Abstract
Chlamydia trachomatis is a bacterial pathogen causing ocular and genital infections in humans. C. trachomatis multiplies exclusively inside host cells within a characteristic vacuole, from where it manipulates host cells by injecting them with type III secretion effector proteins. Here, we identified CteG as the first C. trachomatis effector associated with the Golgi. For this, C. trachomatis strains expressing candidate effectors fused to a double hemagglutinin (2HA) tag were constructed. Then, among these strains, immunofluorescence microscopy revealed that CteG-2HA was delivered into the cytoplasm of infected cells. Between 16–20 h post-infection, CteG-2HA mostly associated with the Golgi; however, CteG-2HA also appeared at the host cell plasma membrane, and at 30 or 40 h post-infection this was its predominant localization. This change in the main localization of CteG-2HA was independent of intact microfilaments or microtubules. Ectopic expression of different regions of CteG (656 amino acid residues) in uninfected cells revealed that its first 100 residues contain a Golgi targeting region. Although a C. trachomatis cteG mutant did not display a defect in intracellular multiplication, CteG induced a vacuolar protein sorting defect when expressed in Saccharomyces cerevisiae. This suggested that CteG might function by subverting host cell vesicular transport.
Introduction
Chlamydia trachomatis serovars are obligate intracellular bacterial pathogens usually causing ocular and genital infections that affect millions of people worldwide and can lead to blindness and sterility. Serovars A-C are normally associated with trachoma1, while serovars D-K are the most common cause of sexually transmitted bacterial infections2. The less common serovars L1–L3 cause lymphogranuloma venereum (LGV), an invasive infection2. The Chlamydia genus includes other species pathogenic for humans (C. pneumoniae and C. psittaci) and other animals (C. abortus, C. avium, C. caviae, C. felis, C. gallinacea, C. muridarum, C. pecorum, C. psittaci, and C. suis)3.
Chlamydia are characterized by a developmental cycle involving an infectious but non-replicative form, the elementary body (EB), and a non-infectious but replicative form, the reticulate body (RB). Adherence of extracellular EBs to host cells leads to invasion and formation of a membrane-bound vacuolar compartment (known as the inclusion) where Chlamydia resides, develops and grows intracellularly4. As with many other Gram-negative bacteria5, the capacity of Chlamydia to subvert host cells largely relies on a type III secretion (T3S) system mediating the transport of effector proteins into host cells6. In general, the biological function of T3S effectors depends on their biochemical activity, timing of delivery and specific subcellular targeting in host cells, and is coordinated with the action of other effectors injected by the same bacterium7,8. In Chlamydia, T3S effectors include the Inc proteins, characterized by a bilobed hydrophobic motif mediating their insertion into the inclusion membrane9,10. The identification of Chlamydia effectors without the bilobed hydrophobic motif is normally more challenging because their primary structure normally lacks other obvious distinguishable features. However, several of these non-Inc C. trachomatis T3S effectors (e.g., TarP, TepP, CT694/TmeA) have been identified and shown to modulate chlamydial invasion and diverse host cell functions4,11–15. There are also Chlamydia effectors, such as deubiquitinating enzymes16,17, which localize within the cytoplasm of host cells and that have not been shown to be T3S substrates, as well as chlamydial T3S substrates secreted into the inclusion lumen18,19. Some of the non-Inc chlamydial effectors localize at the inclusion membrane17,20–22, at the host cell plasma membrane22, or at the host cell nucleus23–25, while others are membrane-associated11,26 or have undefined localization.
In this work, following the identification of candidate chlamydial T3S substrates using Yersinia enterocolitica as a heterologous host27,28, we show that the C. trachomatis CT105 protein (CTL0360 in C. trachomatis serovar L2 strain 434/Bu; L2/434) is delivered into host cells during infection. In infected cells, bacterially-delivered CT105 initially mainly localized at the Golgi complex and then at the plasma membrane. CT105 is the first Chlamydia protein described to localize at the Golgi in infected cells, and we identified a Golgi-targeting region within its first 100 amino acid residues. Using Saccharomyces cerevisae as model, we also show that CT105 can modulate eukaryotic vesicular trafficking.
Results
CT105-2HA is delivered by C. trachomatis into the cytoplasm of infected cells
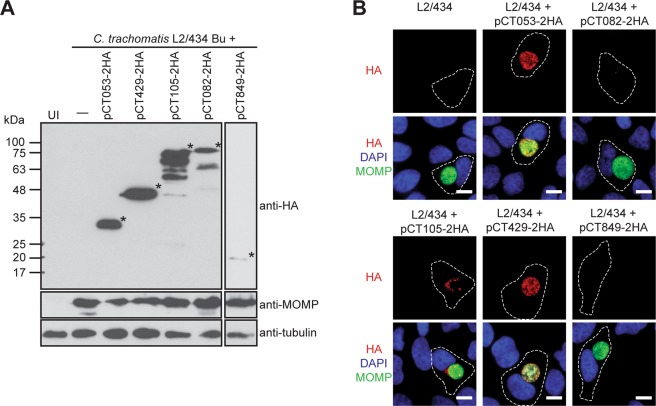
To test if the candidate chlamydial T3S substrates CT053, CT082, CT105, CT429, and CT84927,28 can be transported by Chlamydia into the cytoplasm of host cells, C. trachomatis strain L2/434 was transformed with plasmids encoding these proteins with a double hemagglutinin (2HA) epitope tag at their C-termini. Protein production was confirmed by immunoblotting of extracts of HeLa cells infected for 40 h with C. trachomatis strains harboring plasmids encoding CT053-2HA (predicted molecular mass of 17 kDa), CT082-2HA (60 kDa), CT105-2HA (68 kDa), CT429-2HA (39 kDa), or CT849-2HA (18 kDa) (Figs 1A and S1). The strains producing CT053-2HA, CT082-2HA and CT105-2HA also showed species migrating on SDS-PAGE at a molecular mass different from the one predicted for the full-length proteins (Figs 1A and S1), as previously observed when identical 2HA-tagged versions of the proteins were produced in Y. enterocolitica27,28. Overall, these experiments confirmed that the constructed C. trachomatis strains expressed the expected 2HA-epitope tagged proteins.
Figure 1.
The chlamydial candidate T3S effector CT105 is delivered by C. trachomatis into the cytoplasm of infected cells. HeLa cells were either left uninfected (UI) or infected by C. trachomatis L2/434-derived strains harboring the indicated plasmids, encoding candidate T3S effectors (CT053, CT429, CT105, CT082, CT849) with a 2HA epitope tag at their C-termini. (A) At 40 h p.i., whole cell extracts were analyzed by immunoblotting with antibodies against HA, MOMP (bacterial loading control) and α-tubulin (HeLa loading control) using SuperSignal West Pico detection kit (Thermo Fisher Scientific), or SuperSignal West Femto detection kit (Thermo Fisher Scientific) for CT849-2HA. Asterisks indicate the bands likely corresponding to the full-length proteins. Whole immunoblots are presented in Supplementary Fig. S1. (B) At 20 h p.i., cells were fixed with 4% (w/v) PFA, stained with DAPI (blue), immunolabeled with antibodies against HA (red) and C. trachomatis MOMP (green), and appropriate fluorophore-conjugated secondary antibodies, and imaged by fluorescence microscopy. Scale bars, 10 µm.
To analyze the subcellular localization of CT053-2HA, CT082-2HA, CT105-2HA, CT429-2HA, and CT849-2HA, HeLa cells were infected for 20 or 40 h with C. trachomatis L2/434-derived strains bearing the corresponding encoding plasmids and then analyzed by immunofluorescence microscopy. At 20 p.i. (Fig. 1B) or 40 h p.i. (Supplementary Fig. S2), CT053-2HA, CT082-2HA, CT429-2HA, and CT849-2HA were only detected within the inclusion and/or colocalizing with the C. trachomatis major outer membrane protein (MOMP) signal (Figs 1B and S2). In contrast, CT105-2HA was detected outside of the inclusion, indicating it is bacterially-delivered into the cytoplasm of infected host cells (Figs 1B and S2).
Full-length orthologues of CT105 are only present in C. muridarum and C. suis, and ct105 expression is mostly restricted to C. trachomatis LGV strains
CT105 is a protein of 656 amino acid residues whose sequence does not show significant similarity to other proteins except for potential orthologues in other Chlamydia spp. However, full-length orthologues of CT105 were only found in C. muridarum and C. suis (Supplementary Table S1). In C. suis and in other Chlamydia spp. different open reading frames might encode proteins with some identity (between 30–22%) to only some parts of the amino acid sequence of CT105 (Supplementary Table S1). As previously noted29, analysis of the mRNA levels of ct105 by reverse transcription quantitative PCR (RT-qPCR) in different C. trachomatis strains revealed that the gene is only significantly expressed in LGV strains (serovars L1–L3) (Supplementary Fig. S3). Furthermore, also as previously noted29, considering the promoter region of ct105 based on the transcription start site identified in C. trachomatis LGV strain L2b/UCH-1/proctitis30, C. trachomatis non-LGV strains (serovars A-K) lack 74 nucleotides upstream from the putative −10 region recognized by C. trachomatis σ66 (Supplementary Fig. S4). It has also been previously shown that ct105 is a pseudogene in C. trachomatis ocular strains (serovars A-C)31. Therefore, among C. trachomatis serovars, active CT105 is mostly produced by LGV strains.
Characterization of C. trachomatis expressing plasmid-encoded CT105-2HA
The L2/434 strain harboring pCT105-2HA, was then characterized by comparison to the parental strain. The profile of expression of ct105 during the developmental cycle of C. trachomatis was similar between the two strains (Supplementary Fig. S5). In agreement with previous observations27 (Supplementary Fig. S3), the highest levels of ct105 mRNA were detected at 2 h p.i. (Supplementary Fig. S5). Furthermore, at 2 h p.i., the strain harboring pCT105-2HA showed a 10-fold increase in the mRNA levels of ct105 relative to the parental strain (Supplementary Fig. S5). However, expression of plasmid-encoded CT105-2HA in C. trachomatis had no significant impact on chlamydial growth (Supplementary Fig. S5).
