Abstract
Background
Magnesium (Mg) deficiency causes physiological and molecular responses, already dissected in several plant species. The study of these responses among genotypes showing a different tolerance to the Mg shortage can allow identifying the mechanisms underlying the resistance to this nutritional disorder. To this aim, we compared the physiological and molecular responses (e.g. changes in root metabolome and transcriptome) of two grapevine rootstocks exhibiting, in field, different behaviors with respect to Mg shortage (1103P, tolerant and SO4 susceptible).
Results
The two grapevine rootstocks confirmed, in a controlled growing system, their behavior in relation to the tolerance to Mg deficiency. Differences in metabolite and transcriptional profiles between the roots of the two genotypes were mainly linked to antioxidative compounds and the cell wall constituents. In addition, differences in secondary metabolism, in term of both metabolites (e.g. alkaloids, terpenoids and phenylpropanoids) and transcripts, assessed between 1103P and SO4 suggest a different behavior in relation to stress responses particularly at early stages of Mg deficiency.
Conclusions
Our results suggested that the higher ability of 1103P to tolerate Mg shortage is mainly linked to its capability of coping, faster and more efficiently, with the oxidative stress condition caused by the nutritional disorder.
Electronic supplementary material
The online version of this article (10.1186/s12870-019-1726-x) contains supplementary material, which is available to authorized users.
Keywords: Magnesium deficiency, Grapevine rootstocks, Metabolome and transcriptome
Background
Magnesium (Mg) is an essential macronutrient playing a key role in plant growth and development. It is the most abundant free divalent cation in the cytosol, and it is involved into chlorophyll biosynthesis [1]; Mg is also an essential cofactor for the activity of several enzymes, as for instance RNA polymerases, ATPases, protein kinases, phosphatases, glutathione synthase and carboxylases [2–6]. In addition, Mg is important for protein synthesis being the bridging element for ribosome aggregation [7] and it also represents a regulator of stromal pH [8, 9].
Magnesium concentration in soil solution generally ranges between 0.5 and 8.0 mM [10], depending on soil characteristics and environmental or anthropogenic factors [11]. There are two main reasons causing Mg deficiency: absolute deficiency and cation competition [11]. All these conditions result in a lower accumulation of Mg in plant tissues with a consequent reduction of crop productivity and quality [12].
The physiological effects of Mg deficiency were well described in different species revealing an impact on growth parameters [13–18], sugar accumulation in source leaves [19–21] and antioxidant system [22–28]; however, the responses generally depend on the plant species, age and the system used to induce Mg deficiency.
Plant responses to Mg deficiency have been also analyzed in terms of changes in transcriptome [29–32], proteome [33, 34] and metabolome [35] in root and/or shoot tissues. In addition, miRNAs putatively involved into the responses to Mg deficiency were identified in Citrus leaf and root tissues [36, 37]. As far as root tissues are concerned it has been ascertained that Mg deficiency affected transcripts, proteins and metabolites involved into oxidative stress. In fact, among the long-term responses to Mg shortage in roots of Arabidopsis thaliana was observed an increase of the dehydroascorbate:ascorbate ratio and an upregulation of genes encoding thioredoxin (TRX), glutaredoxin (GRX) and glutathione-S-transferase (GST) [30]. Modulation of proteins and transcripts involved into the same response was also recorded in roots of Citrus sinensis [33] and Citrus reticulata [32], respectively. The shortage of Mg caused also an impairment of other processes such as respiration and metabolism of carbohydrates and energy as suggested by the modulation of hexokinase (HK), pyruvate decarboxylase (PDC), phosphoglycerate kinase (PGK), pyruvate dehydrogenase (PDH) and ATP synthase in Citrus sinensis [33]. In addition, a strong upregulation of a transcripts encoding an ATP synthase subunit β was observed in response to Mg deficiency also in Citrus reticulata [32]. Furthermore, Mg deficiency caused a decrease of organic acids as pyruvic, citric, 2-keto-glutaric, succinic, fumaric and malic in soybean roots [35]. The cell wall and cytoskeleton have been recorded as other targets of Mg shortage. It was in fact shown that changes in the abundance of proteins (e.g. actin 1, villin 3 and tubulin γ-1 chain) [33] and transcripts (cell wall protein-like, α-(1,4)-fucosyltransferase and endo-1,4-β-D-glucanase) [32] occurred in response to this nutritional disorder in Citrus.
Notwithstanding, to the best of our knowledge, the molecular events taking place during Mg starvation in genotypes differing in their tolerance to Mg shortage have not been described so far. The results of similar experiments could help dissecting the processes underlying the different tolerance to Mg deficiency, considering that, at present, the molecular bases accounting for these differences are still lacking. To this aim, we compared the physiological and molecular responses (e.g. changes in metabolome and transcriptome) of two grapevine rootstocks, Paulsen 1103 (1103P) and Selection Oppenheim 4 (SO4), exhibiting different tolerance to Mg deficiency, being tolerant and susceptible, respectively [38]. This kind of approach is particularly relevant to viticulture as grapevine is one of the world’s most economically important and cultivated fruit crop. It is in fact well recognized that Mg-deficiency in grapevine is a widespread nutritional disorder causing serious loss of production [39].
Results
Physiological responses to Mg deficiency of 1103P and SO4
The physiological responses to Mg shortage of the two grapevine rootstocks, SO4 and 1103P, reported as susceptible and tolerant to Mg paucity respectively, were evaluated using microcuttings grown in hydroponics. The experiment was carried out for 14 days by cultivating the plants with or without Mg. After this time, the typical symptoms of Mg deficiency (e.g. the interveinal chlorosis in old leaves) were visible in the leaves of both rootstocks, grown in the absence of Mg (Fig. 1). However, Mg deficiency symptoms were more severe in old leaves of SO4 that showed necrotic spots near the margin of the basal leaves (Fig. 1). This observation was confirmed by the significant decrease in SPAD index values of SO4 plants grown in the absence of Mg (Fig. 2a). In addition, the 14-day Mg shortage period caused a decrease of S/R ratio (Fig. 2b) and a significant increase in shoot sugar accumulation only in the susceptible SO4 rootstock (Fig. 3).
Fig. 1.
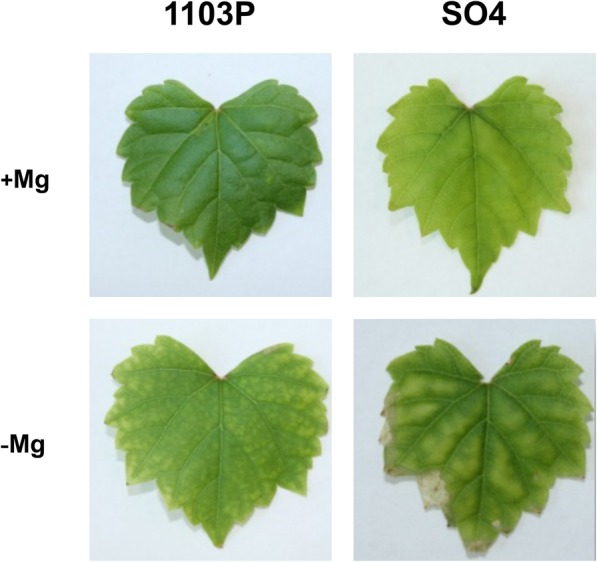
Details of old leaf of SO4 and 1103P cuttings grown for 14 days in presence (+Mg) and absence of Mg (−Mg)
Fig. 2.
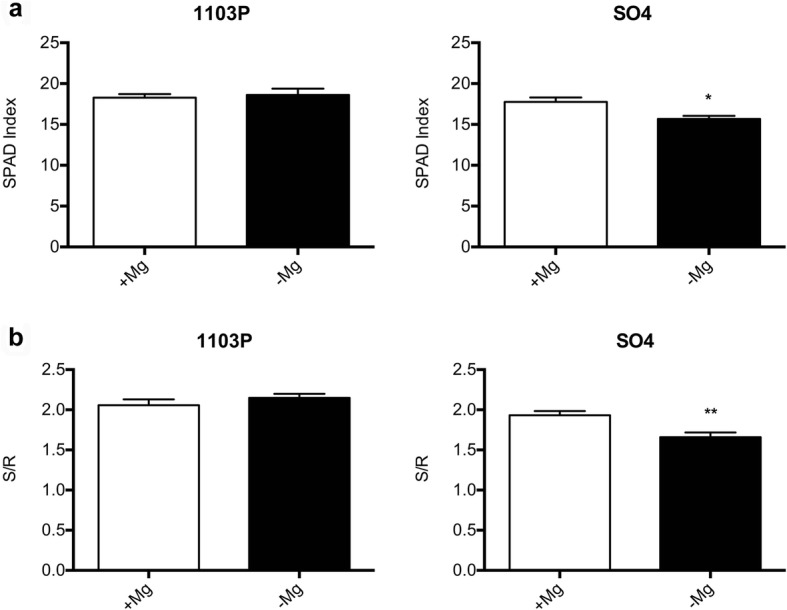
Mean SPAD index (a) and S/R (b) values of SO4 and 1103P microcuttings grown for 14 days in presence (+Mg) and absence of Mg (−Mg). Data were expressed as mean ± SEM of data from three growth independent experiments (n = 3). The statistical significance was determined by means of Student’s t-test. (∗P < 0.05) using the GraphPad InStat Program (version 5.0)
Fig. 3.
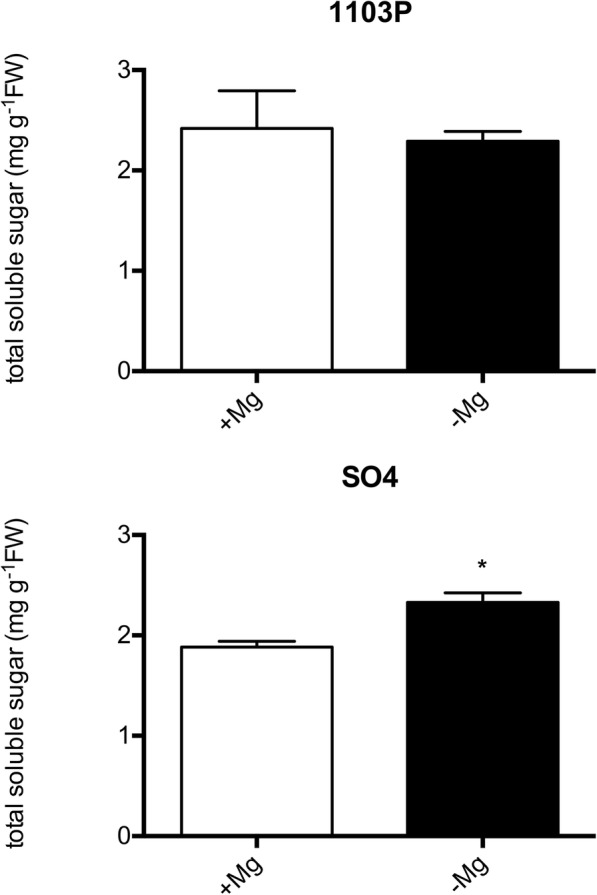
Total shoot soluble sugar concentration of SO4 and 1103P microcuttings grown for 14 days in presence (+Mg) and absence of Mg (−Mg). Data were expressed as mean ± SEM of data from three growth independent experiments (n = 3). The statistical significance was determined by means of Student’s t-test. (∗P < 0.05) using the GraphPad InStat Program (version 5.0)
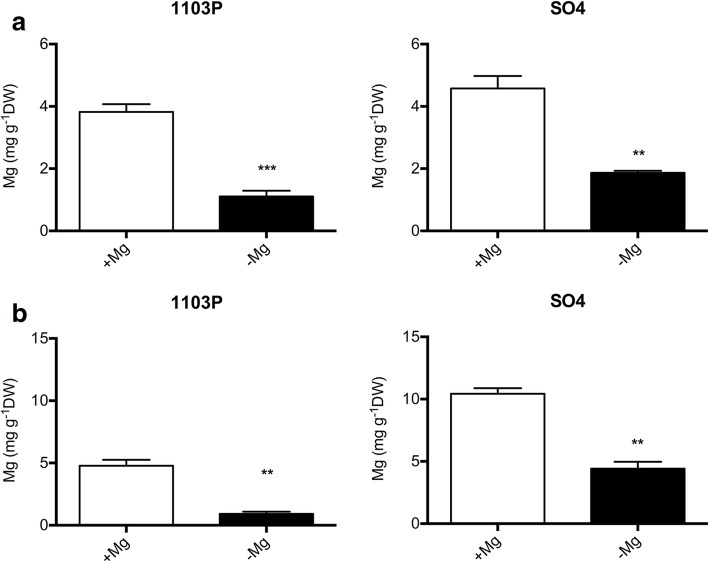
Tissue concentrations of Mg are reported in Fig. 4; a statistically significant decrease in Mg concentration in shoots of both SO4 and 1103P was recorded when plants were grown under macronutrient shortage (Fig. 4a). Similar behavior was observed for the root Mg concentration (Fig. 4b). Interestingly, root Mg levels were higher in SO4 than in 1103P in both nutritional conditions (Fig. 4b).
Fig. 4.
