Abstract
My intention here is to describe the history of the molecular aspects of the antigen processing field from a personal perspective, beginning with the early identification of the species that we now know as MHC class I and MHC class II molecules, to the recognition that their stable surface expression and detection by T cells depends on peptide association, and to the unraveling of the biochemical and cell biological mechanisms that regulate peptide binding. One goal is to highlight the role that serendipity or, more colloquially, pure blind luck, can play in advancing the research enterprise when it is combined with an appropriately receptive mind. This is not intended to be an overarching review, and because of my own work I focus primarily on studies of the human MHC. This means that I neglect the work of many other individuals who made advances in other species, particularly those who produced the many knockout mouse strains used to demonstrate the importance of the antigen processing machinery for initiating immune responses. I apologize in advance to colleagues around the globe whose contributions I deal with inadequately for these reasons, and to those whose foundational work is now firmly established in text books and therefore not cited. So many individuals have worked to advance the field that giving all of them the credit they deserve is almost impossible. I have attempted, while focusing on work from my own laboratory, to point out contemporaneous or sometimes earlier advances made by others. Much of the success of my own laboratory came because we simultaneously worked on both the MHC class I and class II systems and used the findings in one area to inform the other, but mainly it depended on the extraordinary group of students and fellows who have worked on these projects over the years. To those who worked in other areas who are not mentioned here, rest assured that I appreciate your efforts just as much.
Major Histocompatibility Complex (MHC) molecules are currently so familiar that it is difficult to imagine that until the late 1960’s and early 1970’s they were undefined except as the targets for immune responses induced by transplantation. The molecular species recognized by alloantisera and alloreactive T cells were unknown. A number of individuals began to isolate and purify the critical cell surface molecules using their ability to bind alloantisera in a variety of assay techniques. The late Stanley Nathenson, working at Albert Einstein College of Medicine, simplified the process by showing that mouse MHC molecules, or H2 molecules, could be released from cell membranes by cleavage with papain (Shimada A 1967). The late Arnold Sanderson, at the McIndoe Memorial Laboratories in East Grinstead, Sussex, U.K., adapted this to the human system, using papain to release soluble HLA molecules from human spleens, and showed that different gene products could be separated by ion exchange chromatography (SandersonAR 1968).Both investigators identified the purified products as proteins, although for a few years Sanderson held on to the hope that the components recognized by anti HLA antibodies would be the glycans of what proved to be glycoproteins. This early work preceded the eventual division of MHC genes and their products into class I and class II subsets, and the species they purified later proved to be MHC class I molecules, now often abbreviated MHC-I. MHC class II molecules (MHC-II) were characterized later.
I obtained my Ph.D. in the Sanderson laboratory and subsequently took up a postdoctoral fellowship with Jack Strominger at Harvard University where, with another British postdoc, Mervyn Turner, I helped to transfer the papain solubilization and HLA purification technique to Cambridge, MA, using as a source EBV-transformed human B-lymphoblastoid cell lines (BLCL), generously provided by Dean Mann at the NIH, rather than spleens. We continued with the analysis of the papain-released molecules, showing that they were comprised of two subunits, that the larger one was glycosylated and polymorphic while the smaller one was not (Cresswell P 1974a; Cresswell P 1973), and eventually finding, in collaboration with Howard Grey and Ralph Kubo, that the smaller one was β2-microglobulin (β2m) (Cresswell P 1974b; Grey HM 1973). Tim Springer, then a Ph.D. student in the Strominger laboratory, was the first to successfully use detergents to solubilize, purify and characterize full-length MHC-I molecules (Springer TA 1977). In 1973 I left Harvard to begin an independent position at Duke University and later additions to the Strominger group determined the amino acid sequences of papain-solubilized HLA class I molecules, and eventually many laboratories conspired to obtain complete sequences of numerous alleles with the advent of cDNA cloning and sequencing.
In my laboratory at Duke we made rabbit antisera to papain solubilized MHC-I molecules and found that they were not as specific as we hoped. When we used them to immunoprecipitate radiolabeled detergent extracts of BLCL membranes we found that, in addition to the expected MHC-I heavy chain and β2m, two other proteins were identifiable by SDS-PAGE (Cresswell P 1975). These proved to be the α- and β-subunits of MHC-II molecules, probably HLA-DR, which were highly immunogenic minor contaminants in the immunogen. In the Strominger group Robert Humphreys deliberately purified these contaminants and came to similar conclusions (Humphreys RE 1976). We now know that MHC-II molecules are poorly susceptible to papain-mediated cleavage and release. Contemporaneous work by Stanley Nathenson and others determined the subunit structures of the mouse MHC-II molecules.
The function of MHC molecules moved beyond their role in transplantation with the discovery by Peter Doherty and Rolf Zinkernagel that killing of virally-infected cells in vitro by CD8-positive T cells isolated from a virus-infected mouse depended on the expression by the targets of a host MHC-I allele (Zinkernagel RM 1974). This became known as ‘MHC restriction’. Similar work in the guinea pig and in mice showed that this was also true for CD4-positive T cell recognition of antigen presenting cells (APCs) incubated with protein or synthetic peptide antigens: the APCs had to be from the same strain as the immunized animal that was the source of the CD4-positive T cells. The ‘restricting elements’ in this case proved to be MHC-II gene products. The addition of a short peptide derived from an antigen to the APCs could also stimulate CD4-positive T cells, and this, unlike the recognition of intact antigen, was resistant to paraformaldehyde fixation of the APCs. This led to the concept that the antigen had to be partially degraded by the APC to generate an MHC-II-peptide complex that was recognized by the T cell. Purified MHC-I and MHC-II molecules ultimately proved to be associated with endogenous peptides, derived predominantly from cytosolic proteins in the case of MHC-I and external or luminal proteins in the case of MHC-II. The definition of the HLAA2 crystal structure by Pamela Bjorkman and Don Wiley showed that peptides occupied the now classical MHC-I binding groove (Bjorkman PJ 1987), and multiple subsequent MHC-I and MHC-II structures defined the mode of peptide binding for each. Emil Unanue was the first to show that peptides could bind to purified MHC-II molecules in vitro and to calculate a binding affinity (Babbitt BP 1985). These remarkable advances set the stage for what eventually became a major field, the study of the molecular events underlying the generation of MHC-I- and MHC-II-peptide complexes recognized by CD8-positive and CD4-positive T cells, respectively. This became known as antigen processing. I reserve the term antigen presentation to describe subsequent interactions with T cells and will not address that here.
The biochemistry and cell biology of human MHC-II-restricted antigen processing
The discovery of human MHC-II molecules established the initial path for my independent work at Duke. Analysis of immunoprecipitated MHC-II molecules by two-dimensional electrophoresis, prior to their division into HLA-DR, DQ and DP gene products, revealed electrophoretic variability, particularly in the β-subunits, isolated from BLCL that were homozygous for different haplotypes (Markert ML 1980). This work, initiated by Louise Markert, an M.D./Ph.D. student, used BLCL labeled with 35S-methionine. Evident on some of the gels was a string of ‘spots’ with variable charge at around 30–35 kDa. Carolyn Machamer, a Ph.D. student, showed that these spots were derivatives of the invariant chain (Ii), first identified and named by Patricia Jones and Hugh McDevitt at Stanford University (Jones PP 1979), containing different numbers of sialic acid residues on its two N-linked glycans as well as acquired O-linked glycans (Machamer CE 1982). Initially we resisted the abbreviation Ii because its continuous repetition lent a peculiar nautical flair (‘aye aye, sir’) to any sentence containing it, and in fact even the name ‘invariant chain’ was belied by its electrophoretic variability and by the eventual discovery of additional forms of the protein generated by alternative splicing and, in humans but not mice, alternative translational start sites. This results in four human Ii forms, called p33, p35, p41 and p43 (Strubin M 1986). Fortunately for us the addition of sialic acid residues, an event occurring in the late Golgi and the trans-Golgi network, provided the analytical tool required for Carolyn to perform sophisticated pulse-chase analyses that revealed the association of HLA-DR molecules with Ii in the endoplasmic reticulum (ER), the progressive modification of Ii by sialic acid and O-linked glycan addition during transport, and its disappearance prior to expression of the MHC-II molecules themselves on the plasma membrane (Machamer CE 1982). Work by another Ph.D. student, Michael Marks, showed that Ii was trimeric, containing mixtures of the p33, p35, p41 and p43 forms, and Paul Roche, a postdoctoral fellow, showed with Michael that the complex assembled in the ER was a nonamer, with three MHC-II αβ dimers associated with an Ii trimer (Marks MS 1990; Roche PA 1991). Carilee Lamb, a Ph.D. student, showed the progressive association of individual αβ dimers with the Ii trimer (Lamb CA 1992), and later work by Karen Anderson, a Duke M.D./Ph.D. student, showed that the assembly process was facilitated by the lectin chaperone calnexin (Anderson KS 1994). Paul found that association of Ii with MHC-II molecules prevented them from binding antigenic peptides, arguing that one of its functions was to prevent their premature association with peptides or, probably more importantly, unfolded proteins in the ER (Roche PA 1990). Cari Lamb, following in the footsteps of Per Peterson’s laboratory (Lotteau V 1990) and with a major assist from Jonathan Yewdell, showed that late in transport the MHC-II-Ii complex was transferred by targeting signals in the Ii cytoplasmic domain to lysosome-like late endosomal compartments containing internalized influenza virus virions (Lamb CA 1991). Janice Blum, a postdoctoral fellow, showed that the disappearance of Ii was a consequence of its proteolytic degradation within those compartments: addition to cells of protease inhibitors such as leupeptin resulted in the accumulation of MHC-II molecules associated with partial Ii degradation products (Blum JS 1988). Overall, this work, together with the work of other groups, particularly those of Ronald Germain, Per Peterson and Hidde Ploegh, established the now familiar paradigm of MHC-II assembly and intracellular transport, encapsulated and updated in Fig. 1.
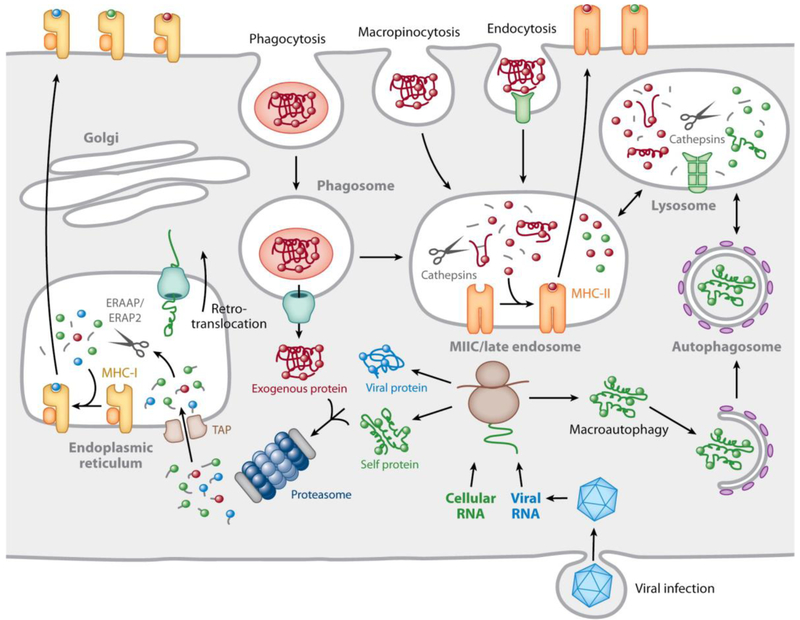
Fig. 1. Mechanisms underlying MHC-II-restricted antigen processing.
MHC-II αβ dimers associate with trimers of the invariant chain (labeled I) in the ER. When the nonameric complex is completely assembled it leaves the ER and moves through the Golgi apparatus before being diverted into the endocytic pathway by a C-terminal cytoplasmic motif in the invariant chain. Late in the endocytic pathway, in the MHC-II compartment or MIIC, exposure to an acidic environment combined with a collection of proteases known as cathepsins results in invariant chain proteolysis, leaving the Class II-associated Invariant chain Peptide (CLIP) in the MHC-II binding groove. Peptides derived from exogenous proteins internalized by endocytosis, micropinocytosis, or phagocytosis, or introduced from the cytosol by autophagy, are also proteolyzed, producing a range of peptides. A subset of these peptides has appropriate sequences for binding to the specific MHC-II alleles expressed by the cell. However, binding does not occur spontaneously. The MHC-II molecule interacts with a homologous non-peptide binding αβ dimer, DM, which is resident in the MIIC, which allosterically modifies the MHC-II binding groove to induce CLIP release and exchange for locally generated peptides. Peptides of sufficiently high affinity avoid subsequent DM-induced release and are expressed on the plasma membrane. A second MHC-II homologue, DO, can bind to DM in the ER and DM/DO complexes also localize to the MIIC but are inactive in inducing CLIP release. DO serves to modulate DM-induced CLIP release and peptide exchange. (reproduced from Blum et al., 2013).
