Abstract
We report the development and initial experimental validation of a computational design procedure aimed at generating enzyme-like protein catalysts called “protozymes.” Our design approach utilizes a “compute and build” strategy that is based on the physical/chemical principles governing protein stability and catalytic mechanism. By using the catalytically inert 108-residue Escherichia coli thioredoxin as a scaffold, the histidine-mediated nucleophilic hydrolysis of p-nitrophenyl acetate as a model reaction, and the ORBIT protein design software to compute sequences, an active site scan identified two promising catalytic positions and surrounding active-site mutations required for substrate binding. Experimentally, both candidate protozymes demonstrated catalytic activity significantly above background. One of the proteins, PZD2, displayed “burst” phase kinetics at high substrate concentrations, consistent with the formation of a stable enzyme intermediate. The kinetic parameters of PZD2 are comparable to early catalytic Abs. But, unlike catalytic Ab design, our design procedure is independent of fold, suggesting a possible mechanism for examining the relationships between protein fold and the evolvability of protein function.
A prominent goal of protein design is the generation of proteins with novel functions, including the catalytic rate enhancement of chemical reactions. The ability to design an enzyme to perform a given chemical reaction has considerable practical application for industry and medicine, particularly for the synthesis of pharmaceuticals (1). Significant progress has been made at enhancing the catalytic properties of existing enzymes through directed evolution (2). In contrast, the design of proteins with novel catalytic properties has met with relatively limited success (3–5). We present a general computational approach for the design of enzyme-like proteins with novel catalytic activities.
The use of transition-state analogs as haptens to elicit catalytic Abs has been the most successful technique to date for generating novel protein catalysts (6). Natural enzymes combine transition-state stabilization with precisely oriented catalytic side chains. Although a reactive hapten has been used in the generation of an Ab with a powerful nucleophile at the active site (7), current catalytic Ab technology does not efficiently select for both catalytic side chains and tight noncovalent affinity in the same molecule. The relationship between the general backbone fold of an enzyme and its catalytic properties is not well understood. This observation is particularly relevant to catalytic Abs that are currently constrained to the Ab fold and which have yet to show catalytic activity on par with natural enzymes.
Rational design efforts have recently succeeded at altering the catalytic reactions of two different enzymes (8, 9). Cyclophilin, a cis-trans isomerase of X-Pro peptide bonds, was engineered into an endopeptidase by grafting a triad of catalytic residues commonly found in serine proteases at the binding cleft (8). In a more complicated design effort, Fersht and coworkers (9) were able to convert indoleglycerol-phosphate synthase into phosphoribosylanthranilate isomerase. The naturally occurring versions of these enzymes share a similar fold and catalyze consecutive reactions in the tryptophan biosynthetic pathway with the product of the isomerase being the substrate for the synthase. On the basis of a comparison of the crystal structures of both natural enzymes, loops were altered in the synthase to resemble the target isomerase, followed by directed evolution. These two successful design efforts both used starting proteins with substantial substrate affinity and altered existing active-site residues to catalyze new reactions.
Compared with altering substrate specificity or catalytic mechanism, one of the most significant challenges of enzyme design utilizes a starting protein scaffold devoid of substrate binding and catalytic activity and introduces residues that convert the scaffold into an enzyme (10). Computational techniques have been used to design novel metal binding sites into proteins (11–13). By leaving one of the primary coordination spheres of the metal unligated by the protein, nascent metalloenzymes with a variety of oxygen redox chemistries have been generated (14, 15). The general design of metalloenzymes to react specifically with more complicated organic molecules has not been demonstrated.
Numerous studies of natural proteins have revealed the basic principles of enzymatic catalysis, including proximity and orientation of substrate molecules, transition-state stabilization, acid-base catalysis, and covalent catalysis (16). The continued improvement in computer speed combined with the development and improvement of computational protein design algorithms (17–19) and force fields (20) makes this an opportune time to apply our understanding of enzyme mechanism to the rational, computational design of enzymes with novel functions. We model a high-energy state along a specified reaction pathway including the interaction of at least one catalytic residue with the high-energy state. The computational techniques described below, which have been incorporated into the orbit (Optimization of Rotamers By Iterative Techniques) protein design software (17), identify catalytic sites within a protein structure as well as mutations necessary to accommodate substrate binding (that is, to build the active site).
