ABSTRACT
miR-203 is a tumor-suppressor microRNA with known functions in cancer metastasis. Here, we explore its normal developmental role in the context of neural crest development. During the epithelial-to-mesenchymal transition of neural crest cells to emigrate from the neural tube, miR-203 displays a reciprocal expression pattern with key regulators of neural crest delamination, Phf12 and Snail2, and interacts with their 3′UTRs. We show that ectopic maintenance of miR-203 inhibits neural crest migration in chick, whereas its functional inhibition using a ‘sponge’ vector or morpholinos promotes premature neural crest delamination. Bisulfite sequencing further shows that epigenetic repression of miR-203 is mediated by the de novo DNA methyltransferase DNMT3B, the recruitment of which to regulatory regions on the miR-203 locus is directed by SNAIL2 in a negative-feedback loop. These findings reveal an important role for miR-203 in an epigenetic-microRNA regulatory network that influences the timing of neural crest delamination.
KEY WORDS: EMT, Neural crest, miR-203, Snail2, DNA methylation, Phf12, Migration
Summary: Maintenance and inhibition of the tumor-suppressor microRNA miR203 reveals an epigenetic miRNA-gene regulatory circuit, conserved in cancer, that controls the timing of neural crest epithelial-to-mesenchymal transition.
INTRODUCTION
The neural crest is a transient embryonic cell population that arises in the neuroectoderm then migrates to the periphery where these cells contribute to diverse derivatives, including craniofacial bone and cartilage, neurons and glia of the peripheral nervous system, pigment cells, and parts of the cardiovascular system (Crane and Trainor, 2006). Neural crest cells (NCCs) emigrate from the forming central nervous system by undergoing an epithelial-mesenchymal transition (EMT), similar to that observed during initiation of tumor metastasis (Kerosuo and Bronner-Fraser, 2012). An evolutionarily conserved gene regulatory network (GRN) regulates NCC EMT and depends upon the coordinated action of transcription factors, including Snai1, Snai2, Zeb2 (also known as Sip1) and Foxd3 (Simoes-Costa and Bronner, 2015). In addition to transcriptional regulators that mediate neural crest EMT, epigenetic mechanisms, including DNA methylation (Hu et al., 2014; Hu et al., 2012) and histone modifications (Strobl-Mazzulla and Bronner, 2012; Strobl-Mazzulla et al., 2010), are also at play. In particular, a repressor complex, comprising SNAIL2 and the epigenetic reader PHF12 represses cadherin 6b (Cad6b) (Strobl-Mazzulla and Bronner, 2012), the downregulation of which is required for initiation of neural crest delamination from the neural tube. These results demonstrate an additional level of fine tuning and an important role for epigenetic regulators during neural crest EMT, raising the intriguing possibility that other yet to be identified factors may be involved.
In recent years, microRNAs (miRNAs) have been shown to be key regulators of EMT in several tumor cells (Diaz-Lopez et al., 2014; Ding, 2014; Xia and Hui, 2012). MicroRNAs are ∼22-nucleotide single-stranded RNAs that negatively regulate gene expression post-transcriptionally (Kloosterman and Plasterk, 2006) by inhibiting translation and/or causing degradation by binding to complementary sequences located at the 3′-untranslated region (3′-UTR) of target mRNAs. Approximately 50% of miRNA genes are embedded in or associated with CpGs islands, and their expression is most often regulated by methylation of cytosines therein (Weber et al., 2007). In several tumor cells, it has been shown that hypermethylation of anti-tumoral miRNAs leads to initiation of EMT (Ahmad et al., 2014; Bonnomet et al., 2010; Diaz-Lopez et al., 2014; Ding, 2014; Kiesslich, Pichler, and Neureiter, 2013; Lujambio et al., 2008; Nelson and Weiss, 2008; Wang et al., 2012; Xia and Hui, 2012). The commonalities between the migration of cancer cells and embryonic neural crest cells (Friedl and Gilmour, 2009; Mayor and Theveneau, 2013; Scarpa and Mayor, 2016) suggest the intriguing possibility that similar epigenetic-microRNA interactions to those functioning in metastasis may be involved in NCC development. As a case in point, we describe in avian embryos that the epigenetic repression of miR-203, which is directed by SNAIL2, enables upregulation of two of its direct targets, Phf12 and Snai2, that are necessary for the delamination of the neural crest from the neural tube. These findings support the idea that a single microRNA, regulating key genes of the EMT process, may ‘throw’ a precise switch that mediates an epithelial-to-mesenchymal transition in neural crest cells.
RESULTS
miR-203 expression is reduced at the time of NCC delamination
Given that PHF12 and SNAIL2 are involved in regulation of NCC EMT (Strobl-Mazzulla and Bronner, 2012), we performed an in silico and literature analysis to investigate miRNAs that might regulate these transcription factors. Interestingly, we found nine and seven families, respectively, of miRNAs in the 3′UTRs of Phf12 and Snal2 with sites conserved across vertebrates (Table S1). We focused on miR-203 for in-depth analysis based on the conservation of deep binding sites across vertebrates (Fig. S1). Importantly, miR-203 has been described to act as tumor suppressor (Benaich et al., 2014; Miao et al., 2014; Zhu et al., 2013) that directly regulates Snai2 expression (Gao et al., 2017; Shi et al., 2015; Xiao et al., 2017; Zhang et al., 2015), the epigenetic repression of which causes metastasis in several tumor cells, including NCC-derived melanoma (Boldrup et al., 2012; Boll et al., 2013; Bu and Yang, 2014; Chen et al., 2012; Chiang et al., 2011; Ding et al., 2013; Furuta et al., 2010; Huang et al., 2014; Ju et al., 2014; Lohcharoenkal et al., 2018; Moes et al., 2012; Zhang et al., 2014; Zhao et al., 2013). Moreover, the mature miR-203 sequence is highly conserved throughout vertebrates, including the basal lamprey (Fig. 1B), suggesting an ancient and conserved function. Thus, like the neural crest, miR-203 is a vertebrate novelty (Heimberg et al., 2010).
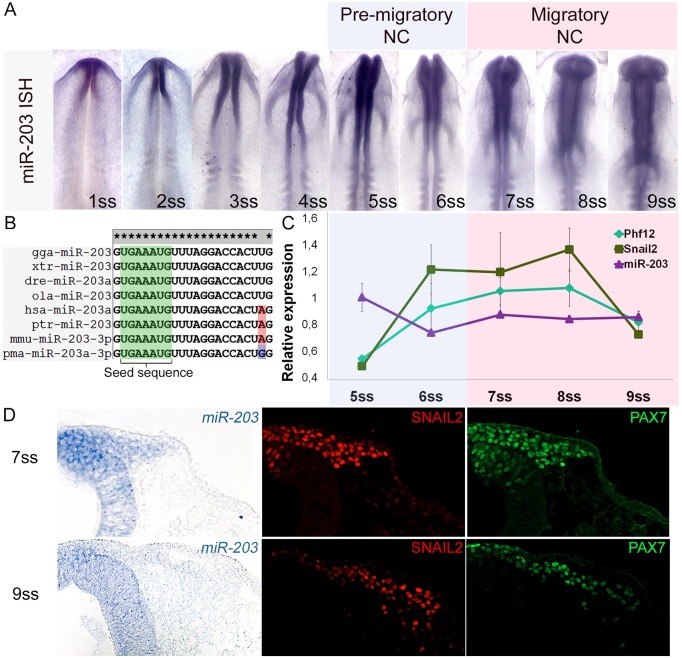
Fig. 1.
miR-203 expression is reduced at the time of NCC delamination. (A) Whole-mount in situ hybridization using LNA-DIG-labeled probes reveals specific expression of mature miR-203 in the neural tube that is reduced from the 5-somite stage (ss) to 9 ss. (B) Conservation analysis of mature miR-203 in vertebrates. Gga (Gallus gallus), xtr (Xenopus tropicalis), dre (Danio rerio), ola (Oryzias latipes), hsa (Homo sapiens), ptr (Pan troflodytes), mmu (Mus musculus) and pma (Petromyzon marinus). (C) RT-qPCR analyses show falling miR-203 expression from 5 to 6 ss in a reciprocal manner to the observed increases in Snai2 and Phf12 expression at the beginning of NCC delamination. (D) Transverse sections of miR-203 in situ hybridization followed by SNAIL2 (red) and PAX7 (green) immunostaining across the dorsal neural tube and delaminating/migrating NCCs.
