Abstract
Mitochondrial DNA (mtDNA) has been one of the most extensively studied molecules in ecological, evolutionary and clinical genetics. In its early application in evolutionary genetics, mtDNA was assumed to be a selectively neutral marker conferring negligible fitness consequences for its host. However, this dogma has been overturned in recent years due to now extensive evidence for non-neutral evolutionary dynamics. Since mtDNA proteins physically interact with nuclear proteins to provide the mitochondrial machinery for aerobic ATP production, among other cell functions, co-variation of the respective genes is predicted to affect organismal fitness. To test this hypothesis we used an mtDNA-nuclear DNA introgression model in Drosophila melanogaster to test the fitness of genotypes in perturbation-reperturbation population cages and in a non-competitive assay for female fecundity. Genotypes consisted of both conspecific and heterospecific mtDNA-nDNA constructs, with either D. melanogaster or D. simulans mtDNAs on two alternative D. melanogaster nuclear backgrounds, to investigate mitonuclear genetic interactions (G x G effects). We found considerable variation between nuclear genetic backgrounds on the selection of mtDNA haplotypes. In addition, there was variation in the selection on mtDNAs pre- and post- reperturbation, demonstrating overall poor repeatability of selection. There was a strong influence of nuclear background on non-competitive fecundity across all the mtDNA species types. In only one of the four cage types did we see a significant fecundity effect between genotypes that could help explain the respective change in genotype frequency over generational time. We discuss these results in the context of G x G interactions and the possible influence of stochastic environments on mtDNA-nDNA selection.
Keywords: mitochondrial DNA, epistasis, fitness, Drosophila, introgression, haplotype, reperturbation cages
During the last half century, studies on mitochondrial biology have revolutionized the fields of metabolism, aging, evolutionary processes and organismal fitness. Seventy years ago it was hypothesized that mitochondria exclusively synthesize adenosine triphosphate (ATP) via oxidative energy metabolism (Ephrussi et al. 1949; Kennedy and Lehninger 1949). Now, the precise roles of mitochondria in the cell are understood to be far more comprehensive, involving many biosynthetic and degradative reactions including metabolism of amino acids, lipids and iron, and programmed cell death (apoptosis) (Tzagoloff 1982; Pfanner et al. 2004). More recently, the role of mitochondrial ‘performance’ and its associated genetic variation has been suggested to underpin a grossly underestimated number of human diseases and fitness-related phenotypes (Schon et al. 2012).
Mitochondrial biogenesis is jointly encoded by both mitochondrial and nuclear genomes. The nuclear DNA encodes ∼1200 mitochondrial genes whereas the mitochondrial DNA (mtDNA) encodes 37 (13 protein coding, 22 transfer RNAs and 2 ribosomal RNAs) (Smeitink et al. 2001). This co-evolved mito-nuclear gene complex provides an interesting target for studies of selection for two main reasons. First, the genome inheritance patterns differ; mtDNAs are maternally inherited, whereas nuclear DNAs (nDNA) are biparentally inherited. Second, mutations in nDNA and mtDNA genes that encode for proteins of the oxidative phosphorylation (OXPHOS) pathway are associated with deleterious phenotypes, including mitochondrial diseases (Wallace 1999; Smeitink et al. 2001), organismal longevity (Rand et al. 2006; Camus et al. 2012; Clancy 2008; De Benedictis et al. 1999), and measures of overall organismal fitness (Ballard and James 2004; Ballard and Rand 2005; Gerber et al. 2001).
Mutations in mtDNA are predicted to have phenotypic consequences on the organism and to date there is good evidence that both point mutations (SNPs) and large scale deletions in mtDNA have been shown to affect the organismal phenotype (Schon et al. 2012; Taylor and Turnbull 2005). Interestingly, mtDNA mutations principally manifest in tissue types with a high metabolic demand, i.e., where ATP production is normally highest (Wallace 2005, 1999). It is therefore not surprising that an organelle that provides ∼90% of ATP to the cell may be sensitive to mutation, and that those mutations confer phenotypic consequences. However, the precise roles of mutations in either genome on the whole organism have been historically difficult to disentangle (Rand et al. 2004).
Given that mtDNA and nDNA have co-evolved over evolutionary time, it is suggested that disruption of co-evolved mito-nuclear gene complexes will be deleterious to organisms, when non-coevolved combinations are compared to coevolved mito-nuclear combinations (Hutter and Rand 1995; Rand et al. 2004). Moreover, the degree of disruption of the co-evolved state is expected to correlate with phenotypic effects although explicit tests of this hypothesis are unresolved across species (but see Clark and Lyckegaard 1988; Montooth et al. 2010; Camus et al. 2012). We would predict that higher numbers of mtDNA mutations from the co-evolved complex would have a greater phenotypic effect than lower numbers and that the magnitude of the phenotypic response to genetic disruption would be correlated with the degree of genetic disruption.
Previous mtDNA-nDNA fitness studies have largely concentrated on either natural genetic variation or within-species introgression in Drosophila (Clark and Lyckegaard 1988; Macrae and Anderson 1988; Fos et al. 1990; Garcia-Martinez et al. 1998; Rand 2001; James and Ballard 2003; Ballard and James 2004; Dowling et al. 2008). Within-species mtDNA-nDNA introgression experiments have demonstrated various effects on fitness, including: (i) haplotype frequency changes in population cages in D. pseudoobscura (Macrae and Anderson 1988), D. melanogaster (Kilpatrick and Rand 1995), and D. simulans (Ballard and James 2004); (ii) changes in nuclear gene expression in D. melanogaster (Innocenti et al. 2011); (iii) aging phenotypes in D. melanogaster (Camus et al. 2012); (iv) sperm competitiveness in D. melanogaster (Yee et al. 2013), but see Friberg and Dowling 2008; (v) development time, survival probability, and organismal activity in D. simulans (James and Ballard 2003); (vi) development time in D. melanogaster (Mossman et al. 2016a); and (vii) female fitness in D. melanogaster (Dowling et al. 2007).
Investigating genetic components of fitness in natural populations is somewhat constrained by the lack of precise control over genotypic variation and the extent of genetic polymorphism and sequence divergence in study populations (Lewontin 1974). Drosophila genetics has proven to be a powerful tool to avoid these issues and to precisely manipulate genetic variation in a phylogenetic framework. For example, heterospecific mtDNA-nDNA introgression can be achieved due to the generation of reproductively viable hybrids between various species pairs. One example of this hybrid viability is between D. melanogaster and D. simulans. Drosophila simulans is a sister species to D. melanogaster and the separate species do not freely interbreed (Sturtevant 1920). However, the offspring arising from a cross between a female D. simulans (C167.4 strain) and a D. melanogaster In(1)AB male are reproductively viable (Davis et al. 1996). The resulting inter-specific hybrids, while not present in nature, provide an opportunity to tease apart: (i) nDNA effects; (ii) mtDNA effects; and (iii) nDNA x mtDNA interactions (epistases), in populations of flies that probably demonstrate greater mtDNA-nDNA sequence divergence (and phenotypic variation) than flies found in natural populations.
There is good reason to introgress genomes between closely-related species. One of the pioneering studies of Drosophila cytoplasmic-nuclear introgression clearly demonstrated that the greatest effects on 2nd chromosome and cytoplasm segregation patterns occurred when genetic material was exchanged between geographically isolated populations, with negligible effects observed when genetic material was exchanged within a population (Clark and Lyckegaard 1988). Introgressing genomes between-species essentially mimics and exaggerates this genetic polymorphism and the potential to detect mtDNA-nDNA interactions. Heterospecific mtDNA-nDNA introgressions in Drosophila have previously revealed mtDNA-nDNA epistases for fitness, including: (i) haplotype frequency changes in population cages of D. persimilis- D. pseudoobscura introgressions (Hutter and Rand 1995); and (ii) development time, bristle size and fecundity (Meiklejohn et al. 2013; Montooth et al. 2010; Mossman et al. 2016a), and aging phenotypes (Rand et al. 2006) in D. simulans - D. melanogaster mtDNA-nDNA introgressions. However, no study to date has investigated the relative fitness of mtDNA haplotypes in D. simulans – D. melanogaster introgressed fruit flies in a competitive context.
Comparisons of mtDNA coding sequence variation reveal an approximately fivefold difference in amino acid divergence within- and between- species in the D. melanogaster- D. simulans clade. For example, there are 18 amino acid substitutions between mtDNA haplotypes within D. melanogaster, and up to 45 amino acid substitutions between mtDNA haplotypes within D. simulans. Furthermore, the between-species comparison demonstrates up to 103 amino acid substitutions between D. melanogaster and D. simulans ((Ballard 2000); and see (Montooth et al. 2010) for all haplotype pairwise substitutions). These pairwise comparisons reveal two key features of this heterospecific introgression model that are important to formulate predictions for the current investigation. First, the degree of amino acid divergence within D. simulans is at least twice that of D. melanogaster. If we assume a greater level of mtDNA polymorphism within a species can potentially confer greater fitness differences than lower levels, then we would predict a priori that there would be greater divergence in phenotypes in the D. simulans clade than in the D. melanogaster clade. Second, the amino acid divergence within either species’ haplotypes is less than half the potential number of differences between species. A second prediction can thus be formulated that if tested, we would expect more phenotypic divergence between clades than within a clade. In view of the fact that in the D. simulans – D. melanogaster introgression model the mtDNA haplotypes are all resident on D. melanogaster nuclear backgrounds, we can further predict that the non-coevolved mtDNA haplotypes (in the D. simulans clade) will demonstrate deleterious phenotypes if compared to D. melanogaster mtDNA haplotypes. Is there any evidence, though, for differential fitness in the D. simulans haplotypes when on their native D. simulans nuclear background?
