Abstract
Aminoacyl-tRNA synthetases (ARSs) are enzymes that ligate their cognate amino acids to tRNAs for protein synthesis. However, recent studies have shown that their functions are expanded beyond protein synthesis through the interactions with diverse cellular factors. In this review, we discuss how ARSs have evolved to expand and control their functions by forming protein assemblies. We particularly focus on a macromolecular ARS complex in eukaryotes, named multi-tRNA synthetase complex (MSC), which is proposed to provide a channel through which tRNAs reach bound ARSs to receive their cognate amino acid and transit further to the translation machinery. Approximately half of the ARSs assemble into the MSC through cis-acting noncatalytic domains attached to their catalytic domains and trans-acting factors. Evolution of the MSC included its functional expansion, during which the MSC interaction network was augmented by additional cellular pathways present in higher eukaryotes. We also discuss MSC components that could be functionally involved in the pathophysiology of tumorigenesis. For example, the activities of some trans-acting factors have tumor-suppressing effects or maintain DNA integrity and are functionally compromised in cancer. On the basis of Gene Ontology analyses, we propose that the regulatory activities of the MSC-associated ARSs mainly converge on five biological processes, including mammalian target of rapamycin (mTOR) and DNA repair pathways. Future studies are needed to investigate how the MSC-associated and free-ARSs interact with each other and other factors in the control of multiple cellular pathways, and how aberrant or disrupted interactions in the MSC can cause disease.
Keywords: aminoacyl tRNA synthetase, protein synthesis, intracellular processing, pathology, cancer biology, Aminoacyl-tRNA synthetases, Cancer, multi-tRNA synthetase complex, network analysis, protein–protein interaction
Introduction
In light of their integral catalytic roles that link the genetic code to protein (1–3), aminoacyl-tRNA synthetases (ARSs)5 are thought to have ancient origins and to have evolved to meet the demands of accurate protein synthesis and complex system development. During evolution, the ARSs have progressively adopted additional domains that mediate interactions with cellular factors involved in functions beyond protein synthesis, while keeping their catalytic domains relatively well conserved (4–6). Thus, these acquired domains appear to be responsible for the functional expansion of the attached enzymes.
Eukaryotic ARSs fall into two groups based on their capability to associate with a multi-tRNA synthetase complex (MSC) or to remain free (7, 8). The mammalian MSC consists of nine cytoplasmic ARSs (glutamyl-, prolyl-, isoleucyl-, leucyl-, methionyl-, glutaminyl-, lysyl-, arginyl-, aspartyl-tRNA synthetase) and three nonenzyme components (ARS-interacting multifunctional proteins (AIMPs) 1–3) (9, 10). Herein, we will refer to the MSC components as “MSC–ARSs,” and those ARSs not localized to the MSC as “free-ARSs.” The formation of the MSC appears to facilitate specific amino acid charging to the incoming tRNAs (11, 12) and delivery of the charged tRNAs to the protein synthesis machinery (13–15).
ARSs first bind ATP and their corresponding amino acid to form an aminoacyl-adenylate, releasing PPi. The aminoacyl-adenylate–enzyme complex then binds the substrate tRNA, and the amino acid is transferred from the amino acid–AMP to either the 2′-OH or the 3′-OH at the 3′-end of tRNA. Once the tRNA is charged, the amino acid is transferred from the tRNA to ribosome onto a growing peptide, guided by the genetic code.
Although the structure and function of the mammalian MSC in its entirety remain enigmatic, a few subcomplex structures recently have been revealed (16, 17). Interestingly, the MSC components dissociate from the complex upon various stresses and stimuli and interact with cellular factors to delineate their noncatalytic functions (5, 6).
Although the MSC is not present in prokaryotes, a primitive form of the complex consisting of glutamyl-(ERS) and methionyl-tRNA synthetase (MRS) and an auxiliary factor (Arc1p) has been detected in Saccharomyces cerevisiae (18). These findings suggest an evolutionary expansion in the size of the MSC and the interaction network among its components throughout evolution. In addition to tRNA charging, the MSC components have been shown to play crucial roles in fundamental biological processes such as transcription, translation, DNA repair, and various signaling pathways in extra- as well as intracellular space (19–21). In this review, we describe some of the MSC components that have been previously shown to positively or negatively control tumorigenesis in different pathways. We also present an evolutionary analysis of the MSC from yeast to mammals to gain a more systematic view of the MSC's role in the control of cancer-related pathways. Considering the significance of the mTOR and DNA-repair pathways in cancer, our review also highlights additional MSC components that could be involved in the regulation of these pathways.
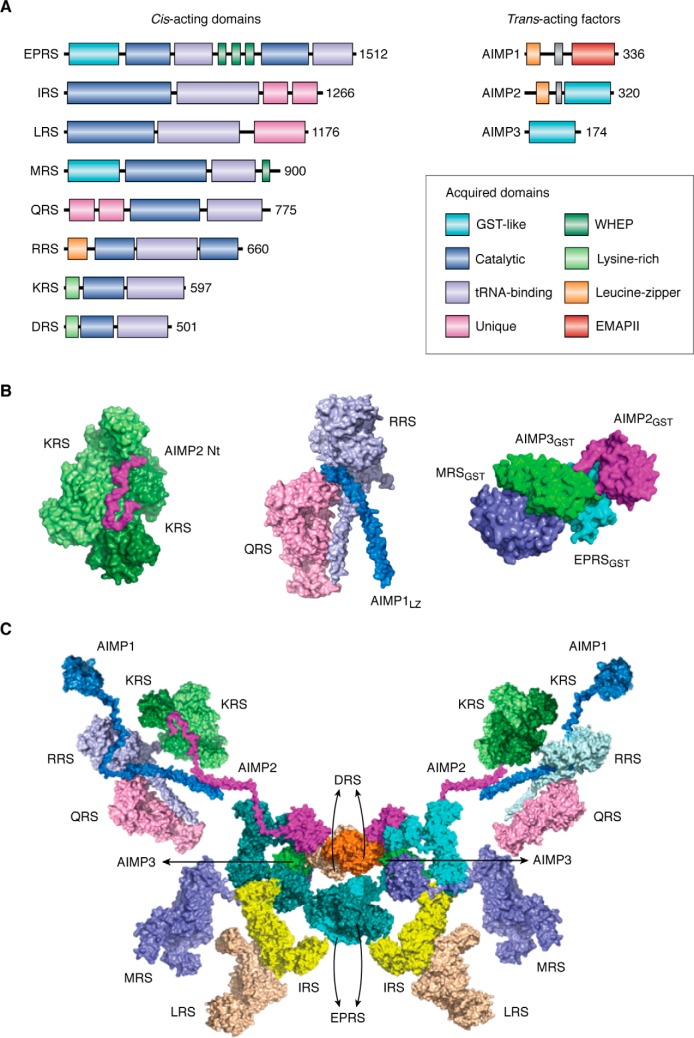
Formation of the MSC
Several ARSs assemble to form the MSC through cis-acting noncatalytic domains attached to their catalytic domains as well as the three trans-acting factors, AIMP1, AIMP2, and AIMP3 (22–26). For instance, GST-homology domains were inserted to the EPRS (glutamyl–prolyl-tRNA synthetase), MRS, AIMP2, and AIMP3, and the WHEP domains were embedded in EPRS and MRS. Leucine zipper motifs are found in the N-terminal regions of RRS, AIMP1, and AIMP2 (Fig. 1A). N-terminal helical motifs were also added to KRS and DRS. Unique additional domains were attached to the N-terminal end of QRS and the C-terminal ends of IRS and LRS. These appended domains are involved in MSC formation, interaction with other proteins, and mediation of new function. Among trans-acting factors, AIMP2 appears to serve as a critical nucleation factor, mediating multiple interactions with many ARS components (27, 28). For this reason, depletion of AIMP2 triggers a massive disintegration of the MSC (22).
Figure 1.
MSC and sub-MSC structures and signaling network functionally related to cancer. A, human MSC components have several appended domains or motifs. The conserved catalytic domains and tRNA recognition domains are shown in dark gray or light gray boxes. GST-like domains are shown in the EPRS, MRS, AIMP2, and AIMP3, whereas the WHEP domains are shown in ERPS and MRS. Leucine–zipper motif is also observed in AIMP1, AIMP2, and RRS. AIMP1 has an EMAPII domain that is involved in several cellular responses. Whereas the DRS and KRS have the lysine-rich domains in the N-terminal region, LRS and IRS have the appended sequences. QRS also has the appended sequences in the C-terminal regions. B, known sub-MSC complex structures. The KRS homodimer (light and dark green) is anchored to the N-terminal peptide region of AIMP2 within the MSC (left) (16). The N-terminal helix of AIMP1 forms the ternary complex with the noncatalytic N-terminal extensions of RRS and the C-terminal core of QRS to assemble a heterotrimeric complex (center) (30). The MRS, AIMP3, EPRS, and AIMP2 are tightly linked through their GST-homology domains (right) (17). C, bisymmetrical model describing one of the possible arrangements of the MSC–ARS/AIMPs is shown as bisymmetrical model, based on the subcomplex and interaction data (17). In this model, homodimerization of DRS and PRS contributes to the bilateral symmetry of the whole complex.
