SUMMARY
Localization of mRNA and small RNAs (sRNA) is important for understanding their function. Fluorescent in situ hybridization (FISH) has been used extensively in animal systems to study sRNA localization and expression. However, current methods for fluorescent in situ detection of sRNA in plant tissues are less developed. Here we report a protocol for sRNA-FISH, for efficient fluorescent detection of sRNAs in plants. sRNA-FISH is suitable for application in diverse plant species and tissue types. The use of Locked Nucleic Acid (LNA) probes and antibodies conjugated with different fluorophores allows the detection of two sRNAs in the same sample. Using this method, we successfully detected co-localization of miR2275 and a 24-nt phasiRNA (phased siRNA) in maize anther tapetal and archesporial cells. We describe how to overcome the common problem of the wide range of autofluorescence in embedded plant tissue using linear spectral unmixing on a laser scanning confocal microscope. For highly autofluorescent samples, we show that multi-photon fluorescence excitation microscopy can be used to separate the target sRNA-FISH signal from background autofluorescence. In contrast to colorimetric in situs, sRNA-FISH signal can be imaged using super-resolution microscopy to examine the sub-cellular localization of sRNAs. We detected maize miR2275 by super-resolution structured illumination microscopy (SR-SIM) and direct stochastic optical reconstruction microscopy (dSTORM). In this study, we describe how we overcame the challenges of adapting fluorescent in situ hybridization for imaging in plant tissue and provide a step-by-step sRNA-FISH protocol for studying sRNAs at the cellular and even subcellular level.
Keywords: Zea mays, Litchi chinensis, Oryza sativa, fluorescent in situ hybridization (FISH), microRNA, sRNA, multi-photon microscopy, immunofluorescence, LNA probes
SIGNIFICANCE STATEMENT
Fluorescence-based microscopy methods are challenging in plants due to a broad range of interfering autofluorescence. Here, we provide a detailed protocol to detect small RNAs by fluorescence in situ hybridization (FISH) that can specifically detect multiple targets and is compatible with advanced imaging technologies, such as super-resolution microscopy.
INTRODUCTION
RNA is information, and localization of this information is critical for its function. Plants and animals have several pathways leading to the production of developmental and functionally important small RNAs (sRNAs) (21 to 24 nt in size). These sRNAs can act in a homology-dependent manner to guide transcriptional and post-transcriptional silencing (Reinhart et al., 2002, Xie et al., 2010, Berezikov, 2011). Recent studies demonstrate cross-kingdom and host-pathogen movement of miRNAs that can regulate gene expression in vitro (Shahid et al., 2018). The subcellular localizations of the components of microRNA (miRNA) and short interfering RNA (siRNA) pathways have been described, including Cajal bodies (CBs) (Fang and Spector, 2007, Pontes and Pikaard, 2008), dicing bodies (D-bodies) (Fang and Spector, 2007), processing bodies (P-bodies) (Pontes and Pikaard, 2008), and more recently described, membrane-bound polysomes (Li et al., 2016). In animal cells, siRNA and miRNA dicing occurs in the nucleus and cytosol (Fang and Spector, 2007). In plant cells, miRNA processing occurs in the nucleus via DCL1 (DICER-LIKE 1) and other miRNA processing proteins (Fang and Spector, 2007).
In the past, precise subcellular and even cellular imaging of production of sRNAs was challenging in plants. Two major methods include green fluorescent protein (GFP)-based sensors and colorimetric in situ hybridization. sRNAs have been detected indirectly using a GFP-based small RNA sensor that detects the silencing of GFP by a small RNA, such as miR156 (Nodine and Bartel, 2010). This method has been used in grafting experiments to detect the shoot-to-root movement of mobile sRNAs that direct transcriptional gene silencing (TGS) (Melnyk et al., 2011). Similar GFP-based small RNA sensors have been used in C. elegans to investigate systemic spreading of gene-silencing between tissues (Winston et al., 2002). These sensors reveal a loss of signal, but lack quantitative and precise spatial resolution. Methods for localization of mRNA by in situ hybridization are routine, but they lack the sensitivity to directly adapting these methods for most sRNAs. A major advance occurred with the advent of locked nucleic acid (LNA) oligonucleotide probes that have improved affinity, sensitivity and specificity (Vester and Wengel, 2004). LNAs have been used in combination with NBT/BCIP (nitro blue tetrazolum and 5-bromo-4-chloro-3-indolyl phosphate) for colorimetric localization of sRNAs in the maize shoot apex (Javelle and Timmermans, 2012), mouse brain (Bak et al., 2008), Drosophila embryos (Rozhkov et al., 2011) and, by us, in maize anthers (Zhai et al., 2015). Using this method, hydrolysis of BCIP by phosphatase produces a blue-colored precipitate at the site of enzymatic activity that is easy to see by transmitted light microscopy but cannot be detected by microscopes that require fluorescence (Trinh le et al., 2007). Whole plant- or tissue-level in situ hybridizations were made possible by using EDC (1-Ethyl-3–3-dimethyl-aminopropyl carbodiimide), a chemical that crosslinks the 5’ end of small RNAs to protein, with a miRNA preserved through the washes and tissue clearing (Ghosh Dastidar et al., 2016). However, these non-fluorescent, colorimetric methods are not easily modified for multiplexed detection, and they are poorly suited for the generation of three-dimensional (3D) images.
Fluorescent in situ hybridization (FISH) has been used extensively in animal systems to study sRNA localization and expression. Obernosterer et al. (2007) used a Fast Red substrate instead of NBT/BCIP for detection of miRNAs in mouse brain. Dual target fluorescence in situ hybridization assays were also used for detecting pathogens in cell cultures (Shah et al., 2017), as well as the specific chromosomal location related to invasive breast tumor (Walker et al., 2013). In higher eukaryotes, the fine structure of RNA processing bodies, including P-bodies, D-bodies, and CBs, have been challenging to observe using conventional microscope because of their small size (300–500 nm in diameter), which is close to the diffraction limit of light (Mito et al., 2016). Meanwhile, super-resolution microscopy, including structured illumination microscopy (SIM) and stochastic optical reconstruction microscopy (STORM), images beyond the diffraction limit of light. Markaki et al. (2013) combined 3D SIM and FISH on cultured mammalian cells to analyze the spatial relations and substructures of nuclear targets. STORM was used to determine the relative distance between nascent transcripts, with a precision of a few tens of nanometers, in combination with RNA-FISH (Larkin and Cook, 2016). The sensitivity and specificity of these methods inspired us to develop a similar technique for plant tissues, in combination with LNA-locked in situ hybridization for detection of sRNA targets.