Cytoplasmic (including mtDNA) micro-injection experiments performed within D. simulans nDNA flies have previously shown differential fitness of D. simulans mtDNA haplotypes, judged as levels of heteroplasmy; a measure of mtDNA competitive exclusion (De Stordeur 1997). Assessed mtDNAs were from the monophyletic siI, siII and siIII haplotypes. Overall, the rank fitness of the haplotypes corresponded to siII>siIII>siI (De Stordeur 1997). In a separate examination using the same mtDNA haplotypes, development times (egg-to-puparium, and egg-to-eclosion) were shown to be longest in siII and siIII flies, and shortest in siI flies (James and Ballard 2003). A follow-up study showed repeatable haplotype frequency changes in perturbation-reperturbation experiments (Ballard and James 2004), corresponding to a rank fitness of siII>siIII>siI; the same as in De Stordeur (1997). There therefore appears to be functional variation between these haplotypes on a D. simulans nuclear background.
The observed fitness effects of alternative mtDNA haplotypes on a D. simulans nDNA background allows us to now ask the question: are the effects associated with mtDNA variation alone, or are they mediated by nuclear genetic variation? Previous studies have explicitly conducted experiments on a fixed nuclear background to eliminate this potential source of fitness variation. The heterospecific mito-nuclear introgression model provides a powerful experimental tool to test for repeatable main effects of mtDNA variation, yet crucially permits a simultaneous test of whether the rank order of mtDNA fitness changes on an alternative nuclear background (to test whether mtDNA selection is repeatable or not). Altered fitness on different nDNA backgrounds would provide good evidence for a mtDNA-nDNA epistasis for fitness.
Here, we tested whether there is any genetic variance for fitness of D. melanogaster and D. simulans mtDNA haplotypes when on D. melanogaster nDNA backgrounds. Our investigation was divided into separate species comparisons, so D. simulans mtDNA haplotypes were competed against each other and D. melanogaster mtDNA haplotypes were competed against each other. We did not compete D. melanogaster mtDNAs against D. simulans mtDNAs. Specifically, we explored the effect of genome introgressions in population perturbation- reperturbation cages. Perturbation cages allow the monitoring of haplotype frequency over discrete, non-overlapping generations of breeding to determine gross estimates of competitiveness, or fitness, between different mtDNA haplotypes (Arnason and Lewontin 1991; Hutter and Rand 1995). By reperturbing the population after a given number of generations it is possible to re-set the populations back to the original equal haplotype frequencies and observe the repeatability of haplotype selection coefficients, pre- and post- perturbation. Perturbation-reperturbation also allows subtle changes in nuclear genetic effects to be detected pre- and post- perturbation and allows delineation of selection and drift processes. Each population cage contained only one nuclear genetic background. In addition to the population cages, we also tested the fitness (fecundity) of introgressed genotypes in a non-competitive context to observe whether the fitness differences observed in population cages could be explained by the numbers of offspring produced by females of known mtDNA-nDNA introgressions.
Materials and Methods
Fly stocks
MtDNA-nDNA introgression flies, in which alternative nuclear DNA backgrounds have been precisely placed on different mtDNA haplotypes using balancer chromosomes, were used for this experiment. We used balancer chromosomes to effectively introgress genomes instead of reciprocal back-crossing because the latter procedure may retain nuclear variants from maternal parents during the back-crossing process (see Montooth et al. 2010). In brief, two nuclear backgrounds were used corresponding to the inbred laboratory strain Oregon R (OreR) and a wild-caught strain from Austria (AutW132, hereafter named Aut), described in Montooth et al. (2010). Both nuclear types were introgressed with either: (i) three D. simulans mtDNA haplotypes separately (mau12, siI, and sm21) corresponding to the monophyletic D. simulans siIII, siI, and siII haplotypes, respectively, or (ii) two D. melanogaster mtDNA haplotypes separately (Zimbabwe 53 (Zm53) and Oregon R (OreR)). These generated flies are viable and have been previously used to research the effects of mito-nuclear epistases on fitness (Montooth et al. 2010). Flies used in this experiment had previously been tetracycline-cleared to eliminate confounding effects of Wolbachia infection.
Perturbation reperturbation cages
To monitor the change in mitochondrial haplotype frequency over time, perturbation reperturbation cages were constructed to house ∼700 flies whose starting haplotype frequency was equal between haplotypes, following a similar procedure to Ballard and James (Ballard and James 2004). In total, the 16 experimental cages comprised of 8 cages with OreR nuclear genomes and 8 cages with Aut nuclear genomes. Each nuclear genome cage type was initiated with either: (a) each of three D. simulans mitochondrial haplotypes (siI, mau12 and sm21: starting frequency 33% each), or (b) each of two D. melanogaster mitochondrial haplotypes (Zm53 and OreR: starting frequency 50% each). Mau12 is a haplotype from D. mauritiana that differs by 1-2bp when compared to the D. simulans siIII haplotype, and is therefore phylogenetically equivalent to siIII. Each mtDNA-nDNA cage trial had four replicates per treatment. A population size ∼700 was targeted throughout to minimize the effects of fluctuating effective population size or genetic drift on mtDNA haplotype frequencies. Population sizes in each cage are reported in the Supplementary Materials Table S2 and Figure S4. Cages were maintained at 25° on a [12hr: 12hr] light: dark cycle. Prior to the experiment, flies from each haplotype were maintained at a controlled density for two generations.
To test the repeatability of haplotype frequency change over time, the experiment was divided into two sections; pre-perturbation and post-perturbation. The experimental food was a standard 2% yeast diet with no additional sprinkled yeast (11% sugar, 2% autolyzed yeast, 5.2% cornmeal, agar 0.79% w/v in water and 0.2% tegosept -methyl 4-hydroxybenzoate, from Sigma (St. Louis, MO, USA). Food (approximately 100ml) was added to a deep-sided petri dish (Fisher Scientific, Pittsburg, PA, USA). In the first generation, cages were seeded with presumably- mated five day old females (233 per genotype in D. simulans cages (699 total) and 350 per genotype in D. melanogaster cages (700 total)). Females were separated from males at ∼4 days old and allowed to recover for one day prior to addition into population cages (when no CO2 knock-down was used). These seeding females were allowed to lay eggs for 5 days. After 5 days of egg laying flies were removed and kept for genotyping (generation 0 estimate). The eggs in the petri dish were allowed to hatch and the adults eclose. After 14 days post egg laying adults were knocked-down using CO2 and transferred to a fresh population cage with a new petri dish and food. The freshly-transferred adults were allowed to lay eggs for 5 days and the process was repeated for subsequent generations. The pre-perturbation stage lasted 9 generations and in the 10th generation 48 presumably mated females were removed from each cage and each female was isolated into an individual vial to establish an isofemale line. The isofemales were allowed to lay eggs for 5 days and were then removed for genotyping. When the haplotypes of the isofemales from each cage were known (see genotyping protocol, below), all isofemale offspring of the same haplotype were pooled and mixed, and then the reperturbation experiment was conducted. The 11th generation (generation 0 post-reperturbation) was founded with equal numbers of individuals from the respective haplotypes of that cage (as in the initial set-up, above). The post-reperturbation episode lasted 13 generations and flies were sampled at generations 0, 6, and 13.
Non-competitive fecundity assay
To measure the fitness of different mtDNA haplotypes in a non-competitive environment, we first isolated 48 presumably mated isofemales from each of 16 cages at generation 10 pre-perturbation (see above) and allowed them to lay eggs, singly in single vials, for 5 days on standard 2% yeast-sugar-cornmeal medium with no added surface yeast (see above). After 5 days of egg laying, the isofemales were genotyped (see below) to determine their mtDNA haplotype. All flies from the same cage type have a known nuclear background and it was not necessary to genotype at nuclear loci. We counted the number of total eclosed offspring from each vial (16 cages × 48 vials = 768 isofemale vials that could be scored for a known haplotype). The number of offspring per vial was scored blind to the mtDNA haplotype identity. Following counting, the eclosed offspring of known haplotype were pooled to re-start the post-perturbation phase of the experiment (see above). The numbers of individuals of each haplotype scored for fecundity therefore represents a highly similar haplotype frequency of the generation nine populations.
DNA extraction
For pre-perturbation experiments flies were sampled at the first two generations (generations 0 and 1) and subsequently every 2nd generation (generation 3, 5, 7, 9). The post perturbation sampling frequency was the first (introduced) generation, then at generations six and thirteen. In total, for D. simulans haplotype cages, 92 flies were sampled per cage following the methods outlined in (Kilpatrick and Rand 1995), allowing up to three positive controls (corresponding to the three D. simulans haplotypes) and one negative control. Ninety three flies were sampled in D. melanogaster haplotype cages, allowing two positive controls (corresponding to the two D. melanogaster haplotypes) and one negative control. Briefly, each digest consisted of standard squish prep with 100μl squish buffer (10 mM Tris (pH 8.2), 1 mM EDTA and 25 mM NaCl and Proteinase K added to a concentration of 200µg/ml (see Gloor et al. 1993). DNA extractions were conducted in 96-well plates and flies were homogenized using a pellet pestle (Kontes, Sigma, St. Louis, MO, USA) and wooden toothpick. Following digestion at 37° for 1hr, the digestion mixture was denatured by boiling at 100° for 5 min. The resulting denatured homogenate was used as the DNA template for PCR.