Among these interactions is the KRS dimer's anchorage to the N-terminal peptide region of AIMP2 within the MSC (Fig. 1B, left) (16). Also, AIMP1 specifically interacts with the noncatalytic N-terminal extensions of RRS and QRS to form a heterotrimeric complex (Fig. 1B, middle) (30). AIMP3 and MRS are connected through their GST-homology domains (Fig. 1B, right) (17). This complex is further extended to form a stable heterotetramer with the GST-homology domains embedded in EPRS and AIMP2 (17).
These subcomplexes are further connected to form the whole MSC (Fig. 1C). For instance, the KRS–AIMP2 subcomplex is linked to the MRS–AIMP3–EPRS–AIMP2 subcomplex via the shared GST-homology domains. There is a report showing that IRS can be anchored to EPRS via its C-terminal added domain (31), and IRS, LRS, and EPRS are interdependent for their cellular stability (32), implying that these three largest enzyme components may be positioned in proximity. All the components of MSC can form a bisymmetric complex via homodimerization of DRS and PRS. One of the potential arrangements of the MSC components is schematically shown in Fig. 1C.
The MSC is thought to provide a channel through which tRNAs transit to the bound ARSs for aminoacylation and also to the translation machinery for protein synthesis. For instance, AIMP1, which is anchored to the N-terminal extension of RRS, can facilitate the delivery of the substrate tRNA to the catalytic cleft of RRS (33). AIMP3, which is specifically bound to MRS, relays the methionylated initiator tRNA to the initiator complex (11).
The MSC is also hypothesized to facilitate the delivery of amino acids to the corresponding enzymes within the complex (12, 34). Elucidation of the detailed mechanisms for the delivery of tRNAs and amino acids awaits further in-depth analysis.
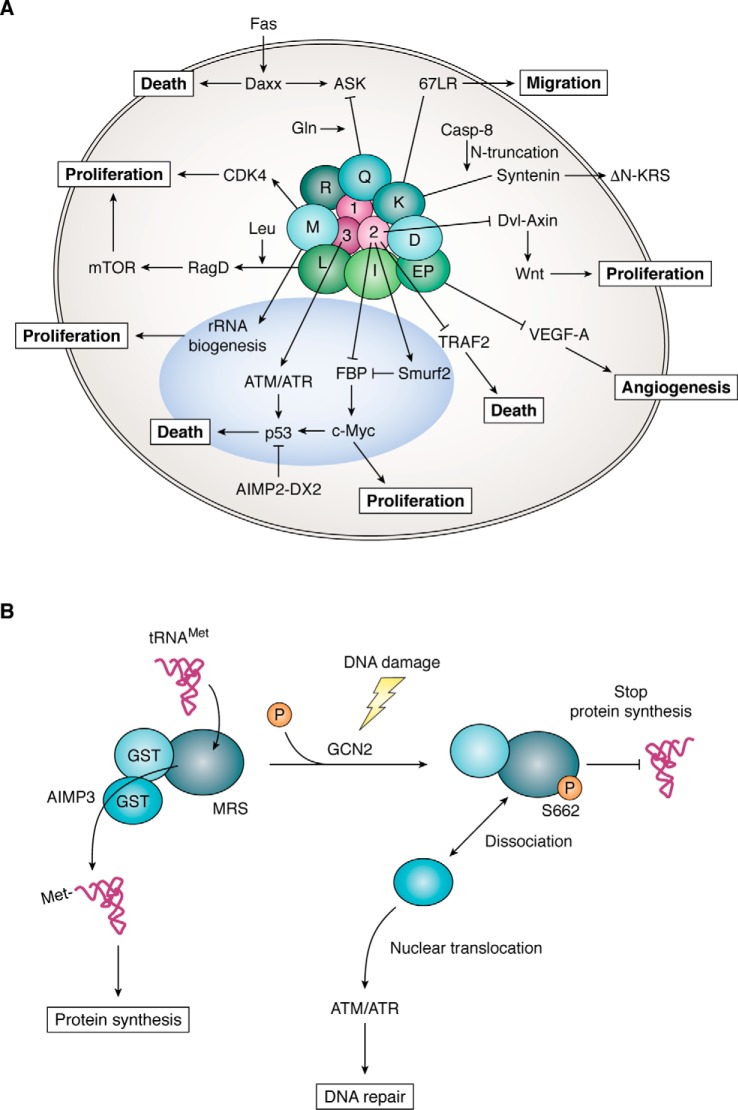
Pathophysiological implications of the MSC in cancer
To explore the evidence linking ARSs with human diseases, we surveyed research articles focusing on the association of ARSs and human diseases published within the past 50 years. Since 1970, reports describing the association of ARSs with cancer are most frequent, implying the potential significance of ARSs in cancer biology (Fig. S1). In recent years, the number of studies implicating ARS with other diseases has rapidly increased as well.
Although the MSC components appear to work together as a complex for protein synthesis, components of the complex can dissociate in response to specific signals and translocate to various cellular locations, where they interact with other proteins to regulate biological processes beyond protein synthesis (4, 35). Among diverse signaling pathways involving MSC components, some of the representative pathways that are functionally related to cancer are schematically shown in Fig. 2A (see figure legend for more details). For instance, AIMP2 exerts a potent tumor-suppressive activity through its interactions with key factors in the TGF-β, TNFα, Wnt, and p53 pathways (36–39). Moreover, cancer cells produce a splicing variant of AIMP2 lacking exon 2 that compromises AIMP2's tumor-suppressive activities (40). For these activities, loss of a single AIMP2 allele enhances the cell and in vivo cancer susceptibility (41). AIMP1 plays multiple roles in both the intracellular and extracellular space. Relevant to tumorigenesis, secreted AIMP1 not only stimulates immune responses but also suppresses tumor vascularization (42, 43). Thus, systemic administration of purified AIMP1 exerts a potent tumor-suppressive activity (44, 45).
Figure 2.
Signaling network of the MSC components related to protein synthesis and cancer. A, cancer-related signaling network mediated by the MSC-forming ARSs and AIMPs. LRS functioning as a leucine sensor interacts with the RagD GTPase to stimulate the mTOR pathway (50, 51). KRS forms a metastasis-promoting interaction with the 67-kDa laminin receptor in the cell membrane (54, 55). Caspase-8 cleaves the N-terminal 12 amino acids of KRS, exposing its PDZ-binding motif at the C terminus. Syntenin binds to the exposed PDZ-binding motif of KRS and facilitates the exosome-mediated secretion of MSC-dissociated KRS (56). Induced by growth stimuli, MRS is translocated to the nucleoli to stimulate rRNA synthesis (15). MRS binds to and stabilizes CDK4 to promote the cell cycle in p16-negative cancers (57). QRS binds to apoptosis signal-regulating kinase 1 (ASK1) to regulate apoptosis in a glutamine-dependent manner (58). EPRS forms the GAIT (interferon γ–activated inhibitor of translation) complex with other cell factors to regulate the expression of VEGF-A mRNA (59). AIMP2 is one of three nonenzymatic factors, and it works as a potent tumor suppressor through multiple pathways, including TGF-β- (36), TNFα- (37), Wnt- (38), and p53 (39)-mediated pathways. AIMP3 is mobilized to the nucleus by DNA damage (46, 47) or via an oncogenic stimulus (21) to activate p53 via ATM/ATR for DNA repair. B, MRS forms a complex with AIMP3 via their GST-homology domains (17). AIMP3 relays methionylated tRNA to the initiation factor to facilitate protein synthesis (11). However, upon DNA damage, MRS is phosphorylated by the activated GCN2 at the serine 662 residue that blocks tRNAMet binding, leading to the inhibition of protein synthesis (48). The dissociated AIMP3 is translocated into nucleus and activates ATM and ATR for DNA repair (21).
Deletion of AIMP3 both in embryonic and adult stages induces severe and lethal DNA damage (46, 47), suggesting the vital role of AIMP3 for maintaining the integrity of cellular DNA. AIMP3 is normally tightly bound to the N-terminal GST-homology domain of MRS via its similar domain and serves as a conduit for the passage of the methionine-charged tRNA to the initiation complex, as mentioned earlier (Fig. 2B) (11). However, it is dissociated from MRS upon DNA damage due to the conformational change of MRS that is phosphorylated by GCN2 at serine 662 (48). The phosphorylated MRS becomes catalytically inactive because it cannot bind tRNAMet anymore. Thus, MRS appears to work as a crucial regulator of translational initiation together with eIF2a as a substrate of GCN2. The AIMP3 dissociated from MRS is then mobilized to the nucleus to activate p53 through ataxia telangiectasia mutated (ATM) and ATM and RAD3-related (ATR) proteins (21, 49). Thus, the MRS–AIMP3 axis functionally coordinates the nuclear DNA replication and repair process with cytosolic protein synthesis.
