ABSTRACT
Adipose tissue is composed of anatomically distinct depots that mediate several important aspects of energy homeostasis. The past two decades have witnessed increased research effort to elucidate the ontogenetic basis of adipose form and function. In this Review, we discuss advances in our understanding of adipose tissue development with particular emphasis on the embryonic patterning of depot-specific adipocyte lineages and adipocyte differentiation in vivo. Micro-environmental cues and other factors that influence cell identity and cell behavior at various junctures in the adipocyte lineage hierarchy are also considered.
KEY WORDS: Adipocyte, Adipocyte lineages, Adipocyte precursors, Adipogenesis, Adipose, Adipose stem cells
Summary: This review discusses the surprisingly complex lineage hierarchy for adipocytes as well micro-environmental and cell-intrinsic factors that influence adipocyte formation in vivo.
Introduction
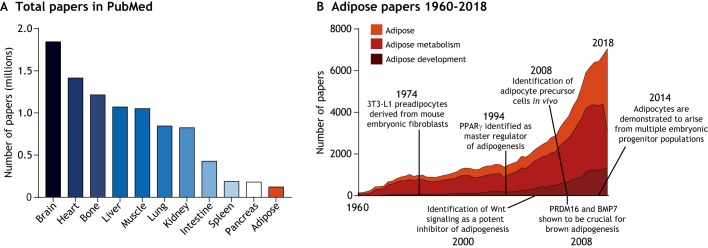
Upwards of 2 billion people worldwide are overweight or obese (Ng et al., 2014), with obesity estimated to account for 147-210 billion dollars in annual healthcare costs in the USA alone (Cawley and Meyerhoefer, 2012). Despite the important implications of increased adipose mass in human health, adipose tissue is remarkably understudied compared with other organs (Fig. 1A). The scarcity of adipose-focused literature can be at least partly explained by a historical lack of interest in the tissue; until the mid-20th century, adipose was considered a metabolically inert connective tissue with no distinguishing functional properties, thus its physiology and ontogeny were largely ignored (Wells, 1940; Wertheimer and Shapiro, 1948). Gradually, however, adipose tissue has garnered greater attention from the research community (Fig. 1B) (Rosen and Spiegelman, 2014) and is now properly recognized as a multi-depot organ with complex regulatory roles in processes ranging from nutrient flux to feeding behavior (Cinti and Vettor, 2009; Frayn, 2002; Herman et al., 2012; Pinto et al., 2004). Indeed, proper adipose tissue mass and function are crucial for maintaining metabolic health.
Fig. 1.
Adipose tissue publication metrics. (A) Adipose tissue has received little research attention compared with other organs/tissues, as illustrated by a bar graph depicting the number of published papers focusing on specific organs/tissues. To construct the graph, each organ/tissue name was typed into the search box on PubMed in January 2019, and the number of items returned for each organ/tissue was plotted. (B) Research investment in adipose has grown, yet is biased toward studies on adipose metabolism. As for the graph in A, the terms ‘adipose’, ‘adipose metabolism’ or ‘adipose development’ were typed into the search box on PubMed. Results by year are shown. Major discoveries in adipose development are also noted along the timeline. The original derivation of 3T3-L1 cells has been described previously (Green and Meuth, 1974). Other major discoveries along the timeline are referred to in the main text.
Advances in our understanding of adipose tissue biology have, unsurprisingly, coincided with a rising prevalence of obesity and related metabolic diseases such as type II diabetes (Ng et al., 2014; Ogurtsova et al., 2017). However, this has resulted in a lopsided research investment that favors studies of the terminal metabolic activities of adipose tissue over those examining the mechanisms responsible for assembling and functionally specifying distinct adipose depots (Fig. 1B, ‘adipose metabolism’ versus ‘adipose development’). Our understanding of adipose tissue development therefore remains exceedingly rudimentary. Nonetheless, seminal in vitro studies in the mid-1990s that identified key transcriptional regulators of adipogenesis (Tontonoz et al., 1994; Yeh et al., 1995) triggered heightened interest in adipose development and laid the intellectual foundation for many important discoveries made in the past 20 years.
In this Review, we describe our current understanding of adipose tissue development, highlighting the lineage hierarchy for adipocytes that populate major adipose depots and the cellular and molecular determinants of adipogenesis in vivo. We critically evaluate the capacity of depot-resident adipocyte precursors to functionally adapt according to systemic and micro-environmental cues, as well as comment on technological breakthroughs that have greatly enhanced the experimental tractability of adipose tissue development.
Distinctive features of white, brown and beige adipocytes
Adipocytes are classified as white, brown or beige (also referred to as brite) according to the unique morphophysiological properties they possess. White adipocytes, which are capable of expanding to well over 100 μm in diameter, are best appreciated as receptacles for lipid storage, but also secrete adipokines that have crucial roles in satiety regulation and whole-body insulin sensitivity (Pinto et al., 2004; Scherer, 2006; Skurk et al., 2007; Steppan et al., 2001; Yamauchi et al., 2002, 2001; Zhang et al., 1994). These cells compose the bulk of white adipose tissue (WAT) mass in both mice and humans, and populate visceral depots in the abdomen as well as subcutaneous depots around the trunk, limbs and face (Fig. 2A) (de Jong et al., 2015; Gesta et al., 2007; Rosen and Spiegelman, 2014; Shen et al., 2003). Brown adipocytes, in contrast, share a similar gene expression profile with myocytes (Timmons et al., 2007), are rich in mitochondria and generate heat through the combustion of various metabolites (e.g. fatty acids and glucose) (Harms and Seale, 2013). Bona fide brown adipocytes have anatomically distinct positions in depots between the scapulae, around the neck and within the chest cavity (Fig. 2A) (Cypess et al., 2009; de Jong et al., 2015; Nedergaard et al., 2007). Beige adipocytes, as their name implies, exhibit properties of both white and brown adipocytes. They possess abundant mitochondria, are thermogenic like brown adipocytes and are located predominately in classical subcutaneous WAT depots in mice (Fig. 2A) (Harms and Seale, 2013; Wu et al., 2012). Beige adipocyte formation in human WAT has not been characterized. However, it has been suggested that human brown adipose depots are more functionally and morphologically similar to murine beige fat, despite having a similar anatomic distribution to brown fat depots in mice (Sharp et al., 2012; Wu et al., 2012).
Fig. 2.
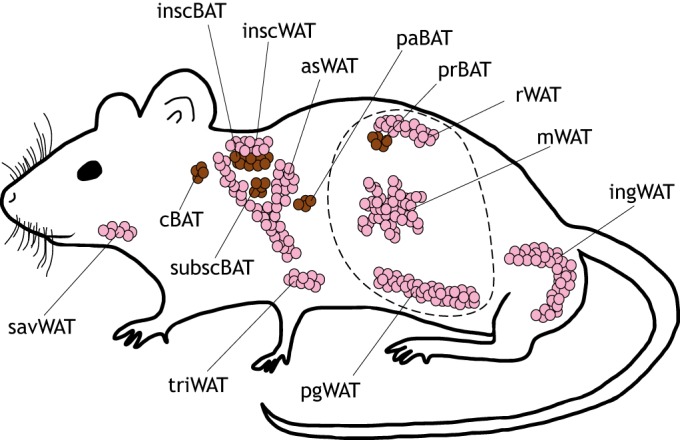
Anatomical distribution of brown and white fat depots in the mouse. Brown fat depots are shown in brown and white fat depots are shown in pink. The visceral cavity is indicated by a dotted line. Notably, the anterior subcutaneous white adipose depot (asWAT) appears to be continuous with the interscapular white adipose depot (inscWAT) under obese conditions. The inscWAT and asWAT overlie the more internal interscapular and subscapular brown depots (inscBAT and subscBAT, respectively). cBAT, cervical brown adipose tissue; ingWAT, inguinal white adipose tissue; mWAT, mesenteric white adipose tissue; paBAT, peri-aortal brown adipose tissue; pgWAT, perigonadal white adipose tissue; prBAT, perirenal brown adipose tissue; rWAT, retroperitoneal white adipose tissue; savWAT, salivary gland white adipose tissue.
The adipocyte lineage tree
Although the adipocyte classifications discussed above help describe the key features of adipocytes, they do not inform us about the ontogenetic relationships among adipocytes. To acquire that knowledge, a fate map must be constructed. As we discuss below, recent technological advances in lineage-tracing and fate-mapping techniques in mice have furthered our understanding of the developmental origins of adipocytes and the relationships between them.
Methods to track cell lineages
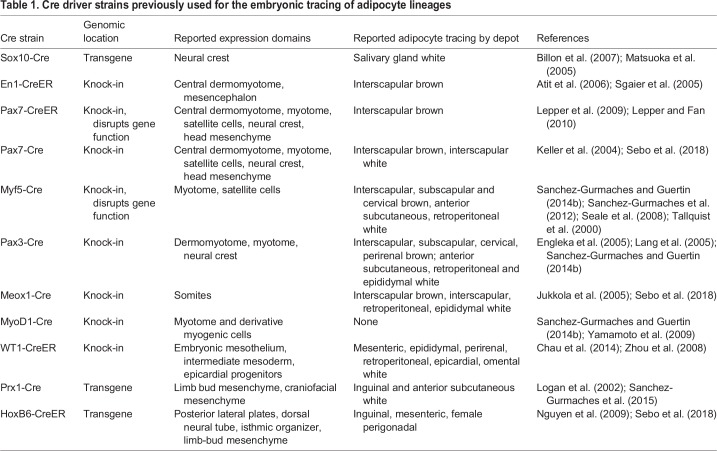
Fate mapping refers to the technique by which a founder cell or cell population is heritably labeled such that the origin of all progeny may be known. The term ‘lineage tracing’ is generally used to refer to single-cell fate maps, although in practice multiple founder cells are typically labeled (Kretzschmar and Watt, 2012). Myriad labeling strategies have been employed since the inception of lineage tracing over one century ago (Conklin, 1905; Stern and Fraser, 2001; Wilson, 1898). These include the use of natural pigment variation among cells of divergent fates (Conklin, 1905) as well as the injection of vital dyes into founder cells (Vogt, 1924). Today, the construction of lineage hierarchies (or trees) occurs mainly via the use of conditional reporter genes, which require activation by genetic approaches such as the Cre-loxP and FLP-FRT systems (Dymecki and Tomasiewicz, 1998; Kretzschmar and Watt, 2012; Orban et al., 1992).