As a first step in analyzing its possible function during NCC development, we examined the expression pattern of miR-203 transcripts using in situ hybridization in early chick embryos. By using locked nucleic acid-digoxigenin-labeled probes, we found that mature miR-203 expression begins during gastrulation by stage 4 (Fig. S2A). During neurulation at the 1-5 somite stage (ss), miR-203 is consistently expressed in the forming neural tube (Fig. 1A). Interestingly, we observed a clear reduction by the 6-8 ss in miR-203 expression in the cranial neural tube, corresponding to the initiation of neural crest emigration. Analyses by stem-loop RT-qPCR confirmed that mature miR-203 expression is reduced from 5-6 ss, coincident with the increase of Snai2 and Phf12 expression at the time of NCC delamination (Fig. 1C). We next performed in situ hybridization against miR-203, followed by immunostaining for PAX7 and SNAIL2 to determine the spatial distribution of these factors as a function of time at three different developmental states (pre-migratory, delaminating and migratory) (Fig. 1D, Fig. S2B). Transverse sections confirm the specific expression of miR-203 in pre-migratory NCC and neural tube at the 4-6 ss (Fig. S2B), whereas transverse sections at 7 ss and 9 ss showed a high level of miR-203 expression in the dorsal neural tube that then dropped in delaminated NCC (Fig. 1D). Taken together, these data suggest that there are dynamic changes in miR-203 at the time of delamination and these are associated with high levels of SNAIL2, consistent with the intriguing possibility that miR-203 has an important role in neural crest EMT.
Overexpression of miR-203 causes loss of migrating NCCs
As miR-203 expression is downregulated as neural crest migration initiates, we investigated whether maintenance of miR-203 expression would prevent neural crest delamination. To achieve this, we designed an overexpression vector in which we cloned the pre-miR-203 sequence, plus a few hundred base pair arms for correct processing, under the control of a CAG promoter (pCAG-203) (Fig. S3A). Overexpression of miR-203 was achieved by electroporation of 3 µg/µl of pCAG-203 and was confirmed by LNA in situ hybridization against miR-203 (Fig. S3B). To demonstrate that the pCAG-203 expresses a functional miR-203, we first used a two-colored sensor vector (Cao et al., 2007) comprising a nuclear-localized destabilized EGFP with a half-life of 4 h (d4EGFPN), driven by a CAG promoter, followed by a 3′UTR containing two copies of a bulged complementary sequence for miR-203. In addition, the vector contains a nuclear-localized monomeric red fluorescent protein (mRFPN) driven by another CAG promoter that serves as an electroporation control (see scheme of pSdmiR-203 in Fig. S3C). Together with the dual sensor pSdmiR-203, we co-electroporated either control empty vector or the vector expressing miR-203 onto each side of the embryo (Fig. S3D). The results show that, on the control side, most of the electroporated cells appeared yellow, owing to expression of d4EGFPN and mRFPN. However, on the experimental side, overexpression of miR-203 causes a strong reduction of d4EGFPN and the cells only express mRFPN (Fig. S3E). These results confirm the functionality of our miR-203 expression construct.
Gain-of-function experiments were conducted by electroporating the miR-203 vector on one side of the embryo (Fig. S3A). Overexpression of miR-203 in pre-migratory NCC causes a clear downregulation of SNAIL2 but had no significant effect on the neural crest specifier gene Foxd3 (Fig. 2A). miR-203 overexpression at later stages (9 ss) causes a drastic reduction in the numbers of migratory neural crest cells identified by in situ hybridization against Sox10 (Fig. S4A) and Tfap2b, and PAX7 immunostaining (Fig. 2C-D). Given the well-known effect of SNAIL2 and PHF12 in repressing the Cad6b gene in pre-migratory neural crest (Strobl-Mazzulla and Bronner, 2012), we performed epistatic rescue experiments using a well-characterized morpholino against Cad6b (Cad6b MO) (Coles et al., 2007) to test the specificity of the effect of miR-203 overexpression on inhibition of NCC migration. First, we showed that co-electroporation of pCAG-203 with control-MO causes SNAIL2 reduction and Cad6B maintenance on the dorsal neural tube (Fig. 2B,B′), in agreement with the lack of migration observed at later stages. Importantly, co-electroporation of pCAG-203 with Cad6b-MO also caused a reduction in SNAIL2 levels, but rescued the Cad6b repression (Fig. 2B,B″) necessary for the neural crest delamination. At later stages, we demonstrated that co-electroporation of pCAG-203 with Cad6b-MO partially rescued the neural crest migratory capacities as shown by in situ hybridization for Tfap2b and immunostaining for PAX7 (Fig. 2C,D). Our results demonstrate that overexpression of miR-203 does not affect expression of FOXD3, one of the earliest neural crest specification markers, suggesting that specification occurs normally. In contrast, there was a clear reduction in SNAIL2 levels and in the maintenance of CAD6B in the dorsal neural tube. Taken together, our results highlight the role of miR-203 in NCC delamination, likely by affecting EMT inducers such as SNAIL2.
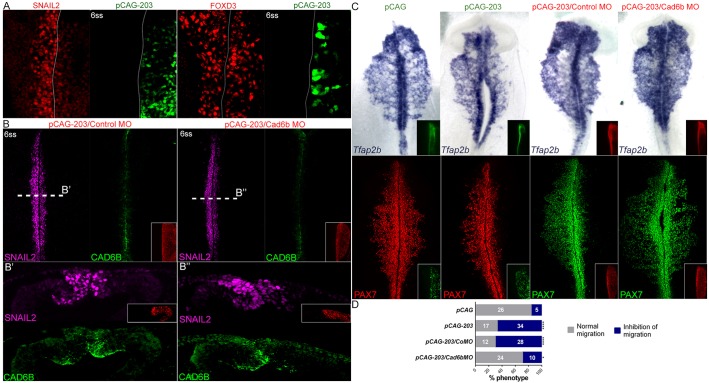
Fig. 2.
miR-203 overexpression prevents NCC delamination without affecting specification. (A) Immunohistochemistry analyses on miR-203-overexpressing embryos shows reduced SNAIL2 immunostaining, but does not affect expression of the early NCC specifier FOXD3. (B) At 6 ss, co-electroporation of pCAG-203 with control MO causes downregulation of SNAIL2 and maintenance of Cad6b in the dorsal neural tube compared with the uninjected site (left). This phenotype is rescued by co-electroporation of pCAG-203 with Cad6b MO where the injected site shows similar expression of Cad6b compared with the control side (right). (B′,B″) Transverse sections at the levels shown in B. (C) In situ hybridization for Tfap2b and immunostaining for PAX7 shows inhibition of NCC migration in the pCAG-203 injected side, compared with the uninjected side of the same embryos and with embryos injected with an empty pCAG vector. This phenotype is partially rescued by the co-electroporation of cadherin 6b morpholino (Cad6b MO). (D) The percentages of embryos showing a phenotype (normal migration versus inhibition of migration) on the side injected with pCAG, pCAG-203, pCAG-203/CoMO and pCAG-203/Cad6bMO. Numbers in the graphs represent the numbers of analyzed embryos. *P<0.05 and ****P<00001 compared with the pCAG vector (contingency table followed by a χ2 test).
