Abstract
Shigella flexneri, an intracellular Gram-negative bacterium causative for shigellosis, employs a type III secretion system to deliver virulence effectors into host cells. One such effector, IcsB, is critical for S. flexneri intracellular survival and pathogenesis, but its mechanism of action is unknown. Here, we discover that IcsB is an 18-carbon fatty acyltransferase catalysing lysine Nε-fatty acylation. IcsB disrupted the actin cytoskeleton in eukaryotes, resulting from Nε-fatty acylation of RhoGTPases on lysine residues in their polybasic region. Chemical proteomic profiling identified about 60 additional targets modified by IcsB during infection, which were validated by biochemical assays. Most IcsB targets are membrane-associated proteins bearing a lysine-rich polybasic region, including members of the Ras, Rho and Rab families of small GTPases. IcsB also modifies SNARE proteins and other non-GTPase substrates, suggesting an extensive interplay between S. flexneri and host membrane trafficking. IcsB is localized on the Shigella-containing vacuole to fatty-acylate its targets. Knockout of CHMP5—one of the IcsB targets and a component of the ESCRT-III complex—specifically affected S. flexneri escape from host autophagy. The unique Nε-fatty acyltransferase activity of IcsB and its altering of the fatty acylation landscape of host membrane proteomes represent an unprecedented mechanism in bacterial pathogenesis.
Bacterial pathogens have sophisticated interplay with their host to promote infection and cause diseases. A critical virulence mechanism commonly used by different pathogens is to secrete effectors or toxins that manipulate various host processes. Many Gram-negative bacteria have evolved a type III secretion system (T3SS) to deliver multiple effectors into host cytosol. These effectors are often endowed with unique and potent biochemical activities to post-translationally modify host proteins1. Shigella flexneri is an enteropathogenic bacterium that causes bacillary dysentery by invasion and spread through the colonic epithelium2. S. flexneri deploys more than a dozen T3SS effectors to modulate host immune responses, cytoskeleton dynamics and vesicle trafficking3,4. Revealing the mechanism of these effectors not only advances the understanding of bacterial infection, but can also lead to the discovery of new enzymatic post-translational modifications. For example, OspF and its homologous effectors in other bacteria harbour an unprecendented phosphothreonine lyase activity that catalyses elimination of the phosphothreonine in MAPKs to block host immunity5–8.
IcsB—one of the first identified Shigella T3SS effectors9—modulates several host cellular processes, including lysis of the protrusions during S. flexneri intercellular spread9,10, evasion of host autophagy11–13 and avoiding septin cage entrapment14. The ΔicsB mutant is deficient in provoking keratoconjunctivitis in infected guinea pigs, indicating its critical role in S. flexneri pathogenicity. Various models are proposed to account for the functional mechanism of IcsB11,13–15, but the host target(s) that can explain its pleiotropic functions are not identified. While bioinformatic analyses have indicated a possible enzyamtic activity in IcsB16, whether and how IcsB employs a catalytic mechanism to modulate its host target(s) remains unknown.
Here, we discover that IcsB is an 18-carbon fatty acyltransferase that can modify RhoGTPases and disrupt their membrane cycling. The modification occurs on the ε -NH2 group of several lysines in the polybasic region (PBR) preceding the prenylated cysteine. Chemical proteomics identify about 60 IcsB substrates, including members of the Ras, Rho and Rab families of small GTPases and other membrane trafficking proteins. We further observe that CHMP5—a substrate of IcsB and a component of the endosomal sorting complex required for transport III (ESCRT-III) complex—is essential for anti-Shigella autophagy. Our work demonstrates that long-chain Nε-fatty acylation, which rarely occurs in eukaryotes, is exploited by the pathogen to alter the acylation landscape of host proteomes and manipulate host defences.
Results
The putative catalytic motif in IcsB is critical for its functioning.
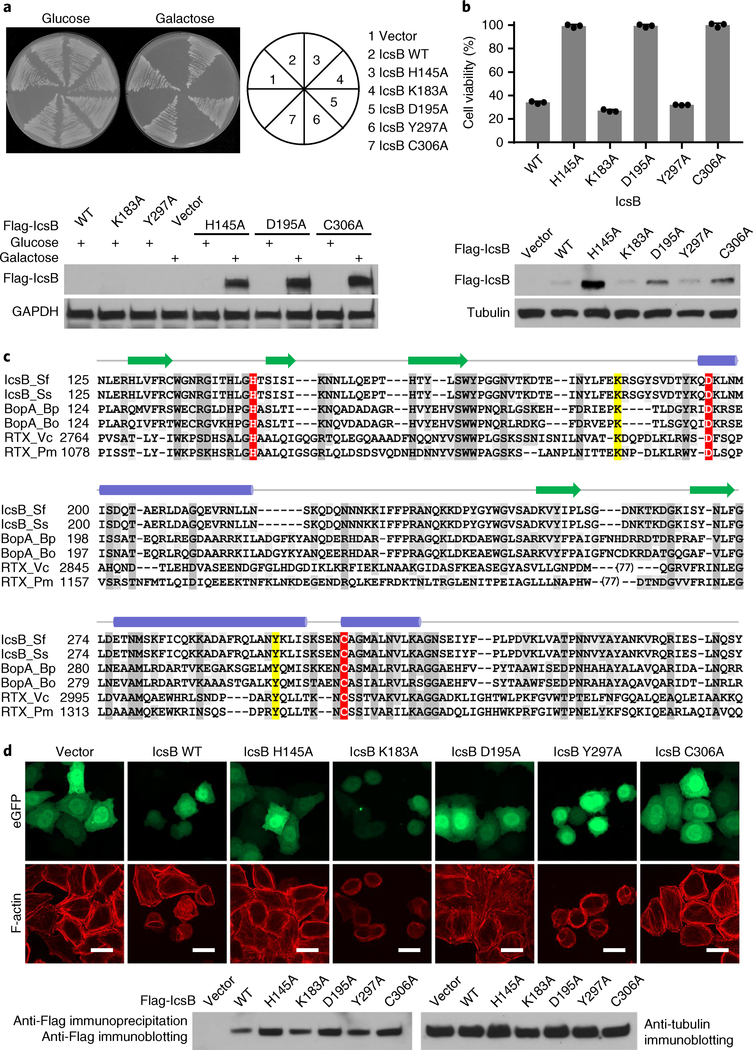
Saccharomyces cerevisiae is a useful model for elucidating the molecular function of bacterial effectors. We found that inducible expression of IcsB could severely block S. cerevisiae growth (Fig. 1a). The cytotoxicity was also observed in IcsB-transfected 293T cells (Fig. 1b). IcsB is present in all four pathogenic Shigella species (S. flexneri, S. sonnei, S. dysenteriae and S. boydii), and shares about 24% sequence homology with the Burkholderia pseudomallei T3SS effector BopA as well as the RID (Rho-inactivation domain) domain of Vibrio species multifunctional autoprocessing repeats-in-toxin (MARTX) toxins (Fig. 1c). The three proteins adopt a similar secondary structure arrangement16 (Fig. 1c). Previous bioinformatics analysis identifies His-145, Asp-195 and Cys-306 in IcsB that are conserved in BopA and RID (Fig. 1c), which may form a catalytic triad resembling that in the circularly permuted papain-like hydrolytic enzymes16,17. Alanine substitution of His-145, Asp-195 or Cys-306 in IcsB diminished its cytotoxicity in yeast, while mutation of two other residues (K183A and Y297A) had no effect (Fig. 1a). Similar findings were obtained in IcsB-transfected 293T cells (Fig. 1b). icsB is critical for S. flexneri intercellular spread, particularly in the absence of virA9,10,15. We confirmed this virulence defect caused by ΔicsB in S. flexneri 2457T (Supplementary Fig. 1a), as well as the requirement of the His–Asp–Cys triad of IcsB (Supplementary Fig. 1b,c). Thus, IcsB and its homologous effectors may function as a papain-like hydrolytic enzyme in modulating host function.
Fig. 1 |. ectopic expression of IcsB is toxic to yeast and disrupts the actin cytoskeleton in mammalian cells, which requires the putative catalytic motif.
a, Effects of IcsB expression on yeast growth. S. cerevisiae strains harbouring an empty vector or an indicated galactose-inducible IcsB expression plasmid were stripped onto the selective media containing glucose or galactose. Cells cultured in the liquid media were subjected to western blot analysis (bottom). b, Effects of IcsB expression on mammalian cell viability. 293T cells were transfected with an empty vector or an indicated IcsB expression plasmid. Cell viability was determined by measuring cytosolic ATP levels normalized to the vector control (mean ± s.d. from three replicates). Bottom: immunoblots of IcsB expression. c, Sequence alignment of the IcsB family. IcsB_Sf and IcsB_Ss are IcsB of S. flexneri and S. sonnei, respectively. BopA_Bp and BopA_Bo are BopA of B. pseudomallei and Burkholderia oklahomensis, respectively. RTX_Vc and RTX_Pm are RIDs of MARTX toxins from Vibrio cholerae and Proteus mirabilis, respectively. In total, 77 amino acids of RTX_Vc and RTX_Pm that are not conserved were omitted from the alignment. The putative catalytic residues and other conserved residues are coloured in red and grey, respectively. The yellow-coloured residues are not required for cytotoxicity of IcsB in yeast and 293T cells. Secondary structures predicted from IcsB sequence are shown above the sequence. Blue ovals represent α helices and green arrows represent β strands. d, Effects of IcsB expression on filamentous actin cytoskeleton structure. HeLa cells were co-transfected with eGFP and an indicated Flag-IcsB expression plasmid. F-actin was stained by rhodamine phalloidin. Scale bars, 20 μm. Total cell lysates were subjected to anti-Flag immunoprecipitation followed by anti-Flag immunoblotting (bottom). The data in a, b and d are representative of three independent experiments.
IcsB disrupts the actin cytoskeleton by inactivating RhoGTPases.
IcsB expression could disrupt the actin cytoskeleton in HeLa cells, as is evident from the loss of filamentous actin fibres and the development of cell-rounding responses (Fig. 1d). Such activity was also blocked by mutation of any residue in the His–Asp–Cys motif. The cytoskeleton phenotype caused by IcsB resembles that of the Yersinia T3SS effector YopT18. YopT is a papain-like cysteine protease that cleaves the carboxy (C)-terminal prenylated cysteine in Rho family small GTPases (RhoA, Rac1 and Cdc42). Notably, the RID of MARTX toxin can also disrupt the actin cytoskeleton by inactivating RhoGTPases19, which requires the His–Asp–Cys motif17 (rather than the His–Tyr–Cys motif proposed in the bioinformatics analysis16). Consistent with these observations, IcsB expression could diminish the interaction of RhoA Q63L (a mutant locked in the GTP-bound form) with its effector domain RBD (Rho-binding domain of Rhotekin), dependent on the Cys-306 (Supplementary Fig. 2a). IcsB also disrupted the interaction between RhoA and Rho GDP-dissociation inhibitor (RhoGDI), which was abolished by mutations of the His–Asp–Cys motif but not Tyr-297 (Supplementary Fig. 2b). Thus, IcsB uses its papain-like activity to inactivate RhoA in mammalian cells.
