Abstract
Antimicrobial peptides (AMPs) have emerged as a promising class of antimicrobial agents that could potentially address the global antibiotic resistance. Generating mirror-like peptides by minimizing dermaseptin family sequences is an effective strategy for designing AMPs. However, the previous research still had some limitations such as lower effectiveness and a narrow spectrum of antibacterial activity. To further expand and hone this strategy, we designed a series of AMPs consisting of the WXMXW-NH2 motif (X represents V, I, F, and W; M represents KAAAKAAAK). The peptides formed α-helices and displayed broad-spectrum antimicrobial activities against eleven types of clinical bacteria including both Gram-negative and Gram-positive bacteria. The optimized peptide WW exhibited high physical rupture by inducing membrane shrinkage, disruption, and lysis. Moreover, WW effectively neutralized endotoxins and inhibited the inflammatory response while having the highest therapeutic index. In conclusion, these results indicated that the peptide WW has potential as a broad-spectrum antimicrobial agent or preservative for overcoming the risk of multidrug resistance in localized or external therapeutic applications.
Keywords: antimicrobial peptide, minimized dermaseptins, antibacterial mechanism, endotoxin neutralization
1. Introduction
The worldwide challenge for infectious diseases remains exceedingly prominent and has become more disturbing due to the rise of antibiotic resistant bacteria, which is one of the main incentives of social, economic, and public health problems [1]. Antimicrobial peptides (AMPs) play an important role in all multicellular organisms as the first line of the host defense [2]. Unlike conventional antibiotics that cause microbial death by acting on specific cellular targets, the majority of AMPs exert their bioactivities via the non-receptor mediated permeabilization of membranes [3]. The physical disruption of the membrane leads to a low probability of the development of drug resistance as a fundamental change in membrane components is considered to be a slow process [4]. Therefore, AMPs have been found to be equally effective against both antibiotic-sensitive and resistant strains and are expected to become potential substitutes for traditional antibiotics.
However, the use of AMPs as therapeutic agents has some limitations due to its long peptide sequences, low efficacy, instability, systemic toxicities, and potential for compromising innate host defense immunity, which has hampered the development and clinical application of AMPs [5]. Numerous methods have been developed in the design of analogous peptides to overcome these barriers such as motif hybridization to increase antimicrobial efficacy and functionality [6], truncation/ substitution to decrease toxicity [7], and de novo design to shorten peptide length and eliminate host defense immunogenicity [8]. Nevertheless, these comprehensive strategies represent only a small part of the available methods to promote the development of AMPs.
In our previous study, “minimized proteins” as an old protein design strategy were newly applied to the design of antimicrobial peptides. According to the dermaseptin family sequence and structure information such as charge, amino acid frequency, hydrophobicity, charge ratio, a cluster of Ala in trimer style, and α-helix structure, we designed mirror-like peptides by minimized theory and selected the peptide KL4A6 (LLKAAAKAAAKLL-NH2), which showed the greatest therapeutic index [9]. However, there are still some limitations for KL4A6 to become a potential therapeutic drug (e.g., narrow spectrum of antibacterial activity and low activity). It has been suggested that tryptophan residues that interact with the membrane at the end of an amphiphilic α-helix will increase antimicrobial activity [10,11], Thus, in this study, short α-helix-folding AMPs (WX) with symmetric sequences, based upon the previous designing principles, consisting of WXMXW (X represents V, I, F, and W; M represents KAAAKAAAK) were designed. The peptide lengths were shortened to 13 residues from 28.75 residues of the dermaseptin family, of which three were cationic amino acids (K) and ten were hydrophobic amino acids. The centrosymmetric structure was maintained to increase biological activity [12,13], while different hydrophobic amino acids were used symmetrically for the replacement of residues in the sequence to perturb microbial cell membranes [4,8,14]. Additionally, the C-terminus of the peptide was aminated to enhance antimicrobial activity and improve stabilization [15].
2. Results and Discussion
2.1. Peptide Design and Characterization
“Minimized proteins” were first proposed in 1997, which represented a potential intermediate step toward the development of drugs targeted to protein–protein interfaces [16]. In fact, the binding surfaces of large protein–protein interactions possess very few residues that confer key functions in the recognition and binding between these proteins. Correspondingly, the interaction between AMP and the bacterial membrane is similar to the interaction between proteins as determinants of the peptide-membrane interaction depend on the physical parameters of AMPs such as charge, amphipathicity, sequence, structure, hydrophobicity, and polar angle [17].