Loss of miR-203 function causes premature NCC delamination
Given that miR-203 is downregulated in migratory NCCs and its overexpression causes defects in their delamination, we next asked whether early loss of miR-203 function would result in premature neural crest delamination. To test this possibility, we adapted a protocol (Kluiver et al., 2012) to generate a ‘sponge’ vector containing repeated miR-203 antisense sequences (pSmiR-203) to sequester endogenous miR-203 (Fig. 3A). A control sponge vector was designed by addition of miR-203 scrambled sequence (pSmiR-scramble). As a second knockdown strategy, we used a morpholino that is complementary to miR-203 (miR-203 MO), thus affecting the processing and the activity of miR-203. The electroporation of miR203 MO resulted in a significant reduction of the mature miR-203 (Fig. S4B). As predicted, electroporation of pSmiR-203 or miR-203 MO resulted in premature NCC migration, when compared with the contralateral uninjected side, analyzed by immunostaining for SNAIL2 and FOXD3, or by in situ hybridization for Sox10 (Fig. 3B). Importantly, no difference in the timing of delamination was observed after electroporation of the pSmiR-scramble or control morpholino (control MO). By categorizing embryos according to their phenotype, premature versus normal migration, we observed a significant increase in the percentage of embryos exhibiting premature NCC migration in pSmiR-203 and miR-203 MO electroporated embryos (Fig. 3C). Notably, reduction of miR-203 function altered not only initiation of NCC delamination, but also shortened the overall length of time during which delamination occurred (Fig. S4C). This is based on the observation that, on the side injected with pSmiR-203, all the Sox10+ NCCs have completed their delamination, compared with the contralateral side where many pre-migratory neural crest cells still persist in the dorsal neural tube. These results clearly confirm that miR-203 controls the temporal regulation of NCC delamination.
Fig. 3.
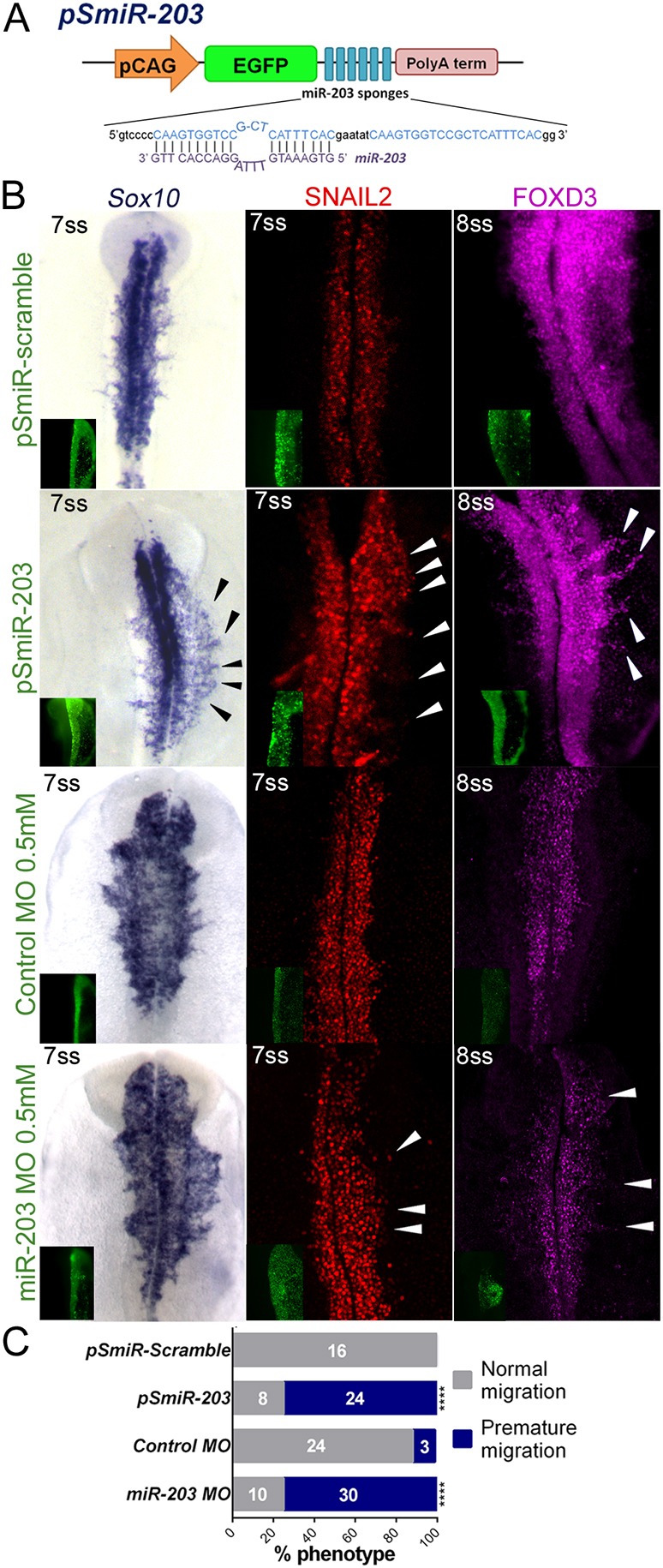
miR-203 sponge vector causes premature NCC delamination. (A) Schematic diagram of the pSmiR-203 sponge vector with bulged miR-203 antisense sequences downstream of the EGFP reporter. (B) Embryos electroporated with pSmiR-203 and miR-203 MO have premature migration of NCCs, as shown by in situ hybridization for Sox10 and immunostaining for SNAIL2 and FOXD3, compared with the uninjected side of the same embryos or injected (right) with pSmiR-scramble vector or control MO. Arrowheads indicate premature migratory NCCs. (C) Quantitation of premature NCC migration in pSmiR-203-, pSmiR-scramble-, control MO- and miR-203 MO-treated embryos. Numbers in the graph represent the analyzed embryos. ****P<00001 (contingency table followed by χ2 test).
miR-203 targets the 3′UTRs of Snail2 and Phf12
To test whether Snail2 and Phf12 genes are direct targets of miR-203, we designed two-colored sensor vectors in which we cloned, downstream of the pCAG and d4EGFPN, the 3′UTRs containing the wild (pUTR-Snail2/Phf12) or mutated (pUTR-mutSnail2/Phf12) miR-203-binding sites (Fig. 4A). Co-electroporation of these vectors (Fig. 4B) demonstrated that overexpression of miR-203 specifically inhibited d4EGFPN expression when it was fused to the 3′UTRs of Snail2 and Phf12 but was uniformly distributed when miR-203-binding sites were mutated (Fig. 4C-D). These results, combined with our gain and loss of miR-203 function, confirm that Snail2 and Phf12 are endogenous targets of the same microRNA: miR-203.
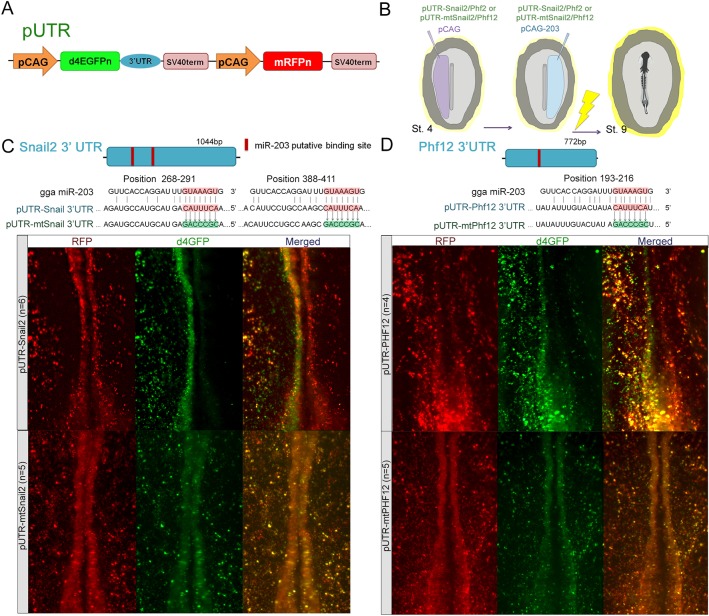
Fig. 4.
