood cells in NH4Cl, a single-cell suspension was obtained by sequential incubation with 0.05% trypsin-EDTA at 37 °C for 5 min and 0.1 mg/ml DNase I (Sigma) for 5 min with gentle pipetting, followed by filtration through 70 μm cell strainers. Cell pellets were resuspended with 5% FBS in PBS and incubated with the following antibodies in 1:200 dilutions. Biotin-conjugated CD31, CD45, TER119 (BD PharMingen, clone MEC 13.3, 30-F11 and TER-119; catalogue # 553371, #55307, # 553672), CD24−PE/cy7, CD29-APC (Biolegend, clone M1/69 and HMb1-1; catalogue #101822 and #102216), Streptavidin−V450 (BD PharMingen). Antibody incubation was performed on ice for 20 min in PBS with 5% fetal bovine serum (FBS; HyClone, Thermo Scientific). Cells were filtered through 40 μm strainers before FACS analysis. All analysis and sorting were performed using a FACSJazz (Becton Dickinson). The purity of sorted population was routinely checked and ensured to be > 95%.
Mammary fat pad transplantation and analysis
Sorted cells were resuspended in 50% Matrigel, PBS with 25% FBS, and 0.04% Trypan Blue (Sigma), and injected in 10 μl volumes into the cleared fat pads of 3-week-old female Nude mice. Reconstituted mammary glands were harvested 6–10 weeks post-surgery. All transplanted cells were labelled with GFP or tdTomato. Outgrowths were detected by a fluorescence dissection microscope (Leica). Outgrowths with more than 10% of the host fat pad filled were scored as positive.
Immunohistochemistry
Whole-mount staining was performed as previously described,5 with minor modifications. In brief, mammary glands were dissected into small pieces, then processed in digestion buffer (RPMI 1640 with 25 mM HEPES, 5% fetal bovine serum, 1% penicillin–streptomycin–glutamine (PSQ), 300 U/ml collagenase III (Worthington)) for 30 min at 37 °C, before fixation in 4% paraformaldehyde and incubation with antibodies. For immunostaining, mammary glands were fixed in 4% paraformaldehyde for 2 h at 4 °C, embedded in OCT and sectioned for 10–12 μm. Tissue sections were incubated with primary antibodies at 4 °C overnight, followed by washes, incubated with secondary antibodies for 2 h at room temperature, and counterstained with DAPI (Life Technologies). For p63 staining, antigen retrival was performed in antigen retrieval buffer (citrate buffer, Ph = 5.7) using pressure cooker before blocking and antibody staining. Confocal images were captured using Leica DM6000 TCS/ SP8 laser confocal scanning microscope. Representative images were shown in the figures.
Quantification of lineage-specific cells and the size of clones
In K8-CreERT2;Rosa26-mTmG mammary glands, clones were defined as clusters of one or more GFP+ cells that contacted each other. A minimum of 3 experimental mice and a minimum of 100 GFP+ clones on more than 20 sections were analysed per time point. For each clone, the number of cells was scored in reference to K14 and K8 expression. Representative clones were documented by confocal imaging.
RNA sequencing and data processing
Approximately 50 cells of Lin−, CD24+, CD29hi, tdTomato+ (normal basal), Lin−, CD24+, CD29hi, GFP+ (LdBC) and Lin−, CD24+, CD29low, GFP+ (normal luminal) were harvested by FACS. Cell samples were lysed in 50 μl of 4 M guanidine isothiocyanate solution (GuSCN; Invitrogen, 15577-018) at 42 °C for 10 min. The volume of the lysate was adjusted to 200 μl by nuclease-free water, and was further concentrated by ethanol precipitation in the presence of 1/10 volume of acetate sodium (pH 5.7, 3 M; Ambion) and 2 μl of carrier glycogen (20 mg/ml; Roche). Total RNA pellets were dissolved in lysis solution and reverse-transcribed and amplified with modified Smart-seq2 PCR technique,64 followed by sequencing on Illumina HiSeq 2500. RNA-seq data can be viewed online under GEO accession number GSE109543.