Polymerase Chain Reaction (PCR)
A PCR product within the mtDNA Cytochrome Oxidase I (COI) gene was amplified using the forward primer 3593 (GAACAGTTCCCGCTTTAGGAG) and reverse primer 4528 (GCAGTTAATCGGACAGCTAATGTCCC). PCRs were conducted in 10μl volumes and reagent concentrations were as follows: 1X PCR Reaction Buffer (containing 1.5mM MgCl2 (Denville Scientific Inc.); 0.1mM of each dNTP (Invitrogen); 5µM of each primer (Invitrogen); 0.1U of Taq polymerase (Denville Scientific Inc.); 1μl squish prep DNA template. Thermocycling conditions were as follows: an initial incubation at 94° (2 min) followed by 30 cycles of 94° (30s), 54° (30s), 72° (45s). A final extension incubation at 72° (8 min) completed the reaction. Positive controls of haplotypes were run on each 96-well plate for the respective Drosophila species haplotypes.
Restriction Fragment Length Polymorphism (RFLP) analysis
Following PCR, PCR fragments were digested to characterize haplotype using a combination of two restriction enzymes. For D. melanogaster haplotypes, the enzyme AluI (Invitrogen) was used in a restriction digest using 10μl of PCR product as reaction template. 6μl of a restriction digest mixture containing 1.5μl 10X digestion buffer (Invitrogen), 0.15μl 1X BSA, 0.2μl AluI restriction enzyme, and 4.15μl H2O was added. For D. simulans haplotypes, a combination of AluI and RsaI enzymes were used to characterize haplotypes. The 10μl PCR product was split into two 5μl sub-samples and each was used as template for alternative enzymes. Each 5μl PCR product was added to 3μl of a digestion mixture containing 0.75μl 10X digestion buffer (Invitrogen), 0.075μl 1X BSA, 0.1μl AluI or RsaI restriction enzyme, and 2.075μl H2O. All restriction digests were incubated at 37° for 5 hr. Post incubation, the independent D. simulans digests from the same sample were pooled for genotype (haplotype) scoring. Restricted PCR products were scored by the same researcher using agarose gel electrophoresis. Restriction patterns were confirmed with positive sample controls and in silico digestions of the known fragment, providing the theoretical restriction pattern. For D. melanogaster haplotypes, frequency estimates were based on genotypes obtained from mean = 83.73 ± 11.63 (1 SD), range= 36-93 individuals. For D. simulans haplotypes, frequency estimates were based on genotypes obtained from mean = 85.63 ± 8.35 (1 SD), range = 50-92 individuals.
We found the starting haplotype frequencies to vary considerably between cages, in spite of equal numbers of representatives being added at the start of the pre- and post-reperturbation experimental phases. This posed a problem for the analyses, since most population cage studies assume the starting frequency to be equal, or exactly according to the ratio of flies from distinct genetic backgrounds, even if that is not a true assumption. Our results and those of (Hutter and Rand 1995; Kilpatrick and Rand 1995) suggest this assumption is not a robust strategy. Instead, we used the estimated frequencies from the flies at the end of the starting generation as the starting frequency, since the target starting frequency was not achieved. This was partly due to flies not surviving the first generation and presumably not contributing equally to the first generation breeders.
Statistical analysis
Statistical analyses were divided into pre- and post-perturbation phases. To estimate the selection coefficient, s, of each cage, we used linear models to regress the natural log of the haplotype frequency (independent variable) against generation (dependent variable) sensu Dykhuizen and Hartl (Dykhuizen and Hartl 1980). To determine if the overall haplotype frequency coefficient (including all four replicate cages) was significantly different from zero, we used general linear mixed-effect models on the natural log of haplotype frequency, with generation fitted as a fixed effect and cage ID fitted as a random effect. Markov Chain Monte Carlo sampling with 5000 iterations was used to calculate 95% confidence intervals (CI). Significance was judged as 95% CIs excluding zero. Log-likelihood ratio tests were performed on models with (mixed effect models: MEM) and without (linear models: LM) the random effect (Cage ID) fitted to determine if the haplotype frequency of cages was repeatable between generations within each haplotype. In other words, we asked whether cages that had relatively high haplotype frequencies in Generation 0, also had relatively high haplotype frequencies in Generations 1, 3, 5 etc., or alternatively, whether there were no repeatable effects between generations. Between generation repeatability for each cage, VR, was estimated as: VR = v/(v+r), where v= random effect variance across cages, and r= random effect residual across cages. The test statistic was 2× the difference in model Log-likelihoods (logLikeLM-logLikeMEM). Chi-squared tests with 1 d.f. on the test statistic gave the associated p-value. Significance was judged as P < 0.05.
For the analysis of the non-competitive fecundity assay, we conducted linear models to test the effects of mtDNA haplotype on offspring number. T-test statistics were used to test the difference between haplotypes compared to the model genotype, and ANOVAs were conducted to test for an overall haplotype effect. All statistical analyses and graphics were performed using the R-package software version 3.1.3 (R Core Team 2018).
Tests of isogenicity of the nuclear backgrounds
We tested the nuclear backgrounds for any nuclear variation using diagnostic tests on RNA-seq libraries from a previous study (Mossman et al. 2016b; Mossman et al. 2017) following a method outlined in (Mossman et al. 2019) . We performed ‘pair’ and ‘trio’ analyses on the .bam files (samtools v.1.1.19 (Li et al. 2009)) in four of the ten genotypes used here (OreR;OreR, siI;OreR, OreR;Aut, siI;Aut) (Figure S1). These genotypes had been maintained in the laboratory for approximately 100 generations at the time of this experiment. We tested for: (i) transcriptome nucleotide variants that segregate between haplotypes within a nuclear background (e.g., siI;OreR vs. OreR;OreR, and siI;Aut vs. OreR;Aut) (pair analysis), and (ii) variants that are present in the transcriptome within each of four genotypes (trio analysis). The pair and trio analyses provide a phred log ratio of genotype likelihoods with and without the pair or trio constraint (CLR: an integer value between 0-255). For the pair analysis we merged the replicate RNA-seq libraries (.bam files) and in the trio analysis we tested the individual replicate libraries against each other, assigning parent and offspring randomly in the trios. Inconsistent genotypes between parents and the offspring are flagged as putative variants and assigned a CLR score. We plotted the CLR of each putative variant against the linearized genome coordinates. The numerical results are described in the Supplementary Materials (Figure S2, Figure S1 and Table S1). Peaks in this nucleotide landscape indicate regions of dissimilarity between the transcriptome sequences. Higher CLR values indicate higher confidence in the difference at that transcriptome site. An extended methodology is described in the Supplementary Materials.
Data availability
Drosophila strains are available upon request. Population cage frequency data and fecundity data are supplied as Supplementary Materials. Supplemental material available at Figshare: https://doi.org/10.25387/g3.7691939.
Results
Pre-perturbation
For each mtDNA-type cage (D. melanogaster or D. simulans), equal numbers of individuals per haplotype were added to their allotted cages. These were 350 flies for each haplotype for the two-haplotype D. melanogaster cages, and 233 flies for each haplotype for the three-haplotype D. simulans cages. The same numbers of flies were introduced pre- and post-perturbation. However, in spite of careful counting of flies to start the population cages, there was considerable variation in the starting frequencies of mtDNA haplotypes at generation 0 (the starting generation) as determined by the PCR-RFLP assay (Figure 1). For all statistical analyses we used the haplotype frequency estimates throughout and not the absolute frequency of flies added. The former values better represent the number of flies that were alive at the end of the 1st generation, since some flies did not survive the 5 days of egg laying, and would therefore not have contributed to the egg population laid in the first 5 days.
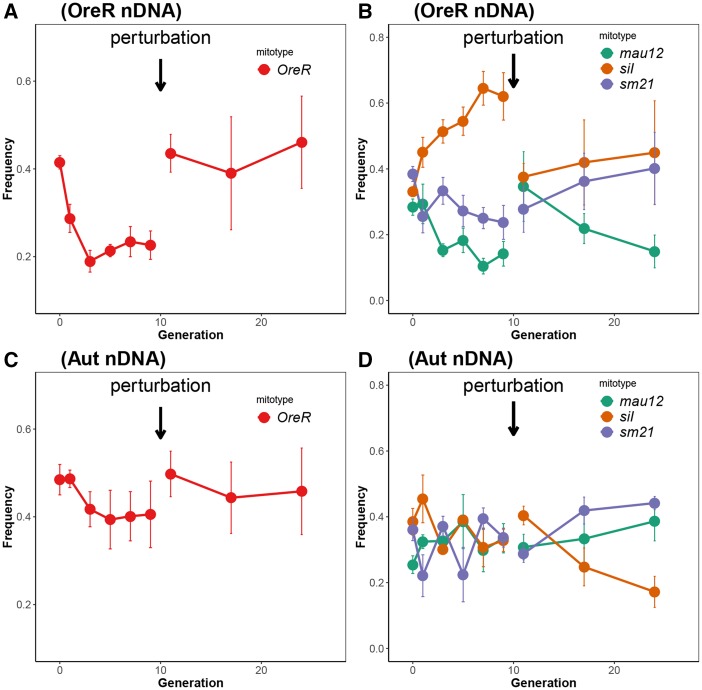
Figure 1.
Frequencies of mtDNAs with different nDNA types as a function of generational time in population cages. (A) shows the pre- and post-perturbation frequency changes for OreR nDNA when the competitor mtDNA types were OreR and Zm53. Only the frequency changes of the OreR haplotype (red line) is shown for clarity. Figure (C) shows the pre- and post-perturbation frequency changes of OreR mtDNAs on the Aut nDNA background. (B) shows the frequencies of D. simulans haplotypes in the pre- and post-perturbation cages on an OreR nuclear background. The figure displays the frequency changes of mau12 (green), siI (orange) and sm21 (purple) haplotypes over generational time. (D) shows the frequency changes of D. simulans haplotypes on an Aut nuclear background. Selection coefficients are reported in Tables 1 and 2. Mean frequencies ± 1 SEM are shown.
