Significance
Complex multicellularity is a major evolutionary innovation in the history of life. Mushroom-forming fungi (Agaricomycetes) represent one of the most diverse complex multicellular clades, yet the genetic bases and evolutionary origins of their multicellular development are hardly known. We used readouts of gene expression in six species to find genes with a dynamic expression during the development of fruiting bodies. Comparisons across species and to 200 fungal genomes identified the gene families with a conserved expression dynamics in multicellular fruiting bodies and their ancient evolutionary origins. These data outline the major multicellularity-related and developmental processes of mushrooms, including the role of transcriptional reprogramming, gene coexpression networks, and alternative splicing, and reveal significant convergence with other complex multicellular lineages.
Keywords: complex multicellularity, evolution, fungi, comparative genomics, fruiting body development
Abstract
The evolution of complex multicellularity has been one of the major transitions in the history of life. In contrast to simple multicellular aggregates of cells, it has evolved only in a handful of lineages, including animals, embryophytes, red and brown algae, and fungi. Despite being a key step toward the evolution of complex organisms, the evolutionary origins and the genetic underpinnings of complex multicellularity are incompletely known. The development of fungal fruiting bodies from a hyphal thallus represents a transition from simple to complex multicellularity that is inducible under laboratory conditions. We constructed a reference atlas of mushroom formation based on developmental transcriptome data of six species and comparisons of 200 whole genomes, to elucidate the core genetic program of complex multicellularity and fruiting body development in mushroom-forming fungi (Agaricomycetes). Nearly 300 conserved gene families and 70 functional groups contained developmentally regulated genes from five to six species, covering functions related to fungal cell wall remodeling, targeted protein degradation, signal transduction, adhesion, and small secreted proteins (including effector-like orphan genes). Several of these families, including F-box proteins, expansin-like proteins, protein kinases, and transcription factors, showed expansions in Agaricomycetes, many of which convergently expanded in multicellular plants and/or animals too, reflecting convergent solutions to genetic hurdles imposed by complex multicellularity among independently evolved lineages. This study provides an entry point to studying mushroom development and complex multicellularity in one of the largest clades of complex eukaryotic organisms.
Fungi represent a diverse lineage of complex multicellular organisms with a unique evolutionary history compared with complex multicellular animals, embryophytes, florideophytes, and laminarean brown algae (1–4). Within the fungal kingdom, complex multicellularity is discussed mostly in the context of fruiting bodies, which are found in at least eight independent lineages (2), of which the Pezizomycotina (Ascomycota) and the Agaricomycetes (Basidiomycota) contain the vast majority of species. The mushroom-forming fungi (Agaricomycetes) comprise 21,000 species and originated 350 million years ago (5), approximately coinciding with the origin of tetrapods. Fruiting bodies of mushroom-forming fungi have immense importance in agriculture, ecology, and medicine; they represent an important and sustainable food source, with favorable medicinal properties (e.g., antitumor, immunomodulatory) (6). Mushroom-forming fungi share a single origin of fruiting body formation that probably dates to the most recent common ancestor of the Agaricomycetes, Dacrymycetes, and Tremellomycetes (2).
Fruiting body development in mushroom-forming fungi has been subject to surprisingly few studies (see, e.g., refs. 7–10), resulting in a paucity of information on the genetic underpinnings of the origins of complex multicellularity in this group (2). During fruiting body development, fungi deploy mechanisms for hypha-to-hypha adhesion, communication (e.g., via cell−cell channels; ref. 11), cell differentiation, and defense, and execute a developmental program that results in a genetically determined shape and size (2, 10). Fruiting bodies shelter and protect reproductive cells and facilitate spore dispersal. Uniquely, complex multicellularity in fungi comprises short-lived reproductive organs, whereas, in animals and plants, it comprises the reproducing individual. Nevertheless, fruiting bodies evolved complexity levels comparable to that of simple animals, with up to 30 morphologically distinguishable cell types described so far (10). Fruiting body development is triggered by changing environmental variables (e.g., nutrient availability), and involves a transition from vegetative mycelium to a complex multicellular fruiting body initial. While the vegetative mycelium is composed of loosely arranged hyphae and shows little differentiation [hence, better regarded as a grade of simple multicellularity (1, 2)], the emergence of a fruiting body initial involves a reprogramming of hyphal branching patterns to form a compact, three-dimensional structure in which hyphae adhere tightly to each other. The initial follows genetically encoded programs to develop species-specific morphologies (9, 10), which, in the Agaricomycetes, ranges from simple crust-like forms (e.g., Phanerochaete) to the most complex toadstools (e.g., Agaricus bisporus). Previous studies identified several developmental genes, including hydrophobins (12), defense-related proteins (13), fungal cell wall (FCW) modifying enzymes (14–17), transcriptional regulators (8, 9, 18) (e.g., mating genes), and light receptors (19) (e.g., white collar complex). Since all these studies focused on a single species, they provide little information on what genes comprise the conserved and species-specific toolkits of multicellularity and development in the Agaricomycetes.
Here we investigate the general evolutionary and functional properties of fruiting body development using comparative transcriptomes of fruiting bodies of complex multicellular Agaricomycetes (mushroom-forming fungi). We sampled RNA from different developmental stages of six species that share a complex multicellular ancestor and represent the levels of fruiting body complexity found in the Agaricomycetes. We combine comparative analyses of developmental transcriptomes with comparisons of 201 whole genomes and focus on conserved developmental functions and complex multicellularity in fruiting bodies.
Results
We obtained fruiting bodies in the laboratory for Coprinopsis cinerea AmutBmut, Schizophyllum commune H4-8, Phanerochaete chrysosporium RP78, and Lentinus tigrinus RLG9953-sp and, from the field, for Rickenella mellea SZMC22713 and profiled gene expression in three to nine developmental stages and tissue types (Fig. 1B). For Armillaria ostoyae C18/9, we used RNA-Seq data of five developmental stages and three tissue types from our previous work (20). Besides, we report the de novo draft genome of R. mellea (Hymenochaetales). The phylogenetically most distant species in our dataset are Rickenella and Coprinopsis, spanning 200 million years of evolution (5) and having a complex multicellular last common ancestor. Species with the most complex fruiting bodies are A. ostoyae, C. cinerea, L. tigrinus (21), and R. mellea form fruiting bodies with cap, stipe, and gills, whereas P. chrysosporium and S. commune produce plesiomorphically and secondarily simple fruiting bodies, respectively. To construct a reference atlas of mushroom development, we performed poly(A)+ RNA-Seq on Illumina platforms, in triplicates (totaling to 120 libraries; Dataset S1). We obtained an average of 60.8 million reads per sample, of which, on average, 83.3% mapped to the genomes (SI Appendix, Fig. S1). For each species, the first and last developmental stages sampled were vegetative mycelium and mature fruiting body at the time of spore release, respectively. This spans all developmental events of fruiting bodies except senescence. We defined two groups of developmentally regulated genes: those that show greater than fourfold change and a fragment per kilobase per million mapped reads of 4 between any two stages of fruiting body development (referred to as “FB development genes”) and that show greater than fourfold increase in expression from vegetative mycelium to the first primordium stage (referred to as “FB-init genes”). These definitions exclude genes that show highest expression in vegetative mycelium and little or no dynamics later on. Using this strategy, we could recover 80% of previously reported developmental genes of Coprinopsis (Dataset S2). To more broadly infer functionalities enriched in mushroom-forming fungi, we analyzed Interpro domain counts across 201 fungal genomes (including 104 Agaricomycetes), which revealed 631 significantly overrepresented domains in mushroom-forming fungi (P 0.01, Fisher exact test Benjamini Hochberg adjusted P values, abbreviated as FET; Datasets S3 and S4).