Shoot (a) and root (b) Mg concentration of SO4 and 1103P microcuttings grown for 14 days in presence (+Mg) and absence of Mg (−Mg). Data were expressed as mean ± SEM of data from three growth independent experiments (n = 3). The statistical significance was determined by means of Student’s t-test. (∗∗P < 0.01, ∗∗∗P < 0.001) using the GraphPad InStat Program (version 5.0)
Metabolic responses to Mg deficiency in roots of SO4 and 1103P
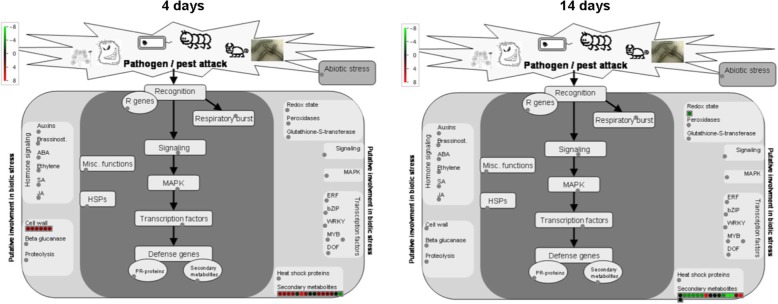
Changes in metabolomic profiles in roots of both grapevine rootstocks were evaluated in relation to the two nutritional conditions (+Mg and -Mg) through GC/MS and UHPLC-ESI/QTOF-MS for primary and secondary metabolites, respectively. The whole list of differentially accumulated metabolites, as selected by Volcano Plot analysis, is provided as supplementary information (Additional file 1: Table S1). The metabolic profiles of 1103P and SO4 microcuttings grown in +Mg, differed for the abundance of 77 and 250 metabolites after 4 and 14 days, respectively (Table 1, Additional file 2: Table S2 and Additional file 3: Table S3). Considering -Mg condition, the two genotypes differed at the two sampling time-points for 60 and 57 compounds, respectively (Table 1, Additional file 2: Tables S2 and Additional file 3: Table S3). By the means of Venn diagram, we identified the differentially abundant metabolites that distinguish the two rootstocks under the different nutritional conditions (+Mg, −Mg) and those that depends on the genetic background only (Fig. 5). In particular, we focused on the 48 and 32 metabolites that were differentially abundant at 4 (Fig. 5a) and 14 days (Fig. 5b) only in the roots of plants grown in -Mg conditions. At 4 days after treatments, 34 metabolites out of 48 were more abundant in 1103P as compared to SO4, whilst 14 were less abundant; at 14 days after treatments, 12 metabolites out of 32 were more abundant in 1103P with respect to SO4, whereas 20 were less represented (Additional file 4: Table S4). Considering the distribution in the main chemical classes, we observed that carbohydrate (41.18%) and terpenoids (11.76%) were the most represented classes for the “more abundant metabolites” in 1103P identified at 4 days. On the other hand, amino acids or their derivatives (28.57%), carbohydrates (14.29%), gibberellin (14.29%) and phenylpropanoids (14.29%) were the most represented classes for the “less abundant metabolites” (Additional file 4: Figure S1). After 14 days, coenzyme A-activated compounds (16.67%), glucosinolates (16.67%) and lipids (16.67%) were the most abundant classes for the metabolites showing higher levels in 1103P relative to SO4, whilst terpenoids (30%), alkaloids (20%) and phenylpropanoids (20%) were the most represented classes considering the “less abundant metabolites” in 1103P (Additional file 4: Figure S2). A selection of these metabolites is reported in Table 2. Considering all the differentially abundant metabolites under Mg deficiency in relation to the stress responses (Fig. 6), a higher level of metabolites belonging to secondary metabolism in the roots of the tolerant 1103P genotype at 4 days was observed. On the other hand, the production of secondary metabolites was delayed in the susceptible rootstock as supported by the major content of secondary compounds recorded in SO4 at 14 days (Additional file 4: Figure S3). In particular, the roots of 1103P produced more alkaloid-like compounds after 4 days, whilst the levels of this class of compounds and terpenoids were reduced at 14 days (Additional file 4: Figure S3). Furthermore, glucosinolates were specifically accumulated in the tolerant genotype 1103P at 14 days (Additional file 4: Figure S3). Differences in the responses to stress between the two rootstocks were also highlighted by the higher production of compounds involved into cell wall metabolism that were more abundant in 1103P after 4 days of Mg deficiency (Fig. 6).
Table 1.
Number of more and less abundant metabolites between 1103P and SO4 at 4 and 14 days
| 1103P + Mg vs SO4 + Mg | 1103P -Mg vs SO4 -Mg | |
|---|---|---|
| 4 days | ||
| More abundant | 40 | 41 |
| Less abundant | 37 | 19 |
| 14 days | ||
| More abundant | 85 | 25 |
| Less abundant | 165 | 32 |
Fig. 5.
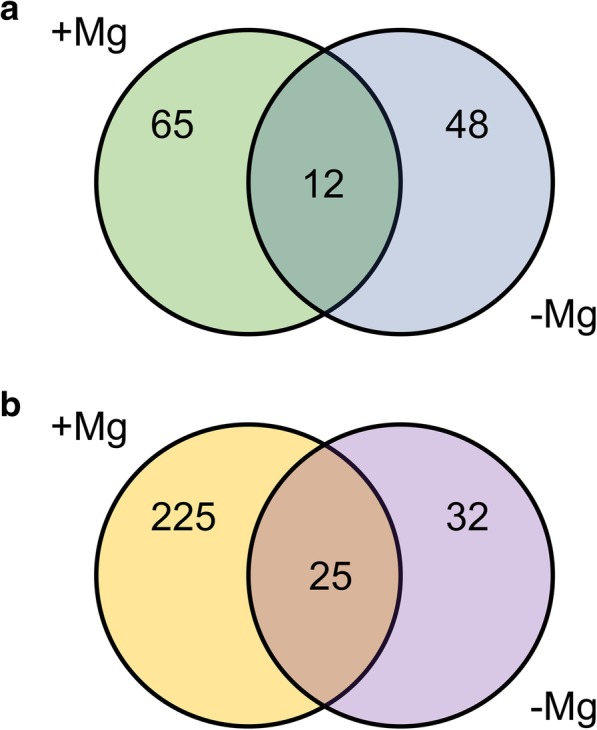
Metabolites differentially abundant in roots between 1103P and SO4 depending on the growth condition. (a) Metabolites specifically modulated at 4 days in presence (+Mg) and absence (−Mg) of Mg and modulated between the two genotypes in both nutritional conditions. (b) Metabolites specifically modulated at 14 days in presence (+Mg) and absence (−Mg) of Mg and modulated between the two genotypes in both nutritional conditions
Table 2.
Selection of metabolites differentially accumulated in roots between 1103P and SO4 at 4 and 14 days of growth under Mg deficiency
| Metabolite | FC |
|---|---|
| 1103P vs SO4 at 4 days | |
| Alkaloids | |
| (S)-corytuberine | 10,080.12 |
| 1,2-dehydroreticulinium | 10,080.12 |
| Norephedrine | 5055.84 |
| Amino acids or their derivatives | |
| Homomethionine | −7.34 |
| L-DOPA | −32,643.51 |
| Carbohydrates | |
| α-D-xylose | 23,691.90 |
| β-D-xylose | 23,691.90 |
| α-L-arabinofuranose | 23,691.90 |
| β-L-arabinopyranose | 23,691.90 |
| Maltohexaose | − 287,939.56 |
| Cofactors | |
| Molybdenum cofactor | 7.38 |
| Gibberellins | |
| Gibberellin A12 | –22.72 |
| Methyl gibberellin A9 | –22.72 |
| Nucleic acid components | |
| ppGpp | −131.55 |
| Phenylpropanoids | |
| Ferulate | 18,227.24 |
| Leachianone G | −7.82 |
| Pelargonidin 3-O-(6-O-malyl)-β-D-glucoside | 16.00a |
| Terpenoids | |
| 1′-hydroxy-γ-carotene | 41,946.68 |
| β-cryptoxanthin | 41,946.68 |
| ε-carotene-3-diol | 41,946.68 |
| Kauralexin B2 | −22.72 |
| β-carotene 15,15′ epoxide | 41,946.68 |
| Vitamins | |
| Thiamin | −8.64 |
| 1103P vs SO4 at 14 days | |
| Alkaloids | |
| (R)-N-methylcoclaurine | −28,523.96 |
| (S)-N-methylcoclaurine | −28,523.96 |
| S-cheilanthifoline | −16.00a |
| Senecionine | − 16.00 |
| Amino acids or their derivatives | |
| Allylcysteine | − 4892.33 |
| Aromatic compounds | |
| L-arogenate | −2.51 |
| Lipids | |
| 1–18:1–2-18:3-phosphatidylcholine | 46,713,708.00 |
| Terpenoids | |
| Loganin | −2.49 |
| (−)-menthol | −39,919.66 |
| (+)-isomenthol | −39,919.66 |
| (+)-neoisomenthol | −39,919.66 |
| (+)-neomenthol | −39,919.66 |
| (S)-(−)-citronellol | −39,919.64 |
| 4,9,13-trimethyltetradeca-2,4,6,8,10,12-hexaene-1,14-dial | 8299.46 |
| Phenylpropanoids | |
| 3,5-dihydroxyanisole | −4.45 |
| 4-hydroxycinnamic acid | −16.00 |
| Feruloyl-CoA | 16.00 |
| Isoliquiritigenin 4′-glucoside | −3.84 |
| p-coumaroyltyramine | −11.51 |
aAccording to Mass Profiler Professional output, FC = 16.00 and FC = − 16.00 denote very high and a very low FC (fold change), respectively
Fig. 6.
Differentially abundant metabolites between 1103P and SO4 linked to stress responses. The Log10(ratio) is shown by the color scale (green indicates a decrease and red an increase in metabolite abundance between 1103P and SO4). The analysis was carried out using MapMan software (https://mapman.gabipd.org/)
Transcriptional responses to Mg deficiency in roots of SO4 and 1103P
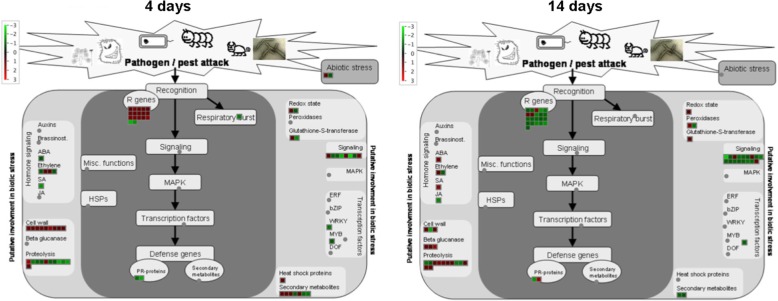
The transcriptional differences between the two rootstocks in relation to Mg nutritional status were identified through microarray analysis. Different comparisons between transcriptional profiles were carried out using Linear Models for MicroArray [40] (adjusted P-value of 0.05 and a |Log2(T/C)| ≥ 1; Table 3 and Additional file 5: Table S5). In particular, 771 and 791 transcripts resulted differentially modulated in roots of 1103P and SO4 after 4 days of growth in presence and absence of Mg, respectively. The transcriptional differences between 1103P and SO4 after 14 days of growth in presence and absence of Mg, consisted in 593 and 699 transcripts. In addition, by comparing the lists of differentially expressed transcripts through Venn diagram (Fig. 7; Additional file 6: Table S6 and Additional file 7: Table S7), we pinpointed transcripts specifically modulated in presence or absence of Mg and commonly modulated under both nutritional conditions after 4 and 14 days. The attention was paid to transcripts modulated only under Mg shortage (219 after 4 days and 361 after 14 days). At 4 days, 115 and 104 transcripts resulted up- and down-regulated, respectively, when the two rootstocks were compared (Additional file 6: Table S6). Considering the second sampling time-point, we identified 128 positively and 233 negatively modulated transcripts between the two genotypes (Additional file 7: Table S7). A Singular Enrichment Analysis (SEA) was performed on transcripts encoding proteins involved in known biological processes using agriGO v2.0 [41]. This analysis showed that the more significant enriched GO terms for the up-regulated transcripts at 4 days were “secondary metabolic process” and “response to stress” (Additional file 8: Figure S4) whilst “cellular homeostasis” was the more enriched term for the down-regulated transcripts (Additional file 8: Figure S5). For the second sampling time-point (14 days), only “secondary metabolic process” resulted significantly enriched for the up-regulated set of transcripts (Additional file 8: Figure S6). Considering the down-regulated transcripts, “secondary metabolic process” and “responses to stimulus” were the highest significant enriched GO terms (Additional file 8: FigureS7). A selection of differentially expressed transcripts between the two rootstocks is reported in Table 4. A further insight into the data were obtained by analyzing the differentially expressed transcripts at both sampling time-points in relation to stress responses (Fig. 8). The tolerant genotype 1103P showed a higher engagement of transcripts involved into secondary metabolism, particularly evident at 4 days of treatment (Fig. 8 and Additional file 8: Figure S8). In addition, the faster response of this rootstock to the Mg shortage was also evident on the basis of a positive modulation of transcripts involved into cell wall metabolism at 4 days as previously observed for the metabolite dataset (Figs. 6, and 8). Furthermore, in the tolerant genotype Mg shortage positively affected R genes at 4 days whilst this response was delayed in the susceptible SO4 (Fig. 8). Conversely, transcripts playing a role in signaling were mostly up-regulated in SO4 roots particularly at 14 days (Fig. 8).