The rationale behind the diversion of MHC-II molecules to the endocytic pathway lies in their function, namely to bind peptides derived from antigenic proteins internalized by antigen presenting cells. The first experiment to provide evidence for the intersection of the MHC-II intracellular transport pathway and the endocytic pathway was provided by an experiment of my own, which showed that neuraminidase covalently linked to transferrin and internalized via the transferrin receptor was capable of removing sialic acid residues from Ii associated with HLA-DR αβ dimers in BLCL (Cresswell 1985). Although the transferrin receptor is not a relevant receptor for antigens, work by Colin Watts and Antonio Lanzavecchia demonstrated that the surface Ig of BLCL specific for tetanus toxoid delivered specific antigen for binding to intracellular HLA-DR molecules (Davidson HW 1991). The compartment where the endocytic pathway intersects the MHC-II transport pathway was defined by Hans Geuze, Jacques Neefjes, Peter Peters and Hidde Ploegh using immunoelectron microscopy and named the MHC class II compartment, or MIIC (Neefjes JJ 1990).
The combination of Ii proteolysis that liberated MHC-II molecules capable of binding peptides, and antigen proteolysis in the same compartment generating the peptides that bind to them, provided an irresistible model to explain antigen processing. However, it proved not to be so simple, as revealed by a mutant BLCL that proved incapable of mediating antigen processing, which was generated by Elizabeth Mellins and the late Donald Pious (Mellins E 1990). Their experiments evolved in parallel with similar experiments by Robert DeMars that began with the goal of uncovering the genetic organization of the human MHC by making a series of deletion mutants within it (Orr HT 1982). More importantly, however, they revealed the existence of MHC-linked genes that regulated both MHC-I- and MHC-II-restricted antigen processing. Although we pursued work on the MHC-I and MHC-II systems simultaneously, I will address the MHC-I story first.
The biochemistry and cell biology of MHC-I-restricted antigen processing
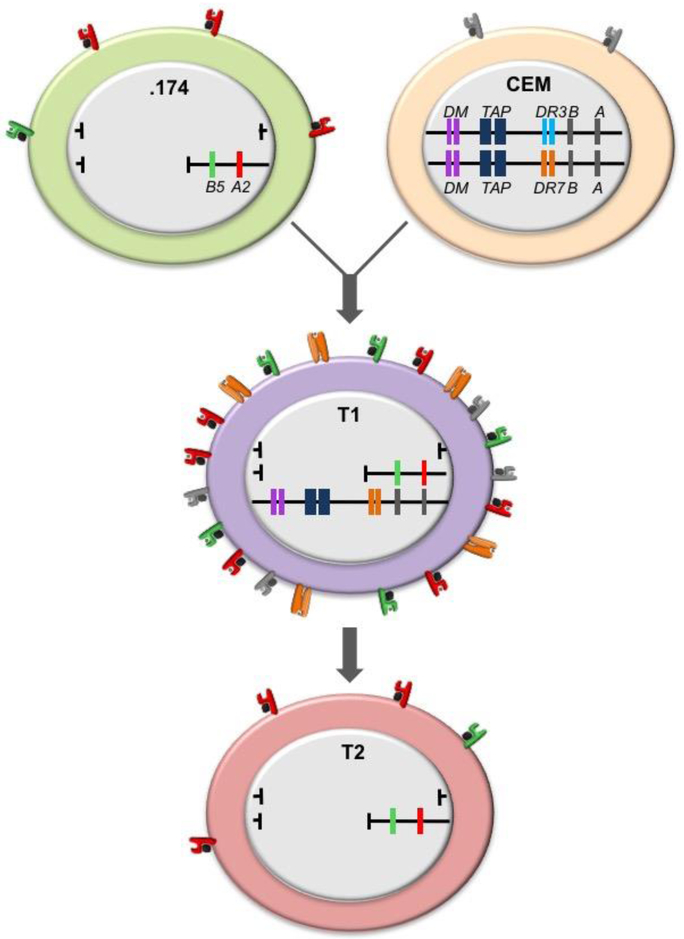
Our involvement with Robert DeMars and his remarkable collection of mutant cell lines arose from previous experiments using somatic cell fusion to examine the regulation of MHC-II expression. David Howell, a Duke M.D./Ph.D. student, had shown that somatic cell fusion of BLCL with MHC-II-negative T lymphoblastoid cell lines (TLCL) resulted in hybrids that expressed the T cell-encoded MHC-II antigens (Howell DN 1983). The DeMars mutants began with a BLCL heterozygous for the HLA complex that was subjected to sequential rounds of gamma irradiation followed by cytotoxic selection using antibodies specific for its HLA-A, B, and DR alleles. Gamma irradiation tends to generate large deletions and one resulting cell line was 721.174, abbreviated to .174, which lacked one complete HLA haplotype and had a deletion of only the HLA class II region in the other (DeMars R 1984). Thus, it had a homozygous deletion of the HLA class II region. We posed a simple question, did the gene responsible for restoring T cell-encoded MHC-II expression in BLCL × TLCL hybrids lie within the deleted HLA class II region? We obtained the .174 cell line, and Russ Salter, a graduate student, fused it with the TLCL CCRF-CEM, which was derived from an acute lymphoblastic leukemia patient (Salter RD 1985). The results of the fusion are illustrated in Fig. 2. If the critical gene was in the HLA class II region a hybrid should be MHC-II negative. To our disappointment the hybrid (.174×CEM.T1, or T1) expressed the T cell-encoded class II allele HLA-DR7, indicating that the gene or genes regulating MHC-II expression were not in the HLA class-II region. However, .174 had a second phenotype: it had retained the genes encoding HLA-A2 and HLA-B5 but surface expression of the former was significantly reduced and surface expression of the latter was virtually eliminated (DeMars R 1984). To our surprise T1 expressed both at normal levels, indicating that a gene or genes regulating MHC class I expression probably lay within the class II region of the MHC. T1 turned out to have spontaneously lost one copy of the MHC-bearing chromosome 6 of the T cell partner. Russ then selected T1 for loss of its T cell-encoded HLA-DR7 allele using a monomorphic anti DR mAb and obtained the now well-known derivative T2, which lacked both T cell-derived copies of chromosome 6. T2, like the parental cell .174, was MHC-II-negative, low in HLA-A2 expression and virtually negative for HLA-B5 (Salter RD 1985). The derivation of T2 is illustrated in Fig.2. This experiment effectively showed that genes within the MHC-II region of the HLA complex regulated HLA class I surface expression, and Russ went on to show that transcription and translation of the MHC-I genes were unaffected and the problem lay at the level of MHCI assembly in the ER (Salter RD 1986).
Fig. 2. Human antigen processing mutant cell lines: the derivation of T2 from 731.174.
721.174 (.174) and T2 are two cell line that focused attention on the MHC-II region of the MHC as a place where genes regulating peptide binding by MHC-I and MHC-II were localized. These were eventually identified as the TAP1 and TAP2 genes for MHC-I and HLA-DMA and HLA-DMB genes for MHC-II. The lines within the individual cells represent the two MHC regions present in each cell. T2 arose from an initial somatic cell fusion between .174 (top left) and the T cell line CCRF-CEM (CEM) (top right). .174 has a complete deletion of the HLA complex on one copy of chromosome 6 and a deletion of the MHC-II region of the HLA complex on the other. It expresses no MHC-II on the cell surface because the coding genes are absent but, although the genes encoding HLA-A2 and -B5 are present, A2 surface expression is lower while B5 expression is dramatically reduced it is minimally expressed at the cell surface. CEM has two intact copies of the HLA complex, but MHC-I expression is very low and MHC-II expression is absent for transcriptional reasons. The initial fusion gave rise to a cell line called T1, where one of the CEM copies of chromosome 6 is missing, but it expresses high levels of HLA-A2 and -B5 encoded by .174 because the TAP genes were introduced from the parental CEM line. HLA-DR7 (orange), encoded but not synthesized by CEM, is also highly expressed, presumably because a transcriptional factor(s) derived from .174 has been introduced. The critical genes for MHC-I function encode the TAP1 and TAP2 (dark blue in the figure). T2 resulted from selection for loss of DR7 expression, which eliminated the remaining CEM-derived chromosome 6 and restored the surface expression phenotype of .174.
While we were pursuing our analysis of T2 cells, Alain Townsend in Oxford, who was already well known for his work with Andrew McMichael showing that CD8+ T cell recognition of influenza virus-infected mouse cells depended on their expression of H2-Kb and Db molecules associated with virus-derived peptides (Townsend AR 1986), was trying to unravel a new mystery. He had found that a mutant mouse T cell line, RMA-S, selected for low MHC-I expression by Klas Karre in his investigations of the role of MHCI in resistance to NK cell killing, could not be killed by CD8+ cytotoxic T cells when infected by influenza virus (Townsend A1 1989). However, like RMA, its parental cell line that has normal MHC-I expression, RMA-S could be killed when incubated with the precise MHC-I-binding peptide for which the CD8+ T cells were specific. This indicated that RMA-S cells were defective in generating MHC-I-peptide complexes from endogenously expressed viral proteins. The phenotype of low MHC-I surface expression shared by RMA-S, .174 and T2 suggested that the human cells might have the same problem, and that our somatic cell genetic work may have localized the critical gene(s) responsible to the MHC. Vincenzo Cerundolo in Alain’s laboratory, using an HLA-A2-restricted cytotoxic human CD8+ T cell clone specific for the influenza virus matrix protein, showed that this was indeed the case (Cerundolo V 1990). When infected, T1 cells were killed by the T cell clone while .174 and T2 cells were not. All could be killed when the precise peptide epitope was added to the cells, when it could bind directly to HLA-A2 at the surface. This was not simply a problem restricted to HLA-A2: numerous human and mouse MHC-I alleles proved to have cell surface expression problems when expressed in T2 cells, although this phenotype was less severe for most mouse alleles. Such cell lines, many produced by Jeff Alexander, a postdoctoral fellow in my laboratory, could not be recognized by virus-specific CD8+ T cells restricted to the specific allele (Alexander J 1989; Anderson KS 1993; Hosken NA 1990). As described earlier the majority of MHC-I-associated peptides had been shown to derive from cytosolic proteins, leading to the question of how they accessed MHC-I molecules. The molecules in .174, T2 and RMA-S proved to lack significant levels of associated endogenous peptides, resulting in impaired assembly with β2m, poor transport out of the ER, and reduced surface expression, suggesting that a lack of available cytosol-derived peptides might be the problem. Surprisingly, surface expression of endogenous HLA-A2 in T2 cells was never as low as the other human alleles that we introduced. This could be accounted for by its association with an alternative source of peptides in the ER, namely those derived from the signal sequences of a subset of secreted proteins. This was shown by Maria Wei, a Duke M.D./Ph.D. student, and Victor Engelhard’s laboratory at the University of Virginia (Henderson RA 1992; Wei ML 1992). Oddly, and probably coincidentally, a dominant peptide was from the signal sequence of calreticulin, which we later found plays an important role in MHC-I assembly (see below). Another was from the lysosomal protein IP-30, which we renamed GILT, for gamma interferon-inducible lysosomal thiol-reductase when B. ‘Arun’ Arunachalam, a postdoctoral fellow in the laboratory, demonstrated its enzymatic activity (Arunachalam B 2000). Later another postdoctoral fellow, Maja Maric, conclusively showed its importance for MHC-II-restricted processing of antigens containing disulfide bonds in a GILT knockout mouse, as illustrated in Fig. 1 (Maric M 2001).
The recognition that MHC-linked genes regulate MHC-I assembly and expression coincided with the intensive application of genomic cloning and sequencing to the MHC. Many investigators were beginning to work their way through the MHC-II region and identifying numerous ‘Really Interesting New Genes’, as John Trowsdale memorably called them. He was joined in this endeavor by Thomas Spies and John Monaco, among others. Two genes, encoding the interferon-inducible LMP2 and LMP7 β-subunits of the proteasome, later shown by Kenneth Rock and Alfred Goldberg to be the cytosolic protease responsible for generating MHC-I-associated peptides or their precursors (Rock KL 1994) proved to not be absolutely essential for MHC-I-restricted processing (Arnold D 1992), but two other genes, each encoding one half of a potential transporter of the ATP Binding Cassette (ABC) family emerged as candidates for peptide delivery into the ER. A variety of names emerged, my favorites being Trowsdale’s RING4 and RING11, but they all eventually coalesced into TAP1 and TAP2, the two subunits of the Transporter associated with Antigen Processing (Deverson EV 1990; Monaco JJ 1990; Spies T 1990; Trowsdale J 1990). Work by a number of investigators showed that reintroduction of TAP2 into RMA-S and TAP1 into another DeMars mutant that had a similar phenotype to .174 repaired their MHC-I antigen processing defect, while .174 and T2 required the introduction of both (Kelly A 1992; Powis SJ 1991; Spies T 1992). In 1991, towards the end of this period of cloning and gene identification, I moved from Duke to Yale University and we continued to work on these problems in my new laboratory and to make inroads into both MHC-I and MHC-II antigen processing mechanisms.
One of my first recruits to Yale, Matthew Androlewicz, a postdoctoral fellow, successfully showed that TAP was indeed a peptide transporter (Androlewicz MJ 1993). He found that a radio-iodinated HLA-A3-binding peptide bound efficiently to HLA-A3 in an HLA-A3-positive BLCL, or RMA cells expressing HLA-A3, when granted access to the cytosol by permeabilizing the plasma membrane with streptolysin O. However, this was not the case for T2 or RMA-S cells expressing HLA-A3. ATP was essential, which we interpreted to reflect the TAP requirement for ATP hydrolysis. Introduction of the peptide into the streptolysin O-permeablized cells also promoted the exit of newly synthesized HLA-A3 molecules from the ER. In follow-up experiments we borrowed an idea from Jacques Neefjes and Gunter Hammerling, who also demonstrated TAP-mediated peptide transport (Neefjes JJ 1993), and used a labeled peptide containing an N-linked glycosylation site that becomes glycosylated upon entry into the ER. Translocation could be quantitated using Concanavilin A-Sepharose to capture the radioactive glycopeptide. Matt used this assay to better define the length and sequence requirements for peptide interaction with TAP and subsequent translocation (Androlewicz MJ 1994a). Using .174 transfectants from Thomas Spies he also demonstrated that peptide translocation via TAP required both TAP1 and TAP2 subunits, and using photoactivatable labeled peptides he showed that the peptide binding site was comprised of cytosolic loops of both TAP1 and TAP2, a finding that was further extended by Gunter Hammerling’s group (Androlewicz MJ 1994b; Nijenhuis M 1996).