IcsB disrupts RhoGTPase membrane cycling by modifying its C-terminal tail.
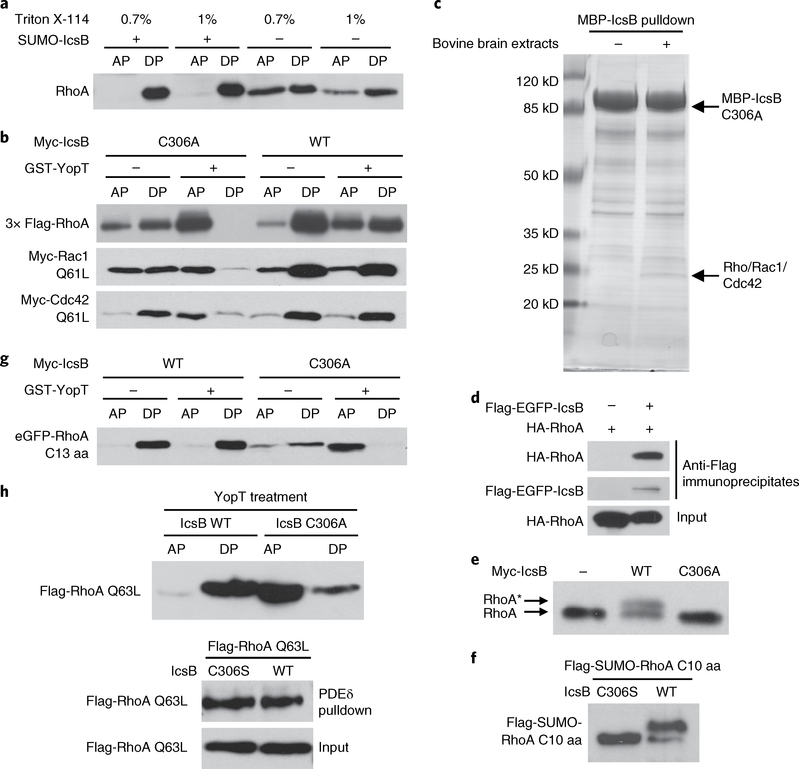
The fact that IcsB could block the RhoA–GTP interaction with RhoGDI indicates that IcsB, like YopT, may modulate RhoGTPase cycling onto and off the membrane. Consistent with this idea, IcsB expression abolished the precipitation of Rac1/Cdc42 Q61L (equivalent to RhoA Q63L) by RhoGDI while it had no effect on their binding to the effector domain PBD (The Rac/Cdc42 (p21)-binding domain of PAK-1 protein) (Supplementary Fig. 2c,d). When the prenylated Cys-190 in RhoA was mutated into alanine, its precipitation by the RBD resisted disruption by IcsB (Supplementary Fig. 2e). These results indicate that IcsB inhibition of RhoA–RBD binding is an indirect result of the alteration of RhoA membrane association. The prenylated RhoGTPase is prone to be partitioned into the detergent phase in the Triton X-114 partitioning assay. Overexpressed RhoA was partitioned into both the aqueous and the detergent phase18, but IcsB treatment of the cell lysates led to RhoA partitioning exclusively into the detergent phase (Fig. 2a). Thus, the action of IcsB increases the hydrophobicity and membrane association of RhoA. YopT treatment abolishes RhoA partitioning into the detergent phase18. Using this assay, we observed that RhoA from IcsB but not IcsB C306A mutant-transfected cells resisted YopT cleavage (Fig. 2b). Similar results were obtained with Rac1 and Cdc42 (Fig. 2b).
Fig. 2 |. IcsB disrupts RhogTPase membrane cycling by modifying its C-terminal tail.
a, Effects of IcsB on the hydrophobicity of RhoA. Cytosolic extracts of RhoA-transfected 293T cells were incubated with purified IcsB and ATP followed by Triton X-114 partitioning. b, Effects of IcsB on RhoGTPase cleavage by YopT. 293T cells were co-transfected with an indicated Rho expression plasmid and IcsB (WT or C306A). Cell lysates were treated with or without YopT, then subjected to Triton X-114 partitioning. c, Identification of RhoGTPases as IcsB-binding proteins. Purified MBP-IcsB C306A was incubated with or without bovine brain extracts and then subjected to amylose bead pulldown. Bound proteins were separated on the SDS-PAGE gel and Coomassie blue staining of the gel is shown. The specific band bound by IcsB C306A contains Rho, Rac and Cdc42, revealed by mass spectrometry. d, Interaction between transfected IcsB and RhoA in 293T cells. Shown are immunoblots of anti-Flag immunoprecipitates and total cell lysates (input). e,f, SDS-PAGE mobility shift of the endogenous RhoA or RhoA C-terminal tail induced by IcsB. 293T cells were transfected with an indicated IcsB expression plasmid alone (e) or co-transfected with a Flag-SUMO-tagged RhoA C-terminal ten-residue tail (C10 aa) (f). Cell lysates were subjected to 15% SDS-PAGE, followed by immunoblotting analyses. The asterisk in (e) indicates the mobility shift of the endogenous RhoA. g, Effects of IcsB on the cleavage of the RhoA C-terminal tail by YopT. 293T cells were co-transfected with an eGFP-tagged RhoA C-terminal 13-residue tail (C13 aa) and IcsB (WT or C306A). Cell lysates were treated with or without YopT, then subjected to Triton X-114 partitioning. h, Effects of IcsB on RhoA precipitation by PDEδ. 293T cells were co-transfected with Flag-RhoA Q63L and an indicated IcsB expression plasmid. Cell lysates were subjected to GST-PDEδ pulldown or YopT digestion followed by Triton X-114 partitioning. Proteins in the aqueous phase (AP) and detergent phase (DP) were subjected to immunoblotting analyses (a, b, g and h). All data (a–h) are representative of three independent experiments.
RhoGTPases are frequently targeted by bacterial pathogens20, which is often mediated by bacterial toxins or effector-catalysed covalent modifications. In vitro pulldown of recombinant MBP (maltose-binding protein)-IcsB C306A from bovine brain extracts identified a specific binding partner of 25 kDa (Fig. 2c), and this band mainly contained Rho, Rac1 and Cdc42, according to mass spectrometry identification. Confirming this observation, RhoA was readily co-immunoprecipitated by IcsB from transfected 293T cells (Fig. 2d). Furthermore, endogenous RhoA in 293T cells often showed a slow migration on a high-concentration sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) gel upon expression of IcsB, but not IcsB C306A (Fig. 2e). Thus, IcsB probably directly modifies RhoGTPases, rendering them more hydrophobic.
The C-terminal tail of RhoA, bearing the prenylated cysteine, is sufficient for recognition and cleavage by YopT18,21. Expression of IcsB, but not IcsB C306A, rendered the enhanced green fluorescent protein (eGFP)-tagged RhoA C-terminal tail resistant to YopT cleavage (Fig. 2f). The tail, when co-expressed with IcsB in 293T cells, exhibited a more evident mobility shift (Fig. 2g). PDEδ, a non-selective prenyl-binding protein, could efficiently pull down RhoA from cell lysates, but this was not altered by IcsB expression (Fig. 2h). Taken together, these data suggest that IcsB probably modifies the C-terminal tail of RhoA without affecting its prenylation.
IcsB is an 18-carbon N-fatty acyltransferase that modifies lysine residues in the C-terminal PBR in RhoGTPases.
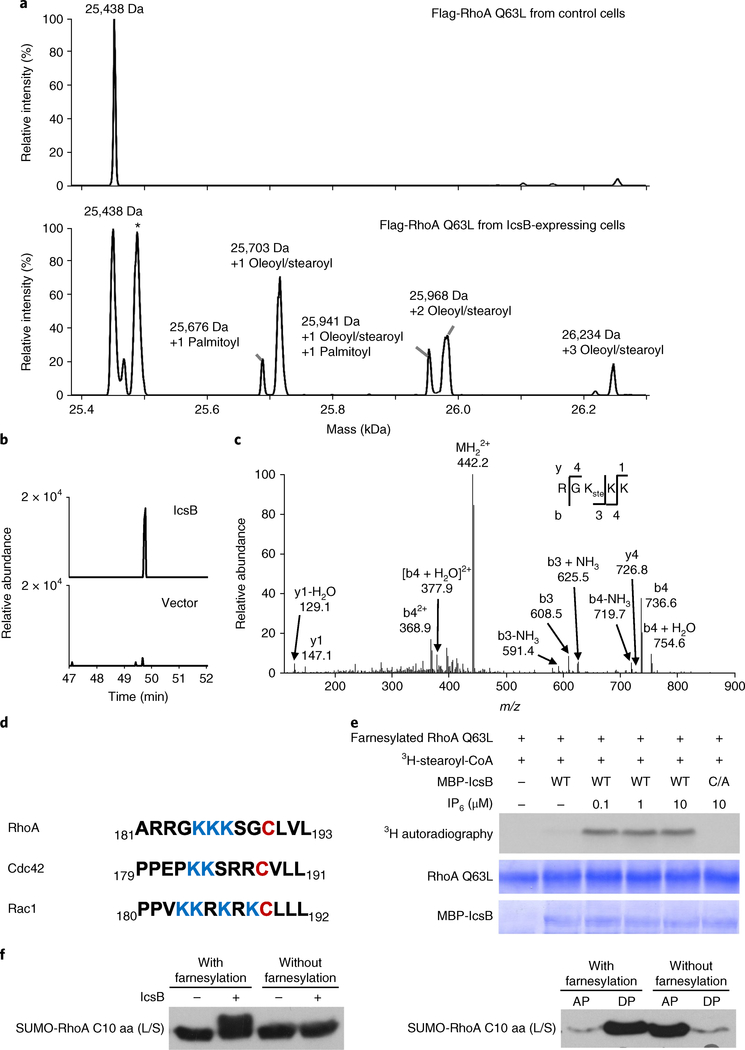
To reveal the modification on RhoA, we affinity-purified Flag-RhoA Q63L from control or IcsB-expressing cells and performed mass spectrometry analyses. Determination of the total molecular mass revealed that RhoA was modified with 1, 2 or 3 18-carbon fatty acyl groups (stearoyl or oleoyl) (Fig. 3a). Tandem mass spectrometry (MS/MS) analysis showed that the acyl groups were conjugated to Lys-185, Lys-186 and Lys-187 in the PBR of RhoA (Fig. 3b–d). The PBR, preceding the prenylated cysteine, has electrostatic interactions with phospholipid head groups in the membrane. Thus, fatty acylation of the PBR increased the hydrophobicity and membrane association of RhoA (Fig. 2a). The modification also explains the resistance of IcsB-modified RhoA to YopT cleavage, as an intact PBR is required for recognition by YopT18,21. According to the crystal structure of RhoGDI complexed with prenylated Cdc42 (ref. 22), fatty acylation of the PBR should disrupt the binding of RhoGDI to RhoGTPases, also explaining our experimental observations (Supplementary Fig. 2b,d).
