Abstract
Background:
SMPD1 (acid-sphingomyelinase, ASMase) variants have been associated with Parkinson’s disease (PD) in recent studies.
Objective:
To further study the role of SMPD1 mutations in PD.
Methods:
SMPD1 was sequenced in three cohorts (Israel Ashkenazi-Jewish cohort, Montreal/Montpellier and New York), including 1,592 PD patients and 975 controls. Additional data was available for 10,709 Ashkenazi-Jewish controls. ASMase activity was measured by a mass-spectrometry-based assay in the New York cohort. α-Synuclein levels were measured in-vitro following CRISPR/Cas9-mediated knock out and siRNA knockdown of SMPD1 in HeLa and BE(2)-M17 cells. Lysosomal localization of ASMase with different mutations was studied, and in-silico analysis of their effect on ASMase structure was performed.
Results:
SMPD1 mutations were associated with PD in the Ashkenazi-Jewish cohort, as 1.4% of PD patients carried the p.L302P or p.fsP330 mutation, compared to 0.37% in 10,709 Ashkenazi-Jewish controls (OR=3.7, 95%CI=1.6–8.2, p=0.0025). In the Montreal/Montpellier cohort, the p.A487V variant was nominally associated with PD (1.5% versus 0.14%, p=0.0065, not significant after correction for multiple comparisons). Among PD patients, reduced ASMase activity was associated with a 3.5–5.8 years earlier onset of PD in the lowest quartile versus the highest quartile of ASMase activity (p=0.01–0.001). We further demonstrated that SMPD1 knockout and knockdown resulted in increased α-synuclein levels in HeLa and BE(2)-M17 dopaminergic cells, and that the p.L302P and p.fsP330 mutations impair the traffic of ASMase to the lysosome.
Conclusions:
Our results support an association between SMPD1 variants, ASMase activity and PD. Furthermore, they suggest that reduced ASMase activity may lead to α-synuclein accumulation.
Keywords: Parkinson’s disease, SMPD1, acid sphingomyelinase, Genetics, α-synuclein
Introduction
Accumulating genetic and biological data suggest an important role for the lysosome and the sphingolipids metabolic pathway in the pathogenesis of Parkinson’s disease (PD).1 The autophagy-lysosomal pathway is responsible for the degradation of α-synuclein, and dysfunction of this pathway may lead to α-synuclein accumulation and to the development of PD.1, 2 Variants in the GBA gene, encoding the lysosomal enzyme glucocerebrosidase (GCase), are among the most common risk factors for PD, found in 3–20% of PD patients from different populations.3–9 Recently, mutations in another lysosomal gene involved in sphingolipid metabolism, SMPD1, which encodes the lysosomal enzyme acid sphingomyelinase (ASMase), has also been associated with an increased risk for PD.10–17 In the Ashkenazi-Jewish population, specific Niemann-Pick type A (NPA)-causing SMPD1 mutations, p.L302P (also called p.L304P) and p.fsP330 (also called p.F333Sfs*52 or c.996delC), were associated with PD in two independent studies.11, 15 Additionally, two studies in Chinese populations and two studies in European populations identified additional SMPD1 mutations and variants associated with PD.12, 13, 16, 17
Both GCase and ASMase hydrolyze sphingolipids in the lysosome and generate a common product, ceramide, suggesting that they may lead to PD in a similar manner.1 ASMase is responsible for the hydrolysis of sphingomyelin into ceramide and phosphorylcholine, and biallelic mutations in SMPD1 lead to NPA or Niemann-Pick type B (NPB),18 the acute neurovisceral and chronic visceral forms, respectively, of ASMase deficiency. NPA is a severe and rapidly progressive disease of infancy, characterized by hepatosplenomegaly, failure to thrive, psychomotor regression, interstitial lung disease and death by early childhood. NPB has a variable age of onset with a slower disease course, and patients can survive into adulthood.1, 19
In the current study we examined the effects of SMPD1 mutations on PD risk and onset in two case-control cohorts. We further studied the effect of SMPD1 mutations on ASMase enzymatic activity, and the association of this activity with risk and clinical characteristics of PD. In addition, we examined whether SMPD1 deficiency affects α-synuclein accumulation in cellular models, and how specific SMPD1 mutations affect the lysosomal localization of ASMase. Lastly, we performed an in-silico analysis of different SMPD1 mutations and their effect on ASMase structure.
Materials and Methods
Full version of the materials and methods can be found in the Supplementary Material online.
Study populations
Basic demographic characteristics of the three cohorts that were analyzed in the current study are described in Supplementary Table 1 and the Supplementary Materials. The study population included three cohorts: a) a cohort of 517 unrelated Ashkenazi Jewish (AJ) PD patients recruited at the Sheba Medical Center (termed “Sheba cohort” hereafter), Israel, who were compared to publicly available data including 10,709 AJ controls, b) a cohort of 525 PD patients and 691 controls of French-Canadian/French origin, all unrelated, collected at the Montreal Neurological Institute (“MTL-F cohort”), Montreal, Canada, and c) a cohort of 550 patients and 284 controls, all unrelated, recruited at Columbia University, New York (NY), USA (“NY cohort”). The AJ PD patient population from the Sheba cohort is independent, with no overlap with previous AJ populations in which SMPD1 was genotyped.11, 15 The Montreal and French cohort (termed hereafter MTL-F cohort) was composed of French-Canadian PD patients and controls who were recruited in Quebec, Canada (n=1027), and French PD patients and controls who were recruited in Montpellier, France (n=189). All French-Canadian and French patients and controls underwent genotyping using a genome-wide association study genotyping array (unpublished data, using the OmniExpress array + NeuroX, Illumina), and principal component analysis was performed. Only patients and controls that segregated together with French ancestry were included in the current study. Furthermore, identity-by-descent (IBD) was performed to exclude individuals with cryptic relatedness. Detailed description of the recruitment and demographics of the NY cohort was previously published.3, 20 All participants signed informed consent forms prior to their enrollment into the study, and the institutional review boards approved the study protocols.
