Abstract
Inflammation plays a significant role in the pathophysiology of depression. However, not all individuals exposed to inflammatory challenge develop depression, and identifying those at risk is necessary to develop targeted monitoring, prevention, and treatment strategies. Within a randomized double-blind placebo-controlled study (n = 115), we examined whether leukocyte transcriptome profiles predicted inflammation-induced depressed mood in volunteers who received low-dose intravenous endotoxin (n = 58; aged 18–50). At baseline, transcription factor (TF) activities were assessed using genome-wide transcriptional profiling of peripheral blood mononuclear cells and promoter-based bioinformatic analyses. Then, participants were administered endotoxin. Self-reported depressed mood was assessed using the Profile of Mood States. Based on extant studies linking transcriptional profiles to depressive disorder, we examined whether post-endotoxin depressed mood is predicted by baseline activity of TFs related to immune activation, sympathetic activation, and glucocorticoid insensitivity: respectively, nuclear factor kappa B (NF-kB), cAMP response element-binding protein (CREB), and glucocorticoid receptor (GR). Twenty-one participants (36%) experienced an increase in depressed mood from baseline to 2 h post endotoxin, when depressive response peaks. Bioinformatics analyses controlling for age, sex, ethnicity, body mass index, and physical sickness response revealed that post-endotoxin depressed mood was predicted by increased baseline activity of TFs related to inflammation (NF-kB) and beta-adrenergic signaling (CREB) and by decreased activity of GR-related TFs (P’s < 0.001). Inflammation-induced depressed mood is predicted by peripheral transcriptome profiles related to immune activation, sympathetic activation, and glucocorticoid insensitivity. With further replication, these stress-related molecular profiles could be used for a novel genomic approach for identifying individuals at high-risk for the inflammatory subtype of depression.
Subject terms: Depression, Transcriptional regulatory elements
Introduction
Inflammation is hypothesized to play an important role in the onset and perpetuation of depression [1, 2]. Indeed, a high proportion of depressed individuals present with elevated inflammation. In the National Health and Nutrition Examination Survey, 47% of individuals with Patient Health Questionnaire (PHQ-9) scores ≥ 10 (i.e., depression) had C-reactive protein > 3 mg/L [3]. Additionally, a poor treatment response to antidepressants such as selective serotonin reuptake inhibitors is found in those with inflammatory disorders, inflammation-related gene variants, and elevated levels of inflammatory biomarkers [4–6]. Thus, inflammation likely has a role in some types depression (i.e., inflammatory subtype) acting possibly as an etiological factor, a correlate of severity, or a moderator of treatment response.
However, not all patients with inflammatory disorders present with depression, and not all individuals exposed to robust inflammatory stimuli such as interferon and endotoxin develop depression [7–11]. One way of identifying individual risk factors for inflammation-related depressive symptoms is to experimentally induce inflammation and quantify variations in affective response. Endotoxin administration is a safe procedure that, at low doses (<1 ng/kg of body weight), induces symptoms such as depressed mood, anhedonia, fatigue, reduced appetite, and cognitive impairment, and has been used as a novel experimental model for inflammation-associated depression [12, 13]. Notably, not all variability in depressive responses to endotoxin is accounted for by differences in the magnitude of inflammatory responses [7–9, 14]. For example, females show greater increases in depressed mood in response to endotoxin as compared to males, even though both sexes show similar increases in inflammation [9, 15]. Defining the factors that account for this variability, i.e., predictors of depressive responses to an inflammatory challenge, may identify individuals at risk of developing depression when exposed to heightened inflammatory states such as infection, inflammatory disorders, cancer, aging, obesity, medical treatments (e.g., interferon), or psychosocial distress. Furthermore, transcriptome profiles that predict depressive symptoms following an inflammatory challenge could be used to identify biological mechanisms underlying inflammation-induced depression, which could inform the development of more precise and targeted strategies for monitoring, prevention, or treatment.
This study sought to identify leukocyte transcriptome profiles that predict inflammation-induced depressed mood by conducting baseline gene expression analyses of peripheral blood mononuclear cells (PBMCs) from healthy volunteers who were subsequently exposed to low-dose intravenous endotoxin.
Examination of transcriptome profile might involve either individual gene transcripts or transcriptional control pathways. In this study, we focused on the latter approach with a priori hypotheses because, compared to small variations in expression of individual downstream gene targets, variations in the activity of upstream transcriptional control pathways might provide more reliable information regarding the prediction of inflammation-induced depression—see Mellon et al. [16] for more background on this approach. Thus, a promoter-based bioinformatics strategy was used to assess the activity of specific transcription factors (TFs) that might contribute to the development of inflammation-induced depressed mood. TFs can bind to closely related DNA promoter sequences, or TF-binding motifs (TFBMs), in the promoter regions of multiple genes to drive coordinated alterations in gene expression. Therefore, examination of promoter sequences of large sets of differentially expressed genes can identify TFBMs that are selectively over-represented among those genes, and thus provide information about transcriptional control pathways that may contribute to inflammation-induced depression.
Several previous studies have linked individual gene transcripts and transcriptional control pathways involved in immune activation to major depressive disorder (MDD) [17–20] and to interferon-induced depression [21]. In particular, the nuclear factor kappa B (NF-kB)/Rel family of TFs, which directly mediates inflammation, has been found to be increased in the periphery and central nervous system of depressed individuals [22, 23]. Glucocorticoid receptor (GR) dysfunction has been identified in MDD [24], and a downregulation of GR-related individual gene transcripts and transcriptional control pathways has been found in depressed subjects [18, 19, 25–29]. The cyclic AMP response element-binding protein (CREB) TF has also been implicated in signaling pathways relevant for pathogenesis and therapy of depression [30, 31]. CREB is downregulated in fibroblasts and neutrophils from patients with MDD [32, 33] and in postmortem brains of suicide victims with a history of depression [34]. Furthermore, CREB is upregulated by chronic antidepressant treatment [35]; increasing CREB levels result in antidepressant-like behaviors in rodent models [35]; and an early increase in peripheral lymphocyte CREB is associated with response to psychotherapy [36]. Promoter-based bioinformatic analyses using PBMCs have revealed a decreased activity of CREB in association with interferon-induced depression [21], although an increased activity of CREB transcriptional control pathway and an increased CREB mRNA expression in peripheral leukocytes have also been found to be associated with MDD [16, 37]. Overall, despite these conflicting findings, more studies have shown a downregulated CREB gene expression in depression.