Although AIMPs show tumor-suppressive activities through their unique mechanisms, specific ARSs in the complex also appear to control cancer-associated pathways. For instance, LRS stimulates the activity of the mTOR by interacting with Ras-related GTP-binding protein D (RagD) to promote cellular protein synthesis and proliferation upon leucine treatment (Fig. 2A) (50–52). KRS is translocated to the nucleus where it mediates transcriptional control (53), and to the cell membrane where it augments laminin-dependent cell migration, which leads to cancer metastasis (54, 55). KRS is also secreted from cancer cells to attract immune cells (56).
Another example of ARS involvement in cancer-associated pathways is MRS, which is translocated to the nucleus to stimulate rRNA synthesis upon growth signals (15). It can also bind and stabilize CDK4 to promote the cell cycle (57). QRS binds to apoptosis signal-regulating kinase 1 (ASK1) to regulate apoptosis in a glutamine-dependent manner (58), and EPRS can repress VEGF-A synthesis at the translational suppression of its mRNA (Fig. 2A) (59, 60).
To test the extent to which expression levels of ARSs are relevant for the survival of cancer patients, we estimated the expression levels of the MSC components from the mRNA-sequencing data generated from 26 types of human cancers available in The Cancer Genome Atlas (TCGA). For each MSC component, we aligned them based on the expression levels in the individual types of cancers (Table S1), and then we divided the patients into the top 25% and bottom 25% expression groups and compared the two groups for survival. Interestingly, ARS and AIMP expression levels appear to significantly affect cancer patient survival, although the relationship varies depending on the type of ARS/AIMPs and cancer. Among the MSC components, reduced survival is correlated with IRS overexpression in liver hepatocellular carcinoma, MRS overexpression in breast-invasive carcinoma, and AIMP1 overexpression in head and neck squamous cell carcinoma (Fig. S2). These data strongly suggest the pathological significance of ARS and AIMP expression for tumorigenesis.
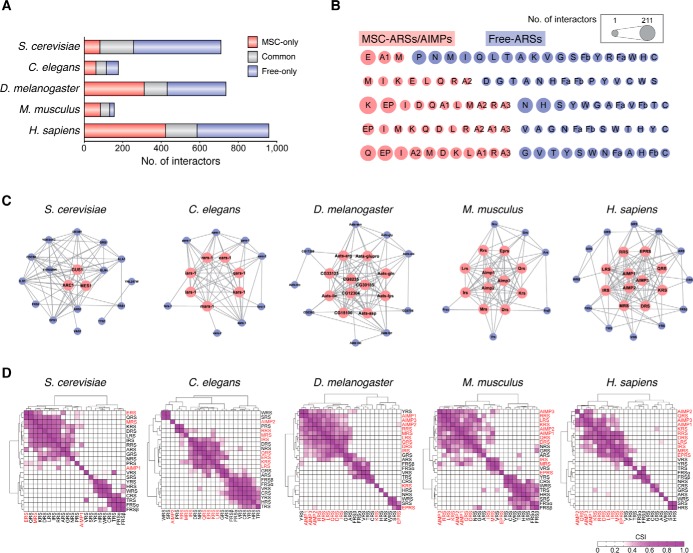
Comparison of ARS networks among different species
Evolution of the MSC included its functional expansion by augmenting the MSC interaction network with additional cellular pathways present in higher eukaryotes. To understand the expansion of the MSC interaction network, we investigated the characteristics of ARS interaction networks in five different species. From 10 interactome databases, we collected protein–protein interaction (PPI) data for ARSs and AIMPs in S. cerevisiae (1718 PPIs), Caenorhabditis elegans (567 PPIs), Drosophila melanogaster (1308 PPIs), Mus musculus (497 PPIs), and Homo sapiens (1963 PPIs) (Table S2). The numbers of ARS/AIMP interactors that participate in these PPIs were 710, 177, 736, 155, and 958 for S. cerevisiae, C. elegans, D. melanogaster, M. musculus and H. sapiens, respectively (Fig. 3A). The numbers of PPIs and ARS/AIMP interactors in C. elegans and M. musculus were smaller than expected, perhaps due to the smaller amount of PPI data available for these species.
Figure 3.
Comparative interactome analysis of ARSs in five species. A, protein–protein interactions of ARSs and AIMPs. The identified ARS interactors were grouped as those specifically interacting with MSC–ARSs/AIMPs (MSC-only), with free-ARSs (free-only), and with both MSC–ARSs/AIMPs and free-ARSs (common). B, number of interactors for individual MSC–ARSs/AIMPs (pink circles) and free-ARSs (purple circles). The circle sizes (see box) represent the interactome size of each ARS or AIMP in five species from yeast (top) to human (bottom). C, networks describing protein–protein interactions (gray edges) among ARSs and AIMPs are shown in five species. Pink and purple nodes denote MSC–ARSs/AIMPs and free-ARSs, respectively. D, hierarchical clustering of ARSs and AIMPs using their scores, called CSIs, which represent the degrees of shared interactors between the pairs of ARSs and AIMPs in each of the five species. Ward linkage and Euclidean distance as the similarity measures were used for the clustering (67, 68). The color bar represents the gradient of CSI scores. MSC–ARSs/AIMPs and free-ARSs were labeled in red and black, respectively.
MSC- and free-ARSs showed a relatively similar number of interactors in most of the species and shared some interacting partners (Fig. 3A). Among MSC–ARSs, EPRS showed a higher number of interactors compared with other components (Fig. 3B). EPRS is a unique, bifunctional tRNA synthetase consisting of the two different catalytic units that ligate Glu and Pro to cognate tRNAs. The two catalytic domains are joined by a noncatalytic linker containing three tandem WHEP domains (31, 61). The large EPRS polypeptide containing the two catalytic units with the GST domain (12) and three WHEP domains (62–64) appears to accommodate many cellular interactors (59–61, 65, 66).
The network size and density of the MSC increases from yeast to mammals, suggesting that the MSC might play a role in the system development of complex organisms (Fig. 3C). We assessed the extent to which each ARS and AIMP pair shared interactors in each species, using the connection specificity index (CSI) measure as described previously (67, 68). Among the five species, the MSC components in higher eukaryotes (D. melanogaster, M. musculus and H. sapiens) showed a higher degree of shared interactors relative to the free ARSs (Fig. 3D), suggesting that they could work in a more coordinated manner.
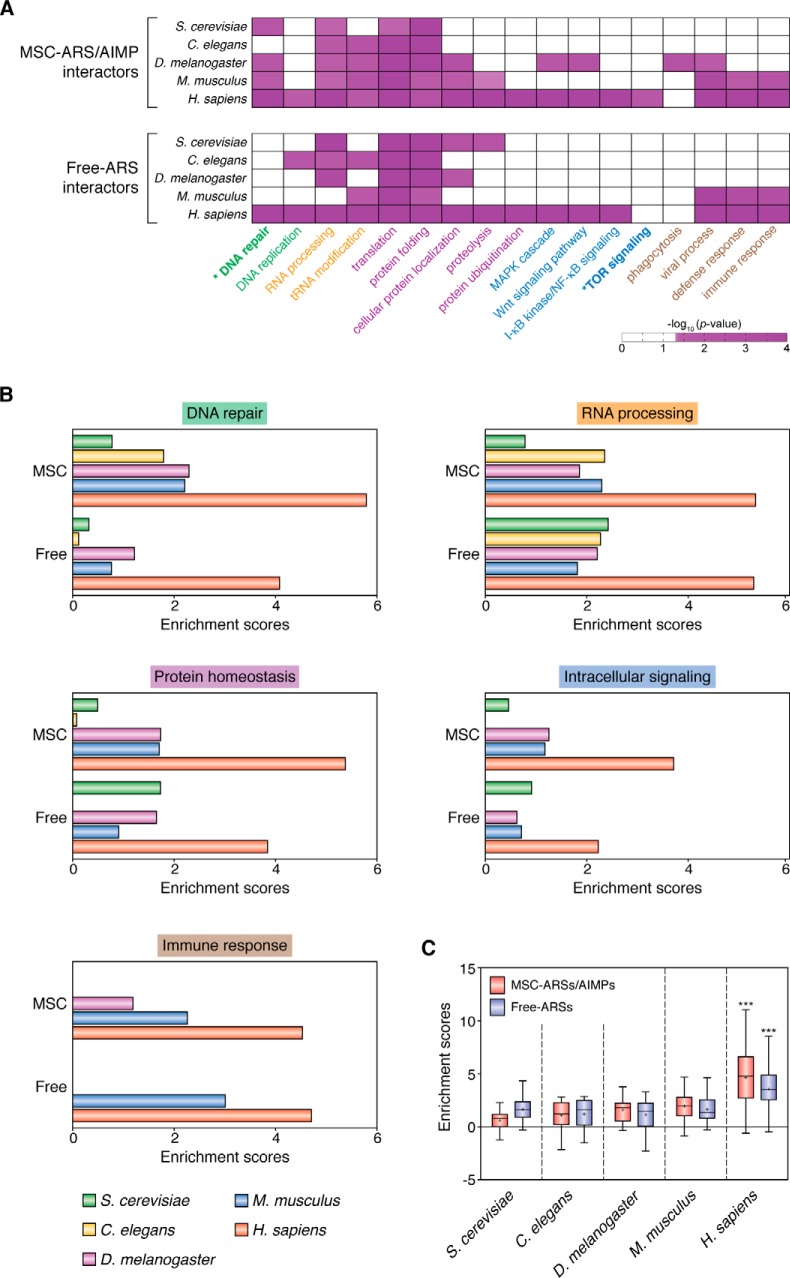
Functional expansion of the MSC throughout evolution
To understand how the MSC- and free-ARSs have expanded their functions beyond their catalytic roles in the five species, we performed enrichment analysis of gene ontology biological processes (GOBPs) for the ARS interactors. Based on the enrichment significance (p values), the following five cellular processes were significantly (p < 0.05) enriched by the interactors of MSC–ARSs in H. sapiens (Fig. 4, A and B). Those include DNA replication (DNA repair and replication), RNA processing (tRNA modification and RNA processing), protein homeostasis (translation, protein localization, and proteolysis), intracellular signaling (mTOR, Wnt, MAPK, and NF-κB signaling), and immune responses (defense response, phagocytosis, and viral process). Interestingly, these processes were similarly enriched by the interactors of free-ARSs in H. sapiens, although free-ARSs showed relatively less shared interactors, compared with MSC–ARSs (Fig. 3D). In the other eukaryotes, fewer processes were enriched by the interactors of the MSC- or free-ARSs with the similar enrichment patterns across the species (Fig. 4A), and the enrichment scores were less significant than those in H. sapiens (Fig. 4, B and C). These data suggest that the MSC- and free-ARS have not separately evolved for different functions (Fig. 4A). Combined together, it appears that noncatalytic functions of the MSC- and free-ARS appear to have commonly expanded toward the five biological processes (Fig. 4, A and B), and this conclusion is most apparent in H. sapiens.