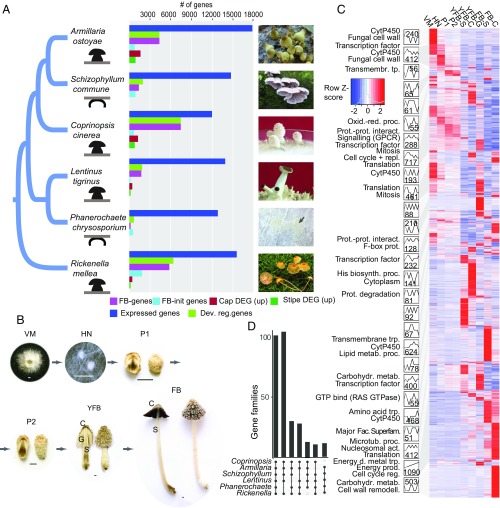
Fig. 1.
Overview of the developmental transcriptomes. (A) The distribution of developmentally regulated genes and significantly differentially expressed genes (DEGs) of cap and stipe in six mushroom-forming species (“up” denotes significantly upregulated). Fruiting bodies are shown on the right (Phanerochaete, K.K.; Rickenella, Bálint Dima; others, L.G.N.). (B) Schematic of the developmental time series data with C. cinerea as an example. C, cap; FB, fruiting body; G, spore producing gills; HN, hyphal knot; P1, stage 1 primordium; P2, stage 2 primordium; S, stipe; VM, vegetative mycelium; YFB, young fruiting body. (C) Analysis of coexpression modules in C. cinerea. Heatmap of 7,475 developmentally expressed genes is arranged based on module assignment, with simplified expression profiles and enriched GO terms (no term means no enriched GO) given for each module (see also Dataset S7). We graphically depict only 27 modules with 50 genes (refer to SI Appendix, Figs. S4–S9 for the complete list of modules and data for other species). The distribution of key developmental genes is given on the right side of the heatmap. (D) UpSetR representation of gene families developmentally regulated in at least five species (see also SI Appendix, Fig. S10).
Dynamic Reprogramming of the Fungal Transcriptome.
We detected 12,003 to 17,822 expressed genes, of which 938 to 7,605 were developmentally regulated in the six species (Fig. 1A and Dataset S5). We found 192 to 7,584 genes that showed significant expression dynamics during fruiting body development (FB development genes). Of developmentally regulated genes, 188 to 1,856 genes were upregulated at fruiting body initiation (FB-init genes), which represents a transition from simple to complex multicellular organization. Only P. chrysosporium had more FB-init genes than FB development genes, which is consistent with its fruiting bodies being among the least complex types in the Agaricomycetes. The number of genes significantly differentially expressed (DEGs) at fruiting body initiation further suggests that the transition to complex multicellularity is associated with a major reprogramming of gene expression (SI Appendix, Fig. S2). The largest numbers of DEGs were observed in cap and gill tissues in all four species with complex fruiting bodies. On the other hand, the expression profiles of stipes changed little relative to primordium stages in Armillaria, Lentinus, and Rickenella, which is explained by the completion of primordial stipe early during development in these species [epinodular development (22)], as opposed to Coprinopsis, in which stipe and cap initials develop simultaneously inside the fruiting body initial [endonodular development (22)]. Many Gene Ontology (GO) terms were partitioned between vegetative mycelium and fruiting body samples (P 0.05, FET). Terms related to FCW, oxidoreductase activity, and carbohydrate metabolism were enriched in FB-init and FB development genes of all six species (SI Appendix, Fig. S3 and Dataset S6), suggesting that cell wall remodeling is a common function during fruiting body development. Other commonly enriched terms cover functions such as DNA replication, transmembrane sugar transport, and ribosome, membrane, and lipid biosynthesis, while many others were specific to single species (SI Appendix, Fig. S3).
To obtain a higher-resolution picture of developmental events, we arranged developmentally regulated genes into coexpression modules using the Short Time-Series Expression Miner (STEM) (23). Developmentally regulated genes grouped into 28 to 40 modules, except Phanerochaete, which had 11. The largest modules in all species contained genes expressed at fruiting body initiation or in early primordia and genes with tissue-specific expression peaks, in young fruiting body caps, gills, stipes, mature fruiting bodies, and stipes or caps (Fig. 1C; only results for C. cinerea are shown; for the other species, see SI Appendix, Figs. S4–S9 and Supplementary Text). Many early-expressed modules show upregulation across multiple stages (hyphal knot, stage 1 and 2 primordia), suggestive of an early expression program overarching multiple primordium stages. Coexpression modules display distinct functional enrichment signatures, as shown in Fig. 1C and Dataset S7. For example, DNA replication and mitosis were characteristic for early-expressed modules, consistent with an early wave of nuclear and cellular division events followed by cell expansion without significant change in cell numbers (9). Growth by cell expansion is a mechanism shared with plants and possibly reflects constraints imposed by independently evolved rigid cell walls in these groups.
Splicing Patterns Associate with Development.
We reconstructed transcript isoforms across developmental stages and tissue types in the six species using region-restricted probabilistic modeling, a strategy developed for gene-dense fungal genomes (24). We found evidence of alternative splicing for 36 to 46% of the expressed genes (Dataset S9), which is significantly higher than what was reported for fungi outside the Agaricomycetes (25, 26) (1 to 8%). This transcript diversity was generated by 6,414 to 13,780 splicing events in the six species. Of the four main types of events, intron retention (44.3 to 60.5%) was the most abundant in all species, followed by alternative 3′ splice site (22.9 to 30.1%), alternative 5′ SS (15.6 to 24.1%), and exon skipping (0.8 to 2.9%) (SI Appendix, Fig. S11A), consistent with observations made on other fungi (25–27). No substantial difference in the proportion of spliced genes and of splicing events was observed across developmental stages, tissue types, or species. Nevertheless, we found that several genes with nearly constant overall expression level had developmentally regulated transcript isoforms (SI Appendix, Fig. S11 B and C). The six species had 159 to 1,278 such genes, the highest number in Rickenella (1,278) and the lowest in Phanerochaete (159) (SI Appendix, Fig. S11D and Dataset S9). Based on their expression dynamics, these transcripts potentially also contribute to development, expanding the space of developmentally regulated genes through alternative splicing.
Conserved Transcriptomic Signatures of Mushroom Development.