Of note, these transcriptional control pathways—NF-kB, GR, and CREB—have also been implicated in the Conserved Transcriptional Response to Adversity (CTRA), which refers to the pattern of gene expression responses that favor proinflammatory antibacterial responses at the expense of antiviral responses when people face threatening or adverse situations [38]. Interferon-related antiviral responses and inflammation-related antibacterial responses derive from distinct gene expression programs that are mediated by separate TF pathways (e.g., interferon response factors vs. NF-kB) and are reciprocally regulated (i.e., upregulation of one induces downregulation of the other) [39]. Under basal conditions, mammalian immune systems show a bias toward strong interferon/antiviral responses (possibly to counteract the enhanced viral colonization of highly social species) [40]. However, in response to extended periods of threat or uncertainty, the sympathetic nervous system (SNS) induces the CTRA and shifts the basal immune cell transcriptome toward proinflammatory responses that would be optimal for defense against wound-related bacterial infection and tissue damage [23, 41]. Under ancestral conditions, this molecular defense response was likely adaptive because physical threats such as an animal attack would cause injuries and tissue damage, situations in which bacterial infections are imminent. However, in the modern world, this physiologic linkage between stress and inflammation may allow other types of adversity such as social isolation and social rejection to promote proinflammatory responses [23].
Both physical and social threats can elicit SNS activation, which results in stimulation of beta-adrenergic receptors and downstream activation of CREB TFs. Chronic social stress has also been found to desensitize the activity of the anti-inflammatory GR [42, 43]. In the context of the CTRA, an increased activity of NF-kB and CREB and a decreased activity of GR would all contribute to increased inflammation and thereby potentially predict depression. Thus, while most depression studies have found associations between decreased CREB activity and depression, the CTRA would link depression to increased CREB activity in circulating leukocytes. In particular, given the coordinated actions of SNS, innate immune system, and hypothalamic pituitary adrenal (HPA) axis according to the notion of the CTRA, increased CREB activity in peripheral blood cells may be an important predisposing factor for the inflammatory subtype of depression.
Based on the aforementioned literature, we examined the following a priori hypotheses in peripheral blood cells. Post-endotoxin depressed mood would be predicted by (1) higher baseline expression of genes regulated by NF-kB; (2) lower baseline expression of genes regulated by GR; and (3) differential expression of genes regulated by CREB (either up- or downregulated, as both have been observed in the literature).
Materials and methods
Participants
One hundred-fifteen healthy participants (mean age 24.2, standard deviation [SD] 6.6 years; 69 females and 46 males) completed a randomized controlled trial of endotoxin administration between March 2011 and August 2013 (Supplemental Material) [9]. Transcriptome data were available for 111 participants, 59 of whom had received endotoxin. One of these 59 participants had missing data on depressed mood. Here we report the data from 58 participants who received endotoxin and had full mood data (age range 18–50; mean age 25.2, SD 7.2 years; 37 females and 21 males) because the aim of this study was to examine transcriptomic predictors of inflammation-induced depressed mood.
Procedures
This randomized controlled trial was conducted at the UCLA Clinical and Translational Research Center. All procedures were approved by the UCLA Institutional Review Board. All subjects provided written consent before participating. Partial results from this trial (NCT01671150) were previously reported, including the moderating roles of sex, preexisting sleep disturbance, and anxiety in endotoxin-induced depressed mood [7, 9, 14] and the enhancement of inflammatory response to endotoxin by sensitivity to social disconnection [44]. No prior study has examined transcriptome profiles as predictors of inflammation-induced depressed mood. Details of the trial protocol are provided in previous reports [7, 9, 44]. Briefly, the participants received low-dose endotoxin (0.8 ng/kg of body weight, Escherichia coli group O:113) as an intravenous bolus, which mimics increases in inflammation found in inflammatory disorders, infections, and also psychological stress [45–47]. As reported previously [7], endotoxin administration led to about 10-fold increases in interleukin-6 levels and 5-fold increases in tumor necrosis factor-α levels (by comparing geometric means), which corresponds to real-world clinical settings such as HIV infection [47] and rheumatoid arthritis [48]. At baseline (prior to endotoxin administration), blood samples were drawn into B-D Biosciences CPT Tubes for subsequent isolation of PBMCs and gene expression profiling. Participants completed a self-report questionnaire on mood at baseline (T0) and then approximately every hour post injection for the next 6 h (T1–T6).
Assessment of depressed mood
Depressed mood was assessed approximately hourly from T0 to T6 using the depression subscale of the short-form Profile of Mood States [49, 50]. Participants rated the extent to which they felt at the moment (“right now”): “unhappy”, “sad”, “blue”, “hopeless”, “discouraged”, “miserable”, “helpless”, and “worthless” on a scale from 0 (not at all) to 4 (extremely). Depressed mood was calculated by summing scores from each of these items at each timepoint. The reliability of the scale was high (α = 0.83). The main outcome measure of this study was “post-endotoxin depressed mood”, a binary variable that was defined as any increase in depressed mood from baseline to T2, when both inflammatory and depressive responses to endotoxin peak. Absence of post-endotoxin depressed mood was defined by no change or decrease in depressed mood from baseline to T2.