Figure 4.
Comparative functional enrichment analysis of ARS interactomes in five species. A, GOBPs enriched by the interactors of MSC–ARSs/AIMPs and free-ARSs in S. cerevisiae, C. elegans, D. melanogaster, M. musculus, and H. sapiens. The five enriched cellular processes are labeled in different colors: 1) DNA replication (DNA repair and replication; green); 2) RNA processing (tRNA modification and RNA processing; orange); 3) protein homeostasis (translation, protein localization, and proteolysis; purple); 4) intracellular signaling (Wnt, MAPK, mTOR, and NF-κB signaling; blue); and 5) immune response (defense response, phagocytosis, and viral process; brown). The color bar represents the gradient of −log10 (p value), where the p value is the significance of the GOBPs being enriched by the interactors, which was computed from DAVID software. For visualization, hierarchical clustering was performed for each group of cellular processes using Ward linkage and Euclidean distance as the similarity measure. B, enrichment scores of the indicated representative processes (DNA repair, RNA processing, protein localization–proteolysis, immune response–viral process, and intracellular signaling) for the five groups of cellular processes enriched by MSC–ARS/AIMP (top) and free-ARS interactors (bottom) in the five species. C, distributions of the enrichment scores of the representative processes by MSC–ARS/AIMP (pink) and free-ARS interactors (purple) in the five species are shown using box plots. Z >2.33, *, p < 0.05, and ***, p < 0.001, two-way ANOVA with Bonferroni's post-hoc correction. See supporting information for methodological details.
Implications of the MSC in mTOR signaling and DNA repair
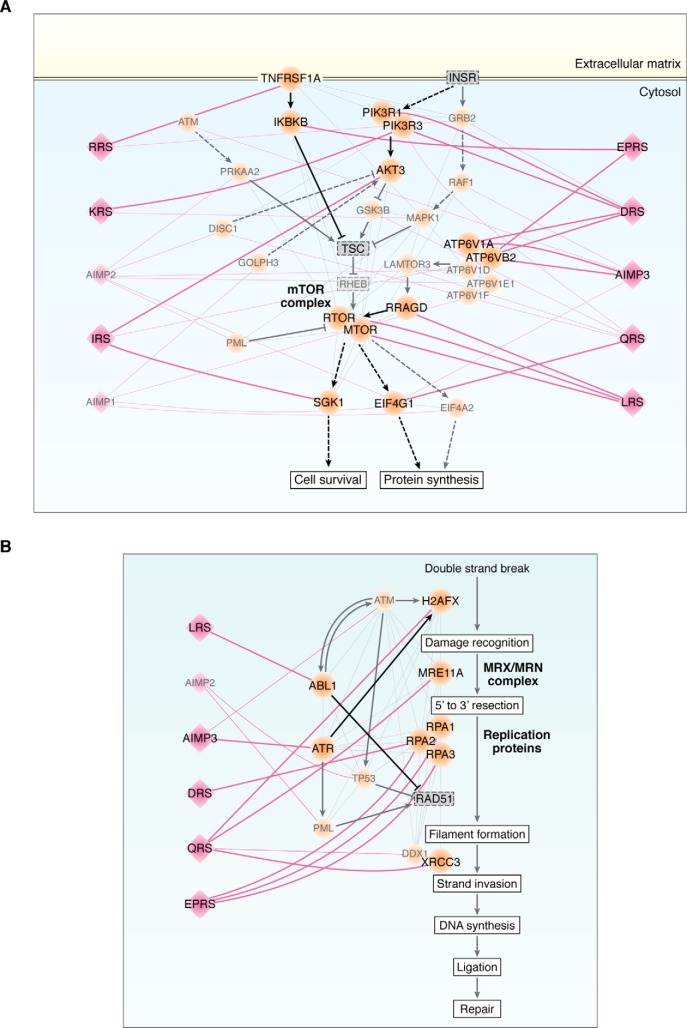
Among the aforementioned five cellular processes, we focused on further examining the mTOR and DNA repair pathways, because these processes are known to play crucial roles in tumorigenesis, and some of the MSC components have been shown to be involved in their control (Fig. 2A). Previously, LRS has been shown to interact with the RagD GTPase to activate the mTORC1 pathway (50, 51). Using the human ARS interactome, we built a network model describing interactions of the MSC components with the factors involved in the mTOR pathway.
For each interacting pair of the MSC component and the factor, we further evaluated the correlations between mRNA levels and patient survival (shown in Fig. S2) and considered the interaction to be potentially active when both interactors have significant mRNA-survival correlations in at least one of 26 cancer types tested. The network model further suggests that in addition to LRS, DRS, RRS, IRS, KRS, QRS, and EPRS also showed potential active interactions with different factors in the MAPK and PI3K–AKT pathways, which are upstream of the mTOR pathway (Fig. 5A). The network model also suggests that IRS and QRS have potential active interactions with downstream molecules in the mTOR pathway (Fig. 5A). In addition, the data revealed active interactions of AIMP3, DRS, and EPRS with different components of the vacuolar H+-ATPases that localize to the phagosome, late endosome, or lysosome membranes (Fig. 5A).
Figure 5.
Systematic association of the MSC components with mTOR and DNA repair pathways. Network models describing interactions of MSC–ARS/AIMPs with molecules involved in the mTOR signaling (A) and HR-based DNA repair pathways (B) in H. sapiens. Pink diamonds and orange circles represent MSC–ARS/AIMPs and their interactors in the pathways, respectively. Pink lines are the interactions between MSC–ARS/AIMPs and their interactors in the two pathways. Gray lines indicate the known interactions among the cellular factors in the pathways. Known activation (arrows) and inhibition (inhibition symbols) information obtained from Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway database and the previous literature are denoted with black lines. Solid and dashed lines are direct and indirect interactions between the molecules, respectively. Active interactions between the pairs of MSC–ARS/AIMPs and the molecules in the pathways with significant mRNA expression–survival correlations are highlighted with thick pink lines. See supporting information for methodological details. ATP6V1A/B2/D/E1/F are the components of ATPases in phagosomes or lysosomes (A). QRS interacts with MRE11, a component of MRN complex involved in the initial processing of double-strand DNA breaks before HR-based DNA repair. EPRS and DRS interact with RPA1/2/3 that prevent ssDNA from winding back before HR-based DNA repair. QRS, LRS, and AIMP3 interact with H2AFX, ABL1, and ATR, respectively, which are involved in RAD51-mediated heteroduplex formation during HR-based DNA repair. QRS interacts with XRCC3 and DDX1 involved in DNA extension during HR-based DNA repair (B).