Our transcriptome data are particularly suited to detecting shared patterns of gene expression across species. We analyzed common functional signals in the six species by estimating the percent of developmentally regulated genes shared by all or subsets of the species based on Markov clustering (28) of protein sequences. We found 100 clusters containing developmentally regulated genes from all six species, and 196 in five species (Fig. 1D and Dataset S10). These are enriched for GO terms related to oxidation−reduction processes, oxidoreductase activity, and carbohydrate metabolism, among others, corresponding to a suite of carbohydrate active enzymes. Of the 100 families shared by six species, 15 can be linked to the FCW, while the remaining families cover diverse cellular functions such as transmembrane transport (6 families), cytochrome p450s (5 families), targeted protein degradation (5 families), or peptidases (3 families). One hundred and four gene families are shared by five species excluding Phanerochaete (Fig. 1D), which comes as no surprise, as this species produces the simple crust-like, fruiting bodies. Besides these highly conserved families, genes containing another 73 InterPro terms are developmentally regulated in six or five species but didn’t group into gene families due to their higher rate of evolution. These include most transcription factors (TFs), kinases, aquaporins, certain peptidase families, and enzymes of primary carbohydrate metabolism (trehalose and mannitol; SI Appendix, Fig. S13), among others (Dataset S10).
Shared developmentally regulated gene families included a conserved suite of CAZymes active on the main chitin and -1,3- and -1,6-glucan polymers as well as minor components of the FCW. These included various glycoside hydrolases (GH), hydrophobins, expansin-like proteins, and cerato-platanins, among others. A large suite of -glucanases, chitinases, laccases, endo--1,4-mannanases, and -1,3-mannosidases were developmentally regulated, many of which are also expanded in Agaricomycetes (SI Appendix, Table S2 and Fig. S12). The expression of glucan-, chitin-, and mannose-active enzymes is consistent with active FCW remodeling during fruiting body formation and recent reports of similar genes upregulated in the fruiting bodies of Lentinula (16, 17, 29), Flammulina (30), and Coprinopsis (31). Kre9/Knh1 homologs are developmentally regulated in all species and are overrepresented in mushroom-forming fungi (P = 1.45 , FET; Dataset S3). This family is involved in -glucan assembly in Saccharomyces and has putative signaling roles through an interaction with MAP kinases (32). Although generally linked to cellulose degradation (33, 34), expansins, lytic polysaccharide monooxygenases, and cellobiose dehydrogenases have recently been shown to target chitin polymers (35, 36) or to be expressed in fruiting bodies of Pycnoporus (37) and Flammulina (38), suggesting a role in fruiting body development. In addition, developmental expression of two alginate lyase-like families (SI Appendix, Table S2) were shared by six species, while that of a -glucuronidase (GH79_1) was shared by four species (Armillaria, Coprinopsis, Rickenella, and Lentinus). The targets of these families in fruiting bodies are currently unknown, yet their conserved expression pattern suggests roles in polysaccharide metabolism during development (39). Comparison across 201 genomes revealed that 24 of these families have undergone expansions in the Agaricomycetes (SI Appendix, Table S2 and Dataset S3). In summary, CAZymes might be responsible for producing fruiting body-specific FCW architectures, conferring adhesive properties to neighboring hyphae or plasticity for growth by cell expansion. We, therefore, suggest that FCW remodeling comprises one of the foundations of the transition to complex multicellularity during the life cycle of fungi.
A significant fraction of conserved developmentally regulated genes carry extracellular secretion signals and were predicted to be glycosylphosphatidylinositol (GPI) anchored (Fig. 2A and SI Appendix, Fig. S14). These include diverse FCW-active proteins, such as laccases (AA1), glucanases (GH5, GH16, Kre9/Knh1 family), and NodB homologs (chitooligosaccharide deacethylases), but also lectins, A1 aspartic peptidases, and sedolisins, among others (Dataset S11). Homologs of rhizobial NodB genes were developmentally regulated in all six species, whereas, in the context of ectomycorrhizal symbioses, they were discussed as potential elicitors of plant immune responses (40). GPI-anchored proteins often mediate adhesion in filamentous and pathogenic fungi (41), but it is not known whether similar mechanisms are at play in fruiting bodies (2). Laccases and glucanases could facilitate adhesion by oxidative cross-linking or other covalent modifications of neighboring hyphal surfaces, although more data are needed on the biochemistry involved. Nevertheless, it seems safe to conclude that FCW-active proteins may bind neighboring hyphae through covalent FCW modifications in fruiting bodies, which would represent a unique adhesion mechanism among complex multicellular organisms. Homologs of the dystroglycan-type cadherin-like domain containing protein of Schizophyllum cerevisiae [Axl2p (42)] were enriched in Agaricomycetes compared with other fungi (P = 1.1 , FET; Dataset S3) and were developmentally regulated in all species (Dataset S11). These proteins share significant sequence similarity with animal cadherins (Blast E value of <10−30) and, although fewer in numbers than in animals, their convergent expansion in complex multicellular fungi and metazoans could indicate recurrent cooption for developmental functions.
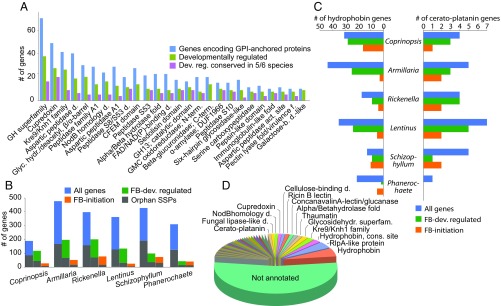
Fig. 2.
Diverse secreted proteins are developmentally regulated in fruiting bodies. (A) The distribution of GPI-anchored secreted protein repertoire, and its developmentally regulated and conserved developmentally regulated subsets. (B) Numbers of all and developmentally regulated SSPs, with orphan genes shaded differently (color legend as in Fig. 4C). (C) Copy number distribution and developmental regulation of hydrophobins and cerato-platanins in the six species. (D) Functional annotation of SSPs in the six species.
Fruiting body secretomes contained a rich suite of genes encoding small secreted proteins (SSPs, 300 amino acids, with extracellular secretion signal). Of the 190 to 477 SSPs predicted in the genomes of the six species, 20 to 61% are developmentally regulated, with 20% being conserved across the six species (Fig. 2B and SI Appendix, Fig. S15). Conserved and annotated genes comprise various FCW-related families, such as hydrophobins, cerato-platanins, cupredoxins, lectins, Kre9/Knh1, GH12 and LysM domain proteins, among others (Fig. 2C and SI Appendix, Fig. S16). Hydrophobins and cerato-platanins are SSPs that self-assemble into a rodlet layer on the cell surface, conferring hydrophobic surfaces to hyphae that hinder soaking of fruiting bodies with water. They are hypothesized to mediate adhesion, the aeration of fruiting bodies (12, 43), or pathogenicity (44). As reported previously (12), most hydrophobin genes are developmentally regulated (Fig. 2D and Dataset S10), and the family is overrepresented in the genomes of mushroom-forming fungi (P , FET; Dataset S3). Cerato-platanins are also expanded (P = 1.56 , FET; Dataset S3) and developmentally regulated (except in Phanerochaete). In addition to conserved genes, 40% of developmentally regulated SSPs had no functional annotations and/or were species-specific orphans (Fig. 2B). This proportion is similar to that observed in ectomycorrhiza-induced SSPs (5, 45) and suggests that species-specific secreted proteins have a role also in fruiting body development. Although their function in fruiting bodies is not known, their role in signaling across partners in ectomycorrhizal (45) and pathogenic interactions (46), or within species (47, 48), raises the possibility that some of the detected SSPs might act as fruiting body effectors. This could also explain the rich SSP repertoires of saprotrophic Agaricomycetes (49).