Cre-loxP-based strategies are most widely employed in mouse models, which are favored for the study of adipose biology, and require a user-defined gene regulatory element to control the expression domain of Cre recombinase (Table 1). In cells where Cre is expressed, it will dock at loxP sites and orchestrate recombination to drive reporter gene expression (Kretzschmar and Watt, 2012; Orban et al., 1992). Importantly, the proper interpretation of any lineage-tracing experiment depends on a thorough knowledge of the founder cell population, which, in the case of constitutive Cre-loxP systems, may include multiple non-overlapping groups of progenitor cells and terminally differentiated cell types. This means that cells marked by reporter gene expression do not necessarily belong to the same lineage tree. Inducible variants of the Cre-loxP system are therefore preferred, given their capacity to limit the time frame of Cre activity and to further restrict the labeled group of founder cells (Feil et al., 1996, 2009). Such methods typically involve either tamoxifen or doxycycline treatment to modulate Cre activity. Of note, it has been reported that tamoxifen causes adverse effects on adipose tissue when administered to adult mice (Hesselbarth et al., 2015; Liu et al., 2015; Ye et al., 2015), and that doxycycline disrupts mitochondrial function in mice and other model systems (Moullan et al., 2015). Thus, when tamoxifen- and doxycycline-dependent Cre-loxP tools are implemented, careful control experiments should be designed and low doses used to minimize unintended effects (Hesselbarth et al., 2015; Jeffery et al., 2016; Moullan et al., 2015). In addition, when using either inducible or constitutive Cre for lineage tracing, multiple Cre models with well-defined expression domains ought to be leveraged in order to control for ‘off-target’ Cre activity, thereby improving the probability of accurately judging the branchpoints and order of succession in a cell lineage tree. For this same reason, it is crucial that the cell populations labeled in any given Cre model are broadly assessed.
Table 1.
Cre driver strains previously used for the embryonic tracing of adipocyte lineages
The use of reporter genes that are appropriate for the progeny cells of interest is also crucial to make an accurate assessment of their developmental origin. Adipocytes present a special case in that traditional soluble reporters, such as Xgal and YFP, fail to appreciably label these cells (Jeffery et al., 2014; Sanchez-Gurmaches and Guertin, 2014b; Sanchez-Gurmaches et al., 2016, 2012). This is likely due to the intracellular lipid engorgement and lack of cytoplasmic volume characteristic of adipocytes. Fortunately, this problem can be circumvented by using cell membrane-localized reporters. The current gold standard for tracing adipocytes is the mTmG system (Berry and Rodeheffer, 2013; Jeffery et al., 2014; Sanchez-Gurmaches et al., 2016). This system drives the ubiquitous expression of membrane-localized tdTomato (mTomato) in the absence of Cre, but in cells where Cre is active, the mTomato element of the mTmG cassette is excised, resulting in the permanent expression of membrane-localized GFP (mGFP) in founder cells and their progeny (Muzumdar et al., 2007).
The germ layer origins of adipocytes
A fundamentally important event in vertebrate embryogenesis is the differentiation of epiblast stem cells into the three germ layers – endoderm, mesoderm and ectoderm – during gastrulation (Hatada and Stern, 1994; Kimmel et al., 1990; Lawson et al., 1991). This marks the first stage of lineage segregation in the embryo proper and sets the stage for organogenesis, with each germ layer giving rise to a defined set of tissues and organs (Tam and Loebel, 2007).
Early understanding of the germ layer origin of adipocytes was based mainly on histological analyses performed by Walther Flemming in the 1870s. In these experiments, Flemming longitudinally observed mouse adipose development and found that at least some adipocytes emerge from a connective tissue anlagen known to be mesodermally derived (Flemming, 1871; Flemming, 1879). Consequently, adipocytes have been considered a predominately mesodermal cell type for well over 100 years. Recent findings (which we discuss below) have verified this notion. However, mesoderm is composed of numerous spatially diminutive progenitor fields, each with unique developmental trajectories. Thus, classifying adipocytes as merely mesodermal cells is an oversimplification. Indeed, from the work of many research groups over the past decade, it is now clear that a remarkably complex lineage hierarchy exists for mesodermally derived adipocytes. Moreover, in 2007, a Cre-dependent lineage-tracing paradigm showed that adipocytes populating the murine salivary gland arise from neural crest cells (Billon et al., 2007), which are an ectodermally derived cell population (Simões-Costa and Bronner, 2015). It remains to be tested whether adipocytes in other craniofacial adipose depots originate from neural crest, or from other ectoderm derivatives. Nonetheless, this finding was an early indicator that adipocytes could have a more complex developmental history than previously thought. Notably, the endoderm, which is the generative tissue for the alimentary canal, pancreas, thyroid, respiratory tract and certain components of the liver and thymus (Gordillo et al., 2015; Gordon and Manley, 2011; Grapin-Botton, 2009; Nilsson and Fagman, 2017), has not been demonstrated to produce adipose tissue.
An introduction to mesoderm anatomy and fate
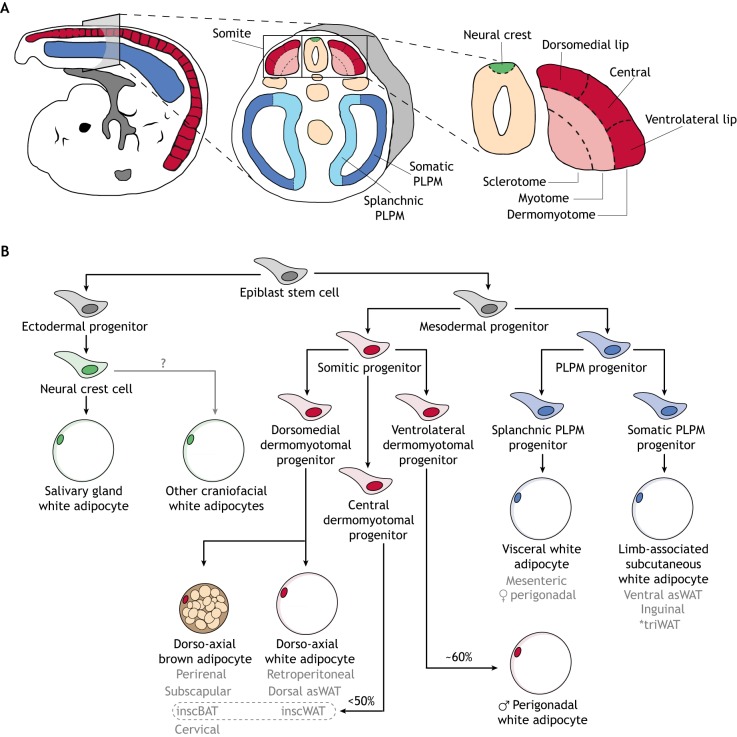
Mesoderm is transiently divided into three subcompartments following gastrulation: the somites, lateral plates and intermediate mesoderm. Somites are the most dorsal of the mesodermal subcompartments, forming in dyadic segments as the embryo elongates posteriorly. Each somite is partitioned further into sclerotome, myotome and dermomyotome (Fig. 3A) (Tajbakhsh and Spörle, 1998). Progenitor cells that compose these somitic domains harbor distinct developmental potentials, with the sclerotome producing cartilaginous and osteoid tissues of the vertebral column, and the myotome producing skeletal muscle (Pang and Thompson, 2011). By contrast, dermomyotomal progenitors exhibit the greatest diversity in cell fates: they are responsible for establishing the myotome (Gros et al., 2004; Pu et al., 2013) and, beyond muscle, give rise to the dermis of the back, as well as adipocytes in several adipose depots (Atit et al., 2006; Lepper and Fan, 2010; Sanchez-Gurmaches and Guertin, 2014a,b; Seale et al., 2008; Sebo et al., 2018).
Fig. 3.
Embryonic patterning of adipocyte lineages. (A) Schematic of an ∼E9.5 mouse embryo. Cross-sectional views show relevant mesodermal and neuro-ectodermal subcompartments. (B) A model adipocyte lineage tree. This tree was constructed based on current lineage-tracing data in mice. Beige adipocytes are not included in the tree, given that it is unclear whether these cells are specified embryonically. *The triceps white adipose depot (triWAT) is not a subcutaneous adipose depot. Rather, it is an internal supra-muscular depot. Grey text indicates specific adipose depots; arrows indicate cell lineage progression; '?' indicates an unknown in the field. inscBAT and inscWAT are outlined as <50% of adipocytes in these depots are derived from progenitors of the central dermomyotome. asWAT, anterior subcutaneous white adipose tissue; inscBAT, interscapular brown adipose tissue; inscWAT, interscapular white adipose tissue; PLPM, posterior lateral plate mesoderm.
Lateral plate mesoderm is composed of two bilaterally symmetric pouches that fill the presumptive abdominal region of the embryo. These pouches are divided into splanchnic and somatic layers (Fig. 3A). The splanchnic layer produces the cardiovascular system and the mesentery, which anchors the intestines to the abdominal wall and suspends them within the body cavity (Brand, 2003; Davis et al., 2008), while the somatic layer is the source of appendicular skeleton and ventral dermis (Newman, 1988; Ohtola et al., 2008). The splanchnic and somatic layers also contribute to adipocytes in visceral and limb-associated subcutaneous adipose depots, respectively (Sanchez-Gurmaches and Guertin, 2014b; Sebo et al., 2018). Finally, intermediate mesoderm, which is sandwiched between the somites and lateral plates and is the smallest of these subcompartments, contributes to the majority of the urogenital tract (Davidson, 2009; Torres et al., 1995) but has not been demonstrated to produce adipocytes.
Dermomyotomal branches of the adipocyte lineage tree
The earliest definitive evidence for the existence of mesodermally -derived adipocytes came from a study showing that tracing of the central dermomyotome using an En1-CreER:lacZ system (Table 1) results in the labeling of interscapular BAT (inscBAT) late in gestation (Atit et al., 2006). These data were corroborated several years later when progenitors expressing Pax7 (a key myogenic transcription factor) in the central dermomyotome were inducibly labeled using a lacZ reporter system (Table 1) and shown to give rise to embryonic inscBAT (Lepper and Fan, 2010). Moreover, progenitors of interscapular brown adipocytes – but not white adipocytes that populate gluteal, inguinal and epididymal depots – have been reported to express Myf5 (Seale et al., 2008), which also functions as a myogenic transcription factor and is known to first be active in the dorsomedial lip of the dermomyotome (Relaix et al., 2005; Tallquist et al., 2000; Teboul et al., 2002). Taken together, these data support the notion that interscapular brown adipocytes arise from dermomyotomal progenitors and thus share a common origin with myocytes in mice.