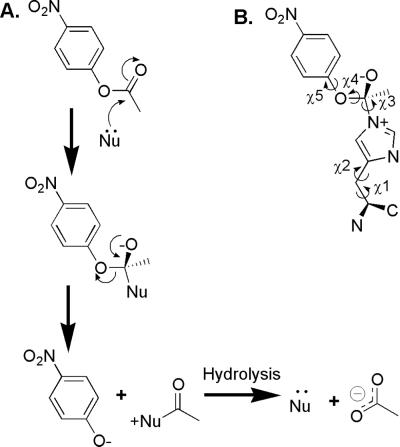
The histidine-mediated nucleophilic hydrolysis of p-nitrophenyl acetate (PNPA) into p-nitrophenol (PNP) and acetate (Fig. 1A) was selected as the target reaction. This reaction has a moderate activation barrier in pH neutral aqueous solution and is similar to the ester and carbonate hydrolysis reactions studied by the first catalytic Abs (21, 22). Nucleophile-catalyzed hydrolysis is commonly achieved by using a catalytic serine or cysteine in natural proteases. Histidine is often used as a general acid or base, but occasionally histidine acts as a nucleophile in phosphate transfer reactions. For nucleophilic catalysis to be effective, the acylated enzyme intermediate must be unstable relative to substrate in buffer to increase the rate of product formation over the uncatalyzed buffer rate. An acyl-histidine intermediate would be less stable than either an acyl-serine or an acyl-cysteine intermediate. Therefore, we modeled histidine as the nucleophile in our design.
Figure 1.
(A) Nucleophile-mediated catalysis of PNPA hydrolysis. (B) High-energy state structure used in the computational active site scan. Labeled dihedral angles were varied to generate the set of high-energy state rotamers used in the design calculations.
Materials and Methods
Molecular Modeling.
For the histidine-PNPA high-energy state rotamers, a backbone-independent rotamer library was generated that included nucleophilic attack by both the Nδ and Nɛ atoms of histidine and attack on both enantiotopic faces of PNPA. The χ1 and χ2 dihedral angles were based on histidine dihedral angles in a survey of protein structures (23) and were expanded by ±1 SD from the reported values. Dihedral angles for χ3 (±30, ±90, and ±150°), χ4 (±60 and 180°), and χ5 (±90°) were based on canonical values for the relevant central atom hybridizations as described in the DREIDING force field (24). Bond lengths and angles as well as additional dihedral angles were optimized by using the DREIDING force field (24). All other side chains were modeled by using a backbone-dependent rotamer library (23) that was expanded about χ1 and χ2 dihedral angles for aromatic residues, expanded about χ1 dihedral angle for aliphatic residues, and unexpanded for polar residues as described (25).
The calculations used an energy function based on the DREIDING force field (24). A Lennard–Jones 12–6 potential with van der Waals radii scaled by 0.9 was applied to all side chain–backbone and side chain–side chain interactions as described (26). An explicit hydrogen bond potential was applied to all groups that contained hydrogen bond donors and/or acceptors (including the high-energy state complex). The 12–10 hydrogen bond potential used a well depth of 8.0 kcal/mol, an equilibrium donor/acceptor distance of 2.8 Å, and a hybridization-dependent angle term as described (27). Electrostatic energies were computed by using a distance-dependent dielectric constant of 40r with partial atomic charges as described (27). Partial atomic charges for the high-energy state rotamers were generated by using the same bond polarization scheme used for the other amino acids and included a net +1 charge on the histidine moiety and a net −1 charge on the high-energy state substrate (Table 2, which is published as supporting information on the PNAS web site, www.pnas.org). A surface area-based atomic solvation potential was selectively applied to the high-energy state rotamers to favor interaction of the substrate with the protein (i.e., only surface area burial of atoms on the high-energy state rotamers was considered). This selective application of the solvation potential, which differs from our previous work, was required to reduce the tendency of generating predominately wild-type amino acid sequence solutions. Surface areas for the solvation potential were calculated by using the Connolly algorithm (28) and the definition of solvent-accessible surface area by Lee and Richards (29) with unscaled atomic radii and an add-on radius of 1.4 Å. A hydrophobic burial benefit of 48 cal/mol/Å2 and a hydrophobic exposure penalty of 76.8 cal/mol/Å2 were used with the two-body decomposition method of Street and Mayo (30). The solvation potential was not applied to polar atoms.
The hydrophobic solvent-accessible surface area of substrate atoms was used to evaluate recognition. For this assessment, solvent-accessible surface area was computed by using the Connolly algorithm (28) and the Lee and Richards definition of solvent-accessible surface area (29) with an add-on radius of 1.4 Å. The solvent-accessible surface area of the substrate was determined by using all atoms in the folded structure as well as by using an unfolded reference that included only the high-energy state rotamer and the local backbone. Active-site designs were ranked by the fraction-accessible surface area defined as the ratio of hydrophobic-accessible surface area in the folded structure and the hydrophobic-accessible surface area in the unfolded reference. Computed designs with zero total solvent-accessible surface area for the substrate atoms were considered inaccessible to substrate and were eliminated from further consideration.
Experimental Validation.