Pre-perturbation D. melanogaster mtDNA haplotype cages
For D. melanogaster mtDNA haplotypes, the frequency of the OreR mtDNA haplotype decreased relative to the Zim53 mtDNA as a function of generational time with selection coefficients (mean ± 1 SE) of -0.052 (0.020) and -0.028 (0.011) for OreR and Aut nuclear types, respectively (Table 1 and Figure 1A & 1C). We report only the selection coefficients of OreR mtDNA because the Zm53 mtDNA haplotype frequency is inversely proportional to that of OreR, and is therefore redundant. The rank order (repeatability, VR) of mtDNA haplotype frequency differed between cages for the OreR nuclear type (P > 0.05) as a function of generation. In other words, there was significant crossing of frequency trajectories between the cages and the cage with the highest frequency at generation 0 was not necessarily the same as the cage with the highest frequency at generations 1, 3, 5, 7 and 9. In contrast, the Aut nuclear type cages showed a significantly repeatable rank order of cages as a function of generational time; cages with relatively high frequency at generation 0 also had relatively high frequency at generations 1,3,5,7 and 9 (P < 0.05, Table 1 and Figure 1C). The rank order of the mean haplotype frequency (fitness) was Zm53>OreR on both nuclear genetic backgrounds (see Figure 1A & 1C).
Table 1. Linear and mixed effect model results for pre- and post-perturbation selection coefficients for D. melanogaster mtDNA cage types. Selection coefficients for each cage (S cage) and the overall haplotype selection coefficient (S Haplotype) are shown. 95% confidence intervals are shown, along with the repeatability (VR) of the rank order of each cage within haplotype (see Materials and Methods). P-values (in parentheses) are from Chi-sq distributions. Bold denotes significance at α=0.05.
| Pre-perturbation | Post-perturbation | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| nDNA | Haplotype | Cage | S cage | S Haplotype | 95% CI | VR | S cage | S Haplotype | 95% CI | VR |
| OreR | OreR | 1 | −0.102 (0.051) | −0.052 (0.020) | −0.092 to −0.012 | <0.001 | −0.046 (0.062) | −0.001 (0.019) | −0.054 to 0.054 | 0.63 |
| 2 | −0.026 (0.030) | (P = 0.91) | 0.007 (0.006) | (P = 0.049) | ||||||
| 3 | −0.054 (0.028) | 0.010 (0.056) | ||||||||
| 4 | −0.028 (0.046) | 0.024 (0.010) | ||||||||
| Aut | OreR | 1 | −0.047 (0.030) | −0.028 (0.011) | −0.055 to −0.003 | 0.55 | −0.027 (0.007) | −0.010 (0.011) | −0.042 to 0.022 | 0.62 |
| 2 | 0.025 (0.010) | (P = 0.003) | −0.042 (0.012) | (P = 0.053) | ||||||
| 3 | −0.019 (0.006) | 0.013 (0.006) | ||||||||
| 4 | −0.071 (0.012) | 0.015 (0.028) | ||||||||
Post-reperturbation D. melanogaster haplotype cages
In contrast to the pre-perturbation cages, there were no significant relationships between haplotype frequency and generational time for D. melanogaster mtDNA haplotypes in either OreR or Aut nuclear backgrounds (P > 0.05 in all cases). In the OreR nuclear genetic background, the rank order of OreR mtDNA frequency was significantly repeatable over time in the pre-perturbation cages. In the post-perturbation cages, however, there was no consistent directionality of selection and therefore no significant pattern that would suggest consistent selection on mtDNA haplotype. The OreR mtDNA haplotype frequency in the Aut nuclear background showed no repeatable rank order over time between cages (Table 1). However, the post-reperturbation cages demonstrated a consistent rank order of mean haplotype frequency, similar to the pre-perturbation cages. In both nuclear genetic backgrounds, Zm53 showed greater mean fitness than OreR (Zm53>OreR) (see Figure 1A & 1C).
Pre-perturbation D. simulans mtDNA haplotype cages
For D. simulans mtDNA haplotypes, there were clear differences between the selection coefficients of mtDNA haplotypes on alternative nuclear genetic backgrounds. For OreR nuclear genetic background, selection coefficients for mau12 (s= -0.106 ± 0.025) and sm21 (s = -0.044 ± 0.02) mtDNAs were negative and significant, whereas the siI (s = 0.062 ± 0.01) mtDNA significantly increased in frequency (Figure 1B and Table 2). All three selection coefficients were significantly different from zero at P < 0.05. The rank orders of haplotype frequency for all except one of the cage sets were non-repeatable, indicating that there was significant crossing over between the norms of reaction of cages within each haplotype between generations (Table 2). For the OreR nuclear type, the rank order of the mean haplotype frequencies at the end of the pre-perturbation episode was siI>sm21>mau12 (see Figure 1B).
Table 2. Linear and mixed effect model results for pre- and post-perturbation selection coefficients for D. simulans mtDNA cage types. Selection coefficients for each cage (S cage) and the overall haplotype selection coefficient (S Haplotype) are shown. 95% confidence intervals are shown, along with the repeatability (VR) of the rank order of each cage within haplotype (see Materials and Methods). P-values (in parentheses) are from Chi-sq distributions. Bold denotes significance at α=0.05.
| Pre perturbation | Post perturbation | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| nDNA | Haplotype | Cage | S cage | S Haplotype | 95% CI | VR | S cage | S Haplotype | 95% CI | VR |
| OreR | mau12(siIII) | 1 | −0.161 (0.057) | −0.106 (0.025) | −0.156 to −0.056 | <0.001 | 0.037 (0.030) | −0.085 (0.040) | −0.185 to 0.009 | 0.37 |
| 2 | −0.155 (0.016) | (P = 1.0) | −0.242 (0.050) | (P = 0.28) | ||||||
| 3 | −0.024 (0.022) | −0.096 (0.007) | ||||||||
| 4 | −0.085 (0.075) | −0.039 (0.016) | ||||||||
| siI (siI) | 1 | 0.058 (0.019) | 0.062 (0.010) | 0.042 to 0.084 | 0.29 | −0.003 (0.001) | −0.000 (0.018) | −0.055 to 0.051 | 0.64 | |
| 2 | 0.079 (0.022) | (P = 0.11) | 0.063 (0.027) | (P = 0.04) | ||||||
| 3 | 0.037 (0.018) | −0.004 (0.033) | ||||||||
| 4 | 0.074 (0.021) | −0.057 (0.002) | ||||||||
| sm21(siII) | 1 | 0.030 (0.046) | −0.044 (0.020) | −0.085 to −0.0004 | 0.28 | −0.008 (0.009) | 0.025 (0.037) | −0.053 to 0.105 | <0.001 | |
| 2 | −0.109 (0.035) | (P = 0.11) | −0.089 (0.044) | (P = 0.85) | ||||||
| 3 | −0.031 (0.014) | 0.136 (0.077) | ||||||||
| 4 | −0.068 (0.034) | 0.062 (0.022) | ||||||||
| Aut | mau12(siIII) | 1 | 0.017 (0.034) | 0.013 (0.019) | −0.026 to 0.052 | <0.001 | 0.023 (0.010) | 0.017 (0.019) | −0.033 to 0.066 | 0.39 |
| 2 | 0.043 (0.052) | (P = 1.0) | 0.006 (0.084) | (P = 0.25) | ||||||
| 3 | 0.041 (0.034) | 0.001 (0.026) | ||||||||
| 4 | −0.050 (0.029) | 0.039 (0.043) | ||||||||
| siI (siI) | 1 | −0.037 (0.030) | −0.029 (0.018) | −0.064 to 0.009 | <0.001 | −0.179 (0.013) | −0.083 (0.024) | −0.143 to −0.018 | 0.48 | |
| 2 | −0.024 (0.030) | (P = 0.72) | −0.048 (0.025) | (P = 0.15) | ||||||
| 3 | −0.074 (0.051) | −0.049 (0.014) | ||||||||
| 4 | 0.019 (0.014) | −0.055 (0.048) | ||||||||
| sm21(siII) | 1 | 0.064 (0.118) | 0.022 (0.034) | −0.051 to 0.090 | 0.10 | 0.037 (0.006) | 0.033 (0.010) | 0.013 to 0.054 | <0.001 | |
| 2 | −0.020 (0.070) | (P = 0.55) | 0.031 (0.021) | (P = 0.84) | ||||||
| 3 | 0.031 (0.046) | 0.050 (0.037) | ||||||||
| 4 | 0.014 (0.025) | 0.015 (0.004) | ||||||||
For the Aut nuclear genetic background, there were no significant relationships between haplotype frequency and generational time across all D. simulans mtDNA haplotypes (P > 0.05 in all cases, Table 2 and Figure 1D). For the Aut nuclear type, the rank order of the mean haplotype frequencies at the end of the pre-perturbation episode was siI=sm21=mau12 (all three haplotypes had a mean frequency of 33%, see Figure 1D).
Post-reperturbation D. simulans haplotype cages
For D. simulans mtDNA haplotypes, the post-reperturbation cages showed clear differences between the OreR nuclear background and the Aut nuclear background. In the OreR nuclear background, there was no evidence of selection on any haplotype, since all the haplotypes demonstrated non-significant selection coefficients (P > 0.05, Table 2). For the OreR nuclear type, the rank order of the mean haplotype frequencies at the end of the post-reperturbation episode was siI>sm21>mau12 (see Figure 1C), the same as in the pre-perturbation experimental episode.