The quality of the reads was evaluated using the FASTQC tool. Raw reads were mapped to mm10 version of mouse genome using Tophat2 v2.0.4 program.65 We calculated fragment per kilobase per million (FPKM) as expression level using Cufflinks v2.0.2 with default parameters.66 Genes with the FPKM > 1.0 in at least one sample across all samples were retained for further analysis. Finally, the expression levels were transformed to logarithmic space by using the log2 (FPKM + 1). Hierarchical clustering and Principal component analysis (PCA) was based on all the expressed genes as described in the RNA-Seq data preprocessing and was performed using FactoMineR package in R.67,68 Differentially expressed genes (DEGs) between each cell groups were identified using RankProd69 with P value < 0.05 and fold change > 1.5. Heatmaps were generated using Cluster 3.0 and JavaTreeView.70 Connection Specificity Index (CSI) analysis employed the PCC (Pearson Correlation Coefficient) as a first-level association index to rank the similarity between nodes, and then a constant of 0.9 was used to define the boundary of interaction-profile similarity.71 Functional enrichment of gene sets with different expression patterns was performed using the Database for Annotation, Visualization and Integrated Discovery v6.8 (DAVID v6.8).69,72 Gene set enrichment analysis (GSEA) was used to examine whether the genes identified as DEGs for each cell type are members of categories with specific signaling pathway. Expression of selected genes was validated and quantitative RT-PCR was performed using SYBR Green gene expression systems for mouse (Applied Biosystems).
Primers used in qPCR analysis
Primers used were as follows.
K14 forward: TGACCATGCAGAACCTCAATGA;
K14 reverse: ATTGGCATTGTCCACGG;
K8 forward: AGGATGAGATCAACAAGCGT;
K8 reverse: CTTCATGGATCTGCCGGA;
ΔNp63 forward: CTGGCAAAACCCTGGAAG;
ΔNp63 reverse: CAACATGTTAGCAGTGAGACTGG.
Esr1 forward: TCCAGCAGTAACGAGAAAGGA
Esr1 reverse: AGCCAGAGGCATAGTCATTGC
Statistical analysis
Student’s t-test was performed when comparing two groups. One-way ANOVA was performed for comparision among more than three groups. Data analysed with One-way ANOVA were specially annotated in figure legends, the others were analysed with t-test. The P value was calculated in Prism for data represented by bar charts, which consisted of results from three independent experiments unless specified otherwise. For all experiments with error bars, the standard deviation (SD) was calculated to indicate the variation within each experiment. The experiments were not randomized. The investigators were not blinded to allocation during experiments and outcome assessment.
Supplementary information
Acknowledgements
We thank Dr. Li Xin for kindly sharing the K8-CreERT2 mouse line and Dr. Fiona Watt for kindly sharing the K14-Lef1Δn mouse line. This work was supported by grants from National Natural Science Foundation of China (31830056, 31861163006, 31625020, 31530045, 31661143043 to Y.A.Z.; 31871456, 31661143042, 91519314, 31630043, 31571513, 31430058 to N.J.), Chinese Academy of Sciences (XDB19000000 and XDA12020349 to Y.A.Z.; XDA16020501 to N.J., XDA16020404 to G.P.), Ministry of Science and Technology of China (2018YFA0108000, 2017YFA0102700, 2015CB964500 to N.J., 2018YFA0107201 to G.P.), and Research Grants Council (N_HKUST628/18 to T.H.C), Shanghai Municipal Science and Technology Commission (17XD1404000 to Y.A.Z.).
Author contributions
W.S. and Y.A.Z. designed the experiments. W.S. and R.Y. performed immunostaining staining, FACS and transplantation experiments. R.W. and G.P. performed RNA sequencing and data analysis, T.C. and N.J. supervised bioinformatics analysis. W.S., W.J. and Q.C.Y. generated mouse strains and conducted genetic experiments. Q.Y. and J.L. redirived and generated the rainbow-reporter mouse strain. W.S., R.W. and Y.A.Z. analyzed the data and wrote the manuscript.
Competing interests
The authors declare no competing interests.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41422-018-0137-0.
