Abstract
Designer nucleases are versatile tools for genome modification and therapy development and have gained widespread accessibility with the advent of clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein (Cas) technology. Prokaryotic RNA-guided nucleases of CRISPR/Cas type, since first being adopted as editing tools in eukaryotic cells, have experienced rapid uptake and development. Diverse modes of delivery by viral and non-viral vectors and ongoing discovery and engineering of new CRISPR/Cas-type tools with alternative target site requirements, cleavage patterns and DNA- or RNA-specific action continue to expand the versatility of this family of nucleases. CRISPR/Cas-based molecules may also act without double-strand breaks as DNA base editors or even without single-stranded cleavage, be it as epigenetic regulators, transcription factors or RNA base editors, with further scope for discovery and development. For many potential therapeutic applications of CRISPR/Cas-type molecules and their derivatives, efficiencies still need to be improved and safety issues addressed, including those of preexisting immunity against Cas molecules, off-target activity and recombination and sequence alterations relating to double-strand-break events. This review gives a concise overview of current CRISPR/Cas tools, applications, concerns and trends.
Key Points
| CRISPR/Cas technology is widely applied for targeted genome modification, with ongoing discovery of new enzymes and improvements to specificity, delivery and efficiency. |
| Ingenious reengineering and reemployment of the basic ribonucleoprotein particles has already created versatile genome disruptors, transcriptional regulators, epigenetic modifiers and base editors. |
| Therapeutic approaches based on CRISPR/Cas technology have raised safety concerns in recent studies, indicating inherent risks, which require full characterization, and avoidable risks, which may be addressed by ongoing refinement of tools and protocols. |
| CRISPR/Cas has proven a disruptive innovation that has changed the conduct of functional studies, conception of disease models and creation of new therapies. |
The Context of Gene Therapy
Gene therapy, the introduction of genetic material for the cure or prevention of disease, has been 39 years in the making since its contentious beginnings [1]. Gene therapy by gene addition, providing an additional, functional copy of a disease-causing defective endogene, has already led to remarkable success for several genetic diseases, including gaining orphan drug approval [2]. Non-viral vectors are usually inefficient as delivery vehicles for gene addition [3], whereas integrating viral vectors impose size restrictions that are unsuitable for many potentially therapeutic genes, a challenge that can be addressed by combinatorial approaches but at the price of reduced efficiency [4–6]. Even in the case of smaller target genes, the size limit of the expression cassette typically remains an obstacle to adding intronic and regulatory sequences required for suitably regulated or high-level gene expression. For example, efficient lentiviral delivery of β-globin with an open reading frame (ORF) of only 441 base pairs is consistently based on size-reduced cassettes that only achieve subphysiological β-globin expression and frequently result in only partially therapeutic treatments for more severe forms of the disease [7], as reviewed in this issue by Ghiaccio et al. [8]. Delivery of short-hairpin RNAs as therapeutic cargo for posttranscriptional regulation overcomes the problem of size but is only of therapeutic interest in specific cases [9, 10] and, like all integrational approaches, still has the inherent risk of insertional mutagenesis [11]. A potentially safer option would be the application of synthetic nucleotides for modulation of gene expression or RNA processing [12], but despite its promise and decades of development, the approach continues to suffer problems with efficiency, systemic delivery, toxicity and the need for chronic application [13–15]. Finally, gene addition by non-integrating vectors even in its most advanced forms is still marred by low efficiency and durability or by adverse events upon repeat application [16–18]. What is more, therapeutic application of each of these approaches has taken years of insight into individual disease and expression mechanisms, and none is universally applicable to loss of function and toxic gain-of-function mutations. Such delays and restrictions in new therapy development for genetic diseases could be avoided if efficient straightforward correction of causative mutations were possible.
Gene Editing 101
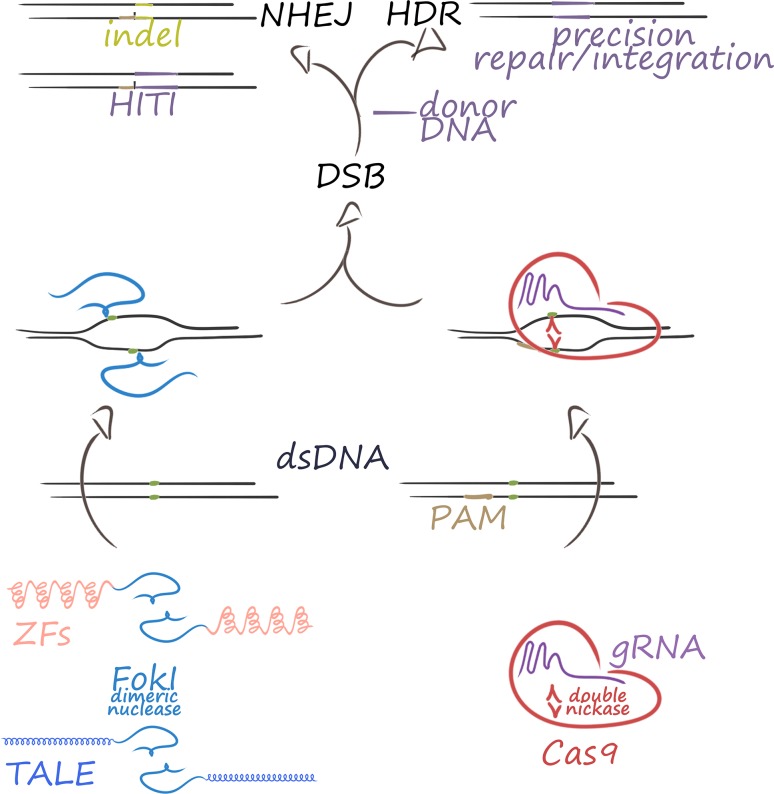
Progress in gene editing technology was thus of potentially critical importance for the gene therapy field. A key factor that would encourage accelerated adoption of the technology to genetic diseases and rare diseases in particular was the establishment of more versatile editing tools, with increasing simplicity of target redesign from meganucleases [19] over zinc finger (ZF) [20] and transcription activator-like effector (TALE) nucleases [21, 22] to clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated (Cas)-type RNA-guided nucleases (RGNs) [23–25] (Fig. 1). A property common to all these genome-editing technologies is the introduction of double-strand breaks (DSBs) at the site of interest in the genome as a trigger of targeted modification. In interphase, the DSB is usually quickly mended by the classical non-homologous end joining (NHEJ) pathway in the absence of a repair template [26], faithfully retaining the original DSB-flanking sequences in the assumed vast majority of ligation events [27]. However, in the continued presence of designer nucleases, the target is recleaved until no longer suitable for recognition, which leads to highly efficient introduction of insertions and/or deletions (indels) at the DSB site as recognizable events, making NHEJ ideal for disruption of endogenous sequences. A major alternative pathway for DSB repair and the one suitable for most therapeutic applications is homology-directed repair (HDR), which can be used to introduce precise changes at the target site. However, HDR is much slower than NHEJ, normally restricted to late S and G2 phases in a cell-type- and locus-dependent fashion [28, 29] and appears to be underrepresented in long-term repopulating hematopoietic stem cells (LT-HSCs) [30–33], a cell population of particular interest for curative treatment of genetic diseases such as rare anemias, immunological disorders and lysosomal storage disorders [34]. While work continues to shift the balance toward HDR (i.e., by ongoing refinement of NHEJ inhibitors, HDR enhancers, cell-cycle regulators, design of donor DNA templates and cell cycle-specific Cas9 expression) [30, 35–40] or to find ways of enriching suitably modified cells [41] to achieve therapeutically relevant efficiencies for DSB-mediated precision editing, the CRISPR/Cas system has emerged as the editing platform of choice and already as the basis for a new generation of editing and regulatory molecules.
Fig. 1.
The three most popular platforms of designer nucleases. Conventional gene editing is based on designer nucleases, of which ZF nucleases, TALE nucleases and CRISPR/Cas9 nucleases are the most popular. Most readily designed for new targets with suitable protospacer adjacent motif (PAM) sequences are CRISPR/Cas-type RGNs. All three classes of nucleases introduce a DSB as a basis for genome modification. ZF and TALE nucleases employ dimeric protein modules containing obligate dimeric FokI nucleases that introduce a staggered cut, whereas CRISPR/Cas9 is most frequently employed as a ribonucleoprotein complex made up of a single guide RNA (sgRNA) and a double-nickase protein component (Cas9). Most RGNs of CRISPR/Cas type introduce blunt-ended DSBs. For any nuclease platform, repair of the DSB by non-homologous end-joining (NHEJ) may achieve insertions and/or deletions (indels) or homology-independent targeted insertions (HITI) at high efficiency. Repair by homology-directed repair (HDR) in the presence of a suitable donor DNA may be used to correct or precisely insert a sequence of interest at the target site. dsDNA double-stranded DNA
CRISPR/Cas Versatility
RGNs (Fig. 2a), initially identified as a prokaryotic adaptive immune system, were the first programmable nuclease system to act as ribonucleoprotein particles (RNPs) and to employ simple Watson–Crick base pairing for target recognition. The corresponding ease of redesigning target specificity at long last allowed speedy and widespread adoption of genome-editing technology for new targets. Demonstration of effective redesign, characterization of two independent strand-specific nickase domains and modification for easier exploitation as genome-editing tools [42, 43] then allowed successful application of CRISPR/Cas9 to mammalian cells [24, 25]. Its versatility and ease of target design, construction and delivery suggested early on that the CRISPR/Cas system may bring about a landmark shift in gene therapy development [44], despite apparent limitations. For instance, the originally employed Streptococcus pyogenes Cas9 (SpCas9) restricted the choice of target sequence by sequence requirement of a DSB-proximal NGG protospacer adjacent motif (PAM) trinucleotide. However, this could be overcome by turning to naturally occurring CRISPR-associated enzymes from other species or by reengineering or evolving Cas9 to achieve alternative or relaxed RGN specificities [45–49]. Of note here are Staphylococcus aureus Cas9, which is particularly small and therefore most suited for adeno-associated virus (AAV)-mediated delivery [50, 51], and the Cas12a (aka Cpf1, CRISPR from Prevotella and Francisella) RGN system. Cpf1 nuclease uses a shorter RNA component more suitable for direct synthesis than that of other CRISPR/Cas systems and creates staggered DSBs 18 base pairs from the TTTV PAM tetranucleotide, with 5ʹ overhangs of 4–5 nucleotides [52–54]. These overhangs may favor directional and HDR-mediated events compared with Cas9-induced DSBs, which typically have blunt ends or single-nucleotide 5ʹ overhangs [55]. Incidentally, the presence of blunt- and sticky-end cutting enzymes in the adaptive CRISPR/Cas system parallels the presence of both types of enzymes in the “innate” prokaryotic defense system of restriction enzymes, which likewise uses DSB induction to fend off intruders and which provides valuable lessons in the dynamics of related repair events [56]. As data accumulate for large-scale application of RGNs across cell types, on-target events surrounding the DSB will become more predictable [57], facilitating designs for particular outcomes, such as frame-shift mutations, at the design stage [58]. As a problem common to all RGNs and designer nucleases, the relatively short recognition sequence, which for RGNs are encoded by the single guide RNA (sgRNA) component of the RNP, mediates on-target cleavage but also off-target cleavage of sequence-similar sites [59]. In this context, employment of mutated Cas9 nickases or heterologous nickases for dimeric target recognition [60, 61] of truncated sgRNAs [62] and of modified high-fidelity Cas9 molecules with minimal unspecific protein-DNA interactions [63, 64] all led to decreasing off-target activity and maintenance of on-target activity. In this vein, programmable Cas9 molecules, such as the recently engineered single-molecule protease-sensing Cas9s [65], may not only expand the possible uses of RGNs but may help reduce any residual off-target activity and its impact. Generally, off-target cleavage is exacerbated by prolonged expression of the designer nuclease and can thus be addressed by highly transient expression of RGNs [66], which will also depend on the delivery method. For instance, RNP-based delivery of CRISPR/Cas allows RNP detection for up to 48 h [66], where delivery as RNAs allows RNA detection for up to 24 h [67] and ongoing RNP action for up to 96 h [68], although this will vary with each specific application. By contrast, plasmid DNA in cells and tissue is highly stable, persists in vivo for several months [69–71] and, moreover, poses the risk of illegitimate integration in the genome [72–74]. The same is true for AAV vectors, which are highly efficient for in vivo delivery but persist as episomes for months [17]. For clinical application, delivery of the RGN components themselves (rather than of possible donor templates) as RNPs or RNAs is therefore highly preferable to delivery as naked DNA or as persistent viral vectors.