The sRNA-FISH we developed is based on the colorimetric sRNA in situ method (Javelle and Timmermans, 2012). We show that sRNA-FISH can be used to assay two sRNA targets in the same sample. Furthermore, plants exhibit strong autofluorescence, which often confounds FISH imaging. We show that for many samples, laser scanning confocal microscopy can be used to spectrally separate autofluorescence from the RNA-FISH signal. For highly autofluorescent samples, multi-photon fluorescence excitation alters the autofluorescence spectra so that it can be easily separated from the sRNA-FISH signal, leading to an increase in the signal-to-noise ratio and specificity. With little adaptation, the sRNA-FISH protocol can be used with super-resolution SIM and STORM for sub-cellular localization of small RNAs.
RESULTS AND DISCUSSION
Development of fluorescent in situ methods to analyze sRNA localization
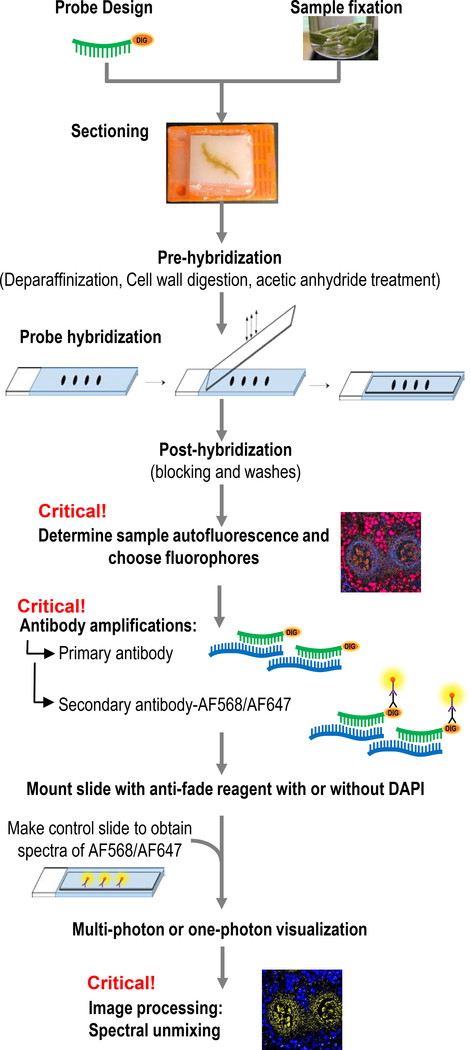
Maize (Zea mays) anthers from early development stages were selected as the model system to develop a sRNA-FISH method. Our method is modified based on the Javelle and Timmermans protocol (2012) for colorimetric sRNA in situ hybridization in plant tissues. Figure 1 provides an overview of the workflow. A detailed step-by-step protocol and reagents list can be found in Supplementary Protocol S1.
Figure 1. Workflow of sRNA-FISH.
Starting with sample preparation and probe design, tissues were fixed, embedded, sectioned and adhered to glass slides. Critical steps include determining sample autofluorescence and choosing antibodies with the right fluorophore combinations. After imaging, linear spectral unmixing is necessary for precise localization of sRNAs.
Samples, in this case maize flower buds, were dissected and immediately fixed to avoid sRNA degradation. The fixative was in PHEM (PIPES, HEPES, EGTA, MgCl2) buffer because it provides better preservation of overall cell structure compared with PBS (phosphate-buffered saline) buffer (see Experimental Procedure and Supplementary Protocol S1). PHEM was originally developed to preserve the microtubule distribution and pericentriolar material (Houliston et al., 1987). Another critical step is vacuum infiltration; it helps the fixative penetrate into tissues with air spaces. For hybridizations, we used LNA-modified probes with a digoxigenin N-hydroxysuccinimide (NHS) ester group added to the 5’ or 3’ end of the probes. LNA probes have high sensitivity and specificity; they are suitable for analysis of short RNA and DNA targets (Vester and Wengel, 2004). After pre-hybridization, probe hybridization, and post-hybridization washing and blocking, probes can either be detected using the colorimetric method or by immunofluorescence methods. Our protocol up until the post-hybridization step can be used for either fluorescent or colorimetric detection. Both methods have their advantages and we compare the methods below. For immunofluorescence detection, samples are incubated with primary antibodies to the probes and secondary antibodies with chosen fluorophores. We tested two primary-secondary antibody combinations. Mouse anti-DIG IgG primary with goat anti-mouse conjugated with Alexa Fluor® 488 (AF488) resulted in background signal in anther cells with either the specific probe or scramble control. In contrast, the combination of sheep anti-Digoxigenin Fab fragment primary and donkey anti-sheep IgG AF568 secondary led to successful in situs in maize tissues. After antibody washing, samples were mounted in SlowFade® Gold or Diamond Antifade Mountant and stored at 4°C until imaging.
Linear spectral unmixing of autofluorescence for specific detection of sRNA-FISH signal
Due to the autofluorescence in paraffin-embedded plant tissue, all confocal and multiphoton microscope images needed to be spectrally unmixed. Laser-scanning confocal microscopes (LSCM) with spectral imaging are becoming increasingly more common (Zimmermann et al., 2014); nearly all microscopes from the major manufacturers can be equipped with this capability. Linear spectral unmixing of fluorophores from autofluorescence is essential for sRNA-FISH protocol described here because nearly all paraffin-embedded plant tissue has a significant amount of autofluorescence that can vary based on species or tissue type. To conduct linear spectral unmixing, two important controls are needed. First, a sample with no secondary antibodies, which have the fluorescence dye conjugate, must be examined with different laser excitations to determine the autofluorescence spectra of the paraffin embedded sample section. The spectra must be saved and will be used later for unmixing. This step is critical, as it provides the spectral information for choosing fluorophores that have the least amount of overlap with the autofluorescence. Second, once a fluorophore is chosen, a solution of the pure secondary antibody conjugated to the fluorophore should be mixed with the mounting media and imaged to obtain pure spectra of the fluorophore. The spectra of the autofluorescence and the pure fluorophore fluorescence can then be used to unmix the tissue autofluorescence from the specific sRNA-FISH signal. All of the laser scanning confocal and multiphoton microscopy images shown here were acquired using spectral detection followed by linear spectral unmixing.