Sequencing and analysis of SMPD1 variants
In the Sheba cohort, two SMPD1 mutations, known to cause NPA and previously shown to be associated with PD were genotyped. The SMPD1 p.L302P (also known as p.L304P) was genotyped using a TaqMan array (assay ID: C____945630_20, ThermoFisher) according to the manufacturer instructions, and was validated by Sanger sequencing. The p.fsP330 mutation (also known as p.F333Sfs*52 or c.996delC) was screened by Sanger sequencing. The Sanger sequencing chromatograms were analyzed using the Genalys 3.3b software. In the NY cohort, sequencing of the entire SMPD1 gene was performed using Sanger sequencing (primer sequences and reagents used for PCR amplification and sequencing are available upon request). In the MTL-F cohort, targeted next generation sequencing of the entire coding region of SMPD1 was performed (Supplementary Table 2 and online supplementary methods).
Acid sphingomyelinase enzymatic activity
Acid sphingomyelinase activity was measured at Sanofi laboratories, and the researchers were blinded to the genetic status of the patients and controls. Dried blood spots were obtained as previously described in the NY cohort.21, 22 In brief, blood samples were collected in a 10 ml EDTA tube. Seventy-five microliters of blood was “spotted” on each of five circles on filter paper (Whatman 903 protein savor card, St. Louis, MI) and dried at room temperature for at least 4 hours. Absorbent filter paper was then stored in a sealed plastic bag with desiccants and a humidity indicator in a −20°C freezer and later shipped to the laboratories at room temperature. Upon receipt, the samples were stored at −80°C before analysis. The detailed procedure of enzymatic activity measurement can be found in the online Supplementary Materials.
Knockout of SMPD1 in Hela cells using CRISPR/Cas9
To generate SMPD1 knockout HeLa cells, two guide RNAs, sg1 and sg2, were cloned into the pSpCas9(BB)-2A-Puro vector (pX459_v2, Addgene 62988).23 Target sequences were (cut site indicated by an asterisk, PAM sequence in parentheses): sg1 AGAGCTGCCCCAGGTCC*GGC (CGG); sg2 CAAAGACATCTCGGAG*CCG (GGG). Further details can be found in the online Supplementary Materials.
Cell cultures, siRNAs and cell transfection
HeLa and BE(2)-M17 cells were obtained from the American Type Culture Collection (ATCC, Manassas, VA) and were maintained at 37°C and 5% CO2. HeLa cells were cultured in Dulbecco’s modified Eagle medium supplemented with 10% heat-inactivated fetal bovine serum (FBS), 2 mM glutamine and 100 units/ml penicillin/streptomycin. BE(2)-M17 cells were cultured in a medium containing 50% of Eagle’s Minimum Essential Medium (EMEM), 50% of HAM-F12 supplemented with 10% heat-inactivated fetal bovine serum (FBS), 2 mM glutamine and 100 units/ml penicillin/streptomycin. Details on siRNAs and cell transfection can be found in the online Supplementary Materials.
Immunoblotting and α-synuclein quantification
Cells were lysed using RIPA lysis buffer with a mixture of protease inhibitors. After adding Laemmli buffer with DTT to the samples, they were boiled for 10 min and separated by SDS-PAGE. Then separated samples were transferred to a nitrocellulose membrane. For α-synuclein, the membrane was incubated with phosphate-buffered saline (PBS) as described previously.24 Membranes were blocked and incubated overnight with primary antibody in PBS-T containing 1% milk. Finally, membranes were washed three times with PBST24 and were developed using ECL substrate.
Immunofluorescence and microscopy
HeLa cells cultured on glass coverslips were transfected for 24 h with plasmids encoding Venus-fused ASMase variants. Cells were then rinsed with PBS, fixed for 10 min at room temperature in 4% paraformaldehyde/PBS, rinsed again with PBS, permeabilized and rinsed in PBS before blocking. Cells were then incubated with mouse anti-LAMP2 primary antibody for 1 h at room temperature in blocking solution, rinsed and incubated with anti-mouse Alexa-555 coupled secondary antibody for 1 h at room temperature. Full details can be found in the online Supplementary Materials.
Molecular cloning
The wild-type human SMPD1 coding sequence was ordered as two synthetic double stranded DNA fragments (gBlocks 1 and 2, Intregrated DNA Technologies) and assembled using Gibson Assembly into the BamHI site of the mVenus N1 plasmid, allowing the addition of a C-terminal fluorescent Venus tag. Using gBlock 2 as a template, we used PCR-based mutagenesis to generate the modified coding sequences associated with the p.L302P, p.fsP330 or p.A487V variants, and further assembled these DNA fragments together with gBlock 1 into the BamHI site of the mVenus N1 plasmid by Gibson Assembly. Constructs were verified by Sanger sequencing.
In silico structural analysis
The atomic coordinates of mouse SMPD1 bound to a lipid and to a bisphosphonate inhibitor were downloaded from the Protein Data Bank. The steric clashes induced by each mutation were evaluated using the “mutagenesis” toolbox in PyMol v. 1.5.
Statistical analysis
Statistical analysis is detailed in the online Supplementary Materials.