The background mutation D26I was included in both protozyme design (PZD) 1 and 2. D26I was predicted by orbit in an independent calculation and results in increased thermodynamic stability similar to the previously reported D26A protein (31). Position 26 is distant in space to the designed active sites in both PZD1 and PZD2.
The genes for PZD1 and PZD2 were constructed by site-directed mutagenesis, using the wild-type thioredoxin gene (Invitrogen) cloned into PET-11A. Protein expression was induced with 0.5 mM IPTG from BL21(DE3) cells grown to mid-log phase. Cells were lysed by sonication and pelleted twice at 20,000 × g for 30 min. The soluble fraction was brought to 60% acetonitrile, pelleted, and rotary-evaporated to half volume. For initial studies, purification was accomplished by reversed-phase HPLC. For subsequent studies with PZD2, all protein samples were additionally purified by ion exchange and size exclusion chromatography to a purity of >99% as judged by silver-stained PAGE.
PZD2 was dialyzed extensively against 10 mM sodium phosphate buffer at pH 6.95. Kinetic experiments at 22°C were started by the addition of substrate dissolved in acetonitrile to buffer solution with and without PZD2 (final protein concentration of 4 μM). Protein concentration was determined by UV absorbance in 6 M guanidinium hydrochloride assuming an extinction coefficient of 12,400 M−1cm−1 at 280 nm. Product concentration was determined by the change in absorbance at 400.5 nm assuming an extinction coefficient for deprotonated PNP of 19,700 M−1cm−1. Final acetonitrile concentration was 1% for all experiments. The steady-state rate of hydrolysis by PZD2 was corrected for the buffer rate. Burst-phase hydrolysis assays were performed at a substrate concentration of 1.6 mM. Protein concentration was 4 μM, with the exception of wild-type thioredoxin. Wild-type thioredoxin was assayed at 50 μM to improve signal to noise and extrapolated to a protein concentration of 4 μM. The dead time for this experiment was ≈30 sec.
The +42 mass unit species detected in the trapping experiment is a result of replacement of a hydrogen (−1) with an acetyl group (+43). PZD2 (100 μM ) in 10 mM Tris at pH 7.0 was reacted with 1.6 mM PNPA to steady-state conditions, and a mass spectrum was acquired. The same protein solution without PNPA was used as a control. Burst-phase kinetics in this buffer system yielded essentially identical results as in phosphate buffer.
Results and Discussion
An important step in the design of a protein catalyst is locating the substrate relative to the protein scaffold. We took advantage of the observation that the vast majority of natural enzymes contain one or more side chains in highly specific geometric relation to substrate. In the case of PNPA hydrolysis catalyzed by histidine, the histidine side chain should be positioned with a nitrogen atom approaching the carbonyl carbon adjacent to the scissile bond. In a procedure reminiscent of transition-state analog synthesis for catalytic Ab design, we modeled a high-energy state of histidine-catalyzed PNPA hydrolysis as a series of side chain rotamers (Fig. 1B). The rotamer model focuses the subsequent search on the relevant phase space where a catalytic side chain can be positioned to interact with substrate atoms. Modeling a high-energy state and optimizing the surrounding protein sequence for binding to the high-energy state may lead to a reduction in activation energy for the reaction and catalytic rate enhancement (32). Substrate affinity is explicitly included in the design because chemical similarity exists between the high-energy state and the substrate.
The well studied protein Escherichia coli thioredoxin (33) was selected as a scaffold because of its favorable expression properties, thermodynamic stability (34), and successful history in computational design (15). Naturally occurring thioredoxin can supply the reducing power for ribonucleotide reductase by way of its disulfide bond, but is essentially catalytically inert with respect to PNPA binding and hydrolysis. The high thermodynamic stability of thioredoxin suggests that it can tolerate destabilizing mutations that are required to build an active site (35). An active site scan was performed to identify favorable positions for the catalytic histidine as well as mutations necessary for substrate recognition and binding. In separate calculations using the orbit computational protein design programs, each position in the protein structure of thioredoxin was modeled by using the set of high-energy state rotamers (Fig. 1B). All other positions in the protein were allowed to choose (with proper consideration for rotamer flexibility) between their wild-type identity and alanine to accommodate the substrate and to build the active site. After computing the optimal solution by using algorithms based on the Dead-End Elimination theorem (36, 37), positions that changed to alanine can be subsequently allowed to change identity to other amino acids to form better interactions with the high-energy state rotamer. The results reported below are based on the first stage calculation that allows only mutation to alanine. The initial limited combinatorial complexity approach provides for tractable calculations and is expected to generate enzyme-like proteins that we call protozymes. As computer power and computational methods become more sophisticated, the need for a two-stage calculation will diminish. All 94 nonglycine nonproline positions, a combinatorial complexity of ≈1026 amino acid sequences that corresponded to 10101 rotamer sequences, were scanned in approximately 2 days on 14 195 MHz R10000 processors (Silicon Graphics, Mountain View, CA) running in parallel. Hydrophobic solvent-accessible surface area of the substrate atoms in the computed high-energy state was used as a measure of substrate recognition to rank designs (resulting from the limited combinatorial complexity calculations) (Table 1). In addition, the simple requirement that the total solvent-accessible surface area of the substrate atoms be greater than zero was used to eliminate designs where the active site would not be accessible.