More recent lineage-tracing studies have revealed some inconsistencies with earlier findings and have produced a surprisingly sophisticated fate map for dermomyotome-derived adipocytes (Fig. 3B). For example, a thorough analysis of Myf5-Cre:mTmG (Table 1) tracing has shown that not all brown adipocytes arise from a Myf5+ lineage. Rather, Myf5lin+ brown adipocytes are restricted to interscapular, subscapular and cervical depots. Furthermore, white adipocytes of the anterior subcutaneous and retroperitoneal depots are Myf5lin+ (Sanchez-Gurmaches and Guertin, 2014b; Sanchez-Gurmaches et al., 2012). These findings were unexpected, as the divergent functional properties of white and brown adipocytes were thought to be explained, at least in part, by their ontogenetic proximity to myocytes (Rosen and Spiegelman, 2014; Seale et al., 2008; Timmons et al., 2007). Instead, emerging data suggest that the allocation of dermomyotomal progenitors to white or brown adipocyte lineages may be better explained by their physical location along the dorsoventral axis of the dermomyotome. Indeed, by conceptually superimposing multiple single-gene fate maps, it is possible to construct a working lineage tree for adipocytes arising from sub-domains of the dermomyotome. The embryonic expression domain for Pax3-Cre includes neural crest cells and the whole dermomyotome (Table 1) (Lang et al., 2005); coupling this Cre with mTmG results in a labeling pattern slightly broader than that observed in the case of Myf5-Cre:mTmG labeling (i.e. perirenal BAT and a subset of male, but not female, perigonadal white adipocytes are also mGFP+). This suggests that the mGFP+ adipocytes in the Pax3-Cre:mTmG system that do not overlap with mGFP+ adipocytes in the Myf5-Cre:mTmG system are derived from either the neural crest or a region of the dermomyotome in which Myf5 is not appreciably expressed, particularly the ventrolateral lip (Teboul et al., 2002).
Interestingly, pan-somite fate mapping using the Meox1-Cre:mTmG system (Table 1) mirrors the adipocyte labeling pattern of Pax3-Cre:mTmG, at least for adipose depots analyzed in both studies (Jukkola et al., 2005; Sanchez-Gurmaches and Guertin, 2014b; Sebo et al., 2018). This labeling pattern includes a subset of adipocytes residing in the male perigonadal fat pad. Thus, these findings suggest that dermomyotomal progenitors of the ventrolateral lip rather than neural crest cells contribute to a proportion of male, but not female, perigonadal adipocytes. Moreover, the sclerotome can probably be excluded as a tissue of origin for adipocytes, as only Meox1-Cre:mTmG labels founder cells here, yet the pattern of mGFP+ adipocyte labeling in this map does not recognizably diverge from that of Pax3-Cre:mTmG labeling (Sanchez-Gurmaches and Guertin, 2014b; Sebo et al., 2018).
Recently, a constitutive Pax7-Cre (Keller et al., 2004) was combined with the mTmG reporter to label adipocytes originating from progenitors of the central dermomyotome (Table 1). This resulted in <50% of interscapular white and brown adipocytes being mGFP+ in mice ranging from 1 day to 10 weeks of age, with no detectable labeling in other depots analyzed (Sebo et al., 2018). These data contrast with a previous study in which intense staining of inscBAT was observed at E16.5 in Pax7-CreERT2:lacZ mouse embryos after induction of Cre activity at E9.5 (Lepper and Fan, 2010). Possible reasons for this discrepancy have been discussed previously (Sebo et al., 2018); these include the use of distinct reporters and Cre strains in the two studies. Moreover, the observation that Pax7+ progenitors contribute to only a subset rather than the majority of interscapular adipocytes is consistent with Myf5-tracing, as Myf5 is expressed along a dorsoventral gradient in the dermomyotome, overlapping with Pax7 in the center and diminishing ventrally (Teboul et al., 2002). This suggests that most interscapular adipocytes arise from the dorsomedial lip of the dermomyotome, with the remainder deriving from the central region (Sebo et al., 2018). Importantly, Pax7+ progenitors that localize to the myotome have been shown to undergo a myogenic developmental trajectory, ultimately producing skeletal muscle fibers (Relaix et al., 2005). This indicates that adipocyte-fated Pax7+ progenitors do not pass through the myotome prior to lineage segregation from myocytes. Indeed, tracing of the myotome and its derivative tissues by MyoD1-Cre:mTmG fails to label any adipocytes, brown or white (Table 1) (Sanchez-Gurmaches and Guertin, 2014b).
To summarize, adipocytes residing in dorso-axial adipose depots are likely to find their origin predominately in the dorsomedial lip of the dermomyotome, with a subset of interscapular adipocytes arising from the central dermomyotome. A proportion of male, but not female, perigonadal adipocytes arise from the ventrolateral lip of the dermomyotome. Finally, evidence to date indicates that adipocyte founder cells do not contribute to the myotome during their developmental history.
Lateral plate branches of the adipocyte lineage tree
Less work has been done to precisely characterize the segregation of lateral plate-derived adipocyte lineages, although a few key experiments have provided insight into this process. The first indication that some adipocytes arise from lateral plate mesoderm came in 2014 when it was reported that several visceral, but not subcutaneous, adipocyte populations differentiated from embryonic mesothelial cells expressing the transcription factor Wilms tumor 1 (WT1) (Chau et al., 2014); these cells are generally thought to descend from splanchnic lateral plate progenitors (Winters et al., 2012). However, it should be noted that WT1+ cells also include derivatives of the intermediate mesoderm (Armstrong et al., 1993). Soon afterwards, it was shown that tracing of the limb bud mesenchyme by Prx1-Cre (Logan et al., 2002) resulted in labeling of limb-associated subcutaneous adipocytes (Krueger et al., 2014; Sanchez-Gurmaches et al., 2015), suggesting these cells are derived from progenitors of the somatic layer of the lateral plates (Durland et al., 2008). A lateral plate origin for adipocytes was recently confirmed through the transient induction of Cre activity at E8.5 in HoxB6-CreERT:mTmG embryos. This marks founder cells in the splanchnic and somatic layers of the posterior domain of lateral plate mesoderm and results in the labeling of adipocytes in ventrolateral adipose depots (i.e. mesenteric, inguinal and female perigonadal) (Sebo et al., 2018). These data suggest that some visceral and subcutaneous adipocytes share a common group of founder cells in the lateral plates, yet these appear to be partitioned into splanchnic and somatic progenitor pools, respectively. These data also confirm that perigonadal adipocytes arise from distinct mesodermal subcompartments in males and females (Fig. 3B), suggesting that the sex-specific anatomy of the perigonadal fat pad in mice is coordinately patterned with internal reproductive organs and, thus, uniquely encoded by male and female genomes.
Our understanding of the adipocyte lineage tree in mice is far more detailed than it was a decade ago. Nonetheless, we still know very little about the mechanisms driving the ramification of these lineages. A functional marker for embryonic interscapular brown adipocyte progenitors has been identified (Wang et al., 2014), although it is unclear whether other depot-specific adipocyte lineages can be molecularly distinguished prenatally, as most adipose tissue does not form until after birth in mice (Jiang et al., 2014). Indeed, it is possible that, for the majority of mouse adipocyte lineages, embryogenesis serves merely to spatially segregate depot-specific progenitor fields. In this way, such fields could be directed to an adipocyte fate by local inductive signals at the outset of depot formation. Consistent with this notion, the formation of adipose tissue in postnatal mice is partitioned both spatially and temporally such that each depot emerges in a stereotyped developmental sequence rather than all at once. For example, inguinal and retroperitoneal adipocytes become morphologically distinguishable soon after pups are delivered, whereas perigonadal adipocytes are not visible until 1 week of age (Birsoy et al., 2011; Han et al., 2011). Human adipose depots are also established sequentially, albeit prior to birth. During the second trimester of gestation, adipose in the head and neck arise, followed by depots in the torso and limbs (Poissonnet et al., 1984, 1988). Thus, adipocyte lineage segregation and adipogenesis are likely to be coordinated developmental events in mice and humans.
Human adipocyte lineage segregation and lipodystrophy
Unsurprisingly, adipocyte lineage segregation is much less understood in humans. However, it is known that several rare adipose disorders (RADs) and lipodystrophies result in depot-specific redistribution and/or loss of fat mass (Herbst, 2012; Herbst et al., 2003; Köbberling and Dunnigan, 1986), raising the possibility that distinct adipocyte lineages are affected in these conditions. Indeed, it has been shown in mice that knocking out phosphatase and tensin homolog (PTEN), an important negative regulator of the insulin signaling pathway, in the Myf5+-lineage results in a dramatic reduction in visceral fat mass and a corresponding increase in interscapular fat mass (Sanchez-Gurmaches et al., 2012). Interestingly, these observations phenocopy a RAD in humans called multiple symmetric lipomatosis (Enzi et al., 2002; Herbst, 2012), suggesting a genetic defect that only affects a subset of adipocyte lineages may be responsible for this disease (Sanchez-Gurmaches et al., 2012).
In line with this, it has been proposed that mosaicism (a condition in which a single organism harbors two or more cell populations with different genotypes) could explain the etiology of at least some idiopathic partial lipodystrophies (Kim et al., 2009). In this case, a given embryonic progenitor cell would acquire a deleterious mutation in a gene important for adipose tissue development or maintenance. This mutation would be passed on to daughter cells, including some but not all adipose-resident cell types, resulting in the failed establishment and/or maintenance of affected adipose depots. Importantly, such mutations need not occur strictly in adipocyte lineages, as it is now clear from mouse studies that proper adipose tissue function and adipogenesis require the activity of multiple adipose-resident cell types (Han et al., 2011; Nishimura et al., 2007).
Adipogenesis in vivo
Adipogenesis refers to the process by which new adipocytes differentiate from precursor cells. Below, we discuss the regulation of adipogenesis both during development and during tissue homeostasis.