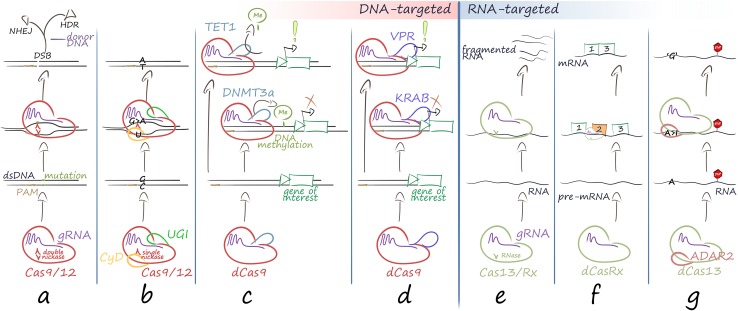
Fig. 2.
CRISPR/Cas9-based tools for therapy development. CRISPR/Cas ribonucleoprotein complexes already offer up an array of molecular tools, which all exploit sequence-specific recognition of DNA or RNA molecules by the sgRNA component of the complex. The following are important categories of CRISPR/Cas-based tools. aRNA-guided nucleases introduce a DSB to trigger repair by NHEJ or HDR. NHEJ introduces indels or can be exploited for homology-independent targeted integration of donor sequences, whereas HDR allows precise repair and homology-based insertion of donor sequences. bDesigner base editors (BEs) exploit chemical conversion of DNA bases to introduce permanent sequence changes without DSB. Shown is the conversion of C to U, the correction of the complementary base from G to A triggered by the Cas9 nickase activity, and the eventual establishment of a T-A base pair after DNA replication. cEpigenome regulators incorporate or recruit DNA or histone-modifying enzymes, such as the DNMT3 DNA methyltransferase or the TET1 DNA demethylase, which add or remove a methyl group (Me) from proximal CpG sequences. dTranscriptional regulators incorporate or recruit transcriptional activators or repressor for nonpermanent modulation of target gene expression. ePosttranscriptional regulators, such as Cas13, CasRx and derivatives, are mostly based on the Cas13 family of RNA-guided RNases and perform targeted RNA cleavage without sequence restriction to cognate PAM sites. fDeactivated CasRx may bind to cis-regulatory elements of pre-messenger RNA (pre-mRNA) and effect alternative splicing. gRNA BEs, in analogy to DNA BEs, contain a Cas13 fusion with an ADAR2 domain, which performs adenosine-to-inosine conversion. This A > I conversion is read as an A > G base change during translation and may be exploited to achieve codon changes or the removal of premature stop codons in the open reading frame of mRNAs. In contrast to DNA-targeting Cas molecules, RNA-targeting Cas molecules are not restricted to target sequences with a corresponding PAM sequence. For clarity, Cpf1, with its staggered DSB, RNA BEs and a plethora of additional tools, such as many additional transcriptional regulators and epigenome regulators [45] and different flavors of paired nickases (two-component RGNs), are not shown. Exclamation mark activation of expression; Red cross deactivation of expression; STOP sign translation termination (nonsense) codon, dCas9 deactivated Cas9 without endonuclease activity, dsDNA double-stranded DNA, catalytic domains for functional expansion of the RGN complex: CyD cytidine deaminase domain for C > U conversion in the ssDNA loopout, currently with precision of ≤ 2 bp, UGI uracil DNA glycosylase inhibitor domain to prevent base excision repair and removal of base edit, DNMT3a catalytic domain of DNA methyltransferase 3 alpha for DNA methylation and potentially persistent repression of gene expression for affected promoters [82], TET1 catalytic domain of Ten-Eleven Translocation dioxygenase 1 (TET) for DNA demethylation and potentially persistent transcriptional activation of affected promoters [84], VPR VP64 (four tandem repeats of herpes simplex virus VP16) linked to p65 (the transactivation domain of nuclear factor [NF]-κB) and Rta (the Epstein-Barr virus transcriptional activation domain) for broad and potent transcriptional activation of affected promoters [174], KRAB catalytic domain of Krüppel-associated box epigenetic repressor [175], ADAR2 adenosine deaminase acting on RNA 2 [101]
CRISPR/Cas Beyond the Double-Strand Break
CRISPR/Cas molecules form the basis for new classes of sequence-targeted DNA modifiers by modular combination of their RNP components with additional functional domains. These molecules currently include designer base editors (BEs) [75–81], epigenome modifiers [82–86] and transcriptional regulators [87]. BEs (Fig. 2b) exploit CRISPR/Cas9-mediated target recognition and Cas9 fusion with nucleotide-specific chemical modifiers plus auxiliary components to achieve chemical conversion of DNA bases. To date, conversion of cytosine to uracil (read as thymine; by cytidine deaminase) [76, 78–80] and of adenine to inosine (read as guanine; by artificially evolved adenine deaminase) [81] has been demonstrated. Triggered by nickase activity of a mutated Cas9 and made more efficient by inhibiting base excision repair of the altered base, the conversion is then copied over to the opposite strand to create a double-stranded sequence change without DSB and thereby with minimal chance of recombination events and at potentially high efficiency [75]. For functional analyses without DSB induction, cytidine-deaminase-based BEs alone would be able to produce functional knockouts by introduction of stop codons in 98.6% of known protein-coding genes in the human genome [88]. Recently, a novel Cpf1-based cytidine deaminase BE was developed, which expands the targeting range for base editing from the G/C-rich PAM of Cas9 to the T/A-rich PAM of Cpf1, at low frequency of indel formation, non-C-to-T substitutions and off-target editing [89]. BEs therefore have great therapeutic potential for the correction of single-base mutations, especially in the case of adenine-deaminase BEs that have the capacity to correct G > A mutations, which account for approximately half of all known pathogenic single nucleotide polymorphisms [81]. However, BEs are restricted to catalyzing base transitions, i.e., to the conversion of a purine to another purine or of a pyrimidine to another pyrimidine. This inability to catalyze base transversions, i.e., substitutions of purines with pyrimidines and vice versa, combined with PAM restrictions and a limited editing window, therefore constrains their use to only a fraction of all pathogenic single-nucleotide mutations. Epigenome modifiers (Fig. 2c) can modify the accessibility of target DNA and contain fusions of nuclease-deactivated Cas9 (dCas9) with functional enzyme domains or with a binding domain recruiting such enzymes, including histone methylases [82, 83], histone acetyltransferases [82, 83, 86], DNA methylases [82] and DNA demethylases [84]. In transcriptional regulators (Fig. 2d), the dCas9 or sgRNA component of RGNs is fused to transcriptional activator or repressor domains [45] and can be used to regulate gene expression of specific target genes or even gene networks [90, 91]. Application of CRISPR/dCas9 alone can also lead to repression of gene transcription by interfering with the successful formation of the RNA polymerase complex machinery at the initiation step or by disrupting the elongation stage, both via steric hindrance [85, 92]. Modification of protein or RNA components of the RGN allows regulated modulation of transcription [93, 94] and even differential ligand-induced regulation of multiple genes [94]. Sufficiently small artificial transcriptional regulators of this type can be delivered by AAV vectors, which has achieved activity in target cells for over 5 months [95]. However, action of CRISPR/Cas RNPs is not restricted to DNA. Analyses of CRISPR/Cas molecules as a prokaryotic defense system had already indicated targeted RNA cleavage by certain Cas molecules [96] 5 years before CRISPR/Cas9 was also harnessed for targeted cleavage of eukaryotic messenger RNA (mRNA) in 2014 by provision of mRNA-complementary PAM-containing oligonucleotides [97]. Discovery and application of new RNA-targeting CRISPR/Cas molecules continues [98–100] and has recently led to the discovery of the RNA-editing Cas13 family [99–101] and the engineering of CasRx, which shows potent and specific RNA cleavage in human cells and, at a small size of 930 amino acids, is suitable for delivery by AAV vectors [102] to primary cells and for in vivo delivery (Fig. 2e). In addition to the knockdown activity of RNA-binding Cas complex, utilization of a catalytically inactivated CasRx guided by single or multiple guide RNA (gRNA) has the capacity to regulate pre-mRNA splicing (i.e., exon excision) and act as splice effectors (Fig. 2f) [102]. Moreover, and in analogy to designer DNA BEs, modification of Cas13-type molecules can be exploited to edit RNA and convert adenosine to inosine (read as guanosine), for instance to repair premature stop codons, without PAM sequence constraints on the target sequence [101] (Fig. 2g).
Brief Instructions for the Toolkit
The current momentum of novel discovery and developments of and around CRISPR/Cas suggests that the tools we have in hand after barely 6 years of research into tailored RGNs are all but the tip of the iceberg. However, these tools already provide a veritable arsenal of possible treatment strategies for genetic diseases. Based on HDR with short donor templates or by base editing, we can fix causative point mutations, activate positive disease modifiers of disease severity and introduce stop codons into gain-of-function toxic ORFs or into negative modifiers. Based on HDR in combination with gene addition, we can minimize insertional mutagenesis by targeting large expression cassettes into inert genomic loci, such as the paradigmatic safe-harbor AAVS1 locus inside the PPP1R12C gene in the case of hematopoietic stem and progenitor cells. Alternatively, we can target gene fragments or complementary DNA (cDNA) into endogenous loci to replace defective sequences and achieve physiological expression from the resulting chimeric expression cassette. Based on NHEJ, we can disrupt toxic mutated genes or regulatory sequences of disease modifiers or, using pairs of RGNs, excise and rearrange parts of the genome. Based on epigenetic and transcriptional regulators, we can boost or dampen expression, respectively, of positive and negative disease modifiers or directly counteract the effect of causative mutations in regulatory regions. For transcriptional regulators, this can already be done in a regulated and multiplexed fashion for several regulatory targets [94]. Finally, the recent addition to the CRISPR/Cas-based toolkit of posttranscriptional regulators that perform targeted RNA cleavage allows the removal of toxic RNAs or accelerated turnover of normal RNAs to alter the transcriptome and cell state [83].
Approaches acting at the level of genomic DNA introduce permanent changes and are best performed using one-off transient RGN applications to limit off-target effects. Conversely, transcriptional and posttranscriptional modifiers need to act continuously and thus require one-off permanent RGN delivery or repeated transient application over time. For epigenetic modifiers, long-term maintenance and thus the need for reapplication of epigenetic reprogramming depend on the nature and locus of the modification [85]. Delivery of CRISPR/Cas components may be performed by a plethora of methods [68, 103], viral delivery (as AAV or lentivirus [LV]) and electroporation (as RNP or RNAs) being amongst the most widely applicable. Transient delivery may be achieved by electroporation of RNPs or RNAs or by transduction with integration-defective viral vectors, whereas permanent delivery would rely on integrating LVs or, at lower efficiency but also lower risk of insertional mutagenesis, by employing an additional designer nuclease for integration into AAVS1 or other suitable loci of choice. Either methodology may be employed for ex vivo and in vivo delivery of the CRISPR/Cas complex, with a preference of DNA-free delivery by electroporation for ex vivo approaches for reasons of biosafety, and of AAV-mediated delivery for in vivo approaches for reasons of efficiency in a range of tissues [18]. Where donor templates or tags for integration at DSBs are required, those may be delivered as either AAV vectors or as naked nucleic acids or analogs, depending on insert size and type of application [18, 103–105].