Results
Association of SMPD1 mutations with Parkinson’s disease
As the three cohorts (Sheba cohort, MTL-F cohort and NY cohort) were of different origins, they were analyzed separately. Table 1 details the frequencies of the p.L302P and p.fsP330 SMPD1 mutations in the Sheba cohort and in previous studies in AJ populations.11, 15 These populations are independent, and patients do not overlap. The frequency of each mutation was compared to the frequency in publicly available control population of 10,709 individuals of AJ origin. A combined analysis for each mutation and for both mutations combined was further performed. In the Sheba cohort alone, the p.L302P mutation was associated with PD, and the p.fsP330 mutation was not, likely due to reduced power. In the combined analysis of the current AJ population together with previously published AJ populations (Table 1, total number of patients of 1,742 AJ PD patients), carriers of the p.L302P mutation had an OR of 9.0 (95% CI 4.2–19.5, p<0.0001), and carriers of the p.fsP330 had an OR of 2.8 (95% CI 1.4–5.3, p=0.003). In the MTL-F cohort, a total of 29 different SMPD1 variants that affect the coding sequence or alternative splicing were identified (Supplementary Table 3). Among PD patients, 5.3% (n=28) carried a rare SMPD1 variant compared to 2.9% (n=20) among controls (OR=1.88, 95% CI 1.05–3.39, p=0.037). After adjustment for age and sex, this association was not significant, which is likely due to the patients with SMPD1 variants having a trend towards an earlier AAE (Mann-Whitney U test, p=0.079). The association was mainly driven by a single variant, p.A487V, found in eight (1.5%) patients and in one (0.14%) control (OR=10.68, 95%CI 1.3–85.6, p=0.0065, not significant after correction for multiple comparisons). In the NY cohort (Table 2), the p.A487V variant was found in three patients (0.55%) and in one control (0.35%, p=1.0). Combining the two cohorts, the p.A487V was found in 11 (1.0%) patients and 2 (0.2%) controls (OR = 5.03, 95%CI 1.11–22.75, p=0.024). None of the SMPD1 p.A487V mutation carriers was a carrier of GBA or the LRRK2 G2019S mutations. The frequency of rare SMPD1 variants in the NY cohort was not significantly different between patients and controls, 3.8% versus 5.3%, p=0.34. We further compared the frequency of rare variants that had ASMase reduced activity in patients and controls. Although these variants (p.R291H, p.P331A, p.R378H, p.A487V, p.G492S, p.R498L, p.E517V, p.G530A, c.1829delCGG, p.R610H) were more frequent in patients (n=12/550, 2.2%) than in controls (n=4/285, 1.4%), these differences did not reach statistical significance. Table 2 details the mutations identified in the NY cohort and their enzymatic activity. Interestingly, the p.V114M variant that did not have reduced ASMase activity in our study, has been previously identified in Niemann-Pick patients.18 This variant was identified in three patients from the MTL-F and NY cohorts, but no carriers were found in the control populations.
Table 1.
Association between SMPD1 mutations in Ashkenazi Jewish PD patient from the current study and previously published studies.
| p.L302P | p.fsP330 | p.L302P and p.fsP330 combined | ||||
|---|---|---|---|---|---|---|
| Study | frequency | p value, OR (95% CI)a | frequency | p value, OR (95% CI)a | frequency | P value, OR (95% CI)a |
| Current study Sheba cohort PD patients | 4/517 (0.8%) | 0.0005, 7.6 (2.4–23.9) | 3/517 (0.6%) | 0.39, 2.1 (0.7–7.1) | 7/517 (1.4%) | 0.0025, 3.7 (1.6–8.2) |
| Dagan 2015 PD patients | 3/287 (1.0%) | 0.0003, 10.3 (2.9–37.0) | 5/287 (1.7%) | <0.0001, 6.5 (2.5–17.0) | 8/287 (2.8%) | <0.0001, 7.6 (3.5–16.5) |
| Gan-Or 2013 PD patients | 9/938 (1.0%) | <0.0001, 9.4 (3.9–22.8) | 5/938 (0.5%) | 0.3, 2.0 (0.8–5.1) | 14/938 (1.5%) | <0.0001, 4.0 (2.2–7.5) |
| Combined PD patients | 16/1742 (0.9%) | <0.0001, 9.0 (4.2–19.5) | 13/1742 (0.7) | 0.003, 2.8 (1.4–5.3) | 29/1742 (1.7%) | <0.0001, 4.5 (2.8–7.3) |
| Controls | 11/10709 (0.1%) | - | 29/10709 (0.3%) | - | 40/10709 (0.4%) | - |
The table presents the frequencies of the p.L302P and p.fsP330 mutations in the current and previous studies, all in Ashkenazi Jewish PD patients, and compares them to the frequencies in publicly available data from 10,709 controls. OR, odds ratio; CI; confidence interval; PD, Parkinson’s disease
Compared to the control population at the bottom row.
Table 2.
SMPD1 variants and their associated ASMase enzymatic activity in PD patients and controls in the NY cohort
| SMPD1 Variant | Pathogenicitya | Patients (n=550) | Controls (n=284) | ASMase activityb (combined PD and controls) | p value, comparing ASMase activity in carriers vs. non-carries’ |
|---|---|---|---|---|---|
| Rare variants | |||||
| p.Q19R | Unknown | 4 (0.7%) | 2 (0.7%) | 6.31 ± 2.67 | 0.13 |
| p.V114M | Probably pathogenic | 2 (0.4%) | 0 (0%) | 5.04 ± 1.45 | 0.59 |
| p.P187S | Unknown | 1 (0.2%) | 3 (1.1%) | 5.70 ± 1.43 | 0.13 |
| p.G269S | Unknown | 0 (0%) | 1 (0.4%) | 4.59 | NA |
| p.R291H | Probably pathogenic | 1 (0.2%) | 0 (0%) | 3.45 | NA |
| p.P331A | Unknown | 1 (0.2%) | 0 (0%) | 2.76 | NA |
| p.R378H | Probably pathogenic | 0 (0%) | 1 (0.4%) | 2.22 | NA |
| p.R389C | Unknown | 1 (0.2%) | 1 (0.4%) | 7.51 ± 3.03 | 0.08 |
| p.W393G | Probably pathogenic | 3 (0.5%) | 3 (1.1%) | 5.14 ± 1.77 | 0.54 |
| p.A487V | Unknown | 3 (0.5%) | 1 (0.4%) | 2.85 ± 0.59 | 0.01 |
| p.G492S | Probably pathogenic | 1 (0.2%) | 0 (0%) | 2.16 | NA |
| p.R498L | Probably pathogenic | 2 (0.4%) | 0 (0%) | 3.32 ± 1.20 | 0.20 |
| p.E517V | Probably pathogenic | 1 (0.2%) | 2 (0.7%) | 2.38 ± 0.47 | 0.007 |
| p.G530A | Unknown | 1 (0.2%) | 0 (0%) | 3.00 | NA |
| c.1829delCGG | Unknown | 1 (0.2%) | 0 (0%) | 2.14 | NA |
| p.M613I | Unknown | 0 (0%) | 1 (0.4%) | 7.61 | NA |
| p.R610H | Unknown | 1 (0.2%) | 0 (0%) | 2.14 | NA |
| Common variants | |||||
| p.G508R | Not pathogenic | G/G 345 (62.7%) | G/G 196 (69.0%) | 4.96 ± 1.73 | |
| G/R 186 (33.8%) | G/R 80 (28.2%) | 4.09 ± 1.37 | |||
| R/R 19 (3.5%) | R/R 8 (2.8%) | 3.56 ± 1.18 | p<0.001c | ||
| Non-carriersd | 529 (96.2%) | 269 (94.7%) | 4.64 ± 1.64 | ||
ASMase, acid sphingomyelinase
A pathogenic variant that may lead to Neimann-Pick type A or B, based on a recently published database of pathogenic mutations.18
ASMase enzymatic activity is measured in μmol/l/h units.