Bodo Ortmann was also an early postdoctoral recruit to Yale, and he made the first observation that led to the eventual definition of the peptide loading complex, or PLC, that facilitates peptide association with MHC-I molecules in the ER. Using digitonin as a detergent he demonstrated that solubilized TAP molecules physically associate with peptide-free MHC-I-β2m dimers (Ortmann B 1994). A similar observation was made in the mouse system by David Williams’ laboratory in Toronto (Suh WK 1994). Notably, Bodo also observed an unidentified component, a glycoprotein of approximately 48kDa, associated with MHC-I-TAP complexes. Calnexin, the transmembrane chaperone that Karen Anderson showed to be important in the assembly of the MHC-II-Ii complex, was found to associate with free HLA class I heavy chains but not with the assembled PLC, setting up the scenario that calnexin helped with the folding of the class I heavy chain, facilitating its association with β2m, and that the MHC-I-β2m dimers then associated with TAP. The latter observation provoked Bhanu Sadasivan, my first Yale Ph.D. student, to look for a possible role for calreticulin, a soluble homologue of calnexin, in MHC-I assembly. Bhanu observed that calreticulin associated with MHC-I-TAP complexes but did not associate with TAP in cells lacking MHC-I-β2m dimers (Sadasivan B 1996). She also found that adding a conventional detergent, such as Triton X-100, to MHC-I-TAP complexes released a subcomplex containing MHC-I-β2m dimers, calreticulin, and the 48kDa species identified by Bodo, which we named tapasin, for TAP-associated protein. The name famously arose in a discussion with Gunter Hammerling as we walked to a tapas bar in Madrid. The detergent-dependent interaction of the sub-complex with TAP led to the proposal that tapasin was a bridge that linked MHC-I-β2m to TAP, fleshing out the description of the PLC. We determined the N-terminal amino acid sequence of purified tapasin by classical Edman sequencing, and this allowed us to make a useful rabbit-anti-peptide antibody specific for it as well as providing a handle for the eventual cloning of a tapasin cDNA by James Copeman, a postdoctoral fellow in the laboratory.
We had a major assist in defining the role of tapasin in the PLC from yet another BLCL mutant from Robert DeMars. This cell line, called .220, had properties similar to TAP-negative cells but not as profound. MHC-I alleles generally had significantly lower surface expression when expressed in this cell line but some were more affected than others (Greenwood R 1994).Thomas Spies’ laboratory in Seattle showed that in these cells MHC-I-β2m dimers failed to interact with TAP (Grandea AG 1995). A collaborative venture between my laboratory and the Spies and Trowsdale laboratories established that these cells lacked tapasin, that this led to the impaired cell surface expression of MHC-I, and that CD8+ T cells failed to recognize HLA-B8-expressing .220 cells when they were infected by influenza virus (Ortmann B 1997). In this paper we also presented the amino acid sequence of tapasin based on the cDNA clone generated by James Copeman but we were pipped at the post by Ping Wang in Sweden, who published the sequence shortly before us (Li S 1997). This is why the gene name for tapasin is TAPBP, for TAP-binding protein. TAPBP is MHC linked but lies outside the homozygous deletion in .174 and T2. The final component of the PLC, ERp57, was simultaneously identified by Eric Hughes, a Yale M.D./Ph.D. student, Gunther Hammerling’s laboratory in Heidelberg, and Simon Powis’s laboratory in Aberdeen (Hughes EA 1998; Lindquist JA 1998; Morrice NA 1998). ERp57 is a member of the protein disulfide isomerase (PDI) family involved in glycoprotein folding in the ER, which we all discovered as a protein co-immunoprecipitated with TAP.
Back to MHC-II
Now I return to the problem posed by the mutant cell lines developed by Mellins and Pious that could not mediate MHC-II-restricted antigen processing (Mellins E 1990). They developed mutants using the same selection principle as DeMars, namely an initial selection of a heterozygous BLCL followed by additional selections using anti HLA class II-specific antibodies after further rounds of mutagenesis. This suggested that the mutated gene in the cell line that failed to mediate MHC-II processing might lie within the HLA complex, perhaps in the deletion common to .174 and T2. To test this, Janice Riberdy, a Duke Ph.D. student who made the transition to Yale with me, expressed the genes encoding the α– and β-subunits of the mouse MHC-II molecule I-Ak in T2 cells. Janice found that T2.I-Ak cells incubated with lysozyme failed to stimulate an I-Ak-restricted, lysozyme-specific mouse T cell hybridoma, while addition of the defined peptide epitope to cells resulted in normal stimulation (Riberdy JM 1992a). T1 cells expressing I-Ak processed and presented lysozyme normally, indicating that a critical antigen processing gene did indeed reside in the class II region of the MHC. One antibody used for selection by Elizabeth Mellins was a DR3-specific mAb called 16.23, which curiously lost reactivity for the antigen processing mutants even though they retained the DR3 allele. Janice made DR3-expressing T2 cells and found that they were also non-reactive with 16.23 (Riberdy JM 1992a). These experiments, together with those from the Mellins laboratory, suggested that 16.23 reactivity might be peptide dependent, and consistent with this DR3 αβ dimers purified from T2.DR3 dissociated in SDS. Instability in SDS at room temperature is a characteristic of MHC-II molecules containing no peptides or associated with low affinity peptides. Together with John Newcombe, a postdoctoral fellow, Janice purified the DR3 molecules from T2.DR3 to identify the associated peptides. In one of the most startling results of my scientific career, they all proved to be overlapping peptides derived from a short region of the invariant chain (Riberdy JM 1992b). Similar findings were reported by Alex Sette, Robert DeMars and their colleagues (Sette A 1992). When writing our manuscript I became irritated by continuously having to write ‘Class II-associated Invariant chain Peptides’ and settled on the acronym CLIP, a name that still survives.
The above findings gave rise to a model in which three MHC-II αβ dimers associate with Ii trimers in the ER and a specific sequence, the CLIP region, is embedded in the peptide binding groove. This blocks the binding of other peptides or unfolded proteins in the ER. After assembly, the MHC-II-Ii complexes traverse the Golgi and are diverted, either in the Trans Golgi Network (TGN) or at the cell surface, into the endocytic pathway and enter MIICs, where Ii is subjected to proteolysis by lysosomal proteases and CLIP remains in the binding groove. A critical molecule, missing in the Mellins and Pious mutants and .174 and T2, encoded in the MHC class II region, induces CLIP dissociation and allows peptides generated by lysosomal proteolysis to replace it. There followed a highly competitive race to identify the critical molecule. The answer, from experiments by Mellins and Pious, now working independently, was that the MHC-II homolog HLA-DM was the critical factor (Fling SP 1994; Morris P 1994). HLA-DM is a non-peptide binding MHC-II homologue consisting of α- and β-subunits encoded within the MHC, and depending on the individual mutant cell line the introduction of the DMA gene, the DMB gene, or both, restored antigen processing. Lisa Denzin, then a postdoctoral fellow in my laboratory, showed that the introduction of both genes, together encoding a complete HLA-DM molecule, was required to repair the defect in T2 cells (Denzin LK 1994). DM expression in HLA-DR3- or DR11-positive T2 cells restored normal MHC-II intracellular transport, CLIP dissociation and SDS-stable MHC-II-peptide complex formation. Lisa also showed that DM αβ dimers reside in the endocytic compartment transiently containing αβIi complexes, or MIIC. This observation suggested how HLA-DM might work. If it is in the same compartment where αβCLIP complexes are generated and where CLIP is exchanged for locally generated peptides, DM might interact with the complexes, mediate CLIP dissociation, and allow other peptides to bind. This proved to be the case, as illustrated in Fig. 1.
In our own experiments that addressed this, we affinity purified HLA-DM from human BLCL using a mAb we had raised to it. Simply mixing HLA-DR3 or H2-I-Ab αβCLIP complexes, purified from T2.DR3 or T2.I-Ab cells, with a known specific binding peptide plus HLA-DM at a mildly acidic pH resulted in CLIP dissociation and formation of SDS-stable MHC-II αβ dimers (Denzin LK 1995). This was inhibited by the addition of CerCLIP.1, a mAb specific for the N-terminus of CLIP that reacted with MHC-II-CLIP complexes, suggesting a physical interaction between the MHC-II-CLIP complexes and HLA-DM. Also consistent with this was a fascinating antigen processing-defective cell line made by Elizabeth Mellins in which mutagenesis had randomly introduced an N-linked glycan into the DR α-chain in a position on the same side of the DR molecule as the N-terminal end of the associated peptides (Mellins E 1994).Overall, the results from my laboratory, the Mellins laboratory, and Peter Jensen’s laboratory during this period were all consistent, revealing a remarkable peptide exchange mechanism caused by a physical interaction of HLA-DM with MHC-II-CLIP, illustrated in Fig. 1 (Denzin LK 1995; Sherman MA 1995; Sloan VS 1995). Furthermore, the exchange process didn’t end there. DM proved to be an editor of peptide binding: Gunter Hammerling’s group showed that, following CLIP dissociation, additional cycles of HLA-DM/MHC-II-peptide interaction displace low affinity peptides at the expense of high affinity ones, resulting in the eventual accumulation of very stable MHC-II-peptide complexes that are exported to the cell surface (Kropshofer H 1996).
One amusing aspect of our peptide exchange experiments was that before we studied DM we had already described low pH-induced in vitro displacement of CLIP from HLA-DR3-CLIP and its replacement by a DR3-binding peptide, actually in the paper that identified CLIP (Riberdy JM 1992b). Bizarrely, this proved to depend on the presence of the detergent N-octyl glucoside, used during purification of the DR3-CLIP complex. Lisa Denzin showed that even N-octyl glucoside-mediated peptide replacement was accelerated by the addition of purified HLA-DM (Denzin LK 1995). She also found that CLIP dissociation from I-Ab-CLIP complexes was not induced by N-octyl glucoside. How this particular detergent induces CLIP dissociation is not clear, although experiments by my Duke M.D./Ph.D. student Ravi Avva, also a transplant to Yale, suggested that it was a function of the short hydrocarbon chain (Avva RR 1994). Why it does not occur with IAb-CLIP is also unclear. However, the mechanism of DM-induced dissociation was revealed much later by Kai Wucherpfennig’s group (Pos W 2012). They solved the structure of a C-terminally tethered complex of DR1 and DM αβ dimers and showed that DM association stabilizes an open, normally transient, peptide-free form of the DR peptide binding groove. Conformational switching between this stabilized open form and the peptide-occupied form ends when the bound peptide is of a sufficiently high affinity that the DM-mediated conformational reversal does not occur. While in my laboratory Lisa Denzin also demonstrated that another MHC-II homologue encoded in the MHC, HLA-DO, expressed predominantly in B cells, thymic epithelium and some dendritic cells, was an inhibitor of HLA-DM function (Denzin LK 1997). This extended earlier experiments by Lars Karlsson which showed that DO formed complexes with DM that accumulated in MIICs, and that this complex formation was required for proper DO assembly and exit from the ER, as illustrated in Fig. 1 (Liljedahl M 1996). DM/DO complexes failed to induce CLIP dissociation from DR3/CLIP complexes. Updated discussions of HLA-DM and DO are presented in Lisa’s review in this issue and I will leave additional details of the DM-DO interaction and its biological significance to her.
Back to MHC-I
Our understanding of the complex processes in the ER that regulate peptide binding to MHC-I molecules is encapsulated in Fig. 3. The evolution of this understanding, again from a personal perspective, began with the discovery of components other than TAP and the MHC-I molecules that play important roles. When the PLC was initially defined the peptide transporter TAP was the only component with a known function. The precise roles of tapasin, calreticulin and ERp57, were not understood. Tapasin was required for the interaction of MHC heavy chain-β2m dimers with TAP, leading to the hypothesis that ensuring proximity was its function: peptides didn’t have far to go to find an empty MHC-I molecule after entry into the ER. Doubt was cast on this simple model by Paul Lehner, a postdoctoral fellow, who showed that tapasin lacking its transmembrane region, and therefore soluble, could restore HLA-B8 cell surface expression and antigen processing function in tapasin-negative .220 cells even though MHC-I association with TAP was not detectable (Lehner PJ 1998). Parenthetically, he also found that, in that absence of tapasin, TAP expression was reduced, a finding also made by the Hammerling group working in the mouse system (Garbi N 2003), and shown to be a function of the C-terminal region of tapasin by Naveen Bangia (Bangia N 1999). Much later, Ralf Leonhardt expanded on this when he moved to my laboratory as a postdoctoral fellow, showing that it reflects a chaperone function in TAP assembly that involves the interaction of the tapasin transmembrane domain with one of the transmembrane domains (TM9) in the core region of TAP1 essential for peptide transport (Leonhardt RM 2014). This specific interaction is lost when TAP2 binds to TAP1, but two tapasin molecules remain associated with the TAP1/TAP2 heterodimer, interacting with the N-terminal regions of the two subunits that are not required for the peptide transport function.