Comparison of sRNA-FISH and colorimetric detection of the same targets
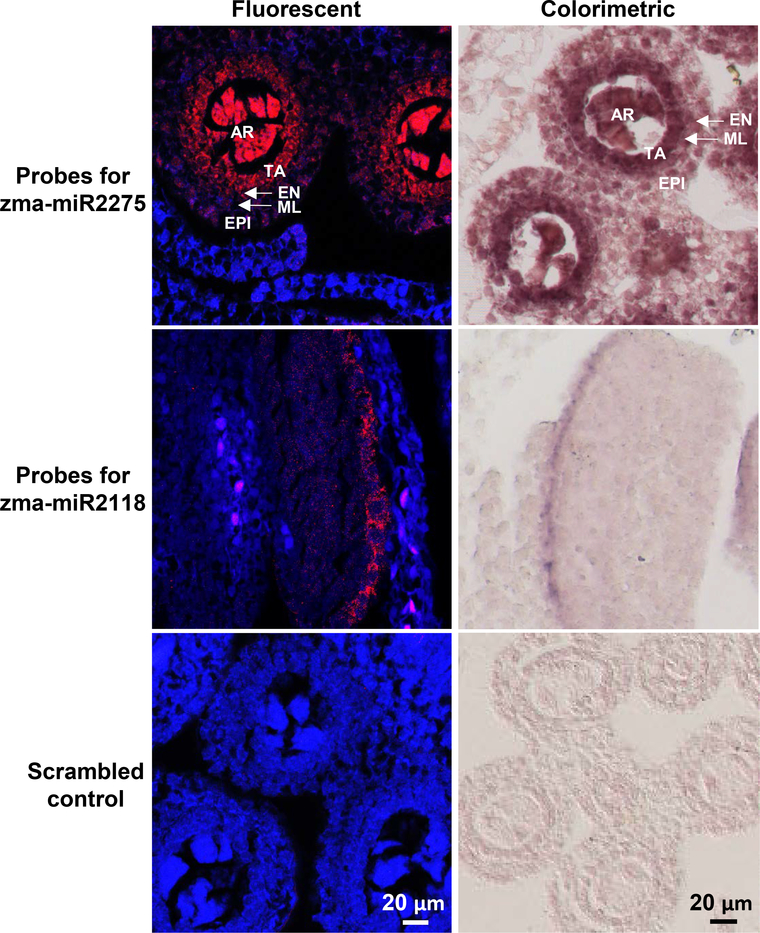
In order to examine the specificity and sensitivity of sRNA-FISH, we compared our method to the colorimetric method. miR2275 and miR2118 are abundant miRNAs that are important for anther development, with a maximum observed abundance in fertile maize anthers of 12,100 transcripts per 10 million reads (TP10M) for miR2275 in 1 mm anthers, and 11,433 TP10M for miR2118 in 0.4 mm anthers (Zhai et al., 2015). Both miRNAs have known localization patterns, with miR2275 abundant in maize anthers of 1.0 to 2.0 mm and localized to the tapetal layer and archesporial cells (Zhai et al., 2015). miR2118 is enriched in pre-meiotic maize anthers and mainly localizes to the epidermal cell layer (Zhai et al., 2015). We designed probes for both miR2275 and miR2118 (probes sequences are listed in Table 1). sRNA-FISH and colorimetric methods both showed the same localization of miR2275 and miR2118 for 1.5 mm and 0.4 mm maize anthers respectively (Figure 2). For miR2275, we observed strong signal (red in fluorescent and dark color in colorimetric) in the tapetal layer and archesporial cells using both methods. For miR2118, we observed signal mainly in the epidermal layer of early-stage anthers, also using both methods. The specific signal in the different locations for miR2275 and miR2118 probes were detectable compared to no signal for the scrambled probe control. The traditional colorimetric in situ method provides more signal amplification, which can be seen for miR2118 detection. It is the best method when maximum sensitivity is required for tissue-level expression studies of one sRNA target. FISH is a powerful strategy to overcome the limitations of colorimetric-based detection schemes (Clay and Ramakrishnan, 2005, Barroso-Chinea et al., 2007). Specifically, our sRNA-FISH method is a better choice when more than one sRNA target needs to be detected simultaneously and when cellular and subcellular localization of sRNAs is required. Below we describe how our basic sRNA-FISH method can be applied for the detection of two sRNA targets or super-resolution localization of sRNAs.
Table 1:
Information on probes used in this study
| miRNA | miRNA abundance (TP10M) | Probe sequence | Probe Tm (°C) | Hybridization temperature (°C) | Probe concentration |
|---|---|---|---|---|---|
| zma-miR2118 | 4,064 | /5DigN/TAGGAATGGGAGGCATCGGGAA | 86 | 53.3 | 10 nM |
| 24-phasiRNA | 16,318 | GGCCAAGGTCCGGTCCAACAACT/36-FAM/ | 87 | 53.3 | 10 nM |
| zma-miR2275 | 12,100 | GGTTCAGTTGCCTTAATCCT/3DigN/ | 88 | 53.3 | 250 nM |
| 24-phasiRNA for litchi | 837 | TCTCATGTTTTCTCATAGGTCT/3DigN/ | 83 | 53.3 | 10 nM |
| Osa-miR1874 | 59,000 | ATCGGGTTACACCTCCATCCATA/3Dig_N/ | 86 | 53.3 | 100 nM |
| Scrambled control | NA | /5DigN/GTGTAACACGTCTATACGCCCA | 87 | 53.3 | 10 nM |
Footnote: Tm, melting temperature; 5DigN, 5’ Digoxigenin NHS Ester
Figure 2. Comparison of sRNA-FISH and traditional non-fluorescent in situ hybridization.
Probes reverse complement with zma-miR2275, zma-miR2118 and scrambled control were used to hybridize to maize anthers (~1 mm and ~0.4 mm in length respectively). The fluorescent micrographs were taken with laser scanning confocal microscopy. miR2275 localized mostly in tapetal and archesporial cells (top row, red), and miR2118 localized to cells in the epidermal layer (middle row, red). No signal was detected in the scramble control (bottom row). The background (blue) was spectrally unmixed from AF568 fluorescence. The micrographs of the non-fluorescent, colorimetric in situs were taken with bright-field microscopy and have the same localization pattern. EPI, epidermis; EN, endothecium; ML, middle layer; TA, tapetal layer; AR, archesporial cells. Scale bars = 20 μm for all images.