In extracts of HeLa cells infected by C. trachomatis L2/434 harboring pCT105-2HA, the 2HA-tagged protein could be detected by immunoblotting from 16 to 40 h p.i. (Supplementary Fig. S6). In addition to the band likely corresponding to full-length CT105-2HA (68 kDa), several faster migrating species of a lower molecular mass were consistently observed between 20 to 40 h p.i. (Supplementary Fig. S6). Similar observations were made by immunoblotting of extracts of HeLa cells infected by strain L2/434 harboring a plasmid (pTet-CT105-2HA) where production of CT105-2HA is driven by the tetracycline promoter (Ptet) (Supplementary Fig. S6). This indicates that plasmid-encoded CT105-2HA is modified and/or degraded as the C. trachomatis developmental cycle progresses. Immunoblotting of extracts of HeLa cells infected by L2/434 harboring either pCT105-2HA or pTet-CT105-2HA, or of corresponding Chlamydia-enriched extracts, suggested that this modification and/or degradation of CT105-2HA occurs within the bacterium (Supplementary Fig. S6).
CT105-2HA localizes at the Golgi complex in C. trachomatis-infected cells
To analyze in further detail the localization of CT105-2HA during infection, HeLa cells were infected for different times by C. trachomatis L2/434 harboring pCT105-2HA and then analysed by immunofluorescence microscopy. At 2, 4, and 8 h p.i., CT105-2HA could be detected colocalizing with the signal for C. trachomatis Hsp60 (Supplementary Fig. S7), which suggested its presence within the bacterial cells. At 16 and 20 h p.i., CT105-2HA was detected outside of the inclusion in approximately 37% and 90% of the infected cells, respectively (Supplementary Fig. S7); at 30 and 40 h p.i., the protein was found outside of the inclusion in all infected cells (Supplementary Fig. S7). While at 16, 20, and 30 h p.i. CT105-2HA was detected in the host cell cytoplasm concentrated at one of the sides of the inclusion, at 30 h p.i. the protein was also detected at the periphery of the infected cells (Supplementary Fig. S7). Furthermore, at 40 h p.i., CT105-2HA was mostly seen at the periphery of the infected cells (Supplementary Fig. S7).
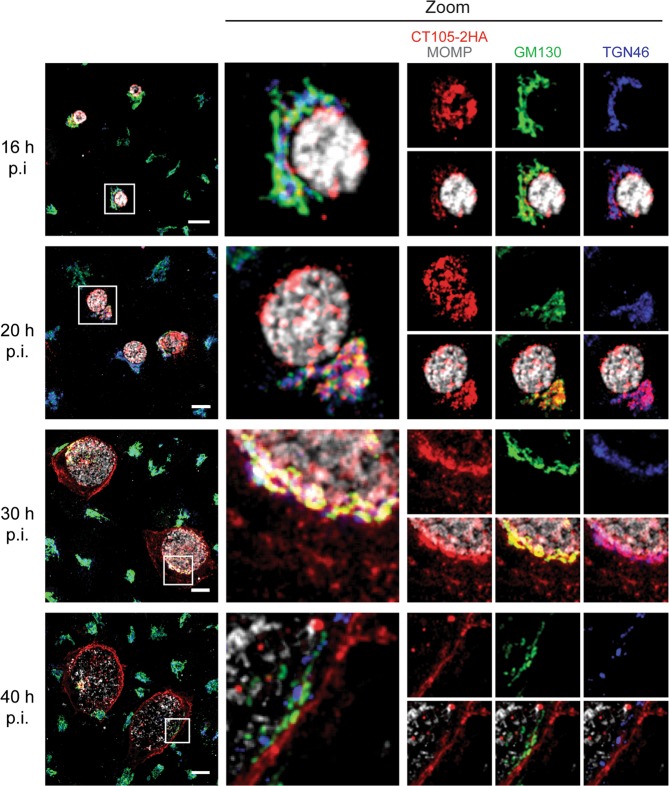
The accumulation of CT105-2HA near the inclusion in the cytoplasm of infected cells suggested it could localize at the Golgi complex. This immunofluorescence signal of CT105-2HA also appeared less compact at longer times of infection, which was evocative of the Golgi fragmentation seen in C. trachomatis infected-cells32. To analyze this, HeLa cells infected by C. trachomatis L2/434 harboring pCT105-2HA were fixed at 16, 20, 30, or 40 h p.i. and then analyzed by immunofluorescence microscopy. This revealed that CT105-2HA localized in the Golgi region at 16, 20, and 30 h p.i., and that this was much less evident at 40 h p.i. (Fig. 2). There was not a perfect colocalization of CT105-2HA with cis-Golgi (GM130) and TGN (TGN46) markers, but the immunofluorescence signal of CT105-2HA near the inclusion accompanied the dispersion of the Golgi complex during infection (Fig. 2).
Figure 2.
CT105 colocalizes with the Golgi complex in C. trachomatis-infected cells. HeLa cells were infected by C. trachomatis L2/434 encoding CT105-2HA. At the indicated times p.i., cells were fixed with 4% (w/v) PFA, immunolabeled with antibodies against HA (red), C. trachomatis MOMP (gray), GM130 (green), and TGN46 (blue), and appropriate fluorophore-conjugated secondary antibodies. In the area delimited by a white square (left-side panels) images were zoomed (middle and right-side panels). Immunolabeled cells were examined by confocal fluorescence microscopy, and images correspond to single z sections. Scale bars, 10 µm.
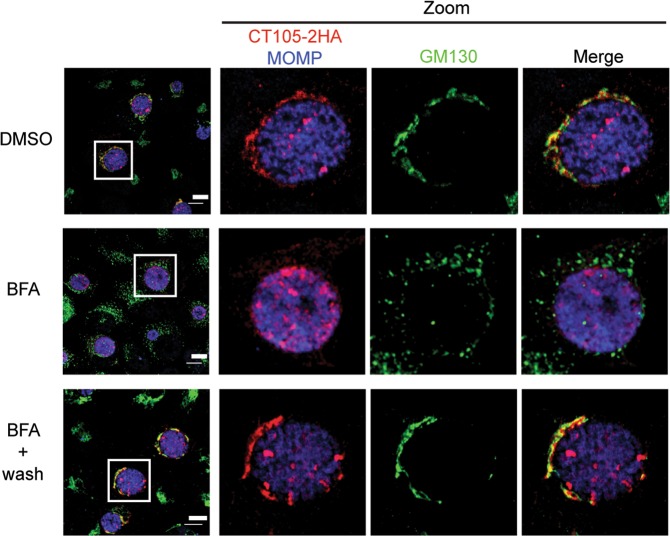
To further confirm the association of CT105-2HA with the Golgi, HeLa cells infected by the L2/434 strain harboring pCT105-2HA were treated with Brefeldin A (BFA), which induces reversible Golgi fragmentation33. For this, at 19 h p.i., the infected cells were either incubated for 1 h with BFA or with dimethyl sulfoxide (DMSO)-solvent alone and then fixed. In addition, the infected cells were incubated for 1 h with BFA, washed out from the BFA, incubated for 1 h and then fixed. Immunofluorescence microscopy revealed that in the DMSO-treated infected cells, CT105-2HA accumulated in the Golgi region (Fig. 3), similarly to untreated cells infected for 16 or 20 h p.i. (Fig. 2). In cells treated with BFA, the GM130-labeled Golgi fragmented into small vesicles and the immunofluorescence signal of CT105-2HA became dispersed and mostly below the limit of detection (Fig. 3). In cells treated with BFA and subsequently washed-out of the drug, both the GM130-labeled Golgi and CT105-2HA appeared mostly compact and near the inclusion (Fig. 3). Overall, this showed that CT105-2HA associates with the Golgi, at 16–20 h p.i., upon the initial detection of its delivery by C. trachomatis into the cytoplasm of infected host cells.
Figure 3.
CT105 associates with the Golgi complex in C. trachomatis-infected cells. HeLa cells were infected by C. trachomatis L2/434 encoding CT105-2HA. At 19 h p.i., cells were treated for 1 h with DMSO or 1 µg/ml BFA. Then, the cells were either fixed with 4% (w/v) PFA (upper and middle panels) or washed with complete medium lacking BFA and incubated for an additional 1 h and then fixed with 4% (w/v) PFA (lower panels). Fixed cells were immunolabeled with antibodies against HA (red), C. trachomatis MOMP (gray), and GM130 (green), and appropriate fluorophore-conjugated secondary antibodies. In the area delimited by a white square (left-side panels) images were zoomed (right-side panels). All immunolabeled cells were examined by confocal fluorescence microscopy, and images correspond to single z sections. Scale bars, 10 µm.