Snail2 and Phf12 3′UTRs are direct targets of miR-203. (A) Schematic diagram of a dual-colored sensor vector containing wild or mutated (mt) 3′UTRs from Snail2 (pUTR-Snail2) and Phf12 (pUTR-Phf12). pCAG, chick β-actin promoter; d4EGFPN, nuclear-localized destabilized EGFP with a half-life of 4 h; mRFPN, nuclear-localized monomeric red fluorescent protein. (B) Diagram of electroporation assays for 3′UTR sensor experiments. Electroporation of sensor vector containing the 3′UTRs for (C) Snail2 (pUTR-Snail2) or (D) Phf12 (pUTR-Phf12) together with miR-203-overexpressing vector (pCAG-203) causes a consistent reduction on the d4EGFPN expression (right side) compared with the control side (left). (C,D) Mutation of miR-203-binding sites in the 3′UTRs of Snail2 (pUTR-mtSnail2) or Phf12 (pUTR-mtPhf12); most of the electroporated cells are yellow, expressing both d4EGFPN and mRFPN, even when miR-203 is overexpressed (right side).
The miR-203 locus is highly methylated on pre-migratory NCC
Finally, we asked how miR-203 is turned down in the delaminating neural crest. In several metastatic tumor cells, the miR-203 locus is epigenetically silenced by DNA methylation (Chim et al., 2011a; Diao et al., 2014; Furuta et al., 2010; Huang et al., 2014; Taube et al., 2013; Zhang et al., 2013, 2011), raising the intriguing possibility that the same may be true in the pre-migratory neural crest. Consistent with this possibility, we found, using in silico analysis, that the miR-203 locus is embedded in a CpG island (Fig. 5A). On this basis, we selected two genomic regions, one at the putative proximal promoter (region 1) and the other at the beginning of the pri-miR-203 (region 2), to analyze the DNA methylation abundance on pre-migratory (PM-NCC) or migratory (M-NCC) neural crest cells and the ventral neural tube (NT). By using bisulfite conversion, we observed high enrichment of methylated CpGs in region 2, but not in region 1, in the PM-NCCs compared with the low abundance detected on the M-NCCs and NT (Fig. 5B,C). These significant differences are clearly evident when comparing the total percentage of methylated CpGs (Fig. 5D,E) from the different samples. We found that 18.4% and 28.9% of the CpGs are methylated in region 1 and 2 in PM-NCCs, compared with 9.5% and 6.1% on the NT, and 5.3% and 7% detected on the M-NCCs, respectively. These results suggest that the decreased expression of miR-203 in pre-migratory NC may be the consequence of hypermethylation of its locus.
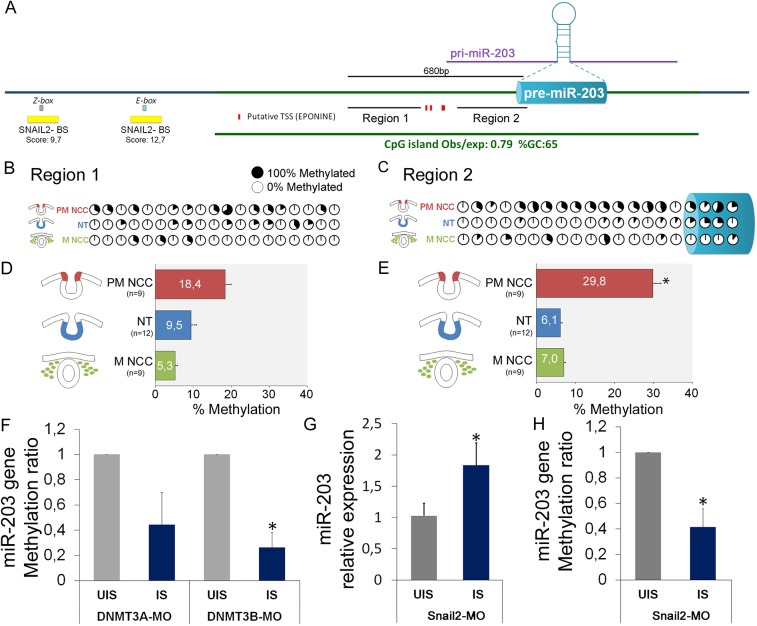
Fig. 5.
SNAIL2 and DNMT3B are required for DNA methylation of the miR-203 locus on premigratory NCC. (A) miR-203 genomic context. The CpG island content (green bar) for the miR-203 gene was identified using the UCSC genome browser (genome.ucsc.edu/) and Ensembl (www.ensembl.org/). The putative transcriptional start sites (TSS) (Tobiasson et al., 2017) were obtained using Eponine (www.sanger.ac.uk/science/tools/eponine). The putative binding sites for SNAIL2 were mapped using JASPAR 2018 (jaspar.genereg.net/). We also show the two regions used in our bisulfite sequencing and the pre-miR-203 accordingly to miRbase (www.mirbase.org/). (B,C) Bisulfite sequencing profiles of CpG site methylation in region 1 (B) and region 2 (C) were analyzed on pre-migratory NCCs (PM-NCCs), migratory NCCs (M-NCCs) and ventral neural tube (NT). The percentages of methylated CpG sites are shown with filled (100% methylated) and open (0% methylated) circles. (D,E) Bar graphs show the total percentages of methylated CpG sites on the different regions for the three analyzed samples in region 1 (D) and in region 2 (E). We found a higher percentage of methylated CpGs in PM-NCCs compared with other samples. Number in brackets indicate the number of analyzed sequences. *P<0.05 (ANOVA). (F) Morpholino-mediated loss of DNMT3A (DNMT3A-MO) and DNMT3B (DNMT3B-MO) function results in a reduction of methylated CpG sites on the injected side (IS) compared with the uninjected side (UIS) of the same group of embryos. Independent clones (n=10) were sequence out of two samples containing six half-electroporated embryos. (G,H) Morpholino-mediated loss of SNAIL2 (SNAIL2-MO) function maintains an elevated level of miR-203 expression (G) and reduced CpGs methylation in region 2 (H) on the IS compared with the UIS of the same group of embryos (n=6). *P<0.05 (Student's t-test).
DNMT3B and SNAIL2 are required for miR-203 methylation
De novo DNA methyltransferases DNMT3A and DNMT3B are both involved in neural crest development (Hu et al., 2014, 2012). To examine the possibility that DNA methylation may be involved in regulating miR-203 expression, we examined the effects of loss of DNMT3A and DNMT3B on miR-203 expression. To achieve this, we performed bisulfite sequencing after loss-of-function experiments with previously characterized fluorescein-tagged morpholino oligonucleotides: DNMT3A-MO and DNMT3B-MO (Hu et al., 2014, 2012). After unilateral injection and electroporation, dorsal neural tubes from 6 ss embryos were dissected and bisulfite converted to analyze the methylation abundance in region 2 of the miR-203 locus (Fig. 5F, Fig. S5). The results show that loss of function of either of the two DNMTs consistently decreased the abundance of methylated CpGs in the region 2. However, depletion of DNMT3B, but not DNMT3A, resulted in significant reduction in the methylation of miR-203.