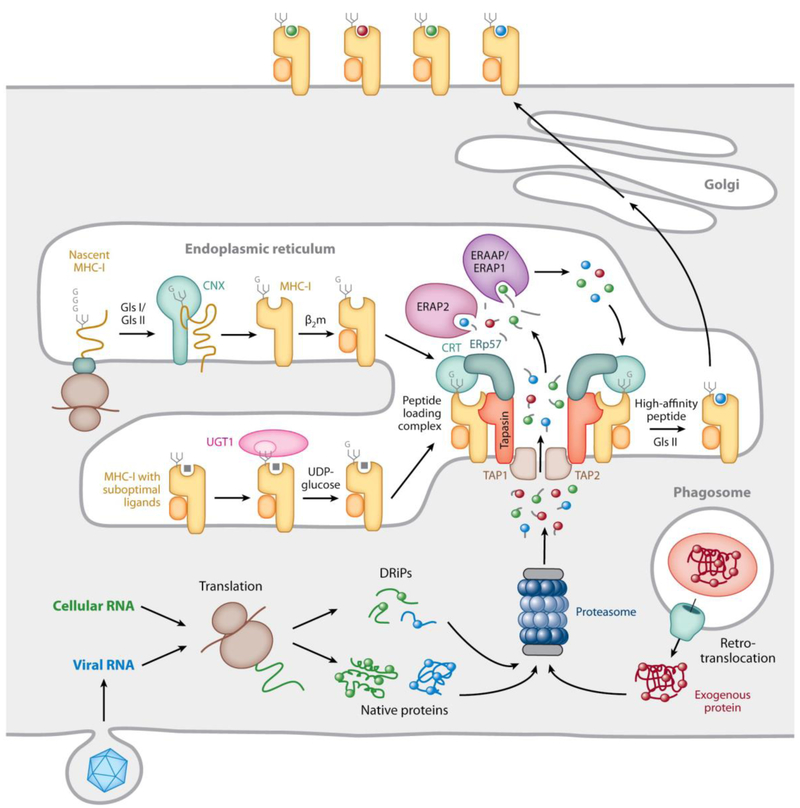
Fig. 3. Mechanisms underlying MHC-I-restricted antigen processing.
MHC-I-β2m dimers assemble in the ER with the help of the lectin chaperone calnexin (CNX in the figure) before incorporation into the peptide loading complex (PLC). This involves non-covalent interactions of the MHC-I-β2m dimer with calreticulin (CRT in the figure), via a monoglucosylated N-linked glycan at position 86 of the MHC-I heavy chain, and with tapasin, two copies of which are non-covalently associated with the TAP1/TAP2 heterodimeric transporter. The MHC-I/calreticulin/tapasin interactions are cooperatively further stabilized by calreticulin association with ERp57, which is itself disulfide-linked to tapasin (see Fig. 4). Peptides generated by proteasomal degradation from cellular or pathogen-derived cytosolic proteins, perhaps prematurely terminated or otherwise defective ribosomal products (DRiPs), are translocated via TAP into the ER, where they can be N-terminally trimmed by the ER aminopeptidases ERAP1 (ERAAP in mice) and ERAP2. Tapasin is assumed to maintain the MHC-I binding groove in an open, peptide-receptive state within the PLC. Peptides of the appropriate sequence and length can bind to the MHC-I molecules and if they are of a sufficiently high affinity the peptide-occupied MHC-I-β2m dimers may dissociate from the PLC. The terminal glucose of the N-linked glycan can be removed by the enzyme glucosidase II and if the peptide is suboptimal, i.e. unable to maintain the properly folded conformation of the MHC-I molecule, the glycan can be re-glucosylated by the enzyme UDP-glucose glycoprotein transferase-1 (UGT1), allowing the MHC-I-β2m dimers to maintain interaction with the PLC or to re-enter it. The end products of these complex quality control processes are stable, surface expressed MHC-I-peptide complexes. Illustrated in the lower left of the figure is the process thought to initiate the cytosolic pathway of cross-presentation: proteins internalized by phagocytosis are extruded into the cytosol, where they meet the same proteolytic fate as endogenously synthesized proteins. (reproduced from Blum et al., 2013).
The finding that MHC-I expression is generally lower on tapasin-negative cells and that these molecules are less stable led to the suggestion by many that tapasin is an editor of peptide binding to MHC-I in the same way that DM is an editor of peptide binding to MHC-II. Data from Victor Engelhard’s laboratory in Virginia comparing Paul Lehner’s .220.B8 cells expressing full length or soluble tapasin suggested that, rather than being an editor that promotes binding of high affinity peptides, it might be a more general facilitator of peptide binding (Zarling AL 2003). However, the idea that tapasin was the DM equivalent for MHC-I persisted and led to attempts to demonstrate peptide editing biochemically, as was done for HLA-DM and MHC-II. Pamela Wearsch, a postdoctoral fellow, took on this project, expressing soluble tapasin in insect cells and adding it to detergent extracts from tapasin-negative, HLA-B8-positive .220 cells to see if it would promote peptide binding to assembling HLA-B8 molecules. This project was initially extremely frustrating and yielded ambiguous results. However, eventually it was rescued with the help of David Peaper, a Yale M.D./Ph.D. student working in the laboratory at the same time, who showed that tapasin within the PLC was a heterodimer with ERp57 (Peaper DR 2005).
David Peaper’s work derived from initial observations by Tobias Dick, a postdoctoral fellow, who found that a fraction of the tapasin in the PLC was associated with ERp57 by a disulfide bond (Dick TP 2002). Detecting the interaction required pretreatment of cells with the sulfhydryl reactive reagent N-ethyl maleimide prior to detergent extraction, and initially we thought that redox reactions involving ERp57 might be directly involved in peptide loading. Tobias found that the linkage involved cysteine 95 of tapasin, which is not involved in an intrachain disulfide bond, and the N-terminal cysteine of the most N-terminal of the two ‘CXXC’ active site motifs present in the thioredoxin-like domains in ERp57 responsible for its reductase activity. ERp57 has a four domain structure and the active site-containing domains are at the N- and C-terminal ends of the molecule. The general thioredoxin reduction reaction involves an attack by the thiol of the first cysteine of the CXXC motif on a disulfide bond in a substrate, leading to a transient enzyme-substrate disulfide bond. This is reduced by an attack of the second cysteine thiol on the first, a reaction known as the escape pathway (Walker KW 1997). N-ethyl maleimide stabilizes the ERp57-tapasin disulfide bond by reacting with the second cysteine (Cys60) in the CXXC motif to block the escape pathway. The extension of the work by David Peaper arose from experiments using an alternative to N-ethyl maleimide called methyl methanethiosulfonate (MMTS), which, unlike N-ethyl maleimide, reacts with the thiol group of a cysteine residue to form a reversible disulfide bond. The use of MMTS began with an experiment of my own. I thought that a reversible reagent might allow a better analysis of disulfide rearrangements that we still hoped to see in the MHC-I molecules themselves, but I was startled to find that when cells were treated with MMTS virtually all of the tapasin in the PLC was disulfide linked to ERp57. David followed this up energetically, demonstrating among other things that PLCs contain tapasin-ERp57 heterodimers even when no MHCI molecules are present, that they form during the assembly of the PLC, and that their formation is independent of the presence of TAP. Perhaps the oddest aspect of the disulfide linkage is that does not survive denaturation in SDS, even in the absence of reducing agents. Denaturation activates the escape pathway, resulting in the spontaneous dissociation of tapasin from ERp57 (Peaper DR 2005).
Pamela applied these findings to her MHC-I peptide editing experiments, making soluble disulfide-linked tapasin-ERp57 heterodimers rather than free tapasin molecules by co-expressing the two components in insect cells. These proved to be more stable if Cys60 was mutated to alanine, eliminating the possible engagement of the escape pathway. Pamela found that addition of tapasin-ERp57 dimers to digitonin extracts of tapasin-negative .220.B8 cells, pulse-labeled with 35S-methionine, promoted binding of B8-specific peptides to labeled, newly synthesized, HLA-B8-β2m heterodimers (Wearsch PA 2007). In a beautiful series of experiments she showed that the ability of peptides to compete for binding, generally proportional to their affinity for B8, was enhanced by the presence of tapasin-ERp57 according to their affinity. Higher affinity peptides became better competitors while low affinity ones did not, which is essentially the definition of peptide editing. Marlene Bouvier, then at the University of Connecticut, showed that soluble tapasin would induce peptide exchange by MHC-I molecules if association was forced by the addition to each of a C-terminal leucine zipper, demonstrating that the tapasin component of the heterodimer was the actual peptide editor (Chen M 2007). In collaboration with Karin Reinisch at Yale, Pamela and Gang Dong, a postdoctoral fellow in the Reinisch laboratory, succeeded in crystallizing and solving the structure of the tapasin-ERp57 dimer, which showed that tapasin naturally suppresses the ERp57 escape pathway by non-covalent elements of the interaction. They also performed an extensive mutagenesis screen that delineated the surface on tapasin that interacts with MHC-I molecules (Dong G 2009). The structure of the tapasin-ERp57 dimer, and an illustration of the mutants that defined the interaction surface, are illustrated in Fig. 4, panels A and B.
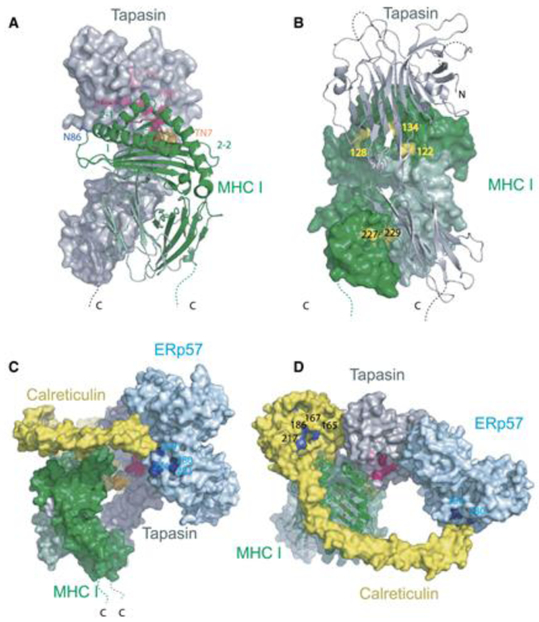
Fig. 4. Interactions within the MHC-I Peptide Loading Complex (PLC).
Panels A and B represent the structure of tapasin (grey) and its predicted interaction with an HLA class I molecule (green). In panel A tapasin is depicted in a space filling model and the multi residue mutations of tapasin that defined its interaction with MHC-I (TN3, TN4, TN6 and TN7) are indicated in pink, red and orange. The HLA class I molecule is presented as a ribbon diagram and position 86, the N-glycosylation site at the end of the α1 helix is indicated. In panel B tapasin is presented as a ribbon diagram and the HLA molecule is depicted as a space filling model. Published mutations that were defined as interfering with the MHC-I/PLC interaction are indicated. Panels C and D represent different views of an inferred model of the subcomplex of the PLC that includes MHC-I (green), tapasin (grey), calreticulin (yellow) and ERp57 (light blue). The pink, red and orange areas represent the MHC-I interaction sites on tapasin depicted in panel A. In both panels C and D the residue numbers in light blue define the site on ERp57 where the extended proline-rich P domain of calreticulin interacts based on mutagenesis. In panel D the residue numbers in dark blue define the site on calreticulin that binds to the monoglucosylated N-linked glycan on the MHC-I heavy chain. The MHC-I peptide binding groove as depicted is derived from a crystallographic structure that contains a bound peptide. Within the PLC this structure is undoubtedly modified, probably with a displaced segment of the α2 helix, as observed in the TAPBPR/MHC-I structures discussed in the text. (reproduced with permission from Dong, Wearsch et al., 2009).
The theoretical attraction of the physical association of ERp57 with tapasin is that it normally cooperates with calreticulin (or calnexin) in the folding of newly synthesized glycoproteins in the ER (ParodiAJ. 2000; WilliamsDB. 2006).One branch of the N-linked glycans on newly synthesized glycoproteins naturally terminates in three glucose residues, and the enzymatic removal of two of them provides the monoglucosylated glycan that is the ligand for calreticulin binding. Calreticulin also interacts with ERp57, which focuses its oxidoreductase activity on the associated glycoprotein and promotes the formation of disulfide bonds. Reiteration of this process constitutes a folding cycle in which the glucose residue of the glycan is removed by the ER enzyme glucosidase II and, if the glycoprotein remains improperly folded, it is recognized by the enzyme UDP-glucose glycoprotein transferase I (UGT1) and re-glucosylated, allowing it to rebind calreticulin. UGT1 has chaperone-like properties, and if the glycoprotein is properly folded it is no longer a UGT1 substrate. This folding cycle had obvious attractions for the regulation of MHC-I peptide loading, as did the potential for disulfide isomerization mediated by ERp57. However, David Peaper showed that the oxidoreductase activity of ERp57 is not required for the peptide loading function: mutagenesis of all the active site the tapasin-ERp57 linkage, had no discernable effect on cellular MHC-I assembly and peptide loading (Peaper DR 2008). This suggested that ERp57 has a structural rather than catalytic role in the PLC. Pamela Wearsch confirmed this in a cell free system and showed that recombinant HLA-B8 molecules could associate with purified calreticulin and bind to tapasin-ERp57 heterodimers, forming a stable complex (Wearsch PA 2011). The HLA-B8 molecules had to have mono-glucosylated glycans, which she achieved by making them in insect cells, stabilizing them during purification by adding a peptide of moderate affinity, and separating those that had mono-glucosylated glycans from those that did not by affinity purification with recombinant calreticulin.