CT105-2HA localizes at the host cell plasma membrane in C. trachomatis-infected cells
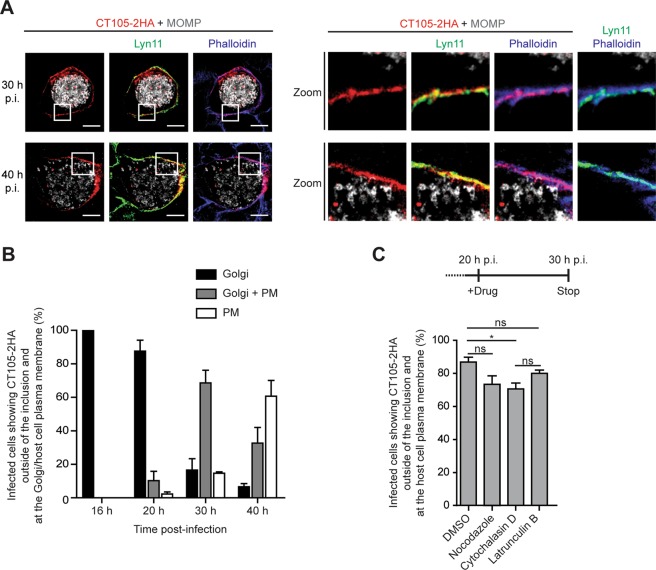
CT105-2HA was also detected in the periphery of host cells infected by C. trachomatis at 30 or 40 h p.i. (Supplementary Fig. S7), which suggested it could localize at the plasma membrane. To analyze this, HeLa cells were infected for 30 and 40 h p.i. by C. trachomatis L2/434 harboring CT105-2HA and transfected with a plasmid encoding a palmitoylated/myristoylated peptide from Lyn kinase (Lyn11) [targeting proteins to the plasma membrane34] fused to monomeric EGFP35 (Lyn11-mEGFP). The cells were then fixed and analyzed by immunofluorescence microscopy. Although CT105-2HA did not show a perfect colocalization with either Lyn11-mEGFP or with phalloidin-stained cortical actin (Fig. 4A), this showed that at 30 and 40 h p.i., CT105-2HA localizes at the host cell plasma membrane.
Figure 4.
CT105 localizes at plasma membrane in C. trachomatis-infected cells. HeLa cells were infected by C. trachomatis L2/434 encoding pCT105-2HA. (A) Cells were also transfected with a plasmid encoding the plasma membrane marker Lyn11-mEGFP (green). At 30 or 40 h p.i., cells were fixed with 4% (w/v) PFA, stained with phalloidin (blue), and immunolabeled with antibodies against HA (red) and C. trachomatis MOMP (gray), and appropriate fluorophore-conjugated secondary antibodies. Cells were imaged by confocal fluorescence microscopy and images correspond to single z sections. In the area delimited by a white square (left-side panels) images were zoomed (right-side panels). Scale bars, 10 µm. (B) Infected cells were fixed with 4% (w/v) PFA at 16, 20, 30 or 40 h p.i. and immunolabeled with antibodies against HA, C. trachomatis MOMP, and GM130, and appropriate fluorophore-conjugated secondary antibodies. Fluorescence microscopy was then used to enumerate cells showing CT105-2HA only at the Golgi, only at the plasma membrane (PM), or both at the Golgi and at the plasma membrane (Golgi + PM). Data are mean ± standard error of the mean (SEM) of three independent experiments (n ≥ 25). (C) At 20 h p.i., infected cells were treated with DMSO, 1 µg/ml nocodazole, 2 µM cytochalasin D or 500 nM latrunculin B. At 30 h p.i., the cells were fixed with 4% (w/v) PFA and labeled with immunolabeled with antibodies against HA, C. trachomatis MOMP, and appropriate fluorophore-conjugated secondary antibodies. Fluorescence microscopy was used to enumerate cells showing CT105-2HA at the periphery of the cell/plasma membrane. Data are mean ± SEM of three independent experiments (n = 50). P-values were obtained by one-way ANOVA and Bonferroni post-test analyses; *statistical significant (P < 0.05); ns, not significant.
The number of infected cells showing CT105-2HA outside of the inclusion and at the Golgi or at the plasma membrane were then enumerated (Fig. 4B), which confirmed that in C. trachomatis-infected cells, initial delivery of CT105-2HA into the host cell cytoplasm (first detected at 16 h p.i.) results in its accumulation at the Golgi region, but as infection progresses this localization of the protein becomes less frequent and CT105-2HA accumulates more often at the periphery of the cell. These observations were recapitulated by the analysis of HeLa cells infected by C. trachomatis L2/434 harboring pTet-CT105-2HA (Supplementary Fig. S8). To test if CT105-2HA was transported from the Golgi to the plasma membrane in vesicles associated with the microtubule and/or actin cytoskeleton, HeLa cells were infected with C. trachomatis harboring pCT105-2HA. At 20 h p.i., when CT105-2HA mostly localizes at the Golgi region (Fig. 4B), the cells were incubated in the presence of DMSO solvent as a control, cytochalasin D or latrunculin B (to inhibit actin polymerization by different mechanisms), or nocodazole (to depolymerize microtubules) (Fig. 4C). The cells were then fixed at 30 h p.i., when CT105-2HA mostly localizes at the plasma membrane (Fig. 4B), and analyzed by immunofluorescence microscopy. We observed a statistically significant difference in the localization of CT105-2HA at the cell periphery when microfilaments were disrupted with cytochalasin D (Fig. 4C). However, the small difference relative to DMSO-treated cells, and the absence of a significantly different effect upon disruption of the microfilaments with latrunculin B (Fig. 4C), suggests this should have no biological significance. Furthermore, disruption of the microtubule network also did not significantly affect the localization of CT105-2HA at the cell periphery (Fig. 4C). Therefore, CT105-2HA changes its predominant localization during infection of host cells by C. trachomatis from the Golgi to the plasma membrane, but this is independent of intact host cell microfilaments and microtubules.
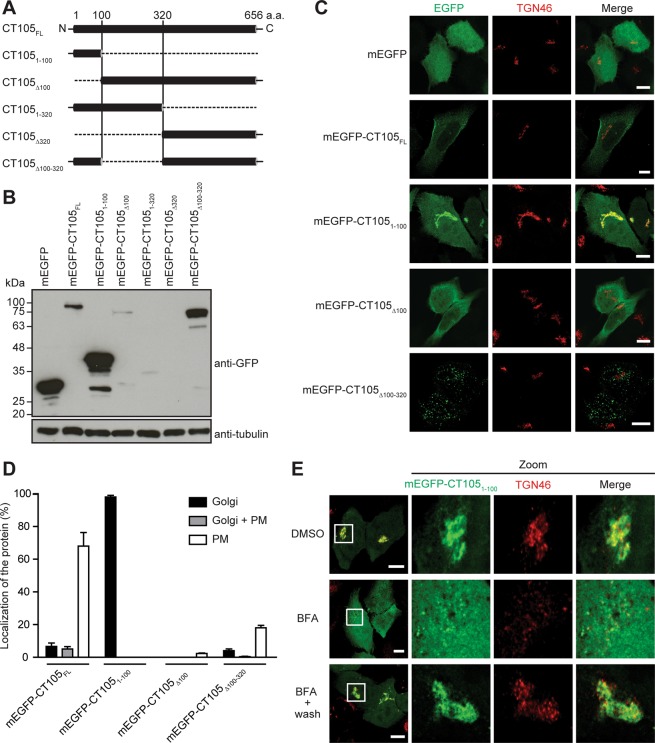
The first 100 amino acid residues of CT105 are sufficient to target the protein to the Golgi in uninfected mammalian cells
To map the regions of CT105 that could determine its different localizations within the cytoplasm of mammalian cells, HeLa cells were first transfected for 24 h with plasmids encoding full length CT105 fused to the C- or N-termini of monomeric EGFP (mEGFP-CT105FL and CT105FL-mEGFP, respectively) or with C-terminal 2HA-epitope tag. Immunoblotting of whole cell extracts confirmed the production of proteins of the predicted molecular mass (Supplementary Fig. S9). Furthermore, fluorescence microscopy showed that mEGFP-CT105FL, CT105FL-mEGFP, or CT105-2HA localized predominantly at the periphery of the cell (Supplementary Fig. S9), recapitulating a main localization of CT105-2HA in cells infected by C. trachomatis for 30 h or 40 h (Fig. 4B). Next, HeLa cells were transfected with plasmids encoding mEGFP-CT105FL (predicted molecular mass of 96 kDa) and different truncated versions of CT105 fused to the C-terminus of mEGFP: mEGFP-CT1051–100 (38 kDa), mEGFP-CT105Δ100 (86 kDa) mEGFP-CT1051–320 (60 kDa), mEGFP-CT105Δ320 (65 kDa), or mEGFP-CT105Δ100–320 (74 kDa) (Fig. 5A). As the primary structure of CT105 revealed no predicted transmembrane domains or other obvious targeting motifs, these truncations were designed only considering the predicted secondary structure of CT105 [deduced using JPred436]. Immunoblotting of whole cell extracts confirmed the production of mEGFP-CT105FL, mEGFP-CT1051–100, mEGFP-CT105Δ100, or mEGFP-CT105Δ100–320 proteins of the predicted molecular mass (Fig. 5B). The production of mEGFP-CT1051–320 and mEGFP-CT105Δ320 proteins of the predicted molecular mass was not detected (Fig. 5B), and they were not further analyzed. HeLa cells ectopically expressing mEGFP-CT105FL, mEGFP-CT1051–100, mEGFP-CT105Δ100, or mEGFP-CT105Δ100–320 were fixed and analyzed by immunofluorescence microscopy for localization of the fusion protein at the cell periphery (as an indication of plasma membrane localization) and at the Golgi. The mEGFP-CT105FL protein localized only at the cell periphery in 68 ± 8% of the transfected cells, and only at the Golgi in 7 ± 2% of the transfected cells (Fig. 5C,D). However, the fluorescent signal was generally weak in the Golgi region (Fig. 5C). Strikingly, mEGFP-CT1051–100 localized at the Golgi region in almost all transfected cells (98 ± 1%) and was not detected at the cell periphery (Fig. 5C,D). In contrast, mEGFP-CT105Δ100 was mostly cytosolic and was never seen at the Golgi complex and only seldom (2 ± 0%) at the cell periphery (Fig. 5C,D). mEGFP-CT105Δ100–320 showed a punctate/vesicular appearance with no marked localization at the Golgi (4 ± 1%) or cell periphery (18 ± 1%), particularly when compared with mEGFP-CT105Δ100 or mEGFP-CT105FL (Fig. 5C,D).