Applying sRNA-FISH to detect and co-localize two sRNAs in the same sample
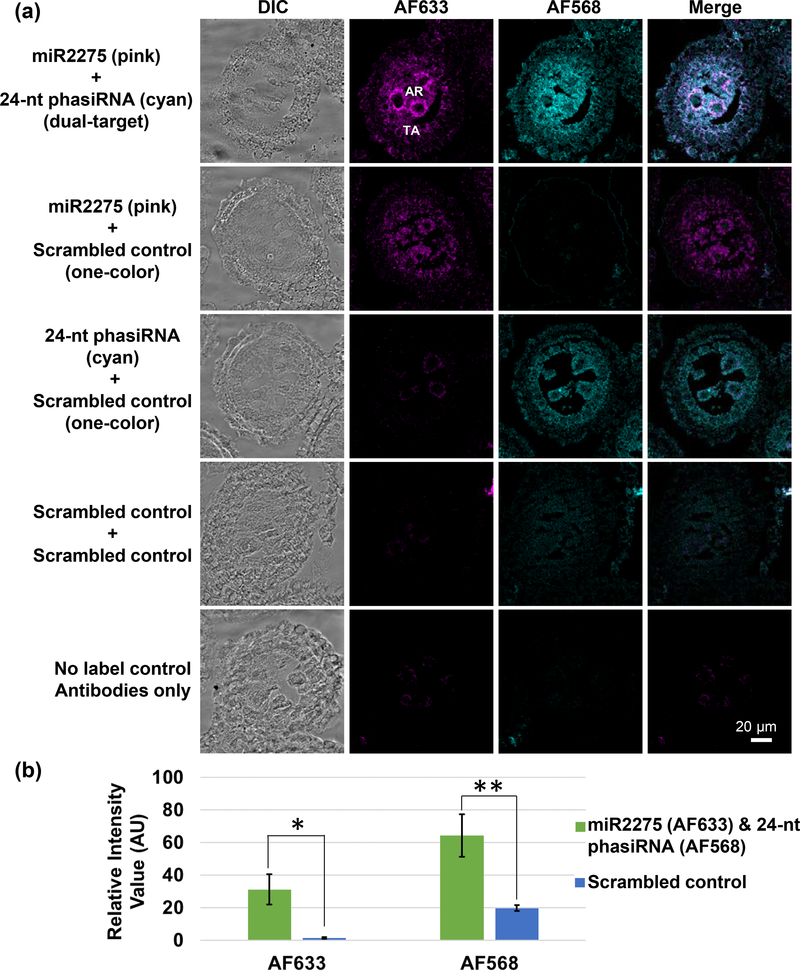
To extend our in situ method from one sRNA target to two sRNA targets, we included LNA probes conjugated with 3’ 6-carboxyfluorescein (FAM-), which is an NHS ester-modified single isomer derivative of fluorescein. The FAM fluorescence was not detected directly (quenched by the hybridization protocol), but rather, was amplified and detected with anti-fluorescein rabbit antibody and donkey Fab anti-rabbit IgG secondary conjugated with Alexa Fluor® 568. To test this method, we designed two LNA probes to miR2275 and its 24-nt phasiRNA. The probe for a miR2275 was conjugated with 3’-DIG and the probe for 24-nt phasiRNA was conjugated to 3’ FAM. They were hybridized to 1.5 mm maize anther sections because the sRNA targets are both present and abundant at this stage (Zhai et al., 2015). Using both DIG- and fluorescein-labeled probes in the same hybridization experiment, we were able to detect specific fluorescence for both miR2275 and the 24-nt phasiRNA compared to the scrambled LNA probe controls (Figure 3). The 24-nt phasiRNA (Figure 3, cyan) and miR2275 (Figure 3, magenta) co-localized to the tapetal layer and archesporial cells.
Figure 3. Dual-target sRNA-FISH for maize anthers.
(a) sRNA-FISH detected both zma-miR2275 (detected in the AF633 channel; magenta) and the 24 nt phasiRNA (detected in the AF568 channel; cyan) in the tapetal layer and archesporial cells. Each image was collected in spectra mode with laser scanning confocal microscopy and then spectrally unmixed using Zen Software. Bright-field and merged images were also shown for each image. TA, tapetal layer; AR, archesporial cells. Scale bars = 20 μm for all images. (b) Quantification of the AF633 and AF568 signal intensity in dual-target sRNA FISH and controls. (Significance level: < 0.05, *; < 0.01, **).
Use of multi-photon excitation to mitigate plant autofluorescence
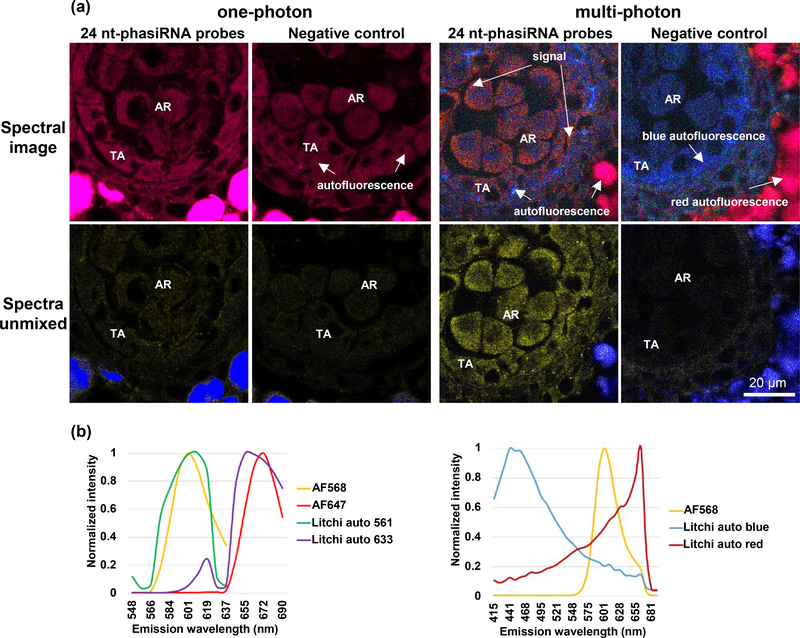
Above, we have shown that our sRNA-FISH method can be used with LSCM for both single and dual target sRNA detection in maize anthers. However, LSCM uses one-photon excitation that can exhibit much higher levels of autofluorescence in certain types of plant tissue and species. Imaging is dependent on having a high signal-to-background ratio; and, these high levels of autofluorescence can prevent the detection of weaker signals below the autofluorescence level. For example, we were unable to use one-photon detection of sRNA-FISH in litchi (Litchi chinensis Sonn.) anthers because the autofluorescence with either 561 nm or 633 nm laser excitation was similar to the emission spectrum of AF568 or AF647 fluorophores (Figure 4). This was evident in both the lambda-coded spectral image that depicts true color and the graphed spectra of the autofluorescence and AF dyes (Figure 4a). In comparison, using multi-photon excitation, we were able to overcome this autofluorescence issue. Imaging with 745 nm multi-photon excitation exhibited blue-to-green autofluorescence that was well separated from the distinct orange fluorescent signal of AF568. The 745 nm multi-photon excitation also exhibited a second autofluorescence peak around 655 nm that was mainly outside of the anther. The emission peak of an AF568-conjugated secondary antibody (603 nm) falls between the two autofluorescence spectra, making it possible to linear spectrally unmix it from the two autofluorescence peaks (Figure 4). We also tested this approach in rice (Oryza sativa) endosperm, another highly autofluorescent tissue type. The exact same area of the endosperm section was imaged with either one-photon or multi-photon excitation (Supplementary Figure 1). sRNA-FISH of osa-miR1874 showed numerous discrete spots of signal by multi-photon excitation and many of these spots could not be spectrally unmixed from the autofluorescence using one-photon excitation. In summary, multi-photon excitation microscopy can be used to overcome the high autofluorescence levels of plant samples that cannot be imaged with one-photon LSCM.
Figure 4. Comparison of one-photon and multi-photon excitation in litchi anthers.
(a) Images show 24 nt-phasiRNA localization in litchi, stage IV anthers. In images acquired with one-photon excitation, the spectra from strong autofluorescence (red) overlaps with AF647 signal. In images acquired with multi-photon excitation, distinct AF568 can be spectrally unmixed from background autofluorescence (both red and blue). TA, tapetal layer; AR, archesporial cells. Scale bar = 20 μm for all images. (b) Spectra profile of AF568/AF647 and litchi anther autofluorescence using one-photon and multi-photon excitation. Both AF568 and AF647 spectra are very close to background autofluorescence using one-photon excitation. AF568 has a distinct spectra compared with background autofluorescence using multi-photon excitation.