Targeted Protein Degradation Shows Striking Expansion in Mushrooms.
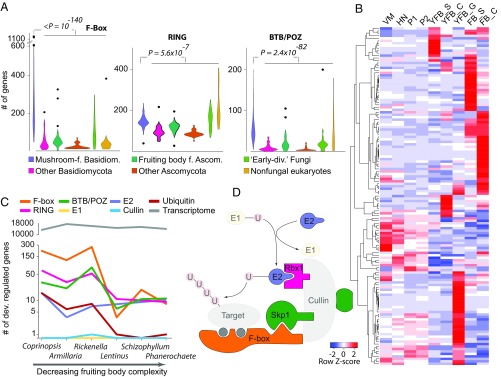
We found a strong signal for developmental expression of components of the E3 ubiquitin ligase complex. Several genes encoding F-box proteins and RING-type zinc-finger and BTB/POZ domain proteins are developmentally regulated in all species, often displaying tissue or developmental stage-specific expression peaks (Fig. 3 and SI Appendix, Figs. S17–S19). These gene families are also strongly overrepresented in the genomes of mushroom-forming fungi compared with related filamentous fungi and yeasts (Fig. 3A). For example, while yeasts and filamentous fungi possess 20 and 60 to 90 F-box proteins (50), respectively, mushroom-forming fungi have 67 to 1,199 copies (mean: 274), comparable to the numbers seen in higher plants (51) and resulting predominantly from recent tandem duplications (SI Appendix, Fig. S20). They mostly showed a single peak in expression, and many of them were upregulated at fruiting body initiation or in caps, gills, and stipes (Fig. 3B). The numbers of developmentally regulated F-box, RING-type zinc-finger, and BTB/POZ domain containing genes found in the six species show a good correlation with fruiting body complexity but a poor correlation with the number of expressed genes (Fig. 3C), suggesting a link between the expansion of these genes and the evolution of complex fruiting body morphologies. These genes define the target specificity of E3 ubiquitin ligases (50, 52), which enables a tight regulation of selective proteolysis during development (51). In plants, F-box proteins can also act as transcriptional regulators (53), although this is yet to be proven in fungi. On the other hand, ubiquitin-conjugating (E2) enzymes are developmentally regulated only in Coprinopsis, Armillaria, and Rickenella, whereas ubiquitin-activating (E1) enzymes, cullins, SKP1, and HECT-type ubiquitin ligases are neither developmentally regulated nor significantly overrepresented in mushroom-forming fungi (Fig. 3D; P 0.05, FET). With the exception of Coprinopsis, we did not detect specific expression patterns of neddylation and deneddylation genes as reported for the Ascomycota (54). Protein ubiquitylation also has a connection with programmed cell death (PCD), which seems to be limited to certain tissue types in fruiting bodies (55). We did not observe an expansion or systematic upregulation of PCD-related genes in the Agaricomycetes, suggesting that the expansion of F-box proteins is independent of the evolution of PCD in fruiting body-forming fungi. Taken together, we observed a striking expansion and distinctive expression patterns of genes that define target specificity of the E3 ubiquitin ligase complex (F-box, RING, and BTB/POZ proteins) in Agaricomycetes. This parallels F-box gene expansion in plants which, combined with their widespread role in development (51, 56) similarly to the apoptotic-like cell death and SUMOylation-related proteins, suggests that they likely have key roles in complex multicellular development in mushroom-forming fungi.
Fig. 3.
Expansion and developmental regulation of selective protein degradation pathways. (A) Copy number distribution of F-box, RING-type zinc finger, and BTB/POZ domain proteins showing their enrichment in mushroom-forming fungi compared with other analyzed organisms among 200 genomes. (B) Heatmap of developmentally regulated F-box proteins in C. cinerea. Genes were hierarchically clustered based on gene expression similarity using average linkage clustering. (C) The correlation between morphological complexity of fruiting bodies and the number of developmentally regulated elements of the E3 ligase complex. (D) Outline of the E3 ubiquitin ligase complex highlighting members expanded and developmentally regulated (solid) in mushroom-forming fungi. Transparent members are not expanded nor developmentally regulated.
Key Multicellularity-Related Genes Are Developmentally Regulated in Fruiting Bodies.
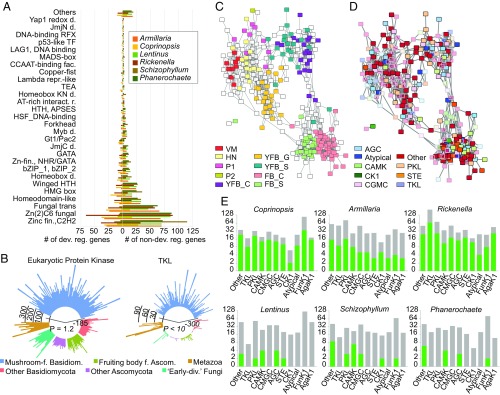
Complex multicellularity in fungi is implemented by the reprogramming of hyphal branching patterns, followed by their adhesion and differentiation (2). This assumes mechanisms for cell-to-cell communication, adhesion, differentiation, and defense. We examined the expression dynamics of gene families related to these traits, including TFs, protein kinases, adhesion and defense-related genes. Like other complex multicellular lineages, mushroom-forming fungi make extensive use of TFs in development. To identify development-related TFs, we manually curated TF candidate genes to exclude ones that nonspecifically bind DNA. The resulting TFomes contain 278 to 408 genes, of which 4.5 to 64% were developmentally regulated (SI Appendix, Figs. S21 and S22). These were dominated by C2H2 and Zn(2)C6 fungal type, fungal trans, and homeodomain-like TFs (Fig. 4A). Although TF families were usually not conserved, we found five TF families that contained developmentally regulated genes from five or six species (Dataset S10). These included C2H2-type zinc fingers [including c2h2 of Schizophyllum (18, 57)], and Zn(2)-C6 fungal-type and homeobox TFs [containing hom1 of Schizophyllum (18, 57)]. Two clusters of C2H2 and homeobox TFs showed expression peaks in stipes of Coprinopsis, Lentinus, Armillaria, and Rickenella, confirming previous reports of Hom1 expression in Coprinopsis (58) and Schizophyllum (57, 59). Members of the light receptor white collar complex were developmentally regulated in all species except Phanerochaete, mostly showing a significant increase in expression at initiation. However, these genes did not group into one family in the clustering, which was a common pattern for TF families, perhaps caused by their high rate of sequence evolution.
Fig. 4.
Expression and copy number distribution of TF and kinase genes. (A) TF family distribution and the proportions of developmentally regulated (left) versus not regulated (right) genes across six species. (B) Circular bar diagram of eukaryotic protein kinase (Left) and TKL (Right) repertoires of mushroom-forming fungi versus in other fungal groups and Metazoa. P values of overrepresentation in mushroom-forming fungi are given for both plots. (C and D) Coexpression network of 306 expressed kinases of C. cinerea overlaid with (C) expression peak and (D) kinase classification. Pairwise correlation coefficients of expression profile similarity were calculated for each pair of kinase genes, and networks were visualized with a correlation coefficient cutoff of 0.825. (E) Kinome (gray) and developmentally regulated kinome (green) repertoires of the six species split by kinase classification. The families Funk1 and Agak1 of the “Other” group are shown separately. Axes are transformed to scale.