Table 1.
Top 10 designs from active site scan
| Design | Catalytic His position | Fraction hydrophobic exposure | Active site mutations |
|---|---|---|---|
| PZD1 | 12 | 0.11 | F12H Y70A |
| PZD2 | 17 | 0.15 | F12A L17H Y70A |
| PZD3 | 86 | 0.29 | V86H I38A L42A L99A |
| PZD4 | 72 | 0.34 | I72H L79A |
| PZD5 | 66 | 0.34 | T66H F12A Y70A |
| PZD6 | 6 | 0.36 | None |
| PZD7 | 39 | 0.37 | A39H K57A |
| PZD8 | 91 | 0.39 | V91H T77A |
| PZD9 | 49 | 0.39 | Y49H K52A |
| PZD10 | 77 | 0.43 | T77H L79A T89A |
Ranked based on hydrophobic surface area burial of substrate atoms in the high-energy state complex. The top two designs, PZD1 and PZD2, were experimentally tested for catalytic activity.
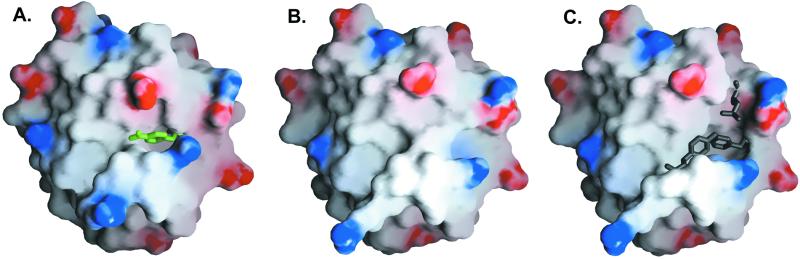
The top two sequences from the limited combinatorial complexity active site scan were selected for experimental analysis. PZD1 contains two mutations (F12H and Y70A) required to introduce the catalytic histidine and to build the active site, whereas PZD2 contains three mutations (F12A, L17H, and Y70A). Experimentally, both proteins demonstrated catalytic hydrolysis of PNPA at a rate significantly above background. Based on preliminary kinetic experiments at high substrate concentration, PZD2 was approximately twice as effective a catalyst as PZD1 and was selected for further analysis. The designed structure of PZD2 computed by orbit shows the substrate atoms in a cleft (Fig. 2A). This cleft is not present in the wild-type thioredoxin x-ray structure (ref. 38; Fig. 2B), indicating that the space-creating mutations (F12A and Y70A) are necessary to create the putative substrate-binding site (Fig. 2C).
Figure 2.
Molecular surfaces (43) focusing on the active site of PZD2 with substrate atoms in green (A) and the corresponding region in the x-ray crystal structure (38) of the wild-type scaffold (B and C). An active-site cleft is present in the design of PZD2 that is largely filled in the wild-type structure. Wild-type residues that were mutated to create the active site are shown in C (F12, L17, and Y70). In the design of PZD2, all side chains were allowed to change geometry, resulting in a slightly different surface compared with that of the wild-type protein.
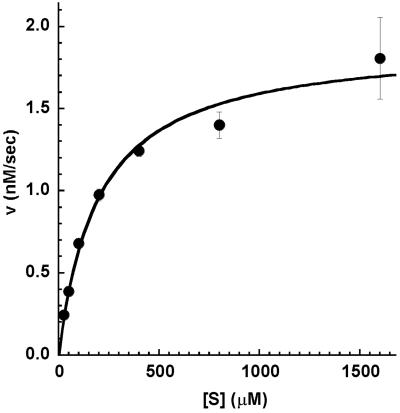
The rate of PNPA hydrolysis by PZD2 at 22°C in 10 mM sodium phosphate buffer at pH 6.95 was experimentally determined over a range of substrate concentrations with standard Briggs–Haldane steady-state treatment (Fig. 3). The reaction velocity of PZD2 demonstrated saturation kinetics with respect to increasing substrate concentration (Fig. 4), indicating that the molecule is acting as an enzyme-like protein with both substrate affinity and catalytic rate enhancement. Hofstee analysis of the data gives a Km of 170 ± 20 μM, kcat of 4.6 ± 0.2 × 10−4 sec−1, and kcat/kuncat of 180 where kuncat (2.5 × 10−6 sec−1) is the rate of hydrolysis under the same conditions. The kinetics of PZD2 are comparable to those of the first catalytic Abs: Km of 208 μM and kcat/kuncat of 770 for MOPC167 (21) and Km of 1.9 μM and kcat/kuncat of 960 for 6D4 (22).