Development of sRNA-FISH for detecting sub-cellular localization of sRNAs
Introducing fluorescence to in situ hybridization enabled sub-cellular localization of RNA. However, the diffraction limit of light restricts the resolution of LSCM to ~200 nm in the focal plane (x,y) and ~450 nm in the optical (z) axis, making details of the subcellular structures and RNA assemblies unresolvable (Hell, 2007, Huang et al., 2016). A multitude of super-resolution light microscopy techniques have emerged that can surpass the diffraction limit of light (Rust et al., 2006, Huang et al., 2010). Here, we examined if two super-resolution approaches, SR-SIM and direct STORM (dSTORM), can be applied to sRNA-FISH. SR-SIM utilizes Moiré fringes to render otherwise unresolvable high-resolution information (Gustafsson, 2005). The high-frequency information in each SR-SIM image is used to double the resolution about 100 nm (Kner et al., 2009). Probes against miR2275 were used to detect miR2275 expression in pre-meiotic stage anthers of maize. The result was very similar to fluorescent and colorimetric detection of miR2275 expression in the same stage of maize anthers – that is, Using SR-SIM, miR2275 is also mainly localized to the tapetal layer and archesporial cells. We also detected miR2275 in secondary parietal cells, which later give rise to the middle layer and tapetal cells (Kelliher et al., 2014) (Figure 5). The original STORM method used two dyes and required conjugating the dyes to the same antibody. In contrast, dSTORM uses only one dye and commercially available secondary antibodies are readily available (Heilemann et al., 2008). Both methods require driving the molecules into a dark state and detecting single molecules that stochastically enter the fluorescent state; a Gaussian fit of fluorescence of spatially separated single molecules can be used to determine their location to approximately 20 nm (Hell, 2007). That single molecule detection is also the key to capturing a dSTORM image without linear spectral unmixing. A single AF647 dye molecule emits significantly more photons than autofluorescence. Filtering based on the high photon counts of AF647 was used to minimize background autofluorescence without linear spectral unmixing. Using dSTORM (Figure 6), we were able to obtain <20 nm localization precision of miR2275 at the subcellular level (Supplementary Table 1), although the size of the antibodies will decrease the actual resolution. With the super-resolution of dSTORM images, the detailed localization of each sRNA can be achieved. In Figure 6b, we were able to capture the precise localization of miR2275 around and in the nucleus and in the cytosol. These localization events as well as brightness and precision radius were documented with exact positions as x and y coordinates (Supplementary Table 1).
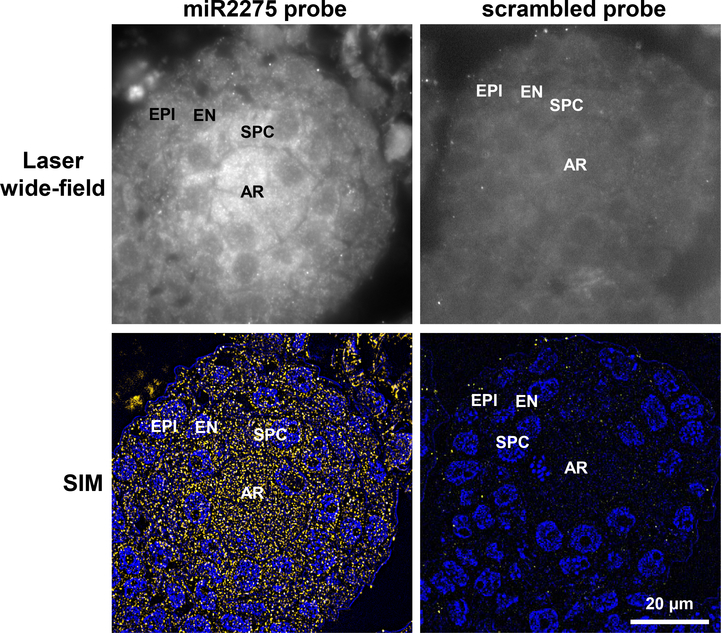
Figure 5. Localization of miR2275 in premeiotic maize anthers using SIM.
Top left panel: Laser wide-field images shown miR2275 is detected in the archesporial cells and secondary parietal cells; the latter give rise to the middle layer and tapetum. Bottom left panel: detection of miR2275 using super-resolution structured illumination. miR2275 is localized to archesporial and secondary parietal cells. Right panels are images of the scrambled probe control. AR, archesporial cells; SPC, secondary parietal cells; EN, endothecium; EPI, epidermis. Scale bar = 20 μm for all images.
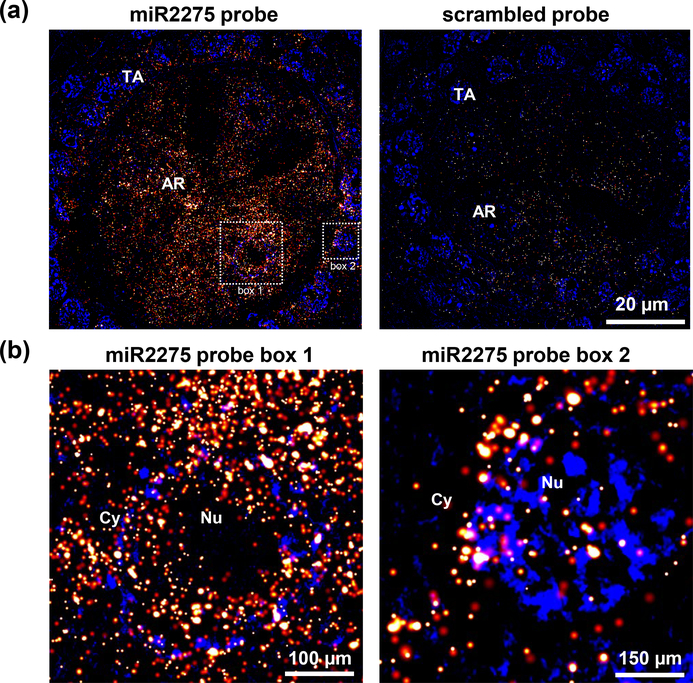
Figure 6. Localization of miR2275 in premeiotic maize anthers using dSTORM.
(a) miR2275 was detected in the tapetal layer and archesporial cells. In comparison, scrambled probe yield very low signal. (b) Higher magnification images of box #1 and 2 showing localization around nucleus (Nu), and in the cytosol (Cy). TA, tapetal layer; AR, archesporial cells.