The requirement for the terminal glucose on the glycan was consistent with a cooperative binding model in which MHC-I-β2m dimers in the PLC associate with tapasin by a protein-protein interaction, indicated by a number of MHC-I mutagenesis studies and confirmed by our tapasin mutagenesis studies, and calreticulin interacts noncovalently with ERp57 while simultaneously associating with MHC-I molecules via the conserved N-linked glycan at the end of the peptide binding groove. Karin Reinisch generated a plausible structural model of the sub-complex of the PLC (lacking TAP) (Fig. 4, panels B and C) based on these interactions, which is gratifyingly consistent with the structure of the entire PLC recently solved by Robert Tampe’s group by cryoelectron microscopy (Blees A 2017).
Also compatible with the Tampe structure was our analysis of the stoichiometry of the PLC by my Ph.D.student Michaela Panter (Panter MS 2012). In collaboration with Ankur Jain in Taekjip Ha’s laboratory, then at the University of Illinois, she used a single molecule approach to show that it contained two tapasin-ERp57 heterodimers per TAP molecule, consistent with the work of Michael Knittler (Rufer E. 2007). She also found that the number of MHC-I-β2m dimers varied depending on peptide supply. If peptide supply was limited by proteasome inhibition or the use of viral TAP inhibitors there were two, equaling the number of tapasin-ERp57 heterodimers, but under steady state conditions PLCs with a single MHC-I-β2m dimer were present, consistent with idea that the flux of peptides through TAP and peptide binding by the associated MHC-I-β2m dimers combine to determine their rate of dissociation.
Peptide editing within the PLC must take place in the context of the cooperative interactions described above. Indeed, the absence of calreticulin or ERp57, or disruption of various interactions by mutagenesis, impairs binding of MHC-I-β2m dimers to the PLC. This leads to their association with low-affinity peptides and reduced expression on the cell surface. For example, Najla Arshad, a postdoc in the lab, studied the effects on antigen processing of frameshift mutations in calreticulin that drive a subset of myeloid malignancies (Arshad N 2018). These mutations alter the C-terminal amino acid sequence of calreticulin and lead to ineffectual association with the PLC. Najla found that, while the mutant calreticulin can interact with MHC-I in a cell-free system, its association with ERp57 is reduced. Examination of the Tampe cryo-EM structure of the PLC suggests that the mutant C-terminal sequence disrupts an interaction between calreticulin and tapasin, underscoring the importance of precise interactions between the different PLC components for effective MHC-I peptide loading.
The probable mechanism of action of tapasin recently emerged from an unexpected quarter, when groups respectively led by Robert Tampe and David Margulies solved the structures of H2-Db and H2-Dd molecules associated with human TABPR (for TAPBP-Related) (Jiang J 2017; Thomas C 2017). TAPBPR is a tapasin homologue identified by John Trowsdale’s laboratory that is neither encoded in the MHC nor a component of the PLC (Teng MS 2002). While the biological role of TAPBR remains unclear, work by led by Louise Boyle in Cambridge showed that it interacts with MHC-I molecules via an interface virtually identical to that involved in the tapasin-MHC-I interaction (see Fig. 4) and that it catalyzes peptide exchange (Boyle LH 2013; Hermann C 2015). Similar data was obtained by the Margulies laboratory (Morozov GI 2016). The structures of the MHC-I-TAPBPR complexes suggest a mechanism of action very similar to that by which DM mediates peptide exchange by MHC-II molecules. The peptide binding groove is widened relative to the structure with a bound peptide and a segment of one of the α-helices is shifted by its interaction with TAPBPR, reducing the interactions available to a peptide. The H2-Db-TAPBPR structure also shows that a loop (dubbed the scoop loop) within TAPBPR is actually inserted into the peptide binding groove where the C-terminus of the associated peptide normally sits. The loop region is shared by tapasin but is disordered and invisible in the tapasin-ERp57 structure, suggesting that a similar interaction with the peptide binding groove may induce order within this loop when MHC-I molecules associate with the PLC. The structures are consistent with an editing mechanism in which peptide-bound MHC-I and TAPBPR-bound MHC-I have alternative conformations of the peptide binding groove, and interactions between peptide-occupied MHC-I and TAPBPR induce peptide dissociation until a peptide of sufficiently high affinity stabilizes the binding groove such that TAPBPR cannot bind.
In the case of tapasin-mediated peptide exchange the editing occurs in the context of the PLC. One presumes that within the PLC the MHC-I structure can oscillate between an open, peptide-free, form that binds tapasin and a peptide-bound form that cannot. In our biochemical experiments we used soluble components, so TAP as well as membrane integration of tapasin and MHC-I molecules are missing from the equation (Wearsch PA 2011). Although glucosidase II could not remove the glucose residue on the MHC-I glycan within the complex, presumably because calreticulin blocks access, a high affinity peptide would bind to mono-glucosylated, but not non-glucosylated, HLA-B8 molecules in the presence of calreticulin and the tapasin-ERp57 dimer. Furthermore, purified recombinant UGT-I could reglucosylate HLA-B8 molecules that were associated with a sub-optimal, low affinity, peptide but not those associated with a high affinity one. This suggests that UGT-1-mediated glucosylation can drive the re-association of MHC-I molecules with calreticulin and in turn with the tapasin-ERp57 dimer, indicating that UGT-1 provides a secondary mechanism regulating the affinity of bound peptides, illustrated in Fig. 4. Consistent with this, Wei Zhang, a Ph.D. student in the laboratory, demonstrated that mouse embryonic fibroblasts that lack UGT1 have a defect in MHC-I assembly and transport and MHC-I cell surface expression is reduced (Zhang W 2011). Still unanswered questions are whether complete MHC-I dissociation from the PLC occurs during peptide exchange and whether glucosidase II-mediated glucose removal and UGT1-mediated re-glucosylation occur within the PLC. The enzymes may act on MHC-I molecules that have dissociated from it. In the context of the complete PLC, transmembrane interactions may serve to constrain lateral diffusion of MHC-I molecules away from the complex during peptide editing. A significant fraction of the total MHC-I in the ER at steady state is not PLC-associated, but whether this is in equilibrium with that in the PLC remains unclear.
Cross-presentation
In general, the pattern that peptides derived from endogenous cytosolic proteins are associated with MHC-I and peptides from external or luminal proteins are associated with MHC-II matches the nature of the antigens they present to T cells. There are occasional exceptions, but the fundamental principle that MHC-II presents exogenous or luminal antigens while MHC-I presents antigens expressed in the cytosol has held up very well. One exception is that MHC-II can present peptides derived from cytosolic proteins that enter lysosomes via chaperone-mediated or conventional autophagy (Paludan C 2005), as indicated in Fig. 1, but the phenomenon that most upends the paradigm, and requires a novel cell biological mechanism to explain it, is antigen cross-presentation. This was originally defined as cross-priming by Michael Bevan (BevanMJ 1976). He established in bone marrow transplantation experiments that minor histocompatibility antigens, which are proteins that exhibit allelic polymorphism and are expressed by the recipient, can prime CD8+ T cell responses by the donor even though they are not expressed on the transplanted hematopoetic cells. This suggests that peptides derived from such antigens can somehow find their way onto MHC-I molecules expressed on antigen presenting cells (APCs) derived from the transplanted bone marrow. It is now well established that many exogenous protein antigens can be processed by APCs, particularly by a specific subset of dendritic cells (DCs), now referred to as the cDC1 subset, and stimulate antigen specific CD8+ T cells, and that in the case of viral infections this is generally required to prime a CD8 T cell response (Sigal LJ 1999).
Our own interest in cross-presentation stemmed less from immunological questions than from cell biological ones. How do peptides derived from exogenous antigens encounter and bind MHC-I molecules? Two fundamental mechanisms have been proposed, generally called the vacuolar and the cytosolic pathways, and evidence exists in support of both. The former holds that the proteins are degraded into peptides in the endocytic system, much as they are for the MHC-II processing pathway, and that the peptides bind to recycling MHC-I molecules. The latter holds that DCs, in particular, are capable of translocating external proteins, or large fragments of them, into the cytosol and that proteasomal degradation generates peptides that are translocated via TAP to bind MHC-I molecules. The theoretical advantage of the cytosolic pathway is that the peptides generated would be the same as those generated by normal cells. In the case of a viral infection, for example, cross-presenting DCs that are not infected could process internalized viral antigens in the cytosol and generate the same spectrum of peptides produced in the virally infected cell, meaning that the virus-specific CD8-positive T cells generated by cross-priming could see the infected cells and kill them. For the vacuolar, or lysosomal, proteases in DCs to produce the same peptides as the proteasomes in infected cells would be largely a matter of chance.
Ovalbumin (OVA), became the favorite antigen for many investigators studying cross-presentation in the mouse because a) as pointed out by Michael Bevan, it is cheap, and b) because of the availability of a robust H2-Kb-restricted OVA-specific hybridoma, B3Z, developed in Nilabh Shastri’s laboratory. The existence of the OT-1 transgenic mouse expressing a T cell receptor specific for the same epitope in its CD8-positive T cells makes It even more attractive. No similarly useful human CD8+ T cell line exists, but Anne Ackerman, a Ph.D. student in my laboratory, found that a monocytic human cell line, KG-1, which can be differentiated into a DC-like cell in culture, could cross-present OVA when transfected with H2-Kb (Ackerman AL 2003a). KG-1 became a useful tool that allowed us to do experiments without generating the large numbers of DCs that would be required for biochemical and cell biological studies. It is worth noting that, although DCs are undoubtedly the critical cells in vivo, other cells are certainly capable of cross-presentation in vitro. Alessandra Giodini, a Ph.D. student in my laboratory, showed that even HEK 293 T cells could do it when equipped with H2-Kb plus an Fc receptor to allow phagocytosis of OVA immune complexes (Giodini A 2009). The cDC1 subset, defined as CD8-positive DCs in mice, are superior largely because they have adapted their endocytic and phagocytic systems to minimize premature proteolysis of internalized antigens (Savina A 2007). Mohammed Samie, a postdoctoral fellow in my laboratory, showed that the transcription factor TFEB, which stimulates the expression of numerous lysosomal proteases as well as the vacuolar ATPase responsible for lysosomal acidification, plays a major role in regulating the ability of cells to mediate cross-presentation (Samie M 2015).
An important, and at the time contentious, finding that stimulated many investigators working on cross-presentation came from Michel Desjardins in Montreal, who found that early phagosomes contain proteins derived from the ER (Gagnon E 2002).This suggested that the phagosome itself could be the site where MHC-I molecules might bind cytosolic peptides imported by TAP. Anne Ackerman was able to show that purified phagosomes from KG-1 cells contain PLC components (TAP, tapasin, calreticulin and ERp57) and that they could import and glycosylate a radioiodinated peptide containing a glycosylation sequence, just as we had shown for SLO-permeabilized cells in our earlier experiments, indicating that the ER glycosylation machinery was present(Ackerman AL 2003b). She also showed that import of an HLAA3-binding peptide into purified phagosomes from HLA-A3-positive human DCs could induce the dissociation of HLA-A3 from PLCs within the phagosomes. Crucially, entry of peptides into the phagosome was blocked by the addition of ICP47, a Herpes simplex virus immune evasion protein that inhibits TAP function by binding to its cytosolic face. At the time a criticism of Michel Desjardin’s work was that purified phagosomes might be contaminated by small amounts of ER that could be picked up by the mass spectrometry-based proteomics analysis that he used to identify the ER components. Cautious individuals might argue, and indeed did argue, that our own data could have the same problem; small sealed ER-derived vesicles present in the phagosomal preparation could be internalizing peptides via TAP and confounding the results. Anne addressed this in two ways. First, she showed that latex beads coated with an anti tapasin antibody could bind tapasin associated with immature MHC-I molecules upon phagocytosis. Second, she used a crucial reagent provided by Robert Tampe’s laboratory; a purified, soluble version of the TAP inhibitor US6, which is encoded by human cytomegalovirus and, unlike ICP47, functions on the luminal side of the ER. When soluble US6 was added to KG-1 cells expressing Kb it blocked cross-presentation directly. Not only that, it induced downregulation of surface Kb molecules in KG-1.Kb cells and also surface HLA class I molecules in human DCs, suggesting that a major fraction of the MHC-I molecules in this cell were unstable because internalized US6 impeded their access to peptides (Ackerman AL 2003b). All of these experiments, together with similar work from Sebastian Amigorena’s group as well as Michel Desjardin’s own group, made a strong argument that DC phagosomes were potential cross-presenting organelles (Allan RS 2003; Houde M 2003).