Transcription factors specifically bind to target DNAs to recruit and guide DNA methyltransferases to specific genomic sites (Siegfried and Simon, 2010). During tumor metastasis, SNAIL2 binds to the miR-203 promoter to inhibit its transcription (Ding et al., 2013), although the mechanism underlying this repression is unknown. Our bioinformatic analysis reveals several SNAIL2-binding sites ∼1 kb upstream of the pre-miR-203 (Fig. 5A, Table S2), two of which have a high binding score (>9) accordingly to JASPAR 2018 (http://jaspar.genereg.net/). To test the effects of SNAIL2 loss of function, we electroporated a previously characterized Snail2 morpholino (Snail2-MO) (Taneyhill et al., 2007) unilaterally and dissected half dorsal neural tubes from 6 ss embryos for analysis of miR-203 expression and DNA methylation. Interestingly, the results show that Snail2 knockdown causes a significant upregulation of miR-203 expression (Fig. 5G). Consistent with this finding, we found a significant reduction in the abundance of methylated CpGs in region 2 of the miR-203 locus (Fig. 5H). These results suggest that SNAIL2 is involved in the epigenetic repression of miR-203 in the pre-migratory neural crest.
DISCUSSION
There is accumulating evidence for the importance of microRNAs in normal development as well as in several diseases, including tumor metastasis. Our study highlights the key role of a single microRNA, miR-203, in regulating the timing of initiation of the EMT program in neural crest cells. The results show that miR-203 gain and loss of function cause reduction or premature NCC delamination, respectively. We further show that the repression of miR-203 occurs via high levels of DNA methylation of the miR-203 locus by the DNMT3B, the specific activity of which is directed by SNAIL2. In this scenario, repression of miR-203 is directed by SNAIL2 in a feedback loop that enables expression of two direct targets of miR-203: Phf12 and Snai2, which in turn are necessary for neural crest delamination. Taken together, the results reveal for the first time an epigenetic-miRNA-gene regulatory circuit that controls the timing of neural crest delamination (Fig. 6). These findings support the idea that a single microRNA may ‘throw the switch’ from an epithelial to a mesenchymal state in the neural crest and thus stabilize the core gene regulatory networks in these two states.
Fig. 6.
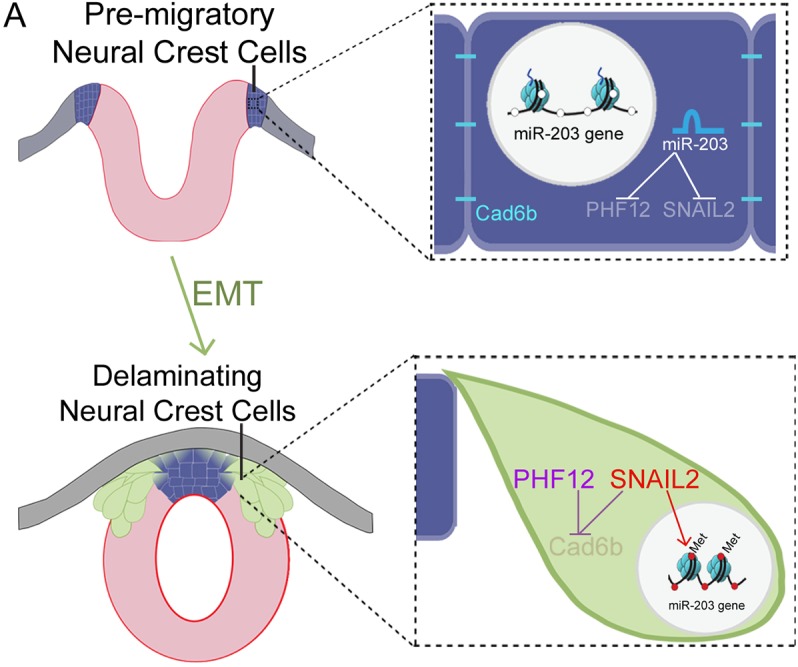
Hypothetical model. Our results show that, during NCC specification, miR-203 is highly expressed and prevents the initial accumulation of SNAIL2 and PHF12. Prior to NCC delamination, the accumulation of SNAIL2 causes epigenetic silencing of miR-203, mediated by DNMT3B. This silencing enables rapid concomitant upregulation of both Snail2 and Phf12, resulting in Cad6b repression at the beginning of the NCC epithelial-to-mesenchymal transition.
There is increasing evidence to suggest that miRNAs often act as fine-tuning regulators rather than as primary gene regulators (Hornstein and Shomron, 2006). Accordingly, we postulate that miR-203 may act by shifting the levels of SNAIL2 and PHF12 to prevent premature NCC delamination. A similar epigenetic-miRNA-mediated control of the core transcription factors necessary for EMT (EMT-TFs) has been also described in cancer cells (Ding, 2014; Guttilla et al., 2012; Kiesslich et al., 2013; Wright et al., 2010; Xia and Hui, 2012). Interestingly, some miRNAs and EMT-TFs form a tightly interconnected feedback loop that controls epithelial cell plasticity (Ding, 2014; Moes et al., 2012; Wellner et al., 2009), similar to our observation that SNAIL2 directs the epigenetic repression of miR-203. These feedback loops provide self-reinforcing signals and robustness to maintain the epithelial or mesenchymal cell state in response to different environmental cues. miR-203 is highly conserved from lamprey to human and appears to be an evolutionary novelty in vertebrates (Heimberg et al., 2010).
In the avian neural crest, EMT has two discrete steps: detachment from the neural tube; and subsequent separation of NCCs from the epidermis and from each other. Here, we show that the regulation of Snail2/Phf12 by miR-203 is required for the first step of EMT. Another transcription factor, SIP1, has been shown to be important for completion of the second step of mesenchymalization by mediating a cadherin switch (Rogers et al., 2013). Thus, there are multiple events, both transcriptional and post-transcriptional, that influence neural crest EMT. As case in point, we have recently shown that the secreted Wnt effector draxin influences the timing of EMT, by directly or indirectly controlling Snail2 expression (Hutchins and Bronner, 2017). Thus, it will be interesting in the future to explore the possible connection between miR-203 and draxin in regulating the timing of neural crest EMT. The present data demonstrate that miR-203 is a key regulator of NCC delamination by a reversible epigenetic-miRNA-mediated process. Given that the EMT program is a highly conserved process, involving similar transcription factors in both embryonic and cancer cells, these finding open new avenues for understanding normal and pathological development, as well as tumor metastasis.
MATERIALS AND METHODS
Chick embryos
Fertilized White Leghorn chick embryos were obtained from chicken farmers local to Chascomús, Argentina.
RNA preparation and RT-qPCR
RNA was prepared from individual chick embryos (n=6) using the isolation kit RNAqueous-Micro (Ambion) following the manufacturer's instructions. The RNA was treated with amplification grade DNaseI (Invitrogen) and then reverse transcribed to cDNA using a reverse transcription kit (SuperScript II; Invitrogen) with stem-loop-miRNA-specific (Chen et al., 2005) primers and random hexamers. QPCRs were performed using a 96-well plate qPCR machine (Stratagen) with SYBR green with ROX (Roche). Normalization control genes for qPCR were: for miR-203, miR-16 (Lardizábal et al., 2012); and for Snail2 and Phf12, Hprt1 (Simoes-Costa and Bronner, 2016). For a complete list of primers see Table S3.
Bisulfite sequencing
Samples were obtained by dissecting nine embryos at 6 ss to isolate the pre-migratory NCCs (PM-NCC), dorsal neural tube and ventral neural tube (NT). In addition, we dissected 13 embryos at 11-13 ss to obtain the migratory NCCs (M-NCCs). For the morpholino-mediated loss of DNMT3A, DNMT3B and SNAIL2 experiment, eight dorsal neural tubes from the injected and uninjected sides were dissected. For a complete list of morpholinos, see Table S3. All the tissues were lysed and bisulfite-converted with the EpiTect Plus Bisulfite Conversion Kit (Qiagen) following the manufacturer's instructions. The regulatory regions of miR-203 were amplified by using two sets of nested primers (see Table S3) from the bisulfite-converted DNA. The obtained products were gel purified and cloned into the pGEM-T Easy Vector (Promega). Twenty individual clones were sequenced and analyzed.