Fig. 3 |. IcsB is an 18-carbon N-fatty acyltransferase that modifies lysine residues in the C-terminal PBR of RhogTPases.
a, Mass spectrometry identification RhoA modification by IcsB in transfected 293T cells. Flag-RhoA Q63L was purified using the anti-Flag affinity resin. The total molecular mass was determined by electrospray ionization mass spectrometry. The asterisk denotes RhoA Q63L without the prenylation modification. b, Extracted ion chromatograms of Flag-RhoA Q63L purified from transfected 293T cells. Shown are graphs of the stearoylated peptide RGKKK (m/z = 441.8). c, MS/MS mass spectrum of the stearoylated peptide RGKKK in IcsB-modified Flag-RhoA Q63L purified from 293T cells. b and y fragments in (c) are obtained from the MS/MS spectra of the fatty acylated peptide (RGK(ste)KK). d, Alignment of the RhoA, Cdc42 and Rac1 C-terminal 13-residue sequences. The prenylated cysteine residues are in red, and the PBR lysines are in blue. e, In vitro reconstitution of RhoA stearoylation by IcsB. Purified farnesylated RhoA was incubated with MBP-IcsB, 3H-stearoyl-CoA and the indicated concentration of IP6. The reactions were stopped by adding the SDS sample buffer, and subjected to SDS-PAGE followed by Coomassie blue staining and 3H autoradiography. f, Requirement of RhoA prenylation for fatty acylation by IcsB. The farnesylated or unmodified Flag-SUMO-RhoA C-terminal 10-residue (C10 aa) tail was incubated with SUMO-IcsB, ATP and cell extracts. The reactions were stopped by adding the SDS sample buffer, then subjected to 15% SDS-PAGE followed by anti-Flag immunoblotting (left). Cell lysates were also subjected to Triton X-114 partitioning (AP, aqueous phase; DP, detergent phase; right). L/S is an L193S mutation that switches the native geranylgeranylation to farnesylation. Data in a–c, e and f are representative of three independent experiments.
We attempted to reconstitute IcsB fatty acylation of RhoA in vitro. Purified farnesylated RhoA (the L193S mutant was used to switch the native geranylgeranylation to farnesylation), obtained from coexpression with the farnesyltransferase (FTase) in Escherichia coli, was incubated with recombinant MBP-IcsB in the presence of H3-stearoyl-CoA and inositol hexaphosphate (IP6). Autoradiography showed that the GTPase was readily modified by the stearoyl group, which did not occur with IcsB C306A (Fig. 3e). We also performed the same reaction using cold stearoyl-CoA. Mass spectrometry then revealed that the molecular mass of RhoA was increased by 535 and 801 Da (Supplementary Fig. 3a), corresponding to the addition of 2 and 3 stearoyl groups, respectively. MS/MS analyses confirmed the modification on Lys-185, Lys-186 and Lys-187 (Supplementary Fig. 3b,c). The Yersinia T3SS effector YopJ, which also bears a papainlike hydrolytic triad, acetylates serine and threonine residues in MAPK kinases23. Different from YopJ, IcsB is an 18-carbon fatty acyltransferase that modifies lysine residues. IcsB-catalysed modification also differs from the known long-chain fatty acylation that generally occurs on a cysteine or the amino (N) terminus of a glycine residue.
We found that the C-terminal tail of RhoA expressed in the absence of the FTase resisted modification by IcsB and was therefore only partitioned into the aqueous phase (Fig. 3f). Consistently, IcsB did not affect the activation state of non-prenylated RhoA C190A (Supplementary Fig. 2e). Thus, prenylation of RhoA is required for its recognition by IcsB. Moreover, IcsB-catalysed H3-stearoyl labelling of RhoA was barely detectable in the absence of IP6 (Fig. 3e). This property echoes several other bacterial toxins, including the autoprocessing cysteine protease domain of the large clostridial cytotoxins TcdA and TcdB24 and the Vibrio MARTX toxin25, and a cysteine protease domain-like T3SS effector from V. parahaemolyticus26, as well as the YopJ family of acetyltransferase effectors27. A previous study reported a cholesterol-binding region in IcsB12, but cholesterol was not required for IcsB stearoylation of RhoA in vitro.
Stearoylome profiling reveals that IcsB modifies multiple host proteins.
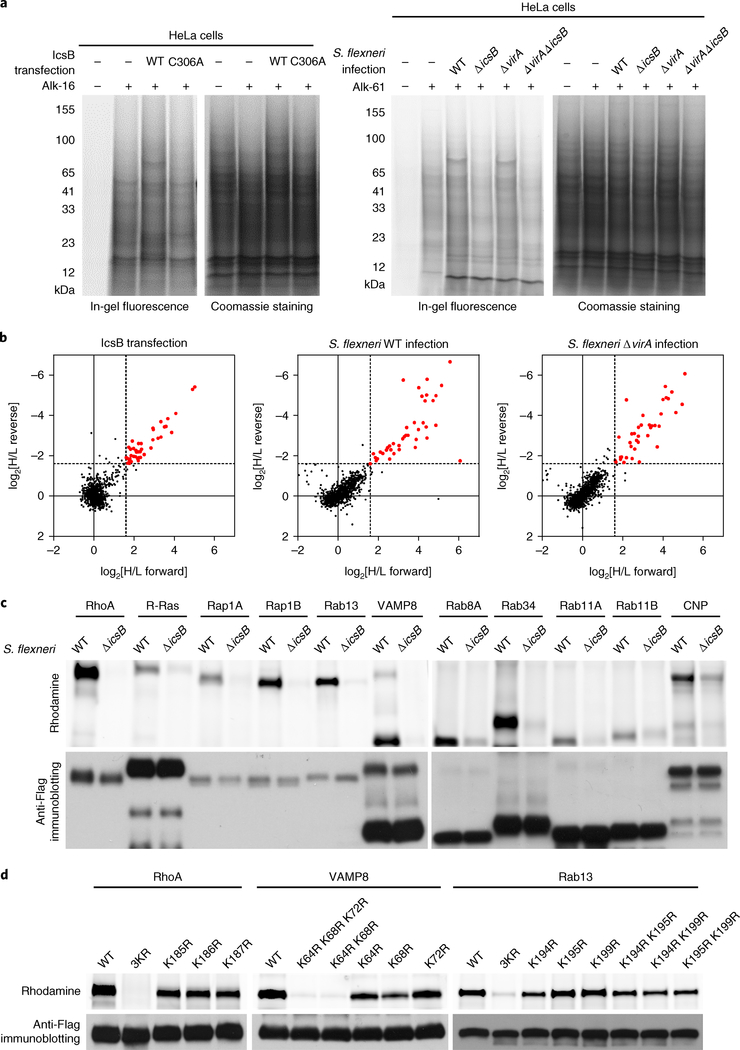
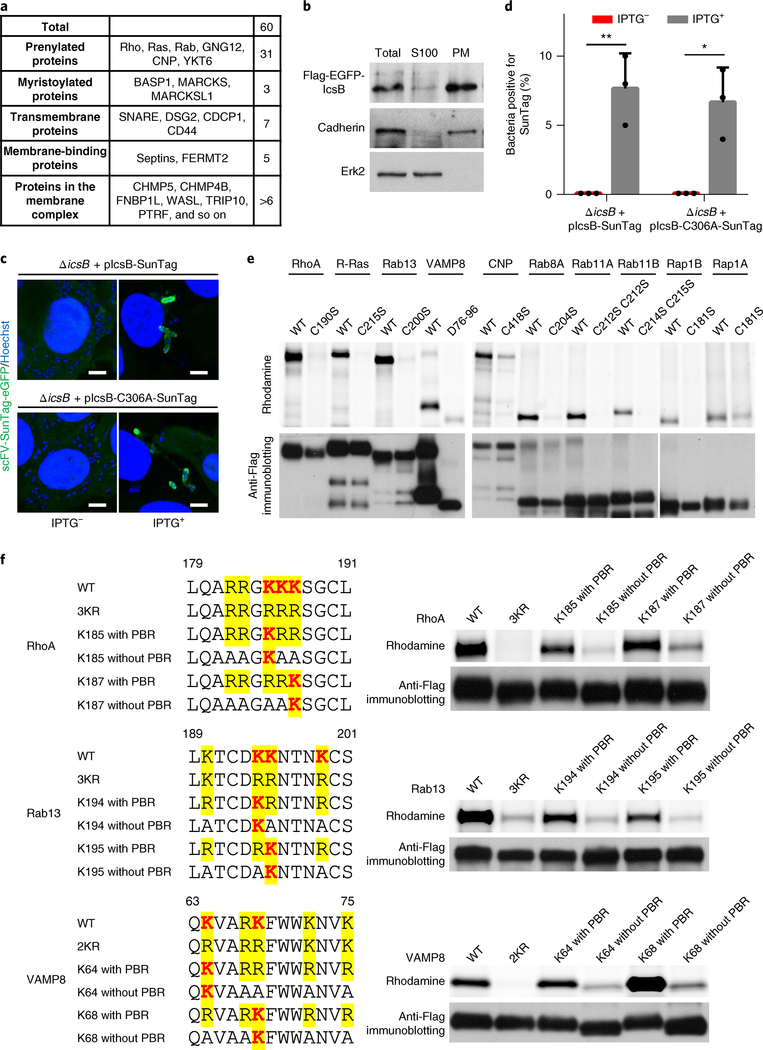
The fact that IcsB can target the prenylated C-terminal tail of RhoA without full-length RhoA indicates that IcsB may modify other host proteins. We therefore employed bioorthogonal chemical proteomics28,29 to profile IcsB-fatty-acylated proteins in living cells. Specifically, HeLa cells were labelled with alk-16—an alkyne analogue of stearic acid that can be metabolized and covalently installed onto cellular proteins30,31. Alkyne-labelled proteins in cell lysates were then subjected to the click-chemistry reaction with azide-rhodamine for in-gel fluorescence, or azide-biotin for streptavidin enrichment and mass spectrometry. In the pilot in-gel fluorescence assay, several abundant stearoylated proteins were detected specifically in the lysates of cells expressing wild-type (WT) IcsB (Fig. 4a). To quantify the differences in stearoylated proteins between WT IcsB and IcsB C306A-expressing cells, double SILAC (stable isotope labelling with amino acids in cell culture) proteomic analyses were also performed (Supplementary Fig. 4a). The quantification identified about 60 proteins that were stearoylated in an IcsB-dependent manner (Fig. 4b, Table 1 and Supplementary Table 1), most of which belong to the Ras superfamily of small GTPases, including the Ras family (R-Ras, K-Ras, H-Ras, Rap1B, RalA, RalB and RheB), Rho family (Rac1, Rac2, RhoA, RhoC and RhoG) and Rab family (Rab1A, Rab5B, Rab8A, Rab10, Rab11A, Rab11B, Rab13, Rab22A, Rab23, Rab34 and Rab35). The list also contains non-small GTPase proteins, including SNARE proteins (YKT6, VAMP3, VAMP8, SEC22B and BNIP1/SEC20), N-myristoylated proteins (MARCKS, MARCKSL1 and BASP1), septins (SEPT7 and SEPT11) and some other proteins. Gene Ontology-based clustering analysis suggested that these proteins regulate the actin cytoskeleton, membrane trafficking, cell–cell adherens junctions, and endocytosis and endosomal recycling (Supplementary Fig. 4b).