A second consequence of the recruitment of ER components to the phagosome that is potentially relevant to cross-presentation is that a defined mechanism exists that can extrude proteins from the ER lumen into the cytosol where they are degraded by the proteasome. This is known as ER-associated degradation, or ERAD, and is used to dispose of misfolded proteins (Meusser B 2005). We and others jumped in enthusiastically to ask if ERAD components recruited to the phagosome were involved in cross-presentation. Initially we thought the answer was yes. Experiments in favor of the idea were performed again by Anne Ackerman, together with Alessandra Giodini (Ackerman AL 2006). First, they reinforced the idea that ER components were imported into phagosomes by showing that a peptide containing a glycosylation site and bound to latex beads was glycosylated upon phagocytosis by DCs. They then designed experiments to eliminate the idea that the vacuolar pathway explained cross-presentation in KG-1 cells by showing that proteins were indeed extruded into the cytosol from the phagosomes. They found that, just like US6, ICP47, the TAP inhibitor that functions at the cytosolic face of TAP, could be internalized by KG-1.Kb cells and block cross-presentation, as well as inducing downregulation of surface MHC-I. This implied that ICP47 enters the cytosol to block TAP. At the time, a candidate for an ERAD channel was Sec61, the channel used to introduce newly synthesized secreted and transmembrane proteins into the ER during synthesis on membrane associated ribosomes, essentially operating in reverse. Export of peptides from the ER via Sec61 was reported to be blocked by exotoxin A from Pseudomonas aeruginosa, which itself uses the ERAD pathway to enter the cytosol. Exotoxin A proved to be capable of blocking cross-presentation of OVA under conditions where it did not block the presentation of OVA expressed in the cytosol, an essential control because exotoxin A is a protein synthesis inhibitor.
Another requirement for ERAD that we sought to implicate in cross-presentation is that proteins destined for disposal often require chaperone-mediated unfolding and reduction of disulfide bonds in order to traverse the ER membrane into the cytosol (Brodsky JL 2009). We wondered whether this might be true for cross-presented proteins, particularly viral glycoproteins which are often tightly folded and contain multiple disulfide bonds. Reshma Singh, a postdoctoral fellow, asked whether GILT, the lysosomal thiol reductase we had shown to be important for the processing of MHC-II-restricted antigens containing disulfide bonds (see Fig.1), might also facilitate the cross-presentation of such an antigen (Singh R 2010). Reshma chose herpes simplex virus (HSV-1) glycoprotein B (gB) as an antigen because its trimeric structure was established and it contains multiple intrachain disulfide bonds, and an H2-Kb-restricted epitope had been defined. Reshma found using our GILT knockout mice that priming of CD8-positive T cells to gB upon HSV-1 infection was GILT-dependent. When infected cell debris or purified soluble gB was added to bone marrow-derived DCs derived from wildtype or GILT knockout mice, processing and recognition by a gB-specific, Kb-restricted, T cell hybridoma also proved to be GILT-dependent. Processing was both proteasome and TAP-dependent. Crucially, the GILT requirement was lost when the purified antigen was pre-reduced prior to its addition to the DCs. These findings are consistent with a requirement for reduction and unfolding of the glycoprotein prior to translocation into the cytosol. Further evidence for a role for unfolding was provided by Alessandra Giodini, looking at the requirements for the cytosolic entry of internalized luciferases in KG-1 cells (Giodini A 2008). She determined that the cytosolic chaperone Hsp90 was required to restore activity by refolding the denatured enzymes upon access to the cytosol.
A major guiding force for our early cross-presentation experiments was the work of individuals developing techniques to reconstitute ERAD in cell free systems (see, for example, (Ackerman AL 2006; Shamu CE 1999)). Tom Rapoport’s laboratory was able to take isolated microsomes (ER) containing an ERAD substrate, add cytosol and an ATP regenerating system, and observe that the substrate was translocated out of the microsomes into the added cytosol (Ye Y 2001). We decided to follow this example, taking KG-1 phagosomes containing an enzyme, adding the same cytosol and ATP regenerating system, and asking if we could observe the release of the enzyme. Chris Norbury and Colin Watts had previously demonstrated that horse radish peroxidase could be released into the cytosol of intact macrophages following micropinocytosis (Norbury CC 1995), but for us firefly luciferase proved to be a better choice. Anne and Alessandra found that phagosomes isolated from KG-1 cells that had internalized latex beads coated with luciferase could export the enzyme, and that this was cytosol and ATP dependent (Ackerman AL 2006). A compelling experiment showed that a purified cytosolic component essential for ERAD could substitute for cytosol to mediate luciferase release from the phagosome. This was the AAA ATPase p97, which hydrolyses ATP to provide the force required to extract proteins from the ERAD channel (Ye Y 2001). A dominant negative version of p97 with mutated ATPase sites traps ERAD substrates in the ER lumen. This mutant was unable to induce the release of luciferase from purified phagosomes and furthermore it blocked the release mediated by the addition of purified cytosol and ATP. Finally, expression of the dominant negative p97 in KG-1.Kb cells blocked its ability to cross-present OVA, without compromising their ability to mediate conventional processing of virally-encoded cytosolic OVA.
All of the above data, combined with work by other groups, seemed to provide a compelling case that the ERAD mechanism had been adapted to mediate cross-presentation. However, major strides were simultaneously being made in the identification of the channel (or channels) involved in ERAD. Sec61 fell out of favor, and other ER membrane proteins, including multi-pass transmembrane proteins such as Derlin-1 and the ubiquitin ligase Hrd1, came to the fore. Because our data suggested that cross-presentation used ERAD components, Jeff Grotzke, a postdoctoral fellow, devised a clever siRNA screen for ERAD components that used as a substrate a glycosylated ‘split’ version of the fluorescent protein Venus, where the glycosylated half was provided with a signal sequence while other half was expressed in the cytosol (Grotzke JE 2013). Retrotranslocation of the mis-folded ER-targeted half into the cytosol resulted in its deglycosylation by the cytosolic enzyme N-glycosidase-1 (Ngly-1) and the two halves, each equipped with leucine zippers, then combined to make a complete molecule. Deglycosylation by N-Gly-1 converts the glycan-coupled asparagine residue to an aspartic acid, and the mutant was designed so that this switch of amino acids enhanced the fluorescent signal, particularly when proteasomal degradation was blocked by the specific inhibitor, epoxomicin. The assumption behind the screen was that the loss of any component that was required to move the ER-targeted fragment into the cytosol would block the fluorescence signal.
Jeff used HEK 293T cells for the ERAD screen because Alessandra Giodini had found that these cells, when transfected with H2-Kb, were perfectly capable of mediating cross-presentation of OVA as well as allowing luciferase to enter the cytosol from phagosomes, so any component required for both ERAD and cross-presentation was likely to be present (Giodini A 2009). Unfortunately, however, the screen did not identify any novel components, although it did identify numerous known ERAD factors, including Hrd1. Jeff then asked if siRNA-mediated knock down of any of these species in mouse bone marrow-derived DCs reduced their ability to cross-present OVA and the answer was no, with the exception of p97 (summarized in (Grotzke JE 2015). We are left then, with a definite demonstration that external proteins can enter the cytosol of DCs and that p97 is important for the process, but we do not know, at the time of writing, the exact mechanism that allows them to cross the phagosomal membrane. ERAD could still be involved if more than one mechanism can mediate translocation, but this remains to be determined. Qiao Lu, a Ph.D. student, recently devised an excellent substrate to detect the translocation event, which is a glycosylated derivative of Renilla luciferase that is inactive until the glycan is removed by cytosolic N-glycosidase-1 (Lu Q 2018). DCs and HEK 293T cells that take up the enzyme by phagocytosis become luciferase-positive after the enzyme is translocated to the cytosol. This may provide the basis for a screen to identify components required for the dislocation event. However, the problem has proved remarkably difficult to solve, as indicated by the limited information on the process incorporated into Fig. 3, and new approaches and ideas are likely to be needed.
Summary
Writing this manuscript served as a reminder of how far our understanding of the mechanisms of antigen processing has come during my career. My delight in the obvious progress is somewhat tempered by the sobering realization that this period covers almost half a century. Nevertheless, because of the efforts of many investigators the surface molecules causing allograft rejection have been identified as the products of the massively polymorphic MHC-I and MHC-II genes and the molecular and cell biological processes used by these molecules to bind peptides and facilitate the adaptive immune response via T cell stimulation are exceptionally well defined. The level of understanding is such that clinical outcomes associated with these principles beyond transplantation are likely, including therapeutic interventions for cancer and autoimmunity. However, there is still much to learn and I anticipate being involved in the learning process for a considerable time to come.
Acknowledgements
As stated at the outset, all progress would have been impossible without the contributions of the many students and postdoctoral fellows who have passed through my laboratory. Their hard work, creativity, dedication and love of science made it all possible. My own mentors, the late Arnold Sanderson, Jack Strominger and, in particular, my role model at Duke, the late Bernard Amos, were all critical in their different ways to the development of my career. I also would like to express my appreciation to the dedicated technicians who made things work over the years and to the administrative staff who compensated for my organizational inadequacies. And of course, without twenty-five years of funding from the Howard Hughes Medical Institute and continuous funding since 1974 from the National Institute of Allergy and Infectious Diseases, my life would have taken quite a different turn. Finally, if this work seems far removed from an exercise in modesty, I would like all blame to be directed at Martin Flajnik for generously extending the invitation.
REFERENCES
- Ackerman AL, Cresswell P (2003a) Regulation of MHC class I transport in human dendritic cells and the dendritic-like cell line KG-1. J. Immunol 170:4178–4188 [DOI] [PubMed] [Google Scholar]
- Ackerman AL, Giodini A, Cresswell P. (2006) A role for the endoplasmic reticulum protein retrotranslocation machinery during crosspresentation by dendritic cells. Immunity:607–617 [DOI] [PubMed] [Google Scholar]
- Ackerman AL, Kyritsis C, Tampé R, Cresswell P. (2003b) Early phagosomes in dendritic cells form a cellular compartment sufficient for cross presentation of exogenous antigens. Proc Natl Acad Sci U S A. 100:12889–12894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander J, Payne AJ, Murray R, Frelinger JA, Cresswell P. (1989) Differential transport requirements of HLA and H-2 class I glycoproteins. Immunogenetics 29:380–388 [DOI] [PubMed] [Google Scholar]
- Allan RS, Zueva E, Cammas F, Schreiber HA, Masson V, Belz GT, Roche D, Maison C, Quivy JP, Almouzni G, Amigorena S. (2003) ER-phagosome fusion defines an MHC class I cross-presentation compartment in dendritic cells. Nature 425:397–412 [DOI] [PubMed] [Google Scholar]
- Anderson KS, Alexander J, Wei M, Cresswell P. (1993) Intracellular transport of class I MHC molecules in antigen processing mutant cell lines. J. Immunol 151:3407–3419 [PubMed] [Google Scholar]
- Anderson KS, Cresswell P. (1994) A role for calnexin (IP90) in the assembly of class II MHC molecules. EMBO J. 13:675–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Androlewicz MJ, Anderson KS, Cresswell P. (1993) Evidence that transporters associated with antigen processing translocate a major histocompatibility complex class I-binding peptide into the endoplasmic reticulum in an ATP-dependent manner. Proc Natl Acad Sci U S A. 90:9130–9134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Androlewicz MJ, Cresswell P. (1994a) Human transporters associated with antigen processing possess a promiscuous peptide-binding site. Immunity 1:7–17 [DOI] [PubMed] [Google Scholar]