Both ANOVA and the non-parametric Kruskal-Wallis tests were performed; in both p<0.001.
Non-carriers of rare SMPD1 mutations, not including the p.G508R variant.
ASMase enzymatic activity is associated with specific SMPD1 variants and the age at onset of PD
Among controls, there was no association between age and sex and ASMase activity (p=0.11 for age, p=0.46 for sex). There was no difference between the average enzymatic activity in patients and controls (4.64 μmol/l/h ± 1.68 μmol/l/h versus 4.62 μmol/l/h ± 1.64 μmol/l/h, respectively, p=0.84). ASMase enzymatic activity was not different between GBA variant/mutation carriers and non-carriers (4.67 μmol/l/h ± 1.78 μmol/l/h versus 4.59 μmol/l/h ± 1.65 μmol/l/h, respectively, p=0.69). Interestingly, it was previously reported that GCase activity is increased in PD patients who carry the LRRK2 p.G2019S mutation.3 Therefore, we examined the ASMase activity in carriers of LRRK2 mutations. Among PD patients, carriers of the LRRK2 p.G2019S mutation (n=36) had an average ASMase activity of 5.20 μmol/l/h ± 1.84 compared to 4.59 μmol/l/h ± 1.65 among non-carriers (n=426, p=0.033, t-test). When including controls in this analysis, the results remained similar, 5.23 μmol/l/h ± 1.79 compared to 4.61 μmol/l/h ± 1.64, respectively (n=38 and n=685, respectively, p=0.025, t-test).
Among PD patients without GBA or LRRK2 p.G2019S mutations, reduced ASMase enzymatic activity was associated with an earlier AAO (B=1.419, β=0.194, 95% CI for B 0.80–2.04, p<0.001, i.e., for each 1 μmol/l/h reduction age at onset was younger by 1.4 years). Further adjustment to GCase activity yielded similar results (B=1.470, β=0.203, 95% CI for B 0.83–2.11, p<0.001). We further divided the cohort to four groups, based on the ASMase enzymatic activity quartiles in controls (Table 3). First, the entire cohort was analyzed, followed by analysis of late-onset PD only (AAO≥50 years), and further exclusion of carriers of GBA and LRRK2 p.G2019S mutations that can also affect the AAO. In all analyses, there was a decrease in AAO with the decrease of enzymatic activity, with significant differences of 3.5–5.8 years between the first and last quartiles in the different analyses (Table 3, p≤0.01 for all comparisons). Of note, when dividing the patients group into quartiles based on the ASMase activity values in patients only, or patients and controls combined, nearly identical and statistically significant results were obtained. Other demographics and disease characteristics were not different among the quartiles (Supplementary Table 4) among PD patients with late-onset PD who do not carry GBA or LRRK2 mutations. Similar results were obtained when early-onset PD and GBA and LRRK2 mutation carriers were included.
Table 3.
ASMase activity quartilesa and the age at onset of PD.
| Quartile 1 | Quartile 2 | Quartile 3 | Quartile 4 | p valueb | |
|---|---|---|---|---|---|
| ASMase activity range (μmol/l/h) | <3.485 | 3.485–4.46 | 4.46–5.64 | >5.64 | |
| All patients | |||||
| Nc | 134 | 148 | 146 | 120 | |
| Average AAO ± SD | 58.6 ± 10.3 | 58.2 ± 12.2 | 58.2 ± 12.4 | 62.2 ± 11.0 | p=0.01 |
| All patients with late-onset PD (AAO ≥ 50) | |||||
| N | 115 | 117 | 111 | 104 | |
| Average AAO ± SD | 61.5 ± 7.2 | 63.0 ± 7.5 | 63.5 ± 8.3 | 65.0 ± 8.8 | p=0.01 |
| All patients excluding GBA and LRRK2 mutation carriers | |||||
| N | 109 | 119 | 113 | 89 | |
| Average AAO ± SD | 57.4 ± 10.4 | 58.2 ± 12.5 | 58.1 ± 12.9 | 63.2 ± 11.2 | p=0.003 |
| All patients with late-onset PD, excluding GBA and LRRK2 mutation carriers | |||||
| N | 91 | 93 | 85 | 80 | |
| Average AAO ± SD | 60.7 ± 7.1 | 63.4 ± 7.6 | 64.0 ± 8.2 | 65.5 ± 9.1 | p=0.001 |
ASMase, acid sphingomyelinase; N, number; AAO, age at onset; SD, standard deviation.
Quartiles were determined according to ASMase activity in controls, however nearly identical results were calculated when the quartiles were determined according to patients only, or patients and controls combined.
Analysis of variance (ANOVA).
Data on AAO was not available for one patient.