Figure 3.
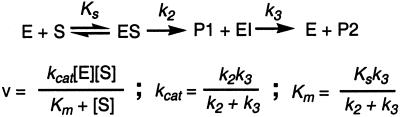
Kinetic model used to analyze the activity of PZD2.
Figure 4.
Velocity vs. substrate concentration for the hydrolysis of PNPA by PZD2.
De novo-designed peptides have recently been designed which catalyze PNPA hydrolysis by a second-order mechanism (i.e., without substrate saturation). The second-order rates for these peptides were determined under different conditions than those used here, making direct comparisons difficult. At pH 5.1, rates of 0.29 M−1sec−1 and 0.056 M−1sec−1 have been reported (39, 40). Extrapolation based on the pH rate profiles (39) reported for the hydrolysis of similarly activated esters suggests rates of ≈0.7 and 0.2 M−1sec−1 for PNPA hydrolysis at pH 7, respectively. The second-order rate of hydrolysis by 4-methyl imidazole (determined under the conditions used in the analysis of PZD2) is 0.11 M−1sec−1. Computing a second-order rate of hydrolysis for PZD2 as kcat/Km gives 2.7 M−1sec−1, which is a factor of 25 better than catalysis by 4-methyl imidazole and about an order of magnitude better than the peptide systems. It is extremely important to note, however, that the lack of substrate saturation in the second-order systems complicates this analysis because substrate affinity was determined for PZD2. Substrate affinity is central to enzyme catalysis and clearly will be important in the design of protein-based catalysts for more complicated reactions.
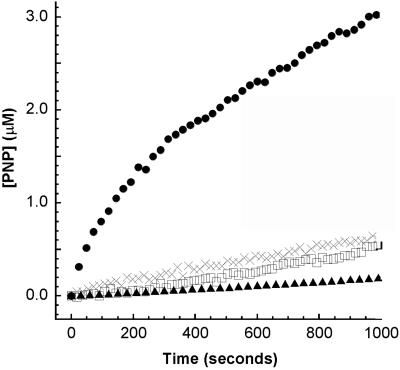
At high substrate concentrations, an initial “burst” phase is a common feature of natural enzymes and is a consequence of a kinetic bottleneck on the reaction pathway. PZD2 displays burst-phase kinetics at high substrate concentration (Fig. 5) consistent with the formation of an enzyme intermediate. The observed burst-phase kinetics could also be a result of slow product release; however, this is unlikely given the kinetic parameters of PZD2. Based on the burst-phase data, we estimate Ks = 1 mM, k2 = 3 × 10−3 sec−1, and k3 = 8 × 10−4 sec−1. Analysis of the magnitude of the burst indicates that ≈30% of PZD2 is in an active state. Using the burst magnitude as a metric of the enzyme concentration indicates a kcat of 1.5 ± 0.1 × 10−3 sec−1, approximately 3 times larger than the kinetic analysis with enzyme concentrations determined from protein UV absorbance.
Figure 5.
Buffer-corrected hydrolysis of PNPA by PZD2 (●), PZD2 H17A (□), wild-type thioredoxin (▴), and wild-type L17H/D26I (×). Data are shown for high substrate concentration and equivalent low protein concentration.
Wild-type thioredoxin is essentially inactive, but does show weak second-order PNPA hydrolysis consistent with its single surface-exposed histidine at position 6. Mutation of the designed catalytic histidine to alanine in PZD2 (H17A) results in a protein with catalytic activity similar to wild-type thioredoxin, indicating that the designed catalytic histidine at position 17 is necessary for the enzyme-like activity in PZD2 (Fig. 5). Mutation of the two other active site residues in PZD2 back to their wild-type identities (A12F and A70Y) also results in a protein with activity similar to wild type. Additionally, at pH 5.7 the activity of PZD2 is almost entirely eliminated, consistent with protonation of the catalytic histidine (data not shown). The kinetic and mutational evidence strongly indicate that PZD2 is working as designed with H17 acting as a catalytic nucleophile, and the space-creating mutations (F12A and Y70A) forming a binding site for substrate.