We also built a network model describing interactions of the MSC components with molecules involved in DNA repair (Fig. 5B). Among the various DNA repair mechanisms, we focused on the homologous recombination (HR)–based DNA repair pathway because it included the largest number of the ARS interactors. AIMP3 was shown to facilitate DNA repair through homologous DNA recombination (46, 47), although its additional role in nonhomologous DNA recombination is not excluded. The network model revealed four instances in which MSC–ARSs/AIMPs interact with core molecules in the HR-based DNA repair pathway (Fig. 5B). For example, LRS, AIMP2, DRS, QRS, or EPRS showed potentially active interactions with H2AFX, MRE11A, RAP1/2/3, or XRCC3 throughout the HR-based DNA repair pathway, as well as ABL1 or ATR, the modulators of the pathway. Together, these data suggest that the MSC components are functionally linked to the mTORC1 and HR-based DNA repair pathways, systematically interacting with the core, upstream, downstream, and/or modulating factors in the pathways. The detailed working mechanisms for the individual interactions in the network models need further investigation.
ARS and disease
Having been equipped with both the catalytic activities for protein synthesis and additional arms to mediate diverse molecular interactions, ARSs are uniquely positioned to be coordinators of system development, intrinsic defense mechanisms, and homeostasis. Much of their functional significance, aberrant expression, mutations, splicing variant formation, and secretion of ARSs can result in diverse pathological symptoms. For instance, mutations in GRS, YRS, AlaRS, HRS, KRS, and MRS are associated with Charcot-Marie-Tooth disease, a peripheral neuropathy (69–77). The uncontrolled extracellular localization of several different ARSs can elicit the formation of autoantibodies that provokes the immune system, resulting in antisynthetase syndrome, an autoimmune disease (78–84).
Aminoacyl-tRNA biosynthesis has been shown to be highly up-regulated in cancer metabolism (85), perhaps because of the increased demand for protein synthesis in cancer. Accordingly, higher activities of ARSs are expected to be present in cancer, and indeed, increased expression of ARSs is frequently observed in many types of cancers. Although ARSs can be catalytically involved in protein synthesis and cancer metabolism, each ARS or AIMP may be specifically involved in a certain type of cancer through noncatalytic mechanisms. Mutations in the ARS/AIMP-encoding genes have not been found as frequently as those in the well-known oncogenes. Nonetheless, the proline 42 to alanine mutation of human cytoplasmic GRS was found in nearly 40% of adenoid cystic carcinoma patients (29), although whether these mutations are causal awaits further investigation. Cancer-specific variant formation and protein–protein interactions of ARSs and AIMPs also deserve attention. As mentioned earlier, AIMP2–DX2, a splicing variant that lacks exon 2 of AIMP2, is expressed in various cancers, compromising the tumor-suppressive activities of AIMP2 (40, 41). In p16-negative cancers, MRS appears to bind and stabilize CDK4, resulting in the promotion of the cell cycle (57). In light of the ARS connection to cancer, their cancer-specific expression, variant formation, and protein–protein interactions represent promising novel targets for developing anti-cancer therapeutics. Moreover, the immune modulation activities of the secreted ARSs and AIMPs suggest they may serve as a novel resource for cancer immunotherapy and vaccines.
Perspectives
Despite MSC's significant role in protein synthesis and system control, understanding its structural features and dynamic nature is still in its infancy. In light of the multidimensional functionality of the MSC components, the tight regulation of their catalytic and noncatalytic activities is critical to prevent pathogenesis resulting from uncontrolled behaviors. In this context, MSC formation provides an effective mechanism for tight yet dynamic control of its components, which are constitutively expressed and ubiquitous in all cells. While still working in protein synthesis as catalysts, a subset of ARSs can exist within the MSC where they are poised for rapid response to incoming stresses and stimuli.
The network analysis of ARSs revealed intriguing features of the MSC. First, although MSC components are more tightly bound to each other than free-ARSs, they may communicate with free-ARSs (Fig. 3C). Second, MSC–ARSs interact with the same cellular factors to a higher degree than with free-ARSs (Fig. 3D). Third, beyond their catalytic roles in protein synthesis, the regulatory activities of MSC–ARSs mainly converge on five biological processes (Fig. 4A) as follows: DNA repair; RNA processing; protein homeostasis; intracellular signaling; and immune responses. Interestingly, these processes are positively or negatively implicated in tumorigenesis. Fourth, the MSC components may be involved in these processes as components of subcomplexes or modules involving multiple components rather than as a single regulatory factor. Future in-depth and systematic investigations will be necessary to understand how the MSC components communicate with each other and with free-ARSs and diverse cellular factors.
Supplementary Material
Acknowledgment
We thank Dr. Bum Sik Kang at Kyungbook National University for the kind help to make the whole MSC model.
This work was supported by Global Frontier Project Grant NRF-M1AXA002-2010-0029785 and the Institute for Basic Science Grant IBS-R013-G1 funded by Korean Ministry of Science, ICT, and Future Planning. This is the fourth article in the “tRNAs and aminoacyl-tRNA synthetases in human disease” JBC Reviews series. The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1 and S2.
- ARS
- aminoacyl-tRNA synthetase
- MSC
- multi-tRNA synthetase complex
- CSI
- connection specificity index
- mTOR
- mammalian target of rapamycin
- RRS
- arginyl-tRNA synthetase
- EPRS
- glutamyl–prolyl-tRNA synthetase
- QRS
- glutaminyl-tRNA synthetase
- DRS
- aspartyl-tRNA synthetase
- PRS
- prolyl-tRNA synthetase
- LRS
- leucyl-tRNA synthetase
- IRS
- isoleucyl-tRNA synthetase
- ERS
- glutamyl-tRNA
- KRS
- lysyl-tRNA synthetase
- QRS
- glutaminyl-tRNA
- AIMP
- ARS-interacting multifunctional protein
- VEGF
- vascular endothelial growth factor
- TGF-β
- transforming growth factor-β
- TNFα
- tumor necrosis factor-α
- ATM
- ataxia telangiectasia-mutated
- ATR
- ATM and Rad3-related
- GST
- glutathione S-transferase
- ANOVA
- analysis of variance
- HR
- homologous recombination
- MRS
- methionyl-tRNA synthetase
- GOBP
- gene ontology biological process
- PPI
- protein–protein interaction.
References
- 1. McClain W. H. (1993) Rules that govern tRNA identity in protein synthesis. J. Mol. Biol. 234, 257–280 10.1006/jmbi.1993.1582 [DOI] [PubMed] [Google Scholar]
- 2. Swanson R., Hoben P., Sumner-Smith M., Uemura H., Watson L., and Söll D. (1988) Accuracy of in vivo aminoacylation requires proper balance of tRNA and aminoacyl-tRNA synthetase. Science 242, 1548–1551 [DOI] [PubMed] [Google Scholar]
- 3. Delarue M. (1995) Aminoacyl-tRNA synthetases. Curr. Opin. Struct. Biol. 5, 48–55 10.1016/0959-440X(95)80008-O [DOI] [PubMed] [Google Scholar]
- 4. Kim S., You S., and Hwang D. (2011) Aminoacyl-tRNA synthetases and tumorigenesis: more than housekeeping. Nat. Rev. Cancer 11, 708–718 10.1038/nrc3124 [DOI] [PubMed] [Google Scholar]
- 5. Lee S. W., Cho B. H., Park S. G., and Kim S. (2004) Aminoacyl-tRNA synthetase complexes: beyond translation. J. Cell Sci. 117, 3725–3734 10.1242/jcs.01342 [DOI] [PubMed] [Google Scholar]
- 6. Guo M., Yang X. L., and Schimmel P. (2010) New functions of tRNA synthetases beyond translation. Nat. Rev. Mol. Cell Biol. 11, 668–674 10.1038/nrm2956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Quevillon S., and Mirande M. (1996) The p18 component of the multisynthetase complex shares a protein motif with the β and γ subunits of eukaryotic elongation factor 1. FEBS Lett. 395, 63–67 10.1016/0014-5793(96)01005-8 [DOI] [PubMed] [Google Scholar]