References
- 1.Sreekumar A, Roarty K, Rosen JM. The mammary stem cell hierarchy: a looking glass into heterogeneous breast cancer landscapes. Endocr. Relat. Cancer. 2015;22:T161–T176. doi: 10.1530/ERC-15-0263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Visvader JE, Stingl J. Mammary stem cells and the differentiation hierarchy: current status and perspectives. Genes Dev. 2014;28:1143–1158. doi: 10.1101/gad.242511.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stingl J, et al. Purification and unique properties of mammary epithelial stem cells. Nature. 2006;439:993–997. doi: 10.1038/nature04496. [DOI] [PubMed] [Google Scholar]
- 4.Shackleton M, et al. Generation of a functional mammary gland from a single stem cell. Nature. 2006;439:84–88. doi: 10.1038/nature04372. [DOI] [PubMed] [Google Scholar]
- 5.Rios AC, Fu NY, Lindeman GJ, Visvader JE. In situ identification of bipotent stem cells in the mammary gland. Nature. 2014;506:322–327. doi: 10.1038/nature12948. [DOI] [PubMed] [Google Scholar]
- 6.Wang D, et al. Identification of multipotent mammary stem cells by protein C receptor expression. Nature. 2015;517:81–84. doi: 10.1038/nature13851. [DOI] [PubMed] [Google Scholar]
- 7.Van Keymeulen A, et al. Distinct stem cells contribute to mammary gland development and maintenance. Nature. 2011;479:189–193. doi: 10.1038/nature10573. [DOI] [PubMed] [Google Scholar]
- 8.van Amerongen R, Bowman AN, Nusse R. Developmental stage and time dictate the fate of Wnt/beta-catenin−responsive stem cells in the mammary gland. Cell Stem Cell. 2012;11:387–400. doi: 10.1016/j.stem.2012.05.023. [DOI] [PubMed] [Google Scholar]
- 9.Prater MD, et al. Mammary stem cells have myoepithelial cell properties. Nat. Cell Biol. 2014;16:942–950. doi: 10.1038/ncb3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Visser KE, et al. Developmental stage-specific contribution of LGR5(+) cells to basal and luminal epithelial lineages in the postnatal mammary gland. J. Pathol. 2012;228:300–309. doi: 10.1002/path.4096. [DOI] [PubMed] [Google Scholar]
- 11.Wuidart A, et al. Quantitative lineage tracing strategies to resolve multipotency in tissue-specific stem cells. Genes Dev. 2016;30:1261–1277. doi: 10.1101/gad.280057.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davis FM, et al. Single-cell lineage tracing in the mammary gland reveals stochastic clonal dispersion of stem/progenitor cell progeny. Nat. Commun. 2016;7:13053. doi: 10.1038/ncomms13053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brisken C, O’Malley B. Hormone action in the mammary gland. Cold Spring Harb. Perspect. Biol. 2010;2:a003178. doi: 10.1101/cshperspect.a003178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clarke RB, Howell A, Potten CS, Anderson E. Dissociation between steroid receptor expression and cell proliferation in the human breast. Cancer Res. 1997;57:4987–4991. [PubMed] [Google Scholar]
- 15.Shyamala G, et al. Cellular expression of estrogen and progesterone receptors in mammary glands: regulation by hormones, development and aging. J. Steroid Biochem Mol. Biol. 2002;80:137–148. doi: 10.1016/S0960-0760(01)00182-0. [DOI] [PubMed] [Google Scholar]
- 16.Petersen OW, Hoyer PE, van Deurs B. Frequency and distribution of estrogen receptor-positive cells in normal, nonlactating human breast tissue. Cancer Res. 1987;47:5748–5751. [PubMed] [Google Scholar]
- 17.Lim E, et al. Aberrant luminal progenitors as the candidate target population for basal tumor development in BRCA1 mutation carriers. Nat. Med. 2009;15:907–913. doi: 10.1038/nm.2000. [DOI] [PubMed] [Google Scholar]
- 18.Asselin-Labat ML, et al. Steroid hormone receptor status of mouse mammary stem cells. J. Natl Cancer Inst. 2006;98:1011–1014. doi: 10.1093/jnci/djj267. [DOI] [PubMed] [Google Scholar]