Table 3.
Number of up- and down-regulated transcripts between 1103P and SO4 at 4 and 14 days
| 1103P + Mg vs SO4 + Mg | 1103P -Mg vs SO4 -Mg | |
|---|---|---|
| 4 days | ||
| Up-regulated | 412 | 421 |
| Down-regulated | 359 | 370 |
| 14 days | ||
| Up-regulated | 183 | 247 |
| Down-regulated | 410 | 452 |
Fig. 7.
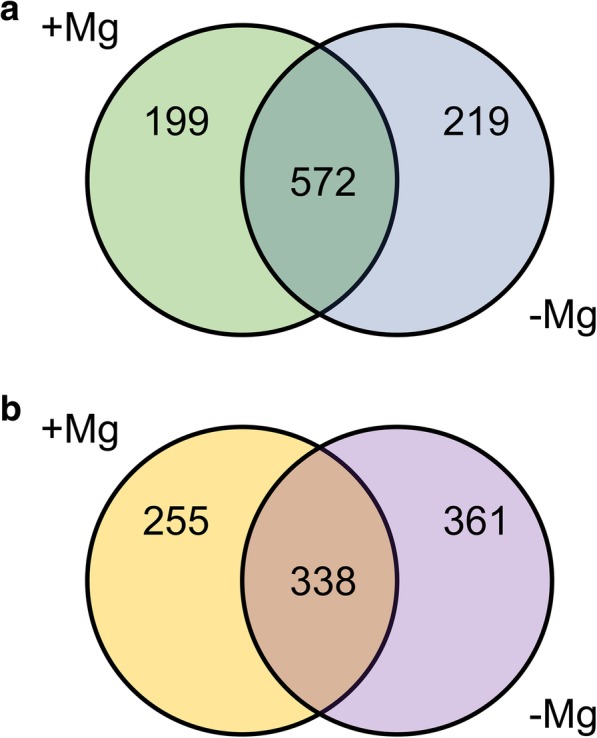
Transcripts differentially expressed in roots between 1103P and SO4 depending on the growth condition. (a) Transcripts specifically modulated at 4 days in presence (+Mg) and absence (−Mg) of Mg and modulated between the two genotypes in both nutritional conditions. (b) Transcripts specifically modulated at 14 days in presence (+Mg) and absence (−Mg) of Mg and modulated between the two genotypes in both nutritional conditions
Table 4.
Selection of transcripts differentially expressed in roots between 1103P and SO4 at 4 and 14 days of growth under Mg deficiency
| V1 Genome ID | Functional annotation | Log2(ratio) |
|---|---|---|
| 1103P vs SO4 at 4 days | ||
| Cell wall organization and biogenesis | ||
| VIT_00s0414g00010 | cellulose synthase CSLE1 | 1.19 |
| VIT_00s0469g00040 | cellulose synthase CSLE1 | 1.30 |
| VIT_00s0531g00060 | cellulose synthase CSLE1 | 1.01 |
| VIT_00s1213g00010 | cellulose synthase CSLE1 | 1.23 |
| VIT_00s1213g00020 | cellulose synthase CSLE1 | 1.19 |
| VIT_01s0127g00850 | polygalacturonase BURP | 1.02 |
| VIT_04s0044g01010 | pectinesterase family | 1.03 |
| VIT_05s0020g02540 | endo-1,3;1,4-beta-D-glucanase precursor | 1.11 |
| VIT_10s0116g00590 | pectinesterase family | 1.04 |
| VIT_12s0059g00960 | cellulose synthase CSLB04 | 1.71 |
| VIT_12s0059g00990 | cellulose synthase CSLB04 | 1.06 |
| VIT_12s0059g00990 | cellulose synthase CSLB04 | 1.09 |
| Cellular process | ||
| VIT_09s0002g06470 | CYP81B2v1 | −1.16 |
| VIT_09s0002g06480 | CYP81B2v1 | −1.00 |
| Establishment of localization | ||
| VIT_08s0007g00030 | cation/hydrogen exchanger 20 (CHX20) | −1.27 |
| VIT_11s0103g00010 | potassium-sodium symporter HKT2 | 1.23 |
| Homostatic process | ||
| VIT_06s0004g07040 | glutaredoxin | −1.08 |
| Regulation of biological process | ||
| VIT_00s0684g00020 | calmodulin binding IQD20 (IQ-domain 20) | −1.03 |
| Regulation of molecular function | ||
| VIT_01s0010g03930 | WRKY transcription Factor (VvWRKY03) | −1.39 |
| VIT_10s0003g00870 | TCP family transcription factor 4 | 1.38 |
| VIT_19s0014g03300 | NAC domain-containing protein (VvNAC18) | −1.27 |
| VIT_19s0027g00870 | NAC domain-containing protein (VvNAC30) | −1.57 |
| VIT_19s0090g00590 | MYB domain protein 101 | −1.07 |
| Response to stress | ||
| VIT_00s0370g00070 | epoxide hydrolase | 1.27 |
| VIT_01s0150g00450 | respiratory burst oxidase protein D (RBOHD) | −1.45 |
| VIT_05s0077g02020 | epoxide hydrolase | −1.26 |
| Secondary metabolic process | ||
| VIT_00s0724g00010 | pinene synthase | −1.01 |
| VIT_06s0004g05670 | glutathione S-transferase 25 | −1.01 |
| VIT_15s0046g03580 | (+)-neomenthol dehydrogenase | 1.06 |
| VIT_16s0100g01190 | stilbene synthase (VvSTS47) | −1.39 |
| VIT_18s0001g04110 | (−)-germacrene D synthase (VvTPS05) | 1.10 |
| VIT_18s0001g11430 | flavonoid 3-monooxygenase | −1.96 |
| VIT_19s0093g00320 | glutathione S-transferase (VvGST1) | −1.65 |
| VIT_19s0135g00180 | CYP72A59 | −1.10 |
| 1103P vs SO4 at 14 days | ||
| Cell wall organization and biogenesis | ||
| VIT_04s0044g01010 | pectinesterase family | 1.15 |
| VIT_05s0020g02540 | endo-1,3;1,4-beta-D-glucanase precursor | 1.06 |
| VIT_09s0002g06380 | polygalacturonase GH28 | 1.12 |
| Cellular process | ||
| VIT_15s0048g01680 | CYP704G7 | −1.35 |
| VIT_15s0048g01690 | CYP86A1 | −1.28 |
| VIT_18s0001g11490 | CYP82C1p | −1.51 |
| VIT_18s0001g11560 | CYP82A3 | −1.20 |
| Establishment of localization | ||
| VIT_01s0011g03020 | HAK5 (High affinity K+ transporter 5) | −1.08 |
| VIT_04s0008g04830 | ABC Transporter (VvPDR31 - VvABCG61) | −1.49 |
| VIT_04s0023g02530 | non-intrinsic ABC protein 12 | −1.03 |
| VIT_08s0007g03710 | ABC Transporter (VvPDR23 - VvABCG53) | 1.10 |
| VIT_09s0002g03550 | ABC Transporter (VvPDR3 - VvABCG33) | −1.92 |
| VIT_09s0002g05380 | ABC transporter g family pleiotropic drug resistance 12 PDR12 | −1.43 |
| VIT_09s0002g05470 | ABC transporter g family pleiotropic drug resistance 12 PDR12 | −1.57 |
| VIT_09s0018g00900 | ABC transporter g family pleiotropic drug resistance 12 PDR12 | −1.54 |
| VIT_13s0106g00580 | potassium channel AKT1 | −1.54 |
| VIT_14s0068g00210 | ABC Transporter (VvMDR16 - VvABCB16) | −1.18 |
| VIT_18s0001g10650 | ABC transporter g family pleiotropic drug resistance 12 PDR12 | −1.22 |
| Homostatic process | ||
| VIT_04s0023g02800 | glutaredoxin | 1.03 |
| Lipid metabolic process | ||
| VIT_13s0064g01480 | lipoxygenase LOX1 | −1.59 |
| VIT_13s0064g01490 | lipoxygenase | −1.30 |
| Regulation of molecular function | ||
| VIT_01s0127g00640 | bHLH | −1.15 |
| VIT_03s0167g00070 | putative MADS-box Short Vegetal Phase 5 | 1.18 |
| Response to endogenous stimulus | ||
| VIT_04s0008g06000 | VvERF045 | 1.04 |
| VIT_04s0079g00160 | auxin response factor 8 | −1.06 |
| Response to stress | ||
| VIT_00s0120g00010 | peroxisomal biogenesis factor 11 (PEX11C) | −1.09 |
| VIT_00s0370g00060 | epoxide hydrolase 2 | 1.19 |
| VIT_12s0059g02410 | peroxidase | 1.43 |
| VIT_13s0067g02360 | peroxidase, class III | −1.35 |
| Secondary metabolic process | ||
| VIT_00s0444g00010 | laccase | −1.17 |
| VIT_00s0704g00020 | 5-alpha-taxadienol-10-beta-hydroxylase | 1.04 |
| VIT_00s0731g00010 | laccase | −1.18 |
| VIT_00s1212g00020 | laccase | −1.33 |
| VIT_03s0063g01590 | CYP82C4 | 1.33 |
| VIT_06s0004g07650 | taxadien-5-alpha-ol-O-acetyltransferase | −1.26 |
| VIT_06s0009g03050 | flavonoid 3′,5′-hydroxylase | −1.21 |
| VIT_07s0031g01380 | ferulate 5-hydroxylase | −1.43 |
| VIT_12s0028g02700 | isoflavone methyltransferase | −1.69 |
| VIT_13s0067g00380 | pinene synthase | −1.14 |
| VIT_17s0000g05610 | isopiperitenol dehydrogenase | 1.08 |
| VIT_18s0001g01010 | laccase | −1.32 |
| VIT_19s0093g00350 | glutathione S-transferase 25 | 1.09 |
Fig. 8.
Differentially expressed transcripts between 1103P and SO4 linked to stress responses. The Log2(ratio) is shown by the color scale (green indicates a decrease and red an increase in transcript abundance between 1103P and SO4). The analysis was carried out using MapMan software (https://mapman.gabipd.org/)
Discussion
1103P is more tolerant to Mg deficiency than SO4
Physiological and molecular responses to Mg deficiency were widely described in several plant species [11, 42, 43]. The results here presented allowed us to identify the key molecular mechanisms underlying the responses to Mg shortage exhibited by two grapevine genotypes showing a different tolerance to this nutritional disorder. Considering grapevine, no description about the physiological and molecular responses to the shortage of this macronutrient is available, although a different tolerance to Mg deficiency among grapevine rootstocks was reported [38, 44]. Changes in physiological parameters (Figs. 1, 2 and 3) confirmed the appearance of typical symptoms described for other plant species such as chlorosis on older leaves and impairment of carbohydrate partitioning [45, 46]. Interestingly, the Mg-shortage-like changes were more evident for microcuttings of the SO4 (susceptible) rootstock.
The earlier plant response to Mg shortage consists in the impairment of the carbohydrate partitioning between leaves and roots, causing an accumulation of sugars in source leaves and a reduced translocation of them to roots [11]. Our results obtained for SO4 are in line with the increase in leaf glucose and fructose concentrations due to Mg deficiency reported for the first time in bean [13, 14] and later in other species [17, 19, 20, 47–50]. The alteration of carbohydrate partitioning between leaves and roots can cause a decrease in root growth as reported in bean [13, 14]. On the other hand, in other plant species such as sugar beet [19], arabidopsis [20] and rice [17], the Mg deficiency can be responsible for a negative effect on shoot growth as we observed in the susceptible genotypes (Fig. 2b). Verbruggen and Hermans [42] ascribed these contrasting effects on shoots or roots growth to the plant species or to the experimental conditions used for applying Mg shortage (e.g. Mg depletion since germination and/or very young growth stage or plants developed in Mg replete conditions before transfer to Mg deficient solution). Considering these observations, we might hypothesize that our results can be ascribable to the experimental design, in which microcuttings, before the 14-days deficiency period, were first exposed to a diluted NS containing Mg.
The results hereby presented, obtained with microcuttings grown in controlled conditions (i.e. growth chamber, plants in hydroponics), confirmed the difference in the tolerance to Mg deficiency observed in field for these two rootstocks. In both rootstocks, shoot Mg concentration (Fig. 4a) was under the threshold value (about 0.2% of dry weight) recorded for grapevine plants in optimal nutritional conditions and without any deficiency symptoms [51]. However, only the susceptible genotype displayed Mg-shortage responses (Figs. 1, 2 and 3) albeit, under Mg deficiency, shoot and root Mg concentrations were lower in the microcuttings of the tolerant 1103P rootstocks than in the susceptible one (Fig. 4). In grapevine, significant differences in ions bound in the apoplast in a not exchangeable form were reported as influenced by the genotype [52, 53] suggesting the need of an accurate evaluation of Mg distribution in sub-cellular compartments.