Assessment of physical sickness symptoms
Physical symptoms such as headaches, muscle pain, shivering, nausea, breathing difficulties, and fatigue were assessed from T0 to T6. Participants rated the extent to which they felt each symptom on a scale from 0 (no symptoms) to 4 (very severe symptoms). Scores from each of these items were then dichotomized for each timepoint: any reports of 2 (“moderate”) or greater for any of the symptoms were categorized as presence of physical sickness symptoms, zero otherwise. Then, a new variable, “physical sickness response”, was defined as any increase in physical sickness symptoms from baseline to T2. Absence of physical symptom response was defined by no change or decrease in physical sickness symptoms from baseline to T2.
Gene expression profiling
Blood samples were collected immediately prior to endotoxin administration, and genome-wide transcriptional profiling was conducted on PBMCs isolated from 8 mL of blood using density gradient centrifugation (B-D Biosciences CPT Tubes). RNA was extracted (Qiagen QIAcube), tested for suitable mass (Nanodrop ND1000) and integrity (Agilent Bioanalyzer), converted to fluorescent cRNA (Ambion TotalPrep), and hybridized to Illumina Human HT-12 v4 BeadArrays following the manufacturer’s standard protocol in the UCLA Neuroscience Genomics Core Laboratory. All samples were assayed in a single batch and yielded valid results according to standard data quality metrics (e.g., median probe fluorescence intensity > 100 units). Gene expression data on 34,670 assayed transcripts were quantile normalized [51] and log2-transformed for analysis. TF activity was inferred from gene expression data using the TELiS promoter-based bioinformatics analysis [52], with modification to yield individual-level estimates of TF activity suitable for predicting individual differences in depressive symptom response to endotoxin exposure. Briefly, (1) a set of up- and downregulated genes were defined for each individual’s expression profile by first standardizing data for each gene,
where g indexes genes, i indexes individuals, and the mean () and SD (S) operators are applied to the vector of gene expression data across individuals for gene g, with “.” indicating averaging over the substituted i index; collecting then all genes showing Zgi > 1.5 (1.5 SD above the sample-wide average for that gene) or Zgi < −1.5 (1.5 SD below the sample-wide average for that gene) for a given individual; and excluding genes that showed little absolute variability in expression across individuals (Sg < 0.074 log2 expression units) or minimal expression levels ( < 7.04 log2 expression units); (2) promoters of up- and downregulated genes for each individual were then scanned for TFBMs as defined by TRANSFAC position-specific weight matrices for NF-κB/Rel family factors (V$CREL_01, V$NFKAPPAB50_01, V$NFKAPPAB65_01, V$NFKAPPAB_01, V$NFKB_C, and V$NFKB_Q6), the GR (V$GRE_C and V$GR_Q6), and CREB family factors (V$CREBP1CJUN_01, V$CREBP1_01, V$CREBP1_Q2, V$CREB_01, V$CREB_02, V$CREB_Q2, and V$CREB_Q4); and (3) the relative activation of each TF for a given individual was indicated by the log2-transformed ratio of TFBM prevalence in the promoters of up- vs. downregulated genes for that individual (with the total number of up- and downregulated genes serving as a precision weight for that ratio); these TFBM ratios were then used as predictor variables as described below.
Statistical analysis
To evaluate whether activity of a priori selected TFs predicts post-endotoxin depressed mood, we conducted multivariate weighted logistic regression with (log2) TFBM ratios as the predictor variables in their original continuous format and post-endotoxin depressed mood as the outcome variable in the binary format as described above. The numbers of genes declared up- and downregulated for each individual were used as precision weights. The covariates for multivariate analyses included not only age, sex, ethnicity (white vs. non-white), and body mass index (BMI), which are often associated with systemic inflammation and/or depression [53], but also physical sickness response. This highly conservative approach of controlling for physical sickness response as well (since fatigue is one of the diagnostic criteria for MDD) was adopted to ensure that the transcriptomic predictors of post-endotoxin depressed mood identified in this study are independent of the influence from post-endotoxin sickness symptoms. The magnitude of prediction was described in odds ratios (ORs), indicating the odds of developing post-endotoxin depressed mood according to the baseline activity of the pre-selected TFs. Statistical analyses were performed using STATA 14.2 (StataCorp, College Station, TX).
Results
Baseline sample characteristics
As described in Methods, the final analytical sample included 58 participants who were administered endotoxin; 21 of them (36%) evidenced depressed mood in response to endotoxin. In this final analytical sample, those with and without post-endotoxin depressed mood did not significantly differ in age, sex, and ethnicity (Table 1). Those with post-endotoxin depressed mood had significantly lower BMI (23.1 vs. 24.7, P = 0.04).
Table 1.
Sample characteristics at baseline
| Characteristic | Post-endotoxin depressed mood (n = 21) | No post-endotoxin depressed mood (n = 37) | P* |
|---|---|---|---|
| Mean age (SD) | 24.3 (7.1) | 25.7 (7.3) | 0.47 |
| Female (%) | 16 (76.2) | 21 (56.8) | 0.14 |
| White (%) | 7 (33.3) | 12 (32.4) | 0.94 |
| Mean body mass index (SD) | 23.1 (2.9) | 24.7 (2.6) | 0.04 |
| Physical sickness response (%) | 19 (90.5) | 21 (56.8) | 0.008 |
SD standard deviation
*P-values from t-tests when the sample characteristic is a continuous variable and from chi-square tests when the sample characteristic is a categorical variable
Transcriptomic predictors of post-endotoxin depressed mood
To bioinformatically determine whether the baseline activity of the selected TFs predicts the development of post-endotoxin depressed mood, we performed promoter-based bioinformatic assessment of TF activity followed by logistic regression analysis with: (1) individual-level measures of TFBM asymmetry in the promoters of genes that were relatively up- and downregulated in that individual at baseline (prior to endotoxin administration) as the predictor variables; (2) post-endotoxin depressed mood as the outcome variable; and (3) the total number of baseline up- and downregulated genes serving as a precision weight. Multivariate logistic regression models were constructed to compute adjusted ORs, first controlling for age, sex, ethnicity, and BMI and then controlling for all these variables as well as physical sickness response (Table 2). These two multivariate models were constructed and described so that the separate role of physical sickness response in the final multivariate model could be clearly visualized.