Decreased SMPD1 expression leads to accumulation of α-synuclein in cellular models
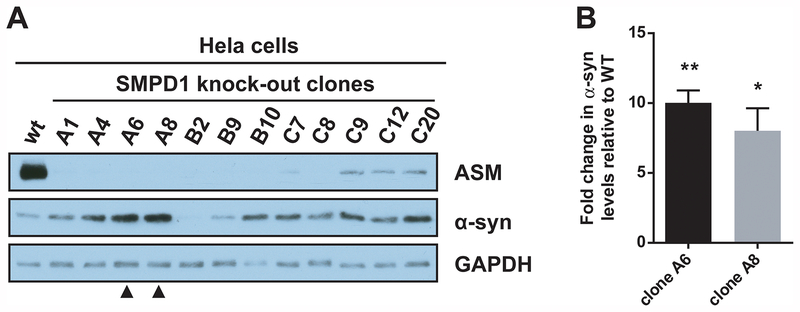
To further investigate the link between ASMase function and PD pathogenesis, a CRISPR/Cas9-mediated knockout of SMPD1 was performed in HeLa cells. Twelve knockout clones were obtained and compared to wild-type cells. Western blot analysis demonstrated almost complete loss of ASMase expression in all clones, and this was correlated in ten out of twelve clones with increased α-synuclein levels (Figure 1A). Precise quantification in two clones (A6 and A8) done in triplicates showed an 8–10-fold increase (Figure 1B). Quantification of all 12 single clones can be seen in Supplementary Figure 1, and 10 out of the 12 clones demonstrated increase levels of α-synuclein. These results were further confirmed using RNA interference against SMPD1 in HeLa cells and in the dopaminergic cell line M17 (Supplementary Figure 2). Regardless of the siRNA used, ASMase expression was reduced and this led to an average increase of about 3-fold of α-synuclein levels.
Figure 1. CRISPR/Cas9-mediated knockout of SMDP1 in Hela cells leads to α-synuclein accumulation.
(A) Twelve knockout clones were obtained using CRISPR/Cas9, and ASMase and α-synuclein expression were compared with wild-type cells by Western blotting. GAPDH was used as a loading control. Clones A6 and A8 (arrowheads) were further used for quantification. (B) Densitometric quantification of α-synuclein levels in clones A6 and A8 is shown. Data are mean ±SEM from 3 independent experiments. ** p<0.01, * p<0.05, paired t-test.
ASMase lysosomal localization is affected by specific SMPD1 variants
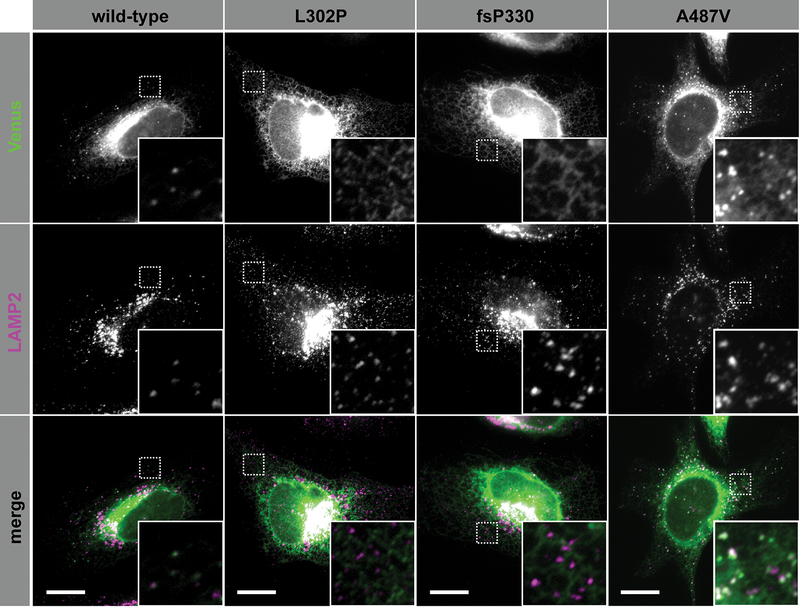
To investigate how SMPD1 variants could affect ASMase activity, we compared the subcellular localization of ASMase with three variants (p.L302P, p.fsP330 or p.A487V) to that of wild-type ASMase. We transfected all four forms of ASMase (fused to a C-terminal Venus tag) into HeLa cells and performed immunocytochemistry against the lysosomal marker LAMP2 (Figure 2). As expected for a protein that travels through the secretory pathway, wild-type ASMase was mainly detected in a perinuclear network reminiscent of the endoplasmic reticulum (ER). In addition, some puncta were observed closer to the periphery, which colocalized with LAMP2 suggesting that our ASMase-Venus fusion was successfully sorted to the lysosome. Strikingly, ASMase with the p.L302P or p.fsP330 mutations failed to reach the lysosomal compartment, and was almost exclusively localized in the ER. The p.A487V variant seemed unaffected.
Figure 2. Some SMPD1 variants are associated with altered lysosomal localization of ASMase.
Hela cells were transfected with plasmids encoding variants of ASMase fused to a C-terminal Venus tag (green), and processed for immunofluorescence using an antibody against the lysosomal marker LAMP2 (magenta) before imaging using an epifluorescence microscope. Scale bar: 10 μm. The insets correspond to a zoom on the areas delineated by a dashed-square. ASMase with the p.L302P or p.P330fs mutations failed to reach the lysosomal compartment, and was almost exclusively localized in the endoplasmic reticulum. Trafficking of ASMase with the p.A487V variant seemed unaffected.
In silico analysis suggests that mutations in SMPD1 disrupt enzymatic domain fold and lipid-binding site
To investigate the effect of SMPD1 variants on the structure and activity of ASMase, we performed in silico structural analysis using the recently published structures of mouse ASMase in the closed and open states, bound to a bisphosphonate inhibitor and a lipid, respectively.25 The p.L302P, p.A487V and p.V114M were modelled, the p.fsP330 was not modeled as it causes early termination of the protein. Superposition of the two structures shows that Leu302 and Ala487 are in the structurally invariant catalytic domain of ASMase, whereas Val114 is located in the lipid-binding domain that undergoes a large conformation change (Supplementary Figure 3A). A proline at position 302 (position 304 in the full transcript) would be incompatible with the helical conformation of Leu304, and thus the mutation L302P would likely unfold the catalytic domain. The mutation A487V would introduce a steric clash in the hydrophobic core of the catalytic domain and would likewise destabilize the domain (Supplementary Figure 3B). The V114M mutation would have two potential effects: the side-chain of Val114 is oriented towards the lipid in the open conformation, and the mutation V114M could interfere with lipid-binding via steric hindrance (Supplementary Figure 3C). On the other hand, the V114M mutation would also introduce severe clashes in the closed state and destabilize this conformation (Supplementary Figure 3D).