Figure 5.
The first 100 amino acids of CT105 contain a Golgi-targeting region. HeLa cells were transfected for 24 h with plasmids encoding monomeric EGFP (mEGFP) or different regions of CT105 fused to the C-terminus of mEGFP (CT105-mEGFP proteins), as indicated. (A) Schematic representation of the mEGFP-CT105 proteins expressed and analyzed. (B) Total extracts of transfected cells were analyzed by immunoblotting with antibodies against GFP and α-tubulin (HeLa loading control) using SuperSignal West Pico detection kit (Thermo Fisher Scientific). (C) Transfected cells were fixed with 4% (w/v) PFA, labeled with antibodies against TGN46 (red), and the appropriate fluorophore-conjugated secondary antibody, and imaged by confocal fluorescence microscopy. Images correspond to single z sections. Scale bars, 10 µm. (D) Cells immunolabeled as described in C were enumerated by fluorescence microscopy for localization of mEGFP-CT105 only at the Golgi, only at the plasma membrane (PM), or both at the Golgi and plasma membrane (Golgi + PM). Data are mean ± standard error of the mean of three independent experiments (n = 100). (E) Cells were treated for 1 h with DMSO or 1 µg/ml BFA. Then, the cells were either fixed with 4% (w/v) PFA (upper and middle panels) or washed with complete medium lacking BFA and incubated for an additional 1 h and then fixed with 4% (w/v) PFA (lower panels). The fixed cells were immunolabeled with antibodies against TGN46 (red), and the appropriate fluorophore-conjugated secondary antibody, and examined by confocal immunofluorescence microscopy. Images correspond to single z sections. In the area delimited by a white square (left-side panels) images were zoomed (right-side panels). Scale bars, 10 µm.
To confirm the association of mEGFP-CT1051–100 with the Golgi, and to analyze its localization within the Golgi complex, HeLa cells that had been transfected for 24 h were either incubated for 1 h with BFA or DMSO solvent, and then fixed. In addition, the transfected cells were incubated for 1 h with BFA, washed out from the BFA, incubated for 1 h, and fixed. Immunofluorescence microscopy revealed that in the control DMSO-treated cells, mEGFP-CT1051–100 localized at the Golgi region, and showed significant colocalization with TGN46 (Fig. 5E) but not with GM130 (Supplementary Fig. S9). BFA-induced Golgi fragmentation resulted in near complete dispersion of the fluorescence signal of mEGFP-CT1051–100 (Figs 5E and S9). However, BFA-induced Golgi fragmentation followed by BFA wash out revealed again a compact Golgi complex and mEGFP-CT1051–100 in that region mostly colocalizing with TGN46 (Fig. 5E) but not with GM130 (Supplementary Fig. S9).
In summary, ectopically expressed full-length CT105 (mEGFP-CT105FL, CT105FL-mEGFP, and CT105-2HA) localized at the cell periphery and mEGFP-CT1051–100 at the Golgi, generally recapitulating the localization of CT105-2HA in infected cells. As mEGFP-CT1051–100 showed a striking association with the TGN in uninfected cells, this indicated that the first 100 amino acids of CT105 contain a Golgi-targeting region.
CT105 is not essential for intracellular multiplication of C. trachomatis in infected tissue culture cells
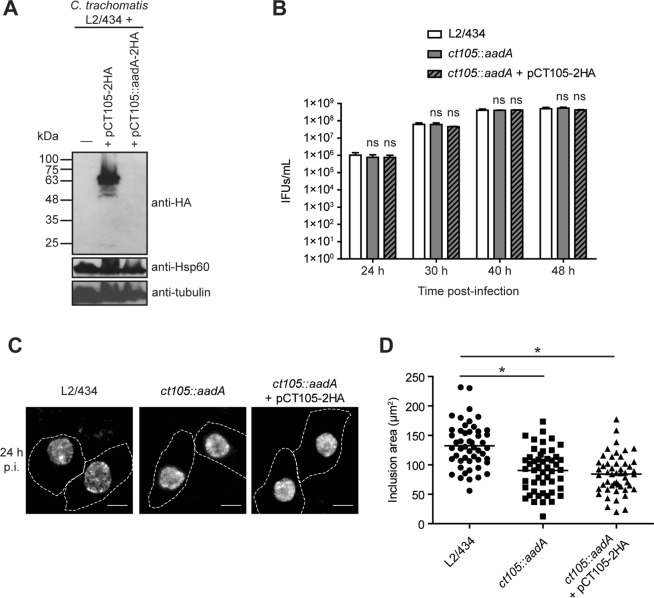
To examine the importance of CT105 during infection of tissue culture cells by C. trachomatis, a L2/434-derived strain where the ct105 gene was inactivated by an insertion between nucleotides 261 and 262 of a modified group II intron containing a spectinomycin-resistance gene (aadA) was constructed (Supplementary Fig. S10). To confirm that insertion of the intron prevented production of CT105, in the absence of a specific anti-CT105 antibody, a C. trachomatis strain harboring a plasmid (pCT105::aadA-2HA) was constructed with the ct105::aadA mutant allele and the sequence encoding 2HA fused to the 3′ end of ct105. HeLa cells were then infected for 40 h with C. trachomatis L2/434 harboring either pCT105-2HA or pCT105::aadA-2HA and analysed by immunoblotting. This confirmed that intron insertion within ct105-2HA prevented production of a protein detectable with anti-HA antibodies (Fig. 6A).
Figure 6.
A C. trachomatis ct105::aadA insertional mutant is not defective for intracellular growth in tissue culture cells. (A,C) trachomatis ct105::aadA insertional mutant was generated in the strain L2/434 by the targeted insertion of a modified group II intron carrying a spectinomycin-resistance gene. (A) Hela cells were infected by C. trachomatis L2/434 harboring a plasmid encoding CT105-2HA or by an identical plasmid carrying a ct105::aadA-2HA mutant allele. Whole cell extracts were analyzed by immunoblotting with antibodies against HA, C. trachomatis Hsp60 (bacterial loading control) and α-tubulin (HeLa loading control) using SuperSignal West Pico detection kit (Thermo Fisher Scientific) for Hsp60 and α-tubulin, or SuperSignal West Femto detection kit (Thermo Fisher Scientific) for HA. (B) HeLa cells were infected for 24, 30, or 40 h by C. trachomatis L2/434 or ct105::aadA mutant at the multiplicity of infection of 1, and recoverable inclusion forming units (IFUs) were enumerated. Data ± standard error of the mean of 3 independent experiments. For each time-point, P-values were obtained by one-way ANOVA and Dunnett post-test analyses relative to the L2/434 parental strain; ns, not significant (P ≥ 0.05). (C) Hela cells were infected for 24 h by C. trachomatis L2/434, ct105::aadA mutant, or ct105:aadA harboring pCT105-2HA. Cells were fixed with methanol, labeled with goat anti-C. trachomatis FITC-conjugated antibody and imaged by fluorescence microscopy. Scale bars, 10 µm. (D) The inclusion area was measured (from images as those depicted in C) for 50 particles randomly chosen from independent images using Fiji software. P-values were obtained by one-way ANOVA and Dunnett post-test analyses; *statistical significant (P < 0.05).
The intracellular growth of C. trachomatis L2/434 (parental strain), ct105::aadA mutant, and ct105::aadA mutant harboring CT105-2HA (complemented strain) was then monitored by quantifying the production of infectious progeny during the chlamydial developmental cycle. Based on this, no significant differences between the three strains were detected (Fig. 6B). However, when HeLa cells infected by the same strains for 24 h were fixed and analysed by immunofluorescence microscopy, this revealed that the inclusion size was smaller in the mutant and complemented strains by comparison to the parental strain (Fig. 6C,D). Because this difference in inclusion size between the mutant and the parental strain was also observed in the complemented strain, the defect is likely not due to the absence of CT105. Probably, intron insertion within ct105 might have caused a polar effect in the neighbouring genes. In summary, this showed that CT105 is not required for C. trachomatis growth in tissue culture cells. In addition, considering the localization of CteG at the Golgi, we analysed if its morphology was altered in HeLa cells infected by the ct105::aadA mutant, by comparison to cells infected by the parental L2/434 strain. However, significant differences in Golgi morphology were not detected (Supplementary Fig. S11).
CT105 induces a vacuolar protein sorting defect when ectopically expressed in S. cerevisiae
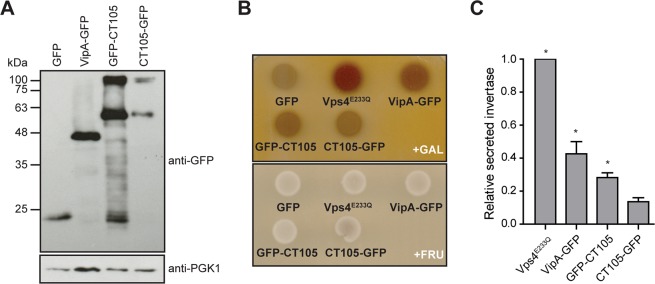
Given the localization of CT105 at the Golgi and plasma membrane in both infected and transfected mammalian cells, we hypothesized that the protein could interfere with eukaryotic vesicular trafficking. To test this, we asked if CT105 could cause a vacuolar protein sorting (VPS) defect in S. cerevisiae37, a eukaryotic model organism that has been often used to study bacterial effector proteins38. The VPS assay consists in a yeast reporter strain NSY01 (Supplementary Table S2) that produces a hybrid protein composed of carboxypeptidase Y and Invertase (CPY-Inv), which normally travels to the yeast vacuole but goes to the cell surface if trafficking is disrupted. As strain NSY01 does not produce endogenous Invertase, an enzyme that hydrolyzes sucrose into glucose and fructose, normal (Vps+) or aberrant (Vps−) trafficking of CPY-Inv can be scored by using an agar overlay solution indicating glucose production at the cell surface by formation of a brown precipitate (Vps+, white colonies; Vps−, brown colonies)37.