Table 2.
Prediction of post-endotoxin depressed mood by baseline transcription factor activity
| Transcription factor | TFBM | Unadjusted OR (95% CI) | P | Model 1Adjusted ORa (95% CI) | P | Model 2Adjusted ORb (95% CI) | P |
|---|---|---|---|---|---|---|---|
| NF-kB/Rel family | V$CREL_01 | 5.38 (4.76–6.08) | <0.001 | 2.73 (2.39–3.12) | <0.001 | 2.39 (2.08–2.75) | <0.001 |
| V$NFKAPPAB50_01 | 4.52 (4.32–4.74) | <0.001 | 11.01 (10.31–11.76) | <0.001 | 14.72 (13.74–15.77) | <0.001 | |
| V$NFKAPPAB65_01 | 1.61 (1.51–1.72) | <0.001 | 1.97 (1.83–2.12) | <0.001 | 1.91 (1.77–2.06) | <0.001 | |
| V$NFKAPPAB_01 | 3.45 (3.19–3.73) | <0.001 | 3.40 (3.14–3.69) | <0.001 | 4.17 (3.82–4.54) | <0.001 | |
| V$NFKB_C | 1.00 (0.95–1.05) | 0.97 | 1.61 (1.53–1.69) | <0.001 | 1.68 (1.60–1.77) | <0.001 | |
| V$NFKB_Q6 | 0.59 (0.55–0.62) | <0.001 | 0.91 (0.86–0.97) | 0.002 | 0.99 (0.93–1.05) | 0.72 | |
| GR | V$GRE_C | 0.67 (0.64–0.70) | <0.001 | 0.61 (0.58–0.64) | <0.001 | 0.61 (0.57–0.65) | <0.001 |
| V$GR_Q6 | 0.29 (0.26–0.31) | <0.001 | 0.28 (0.26–0.31) | <0.001 | 0.30 (0.28–0.33) | <0.001 | |
| CREB | V$CREBP1CJUN_01 | 0.90 (0.86–0.95) | <0.001 | 1.15 (1.09–1.21) | 0.13 | 0.96 (0.91–1.01) | 0.13 |
| V$CREBP1_01 | 0.87 (0.85–0.90) | <0.001 | 0.71 (0.68–0.73) | <0.001 | 0.72 (0.70–0.75) | <0.001 | |
| V$CREBP1_Q2 | 1.16 (1.11–1.22) | <0.001 | 1.61 (1.53–1.70) | <0.001 | 1.67 (1.58–1.76) | <0.001 | |
| V$CREB_01 | 1.95 (1.83–2.07) | <0.001 | 2.46 (2.30–2.63) | <0.001 | 3.51 (3.26–3.79) | <0.001 | |
| V$CREB_02 | 4.29 (3.99–4.62) | <0.001 | 13.05 (11.88–14.33) | <0.001 | 41.01 (36.79–45.72) | <0.001 | |
| V$CREB_Q2 | 0.70 (0.67–0.73) | <0.001 | 0.87 (0.83–0.92) | <0.001 | 1.51 (1.42–1.61) | <0.001 | |
| V$CREB_Q4 | 0.76 (0.72–0.81) | <0.001 | 1.17 (1.09–1.25) | <0.001 | 1.28 (1.18–1.38) | <0.001 |
TFBM transcription factor-binding motif, OR odds ratio, CI confidence interval, NF-kB nuclear factor kappa B, GR glucocorticoid receptor, CREB cyclic AMP response element-binding protein
aMultivariate logistic regression model adjusted for age, sex, ethnicity, and body mass index
bMultivariate logistic regression model adjusted for age, sex, ethnicity, body mass index, and physical sickness response
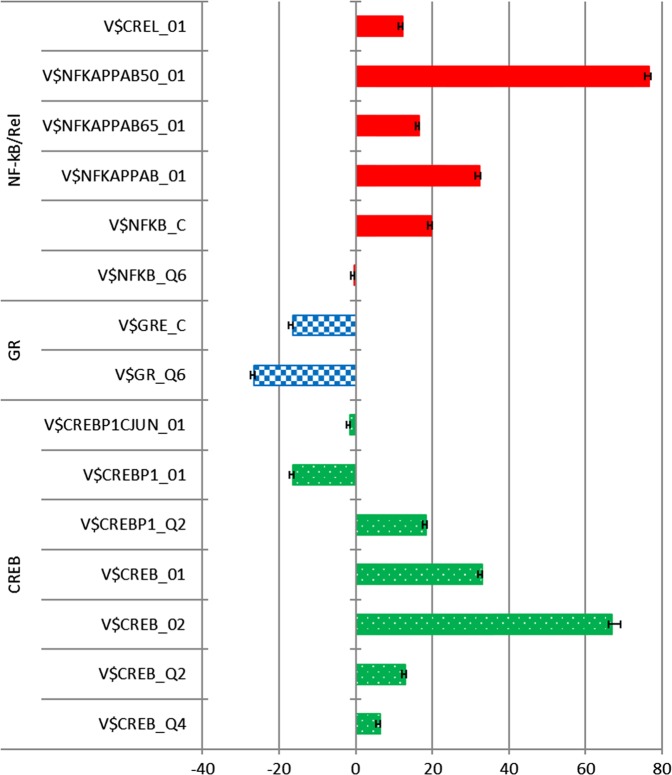
In accordance with the a priori hypotheses, higher activity of NF-kB and lower activity of GR predicted post-endotoxin depressed mood. As shown in Table 2 and Fig. 1, weighted logistic regression controlling for age, sex, ethnicity, BMI, and physical sickness response revealed that for five out of six distinct TFBMs defined for the NF-κB/Rel family, higher TF activity predicted a significantly higher likelihood of post-endotoxin depressed mood as indicated by ORs greater than unity. This was uniformly true for the four NF-kB TFBMs with the strongest biochemical validation (those bearing the _01/_02 suffix from the TRANSFAC database). For both TFBMs assessing GR activity, lower TF activity predicted a higher likelihood of post-endotoxin depressed mood.