In the Aut nuclear background, there were significant relationships between the frequency of siI (s= -0.083 ± 0.024) and sm21 (s= 0.033 ± 0.010) haplotypes and generational time, however, the selection coefficient of the mau12 mtDNA haplotype did not significantly differ from zero (Table 2 and Figures 1D). For the Aut nuclear type, the rank order of the mean haplotype frequencies at the end of the post-reperturbation episode was sm21>mau12>siI (see Figure 1D).
Non-competitive fecundity effects
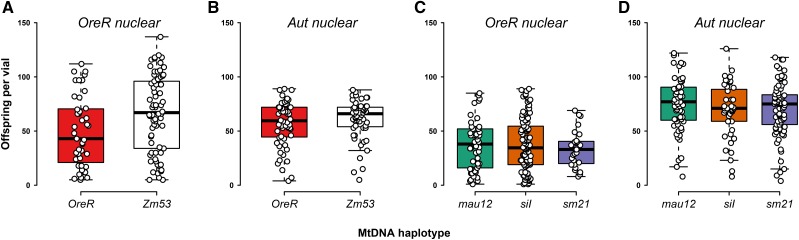
For the analyses of offspring number and haplotype variation for cages of known nuclear type, we found large differences between cage types for fecundity in the generation prior to reperturbation (Tables 3 and 4, Figure 2). In the OreR nuclear background cages, there was a significant difference between the number of offspring between D. melanogaster-type and D. simulans-type mtDNAs. OreR nDNA isofemales with D. melanogaster mtDNAs produced significantly greater offspring than isofemales harboring D. simulans mtDNAs (Table 4). In contrast, isofemales with the Aut nuclear background produced statistically greater numbers of offspring when harboring D. simulans-type mtDNAs (Table 4), and greater numbers than the OreR nuclear background overall. Within the cage types, the results were less dramatic (Table 3 and Figure 2). In OreR nDNA cages there were no statistical differences between the numbers of offspring per isofemale with different D. simulans mtDNA haplotypes (Table 3 and Figure 2C). There was also no effect of mtDNA haplotype on offspring number in the Aut nDNA background with D. simulans mtDNAs (Table 3 and Figure 2D). For the analyses of D. melanogaster type mtDNAs and offspring number, there were clear contrasts between the nuclear backgrounds. In the OreR nDNA background, the Zm53 mtDNA-bearing isofemales produced statistically greater numbers of offspring than the OreR mtDNA-bearing isofemales (Table 3 and Figure 2A). There was no effect of mtDNA haplotype on offspring number when the mtDNAs were on Aut nDNA genetic backgrounds (Table 3 and Figure 2B).
Table 3. MtDNA haplotype effects on isofemale offspring numbers for the four experimental cage types. We report results from models of individual haplotype performance against the model haplotype (mau12 in D. simulans types and OreR in D. melanogaster types) and conducted T-tests. Coefficients (β ± SE), T-test statistics and P-values are shown. We also report the effects of haplotype in an ANOVA analysis, where degrees-of-freedom, sum-of-squares, R2, F statistics and P-values are shown. Bold denotes significance at α=0.05.
| T-test (Individual haplotypes against model) | ANOVA (Haplotype effect) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Experimental mtDNA types | Nuclear type (intercept) | mtDNA type | β (± SE) | T | P | Model term | d.f. | Sum-of-squares | R2 | F | P |
| D. simulans | OreR | siI | 0.247 (3.99) | 0.062 | 0.95 | mtDNA | 2 | 355 | 0.004 | 0.339 | 0.71 |
| 37.18 (mau12) | sm21 | −3.716 (5.40) | −0.69 | 0.49 | Residuals | 171 | 89384 | ||||
| Aut | siI | −4.901 (4.97) | −0.99 | 0.33 | mtDNA | 2 | 776 | 0.007 | 0.624 | 0.54 | |
| 74.42 (mau12) | sm21 | −4.077 (4.34) | −0.94 | 0.35 | Residuals | 175 | 108762 | ||||
| D. melanogaster | OreR | Zm53 | 17.113 (6.651) | 2.57 | 0.01 | mtDNA | 1 | 7801 | 0.057 | 6.620 | 0.01 |
| 49.56 (OreR) | Residuals | 111 | 130810 | ||||||||
| Aut | Zm53 | 4.303 (3.719) | 1.16 | 0.25 | mtDNA | 1 | 514 | 0.012 | 1.339 | 0.25 | |
| 56.68 (OreR) | Residuals | 109 | 41841 | ||||||||
Table 4. Linear model results for isofemale number of offspring eclosed against mtDNA species type. Results for OreR and Aut nuclear types are shown. The model coefficients (β ± SE) are compared against the respective values for D. melanogaster mtDNAs (intercept= 60.16 for OreR nuclear type; intercept= 58.81 for Aut nuclear type). In the OreR nuclear background, isofemales with D. simulans mtDNAs produced significantly fewer offspring than isofemales with D. melanogaster mtDNAs. In the Aut nuclear background, isofemales with D. simulans mtDNAs produced significantly more offspring than isofemales with D. melanogaster mtDNAs (see Figure 3). In both nuclear backgrounds, there was a significant effect of mtDNA species type (D. simulans vs. D. melanogaster) on isofemale offspring numbers. Degrees-of-freedom, R2, F-statistics and P-values are shown. Bold denotes significance at α=0.05.
| T-test (Individual haplotypes against model) | ANOVA (Haplotype effect) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Nuclear type | Term against model | β (± SE) | T | Nuclear type | Model term | d.f. | Sum-of-squares | R2 | F | P |
| OreR | D. simulans mtDNA | −23.44 (3.42) | −6.85 | OreR | mtDNA species | 1 | 37644 | 0.145 | 46.98 | 4.43e-11 |
| Residual | 285 | 228350 | ||||||||
| Aut | D. simulans mtDNA | 12.68 (2.78) | 4.56 | Aut | mtDNA species | 1 | 10998 | 0.067 | 20.78 | 7.63e-6 |
Figure 2.
Mito-nuclear introgression effects on isofemale offspring production in a non-competitive five-day egg-laying period. Panels (A) and (B) show D. melanogaster mtDNAs on OreR and Aut nDNAs, respectively. Panels (C) and (D) show isofemale offspring numbers for D. simulans mtDNAs on OreR and Aut nDNAs, respectively. For comparisons within nDNA types, D. simulans mtDNA isofemales produced significantly more offspring than the D. melanogaster mtDNA types on the Aut background (comparison between (B) and (D); Table 4). D. melanogaster mtDNA-bearing isofemales produced significantly more offspring than the D. simulans mtDNA-bearing isofemales in the OreR nDNA background (comparison between (A) and (C); Table 4). Within a cage type, the only significant difference in offspring numbers were between Zm53 and OreR haplotypes on OreR nDNA (Figure 2A and Table 3).
Nuclear genome variation is present in the genetic stocks
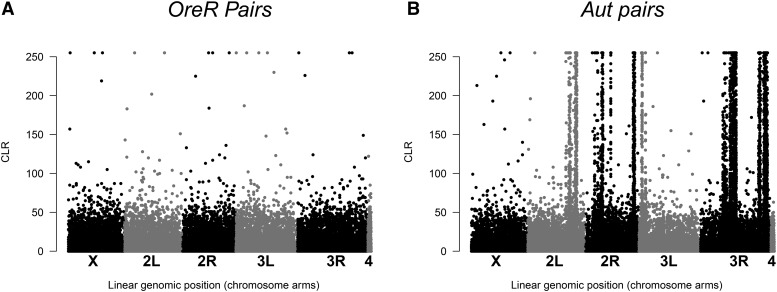
Our pairs and trios analyses suggest there are low levels of nucleotide variation within and between siI;OreR and OreR;OreR (Figures 3 and 4; Table S1). There are regions of the Aut nuclear background that contain clustered variants between the siI;Aut and OreR;Aut haplotypes (Figure 3B). On closer inspection in trio analyses, the between-haplotype variation identified in the pairs analysis is likely caused by SNPs that are segregating solely in the siI;Aut genotype libraries (Figure 4D; Table S1).
Figure 3.
Whole transcriptome SNP variation between haplotypes within nuclear genetic backgrounds (pairs analysis). (A) shows the relative chromosome position of putative transcriptome-wide SNPs on the abscissa against the CLR (confidence) score (see main text) on the ordinal axis for the between-OreR comparison (siI;OreR vs. OreR;OreR). (B) shows the between-Aut nuclear haplotype comparison (siI;Aut vs. OreR;Aut).
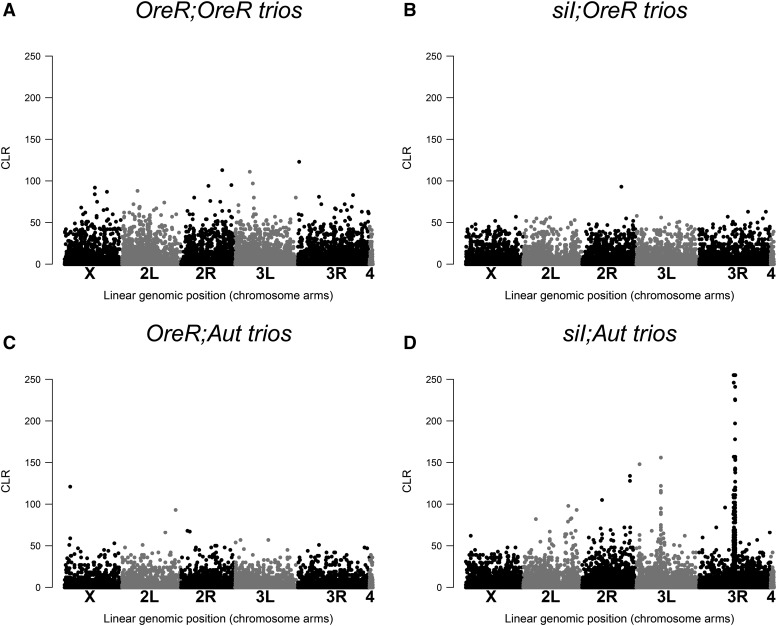
Figure 4.