- 19.Sleeman KE, et al. Dissociation of estrogen receptor expression and in vivo stem cell activity in the mammary gland. J. Cell Biol. 2007;176:19–26. doi: 10.1083/jcb.200604065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stingl J. Estrogen and progesterone in normal mammary gland development and in cancer. Horm. Cancer. 2011;2:85–90. doi: 10.1007/s12672-010-0055-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bergkvist L, Adami HO, Persson I, Hoover R, Schairer C. The risk of breast cancer after estrogen and estrogen-progestin replacement. N. Engl. J. Med. 1989;321:293–297. doi: 10.1056/NEJM198908033210505. [DOI] [PubMed] [Google Scholar]
- 22.Pike MC, Spicer DV, Dahmoush L, Press MF. Estrogens, progestogens, normal breast cell proliferation, and breast cancer risk. Epidemiol. Rev. 1993;15:17–35. doi: 10.1093/oxfordjournals.epirev.a036102. [DOI] [PubMed] [Google Scholar]
- 23.Clemons M, Goss P. Estrogen and the risk of breast cancer. N. Engl. J. Med. 2001;344:276–285. doi: 10.1056/NEJM200101253440407. [DOI] [PubMed] [Google Scholar]
- 24.Chang TH, et al. New insights into lineage restriction of mammary gland epithelium using parity-identified mammary epithelial cells. Breast Cancer Res. 2014;16:R1. doi: 10.1186/bcr3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rodilla V, et al. Luminal progenitors restrict their lineage potential during mammary gland development. PLoS Biol. 2015;13:e1002069. doi: 10.1371/journal.pbio.1002069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Van Keymeulen A, et al. Lineage-restricted mammary stem cells sustain the development, homeostasis, and regeneration of the estrogen receptor positive lineage. Cell Rep. 2017;20:1525–1532. doi: 10.1016/j.celrep.2017.07.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang C, Christin JR, Oktay MH, Guo W. Lineage-biased stem cells maintain estrogen-receptor-positive and -negative mouse mammary luminal lineages. Cell Rep. 2017;18:2825–2835. doi: 10.1016/j.celrep.2017.02.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Elias S, Morgan MA, Bikoff EK, Robertson EJ. Long-lived unipotent Blimp1-positive luminal stem cells drive mammary gland organogenesis throughout adult life. Nat. Commun. 2017;8:1714. doi: 10.1038/s41467-017-01971-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lafkas D, et al. Notch3 marks clonogenic mammary luminal progenitor cells in vivo. J. Cell Biol. 2013;203:47–56. doi: 10.1083/jcb.201307046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bach K, et al. Differentiation dynamics of mammary epithelial cells revealed by single-cell RNA sequencing. Nat. Commun. 2017;8:2128. doi: 10.1038/s41467-017-02001-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pal B, et al. Construction of developmental lineage relationships in the mouse mammary gland by single-cell RNA profiling. Nat. Commun. 2017;8:1627. doi: 10.1038/s41467-017-01560-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guo W, et al. Slug and Sox9 cooperatively determine the mammary stem cell state. Cell. 2012;148:1015–1028. doi: 10.1016/j.cell.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Panciera T, et al. Induction of expandable tissue-specific stem/progenitor cells through transient expression of YAP/TAZ. Cell Stem Cell. 2016;19:725–737. doi: 10.1016/j.stem.2016.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Molyneux G, et al. BRCA1 basal-like breast cancers originate from luminal epithelial progenitors and not from basal stem cells. Cell Stem Cell. 2010;7:403–417. doi: 10.1016/j.stem.2010.07.010. [DOI] [PubMed] [Google Scholar]
- 35.Ling H, Jolicoeur P. Notch-1 signaling promotes the cyclinD1-dependent generation of mammary tumor-initiating cells that can revert to bi-potential progenitors from which they arise. Oncogene. 2013;32:3410–3419. doi: 10.1038/onc.2012.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hein SM, et al. Luminal epithelial cells within the mammary gland can produce basal cells upon oncogenic stress. Oncogene. 2016;35:1461–1467. doi: 10.1038/onc.2015.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Koren S, et al. PIK3CA(H1047R) induces multipotency and multi-lineage mammary tumours. Nature. 2015;525:114–118. doi: 10.1038/nature14669. [DOI] [PubMed] [Google Scholar]