A Lever to Move the Medical World
Based on these CRISPR/Cas-based technologies, therapy development for many human diseases, including infectious diseases, cancers and monogenically inherited diseases, is underway. As regards human infectious diseases, one of the most intensely researched viral diseases is HIV, for which several CRISPR/Cas-based therapeutic approaches are being evaluated. These include antiviral action by elimination of the C-C chemokine receptor type 5 (CCR5) HIV co-receptor [106] and excision of the HIV provirus from latently infected cluster of differentiation (CD)-4+ T cells for complete elimination of the virus [107]. Besides disruption of integrated proviruses as antiviral therapy, CRISPR/Cas can also eliminate episomal viral DNA species, such as the covalently closed circular DNA responsible for relapse after withdrawal of antiviral treatments in hepatitis B and C infections [108, 109]. In cancers, CRISPR/Cas therapeutic application has two main angles. On the one hand, CRISPR/Cas has been used to accelerate development of chimeric antigen receptor (CAR) T cells [110], enhance their action or enhance the action of naturally occurring T cells [111] as an auxiliary factor to therapy. On the other hand, CRISPR/Cas may be used as direct and possibly the primary therapeutic agents, as proposed for noncoding RNAs [112] or as shown with pro-apoptotic effect for cancer cells by knockout of the programmed cell death protein 1 (PD-1) [113]. The latter approach is at the basis of at least seven current clinical trials (ClinicalTrials.gov ID NCT02793856, NCT02863913, NCT02867332, NCT02867345, NCT03044743, NCT03081715, NCT03342547) for various types of cancer and of an additional trial based on PD-1 knockout in combination with gene-edited CAR T-cells (NCT03545815). As regards monogenically inherited disease and reviewed elsewhere in this issue [114], ease of CRISPR/Cas design favors therapy development for an abundance of rare diseases, most of which are currently without a cure. The CRISPR/Cas strategies employed for rare diseases are highly varied and, depending on the disease mechanism and type of causative mutation, may include disruption, precision editing, targeted integration of expression cassettes or of gene fragments, and transcriptional and posttranscriptional regulation. Despite the experimental nature of therapies based on the nascent CRISPR/Cas technology, three clinical trials aiming to treat patients with rare diseases are already registered, all three with a focus on β-hemoglobinopathies (NCT03655678, NCT03728322 and NCT03745287).
Towards future therapies, through the ease of creating large sgRNA libraries and corresponding viral vectors or RNP complexes, CRISPR/Cas technology also greatly facilitates functional genetic screens for forward or reverse genetics [115]. Compared with screens based on knockdown by RNA interference (RNAi), CRISPR/Cas screens have much greater versatility, in that CRISPR/Cas-based transcriptional inactivation screens mimic RNAi-based screens [116], whereas knockout screens allow more reproducible, complete inactivation of targets in coding as well as non-coding parts of the genome [117, 118], and whereas gain-of-function screens additionally allow assaying for the effects of transcriptional activation [118]. In the context of viral infections, genome-wide screens using CRISPR/Cas have thus already achieved identification of additional host-dispensable critical host factor for HIV for future therapy [119], of antiviral host defense mechanisms against flaviviruses [120] and of critical host factors for additional viruses, such as Dengue, hepatitis C, West Nile and Zika viruses [121], with a clinical trial for the identification of norovirus-critical host factors underway (NCT03342547). As examples for cancer research, CRISPR/Cas is employed to validate shortlisted drug-resistance candidates [122, 123], knockout screens have already identified therapeutic targets for BRCA-, KRAS- and MYCN-linked cancers and large B-cell lymphomas [124–128], and a transcriptional activation screen has revealed genes conferring resistance to cancer treatment by BRAF inhibitors [129]. Because high-throughput screens depend on readily scorable phenotypes, such as viral infection, cell death or cell proliferation, screening for nonmalignant genetic diseases with phenotypes that mostly only appear at the organismal level is more difficult. Nevertheless, here CRISPR/Cas screens have already revealed novel genes with therapeutic relevance in β-hemoglobinopathies [128], Parkinson’s disease [130], thromboembolisms [131] and inflammatory responses [132] and have shortlisted genes required for processes as diverse as neuronal fate [133], energy metabolism [134] and ferritinophagy [135], giving pointers to novel disease-related pathways and candidate genes.
CRISPR/Cas-type molecules thus have great potential both as therapeutic tools and for the identification of future therapeutic targets. Combined with the wider importance of CRISPR/Cas technology for therapy development, where it also contributes to several-fold accelerated development of disease models by facilitating the creation of isogenic cell-line [136–138] and animal [138–146] models, CRISPR/Cas technology has already become an essential and ubiquitous component of biomedical research for infectious, malignant and nonmalignant diseases.
Room for Improvement
As versatile and powerful as CRISPR/Cas technology may already be, many aspects of the technology still need to be improved, understood, or both, for therapeutic application. Whereas for research applications, suboptimal aspects of CRISPR/Cas technology will become an inconvenience or a significant cost factor, for therapy these same aspects will prevent regulatory approval for clinical application or may turn a trial into a tragedy. The issues that remain to be addressed may be divided into aspects of safety, efficiency and utility, with some overlap between the categories.
Safety
A key safety concern is that of off-target activity, which is still hard to predict, despite large-scale analysis efforts, and is in part brought about by issues of cell-type- and development-specific genome accessibility, which cannot be fully controlled [147]. The problem of potential off-target activity also affects BEs, which moreover have an editing window of 2–5 nucleotides that limits their use for precision editing of certain target sequences [78, 148]. While off-target activity at least may be reduced by high-fidelity variants or, for DSB-based approaches, by paired CRISPR/Cas nickases or CRISPR/FokI dimeric nucleases, approval for clinical trials will be difficult to obtain without comprehensive experimental analysis of potential off-target effects, so as to minimize the residual risk to patients [149]. Another safety concern is that of pre-existing immunity against Cas molecules in humans [39, 150, 151]. While this issue may be of minor importance for most ex vivo approaches and may be overcome for in vivo approaches by alternative enzymes or transient immunosuppression [39], it nevertheless casts a shadow of doubt over in vivo applications of CRISPR/Cas, which are already at risk of adverse reactions or lowered efficiency by pre-existing immunity to AAV and other delivery vectors [152–154]. Two additional and significant safety concerns are directly linked to the induction of DSBs for genome editing by RGNs and could altogether be prevented by approaches that act independent of DSBs, such as base editing and transcriptional or posttranscriptional regulation. First, effective DSB induction may lead to P53-dependent apoptosis, and recent independent studies have pointed out that selecting for DSB-dependent editing events may thus select for P53-deficient cells and cell populations with elevated cancer risk [155–157]. Such enrichment may be prevented by suppressing P53 activity at the time of editing, and it remains to be shown whether the observations for both studies can be reproduced in clinically relevant cell types. As a second safety concern linked to DSB-dependent editing, even single DSB events have been demonstrated to induce unpredictable indel and interchromosomal recombination events [158]. While future research will show how far such events can be minimized by encouraging speedy and faithful DSB repair, they appear to be inherent to RGN activity and therefore represent a safety risk tied up with their application. Further safety concerns exist regarding the long-term safety of editing events. One such concern arises from the long persistence of AAV vectors in target tissue. A favorite vehicle for in vivo delivery of HDR donor templates, AAV vectors show long-term persistence of circular episomal concatemers and the occurrence of illegitimate recombination and rare random integration events [17, 159], which may represent a significant lifetime risk of insertional mutagenesis for treated patients. Another concern is common to all gene therapy approaches based on permanent modification of stem cells and relates to the natural turnover of stem cells actively contributing to tissue reconstitution. Exemplary work on HSCs and multipotent progenitor cells demonstrates that treated cells experience waves of quiescence and active contribution to repopulation [160, 161]. Disconcertingly, this implies that events of insertional mutagenesis or off-target-related genotoxicity in quiescent cells may remain hidden for an extended period of time until the affected cell may become active and experience pathological clonal expansion.
Efficiency
The frequency of HDR events is a major concern for many applications, particularly owing to bias of HDR events for less basic stem cells. This might partly be addressed by ex vivo selection of genome-edited cells [162], by refinement of additives for the maintenance of stemness in ex vivo procedures and culture [30, 163], by timing DSB induction with suitable phases of the cell cycle, or by improving the selection procedure for enrichment of true stem cells [164], which would also help lower vector requirements and the cost of the procedure. Moreover, Cpf1 appears to have an advantage over Cas9 molecules in general for certain HDR-mediated knock-in or editing events, which might partly relate to the staggered DSB introduced by Cpf1, akin to that produced by TALENs [165, 166]. Additionally, the nature and symmetry of the HDR donor template affect HDR efficiency [166, 167]. Therefore, many factors allow optimization of HDR efficiency, from cell isolation, selection and culture to choice of RGN and donor sequence and type.
Utility
Several aspects of Cas9 are points of practical limitation for their employment. For one, Cas9 molecules are fairly large and difficult to deliver by AAV for systemic application, a problem that is aggravated for Cas9 fusion proteins, such as BEs, that include additional functional domains. Although this may be addressed by split delivery [168], the search continues for smaller Cas-type molecules that would more readily accommodate additional domains and efficient delivery. As a second point, PAM site requirement is an evolutionary necessity of the bacterial adaptive immune system and prevents self-cleavage of the host, so that the search for additional Cas variants with alternative PAM sites continues to extend the choice of target sites. However, true freedom to select the target site of DNA-targeting CRISPR/Cas molecules, albeit at higher likelihood of off-target hits for single-molecule CRISPR/Cas approaches, would only be warranted by protein engineering and removal of PAM restriction altogether, as it already applies for RNA-targeting CRISPR/Cas molecules.
Of Disruptive Technologies
In any field of science, new discovery critically depends upon new technological advances and then, in turn, engenders new technological advances. Sometimes the resulting developments are so profound or their practical implications so wide-ranging that they cause a break with previous industry and scientific practice. A key example of such disruptive technologies in molecular biology is the polymerase chain reaction, with such versatility in its application that it had a rapid impact on virtually every aspect of molecular biology shortly after its publication in 1986 [169]. It then seemed that the discovery of RNAi as an antiviral defense and developmental regulatory system may also find wide-ranging biotechnological application [170]. However, more than 20 years after its first conceptualization [171], RNAi has simply turned into one of several arrows in our quiver for reverse genetics and functional analyses and only gradually is its potential for therapy coming to fruition [172, 173]. Given the RNAi experience, it was therefore hard to gauge early on whether the adoption of a prokaryotic antiviral defense system for genome engineering [25] truly heralded a new revolution in molecular biology or merely a further addition of tools for research and therapy development. However, and as we have summarized here, in the 6 years since its original application to mammalian cells, CRISPR/Cas technology has been adopted so widely and has been modified, functionally extended and adapted for so many purposes, that its utility to molecular biology and genetics has taken on a new quality altogether. In more senses than one, therefore, and going well beyond its application for design and delivery of effective therapeutic tools, CRISPR/Cas technology might prove a truly disruptive technology for many aspects of biomedicine and fundamental research.
Funding
The authors were co-funded by TELETHON Cyprus, the EU Erasmus+ program and the European Union’s Seventh Framework Program for Research, Technological Development and Demonstration under Grant agreement no. 306201 (THALAMOSS).
Conflict of interest
CWL, PP and MK have no conflicts of interest that are directly relevant to the content of this article. No writing assistance was utilized in the production of this manuscript.