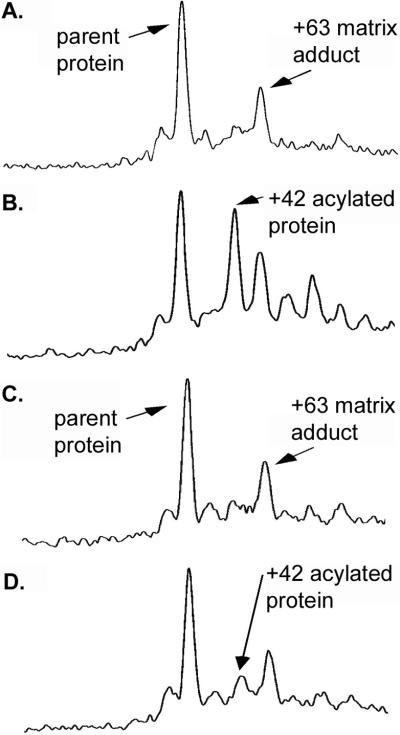
According to its design, reaction of PZD2 with PNPA should yield an acyl-histidine enzyme intermediate, increasing the population of a +42-mass unit species compared with free protein. Mass spectrometry of PZD2 clearly indicates increased population of an acylated species on reaction with PNPA (Fig. 6 A and B). Acylation due to reaction with PNPA is essentially absent for the H17A mutant of PZD2 (Fig. 6 C and D), indicating that acylation depends on H17. From the data it is not possible to distinguish between direct acylation of H17 or H17-mediated (acid/base or acyl transfer) acylation of a different residue.
Figure 6.
Trapping of an acylated intermediate by mass spectrometry. (A) PZD2, (B) PZD2 reacted with substrate, (C) PZD2 H17A, and (D) PZD2 H17A reacted with substrate. A large increase in the population of a +42 species occurs on reaction of PZD2 with substrate, indicating the buildup of an acyl-enzyme intermediate. This +42 species is dramatically reduced for PZD2 H17A where the designed catalytic histidine was mutated to alanine. A small increase in the population of a +42 species is detected in PZD2 H17A on reaction with substrate and is likely the result of acylation at the single surface-exposed histidine at position 6. Consistent with this analysis, a small increase in the population of a double acetylated +84 product is detected on reaction of PZD2 with substrate. A copper matrix adduct (+63) is present (38) in all spectra and in combination with free and acylated protein results in multiple peaks.
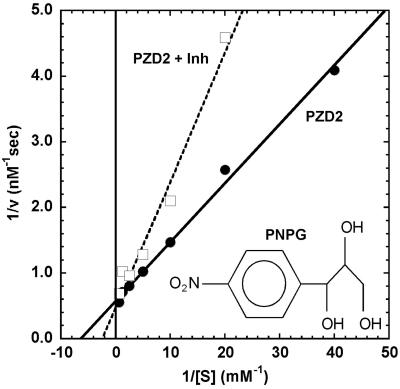
Naturally occurring enzymes are frequently inhibited in a competitive manner by inert compounds resembling the substrate molecule. With this in mind, we assayed the inhibitory effects of p-nitrophenyl glycerol (PNPG) a highly soluble compound with structural similarity to PNPA. A double reciprocal analysis of PZD2 catalyzed PNPA hydrolysis in the presence and absence of inhibitor shows the hallmark features of competitive inhibition (Fig. 7) implying that PNPG is able to bind in the active site and block PNPA binding. A Dixon analysis, varying PNPG concentration at a fixed substrate concentration of 200 μM yielded a Ki of 20 mM. The decreased binding affinity of PNPG relative to PNPA may be due to burial of polar hydroxyl groups against hydrophobic regions in the active site or differences in the steric requirements for PNPG and PNPA. No inhibition was detected with PNP phosphate at concentrations up to 20 mM, suggesting that PZD2 binds preferentially to uncharged nitrophenyl molecules.
Figure 7.
Lineweaver–Burk analysis of PZD2-catalyzed PNPA hydrolysis in the presence (□) and absence (●) of 10 mM PNPG.
To further test the substrate specificity of PZD2, we assayed its kinetics with PNP propionate (PNPP), which has an additional methylene group compared with PNPA. Hofstee analysis indicates within error an identical kcat/kuncat and a Km of 110 ± 20 μM for PNPP, compared with 170 ± 20 μM for PNPA. Inspection of the designed structure of PZD2 with bound PNPA shows that space exists to accommodate the additional methylene of PNPP. Both favorable van der Waals interactions and hydrophobic burial afforded by the relatively open active site likely explain the slightly increased affinity of PZD2 for PNPP compared with PNPA.
Conclusions
The computational design method presented here successfully identified enzyme-like active sites within a protein scaffold essentially devoid of relevant catalytic activity. Although the reaction catalyzed (PNPA hydrolysis) and the level of activity of PZD2 are likely to be insufficient for practical applications, clear paths for improvement exist including further computational optimization and experimentally directed evolution. Computational optimization of the identity of residues proximal in space to the high-energy state complex could result in improved shape and charge complementarity with the high-energy state, improving both substrate affinity and catalytic rate enhancement. Directed evolution, combining mutagenesis and recombination with screening and/or selection, has been an effective method for increasing catalytic activity. The use of computational modeling to focus directed evolution experiments promises to increase the effectiveness of directed evolution methods (41). For the creation and optimization of novel protein catalysts, computational protein design and experimentally directed evolution complement each other well. Use of the design methods presented here to generate proteins with novel catalytic activity and directed evolution to further enhance this activity is a promising path to the production of novel high efficiency protein catalysts.