Electroporation
Chicken embryos were electroporated at stage 4-5 using previously described techniques (Sauka-Spengler and Barembaum, 2008). The vectors and morpholinos were injected by air pressure using a glass micropipette, and platinum electrodes were placed vertically across the chick embryos and electroporated with five pulses of 5.5 V for 50 ms at 100 ms intervals. Embryos were cultured in 0.5 ml albumen in tissue-culture dishes until the desired stages. Embryos were then removed and fixed in 4% PFA and used for immunohistochemistry or in situ hybridization.
In situ hybridization
Whole-mount chick in situ hybridization for mRNAs and for microRNA was performed as described previously (Acloque et al., 2008; Darnell et al., 2006). LNA probes for miR-203 used in the assay were obtained from Exiqon and DIG-labeled using the DIG oligonucleotide 3′ end labeling kit (Roche). After in situ hybridization, some embryos were fixed in 4% PFA in PBS, washed, embedded in gelatin and cryostat sectioned at 14-16 µm. They were photographed using the NIS-Elements Advanced Research software (Nikon) with an Eclipse E600 microscope (Nikon) and processed using Photoshop CS3 (Adobe).
Immunohistochemistry
Whole-mount chick immunohistochemistry was performed as described previously (Taneyhill et al., 2007). Briefly, embryos were fixed for 15 min in 4% paraformaldehyde (PFA) and then permeabilized and blocked in TBS [500 mM Tris-HCl (pH 7.4), 1.5 M NaCl, and 10 mM CaCl2] containing 0.1% Triton X-100 (TBS-T) and 5% FBS for 60 min at room temperature. Primary antibodies were diluted in TBS-T/fetal bovine serum and incubated overnight at 4°C. Primary antibodies used were mouse anti-Snail2 (1:100; supplied by the Developmental Studies Hybridoma Bank) and rabbit anti-FoxD3 (1:300; a gift from P. Labosky, Vanderbilt University Medical Center, Nashville, TN, USA). Secondary antibodies used were goat anti-mouse and anti-rabbit Alexa Fluor 594 (1:500; all obtained from Molecular Probes) diluted in TBS-T/FBS and incubated for 45 min at room temperature. All washes were performed in TBS-T at room temperature. Some embryos were subsequently embedded in gelatin, cryostat sectioned at 12-16 μm, photographed using the NIS-Elements Advanced Research software (Nikon) with an Eclipse E600 microscope (Nikon) and processed using Photoshop CS3 (Adobe).
MicroRNA sponge generation
Oligos designed to generate miR-203 sponge were ordered, phosphorylated and PAGE purified at a 100 nmol scale (see Table S2), and dissolved to 50 mM in STE buffer [100 mM NaCl, 10 mM Tris/HCl, 1 mM EDT (pH 8.0)]. Sense and antisense oligos were mixed at a 1:1 ratio and annealed by incubation at 100°C for 10 min followed by slow cooling. The ‘sponge’ vector was generated by following previously described protocols (Kluiver et al., 2012).
Supplementary Material
Acknowledgements
We thank Dr Xinwei Cao for the two-colored sensor vector and Dr Andrew Pollok for advice on the design of sponge vector. We also thank the Wood-Whelan research fellowships and The Company of Biologists (Development Travelling Fellowship DEV-180502) for travel support for E.S.-V.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: P.H.S.-M.; Methodology: E.S.-V., P.H.S.-M.; Validation: E.S.-V.; Formal analysis: E.S.-V.; Investigation: E.S.-V., P.H.S.-M.; Resources: M.E.B., P.H.S.-M.; Writing - original draft: E.S.-V., P.H.S.-M.; Writing - review & editing: M.E.B., P.H.S.-M.; Visualization: E.S.-V., P.H.S.-M.; Supervision: M.E.B., P.H.S.-M.; Project administration: P.H.S.-M.; Funding acquisition: M.E.B., P.H.S.-M.
Funding
This work was supported by the Fogarty International Center of the National Institutes of Health (R21TW011224 to M.E.B. and P.H.S.-M.) and by the Agencia Nacional de Promoción Científica y Tecnológica (PICT 2016-0747 to P.H.S.-M.). Deposited in PMC for release after 12 months.
Supplementary information
Supplementary information available online at http://dev.biologists.org/lookup/doi/10.1242/dev.171017.supplemental
References
- Acloque H., Wilkinson D. G. and Nieto M. A. (2008). In situ hybridization analysis of chick embryos in whole-mount and tissue sections. Methods Cell Biol. 87, 169-185. 10.1016/S0091-679X(08)00209-4 [DOI] [PubMed] [Google Scholar]
- Ahmad A., Li Y., Bao B., Kong D. and Sarkar F. H. (2014). Epigenetic regulation of miRNA-cancer stem cells nexus by nutraceuticals. Mol. Nutr. Food Res. 58, 79-86. 10.1002/mnfr.201300528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benaich N., Woodhouse S., Goldie S. J., Mishra A., Quist S. R. and Watt F. M. (2014). Rewiring of an epithelial differentiation factor, miR-203, to inhibit human squamous cell carcinoma metastasis. Cell Rep. 9, 104-117. 10.1016/j.celrep.2014.08.062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boldrup L., Coates P. J., Wahlgren M., Laurell G. and Nylander K. (2012). Subsite-based alterations in miR-21, miR-125b, and miR-203 in squamous cell carcinoma of the oral cavity and correlation to important target proteins. J. Carcinog. 11, 18 10.4103/1477-3163.104007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boll K., Reiche K., Kasack K., Mörbt N., Kretzschmar A. K., Tomm J. M., Verhaegh G., Schalken J., von Bergen M., Horn F. et al. (2013). MiR-130a, miR-203 and miR-205 jointly repress key oncogenic pathways and are downregulated in prostate carcinoma. Oncogene 32, 277-285. 10.1038/onc.2012.55 [DOI] [PubMed] [Google Scholar]
- Bonnomet A., Brysse A., Tachsidis A., Waltham M., Thompson E. W., Polette M. and Gilles C. (2010). Epithelial-to-mesenchymal transitions and circulating tumor cells. J. Mammary Gland Biol. Neoplasia 15, 261-273. 10.1007/s10911-010-9174-0 [DOI] [PubMed] [Google Scholar]
- Bu P. and Yang P. (2014). MicroRNA-203 inhibits malignant melanoma cell migration by targeting versican. Exp Ther Med 8, 309-315. 10.3892/etm.2014.1708 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Cao X., Pfaff S. L. and Gage F. H. (2007). A functional study of miR-124 in the developing neural tube. Genes Dev. 21, 531-536. 10.1101/gad.1519207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coles E. G., Taneyhill L. A. and Bronner-Fraser M. (2007). A critical role for Cadherin6B in regulating avian neural crest emigration. Dev. Biol. 312, 533-544. 10.1016/j.ydbio.2007.09.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crane J. F. and Trainor P. A. (2006). Neural crest stem and progenitor cells. Annu. Rev. Cell Dev. Biol. 22, 267-286. 10.1146/annurev.cellbio.22.010305.103814 [DOI] [PubMed] [Google Scholar]
- Chen C., Ridzon D. A., Broomer A. J., Zhou Z., Lee D. H., Nguyen J. T., Barbisin M., Xu N. L., Mahuvakar V. R. and Andersen M. R. (2005). Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 33, e179 10.1093/nar/gni178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H.-Y., Han Z.-B., Fan J.-W., Xia J., Wu J.-Y., Qiu G. Q., Tang H.-M. and Peng Z. H. (2012). miR-203 expression predicts outcome after liver transplantation for hepatocellular carcinoma in cirrhotic liver. Med. Oncol. 29, 1859-1865. 10.1007/s12032-011-0031-9 [DOI] [PubMed] [Google Scholar]