Strain NSY01 was transformed with yeast expression plasmids encoding either CT105 fused to the N-terminus or C-terminus of GFP (CT105-GFP or GFP-CT105, respectively) under the control of a galactose inducible promoter (Fig. 7A). The first controls used were a NSY01 derivative strain that expressed only GFP (Fig. 7A), which leads to a Vps+ phenotype39, and NSY01 derivative strains expressing a dominant-negative form of the yeast ATPase Vps4 (Vps4E233Q)40 or the Legionella pneumophila effector protein VipA (Fig. 7A), both known to cause a Vps− phenotype37,39. In the qualitative colorimetric enzymatic assay in solid media, it was consistently observed that GFP-CT105 or CT105-GFP induced a Vps− phenotype (Fig. 7B). In addition, quantitative analyses in liquid media were performed and the amounts of secreted and total invertase were assessed for each strain. In this assay, both GFP-CT105 and CT105-GFP caused an increase in the levels of secreted invertase (relative to those of the control strain expressing GFP), but the difference was only statistically significant for GFP-CT105 (Fig. 7C). The C-terminal half of CT105 (amino acid residues 351 to 656), but not its N-terminal part (amino acid residues 1-443), was sufficient to induce a VPS defect in S. cerevisiae (Supplementary Fig. S12), indicating that the VPS-inhibitory activity of CT105 depends on its C-terminal region. Moreover, production in yeast of a fusion to GFP of another C. trachomatis type III secreted protein (CT14227) did not induce a VPS defect (Supplementary Fig. S12). Finally, production of GFP-CT105 or CT105-GFP caused no toxicity in S. cerevisiae NSY01 (Supplementary Fig. S12), indicating that the induction of the VPS defect by CT105 was not a consequence of an overall impact on yeast physiology. Thus, ectopic expression of CT105 in S. cerevisiae disrupted vesicular trafficking to the yeast vacuole.
Figure 7.
CT105 induces a vacuolar protein sorting defect in S. cerevisiae. S. cerevisiae reporter strain NSY01 producing CPY-invertase was transformed with plasmids encoding GFP (pKS84), VipA-GFP, GFP-CT105, or CT105-GFP, where their expression can be induced by galactose. A NSY01 derivative strain encoding a dominant-negative form of the yeast ATPase Vps4 (Vps4E233Q) was also used. Yeast strains are listed in Supplementary Table S2. (A) Whole cell extracts of S. cerevisiae NSY01 producing the indicated proteins were analyzed by immunoblotting with antibodies against GFP and PGK1 (yeast loading control), using SuperSignal West Pico detection kit (Thermo Fisher Scientific). (B) S. cerevisiae NSY01 strains encoding the indicated proteins were grown in solid medium in the presence of galactose (inducing conditions, +GAL) or in the presence of fructose (non-inducing conditions; +FRU). The vacuolar protein sorting (VPS) phenotype was analyzed using a sucrose overlay to assess activity of secreted invertase. A vacuolar protein sorting defect (VPS−) leads to the formation of a brown precipitate. (C) S. cerevisiae NSY01 strains encoding the indicated proteins were grown in liquid medium in the presence of galactose (inducing conditions) and the relative activity of secreted invertase was quantified (see Experimental Procedures). Data are mean ± standard error of the mean of five independent experiments. P-values were obtained by one-way ANOVA and Dunnett post-test analyses relative to GFP (0% of relative secreted invertase; not shown); *statistical significant (P < 0.05); ns, not significant.
Discussion
We found that the C. trachomatis CT105 protein is delivered into the cytoplasm of infected host cells. At distinct times of the chlamydial developmental cycle, CT105 accumulates at the Golgi complex (16–30 h p.i.) and/or at the host cell plasma membrane (30–40 h p.i.). Because CT105 can be secreted by the T3S system of Y. enterocolitica27 and can interfere with vesicular trafficking in S. cerevisiae (Figs 7 and S12), this indicates that CT105 is a newly identified C. trachomatis T3S effector. However, thus far, we have no direct evidence for the ability of CT105 to interfere with vesicular trafficking in infected human cells and this must be analyzed directly in future studies. Overall, this work expands the portfolio of known chlamydial effector proteins and reveals the first Chlamydia protein that associates with the Golgi complex. Moreover, it illustrates that a single C. trachomatis effector can have distinct subcellular localizations, and possibly different functions, during the chlamydial developmental cycle. Such dual localization during infection as a likely means to diversify effector function is evocative of the Salmonella SopB effector, which initially localizes at the host cell membrane to mediate bacterial invasion but is then redirected to the Salmonella-containing vacuole to modulate bacterial intracellular growth41.
In addition to CT105, the C. trachomatis effector CT867, a deubiquitinase enzyme known as ChlaDub216 or Cdu217, has been detected at the inclusion membrane and at the host cell plasma membrane at distinct times of infection22. Such localization of CT105 and CT867/ChlaDub2/Cdu2 later in infection might indicate a function in host cell exit. It is unknown how CT105 and CT867/ChlaDub2/Cdu2 are differentially directed to different host cell localizations. In the case of CT105, it is unlikely that vesicular trafficking is involved as disruption of microfilaments and microtubules did not affect its localization at the plasma membrane in infected cells. We envision that host cytosolic CT105 is specifically directed to different subcellular localizations by covalent modifications it might be subjected to and/or by changes in the lipid and/or protein composition of host cellular membranes.
Ectopic expression of CT105 in yeast inhibited protein transport to the vacuole. While it remains to be directly tested, this observation suggested a potential capacity of CT105 to interfere with vesicular trafficking in infected cells. An appealing possibility for the function of CT105 would be that it contributes to nutrient acquisition by the inclusion and/or avoidance of fusion with hydrolytic compartments. However, the C. trachomatis ct105::aadA mutant did not reveal an obvious intracellular growth defect, which is not uncommon amongst C. trachomatis effector gene mutants characterized thus far11,42,43 and might reflect redundancy in effector function or inexistence of adequate infection models7. Furthermore, a putative activity of CT105 on vesicular trafficking in infected cells could also affect cytokine secretion44 or immune signaling45, which would not have an impact on chlamydial intracellular growth in tissue culture cells. Although the ct105 mutant revealed smaller inclusions relative to the parental strain (suggesting a possible defect in vacuolar membrane expansion), this could not be restored by reintroduction of ct105 in trans. The exact reasons for this need to be further analyzed, but the inability to complement intracellular growth-like phenotypes in intron insertional mutants of C. trachomatis was also recently described for ct101/mrcA46, while other specific phenotypes of this mutant strain could be complemented46.
The precise spatial and temporal subcellular targeting of bacterial effector proteins during infection is an essential feature of their function47. In other bacterial pathogens at least three other effector proteins have been shown to localize at the Golgi in infected host cells. These are EspI/NleA from Escherichia coli48, GobX from L. pneumophila49, and SteD from Salmonella enterica50. EspI/NleA interferes with coatomer protein II complex function, inhibits host cell protein secretion51, and disrupts intestinal tight junctions52. In addition, EspI/NleA inhibits NLRP3 inflammasome activation53. GobX displays E3 ubiquitin ligase activity, but how this promotes L. pneumophila infection is unknown49. Finally, SteD interferes with the activity of a host E3 ubiquitin ligase, leading to a reduction of surface-localized major histocompatibility complex class II molecules and suppression of T cell activation50. This illustrates that no specific function can be predicted for CT105 based on its localization at the Golgi.
The first 100 amino acid residues of CT105 are sufficient for Golgi localization when the protein is ectopically expressed in Hela cells. However, this region does not display obvious motifs found in other relevant eukaryotic or bacterial proteins, and the exact molecular determinants of the Golgi localization of CT105 remain to be identified. Other relevant questions are whether the same determinants are responsible for Golgi localization in infected cells and which characteristics of the full-length CT105 protein enable its targeting to the plasma membrane. Other effector proteins that localize at the Golgi in infected cells use distinct mechanisms directing them to the organelle49,50,54,55. In the case of EspI/NleA, Golgi targeting is partially mediated by a Postsynaptic Density95/Disc Large/Zonula Occludens-1 (PDZ) domain on its C-terminus54. CT105 also possesses a putative PDZ domain56, but the relevant residues are not involved in its subcellular localization after ectopic expression in mammalian cells (data not shown).
CT105 displays a striking variability within C. trachomatis serovars, revealing LGV-specific genetic and transcriptomic traits. This suggests that CT105 could have a specific function related with the unique characteristics of infections by these strains, which, unlike ocular and urogenital C. trachomatis strains, can infect mononuclear phagocytes and disseminate into regional lymph nodes29,57. It also indicates that features of the infection by LGV strains should be an evolutionary pressure to maintain an active ct105 gene. Furthermore, among Chlamydia species, full-length orthologues of CT105 are only present in C. muridarum and C. suis, consistent with the fact that these species are the closer relatives of C. trachomatis3.
In summary, we identified CT105 as a T3S effector of C. trachomatis that localizes to the Golgi and plasma membrane in infected cells, and which can disrupt trafficking to the S. cerevisiae vacuole. Because CT105 is the first Chlamydia effector shown to associate with the Golgi complex, we propose to name it CteG (for C. trachomatis effector associated with the Golgi). The identification of CteG also leaves many questions to be addressed related to its function, subcellular targeting mechanisms, diversity, and specificity within C. trachomatis and among Chlamydia species.