Communication among cells by various signaling pathways is paramount to the increase of distinct cell types in evolution. In accordance with the higher complexity of mushroom-forming fungi, their kinomes are significantly larger than those of other fungi (P = 1.1 , FET; Fig. 4B), due to expansions of the eukaryotic protein kinase superfamily. We classified protein kinases into nine groups (60) using published kinome classifications for Coprinopsis (7). The six mushroom kinomes (Dataset S12) have a similar composition, with PKL, CMGC, and CAMK families being most diverse and RGC and tyrosine kinases missing. The Agaricomycetes-specific FunK1 family is expanded significantly, as reported earlier (7). Tyrosine kinase-like (TKL) kinases also show a strong expansion in Agaricomycetes (P ), consistent with observations in Laccaria bicolor (61) and other species (62). Histidine kinases are underrepresented (P = 8.95 ) relative to other fungal groups.
A kinase coexpression network revealed tissue specificity as the main driver of network topology (Fig. 4 C and D), with most kinases showing an expression peak late in development (SI Appendix, Figs. S23–S25). Kinases with early expression peaks are mostly highly expressed through multiple stages, resembling early expressed modules of Coprinopsis (Fig. 1C and SI Appendix, Figs. S23–S25). Many CAMK family members showed expression peaks in cap and gill tissues. However, overlaying the network with classification shows no general enrichment of any family in developmental stages or tissues (Fig 4 C–E), indicating diverse cooption events for development at the highest level of kinase classification. The FunK1 family has been linked to multicellular development, based on an upregulation in fruiting bodies (7). In our species, FunK1 comprises 4 to 33% of the kinome and 0.2 to 35% of the developmentally regulated kinases, although this figure resembles that of other kinase families. Members of the highly expanded TKL family (Fig. 4E) are developmentally regulated in Coprinopsis, Armillaria, and Rickenella, but not in species with simpler morphologies. The expansion and developmental expression of the TKL family in mushroom-forming fungi is a remarkable case of convergence with complex multicellular plants (1, 63) [which is distinct from the TK expansion of animals (64)] and may be related to parallel increases in organismal complexity. However, while most plant TKL genes have receptor-like architectures (65), we found no evidence of extracellular domains or secretion signals in TKL and other Ser/Thr kinase genes of mushroom-forming fungi, suggesting they orchestrate signal transduction via soluble kinases or other mechanisms different from those of multicellular plants and animals.
Fungal immune systems comprise innate chemical defense mechanisms against metazoan predators as well as bacterial and fungal infections (66). We cataloged 11 families of defense effector proteins and their expression to assess the conservation of the defensive arsenal of Agaricomycetes. Genomes of mushroom-forming fungi harbor highly species-specific combinations of defense-related genes encoding pore-forming toxins, cerato-platanins, lectins, and copsins, among others, with most of them being developmentally expressed and upregulated at fruiting body initiation (SI Appendix, Fig. S26). Of the 11 families, only 3 were conserved, and only 1 (thaumatins) was developmentally regulated in all six species, which can display either endoglucanase or antimicrobial activity, depending on the structure of the mature protein. In silico structure prediction identified an acidic cleft in fruiting body expressed thaumatins (SI Appendix, Fig. S26), consistent with an antimicrobial activity. Several defense-related lectins have been reported from fruiting bodies (67), although lectins have been implicated in cell adhesion and signaling too. Agaricomycete genomes encode at least 17 lectin and 2 lectin-like families, of which 7 are significantly overrepresented (P 0.05, FET; Dataset S3). Developmentally regulated lectins belong to nine families, with four to seven families per species, but only ricin B lectins were developmentally regulated in all six species (SI Appendix, Fig. S27). Other lectin families show a patchy phylogenetic distribution, which is also reflected in their expression patterns in fruiting bodies. Several lectins are induced at fruiting body initiation, including all previously reported nematotoxic Coprinopsis lectins (CCL1-2, CGL1-3, and CGL3). Taken together, the defense effector and lectin-encoding arsenal of mushroom-forming fungi shows a patchy phylogenetic distribution, consistent with high gene turnover rates or gains via horizontal gene transfer (66). Accordingly, expressed defense gene sets are highly species-specific, with most of the encoded genes upregulated in fruiting bodies, suggesting that chemical defense is a key fruiting body function.
Most Developmental Gene Families Are Older than Fruiting Body Formation.
We investigated the evolutionary age distribution of developmentally regulated genes using phylostratigraphy (68), and gene tree−species tree reconciliation analyses. We assigned genes to phylogenetic ages, “phylostrata,” by identifying for each gene the most phylogenetically distant species in which a homolog could be detected. The phylostratigraphic profiles of all species show three peaks, corresponding to two major periods of fungal gene origin: the first containing genes shared by all living species, the second containing genes shared by the Dikarya (Ascomycota + Basidiomycota), and the third containing species-specific genes (SI Appendix, Fig. S28). In terms of gene duplications and losses, we observed two major expansion in 292 shared developmentally regulated gene families, one in the most recent common ancestor (mrca) of the Dikarya and the other from the mrca of Agaricomycetes and Dacrymycetes extending to basal nodes of the Agaricomycetes (SI Appendix, Fig. S29). Notably, the expansion in the Agaricomycetes roughly correlates with the origin of fruiting body development in this class. These data suggest that many developmentally regulated genes have homologs in simple multicellular or unicellular organisms: The origin of 83.3% predate the origin of mushroom-forming fungi (SI Appendix, Fig. S28), indicating that several conserved gene families were recruited for fruiting body development during evolution. Nevertheless, Agaricomycetes-specific phylostrata showed a characteristic enrichment for F-box genes, TFs, and protein kinases, indicating an increased rate of origin for these in mushroom-forming fungi (Dataset S8).
Discussion
We charted the transcriptomic landscape of multicellular development in six phylogenetically diverse mushroom-forming species and performed comparative analyses of 200 genomes. We pinpointed nearly 300 conserved gene families, and another 73 gene groups with developmentally dynamic expression in five or more species, as well as 631 domains significantly overrepresented in mushroom-forming fungi. These are enriched in cell wall-modifying enzymes, various secreted proteins (including GPI-anchored and SSPs), components of the ubiquitin ligase complex, kinases, or TFs. Lectins and defense effectors, on the other hand, showed species-specific repertoires, indicating a higher rate of evolutionary turnover. These data provide a framework for elucidating the core genetic program of fruiting body formation and will serve as guideposts for a systems approach to understanding the genetic bases of mushroom development and multicellularity.