The previously designed mirror-like peptide KL4A6 showed the worst cell selectivity at a therapeutic index (TI) of 9.6, with several limitations. In this study, some rational modification strategies were employed in the optimization of synthetic AMPs to enhance therapeutic potentials including tryptophan residues localizing to the end of an amphiphilic α-helix, centrosymmetric structures, and modifications of different hydrophobic amino acids. The measured molecular weights of each peptide were very close to its theoretical value, suggesting that the products corresponded to the designed compositions (Table 1). Each peptide has four positive charges. 3D hydrophobic moment vectors (3D HMs) are useful as qualitative descriptions of peptide helices, which took into account the cooperation between the side chains and some conformational rearrangements within a whole peptide [18]. The length of 3D HMs for WV, WI, WF, and WW were 36.49, 30.30, 35.79, and 47.47 kTÅ/e, respectively, indicating the following 3D HMs order: WW > WF > WV > WI.
Table 1.
Peptides design and key physicochemical parameters.
| Peptides | Sequences | Molecular Mass | Net Charge | 3D Hydrophobic Moment Vectors (20.0) 1 | ||
|---|---|---|---|---|---|---|
| Calculated | Measured | Length (kTÅ/e) | Angle (°) | |||
| KL4A6 [9] | LLKAAAKAAAKLL-NH2 | 1280.69 | 1280.68 | +4 | 18.94 | 109.81 |
| WV | WVKAAAKAAAKVW-NH2 | 1398.74 | 1398.73 | +4 | 36.49 | 30.87 |
| WI | WIKAAAKAAAKIW-NH2 | 1426.79 | 1426.78 | +4 | 30.30 | 141.05 |
| WF | WFKAAAKAAAKFW-NH2 | 1494.83 | 1494.82 | +4 | 35.79 | 86.60 |
| WW | WWKAAAKAAAKWW-NH2 | 1572.87 | 1572.86 | +4 | 47.40 | 84.50 |
1 20.0 is the solvent dielectric constants used for the membrane-water interface.
2.2. Structure Variability of the Peptides
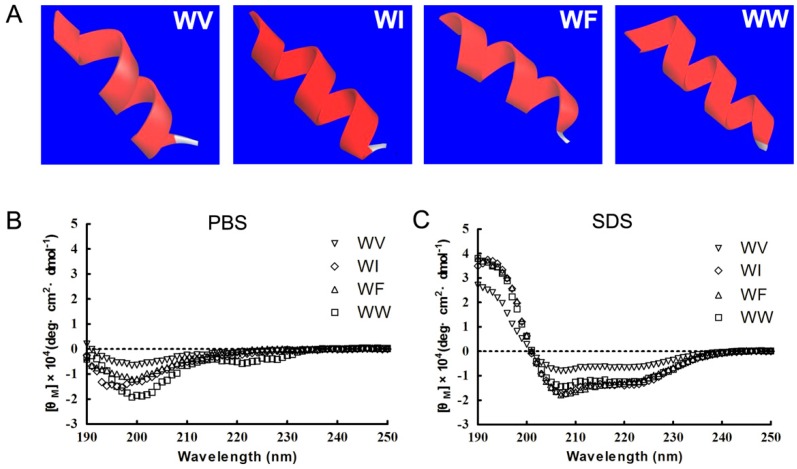
As shown in Figure 1A, the designed peptides showed similar α-helices, which was predicted online by computer-aided I-TASSER. These structures were in accordance with the requirement of the dermaseptin family structures based on minimized peptide theory. Then, circular dichroism (CD) spectra were measured under 10 mM phosphate-buffered saline (PBS) and 30 mM sodium dodecyl sulfate (SDS). In PBS, the CD spectra of WV, WI, and WF adopted a random coil structure with a minimum at ≈200 nm. The spectrum of WW displayed minima at ≈205 and 220 nm, which supports a disordered structure (Figure 1B). However, in the presence of SDS, the secondary structure of the peptides, with minima at ≈205 and 220 nm, was mainly α-helix (Figure 1C), which indicated that the cationic residues could initiate the association of the peptides onto the surface of the SDS micelles and facilitate the folding of the peptides through electrostatic interactions [19]. Moreover, it has been reported that the conformational transformation can enlarge the area of membrane damage as the peptide approaches the bacterial cell membrane [20].