References
- 1.Johnson RS. Gene transfer experiment in humans meets with scant approval. Jama [Internet]. 1980;244:2139–40. http://jama.jamanetwork.com/article.aspx.articleid=372398. Accessed 14 Nov 1980. [PubMed]
- 2.Aiuti A, Roncarolo MG, Naldini L. Gene therapy for ADA-SCID, the first marketing approval of an ex vivo gene therapy in Europe: paving the road for the next generation of advanced therapy medicinal products. EMBO Mol Med [Internet]. 2017;9:737–40. http://www.ncbi.nlm.nih.gov/pubmed/28396566. Accessed 31 May 2017. [DOI] [PMC free article] [PubMed]
- 3.Šimčíková M, Prather KLJ, Prazeres DMF, Monteiro GA. Towards effective non-viral gene delivery vector. Biotechnol Genet Eng Rev [Internet]. 2015;31:82–107. http://www.ncbi.nlm.nih.gov/pubmed/27160661. Accessed 11 May 2018. [DOI] [PubMed]
- 4.Counsell JR, Asgarian Z, Meng J, Ferrer V, Vink CA, Howe SJ, et al. Lentiviral vectors can be used for full-length dystrophin gene therapy. Sci Rep [Internet]. Nature Publishing Group; 2017;7:79. http://www.ncbi.nlm.nih.gov/pubmed/28250438. Accessed 10 May 2018. [DOI] [PMC free article] [PubMed]
- 5.Chamberlain K, Riyad JM, Weber T. Expressing Transgenes that exceed the packaging capacity of adeno-associated virus capsids. Hum Gene Ther Methods [Internet]. Mary Ann Liebert, Inc.; 2016;27:1–12. http://www.ncbi.nlm.nih.gov/pubmed/26757051. Accessed 10 May 2018. [DOI] [PMC free article] [PubMed]
- 6.Canté-Barrett K, Mendes RD, Smits WK, van Helsdingen-van Wijk YM, Pieters R, Meijerink JPP. Lentiviral gene transfer into human and murine hematopoietic stem cells: size matters. BMC Res Notes. 2016;9:312. doi: 10.1186/s13104-016-2118-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bluebird Bio Announces New England Journal of Medicine Publication of Interim Data from Two Phase 1/2 Clinical Studies of LentiGlobinTM Gene Therapy in Patients with Transfusion-Dependent β-Thalassemia. [Internet]. bluebird bio Press Statement (Published 2018-04-18). https://www.businesswire.com/news/home/20180418006407/en/bluebird-bio-Announces-New-England-Journal-Medicine. Accessed 26 Apr 2018.
- 8.Ghiaccio V, Chappell M, Rivella S, Breda L. Gene therapy for beta-hemoglobinopathies: milestones, new therapies and challenges. Mol Diagn Ther. 2019; 10.1007/s40291-019-00383-4. [DOI] [PubMed]
- 9.Brendel C, Guda S, Renella R, Bauer DE, Canver MC, Kim YJ, et al. Lineage-specific BCL11A knockdown circumvents toxicities and reverses sickle phenotype. J Clin Invest. 2016;126:3868–3878. doi: 10.1172/JCI87885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Patsali P, Papasavva P, Stephanou C, Christou S, Sitarou M, Antoniou MN, et al. Short-hairpin rna against aberrant HBBIVSI-110(G > A) mRNA restores β-globin levels in a novel cell model and acts as mono-and combination therapy for β-thalassemia in primary hematopoietic stem cells. Haematologica [Internet]. 2018;103:e419–23. http://www.haematologica.org/lookup/doi/10.3324/haematol.2018.189357. Accessed 17 Jan 2019. [DOI] [PMC free article] [PubMed]
- 11.Cesana D, Ranzani M, Volpin M, Bartholomae C, Duros C, Artus A, et al. Uncovering and dissecting the genotoxicity of self-inactivating lentiviral vectors in vivo. Mol Ther [Internet]. 2014;22:774–85. http://www.ncbi.nlm.nih.gov/pubmed/24441399. [DOI] [PMC free article] [PubMed]
- 12.van Agtmaal EL, André LM, Willemse M, Cumming SA, van Kessel IDG, van den Broek WJAA, et al. CRISPR/Cas9-Induced (CTG-CAG)n Repeat instability in the myotonic dystrophy type 1 locus: implications for therapeutic genome editing. Mol Ther [Internet]. Am Soc Gene Cell Ther. 2017;25:24–43. http://www.ncbi.nlm.nih.gov/pubmed/28129118. Accessed 11 May 2018. [DOI] [PMC free article] [PubMed]
- 13.Jason TLH, Koropatnick J, Berg RW. Toxicology of antisense therapeutics. Toxicol Appl Pharmacol [Internet]. 2004;201:66–83. http://www.ncbi.nlm.nih.gov/pubmed/15519609. Accessed 2018 May 11. [DOI] [PubMed]
- 14.Juliano RL. The delivery of therapeutic oligonucleotides. Nucleic Acids Res [Internet]. 2016;44:6518–48. http://www.ncbi.nlm.nih.gov/pubmed/27084936. Accessed 11 May 2018. [DOI] [PMC free article] [PubMed]
- 15.Philippou S, Mastroyiannopoulos NP, Makrides N, Lederer CW, Kleanthous M, Phylactou LA. Selection and identification of skeletal-muscle-targeted RNA Aptamers. Mol Ther Nucleic Acids [Internet]. 2018;10:199–214. http://www.ncbi.nlm.nih.gov/pubmed/29499933. Accessed 11 May 2018. [DOI] [PMC free article] [PubMed]
- 16.Stavrou EF, Lazaris VM, Giannakopoulos A, Papapetrou E, Spyridonidis A, Zoumbos NC, et al. The β-globin Replicator greatly enhances the potential of S/MAR based episomal vectors for gene transfer into human haematopoietic progenitor cells. Sci Rep [Internet]. 2017;7:40673. http://www.ncbi.nlm.nih.gov/pubmed/28106085. Accessed 11 May 2018. [DOI] [PMC free article] [PubMed]
- 17.Colella P, Ronzitti G, Mingozzi F. Emerging issues in AAV-mediated in vivo gene therapy. Mol Ther Methods Clin Dev [Internet]. American Society of Gene & Cell Therapy; 2018;8:87–104. http://www.ncbi.nlm.nih.gov/pubmed/29326962. Accessed 11 May 2018. [DOI] [PMC free article] [PubMed]
- 18.Lau C-H, Suh Y. In vivo genome editing in animals using AAV-CRISPR system: applications to translational research of human disease. F1000Research [Internet]. 2017;6:2153. http://www.ncbi.nlm.nih.gov/pubmed/29333255. Accessed 13 May 2018. [DOI] [PMC free article] [PubMed]
- 19.Smith J, Grizot S, Arnould S, Duclert A, Epinat JC, Chames P, et al. A combinatorial approach to create artificial homing endonucleases cleaving chosen sequences. Nucleic Acids Res. 2006. [DOI] [PMC free article] [PubMed]
- 20.Kim YG, Cha J, Chandrasegaran S. Hybrid restriction enzymes: zinc finger fusions to Fok I cleavage domain. Proc Natl Acad Sci. 1996. [DOI] [PMC free article] [PubMed]
- 21.Cermak T, Doyle EL, Christian M, Wang L, Zhang Y, Schmidt C, et al. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res [Internet]. 2011;39:e82. http://www.ncbi.nlm.nih.gov/pubmed/21493687. Accessed 16 Mar 2011. [DOI] [PMC free article] [PubMed]
- 22.Christian M, Cermak T, Doyle EL, Schmidt C, Zhang F, Hummel A, et al. Targeting DNA double-strand breaks with TAL effector nucleases. Genetics. 2010. [DOI] [PMC free article] [PubMed]
- 23.Maeder ML, Gersbach CA. Genome-editing technologies for gene and cell therapy. Mol Ther. 2016;24:430–446. doi: 10.1038/mt.2016.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, et al. RNA-guided human genome engineering via Cas9. Science (80-) [Internet]. NIH Public Access; 2013;339:823–6. Available from: http://www.ncbi.nlm.nih.gov/pubmed/23287722. Accessed 10 May 2018. [DOI] [PMC free article] [PubMed]
- 25.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, et al. Multiplex genome engineering using CRISPR/Cas systems. Science (80-). NIH Public Access; 2013;339:819–23. [DOI] [PMC free article] [PubMed]
- 26.Her J, Bunting SF. How cells ensure correct repair of DNA double-strand breaks. J Biol Chem [Internet]. 2018;jbc.TM118.000371. http://www.ncbi.nlm.nih.gov/pubmed/29414795. Accessed 11 May 2018. [DOI] [PMC free article] [PubMed]
- 27.Bétermier M, Bertrand P, Lopez BS. Is non-homologous end-joining really an inherently error-prone process? Jinks-Robertson S, editor. PLoS Genet [Internet]. Public Library of Science; 2014;10:e1004086. http://dx.plos.org/10.1371/journal.pgen.1004086. Accessed 11 May 2018. [DOI] [PMC free article] [PubMed]
- 28.Zaboikin M, Zaboikina T, Freter C, Srinivasakumar N. Non-Homologous End Joining and Homology Directed DNA Repair Frequency of Double-Stranded Breaks Introduced by Genome Editing Reagents. Hu W, editor. PLoS One [Internet]. 2017;12:e0169931. http://www.ncbi.nlm.nih.gov/pubmed/28095454. Accessed 03 May 2018. [DOI] [PMC free article] [PubMed]
- 29.Miyaoka Y, Berman JR, Cooper SB, Mayerl SJ, Chan AH, Zhang B, et al. Systematic quantification of HDR and NHEJ reveals effects of locus, nuclease, and cell type on genome-editing. Sci Rep [Internet]. 2016;6:23549. http://www.ncbi.nlm.nih.gov/pubmed/27030102. Accessed 11 May 2018. [DOI] [PMC free article] [PubMed]
- 30.Genovese P, Schiroli G, Escobar G, Di Tomaso T, Firrito C, Calabria A, et al. Targeted genome editing in human repopulating haematopoietic stem cells. Nature [Internet]. Nature Publishing Group; 2014;510:235–40. http://www.ncbi.nlm.nih.gov/pubmed/24870228. [DOI] [PMC free article] [PubMed]
- 31.Hoban MD, Cost GJ, Mendel MC, Romero Z, Kaufman ML, Joglekar A V., et al. Correction of the sickle cell disease mutation in human hematopoietic stem/progenitor cells. Blood [Internet]. American Society of Hematology; 2015;125:2597–604. http://www.ncbi.nlm.nih.gov/pubmed/25733580. [DOI] [PMC free article] [PubMed]
- 32.DeWitt MA, Magis W, Bray NL, Wang T, Berman JR, Urbinati F, et al. Selection-free genome editing of the sickle mutation in human adult hematopoietic stem/progenitor cells. Sci Transl Med. 2016;8. [DOI] [PMC free article] [PubMed]
- 33.DeWitt M, Magis W, Bray NL, Wang T, Berman JR, Urbinati F, et al. Efficient correction of the sickle mutation in human hematopoietic stem cells using a Cas9 ribonucleoprotein complex. bioRxiv. 2016;036236.
- 34.Steward CG, Jarisch A. Haemopoietic stem cell transplantation for genetic disorders. Arch Dis Child [Internet]. 2005;90:1259–63. http://www.ncbi.nlm.nih.gov/pubmed/16301554. Accessed 13 Apr 2018. [DOI] [PMC free article] [PubMed]
- 35.Richardson CD, Ray GJ, DeWitt MA, Curie GL, Corn JE. Enhancing homology-directed genome editing by catalytically active and inactive CRISPR-Cas9 using asymmetric donor DNA. Nat Biotechnol [Internet]. 2016;34:339–44. http://www.ncbi.nlm.nih.gov/pubmed/26789497. Accessed 11 May 2018. [DOI] [PubMed]
- 36.Gutschner T, Haemmerle M, Genovese G, Draetta GF, Chin L. Post-translational regulation of Cas9 during G1 enhances homology-directed repair. Cell Rep [Internet]. Elsevier Ltd; 2016;14:1555–66. 10.1016/j.celrep.2016.01.019. [DOI] [PubMed]
- 37.Orthwein A, Noordermeer SM, Wilson MD, Landry S, Enchev RI, Sherker A, et al. A mechanism for the suppression of homologous recombination in G1 cells. Nature. 2015. [DOI] [PMC free article] [PubMed]
- 38.Lin S, Staahl BT, Alla RK, Doudna JA. Enhanced homology-directed human genome engineering by controlled timing of CRISPR/Cas9 delivery. Elife. 2014;3:e04766. doi: 10.7554/eLife.04766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Charlesworth CT, Deshpande PS, Dever DP, Dejene B, Gomez-Ospina N, Mantri S, et al. Identification of pre-existing adaptive immunity to Cas9 proteins in humans. bioRxiv [Internet]. Cold Spring Harbor Laboratory; 2018;243345. https://www.biorxiv.org/content/early/2018/01/05/243345. Accessed 18 May 2018.