Developmental adipogenesis
Adipocyte precursors (APs), also called adipocyte stem/progenitor cells, display a unique cell surface marker profile (CD45−CD31−CD34+CD29+SCA1+CD24+/−) and are functionally separable by the presence of CD24. Specifically, CD24 is lost on the AP cell surface as they commit to the adipocyte fate and exit the cell cycle (Berry and Rodeheffer, 2013), yet it is not known whether CD24 plays an active role in adipogenesis. APs residing in a white adipose anlagen (nascent adipose tissue) are highly proliferative prior to their differentiation and lipid filling (Jeffery et al., 2015). The differentiation of these cells is dependent on their crosstalk with various other cell types in the adipose anlagen. For example, disrupting angiogenesis (blood vessel formation) and reducing the number of depot-resident macrophages both result in defective perigonadal adipose development (Han et al., 2011). Consistent with this, it has been shown that adipose-resident endothelial cells are essential structural and functional components of the niche in which APs reside (Tang et al., 2008), although a direct role for macrophages in adipogenesis has not been identified. Thus, the micro-environment of nascent adipose tissue is an important regulator of developmental adipogenesis.
In addition, several transcription factors have been shown to have important cell-intrinsic roles in the process of adipocyte differentiation. Indeed, a core adipogenic transcriptional cascade is well characterized (Farmer, 2006; Lefterova and Lazar, 2009; Rosen and MacDougald, 2006; Siersbæk et al., 2012). Upstream components of this pathway include C/EBP transcription factors that work sequentially to activate PPARγ: the master regulator of adipocyte differentiation. Knocking out or disrupting the function of these genes results in severe lipodystrophy (Moitra et al., 1998; Rosen et al., 1999). A recent study suggested that C/EBPα is not required for developmental adipogenesis (Wang et al., 2015); however, this study knocked out C/EBPα using an adiponectin-Cre, which is active in mature adipocytes but not in APs (Berry and Rodeheffer, 2013; Jeffery et al., 2014). Thus, C/EBPα may be dispensable for maintaining certain aspects of adipocyte identity once these cells have been established. For brown adipogenesis, the core program is adjoined by unique transcriptional co-regulators, including PRDM16 and Ebf2, that physically interact with PPARγ to direct brown adipocyte-specific target gene expression (Rajakumari et al., 2013; Seale et al., 2008). Notably, these factors also play a role in beiging, but it is currently unclear whether beige adipocyte formation is developmentally encoded or whether these cells arise only upon induction in adult adipose tissue.
Secreted proteins are also thought to play important roles in developmental adipogenesis. BMP7, for example, has been shown to be crucial for the induction of brown adipose tissue formation (Tseng et al., 2008), whereas BMP2 and BMP4 have been implicated as positive regulators of white adipogenesis (Huang et al., 2009; Jin et al., 2006). However, the pro-adipogenic activity of BMP2 and BMP4 have not been validated in vivo. Similarly, Wnt10b has been shown to be a potent inhibitor of adipogenesis in cell culture and implantation models (Bennett et al., 2002; Ross et al., 2000), but no transgenic mice have been developed to test the role of Wnt10b in regulating the establishment of specific adipose depots. Interestingly, Wnts and BMPs are well known morphogens that direct the patterning and differentiation of various organs in development (Clevers, 2006; Wagner et al., 2010). It is possible that opposing gradients of these cytokines are present at presumptive adipose depots and act to spatiotemporally constrain depot formation.
Obesogenic adipogenesis
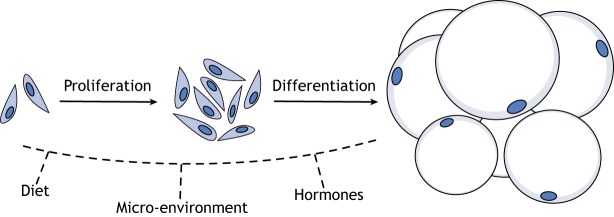
The development of obesity occurs through an increase in adipocyte number by adipogenesis (hyperplasia) (Fig. 4) and/or an increase in the size of existing adipocytes (hypertrophy). Here, we will focus on adipocyte hyperplasia. At the onset of high-fat diet (HFD) feeding, APs in mice exhibit a burst in proliferation and, over the course of 7-8 weeks, differentiate into adipocytes (Jeffery et al., 2015; Vishvanath et al., 2016; Wang et al., 2013). This response is restricted to visceral adipose tissue in males, but occurs in both visceral and subcutaneous fat in females (Jeffery et al., 2016; Vishvanath et al., 2016; Wang et al., 2013). Interestingly, obesogenic adipogenesis appears to take place preferentially in APs that express the pericyte marker, Pdgfrb (Vishvanath et al., 2016), although a functional role for Pdgfrb in this process has not been identified. Furthermore, obesogenic adipogenesis and developmental adipogenesis seem to be initiated by distinct signal transduction pathways, as Akt2 is required for HFD-induced adipocyte hyperplasia but not for the developmental establishment of adipocytes (Jeffery et al., 2015). Akt2 knockout mice display age-dependent lipoatrophy (Garofalo et al., 2003) and an Akt2 mutation in humans (Akt2H274) is associated with reduced adipose tissue mass (George et al., 2004), indicating this gene is required for normal adipose tissue maintenance, in addition to obesogenic adipogenesis. Indeed, it is known that ∼10% of adipocytes are replaced annually in humans under homeostatic conditions (Spalding et al., 2008). Thus, loss of Akt2 function may impair homeostatic adipocyte renewal, resulting in the progressive loss of adipose tissue mass.
Fig. 4.
Adipocyte hyperplasia is regulated by dietary, micro-environmental and hormonal cues. New adipocytes form through the proliferation and differentiation of depot-resident adipocyte precursor cells. This process is combinatorially regulated in vivo. However, the molecular mechanisms by which different cues influence adipocyte hyperplasia are only beginning to be understood.
AP behavior is also regulated by micro-environmental and systemic factors. The reciprocal transplantation of visceral and subcutaneous APs in male mice results in transplanted APs taking on the behavioral properties of host-depot APs. That is, subcutaneous APs transplanted into visceral fat respond to HFD by proliferating, whereas visceral APs transplanted into subcutaneous fat do not (Jeffery et al., 2016). Furthermore, it is known that, in response to HFD, APs in the anterior tip of male perigonadal fat undergo more adipogenesis than those located in the posterior region of the depot. Importantly, this seems to be independent of AP lineage (Sebo et al., 2018), indicating that AP behavior is strongly influenced by the intra-depot micro-environment. Indeed, a newly discovered cell population in the perivascular compartment of adipose tissue has been demonstrated to negatively regulate adipogenesis in cell culture and Matrigel implantation models through a paracrine mechanism (Schwalie et al., 2018). These cells, which are called Aregs and display a novel cell-surface marker profile (Lin−CD29+CD34+SCA1+CD142+ABCG1+), might fine-tune the pattern of adipocyte hyperplasia between and within depots to regulate fat mass and distribution. Sex hormones also play a role in AP activity, as treating male mice with estradiol results in a feminized pattern of AP proliferation upon HFD feeding (Jeffery et al., 2016). Thus, AP behavior is regulated by cues from the diet, circulating hormones and the micro-environment (Fig. 4).
Recently, it was shown that feeding PdgfRβ-Cre:mTmG male mice a Surwit diet (58% kcals from coconut/soybean oil and sucrose) results in high levels of mGFP+ adipocytes in visceral (∼35%) and subcutaneous (∼70%) adipose depots after 4 weeks (Gao et al., 2018). This was interpreted as obesogenic adipogenesis from PdgfRβ+ APs. However, this conclusion conflicts with previous findings, particularly observations indicating that adipocyte hyperplasia is restricted to visceral fat in males and takes approximately 2 months (Jeffery et al., 2015, 2016; Vishvanath et al., 2016; Wang et al., 2013). It is possible that a Surwit diet initiates a pro-adipogenic response that is distinct from that induced by standard lard-based HFDs. The data from Gao et al. can also be explained by the induction of PdgfRβ expression in mature adipocytes; this would be reflected in increased mGFP labeling of adipocytes in the absence of increased PdgfRβ+ AP differentiation. Notably, these possibilities are not mutually exclusive, and further experiments will be required to better understand the effect of a Surwit diet on obesogenic adipogenesis.
Beiging
Beige adipocyte formation, in contrast to white adipocyte formation, in obesity is induced by cold exposure or pharmacological activation of β-adrenergic receptors (Harms and Seale, 2013). Unlike white adipogenesis, beige adipogenesis is rapid and can occur in less than 3 days (Wang et al., 2013). De novo adipogenesis is thought to be the major mechanism by which beige adipocytes arise (Harms and Seale, 2013; Wang et al., 2013), although these cells can also form through the direct conversion of white adipocytes (a process also referred to as transdifferentiation) (Rosenwald et al., 2013; Vitali et al., 2012). It remains unclear whether all white adipocytes are capable of taking on thermogenic properties. However, the depot-specific micro-environment, particularly innervation, is likely to be important for both white/beige interconversion and de novo beige adipogenesis, as higher neurite density in inguinal fat is associated with beiging (Chi et al., 2018) and ablation of sympathetic arborizations in inguinal fat prevents beiging upon cold exposure in this tissue (Jiang et al., 2017). In addition, β3-adrenergic receptor activation has been shown to be dispensable for cold-induced beiging and thermogenesis (de Jong et al., 2017), indicating that beige adipocyte formation can occur by at least two distinct molecular pathways. Interestingly, beige adipocyte formation occurs in 3-week-old mice in the absence of overt cold exposure. These so-called postnatal beige adipocytes subsequently whiten in adulthood and, if exposed to cold, are more likely to become beige than are other depot-resident white adipocytes (Wang et al., 2017). It is unclear whether postnatal beige adipocyte formation is a genetically encoded process or takes place as a result of reduced body temperature after weaning (i.e. separation from parents).