Fig. 4 |. Chemical proteomics reveals that IcsB targets multiple host membrane proteins for lysine Nε-fatty acylation.
a, In-gel fluorescence visualization of the stearoylome profile of IcsB (WT or C306A)-transfected (left) and S. flexneri (WT, ΔicsB, Δ virA or Δ virAΔ icsB)-infected (right) HeLa cells. Coomassie blue staining confirms equal protein loading. b, Scatter plots of SILAC proteomic data. Hits with an abundance increase of more than threefold (log2[H/L > 1.58] in the forward SILAC and log2[H/L < − 1.58] in the reverse SILAC) in the IcsB group relative to the control group are shown in red. H/L represents the ratio between heavy and light label partners in the indicated (forward or reverse) experiment. c, Validation of IcsB modification of the proteomic hits. A total of 11 selected candidate substrates were individually transfected into 293T cells. The cells were infected with S. flexneri WT or ΔicsB in the presence of Alk-16, and subjected to in-gel fluorescence assay. Anti-Flag immunoblotting confirms equal loading of the indicated samples. d, Effects of lysine mutation in the PBR of selected substrates on their modification by IcsB. 293T cells were co-transfected with IcsB and Flag-tagged RhoA, VAMP8 or Rab13, or the indicated lysine mutant. The cells were metabolized with Alk-16 and subjected to in-gel fluorescence assay. Data are representative of two (a and b) or three (c and d) independent experiments.
Table 1 |.
IcsB substrates identified in chemical proteomic screens
| Protein symbol | IcsB transfection | S. flexneri WT infection | S. flexneri ΔvirA infection | Loss of function test | Lipidation | Classification |
|---|---|---|---|---|---|---|
| RHOA, RHOC, RHOG, RAC1 | √ | √ | √ | Knockout | * | Rho family |
| RHOF | √ | √ | Knockout | * | ||
| RAC2 | √ | Not tested | * | |||
| CDC42 | √ | √ | Knockout | * | ||
| H-RAS, K-RAS, R-RAS, RAP1B | √ | √ | √ | Knockout | * | Ras family |
| RALA | √ | √ | √ | Not tested | * | |
| RALB | √ | √ | Not tested | * | ||
| RAP1A | √ | √ | Knockout | * | ||
| RHEB | √ | Not tested | * | |||
| RAB8A, RAB11A, RAB11B, RAB13, | √ | √ | √ | Knockout | * | Rab family |
| RAB22A, RAB23, RAB34, RAB35 | ||||||
| RAB5B | √ | √ | Not tested | * | ||
| RAB6A | √ | Knockout | * | |||
| RAB6B | √ | Knockout | * | |||
| RAB9A | √ | √ | Knockout | * | ||
| RAB39A | √ | Not tested | * | |||
| YKT6 | √ | √ | √ | RNAi (85%) | * | SNARE |
| VAMP3 | √ | RNAi (89%) | ||||
| VAMP8 | √ | √ | √ | Knockout | ||
| BNIP1 | √ | Not tested | ||||
| SEC22B | √ | RNAi (95%) | ||||
| SEPT2 | √ | Not tested | Septin | |||
| SEPT7 | √ | √ | √ | RNAi (70%) | ||
| SEPT9 | √ | Not tested | ||||
| SEPT11 | √ | √ | Knockout | |||
| CHMP5 | √ | √ | √ | Knockout | ||
| CHMP4B | √ | Not tested | ||||
| BASP1 | √ | √ | √ | Not tested | * | |
| MARCKS | √ | √ | √ | Knockout | * | |
| MARCKSL1 | √ | √ | √ | Not tested | * | |
| CNP | √ | √ | √ | Knockout | * | |
| GNG12 | √ | √ | Not tested | * | ||
| CD44 | √ | √ | Knockout | |||
| CDCP1 | √ | √ | Not tested | |||
| DSG2 | √ | √ | Knockout | |||
| FNBP1L | √ | √ | Knockout | |||
| WASL | √ | √ | Not tested | |||
| TRIP10 | √ | √ | Knockout | |||
| FERMT2 | √ | Not tested | ||||
| EPS8 | √ | Not tested | ||||
| EPS8L2 | √ | Not tested | ||||
| S100A13 | √ | Not tested | ||||
| PTRF | √ | Not tested | ||||
| GIPC1 | √ | Not tested | ||||
| FKBP8 | √ | RNAi (95%) | ||||
| FKBP11 | √ | Not tested | ||||
| EPB41L2 | √ | Not tested | ||||
| EPB41L1 | √ | Not tested | ||||
RNAi, RNA interference.
We also performed the proteomic profiling under S. flexneri infection conditions. For this, HeLa cells were infected with the icsB-sufficient or -deficient S. flexneri strains. An evident increase of total stearoylated proteins was observed with icsB-positive infections (Fig. 4a). SILAC quantitative proteomics identified more than 40 proteins that were stearoylated in an icsB-dependent manner (Fig. 4b). The presence or absence of VirA, which affects autophagy of intracellular S. flexneri32, had little influence on the stearoylated proteins identified (Fig. 4b). The list of stearoylated proteins during infection largely overlapped with that of the transfection experiment (Table 1 and Supplementary Table 2). In-gel fluorescence assay of a dozen selected candidates, including small GTPases (Rho, Ras, Rap and Rab) and the non-small GTPase protein VAMP8 showed that they could all be labelled with azide-rhodamine upon icsB-positive infections (Fig. 4c), validating the proteomic data. Thus, IcsB alters the fatty acylation landscape of the host cell proteome to modulate multiple host cellular processes.
Using the alk-16 labelling assay, we found that only mutations of all three lysines (Lys-185, Lys-186 and Lys-187) in the PBR of RhoA could completely block its stearoylation by IcsB in 293T cells (Fig. 4d). Results of a similar nature were obtained with VAMP8 and Rab13—two other IcsB substrates that also have a PBR preceding their C-terminal membrane insertion or association sequences (Fig. 4d).
IcsB is localized on Shigella-containing vacuoles and modifies its substrates there.
The substrates of IcsB are all known to be membrane associated or within a membrane complex (Table 1 and Fig. 5a). Consistently, ectopically expressed IcsB in 293T cells was in the membrane fraction (Fig. 5b). To investigate the localization of IcsB during S. flexneri infection, we used the recently developed SunTag system, which is extremely sensitive in imaging a cellular protein tagged with a GCN4 protein-derived peptide (SunTag)33,34. Specifically, eGFP-fused single-chain anti-SunTag antibody was stably expressed in HeLa cells, and the cells were then infected with S. flexneri expressing IcsB harbouring 24 copies of the SunTag. The 24× SunTag did not affect T3SS-dependent secretion of IcsB (Supplementary Fig. 5a). The data revealed that T3SS-delivered IcsB, before bacterial escape from the vacuole into the cytosol, was localized on S. flexneri-containing vacuole membranes (Fig. 5c,d).
Fig. 5 |. IcsB is localized on Shigella-containing vacuoles and modifies its substrates on the membrane location.
a, Classification of IcsB substrates by their membrane-targeting mechanisms. b, Sub-cellular fractionation of IcsB. 293T cells transfected with Flag-eGFP-IcsB were lysed in hypotonic buffer, and the cell lysates (S10) were ultracentrifuged (100,000g) to obtain the plasma membrane (PM) and the S100 cytoplasmic fraction. Shown are the anti-Flag, anti-cadherin and anti-Erk2 immunoblots. c,d, Localization of IcsB during S. flexneri infection. HeLa cells stably expressing scFV-SunTag-eGFP were infected with S. flexneri Δ icsB complemented with 24× SunTag-fused IcsB (WT or C306A). IPTG was used to induce IcsB expression. Fluorescence images taken at 4 h post-infection are shown in c (scale bars, 3 μm), and percentages of intracellular bacteria containing SunTag-positive signals are shown in d. At least 200 infected cells were examined for each experiment, and data are mean ± s.d. from three replicates. A two-tailed unpaired Student’s t-test was performed (*P < 0.05; **P < 0.01). e,f, Membrane localization of the substrates is required for their modification by IcsB. 293T cells were co-transfected with IcsB and an indicated Flag-tagged substrate. The mutants assayed in e are cysteine mutants devoid of lipid modification or deletion of the transmembrane helix for VAMP8 (D76–96). f, RhoA, Rab13 and VAMP8 mutants with or without the PBR, both of which bear a single modifiable lysine, were assayed. Cells were metabolized with Alk-16 and harvested for in-gel fluorescence assay. Data (b–f) are representative of three independent experiments.