Discussion
The current study provides support for the association between SMPD1, ASMase activity and PD. The results suggest that specific SMPD1 variants are associated with PD, and that reduced ASMase activity is associated with earlier AAO of PD. Using two cellular models, we further demonstrated that SMPD1 knockdown resulted in decreased ASMase levels and led to α-synuclein accumulation. Lastly, we demonstrated that some of the SMPD1 mutations lead to altered lysosomal localization of ASMase, and that in silico analysis suggests deleterious effects on ASMase structure and function. These results suggest that while only a small proportion of PD patients have SMPD1 variants that increase the risk for PD, ASMase activity may have a role in PD. More studies are necessary to replicate our findings and to elucidate the underlying mechanisms responsible for the effects on AAO and α-synuclein accumulation.
Thus far, not including the current study, SMPD1 variants have been associated with PD or synucleinopathy risk in seven independent cohorts: two of Ashkenazi-Jewish origin;11, 15 two Chinese;12, 13 two European;16, 17 and one North-American.10 In these studies, as well as in our cohort, only some of the mutations were associated with PD, while other SMPD1 mutations, including some that can cause Niemann-Pick type A/B, were not associated with PD. The current study independently shows (i.e. using a different population than the two previous AJ populations that were studied) that the two AJ mutations, p.L302P and p.fsP330, are associated with PD, and that both mutations result in lack of lysosomal localization of ASMase. Together with the experiments demonstrating that reduced ASMase levels lead to α-synuclein accumulation, a hypothesis can be made that these mutations increase the risk for PD by causing reduced lysosomal localization of ASMase and as a result, accumulation of α-synuclein due to yet unknown mechanism.
It is important to note that when analyzing the AJ cohorts (Table 1), we have performed a combined analysis of both mutations, and a separate analysis of each mutation, which is important for determining their individual effects on risk. At the same time, a combined analysis, especially when analyzing rare mutations, may be crucial for understanding whether a specific gene is important in PD or not. For example, when the association of GBA with PD was initially described, there were conflicting results due to one study combining all mutations with a positive association and another report on negative association in which the separate effect of only one mutation was examined.26, 27 Subsequent studies that included a combined analysis of various GBA mutations have revealed that mutations in GBA are indeed associated with PD.5, 6, 9 Such combined analyses are also useful for analysis of associations with phenotypes or biomarkers,28, 29 and currently burden analyses of multiple genes from the same pathways are routinely performed.16, 30 Such recent analysis demonstrated that there is a burden of lysosomal storage disease-causing genes in PD, which was mainly driven by GBA but also by SMPD1,16 further supporting the current results.
The role of the third mutation implicated here, p.A487V, is still unclear. In the cellular experiments and in silico models, while there was lack of effect of p.A487V on ASMase localization, the in silico models suggested that it can destabilize the catalytic domain of ASMase. However, it is still possible that the association of this mutation with PD in our cohort is due to chance alone, and further studies are necessary. Interestingly, it is controversial whether the p.A487V variant causes Niemann-Pick A or B.31 Similarly, GBA variants that do not cause Gaucher disease, such as the p.E326K variant,32 are important risk factors for PD.33 These observations may suggest that the mechanisms that lead to PD and accumulation of α-synuclein could be somewhat different than those that lead to either Gaucher or Niemann-Pick types A or B disease. The role of the p.V114M mutation, found in three patients and none in controls, is still unclear, and additional studies are required.
We previously reported that among carriers of the LRRK2 p.G2019S mutation in the NY cohort, the enzymatic activity of GCase was higher than in non-carriers.3 In the current study, we found a similar association between LRRK2 p.G2019S genotype and ASMase activity. The mechanism of both associations is still unknown. The reason for the increased activity is currently unclear. We previously suggested that it could be due to lysosomal compartment expansion or effects on the retromer and turnover of GCase/ASMase.3 If another, possibly compensatory, mechanism also plays a role remains to be determined.
Our study has several limitations. Although we studied more than 2500 individuals, due to the rarity of these mutations, we may have been underpowered to test the association of rare mutations with PD and even larger studies will be needed to more accurately determine the role of SMPD1 variants in PD. Another limitation was that the MTL-F and NY cohorts were not matched for sex and that the MTL-F cohort was not matched for age. However, we adjusted for these differences in the statistical analysis. Of note, the younger age of the control population in the MTL-F cohort is more likely to reduce rather than increase the risk estimates, since it is possible that control carriers of SMPD1 variants will develop PD in the future. For example, the control individual with the p.G504X stop mutation was only 25 years old at recruitment, and could develop PD in the future. An additional limitation is that the ASMase enzymatic activity was only available for the NY cohort. Therefore, the association between reduced ASMase activity and earlier AAO requires replication in other cohorts. Another limitation is that our cellular models represent complete or almost complete dysfunction of ASMase, which is not the case in heterozygous carriers of SMPD1 mutations. However, as can be seen in some of the knockdown and knockout models, there is still residual amount of ASMase, and α-synuclein still accumulates.
The major strength of our study is that we tested the association between SMPD1 and PD in multiple modalities. First, we conducted a genetic correlation. Then, we tested ASMase activity in PD patients and controls, all of whom were sequenced for SMPD1 and GBA and genotyped for the LRRK2 G2019S mutation. We further used cellular models to test a potential mechanism for the association between PD and SMPD1 and performed in silico structural analysis of specific SMPD1 variants. In all modalities, we found suggestive evidence for an association. However, it will be important to further study the functional effects of SMPD1 mutations in additional cohorts and in human-derived neuronal models.
SMPD1, like many other PD-associated genes, is involved in the autophagy lysosomal pathway, and it is possible that PD could be treated by enhancing specific components of this pathway. Our results, together with the previous studies, suggest that SMPD1 could be one of these components.