Figure 1.
The structure of α-helix forming peptides. (A) The 3D structure of the peptides predicted by I-TASSER. The peptides are displayed by a solid ribbon style which was colored online according to secondary structures. red square: α-helix. (B,C) The CD spectra of the peptides in 10 mM PBS (pH 7.4) (B) and 30 mM SDS (C). An average of three scans was recorded for each peptide.
2.3. Antimicrobial Activity
To prove the biological activity of the synthetic peptides, a broad spectrum of model microbes was used in a minimal inhibitory concentration (MIC) assay. As listed in Table 2, the peptides exhibited antibacterial activity against both Gram-negative and Gram-positive pathogens. The geometric means (GM) of the MICs varied from 4.0 to 149.3 μM. Compared with KL4A6, all peptides, with the exception of WV, showed significantly improved antimicrobial activity and achieved the effective inhibition of B. pyocyaneum ATCC 27853, S. typhimurium ATCC 14028, S. pullorum C79-13, S. aureus ATCC 29213, and S. epidermidis ATCC 12228. The peptide WW with the lowest GM MIC value of 4.0 μM displayed approximately 1–28 folds higher antimicrobial activity than KL4A6, indicating the enhanced effectiveness of centrosymmetric Trp modification. It has been previously demonstrated that the Trp residue interacts strongly with the hydrophobic tails of lipids because of its large and bulky indole side chain, which is surrounded by negatively charged π-electron clouds [21], and the Trp residue arranged in the headgroup of the α-helical peptides plays an important role in promoting the biological activity of the peptide [11]. In addition, for the peptide WV, through W residues located at positions 1 and 13 of the sequence, antimicrobial activity was significantly decreased when compared to the peptide KL4A6. One possible explanation might be that Val has strong β-sheet folding propensity and a negative effect on helical propensity [4]. This phenomenon can be confirmed by CD spectra. The CD spectra of WV showed weak CD signals in the PBS and SDS solution.
Table 2.
MICs and therapeutic index of the peptides.
| MIC (μM) | KL4A6 [9] | WV | WI | WF | WW | Melittin |
|---|---|---|---|---|---|---|
| Gram-negative bacteria | ||||||
| E. coli ATCC25922 | 2 | 32 | 8 | 4 | 2 | 1 |
| E. coli UB1005 | 32 | >128 | 8 | 4 | 4 | 2 |
| P. aeruginosa ATCC 27853 | >128 | >128 | 32 | 16 | 8 | 2 |
| S. typhimurium ATCC 14028 | 128 | >128 | 32 | 32 | 8 | 4 |
| S. pullorum C79-13 | >128 | 64 | 8 | 8 | 2 | 1 |
| Gram-positive bacteria | ||||||
| S. aureus ATCC 29213 | >128 | >128 | 32 | 8 | 4 | 1 |
| S. epidermidis ATCC 12228 | 128 | >128 | 32 | 16 | 4 | 0.5 |
| S. faecalis ATCC 29212 | 16 | 64 | 32 | 32 | 4 | 1 |
| B. subtilis CMCC 63501 | 32 | >128 | 8 | 8 | 4 | 1 |
| MHC5(μM) 1 | >256 | >256 | 256 | 256 | 256 | 0.25 |
| GM(μM) 2 | 59.3 | 149.3 | 17.3 | 10.9 | 4.0 | 1.3 |
| Therapeutic index 3 | 8.6 | 3.4 | 14.8 | 23.5 | 64.0 | 0.2 |
1 Minimum hemolysis concentration 5% (MHC5) is defined as the lowest peptide concentration that induces 5% hemolysis. When no detectable hemolytic activity was observed at 256 μM, a value of 512 μM was used to calculate the therapeutic index. 2 Geometric mean (GM) of MIC values for the 10 bacteria were tested. When no detectable antimicrobial activity was observed at 128 μM, a value of 256 μM was used to calculate the therapeutic index. 3 Therapeutic index (TI) was calculated as MHC5/GM.