It has been reported that F4/80−CD19−CD137+TMEM26+ cells in subcutaneous fat preferentially differentiate into beige adipocytes in adults (Wu et al., 2012). However, this cell pool includes endothelial cells (CD31+) and is not enriched for cells with accepted markers of stemness (CD34+SCA1+), highlighting that the identity of beige APs requires further refinement. Importantly, the extent to which depot-resident white and beige AP populations overlap, if at all, is currently unclear. The embryonic founder cells from which beige adipocytes arise are also not known, although it would be surprising if beige and white adipocyte lineages diverged prior to depot formation. Indeed, Prx1-Cre tracing labels both white and beige adipocytes in subcutaneous fat (Sanchez-Gurmaches et al., 2015), indicating lineage divergence has not occurred by the outset of limb formation (E9.5-E11.5) (Logan et al., 2002). Consistent with this notion, it has been reported that at least some beige adipocytes, but not white adipocytes, arise from a common depot-resident progenitor pool with smooth muscle cells (Long et al., 2014). However, lineage-tracing experiments from this study are difficult to interpret due to poor adipocyte labeling. Interestingly, both white and beige adipocytes arise from a PdgfRa+ lineage (Berry and Rodeheffer, 2013; Lee et al., 2012), and depot-resident PdgfRb+ APs can differentiate into adipocytes of either type depending on the mode of stimulation (i.e. HFD or cold) (Vishvanath et al., 2016). Thus, distinguishing white and beige adipocyte lineages merely by the ancestral expression of a single gene may not be sufficient. Indeed, beige adipocytes can switch to a white adipocyte phenotype when cold exposure is abated (Rosenwald et al., 2013) and also retain a beige-like chromatin state poised for thermogenic gene expression under such conditions (Roh et al., 2018). Thus, the concept of distinct white and beige adipocyte lineages may not be valid in all physiologic contexts. Further work will be required to identify the in vivo regulators of beiging and the phenotypic range of adipocytes.
Conclusions and future perspectives
In this Review, we have discussed cellular and molecular features of adipose tissue assembly, focusing on the embryonic segregation of adipocyte lineages and adipogenesis in vivo. It is now apparent that adipocytes have remarkably diverse developmental ancestries, and that adipogenesis is a context-dependent, modifiable cell differentiation program. Yet we know little about how adipocyte-fated embryonic progenitors are allocated to specific depots, nor have any mechanisms been implicated in the temporal regulation of depot formation. Indeed, how the core adipogenic transcriptional program is differentially deployed in development and in response to specific external cues, such as diet and temperature, has only recently become an area of active investigation. Therefore, a great research frontier lies ahead.
Next-generation sequencing methods involving DNA barcodes and single cell transcriptional profiling have emerged as powerful tools for lineage tree reconstruction and the study of cell differentiation (Kalhor et al., 2018; Kumar et al., 2017; McKenna et al., 2016). In addition, intravital imaging approaches have tremendous utility for viewing cell dynamics in live animals in real time (Nishimura et al., 2008). Implementing such methods in the study of adipose tissue will be crucial to gain further insight into how adipose tissue develops and how micro-environmental factors influence adipogenesis in vivo. This has implications for understanding not only the structural emergence and maintenance of adipose tissue, but also how adipose formation is involved more broadly in establishing and integrating systemic metabolic regulatory networks.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Funding
This work was supported by a National Science Foundation Graduate Research Fellowship (DGE1122492 to Z.L.S.), and by the National Institute of Diabetes and Digestive and Kidney Diseases (DK090489 and DK110147 to M.S.R.). Deposited in PMC for release after 12 months.
References
- Armstrong J. F., Pritchard-Jones K., Bickmore W. A., Hastie N. D. and Bard J. B. L. (1993). The expression of the Wilms’ tumour gene, WT1, in the developing mammalian embryo. Mech. Dev. 40, 85-97. 10.1016/0925-4773(93)90090-K [DOI] [PubMed] [Google Scholar]
- Atit R., Sgaier S. K., Mohamed O. A., Taketo M. M., Dufort D., Joyner A. L., Niswander L. and Conlon R. A. (2006). β-catenin activation is necessary and sufficient to specify the dorsal dermal fate in the mouse. Dev. Biol. 296, 164-176. 10.1016/j.ydbio.2006.04.449 [DOI] [PubMed] [Google Scholar]
- Bennett C. N., Ross S. E., Longo K. A., Bajnok L., Hemati N., Johnson K. W., Harrison S. D. and MacDougald O. A. (2002). Regulation of Wnt signaling during adipogenesis. J. Biol. Chem. 277, 30998-31004. 10.1074/jbc.M204527200 [DOI] [PubMed] [Google Scholar]
- Berry R. and Rodeheffer M. S. (2013). Characterization of the adipocyte cellular lineage in vivo. Nat. Cell Biol. 15, 302 10.1038/ncb2696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billon N., Iannarelli P., Monteiro M. C., Glavieux-Pardanaud C., Richardson W. D., Kessaris N., Dani C. and Dupin E. (2007). The generation of adipocytes by the neural crest. Development 134, 2283-2292. 10.1242/dev.002642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birsoy K., Berry R., Wang T., Ceyhan O., Tavazoie S., Friedman J. M. and Rodeheffer M. S. (2011). Analysis of gene networks in white adipose tissue development reveals a role for ETS2 in adipogenesis. Development 138, 4709-4719. 10.1242/dev.067710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand T. (2003). Heart development: molecular insights into cardiac specification and early morphogenesis. Dev. Biol. 258, 1-19. 10.1016/S0012-1606(03)00112-X [DOI] [PubMed] [Google Scholar]
- Cawley J. and Meyerhoefer C. (2012). The medical care costs of obesity: an instrumental variables approach. J. Health Econ. 31, 219-230. 10.1016/j.jhealeco.2011.10.003 [DOI] [PubMed] [Google Scholar]
- Chau Y.-Y., Bandiera R., Serrels A., Martínez-Estrada O. M., Qing W., Lee M., Slight J., Thornburn A., Berry R. and McHaffie S. (2014). Visceral and subcutaneous fat have different origins and evidence supports a mesothelial source. Nat. Cell Biol. 16, 367-375. 10.1038/ncb2922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi J., Wu Z., Choi C. H. J., Nguyen L., Tegegne S., Ackerman S. E., Crane A., Marchildon F., Tessier-Lavigne M. and Cohen P. (2018). Three-dimensional adipose tissue imaging reveals regional variation in beige fat biogenesis and PRDM16-dependent sympathetic neurite density. Cell Metab. 27, 226-236. e223. 10.1016/j.cmet.2017.12.011 [DOI] [PubMed] [Google Scholar]
- Cinti S. and Vettor R. (2009). The adipose organ. In Adipose Tissue and Inflammation (A.B. Awad and P. G. Bradford, eds), pp. 11-31. CRC Press. [Google Scholar]
- Clevers H. (2006). Wnt/β-catenin signaling in development and disease. Cell 127, 469-480. 10.1016/j.cell.2006.10.018 [DOI] [PubMed] [Google Scholar]
- Conklin E. G. (1905). The Organization and Cell-Lineage of the Ascidian Egg. Academy of Natural Sciences, Philadelphia. [Google Scholar]
- Cypess A. M., Lehman S., Williams G., Tal I., Rodman D., Goldfine A. B., Kuo F. C., Palmer E. L., Tseng Y.-H. and Doria A. (2009). Identification and importance of brown adipose tissue in adult humans. N. Engl. J. Med. 360, 1509-1517. 10.1056/NEJMoa0810780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson A. J. (2009). Mouse Kidney Development. [PubMed] [Google Scholar]
- Davis N. M., Kurpios N. A., Sun X., Gros J., Martin J. F. and Tabin C. J. (2008). The chirality of gut rotation derives from left-right asymmetric changes in the architecture of the dorsal mesentery. Dev. Cell 15, 134-145. 10.1016/j.devcel.2008.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong J. M. A., Larsson O., Cannon B. and Nedergaard J. (2015). A stringent validation of mouse adipose tissue identity markers. Am. J. Physiol. Endocrinol. Metab. 308, E1085-E1105. 10.1152/ajpendo.00023.2015 [DOI] [PubMed] [Google Scholar]
- de Jong J. M. A., Wouters R. T., Boulet N., Cannon B., Nedergaard J. and Petrovic N. (2017). The β3-adrenergic receptor is dispensable for browning of adipose tissues. Am. J. Physiol. Endocrinol. Metab. 312, E508-E518. 10.1152/ajpendo.00437.2016 [DOI] [PubMed] [Google Scholar]
- Durland J. L., Sferlazzo M., Logan M. and Burke A. C. (2008). Visualizing the lateral somitic frontier in the Prx1Cre transgenic mouse. J. Anat. 212, 590-602. 10.1111/j.1469-7580.2008.00879.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dymecki S. M. and Tomasiewicz H. (1998). Using Flp-recombinase to characterize expansion of Wnt1-expressing neural progenitors in the mouse. Dev. Biol. 201, 57-65. 10.1006/dbio.1998.8971 [DOI] [PubMed] [Google Scholar]
- Engleka K. A., Gitler A. D., Zhang M., Zhou D. D., High F. A. and Epstein J. A. (2005). Insertion of Cre into the Pax3 locus creates a new allele of Splotch and identifies unexpected Pax3 derivatives. Dev. Biol. 280, 396-406. 10.1016/j.ydbio.2005.02.002 [DOI] [PubMed] [Google Scholar]
- Enzi G., Busetto L., Ceschin E., Coin A., Digito M. and Pigozzo S. (2002). Multiple symmetric lipomatosis: clinical aspects and outcome in a long-term longitudinal study. Int. J. Obes. 26, 253 10.1038/sj.ijo.0801867 [DOI] [PubMed] [Google Scholar]