- 40.Song J, Yang D, Xu J, Zhu T, Chen YE, Zhang J. RS-1 enhances CRISPR/Cas9- and TALEN-mediated knock-in efficiency. Nat Commun [Internet]. Nature Publishing Group; 2016;7:1–7. 10.1038/ncomms10548. [DOI] [PMC free article] [PubMed]
- 41.Dever DP, Bak RO, Reinisch A, Camarena J, Washington G, Nicolas CE, et al. CRISPR/Cas9 beta-globin gene targeting in human haematopoietic stem cells. Nature [Internet]. Nature Publishing Group; 2016;539:384–9. https://www.ncbi.nlm.nih.gov/pubmed/27820943%5Cnhttp://www.nature.com/nature/journal/v539/n7629/pdf/nature20134.pdf. [DOI] [PMC free article] [PubMed]
- 42.Gasiunas G, Barrangou R, Horvath P, Siksnys V. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc Natl Acad Sci [Internet]. 2012;109:E2579–86. http://www.ncbi.nlm.nih.gov/pubmed/22949671. Accessed 10 May 2018. [DOI] [PMC free article] [PubMed]
- 43.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science (80-.). 2012;816–21. [DOI] [PMC free article] [PubMed]
- 44.Cathomen T, Ehl S. Translating the genomic revolution - targeted genome editing in primates. N Engl J Med [Internet]. 2014;370:2342–5. http://www.ncbi.nlm.nih.gov/pubmed/24918378. Accessed 12 June 2014. [DOI] [PubMed]
- 45.Komor AC, Badran AH, Liu DR. CRISPR-based technologies for the manipulation of eukaryotic genomes. Cell [Internet]. 2017;168:20–36. 10.1016/j.cell.2016.10.044. [DOI] [PMC free article] [PubMed]
- 46.Hu JH, Miller SM, Geurts MH, Tang W, Chen L, Sun N, et al. Evolved Cas9 variants with broad PAM compatibility and high DNA specificity. Nature. 2018;556:57–63. doi: 10.1038/nature26155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gao L, Cox DBT, Yan WX, Manteiga JC, Schneider MW, Yamano T, et al. Engineered Cpf1 variants with altered PAM specificities. Nat Biotechnol. 2017;35:789–792. doi: 10.1038/nbt.3900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kleinstiver BP, Prew MS, Tsai SQ, Topkar VV, Nguyen NT, Zheng Z, et al. Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature. 2015;523:481–485. doi: 10.1038/nature14592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tóth E, Czene BC, Kulcsár PI, Krausz SL, Tálas A, Nyeste A, et al. Mb- and FnCpf1 nucleases are active in mammalian cells: activities and PAM preferences of four wild-type Cpf1 nucleases and of their altered PAM specificity variants. Nucleic Acids Res. 2018;46:10272–10285. doi: 10.1093/nar/gky815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Friedland AE, Baral R, Singhal P, Loveluck K, Shen S, Sanchez M, et al. Characterization of Staphylococcus aureus Cas9: a smaller Cas9 for all-in-one adeno-associated virus delivery and paired nickase applications. Genome Biol [Internet]. BioMed Central; 2015;16:257. Available from: http://genomebiology.com/2015/16/1/257. Accessed 13 May 2018. [DOI] [PMC free article] [PubMed]
- 51.Kleinstiver BP, Prew MS, Tsai SQ, Nguyen NT, Topkar VV, Zheng Z, et al. Broadening the targeting range of Staphylococcus aureus CRISPR-Cas9 by modifying PAM recognition. Nat Biotechnol [Internet]. 2015;33:1293–8. http://www.ncbi.nlm.nih.gov/pubmed/26524662. Accessed 13 May 2018. [DOI] [PMC free article] [PubMed]
- 52.Zetsche B, Gootenberg JSS, Abudayyeh OOO, Slaymaker IMM, Makarova KSS, Essletzbichler P, et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell [Internet]. 2015/10/01. Elsevier Inc.; 2015;163:759–71. http://www.cell.com/article/S0092867415012003/fulltext. [DOI] [PMC free article] [PubMed]
- 53.Kleinstiver BP, Tsai SQ, Prew MS, Nguyen NT, Welch MM, Lopez JM, et al. Genome-wide specificities of CRISPR-Cas Cpf1 nucleases in human cells. Nat Biotechnol. 2016;34:869–874. doi: 10.1038/nbt.3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Singh D, Mallon J, Poddar A, Wang Y, Tippana R, Yang O, et al. Real-time observation of DNA target interrogation and product release by the RNA-guided endonuclease CRISPR Cpf1 (Cas12a). Proc Natl Acad Sci [Internet]. 2018;201718686. http://www.ncbi.nlm.nih.gov/pubmed/29735714. Accessed 13 May 2018. [DOI] [PMC free article] [PubMed]
- 55.Jinek M, Jiang F, Taylor DW, Sternberg SH, Kaya E, Ma E, et al. Structures of Cas9 endonucleases reveal RNA-mediated conformational activation. Science (80-). 2014. [DOI] [PMC free article] [PubMed]
- 56.Schipler A, Iliakis G. DNA double-strand-break complexity levels and their possible contributions to the probability for error-prone processing and repair pathway choice. Nucleic Acids Res. 2013;41:7589–7605. doi: 10.1093/nar/gkt556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Allen F, Crepaldi L, Alsinet C, Strong AJ, Kleshchevnikov V, De Angeli P, et al. Mutations generated by repair of Cas9-induced double strand breaks are predictable from surrounding sequence. bioRxiv [Internet]. 2018;400341. Available from: https://www.biorxiv.org/content/early/2018/08/25/400341%0Ahttp://dx.doi.org/10.1101/400341. Accessed 28 Nov 2018.
- 58.Concordet JP, Haeussler M. CRISPOR: Intuitive guide selection for CRISPR/Cas9 genome editing experiments and screens. Nucleic Acids Res. 2018;46:W242–W245. doi: 10.1093/nar/gky354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fu Y, Foden JA, Khayter C, Maeder ML, Reyon D, Joung JK, et al. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat Biotechnol [Internet]. 2013;31:822–6. http://www.ncbi.nlm.nih.gov/pubmed/23792628. [DOI] [PMC free article] [PubMed]
- 60.Ran FA, Hsu PD, Lin C-Y, Gootenberg JS, Konermann S, Trevino A, et al. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell [Internet]. 2013;154:1380–9. http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3856256/. [DOI] [PMC free article] [PubMed]
- 61.Tsai SQ, Wyvekens N, Khayter C, Foden JA, Thapar V, Reyon D, et al. Dimeric CRISPR RNA-guided FokI nucleases for highly specific genome editing. Nat Biotechnol. 2014;32:569–576. doi: 10.1038/nbt.2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fu Y, Sander JD, Reyon D, Cascio VM, Joung JK. Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat Biotechnol [Internet]. 2014/01/28. Nature Research; 2014;32:279–84. http://www.ncbi.nlm.nih.gov/pubmed/24463574. Accessed 25 Jan 2017. [DOI] [PMC free article] [PubMed]
- 63.Kleinstiver BP, Pattanayak V, Prew MS, Tsai SQ, Nguyen NT, Zheng Z, et al. High-fidelity CRISPR–Cas9 nucleases with no detectable genome-wide off-target effects. Nature [Internet]. 2016;529:490–5. http://www.ncbi.nlm.nih.gov/pubmed/26735016. Accessed 25 Mar 2017. [DOI] [PMC free article] [PubMed]
- 64.Chen JS, Dagdas YS, Kleinstiver BP, Welch MM, Sousa AA, Harrington LB, et al. Enhanced proofreading governs CRISPR–Cas9 targeting accuracy. Nature [Internet]. 2017;550:407–10. http://www.ncbi.nlm.nih.gov/pubmed/28931002. Accessed 13 May 2018. [DOI] [PMC free article] [PubMed]
- 65.Oakes BL, Fellmann C, Rishi H, Taylor KL, Ren SM, Nadler DC, et al. CRISPR-Cas9 Circular permutants as programmable scaffolds for genome modification. Cell [Internet]. Elsevier; 2019;176:254–267.e16. http://www.ncbi.nlm.nih.gov/pubmed/30633905. Accessed 30 Jan 2019. [DOI] [PMC free article] [PubMed]
- 66.Liang X, Potter J, Kumar S, Zou Y, Quintanilla R, Sridharan M, et al. Rapid and highly efficient mammalian cell engineering via Cas9 protein transfection. J Biotechnol [Internet]. 2015;208:44–53. https://ac.els-cdn.com/S016816561500200X/1-s2.0-S016816561500200X-main.pdf?_tid = 7457bdc5-90f7-4bbc-90a0-f760940affb3&acdnat = 1526368421_6e21cd70921a282e0bd2a3fb2576b4f3. Accessed 15 May 2018. [DOI] [PubMed]
- 67.Yin H, Song C-Q, Dorkin JR, Zhu LJ, Li Y, Wu Q, et al. Therapeutic genome editing by combined viral and non-viral delivery of CRISPR system components in vivo. Nat Biotechnol [Internet]. 2016 [cited 2018 May 15];34:328–33. http://www.ncbi.nlm.nih.gov/pubmed/26829318. [DOI] [PMC free article] [PubMed]
- 68.Finn JD, Smith AR, Patel MC, Strapps WR, Chang Y, Morrissey Correspondence D V, et al. A Single Administration of CRISPR/Cas9 lipid nanoparticles achieves robust and persistent in vivo genome editing. Cell Rep [Internet]. Intellia Therapeutics, Inc; 2018 [cited 2018 May 15];22:2227–35. 10.1016/j.celrep.2018.02.014. [DOI] [PubMed]
- 69.Dory D, Le Moigne V, Cariolet R, Béven V, Keranflec’h A, Jestin A. Targeted collection of plasmid DNA in large and growing animal muscles 6 weeks after DNA vaccination with and without electroporation. J Immunol Res [Internet]. Hindawi; 2015 [cited 2018 May 15];2015:1–5. http://www.hindawi.com/journals/jir/2015/326825/. [DOI] [PMC free article] [PubMed]
- 70.Jeong Y-S, Kim EJ, Shim C-K, Hou JH, Kim JM, Choi H-G, et al. Modulation of biodistribution and expression of plasmid DNA following mesenchymal progenitor cell-based delivery. J Drug Target [Internet]. Taylor & Francis; 2008 [cited 2018 May 15];16:405–14. http://www.tandfonline.com/doi/full/10.1080/10611860802088713. [DOI] [PubMed]
- 71.Coelho-Castelo A, Trombone A, Rosada R, Santos R, Bonato V, Sartori A, et al. Tissue distribution of a plasmid DNA encoding Hsp65 gene is dependent on the dose administered through intramuscular delivery. Genet Vaccines Ther [Internet]. BioMed Central; 2006 [cited 2018 May 15];4:1. http://gvt-journal.biomedcentral.com/articles/10.1186/1479-0556-4-1. [DOI] [PMC free article] [PubMed]
- 72.Wang Z, Troilo PJ, Wang X, Griffiths TG, Pacchione SJ, Barnum AB, et al. Detection of integration of plasmid DNA into host genomic DNA following intramuscular injection and electroporation. Gene Ther [Internet]. 2004 [cited 2018 May 14];11:711–21. http://www.ncbi.nlm.nih.gov/pubmed/14724672. [DOI] [PubMed]