- 8. Cerini C., Kerjan P., Astier M., Gratecos D., Mirande M., and Sémériva M. (1991) A component of the multisynthetase complex is a multifunctional aminoacyl-tRNA synthetase. EMBO J. 10, 4267–4277 10.1002/j.1460-2075.1991.tb05005.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rho S. B., Kim M. J., Lee J. S., Seol W., Motegi H., Kim S., and Shiba K. (1999) Genetic dissection of protein–protein interactions in multi-tRNA synthetase complex. Proc. Natl. Acad. Sci. U.S.A. 96, 4488–4493 10.1073/pnas.96.8.4488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kaminska M., Havrylenko S., Decottignies P., Le Maréchal P., Negrutskii B., and Mirande M. (2009) Dynamic organization of aminoacyl-tRNA synthetase complexes in the cytoplasm of human cells. J. Biol. Chem. 284, 13746–13754 10.1074/jbc.M900480200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kang T., Kwon N. H., Lee J. Y, Park M. C., Kang E., Kim H. H., Kang T. J., and Kim S. (2012) AIMP3/p18 controls translational initiation by mediating the delivery of charged initiator tRNA to initiation complex. J. Mol. Biol. 423, 475–481 10.1016/j.jmb.2012.07.020 [DOI] [PubMed] [Google Scholar]
- 12. Eswarappa S. M., and Fox P. L. (2013) Citric acid cycle and the origin of MARS. Trends Biochem. Sci. 38, 222–228 10.1016/j.tibs.2013.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nathanson L., and Deutscher M. P. (2000) Active aminoacyl-tRNA synthetases are present in nuclei as a high molecular weight multienzyme complex. J. Biol. Chem. 275, 31559–31562 10.1074/jbc.C000385200 [DOI] [PubMed] [Google Scholar]
- 14. Kyriacou S. V., and Deutscher M. P. (2008) An important role for the multienzyme aminoacyl-tRNA synthetase complex in mammalian translation and cell growth. Mol. Cell 29, 419–427 10.1016/j.molcel.2007.11.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ko Y. G., Kang Y. S., Kim E. K., Park S. G., and Kim S. (2000) Nucleolar localization of human methionyl-tRNA synthetase and its role in ribosomal RNA synthesis. J. Cell Biol. 149, 567–574 10.1083/jcb.149.3.567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ofir-Birin Y., Fang P., Bennett S. P., Zhang H. M., Wang J., Rachmin I., Shapiro R., Song J., Dagan A., Pozo J., Kim S., Marshall A. G., Schimmel P., Yang X. L., Nechushtan H., et al. (2013) Structural switch of lysyl-tRNA synthetase between translation and transcription. Mol. Cell 49, 30–42 10.1016/j.molcel.2012.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cho H. Y., Maeng S. J., Cho H. J., Choi Y. S., Chung J. M., Lee S., Kim H. K., Kim J. H., Eom C. Y., Kim Y. G., Guo M., Jung H. S., Kang B. S., and Kim S. (2015) Assembly of multi-tRNA synthetase complex via heterotetrameric glutathione transferase-homology domains. J. Biol. Chem. 290, 29313–29328 10.1074/jbc.M115.690867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Galani K., Grosshans H., Deinert K., Hurt E. C., and Simos G. (2001) The intracellular location of two aminoacyl-tRNA synthetases depends on complex formation with Arc1p. EMBO J. 20, 6889–6898 10.1093/emboj/20.23.6889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Park S. G., Schimmel P., and Kim S. (2008) Aminoacyl tRNA synthetases and their connections to disease. Proc. Natl. Acad. Sci. U.S.A. 105, 11043–11049 10.1073/pnas.0802862105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yannay-Cohen N., Carmi-Levy I., Kay G., Yang C. M., Han J. M., Kemeny D. M., Kim S., Nechushtan H., and Razin E. (2009) LysRS serves as a key signaling molecule in the immune response by regulating gene expression. Mol. Cell 34, 603–611 10.1016/j.molcel.2009.05.019 [DOI] [PubMed] [Google Scholar]
- 21. Park B. J., Kang J. W., Lee S. W., Choi S. J., Shin Y. K., Ahn Y. H., Choi Y. H., Choi D., Lee K. S., and Kim S. (2005) The haploinsufficient tumor suppressor p18 upregulates p53 via interactions with ATM/ATR. Cell 120, 209–221 10.1016/j.cell.2004.11.054 [DOI] [PubMed] [Google Scholar]
- 22. Kim J. Y., Kang Y. S., Lee J. W., Kim H. J., Ahn Y. H., Park H., Ko Y. G., and Kim S. (2002) p38 is essential for the assembly and stability of macromolecular tRNA synthetase complex: implications for its physiological significance. Proc. Natl. Acad. Sci. U.S.A. 99, 7912–7916 10.1073/pnas.122110199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kaminska M., Shalak V., and Mirande M. (2001) The appended C-domain of human methionyl-tRNA synthetase has a tRNA-sequestering function. Biochemistry 40, 14309–14316 10.1021/bi015670b [DOI] [PubMed] [Google Scholar]
- 24. Francin M., Kaminska M., Kerjan P., and Mirande M. (2002) The N-terminal domain of mammalian lysyl-tRNA synthetase is a functional tRNA-binding domain. J. Biol. Chem. 277, 1762–1769 10.1074/jbc.M109759200 [DOI] [PubMed] [Google Scholar]
- 25. Francin M., and Mirande M. (2003) Functional dissection of the eukaryotic-specific tRNA-interacting factor of lysyl-tRNA synthetase. J. Biol. Chem. 278, 1472–1479 10.1074/jbc.M208802200 [DOI] [PubMed] [Google Scholar]
- 26. Kim S., Landro J. A., Gale A. J., and Schimmel P. (1993) C-terminal peptide appendix in a class I tRNA synthetase needed for acceptor-helix contacts and microhelix aminoacylation. Biochemistry 32, 13026–13031 10.1021/bi00211a011 [DOI] [PubMed] [Google Scholar]
- 27. Kaminska M., Havrylenko S., Decottignies P., Gillet S., Le Maréchal P., Negrutskii B., and Mirande M. (2009) Dissection of the structural organization of the aminoacyl-tRNA synthetase complex. J. Biol. Chem. 284, 6053–6060 10.1074/jbc.M809636200 [DOI] [PubMed] [Google Scholar]
- 28. Quevillon S., Robinson J. C., Berthonneau E., Siatecka M., and Mirande M. (1999) Macromolecular assemblage of aminoacyl-tRNA synthetases: identification of protein–protein interactions and characterization of a core protein. J. Mol. Biol. 285, 183–195 10.1006/jmbi.1998.2316 [DOI] [PubMed] [Google Scholar]
- 29. Stephens P. J., Davies H. R., Mitani Y., Van Loo P., Shlien A., Tarpey P. S., Papaemmanuil E., Cheverton A., Bignell G. R., Butler A. P., Gamble J., Gamble S., Hardy C., Hinton J., Jia M., et al. (2013) Whole exome sequencing of adenoid cystic carcinoma J. Clin. Invest. 123, 2965–2968 10.1172/JCI67201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Fu Y., Kim Y., Jin K. S., Kim H. S., Kim J. H., Wang D., Park M., Jo C. H., Kwon N. H., Kim D., Kim M. H., Jeon Y. H., Hwang K. Y., Kim S., and Cho Y. (2014) Structure of the ArgRS–GlnRS–AIMP1 complex and its implications for mammalian translation. Proc. Natl. Acad. Sci. U.S.A. 111, 15084–15089 10.1073/pnas.1408836111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rho S. B., Lee J. S., Jeong E. J., Kim K. S., Kim Y. G., and Kim S. (1998) A multifunctional repeated motif is present in human bifunctional tRNA synthetase. J. Biol. Chem. 273, 11267–11273 10.1074/jbc.273.18.11267 [DOI] [PubMed] [Google Scholar]
- 32. Han J. M., Lee M. J., Park S. G., Lee S. H., Razin E., Choi E. C., and Kim S. (2006) Hierarchical network between the components of the multi-tRNA synthetase complex: implications for complex formation. J. Biol. Chem. 281, 38663–38667 10.1074/jbc.M605211200 [DOI] [PubMed] [Google Scholar]
- 33. Park S. G., Jung K. H., Lee J. S., Jo Y. J., Motegi H., Kim S., and Shiba K. (1999) Precursor of pro-apoptotic cytokine modulates aminoacylation activity of tRNA synthetase. J. Biol. Chem. 274, 16673–16676 10.1074/jbc.274.24.16673 [DOI] [PubMed] [Google Scholar]
- 34. Eswarappa S. M., Potdar A. A., Sahoo S., Sankar S., and Fox P. L. (2018) Metabolic origin of the fused aminoacyl tRNA synthetase, glutamyl-prolyl tRNA synthetase. J. Biol. Chem. 293, 19148–19156 10.1074/jbc.RA118.004276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kim J. H., Han J. M., and Kim S. (2014) Protein–protein interactions and multi-component complexes of aminoacyl-tRNA synthetases. Top. Curr. Chem. 344, 119–144 10.1007/128_2013_479 [DOI] [PubMed] [Google Scholar]