Materials and Methods
Plasmids, primers, and DNA manipulation
The plasmids used in this work, their main characteristics and construction details, are described in Supplementary Table S3. The DNA primers used in their construction are listed in Supplementary Table S4. Plasmids were constructed and purified using standard molecular biology procedures. The accuracy of the nucleotide sequence of all the inserts in the constructed plasmids was confirmed by DNA sequencing. The backbone plasmids used in this work were: pSVP24719, a derivative of p2TK2-SW258, was used to generate C. trachomatis expression plasmids encoding proteins with a 2HA at their C-termini; pmEGFP-N1 and pmEGFP-C1 [both constructed from pEGFP-N1 (Clontech) and pEGFP-C1 (Clontech), respectively, where the gene encoding EGFP was replaced by a gene encoding mEGFP from pLAMP1-mGFP35], and pEF6/Myc-His A (Thermo Fisher Scientific), were used to generate mammalian transfection plasmids; pYES2-GFP59 and pKS8460, were used to generate yeast expression plasmids encoding fusions to the C- or N-terminus of GFP, respectively. Furthermore, pBOMB4-Tet-mCherry61 and Lyn11-FRB-mCherry62 were used to amplify the Ptet and the nucleotide sequence encoding the Lyn11 peptide, respectively.
Cells lines and transient transfection
HeLa 229 and Vero cells (from the European Collection of Cell Culture; ECACC) were maintained in high-glucose Dulbecco’s modified Eagle Medium (DMEM; Thermo Fisher Scientific) supplemented with heat-inactivated 10% (v/v) fetal bovine serum (FBS; Thermo Fisher Scientific) at 37 °C in a humidified atmosphere of 5% (v/v) CO2. Cells were checked for Mycoplasma by conventional PCR either using the Venor GeM Advance kit (Minerva Biolabs) or as described63. HeLa cells were transfected by using the jetPEI reagent (Polyplus-Transfection) according to the instructions of the manufacturer.
Bacterial strains and growth conditions
E. coli TOP10 (Thermo Fisher Scientific) or NEB 10β (New England Biolabs) were used for construction and purification of plasmids, and E. coli ER2925 (New England Biolabs) was used to amplify and purify plasmids for transformation of C. trachomatis. E. coli strains were routinely grown in liquid or solid lysogeny broth (LB) medium (NZYTech) with the appropriate antibiotics and supplements at 37 °C.
C. trachomatis prototype strains B/Har36, C/TW3, E/Bour, L2/434, and L3/404 (from ATCC) and clinical strains F/CS465-95 and L2b/CS19-08 (from the collection of the Portuguese National Institute of Health) were propagated in HeLa 229 cells using standard techniques64. Transformation of C. trachomatis was done essentially as described by Agaisse and Derré58. The optimal antibiotic concentrations to select transformants were 1 U/ml of penicillin G, or 250 µg/ml of spectinomycin. Once established, the transformed strains were cultured in the presence of 10 U/ml of penicillin, or 500 µg/ml of spectinomycin, and plaque purified using Vero cells, as described65. Infection of mammalian cells by C. trachomatis and quantification of infection progeny was done as previously described19. Infected cells were harvested at the indicated times post-infection and analyzed by immunoblotting or immunofluorescence microscopy.
Construction of a C. trachomatis ct105::aadA mutant strain
A C. trachomatis ct105::aadA mutant was generated using group II intron-based insertional mutagenesis, as previously described66,67. Briefly, intron-insertion sites in the C. trachomatis L2/434 ctl0360 gene (orthologue of ct105 in C. trachomatis strain D/UW3) were identified using the TargeTron algorithm (Sigma). Then, the intron in pDFTT3aadA67 (Supplementary Table S2) was retargeted for ctl0360 using standard molecular biology procedures and the DNA primers listed in Supplementary Table S4. The ctl0360 mutator plasmid (pDFTT296; Supplementary Table S3) was then used to transform C. trachomatis L2/434.
Yeast strains and invertase assays
S. cerevisiae strains (Supplementary Table S2) used in this work were grown as previously described37,39. For the invertase assays, S. cerevisiae expressing GFP fusion proteins were grown in plates with yeast nitrogen base uracil dropout (YNB-Ura) supplemented with 2% (w/v) fructose for 3 days, at 25 °C. Cells were then streaked on YNB-Ura with 2% (w/v) fructose (non-inducing media) or 2% (w/v) galactose (inducing media) and grown for 3 days at 25 °C. Assays for qualitative and quantitative detection of invertase enzymatic activity were performed as described37,68. Relative secreted invertase was normalized by setting GFP as the minimum value and Vps4E233Q as the maximum value.
Antibodies, fluorescent dyes, and drug treatments
The list of antibodies and fluorescent dyes used is described in Supplementary Materials and Methods. To induce Golgi fragmentation, HeLa 229 cells were incubated with 1 µg/ml of BFA (Sigma; stock solution at 5 mg/ml in DMSO) for 1 h and then either fixed immediately or washed with medium without BFA and incubated for 1 h before fixation. To disrupt microtubules, HeLa 229 cells were incubated in the presence of 1 µg/ml nocodazole (stock solution at 5 mg/ml in DMSO; Sigma). To disrupt the actin cytoskeleton, C. trachomatis-infected cells were incubated in the presence of 2 µM cytochalasin D (stock solution at 5 mg/ml in DMSO; Sigma) or 500 nM latrunculin B (stock solution at 1 mg/ml in DMSO; Sigma) in serum-free DMEM.
Immunoblotting
To harvest infected or transfected HeLa cells, they were washed once with phosphate-buffered saline (PBS) and then detached with TrypLE Express (Thermo Fisher Scientific) by incubation during 5 min at 37 °C in a 5% [v/v] CO2 atmosphere. The cells were then collected, pelleted by a brief centrifugation, washed 2 times with ice-cold PBS, and stored as a pellet at −20 °C until use. Prior to SDS-PAGE, the cells were thawed and resuspended in an appropriate volume of SDS-PAGE loading buffer. The proteins were further denatured by an incubation of 5 min at 100 °C, followed by addition of benzonase (Novagen) to destroy DNA and reduce the viscosity of the samples.
To prepare Chlamydia-enriched extracts, the infected HeLa cells were lysed by osmotic shock (15 min in sterile H2O). Lysates were centrifuged at 170 × g for 10 min at 4 °C. The supernatants were then centrifuged at 24,000 × g for 10 min at 4 °C and resulting pellets washed 2 times with ice-cold PBS. The bacteria-enriched pellets were resuspended in an appropriate volume of SDS-PAGE loading buffer and the proteins were further denatured by an incubation of 5 min at 100 °C.
To prepare yeast extracts, cells were grown for 3 days at 30 °C in YNB-Ura plates supplemented with 2% (w/v) fructose and then streaked into YNB-Ura supplemented with 2% (w/v) galactose. Cells corresponding to an optical density at 600 nm (OD600) of 2.5 were used for immunoblotting.
Samples were separated by 12% (v/v) SDS-PAGE and transferred onto 0.2 μm nitrocellulose membranes (Bio-Rad) using Trans-Blot Turbo Transfer System (BioRad). Immunoblot detection was done with SuperSignal West Pico Chemiluminescent Substrate (Thermo Fisher Scientific) or SuperSignal West Femto Maximum Sensitivity Substrate (Thermo Fisher Scientific) (as indicated in figure legends) and exposure to Amersham Hyperfilm ECL (GE Healthcare).
Immunofluorescence microscopy
Infected HeLa cells were fixed either in PBS containing 4% (w/v) paraformaldehyde (PFA) for 10 min at room temperature or in methanol (−20 °C) for 10 min, as indicated in figure legends. For immunostaining, the antibodies were diluted in PBS containing 10% (v/v) horse serum (when fixation was done with PFA, 0.1% (v/v) Triton X-100 was added to allow permeabilization of cells). After immunolabelling, the cells were consecutively washed with PBS and H2O. The coverslips were assembled using Aqua-poly/Mount (Polysciences) on microscopy glass slides, and the cells were examined by conventional fluorescence microscopy or by confocal microscopy. Images were processed and assembled using Fiji software69. Analysis of Golgi redistribution around the C. trachomatis inclusion was performed as previously described70.
RT-qPCR
To quantify the mRNA levels of ct105 during the developmental cycle of C. trachomatis strains B/Har36, C/TW3, E/Bour, F/CS465-95, L2/434, L2b/CS19-08 and L3/404, RT-qPCR measurements were performed as previously described71, using primers listed in Supplementary Table S4. To compare by RT-qPCR the mRNA levels of ct105 during the developmental cycle of C. trachomatis strains L2/434 or L2/434 harboring pCT105-2HA (pSVP264/pCT105-2HA; Supplementary Table S3), a 6-well plate seeded with HeLa 229 cells was inoculated by each strain at a multiplicity of infection of 50 and cells were harvested at the indicated times post infection by trypsinization. Total RNA was isolated using NZY Total RNA Isolation Kit (NZYtech). For each RNA sample, cDNA was generated using iScript cDNA Synthesis Kit (Bio-Rad) accordingly with the manufacturer’s instructions. RT-qPCR was performed using ct105 and 16S primers (Supplementary Table S4) and SsoFast EvaGreen Supermix (Bio-Rad). For each time point, quantitative PCR was performed on each cDNA sample (including an RT-negative sample) using Rotor Gene 6000 (Corbett Life Science). For normalization, ratios to the 16s rRNA transcript were obtained. Data analysis was carried out with Rotor-Gene 6000 software. Unless otherwise indicated, RT-qPCR results were based in three independent experiments.
Statistical analyses
Statistical analyses were done using GraphPad Prism, version 5.04 for Windows, GraphPad Software, San Diego California, USA (www.graphpad.com).