Metabolite profiling suggested a reduced oxidative stress condition in root of 1103P in response to Mg deficiency
It was reported that Mg deficiency could increase the reactive oxygen species (ROS) levels in plant roots [11]. Our metabolite profiling analysis suggests that the tolerant rootstock could better control the oxidative stress maintaining a lower level of ROS in roots through a higher production of antioxidative metabolites, already after 4 days of Mg deficiency. Indeed, roots of 1103P featured a higher concentration of carotenoids, as well as their oxidized forms, and anthocyanins (pelargonidin 3-O-(6-O-malyl)- β-D-glucoside) (Table 2 and Additional file 2: Table S2), which have a potential role in reducing the accumulation of ROS [54, 55]. In addition, Mg deficient 1103P rootstock showed, at both sampling time-points (4 and 14 days), higher levels of the dehydroxyascorbate (Additional file 2: Table S2 and Additional file 3: Table S3), which has antioxidant activity. Consistently, this compound was also reported to increase in Arabidopsis thaliana roots after 1 week of Mg deficiency [30]. A stronger capability of 1103P to contrast ROS is also supported by the higher abundance of molybdenum cofactor (Moco) recorded after 4 days (Table 2). Some Moco-dependent enzymes are involved in sulfite (sulfite oxidase and sulfite reductase) [56] and hydrogen peroxide (xanthine dehydrogenase) [57] detoxification. In addition, the lower thiamin and 5′-diphosphate 3′-diphosphate (ppGpp) levels (Table 2) in 1103P further support the hypothesis that root tissues are less prone to oxidative stress. Thiamin levels increases in response to several stresses among which oxidative stress in Arabidopsis thaliana [58] and Zea mays [59], whilst the plant alarmone seems to play a role in systemic signaling in response to abiotic stimuli [60]. In 1103P, the limited stress-condition of roots is also supported by lower levels of maltose as compared with SO4 (Table 2). Indeed, maltose is involved into the protection of proteins, membranes and photosynthetic electron transport chain; in fact, its levels were shown to increase in Arabidopsis thaliana leaf tissues during temperature shock [61]. The response to antioxidative stress in roots caused by Mg shortage appear to be delayed in the susceptible rootstock in comparison to the tolerant one. In fact, other metabolites (3,5-dihydroxyanisole and allylcysteine, Table 2) involved in oxidative stress responses [62, 63] showed higher levels in SO4 than 1103P only after 14 days. Nevertheless, at the same sampling time-point, 1103P produced higher quantity of phosphatidylcholine (Table 2). It was hypothesized that phosphatidylcholine plays a role in root system development of Arabidopsis thaliana through phosphatidic acid (PA) signaling [64]. PA is a secondary messenger playing a role in membrane-trafficking, in Ca2+ signaling and in the oxidative burst [65].
Other metabolites differentially accumulated in roots under Mg shortage in the two rootstocks are involved into defense responses, cell wall metabolism and gibberellin (GA) metabolism. Regarding the defense responses, we observed a rootstock-specific synthesis of compounds playing a role in these processes. After 4 days, a higher quantity of alkaloids (Table 2) was recorded in 1103P whilst SO4 produced more phytoalexin kauralexin B2 [66], leachianone G (intermediate of sophoraflavanone G synthesis) [67, 68], homomethionine (precursor of glucosinolates) [69] and the allelopathy L-3,4- dihydroxyphenylalanine (L-DOPA) [70]. The analysis of metabolites involved in responses to abiotic and/or biotic stresses, confirmed that the susceptible rootstock (SO4) has a delayed reaction in comparison to the tolerant one (Fig. 6 and Additional file 4: Figure S3). We observed that only at 14 days SO4 accumulated the higher levels of defense compounds (Table 2) such as alkaloids and their precursors, phenolic compounds and their precursors and terpenoids [71–78].
The microcuttings of the tolerant rootstock could respond to the Mg shortage through a positive modulation of monosaccharides (e.g. β-L-arabinopyranose, α-L-arabinofuranose, α-D-xylose, β-D-xylose, Table 2) involved into the synthesis of cell wall xyloglucans and pectins [79]. These results agree with recent reports concerning an increase in arabinose and xylose observed in Vitis vinifera L. calli in response to N-, P- and S-deficiency [80]. In undifferentiated and differentiated tissues of grapevine, N and P deficiencies caused a decrease in cellulose that can be compensated, in order to reinforce the cell wall, by an increase in pectin with a lower level of methylesterification and a higher quantity of arabinose-containing polysaccharides, putatively tightly linked to the cell wall [81]. It is thus conceivable that Mg shortage could determine a similar response into cell wall of roots, phenomenon that is more evident in the tolerant genotype. In addition, the higher levels of ferulate measured in Mg deficient 1103P (Table 2) suggested matrix reinforcement through cross-linking caused by esterification with arabinoxylans. This phenomenon was also described for grasses under salt stress [82]. In addition, a decrease of cell wall loosening controlled by gibberellin (GA) in root elongation zone [83] could occur as suggested by a lower level of intermediates (GA12 and methyl-GA9; Table 2) of the biosynthesis of bioactive GA in 1103P [84]. Higher quantity of these intermediates in SO4 roots under Mg shortage can be linked to a significant reduction in S/R ratio (Fig. 2b) in this rootstock.
Transcriptional analysis reinforces the hypothesis that SO4 has a delayed response to Mg deficiency
The analysis of the differentially expressed transcripts (Additional file 6: Table S6 and Additional file 7: Table S7) in roots under Mg deficiency confirmed the different physiological and metabolic behavior of 1103P and SO4, as mentioned above. Among these responses, the tolerant rootstock exhibited a lower oxidative stress in the first phase of the response to Mg deficiency (e.g. 4 days). This observation is supported by a down-regulation in 1103P roots of a transcript encoding a Respiratory burst oxidase protein D at 4 days (RBOH D). Plant RBOHs are involved into the production reactive oxygen intermediates (ROI) [85]. In addition, after 14 days we observed in 1103P roots the down-regulation of the peroxisomal biogenesis factor 11 transcript (PEX11C; Table 4). It was reported that OsPEX11 play a role in response to salt stress through the modulation of cation transporters and increasing the activity of antioxidant enzymes [86]. Nevertheless, we did not observe a clear genotype-specific transcriptional behavior of genes encoding enzymes (e.g. epoxide hydrolase, EH; GRX; GST; lipoxygenase, LOX; peroxidase, POX; Table 4 and Fig. 8) involved into the responses to oxidative stress at 4 and 14 days of Mg deficiency [87–90]. On the other hand, we observed a lower level of phenolics in 1103P root tissues under Mg shortage in particular at 14 days (Table 2). Phenolic compounds are involved in the non-enzymatic antioxidative responses in plant tissues [91]. Transcriptional data are in line with this assumption, showing a main down-regulation of transcripts encoding for enzymes involved into phenolic biosynthesis such as flavonoid 3-monooxygenase (F3H), isoflavone methyltransferase (IMT), ferulate 5-hydroxylase (F5H), flavonoid 3′,5′-hydroxylase (F3’5’H), laccase and stilbene synthase (STS) and transcripts encoding cytochrome P450 proteins (CYP) (Table 4, Additional file 8: Figure S8). Considering terpene biosynthesis, our transcriptional analysis (Table 4) suggested that 1103P, compared to SO4, selectively modulated transcripts involved into the biosynthesis of some monoterpenes, sesquiterpene and diterpene in response to Mg shortage. A lower level of monoterpene was measured in 1103P roots, in particular at 14 days (Table 2). At the same time, a strong repression of transcripts encoding ATP-binding cassette (ABC) transporters was measured (Table 4). ABC transporters could play a role into the export of secondary metabolites such as terpenoids, alkaloids, and phenolics to the cell surface and/or in the accumulation into the vacuole of their non-toxic precursors [92]. This transcriptional behavior can explain the lower synthesis of these classes of compounds in 1103P at 14 days (Table 2).
Considering the cell wall metabolism, transcripts encoding cellulose synthase (CEL), endo-1,3;1,4-beta-D-glucanase (GH), pectinesterese (PE) and polygalacturonase (PG) were up-regulated in 1103P as compared with SO4 mainly at 4 days (Table 4). These results partially fit with the previously hypothesized changes in cell wall structure consisting in a decrease of cellulose content and its stiffness through an increase in pectin with a lower level of methylesterification and a higher quantity of arabinose-containing polysaccharides putatively tightly linked to the cell wall as described in response to mineral deficiency in grapevine [81]. Although the N and P deficiencies caused in grapevine a decrease in cellulose, it was reported an up-regulation of transcripts for CELs [81]. This lack of correlation could also occur in the response to Mg shortage of 1103P, exhibiting higher levels of expression of CELs and GH genes involved in cellulose biosynthesis (Table 4). The up-regulation of PE transcripts was in line with the hypothesized increase in methylesterification of pectin [81].
Considering cellular responses after the perception of low Mg, it was reported an increase of ROS and cytosolic calcium (Ca2+) concentration during the elongation of root hairs in response to Mg shortage in Arabidopsis plants [31, 93]. The down-regulation of a transcript encoding a Calmodulin binding IQD20 protein (Table 4) in 1103P in comparison with SO4 at 4 days further supports the hypothesis that the tolerant genotype can counteract more efficiently the oxidative stress caused by Mg deficiency by limiting ROS development and modifying the root apparatus. In fact, IQD proteins could play a role in auxin and Ca2+ signaling in order to regulate plant growth and development [94]. In addition, the repression of an auxin response factor (ARF; Table 4) was in line with this hypothesis since it was proved that the expression of IQD genes is regulated by auxin though ARFs [94].
The processes linked to Mg uptake and redistribution could be a crucial point for the different capability of plants to cope with Mg shortage. The transporters MRS2/MGT have been often related to Mg uptake and, in particular, some members (i.e. AtMGT6) of the family have been suggested to be involved in the acquisition of the nutrient in shortage condition [95]. However, in Mg deficiency we did not observe a differential expression of MRS2/MGT transcripts between the two rootstocks. Nevertheless, a protein belonging to this transporter family (Additional file 7: Table S7, VIT_07s0031g02820) was shown to be up-regulated in 1103P plants grown in the presence of Mg for 14 days. This grapevine transcript encodes a protein showing the highest homology with the AtMRS2–10 (AtMGT1; AT1G80900.1, 40% identity) that is described as a high affinity Mg2+ transporter localized on plasma membrane and expressed in particular in root hair and in elongation zone [95, 96]. However, as recently highlighted [96], the Mg transport mediated by this protein is induced by Al toxicity but not by Mg deficiency. Indeed, other transport mechanisms could be involved in the modulation of Mg uptake in response to Mg deficiency [96]; it was in fact reported that the high-affinity K+ transporter OsHKT2;4 can accomplish a Mg2+ and Ca2+ transport that is impaired by K+ oversupply [97]. Interestingly, we observed the up-regulation of a HKT2 transcript in roots of 1103P as compared to SO4 under 4-day Mg deficiency (Table 4) and, at 14 days, the down-regulation of a transcript encoding a different putative HKT (Table 4), which shows the highest amino acid homology with AtHKT5 (AT4G13420.1, 60%). The AtAKT1 channel, AtHAK5 and AtKUP7 transporters are considered the major Arabidopsis components for K+ uptake at the root-soil interface [98]. In addition, a transcript putatively involved in the intracellular K+ homeostasis (e.g. Cation/H+ exchanger) [99] and a transcript encoding for a K+ channel (AKT1) mediating the low-affinity transport in plant roots [100] were repressed in 1103P at 4 and 14 days of treatment, respectively (Table 4). Overall, these results suggest that 1103P could positively modulate the component putatively involved in Mg uptake in 1103P (HKT2, VIT_11s0103g00010) with a concomitant down-regulation of the genes putatively responsible for K+ uptake and homeostasis.
Our results underlined a differential expression between the two rootstocks under Mg shortage of transcripts encoding several transcription factors that could be involved in the regulation of the different response to Mg deficiency. At both times, transcripts encoding transcription factors were mainly down-regulated in 1103P as compared with SO4 (Additional file 6: Table S6 and Additional file 7: Table S7). In particular, at 4 days we observed a repression of transcripts encoding a MYB protein showing amino acid homology to AtMYB101 (81% identity) among the Arabidopsis thaliana MYBs. AtMYB101 together with AtMYB33 and AtMYB65 was reported to be negatively regulated by miR159 that is described as a post transcriptional negative regulator of primary root growth in A. thaliana [101]. This transcriptional behavior suggests a different response of root apparatus to Mg deficiency between 1103P and SO4. A similar transcriptional behavior was recorded for one and two transcripts encoding a WRKY protein and two NAC proteins respectively (Table 4). The plant transcription factors WRKYs regulates in plants the responses to several biotic and abiotic stresses, among which nutritional stress as tolerance to low phosphate (Pi) condition [102, 103]. The grapevine WRKY transcript encodes a protein showing the highest homology among the A. thaliana a WRKYs with AtWRKY75 (84% amino acid identity), a negative regulator of root growth, whose expression was induced under phosphate (Pi) starvation [104]. Concerning NAC proteins, it was reported their involvement in responses to several abiotic stress [105]. Only a TEOSINTE BRANCHED 1, CYCLOIDEA, PCF1 (TCP) transcripts was more expressed at 4 days of deficiency in 1103P as compared to SO4 (Table 4). Members of TCP family are involved into control of plants growth in response to environmental signals such as red to far-red light ratio, high light stress, salt stress or abundance of nutrients (e.g. nitrate) [106]. After 14 days of deficiency other genes encoding transcription factors could play a role in the differential regulation of the late responses to this stress condition between the two grapevine roots. We observed in 1103P a minor expression level of a basic/helix-loop-helix (bHLH) whilst an opposite transcriptional profile was recorded for transcripts encoding an ethylene transcription factor (VvERF45) and a MADS-box protein (Table 4).