- 73.Schnepp BC, Jensen RL, Chen C-L, Johnson PR, Clark KR. Characterization of Adeno-Associated Virus Genomes Isolated from Human Tissues. J Virol [Internet]. 2005 [cited 2018 May 14];79:14793–803. http://jvi.asm.org/cgi/doi/10.1128/JVI.79.23.14793-14803.2005. [DOI] [PMC free article] [PubMed]
- 74.Chandler RJ, Sands MS, Venditti CP. Recombinant adeno-associated viral integration and genotoxicity: insights from animal models. Hum Gene Ther [Internet]. 2017 [cited 2018 May 14];28:314–22. http://online.liebertpub.com/doi/10.1089/hum.2017.009. [DOI] [PMC free article] [PubMed]
- 75.Kim J-S. Precision genome engineering through adenine and cytosine base editing. Nat Plants [Internet]. 2018 [cited 2018 Apr 18];4:148–51. http://www.nature.com/articles/s41477-018-0115-z. [DOI] [PubMed]
- 76.Rees HA, Komor AC, Yeh W-H, Caetano-Lopes J, Warman M, Edge ASB, et al. Improving the DNA specificity and applicability of base editing through protein engineering and protein delivery. Nat Commun [Internet]. 2017 [cited 2017 Oct 10];8:15790. http://www.nature.com/doifinder/10.1038/ncomms15790. [DOI] [PMC free article] [PubMed]
- 77.Austin CP, Cutillo CM, Lau LPL, Jonker AH, Rath A, Julkowska D, et al. Future of rare diseases research 2017–2027: an IRDiRC perspective. Clin Transl Sci [Internet]. Wiley-Blackwell; 2018 [cited 2018 May 9];11:21–7. http://www.ncbi.nlm.nih.gov/pubmed/28796445. [DOI] [PMC free article] [PubMed]
- 78.Plosky BS. CRISPR-Mediated Base Editing without DNA Double-Strand Breaks. Mol Cell [Internet]. Elsevier Inc.; 2016;62:477–8. 10.1016/j.molcel.2016.05.006. [DOI] [PubMed]
- 79.Li G, Liu Y, Zeng Y, Li J, Wang L, Yang G, et al. Highly efficient and precise base editing in discarded human tripronuclear embryos. Protein Cell. 2017;1–4. [DOI] [PMC free article] [PubMed]
- 80.Wang L, Xue W, Yan L, Li X, Wei J, Chen M, et al. Enhanced base editing by co-expression of free uracil DNA glycosylase inhibitor. Cell Res [Internet]. 2017;1–4. http://www.nature.com/doifinder/10.1038/cr.2017.111. [DOI] [PMC free article] [PubMed]
- 81.Gaudelli NM, Komor AC, Rees HA, Packer MS, Badran AH, Bryson DI, et al. Programmable base editing of A•T to G•C in genomic DNA without DNA cleavage. Nature [Internet]. 2017 [cited 2017 Dec 10];551:464–71. http://www.ncbi.nlm.nih.gov/pubmed/29160308. [DOI] [PMC free article] [PubMed]
- 82.O’Geen H, Ren C, Nicolet CM, Perez AA, Halmai J, Le VM, et al. DCas9-based epigenome editing suggests acquisition of histone methylation is not sufficient for target gene repression. Nucleic Acids Res [Internet]. 2017 [cited 2018 May 13];45:9901–16. http://academic.oup.com/nar/article/45/17/9901/3923391. [DOI] [PMC free article] [PubMed]
- 83.Klann TS, Black JB, Chellappan M, Safi A, Song L, Hilton IB, et al. CRISPR-Cas9 epigenome editing enables high-throughput screening for functional regulatory elements in the human genome. Nat Biotechnol [Internet]. 2017 [cited 2018 May 13];35:561–8. http://www.nature.com/doifinder/10.1038/nbt.3853. [DOI] [PMC free article] [PubMed]
- 84.Choudhury SR, Cui Y, Lubecka K, Stefanska B, Irudayaraj J. CRISPR-dCas9 mediated TET1 targeting for selective DNA demethylation at BRCA1 promoter. Oncotarget [Internet]. 2016 [cited 2018 May 13];7:46545–56. http://www.oncotarget.com/fulltext/10234. [DOI] [PMC free article] [PubMed]
- 85.Lo A, Qi L. Genetic and epigenetic control of gene expression by CRISPR–Cas systems. F1000Research [Internet]. 2017 [cited 2018 May 13];6:747. https://f1000research.com/articles/6-747/v1. [DOI] [PMC free article] [PubMed]
- 86.Hilton IB, D’Ippolito AM, Vockley CM, Thakore PI, Crawford GE, Reddy TE, et al. Epigenome editing by a CRISPR-Cas9-based acetyltransferase activates genes from promoters and enhancers. Nat Biotechnol [Internet]. 2015 [cited 2018 May 13];33:510–7. http://www.ncbi.nlm.nih.gov/pubmed/25849900. [DOI] [PMC free article] [PubMed]
- 87.Vora S, Tuttle M, Cheng J, Church G. Next stop for the CRISPR revolution: RNA-guided epigenetic regulators. FEBS J [Internet]. 2016 [cited 2018 May 10];283:3181–93. http://www.ncbi.nlm.nih.gov/pubmed/27248712. [DOI] [PubMed]
- 88.Billon P, Bryant EE, Joseph SA, Nambiar TS, Hayward SB, Rothstein R, et al. CRISPR-mediated base editing enables efficient disruption of eukaryotic genes through induction of STOP codons. Mol Cell [Internet]. Elsevier Inc.; 2017;67:1068–1079.e4. 10.1016/j.molcel.2017.08.008. [DOI] [PMC free article] [PubMed]
- 89.Li X, Wang Y, Liu Y, Yang B, Wang X, Wei J, et al. Base editing with a Cpf1-cytidine deaminase fusion. Nat Biotechnol. 2018;36:324–327. doi: 10.1038/nbt.4102. [DOI] [PubMed] [Google Scholar]
- 90.Lowder LG, Malzahn A, Qi Y. Plant Gene Regulation Using Multiplex CRISPR-dCas9 Artificial Transcription Factors. Methods Mol Biol [Internet]. 2018 [cited 2018 May 13]. p. 197–214. http://www.ncbi.nlm.nih.gov/pubmed/28986912. [DOI] [PubMed]
- 91.Tak YE, Kleinstiver BP, Nuñez JK, Hsu JY, Horng JE, Gong J, et al. Inducible and multiplex gene regulation using CRISPR–Cpf1-based transcription factors. Nat Methods [Internet]. 2017 [cited 2018 May 13];14:1163–6. http://www.ncbi.nlm.nih.gov/pubmed/29083402. [DOI] [PMC free article] [PubMed]
- 92.Larson MH, Gilbert LA, Wang X, Lim WA, Weissman JS, Qi LS. CRISPR interference (CRISPRi) for sequence-specific control of gene expression. Nat Protoc. 2013;8:2180–2196. doi: 10.1038/nprot.2013.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Liu Y, Zhan Y, Chen Z, He A, Li J, Wu H, et al. Directing cellular information flow via CRISPR signal conductors. Nat Methods [Internet]. Nature Publishing Group; 2016 [cited 2018 May 13];13:938–44. http://www.nature.com/articles/nmeth.3994. [DOI] [PubMed]
- 94.Ferry QR, Lyutova R, Fulga TA. Rational design of inducible CRISPR guide RNAs for de novo assembly of transcriptional programs. Nat Commun [Internet]. Nature Publishing Group; 2017 [cited 2018 May 10];8:14633. http://www.nature.com/doifinder/10.1038/ncomms14633. [DOI] [PMC free article] [PubMed]
- 95.Thakore PI, Kwon JB, Nelson CE, Rouse DC, Gemberling MP, Oliver ML, et al. RNA-guided transcriptional silencing in vivo with S. aureus CRISPR-Cas9 repressors. Nat Commun [Internet]. 2018 [cited 2018 May 13];9:1674. http://www.ncbi.nlm.nih.gov/pubmed/29700298. [DOI] [PMC free article] [PubMed]
- 96.Hale CR, Zhao P, Olson S, Duff MO, Graveley BR, Wells L, et al. RNA-guided RNA cleavage by a CRISPR RNA-Cas protein complex. Cell [Internet]. 2009 [cited 2018 May 10];139:945–56. http://www.ncbi.nlm.nih.gov/pubmed/19945378. [DOI] [PMC free article] [PubMed]
- 97.O’Connell MR, Oakes BL, Sternberg SH, East-Seletsky A, Kaplan M, Doudna JA. Programmable RNA recognition and cleavage by CRISPR/Cas9. Nature [Internet]. 2014 [cited 2018 May 10];516:263–6. http://www.ncbi.nlm.nih.gov/pubmed/25274302. [DOI] [PMC free article] [PubMed]
- 98.Aman R, Ali Z, Butt H, Mahas A, Aljedaani F, Khan MZ, et al. RNA virus interference via CRISPR/Cas13a system in plants. Genome Biol [Internet]. 2018 [cited 2018 May 13];19:1. http://www.ncbi.nlm.nih.gov/pubmed/29301551. [DOI] [PMC free article] [PubMed]
- 99.Yan WX, Chong S, Zhang H, Makarova KS, Koonin E V., Cheng DR, et al. Cas13d Is a Compact RNA-Targeting Type VI CRISPR Effector Positively Modulated by a WYL-Domain-Containing Accessory Protein. Mol Cell [Internet]. 2018 [cited 2018 May 13];70:327–339.e5. http://www.ncbi.nlm.nih.gov/pubmed/29551514. [DOI] [PMC free article] [PubMed]
- 100.Liu L, Li X, Ma J, Li Z, You L, Wang J, et al. The molecular architecture for RNA-guided RNA cleavage by Cas13a. Cell [Internet]. 2017 [cited 2018 May 13];170:714–726.e10. http://www.ncbi.nlm.nih.gov/pubmed/28757251. [DOI] [PubMed]
- 101.Cox DBT, Gootenberg JS, Abudayyeh OO, Franklin B, Kellner MJ, Joung J, et al. RNA editing with CRISPR-Cas13. Science [Internet]. NIH Public Access; 2017 [cited 2018 May 14];358:1019–27. http://www.ncbi.nlm.nih.gov/pubmed/29070703. [DOI] [PMC free article] [PubMed]
- 102.Konermann S, Lotfy P, Brideau NJ, Oki J, Shokhirev MN, Hsu PD. Transcriptome engineering with RNA-targeting type VI-D CRISPR effectors. Cell [Internet]. 2018 [cited 2018 May 11];173:665–676.e14. http://www.ncbi.nlm.nih.gov/pubmed/29551272. [DOI] [PMC free article] [PubMed]
- 103.Liu C, Zhang L, Liu H, Cheng K. Delivery strategies of the CRISPR-Cas9 gene-editing system for therapeutic applications. J Control Release [Internet]. 2017 [cited 2018 May 13];266:17–26. http://linkinghub.elsevier.com/retrieve/pii/S0168365917308453. [DOI] [PMC free article] [PubMed]
- 104.Liang X, Potter J, Kumar S, Ravinder N, Chesnut JD. Enhanced CRISPR/Cas9-mediated precise genome editing by improved design and delivery of gRNA, Cas9 nuclease, and donor DNA. J Biotechnol [Internet]. 2017 [cited 2018 May 11];241:136–46. http://www.ncbi.nlm.nih.gov/pubmed/27845164. [DOI] [PubMed]
- 105.Gaj T, Staahl BT, Rodrigues GMC, Limsirichai P, Ekman FK, Doudna JA, et al. Targeted gene knock-in by homology-directed genome editing using Cas9 ribonucleoprotein and AAV donor delivery. Nucleic Acids Res [Internet]. 2017 [cited 2018 May 14];45:e98–e98. http://www.ncbi.nlm.nih.gov/pubmed/28334779. [DOI] [PMC free article] [PubMed]
- 106.Xu L, Yang H, Gao Y, Chen Z, Xie L, Liu Y, et al. CRISPR/Cas9-mediated CCR5 ablation in human hematopoietic stem/progenitor cells confers HIV-1 resistance in vivo. Mol Ther [Internet]. Elsevier Ltd.; 2017;25:1782–9. 10.1016/j.ymthe.2017.04.027. [DOI] [PMC free article] [PubMed]
- 107.Kaminski R, Chen Y, Fischer T, Tedaldi E, Napoli A, Zhang Y, et al. Elimination of HIV-1 genomes from human T-lymphoid cells by CRISPR/Cas9 gene editing. Sci Rep [Internet]. Nature Publishing Group; 2016;6:1–14. 10.1038/srep22555.