2.4. Cytotoxicity and Therapeutic Index
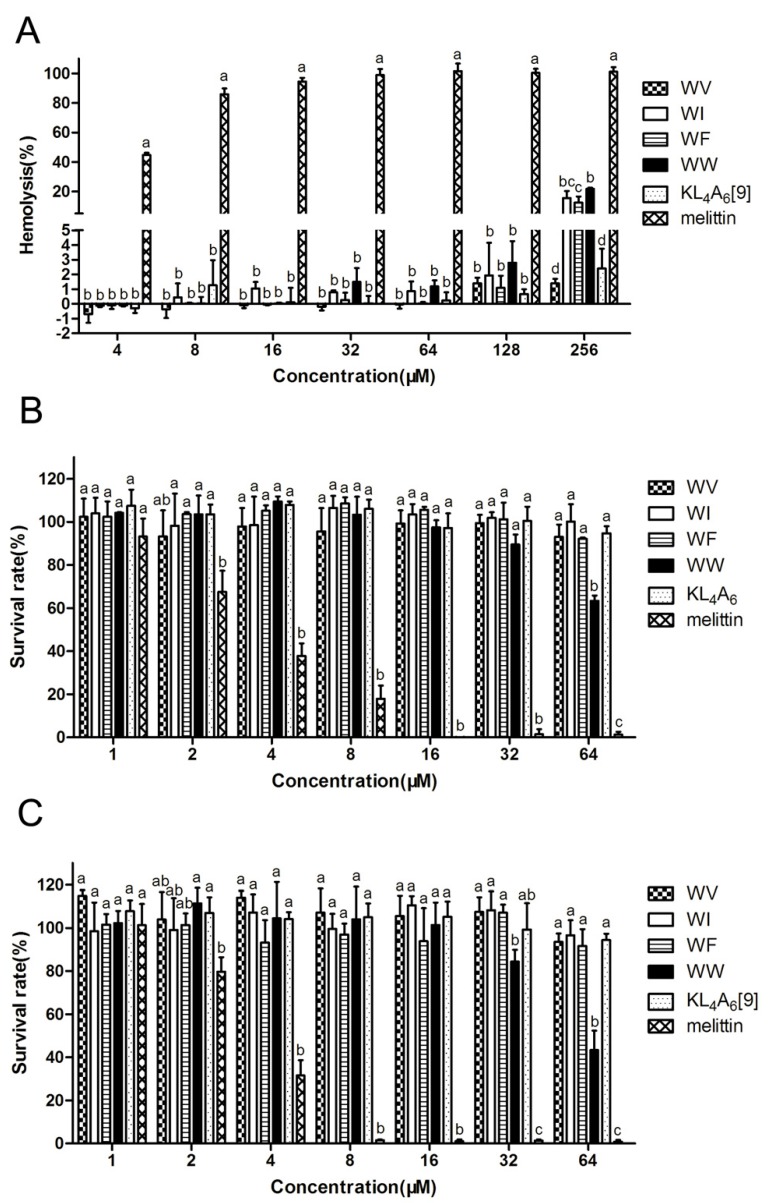
According to Table 2 and Figure 2A, dose-response studies revealed that hemolytic activity was relative to different hydrophobic residues in WX peptides at the higher concentration. On the whole, the results showed that four WX peptides had remarkably lower cytotoxicity and hemolysis when compared with melittin (the control). At the highest concentration (256 μM), the hemolysis of the peptides was as follows: WW > WI > WF > WV, and was higher than that of the peptide KL4A6 [9], suggesting an important role of the tryptophan residues residing at the end of an amphophilic alpha-helix for hemolytic activity. The results showed that the cell survival rate of the murine macrophage cell line (RAW264.7) at a peptide concentration of 64 μM was as follows: WW (63%) < WF (92%) < WV (93%) < WI (100%) (Figure 2B). Similarly, the cell survival rate of human embryonic kidney cells (HEK293T) at a peptide concentration of 64 μM was WW (43%) < WF (91%) < WV (93%) < WI (96%) (Figure 2C). It should also be noted that WW was significantly more toxic to both cells than KL4A6 and the other three WX-series peptides at 64 μM. Thus, the modification of aromatic residues such as Phe and Trp had higher hemolytic activity and cytotoxicity than that of Ile or Val. This was related to the fact that aromatic residues are considered to have a pivotal function on the partition by anchoring the peptide to the membrane [11]. However, taking antimicrobial efficacy into consideration, WW showed the greatest selectivity toward bacteria over mammalian cells with a TI value of 64.0. Therefore, WW was selected to explore the mechanism of action.