Conclusion
Our results allowed obtaining for the first time a picture of the different molecular responses exhibited by two contrasting genotypes with respect to the tolerance towards Mg shortage. In particular, data here presented clearly indicate that the ability of 1103P to tolerate Mg deficiency mainly relies on the early control of ROS level, thus reducing the oxidative stress in roots. In comparison with SO4, the molecular events playing a role in these responses are in fact faster both in terms of metabolites and transcripts involved. Although the data concerning the capability of acquiring cation nutrients more efficiently by the tolerant rootstock are somehow difficult to be interpreted, it seems most likely that the tolerance mechanism might rely on a tight control of cations ratios and homeostasis. Further research is indeed necessary to understand if the responses here highlighted could be generalized also to other plant species.
Methods
Plant material and sampling
Two-week-old grapevine in vitro propagated microcuttings (provided by Vitroplant s.r.l.) of SO4 and 1103P rootstocks were hydroponically grown into 2-L plastic pots for two weeks using a dilute nutrient solution (NS) containing: 200 μM Ca(NO3)2, 50 μM MgSO4, 70 μM K2SO4, 10 μM KCl, 10 μM KH2PO4, 0.1 μM H3BO3, 0.05 μM MnSO4, 0.02 μM CuSO4, 0.05 μM ZnSO4, 0.001 μM (NH4)6Mo7O24 and 10 μM Fe-EDTA. The pH of the solution was adjusted to 6.0 ± 0.1 with KOH. The NS was changed every week and the microcuttings were grown in a growth chamber (Photosynthetically Active Radiation, PAR, 200 μmol m− 2 s− 1, 16/8 h day/night regime, temperature 24 °C). After two weeks, for the Mg starvation experiment a NS with the following composition was set-up: 1000 μM Ca(NO3)2, 350 μM K2SO4, 50 μM KCl, 50 μM KH2PO4, 0.5 μM H3BO3, 0.25 μM MnSO4, 0.1 μM CuSO4, 0.25 μM ZnSO4, 0.005 μM (NH4)6Mo7O24 and 50 μM Fe-EDTA. For both rootstocks, half of the microcuttings were grown in this new NS containing 250 μM MgSO4 (+Mg) and the other half in a Mg-free NS in which MgSO4 was replaced with an equivalent mole of CaSO4 (−Mg). Three independent experiments (biological replicates) were performed growing 24 microcuttings (6 for each pot) for each treatment (SO4 + Mg, SO4 -Mg, 1103P + Mg and 1103P -Mg). For each biological replicate, root tissues from six plants were collected and pooled for each sample (SO4 + Mg, SO4 -Mg, 1103P + Mg and 1103P -Mg) after 4 and 14 days of growth. Samples were ground with liquid nitrogen and used for microarray analysis and Real time RT-PCR analysis, GC-MS analysis, UPLC-MS analysis.
Shoot to root (S/R) ratio and SPAD index were determined at the end (14 days) of each independent experiment using four plants (technical replicates). S/R was calculated measuring the fresh root and shoot weights of each plant. For the same plant a mean SPAD index was calculated using the SPAD values of all the leaves. For each leaf the SPAD value was expressed as the mean of five measurements performed using a SPAD-502 Plus Chlorophyll meter® (Konica Minolta). For each of the three independent experiments (biological replicates, n = 3) the mean value of S/R and SPAD index was calculated using the data its technical replicates. Data were expressed as mean ± SEM (n = 3, biological replicates).
Soluble sugars quantification
Shoot total soluble sugars were quantified using the Sucrose/D-Fructose/D-Glucose Assay kit (Megazyme, USA) according to the manufacturer’s protocol using leaf tissues of three plants (technical replicates) for each independent experiment (biological replicates) sampled at the end of the growth (after 14 days). For each of the three independent experiments (biological replicates, n = 3) the mean value was calculated using the data of their technical replicates. Data were expressed as mean ± SEM (n = 3, biological replicates).
Mg quantification
Magnesium concentrations were determined through ICP-MS analysis using three plants (technical replicates) for each independent experiment (biological replicates) sampled at the end of the growth (after 14 days). Roots and shoots were separated, washed three times in deionized water and two time with ultrapure water (milliQ, 18.2 MΩ cm) and dried in oven at 60 °C for 72 h. Samples (5 mg) were weighted and digested in a TFM microsampling insert using 250 μl of 69% ultrapure HNO3. Three inserts were put into 100-ml oven vessel containing 10 ml of water (milliQ, 18.2 MΩ cm) and 1 ml of 30% H2O2. In addition, 5 mg of the following reference material were digested: NIST 1515 (apple leaves). Digestion reaction was carried out using a microwave oven (Milestone StartD® microwave). A 20-min ramping period was used to reach a digestion temperature of 180 °C, which thereupon was maintained for 20 min. Samples were then diluted with water (milliQ, 18.2 MΩ cm) to a final concentration of 2% HNO3. Sample Mg concentrations were determined by using the Agilent 7500cx ICP-MS (Agilent). The instrument was tuned using tuning solution (Agilent tuning solution 1 ppb) in a standard mode checking the sensitivity of masses 7Li, 89Y, and 205Tl and the oxide and double charged ion levels (< 2%). Mg was quantified using a Mg standard solution (Romil PrimAg-plus). For each of the three independent experiments (biological replicates, n = 3) the mean value was calculated using the data of their technical replicates. Data were expressed as mean ± SEM (n = 3, biological replicates).
GC/MS metabolomic analysis
An amount of 150 mg of grinded root tissue was weighted in 2 mL tube and used for extraction in 80% methanol. After vortexing, the samples were sonicated for 5 min and centrifuged at 11000 g for 10 min at 4 °C. The following GC/MS metabolomic analysis was carried out according to Lisec et al. [107]. A volume of 200 μL of supernatant of each sample was added with 5 μL of internal standard (norvaline 2 mM) and then dried in a SpeedVac™ (Thermo Scientific, Waltham, MA, USA). For derivatization, each sample was added with 3 μL of a second internal standard (myristic acid 10 mM), then with 60 μL of methoxyamine in pyridine (20 mg/ml), incubated at 37 °C with shaking for 30 min and then without shaking for 3 h, in order to protect carbonyl groups. Thereafter, the samples were transferred into vials and added with 90 μL of N-methyl-N-(trimethylsilyl) trifluoroacetamide (MSTFA) with 1% trimethylchlorosilane, as derivatization agent.
The tubes were incubated in ThermoMixer at 400 rpm, for one hour at 55 °C and then transferred into vials to be analyzed.
An Agilent 6890 gas chromatograph equipped with a 30 m DB-5MS capillary column and coupled to an Agilent 5977 quadrupole mass spectrometer was used. Derivatized extracts (1 μl) were injected in splitless mode (250 °C), using helium as carrier gas (1 ml/min). A GC oven program, starting at 100 °C (hold for 2 min) up to 325 °C at 10 °C/min, was adopted. A fatty acids methyl ester mixture was used for retention time locking purposes (FAME mix, Agilent Technologies). Features were deconvoluted using the software “Unknown Analysis” (Agilent Technologies) and identification was based on spectral comparison against the commercially available database Fiehn Library (Agilent Technologies).
UHPLC-ESI/QTOF-MS metabolomic analysis
An aliquot of the extract produced for GC/MS was filtered through a 0.22 μm cellulose membrane and transferred to an amber vial for a complementary untargeted metabolomic analysis. Plant metabolites were screened using UHPLC chromatography coupled to a quadrupole-time-of-flight mass spectrometer (UHPLC/ QTOF-MS) through a JetStream Electrospray ionization system. In detail, a 1290 series liquid chromatograph, equipped with a binary pump, was interfaced to a G6550 iFunnel mass spectrometer (all from Agilent Technologies).
Instrumental conditions were optimized for plant metabolomics in previous experiments [108]. Briefly, chromatographic separation was achieved using a mixture of methanol and water as mobile phase and an Agilent Zorbax Eclipse-plus column (75 × 2.1 mm i.d., 1.8 μm). The gradient started from 5 to 90% methanol within 35 min, with a flow of 220 μL min− 1. Injection volume was 3.5 μL and the QTOF was run in positive polarity and SCAN mode (100–1200 m/z+ range, 0.8 spectra s− 1). Compounds deconvolution, mass and retention time alignment, as well as filtering (mass accuracy < 5 ppm, filter-by-frequerncy with 100% of detection within at least one condition) were carried out in Profinder B.06 (Agilent Technologies). Compounds annotation was based on accurate mass, isotope spacing and isotope ratio, versus the database exported from PlantCyc 9.5 [109]. Molecular features were subjected to a recursive analysis workflow using Mass Profiler Professional B12.06 (from Agilent Technologies) after the initial deconvolution. The peak area of each compound identified after recursive analysis with an identification score above 85/100 was extracted from the total ion current and exported for statistics and data interpretation.
Therefore, compounds identification was carried out as Level 2 (putatively annotated compounds), according to COSMOS Metabolomics Standards Initiative (http://cosmos-fp7.eu/).
Interpretation of either GC/MS and UHPLC-ESI/QTOF-MS metabolomics was carried out in Mass Profiler Professional B.12.06 (Agilent Technologies). Compounds’ abundance was normalized at the 75th percentile and baselined to the median. Thereafter, multivariate ANOVA (P < 0.05, Benjamini–Hochberg multiple testing correction) and fold-change analysis (cut-off = 2) were combined into volcano plots.
Microarray analysis
The total RNA was extracted from 100 mg of root tissues using the Spectrum Plant Total RNA kit (Sigma-Aldrich). RNA quality and quantity were verified using a Nanodrop 2000 instrument (Termo Scientific) and a Bioanalyzer Chip RNA 6000 series II (Agilent Technologies). The cDNA synthesis, labelling, hybridization and washing procedures were carried out according to the NimbleGen Arrays User’s Guide (version 3.2). Each sample was hybridized to one sub-array of a NimbleGen microarray 090818 Vitis exp. HX12 (Roche, NimbleGen; GPL13936) containing probes targeted to 29,549 predicted grapevine transcripts. The microarrays were scanned using an Axon GenePix 4400A (Molecular Devices) at 532 nm (Cy3 absorption peak) and GenePix Pro7 software (Molecular Devices) following the manufacturer’s instructions. A NimbleScan v2.5 software (Roche) was used for images analyses. This software produces Pair Files containing the raw signal intensity data for each probe and Calls Files with normalized expression data (quantile normalization) [110] derived from the probe summarization, performed through RMA analysis [111]. Analysis of normalized data (Calls Files) was performed using the open source software of the Bioconductor project [112] (http://www.bioconductor.org) with the statistical R programming language [113] (http://www.r-project.org). Differentially expressed probes were identified by Linear Models for MicroArrays [40] using LIMMA package and applying Bayesian correction, adjusted P-value of 0.05 and a |Log2(T/C)| ≥ 1. All microarray expression data are available at the GEO (http://www.ncbi.nlm.nih.gov/geo) under the series entry (GSE117849). Transcript annotation of the V1 version is reported by Fasoli et al. [114].
Real time RT-PCR analysis
Microarray data were validated by quantitative RT-PCR experiments analyzing the expression profile of 8 transcripts (Additional file 9: Table S8). Primer sequences were reported in Additional file 9: Table S9. Transcripts encoding tubulin beta chain (VIT_08s0040g00980) and elongation factor 1-alpha (VIT_06s0004g03220) were used to normalize data. DNA traces were removed with DNase I treatment (RQ1 RNase-Free DNase, Promega) according to the manufacturer’s procedure from total RNA samples used in microarray experiments. Reverse transcription reactions were performed using 1 μg of total RNA and the ImProm-II Reverse Transcriptase (Promega) and Real-time RT-PCR reactions were performed using FastSYBR® Green Master Mix (Applied Biosystems) according to the manufacturer’s protocols. The reactions were performed using a StepOnePlus™ (Applied Biosystems) with a final volume of 10 μL, the primer concentration of 350 nm and 1 μL of a 1:4 solution of cDNA. The thermal profile was 95 °C for 20s, 40 cycle of 95 °C for 3 s and 60 °C for 30s. PCR reaction efficiencies were calculated with the LinRegPCR program [115]. For each transcript, two mean normalized expression values (MNE) [116] were calculated using separately the two housekeeping transcripts for each sample (SO4 + Mg, SO4 -Mg, 1103P + Mg and 1103P -Mg). A final mean normalized expression value was calculated using a geometric mean of the two normalized expression values obtained for each transcript [117] and each sample. These normalized expression values were than used to calculated fold change (FC) values between samples (Additional file 7: Table S7).
MapMan analysis
Differentially abundant metabolites and differentially expressed transcripts were mapped on biotic stress overview and secondary metabolism overview using MapMan tool [118] (https://mapman.gabipd.org/). Concerning metabolites, a mapping file (Additional file 10: Table S10) was built assigning a bin code at each metabolite using information retrieved from KEGG (KEGG compound) (https://www.genome.jp/kegg/) and PlantCyc [109] databases. In the case of transcripts, the analysis was carried out using the mapping file for the grapevine Nimblegen chip (vvi_Nimblegen_probe_2018-05-25_mapping.txt) (http://protein.gomapman.org/).