Many of the IcsB substrates, including the Ras superfamily of small GTPases, rely on their C-terminal cysteine prenylation to be membrane associated. Serine substitution of the cysteine in representative members of the Rho, Ras, Rab and Rap families abolished their stearoylation by transfected IcsB in 293T cells (Fig. 5e). Similar results were obtained with a prenylated non-GTPase substrate CNP, a microtubule-associated 2′, 3′ -cyclic nucleotide-3′ -phosphodiesterase (Fig. 5e). Deletion of the C-terminal transmembrane domain in VAMP8 also diminished its stearoylation by IcsB (Fig. 5e). The membrane insertion motif in these IcsB substrates is often preceded by a PBR (Fig. 5f). We generated PBR-mutated RhoA, Rab13 and VAMP8 containing only one lysine, with all other lysines mutated into arginine (with PBR) or with all other lysines or arginines mutated into alanine (without PBR). Mutations to alanine abolish or reduce the membrane association. When subjected to IcsB-catalysed stearoylation in cells, the one-lysine-with-PBR mutants were still modified by IcsB, in contrast with their cognate-without-PBR mutants (Fig. 5f). Thus, membrane localization of IcsB targets is required for their encounter with and subsequent fatty acylation by IcsB from S. flexneri.
Fatty acylation of CHMP5 by IcsB is important for Shigella evasion of autophagy.
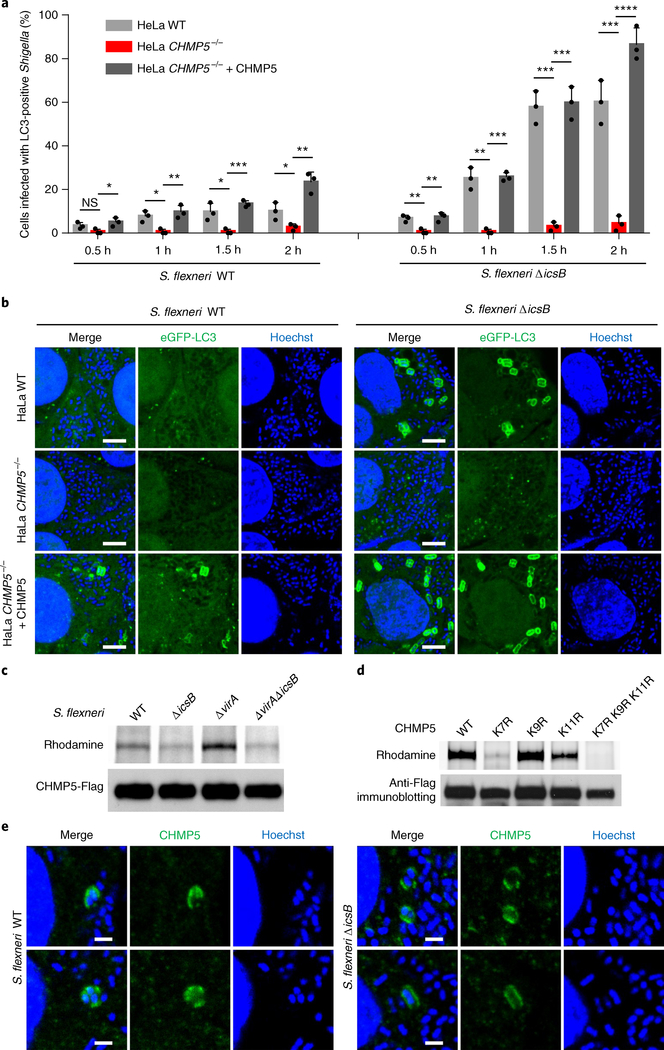
Loss of icsB in S. flexneri results in trapping of the bacterium into the autophagosome11–13,15. We confirmed that few WT S. flexneri were positive for the autophagosome marker eGFP-LC3 while nearly 80% of ΔicsB -infected cells contained eGFP-LC3-decorated bacteria (Supplementary Fig. 5b,c). The increased autophagy of S. flexneri ΔicsB was suppressed by expression of WT IcsB, but not its acyltransferase-inactive H145A, D195A and C306A mutants in the bacteria (Supplementary Fig. 5b,c). As expected, eGFP-LC3 decoration of S. flexneri ΔicsB was diminished in ATG16−/− or ATG5−/− HeLa cells (Supplementary Fig. 5d,e). It is necessary to note that the impact of IcsB deficiency on anti-Shigella autophagy was not that robust, particularly when the percentage of intracellular bacteria decorated with eGFP-LC3 was counted. Thus, the overall relevance of IcsB inhibition of autophagy to Shigella pathogenesis requires further investigation, which is consistent with our finding of IcsB targeting of multiple different host proteins.
To investigate whether any IcsB substrate is involved in S. flexneri autophagy, knockout or knockdown analyses in HeLa cells were performed with nearly all the substrates identified from the proteomic screen. These extensive analyses (Table 1) only identified one gene, CHMP5 (charged MVB protein 5), whose deficiency could inhibit eGFP-LC3 decoration of S. flexneri ΔicsB (Fig. 6a,b and Supplementary Fig. 6a,b). The defective autophagy of S. flexneri ΔicsB was restored by re-expression of CHMP5 in the knockout cells. Consistent with the functional data, CHMP5 was found to be stearoylated by IcsB in S. flexneri-infected 293T cells (Fig. 6c). The N-terminal region in CHMP5 contains three lysines (Lys-7, 9 and 11) and one arginine (Arg-3), resembling the PBR in other IcsB substrates. Mutation of Lys-7 or the three lysines together severely inhibited or completely blocked CHMP5 stearoylation by IcsB, respectively (Fig. 6d), whereas mutation of other lysine residues in CHMP5 showed no effects (Supplementary Fig. 7a). Thus, IcsB mainly targets Lys-7 in CHMP5 for stearoylation, resulting in suppression of antibacterial autophagy in host cells. CHMP5 is a component of the ESCRT-III complex35,36. CHMP5−/− did not affect S. flexneri invasion into HeLa cells (Supplementary Fig. 6c,d). Thus, CHMP5 is probably involved in regulating the trafficking and property of S. flexneri-containing vacuoles, which indirectly affects development of the autophagic response to S. flexneri. Supporting this notion, CHMP5−/− in HeLa cells disrupted autophagosome formation in response to both ΔicsB and WT S. flexneri infection (Fig. 6a,b). Endogenous CHMP5 could be identified on Shigella-containing vacuoles (Fig. 6e).
Fig. 6 |. Fatty acylation of CHMP5 by IcsB is important for Shigella escape from host autophagy.
a,b, Effects of CHMP5 deficiency on bacterial autophagosome formation in response to S. flexneri infection. Indicated HeLa cells expressing eGFP-LC3 were infected with S. flexneri WT or ΔicsB. a, Percentages of infected cells containing LC3-positive S. flexneri at indicated time points after infection. b, Representative fluorescence images taken at 2 h post-infection (scale bars, 3 μm). At least 200 infected cells were examined for each experiment and the data are means ± s.d. from three replicates. A two-tailed unpaired Student’s t-test was performed (*P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001; NS, 0.0734). c, Stearoylation of CHMP5 by IcsB during S. flexneri infection. 293T cells stably expressing Flag-CHMP5 were infected with S. flexneri WT, ΔicsB, Δ virA or Δ virAΔ icsB in the presence of Alk-16. Cells were harvested 2 h after infection for in-gel fluorescence assay. d, Effects of lysine mutation on CHMP5 stearoylation by IcsB. 293T cells were transfected with IcsB and Flag-CHMP5 (WT or an indicated K to R mutant). Cells were metabolized with Alk-16 for 24 h and subjected to in-gel fluorescence assay. e, Localization of endogenous CHMP5 by immunofluorescence staining in S. flexneri-infected HeLa cells (scale bars, 1 μm). Data shown in a–e are representative of three independent experiments.
We also infected CHMP5−/− HeLa cells with other bacterial pathogens, including Salmonella typhimurium, Yersinia pseudotuberculosis (the effector-less Δ HEMOJ(T) IP2666 strain) or Listeria monocytogenes, which are known to be targeted by host autophagy. Interestingly, eGFP-LC3 decoration of these bacteria was not affected by the loss of CHMP5 (Supplementary Fig. 7b,c). This agrees with the hypothesis that CHMP5 and the ESCRT-III complex may have a differential role in endocytosis-mediated entry for different bacteria. Thus, CHMP5 functions specifically in anti-Shigella autophagy, and its fatty acylation by IcsB contributes to Shigella escape from host autophagy.
Discussion
In summary, we show that the T3SS effector IcsB from S. flexneri targets a large set of host proteins for fatty acylation during infection. This property differs from other bacterial effectors that often act on one or a few host proteins. Most IcsB targets, such as the small GTPases, bear the CAAX-mediated prenylation. IcsB modifies RhoGTPases and disrupts their membrane cycling, which causes cell rounding in transfected cells, but not in S. flexneri-infected cells. This is different from the RID of MARTX toxin whose inactivation of Rho causes cell rounding during infection19. It is possible that IcsB modification of RhoGTPases is not extensive enough to generate a gross cytoskeleton phenotype. Thus, the functional consequence of IcsB modification of its other targets during infection is a subject of future studies. Despite this, IcsB appears to be a master effector that alters the fatty acylation landscape of host membrane proteomes during S. flexneri infection.
Autophagy is important for host defence against bacterial infections37. Different bacterial pathogens enter the host cell through different routes with distinct trafficking routes. Successful pathogens have evolved effectors to hijack different steps in bacterial autophagy. The Legionella pneumophila RavZ effector cleaves and deconjugates LC3-PE (LC3-phosphatidylethanolamine conjugate)38. S. flexneri VirA inactivates Rab1 at the endoplasmic reticulum–Golgi exit sites to block autophagosome membrane formation32. We show that IcsB modifies the endosomal ESCRT-III complex protein CHMP5, and CHMP5 deficiency suppresses anti-Shigella autophagy. These results suggest that IcsB functions during the trafficking of endosome-derived Shigella-containing vacuoles, and its suppression of autophagy is an indirect result of CHMP5 inactivation. CHMP5 deficiency does not affect autophagy of other intracellular bacteria, consistent with the distinct endosomal machineries exploited by different bacteria.
Fatty acylation modification is present in all kingdoms of life, and long-chain fatty acylation is commonly manifested as N-myristoylation and S-palmitoylation of the N-terminal glycine and cysteine residues, respectively39–41. Lysine Nε-fatty acylation has only been found on aquaporin-0 (ref. 42) and some secreted cytokines40,43,44, but their functional significance is unclear. IcsB catalyses Nε-fatty acylation to modulate multiple host cellular processes. IcsB adopts a papain-like fold that is common in the protein structure space. It is not unexpected that Nε-fatty acylation might be widely used in regulating eukaryotic biology. A recent study shows that SIRT6 can defatty-acylate PBR lysines in R-Ras245. While our manuscript was under revision, a follow-up study reported that the RID of MARTX toxin also functions as a long-chain Nε-fatty acyltransferase to modify PBR lysine residues of RhoGTPase46, which explains the known functions of RID, such as inducing host cell rounding and inhibiting phagocytosis17,19,47.