- 38.Van Keymeulen A, et al. Reactivation of multipotency by oncogenic PIK3CA induces breast tumour heterogeneity. Nature. 2015;525:119–123. doi: 10.1038/nature14665. [DOI] [PubMed] [Google Scholar]
- 39.Zhang L, et al. Targeting CreER(T2) expression to keratin 8-expressing murine simple epithelia using bacterial artificial chromosome transgenesis. Transgenic Res. 2012;21:1117–1123. doi: 10.1007/s11248-012-9598-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Choi N, Zhang B, Zhang L, Ittmann M, Xin L. Adult murine prostate basal and luminal cells are self-sustained lineages that can both serve as targets for prostate cancer initiation. Cancer Cell. 2012;21:253–265. doi: 10.1016/j.ccr.2012.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yu Qing, Verheyen Esther, Zeng Yi. Mammary Development and Breast Cancer: A Wnt Perspective. Cancers. 2016;8(7):65. doi: 10.3390/cancers8070065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Markopoulos C, Berger U, Wilson P, Gazet JC, Coombes RC. Oestrogen receptor content of normal breast cells and breast carcinomas throughout the menstrual cycle. Br. Med J. 1988;296:1349–1351. doi: 10.1136/bmj.296.6633.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Press MF, Nousek-Goebl N, King WJ, Herbst AL, Greene GL. Immunohistochemical assessment of estrogen receptor distribution in the human endometrium throughout the menstrual cycle. Lab Invest. 1984;51:495–503. [PubMed] [Google Scholar]
- 44.Asselin-Labat ML, et al. Control of mammary stem cell function by steroid hormone signalling. Nature. 2010;465:798–802. doi: 10.1038/nature09027. [DOI] [PubMed] [Google Scholar]
- 45.Ellison-Zelski SJ, Solodin NM, Alarid ET. Repression of ESR1 through actions of estrogen receptor alpha and Sin3A at the proximal promoter. Mol. Cell Biol. 2009;29:4949–4958. doi: 10.1128/MCB.00383-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Niemann C, Owens DM, Hulsken J, Birchmeier W, Watt FM. Expression of DeltaNLef1 in mouse epidermis results in differentiation of hair follicles into squamous epidermal cysts and formation of skin tumours. Development. 2002;129:95–109. doi: 10.1242/dev.129.1.95. [DOI] [PubMed] [Google Scholar]
- 47.Harada N, et al. Intestinal polyposis in mice with a dominant stable mutation of the beta-catenin gene. EMBO J. 1999;18:5931–5942. doi: 10.1093/emboj/18.21.5931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lindley LE, et al. The WNT-controlled transcriptional regulator LBH is required for mammary stem cell expansion and maintenance of the basal lineage. Development. 2015;142:893–904. doi: 10.1242/dev.110403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chakrabarti R, et al. DeltaNp63 promotes stem cell activity in mammary gland development and basal-like breast cancer by enhancing Fzd7 expression and Wnt signalling. Nat. Cell Biol. 2014;16:1004–1015. doi: 10.1038/ncb3040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gu B, Watanabe K, Sun P, Fallahi M, Dai X. Chromatin effector Pygo2 mediates Wnt-notch crosstalk to suppress luminal/alveolar potential of mammary stem and basal cells. Cell Stem Cell. 2013;13:48–61. doi: 10.1016/j.stem.2013.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bouras T, et al. Notch signaling regulates mammary stem cell function and luminal cell-fate commitment. Cell Stem Cell. 2008;3:429–441. doi: 10.1016/j.stem.2008.08.001. [DOI] [PubMed] [Google Scholar]
- 52.Wagner KU, et al. An adjunct mammary epithelial cell population in parous females: its role in functional adaptation and tissue renewal. Development. 2002;129:1377–1386. doi: 10.1242/dev.129.6.1377. [DOI] [PubMed] [Google Scholar]