- 108.Moyo B, Bloom K, Scott T, Ely A, Arbuthnot P. Advances with using CRISPR/Cas-mediated gene editing to treat infections with hepatitis B virus and hepatitis C virus. Virus Res [Internet]. 2018 [cited 2018 Nov 29];244:311–20. http://www.ncbi.nlm.nih.gov/pubmed/28087399. [DOI] [PubMed]
- 109.Seeger C, Sohn JA. Targeting Hepatitis B Virus with CRISPR/Cas9. Mol Ther - Nucleic Acids [Internet]. American Society of Gene & Cell Therapy; 2014;3:e216. Available from: 10.1038/mtna.2014.68. [DOI] [PMC free article] [PubMed]
- 110.Ren J, Zhao Y. Advancing chimeric antigen receptor T cell therapy with CRISPR/Cas9. Protein Cell. 2017;8:634–643. doi: 10.1007/s13238-017-0410-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Jung I-Y, Kim Y-Y, Yu H-S, Lee M, Kim S, Lee J. CRISPR/Cas9-mediated knockout of DGK improves antitumor activities of human T cells. Cancer Res [Internet]. 2018 [cited 2018 Nov 29];78:4692–703. http://www.ncbi.nlm.nih.gov/pubmed/29967261. [DOI] [PubMed]
- 112.Yang J, Meng X, Pan J, Jiang N, Zhou C, Wu Z, et al. CRISPR/Cas9-mediated noncoding RNA editing in human cancers. RNA Biol [Internet]. 2018 [cited 2018 Nov 29];15:35–43. http://www.ncbi.nlm.nih.gov/pubmed/29028415. [DOI] [PMC free article] [PubMed]
- 113.Zhao Z, Shi L, Zhang W, Han J, Zhang S, Fu Z, et al. CRISPR knock out of programmed cell death protein 1 enhances anti-tumor activity of cytotoxic T lymphocytes. Oncotarget [Internet]. 2018;9:5208–15. http://www.ncbi.nlm.nih.gov/pubmed/29435173%0Ahttp://www.pubmedcentral.nih.gov/articlerender.fcgi?artid = PMC5797044. [DOI] [PMC free article] [PubMed]
- 114.Papasavva P, Kleanthous M, Lederer CW. Rare opportunities: CRISPR/Cas-based therapy development for rare genetic diseases. Mol Diagn Ther. 2019. 10.1007/s40291-019-00392-3 [DOI] [PMC free article] [PubMed]
- 115.Gurumurthy CB, Grati M, Ohtsuka M, Schilit SLP, Quadros RM, Liu XZ. CRISPR: a versatile tool for both forward and reverse genetics research. Hum Genet [Internet]. 2016 [cited 2018 Dec 3];135:971–6. http://www.ncbi.nlm.nih.gov/pubmed/27384229. [DOI] [PMC free article] [PubMed]
- 116.Gilbert LA, Horlbeck MA, Adamson B, Villalta JE, Chen Y, Whitehead EH, et al. Genome-scale CRISPR-mediated control of gene repression and activation. Cell [Internet]. Elsevier Inc.; 2014;159:647–61. 10.1016/j.cell.2014.09.029. [DOI] [PMC free article] [PubMed]
- 117.Wang T, Wei JJ, Sabatini DM, Lander ES. Genetic screens in human cells using the CRISPR-Cas9 system. Science (80-) [Internet]. 2014 [cited 2017 Aug 7];343:80–4. http://www.ncbi.nlm.nih.gov/pubmed/24336569. [DOI] [PMC free article] [PubMed]
- 118.Joung J, Konermann S, Gootenberg JS, Abudayyeh OO, Platt RJ, Brigham MD, et al. Genome-scale CRISPR-Cas9 knockout and transcriptional activation screening. Nat Protoc [Internet]. 2017 [cited 2017 Aug 7];12:828–63. http://www.ncbi.nlm.nih.gov/pubmed/28333914. [DOI] [PMC free article] [PubMed]
- 119.Park RJ, Wang T, Koundakjian D, Hultquist JF, Lamothe-Molina P, Monel B, et al. A genome-wide CRISPR screen identifies a restricted set of HIV host dependency factors. Nat Genet [Internet]. 2017 [cited 2018 Nov 29];49:193–203. http://www.ncbi.nlm.nih.gov/pubmed/27992415. [DOI] [PMC free article] [PubMed]
- 120.Richardson RB, Ohlson MB, Eitson JL, Kumar A, McDougal MB, Boys IN, et al. A CRISPR screen identifies IFI6 as an ER-resident interferon effector that blocks flavivirus replication. Nat Microbiol [Internet]. 2018 [cited 2018 Nov 29];3:1214–23. http://www.ncbi.nlm.nih.gov/pubmed/30224801. [DOI] [PMC free article] [PubMed]
- 121.Puschnik AS, Majzoub K, Ooi YS, Carette JE. A CRISPR toolbox to study virus–host interactions. Nat Rev Microbiol [Internet]. 2017 [cited 2018 Nov 29];15:351–64. http://www.ncbi.nlm.nih.gov/pubmed/28420884. [DOI] [PMC free article] [PubMed]
- 122.Neggers JE, Vercruysse T, Jacquemyn M, Vanstreels E, Baloglu E, Shacham S, et al. Identifying drug-target selectivity of small-molecule CRM1/XPO1 inhibitors by CRISPR/Cas9 genome editing. Chem Biol [Internet]. Elsevier Ltd; 2015;22:107–16. 10.1016/j.chembiol.2014.11.015. [DOI] [PubMed]
- 123.Smurnyy Y, Cai M, Wu H, McWhinnie E, Tallarico JA, Yang Y, et al. DNA sequencing and CRISPR-Cas9 gene editing for target validation in mammalian cells. Nat Chem Biol [Internet]. 2014 [cited 2018 Nov 29];10:623–5. http://www.ncbi.nlm.nih.gov/pubmed/24929529. [DOI] [PubMed]
- 124.He YJ, Meghani K, Caron M-C, Yang C, Ronato DA, Bian J, et al. DYNLL1 binds to MRE11 to limit DNA end resection in BRCA1-deficient cells. Nature [Internet]. 2018 [cited 2018 Nov 29];563:522–6. http://www.ncbi.nlm.nih.gov/pubmed/30464262. [DOI] [PMC free article] [PubMed]
- 125.Yau EH, Kummetha IR, Lichinchi G, Tang R, Zhang Y, Rana TM. Genome-wide CRISPR screen for essential cell growth mediators in mutant KRAS colorectal cancers. Cancer Res [Internet]. 2017 [cited 2018 Nov 29];77:6330–9. http://www.ncbi.nlm.nih.gov/pubmed/28954733. [DOI] [PMC free article] [PubMed]
- 126.Dompe N, Klijn C, Watson SA, Leng K, Port J, Cuellar T, et al. A CRISPR screen identifies MAPK7 as a target for combination with MEK inhibition in KRAS mutant NSCLC. Ahmad A, editor. PLoS One [Internet]. 2018 [cited 2018 Nov 29];13:e0199264. http://www.ncbi.nlm.nih.gov/pubmed/29912950. [DOI] [PMC free article] [PubMed]
- 127.Chen L, Alexe G, Dharia NV, Ross L, Iniguez AB, Conway AS, et al. CRISPR-Cas9 screen reveals a MYCN-amplified neuroblastoma dependency on EZH2. J Clin Invest [Internet]. 2017 [cited 2018 Nov 29];128:446–62. http://www.ncbi.nlm.nih.gov/pubmed/29202477. [DOI] [PMC free article] [PubMed]
- 128.Reddy A, Zhang J, Davis NS, Moffitt AB, Love CL, Waldrop A, et al. Genetic and functional drivers of diffuse large B cell lymphoma. Cell [Internet]. 2017 [cited 2018 Nov 29];171:481–494.e15. http://www.ncbi.nlm.nih.gov/pubmed/28985567. [DOI] [PMC free article] [PubMed]
- 129.Konermann S, Brigham MD, Trevino AE, Joung J, Abudayyeh OO, Barcena C, et al. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature [Internet]. 2015 [cited 2018 Nov 29];517:583–8. http://www.nature.com/articles/nature14136. [DOI] [PMC free article] [PubMed]
- 130.Potting C, Crochemore C, Moretti F, Nigsch F, Schmidt I, Manneville C, et al. Genome-wide CRISPR screen for PARKIN regulators reveals transcriptional repression as a determinant of mitophagy. Proc Natl Acad Sci [Internet]. 2017;201711023. http://www.pnas.org/lookup/doi/10.1073/pnas.1711023115. [DOI] [PMC free article] [PubMed]
- 131.Westrick RJ, Tomberg K, Siebert AE, Zhu G, Winn ME, Dobies SL, et al. Sensitized mutagenesis screen in Factor V Leiden mice identifies thrombosis suppressor loci. Proc Natl Acad Sci [Internet]. 2017;114:201705762. http://www.pnas.org/lookup/doi/10.1073/pnas.1705762114. [DOI] [PMC free article] [PubMed]
- 132.Schmid-Burgk JL, Chauhan D, Schmidt T, Ebert TS, Reinhardt J, Endl E, et al. A genome-wide CRISPR (clustered regularly interspaced short palindromic repeats) screen identifies NEK7 as an essential component of NLRP3 inflammasome activation. J Biol Chem. 2016;291:103–109. doi: 10.1074/jbc.C115.700492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Liu Y, Yu C, Daley TP, Wang F, Cao WS, Bhate S, et al. CRISPR Activation Screens Systematically Identify Factors that Drive Neuronal Fate and Reprogramming. Cell Stem Cell [Internet]. 2018 [cited 2018 Nov 29];23:758–771.e8. http://www.ncbi.nlm.nih.gov/pubmed/30318302. [DOI] [PMC free article] [PubMed]
- 134.Mendelsohn BA, Bennett NK, Darch MA, Yu K, Nguyen MK, Pucciarelli D, et al. A high-throughput screen of real-time ATP levels in individual cells reveals mechanisms of energy failure. Tian R, editor. PLOS Biol [Internet]. 2018 [cited 2018 Nov 29];16:e2004624. http://www.ncbi.nlm.nih.gov/pubmed/30148842. [DOI] [PMC free article] [PubMed]
- 135.Goodwin JM, Dowdle WE, DeJesus R, Wang Z, Bergman P, Kobylarz M, et al. Autophagy-Independent Lysosomal Targeting Regulated by ULK1/2-FIP200 and ATG9. Cell Rep [Internet]. ElsevierCompany.; 2017;20:2341–56. 10.1016/j.celrep.2017.08.034. [DOI] [PMC free article] [PubMed]
- 136.Miyamoto T, Akutsu SN, Matsuura S. Updated summary of genome editing technology in human cultured cells linked to human genetics studies review-article. J Hum Genet [Internet]. Springer US; 2018 [cited 2018 May 10];63:133–43. 10.1038/s10038-017-0349-z. [DOI] [PubMed]
- 137.Wang G, McCain ML, Yang L, He A, Pasqualini FS, Agarwal A, et al. Modeling the mitochondrial cardiomyopathy of Barth syndrome with induced pluripotent stem cell and heart-on-chip technologies. Nat Med [Internet]. 2014 [cited 2018 May 14];20:616–23. http://www.nature.com/articles/nm.3545. [DOI] [PMC free article] [PubMed]
- 138.Dow LE. Modeling Disease In Vivo With CRISPR/Cas9. Trends Mol Med [Internet]. Elsevier; 2015 [cited 2018 Nov 29];21:609–21. http://www.ncbi.nlm.nih.gov/pubmed/26432018. [DOI] [PMC free article] [PubMed]
- 139.Fujii W, Kawasaki K, Sugiura K, Naito K. Efficient generation of large-scale genome-modified mice using gRNA and CAS9 endonuclease. Nucleic Acids Res [Internet]. 2013 [cited 2018 May 15];41:e187–e187. http://www.ncbi.nlm.nih.gov/pubmed/23997119. [DOI] [PMC free article] [PubMed]
- 140.Martinez-Lage M. CRISPR/Cas9 Technology: applications and human disease modeling. Prog Mol Biol Transl Sci [Internet]. Academic Press; 2017 [cited 2018 May 11];152:23–48. https://www.sciencedirect.com/science/article/pii/S1877117317301369?via%3Dihub. [DOI] [PubMed]
- 141.Chen Y, Niu Y, Ji W. Genome editing in nonhuman primates: approach to generating human disease models. J Intern Med [Internet]. 2016 [cited 2018 May 11];280:246–51. http://www.ncbi.nlm.nih.gov/pubmed/27114283. [DOI] [PubMed]
- 142.Pelletier S, Gingras S, Green DR. Mouse genome engineering via CRISPR-Cas9 for study of immune function. Immunity [Internet]. Elsevier; 2015 [cited 2018 May 11];42:18–27. http://www.ncbi.nlm.nih.gov/pubmed/25607456. [DOI] [PMC free article] [PubMed]
- 143.Tu Z, Yang W, Yan S, Guo X, Li X-J. CRISPR/Cas9: a powerful genetic engineering tool for establishing large animal models of neurodegenerative diseases. Mol Neurodegener [Internet]. 2015 [cited 2018 May 11];10:35. http://www.ncbi.nlm.nih.gov/pubmed/26238861. [DOI] [PMC free article] [PubMed]
- 144.Wang H, Yang H, Shivalila CS, Dawlaty MM, Cheng AW, Zhang F, et al. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell [Internet]. NIH Public Access; 2013 [cited 2018 May 11];153:910–8. http://www.ncbi.nlm.nih.gov/pubmed/23643243. [DOI] [PMC free article] [PubMed]
- 145.Siggs OM, Popkin DL, Krebs P, Li X, Tang M, Zhan X, et al. Mutation of the ER retention receptor KDELR1 leads to cell-intrinsic lymphopenia and a failure to control chronic viral infection. Proc Natl Acad Sci [Internet]. 2015 [cited 2018 May 14];112:E5706–14. http://www.ncbi.nlm.nih.gov/pubmed/26438836. [DOI] [PMC free article] [PubMed]
- 146.Kimura Y, Hisano Y, Kawahara A, Higashijima S. Efficient generation of knock-in transgenic zebrafish carrying reporter/driver genes by CRISPR/Cas9-mediated genome engineering. Sci Rep [Internet]. 2015 [cited 2018 May 15];4:6545. http://www.ncbi.nlm.nih.gov/pubmed/25293390. [DOI] [PMC free article] [PubMed]
- 147.Allen F, Crepaldi L, Alsinet C, Strong AJ, Kleshchevnikov V, De Angeli P, et al. Predicting the mutations generated by repair of Cas9-induced double-strand breaks. Nat Biotechnol [Internet]. 2018 [cited 2018 Nov 28]; http://www.nature.com/doifinder/10.1038/nbt.4317. [DOI] [PMC free article] [PubMed]
- 148.Li X, Wang Y, Liu Y, Yang B, Wang X, Wei J, et al. Base editing with a Cpf1–cytidine deaminase fusion. Nat Biotechnol [Internet]. 2018 [cited 2018 May 13];36:324–7. http://www.ncbi.nlm.nih.gov/pubmed/29553573. [DOI] [PubMed]
- 149.European Medicines Agency Committee for Advanced Therapies (CAT). Guideline on quality, non-clinical and clinical aspects of 4 medicinal products containing genetically modified cells—Revision 1 [Internet]. 2018. http://www.ema.europa.eu/contact.