Whole transcriptome SNP variation within mitonuclear genotypes (trios analysis). The trio analyses results are shown for OrR;OreR (A), siI;OreR (B), OreR;Aut (C), and siI;Aut (D). See Supplementary Materials for numerical estimates across CLR values (Figures S2, S3 and Table S1). The siI;Aut genotype shows evidence of some SNP variation, particularly in the 3R chromosome arm.
Discussion
The pre- and post-perturbation selection coefficient results suggest that population cages showed several alternative modes of changing mtDNA haplotype frequencies as a function of generational time. For the D. melanogaster mtDNA haplotypes, both of the pre-perturbation cage treatments showed frequency changes consistent with a selection regime. However, in the post-perturbation phase of the experiment, these cages no longer showed consistent and unidirectional mtDNA haplotype frequency changes with time, suggesting mtDNA frequencies were changing by a more neutral process such as genetic drift. In contrast, the D. simulans cages showed an alternative profile, that was also nuclear genetic background-specific. In the OreR nDNA background, the pre-perturbation cages showed evidence of unidirectional change, and the selection coefficients for each haplotype were significantly different from zero. Following reperturbation, these cages no longer showed a unidirectional and significant selection coefficient, consistent with a neutral process. In the Aut nuclear background, the pre-perturbation populations showed evidence consistent with neutral processes, whereas post-perturbation, two of the three cage types showed consistent and unidirectional selection coefficients. We found female fecundity differences between haplotypes on alternative nuclear backgrounds could only partly explain haplotype frequency changes in the population cages. We discuss these results in the context of female fecundity, the differences pre- and post-reperturbation, and the implications of G x G and G x G x E interactions affecting fitness in population cages. We finally discuss whether there was evidence to support our initial predictions based on the first principles of phylogenetic relatedness of haplotypes and its possible phenotypic consequences.
Female fecundity - a trait driving frequency changes in population cages?
There was considerable variation in the number of offspring produced in the non-competitive cage fecundity assays. The results suggest two main effects of nuclear and mtDNA co-variation. First, with OreR nuclear backgrounds, the greatest numbers of offspring were produced with the same species mtDNAs. That is, isofemales with OreR nDNA and D. melanogaster mtDNAs were statistically more fecund than isofemales with D. simulans mtDNAs, consistent with our co-adapted prediction. The opposite effect was evident with Aut nuclear genetic backgrounds; D. simulans mtDNA haplotypes produced significantly greater numbers of offspring than individuals with the co-evolved D. melanogaster mtDNAs, suggesting the nuclear variant modified the mtDNA fecundity results in an epistatic manner. We were surprised that the effects of D. simulans mtDNAs were advantageous in a D. melanogaster nuclear background, in spite of originating in a separate species and contrary to our prediction. However, we have previously recognized this effect in some of these genotypes for egg production (Meiklejohn et al. 2013). Comparisons between nuclear backgrounds in Figures 2A and 2B suggest that OreR mtDNA haplotypes perform more variably on OreR nDNA, because in the Aut nuclear background there is no significant difference between haplotypes (Table 3). Overall, we found that the population cage that demonstrated the greatest deviation from haplotype equality at an early stage of the pre-perturbation experiment was the OreR nuclear background with D. melanogaster mtDNA haplotypes. We found a concurrent significant difference between the performance of the D. melanogaster mtDNA haplotypes in offspring numbers, suggesting that only in cages where there are clear and statistical differences in fecundity between the competing genotypes, do we see clear signatures of selection, which is perhaps predictable.
Inconsistent cage behavior pre-and post-perturbation
Compared to the Ballard and James (2004) study, which found the fitness differences between haplotypes in pre-perturbation phase to be extensive and repeatable (in D. simulans nDNA backgrounds), we did not find the same haplotype effects when the D. simulans haplotypes are resident on D. melanogaster nDNA genetic backgrounds. In the Ballard and James study (2004), the siII haplotype showed the strongest increase in frequency, both pre- and post-perturbation, whereas in the present study, there was no significant increase in siII haplotype frequency on the Aut D. melanogaster nuclear background. In fact, on an OreR D. melanogaster nuclear background, the siII haplotype significantly decreased over time pre-perturbation, demonstrating the opposite effect and therefore suggesting a mtDNA-nDNA epistasis for fitness that is mediated by the D. melanogaster nuclear variant. Although we selected the same general haplotypes to test for repeatable fitness effects in different nuclear genetic backgrounds, there are obviously ‘species’ genetic variants between D. simulans and D. melanogaster nuclear backgrounds that confer fitness effects when in their native (co-evolved) D. simulans mtDNA-nDNA configuration. We further identified SNP variation within the siI;Aut genotype. These genetic differences could partly explain why we did not observe the same rank order of fitness of D. simulans haplotypes in both of the D. melanogaster nuclear backgrounds tested here. Furthermore, sporadic mtDNA-nDNA epistases may have been generated between generations at variable transcriptome sites and these unpredictable events are consistent with no overall signature of selection.
Why are there differences between mtDNA haplotype competitiveness depending on the nuclear (species) background? While we used balancer chromosomes to precisely place isogenic nuclear chromosomes on the mtDNAs (see Montooth et al. 2010), we still observed an effect of erosion of potentially beneficial (linked) markers between the OreR nuclear background and Zm53 D. melanogaster mtDNAs with generational time (see Figure 1A). In the same cage type (OreR nuclear) the Zm53 mtDNA frequency increased rapidly in the preliminary generations pre-perturbation, but formed an asymptote around generation four, an effect similar to other population cage frequency changes (Kilpatrick and Rand 1995). For the following generations, there was no apparent change in frequency and the haplotype did not fix in any of the four replicate populations. The results from the present population cage study are supported by the differences we observed between mito-nuclear types for offspring numbers (see above). In another population cage experiment (Kilpatrick and Rand 1995) the effect of decelerating haplotype frequency change could be explained by the decay of spurious linkage disequilibrium (LD: mtDNA-nDNA association) during the early generations of the experiment. Essentially, the ‘conditional hitchhiking’ in Kilpatrick and Rand (1995) arose through initial association between hybrid nDNA, and mtDNA variants. In theory each generation post- hybridization saw an erosion of LD by a factor of r = 0.5 on average, which was evident in the data, hinting that a nuclear genetic component was contributing to the main haplotype frequency change effect. In the same study any residual LD between mtDNA and nDNA, which was present in the pre-perturbation phase, was not sufficient to cause haplotype frequencies to significantly change post-repurturbation. In the present study we aimed to use balancer chromosome introgression to minimize this occurrence but we did identify some genetic variation within mitonuclear genotypes and between haplotypes within a nuclear background (Aut) that had persisted or had been generated since the genotype construction ∼100 generations prior to the transcriptome analyses. Despite careful genotype construction, the deleterious mutation rate of U∼1-1.2 per diploid genome per generation (Haag-Liautard et al. 2007; Keightley et al. 2009; Charlesworth et al. 2004) all but ensures that any experimental genetic study will have fitness-related LD that may decay over time.
Relaxed selection in the experimental procedure?
Interestingly, in D. simulans nuclear backgrounds, siI females have a significantly shorter egg-to-puparium development time than siII or siIII females (James and Ballard 2003), which would presumably confer a competitive advantage in a population cage. However, siI haplotype frequencies are dramatically reduced as a function of generational time in population cages (Ballard and James 2004), suggesting that development time is not a trait closely coupled to competitive advantage in population cages and is more likely linked to a lower probability of survival in this haplotype (James and Ballard 2003).
A more clear effect of female fecundity driving frequency change was observed by Hutter and Rand (1995), where a population cage experiment of mito-nuclear introgressed D. pseudoobscura and D. persimilis revealed egg production rate differences between genotypes could explain the competitive exclusion of the non-co-evolved D. persimilis mtDNA when it was introgressed with D. pseudoobscura nuclear DNA; the co-evolved D. pseudoobscura mtDNA-nDNA flies had a significant fertility advantage. The discrepancy between haplotype ‘competitiveness’ and female fertility measures in James and Ballard (2003) and Hutter and Rand’s (1995) studies suggest large fitness differences may be required to demonstrate significant changes in haplotype frequencies. Alternatively, non-competitive fitness assays may poorly predict the competitiveness of haplotypes in the population cage environment. The present study also found statistical differences in female fecundity, although we could not explain the differences in haplotype frequency changes by female fecundity alone.