Overall, fluorescent in situ hybridization in plant tissue is challenging, and in this work, we present a sRNA-FISH protocol that can use many of the advantages of fluorescence, including multiple probes to detect multiple targets in the same sample and high resolution localization at the cellular and subcellular level. Another challenge when working with plant tissue is the amount of autofluorescence in many tissues. Multi-photon excitation with a 745 nm laser results in two distinct autofluorescence peaks in most of the plants we tested, including, rice and litchi; this gives us a wide spectral window for selecting fluorophores. Single-photon excitation can be used for tissues that exhibit less autofluorescence, such as maize anthers, and LSCM systems are more readily accessible. Here we demonstrate that our sRNA-FISH method can be applied to a wide variety of plant tissue types and species. It complements the traditional colorimetric ISH method that has much higher signal amplification. Our detailed step-by-step protocol provided in the Supplementary Protocol file is identical for colorimetric ISH and sRNA-FISH up until the step of antibody incubation (step 25), and it can be used to compare the two methods. We have found that the colorimetric ISH method should be applied when aiming towards maximum sensitivity at the tissue-to-organ level. However, these non-fluorescent, colorimetric methods are not easily modified for multiplexed detection. In contrast, the sRNA-FISH method can be easily adapted to dual-detection of two RNA targets. The fact that sRNA-FISH requires antibody amplification complicates its use for absolute copy number quantification of sRNAs. A bioinformatically designed, oligonucleotide-based technology called OligoPAINT, enables single-molecule super-resolution imaging of chromosomes without antibody amplification (Beliveau et al., 2015). Adaptation of a similar method to small RNAs should yield fine resolution imaging method with precise number of fluorophores or binding sites.
Finally, the cellular mechanism of RNA transport and its role in small RNA biogenesis is still unclear. Nucleolus-associated CBs in plants have been implicated as sites of siRNA and miRNA biogenesis (Pontes and Pikaard, 2008). DCL1 and DCL3-AGO4 (Argonaute protein 4) siRNA processing centers can be located in highly dynamic CBs (Pontes and Pikaard, 2008). However, DCL1 and HYL1 (dsRNA-binding domain-like superfamily protein 1) can function in a non-CB-dependent manner when localized to D-bodies, suggesting D-bodies have a role in pri-miRNA processing and miRNA biogenesis (Fang and Spector, 2007). For both animals and plants, P-bodies contain all the components for miRNA-directed cleavage: Argonaute proteins and miRNAs forming the RNA-induced silencing complex (RISC), and untranslated mRNA (Liu et al., 2005). Cellular fractionation and molecular analysis of phased siRNA (phasiRNA) biogenesis showed that phasiRNAs are generated from miRNA-targeted transcripts on membrane-bound polysomes, and the miRNAs are recruited to the membrane in an AGO1-dependent manner (Li et al., 2016). In the future, sRNA-FISH may be combined with other, compatible fluorescence-based methods to detect other RNA and protein targets. The ability to localize small RNAs, potentially other RNA targets including mRNA transcription and sRNA precursors, together with proteinaceous biogenesis components all at a subcellular level may yield breakthroughs in our understanding of RNAs.
EXPERIMENTAL PROCEDURES
Plant material
Maize samples were kindly provided by the Walbot lab (Stanford University). Anthers of the W23 inbred line were grown in Stanford, CA under greenhouse conditions. Anthers were dissected and measured using a micrometer as previously described (Kelliher and Walbot, 2011). Rice samples were provided by Dr. Yuanlong Liu from the Donald Danforth Plant Science Center. Litchi samples were kindly provided by Dr. Rui Xia in South China Agricultural University (China). Arabidopsis (Col-0) were grown on 0.5 MS/0.8% agar (MS agar) plates in controlled-environment chambers under 16 h light – 8 h dark, 23°C temperature.
Probe design
LNA-modified oligonucleotide probes were synthesized by Exiqon (Exiqon A/S, Vedbaek, Denmark). This probe design webpage can be found at: https://www.qiagen.com/us/shop/pcr/primer-sets/custom-lna-oligonucleotides/#orderinginformation. The input requires specific sRNA sequences and species. The output contains probe sequences, TM, and molecular weight. The LNA position information will not be released. Scrambled control probes were directly ordered from Exqion (Cat. No. YD00699004). All probes sequences are listed in Table 1.
Sample preparation
Detailed protocol of sample preparation, including fixation and embedding, can be found in Supplementary Protocol S1. Briefly, anthers were dissected and fixed in a 20 ml glass vial using 4% paraformaldehyde in 1xPHEM buffer (5 mM HEPES, 60 mM PIPES, 10 mM EGTA, 2 mM MgCl2 pH 7). Fixation was done in a vacuum chamber at 0.08 MPa for 3 times, 15 min each. After fixation, samples were sent for paraffin embedding at histology lab from Nemours/Alfred I. duPont Hospital for Children (Wilmington, DE). Samples were sectioned using a paraffin microtome and dried on Fisherbrand- Tissue Path SuperFrost- Plus Gold Slides (Thermo Fisher Scientific cat# 15–188-48).
Fluorescent in situ hybridization
Fluorescent in situ hybridization was modified from the protocol by Javelle et al. (2012) by replacing the antibody with primary anti-Digoxigenin Fab fragment (Sigma-Aldrich cat# 11214667001) and secondary donkey anti-sheep IgG (H+L) AF647, AF568 or AF633 (Thermo Fisher Scientific cat# A-21448, A-21099, and A-21100). Two color in situ hybridization, 6-FAM- labeled probe was amplified with anti-fluorescein (Abcam ab19491) from rabbit and donkey F(ab’)2 anti-rabbit IgG H&L (Alexa Fluor® 568) (Abcam ab 175694). For a detailed protocol, please refer to the Supplementary Protocol. Briefly, samples were de-paraffin using Histo-Clear (Fisher scientific, 50–899-90147) and re-hydrate by going through an ethanol series of 95, 80, 70, 50, 30, 10% (vol/vol) (30 sec each) and water (1min) at room temperature. After protease (Sigma, P5147) digestion (20 min, 37°C), samples were treated with 0.2% glycine (Sigma-Aldrich, G8898) for 2 min, followed by a TEA treatment (Triethanolamine; Sigma-Aldrich, 90279) HCl and acetic anhydride (Sigma-Aldrich, A6404). After two washes in 1xPBS buffer, samples were de-hydrated and then hybridized with probes overnight at 53.3°C. 10 ml of hybridization buffer contains 875 μl of nuclease-free H2O, 1.25 ml in situ hybridization salts, 5 ml of deionized formamide, 2.5 ml of 50% (wt/vol) dextran sulfate, 250 μl of 50x Denhardt’s solution, and 125 μl of 100 mg/ml tRNA. Hybridized slides were then washed twice using 0.2x SSC buffer (saline-sodium citrate), blocked in 1x blocking buffer (1% blocking reagent in 1xTBS buffer), and 1x washing buffer (1% wt/vol BSA; Sigma-Aldrich-Aldrich, A7906) and 0.3% Triton x-100 in 1xTBS buffer) for 1 hour each. Samples were then incubated with primary antibody overnight at 4°C followed by 4x washes in 1x washing buffer, 15 min each. Samples were then incubated with a secondary antibody overnight at 4°C followed by 4x washes in 1x washing buffer, 15 min each. After final wash in 1xTBS buffer, samples were mounted using SlowFade- Gold Antifade Mountant (ThermoFisher Scientific, S36936) or SlowFade- Diamond Antifade Mountant (ThermoFisher Scientific, S36967). The other antibody combination we tested, but resulted in nonspecific background plant tissue, was mouse anti-DIG IgG primary (Sigma-Aldrich, #11333062910) with goat anti-mouse conjugated with Alexa Fluor® 488 (AF488) secondary (Thermo Fisher Scientific, #A11017).