- Chiang Y., Song Y., Wang Z., Chen Y., Yue Z., Xu H., Xing C. and Liu Z. (2011). Aberrant expression of miR-203 and its clinical significance in gastric and colorectal cancers. J. Gastrointest. Surg. 15, 63-70. 10.1007/s11605-010-1367-8 [DOI] [PubMed] [Google Scholar]
- Chim C. S., Wong K. Y., Leung C. Y., Chung L. P., Hui P. K., Chan S. Y. and Yu L. (2011b). Epigenetic inactivation of the hsa-miR-203 in haematological malignancies. J. Cell. Mol. Med. 15, 2760-2767. 10.1111/j.1582-4934.2011.01274.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell D. K., Kaur S., Stanislaw S., Konieczka J. H., Yatskievych T. A. and Antin P. B. (2006). MicroRNA expression during chick embryo development. Dev. Dyn. 235, 3156-3165. 10.1002/dvdy.20956 [DOI] [PubMed] [Google Scholar]
- Diao Y., Guo X., Jiang L., Wang G., Zhang C., Wan J., Jin Y. and Wu Z. (2014). miR-203, a tumor suppressor frequently down-regulated by promoter hypermethylation in rhabdomyosarcoma. J. Biol. Chem. 289, 529-539. 10.1074/jbc.M113.494716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Lopez A., Moreno-Bueno G. and Cano A. (2014). Role of microRNA in epithelial to mesenchymal transition and metastasis and clinical perspectives. Cancer Manag. Res. 6, 205-216. 10.2147/CMAR.S38156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding X.-M. (2014). MicroRNAs: regulators of cancer metastasis and epithelial-mesenchymal transition (EMT). Chin. J. Cancer 33, 140-147. 10.5732/cjc.013.10094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding X., Park S. I., McCauley L. K. and Wang C.-Y. (2013). Signaling between transforming growth factor beta (TGF-beta) and transcription factor SNAI2 represses expression of microRNA miR-203 to promote epithelial-mesenchymal transition and tumor metastasis. J. Biol. Chem. 288, 10241-10253. 10.1074/jbc.M112.443655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedl P. and Gilmour D. (2009). Collective cell migration in morphogenesis, regeneration and cancer. Nat. Rev. Mol. Cell Biol. 10, 445-457. 10.1038/nrm2720 [DOI] [PubMed] [Google Scholar]
- Furuta M., Kozaki K.-I., Tanaka S., Arii S., Imoto I. and Inazawa J. (2010). miR-124 and miR-203 are epigenetically silenced tumor-suppressive microRNAs in hepatocellular carcinoma. Carcinogenesis 31, 766-776. 10.1093/carcin/bgp250 [DOI] [PubMed] [Google Scholar]
- Gao P., Wang S., Jing F., Zhan J. and Wang Y. (2017). microRNA-203 suppresses invasion of gastric cancer cells by targeting ERK1/2/Slug/ E-cadherin signaling. Cancer Biomark. 19, 11-20. 10.3233/CBM-160167 [DOI] [PubMed] [Google Scholar]
- Guttilla I. K., Adams B. D. and White B. A. (2012). ERalpha, microRNAs, and the epithelial-mesenchymal transition in breast cancer. Trends Endocrinol. Metab. 23, 73-82. 10.1016/j.tem.2011.12.001 [DOI] [PubMed] [Google Scholar]
- Heimberg A. M., Cowper-Sal-lari R., Semon M., Donoghue P. C. and Peterson K. J. (2010). microRNAs reveal the interrelationships of hagfish, lampreys, and gnathostomes and the nature of the ancestral vertebrate. Proc. Natl. Acad. Sci. USA 107, 19379-19383. 10.1073/pnas.1010350107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornstein E. and Shomron N. (2006). Canalization of development by microRNAs. Nat. Genet. 38 Suppl., S20-S24. 10.1038/ng1803 [DOI] [PubMed] [Google Scholar]
- Hu N., Strobl-Mazzulla P. H., Simoes-Costa M., Sánchez-Vásquez E. and Bronner M. E. (2014). DNA methyltransferase 3B regulates duration of neural crest production via repression of Sox10. Proc. Natl. Acad. Sci. USA 111, 17911-17916. 10.1073/pnas.1318408111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu N., Strobl-Mazzulla P., Sauka-Spengler T. and Bronner M. E. (2012). DNA methyltransferase3A as a molecular switch mediating the neural tube-to-neural crest fate transition. Genes Dev. 26, 2380-2385. 10.1101/gad.198747.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y.-W., Kuo C.-T., Chen J.-H., Goodfellow P. J., Huang T. H.-M., Rader J. S. and Uyar D. S. (2014). Hypermethylation of miR-203 in endometrial carcinomas. Gynecol. Oncol. 133, 340-345. 10.1016/j.ygyno.2014.02.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchins E. J. and Bronner M. E. (2018). Draxin acts as a molecular rheostat of canonical Wnt signaling to control cranial neural crest EMT. J. Cell Biol. 217, 3683-3697. 10.1083/jcb.201709149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju S.-Y., Chiou S.-H. and Su Y. (2014). Maintenance of the stemness in CD44(+) HCT-15 and HCT-116 human colon cancer cells requires miR-203 suppression. Stem Cell Res. 12, 86-100. 10.1016/j.scr.2013.09.011 [DOI] [PubMed] [Google Scholar]
- Kerosuo L. and Bronner-Fraser M. (2012). What is bad in cancer is good in the embryo: importance of EMT in neural crest development. Semin. Cell Dev. Biol. 23, 320-332. 10.1016/j.semcdb.2012.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiesslich T., Pichler M. and Neureiter D. (2013). Epigenetic control of epithelial-mesenchymal-transition in human cancer. Mol. Clin. Oncol. 1, 3-11. 10.3892/mco.2012.28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloosterman W. P. and Plasterk R. H. (2006). The diverse functions of microRNAs in animal development and disease. Dev. Cell 11, 441-450. 10.1016/j.devcel.2006.09.009 [DOI] [PubMed] [Google Scholar]
- Kluiver J., Gibcus J. H., Hettinga C., Adema A., Richter M. K. S., Halsema N., Slezak-Prochazka I., Ding Y., Kroesen B.-J. and van den Berg A. (2012). Rapid generation of microRNA sponges for microRNA inhibition. PLoS ONE 7, e29275 10.1371/journal.pone.0029275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lardizábal M. N., Nocito A. L., Daniele S. M., Ornella L. A., Palatnik J. F. and Veggi L. M. (2012). Reference genes for real-time PCR quantification of microRNAs and messenger RNAs in rat models of hepatotoxicity. PLoS ONE 7, e36323 10.1371/journal.pone.0036323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohcharoenkal W., Das Mahapatra K., Pasquali L., Crudden C., Kular L., Akkaya Ulum Y. Z., Zhang L., Xu Landén N., Girnita L., Jagodic M., et al. (2018). Genome-wide screen for microRNAs reveals a role for miR-203 in melanoma metastasis. J. Invest. Dermatol. 138, 882-892. 10.1016/j.jid.2017.09.049 [DOI] [PubMed] [Google Scholar]
- Lujambio A., Calin G. A., Villanueva A., Ropero S., Sanchez-Cespedes M., Blanco D., Montuenga L. M., Rossi S., Nicoloso M. S., Faller W. J., et al. (2008). A microRNA DNA methylation signature for human cancer metastasis. Proc. Natl. Acad. Sci. USA 105, 13556-13561. 10.1073/pnas.0803055105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayor R. and Theveneau E. (2013). The neural crest. Development 140, 2247-2251. 10.1242/dev.091751 [DOI] [PubMed] [Google Scholar]