Methods
Plasmids, antibodies and reagents.
DNA for the icsB gene was amplified from the genomic DNA of the S. flexneri 2a strain 2457T. For recombinant expression in E. coli, icsB DNA was constructed into the pSUMO vector with an N-terminal SUMO (small ubiquitin-like modifier) tag and the pMal vector with an N-terminal MBP tag. For expression in mammalian cells, icsB DNA was cloned into the pCS2 vector with an N-terminal 3× Flag tag or a modified peGFP-C1 or pLKO1 vector containing a Flag tag preceding eGFP. ipgA DNA was cloned into the pCS2 vector with an N-terminal RFP (red fluorescent protein) or BFP (blue fluorescent protein) tag. For complementary expression in S. flexneri, icsB was cloned into the pME6032 vector under the control of a tac promoter. For expression in yeast, icsB and ipgA were cloned into the p414 and p413-Gal vector, respectively, and protein expression was induced by galactose in the media. eGFP-tagged 13 C-terminal residues of RhoA were generated using a standard PCR cloning strategy. RhoA was cloned into pET21a with an N-terminal Flag-6× His tandem tag for recombinant bacterial expression. Complementary DNAs for the α and β subunits of human FTase were amplified from the pRSF-FTase α/β plasmid48 and constructed into the pACYCDuet vector with an N-terminal 6× His-SUMO tag and an Ulp1 protease digestion site in the FTase α subunit. 24× SunTag and single-chain variable fragment (scFV)-SunTag-eGFP sequences were synthesized at our gene synthesis facility and cloned into the pME6032 and FUIPW vectors, respectively. Point mutations were generated by QuickChange Site-Directed Mutagenesis. All plasmids were verified by DNA sequencing.
Antibodies for CHMP5 (F-7; sc-374338), RhoA (sc-418), eGFP (sc-8334), HA (Y-11; sc-805) and Erk2 (C-14; sc-154) were purchased from Santa Cruz Biotechnology. Antibodies for Myc (9E10) were from Covance. Anti-Flag M2 mouse monoclonal antibody (F4049) and anti-Flag rabbit polyclonal antibody (F7425) were from Sigma–Aldrich. Anti-pan cadherin antibody (ab6529) was from Abcam. Streptavidin-biotinylated horseradish peroxidase complex was from GE Healthcare. All the yeast culture and transformation reagents were from BD Biosciences. Cell culture reagents were Invitrogen products. All other chemicals and reagents used were Sigma–Aldrich products unless otherwise noted.
Yeast, cell culture, bacterial strains and infection.
Culture and transformation of the S. cerevisiae W303a strain and assays of IcsB toxicity in yeast cells were performed as described recently49. Expression of IcsB and IpgA proteins was induced using 2% galactose as the carbon source in the medium. 293T and HeLa cells were obtained from the American Type Culture Collection. All cell lines were tested to be mycoplasma-free by PCR analyses. The identity of the cells was frequently checked by assessing their morphological features, but has not been authenticated by the short tandem repeat profiling. 293T and HeLa cells were maintained in DMEM supplemented with 10% foetal bovine serum (FBS) and 2 mM L-glutamine at 37 °C in a humidified 5% CO2 incubator. Transient transfection was performed using the VigoFect (Vigorus) or jetPRIME (Polyplus) reagents following the manufacturers’ instructions.
S. flexneri 2a strain 2457T was used in this study. Afimbrial adhesin was transformed into S. flexneri to facilitate the infection. ΔicsB, ΔvirA and ΔvirAΔicsB mutant strains were constructed using the suicide vector pCVD442. For bacterial infection, S. flexneri 2457T, S. typhimurium SL1344 and Y. pseudotuberculosis IP2666 (ΔHEMOJ(T)) were cultured overnight at 37 °C in 2× YT broth with shaking. Before infection, bacteria cultures were diluted by 1:100 in fresh 2× YT broth, and incubated in a shaker until the absorbance at a wavelength of 600 nm reached 1.5. L. monocytogenes EGD strain was cultured overnight at 30 °C in brain heart infusion broth with shaking until the absorbance at a wavelength of 600 nm reached 1.5. For fluorescence microscopy, HeLa cells were seeded on glass coverslips in 24-well plates and cultured for 16 h before infections with the indicated multiplicity of infection (MOI) values. The infection was facilitated by centrifugation at 800g for 5 min at room temperature followed by another hour of incubation at 37 °C in a 5% CO2 incubator. Cells were subsequently washed three times with phosphate-buffered saline (PBS) and fresh DMEM containing 100 μg ml−1 gentamycin (to kill extracellular bacteria). After another 2 h of incubation, infected cells were washed three times with PBS, then subjected to immunofluorescence staining. Cells cultured on the coverslips were fixed with 4% paraformaldehyde for 30 min at room temperature, permeabilized with 0.1% Triton X-100, blocked with bovine serum albumin or FBS, then incubated with the primary antibody for one hour and the secondary antibody for another hour. Rhodamine-labelled phalloidin was used to stain the F-actin structures.
Plaque formation assay.
HeLa cells were seeded onto 6-well plates and cultured for 24 h before the infection (MOI = 0.05). Bacteria were cultured at 37 °C in 2× YT broth containing 100 μg ml−1 streptomycin with shaking at 250 r.p.m. until the absorbance at a wavelength of 600 nm reached 2.0. The infection was facilitated by centrifugation at 800g for 5 min at room temperature, and proceeded for 1.5 h at 37 °C in a 5% CO2 incubator. Cells were subsequently washed with PBS and incubated in 2 ml fresh DMEM containing 10% FBS, 2 mM L-glutamine, 100 μg ml−1 gentamycin and 0.5% (w/v) agarose. For S. flexneri ΔvirAΔicsB harbouring pME6032-icsB-Flag or other catalytically inactive mutants, bacteria were cultured at 37 °C in 2× YT broth containing 100 μg ml−1 streptomycin, 25 μg ml−1 tetracycline and 200 μM isopropyl β-D-1-thiogalactopyranoside (IPTG). The infected cells were incubated in fresh DMEM containing 10% FBS, 2 mM l-glutamine, 100 μg ml−1 gentamycin, 100 μM IPTG and 0.5% (w/v) agarose. Afimbrial adhesin transformed into S. flexneri was not recommended for this assay. The plaque area (A) was defined as A=π(d∕2)2, where d was obtained by measuring the diameter (d) of a plaque. The average plaque area was calculated by analysing at least 15 plaques per group.
Protein expression and purification.
Recombinant expression in E. coli BL21 cells was induced by 0.4 mM IPTG overnight at 25 °C unless otherwise noted. Expression and purification of GST-RBD and GST (glutathione S-transferase)-PBD proteins were previously described50. GST-tagged proteins were purified by Glutathione Sepharose resin (GE Healthcare). 6× His tag-fused proteins were purified by Ni-NTA resin (Qiagen). MBP-tagged proteins were purified by the amylose resin (NEB). The proteins were further purified by ion exchange chromatography followed by gel filtration chromatography. All the proteins were concentrated and stored at −20 °C with 20% glycerol in the final volume. The protein concentration was determined by Coomassie blue staining of SDS-PAGE gels using bovine serum albumin standards.
Immunoprecipitation, GST pulldown and western blot analysis.
For immunoprecipitation, approximately 1 × 107 cells were collected 24 h after transfection, and lysed in 1 ml of cell lysis buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1 mM EDTA and 0.5% Triton X-100) supplemented with protease inhibitor cocktail (Roche) for 15 min on ice. The cell lysates were centrifuged at 13,000 r.p.m. for 15 min to collect the supernatants. Then, 20 μl of Anti-Flag M2 agarose affinity gel (F2426; Sigma) was added to the supernatants. After 2 h rotation at 4 °C, the beads were washed three times with the cell lysis buffer. For GST pulldown assay, cell lysates were incubated with about 30 μg of GST-RBD/PBD/GDI proteins pre-bound on the glutathione beads. After 1 h rotation at 4 °C, the GST beads were collected and washed three times with the cell lysis buffer. Protein bound on the beads was eluted by boiling in 1× sodium dodecyl sulfate (SDS) sample buffer or by adding 300 ng μl−1 Flag peptides to the lysis buffer before immunoblotting analysis, as indicated.
YopT cleavage and Triton X-114 partition assay.
Indicated cells were lysed in an ice-cold buffer containing 50 mM HEPES (pH 7.4), 150 mM NaCl, 1 mM dithiothreitol (DTT) and 1% Triton X-114 supplemented with the protease inhibitor cocktail. Then, 20 μl of clear lysates were incubated with 2 μg of recombinant GST-YopT protein18 at 30 °C for 1 h. The total lysates were then centrifuged at 13,000 r.p.m. for 10 min and partitioned into the aqueous phase and detergent phase followed by immunoblotting analysis as indicated. For the reconstitution assay, the reactions were supplemented with 1% Triton X-114 (final concentration) and treated with GST-YopT for 1 h before performing the phase partition assay.
In vitro acyltransferase assay.
To prepare the prenylated substrates, equal molar amounts of human FTase complex and Flag-6× His-RhoA or SUMO-RhoA C-terminal 13-residue tail proteins (L193S version) were mixed in a reaction buffer containing 25 mM HEPES (pH 7.4), 20 μM farnesyl pyrophosphate, 5 mM MgCl2, 5 μM ZnCl2 and 2 mM DTT. The reactions were carried out at 30 °C for 3 h, then loaded onto a HiTrap Q column (GE Healthcare) to collect farnesylated substrate or FTase complex. The farnesylation modification was confirmed by matrixassisted laser desorption/ionization mass spectrometry analysis. To assay the fatty acyltransferase activity of IcsB, 4 μg of farnesylated substrates was incubated with 2 μg of purified IcsB protein in the presence of 0.5 μCi of 3H-stearoyl coenzyme A (Moravek Biochemicals) at 30 °C for 1 h in the reaction buffer containing 40 mM HEPES (pH 7.4), 50 mM NaCl, 1 mM MgCl2, 10 μM ZnCl2, 0.5 mM DTT and 10 μM IP6. The reactions were stopped by adding 5× SDS sample buffer. Protein samples were separated on 15% SDS-PAGE gel and transferred onto polyvinylidene difluoride membrane followed by Coomassie blue staining. Incorporation of the 3H-stearoyl group was visualized by 3H autoradiography.
Mass spectrometry analyses of RhoA modification by IcsB.