- Androlewicz MJ, Ortmann B, van Endert PM, Spies T, Cresswell P. (1994b) Characteristics of peptide and major histocompatibility complex class I/beta 2-microglobulin binding to the transporters associated with antigen processing (TAP1 and TAP2). Proc Natl Acad Sci U S A. 91:12716–12720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold D, Driscoll J, Androlewicz M, Hughes E, Cresswell P, Spies T. (1992) Proteasome subunits encoded in the MHC are not generally required for the processing of peptides bound by MHC class I molecules. Nature 360:171–174 [DOI] [PubMed] [Google Scholar]
- Arshad N, Cresswell P. (2018) Tumor-associated calreticulin variants functionally compromise the peptide loading complex and impair its recruitment of MHC-I. J Biol Chem. 293:9555–9569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arunachalam B, Phan U, Geuze HJ, Cresswell P. (2000) Enzymatic reduction of disulfide bonds in lysosomes: characterization of a gamma-interferon-inducible lysosomal thiol reductase (GILT). Proc Natl Acad Sci U S A. 97:745–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avva RR, Cresswell P. (1994) In vivo and in vitro formation and dissociation of HLA-DR complexes with invariant chain-derived peptides. Immunity 1:763–774 [DOI] [PubMed] [Google Scholar]
- Babbitt BP, Allen PM, Matsueda G, Haber E, Unanue ER. (1985) Binding of immunogenic peptides to Ia histocompatibility molecules. Nature 317:359–361 [DOI] [PubMed] [Google Scholar]
- Bangia N, Lehner P, Hughes EA, Surman M, Cresswell P. (1999) The N-terminal region of tapasin is required to stabilize the MHC class I loading complex. Eur. J. Immunol 29:1858–1870 [DOI] [PubMed] [Google Scholar]
- Bevan MJ. (1976) Cross-priming for a secondary cytotoxic response to minor H antigens with H-2 congenic cells which do not cross-react in the cytotoxic assay. J Exp Med. 143:1283–1288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjorkman PJ, Saper M, Samraoui B, Bennett WS, Strominger JL, Wiley DC. (1987) Structure of the human class I histocompatibility antigen, HLA-A2. Nature 329:506–512 [DOI] [PubMed] [Google Scholar]
- Blees A, Januliene D, Hofmann T, Koller N, Schmidt C, Trowitzsch S, Moeller A, Tampé R. (2017) Structure of the human MHC-I peptide-loading complex. Nature 551:525–528 [DOI] [PubMed] [Google Scholar]
- Blum JS, Cresswell P. (1988) Role for intracellular proteases in the processing and transport of class II HLA antigens. Proc Natl Acad Sci U S A. 85:3978–3979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle LH, Hermann C, Boname JM, Porter KM, Patel PA, Burr ML, Duncan LM, Harbour ME, Rhodes DA, Skjødt K, Lehner PJ, Trowsdale J. (2013) Tapasin-related protein TAPBPR is an additional component of the MHC class I presentation pathway. Proc Natl Acad Sci U S A. 110:3465–3470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky JL, Wojcikiewicz R. (2009) Substrate-specific mediators of ER associated degradation (ERAD). Curr Opin Cell Biol. 21:516–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerundolo V, Alexander J, Anderson K, Lamb C, Cresswell P, McMichael A, Gotch F, Townsend A. (1990) Presentation of viral antigen controlled by a gene in the major histocompatibility complex. Nature 345:449–452 [DOI] [PubMed] [Google Scholar]
- Chen M, Bouvier M. (2007) Analysis of interactions in a tapasin/class I complex provides a mechanism for peptide selection. EMBO J. 26:1681–1690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cresswell P (1985) Intracellular class II HLA antigens are accessible to transferrin-neuraminidase conjugates internalized by receptor-mediated endocytosis. Proc Natl Acad Sci U S A. 82:8188–8192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cresswell P, Geier SG. (1975) Antisera to human B-lymphocyte membrane glycoproteins block stimulation in mixed lymphocyte culture. Nature 257:147–9 [DOI] [PubMed] [Google Scholar]
- Cresswell P, Robb R, Turner MJ, Strominger JL. (1974a) Papain-solubilized HL-A antigens. Chromatographic and electrophoretic studies of the two subunits from different specificities. J Biol Chem 249:2828–2832 [PubMed] [Google Scholar]
- Cresswell P, Springer T, Strominger JL, Turner MJ, Grey HM, Kubo RT. (1974b) Immunological identity of the small subunit of HL-A antigens and beta2-microglobulin and its turnover on the cell membrane. Proc Natl Acad Sci U S A 71:2123–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cresswell P, Turner MJ, Strominger JL. (1973) Papain-solubilized HL-A antigens from cultured human lymphocytes contain two peptide fragments. Proc Natl Acad Sci U S A 70:1603–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson HW, Reid PA, Lanzavecchia A, Watts C. (1991) Processed antigen binds to newly synthesized MHC class II molecules in antigen-specific B lymphocytes. Cell 67:105–16 [DOI] [PubMed] [Google Scholar]
- DeMars R, Chang CC, Shaw S, Reitnauer PJ, Sondel PM. (1984) Homozygous deletions that simultaneously eliminate expressions of class I and class II antigens of EBV-transformed B-lymphoblastoid cells. I. Reduced proliferative responses of autologous and allogeneic T cells to mutant cells that have decreased expression of class II antigens. Human Immunology 11:77–97 [DOI] [PubMed] [Google Scholar]
- Denzin LK, Cresswell P. (1995) HLA-DM induces CLIP dissociation from MHC class II alpha beta dimers and facilitates peptide loading. Cell 82:1550165. [DOI] [PubMed] [Google Scholar]
- Denzin LK, Robbins N, Carboy-Newcomb C, Cresswell P. (1994) Assembly and intracellular transport of HLA-DM and correction of the class II antigen-processing defect in T2 cells. Immunity 1:595–606 [DOI] [PubMed] [Google Scholar]
- Denzin LK, Sant’Angelo DB, Hammond C, Surman MJ, Cresswell P (1997) Negative regulation by HLA-DO of MHC class II-restricted antigen processing. Science 278:106–109 [DOI] [PubMed] [Google Scholar]
- Deverson EV, Gow IR, Coadwell WJ, Monaco JJ, Butcher GW, Howard JC. (1990) MHC class II region encoding proteins related to the multidrug resistance family of transmembrane transporters. Nature 348:738–741 [DOI] [PubMed] [Google Scholar]
- Dick TP, Bangia N, Peaper DR, Cresswell P. (2002) Disulfide bond isomerization and the assembly of MHC class I-peptide complexes. Immunity 16:87–98 [DOI] [PubMed] [Google Scholar]
- Dong G Wearsch PA, Peaper DR, Cresswell P, Reinisch KM (2009) Insights into MHC class I peptide loading from the structure of the tapasin-ERp57 thiol oxidoreductase heterodimer. Immunity 30:21–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fling SP, Arp B, Pious D. (1994) HLA-DMA and -DMB genes are both required for MHC class II/peptide complex formation in antigen-presenting cells. Nature 368:554–558 [DOI] [PubMed] [Google Scholar]
- Gagnon E, Duclos S, Rondeau C, Chevet E, Cameron PH, Steele-Mortimer O, Paiement J, Bergeron JJ, Desjardins M. (2002) Endoplasmic reticulum-mediated phagocytosis is a mechanism of entry into macrophages. Cell 110:119–131 [DOI] [PubMed] [Google Scholar]
- Garbi N, Tiwari N, Momburg F, Hämmerling GJ. (2003) A major role for tapasin as a stabilizer of the TAP peptide transporter and consequences for MHC class I expression. Eur. J. Immunol 33:264–273 [DOI] [PubMed] [Google Scholar]
- Giodini A, Cresswell P. (2008) Hsp90-mediated cytosolic refolding of exogenous proteins internalized by dendritic cells. EMBO J. 27:201–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giodini A, Rahner C, Cresswell P. (2009) Receptor-mediated phagocytosis elicits cross-presentation in nonprofessional antigen-presenting cells. Proc Natl Acad Sci U S A. 106:3324–3329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandea AG, Androlewicz MJ, Athwal RS, Geraghty DE, Spies T. (1995) Dependence of peptide binding by MHC class I molecules on their interaction with TAP. Science 270:105–108 [DOI] [PubMed] [Google Scholar]
- Greenwood R, Shimuzu Y, Sekhon GS, DeMars R. (1994) Novel allele-specific, post-translational reduction in HLA class I surface expression in a mutant human B cell line. J. Immunol 153:5525–5536 [PubMed] [Google Scholar]
- Grey HM, Kubo R, Colon SM, Poulik MD, Cresswell P, Springer T, Turner M, Strominger JL. (1973) The small subunit of HL-A antigens is beta 2-microglobulin. J Exp Med. 138:1608–1612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grotzke JE, Cresswell P. (2015) Are ERAD components involved in cross-presentation? Mol Immunol. 68:112–115 [DOI] [PubMed] [Google Scholar]
- Grotzke JE, Lu Q, Cresswell P. (2013) Deglycosylation-dependent fluorescent proteins provide unique tools for the study of ER-associated degradation. Proc Natl Acad Sci U S A. 110:3393–3398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson RA, Michel H, Sakaguchi K, Shabanowitz J, Appella E, Hunt DF, Engelhard VH. (1992) HLA-A2.1-associated peptides from a mutant cell line: a second pathway of antigen presentation. Science 255:1264–1266 [DOI] [PubMed] [Google Scholar]
- Hermann C, van Hateren A, Trautwein N, Neerincx A, Duriez PJ, Stevanović S, Trowsdale J, Deane JE, Elliott T, Boyle LH (2015) TAPBPR alters MHC class I peptide presentation by functioning as a peptide exchange catalyst. Elife DOI: 10.7554/eLife.09617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosken NA, Bevan MJ. (1990) Defective presentation of endogenous antigen by a cell line expressing class I molecules. Science 248:367–370 [DOI] [PubMed] [Google Scholar]
- Houde M, Bertholet S, Gagnon E, Brunet S, Goyette G, Laplante A, Princiotta MF, Thibault P, Sacks D, Desjardins M. (2003) Phagosomes are competent organelles for antigen cross-presentation. Nature 425:402–406 [DOI] [PubMed] [Google Scholar]
- Howell DN, Cresswell P. (1983) Expression of T-lymphoblast-encoded HLA-DR antigens on human T-B lymphoblast hybrids. Immunogenetics 17:411–425 [DOI] [PubMed] [Google Scholar]
- Hughes EA, Cresswell P. (1998) The thiol oxidoreductase ERp57 is a component of the MHC class I peptide-loading complex. Current Biology 8:709–712 [DOI] [PubMed] [Google Scholar]
- Humphreys RE, McCune J, Chess L, Herrman HC, Malenka DJ, Mann DL, Parham P, Schlossman SF, Strominger JL. (1976) Isolation and immunologic characterization of a human. B-lymphocyte-specific, cell surface antigen. J Exp Med 144:98–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J, Natarajan K, Boyd LF, Morozov GI, Mage MG, Margulies DH. (2017) Crystal structure of a TAPBPR-MHC I complex reveals the mechanism of peptide editing in antigen presentation. Science 358:1064–1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones PP, Murphy D, Hewgill D, McDevitt HO. (1979) Detection of a common polypeptide chain in I--A and I--E sub-region immunoprecipitates. Mol Immunol. 16:51–60 [DOI] [PubMed] [Google Scholar]
- Kelly A, Powis S, Kerr LA, Mockridge I, Elliott T, Bastin J, Uchanska-Ziegler B, Ziegler A, Trowsdale J, Townsend A. (1992) Assembly and function of the two ABC transporter proteins encoded in the human major histocompatibility complex. Nature 355:641–644 [DOI] [PubMed] [Google Scholar]
- Kropshofer H, Vogt A, Moldenhauer G, Hammer J, Blum JS, Hämmerling GJ. (1996) Editing of the HLA-DR-peptide repertoire by HLA-DM. EMBO J. 15:6144–6154 [PMC free article] [PubMed] [Google Scholar]
- Lamb CA, Cresswell P. (1992) Assembly and transport properties of invariant chain trimers and HLA-DR-invariant chain complexes. J Immunol. 148:3478–3482 [PubMed] [Google Scholar]
- Lamb CA, Yewdell J, Bennink JR, Cresswell P (1991) Invariant chain targets HLA class II molecules to acidic endosomes containing internalized influenza virus. Proc Natl Acad Sci U S A. 88:5998–6002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehner PJ, Surman M, Cresswell P. (1998) Soluble tapasin restores MHC class I expression and function in the tapasin-negative cell line .220. Immunity 8:221–231 [DOI] [PubMed] [Google Scholar]
- Leonhardt RM, Abrahim P, Mitchell SM, Cresswell P. (2014) Three tapasin docking sites in TAP cooperate to facilitate transporter stabilization and heterodimerization. J. Immunol 192:2480–2494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Sjogrenn H-O, Hellman U, Pettersson RF, and Wang P. (1997) Cloning and functional characterization of a subunit of the transporter associated with antigen processing. Proc Natl Acad Sci U S A 94:8708–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liljedahl M, Kuwana T, Fung-Leung WP, Jackson MR, Peterson PA, Karlsson L. (1996) HLA-DO is a lysosomal resident which requires association with HLADM for efficient intracellular transport. EMBO J. 15:4817–4824 [PMC free article] [PubMed] [Google Scholar]
- Lindquist JA, Jensen ON, Mann M, Hämmerling GJ. (1998) ER-60, a chaperone with thiol-dependent reductase activity involved in MHC class I assembly. EMBO J. 17:2186–2195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotteau V, Teyton L, Peleraux A, Nilsson T, Karlsson L, Schmid SL, Quaranta V, Peterson PA. (1990) Intracellular transport of class II MHC molecules directed by invariant chain. Nature 1990:600–605 [DOI] [PubMed] [Google Scholar]
- Lu Q, Grotzke J, Cresswell P. (2018) A novel probe to assess cytosolic entry of exogenous proteins. Nature Communications 9:3104. doi: 10.1038/s41467-018-05556-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machamer CE, Cresswell P. (1982) Biosynthesis and glycosylation of the invariant chain associated with HLA-DR antigens. J Immunol. 129:2564–2569 [PubMed] [Google Scholar]