Fig. 1.
Magnitude and direction of the associations between baseline transcription factor activity and post-endotoxin depressed mood. Bars represent the Z-scores derived from the multivariate weighted logistic regression analyses adjusted for age, sex, body mass index, ethnicity, and physical sickness response. Positive and negative scores indicate, respectively, increased and decreased activity of the transcription factors. Error bars indicate standard errors. NF-kB nuclear factor kappa B, GR glucocorticoid receptor, CREB cyclic AMP response element-binding protein
Contrary to the hypothesis derived from much of the previous depression literature, but consistent with the CTRA hypothesis, bioinformatic indications of higher CREB activity predicted post-endotoxin depressed mood. As shown in Table 2 and Fig. 1, results for five out of seven CREB TFBMs indicated the prediction of post-endotoxin depressed mood by an increased activity of CREB.
Most of the associations tested here were highly statistically significant (P’s < 0.001). Furthermore, these associations remained significant even after the conservative Bonferroni correction of statistical significance at P < 0.003 (i.e., P < 0.05 divided by 15 as we tested 15 variant models involving alternative specifications of 3 TF predictor hypotheses).
Transcriptomic predictors of post-endotoxin cytokine response are provided in Supplemental Material.
Discussion
In this human experimental study, inflammation-induced depressed mood was predicted by baseline leukocyte transcriptome profiles related to immune activation/inflammation, SNS activation, and glucocorticoid insensitivity; more specifically, higher activity of NF-kB and CREB and lower activity of GR. In other words, individuals with baseline peripheral blood gene expression profiles showing transcriptomic indications of immune activation/inflammation, sympathetic activation, and glucocorticoid insensitivity were more likely to develop depressed mood in response to administration of an experimental challenge with endotoxin.
While increased activity of NF-kB and decreased activity of GR are associated with depression [18, 19, 22, 23, 25–29], the direction of the association between CREB and depression has been less consistent, with either decreased [21, 32–34] or increased [16, 37] CREB activities in association with depression. Furthermore, depending on the brain region involved, increased CREB activity can either reduce or increase depressive-like behavior in animal models. While antidepressant treatments increase CREB activity in hippocampus, a sustained elevation of CREB activity in nucleus accumbens produces anhedonia [54]. Regarding the CREB activity in peripheral blood cells, some studies have linked decreased CREB activity to depression [32, 33], whereas other studies have linked increased CREB activity to depression [16, 37]. Our results are consistent with the latter findings.
Whereas previous studies have focused on the association between CREB activity and depression, the current study examined the activity of the CREB transcription control pathway in peripheral leukocytes as a predictor of inflammation-induced depressed mood. Therefore, although this study does not provide any direct evidence regarding the etiological association between CREB activity and depression per se, it provides some insight regarding the risk prediction potential for CREB in inflammation-induced depression. Greater peripheral activity of CREB may predict depressive responses to an inflammatory challenge, which may occur in the context of physiological or psychological stress. Predisposing stress may be expressed in the form of increased peripheral beta-adrenergic activity, and therefore increased CREB activity, which would be consistent with the concept of the CTRA. In line with this possibility, preexisting anxiety symptoms serve as a vulnerability factor for post-endotoxin depressive symptoms [14]. Furthermore, the current experimental study design could have introduced some element of psychological stress in addition to inflammatory stress (i.e., endotoxin) because the study participants may have experienced anticipatory anxiety and SNS activation at baseline due to the anticipation of receiving endotoxin vs. placebo. Thus, those who were vulnerable to anticipation stress could have had an additional upregulation of CREB at baseline beyond their preexisting stress and CREB activation. Personality traits of anticipatory anxiety, longstanding adversities, transient psychological stress, and concurrent enhancement of CREB activity could all contribute to inflammation-induced depression.
Adversities in the form of physical, physiological, psychological, or social stress may activate SNS and also trigger innate immune responses with the elevation of catecholamines and inflammatory mediators. Prolonged stress may lead to a state of glucocorticoid insensitivity. Furthermore, catecholamines acting through alpha- and beta-adrenergic receptors can activate inflammatory signaling pathways, including NF-kB, and increase cytokine expression in the brain and periphery [2]. Glucocorticoid insensitivity may result in a reduced cellular response to cortisol, which is one of the most potent anti-inflammatory hormones, despite the continued presence of stressful factors (and continued cortisol output). Thus, the combined neuroendocrine actions of SNS and HPA axis may lead to a state of chronic systemic inflammation, predisposing individuals to inflammation-associated conditions including depression. More specifically, according to the sequence of events corresponding to the current experimental study design, some individuals may be found in a state of sympathetic activation, heightened inflammation, and glucocorticoid insensitivity for a variety of reasons such as longstanding adversities; and when they face physiological or psychosocial stress that induces an acute proinflammatory state, they may develop depressive symptoms or disorders.