- 36. Kim M. J., Park B. J., Kang Y. S., Kim H. J., Park J. H., Kang J. W., Lee S. W., Han J. M., Lee H. W., and Kim S. (2003) Downregulation of FUSE-binding protein and c-myc by tRNA synthetase cofactor p38 is required for lung cell differentiation. Nat. Genet. 34, 330–336 10.1038/ng1182 [DOI] [PubMed] [Google Scholar]
- 37. Choi J. W., Kim D. G., Park M. C., Um J. Y., Han J. M., Park S. G., Choi E. C., and Kim S. (2009) AIMP2 promotes TNFα-dependent apoptosis via ubiquitin-mediated degradation of TRAF2. J. Cell Sci. 122, 2710–2715 10.1242/jcs.049767 [DOI] [PubMed] [Google Scholar]
- 38. Yum M. K., Kang J. S., Lee A. E., Jo Y. W., Seo J. Y., Kim H. A., Kim Y. Y., Seong J., Lee E. B., Kim J. H., Han J. M., Kim S., and Kong Y. Y. (2016) AIMP2 controls intestinal stem cell compartments and tumorigenesis by modulating Wnt/β-catenin signaling. Cancer Res. 76, 4559–4568 10.1158/0008-5472.CAN-15-3357 [DOI] [PubMed] [Google Scholar]
- 39. Han J. M., Park B. J., Park S. G., Oh Y. S., Choi S. J., Lee S. W., Hwang S. K., Chang S. H., Cho M. H., and Kim S. (2008) AIMP2/p38, the scaffold for the multi-tRNA synthetase complex, responds to genotoxic stresses via p53. Proc. Natl. Acad. Sci. U.S.A. 105, 11206–11211 10.1073/pnas.0800297105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Choi J. W., Kim D. G., Lee A. E., Kim H. R., Lee J. Y., Kwon N. H., Shin Y. K., Hwang S. K., Chang S. H., Cho M. H., Choi Y. L., Kim J., Oh S. H., Kim B., Kim S. Y., et al. (2011) Cancer-associated splicing variant of tumor suppressor AIMP2/p38: pathological implication in tumorigenesis. PLoS Genet. 7, e1001351 10.1371/journal.pgen.1001351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Choi J. W., Um J. Y., Kundu J. K., Surh Y. J., and Kim S. (2009) Multidirectional tumor-suppressive activity of AIMP2/p38 and the enhanced susceptibility of AIMP2 heterozygous mice to carcinogenesis. Carcinogenesis 30, 1638–1644 10.1093/carcin/bgp170 [DOI] [PubMed] [Google Scholar]
- 42. Park S. G., Kang Y. S., Ahn Y. H., Lee S. H., Kim K. R., Kim K. W., Koh G. Y., Ko Y. G., and Kim S. (2002) Dose-dependent biphasic activity of tRNA synthetase-associating factor, p43, in angiogenesis. J. Biol. Chem. 277, 45243–45248 10.1074/jbc.M207934200 [DOI] [PubMed] [Google Scholar]
- 43. Liang D., Tian L., You R., Halpert M. M., Konduri V., Baig Y. C., Paust S., Kim D., Kim S., Jia F., Huang S., Zhang X., Kheradmand F., Corry D. B., et al. (2017) AIMp1 potentiates TH1 polarization and is critical for effective antitumor and antiviral immunity. Front. Immunol. 8, 1801 10.3389/fimmu.2017.01801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lee Y. S., Han J. M., Kang T., Park Y. I., Kim H. M., and Kim S. (2006) Antitumor activity of the novel human cytokine AIMP1 in an in vivo tumor model. Mol. Cells 21, 213–217 [PubMed] [Google Scholar]
- 45. Han J. M., Myung H., and Kim S. (2010), Antitumor activity and pharmacokinetic properties of ARS-interacting multi-functional protein 1 (AIMP1/p43). Cancer Lett. 287, 157–164 10.1016/j.canlet.2009.06.005 [DOI] [PubMed] [Google Scholar]
- 46. Kim S. M., Jeon Y., Kim D., Jang H., Bae J. S., Park M. K., Kim H., Kim S., and Lee H. (2018) AIMP3 depletion causes genome instability and loss of stemness in mouse embryonic stem cells. Cell Death Dis. 9, 972 10.1038/s41419-018-1037-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kim D., Kim S., Oh Y., Park S., Jeon Y., Kim H., Lee H., and Kim S. (2018) AIMP3 deletion induces acute radiation syndrome-like phenotype in mice. Sci. Rep. 8, 15025 10.1038/s41598-018-33303-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kwon N. H., Kang T., Lee J. Y., Kim H. H., Kim H. R., Hong J., Oh Y. S., Han J. M., Ku M. J., Lee S. Y., and Kim S. (2011) Dual role of methionyl-tRNA synthetase in the regulation of translation and tumor suppressor activity of aminoacyl-tRNA synthetase-interacting multifunctional protein-3. Proc. Natl. Acad. Sci. U.S.A. 108, 19635–19640 10.1073/pnas.1103922108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Park B. J., Oh Y. S., Park S. Y., Choi S. J., Rudolph C., Schlegelberger B., and Kim S. (2006) AIMP3 haploinsufficiency disrupts oncogene-induced p53 activation and genomic stability. Cancer Res. 66, 6913–6918 10.1158/0008-5472.CAN-05-3740 [DOI] [PubMed] [Google Scholar]
- 50. Han J. M., Jeong S. J., Park M. C., Kim G., Kwon N. H., Kim H. K., Ha S. H., Ryu S. H., and Kim S. (2012) Leucyl-tRNA synthetase is an intracellular leucine sensor for the mTORC1-signaling pathway. Cell 149, 410–424 10.1016/j.cell.2012.02.044 [DOI] [PubMed] [Google Scholar]
- 51. Bonfils G., Jaquenoud M., Bontron S., Ostrowicz C., Ungermann C., and De Virgilio C. (2012) Leucyl-tRNA synthetase controls TORC1 via the EGO complex. Mol. Cell 46, 105–110 10.1016/j.molcel.2012.02.009 [DOI] [PubMed] [Google Scholar]
- 52. Kim J. H., Lee C., Lee M., Wang H., Kim K., Park S. J., Yoon I., Jang J., Zhao H., Kim H. K., Kwon N. H., Jeong S. J., Yoo H. C., Kim J. H., Yang J. S., et al. (2017) Control of leucine-dependent mTORC1 pathway through chemical intervention of leucyl-tRNA synthetase and RagD interaction. Nat. Commun. 8, 732 10.1038/s41467-017-00785-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Lee Y. N., Nechushtan H., Figov N., and Razin E. (2004) The function of lysyl-tRNA synthetase and Ap4A as signaling regulators of MITF activity in FcϵRI-activated mast cells. Immunity 20, 145–151 10.1016/S1074-7613(04)00020-2 [DOI] [PubMed] [Google Scholar]
- 54. Kim D. G., Choi J. W., Lee J. Y., Kim H., Oh Y. S., Lee J. W., Tak Y. K., Song J. M., Razin E., Yun S. H., and Kim S. (2012) Interaction of two translational components, lysyl-tRNA synthetase and p40/37LRP, in plasma membrane promotes laminin-dependent cell migration. FASEB J. 26, 4142–4159 10.1096/fj.12-207639 [DOI] [PubMed] [Google Scholar]
- 55. Kim D. G., Lee J. Y., Kwon N. H., Fang P., Zhang Q., Wang J., Young N. L., Guo M., Cho H. Y., Mushtaq A. U., Jeon Y. H., Choi J. W., Han J. M., Kang H. W., Joo J. E., et al. (2014) Chemical inhibition of prometastatic lysyl-tRNA synthetase-laminin receptor interaction. Nat. Chem. Biol. 10, 29–34 10.1038/nchembio.1381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kim S. B., Kim H. R., Park M. C., Cho S., Goughnour P. C., Han D., Yoon I., Kim Y., Kang T., Song E., Kim P., Choi H., Mun J. Y., Song C., Lee S., et al. (2017) Caspase-8 controls the secretion of inflammatory lysyl-tRNA synthetase in exosomes from cancer cells. J. Cell Biol. 216, 2201–2216 10.1083/jcb.201605118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kwon N. H., Lee J. Y., Ryu Y., Kim C., Kong J., Oh Y., Kang B. S., Ahn H. W., Ahn S. G., Jeong J., Kim H. K., Kim J. H., Han D. Y., Park M. C., Kim D., et al. (2018) Stabilization of cyclin-dependent kinase 4 by methionyl-tRNA synthetase in p16 INK4a-negative cancer. ACS Pharmacol. Transl. Sci. 1, 21–31 10.1021/acsptsci.8b00001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Ko Y. G., Kim E. Y., Kim T., Park H., Park H. S., Choi E. J., and Kim S. (2001) Glutamine-dependent anti-apoptotic interaction of human glutaminyl-tRNA synthetase with apoptosis signal-regulating kinase 1. J. Biol. Chem. 276, 6030–6036 10.1074/jbc.M006189200 [DOI] [PubMed] [Google Scholar]
- 59. Yao P., Potdar A. A., Arif A., Ray P. S., Mukhopadhyay R., Willard B., Xu Y., Yan J., Saidel G. M., and Fox P. L. (2012) Coding region polyadenylation generates a truncated tRNA synthetase that counters translation repression. Cell 149, 88–100 10.1016/j.cell.2012.02.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Arif A., Jia J., Moodt R. A., DiCorleto P. E., and Fox P. L. (2011) Phosphorylation of glutamyl-prolyl tRNA synthetase by cyclin-dependent kinase 5 dictates transcript-selective translational control. Proc. Natl. Acad. Sci. U.S.A. 108, 1415–1420 10.1073/pnas.1011275108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Jia J., Arif A., Ray P. S., and Fox P. L. (2008) WHEP domains direct noncanonical function of glutamyl-prolyl tRNA synthetase in translational control of gene expression. Mol. Cell 29, 679–690 10.1016/j.molcel.2008.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Jeong E. J., Hwang G. S., Kim K. H., Kim M. J., Kim S., and Kim K. S. (2000) Structural analysis of multifunctional peptide motifs in human bifunctional tRNA synthetase: identification of RNA-binding residues and functional implications for tandem repeats. Biochemistry 39, 15775–15782 10.1021/bi001393h [DOI] [PubMed] [Google Scholar]