- Maric M, Arunachalam B, Phan UT, Dong C, Garrett WS, Cannon KS, Alfonso C, Karlsson L, Flavell RA, Cresswell P. (2001) Defective antigen processing in GILT-free mice. Science 294:1361–1365 [DOI] [PubMed] [Google Scholar]
- Markert ML, Cresswell P. (1980) Polymorphism of human B-cell alloantigens: evidence for three loci within the HLA system. Proc Natl Acad Sci U S A. 77:6101–6104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks MS, Blum JS, Cresswell P. (1990) Invariant chain trimers are sequestered in the rough endoplasmic reticulum in the absence of association with HLA class II antigens. J Cell Biol. 111:839–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellins E, Cameron P, Amaya M, Goodman S, Pious D, Smith L, Arp B. (1994) A mutant human histocompatibility leukocyte antigen DR molecule associated with invariant chain peptides. J. Exp. Med 179:541–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellins E, Smith L, Arp B, Cotner T, Celis E, Pious D. (1990) Defective processing and presentation of exogenous antigens in mutants with normal HLA class II genes. Nature 343:71–74 [DOI] [PubMed] [Google Scholar]
- Meusser B, Hirsch C, Jarosch E, Sommer T. (2005) ERAD: the long road to destruction. Nature Cell Biol. 8:766–772 [DOI] [PubMed] [Google Scholar]
- Monaco JJ, Cho S, Attaya M. (1990) Transport protein genes in the murine MHC: possible implications for antigen processing. Science 250:1723–1726 [DOI] [PubMed] [Google Scholar]
- Morozov GI, Zhao H, Mage MG, Boyd LF, Jiang J, Dolan MA, Venna R, Norcross MA, McMurtrey CP, Hildebrand W, Schuck P, Natarajan K, Margulies DH. (2016) Interaction of TAPBPR, a tapasin homolog, with MHC-I molecules promotes peptide editing. Proc Natl Acad Sci U S A. 2016:1006–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrice NA, Powis SJ. (1998) A role for the thiol-dependent reductase ERp57 in the assembly of MHC class I molecules. Current Biology 8:713–716 [DOI] [PubMed] [Google Scholar]
- Morris P, Shaman J, Attaya M, Amaya M, Goodman S, Bergman C, Monaco JJ, Mellins E. (1994) An essential role for HLA-DM in antigen presentation by class II major histocompatibility molecules. Nature 368:551–554 [DOI] [PubMed] [Google Scholar]
- Neefjes JJ, Momburg F, Hämmerling GJ. (1993) Selective and ATP-dependent translocation of peptides by the MHC-encoded transporter. Science 261:769–771 [DOI] [PubMed] [Google Scholar]
- Neefjes JJ, Stollorz V, Peters PJ, Geuze HJ, Ploegh HL. (1990) The biosynthetic pathway of MHC class II but not class I molecules intersects the endocytic route. Cell 61:171–183 [DOI] [PubMed] [Google Scholar]
- Nijenhuis M, Hammerling G. (1996) Multiple regions of the transporter associated with antigen processing (TAP) contribute to its peptide binding site. J Immunol. 157:5467–5477 [PubMed] [Google Scholar]
- Norbury CC, Hewlett L, Prescott AR, Shastri N, Watts C. (1995) Class I MHC presentation of exogenous soluble antigen via macropinocytosis in bone marrow macrophages. Immunity 3:783–791 [DOI] [PubMed] [Google Scholar]
- Orr HT, Bach F, Ploegh HL, Strominger JL, Kavathas P, DeMars R. (1982) Use of HLA loss mutants to analyse the structure of the human major histocompatibility complex. Nature 296:454–456 [DOI] [PubMed] [Google Scholar]
- Ortmann B, Androlewicz MJ, Cresswell P. (1994) MHC class I/beta 2-microglobulin complexes associate with TAP transporters before peptide binding. Nature 368:864–867 [DOI] [PubMed] [Google Scholar]
- Ortmann B, Copeman J, Lehner PJ, Sadasivan B, Herberg JA, Grandea AG, Riddell SR, Tampé R, Spies T, Trowsdale J, Cresswell P. (1997) A critical role for tapasin in the assembly and function of multimeric MHC class I-TAP complexes. Science 277:1306–1309 [DOI] [PubMed] [Google Scholar]
- Paludan C, Schmid D, Landthaler M, Vockerodt M, Kube D, Tuschl T, Münz C. (2005) Endogenous MHC class II processing of a viral nuclear antigen after autophagy. Science 307:593–596 [DOI] [PubMed] [Google Scholar]
- Panter MS, Jain A, Leonhardt RM, Ha T, Cresswell P (2012) Dynamics of major histocompatibility complex class I association with the human peptide-loading complex. J Biol Chem. 2012:31172–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parodi AJ. (2000) Protein glucosylation and its role in protein folding. Ann. Revi. Biochem 69:69–93. [DOI] [PubMed] [Google Scholar]
- Peaper DR, Cresswell P. (2008) The redox activity of ERp57 is not essential for its functions in MHC class I peptide loading. Proc Natl Acad Sci U S A. 105:10477–10482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peaper DR, Wearsch PA, Cresswell P. (2005) Tapasin and ERp57 form a stable disulfide-linked dimer within the MHC class I peptide-loading complex. EMBO J. 24:3613–3623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pos W, Sethi D, Call MJ, Schulze MS, Anders AK, Pyrdol J, Wucherpfennig KW. (2012) Crystal structure of the HLA-DM-HLA-DR1 complex defines mechanisms for rapid peptide selection. Cell 151:1557–1568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powis SJ Townsend AR, Deverson EV, Bastin J, Butcher GW, Howard JC. (1991) Restoration of antigen presentation to the mutant cell line RMA-S by an MHC-linked transporter. Nature 354:528–531 [DOI] [PubMed] [Google Scholar]
- Riberdy JM, Cresswell P. (1992a) The antigen-processing mutant T2 suggests a role for MHC-linked genes in class II antigen presentation. J. Immunol 148:2586–2590 [PubMed] [Google Scholar]
- Riberdy JM, Newcombe JR, Surman MJ, Barbosa JA, Cresswell P. (1992b) HLA-DR molecules from an antigen-processing mutant cell line are associated with invariant chain peptides. Nature 360:474–477 [DOI] [PubMed] [Google Scholar]
- Roche PA, Cresswell P. (1990) Invariant chain association with HLA-DR molecules inhibits immunogenic peptide binding. Nature 345:615–618 [DOI] [PubMed] [Google Scholar]
- Roche PA, Marks MS, Cresswell P. (1991) Formation of a nine-subunit complex by HLA class II glycoproteins and the invariant chain. Nature 354:392–394 [DOI] [PubMed] [Google Scholar]
- Rock KL, Gramm C, Rothstein L, Clark K, Stein R, Dick L, Hwang D, Goldberg AL (1994) nhibitors of the proteasome block the degradation of most cell proteins and the generation of peptides presented on MHC class I molecules. Cell 78:761–767 [DOI] [PubMed] [Google Scholar]
- Rufer E, Leonhardt RM, Knittler MR (2007) Molecular architecture of the TAP-associated MHC class I peptide-loading complex. J. Immunol 179:5717–5727 [DOI] [PubMed] [Google Scholar]
- Sadasivan B, Lehner PJ, Ortmann B, Spies T, Cresswell P. (1996) Roles for calreticulin and a novel glycoprotein, tapasin, in the interaction of MHC class I molecules with TAP. Immunity 5:103–104 [DOI] [PubMed] [Google Scholar]
- Salter RD, Cresswell P. (1986) Impaired assembly and transport of HLA-A and -B antigens in a mutant TxB cell hybrid. EMBO J. 5:943–949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salter RD, Howell DN, Cresswell P. (1985) Genes regulating HLA class I antigen expression in T-B lymphoblast hybrids. Immunogenetics 21:235–246 [DOI] [PubMed] [Google Scholar]
- Samie M, Cresswell P. (2015) The transcription factor TFEB acts as a molecular switch that regulates exogenous antigen-presentation pathways. Nat Immunol 16:729–736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderson AR. (1968) HL-A substances from human spleens. Nature 220:192–195 [DOI] [PubMed] [Google Scholar]
- Savina A, Amigorena S. (2007) Phagocytosis and antigen presentation in dendritic cells. Immunol Rev 219:143–156 [DOI] [PubMed] [Google Scholar]
- Sette A, Ceman S, Kubo RT, Sakaguchi K, Appella E, Hunt DF, Davis TA, Michel H, Shabanowitz J, Rudersdorf R, et al. (1992) Invariant chain peptides in most HLA-DR molecules of an antigen-processing mutant. Science 258:1801–1804 [DOI] [PubMed] [Google Scholar]
- Singh R, Cresswell P. (2010) Defective cross-presentation of viral antigens in GILT-free mice. Science 328:1394–1398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamu CE, Story C, Rapoport TA, Ploegh HL. (1999) The pathway of US11-dependent degradation of MHC class I heavy chains involves a ubiquitin-conjugated intermediate. J Cell Biol. 147:45–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman MA, Weber DA, Jensen PE. (1995) DM enhances peptide binding to class II MHC by release of invariant chain-derived peptide. Immunity 3:197–205 [DOI] [PubMed] [Google Scholar]
- Shimada A, Nathenson SG (1967) Solubilization of membrane H-2 isoantigens: chromatographic separation of specificities determined by a single H-2 genotype. Biochem Biophys Res Commun 29:828–33 [DOI] [PubMed] [Google Scholar]
- Sigal LJ, Crotty S, Andino R, Rock KL (1999) Cytotoxic T-cell immunity to virus-infected non-haematopoietic cells requires presentation of exogenous antigen. Nature 398:77–80 [DOI] [PubMed] [Google Scholar]
- Sloan VS, Cameron P, Porter G, Gammon M, Amaya M, Mellins E, Zaller DM. (1995) Mediation by HLA-DM of dissociation of peptides from HLA-DR. Nature 375:802–806 [DOI] [PubMed] [Google Scholar]
- Spies T, Bresnahan M, Bahram S, Arnold D, Blanck G, Mellins E, Pious D, DeMars R. (1990) A gene in the human major histocompatibility complex class II region controlling the class I antigen presentation pathway. Nature 348:744–747 [DOI] [PubMed] [Google Scholar]
- Spies T, Cerundolo V, Colonna M, Cresswell P, Townsend A, DeMars R (1992) Presentation of viral antigen by MHC class I molecules is dependent on a putative peptide transporter heterodimer. Nature 355:644–666 [DOI] [PubMed] [Google Scholar]
- Springer TA, Mann DL, DeFranco AL, Strominger JL. (1977) Detergent solubilization, purification, and separation of specificities of HLA antigens from a cultured human lymphoblastoid line, RPMI 4265. J Biol Chem. 252:4682–4693 [PubMed] [Google Scholar]
- Strubin M, Berte C, Mach B. (1986) Alternative splicing and alternative initiation of translation explain the four forms of the Ia antigen-associated invariant chain. EMBO J. 5:3483–3488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh WK, Cohen-Doyle M, Fruh K, Wang K, Peterson PA, Williams DB. (1994) Interaction of MHC class I molecules with the transporter associated with antigen processing. Science 264:1322–1326 [DOI] [PubMed] [Google Scholar]
- Teng MS, Stephens R, Du Pasquier L, Freeman T, Lindquist JA, Trowsdale J. (2002) A human TAPBP (TAPASIN)-related gene, TAPBP-R. Eur J Immunol. 32:1059–1068 [DOI] [PubMed] [Google Scholar]
- Thomas C, Tampe R. (2017) Structure of the TAPBPR-MHC I complex defines the mechanism of peptide loading and editing. Science 358:1060–1064 [DOI] [PubMed] [Google Scholar]
- Townsend AR, Ohlen C, Bastin J, Ljunggren HG, Foster L, Kärre K. (1989) Association of class I major histocompatibility heavy and light chains induced by viral peptides. Nature 340:443–448 [DOI] [PubMed] [Google Scholar]
- Townsend AR, Rothbard J, Gotch FM, Bahadur G, Wraith D, McMichael AJ. (1986) The epitopes of influenza nucleoprotein recognized by cytotoxic T lymphocytes can be defined with short synthetic peptides. Cell 44:959–968 [DOI] [PubMed] [Google Scholar]
- Trowsdale J, Hanson I, Mockridge I, Beck S, Townsend A, Kelly A (1990) Sequences encoded in the class II region of the MHC related to the ‘ABC’ superfamily of transporters. Nature 348:741–744 [DOI] [PubMed] [Google Scholar]
- Walker KW, Gilbert H. (1997) Scanning and escape during protein-disulfide isomerase-assisted protein folding. J Biol Chem. 272:8845–8848 [DOI] [PubMed] [Google Scholar]
- Wearsch PA, Cresswell P. (2007) Selective loading of high-affinity peptides onto major histocompatibility complex class I molecules by the tapasin-ERp57 heterodimer. Nature Immunology 8:873–881 [DOI] [PubMed] [Google Scholar]
- Wearsch PA, Peaper DR, Cresswell P. (2011) Essential glycan-dependent interactions optimize MHC class I peptide loading. Proc Natl Acad Sci U S A. 108:4950–4955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei ML, Cresswell P. (1992) HLA-A2 molecules in an antigen-processing mutant cell contain signal sequence-derived peptides. Nature 356:443–446 [DOI] [PubMed] [Google Scholar]
- Williams DB. (2006) Beyond lectins: the calnexin/calreticulin chaperone system of the endoplasmic reticulum. J Cell Sci. 119:615–623 [DOI] [PubMed] [Google Scholar]
- Ye Y, Meyer HH, Rapoport TA (2001) The AAA ATPase Cdc48/p97 and its partners transport proteins from the ER into the cytosol. Nature 414:652–656 [DOI] [PubMed] [Google Scholar]
- Zarling AL, Luckey CJ, Marto JA, White FM, Brame CJ, Evans AM, Lehner PJ, Cresswell P, Shabanowitz J, Hunt DF, Engelhard VH. (2003) Tapasin is a facilitator, not an editor, of class I MHC peptide binding. J. Immunol 171:5287–5295 [DOI] [PubMed] [Google Scholar]
- Zhang W, Wearsch PA, Zhu Y, Leonhardt RM, Cresswell P. (2011) A role for UDP-glucose glycoprotein glucosyltransferase in expression and quality control of MHC class I molecules. Proc Natl Acad Sci U S A 108:4956–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinkernagel RM, Doherty P (1974) Restriction of in vitro T cell-mediated cytotoxicity in lymphocytic choriomeningitis within a syngeneic or semiallogeneic system. Nature 248:701–702 [DOI] [PubMed] [Google Scholar]