The current study presents several strengths. It is a highly controlled human experimental study using an innovative inflammatory challenge model of depression (i.e., post-endotoxin depressed mood) and a novel transcriptomic approach. Furthermore, the study hypotheses were formulated based on an integrative theoretical concept of the CTRA, and the findings were both consistent with this concept and highly statistically significant. The following limitations should be considered. First, although this experimental model of depression using endotoxin is a unique opportunity for studying depression and has been used in several previous studies [12], post-endotoxin depressive symptoms cannot be equated with clinical depression. Future clinical studies will be required to corroborate and translate the current findings into clinical practice. Second, the study participants were healthy and mostly young, which may limit the external validity of the findings to a broader sphere of the general population including mentally or physically ill individuals and older adults. Lastly, the association between baseline TF measures and depressive symptom responses is fundamentally correlational (even though they are measured at different time points), so the causal relationships involved and their biological mechanisms remain to be clarified in future research.
The current study suggests that CTRA- and stress-related molecular profiles may hold potential as predictive biomarkers for inflammation-induced depressive symptoms. These findings also underscore the potential utility of targeting inflammation-related biological processes in future efforts to prevent depression. Independent replication of these findings in prospective cohort studies of clinical depression will be an important advance in this endeavor. With further replication, these transcriptome profiles could be used as a novel genomic approach for identifying individuals at high risks for depression.
Funding and disclosure
This research was funded by an R01 from the National Institute of Mental Health to NIE (5R01MH091352). The first author (JHC) was supported by K23AG049085, NIH/NCATS UCLA CTSI Grant KL2TR000122, and NARSAD Young Investigator Grant from the Brain and Behavior Research Foundation.
Supplementary information
Acknowledgements
We thank Anthony Suffredini, MD, at the National Institutes of Health, Warren Grant Magnuson Clinical Center, for providing the standard reference endotoxin and the staff of the UCLA Clinical and Translational Research Center for providing technical assistance.
Competing interests
The authors declare no competing interests.
Footnotes
Clinical Trial Registration: ClinicalTrials.gov NCT01671150
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information accompanies this paper at (10.1038/s41386-019-0316-9).
References
- 1.Dantzer R, O’Connor JC, Freund GG, Johnson RW, Kelley KW. From inflammation to sickness and depression: when the immune system subjugates the brain. Nat Rev. 2008;9:46–56. doi: 10.1038/nrn2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Miller AH, Maletic V, Raison CL. Inflammation and its discontents: the role of cytokines in the pathophysiology of major depression. Biol Psychiatry. 2009;65:732–41. doi: 10.1016/j.biopsych.2008.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rethorst CD, Bernstein I, Trivedi MH. Inflammation, obesity, and metabolic syndrome in depression: analysis of the 2009-2010 National Health and Nutrition Examination Survey (NHANES) J Clin Psychiatry. 2014;75:e1428–32. doi: 10.4088/JCP.14m09009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barnes J, Mondelli V, Pariante CM. Genetic contributions of inflammation to depression. Neuropsychopharmacology. 2017;42:81–98. doi: 10.1038/npp.2016.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Strawbridge R, Arnone D, Danese A, Papadopoulos A, Herane Vives A, Cleare AJ. Inflammation and clinical response to treatment in depression: a meta-analysis. Eur Neuropsychopharmacol. 2015;25:1532–43. doi: 10.1016/j.euroneuro.2015.06.007. [DOI] [PubMed] [Google Scholar]
- 6.Vogelzangs N, Beekman AT, van Reedt Dortland AK, Schoevers RA, Giltay EJ, de Jonge P, et al. Inflammatory and metabolic dysregulation and the 2-year course of depressive disorders in antidepressant users. Neuropsychopharmacology. 2014;39:1624–34. doi: 10.1038/npp.2014.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cho HJ, Eisenberger NI, Olmstead R, Breen EC, Irwin MR. Preexisting mild sleep disturbance as a vulnerability factor for inflammation-induced depressed mood: a human experimental study. Transl Psychiatry. 2016;6:e750. doi: 10.1038/tp.2016.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eisenberger NI, Inagaki TK, Mashal NM, Irwin MR. Inflammation and social experience: an inflammatory challenge induces feelings of social disconnection in addition to depressed mood. Brain Behav Immun. 2010;24:558–63. doi: 10.1016/j.bbi.2009.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moieni M, Irwin MR, Jevtic I, Olmstead R, Breen EC, Eisenberger NI. Sex differences in depressive and socioemotional responses to an inflammatory challenge: implications for sex differences in depression. Neuropsychopharmacology. 2015;40:1709–16. doi: 10.1038/npp.2015.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Capuron L, Raison CL, Musselman DL, Lawson DH, Nemeroff CB, Miller AH. Association of exaggerated HPA axis response to the initial injection of interferon-alpha with development of depression during interferon-alpha therapy. Am J Psychiatry. 2003;160:1342–5. doi: 10.1176/appi.ajp.160.7.1342. [DOI] [PubMed] [Google Scholar]