- 63. Ray P. S., and Fox P. L. (2014) Origin and evolution of glutamyl-prolyl-tRNA synthetase WHEP domains reveal evolutionary relationships within holozoa. PLoS ONE 9, e98493 10.1371/journal.pone.0098493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Ray P. S., Sullivan J. C., Jia J., Francis J., Finnerty J. R., and Fox P. (2011) L evolution of function of a fused metazoan tRNA synthetase. Mol. Biol. Evol. 28, 437–447 10.1093/molbev/msq246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Arif A., Jia J., Mukhopadhyay R., Willard B., Kinter M., and Fox P. L. (2009) Two-site phosphorylation of EPRS coordinates multimodal regulation of noncatalytic translational control activity. Mol. Cell 35, 164–180 10.1016/j.molcel.2009.05.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Arif A., Terenzi F., Potdar A. A., Jia J., Sacks J., China A., Halawani D., Vasu K., Li X., Brown J. M., Chen J., Kozma S. C., Thomas G., and Fox P. L. (2017) EPRS is a critical mTORC1-S6K1 effector that influences adiposity in mice. Nature 542, 357–361 10.1038/nature21380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Green R. A., Kao H. L., Audhya A., Arur S., Mayers J. R., Fridolfsson H. N., Schulman M., Schloissnig S., Niessen S., Laband K., Wang S., Starr D. A., Hyman A. A., Schedl T., Desai A., et al. (2011) A high-resolution C.elegans essential gene network based on phenotypic profiling of a complex tissue. Cell 145, 470–482 10.1016/j.cell.2011.03.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Fuxman Bass J. I., Diallo A., Nelson J., Soto J. M., Myers C. L., and Walhout A. J. (2013) Using networks to measure similarity between genes: association index selection. Nat. Methods 10, 1169–1176 10.1038/nmeth.2728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Seburn K. L., Nangle L. A., Cox G. A., Schimmel P., and Burgess R. W. (2006) An active dominant mutation of glycyl-tRNA synthetase causes neuropathy in a Charcot-Marie-Tooth 2D mouse model. Neuron 51, 715–726 10.1016/j.neuron.2006.08.027 [DOI] [PubMed] [Google Scholar]
- 70. Antonellis A., Ellsworth R. E., Sambuughin N., Puls I., Abel A., Lee-Lin S. Q., Jordanova A., Kremensky I., Christodoulou K., Middleton L. T., Sivakumar K., Ionasescu V., Funalot B., Vance J. M., Goldfarb L. G., et al. (2003) Glycyl-tRNA synthetase mutations in Charcot-Marie-Tooth disease type 2D and distal spinal muscular atrophy type V. Am. J. Hum. Genet. 72, 1293–1299 10.1086/375039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Jordanova A., Irobi J., Thomas F. P., Van Dijck P., Meerschaert K., Dewil M., Dierick I., Jacobs A., De Vriendt E., Guergueltcheva V., Rao C. V., Tournev I., Gondim F. A., D'Hooghe M., Van Gerwen V., et al. (2006) Disrupted function and axonal distribution of mutant tyrosyl-tRNA synthetase in dominant intermediate Charcot-Marie-Tooth neuropathy. Nat. Genet. 38, 197–202 10.1038/ng1727 [DOI] [PubMed] [Google Scholar]
- 72. Nangle L. A., Zhang W., Xie W., Yang X. L., and Schimmel P. (2007) Charcot-Marie-Tooth disease-associated mutant tRNA synthetases linked to altered dimer interface and neurite distribution defect. Proc. Natl. Acad. Sci. U.S.A. 104, 11239–11244 10.1073/pnas.0705055104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. McLaughlin H. M., Sakaguchi R., Giblin W., NISC Comparative Sequencing Program, Wilson T. E., Biesecker L., Lupski J. R., Talbot K., Vance J. M., Züchner S., Lee Y. C., Kennerson M., Hou Y. M., Nicholson G., and Antonellis A. (2012) A recurrent loss-of-function alanyl-tRNA synthetase (AARS) mutation in patients with Charcot-Marie-Tooth disease type 2N (CMT2N). Hum. Mutat. 33, 244–253 10.1002/humu.21635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Vester A., Velez-Ruiz G., McLaughlin H. M., NISC Comparative Sequencing Program, Lupski J. R., Talbot K., Vance J. M., Züchner S., Roda R. H., Fischbeck K. H., Biesecker L. G., Nicholson G., Beg A. A., and Antonellis A. (2013) A loss-of-function variant in the human histidyl-tRNA synthetase (HARS) gene is neurotoxic in vivo. Hum. Mutat. 34, 191–199 10.1002/humu.22210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Abbott J. A., Meyer-Schuman R., Lupo V., Feely S., Mademan I., Oprescu S. N., Griffin L. B., Alberti M. A., Casasnovas C., Aharoni S., Basel-Vanagaite L., Züchner S., De Jonghe P., Baets J., Shy M. E., Espinós C., et al. (2018) Substrate interaction defects in histidyl-tRNA synthetase linked to dominant axonal peripheral neuropathy. Hum. Mutat. 39, 415–432 10.1002/humu.23380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. McLaughlin H. M., Sakaguchi R., Liu C., Igarashi T., Pehlivan D., Chu K., Iyer R., Cruz P., Cherukuri P. F., Hansen N. F., Mullikin J. C., NISC Comparative Sequencing Program, Biesecker L. G., Wilson T. E., Ionasescu V., et al. (2010) Compound heterozygosity for loss-of-function lysyl-tRNA synthetase mutations in a patient with peripheral neuropathy. Am. J. Hum. Genet. 87, 560–566 10.1016/j.ajhg.2010.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Gonzalez M., McLaughlin H., Houlden H., Guo M., Yo-Tsen L., Hadjivassilious M., Speziani F., Yang X. L., Antonellis A., Reilly M. M., Züchner S., and Inherited Neuropathy Consortium (2013) Exome sequencing identifies a significant variant in methionyl-tRNA synthetase (MARS) in a family with late-onset CMT2. J. Neurol. Neurosurg. Psychiatry 84, 1247–1249 10.1136/jnnp-2013-305049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Witt L. J., Curran J. J., and Strek M. E. (2016) The diagnosis and treatment of antisynthetase syndrome. Clin. Pulm. Med. 23, 218–226 10.1097/CPM.0000000000000171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Nishikai M., and Reichlin M. (1980) Heterogeneity of precipitating antibodies in polymyositis and dermatomyositis. Characterization of the Jo-1 antibody system. Arthritis Rheum. 23, 881–888 10.1002/art.1780230802 [DOI] [PubMed] [Google Scholar]
- 80. Targoff I. N. (1990) Autoantibodies to aminoacyl-transfer RNA synthetases for isoleucine and glycine. Two additional synthetases are antigenic in myositis. J. Immunol. 144, 1737–1743 [PubMed] [Google Scholar]
- 81. Hirakata M., Suwa A., Nagai S., Kron M. A., Trieu E. P., Mimori T., Akizuki M., and Targoff I. N. (1999) Anti-KS: identification of autoantibodies to asparaginyl-transfer RNA synthetase associated with interstitial lung disease. J. Immunol. 162, 2315–2320 [PubMed] [Google Scholar]
- 82. Betteridge Z., Gunawardena H., North J., Slinn J., and McHugh N. (2007), Anti-synthetase syndrome: a new autoantibody to phenylalanyl transfer RNA synthetase (anti-Zo) associated with polymyositis and interstitial pneumonia. Rheumatology 46, 1005–1008 10.1093/rheumatology/kem045 [DOI] [PubMed] [Google Scholar]
- 83. Hashish L., Trieu E. P., Sadanandan P., and Targoff I. N. (2005) Identification of autoantibodies to tyrosyl-tRNA synthetase in dermatomyositis with features consistent with anti-synthetase syndrome (abstract). Arthritis Rheum. 52, S312 [Google Scholar]
- 84. Labirua-Iturburu A., Selva-O'Callaghan A., Vincze M., Dankó K., Vencovsky J., Fisher B., Charles P., Dastmalchi M., and Lundberg I. E. (2012) Anti-PL-7 (anti-threonyl-tRNA synthetase) antisynthetase syndrome: clinical manifestations in a series of patients from a European multicenter study (EUMYONET) and review of the literature. Medicine 91, 206–211 10.1097/MD.0b013e318260977c [DOI] [PubMed] [Google Scholar]
- 85. Hu J., Locasale J. W., Bielas J. H., O'Sullivan J., Sheahan K., Cantley L. C., Vander Heiden M. G., and Vitkup D. (2013) Heterogeneity of tumor-induced gene expression changes in the human metabolic network. Nat. Biotechnol. 31, 522–529 10.1038/nbt.2530 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.