Figure 2.
Cytotoxicity of the peptides against human red blood cells (hRBCs) (A), RAW 264.7 (B), and HEK293T (C). Results were given as the mean ± standard deviation (SD) of three independent trials. Differences between groups exposed to the same concentration were determined by one-way ANOVA followed by Turkey’s post-hoc analysis (n = 3). The values with different superscripts (a, b, c, d) indicate a significant difference (p < 0.05).
2.5. Mechanism of Action of the Peptides
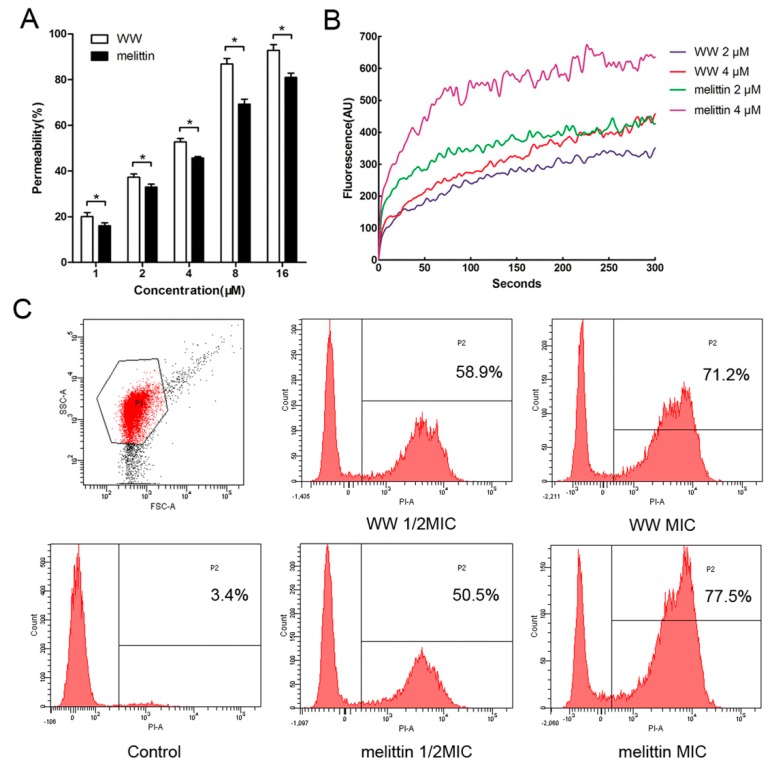
Compared to conventional antibiotics, most AMPs kill bacteria through the disruption of the microbial membrane [22,23]. Gram-positive bacteria and Gram-negative bacteria share the commonality of having a cytoplasmic membrane. Gram-negative bacteria are surrounded by an extra outer membrane (OM) containing lipopolysaccharide (LPS) as the major lipid component, which performs the crucial role of compromising the exchange of material required for sustaining life [24]. Figure 3A shows that the dose-response relationship between OM permeability and concentration (1–16 μM) of the peptides. At a concentration of 16 μM, the two peptides caused an outer membrane permeability greater than 75%. Interestingly, the outer membrane permeability of WW was stronger than those of melittin at the same concentration, even though WW had a lower antibacterial activity against E. coli UB1005 than melittin.
Figure 3.
The influence of WW and melittin on the outer membrane permeability (A), cytoplasmic membrane depolarization (B), and membrane integrity (C). Results in (A) are given as the mean ± standard deviation (SD) of three independent experiments and the difference (* p < 0.05) between two groups exposed to the same peptide concentration was analyzed by the Student’s t test. The results in (B) were expressed as an average of three independent trials. The data in (C) are representative of three independent experiments.