Supplementary Material
Acknowledgements
We thank the patients and controls for their participation in this study. This work was financially supported by the Michael J. Fox Foundation. The cohort from Columbia, NY, was funded by Parkinson’s Disease Foundation and the NIH (K02NS080915, and UL1 TR000040). BV is funded by a postdoctoral fellowship from the Canadian Institutes of Health Research (CIHR) Fellowship program (MFE 152571). This research was undertaken thanks in part to funding from the Canada First Research Excellence Fund, awarded to McGill University for the Healthy Brains for Healthy Lives initiative. GAR holds a Canada Research Chair in Genetics of the Nervous System and the Wilder Penfield Chair in Neurosciences. The access to part of the participants for this research has been made possible thanks to the Quebec Parkinson Network (http://rpq-qpn.ca/en/). We thank Judy Hull, Daniel Rochefort, Pascale Hince, Helene Catoire, Patricia Lanzano, Cynthia Bourassa, Cathy Mirarchi and Vessela Zaharieva for their assistance.
Funding sources: This work was financially supported by the Michael J. Fox Foundation, the Canadian Consortium on Neurodegeneration in Aging (CCNA). The cohort from Montreal was funded by the Canadian Institutes for Health Research (CIHR) and Fonds de Recherche du Québec – Santé (FRQS). The cohort from Columbia, NY, was funded by Parkinson’s Disease Foundation, the NIH (K02NS080915, and UL1 TR000040) and the Brookdale foundation.
Full financial disclosure for the previous 12 months:
Roy N. Alcalay: Received research funding from the NIH, Parkinson’s Foundation and the Michael J Fox Foundation. Consultation fees from Genzyme/Sanofi, Biogen, Denali and Prophase Victoria Mallett: Nothing to disclose Benoît Vanderperre: Recieved Postdoctoral fellowship from the CIHR Fellowship program (MFE 152571). Received research funding from the Canada First Research Excellence Fund, awarded to McGill University for the Healthy Brains for Healthy Lives initiative. Omid Tavassoly: Received Basic Research Postdoctoral Fellowship from Parkinson Society Canada (PSC). Received research grants from the W. Garfield Weston Foundation (Rapid Response: Canada-2016 & 2017) Yves Dauvilliers: Received funds for seminars, board engagements and travel to conferences by UCB Pharma, Jazz, Theranexus, Flamel and Bioprojet Richard Y.J. Yu: Nothing to disclose.
Jennifer A. Rusky: Nothing to disclose.
Claire S. Leblond: Nothing to disclose.
Amirthagowri Ambalavanan: Nothing to disclose.
Sandra B. Laurent: Nothing to disclose.
Dan Spiegelman: Nothing to disclose.
Alexandre Dionne-Laporte: Nothing to disclose.
Christopher Liong: Nothing to disclose.
Oren A. Levy: Advisory board and honorarium from Abbvie; Consulting fee from Schlesinger; Grant support from Michael J. Fox Foundation, Parkinson Foundation, APDA
Stanley Fahn: Consulting and Advisory Board Membership with honoraria: Merz Pharma, Genervon Biotechnology; PixarBio; Lundbeck Pharma. Grants/Research Support: 69Genervon Biotechnology
Cheryl Waters: Advisory Board and honoraria from US World Meds, Acadia, Adamas
Sheng-Han Kuo: Nothing to disclose.
Wendy K. Chung: Nothing to disclose.
Blair Ford: Nothing to disclose.
Karen S. Marder: Received research funding from the NIH (NS036630, 1U01NS100600 1UL1TR001873 1U10NS077267), Michael J Fox, Parkinson Disease Foundation, CHDI, HDSA, Site Investigator (TEVA, Vaccinex).
Un Jung Kang: Received research funding from the NIH (R01 NS101982, R03 NS096494), DoD PR161817, Michael J Fox Foundation for Parkinson’s Research, and the Parkinson’s Foundation
Sharon Hassin-Baer: Nothing to disclose.
Lior Greenbaum: Nothing to disclose.
Jean-Francois Trempe: Nothing to disclose.
Pavlina Wolf: Employee of Sanofi.
Petra Oliva: Employee of Sanofi.
Xiaokui Kate Zhang: Employee of Sanofi.
Lorraine N. Clark: Nothing to disclose.
Melanie Langlois: Advisory board and honorarium from Abbvie, Allergan
Patrick A. Dion: Received research funding from Brain Canada-ALS Canada Hudson Translational Program, International Essential Tremor Foundation.
Edward A. Fon: Grants: CIHR, Michael J Fox Foundation, ALS Canada-Brain Canada Hudson Team grant, CQDM, Brain Canada, Canadian Consortium on Neurodegeneration in Aging (CCNA), and National Parkinson Foundation (NPF)
Nicolas Dupre: Received research funding from CIHR, FRQS (RPQ), CHUdeQ-UL foundation, AQNF, Actelion Pharmaceuticals, Genzyme/Sanofi
Guy A. Rouleau: Received research funding from the CIHR Foundation Grant, Brain Canada-ALS Canada Hudson Translational Program, International Essential Tremor Foundation.
Ziv Gan-Or: Consulting for Genzyme/Sanofi, Lysosomal Therapeutics Inc, Denali, Idorsia and Prevail Therapeutics.
Footnotes
Conflict of interest disclosure: PW, PO and XKZ are employees of Sanofi. Sanofi is performing clinical trials targeting acid sphingomyelinase deficiency. RNA, ND and ZGO have received consultancy fees or research funding from Sanofi on topics not related to the current study. All other authors report no conflict of interests.