Complex multicellularity evolved in five lineages, of which plants, animals, and fungi are the most diverse (1, 2). At the broadest level of comparison, all these lineages evolved solutions to cell adhesion, communication, long-range transport, and differentiation, although the exact mechanisms often differ among lineages (1–3). As in animals and plants, protein kinases, putative adhesive proteins, defense effectors, and certain TFs have expanded repertoires in mushroom-forming fungi and show developmentally dynamic expression patterns. Examples for convergent expansions in mushroom-forming fungi, plants, and/or animals include TKL family kinases, F-box proteins, and cadherin-like proteins, indicating that ancient eukaryotic gene families with apt biochemical properties have been repeatedly coopted for complex multicellularity during evolution. F-box proteins showed the largest expansion in Agaricomycetes across all gene families and were the largest developmentally regulated family, along with RING-type and BTB domain proteins. Among fruiting body-forming fungi (2, 54), Agaricomycetes share some similarity with the Pezizomycotina (Ascomycota), many of which also produce macroscopic fruiting bodies. For example, laccases, lectins, several TFs, and signal transduction systems have also been implicated in fruiting body formation in the Pezizomycotina, although, at the moment, it is unclear whether the Pezizomycotina shares a complex multicellular ancestor with the Agaricomycetes (2). Comparisons of developmental genes and transcriptomes across the Agaricomycetes and the Pezizomycotina will be necessary to elucidate whether these two groups share a single origin of or represent independent acquisitions of complex multicellularity.
Mushroom-forming fungi also show several unique solutions for multicellularity, as expected based on their independent evolutionary origin. These are, in part, explained by the very nature of fungi: Complex multicellularity comprises the reproductive phase of the life cycle (except in sclerotia and rhizomorphs) and so mechanisms have evolved for sensing when fruiting body formation is optimal (e.g., nutrient availability, light). Mushroom development can be partitioned into an early phase of cell proliferation and differentiation and a growth phase of rapid increase in cell size, a division evident on our gene coexpression profiles as well. Broadly speaking, this is similar to the development of fleshy plant fruits, although mechanisms are likely to be different.
This work has provided a glimpse into the core genetic toolkit of complex multicellularity in mushroom-forming fungi. Our comparative transcriptomic and genomic analyses revealed several gene families with conserved developmental expression in fruiting bodies, with scope to increase the resolution both phylogenetically and among cell types (e.g., by single-cell RNA-Seq). Such data should help define the conserved genetic programs underlying multicellularity in mushroom-forming fungi, and uncover the evolutionary origins of a major complex multicellular lineage in the eukaryotes.
Materials and Methods
For additional methods, see SI Appendix. We analyzed developmental transcriptomes of C. cinerea AmutBmut, S. commune H4-8, P. chrysosporium RP78, R. mellea SZMC22713, L. tigrinus RLG9953-sp, and A. ostoyae C18/9. Gene expression profiling was used to identify developmentally regulated genes; these were characterized by Interpro domains, and their enrichment was analyzed across 201 fungal genomes. To reconstruct transcript isoforms, we used region-restricted probabilistic modeling (24). Genes encoding putative carbohydrate-active enzymes were annotated using the CAZy pipeline. Prediction of GPI-anchored proteins was performed using Pred-GPI. TFs were identified based on Interpro domains with sequence-specific DNA-binding activity. Kinases were predicted based on InterPro domain composition. We excluded classical kinases involved in metabolism. Prediction of SSPs was performed using a modified version of the pipeline of Pellegrin et al. (49). Developmentally regulated genes were clustered into coexpression modules using STEM v1.3.11 (23). Phylostratigraphy and gene tree−species tree reconciliation analysis was performed to investigate the evolutionary age distribution of developmentally regulated genes.
Data Availability
Genome assembly and annotation of R. mellea was deposited in the National Center for Biotechnology Information BioProject database (accession no. PRJNA334780). A Gene Expression Omnibus (GEO) archive of the sequenced transcriptome libraries was deposited in the NCBI’s GEO Archive at www.ncbi.nlm.nih.gov/geo under accession GSE125200 (individual accessions: GSE125184, Coprinopsis; GSE125190, Lentinus; GSE125195, Rickenella; GSE125198, Schizophyllum; GSE125199, Phanerochaete).
Supplementary Material
Acknowledgments
The authors thank Daniel Cullen and Jill Gaskell (US Department of Agriculture) for the Phanerochaete strain used in this study. This work was supported by Momentum Program of the Hungarian Academy of Sciences Contract LP2014/12, by National Research, Development and Innovation Office Grant GINOP-2.3.2-15-2016-00001, and by the European Research Council under the Horizon 2020 research and innovation programme Grant Agreements 758161 and 716132. The work by the US Department of Energy (DOE) Joint Genome Institute, a DOE Office of Science User Facility, is supported by the Office of Science of the US DOE under Contract DE-AC02-05CH11231.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Genome assembly and annotation of Rickenella mellea was deposited in the National Center for Biotechnology Information BioProject database (accession no. PRJNA334780). A Gene Expression Omnibus (GEO) archive of the sequenced transcriptome libraries was deposited in the NCBI’s GEO Archive at www.ncbi.nlm.nih.gov/geo (accession no. GSE125200).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1817822116/-/DCSupplemental.
References
- 1.Knoll AH. The multiple origins of complex multicellularity. Earth Planet Sci. 2011;39:217–239. [Google Scholar]
- 2.Nagy LG, Kovács GM, Krizsán K. Complex multicellularity in fungi: Evolutionary convergence, single origin, or both? Biol Rev. 2018;93:1778–1794. doi: 10.1111/brv.12418. [DOI] [PubMed] [Google Scholar]
- 3.Cock JM, et al. The Ectocarpus genome and the independent evolution of multicellularity in brown algae. Nature. 2010;465:617–621. doi: 10.1038/nature09016. [DOI] [PubMed] [Google Scholar]
- 4.Sebé-Pedrós A, Degnan BM, Ruiz-Trillo I. The origin of Metazoa: A unicellular perspective. Nat Rev Genet. 2017;18:498–512. doi: 10.1038/nrg.2017.21. [DOI] [PubMed] [Google Scholar]
- 5.Kohler A, et al. 2015. Convergent losses of decay mechanisms and rapid turnover of symbiosys genes in mycorrhizal mutualists. Nat Genet, 47:410–415.