- Farmer S. R. (2006). Transcriptional control of adipocyte formation. Cell Metab. 4, 263-273. 10.1016/j.cmet.2006.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feil R., Brocard J., Mascrez B., LeMeur M., Metzger D. and Chambon P. (1996). Ligand-activated site-specific recombination in mice. Proc. Natl Acad. Sci. USA 93, 10887-10890. 10.1073/pnas.93.20.10887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feil S., Valtcheva N. and Feil R. (2009). Inducible cre mice. In Gene Knockout Protocols, pp. 343-363. Springer. [DOI] [PubMed] [Google Scholar]
- Flemming W. (1871). On the formation and regression of fat cells in connective tissue with comment on the structure of the latter. Arch. R. mikr. Anat. 7, 32 10.1007/BF02956045 [DOI] [Google Scholar]
- Flemming W. (1879). Beitrage zur Kenntniss der Zelle und ihrer Lebenserscheinungen. Arch. Mikrosk. Anat. 16, 302-436. 10.1007/BF02956386 [DOI] [Google Scholar]
- Frayn K. (2002). Adipose tissue as a buffer for daily lipid flux. Diabetologia 45, 1201-1210. 10.1007/s00125-002-0873-y [DOI] [PubMed] [Google Scholar]
- Gao Z., Daquinag A. C., Su F., Snyder B. and Kolonin M. G. (2018). PDGFRα/PDGFRβ signaling balance modulates progenitor cell differentiation into white and beige adipocytes. Development 145, dev155861 http://dev.biologists.org/content/145/1/dev155861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garofalo R. S., Orena S. J., Rafidi K., Torchia A. J., Stock J. L., Hildebrandt A. L., Coskran T., Black S. C., Brees D. J. and Wicks J. R. (2003). Severe diabetes, age-dependent loss of adipose tissue, and mild growth deficiency in mice lacking Akt2/PKBβ. J. Clin. Invest. 112, 197-208. 10.1172/JCI16885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- George S., Rochford J. J., Wolfrum C., Gray S. L., Schinner S., Wilson J. C., Soos M. A., Murgatroyd P. R., Williams R. M. and Acerini C. L. (2004). A family with severe insulin resistance and diabetes due to a mutation in AKT2. Science 304, 1325-1328. 10.1126/science.1096706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gesta S., Tseng Y.-H. and Kahn C. R. (2007). Developmental origin of fat: tracking obesity to its source. Cell 131, 242-256. 10.1016/j.cell.2007.10.004 [DOI] [PubMed] [Google Scholar]
- Gordillo M., Evans T. and Gouon-Evans V. (2015). Orchestrating liver development. Development 142, 2094-2108. 10.1242/dev.114215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon J. and Manley N. R. (2011). Mechanisms of thymus organogenesis and morphogenesis. Development 138, 3865-3878. 10.1242/dev.059998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grapin-Botton A. (2009). Endoderm specification (November 30, 2008), StemBook, ed. The Stem Cell Research Community, StemBook, doi/10.3824/stembook.1.30.1, http://www.stembook.org [Google Scholar]
- Green H. and Meuth M. (1974). An established pre-adipose cell line and its differentiation in culture. Cell 3.2, 127-133. 10.1016/0092-8674(74)90116-0 [DOI] [PubMed] [Google Scholar]
- Gros J., Scaal M. and Marcelle C. (2004). A two-step mechanism for myotome formation in chick. Dev. Cell 6, 875-882. 10.1016/j.devcel.2004.05.006 [DOI] [PubMed] [Google Scholar]
- Han J., Lee J.-E., Jin J., Lim J. S., Oh N., Kim K., Chang S.-I., Shibuya M., Kim H. and Koh G. Y. (2011). The spatiotemporal development of adipose tissue. Development 138, 5027-5037. 10.1242/dev.067686 [DOI] [PubMed] [Google Scholar]
- Harms M. and Seale P. (2013). Brown and beige fat: development, function and therapeutic potential. Nat. Med. 19, 1252 10.1038/nm.3361 [DOI] [PubMed] [Google Scholar]
- Hatada Y. and Stern C. D. (1994). A fate map of the epiblast of the early chick embryo. Development 120, 2879-2889. [DOI] [PubMed] [Google Scholar]
- Herbst K. L. (2012). Rare adipose disorders (RADs) masquerading as obesity. Acta Pharmacol. Sin. 33, 155 10.1038/aps.2011.153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbst K. L., Tannock L. R., Deeb S. S., Purnell J. Q., Brunzell J. D. and Chait A. (2003). Köbberling type of familial partial lipodystrophy: an underrecognized syndrome. Diabetes Care 26, 1819-1824. 10.2337/diacare.26.6.1819 [DOI] [PubMed] [Google Scholar]
- Herman M. A., Peroni O. D., Villoria J., Schön M. R., Abumrad N. A., Blüher M., Klein S. and Kahn B. B. (2012). A novel ChREBP isoform in adipose tissue regulates systemic glucose metabolism. Nature 484, 333 10.1038/nature10986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hesselbarth N., Pettinelli C., Gericke M., Berger C., Kunath A., Stumvoll M., Blüher M. and Klöting N. (2015). Tamoxifen affects glucose and lipid metabolism parameters, causes browning of subcutaneous adipose tissue and transient body composition changes in C57BL/6NTac mice. Biochem. Biophys. Res. Commun. 464, 724-729. 10.1016/j.bbrc.2015.07.015 [DOI] [PubMed] [Google Scholar]
- Huang H., Song T.-J., Li X., Hu L., He Q., Liu M., Lane M. D. and Tang Q.-Q. (2009). BMP signaling pathway is required for commitment of C3H10T1/2 pluripotent stem cells to the adipocyte lineage. Proc. Natl Acad. Sci. USA 106, 12670-12675. 10.1073/pnas.0906266106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffery E., Berry R., Church C. D., Yu S., Shook B. A., Horsley V., Rosen E. D. and Rodeheffer M. S. (2014). Characterization of Cre recombinase models for the study of adipose tissue. Adipocyte 3, 206-211. 10.4161/adip.29674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffery E., Church C. D., Holtrup B., Colman L. and Rodeheffer M. S. (2015). Rapid depot-specific activation of adipocyte precursor cells at the onset of obesity. Nat. Cell Biol. 17, 376 10.1038/ncb3122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffery E., Wing A., Holtrup B., Sebo Z., Kaplan J. L., Saavedra-Peña R., Church C. D., Colman L., Berry R. and Rodeheffer M. S. (2016). The adipose tissue microenvironment regulates depot-specific adipogenesis in obesity. Cell Metab. 24, 142-150. 10.1016/j.cmet.2016.05.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y., Berry D. C., Tang W. and Graff J. M. (2014). Independent stem cell lineages regulate adipose organogenesis and adipose homeostasis. Cell Rep. 9, 1007-1022. 10.1016/j.celrep.2014.09.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H., Ding X., Cao Y., Wang H. and Zeng W. (2017). Dense intra-adipose sympathetic arborizations are essential for cold-induced beiging of mouse white adipose tissue. Cell Metab. 26, 686-692. e683. 10.1016/j.cmet.2017.08.016 [DOI] [PubMed] [Google Scholar]
- Jin W., Takagi T., Kanesashi S.-N., Kurahashi T., Nomura T., Harada J. and Ishii S. (2006). Schnurri-2 controls BMP-dependent adipogenesis via interaction with Smad proteins. Dev. Cell 10, 461-471. 10.1016/j.devcel.2006.02.016 [DOI] [PubMed] [Google Scholar]
- Jukkola T., Trokovic R., Maj P., Lamberg A., Mankoo B., Pachnis V., Savilahti H. and Partanen J. (2005). Meox1Cre: a mouse line expressing Cre recombinase in somitic mesoderm. Genesis 43, 148-153. 10.1002/gene.20163 [DOI] [PubMed] [Google Scholar]
- Kalhor R., Kalhor K., Mejia L., Leeper K., Graveline A., Mali P. and Church G. M. (2018). Developmental barcoding of whole mouse via homing CRISPR. Science 361, eaat9804 10.1126/science.aat9804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller C., Hansen M. S., Coffin C. M. and Capecchi M. R. (2004). Pax3: Fkhr interferes with embryonic Pax3 and Pax7 function: implications for alveolar rhabdomyosarcoma cell of origin. Genes Dev. 18, 2608-2613. 10.1101/gad.1243904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M., Oh Y., Hong S. P., Jeon S.-Y. and Lee W.-S. (2009). Does segmental lipodystrophy represent mosaicism of inherited lipodystrophy? J. Am. Acad. Dermatol. 60, 519-521. 10.1016/j.jaad.2008.06.021 [DOI] [PubMed] [Google Scholar]
- Kimmel C. B., Warga R. M. and Schilling T. F. (1990). Origin and organization of the zebrafish fate map. Development 108, 581-594. [DOI] [PubMed] [Google Scholar]
- Köbberling J. and Dunnigan M. G. (1986). Familial partial lipodystrophy: two types of an X linked dominant syndrome, lethal in the hemizygous state. J. Med. Genet. 23, 120-127. 10.1136/jmg.23.2.120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretzschmar K. and Watt F. M. (2012). Lineage tracing. Cell 148, 33-45. 10.1016/j.cell.2012.01.002 [DOI] [PubMed] [Google Scholar]
- Krueger K. C., Costa M. J., Du H. and Feldman B. J. (2014). Characterization of Cre recombinase activity for in vivo targeting of adipocyte precursor cells. Stem Cell Rep. 3, 1147-1158. 10.1016/j.stemcr.2014.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P., Tan Y. and Cahan P. (2017). Understanding development and stem cells using single cell-based analyses of gene expression. Development 144, 17-32. 10.1242/dev.133058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang D., Lu M. M., Huang L., Engleka K. A., Zhang M., Chu E. Y., Lipner S., Skoultchi A., Millar S. E. and Epstein J. A. (2005). Pax3 functions at a nodal point in melanocyte stem cell differentiation. Nature 433, 884 10.1038/nature03292 [DOI] [PubMed] [Google Scholar]
- Lawson K. A., Meneses J. J. and Pedersen R. (1991). Clonal analysis of epiblast fate during germ layer formation in the mouse embryo. Development 113, 891-911. [DOI] [PubMed] [Google Scholar]
- Lee Y.-H., Petkova A. P., Mottillo E. P. and Granneman J. G. (2012). In vivo identification of bipotential adipocyte progenitors recruited by β3-adrenoceptor activation and high-fat feeding. Cell Metab. 15, 480-491. 10.1016/j.cmet.2012.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefterova M. I. and Lazar M. A. (2009). New developments in adipogenesis. Trends Endocrinol. Metab. 20, 107-114. 10.1016/j.tem.2008.11.005 [DOI] [PubMed] [Google Scholar]