References
- 1.Gan-Or Z, Dion PA, Rouleau GA. Genetic perspective on the role of the autophagy-lysosome pathway in Parkinson disease. Autophagy 2015;11(9):1443–1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cuervo AM, Stefanis L, Fredenburg R, Lansbury PT, Sulzer D. Impaired degradation of mutant alpha-synuclein by chaperone-mediated autophagy. Science 2004;305(5688):1292–1295. [DOI] [PubMed] [Google Scholar]
- 3.Alcalay RN, Levy OA, Waters CC, et al. Glucocerebrosidase activity in Parkinson’s disease with and without GBA mutations. Brain 2015;138(Pt 9):2648–2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clark LN, Ross BM, Wang Y, et al. Mutations in the glucocerebrosidase gene are associated with early-onset Parkinson disease. Neurology 2007;69(12):1270–1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gan-Or Z, Amshalom I, Kilarski LL, et al. Differential effects of severe vs mild GBA mutations on Parkinson disease. Neurology 2015;84(9):880–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gan-Or Z, Giladi N, Rozovski U, et al. Genotype-phenotype correlations between GBA mutations and Parkinson disease risk and onset. Neurology 2008;70(24):2277–2283. [DOI] [PubMed] [Google Scholar]
- 7.Lesage S, Anheim M, Condroyer C, et al. Large-scale screening of the Gaucher’s disease-related glucocerebrosidase gene in Europeans with Parkinson’s disease. Hum Mol Genet 2011;20(1):202–210. [DOI] [PubMed] [Google Scholar]
- 8.Neumann J, Bras J, Deas E, et al. Glucocerebrosidase mutations in clinical and pathologically proven Parkinson’s disease. Brain 2009;132(Pt 7):1783–1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sidransky E, Nalls MA, Aasly JO, et al. Multicenter analysis of glucocerebrosidase mutations in Parkinson’s disease. N Engl J Med 2009;361(17):1651–1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clark LN, Chan R, Cheng R, et al. Gene-wise association of variants in four lysosomal storage disorder genes in neuropathologically confirmed Lewy body disease. PLoS One 2015;10(5):e0125204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dagan E, Schlesinger I, Ayoub M, et al. The contribution of Niemann-Pick SMPD1 mutations to Parkinson disease in Ashkenazi Jews. Parkinsonism Relat Disord 2015;21(9):1067–1071. [DOI] [PubMed] [Google Scholar]
- 12.Deng S, Deng X, Song Z, et al. Systematic Genetic Analysis of the SMPD1 Gene in Chinese Patients with Parkinson’s Disease. Mol Neurobiol 2015. [DOI] [PubMed] [Google Scholar]
- 13.Foo JN, Liany H, Bei JX, et al. Rare lysosomal enzyme gene SMPD1 variant (p.R591C) associates with Parkinson’s disease. Neurobiol Aging 2013;34(12):2890 e2813–2895. [DOI] [PubMed] [Google Scholar]
- 14.Gan-Or Z, Orr-Urtreger A, Alcalay RN, Bressman S, Giladi N, Rouleau GA. The emerging role of SMPD1 mutations in Parkinson’s disease: Implications for future studies. Parkinsonism Relat Disord 2015;21(10):1294–1295. [DOI] [PubMed] [Google Scholar]
- 15.Gan-Or Z, Ozelius LJ, Bar-Shira A, et al. The p.L302P mutation in the lysosomal enzyme gene SMPD1 is a risk factor for Parkinson disease. Neurology 2013;80(17):1606–1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Robak LA, Jansen IE, van Rooij J, et al. Excessive burden of lysosomal storage disorder gene variants in Parkinson’s disease. Brain 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ylonen S, Siitonen A, Nalls MA, et al. Genetic risk factors in Finnish patients with Parkinson’s disease. Parkinsonism Relat Disord 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zampieri S, Filocamo M, Pianta A, et al. SMPD1 Mutation Update: Database and Comprehensive Analysis of Published and Novel Variants. Hum Mutat 2015. [DOI] [PubMed] [Google Scholar]
- 19.Schuchman EH, Wasserstein MP. Types A and B Niemann-Pick disease. Best Pract Res Clin Endocrinol Metab 2015;29(2):237–247. [DOI] [PubMed] [Google Scholar]
- 20.Sakanaka K, Waters CH, Levy OA, et al. Knowledge of and interest in genetic results among Parkinson disease patients and caregivers. J Genet Couns 2014;23(1):114–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Olivova P, Cullen E, Titlow M, et al. An improved high-throughput dried blood spot screening method for Gaucher disease. Clin Chim Acta 2008;398(1–2):163–164. [DOI] [PubMed] [Google Scholar]
- 22.Reuser AJ, Verheijen FW, Bali D, et al. The use of dried blood spot samples in the diagnosis of lysosomal storage disorders--current status and perspectives. Mol Genet Metab 2011;104(1–2):144–148. [DOI] [PubMed] [Google Scholar]
- 23.Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc 2013;8(11):2281–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee BR, Kamitani T. Improved immunodetection of endogenous alpha-synuclein. PLoS One 2011;6(8):e23939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gorelik A, Illes K, Heinz LX, Superti-Furga G, Nagar B. Crystal structure of mammalian acid sphingomyelinase. Nat Commun 2016;7:12196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aharon-Peretz J, Rosenbaum H, Gershoni-Baruch R. Mutations in the glucocerebrosidase gene and Parkinson’s disease in Ashkenazi Jews. N Engl J Med 2004;351(19):1972–1977. [DOI] [PubMed] [Google Scholar]
- 27.Zimran A, Neudorfer O, Elstein D. The glucocerebrosidase gene and Parkinson’s disease in Ashkenazi Jews. N Engl J Med 2005;352(7):728–731; author reply 728–731. [PubMed] [Google Scholar]
- 28.Lerche S, Schulte C, Srulijes K, et al. Cognitive impairment in Glucocerebrosidase (GBA)-associated PD: Not primarily associated with cerebrospinal fluid Abeta and Tau profiles. Mov Disord 2017;32(12):1780–1783. [DOI] [PubMed] [Google Scholar]
- 29.Brockmann K, Srulijes K, Pflederer S, et al. GBA-associated Parkinson’s disease: reduced survival and more rapid progression in a prospective longitudinal study. Mov Disord 2015;30(3):407–411. [DOI] [PubMed] [Google Scholar]
- 30.Gaare JJ, Nido GS, Sztromwasser P, et al. Rare genetic variation in mitochondrial pathways influences the risk for Parkinson’s disease. Mov Disord 2018;33(10):1591–1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rhein C, Naumann J, Muhle C, et al. The Acid Sphingomyelinase Sequence Variant p.A487V Is Not Associated With Decreased Levels of Enzymatic Activity. JIMD Rep 2013;8:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Duran R, Mencacci NE, Angeli AV, et al. The glucocerobrosidase E326K variant predisposes to Parkinson’s disease, but does not cause Gaucher’s disease. Mov Disord 2013;28(2):232–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pankratz N, Beecham GW, DeStefano AL, et al. Meta-analysis of Parkinson’s disease: identification of a novel locus, RIT2. Ann Neurol 2012;71(3):370–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.