- 150.Wagner DL, Amini L, Wendering DJ, Burkhardt L-M, Akyüz L, Reinke P, et al. High prevalence of Streptococcus pyogenes Cas9-reactive T cells within the adult human population. Nat Med. 2018. [DOI] [PubMed]
- 151.Wagner DL, Amani L, Wendering DJ, Reinke P, Volk H-D, Schmueck-Henneresse M. High prevalence of S. pyogenes Cas9-specific T cell sensitization within the adult human population-A balanced effector/regulatory T cell response. bioRxiv. 2018;295139.
- 152.Sakurai H, Kawabata K, Sakurai F, Nakagawa S, Mizuguchi H. Innate immune response induced by gene delivery vectors. Int J Pharm [Internet]. 2008 [cited 2018 Dec 3];354:9–15. http://www.ncbi.nlm.nih.gov/pubmed/17640834. [DOI] [PubMed]
- 153.Bessis N, GarciaCozar FJ, Boissier M-C. Immune responses to gene therapy vectors: influence on vector function and effector mechanisms. Gene Ther [Internet]. 2004 [cited 2018 Dec 3];11:S10–7. http://www.ncbi.nlm.nih.gov/pubmed/15454952. [DOI] [PubMed]
- 154.Vandamme C, ADJALI O, Mingozzi F. Unraveling the complex story of immune responses to AAV vectors trial after trial. Hum Gene Ther [Internet]. 2017;28:hum.2017.150. http://online.liebertpub.com/doi/10.1089/hum.2017.150. [DOI] [PMC free article] [PubMed]
- 155.Ihry RJ, Worringer KA, Salick MR, Frias E, Ho D, Theriault K, et al. P53 inhibits CRISPR-Cas9 engineering in human pluripotent stem cells. Nat Med [Internet]. Springer US; 2018;24:939–46. 10.1038/s41591-018-0050-6. [DOI] [PubMed]
- 156.Mohrin M, Bourke E, Alexander D, Warr MR, Barry-Holson K, Le Beau MM, et al. Hematopoietic Stem Cell Quiescence Promotes Error-Prone DNA Repair and Mutagenesis. Cell Stem Cell [Internet]. NIH Public Access; 2010 [cited 2018 May 3];7:174–85. http://www.ncbi.nlm.nih.gov/pubmed/20619762. [DOI] [PMC free article] [PubMed]
- 157.Haapaniemi E, Botla S, Persson J, Schmierer B, Taipale J. CRISPR-Cas9 genome editing induces a p53-mediated DNA damage response. Nat Med [Internet]. Springer US; 2018;24:927–30. 10.1038/s41591-018-0049-z. [DOI] [PubMed]
- 158.Kosicki M, Tomberg K, Bradley A. Repair of double-strand breaks induced by CRISPR–Cas9 leads to large deletions and complex rearrangements. Nat Biotechnol [Internet]. 2018; http://www.nature.com/doifinder/10.1038/nbt.4192. [DOI] [PMC free article] [PubMed]
- 159.Tenenbaum L, Lehtonen E, Monahan PE. Evaluation of risks related to the use of adeno-associated virus-based vectors. Curr Gene Ther. 2003;3:545–565. doi: 10.2174/1566523034578131. [DOI] [PubMed] [Google Scholar]
- 160.Biasco L, Pellin D, Scala S, Dionisio F, Basso-Ricci L, Leonardelli L, et al. In vivo tracking of human hematopoiesis reveals patterns of clonal dynamics during early and steady-state reconstitution phases. Cell Stem Cell [Internet]. The Author(s); 2015;19:107–19. 10.1016/j.stem.2016.04.016. [DOI] [PMC free article] [PubMed]
- 161.Scala S, Basso-Ricci L, Dionisio F, Pellin D, Giannelli S, Salerio FA, et al. Dynamics of genetically engineered hematopoietic stem and progenitor cells after autologous transplantation in humans. Nat Med [Internet]. Springer US; 2018;24:1683–90. http://www.nature.com/articles/s41591-018-0195-3. [DOI] [PubMed]
- 162.Voit RA, Hendel A, Pruett-Miller SM, Porteus MH. Nuclease-mediated gene editing by homologous recombination of the human globin locus. Nucleic Acids Res [Internet]. 2013/10/26. 2014;42:1365–78. http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3902937/pdf/gkt947.pdf. [DOI] [PMC free article] [PubMed]
- 163.Zonari E, Desantis G, Petrillo C, Boccalatte FE, Lidonnici MR, Kajaste-Rudnitski A, et al. Efficient ex vivo engineering and expansion of highly purified human hematopoietic stem and progenitor cell populations for gene therapy. Stem Cell Rep [Internet]. ElsevierCompany.; 2017;8:977–90. 10.1016/j.stemcr.2017.02.010. [DOI] [PMC free article] [PubMed]
- 164.Improving Gene Therapy efficiency through the enrichment of human hematopoietic stem cells. Mol Ther. American Society of Gene & Cell Therapy; 2017;25:2163–75. [DOI] [PMC free article] [PubMed]
- 165.He Z, Proudfoot C, Whitelaw CBA, Lillico SG. Comparison of CRISPR/Cas9 and TALENs on editing an integrated EGFP gene in the genome of HEK293FT cells. Springerplus. Springer International Publishing; 2016;5. [DOI] [PMC free article] [PubMed]
- 166.Wang Y, Liu KI, Sutrisnoh NAB, Srinivasan H, Zhang J, Li J, et al. Systematic evaluation of CRISPR-Cas systems reveals design principles for genome editing in human cells. Genome Biol. 2018;19:1–16. doi: 10.1186/s13059-017-1381-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Song F, Stieger K. Optimizing the DNA Donor Template for Homology-Directed Repair of Double-Strand Breaks. Mol Ther - Nucleic Acids [Internet]. Elsevier Ltd.; 2017;7:53–60. 10.1016/j.omtn.2017.02.006. [DOI] [PMC free article] [PubMed]
- 168.Villiger L, Grisch-Chan HM, Lindsay H, Ringnalda F, Pogliano CB, Allegri G, et al. Treatment of a metabolic liver disease by in vivo genome base editing in adult mice. Nat Med [Internet]. 2018;24:1519–25. http://www.ncbi.nlm.nih.gov/pubmed/30297904. Accessed 2018 Dec 3. [DOI] [PubMed]
- 169.Mullis K, Faloona F, Scharf S, Saiki R, Horn G, Erlich H. Specific enzymatic amplification of DNA in vitro: the polymerase chain reaction. Cold Spring Harb Symp Quant Biol [Internet]. 1986;51 Pt 1:263–73. http://www.ncbi.nlm.nih.gov/pubmed/3472723. Accessed 2018 Nov 28. [DOI] [PubMed]
- 170.Bosher JM, Labouesse M. RNA interference: genetic wand and genetic watchdog. Nat Cell Biol [Internet]. 2000;2:E31–6. http://www.ncbi.nlm.nih.gov/pubmed/10655601. Accessed 2018 Nov 29. [DOI] [PubMed]
- 171.Baulcombe DC. RNA as a target and an initiator of post-transcriptional gene silencing in transgenic plants. Plant Mol Biol [Internet]. 1996/10/01. 1996;32:79–88. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd = Retrieve&db = PubMed&dopt = Citation&list_uids = 8980475. Accessed 2018 Nov 29. [DOI] [PubMed]
- 172.Ledford H. Gene-silencing technology gets first drug approval after 20-year wait. Nature [Internet]. 2018;560:291–2. http://www.ncbi.nlm.nih.gov/pubmed/30108348. [DOI] [PubMed]
- 173.RNA interference therapies could be on the cusp of success. Pharm J [Internet]. 2018. http://www.pharmaceutical-journal.com/news-and-analysis/features/rna-interference-therapies-could-be-on-the-cusp-of-success/20204853.article. Accessed 2018 Nov 28.
- 174.Chavez A, Scheiman J, Vora S, Pruitt BW, Tuttle M, P R Iyer E, et al. Highly efficient Cas9-mediated transcriptional programming. Nat Methods [Internet]. 2015;12:326–8. http://www.nature.com/articles/nmeth.3312. Accessed 2018 May 19. [DOI] [PMC free article] [PubMed]
- 175.Moreno AM, Fu X, Zhu J, Katrekar D, Shih Y-R V, Marlett J, et al. < em > In situ </em > gene therapy via AAV-CRISPR-Cas9 mediated targeted gene regulation. Mol Ther [Internet]. Elsevier Ltd.; 2018;26:1–10. 10.1016/j.ymthe.2018.04.017. [DOI] [PMC free article] [PubMed]