- Miao L., Xiong X., Lin Y., Cheng Y., Lu J., Zhang J. and Cheng N. (2014). miR-203 inhibits tumor cell migration and invasion via caveolin-1 in pancreatic cancer cells. Oncol. Lett. 7, 658-662. 10.3892/ol.2014.1807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moes M., Le Béchec A., Crespo I., Laurini C., Halavatyi A., Vetter G., del Sol A. and Friederich E. (2012). A novel network integrating a miRNA-203/SNAI1 feedback loop which regulates epithelial to mesenchymal transition. PLoS ONE 7, e35440 10.1371/journal.pone.0035440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson K. M. and Weiss G. J. (2008). MicroRNAs and cancer: past, present, and potential future. Mol. Cancer Ther. 7, 3655-3660. 10.1158/1535-7163.MCT-08-0586 [DOI] [PubMed] [Google Scholar]
- Rogers C. D., Saxena A. and Bronner M. E. (2013). Sip1 mediates an E-cadherin-to-N-cadherin switch during cranial neural crest EMT. Cell Biol. 203, 835-847. 10.1083/jcb.201305050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauka-Spengler T. and Barembaum M. (2008). Chapter 12 gain- and loss-of-function approaches in the chick embryo. Methods Cell Biol. 87, 237-256. 10.1016/s0091-679x(08)00212-4 [DOI] [PubMed] [Google Scholar]
- Scarpa E. and Mayor R. (2016). Collective cell migration in development. J. Cell Biol. 212, 143-155. 10.1083/jcb.201508047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y., Tan Y. J., Zeng D. Z., Qian F. and Yu P. W. (2015). miR-203 suppression in gastric carcinoma promotes Slug-mediated cancer metastasis. Tumour Biol. 37, 15483-15488. 10.1007/s13277-015-3765-8 [DOI] [PubMed] [Google Scholar]
- Siegfried Z. and Simon I. (2010). DNA methylation and gene expression. Wiley Interdiscip. Rev. Syst. Biol. Med. 2, 362-371. 10.1002/wsbm.64 [DOI] [PubMed] [Google Scholar]
- Simoes-Costa M. and Bronner M. E. (2015). Establishing neural crest identity: a gene regulatory recipe. Development 142, 242-257. 10.1242/dev.105445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simoes-Costa M. and Bronner M. E. (2016). Reprogramming of avian neural crest axial identity and cell fate. Science 352, 1570-1573. 10.1126/science.aaf2729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strobl-Mazzulla P. H. and Bronner M. E. (2012). A PHD12-Snail2 repressive complex epigenetically mediates neural crest epithelial-to-mesenchymal transition. J. Cell Biol. 198, 999-1010. 10.1083/jcb.201203098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strobl-Mazzulla P. H., Sauka-Spengler T. and Bronner-Fraser M. (2010). Histone demethylase JmjD2A regulates neural crest specification. Dev. Cell 19, 460-468. 10.1016/j.devcel.2010.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taneyhill L. A., Coles E. G. and Bronner-Fraser M. (2007). Snail2 directly represses cadherin6B during epithelial-to-mesenchymal transitions of the neural crest. Development 134, 1481-1490. 10.1242/dev.02834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taube J. H., Malouf G. G., Lu E., Sphyris N., Vijay V., Ramachandran P. P., Ueno K. R., Gaur S., Nicoloso M. S., Rossi S., et al. (2013). Epigenetic silencing of microRNA-203 is required for EMT and cancer stem cell properties. Sci. Rep. 3, 2687 10.1038/srep02687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobiasson M., Abdulkadir H., Lennartsson A., Katayama S., Marabita F., De Paepe A., Karimi M., Krjutskov K., Einarsdottir E., Grövdal M., et al. (2017). Comprehensive mapping of the effects of azacitidine on DNA methylation, repressive/permissive histone marks and gene expression in primary cells from patients with MDS and MDS-related disease. Oncotarget 8, 28812-28825. 10.18632/oncotarget.15807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Zhao J., Huang J., Tang H., Yu S. and Chen Y. (2012). The regulatory roles of miRNA and methylation on oncogene and tumor suppressor gene expression in pancreatic cancer cells. Biochem. Biophys. Res. Commun. 425, 51-57. 10.1016/j.bbrc.2012.07.047 [DOI] [PubMed] [Google Scholar]
- Weber B., Stresemann C., Brueckner B. and Lyko F. (2007). Methylation of human microRNA genes in normal and neoplastic cells. Cell Cycle 6, 1001-1005. 10.4161/cc.6.9.4209 [DOI] [PubMed] [Google Scholar]
- Wellner U., Schubert J., Burk U. C., Schmalhofer O., Zhu F., Sonntag A., Waldvogel B., Vannier C., Darling D., zur Hausen A., et al. (2009). The EMT-activator ZEB1 promotes tumorigenicity by repressing stemness-inhibiting microRNAs. Nat. Cell Biol. 11, 1487-1495. 10.1038/ncb1998 [DOI] [PubMed] [Google Scholar]
- Wright J. A., Richer J. K. and Goodall G. J. (2010). microRNAs and EMT in mammary cells and breast cancer. J. Mammary Gland Biol. Neoplasia 15, 213-223. 10.1007/s10911-010-9183-z [DOI] [PubMed] [Google Scholar]
- Xia H. and Hui K. M. (2012). MicroRNAs involved in regulating epithelial-mesenchymal transition and cancer stem cells as molecular targets for cancer therapeutics. Cancer Gene Ther. 19, 723-730. 10.1038/cgt.2012.58 [DOI] [PubMed] [Google Scholar]
- Xiao J.-N., Yan T.-H., Yu R.-M., Gao Y., Zeng W.-L., Lu S.-W., Que H.-X., Liu Z.-P. and Jiang J. H. (2017). Long non-coding RNA UCA1 regulates the expression of Snail2 by miR-203 to promote hepatocellular carcinoma progression. J. Cancer Res. Clin. Oncol. 143, 981-990. 10.1007/s00432-017-2370-1 [DOI] [PubMed] [Google Scholar]
- Zhang F., Yang Z., Cao M., Xu Y., Li J., Chen X., Gao Z., Xin J., Zhou S., Zhou Z., et al. (2014). MiR-203 suppresses tumor growth and invasion and down-regulates MiR-21 expression through repressing Ran in esophageal cancer. Cancer Lett. 342, 121-129. 10.1016/j.canlet.2013.08.037 [DOI] [PubMed] [Google Scholar]
- Zhang J., Zhou Y., Wu Y.-J., Li M.-J., Wang R.-J., Huang S.-Q., Gao R.-R., Ma L., Shi H.-J. and Zhang J. (2013). Hyper-methylated miR-203 dysregulates ABL1 and contributes to the nickel-induced tumorigenesis. Toxicol. Lett. 223, 42-51. 10.1016/j.toxlet.2013.08.007 [DOI] [PubMed] [Google Scholar]
- Zhang K., Dai L., Zhang B., Xu X., Shi J., Fu L., Chen X., Li J. and Bai Y. (2015). miR-203 Is a Direct Transcriptional Target of E2F1 and Causes G1 Arrest in Esophageal Cancer Cells. J. Cell. Physiol. 230, 903-910. 10.1002/jcp.24821 [DOI] [PubMed] [Google Scholar]
- Zhang Z., Zhang B., Li W., Fu L., Fu L., Zhu Z. and Dong J.-T. (2011). Epigenetic silencing of miR-203 upregulates SNAI2 and contributes to the invasiveness of malignant breast cancer cells. Genes Cancer 2, 782-791. 10.1177/1947601911429743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S., Yao D.-S., Chen J.-Y. and Ding N. (2013). Aberrant expression of miR-20a and miR-203 in cervical cancer. Asian Pac. J. Cancer Prev. 14, 2289-2293. 10.7314/APJCP.2013.14.4.2289 [DOI] [PubMed] [Google Scholar]
- Zhu X., Er K., Mao C., Yan Q., Xu H., Zhang Y., Zhu J., Cui F., Zhao W. and Shi H. (2013). miR-203 suppresses tumor growth and angiogenesis by targeting VEGFA in cervical cancer. Cell. Physiol. Biochem. 32, 64-73. 10.1159/000350125 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.