The relationship between protein fold and function is a fundamental question in enzymology. From numerous structural studies, insight has been gained into the properties of the α/β barrel fold that have facilitated its evolution to catalyze many different chemical reactions (42). With the methods presented here, we may now address the evolvability of essentially any fold through the computational design of novel enzymatic function followed by directed evolution. The comparison of an Ab fold to an α/β barrel fold, for example, may prove particularly insightful. Studies of this type may advance our understanding of the functional potential of protein folds, provide insight into natural evolution, and aid in the selection of scaffolds for future design efforts.
Acknowledgments
We acknowledge F. S. Lee and D. Bökenkamp for their important early efforts and P. Strop and R. A. Olofson for helpful discussions. We thank G. Hathaway and the Protein/Peptide Micro Analytical Laboratory for mass spectra. This work was supported by the Howard Hughes Medical Institute and the Ralph M. Parsons Foundation (S.L.M.), the Helen G. and Arthur McCallum Foundation, the Evelyn Sharp Graduate Fellowship, and by a training grant from the National Institutes of Health (D.N.B.).
Abbreviations
- PNP
p-nitrophenyl
- PNPA
PNP acetate
- PZD
protozyme design
References
- 1.Liese A, Filho M V. Curr Opin Biotechnol. 1999;10:595–603. doi: 10.1016/s0958-1669(99)00040-3. [DOI] [PubMed] [Google Scholar]
- 2.Arnold F H, Volkov A A. Curr Opin Chem Biol. 1999;3:54–59. doi: 10.1016/s1367-5931(99)80010-6. [DOI] [PubMed] [Google Scholar]
- 3.Corey M J, Corey E. Proc Natl Acad Sci USA. 1996;93:11428–11434. doi: 10.1073/pnas.93.21.11428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Petsko G A. Nature (London) 2000;403:606–607. doi: 10.1038/35001176. [DOI] [PubMed] [Google Scholar]
- 5.Baltzer L, Nilsson J. Curr Opin Biotechnol. 2001;12:355–360. doi: 10.1016/s0958-1669(00)00227-5. [DOI] [PubMed] [Google Scholar]
- 6.Hilvert D. Annu Rev Biochem. 2000;69:751–793. doi: 10.1146/annurev.biochem.69.1.751. [DOI] [PubMed] [Google Scholar]
- 7.Wagner J, Lerner R A, Barbas C F., 3rd Science. 1995;270:1797–1800. doi: 10.1126/science.270.5243.1797. [DOI] [PubMed] [Google Scholar]
- 8.Quemeneur E, Moutiez M, Charbonnier J B, Menez A. Nature (London) 1998;391:301–304. doi: 10.1038/34687. [DOI] [PubMed] [Google Scholar]
- 9.Altamirano M M, Blackburn J M, Aguayo C, Fersht A R. Nature (London) 2000;403:617–622. doi: 10.1038/35001001. [DOI] [PubMed] [Google Scholar]
- 10.Nixon A E, Firestine S M, Salinas F G, Benkovic S J. Proc Natl Acad Sci USA. 1999;96:3568–3571. doi: 10.1073/pnas.96.7.3568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hellinga H W, Richards F M. J Mol Biol. 1991;222:763–785. doi: 10.1016/0022-2836(91)90510-d. [DOI] [PubMed] [Google Scholar]
- 12.Robertson D E, Farid R S, Moser C C, Urbauer J L, Mulholland S E, Pidikiti R, Lear J D, Wand A J, DeGrado W F, Dutton P L. Nature (London) 1994;368:425–432. doi: 10.1038/368425a0. [DOI] [PubMed] [Google Scholar]
- 13.Klemba M, Gardner K H, Marino S, Clarke N D, Regan L. Nat Struct Biol. 1995;2:368–373. doi: 10.1038/nsb0595-368. [DOI] [PubMed] [Google Scholar]
- 14.Pinto A L, Hellinga H W, Caradonna J P. Proc Natl Acad Sci USA. 1997;94:5562–5567. doi: 10.1073/pnas.94.11.5562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Benson D E, Wisz M S, Hellinga H W. Proc Natl Acad Sci USA. 2000;97:6292–6297. doi: 10.1073/pnas.97.12.6292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fersht A. Enzyme Structure and Mechanism. New York: Freeman; 1985. [Google Scholar]