To prepare RhoA samples for total molecular weight measurement by mass spectrometry, 293T cells cultured in 15 cm dishes were transfected with 3× Flag-RhoA Q63L alone or co-transfected with IcsB. Transfected cells were harvested and resuspended in ice-cold buffer containing 25 mM HEPES (pH 7.4), 150 mM NaCl, 1% octyl β-glucopyranoside and the protease inhibitor cocktail. The collected cells were lysed by sonication, and the soluble cleared lysates were incubated with anti-Flag M2 beads for 4 h. The beads were washed extensively with the buffer containing 25 mM HEPES (pH 7.4), 500 mM NaCl and 1% octyl β-glucopyranoside. The bound proteins were eluted by 300 ng μl−1 Flag peptide in the elution buffer containing 25 mM HEPES (pH 7.4), 150 mM NaCl and 0.1% octyl β-glucopyranoside. For mass spectrometry, the eluted RhoA protein was loaded onto a homemade capillary column (75 μm inner diameter; 6 cm long) packed with POROS R1 medium (Applied Biosystems). Subsequently, the proteins were eluted by an Agilent 1100 binary pump system with the following gradient: 0–80% B in 30 min (A = 0.1 M acetic acid in water; B = 0.1 M acetic acid in 80% acetonitrile/water), then sprayed into a QSTAR XL mass spectrometer (AB Sciex) equipped with a Nano Electrospray ion source. The instrument was acquired in MS mode under 2,100 volts of spray voltage. The protein charge envelope was averaged across the corresponding protein elution peaks and deconvoluted into the uncharged form using the BioAnalyst software provided by the manufacturer. To identify the exact lipid modification, the above purified RhoA was subjected to trypsin digestion as described previously51. The resulting peptides were separated on an EASY-nLC 1000 system (Thermo Fisher Scientific). Peptides eluted from the capillary column were applied directly onto a hybrid LTQ-Orbitrap Elite mass spectrometer by electrospray (Thermo Fisher Scientific) for mass spectrometry (MS) and MS/MS analyses.
In-gel fluorescence assay.
293T cells grown at the confluency of 60–70% in 10 cm dishes were co-transfected with Flag-tagged IcsB substrates and IcsB-WT or IcsB C306A. Some 4 h after transfection, the media were replaced with fresh media containing 50 μM Alk-16. To examine substrate modification during infection, 293T transfected with Flag-tagged IcsB substrates for 24 h were infected with the indicated S. flexneri strains (MOI = 100) in the presence of 50 μM Alk-16. Some 28 h after the co-transfection, or 2 h after the infection, the cells were harvested and the substrates were subjected to anti-Flag immunoprecipitation. The immunoprecipitates were washed once with the cell lysis buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1 mM EDTA and 0.5% Triton X-100) and another three times with the wash buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl). In-gel click reactions with 100 μM azido-rhodamine in the buffer containing 25 mM HEPES (pH 7.4), 1 mM CuSO4, 1 mM TCEP, 100 μM TBTA and 150 mM NaCl were then performed (4 h, room temperature). The samples were then heated at 95 °C for 20 min in the presence of freshly prepared NH2OH (0.75 M) before being loaded onto 4−20% Tris-HCl gels (Bio-Rad) for SDS-PAGE separation. Gels were scanned on the Bio-Rad ChemiDoc Imaging System.
Chemical proteomic profiling of IcsB-modified proteins.
HeLa cells were grown in DMEM supplemented with 4.5 gl−1 D-glucose, 110 mg l−1 sodium pyruvate and 10% FBS (HyClone; Thermo Fisher Scientific). For the SILAC experiments, cells were cultured in arginine- and lysine-deficient DMEM (Thermo Fisher Scientific) supplemented with 10% dialysed FBS. For the culture of ‘light’-labelled cells, SILAC medium containing 12C6-L-lysine-2HCl (Lys0; 0.80 mM; Sigma) and 12C6-l-arginine-HCl (Arg0; 0.40 mM; Sigma) was used. For the culture of ‘heavy’labelled cells, SILAC medium containing 13C6,15N2-L-lysine-2HCl (Lys8; 0.80 mM; Cambridge Isotope) and 13C6,15N4-L-arginine-HCl (Arg10; 0.40 mM; Cambridge Isotope) was used. After seven cell doublings, the incorporation of the heavy isotopes was estimated to be > 98% as determined by liquid chromatography MS/MS (LC-MS/MS) analysis.
HeLa cells were transfected with the indicated eGFP-IcsB plasmid for 6 h and labelled with DMSO or Alk-16 (25 μM) overnight. Alternatively, HeLa cells labelled with Alk-16 (50 μM) for 1 h were infected with S. flexneri WT or Δ icsB for 1.5 h. The media were then replaced with fresh media containing 100 μg ml−1 gentamicin and 50 μM Alk-16. Infected cells were further incubated for 1 h, then washed with PBS twice. For both assays, cells were lysed with 4% SDS (50 mM triethanolamine with 150 mM NaCl, pH 7.4) with vigorous vortexing and sonication. The cell lysates were centrifuged at 16,000g for 20 min at room temperature to remove cellular debris. Protein concentrations were determined using the BCA assay (Pierce). For in-gel fluorescence visualization, whole cell lysates (50 μg) diluted with SDS lysis buffer (1 mg ml−1) were reacted with a freshly prepared click-chemistry reaction cocktail containing azido-rhodamine, TCEP, TBTA and CuSO4 for 1 h at room temperature30. The reactions were terminated by the addition of ice-cold methanol (1 ml). The mixtures were placed at −20 °C overnight, then centrifuged at 18,000g for 20 min at 4 °C. The protein pellets were washed with ice-cold MeOH twice, air-dried for 10 min, resuspended in 35 μl of SDS lysis buffer, and then diluted with 12.5 μl of 4× reducing SDS sample buffer and 2.5 μl of 2-mercaptoethanol. The samples were heated for 5 min at 95 °C in the presence of freshly prepared NH2OH (0.75 M) before being loaded onto 4−20% Tris-HCl gels (Bio-Rad) for SDS-PAGE separation. Gels were de-stained in 40% methanol and 10% acetic acid for at least 1 h, then scanned on a GE Healthcare Typhoon 9400 variable mode imager with excitation and emission at 532 and 580 nm, respectively. The gels were then also stained with Coomassie Brilliant Blue (Bio-Rad).
For the SILAC proteomics studies, ‘heavy’- and ‘light’-labelled cells were transfected with eGFP-tagged WT or the C306A mutant IcsB in the ‘forward’ experiment and vice versa in the ‘reverse’ experiment. For the infection assay, ‘heavy’- and ‘light’-labelled cells were infected with S. flexneri WT or ΔicsB strain in the ‘forward’ experiment and vice versa in the ‘reverse’ experiment. Cells were labelled with Alk-16 and lysed with 4% SDS as described above. After estimating the protein concentration, ‘heavy’-labelled cell lysates (1 mg) were mixed with ‘light’-labelled cell lysates (1 mg) and treated with freshly prepared NH2OH (0.75 M) at room temperature with rotation for 1 h. Proteins were then precipitated with ice-cold MeOH, resuspended in 4% SDS and clicked with azido-biotin using CuAAC reactions as above described. Methanol-precipitated and washed protein pellets were again resuspended in 4% SDS. Equal amounts of each protein were diluted 1/4 by volume with 50 mM triethanolamine buffer (final 1% SDS). Then, 60 μl of prewashed streptavidin agarose beads (Invitrogen) were added to each sample. The protein and bead mixtures were incubated for 1 h at room temperature on a nutating mixer. The beads were then washed once with PBS containing 0.2% (w/v) SDS, three times with PBS and twice with 250 mM ammonium bicarbonate. Beads were resuspended in 500 μl of 8 M urea, reduced with 10 mM DTT for 30 min, then alkylated with 50 mM iodoacetamide in the dark for another 30 min. Finally, the beads were washed with 25 mM ammonium bicarbonate and digested with 0.5 μg of trypsin at 37 °C overnight. The supernatants were collected, dried and pre-fractionated with SCX StageTips for LC-MS analysis.
LC-MS/MS analysis for proteomic profiling was carried out at the Proteomics Resource Center at The Rockefeller University, New York, NY, USA. LC-MS analysis was performed with a Dionex 3000 nano-HPLC coupled to an Orbitrap XL mass spectrometer (Thermo Fisher Scientific). Peptide samples were loaded onto a C18 reverse-phase column (75 μm diameter, 15 cm length). A 180 min gradient increasing from 95% buffer A (water with 0.1% formic acid) and 5% buffer B (acetonitrile with 0.1% formic acid) to 75% buffer B in 133 min was used at 0.2 μl min−1 to inject the samples into the mass spectrometer. The Orbitrap XL was operated in top-8-CID-mode with MS spectra measured at a resolution of 60,000 FWHM (full width at half maximum) at m/z = 400. One full MS scan (300–2,000 molecular weight) was followed by three data-dependent scans of the tth nth most intense ions with dynamic exclusion enabled. Peptides fulfilling a Percolator-calculated 1% false discovery rate threshold were reported. Acquired tandem MS spectra were extracted and quantified using MaxQuant software52. The search results from MaxQuant were analysed by Perseus (http://www.perseus-framework.org). Enzyme specificity was set to trypsin, allowing two missed cleavages. Carbamidomethylation of cysteine was set as a fixed modification, while methionine oxidation and N-terminal acetylation were set as variable modifications. Mass deviation for MS/MS peaks was set at a maximum of 0.5 m/z units, and maximum false discovery rates were set to 0.01, both at the peptide and the protein levels. Peak lists generated by MaxQuant were searched with Andromeda against the UniProt complete human database concatenated with common known contaminants. Only unique and razor peptides were used for quantification with a minimum of two ratio counts. The ‘re-quantify’ feature of MaxQuant was used to correct the quantification of proteins with high ratios. Before analysis, known contaminants and reverse hits were removed.
In this study, no statistical methods were used to predetermine the sample size. The experiments were not randomized, and the investigators were not blinded to allocation during the experiments and outcome assessment.
Supplementary Material
Acknowledgements
We thank R. Isberg for providing Yersinia strains, G. Praefcke for the pRSF-FTase plasmid, and the Proteomics Resource Center at The Rockefeller University for mass spectrometry analysis. We also thank members of the Shao laboratory for technical assistance and stimulating discussions. This work was supported by the Basic Science Center Project of the National Natural Science Foundation of China (81788101), National Key Research and Development Program of China (2017YFA0505900 and 2016YFA0501500) and Strategic Priority Research Program of the Chinese Academy of Sciences (XDB08020202) to F.S. H.C.H. acknowledges support from NIH-NIGMS grant R01 GM087544. The research was also supported in part by an International Early Career Scientist grant from the Howard Hughes Medical Institute and Beijing Scholar Program to F.S.
Footnotes
Competing interests
The authors declare no competing interests.
Reporting Summary. Further information on experimental design is available in the Nature Research Reporting Summary linked to this article.