- 6.Kalaras MD, Richie JP, Calcagnotto A, Beelman RB. Mushrooms: A rich source of the antioxidants ergothioneine and glutathione. Food Chem. 2017;233:429–433. doi: 10.1016/j.foodchem.2017.04.109. [DOI] [PubMed] [Google Scholar]
- 7.Stajich JE, et al. Insights into evolution of multicellular fungi from the assembled chromosomes of the mushroom Coprinopsis cinerea (Coprinus cinereus) Proc Natl Acad Sci USA. 2010;107:11889–11894. doi: 10.1073/pnas.1003391107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ohm RA, et al. Genome sequence of the model mushroom Schizophyllum commune. Nat Biotechnol. 2010;28:957–963. doi: 10.1038/nbt.1643. [DOI] [PubMed] [Google Scholar]
- 9.Kües U. Life history and developmental processes in the basidiomycete Coprinus cinereus. Microbiol Mol Biol Rev. 2000;64:316–353. doi: 10.1128/mmbr.64.2.316-353.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kües U, Navarro-González M. How do Agaricomycetes shape their fruiting bodies? 1. Morphological aspects of development. Fungal Biol Rev. 2015;29:63–97. [Google Scholar]
- 11.Van Peer AF, et al. The septal pore cap is an organelle that functions in vegetative growth and mushroom formation of the wood-rot fungus Schizophyllum commune. Environ Microbiol. 2009;12:833–844. doi: 10.1111/j.1462-2920.2009.02122.x. [DOI] [PubMed] [Google Scholar]
- 12.Bayry J, Aimanianda V, Guijarro JI, Sunde M, Latgé JP. Hydrophobins-unique fungal proteins. PLoS Pathog. 2012;8:e1002700. doi: 10.1371/journal.ppat.1002700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Plaza DF, Lin CW, van der Velden NS, Aebi M, Künzler M. Comparative transcriptomics of the model mushroom Coprinopsis cinerea reveals tissue-specific armories and a conserved circuitry for sexual development. BMC Genomics. 2014;15:492. doi: 10.1186/1471-2164-15-492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Buser R, Lazar Z, Käser S, Künzler M, Aebi M. Identification, characterization, and biosynthesis of a novel N-glycan modification in the fruiting body of the basidiomycete Coprinopsis cinerea. J Biol Chem. 2010;285:10715–10723. doi: 10.1074/jbc.M109.076075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ohga S, Cho N-S, Thurston CF, Wood DA. Transcriptional regulation of laccase and cellulase in relation to fruit body formation in the mycelium of Lentinula edodes on a sawdust-based substrate. Mycoscience. 2000;41:149–153. [Google Scholar]
- 16.Konno N, Sakamoto Y. An endo--1,6-glucanase involved in Lentinula edodes fruiting body autolysis. Appl Microbiol Biotechnol. 2011;91:1365–1373. doi: 10.1007/s00253-011-3295-2. [DOI] [PubMed] [Google Scholar]
- 17.Sakamoto Y, et al. Lentinula edodes tlg1 encodes a thaumatin-like protein that is involved in lentinan degradation and fruiting body senescence. Plant Physiol. 2006;141:793–801. doi: 10.1104/pp.106.076679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ohm RA, de Jong JF, de Bekker C, Wösten HAB, Lugones LG. Transcription factor genes of Schizophyllum commune involved in regulation of mushroom formation. Mol Microbiol. 2011;81:1433–1445. doi: 10.1111/j.1365-2958.2011.07776.x. [DOI] [PubMed] [Google Scholar]
- 19.Corrochano LM. Fungal photoreceptors: Sensory molecules for fungal development and behaviour. Photochem Photobiol Sci. 2007;6:725–736. doi: 10.1039/b702155k. [DOI] [PubMed] [Google Scholar]
- 20.Sipos G, et al. Genome expansion and lineage-specific genetic innovations in the forest pathogenic fungi Armillaria. Nat Ecol Evol. 2017;1:1931–1941. doi: 10.1038/s41559-017-0347-8. [DOI] [PubMed] [Google Scholar]
- 21.Wu B, et al. Genomics and development of Lentinus tigrinus: A white-rot wood-decaying mushroom with dimorphic fruiting bodies. Genome Biol Evol. 2018;10:3250–3261. doi: 10.1093/gbe/evy246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Clémencon H, Emmett V, Emmett EE. Cytology and Plectology of the Hymenomycetes with 12 Tables. Cramer Gebr Borntraeger Verl Buchh; Stuttgart: 2012. [Google Scholar]
- 23.Ernst J, Bar-Joseph Z. STEM: A tool for the analysis of short time series gene expression data. BMC Bioinformatics. 2006;7:191. doi: 10.1186/1471-2105-7-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gehrmann T, et al. Schizophyllum commune has an extensive and functional alternative splicing repertoire. Sci Rep. 2016;6:33640. doi: 10.1038/srep33640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xie B-B, et al. Deep RNA sequencing reveals a high frequency of alternative splicing events in the fungus Trichoderma longibrachiatum. BMC Genomics. 2015;16:54. doi: 10.1186/s12864-015-1251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang B, et al. Survey of the transcriptome of Aspergillus oryzae via massively parallel mRNA sequencing. Nucleic Acids Res. 2010;38:5075–5087. doi: 10.1093/nar/gkq256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gordon SP, et al. Widespread polycistronic transcripts in fungi revealed by single-molecule mRNA sequencing. PLoS One. 2015;10:e0132628. doi: 10.1371/journal.pone.0132628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Enright AJ, Van Dongen S, Ouzounis CA. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 2002;30:1575–1584. doi: 10.1093/nar/30.7.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sakamoto Y, Nakade K, Konno N. Endo--1,3-Glucanase GLU1, from the fruiting body of Lentinula edodes, belongs to a new glycoside hydrolase family. Appl Environ Microbiol. 2011;77:8350–8354. doi: 10.1128/AEM.05581-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fukuda K, et al. Purification and characterization of a novel exo-beta-1,3-1,6-glucanase from the fruiting body of the edible mushroom Enoki (Flammulina velutipes) Biosci Biotechnol Biochem. 2008;72:3107–3113. doi: 10.1271/bbb.80213. [DOI] [PubMed] [Google Scholar]
- 31.Zhou Y, Zhang W, Liu Z, Wang J, Yuan S. Purification, characterization and synergism in autolysis of a group of 1,3--glucan hydrolases from the pilei of Coprinopsis cinerea fruiting bodies. Microbiol. 2015;161:1978–1989. doi: 10.1099/mic.0.000143. [DOI] [PubMed] [Google Scholar]
- 32.Szeto CY, Leung GS, Kwan HS. Le.MAPK and its interacting partner, Le.DRMIP, in fruiting body development in Lentinula edodes. Gene. 2007;393:87–93. doi: 10.1016/j.gene.2007.01.030. [DOI] [PubMed] [Google Scholar]
- 33.Kersten P, Cullen D. Copper radical oxidases and related extracellular oxidoreductases of wood-decay Agaricomycetes. Fungal Genet Biol. 2014;72:124–130. doi: 10.1016/j.fgb.2014.05.011. [DOI] [PubMed] [Google Scholar]
- 34.Rytioja J, et al. Plant-polysaccharide-degrading enzymes from basidiomycetes. Microbiol Mol Biol Rev. 2014;78:614–649. doi: 10.1128/MMBR.00035-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tovar-Herrera OE, et al. A novel expansin protein from the white-rot fungus Schizophyllum commune. PLoS One. 2015;10:e0122296. doi: 10.1371/journal.pone.0122296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lenfant N, et al. A bioinformatics analysis of 3400 lytic polysaccharide oxidases from family AA9. Carbohydr Res. 2017;448:166–174. doi: 10.1016/j.carres.2017.04.012. [DOI] [PubMed] [Google Scholar]