We favor the hypothesis that selection on female life history traits has been relaxed in the current study. This has potentially allowed low fitness genotypes to co-exist with high fitness genotypes in cages when there is no significant fecundity difference between mtDNA haplotypes (as observed for all cages except OreR nuclear DNA with D. melanogaster mtDNAs). For example, if there was a significantly greater developmental time in one haplotype - as evidenced in the w501 D. simulans mtDNA in Montooth et al. (2010) and Meiklejohn et al. (2013)- that haplotype would shift to low frequency rapidly if the experimental transition between generations was before eclosion or reproductive maturation in that female’s mtDNA haplotype. On the other hand, allowing females to lay eggs for five days instead of three or four, we may have effectively relaxed the selection for early development and fecundity, and therefore potentially greater (and possibly equal) offspring numbers could be produced across all female haplotypes, providing no consistent evidence of a selection process. We kept the generation time constant in an effort to minimize any stochastic variation in egg laying between generations that could introduce density-induced effects on fitness (Clark and Feldman 1981). However, it is possible that the Aut nDNA flies, which have generally higher fecundity, laid more eggs in the 5 day laying period. We found that the population sizes in cages did fluctuate over time and this may have resulted in variable larval densities and thus variable larval competition, possibly mitigating any haplotype advantage of faster development.
G X G
The earliest investigations of mtDNA selection suggested that mtDNA behaves as a selectively neutral genetic marker (reviewed in (Rand 2001)) and (Ballard and Whitlock 2004)). Recent elegant studies have challenged this dogma and shown quite clearly strong non-neutral components of mtDNA selection (e.g., Ballard and James 2004). The present study adds further evidence for the complexity of mtDNA evolutionary dynamics and suggests that any selection advantage for fitness of a given mtDNA haplotype is dependent on the nDNA variant it is inherited with (in the case of ostensibly isogenic nDNA). Furthermore, we suggest any mtDNA haplotype selection is also dependent upon residual genetic variation within a population of nDNAs, which can be uncoupled through generational time via the erosion of LD between beneficial or deleterious mtDNA and nDNA variants (sensu conditional hitchhiking: (Kilpatrick and Rand 1995)). For example, the same D. simulans haplotypes used in Ballard and James (2004), when present on alternative D. melanogaster nDNAs, do not demonstrate repeatable selection either between nDNA types or pre- and post-repurturbation. In fact, we found that the rank order of the mean haplotype fitness was variable both between nDNA types and between pre- and post-perturbation experimental phases. This suggests that mtDNA x nDNA interactions are important to reveal or conceal fitness advantages associated with mtDNA selection, and that repurturbation can modify previously evident haplotype selection or neutrality.
G X G X E
The present study determined gross measures of fitness between mtDNA haplotypes using the change in haplotype frequency as a proxy of ‘fitness’. It is possible that different population cages (e.g., OreR nDNA vs. Aut nDNA) can modify their environment in different ways thus providing alternative environmentally-based selection landscapes (Dobzhansky and Spassky 1944). We found some evidence for this possibility in the Aut nDNA background, in which females are statistically more fecund than OreR nDNA females, probably altering the larval developmental environment. In the Aut nDNA background, D. simulans haplotypes demonstrated clear differences in selection coefficients pre- and post reperturbation. In the pre-perturbation phase, haplotypes showed dramatic stochastic fluctuating changes in frequency between generations whereas the post-perturbation phase showed more directional changes consistent with selection (Table 2 and Figure 1C). This may have arisen through nDNA variation being present and maintained pre-perturbation, essentially reducing the selection on mtDNA haplotype. One way nDNA variation may have been maintained is via a stochastic larval environment between generations. Alternatively, any nDNA variation (which was likely present in our cages) at mtDNA-interacting loci could have overridden the main mtDNA haplotype effect. Following reperturbation, residual nDNA variation, if greatly reduced through a genetic bottleneck, may have sensitized the mtDNA-nDNA gene complex to selection. In contrast, the OreR nDNA cages showed directional haplotype frequency change pre- perturbation, then no directional change post- perturbation. This may again be explained by gene x gene x environment interactions (G x G x E) (Arnqvist et al. 2010), which are known to be pervasive modifiers of fitness (Mossman et al. 2016a). We aimed to minimize environmental variation by maintaining a constant population size throughout and a consistent environment for egg laying, although the population sizes were evidently variable across generations. Therefore, larval density may have been considerably variable between nDNA types with a resulting environmental interaction modifying any main effect of mtDNA or nDNA-mtDNA epistasis for fitness. We were not able to assess the egg to larval to puparium to adult survival parameters in this investigation to test this hypothesis.
Any evidence to support phylogenetic predictions?
We formulated three basic predictions about expected fitness in the mtDNA-nDNA introgressed flies. These were: (i) there would be greater divergence in phenotypes in the D. simulans clade than in the D. melanogaster clade; (ii) there would be more phenotypic divergence between clades than within a clade; and (iii) the non-coevolved mtDNA haplotypes (in the D. simulans clade) will demonstrate deleterious phenotypes if compared to D. melanogaster mtDNA haplotypes.
Overall, we found some support for these prediction, although due to the inconsistent behavior of cages in pre- and post-reperturbation episodes of the experiment, we have to conclude that fitness as measured in population cages is a somewhat labile trait. Likewise, the female fecundity assays showed support for the predictions in some cases and contrary evidence in other cases. For female fecundity our first prediction is largely unsupported. We found the only significant difference in fecundity within a cage type to be between D. melanogaster haplotypes, and not the predicted D. simulans, with its greater level of mtDNA genetic polymorphism. For our second prediction we found some evidence of greater offspring number divergence between clades than within clades; in both nuclear types there were significant differences in offspring numbers that was associated with mtDNA species, whereas only one of the four cages demonstrated within-species differences. Our third prediction is supported in the OreR nuclear background as D. melanogaster mtDNA haplotypes performed better than D. simulans haplotypes. However, the Aut nuclear background reversed this effect, and D. simulans haplotypes outperformed the D. melanogaster (co-evolved) counterparts, contrary to our prediction. Generally, the largest effect on offspring numbers across all treatments was the variation associated with alternative nuclear background on D. simulans haplotypes.
In conclusion, there were no consistent patterns of frequency change over generational time for introgressed mito-nuclear genotypes. We did not observe patterns similar to those in previous studies which used the same haplotypes (D. simulans) but on alternative (conspecific D. simulans) nuclear backgrounds. We did find that the one cage that showed a strong and significant change in haplotype frequency was the same cage that possessed haplotypes with significantly different female fecundity in a non-competitive context. We suggest this fecundity difference has large implications for the strength of frequency change, and that competing haplotypes with very similar female fecundity will likely result in non-repeatable selection coefficients. Broadly speaking, genotype effects are likely to be sensitive to both interacting genes and the environments which they are in dialogue with and as a main consequence genetic variants may not display the same fitness effects under all experimental conditions. Furthermore, while mtDNA polymorphisms confer fitness effects in some nuclear backgrounds, they may display neutral effects when paired with alternative nDNAs.
Acknowledgments
We thank A-M Hernandez, reviewers, and the Editors for constructive comments which greatly improved the manuscript. Supported by NIH grants R01GM067862 from NIGMS, and R01AG027849 from NIA. This work was conducted using computational resources and services at the Center for Computation and Visualization, Brown University.
Footnotes
Supplemental material available at Figshare: https://doi.org/10.25387/g3.7691939.