Image acquisition
Bright-field images of colorimetric in situs were acquired on a Zeiss Axioplan 2 using an Axiocam MR color camera. Spectral imaging was conducted on a Carl Zeiss LSM 880 laser scanning microscopy capable of both LSCM and multiphoton microscopy. The Zen software (v2.3; Carl Zeiss) was used for both acquisition of spectral images and linear spectral unmixing. Spectral data for pure Alexa Fluor® fluorophores were used as positive controls, and non-labeled samples were used to obtain autofluorescence spectra for linear spectral unmixing. Brightness and contrast of images in the same figure panel were adjusted equally and linearly in Zen software (Carl Zeiss).
Data quantification
For quantification, the intensity value of each image was calculated using ImageJ (Schindelin et al., 2012). Five replicates for each images were used for calculating the significance. P-value was calculated in Microsoft Excel using t-test assuming equal variance.
SR-SIM imaging
SR-SIM images were acquired on a Zeiss Elyra PS.1 with a Plan-Apochromat 63x/1.4 oil objective. The raw data for each channel was collected as 25 images in which the structured illumination was shifted to five different phases and 5 different rotations. For each field of view, a 5-slice z stack was taken and then maximum intensity images was generated using the stacks with Zen software (Carl Zeiss). The 642 nm laser (10%) was used to excite the AF633, and 405 laser (20%) was used to exite DAPI. After image subsets are obtained for each of the five grid orientations, the collection was analyzed in Zen (Carl Zeiss). The images were processed using the same parameters, which are as follows: Theoretical Point Spread Function; Baseline Cut display; SR-Frequency weighting (1); Noise filter (−1); Sectioning (100%, 83%, 83%).
dSTORM Imaging
dSTORM images were taken using a Zeiss Elyra PS.1 super-resolution microscope with a Plan-Apochromat 100x/1.46 oil objective with 642 nm (100%) and 405 (10%) laser excitation. Anther samples were sectioned and dried on Medium Density wide Spectral Band Coverglass (600+/− 100 nm fiducials, density #(100 μm)2 51–150) (HESTZIG LLC.) coated with poly-l-lysine (Sigma). After sRNA fluorescent in situ hybridization, sample then mounted in a dSTORM imaging buffer. The dSTORM buffer was made by mixing three buffers immediately before use: solution A (containing 30 nM Tris/Cl pH 8.5, 1 mM EDTA, 6.25 uM glucose oxidase, and 2.5 μM catalase for oxygen scavenging); solution B containing (250 mM cysteamine-HCL, pH 3); and solution C (containing 250 mM glucose in water). Sample were sealed in a magnetic CF chamber (Chamlide) during imaging. Images were taken with a exposure time of 100 ms, an EMCCD Gain 30, and 40,000 frames in total. Images were aligned using fiducial-based algorithm. After image subsets were obtained for each image, the raw data was analyzed in Zen software (Carl Zeiss). The images were processed using the same parameters, which are listed below: Discard overlapping molecules; Peak Mask Size (9); Peak Intensity to Noise (8.0); Fit model (x,y 2D Gauss Fit); Average before localization using Drift Correction (Min On time 10%, Capture Radium 5.0 pixel); Grouping using OFF Gap 10% with a Capture Radium of 1.0 pixel. After the dSTORM images were generated, these filters were applied to each image: Number Photons (500–5000) and Chi Square (0.5–100.00).
Supplementary Material
Comparison of one-photon and multi-photon excitation on imaging resolution in rice endosperm amyloplast.
Coordinates of miR2275 localization detected by sRNA-FISH and dSTORM imaging.
Detailed protocol for sRNA-FISH in plant tissues.
ACKNOWLEDGEMENTS
This project was supported by the US NSF Plant Genome Research Program, awards 1649424 andv1754097. We would like to thank Marie Javelle and Marja Timmermans for their guide on in situ hybridization using LNA probes, members of the Meyers and Caplan labs for help and support. Microscopy equipment was acquired with a shared instrumentation grant (S10 OD016361) and access was supported by the NIH-NIGMS (P20 GM103446), the NSF (IIA-1301765) and the State of Delaware.
Footnotes
CONFLICTS OF INTEREST
The authors declare no conflicts of interest.
REFERENCES
- Bak M, Silahtaroglu A, Moller M, Christensen M, Rath MF, Skryabin B, Tommerup N and Kauppinen S (2008) MicroRNA expression in the adult mouse central nervous system. RNA, 14, 432–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barroso-Chinea P, Aymerich MS, Castle MM, Perez-Manso M, Tunon T, Erro E and Lanciego JL (2007) Detection of two different mRNAs in a single section by dual in situ hybridization: a comparison between colorimetric and fluorescent detection. J Neurosci Meth, 162, 119–128. [DOI] [PubMed] [Google Scholar]
- Beliveau BJ, Boettiger AN, Avendano MS, Jungmann R, McCole RB, Joyce EF, Kim-Kiselak C, Bantignies F, Fonseka CY, Erceg J, Hannan MA, Hoang HG, Colognori D, Lee JT, Shih WM, Yin P, Zhuang X and Wu CT (2015) Single-molecule super-resolution imaging of chromosomes and in situ haplotype visualization using Oligopaint FISH probes. Nat Commun, 6, 7147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berezikov E (2011) Evolution of microRNA diversity and regulation in animals. Nat Rev Gen, 12, 846–860. [DOI] [PubMed] [Google Scholar]
- Clay H and Ramakrishnan L (2005) Multiplex fluorescent in situ hybridization in zebrafish embryos using tyramide signal amplification. Zebrafish, 2, 105–111. [DOI] [PubMed] [Google Scholar]
- Fang Y and Spector DL (2007) Identification of nuclear dicing bodies containing proteins for microRNA biogenesis in living Arabidopsis plants. Current Biology, 17, 818–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh Dastidar M, Mosiolek M, Bleckmann A, Dresselhaus T, Nodine MD and Maizel A (2016) Sensitive whole mount in situ localization of small RNAs in plants. Plant J, 88, 694–702. [DOI] [PubMed] [Google Scholar]
- Gustafsson MGL (2005) Nonlinear structured-illumination microscopy: Wide-field fluorescence imaging with theoretically unlimited resolution. Proc Natl Acad Sci USA, 102, 13081–13086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heilemann M, van de Linde S, Schuttpelz M, Kasper R, Seefeldt B, Mukherjee A, Tinnefeld P and Sauer M (2008) Subdiffraction-resolution fluorescence imaging with conventional fluorescent probes. Angewandte Chemie, 47, 6172–6176. [DOI] [PubMed] [Google Scholar]
- Hell SW (2007) Far-field optical nanoscopy. Science, 316, 1153–1158. [DOI] [PubMed] [Google Scholar]