- Lepper C. and Fan C.-M. (2010). Inducible lineage tracing of Pax7-descendant cells reveals embryonic origin of adult satellite cells. Genesis 48, 424-436. 10.1002/dvg.20630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepper C., Conway S. J. and Fan C.-M. (2009). Adult satellite cells and embryonic muscle progenitors have distinct genetic requirements. Nature 460, 627 10.1038/nature08209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Zou P., Zheng L., Linarelli L. E., Amarell S., Passaro A., Liu D. and Cheng Z. (2015). Tamoxifen reduces fat mass by boosting reactive oxygen species. Cell Death Dis. 6, e1586 10.1038/cddis.2014.553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan M., Martin J. F., Nagy A., Lobe C., Olson E. N. and Tabin C. J. (2002). Expression of Cre Recombinase in the developing mouse limb bud driven by a Prxl enhancer. Genesis 33, 77-80. 10.1002/gene.10092 [DOI] [PubMed] [Google Scholar]
- Long J. Z., Svensson K. J., Tsai L., Zeng X., Roh H. C., Kong X., Rao R. R., Lou J., Lokurkar I. and Baur W. (2014). A smooth muscle-like origin for beige adipocytes. Cell Metab. 19, 810-820. 10.1016/j.cmet.2014.03.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka T., Ahlberg P. E., Kessaris N., Iannarelli P., Dennehy U., Richardson W. D., McMahon A. P. and Koentges G. (2005). Neural crest origins of the neck and shoulder. Nature 436, 347 10.1038/nature03837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A., Findlay G. M., Gagnon J. A., Horwitz M. S., Schier A. F. and Shendure J. (2016). Whole-organism lineage tracing by combinatorial and cumulative genome editing. Science 353, aaf7907 10.1126/science.aaf7907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moitra J., Mason M. M., Olive M., Krylov D., Gavrilova O., Marcus-Samuels B., Feigenbaum L., Lee E., Aoyama T. and Eckhaus M. (1998). Life without white fat: a transgenic mouse. Genes Dev. 12, 3168-3181. 10.1101/gad.12.20.3168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moullan N., Mouchiroud L., Wang X., Ryu D., Williams E. G., Mottis A., Jovaisaite V., Frochaux M. V., Quiros P. M. and Deplancke B. (2015). Tetracyclines disturb mitochondrial function across eukaryotic models: a call for caution in biomedical research. Cell Rep. 10, 1681-1691. 10.1016/j.celrep.2015.02.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muzumdar M. D., Tasic B., Miyamichi K., Li L. and Luo L. (2007). A global double-fluorescent Cre reporter mouse. Genesis 45, 593-605. 10.1002/dvg.20335 [DOI] [PubMed] [Google Scholar]
- Nedergaard J., Bengtsson T. and Cannon B. (2007). Unexpected evidence for active brown adipose tissue in adult humans. Am. J. Physiol. Endocrinol. Metab. 293, E444-E452. 10.1152/ajpendo.00691.2006 [DOI] [PubMed] [Google Scholar]
- Newman S. A. (1988). Lineage and pattern in the developing vertebrate limb. Trends Genet. 4, 329-332. 10.1016/0168-9525(88)90051-0 [DOI] [PubMed] [Google Scholar]
- Ng M., Fleming T., Robinson M., Thomson B., Graetz N., Margono C., Mullany E. C., Biryukov S., Abbafati C. and Abera S. F. (2014). Global, regional, and national prevalence of overweight and obesity in children and adults during 1980–2013: a systematic analysis for the global burden of disease study 2013. Lancet 384, 766-781. 10.1016/S0140-6736(14)60460-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen M. T., Zhu J., Nakamura E., Bao X. and Mackem S. (2009). Tamoxifen-dependent, inducible Hoxb6CreERT recombinase function in lateral plate and limb mesoderm, CNS isthmic organizer, posterior trunk neural crest, hindgut, and tailbud. Dev. Dyn. 238, 467-474. 10.1002/dvdy.21846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson M. and Fagman H. (2017). Development of the thyroid gland. Development 144, 2123-2140. 10.1242/dev.145615 [DOI] [PubMed] [Google Scholar]
- Nishimura S., Manabe I., Nagasaki M., Hosoya Y., Yamashita H., Fujita H., Ohsugi M., Tobe K., Kadowaki T. and Nagai R. (2007). Adipogenesis in obesity requires close interplay between differentiating adipocytes, stromal cells, and blood vessels. Diabetes 56, 1517-1526. 10.2337/db06-1749 [DOI] [PubMed] [Google Scholar]
- Nishimura S., Manabe I., Nagasaki M., Seo K., Yamashita H., Hosoya Y., Ohsugi M., Tobe K., Kadowaki T. and Nagai R. (2008). In vivo imaging in mice reveals local cell dynamics and inflammation in obese adipose tissue. J. Clin. Invest. 118, 710-721. 10.1172/JCI33328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogurtsova K., da Rocha Fernandes J., Huang Y., Linnenkamp U., Guariguata L., Cho N., Cavan D., Shaw J. and Makaroff L. (2017). IDF Diabetes Atlas: global estimates for the prevalence of diabetes for 2015 and 2040. Diabetes Res. Clin. Pract. 128, 40-50. 10.1016/j.diabres.2017.03.024 [DOI] [PubMed] [Google Scholar]
- Ohtola J., Myers J., Akhtar-Zaidi B., Zuzindlak D., Sandesara P., Yeh K., Mackem S. and Atit R. (2008). β-Catenin has sequential roles in the survival and specification of ventral dermis. Development 135, 2321-2329. 10.1242/dev.021170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orban P. C., Chui D. and Marth J. D. (1992). Tissue-and site-specific DNA recombination in transgenic mice. Proc. Natl Acad. Sci. USA 89, 6861-6865. 10.1073/pnas.89.15.6861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang D. and Thompson D. N. P. (2011). Embryology and bony malformations of the craniovertebral junction. Child's Nerv. Syst. 27, 523-564. 10.1007/s00381-010-1358-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto S., Roseberry A. G., Liu H., Diano S., Shanabrough M., Cai X., Friedman J. M. and Horvath T. L. (2004). Rapid rewiring of arcuate nucleus feeding circuits by leptin. Science 304, 110-115. 10.1126/science.1089459 [DOI] [PubMed] [Google Scholar]
- Poissonnet C. M., Burdi A. R. and Garn S. M. (1984). The chronology of adipose tissue appearance and distribution in the human fetus. Early Hum. Dev. 10, 1-11. 10.1016/0378-3782(84)90106-3 [DOI] [PubMed] [Google Scholar]
- Poissonnet C. M., LaVelle M. and Burdi A. R. (1988). Growth and development of adipose tissue. J. Pediatr. 113, 1-9. 10.1016/S0022-3476(88)80520-1 [DOI] [PubMed] [Google Scholar]
- Pu Q., Abduelmula A., Masyuk M., Theiss C., Schwandulla D., Hans M., Patel K., Brand-Saberi B. and Huang R. (2013). The dermomyotome ventrolateral lip is essential for the hypaxial myotome formation. BMC Dev. Biol. 13, 37 10.1186/1471-213X-13-37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajakumari S., Wu J., Ishibashi J., Lim H.-W., Giang A.-H., Won K.-J., Reed R. R. and Seale P. (2013). EBF2 determines and maintains brown adipocyte identity. Cell Metab. 17, 562-574. 10.1016/j.cmet.2013.01.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Relaix F., Rocancourt D., Mansouri A. and Buckingham M. (2005). A Pax3/Pax7-dependent population of skeletal muscle progenitor cells. Nature 435, 948 10.1038/nature03594 [DOI] [PubMed] [Google Scholar]
- Roh H. C., Tsai L. T., Shao M., Tenen D., Shen Y., Kumari M., Lyubetskaya A., Jacobs C., Dawes B. and Gupta R. K. (2018). Warming induces significant reprogramming of beige, but not brown, adipocyte cellular identity. Cell Metab. 27, 1121-1137.e5. 10.1016/j.cmet.2018.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen E. D. and MacDougald O. A. (2006). Adipocyte differentiation from the inside out. Nat. Rev. Mol. Cell Biol. 7, 885 10.1038/nrm2066 [DOI] [PubMed] [Google Scholar]
- Rosen E. D. and Spiegelman B. M. (2014). What we talk about when we talk about fat. Cell 156, 20-44. 10.1016/j.cell.2013.12.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen E. D., Sarraf P., Troy A. E., Bradwin G., Moore K., Milstone D. S., Spiegelman B. M. and Mortensen R. M. (1999). PPARγ is required for the differentiation of adipose tissue in vivo and in vitro. Mol. Cell 4, 611-617. 10.1016/S1097-2765(00)80211-7 [DOI] [PubMed] [Google Scholar]
- Rosenwald M., Perdikari A., Rülicke T. and Wolfrum C. (2013). Bi-directional interconversion of brite and white adipocytes. Nat. Cell Biol. 15, 659 10.1038/ncb2740 [DOI] [PubMed] [Google Scholar]
- Ross S. E., Hemati N., Longo K. A., Bennett C. N., Lucas P. C., Erickson R. L. and MacDougald O. A. (2000). Inhibition of adipogenesis by Wnt signaling. Science 289, 950-953. 10.1126/science.289.5481.950 [DOI] [PubMed] [Google Scholar]
- Sanchez-Gurmaches J. and Guertin D. A. (2014a). Adipocyte lineages: tracing back the origins of fat. Biochim. Biophy. Acta Mol. Basis Dis. 1842, 340-351. 10.1016/j.bbadis.2013.05.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Gurmaches J. and Guertin D. A. (2014b). Adipocytes arise from multiple lineages that are heterogeneously and dynamically distributed. Nat. Commun. 5, 4099 10.1038/ncomms5099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Gurmaches J., Hung C.-M., Sparks C. A., Tang Y., Li H. and Guertin D. A. (2012). PTEN loss in the Myf5 lineage redistributes body fat and reveals subsets of white adipocytes that arise from Myf5 precursors. Cell Metab. 16, 348-362. 10.1016/j.cmet.2012.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Gurmaches J., Hsiao W.-Y. and Guertin D. A. (2015). Highly selective in vivo labeling of subcutaneous white adipocyte precursors with Prx1-Cre. Stem Cell Rep. 4, 541-550. 10.1016/j.stemcr.2015.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Gurmaches J., Hung C.-M. and Guertin D. A. (2016). Emerging complexities in adipocyte origins and identity. Trends Cell Biol. 26, 313-326. 10.1016/j.tcb.2016.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherer P. E. (2006). Adipose tissue: from lipid storage compartment to endocrine organ. Diabetes 55, 1537-1545. 10.2337/db06-0263 [DOI] [PubMed] [Google Scholar]
- Schwalie P. C., Dong H., Zachara M., Russeil J., Alpern D., Akchiche N., Caprara C., Sun W., Schlaudraff K.-U. and Soldati G. (2018). A stromal cell population that inhibits adipogenesis in mammalian fat depots. Nature 559, 103-108. [DOI] [PubMed] [Google Scholar]