Additional files
Table S1. Metabolites differentially abundant in roots between 1103P and SO4 at 4 and 14 days of growth in presence (+Mg) and absence (-Mg) of Mg identified by the comparison of profile obtained by QTOF and GC-MS analyses. Description, p(Corr) and fold change (FC) were reported. (XLSX 32 kb)
Table S2. List of metabolites differentially abundant in roots at 4 days between 1103P and SO4 depending on the growth condition. Metabolites specifically modulated at 4 days in presence (+Mg) and absence (-Mg) of Mg and modulated between the two genotypes in both nutritional conditions. Identity of compound, p(Corr) and FC were reported. (XLSX 16 kb)
Table S3. List of metabolites differentially abundant in roots at 14 days between 1103P and SO4 depending on the growth condition. Metabolites specifically modulated at 14 days in presence (+Mg) and absence (-Mg) of Mg and modulated between the two genotypes in both nutritional conditions. Identity of compound, p(Corr) and FC were reported. (XLSX 22 kb)
Table S4. More and less abundant metabolites in 1103P relative to SO4 at 4 and 14 days; metabolites were grouped in main chemical classes. Figures S1-S3. (PDF 801 kb)
Table S5. Differentially expressed transcripts identified by each comparison of transcriptional profiles. Probe ID, adjusted p-value, Log2(ratio) and Functional annotation were reported for each transcript. (XLSX 163 kb)
Table S6. List of transcripts differentially expressed in roots at 4 days between 1103P and SO4 depending on the growth condition. Transcripts specifically modulated at 4 days in presence (+Mg) and absence (-Mg) of Mg and modulated between the two genotypes in both nutritional conditions. Probe ID, adjusted p-value, Log2(ratio), Functional annotation and Gene Ontology term were reported for each transcript. (XLSX 87 kb)
Table S7. List of transcripts differentially expressed in roots at 14 days between 1103P and SO4 depending on the growth condition. Transcripts specifically modulated at 14 days in presence (+Mg) and absence (-Mg) of Mg and modulated between the two genotypes in both nutritional conditions. Probe ID, adjusted p-value, Log2(ratio), Functional annotation and Gene Ontology term were reported for each transcript. (XLSX 85 kb)
Figures S4-S8. (PDF 958 kb)
Table S8. Real-time RT-PCR of a set of transcripts differentially expressed between roots of 1103P and SO4 at 4 and 14 days of growth in presence (+Mg) and absence (-Mg) resulted differentially expressed by microarray analysis. Probe ID, V1 transcript ID, description, microarray fold change (FC) value and RT-PCR data (means ± SE of three biological replicates) were reported. Table S9. Sequence of forward and reverse primers used in Real-time RT-PCR experiments. Probe ID, V1 transcript ID, description, 5’-3’ nucleotide sequence of forward and reverse primers were reported. (PDF 1310 kb)
Table S10. Mapping file of the differentially abundant metabolites identified at 4 and 14 days used to carry out the MapMan analyses. Bin code, name, identifier and type were reported for each metabolite. (XLSX 11 kb)
Acknowledgements
Not applicable.
Funding
The work was supported by JP 2010 UniVR-UIV and JP 2016 UniVR-FCP. The publication costs of the article were funded by JP 2016 UniVR-FCP. The funders have no role in the study design, data analysis and interpretation, and manuscript writing, but just provide the financial.
Availability of data and materials
All microarray expression data are available at the GEO under the series entry (GSE117849).
Authors’ contributions
ZV, AZ and TP planned and designed the research; ON produced plant material; SL, LL and DS performed the experiments, SL, AZ and LL collected and analysed the data; SL, AZ and ZV interpreted the results, AZ and ZV wrote the manuscript. All authors have read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Sonia Livigni, Email: sonia.livigni@gmail.com.
Luigi Lucini, Email: luigi.lucini@unicatt.it.
Davide Sega, Email: davide.sega@univr.it.
Oriano Navacchi, Email: orianonavacchi@vitroplant.it.
Tiziana Pandolfini, Email: tiziana.pandolfini@univr.it.
Anita Zamboni, Email: anita.zamboni@univr.it.
Zeno Varanini, Email: zeno.varanini@univr.it.
References
- 1.Kobayashi K, Mochizuki N, Yoshimura N, Motohashi K, Hisabori T, Masuda T. Functional analysis of Arabidopsis thaliana isoforms of the mg-chelatase CHLI subunit. Photochem Photobiol Sci. 2008;7:1188–1195. doi: 10.1039/b802604c. [DOI] [PubMed] [Google Scholar]
- 2.Hallenbeck PC, Walsh DA. Autophosphorylation of phosphorylase kinase. Divalent metal cation and nucleotide dependency. J Biol Chem. 1983;258:13493–13501. [PubMed] [Google Scholar]
- 3.Rea AP, Sanders D. Tonoplast energization: two H+ pumps, one membrane. Physiol Plant. 1987;71:131–141. doi: 10.1111/j.1399-3054.1987.tb04630.x. [DOI] [Google Scholar]
- 4.Hell R, Bergmann L. Glutathione synthetase in tobacco suspension cultures: catalytic properties and localization. Physiol Plant. 1988;72:70–76. doi: 10.1111/j.1399-3054.1988.tb06624.x. [DOI] [Google Scholar]
- 5.Maeshima M. H+-translocating inorganic pyrophosphatase of plant vacuoles. Inhibition by Ca2+, stabilization by Mg2+ and immunological comparison with other inorganic pyrophosphatases. Eur J Biochem. 1991;196:11–17. doi: 10.1111/j.1432-1033.1991.tb15779.x. [DOI] [PubMed] [Google Scholar]
- 6.Portis R. Regulation of ribulose 1,5-bisphosphate carboxylase oxygenase activity. Annu Rev Plant Physiol Plant Mol Biol. 1992;43:415–437. doi: 10.1146/annurev.pp.43.060192.002215. [DOI] [Google Scholar]
- 7.Cammarano P, Felsani A, Gentile M, Gualerzi C, Romeo C, Wolf G. Formation of active hybrid 80-S particles from subunits of pea seedlings and mammalian liver ribosomes. Biochim Biophys Acta. 1972;281:625–642. doi: 10.1016/0005-2787(72)90160-8. [DOI] [PubMed] [Google Scholar]
- 8.Gardemann A, Schimkat D, Heldt W. Control of CO2 fixation regulation of stromal fructose-1,6-bisphosphatase in spinach by pH and Mg2+ concentration. Planta. 1986;168:536–545. doi: 10.1007/BF00392274. [DOI] [PubMed] [Google Scholar]
- 9.Wu W, Peters J, Berkowitz GA. Surface charge-mediated effects of Mg2+ on K+ flux across the chloroplast envelope are associated with regulation of stromal pH and photosynthesis. Plant Physiol. 1991;97:580–587. doi: 10.1104/pp.97.2.580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barber SA. Soil nutrient availability: a mechanistic approach. New York: Wiley; 1995. [Google Scholar]
- 11.Gransee A, Führs H. Magnesium mobility in soils as a challenge for soil and plant analysis, magnesium fertilization and root uptake under adverse growth conditions. Plant Soil. 2013;368:5–21. doi: 10.1007/s11104-012-1567-y. [DOI] [Google Scholar]
- 12.Cakmak I. Magnesium in crop production, food quality and human health. Plant Soil. 2013;368:1–4. doi: 10.1007/s11104-013-1781-2. [DOI] [Google Scholar]
- 13.Cakmak I, Hengeler C, Marschner H. Partitioning of shoot and root dry matter and carbohydrates in bean plants suffering from phosphorus, potassium and magnesium deficiency. J Exp Bot. 1994;45:1245–1250. doi: 10.1093/jxb/45.9.1245. [DOI] [Google Scholar]
- 14.Cakmak I, Hengeler C, Marschner H. Changes in phloem export of sucrose in leaves in response to phosphorus, potassium and magnesium deficiency in bean plants. J Exp Bot. 1994;45:1251–1257. doi: 10.1093/jxb/45.9.1251. [DOI] [Google Scholar]
- 15.Ericsson T, Kahr M. Growth and nutrition of birch seedlings at varied relative addition rates of magnesium. Tree Physiol. 1995;15:85–93. doi: 10.1093/treephys/15.2.85. [DOI] [PubMed] [Google Scholar]
- 16.Sun OJ, Payn TW. Magnesium nutrition and photosynthesis in Pinus radiata: clonal variation and influence of potassium. Tree Physiol. 1999;19:535–540. doi: 10.1093/treephys/19.8.535. [DOI] [PubMed] [Google Scholar]
- 17.Ding Y, Luo W, Xu G. Characterisation of magnesium nutrition and interaction of magnesium and potassium in rice. Ann App Biol. 2006;149:111–123. doi: 10.1111/j.1744-7348.2006.00080.x. [DOI] [Google Scholar]
- 18.Huang Jing-Hao, Xu Jing, Ye Xin, Luo Tu-Yan, Ren Li-Hua, Fan Guo-Cheng, Qi Yi-Ping, Li Qiang, Ferrarezi Rhuanito S., Chen Li-Song. Magnesium deficiency affects secondary lignification of the vascular system in Citrus sinensis seedlings. Trees. 2018;33(1):171–182. doi: 10.1007/s00468-018-1766-0. [DOI] [Google Scholar]
- 19.Hermans C, Johnson GN, Strasser RJ, Verbruggen N. Physiological characterization of mg deficiency in sugar beet: acclimation to low magnesium differentially affects photosystems I and II. Planta. 2004;220:344–355. doi: 10.1007/s00425-004-1340-4. [DOI] [PubMed] [Google Scholar]
- 20.Hermans C, Verbruggen N. Physiological characterization of mg deficiency in Arabidopsis thaliana. J Exp Bot. 2005;56:2153–2161. doi: 10.1093/jxb/eri215. [DOI] [PubMed] [Google Scholar]
- 21.Li CP, Qi YP, Zhang J, Yang LT, Wang DH, Ye X, Lai NW, Tan LL, Lin D, Chen LS. Magnesium-deficiency-induced alterations of gas exchange, major metabolites and key enzymes differ among roots, and lower and upper leaves of Citrus sinensis seedlings. Tree Physiol. 2017;37:1564–1581. doi: 10.1093/treephys/tpx067. [DOI] [PubMed] [Google Scholar]
- 22.Cakmak I, Marschner H. Magnesium deficiency and high light intensity enhance activities of superoxide dismutase, ascorbate peroxidase, and glutathione reductase in bean leaves. Plant Physiol. 1992;98:1222–1227. doi: 10.1104/pp.98.4.1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Candan N, Tarhan L. Relationship among chlorophyll-carotenoid content, antioxidant enzyme activities and lipid peroxidation levels by Mg2+ deficiency in the Mentha pulegium leaves. Plant Physiol Biochem. 2003;41:35–40. doi: 10.1016/S0981-9428(02)00006-2. [DOI] [Google Scholar]
- 24.Anza M, Riga P, Garbisua C. Time course of antioxidant responses of Capsicum annuum subjected to a progressive magnesium deficiency. Ann App Biol. 2005;146:123–134. doi: 10.1111/j.1744-7348.2005.04023.x. [DOI] [Google Scholar]
- 25.Tewari PK, Kumar P, Sharma PN. Magnesium deficiency induced oxidative stress and antioxidant responses in mulberry plants. Sci Hortic. 2006;108:7–14. doi: 10.1016/j.scienta.2005.12.006. [DOI] [Google Scholar]
- 26.Yang GH, Yang LT, Jiang HX, Li Y, Wang P, Chen LS. Physiological impacts of magnesium-deficiency in Citrus seedlings: photosynthesis, antioxidant system and carbohydrates. Trees. 2012;26:1237–1250. doi: 10.1007/s00468-012-0699-2. [DOI] [Google Scholar]
- 27.da Silva DM, Brandão IR, Alves JD, de Santos MO, de Souza KRD, de Silveira HRO. Physiological and biochemical impacts of magnesium-deficiency in two cultivars of coffee. Plant Soil. 2014;382:133–150. doi: 10.1007/s11104-014-2150-5. [DOI] [Google Scholar]
- 28.Tang N, Li Y, Chen LS. Magnesium deficiency–induced impairment of photosynthesis in leaves of fruiting Citrus reticulata trees accompanied by up-regulation of antioxidant metabolism to avoid photo-oxidative damage. J Plant Nutr Soil Sci. 2012;175:784–793. doi: 10.1002/jpln.201100329. [DOI] [Google Scholar]
- 29.Hermans C, Vuylsteke M, Coppens F, Craciun A, Inzé D, Verbruggen N. Early transcriptomic changes induced by magnesium deficiency in Arabidopsis thaliana reveal the alteration of circadian clock gene expression in roots and the triggering of abscisic acid-responsive genes. New Phytol. 2010;187:119–131. doi: 10.1111/j.1469-8137.2010.03258.x. [DOI] [PubMed] [Google Scholar]
- 30.Hermans C, Vuylsteke M, Coppens F, Cristescu SM, Harren FJ, Inzé D, et al. Systems analysis of the responses to long-term magnesium deficiency and restoration in Arabidopsis thaliana. New Phytol. 2010;187:132–144. doi: 10.1111/j.1469-8137.2010.03257.x. [DOI] [PubMed] [Google Scholar]