Thus, upon the permeabilization of the outer membrane, the electrical potential changes of the cytoplasmic membrane were further measured by 3,3′-dipropylthiadicarbo-cyanine iodide (diSC3-5). The bacterial cytoplasmic membrane consists predominantly of hydroxylated phospholipids (e.g., cardiolipin, phosphatidylglycerol, phosphatidylserine), representing a negative net charge at physiological pH [25], which enhance the binding of the cationic AMPs. Upon membrane absorption and binding, the peptide was then inserted into the lipid bilayer, resulting in membrane permeabilization and pore/ion channel formation, simultaneously concomitant with the membrane potential collapse. As shown in Figure 3B, these two peptides induced a sustained release of diSC3-5 over a period of 300 s, and every peptide produced dose-dependent results at the final time of 300 s. Melittin induced a higher and faster increase of fluorescence than those of WW at 2 μM and 4 μM, respectively. Considering the moderate OM permeabilization of melittin, we reasoned that the stronger antimicrobial activity against E. coli UB1005 by melittin than WW may be due to its higher cytoplasmic membrane depolarization.
Furthermore, fluorescence-assisted cell sorting (FACScan) analysis was performed to determine the membrane integrity of E. coli treated with WW or melittin as the fluorescent probe propidium iodide (PI) can incorporate and stain nucleic acids in cells with damaged membranes [26]. The data were then analyzed once and displayed as two-parameters, complexity (Side Scatter, SSC) and cell size (Forward Scatter, FSC) before the fluorescence frequency distribution histogram of the E. coli population was obtained (P1). As shown in Figure 3C, the x-axis represents the fluorescent emission of PI, and the count (y-axis) of PI-positive cells was measured for each peptide concentration. The data showed that the percentage of PI-positive cells with membranes damaged by WW was 58.9% (1/2 × MIC) and 71.2% (1 × MIC). The percentage of PI-positive cells treated with melittin was 50.5% (1/2 × MIC) and 77.5% (1 × MIC). This observation indicated that WW has the ability to permeabilize the cytoplamic membranes of bacteria, similar to melittin, which results in cell death.
2.6. Membrane Morphological Analysis
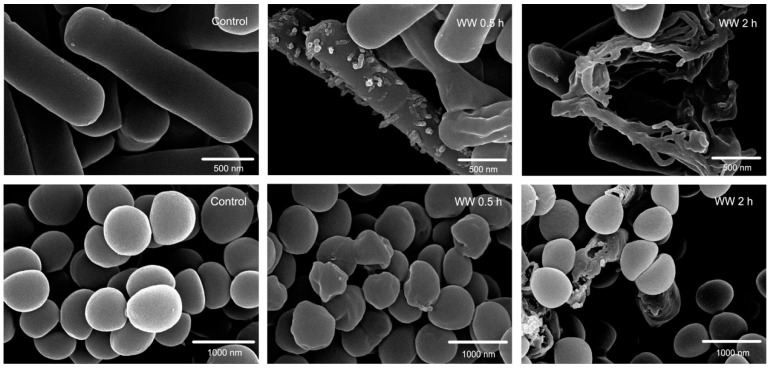
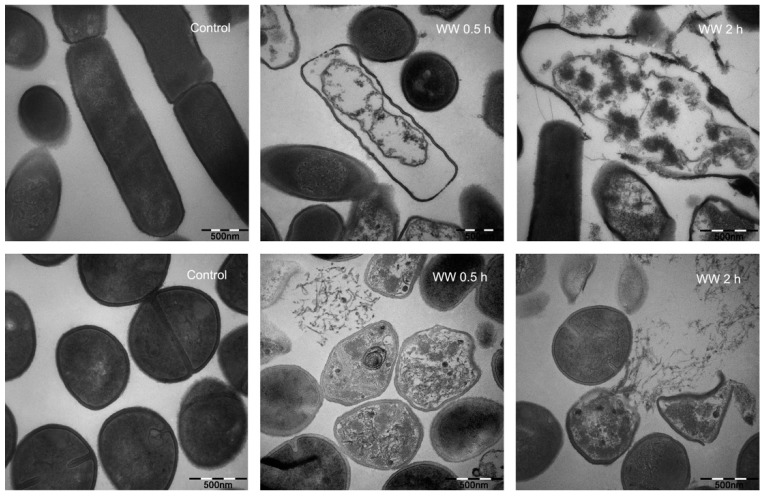
Based on the rapid permeabilization and depolarization of microbial membranes, we visualized the bacterial membrane damage by field emission scanning electron microscopy (FE-SEM) and transmission electron microscopy (TEM). Figure 4 and Figure 5 demonstrate that membrane morphology of the cells treated with the peptides were all obviously different from the control (no peptide). Under FE-SEM, the controls showed a smooth and intact surface. After treatment with the peptide WW for 30 min, the membrane surface morphology and integrity of both the Gram-negative E. coli and Gram-positive S. aureus were observed as a rough, shrunken, and slight leakage of cellular cytoplasmic content to cell lysis, resulting in the formation of visible membrane atrophy and corrugation. After 2 h of WW treatment, significant membrane rupture occurred in the bacteria, some of which even appeared to be completely dissolved. Similarly, after 30 min, peptide WW induced significant morphological and intracellular changes of E. coli and S. aureus when compared to a control with a smooth cell membrane and homogeneous cytoplasm under TEM, and caused severe membrane lysis and plasmolysis after 2 h. This observation is consistent with the documented membrane lysis mechanism of KL4A6 [9] and some other natural or synthetic AMPs [27].