- 17.Dahiyat B I, Mayo S L. Science. 1997;278:82–87. doi: 10.1126/science.278.5335.82. [DOI] [PubMed] [Google Scholar]
- 18.Harbury P B, Plecs J J, Tidor B, Alber T, Kim P S. Science. 1998;282:1462–1467. doi: 10.1126/science.282.5393.1462. [DOI] [PubMed] [Google Scholar]
- 19.Street A G, Mayo S L. Struct Fold Des. 1999;7:R105–R109. doi: 10.1016/s0969-2126(99)80062-8. [DOI] [PubMed] [Google Scholar]
- 20.Gordon D B, Marshall S A, Mayo S L. Curr Opin Struct Biol. 1999;9:509–513. doi: 10.1016/s0959-440x(99)80072-4. [DOI] [PubMed] [Google Scholar]
- 21.Pollack S J, Jacobs J W, Schultz P G. Science. 1986;234:1570–1573. doi: 10.1126/science.3787262. [DOI] [PubMed] [Google Scholar]
- 22.Tramontano A, Janda K D, Lerner R A. Science. 1986;234:1566–1570. doi: 10.1126/science.3787261. [DOI] [PubMed] [Google Scholar]
- 23.Dunbrack R L, Jr, Karplus M. J Mol Biol. 1993;230:543–574. doi: 10.1006/jmbi.1993.1170. [DOI] [PubMed] [Google Scholar]
- 24.Mayo S L, Olafson B D, Goddard W A., 3rd J Phys Chem. 1990;94:8897–8909. [Google Scholar]
- 25.Dahiyat B I, Sarisky C A, Mayo S L. J Mol Biol. 1997;273:789–796. doi: 10.1006/jmbi.1997.1341. [DOI] [PubMed] [Google Scholar]
- 26.Dahiyat B I, Mayo S L. Proc Natl Acad Sci USA. 1997;94:10172–10177. doi: 10.1073/pnas.94.19.10172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dahiyat B I, Gordon D B, Mayo S L. Protein Sci. 1997;6:1333–1337. doi: 10.1002/pro.5560060622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Connolly M L. Science. 1983;221:709–713. doi: 10.1126/science.6879170. [DOI] [PubMed] [Google Scholar]
- 29.Lee B, Richards F M. J Mol Biol. 1971;55:379–400. doi: 10.1016/0022-2836(71)90324-x. [DOI] [PubMed] [Google Scholar]
- 30.Street A G, Mayo S L. Fold Des. 1998;3:253–258. doi: 10.1016/S1359-0278(98)00036-4. [DOI] [PubMed] [Google Scholar]
- 31.Gleason F K. Protein Sci. 1992;1:609–616. doi: 10.1002/pro.5560010507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pauling L. Am Sci. 1948;36:51–58. [PubMed] [Google Scholar]
- 33.Holmgren A. Annu Rev Biochem. 1985;54:237–271. doi: 10.1146/annurev.bi.54.070185.001321. [DOI] [PubMed] [Google Scholar]
- 34.Ladbury J E, Wynn R, Hellinga H W, Sturtevant J M. Biochemistry. 1993;32:7526–7530. doi: 10.1021/bi00080a026. [DOI] [PubMed] [Google Scholar]
- 35.Hellinga H W, Wynn R, Richards F M. Biochemistry. 1992;31:11203–11209. doi: 10.1021/bi00160a034. [DOI] [PubMed] [Google Scholar]
- 36.Desmet J, Maeyer M D, Hazes B, Lasters I. Nature (London) 1992;356:539–542. doi: 10.1038/356539a0. [DOI] [PubMed] [Google Scholar]
- 37.Pierce N A, Spriet J A, Desmet J, Mayo S L. J Comput Chem. 2000;21:999–1009. [Google Scholar]
- 38.Katti S K, LeMaster D M, Eklund H. J Mol Biol. 1990;212:167–184. doi: 10.1016/0022-2836(90)90313-B. [DOI] [PubMed] [Google Scholar]
- 39.Broo K S, Brive L, Ahlberg P, Baltzer L. J Am Chem Soc. 1997;119:11362–11372. [Google Scholar]
- 40.Nilsson J, Baltzer L. Chemistry. 2000;6:2214–2220. doi: 10.1002/1521-3765(20000616)6:12<2214::aid-chem2214>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 41.Voigt C A, Mayo S L, Arnold F H, Wang Z. Proc Natl Acad Sci USA. 2001;98:3778–3783. doi: 10.1073/pnas.051614498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reardon D, Farber G K. FASEB J. 1995;9:497–503. doi: 10.1096/fasebj.9.7.7737457. [DOI] [PubMed] [Google Scholar]
- 43.Nicholls A, Sharp K A, Honig B. Proteins. 1991;11:281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]