- Houliston E, Pickering SJ and Maro B (1987) Redistribution of microtubules and pericentriolar material during the development of polarity in mouse blastomeres. J Cell Biol 104, 1299–1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B, Babcock H and Zhuang X (2010) Breaking the diffraction barrier: super-resolution imaging of cells. Cell, 143, 1047–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang F, Sirinakis G, Allgeyer ES, Schroeder LK, Duim WC, Kromann EB, Phan T, Rivera-Molina FE, Myers JR, Irnov I, Lessard M, Zhang Y, Handel MA, Jacobs-Wagner C, Lusk CP, Rothman JE, Toomre D, Booth MJ and Bewersdorf J (2016) Ultra-high resolution 3D imaging of Whole Cells. Cell, 166, 1028–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javelle M and Timmermans MCP (2012) In situ localization of small RNAs in plants by using LNA probes. Nat Protoc, 7, 533–541. [DOI] [PubMed] [Google Scholar]
- Kelliher T, Egger RL, Zhang H and Walbot V (2014) Unresolved issues in pre-meiotic anther development. Front Plant Sci, 5, 347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelliher T and Walbot V (2011) Emergence and patterning of the five cell types of the Zea mays anther locule. Dev Biol, 350, 32–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kner P, Chhun BB, Griffis ER, Winoto L and Gustafsson MG (2009) Super-resolution video microscopy of live cells by structured illumination. Nat Methods, 6, 339–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin JD and Cook PR (2016) Super-resolution measurement of distance between transcription sites using RNA FISH with intronic probes. Methods, 98, 150–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Le B, Ma X, Li S, You C, Yu Y, Zhang B, Liu L, Gao L, Shi T, Zhao Y, Mo B, Cao X and Chen X (2016) Biogenesis of phased siRNAs on membrane-bound polysomes in Arabidopsis. Elife, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Valencia-Sanchez MA, Hannon GJ and Parker R (2005) MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nat Cell Biol, 7, 719–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markaki Y, Smeets D, Cremer M and Schermelleh L (2013) Fluorescence in situ hybridization applications for super-resolution 3D structured illumination microscopy. Methods Mol Biol, 950, 43–64. [DOI] [PubMed] [Google Scholar]
- Melnyk CW, Molnar A, Bassett A and Baulcombe DC (2011) Mobile 24 nt small RNAs direct transcriptional gene silencing in the root meristems of Arabidopsis thaliana. Curr Biol, 21, 1678–1683. [DOI] [PubMed] [Google Scholar]
- Mito M, Kawaguchi T, Hirose T and Nakagawa S (2016) Simultaneous multicolor detection of RNA and proteins using super-resolution microscopy. Methods, 98, 158–165. [DOI] [PubMed] [Google Scholar]
- Nodine MD and Bartel DP (2010) MicroRNAs prevent precocious gene expression and enable pattern formation during plant embryogenesis. Gene Dev, 24, 2678–2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obernosterer G, Martinez J and Alenius M (2007) Locked nucleic acid-based in situ detection of microRNAs in mouse tissue sections. Nat Protoc, 2, 1508–1514. [DOI] [PubMed] [Google Scholar]
- Pontes O and Pikaard CS (2008) siRNA and miRNA processing: new functions for Cajal bodies. Curr Opin Gen Dev, 18, 197–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart BJ, Weinstein EG, Rhoades MW, Bartel B and Bartel DP (2002) MicroRNAs in plants. Gene Dev, 16, 1616–1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozhkov NV, Zelentsova ES, Shostak NG and Evgen’ev MB (2011) Expression of Drosophila virilis retroelements and role of small RNAs in their intrastrain transposition. Plos One, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rust MJ, Bates M and Zhuang X (2006) Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM). Nat Methods, 3, 793–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, White DJ, Hartenstein V, Eliceiri K, Tomancak P and Cardona A (2012) Fiji: an open-source platform for biological-image analysis. Nat Methods, 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah J, Weltman H, Narciso P, Murphy C, Poruri A, Baliga S, Sharon L, York M, Cunningham G, Miller S, Caviedes L, Gilman R, Desmond E and Ramasamy R (2017) Dual color fluorescence in situ hybridization (FISH) assays for detecting Mycobacterium tuberculosis and Mycobacterium avium complexes and related pathogens in cultures. Plos One, 12, e0174989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahid S, Kim G, Johnson NR, Wafula E, Wang F, Coruh C, Bernal-Galeano V, Phifer T, dePamphilis CW, Westwood JH and Axtell MJ (2018) MicroRNAs from the parasitic plant Cuscuta campestris target host messenger RNAs. Nature, 553, 82–85. [DOI] [PubMed] [Google Scholar]
- Trinh le A, McCutchen MD, Bonner-Fraser M, Fraser SE, Bumm LA and McCauley DW (2007) Fluorescent in situ hybridization employing the conventional NBT/BCIP chromogenic stain. BioTechniques, 42, 756–759. [DOI] [PubMed] [Google Scholar]
- Vester B and Wengel J (2004) LNA (locked nucleic acid): high-affinity targeting of complementary RNA and DNA. Biochemistry, 43, 13233–13241. [DOI] [PubMed] [Google Scholar]
- Walker LC, McDonald M, Wells JE, Harris GC, Robinson BA and Morris CM (2013) Dual-Color Fluorescence In Situ Hybridization Reveals an Association of Chromosome 8q22 but Not 8p21 Imbalance with High Grade Invasive Breast Carcinoma. Plos One, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winston WM, Molodowitch C and Hunter CP (2002) Systemic RNAi in C. elegans requires the putative transmembrane protein SID-1. Science, 295, 2456–2459. [DOI] [PubMed] [Google Scholar]
- Xie ZX, Khanna K and Ruan SL (2010) Expression of microRNAs and its regulation in plants. Semin Cell Dev Biol, 21, 790–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai JX, Zhang H, Arikit S, Huang K, Nan GL, Walbot V and Meyers BC (2015) Spatiotemporally dynamic, cell-type-dependent premeiotic and meiotic phasiRNAs in maize anthers. Proc Natl Acad Sci USA, 112, 3146–3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann T, Marrison J, Hogg K and O’Toole P (2014) Clearing up the signal: spectral imaging and linear unmixing in fluorescence microscopy. Methods Mol Biol, 1075, 129–148. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Comparison of one-photon and multi-photon excitation on imaging resolution in rice endosperm amyloplast.
Coordinates of miR2275 localization detected by sRNA-FISH and dSTORM imaging.
Detailed protocol for sRNA-FISH in plant tissues.