- 53.Matulka LA, Triplett AA, Wagner KU. Parity-induced mammary epithelial cells are multipotent and express cell surface markers associated with stem cells. Dev. Biol. 2007;303:29–44. doi: 10.1016/j.ydbio.2006.12.017. [DOI] [PubMed] [Google Scholar]
- 54.Boulanger CA, Wagner KU, Smith GH. Parity-induced mouse mammary epithelial cells are pluripotent, self-renewing and sensitive to TGF-beta1 expression. Oncogene. 2005;24:552–560. doi: 10.1038/sj.onc.1208185. [DOI] [PubMed] [Google Scholar]
- 55.Keller PJ, et al. Defining the cellular precursors to human breast cancer. Proc. Natl Acad. Sci. USA. 2012;109:2772–2777. doi: 10.1073/pnas.1017626108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ince TA, et al. Transformation of different human breast epithelial cell types leads to distinct tumor phenotypes. Cancer Cell. 2007;12:160–170. doi: 10.1016/j.ccr.2007.06.013. [DOI] [PubMed] [Google Scholar]
- 57.Gastaldi S, et al. Met signaling regulates growth, repopulating potential and basal cell-fate commitment of mammary luminal progenitors: implications for basal-like breast cancer. Oncogene. 2013;32:1428–1440. doi: 10.1038/onc.2012.154. [DOI] [PubMed] [Google Scholar]
- 58.Janerich DT, Hoff MB. Evidence for a crossover in breast cancer risk factors. Am. J. Epidemiol. 1982;116:737–742. doi: 10.1093/oxfordjournals.aje.a113462. [DOI] [PubMed] [Google Scholar]
- 59.Schedin P. Pregnancy-associated breast cancer and metastasis. Nat. Rev. Cancer. 2006;6:281–291. doi: 10.1038/nrc1839. [DOI] [PubMed] [Google Scholar]
- 60.MacMahon B, et al. Age at first birth and breast cancer risk. Bull. World Health Organ. 1970;43:209–221. [PMC free article] [PubMed] [Google Scholar]
- 61.Muzumdar MD, Tasic B, Miyamichi K, Li L, Luo L. A global double-fluorescent Cre reporter mouse. Genesis. 2007;45:593–605. doi: 10.1002/dvg.20335. [DOI] [PubMed] [Google Scholar]
- 62.Rinkevich Y, Lindau P, Ueno H, Longaker MT, Weissman IL. Germ-layer and lineage-restricted stem/progenitors regenerate the mouse digit tip. Nature. 2011;476:409–413. doi: 10.1038/nature10346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Huelsken J, Vogel R, Erdmann B, Cotsarelis G, Birchmeier W. beta-Catenin controls hair follicle morphogenesis and stem cell differentiation in the skin. Cell. 2001;105:533–545. doi: 10.1016/S0092-8674(01)00336-1. [DOI] [PubMed] [Google Scholar]
- 64.Chen J, et al. Spatial transcriptomic analysis of cryosectioned tissue samples with Geo-seq. Nat. Protoc. 2017;12:566–580. doi: 10.1038/nprot.2017.003. [DOI] [PubMed] [Google Scholar]
- 65.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kim D, et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li YY, et al. Transcriptome analysis reveals determinant stages controlling human embryonic stem cell commitment to neuronal cells. J. Biol. Chem. 2017;292:19590–19604. doi: 10.1074/jbc.M117.796383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sozen B, et al. Self-assembly of embryonic and two extra- embryonic stem cell types into gastrulating embryo-like structures. Nat. Cell Biol. 2018;20:979. doi: 10.1038/s41556-018-0147-7. [DOI] [PubMed] [Google Scholar]
- 69.Hong FX, et al. RankProd: a bioconductor package for detecting differentially expressed genes in meta-analysis. Bioinformatics. 2006;22:2825–2827. doi: 10.1093/bioinformatics/btl476. [DOI] [PubMed] [Google Scholar]
- 70.Saldanha AJ. Java Treeview-extensible visualization of microarray data. Bioinformatics. 2004;20:3246–3248. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- 71.Yang X, et al. TGFbeta signaling hyperactivation-induced tumorigenicity during the derivation of neural progenitors from mouse ESCs. J. Mol. Cell Biol. 2018;10:216–228. doi: 10.1093/jmcb/mjy013. [DOI] [PubMed] [Google Scholar]
- 72.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.