Data availability. The data that support the findings of this study are included in this published article along with its Supplementary Information files, and are also available from the corresponding author upon request.
Additional information
Supplementary information is available for this paper at https://doi.org/10.1038/s41564-018-0215-6.
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Cui J & Shao F Biochemistry and cell signaling taught by bacterial effectors. Trends Biochem. Sci 36, 532–540 (2011). [DOI] [PubMed] [Google Scholar]
- 2.Carayol N & Tran Van Nhieu G The inside story of Shigella invasion of intestinal epithelial cells. Cold Spring Harb. Perspect. Med 3, a016717 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ashida H, Mimuro H & Sasakawa C Shigella manipulates host immune responses by delivering effector proteins with specific roles. Front Immunol. 6, 219 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mellouk N & Enninga J Cytosolic access of intracellular bacterial pathogens: the Shigella paradigm. Front. Cell. Infect. Microbiol 6, 35 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li H et al. The phosphothreonine lyase activity of a bacterial type III effector family. Science 315, 1000–1003 (2007). [DOI] [PubMed] [Google Scholar]
- 6.Zhu Y et al. Structural insights into the enzymatic mechanism of the pathogenic MAPK phosphothreonine lyase. Mol. Cell 28, 899–913 (2007). [DOI] [PubMed] [Google Scholar]
- 7.Mazurkiewicz P et al. SpvC is a Salmonella effector with phosphothreonine lyase activity on host mitogen-activated protein kinases. Mol. Microbiol 67, 1371–1383 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goto Y et al. Discovery of unique lanthionine synthetases reveals new mechanistic and evolutionary insights. PLoS Biol. 8, e1000339 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Allaoui A, Mounier J, Prevost MC, Sansonetti PJ & Parsot C icsB: a Shigella flexneri virulence gene necessary for the lysis of protrusions during intercellular spread. Mol. Microbiol 6, 1605–1616 (1992). [DOI] [PubMed] [Google Scholar]
- 10.Ogawa M, Suzuki T, Tatsuno I, Abe H & Sasakawa C IcsB, secreted via the type III secretion system, is chaperoned by IpgA and required at the post-invasion stage of Shigella pathogenicity. Mol. Microbiol 48, 913–931 (2003). [DOI] [PubMed] [Google Scholar]
- 11.Ogawa M et al. Escape of intracellular Shigella from autophagy. Science 307, 727–731 (2005). [DOI] [PubMed] [Google Scholar]
- 12.Kayath CA et al. Escape of intracellular Shigella from autophagy requires binding to cholesterol through the type III effector, IcsB. Microbes Infect 12, 956–966 (2010). [DOI] [PubMed] [Google Scholar]
- 13.Baxt LA & Goldberg MB Host and bacterial proteins that repress recruitment of LC3 to Shigella early during infection. PLoS ONE 9, e94653 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mostowy S et al. Entrapment of intracytosolic bacteria by septin cage-like structures. Cell Host Microbe 8, 433–444 (2010). [DOI] [PubMed] [Google Scholar]
- 15.Campbell-Valois FX, Sachse M, Sansonetti PJ & Parsot C Escape of actively secreting Shigella flexneri from ATG8/LC3-positive vacuoles formed during cell-to-cell spread is facilitated by IcsB and VirA. mBio 6, e02567–14 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pei J & Grishin NV The Rho GTPase inactivation domain in Vibrio cholerae MARTX toxin has a circularly permuted papain-like thiol protease fold. Proteins 77, 413–419 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ahrens S, Geissler B & Satchell KJ Identification of a His–Asp–Cys catalytic triad essential for function of the Rho inactivation domain (RID) of Vibrio cholerae MARTX toxin. J. Biol. Chem 288, 1397–1408 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shao F, Merritt PM, Bao Z, Innes RW & Dixon JE A Yersinia effector and a Pseudomonas avirulence protein define a family of cysteine proteases functioning in bacterial pathogenesis. Cell 109, 575–588 (2002). [DOI] [PubMed] [Google Scholar]
- 19.Sheahan KL & Satchell KJ Inactivation of small Rho GTPases by the multifunctional RTX toxin from Vibrio cholerae. Cell. Microbiol 9, 1324–1335 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lemichez E & Aktories K Hijacking of Rho GTPases during bacterial infection. Exp. Cell Res 319, 2329–2336 (2013). [DOI] [PubMed] [Google Scholar]
- 21.Shao F et al. Biochemical characterization of the Yersinia YopT protease: cleavage site and recognition elements in Rho GTPases. Proc. Natl Acad. Sci. USA 100, 904–909 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hoffman GR, Nassar N & Cerione RA Structure of the Rho family GTP-binding protein Cdc42 in complex with the multifunctional regulator RhoGDI. Cell 100, 345–356 (2000). [DOI] [PubMed] [Google Scholar]
- 23.Mukherjee S et al. Yersinia YopJ acetylates and inhibits kinase activation by blocking phosphorylation. Science 312, 1211–1214 (2006). [DOI] [PubMed] [Google Scholar]
- 24.Lupardus PJ, Shen A, Bogyo M & Garcia KC Small molecule-induced allosteric activation of the Vibrio cholerae RTX cysteine protease domain. Science 322, 265–268 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Prochazkova K & Satchell KJ Structure–function analysis of inositol hexakisphosphate-induced autoprocessing of the Vibrio cholerae multifunctional autoprocessing RTX toxin. J. Biol. Chem 283, 23656–23664 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Calder T et al. Vibrio type III effector VPA1380 is related to the cysteine protease domain of large bacterial toxins. PLoS ONE 9, e104387 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mittal R, Peak-Chew SY, Sade RS, Vallis Y & McMahon HT The acetyltransferase activity of the bacterial toxin YopJ of Yersinia is activated by eukaryotic host cell inositol hexakisphosphate. J. Biol. Chem 285, 19927–19934 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hang HC & Linder ME Exploring protein lipidation with chemical biology. Chem. Rev 111, 6341–6358 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grammel M & Hang HC Identification of lysine acetyltransferase substrates using bioorthogonal chemical proteomics. Methods Mol. Biol 981, 201–210 (2013). [DOI] [PubMed] [Google Scholar]
- 30.Charron G et al. Robust fluorescent detection of protein fatty-acylation with chemical reporters. J. Am. Chem. Soc 131, 4967–4975 (2009). [DOI] [PubMed] [Google Scholar]
- 31.Wilson JP, Raghavan AS, Yang YY, Charron G & Hang HC Proteomic analysis of fatty-acylated proteins in mammalian cells with chemical reporters reveals S-acylation of histone H3 variants. Mol. Cell Proteom 10, M110.001198 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dong N et al. Structurally distinct bacterial TBC-like GAPs link Arf GTPase to Rab1 inactivation to counteract host defenses. Cell 150, 1029–1041 (2012). [DOI] [PubMed] [Google Scholar]
- 33.Tanenbaum ME, Gilbert LA, Qi LS, Weissman JS & Vale RD A protein-tagging system for signal amplification in gene expression and fluorescence imaging. Cell 159, 635–646 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li P et al. Ubiquitination and degradation of GBPs by a Shigella effector to suppress host defence. Nature 551, 378–383 (2017). [DOI] [PubMed] [Google Scholar]
- 35.Shim JH et al. CHMP5 is essential for late endosome function and down-regulation of receptor signaling during mouse embryogenesis. J. Cell Biol 172, 1045–1056 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ward DM et al. The role of LIP5 and CHMP5 in multivesicular body formation and HIV-1 budding in mammalian cells. J. Biol. Chem 280, 10548–10555 (2005). [DOI] [PubMed] [Google Scholar]
- 37.Shibutani ST, Saitoh T, Nowag H, Munz C & Yoshimori T Autophagy and autophagy-related proteins in the immune system. Nat. Immunol 16, 1014–1024 (2015). [DOI] [PubMed] [Google Scholar]
- 38.Choy A et al. The Legionella effector RavZ inhibits host autophagy through irreversible Atg8 deconjugation. Science 338, 1072–1076 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Resh MD Fatty acylation of proteins: the long and the short of it. Prog. Lipid Res 63, 120–131 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hannoush RN Synthetic protein lipidation. Curr. Opin. Chem. Biol 28, 39–46 (2015). [DOI] [PubMed] [Google Scholar]
- 41.Nadolski MJ & Linder ME Protein lipidation. FEBS J. 274, 5202–5210 (2007). [DOI] [PubMed] [Google Scholar]
- 42.Schey KL, Gutierrez DB, Wang Z, Wei J & Grey AC Novel fatty acid acylation of lens integral membrane protein aquaporin-0. Biochemistry 49, 9858–9865 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stevenson FT, Bursten SL, Fanton C, Locksley RM & Lovett DH The 31-kDa precursor of interleukin 1 alpha is myristoylated on specific lysines within the 16-kDa N-terminal propiece. Proc. Natl Acad. Sci. USA 90, 7245–7249 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jiang H et al. SIRT6 regulates TNF-alpha secretion through hydrolysis of long-chain fatty acyl lysine. Nature 496, 110–113 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang X, Spiegelman NA, Nelson OD, Jing H & Lin H SIRT6 regulates Ras-related protein R-Ras2 by lysine defatty-acylation. eLife 6, e25158 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhou Y et al. Nε-fatty acylation of Rho GTPases by a MARTX toxin effector. Science 358, 528–531 (2017). [DOI] [PubMed] [Google Scholar]
- 47.Dolores JS, Agarwal S, Egerer M & Satchell KJ Vibrio cholerae MARTX toxin heterologous translocation of beta-lactamase and roles of individual effector domains on cytoskeleton dynamics. Mol. Microbiol 95, 590–604 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fres JM, Muller S & Praefcke GJ Purification of the CaaX-modified, dynamin-related large GTPase hGBP1 by coexpression with farnesyltransferase. J. Lipid Res 51, 2454–2459 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dong N et al. Modulation of membrane phosphoinositide dynamics by the phosphatidylinositide 4-kinase activity of the Legionella LepB effector. Nat. Microbiol 2, 16236 (2016). [DOI] [PubMed] [Google Scholar]
- 50.Pellegrin S & Mellor H Rho GTPase activation assays. Curr. Protoc. Cell Biol 38, 14.8.1–14.8.19 (2008). [DOI] [PubMed] [Google Scholar]
- 51.Hu M, Liu Y, Yu K & Liu X Decreasing the amount of trypsin in in-gel digestion leads to diminished chemical noise and improved protein identifications. J. Proteom 109, 16–25 (2014). [DOI] [PubMed] [Google Scholar]
- 52.Cox J & Mann M MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol 26, 1367–1372 (2008). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.