Supplementary information
Acknowledgements
We are grateful to Ana Luzia Trindade and to Catarina Milho for the construction of plasmids, and to Agathe Subtil and Guangming Zhong for the gift of antibodies. This work was supported by Fundação para a Ciência e a Tecnologia (FCT/MCTES) through grants PTDC/IMI-MIC/1300/2014 and PTDC/BIA-MIC/28503/2017, and by the Applied Molecular Biosciences Unit (UCIBIO), which is financed by national funds from FCT/MCTES (UID/Multi/04378/2019) and co-financed by the European Regional Development Fund (ERDF) under the PT2020 Partnership Agreement (POCI-01-0145-FEDER-007728). SVP was supported by PhD fellowship PD/BD/52210/2013 within the scope of the PhD program Molecular Biosciences (PD/00133/2012) funded by FCT/MCTES. ISP was supported by PhD fellowship SFRH/BD/129756/2017. Work by DJF was funded by grant R21AI115238 from the National Institutes of Health.
Author Contributions
S.V.P., J.P.G., D.J.F. and L.J.M. conceived the experiments. S.V.P., C.E.K., V.B., I.S.P. and L.J.M. conducted the experiments. S.V.P., V.B., I.S.P., J.P.G., D.J.F. and L.J.M. analyzed the results. S.V.P. and L.J.M. wrote the manuscript, which was revised by all authors.
Data Availability
All data generated or analyzed during this study are included in this published article (and its Supplementary Information files).
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-42647-3.
References
- 1.Taylor HR, Burton MJ, Haddad D, West S, Wright H. Trachoma. Lancet. 2014 doi: 10.1016/S0140-6736(13)62182-0. [DOI] [PubMed] [Google Scholar]
- 2.O’Connell CM, Ferone ME. Chlamydia trachomatis Genital Infections. Microb Cell. 2016;3:390–403. doi: 10.15698/mic2016.09.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sachse K, et al. Emendation of the family Chlamydiaceae: proposal of a single genus, Chlamydia, to include all currently recognized species. Syst Appl Microbiol. 2015;38:99–103. doi: 10.1016/j.syapm.2014.12.004. [DOI] [PubMed] [Google Scholar]
- 4.Elwell C, Mirrashidi K, Engel J. Chlamydia cell biology and pathogenesis. Nat Rev Microbiol. 2016;14:385–400. doi: 10.1038/nrmicro.2016.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Galan JE, Lara-Tejero M, Marlovits TC, Wagner S. Bacterial type III secretion systems: specialized nanomachines for protein delivery into target cells. Annu Rev Microbiol. 2014;68:415–438. doi: 10.1146/annurev-micro-092412-155725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mueller KE, Plano GV, Fields KA. New frontiers in type III secretion biology: the Chlamydia perspective. Infect Immun. 2014;82:2–9. doi: 10.1128/IAI.00917-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Galan JE. Common themes in the design and function of bacterial effectors. Cell Host Microbe. 2009;5:571–579. doi: 10.1016/j.chom.2009.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hicks SW, Galan JE. Exploitation of eukaryotic subcellular targeting mechanisms by bacterial effectors. Nat Rev Microbiol. 2013;11:316–326. doi: 10.1038/nrmicro3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rockey DD, Scidmore MA, Bannantine JP, Brown WJ. Proteins in the chlamydial inclusion membrane. Microbes Infect. 2002;4:333–340. doi: 10.1016/S1286-4579(02)01546-0. [DOI] [PubMed] [Google Scholar]
- 10.Li Z, et al. Characterization of fifty putative inclusion membrane proteins encoded in the Chlamydia trachomatis genome. Infect Immun. 2008;76:2746–2757. doi: 10.1128/IAI.00010-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McKuen, M. J., Mueller, K. E., Bae, Y. S. & Fields, K. A. Fluorescence-Reported Allelic Exchange Mutagenesis Reveals a Role for Chlamydia trachomatis TmeA in Invasion That Is Independent of Host AHNAK. Infect Immun85, 10.1128/IAI.00640-17 (2017). [DOI] [PMC free article] [PubMed]
- 12.Carpenter, V., Chen, Y. S., Dolat, L. & Valdivia, R. H. The Effector TepP Mediates Recruitment and Activation of Phosphoinositide 3-Kinase on Early Chlamydia trachomatis Vacuoles. mSphere2, 10.1128/mSphere.00207-17 (2017). [DOI] [PMC free article] [PubMed]
- 13.Vromman F, Perrinet S, Gehre L, Subtil A. The DUF582 Proteins of Chlamydia trachomatis Bind to Components of the ESCRT Machinery, Which Is Dispensable for Bacterial Growth In vitro. Frontiers in cellular and infection microbiology. 2016;6:123. doi: 10.3389/fcimb.2016.00123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thwaites TR, Pedrosa AT, Peacock TP, Carabeo RA. Vinculin Interacts with the Chlamydia Effector TarP Via a Tripartite Vinculin Binding Domain to Mediate Actin Recruitment and Assembly at the Plasma Membrane. Frontiers in cellular and infection microbiology. 2015;5:88. doi: 10.3389/fcimb.2015.00088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Parrett CJ, Lenoci RV, Nguyen B, Russell L, Jewett TJ. Targeted Disruption of Chlamydia trachomatis Invasion by in Trans Expression of Dominant Negative Tarp Effectors. Frontiers in cellular and infection microbiology. 2016;6:84. doi: 10.3389/fcimb.2016.00084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Misaghi S, et al. Chlamydia trachomatis-derived deubiquitinating enzymes in mammalian cells during infection. Mol Microbiol. 2006;61:142–150. doi: 10.1111/j.1365-2958.2006.05199.x. [DOI] [PubMed] [Google Scholar]
- 17.Fischer, A. et al. Chlamydia trachomatis-containing vacuole serves as deubiquitination platform to stabilize Mcl-1 and to interfere with host defense. eLife6, 10.7554/eLife.21465 (2017). [DOI] [PMC free article] [PubMed]
- 18.Gehre L, et al. Sequestration of host metabolism by an intracellular pathogen. eLife. 2016;5:e12552. doi: 10.7554/eLife.12552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.da Cunha M, Pais SV, Bugalhao JN, Mota LJ. The Chlamydia trachomatis type III secretion substrates CT142, CT143, and CT144 are secreted into the lumen of the inclusion. PLoS One. 2017;12:e0178856. doi: 10.1371/journal.pone.0178856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fling SP, et al. CD8+ T cells recognize an inclusion membrane-associated protein from the vacuolar pathogen Chlamydia trachomatis. Proc Natl Acad Sci USA. 2001;98:1160–1165. doi: 10.1073/pnas.98.3.1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fields KA, Hackstadt T. Evidence for the secretion of Chlamydia trachomatis CopN by a type III secretion mechanism. Mol Microbiol. 2000;38:1048–1060. doi: 10.1046/j.1365-2958.2000.02212.x. [DOI] [PubMed] [Google Scholar]
- 22.Wang, X., Hybiske, K. & Stephens, R. S. Direct visualization of the expression and localization of chlamydial effector proteins within infected host cells. Pathogens and disease76, 10.1093/femspd/fty011 (2018). [DOI] [PMC free article] [PubMed]
- 23.Pennini ME, Perrinet S, Dautry-Varsat A, Subtil A. Histone methylation by NUE, a novel nuclear effector of the intracellular pathogen Chlamydia trachomatis. PLoS Pathog. 2010;6:e1000995. doi: 10.1371/journal.ppat.1000995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Muschiol S, et al. Identification of a family of effectors secreted by the type III secretion system that are conserved in pathogenic Chlamydiae. Infect Immun. 2011;79:571–580. doi: 10.1128/IAI.00825-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hobolt-Pedersen AS, Christiansen G, Timmerman E, Gevaert K, Birkelund S. Identification of Chlamydia trachomatis CT621, a protein delivered through the type III secretion system to the host cell cytoplasm and nucleus. FEMS Immunol Med Microbiol. 2009;57:46–58. doi: 10.1111/j.1574-695X.2009.00581.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bullock HD, Hower S, Fields KA. Domain Analyses Reveal That Chlamydia trachomatis CT694 Protein Belongs to the Membrane-localized Family of Type III Effector Proteins. J Biol Chem. 2012;287:28078–28086. doi: 10.1074/jbc.M112.386904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.da Cunha M, et al. Identification of type III secretion substrates of Chlamydia trachomatis using Yersinia enterocolitica as a heterologous system. BMC Microbiol. 2014;14:40. doi: 10.1186/1471-2180-14-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pais SV, Milho C, Almeida F, Mota LJ. Identification of novel type III secretion chaperone-substrate complexes of Chlamydia trachomatis. PLoS One. 2013;8:e56292. doi: 10.1371/journal.pone.0056292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Borges V, Gomes JP. Deep comparative genomics among Chlamydia trachomatis lymphogranuloma venereum isolates highlights genes potentially involved in pathoadaptation. Infect Genet Evol. 2015;32:74–88. doi: 10.1016/j.meegid.2015.02.026. [DOI] [PubMed] [Google Scholar]
- 30.Albrecht M, Sharma CM, Reinhardt R, Vogel J, Rudel T. Deep sequencing-based discovery of the Chlamydia trachomatis transcriptome. Nucleic Acids Res. 2010;38:868–877. doi: 10.1093/nar/gkp1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Borges V., Nunes A., Ferreira R., Borrego M. J., Gomes J. P. Directional Evolution of Chlamydia trachomatis towards Niche-Specific Adaptation. Journal of Bacteriology. 2012;194(22):6143–6153. doi: 10.1128/JB.01291-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Heuer D, et al. Chlamydia causes fragmentation of the Golgi compartment to ensure reproduction. Nature. 2009;457:731–735. doi: 10.1038/nature07578. [DOI] [PubMed] [Google Scholar]