- 11.Capuron L, Ravaud A, Miller AH, Dantzer R. Baseline mood and psychosocial characteristics of patients developing depressive symptoms during interleukin-2 and/or interferon-alpha cancer therapy. Brain Behav Immun. 2004;18:205–13. doi: 10.1016/j.bbi.2003.11.004. [DOI] [PubMed] [Google Scholar]
- 12.DellaGioia N, Hannestad J. A critical review of human endotoxin administration as an experimental paradigm of depression. Neurosci Biobehav Rev. 2010;34:130–43. doi: 10.1016/j.neubiorev.2009.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reichenberg A, Yirmiya R, Schuld A, Kraus T, Haack M, Morag A, et al. Cytokine-associated emotional and cognitive disturbances in humans. Arch Gen Psychiatry. 2001;58:445–52. doi: 10.1001/archpsyc.58.5.445. [DOI] [PubMed] [Google Scholar]
- 14.Irwin MR, Cole S, Olmstead R, Breen EC, Cho HJ, Moieni M, et al. Moderators for depressed mood and systemic and transcriptional inflammatory responses: a randomized controlled trial of endotoxin. Neuropsychopharmacology. 2018;44:635–41. [DOI] [PMC free article] [PubMed]
- 15.Eisenberger NI, Inagaki TK, Rameson LT, Mashal NM, Irwin MR. An fMRI study of cytokine-induced depressed mood and social pain: the role of sex differences. NeuroImage. 2009;47:881–90. doi: 10.1016/j.neuroimage.2009.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mellon SH, Wolkowitz OM, Schonemann MD, Epel ES, Rosser R, Burke HB, et al. Alterations in leukocyte transcriptional control pathway activity associated with major depressive disorder and antidepressant treatment. Transl Psychiatry. 2016;6:e821. doi: 10.1038/tp.2016.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Galecki P, Galecka E, Maes M, Chamielec M, Orzechowska A, Bobinska K, et al. The expression of genes encoding for COX-2, MPO, iNOS, and sPLA2-IIA in patients with recurrent depressive disorder. J Affect Disord. 2012;138:360–6. doi: 10.1016/j.jad.2012.01.016. [DOI] [PubMed] [Google Scholar]
- 18.Grosse L, Carvalho LA, Wijkhuijs AJ, Bellingrath S, Ruland T, Ambree O, et al. Clinical characteristics of inflammation-associated depression: monocyte gene expression is age-related in major depressive disorder. Brain Behav Immun. 2015;44:48–56. doi: 10.1016/j.bbi.2014.08.004. [DOI] [PubMed] [Google Scholar]
- 19.Hepgul N, Cattaneo A, Zunszain PA, Pariante CM. Depression pathogenesis and treatment: what can we learn from blood mRNA expression? BMC Med. 2013;11:28. doi: 10.1186/1741-7015-11-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yi Z, Li Z, Yu S, Yuan C, Hong W, Wang Z, et al. Blood-based gene expression profiles models for classification of subsyndromal symptomatic depression and major depressive disorder. PLoS ONE. 2012;7:e31283. doi: 10.1371/journal.pone.0031283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Felger JC, Cole SW, Pace TW, Hu F, Woolwine BJ, Doho GH, et al. Molecular signatures of peripheral blood mononuclear cells during chronic interferon-alpha treatment: relationship with depression and fatigue. Psychol Med. 2012;42:1591–603. doi: 10.1017/S0033291711002868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Malki K, Pain O, Tosto MG, Du Rietz E, Carboni L, Schalkwyk LC. Identification of genes and gene pathways associated with major depressive disorder by integrative brain analysis of rat and human prefrontal cortex transcriptomes. Transl Psychiatry. 2015;5:e519. doi: 10.1038/tp.2015.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Savitz J, Frank MB, Victor T, Bebak M, Marino JH, Bellgowan PS, et al. Inflammation and neurological disease-related genes are differentially expressed in depressed patients with mood disorders and correlate with morphometric and functional imaging abnormalities. Brain Behav Immun. 2013;31:161–71. doi: 10.1016/j.bbi.2012.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pariante CM, Miller AH. Glucocorticoid receptors in major depression: relevance to pathophysiology and treatment. Biol Psychiatry. 2001;49:391–404. doi: 10.1016/S0006-3223(00)01088-X. [DOI] [PubMed] [Google Scholar]
- 25.Cattaneo A, Gennarelli M, Uher R, Breen G, Farmer A, Aitchison KJ, et al. Candidate genes expression profile associated with antidepressants response in the GENDEP study: differentiating between baseline ‘predictors’ and longitudinal ‘targets’. Neuropsychopharmacology. 2013;38:377–85. doi: 10.1038/npp.2012.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Frodl T, Carballedo A, Hughes MM, Saleh K, Fagan A, Skokauskas N, et al. Reduced expression of glucocorticoid-inducible genes GILZ and SGK-1: high IL-6 levels are associated with reduced hippocampal volumes in major depressive disorder. Transl Psychiatry. 2012;2:e88. doi: 10.1038/tp.2012.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matsubara T, Funato H, Kobayashi A, Nobumoto M, Watanabe Y. Reduced glucocorticoid receptor alpha expression in mood disorder patients and first-degree relatives. Biol Psychiatry. 2006;59:689–95. doi: 10.1016/j.biopsych.2005.09.026. [DOI] [PubMed] [Google Scholar]
- 28.Menke A, Klengel T, Rubel J, Bruckl T, Pfister H, Lucae S, et al. Genetic variation in FKBP5 associated with the extent of stress hormone dysregulation in major depression. Genes Brain Behav. 2013;12:289–96. doi: 10.1111/gbb.12026. [DOI] [PubMed] [Google Scholar]
- 29.Carvalho LA, Bergink V, Sumaski L, Wijkhuijs J, Hoogendijk WJ, Birkenhager TK, et al. Inflammatory activation is associated with a reduced glucocorticoid receptor alpha/beta expression ratio in monocytes of inpatients with melancholic major depressive disorder. Transl Psychiatry. 2014;4:e344. doi: 10.1038/tp.2013.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gass P, Riva MA. CREB, neurogenesis and depression. Bioessays. 2007;29:957–61. doi: 10.1002/bies.20658. [DOI] [PubMed] [Google Scholar]