- 37.Temp U, Eggert C. Novel interaction between laccase and cellobiose dehydrogenase during pigment synthesis in the white rot fungus Pycnoporus cinnabarinus. Appl Environ Microbiol. 1999;65:389–395. doi: 10.1128/aem.65.2.389-395.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Osińska-Jaroszuk M, et al. Complex biochemical analysis of fruiting bodies from newly isolated Polish Flammulina velutipes strains. Polish J Microbiol. 2016;65:295–305. doi: 10.5604/17331331.1215609. [DOI] [PubMed] [Google Scholar]
- 39.Ji J, Moore D. Glycogen metabolism in relation to fruit body maturation in Coprinus cinereus. Mycol Res. 1993;97:283–289. [Google Scholar]
- 40.Garcia K, Delaux P-M, Cope KR, Ané J-M. Molecular signals required for the establishment and maintenance of ectomycorrhizal symbioses. New Phytol. 2015;208:79–87. doi: 10.1111/nph.13423. [DOI] [PubMed] [Google Scholar]
- 41.Dranginis AM, Rauceo JM, Coronado JE, Lipke PN. A biochemical guide to yeast adhesins: Glycoproteins for social and antisocial occasions. Microbiol Mol Biol Rev. 2007;71:282–294. doi: 10.1128/MMBR.00037-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dickens NJ, Beatson S, Ponting CP. Cadherin-like domains in -dystroglycan, /-sarcoglycan and yeast and bacterial proteins. Curr Biol. 2002;12:R197–R199. doi: 10.1016/s0960-9822(02)00748-0. [DOI] [PubMed] [Google Scholar]
- 43.Lugones LG, et al. Hydrophobins line air channels in fruiting bodies of Schizophyllum commune and Agaricus bisporus. Mycol Res. 1999;103:635–640. [Google Scholar]
- 44.Gaderer R, Bonazza K, Seidl-Seiboth V. Cerato-platanins: A fungal protein family with intriguing properties and application potential. Appl Microbiol Biotechnol. 2014;98:4795–4803. doi: 10.1007/s00253-014-5690-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Martin F, Kohler A, Murat C, Veneault-Fourrey C, Hibbett DS. Unearthing the roots of ectomycorrhizal symbioses. Nat Rev Microbiol. 2016;14:760–773. doi: 10.1038/nrmicro.2016.149. [DOI] [PubMed] [Google Scholar]
- 46.Stergiopoulos I, de Wit PJGM. Fungal effector proteins. Annu Rev Phytopathol. 2009;47:233–263. doi: 10.1146/annurev.phyto.112408.132637. [DOI] [PubMed] [Google Scholar]
- 47.Feldman D, Kowbel DJ, Glass NL, Yarden O, Hadar Y. A role for small secreted proteins (SSPs) in a saprophytic fungal lifestyle: Ligninolytic enzyme regulation in Pleurotus ostreatus. Sci Rep. 2017;7:14553. doi: 10.1038/s41598-017-15112-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang L, Tian X, Gyawali R, Lin X. Fungal adhesion protein guides community behaviors and autoinduction in a paracrine manner. Proc Natl Acad Sci USA. 2013;110:11571–11576. doi: 10.1073/pnas.1308173110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pellegrin C, Morin E, Martin FM, Veneault-Fourrey C. Comparative analysis of secretomes from ectomycorrhizal fungi with an emphasis on small-secreted proteins. Front Microbiol. 2015;6:1278. doi: 10.3389/fmicb.2015.01278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu TB, Xue C. The ubiquitin-proteasome system and F-box proteins in pathogenic fungi. Mycobiology. 2011;39:243–248. doi: 10.5941/MYCO.2011.39.4.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xu G, Ma H, Nei M, Kong H. Evolution of F-box genes in plants: Different modes of sequence divergence and their relationships with functional diversification. Proc Natl Acad Sci USA. 2009;106:835–840. doi: 10.1073/pnas.0812043106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Metzger MB, Pruneda JN, Klevit RE, Weissman AM. RING-type E3 ligases: Master manipulators of E2 ubiquitin-conjugating enzymes and ubiquitination. Biochim Biophys Acta-Mol Cel Res. 2014;1843:47–60. doi: 10.1016/j.bbamcr.2013.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chae E, Tan QK-G, Hill TA, Irish VF. An Arabidopsis F-box protein acts as a transcriptional co-factor to regulate floral development. Development. 2008;135:1235–1245. doi: 10.1242/dev.015842. [DOI] [PubMed] [Google Scholar]
- 54.Pöggeler S, Nowrousian M, Teichert I, Beier A, Kück U. Physiology and Genetics. Springer Int; Cham, Switzerland: 2018. Fruiting-body development in ascomycetes; pp. 1–56. [Google Scholar]
- 55.Lu BC. Cell degeneration and gill remodelling during basidiocarp development in the fungus Coprinus cinereus. Can J Bot. 1991;69:1161–1169. [Google Scholar]
- 56.Dharmasiri N, et al. Plant development is regulated by a family of auxin receptor F box proteins. Dev Cel. 2005;9:109–119. doi: 10.1016/j.devcel.2005.05.014. [DOI] [PubMed] [Google Scholar]
- 57.Pelkmans JF, et al. The transcriptional regulator c2h2 accelerates mushroom formation in Agaricus bisporus. Appl Microbiol Biotechnol. 2016;100:7151–7159. doi: 10.1007/s00253-016-7574-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Muraguchi H, et al. Strand-specific RNA-seq analyses of fruiting body development in Coprinopsis cinerea. PLoS One. 2015;10:e0141586. doi: 10.1371/journal.pone.0141586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pelkmans JF, et al. Transcription factors of Schizophyllum commune involved in mushroom formation and modulation of vegetative growth. Sci Rep. 2017;7:310. doi: 10.1038/s41598-017-00483-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 61.Kosti I, Mandel-Gutfreund Y, Glaser F, Horwitz BA. Comparative analysis of fungal protein kinases and associated domains. BMC Genomics. 2010;11:133. doi: 10.1186/1471-2164-11-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhao Z, Jin Q, Xu JR, Liu H. Identification of a fungi-specific lineage of protein kinases closely related to tyrosine kinases. PLoS One. 2014;9:e89813. doi: 10.1371/journal.pone.0089813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lehti-Shiu MD, Shiu S-H. Diversity, classification and function of the plant protein kinase superfamily. Philos Trans R Soc B Biol Sci. 2012;367:2619–2639. doi: 10.1098/rstb.2012.0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Suga H, Torruella G, Burger G, Brown MW, Ruiz-Trillo I. Earliest Holozoan expansion of phosphotyrosine signaling. Mol Biol Evol. 2014;31:517–528. doi: 10.1093/molbev/mst241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shiu SH, Bleecker AB. Plant receptor-like kinase gene family: Diversity, function, and signaling. Sci Signaling. 2001;2001:re22. doi: 10.1126/stke.2001.113.re22. [DOI] [PubMed] [Google Scholar]
- 66.Künzler M. Hitting the sweet spot-glycans as targets of fungal defense effector proteins. Molecules. 2015;20:8144–8167. doi: 10.3390/molecules20058144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schubert M, et al. Plasticity of the -trefoil protein fold in the recognition and control of invertebrate predators and parasites by a fungal defence system. PLoS Pathog. 2012;8:e1002706. doi: 10.1371/journal.ppat.1002706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Domazet-Lošo T, Brajković J, Tautz D. A phylostratigraphy approach to uncover the genomic history of major adaptations in metazoan lineages. Trends Genet. 2007;23:533–539. doi: 10.1016/j.tig.2007.08.014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Genome assembly and annotation of R. mellea was deposited in the National Center for Biotechnology Information BioProject database (accession no. PRJNA334780). A Gene Expression Omnibus (GEO) archive of the sequenced transcriptome libraries was deposited in the NCBI’s GEO Archive at www.ncbi.nlm.nih.gov/geo under accession GSE125200 (individual accessions: GSE125184, Coprinopsis; GSE125190, Lentinus; GSE125195, Rickenella; GSE125198, Schizophyllum; GSE125199, Phanerochaete).