- 33.Lippincott-Schwartz J, Yuan LC, Bonifacino JS, Klausner RD. Rapid redistribution of Golgi proteins into the ER in cells treated with brefeldin A: evidence for membrane cycling from Golgi to ER. Cell. 1989;56:801–813. doi: 10.1016/0092-8674(89)90685-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Inoue T, Heo WD, Grimley JS, Wandless TJ, Meyer T. An inducible translocation strategy to rapidly activate and inhibit small GTPase signaling pathways. Nat Methods. 2005;2:415–418. doi: 10.1038/nmeth763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Falcon-Perez JM, Nazarian R, Sabatti C, Dell’Angelica EC. Distribution and dynamics of Lamp1-containing endocytic organelles in fibroblasts deficient in BLOC-3. J Cell Sci. 2005;118:5243–5255. doi: 10.1242/jcs.02633. [DOI] [PubMed] [Google Scholar]
- 36.Drozdetskiy A, Cole C, Procter J, Barton GJ. JPred4: a protein secondary structure prediction server. Nucleic Acids Res. 2015;43:W389–394. doi: 10.1093/nar/gkv332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shohdy N, Efe JA, Emr SD, Shuman HA. Pathogen effector protein screening in yeast identifies Legionella factors that interfere with membrane trafficking. Proc Natl Acad Sci USA. 2005;102:4866–4871. doi: 10.1073/pnas.0501315102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Curak J, Rohde J, Stagljar I. Yeast as a tool to study bacterial effectors. Curr Opin Microbiol. 2009;12:18–23. doi: 10.1016/j.mib.2008.11.004. [DOI] [PubMed] [Google Scholar]
- 39.Franco IS, Shohdy N, Shuman HA. The Legionella pneumophila Effector VipA Is an Actin Nucleator That Alters Host Cell Organelle Trafficking. PLoS Pathog. 2012;8:e1002546. doi: 10.1371/journal.ppat.1002546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Babst M, Wendland B, Estepa EJ, Emr SD. The Vps4p AAA ATPase regulates membrane association of a Vps protein complex required for normal endosome function. EMBO J. 1998;17:2982–2993. doi: 10.1093/emboj/17.11.2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Patel JC, Hueffer K, Lam TT, Galan JE. Diversification of a Salmonella virulence protein function by ubiquitin-dependent differential localization. Cell. 2009;137:283–294. doi: 10.1016/j.cell.2009.01.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weber MM, et al. Absence of Specific Chlamydia trachomatis Inclusion Membrane Proteins Triggers Premature Inclusion Membrane Lysis and Host Cell Death. Cell reports. 2017;19:1406–1417. doi: 10.1016/j.celrep.2017.04.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stanhope R, Flora E, Bayne C, Derre I. IncV, a FFAT motif-containing Chlamydia protein, tethers the endoplasmic reticulum to the pathogen-containing vacuole. Proc Natl Acad Sci USA. 2017;114:12039–12044. doi: 10.1073/pnas.1709060114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Murray RZ, Stow JL. Cytokine Secretion in Macrophages: SNAREs, Rabs, and Membrane Trafficking. Front Immunol. 2014;5:538. doi: 10.3389/fimmu.2014.00538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gleeson PA. The role of endosomes in innate and adaptive immunity. Semin Cell Dev Biol. 2014;31:64–72. doi: 10.1016/j.semcdb.2014.03.002. [DOI] [PubMed] [Google Scholar]
- 46.Nguyen PH, Lutter EI, Hackstadt T. Chlamydia trachomatis inclusion membrane protein MrcA interacts with the inositol 1,4,5-trisphosphate receptor type 3 (ITPR3) to regulate extrusion formation. PLoS Pathog. 2018;14:e1006911. doi: 10.1371/journal.ppat.1006911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hicks SW, Charron G, Hang HC, Galan JE. Subcellular targeting of Salmonella virulence proteins by host-mediated S-palmitoylation. Cell Host Microbe. 2011;10:9–20. doi: 10.1016/j.chom.2011.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gruenheid S, et al. Identification and characterization of NleA, a non-LEE-encoded type III translocated virulence factor of enterohaemorrhagic Escherichia coli O157:H7. Mol Microbiol. 2004;51:1233–1249. doi: 10.1046/j.1365-2958.2003.03911.x. [DOI] [PubMed] [Google Scholar]
- 49.Lin YH, et al. Host Cell-catalyzed S-Palmitoylation Mediates Golgi Targeting of the Legionella Ubiquitin Ligase GobX. J Biol Chem. 2015;290:25766–25781. doi: 10.1074/jbc.M115.637397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bayer-Santos E, et al. The Salmonella Effector SteD Mediates MARCH8-Dependent Ubiquitination of MHC II Molecules and Inhibits T Cell Activation. Cell Host Microbe. 2016;20:584–595. doi: 10.1016/j.chom.2016.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim J, et al. The bacterial virulence factor NleA inhibits cellular protein secretion by disrupting mammalian COPII function. Cell Host Microbe. 2007;2:160–171. doi: 10.1016/j.chom.2007.07.010. [DOI] [PubMed] [Google Scholar]
- 52.Thanabalasuriar A, et al. The bacterial virulence factor NleA is required for the disruption of intestinal tight junctions by enteropathogenic Escherichia coli. Cell Microbiol. 2010;12:31–41. doi: 10.1111/j.1462-5822.2009.01376.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yen H, Sugimoto N, Tobe T. Enteropathogenic Escherichia coli Uses NleA to Inhibit NLRP3 Inflammasome Activation. PLoS Pathog. 2015;11:e1005121. doi: 10.1371/journal.ppat.1005121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lee SF, et al. A C-terminal class I PDZ binding motif of EspI/NleA modulates the virulence of attaching and effacing Escherichia coli and Citrobacter rodentium. Cell Microbiol. 2008;10:499–513. doi: 10.1111/j.1462-5822.2007.01065.x. [DOI] [PubMed] [Google Scholar]
- 55.Thanabalasuriar A, et al. Sec. 24 interaction is essential for localization and virulence-associated function of the bacterial effector protein NleA. Cell Microbiol. 2012;14:1206–1218. doi: 10.1111/j.1462-5822.2012.01789.x. [DOI] [PubMed] [Google Scholar]
- 56.Yi CR, et al. Systematic analysis of bacterial effector-postsynaptic density 95/disc large/zonula occludens-1 (PDZ) domain interactions demonstrates Shigella OspE protein promotes protein kinase C activation via PDLIM proteins. J Biol Chem. 2014;289:30101–30113. doi: 10.1074/jbc.M114.595868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Thomson NR, et al. Chlamydia trachomatis: genome sequence analysis of lymphogranuloma venereum isolates. Genome Res. 2008;18:161–171. doi: 10.1101/gr.7020108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Agaisse H, Derre I. A C. trachomatis cloning vector and the generation of C. trachomatis strains expressing fluorescent proteins under the control of a C. trachomatis promoter. PLoS One. 2013;8:e57090. doi: 10.1371/journal.pone.0057090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rodriguez-Escudero I, et al. Reconstitution of the mammalian PI3K/PTEN/Akt pathway in yeast. Biochem J. 2005;390:613–623. doi: 10.1042/BJ20050574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.de Felipe KS, et al. Legionella eukaryotic-like type IV substrates interfere with organelle trafficking. PloS Pathog. 2008;4:e1000117. doi: 10.1371/journal.ppat.1000117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bauler LD, Hackstadt T. Expression and targeting of secreted proteins from Chlamydia trachomatis. J Bacteriol. 2014;196:1325–1334. doi: 10.1128/JB.01290-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hammond GR, et al. PI4P and PI(4,5)P2 are essential but independent lipid determinants of membrane identity. Science. 2012;337:727–730. doi: 10.1126/science.1222483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Uphoff CC, Drexler HG. Detecting mycoplasma contamination in cell cultures by polymerase chain reaction. Methods Mol Biol. 2011;731:93–103. doi: 10.1007/978-1-61779-080-5_8. [DOI] [PubMed] [Google Scholar]
- 64.Scidmore MA. Cultivation and Laboratory Maintenance of Chlamydia trachomatis. Curr Protoc Microbiol Chapter. 2005;11(Unit 11A):11. doi: 10.1002/9780471729259.mc11a01s00. [DOI] [PubMed] [Google Scholar]
- 65.Nguyen, B. D. & Valdivia, R. H. Forward genetic approaches in Chlamydia trachomatis. Journal of visualized experiments: JoVE, e50636, 10.3791/50636 (2013). [DOI] [PMC free article] [PubMed]
- 66.Johnson CM, Fisher DJ. Site-specific, insertional inactivation of incA in Chlamydia trachomatis using a group II intron. PloS One. 2013;8:e83989. doi: 10.1371/journal.pone.0083989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Key CE, Fisher DJ. Use of Group II Intron Technology for Targeted Mutagenesis in Chlamydia trachomatis. Methods Mol Biol. 2017;1498:163–177. doi: 10.1007/978-1-4939-6472-7_11. [DOI] [PubMed] [Google Scholar]
- 68.Darsow T, Odorizzi G, Emr SD. Invertase fusion proteins for analysis of protein trafficking in yeast. Methods Enzymol. 2000;327:95–106. doi: 10.1016/S0076-6879(00)27270-4. [DOI] [PubMed] [Google Scholar]
- 69.Schindelin J, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wesolowski, J. et al. Chlamydia Hijacks ARF GTPases To Coordinate Microtubule Posttranslational Modifications and Golgi Complex Positioning. mBio8, 10.1128/mBio.02280-16 (2017). [DOI] [PMC free article] [PubMed]
- 71.Almeida F, et al. Polymorphisms in Inc Proteins and Differential Expression of inc Genes among Chlamydia trachomatis Strains Correlate with Invasiveness and Tropism of Lymphogranuloma Venereum Isolates. J Bacteriol. 2012;194:6574–6585. doi: 10.1128/JB.01428-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article (and its Supplementary Information files).