- Seale P., Bjork B., Yang W., Kajimura S., Chin S., Kuang S., Scime A., Devarakonda S., Conroe H. M. and Erdjument-Bromage H. (2008). PRDM16 controls a brown fat/skeletal muscle switch. Nature 454, 961 10.1038/nature07182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebo Z. L., Jeffery E., Holtrup B. and Rodeheffer M. S. (2018). A mesodermal fate map for adipose tissue. Development 145, dev166801 10.1242/dev.166801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sgaier S. K., Millet S., Villanueva M. P., Berenshteyn F., Song C. and Joyner A. L. (2005). Morphogenetic and cellular movements that shape the mouse cerebellum: insights from genetic fate mapping. Neuron 45, 27-40. [DOI] [PubMed] [Google Scholar]
- Sharp L. Z., Shinoda K., Ohno H., Scheel D. W., Tomoda E., Ruiz L., Hu H., Wang L., Pavlova Z. and Gilsanz V. (2012). Human BAT possesses molecular signatures that resemble beige/brite cells. PLoS ONE 7, e49452 10.1371/journal.pone.0049452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen W., Wang Z., Punyanita M., Lei J., Sinav A., Kral J. G., Imielinska C., Ross R. and Heymsfield S. B. (2003). Adipose tissue quantification by imaging methods: a proposed classification. Obes. Res. 11, 5-16. 10.1038/oby.2003.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siersbæk R., Nielsen R. and Mandrup S. (2012). Transcriptional networks and chromatin remodeling controlling adipogenesis. Trends Endocrinol. Metab. 23, 56-64. 10.1016/j.tem.2011.10.001 [DOI] [PubMed] [Google Scholar]
- Simões-Costa M. and Bronner M. E. (2015). Establishing neural crest identity: a gene regulatory recipe. Development 142, 242-257. 10.1242/dev.105445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skurk T., Alberti-Huber C., Herder C. and Hauner H. (2007). Relationship between adipocyte size and adipokine expression and secretion. J. Clin. Endocrinol. Metab. 92, 1023-1033. 10.1210/jc.2006-1055 [DOI] [PubMed] [Google Scholar]
- Spalding K. L., Arner E., Westermark P. O., Bernard S., Buchholz B. A., Bergmann O., Blomqvist L., Hoffstedt J., Näslund E. and Britton T. (2008). Dynamics of fat cell turnover in humans. Nature 453, 783 10.1038/nature06902 [DOI] [PubMed] [Google Scholar]
- Steppan C. M., Bailey S. T., Bhat S., Brown E. J., Banerjee R. R., Wright C. M., Patel H. R., Ahima R. S. and Lazar M. A. (2001). The hormone resistin links obesity to diabetes. Nature 409, 307 10.1038/35053000 [DOI] [PubMed] [Google Scholar]
- Stern C. D. and Fraser S. E. (2001). Tracing the lineage of tracing cell lineages. Nat. Cell Biol. 3, E216 10.1038/ncb0901-e216 [DOI] [PubMed] [Google Scholar]
- Tajbakhsh S. and Spörle R. (1998). Somite development: constructing the vertebrate body. Cell 92, 9-16. 10.1016/S0092-8674(00)80894-6 [DOI] [PubMed] [Google Scholar]
- Tallquist M. D., Weismann K. E., Hellstrom M. and Soriano P. (2000). Early myotome specification regulates PDGFA expression and axial skeleton development. Development 127, 5059-5070. [DOI] [PubMed] [Google Scholar]
- Tam P. P. L. and Loebel D. A. F. (2007). Gene function in mouse embryogenesis: get set for gastrulation. Nat. Rev. Genet. 8, 368 10.1038/nrg2084 [DOI] [PubMed] [Google Scholar]
- Tang W., Zeve D., Suh J. M., Bosnakovski D., Kyba M., Hammer R. E., Tallquist M. D. and Graff J. M. (2008). White fat progenitor cells reside in the adipose vasculature. Science 322, 583-586. 10.1126/science.1156232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teboul L., Hadchouel J., Daubas P., Summerbell D., Buckingham M. and Rigby P. W. (2002). The early epaxial enhancer is essential for the initial expression of the skeletal muscle determination gene Myf5 but not for subsequent, multiple phases of somitic myogenesis. Development 129, 4571-4580. [DOI] [PubMed] [Google Scholar]
- Timmons J. A., Wennmalm K., Larsson O., Walden T. B., Lassmann T., Petrovic N., Hamilton D. L., Gimeno R. E., Wahlestedt C. and Baar K. (2007). Myogenic gene expression signature establishes that brown and white adipocytes originate from distinct cell lineages. Proc. Natl Acad. Sci. USA 104, 4401-4406. 10.1073/pnas.0610615104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tontonoz P., Hu E. and Spiegelman B. M. (1994). Stimulation of adipogenesis in fibroblasts by PPARγ2, a lipid-activated transcription factor. Cell 79, 1147-1156. 10.1016/0092-8674(94)90006-X [DOI] [PubMed] [Google Scholar]
- Torres M., Gómez-Pardo E., Dressler G. R. and Gruss P. (1995). Pax-2 controls multiple steps of urogenital development. Development 121, 4057-4065. [DOI] [PubMed] [Google Scholar]
- Tseng Y.-H., Kokkotou E., Schulz T. J., Huang T. L., Winnay J. N., Taniguchi C. M., Tran T. T., Suzuki R., Espinoza D. O. and Yamamoto Y. (2008). New role of bone morphogenetic protein 7 in brown adipogenesis and energy expenditure. Nature 454, 1000 10.1038/nature07221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vishvanath L., MacPherson K. A., Hepler C., Wang Q. A., Shao M., Spurgin S. B., Wang M. Y., Kusminski C. M., Morley T. S. and Gupta R. K. (2016). Pdgfrβ+ mural preadipocytes contribute to adipocyte hyperplasia induced by high-fat-diet feeding and prolonged cold exposure in adult mice. Cell Metab. 23, 350-359. 10.1016/j.cmet.2015.10.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitali A., Murano I., Zingaretti M. C., Frontini A., Ricquier D. and Cinti S. (2012). The adipose organ of obesity-prone C57BL/6J mice is composed of mixed white and brown adipocytes. J. Lipid Res. 53, 619-629. 10.1194/jlr.M018846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt W. (1924). Sitzungsber. Ges. Morph. Physiol. Munchen 35, 22-32. [Google Scholar]
- Wagner D. O., Sieber C., Bhushan R., Börgermann J. H., Graf D. and Knaus P. (2010). BMPs: from bone to body morphogenetic proteins. Sci. Signal. 3, mr1 10.1126/scisignal.3107mr1 [DOI] [PubMed] [Google Scholar]
- Wang Q. A., Tao C., Gupta R. K. and Scherer P. E. (2013). Tracking adipogenesis during white adipose tissue development, expansion and regeneration. Nat. Med. 19, 1338 10.1038/nm.3324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Kissig M., Rajakumari S., Huang L., Lim H.-W., Won K.-J. and Seale P. (2014). Ebf2 is a selective marker of brown and beige adipogenic precursor cells. Proc. Natl Acad. Sci. USA 111, 14466-14471. 10.1073/pnas.1412685111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q. A., Tao C., Jiang L., Shao M., Ye R., Zhu Y., Gordillo R., Ali A., Lian Y., Holland W. L. et al. (2015). Distinct regulatory mechanisms governing embryonic versus adult adipocyte maturation. Nat. Cell Biol. 17, 1099 10.1038/ncb3217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Paulo E., Wu D., Wu Y., Huang W., Chawla A. and Wang B. (2017). Adipocyte liver kinase b1 suppresses beige adipocyte renaissance through class IIa histone deacetylase 4. Diabetes 66, 2952-2963. 10.2337/db17-0296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells H. G. (1940). Adipose tissue, a neglected subject. JAMA 114, 2177-2183. 10.1001/jama.1940.02810220001001 [DOI] [Google Scholar]
- Wertheimer E. and Shapiro B. (1948). The physiology of adipose tissue. Physiol. Rev. 28, 451-464. 10.1152/physrev.1948.28.4.451 [DOI] [PubMed] [Google Scholar]
- Wilson E. (1898). Biological lectures from the marine biological laboratory, woods hole, massachusetts. Boston: Ginn & Co 21-42. [Google Scholar]
- Winters N. I., Thomason R. T. and Bader D. M. (2012). Identification of a novel developmental mechanism in the generation of mesothelia. Development 139, 2926-2934. 10.1242/dev.082396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Boström P., Sparks L. M., Ye L., Choi J. H., Giang A.-H., Khandekar M., Virtanen K. A., Nuutila P. and Schaart G. (2012). Beige adipocytes are a distinct type of thermogenic fat cell in mouse and human. Cell 150, 366-376. 10.1016/j.cell.2012.05.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto M., Shook N. A., Kanisicak O., Yamamoto S., Wosczyna M. N., Camp J. R. and Goldhamer D. J. (2009). A multifunctional reporter mouse line for Cre-and FLP-dependent lineage analysis. Genesis 47, 107-114. 10.1002/dvg.20474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamauchi T., Kamon J., Waki H., Terauchi Y., Kubota N., Hara K., Mori Y., Ide T., Murakami K. and Tsuboyama-Kasaoka N. (2001). The fat-derived hormone adiponectin reverses insulin resistance associated with both lipoatrophy and obesity. Nat. Med. 7, 941 10.1038/90984 [DOI] [PubMed] [Google Scholar]
- Yamauchi T., Kamon J., Minokoshi Y. A., Ito Y., Waki H., Uchida S., Yamashita S., Noda M., Kita S. and Ueki K. (2002). Adiponectin stimulates glucose utilization and fatty-acid oxidation by activating AMP-activated protein kinase. Nat. Med. 8, 1288 10.1038/nm788 [DOI] [PubMed] [Google Scholar]
- Ye R., Wang Q. A., Tao C., Vishvanath L., Shao M., McDonald J. G., Gupta R. K. and Scherer P. E. (2015). Impact of tamoxifen on adipocyte lineage tracing: inducer of adipogenesis and prolonged nuclear translocation of Cre recombinase. Mol. Metab. 4, 771-778. 10.1016/j.molmet.2015.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh W.-C., Cao Z., Classon M. and McKnight S. L. (1995). Cascade regulation of terminal adipocyte differentiation by three members of the C/EBP family of leucine zipper proteins. Genes Dev. 9, 168-181. 10.1101/gad.9.2.168 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Proenca R., Maffei M., Barone M., Leopold L. and Friedman J. M. (1994). Positional cloning of the mouse obese gene and its human homologue. Nature 372, 425 10.1038/372425a0 [DOI] [PubMed] [Google Scholar]
- Zhou B., Ma Q., Rajagopal S., Wu S. M., Domian I., Rivera-Feliciano J., Jiang D., von Gise A., Ikeda S., Chien K. R. et al. (2008). Epicardial progenitors contribute to the cardiomyocyte lineage in the developing heart. Nature 454, 109 10.1038/nature07060 [DOI] [PMC free article] [PubMed] [Google Scholar]