- 31.Niu Y, Jin G, Zhang YS. Root development under control of magnesium availability. Plant Signal Behav. 2014;37:2795–2813. doi: 10.4161/psb.29720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jin XL, Ma CL, Yang LT, Chen LS. Alterations of physiology and gene expression due to long-term magnesium-deficiency differ between leaves and roots of Citrus reticulata. J Plant Physiol. 2016;198:103–115. doi: 10.1016/j.jplph.2016.04.011. [DOI] [PubMed] [Google Scholar]
- 33.Peng HY, Qi YP, Lee J, Yang LT, Guo P, Jiang HX, et al. Proteomic analysis of Citrus sinensis roots and leaves in response to long-term magnesium-deficiency. BMC Genomics. 2015;16:253. doi: 10.1186/s12864-015-1462-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Billard V, Maillard A, Coquet L, Jouenne T, Cruz F, Garcia-Mina JM, et al. Mg deficiency affects leaf mg remobilization and the proteome in Brassica napus. Plant Physiol Biochem. 2016;107:337–343. doi: 10.1016/j.plaphy.2016.06.025. [DOI] [PubMed] [Google Scholar]
- 35.Yang N, Jiang J, Xie H, Bai M, Xu Q, Wang X, et al. Metabolomics reveals distinct carbon and nitrogen metabolic responses to magnesium deficiency in leaves and roots of soybean [Glycine max (Linn.) Merr.]. Front. Plant Sci. 2017;8:2091. doi: 10.3389/fpls.2017.02091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ma CL, Qi YP, Liang WW, Yang LT, Lu YB, Guo P, et al. MicroRNA regulatory mechanisms on Citrus sinensis leaves to magnesium-deficiency. Front Plant Sci. 2016;7:201. doi: 10.3389/fpls.2016.00201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liang WW, Huang JH, Li CP, Yang LT, Ye X, Lin D, et al. MicroRNA-mediated responses to long-term magnesium-deficiency in Citrus sinensis roots revealed by Illumina sequencing. BMC Genomics. 2017;18:657. doi: 10.1186/s12864-017-3999-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Keller M. Botany and anatomy. In: Keller M, editor. The science of grapevine: anatomy and physiology, 2nd ed. London: Elsevier/Academic Press; 2015. pp. 1–57. [Google Scholar]
- 39.Keller M. Environmental constraints and stress physiology. In: Keller M, editor. The science of grapevine: anatomy and physiology, 2nd ed. Elsevier/Academic Press: London; 2015. pp. 267–341. [Google Scholar]
- 40.Smyth GK. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3:article 3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- 41.Tian T, Liu Y, Yan H, You Q, Yi X, Du Z, Xu W, Su Z. agriGO v2.0: a GO analysis toolkit for the agricultural community, 2017 update. Nucleic Acids Res. 2017;45:W122–W129. doi: 10.1093/nar/gkx382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Verbruggen N, Hermans C. Physiological and molecular responses to magnesium nutritional imbalance in plants. Plant Soil. 2013;368:87–99. doi: 10.1007/s11104-013-1589-0. [DOI] [Google Scholar]
- 43.Farhat N, Elkhouni A, Zorrig W, Smaoui A, Abdelly C, Rabhi M. Effects of magnesium deficiency on photosynthesis and carbohydrate partitioning. Acta Physiol Plant. 2016;38:145. doi: 10.1007/s11738-016-2165-z. [DOI] [Google Scholar]
- 44.Garcia M, Gallego P, Daverade C, Ibrahim H. Effects of three rootstocks on grape (Vitis vinifera L.) cv. Négrette, grown hydroponically. I. Potassium, calcium and magnesium nutrition. South Afr J Enol Viticult. 2001;22:101–103. [Google Scholar]
- 45.Cakmak I, Kirkby EA. Role of magnesium in carbon partitioning and alleviating photooxidative damage. Physiol Plant. 2008;133:692–704. doi: 10.1111/j.1399-3054.2007.01042.x. [DOI] [PubMed] [Google Scholar]
- 46.Hawkesford M, Horst W, Kichey T, Lambers H, Schjoerring J, Møller SI, et al. Function of macronutrients. In: Marschner P, et al., editors. Marschner’s mineral nutrition of higher plants, 3rd ed. London: Elsevier/Academic Press; 2012. pp. 135–189. [Google Scholar]
- 47.Mehne-Jakobs B. The influence of magnesium deficiency on carbohydrate concentrations in Norway spruce (Picea abies) needles. Tree Physiol. 1995;15:577–584. doi: 10.1093/treephys/15.9.577. [DOI] [PubMed] [Google Scholar]
- 48.Fischer ES, Lohaus G, Heineke D, Heldt HW. Magnesium deficiency results in accumulation of carbohydrates and amino acids in source and sink leaves of spinach. Physiol Plant. 1998;102:16–20. doi: 10.1034/j.1399-3054.1998.1020103.x. [DOI] [PubMed] [Google Scholar]
- 49.Hermans C, Bourgis F, Faucher M, Strasser RJ, Delrot S, Verbruggen N. Magnesium deficiency in sugar beets alters sugar partitioning and phloem loading in young mature leaves. Planta. 2005;220:541–549. doi: 10.1007/s00425-004-1376-5. [DOI] [PubMed] [Google Scholar]
- 50.Hermans C, Hammond JP, White PJ, Verbruggen N. How do deficiencies of essential mineral elements alter biomass allocation? Trends Plant Sci. 2006;11:610–617. doi: 10.1016/j.tplants.2006.10.007. [DOI] [PubMed] [Google Scholar]
- 51.Wolf TK, Haeseler CW, Bergman EL. Growth and foliar elemental composition Seyval blanc grapevines as affected by four nutrient solution concentration of nitrogen, potassium and magnesium. Am J Enol Vitic. 1983;34:271–277. [Google Scholar]
- 52.Maggioni A, Varanini Z. Free space binding and uptake of ions by excised roots of grapevines. In: Sari MR, Loughman BC, editors. Genetic aspects of plant nutrition. The Hague/Boston/Lancaster: Martinus Nijhoff/Dr W. Junk Publishers; 1983. pp. 133–137. [Google Scholar]
- 53.Marastoni L, Sandri M, Pii Y, Valentinuzzi F, Brunetto G, Cesco S, Mimmo T. Synergism and antagonisms between nutrients induced by copper toxicity in grapevine rootstocks: Monocropping vs intercropping. Chemosphere. 2019;214:563–578. doi: 10.1016/j.chemosphere.2018.09.127. [DOI] [PubMed] [Google Scholar]
- 54.Nakabayashi R, Yonekura-Sakakibara K, Urano K, Suzuki M, Yamada Y, Nishizawa T, et al. Enhancement of oxidative and drought tolerance in Arabidopsis by overaccumulation of antioxidant flavonoids. Plant J. 2014;77:367–379. doi: 10.1111/tpj.12388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nisar N, Li L, Lu S, Khin NC, Pogson BJ. Carotenoid metabolism in plants. Mol Plant. 2015;8:68–82. doi: 10.1016/j.molp.2014.12.007. [DOI] [PubMed] [Google Scholar]
- 56.Yarmolinsky D, Brychkova G, Fluhr R, Sagi M. Sulfite reductase protects plants against sulfite toxicity. Plant Physiol. 2013;161:725–743. doi: 10.1104/pp.112.207712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ma X, Wang W, Bittner F, Schmidt N, Berkey R, Zhang L, et al. Dual and opposing roles of xanthine dehydrogenase in defense-associated reactive oxygen species metabolism in Arabidopsis. Plant Cell. 2016;28:1108–1126. doi: 10.1105/tpc.15.00880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tunc-Ozdemir M, Miller G, Song L, Kim J, Sodek A, Koussevitzky S, et al. Thiamin confers enhanced tolerance to oxidative stress in Arabidopsis. Plant Physiol. 2009;151:421–432. doi: 10.1104/pp.109.140046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rapala-Kozik M, Kowalska E, Ostrowska K. Modulation of thiamine metabolism in Zea mays seedlings under conditions of abiotic stress. J Exp Bot. 2008;59:4133–4143. doi: 10.1093/jxb/ern253. [DOI] [PubMed] [Google Scholar]
- 60.Takahashi K, Kasai K, Ochi K. Identification of the bacterial alarmone guanosine 5′-diphosphate 3′-diphosphate (ppGpp) in plants. PNAS. 2004;10:4320–4324. doi: 10.1073/pnas.0308555101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kaplan F, Guy CL. β-Amylase induction and the protective role of maltose during temperature shock. Plant Physiol. 2004;135:1674–1684. doi: 10.1104/pp.104.040808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jovanovic SV, Hara Y, Steenken S, Simic MG. Antioxidant potential of Gallocatechins. A pulse radiolysis and laser photolysis study. J Am Chem Soc. 1995;117:9881–9888. doi: 10.1021/ja00144a014. [DOI] [Google Scholar]
- 63.Colín-González AL, Santana RA, Silva-Islas CA, Chánez-Cárdenas ME, Santamaría A, Maldonado PD. The antioxidant mechanisms underlying the aged garlic extract- and S-allylcysteine-induced protection. Oxidative Med Cell Longev. 2012;2012:907162. doi: 10.1155/2012/907162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cruz-Ramírez A, López-Bucio J, Ramírez-Pimentel G, Zurita-Silva A, Sánchez-Calderon L, Ramírez-Chávez E, et al. The xipotl mutant of Arabidopsis reveals a critical role for phospholipid metabolism in root system development and epidermal cell integrity. Plant Cell. 2004;16:2020–2034. doi: 10.1105/tpc.103.018648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Canonne J, Froidure-Nicolas S, Rivas S. Phospholipases in action during plant defense signaling. Plant Signal Behav. 2011;6:13–18. doi: 10.4161/psb.6.1.14037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ejike CECC, Gong M, Udenigwe CC. Phytoalexins from the Poaceae: biosynthesis, function and prospects in food preservation. Food Res Int. 2013;52:167–177. doi: 10.1016/j.foodres.2013.03.012. [DOI] [Google Scholar]
- 67.Zhao P, Inoue K, Kouno I, Yamamoto H. Characterization of leachianone G 2″-dimethylallyltransferase, a novel prenyl side-chain elongation enzyme for the formation of the lavandulyl group of sophoraflavanone G in Sophora flavescens Ait. Cell suspension cultures. Plant Physiol. 2003;133:1306–1313. doi: 10.1104/pp.103.025213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kim CS, Park SN, Ahn SJ, Seo YW, Lee YJ, Lim YK, et al. Antimicrobial effect of sophoraflavanone G isolated from Sophora flavescens against mutans streptococci. Anaerobe. 2013;19:17–21. doi: 10.1016/j.anaerobe.2012.11.003. [DOI] [PubMed] [Google Scholar]
- 69.Frisch T, Motawia MS, Olsen CE, Agerbirk N, Møller BL, Bjarnholt N. Diversified glucosinolate metabolism: biosynthesis of hydrogen cyanide and of the hydroxynitrile glucoside alliarinoside in relation to sinigrin metabolism in Alliaria petiolata. Front Plant Sci. 2015;6:926. doi: 10.3389/fpls.2015.00926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Soares AR, Marchiosi R, Siqueira-Soares RC, Barbosa de Lima R, Dantas dos Santos W, Ferrarese-Filho O. The role of L-DOPA in plants. Plant Signal Behav. 2014;9:e28275. doi: 10.4161/psb.28275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hartmann T, Dierich B. Chemical diversity and variation of pyrrolizidine alkaloids of the senecionine type: biological need or coincidence? Planta. 1998;206:443–451. doi: 10.1007/s004250050420. [DOI] [Google Scholar]
- 72.Pearce G, Marchand PA, Griswold J, Lewis NG, Ryan CA. Accumulation of feruloyltyramine and p-coumaroyltyramine in tomato leaves in response to wounding. Phytochemistry. 1998;47:659–664. doi: 10.1016/S0031-9422(97)00620-1. [DOI] [Google Scholar]
- 73.Ikezawa N, Iwasa K, Sato F. Molecular cloning and characterization of methylenedioxy bridge-forming enzymes involved in stylopine biosynthesis in Eschscholzia californica. FEBS J. 2007;274:1019–1035. doi: 10.1111/j.1742-4658.2007.05652.x. [DOI] [PubMed] [Google Scholar]
- 74.Kai K, Mizutani M, Kawamura N, Yamamoto R, Tamai M, Yamaguchi H, et al. Scopoletin is biosynthesized via ortho-hydroxylation of feruloyl CoA by a 2-oxoglutarate-dependent dioxygenase in Arabidopsis thaliana. Plant J. 2008;55:989–999. doi: 10.1111/j.1365-313X.2008.03568.x. [DOI] [PubMed] [Google Scholar]
- 75.Chokchaisiri R, Suaisom C, Sriphota S, Chindaduang A, Chuprajob T, Suksamrarn A. Bioactive flavonoids of the flowers of Butea monosperma. Chem Pharm Bull. 2009;57:428–432. doi: 10.1248/cpb.57.428. [DOI] [PubMed] [Google Scholar]
- 76.Nerio LS, Olivero-Verbel J, Stashenko E. Repellent activity of essential oils: a review. Bioresour Technol. 2010;101:372–378. doi: 10.1016/j.biortech.2009.07.048. [DOI] [PubMed] [Google Scholar]
- 77.Miettinen K, Dong L, Navrot N, Schneider T, Burlat V, Pollier J, et al. The secoiridoid pathway from Catharanthus roseus. Nat Commun. 2013;5:3606. doi: 10.1038/ncomms4606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Funayama S, Cordell GA. Alkaloids: a treasury of poisons and medicines. Boston: Academic Press, Boston; 2015. [Google Scholar]