- 31.Sulser F. The role of CREB and other transcription factors in the pharmacotherapy and etiology of depression. Ann Med. 2002;34:348–56. doi: 10.1080/078538902320772106. [DOI] [PubMed] [Google Scholar]
- 32.Manier DH, Shelton RC, Ellis TC, Peterson CS, Eiring A, Sulser F. Human fibroblasts as a relevant model to study signal transduction in affective disorders. J Affect Disord. 2000;61:51–8. doi: 10.1016/S0165-0327(99)00190-1. [DOI] [PubMed] [Google Scholar]
- 33.Ren X, Dwivedi Y, Mondal AC, Pandey GN. Cyclic-AMP response element binding protein (CREB) in the neutrophils of depressed patients. Psychiatry Res. 2011;185:108–12. doi: 10.1016/j.psychres.2010.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dwivedi Y, Rizavi HS, Shukla PK, Lyons J, Faludi G, Palkovits M, et al. Protein kinase A in postmortem brain of depressed suicide victims: altered expression of specific regulatory and catalytic subunits. Biol Psychiatry. 2004;55:234–43. doi: 10.1016/j.biopsych.2003.11.003. [DOI] [PubMed] [Google Scholar]
- 35.Blendy JA. The role of CREB in depression and antidepressant treatment. Biol Psychiatry. 2006;59:1144–50. doi: 10.1016/j.biopsych.2005.11.003. [DOI] [PubMed] [Google Scholar]
- 36.Koch JM, Hinze-Selch D, Stingele K, Huchzermeier C, Goder R, Seeck-Hirschner M, et al. Changes in CREB phosphorylation and BDNF plasma levels during psychotherapy of depression. Psychother Psychosom. 2009;78:187–92. doi: 10.1159/000209350. [DOI] [PubMed] [Google Scholar]
- 37.Iga J, Ueno S, Yamauchi K, Numata S, Kinouchi S, Tayoshi-Shibuya S, et al. Altered HDAC5 and CREB mRNA expressions in the peripheral leukocytes of major depression. Prog Neuropsychopharmacol Biol Psychiatry. 2007;31:628–32. doi: 10.1016/j.pnpbp.2006.12.014. [DOI] [PubMed] [Google Scholar]
- 38.Slavich GM, Cole SW. The emerging field of human social genomics. Clin Psychol Sci. 2013;1:331–48. doi: 10.1177/2167702613478594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Amit I, Garber M, Chevrier N, Leite AP, Donner Y, Eisenhaure T, et al. Unbiased reconstruction of a mammalian transcriptional network mediating pathogen responses. Science. 2009;326:257–63. doi: 10.1126/science.1179050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nunn C, Altizer S, Altizer SM. Infectious diseases in primates: behavior, ecology and evolution. Oxford University Press; Oxford. 2006.
- 41.Irwin MR, Cole SW. Reciprocal regulation of the neural and innate immune systems. Nat Rev Immunol. 2011;11:625. doi: 10.1038/nri3042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miller G, Chen E, Cole SW. Health psychology: developing biologically plausible models linking the social world and physical health. Annu Rev Psychol. 2009;60:501–24. doi: 10.1146/annurev.psych.60.110707.163551. [DOI] [PubMed] [Google Scholar]
- 43.Weber MD, Godbout JP, Sheridan JF. Repeated social defeat, neuroinflammation, and behavior: monocytes carry the signal. Neuropsychopharmacology. 2017;42:46–61. doi: 10.1038/npp.2016.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moieni M, Irwin MR, Jevtic I, Breen EC, Cho HJ, Arevalo JM, et al. Trait sensitivity to social disconnection enhances pro-inflammatory responses to a randomized controlled trial of endotoxin. Psychoneuroendocrinology. 2015;62:336–42. doi: 10.1016/j.psyneuen.2015.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ishihara K, Hirano T. IL-6 in autoimmune disease and chronic inflammatory proliferative disease. Cytokine Growth Factor Rev. 2002;13:357–68. doi: 10.1016/S1359-6101(02)00027-8. [DOI] [PubMed] [Google Scholar]
- 46.Steptoe A, Hamer M, Chida Y. The effects of acute psychological stress on circulating inflammatory factors in humans: a review and meta-analysis. Brain Behav Immun. 2007;21:901–12. doi: 10.1016/j.bbi.2007.03.011. [DOI] [PubMed] [Google Scholar]
- 47.Breen EC, Rezai AR, Nakajima K, Beall GN, Mitsuyasu RT, Hirano T, et al. Infection with HIV is associated with elevated IL-6 levels and production. J Immunol. 1990;144:480–4. [PubMed] [Google Scholar]
- 48.Mangge H, Kenzian H, Gallistl S, Neuwirth G, Liebmann P, Kaulfersch W, et al. Serum cytokines in juvenile rheumatoid arthritis. Correlation with conventional inflammation parameters and clinical subtypes. Arthritis Rheum. 1995;38:211–20. doi: 10.1002/art.1780380209. [DOI] [PubMed] [Google Scholar]
- 49.Baker F, Denniston M, Zabora J, Polland A, Dudley WN. A POMS short form for cancer patients: psychometric and structural evaluation. Psychooncology. 2002;11:273–81. doi: 10.1002/pon.564. [DOI] [PubMed] [Google Scholar]
- 50.McNair DM, Lorr M, Droppleman LF. Manual for the profile of mood states. San Diego, CA: Education and Industrial Testing Service; 1971. [Google Scholar]
- 51.Gallon S, Loubes JM, Maza E. Statistical properties of the quantile normalization method for density curve alignment. Math Biosci. 2013;242:129–42. doi: 10.1016/j.mbs.2012.12.007. [DOI] [PubMed] [Google Scholar]
- 52.Cole SW, Yan W, Galic Z, Arevalo J, Zack JA. Expression-based monitoring of transcription factor activity: the TELiS database. Bioinformatics. 2005;21:803–10. doi: 10.1093/bioinformatics/bti038. [DOI] [PubMed] [Google Scholar]
- 53.O’Connor MF, Bower JE, Cho HJ, Creswell JD, Dimitrov S, Hamby ME, et al. To assess, to control, to exclude: effects of biobehavioral factors on circulating inflammatory markers. Brain Behav Immun. 2009;23:887–97. doi: 10.1016/j.bbi.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Carlezon WA, Jr., Duman RS, Nestler EJ. The many faces of CREB. Trends Neurosci. 2005;28:436–45. doi: 10.1016/j.tins.2005.06.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.