Communicating editor: R. Kulathinal
Literature Cited
- Arnason E., Lewontin R. C., 1991. Perturbation-reperturbation test of selection vs. hitchhiking of the 2 major alleles of esterase-5 in Drosophila pseudoobscura. Genetics 129: 145–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnqvist G., Dowling D. K., Eady P., Gay L., Tregenza T., et al. , 2010. Genetic architecture of metabolic rate: environment specific epistasis between mitochondrial and nuclear genes in an insect. Evolution 64: 3354–3363. 10.1111/j.1558-5646.2010.01135.x [DOI] [PubMed] [Google Scholar]
- Ballard J. W. O., 2000. Comparative genomics of mitochondrial DNA in members of the Drosophila melanogaster subgroup. J. Mol. Evol. 51: 48–63. 10.1007/s002390010066 [DOI] [PubMed] [Google Scholar]
- Ballard J. W. O., James A. C., 2004. Differential fitness of mitochondrial DNA in perturbation cage studies correlates with global abundance and population history in Drosophila simulans. Proc. Biol. Sci. 271: 1197–1201. 10.1098/rspb.2004.2709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballard J. W. O., Rand D. M., 2005. The population biology of mitochondrial DNA and its phylogenetic implications. Annu. Rev. Ecol. Evol. Syst. 36: 621–642. 10.1146/annurev.ecolsys.36.091704.175513 [DOI] [Google Scholar]
- Ballard J. W. O., Whitlock M. C., 2004. The incomplete natural history of mitochondria. Mol. Ecol. 13: 729–744. 10.1046/j.1365-294X.2003.02063.x [DOI] [PubMed] [Google Scholar]
- Camus M. F., Clancy D. J., Dowling D. K., 2012. Mitochondria, Maternal Inheritance, and Male Aging. Curr. Biol. 22: 1717–1721. 10.1016/j.cub.2012.07.018 [DOI] [PubMed] [Google Scholar]
- Charlesworth B., Borthwick H., Bartolomé C., Pignatelli P., 2004. Estimates of the genomic mutation rate for detrimental alleles in Drosophila melanogaster. Genetics 167: 815–826. 10.1534/genetics.103.025262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clancy D. J., 2008. Variation in mitochondrial genotype has substantial lifespan effects which may be modulated by nuclear background. Aging Cell 7: 795–804. 10.1111/j.1474-9726.2008.00428.x [DOI] [PubMed] [Google Scholar]
- Clark A. G., Feldman M. W., 1981. Density-Dependent Fertility Selection in Experimental Populations of Drosophila melanogaster. Genetics 98: 849–869. [PMC free article] [PubMed] [Google Scholar]
- Clark A. G., Lyckegaard E. M. S., 1988. Natural-selection with nuclear and cytoplasmic transmission. 3. Joint analysis of segregation and mtDNA in Drosophila melanogaster. Genetics 118: 471–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis A. W., Roote J., Morley T., Sawamura K., Herrmann S., et al. , 1996. Rescue of hybrid sterility in crosses between D-melanogaster and D-simulans. Nature 380: 157–159. 10.1038/380157a0 [DOI] [PubMed] [Google Scholar]
- De Benedictis G., Rose G., Carrieri G., Luca M. D., Falcone E., et al. , 1999. Mitochondrial DNA inherited variants are associated with successful aging and longevity in humans. FASEB J. 13: 1532–1536. 10.1096/fasebj.13.12.1532 [DOI] [PubMed] [Google Scholar]
- De Stordeur E., 1997. Nonrandom partition of mitochondria in heteroplasmic Drosophila. Heredity 79: 615–623. 10.1038/hdy.1997.207 [DOI] [PubMed] [Google Scholar]
- Dobzhansky T., Spassky B., 1944. Genetics of natural populations. Xi. Manifestation of genetic variants in Drosophila pseudoobscura in different environments. Genetics 29: 270–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowling D. K., Friberg U., Hailer F., Arnqvist G., 2007. Intergenomic epistasis for fitness: Within-population interactions between cytoplasmic and nuclear genes in Drosophila melanogaster. Genetics 175: 235–244. 10.1534/genetics.105.052050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowling D. K., Friberg U., Lindell J., 2008. Evolutionary implications of non-neutral mitochondrial genetic variation. Trends Ecol. Evol. 23: 546–554. 10.1016/j.tree.2008.05.011 [DOI] [PubMed] [Google Scholar]
- Dykhuizen D., Hartl D. L., 1980. Selective neutrality of 6pgd allozymes in Escherichia coli and the effects of genetic background. Genetics 96: 801–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ephrussi B., Hottinguer H., Chimenes A. M., 1949. Action de lacriflavine sur les levures. 1. La mutation petite colonie. Ann. Inst. Pasteur (Paris) 76: 351–367. [Google Scholar]
- Fos M., Dominguez M. A., Latorre A., Moya A., 1990. Mitochondrial DNA evolution in experimental populations of Drosophila subobscura. Proc. Natl. Acad. Sci. USA 87: 4198–4201. 10.1073/pnas.87.11.4198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friberg U., Dowling D. K., 2008. No evidence of mitochondrial genetic variation for sperm competition within a population of Drosophila melanogaster. J. Evol. Biol. 21: 1798–1807. 10.1111/j.1420-9101.2008.01581.x [DOI] [PubMed] [Google Scholar]
- Garcia-Martinez J., Castro J. A., Ramon M., Latorre A., Moya A., 1998. Mitochondrial DNA haplotype frequencies in natural and experimental populations of Drosophila subobscura. Genetics 149: 1377–1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber A. S., Loggins R., Kumar S., Dowling T. E., 2001. Does Nonneutral Evolution Shape Observed Patterns of DNA Variation in Animal Mitochondrial Genomes? Annu. Rev. Genet. 35: 539–566. 10.1146/annurev.genet.35.102401.091106 [DOI] [PubMed] [Google Scholar]
- Gloor G. B., Preston C. R., Johnson-Schlitz D. M., Nassif N. A., Phillis R. W., et al. , 1993. Type I repressors of P element mobility. Genetics 135: 81–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haag-Liautard C., Dorris M., Maside X., Macaskill S., Halligan D. L., et al. , 2007. Direct estimation of per nucleotide and genomic deleterious mutation rates in Drosophila. Nature 445: 82–85 (erratum: Nature 453: 128). 10.1038/nature05388 [DOI] [PubMed] [Google Scholar]
- Hutter C. M., Rand D. M., 1995. Competition between mitochondrial haplotypes in distinct nuclear genetic environments – Drosophila pseudoobscura vs. D. persimilis. Genetics 140: 537–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innocenti P., Morrow E. H., Dowling D. K., 2011. Experimental Evidence Supports a Sex-Specific Selective Sieve in Mitochondrial Genome Evolution. Science 332: 845–848. 10.1126/science.1201157 [DOI] [PubMed] [Google Scholar]
- James A. C., Ballard J. W. O., 2003. Mitochondrial genotype affects fitness in Drosophila simulans. Genetics 164: 187–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keightley P. D., Trivedi U., Thomson M., Oliver F., Kumar S., et al. , 2009. Analysis of the genome sequences of three Drosophila melanogaster spontaneous mutation accumulation lines. Genome Res. 19: 1195–1201. 10.1101/gr.091231.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy E. P., Lehninger A. L., 1949. Oxidation of fatty acids and tricarboxylic acid cycle intermediates by isolated rat liver mitochondria. J. Biol. Chem. 179: 957–972. [PubMed] [Google Scholar]
- Kilpatrick S. T., Rand D. M., 1995. Conditional hitchhiking of mitochondrial-DNA - frequency-shifts of Drosophila melanogaster mtDNA variants depend on nuclear genetic background. Genetics 141: 1113–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewontin R. C., 1974. The Genetic Basis of Evolutionary Change, Columbia University Press, New York. [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., et al. , 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macrae A. F., Anderson W. W., 1988. Evidence for non-neutrality of mitochondrial-DNA haplotypes in Drosophila-pseudoobscura. Genetics 120: 485–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meiklejohn C. D., Holmbeck M. A., Siddiq M. A., Abt D. N., Rand D. M., et al. , 2013. An Incompatibility between a Mitochondrial tRNA and Its Nuclear-Encoded tRNA Synthetase Compromises Development and Fitness in Drosophila. PLoS Genet. 9: e1003238 10.1371/journal.pgen.1003238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montooth K. L., Meiklejohn C. D., Abt D. N., Rand D. M., 2010. Mitochondrial-nuclear epistasis affects fitness within species but does not contribute to fixed incompatibilities between species of Drosophila. Evolution 64: 3364–3379. 10.1111/j.1558-5646.2010.01077.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mossman J. A., Biancani L. M., Zhu C.-T., Rand D. M., 2016a Mitonuclear Epistasis for Development Time and its Modification by Diet in Drosophila. Genetics 203: 463–484. 10.1534/genetics.116.187286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mossman J. A., Mabeza R. M. S., Blake E., Mehta N., Rand D. M., 2019. Age of both parents influences reproduction and egg dumping behavior in Drosophila melanogaster. J. Hered. 10.1093/jhered/esz009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mossman J. A., Tross J. G., Jourjine N. A., Li N., Wu Z., et al. , 2017. Mitonuclear Interactions Mediate Transcriptional Responses to Hypoxia in Drosophila. Mol. Biol. Evol. 34: 447–466. 10.1093/molbev/msw246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mossman J. A., Tross J. G., Li N., Wu Z., Rand D. M., 2016b Mitochondrial-Nuclear Interactions Mediate Sex-Specific Transcriptional Profiles in Drosophila. Genetics 204: 613–630. 10.1534/genetics.116.192328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfanner N., Wiedemann N., Meisinger C., 2004. Double Membrane Fusion. Science 305: 1723–1724. 10.1126/science.1104244 [DOI] [PubMed] [Google Scholar]
- R Core Team , 2018. R: A language and environment for statistical computing, R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- Rand D. M., 2001. The units of selection on mitochondrial DNA. Annu. Rev. Ecol. Syst. 32: 415–448. 10.1146/annurev.ecolsys.32.081501.114109 [DOI] [Google Scholar]
- Rand D. M., Fry A., Sheldahl L., 2006. Nuclear-mitochondrial epistasis and Drosophila aging: Introgression of Drosophila simulans mtDNA modifies longevity in D-melanogaster nuclear backgrounds. Genetics 172: 329–341. 10.1534/genetics.105.046698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rand D. M., Haney R. A., Fry A. J., 2004. Cytonuclear coevolution: the genomics of cooperation. Trends Ecol. Evol. 19: 645–653. 10.1016/j.tree.2004.10.003 [DOI] [PubMed] [Google Scholar]
- Schon E. A., DiMauro S., Hirano M., 2012. Human mitochondrial DNA: roles of inherited and somatic mutations. Nat. Rev. Genet. 13: 878–890. 10.1038/nrg3275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smeitink J., van den Heuvel L., DiMauro S., 2001. The genetics and pathology of oxidative phosphorylation. Nat. Rev. Genet. 2: 342–352. 10.1038/35072063 [DOI] [PubMed] [Google Scholar]
- Sturtevant A. H., 1920. Genetic studies on drosophila simulans. I. Introduction. Hybrids with Drosophila melanogaster. Genetics 5: 488–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor R. W., Turnbull D. M., 2005. Mitochondrial DNA mutations in human disease. Nat. Rev. Genet. 6: 389–402. 10.1038/nrg1606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzagoloff A., 1982. Mitochondria, Plenum Press, New York. [Google Scholar]
- Wallace D. C., 1999. Mitochondrial diseases in man and mouse. Science 283: 1482–1488. 10.1126/science.283.5407.1482 [DOI] [PubMed] [Google Scholar]
- Wallace D. C., 2005. A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: A dawn for evolutionary medicine. Annu. Rev. Genet. 39: 359–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yee W. K. W., Sutton K. L., Dowling D. K., 2013. In vivo male fertility is affected by naturally occurring mitochondrial haplotypes. Curr. Biol. 23: R55–R56. 10.1016/j.cub.2012.12.002 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Drosophila strains are available upon request. Population cage frequency data and fecundity data are supplied as Supplementary Materials. Supplemental material available at Figshare: https://doi.org/10.25387/g3.7691939.