- 79.Harris PJ, Stone BA. Chemistry and molecular Organization of Plant Cell Walls. In: Himmel ME, editor. Biomass recalcitrance: deconstructing the plant Cell Wall for bioenergy. Oxford: Blackwell Publishing Ltd; 2008. pp. 61–93. [Google Scholar]
- 80.Fernandes JC, García-Angulo P, Goulao LF, Acebes JL, Amâncio S. Mineral stress affects the cell wall composition of grapevine (Vitis vinifera L.) callus. Plant Sci. 2013;205-206:111–120. doi: 10.1016/j.plantsci.2013.01.013. [DOI] [PubMed] [Google Scholar]
- 81.Goulao LF, Fernandes JC, Amâncio S. How the depletion in mineral major elements affects grapevine (Vitis vinifera L.) primary cell wall. Front Plant Sci. 2017;8:1439. doi: 10.3389/fpls.2017.01439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Uddin MN, Hanstein S, Faust F, Eitenmüller PT, Pitann B, Schubert S. Diferulic acids in the cell wall may contribute to the suppression of shoot growth in the first phase of salt stress in maize. Phytochemistry. 2014;102:126–136. doi: 10.1016/j.phytochem.2014.02.014. [DOI] [PubMed] [Google Scholar]
- 83.Somssich M, Khan GA, Persson S. Cell Wall heterogeneity in root development of Arabidopsis. Front Plant Sci. 2016;7:1242. doi: 10.3389/fpls.2016.01242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hedden P, Kamiya Y. Gibberellin biosynthesis: Enzymes, genes and their regulation. Annu Rev Plant Physiol Plant Mol Biol. 1997;48:431–460. doi: 10.1146/annurev.arplant.48.1.431. [DOI] [PubMed] [Google Scholar]
- 85.Torres MA, Dangl JL. Functions of the respiratory burst oxidase in biotic interactions, abiotic stress and development. Curr Opin Plant Biol. 2005;8:397–403. doi: 10.1016/j.pbi.2005.05.014. [DOI] [PubMed] [Google Scholar]
- 86.Cui P, Liu H, Islam F, Li L, Farooq MA, Ruan S, Zhou W. OsPEX11, a Peroxisomal biogenesis factor 11, Contributes to Salt Stress Tolerance in Oryza sativa. Front Plant Sci. 2016;7:1357. doi: 10.3389/fpls.2016.01357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Guo A, Durner J, Klessig DF. Characterization of a tobacco epoxide hydrolase gene induced during the resistance response to TMV. Plant J. 1998;15:647–656. doi: 10.1046/j.1365-313x.1998.00241.x. [DOI] [PubMed] [Google Scholar]
- 88.Dixon DP, Lapthorn A, Edwards R. Plant glutathione transferases. Genome Biol. 2002;3:reviews3004. doi: 10.1186/gb-2002-3-3-reviews3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lim CW, Han SW, Hwang IS, Kim DS, Hwang BK, Lee SC. The pepper lipoxygenase CaLOX1 plays a role in osmotic, drought and high salinity stress response. Plant Cell Physiol. 2017;56:930–942. doi: 10.1093/pcp/pcv020. [DOI] [PubMed] [Google Scholar]
- 90.Noctor G, Reichheld JP, Foyer CH. ROS-related redox regulation and signaling in plants. Semin Cell Dev Biol. 2017;80:3–12. doi: 10.1016/j.semcdb.2017.07.013. [DOI] [PubMed] [Google Scholar]
- 91.Kubalt K. The role of phenolic compounds in plant resistance. Biotech Food Sci. 2016;80:97–102. [Google Scholar]
- 92.Hwang JU, Song WY, Hong D, Ko D, Yamaoka Y, Jang S, et al. Plant ABC transporters enable many unique aspects of a terrestrial Plant's lifestyle. Mol Plant. 2016;9:338–355. doi: 10.1016/j.molp.2016.02.003. [DOI] [PubMed] [Google Scholar]
- 93.Niu Y, Chai R, Liu L, Jin G, Liu M, Tang C, et al. Magnesium availability regulates the development of root hairs in Arabidopsis thaliana (L.) Heynh. Plant Cell Environ. 2014;37:2795–2813. doi: 10.1111/pce.12362. [DOI] [PubMed] [Google Scholar]
- 94.Bürstenbinder K, Mitra D, Quegwer J. Functions of IQD proteins as hubs in cellular calcium and auxin signaling: a toolbox for shape formation and tissue-specification in plants? Plant Signal Behav. 2017;12:e1331198. doi: 10.1080/15592324.2017.1331198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Gebert M, Meschenmoser K, Svidová S, Weghuber J, Schweyen R, Eifler K, et al. A root-expressed magnesium transporter of the MRS2/MGT gene family in Arabidopsis thaliana allows for growth in low-Mg2+ environments. Plant Cell. 2009;21:4018–4030. doi: 10.1105/tpc.109.070557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Guo W, Nazimc H, Lianga Z, Yanga D. Magnesium deficiency in plants: An urgent problem. Crop J. 2016;4:83–91. doi: 10.1016/j.cj.2015.11.003. [DOI] [Google Scholar]
- 97.Horie T, Brodsky DE, Costa A, Kaneko T, Lo Schiavo F, Katsuhara M, et al. K+ transport by the OsHKT2;4 transporter from rice with atypical Na+ transport properties and competition in permeation of K+ over Mg2+ and Ca2+ ions. Plant Physiol. 2011;156:1493–1507. doi: 10.1104/pp.110.168047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wang Y, Wu W-H. Regulation of potassium transport and signaling in plants. Curr Opin Plant Biol. 2017;39:123–128. doi: 10.1016/j.pbi.2017.06.006. [DOI] [PubMed] [Google Scholar]
- 99.Padmanaban S, Chanroj S, Kwak JM, Li X, Ward JM, Sze H. Participation of endomembrane cation/H+ exchanger AtCHX20 in osmoregulation of guard cells. Plant Physiol. 2007;144:82–93. doi: 10.1104/pp.106.092155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Szczerba MW, Britto DT, Kronzucker HJ. K+ transport in plants: physiology and molecular biology. J Plant Physiol. 2009;166:447–466. doi: 10.1016/j.jplph.2008.12.009. [DOI] [PubMed] [Google Scholar]
- 101.Xue T, Liu Z, Dai X, Xiang F. Primary root growth in Arabidopsis thaliana is inhibited by the miR159 mediated repression of MYB33, MYB65 and MYB101. Plant Sci. 2017;262:182–189. doi: 10.1016/j.plantsci.2017.06.008. [DOI] [PubMed] [Google Scholar]
- 102.Chen YF, Li LQ, Kong YH, Wang H, Wu WH. The WRKY6 transcription factor modulates PHOSPHATE 1 expression in response to low pi stress in Arabidopsis. Plant Cell. 2009;21:3554–3566. doi: 10.1105/tpc.108.064980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Jiang J, Ma S, Ye N, Jiang M, Cao J, Zhang J. WRKY transcription factors in plant responses to stresses. J Integr Plant Biol. 2017;59:86–101. doi: 10.1111/jipb.12513. [DOI] [PubMed] [Google Scholar]
- 104.Devaiah BN, Karthikeyan AS, Raghothama KG. WRKY75 transcription factor is a modulator of phosphate acquisition and root development in Arabidopsis. Plant Physiol. 2007;143:1789–1801. doi: 10.1104/pp.106.093971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Shao H, Wang H, Tang X. NAC transcription factors in plant multiple abiotic stress responses: progress and prospects. Front Plant Sci. 2015;6:902. doi: 10.3389/fpls.2015.00902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Danisman S. TCP transcription factors at the interface between environmental challenges and the plant’s growth responses. Front Plant Sci. 2016;7:1930. doi: 10.3389/fpls.2016.01930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lisec J, Schauer N, Kopka J, Willmitzer L, Fernie AR. Gas chromatography mass spectrometry-based metabolite profiling in plants. Nat Prot. 2006;1:387–396. doi: 10.1038/nprot.2006.59. [DOI] [PubMed] [Google Scholar]
- 108.Lucini L, Borgognone D, Rouphael Y, Cardarelli M, Bernardi J, Colla G. Mild potassium chloride stress alters the mineral composition, hormone network, and phenolic profile in artichoke leaves. Front Plant Sci. 2016;7:948. doi: 10.3389/fpls.2016.00948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Plant Metabolic Network, http://www.plantcyc.org.
- 110.Bolstad B.M., Irizarry R.A, Astrand M., Speed T.P. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19(2):185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 111.Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ihaka R, Gentleman R. R: a language for data analysis and graphics. J Comput Graph Stat. 1996;5:299–314. [Google Scholar]
- 114.Fasoli M, Dal Santo S, Zenoni S, Tornielli GB, Farina L, Zamboni A, et al. The grapevine expression atlas reveals a deep transcriptome shift driving the entire plant into a maturation program. Plant Cell. 2012;24:3489–3505. doi: 10.1105/tpc.112.100230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ramakers C, Ruijter JM, Deprez RHL, Moorman AFM. Assumption free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci Lett. 2003;339:62–66. doi: 10.1016/S0304-3940(02)01423-4. [DOI] [PubMed] [Google Scholar]
- 116.Simon P. Q-Gene: processing quantitative real-time RT–PCR data. Bioinformatics. 2003;19:1439–1440. doi: 10.1093/bioinformatics/btg157. [DOI] [PubMed] [Google Scholar]
- 117.Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3:research0034.1. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Thimm O, Bläsing O, Gibon Y, Nagel A, Meyer S, Krüger P, Selbig J, Müller LA, Rhee SY, Stitt M. MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 2004;3:914–939. doi: 10.1111/j.1365-313X.2004.02016.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Metabolites differentially abundant in roots between 1103P and SO4 at 4 and 14 days of growth in presence (+Mg) and absence (-Mg) of Mg identified by the comparison of profile obtained by QTOF and GC-MS analyses. Description, p(Corr) and fold change (FC) were reported. (XLSX 32 kb)
Table S2. List of metabolites differentially abundant in roots at 4 days between 1103P and SO4 depending on the growth condition. Metabolites specifically modulated at 4 days in presence (+Mg) and absence (-Mg) of Mg and modulated between the two genotypes in both nutritional conditions. Identity of compound, p(Corr) and FC were reported. (XLSX 16 kb)
Table S3. List of metabolites differentially abundant in roots at 14 days between 1103P and SO4 depending on the growth condition. Metabolites specifically modulated at 14 days in presence (+Mg) and absence (-Mg) of Mg and modulated between the two genotypes in both nutritional conditions. Identity of compound, p(Corr) and FC were reported. (XLSX 22 kb)
Table S4. More and less abundant metabolites in 1103P relative to SO4 at 4 and 14 days; metabolites were grouped in main chemical classes. Figures S1-S3. (PDF 801 kb)
Table S5. Differentially expressed transcripts identified by each comparison of transcriptional profiles. Probe ID, adjusted p-value, Log2(ratio) and Functional annotation were reported for each transcript. (XLSX 163 kb)
Table S6. List of transcripts differentially expressed in roots at 4 days between 1103P and SO4 depending on the growth condition. Transcripts specifically modulated at 4 days in presence (+Mg) and absence (-Mg) of Mg and modulated between the two genotypes in both nutritional conditions. Probe ID, adjusted p-value, Log2(ratio), Functional annotation and Gene Ontology term were reported for each transcript. (XLSX 87 kb)
Table S7. List of transcripts differentially expressed in roots at 14 days between 1103P and SO4 depending on the growth condition. Transcripts specifically modulated at 14 days in presence (+Mg) and absence (-Mg) of Mg and modulated between the two genotypes in both nutritional conditions. Probe ID, adjusted p-value, Log2(ratio), Functional annotation and Gene Ontology term were reported for each transcript. (XLSX 85 kb)
Figures S4-S8. (PDF 958 kb)
Table S8. Real-time RT-PCR of a set of transcripts differentially expressed between roots of 1103P and SO4 at 4 and 14 days of growth in presence (+Mg) and absence (-Mg) resulted differentially expressed by microarray analysis. Probe ID, V1 transcript ID, description, microarray fold change (FC) value and RT-PCR data (means ± SE of three biological replicates) were reported. Table S9. Sequence of forward and reverse primers used in Real-time RT-PCR experiments. Probe ID, V1 transcript ID, description, 5’-3’ nucleotide sequence of forward and reverse primers were reported. (PDF 1310 kb)
Table S10. Mapping file of the differentially abundant metabolites identified at 4 and 14 days used to carry out the MapMan analyses. Bin code, name, identifier and type were reported for each metabolite. (XLSX 11 kb)
Data Availability Statement
All microarray expression data are available at the GEO under the series entry (GSE117849).