Figure 4.
Scanning electron micrographs of E. coli 25922 and S. aureus 29213 treated with WW at 1 × MIC. All data are representative of three independent experiments.
Figure 5.
Transmission electron micrographs of E. coli 25922 and S. aureus 29213 treated with WW at 1 × MIC. All data are representative of three independent experiments.
2.7. Endotoxin Neutralization Assay
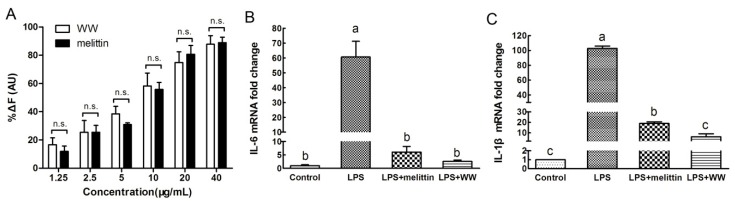
As we know, LPS, as a major component of the outer membrane of Gram-negative bacteria, can increase the expression of pro-inflammatory cytokines by binding to its receptors, thereby triggering a systemic inflammatory response [28]. With the continuous advancement of the properties and performance of antimicrobial peptides in the past two decades, in addition to classical antibacterial activity, they can also be used as antitoxin drugs to bind and isolate LPS [29]. In this study, we first assessed the binding activity of WW to LPS (100 ng/mL) by using a fluorescence-based displacement assay with BODIPY-TR cadaverine (BC). As shown in Figure 6A, both WW and melittin displayed a concentration-dependent increase in the fluorescent intensities. At all concentrations, the %ΔF (AU) caused by WW and melittin did not show significant differences. However, at low concentrations (1.25, 2.5, 5, 10 μg/mL), the changes in fluorescence intensity caused by WW were slightly higher than that of melittin. As the peptide concentration increased, the binding of melittin to LPS showed a stronger increase than WW.
Figure 6.
(A) Peptide binding affinity to LPS from E. coli 0111:B4. Results in Figure 6A are given as mean ± standard deviation (SD) of three independent experiments and the difference (n.s., not significant at p < 0.05) between two groups exposed to the same peptide concentration was analyzed by the Student’s t test. (B,C) WW inhibited the LPS-induced gene expression of pro-inflammatory cytokine effects in the murine macrophage cell line RAW264.7. Results are given as the mean ± SD. Differences between groups were determined by one-way ANOVA followed by Turkey’s post-hoc analysis (n = 3). The values with different superscripts (a, b, c) indicated a significant difference (p < 0.05).
Subsequently, the gene expression of pro-inflammatory cytokines (e.g., interleukin-6(IL-6) and interleukin-1β(IL-1β)) in macrophages was measured to determine the inhibitory effect of WW on the LPS-induced inflammatory response. As shown in Figure 6B,C, untreated cells and LPS-alone (100 ng·mL−1) treated cells served as the negative and positive controls, respectively. After stimulation with LPS, the gene expression of IL-6 and IL-1β from the murine macrophages were robustly increased. However, in the presence of WW or melittin, the gene expression of pro-inflammatory factors in cells treated with LPS were markedly decreased. WW was more effective than melittin to attenuate the mRNA expression of IL-1β. Thus, the peptide WW has the potential to maintain the balance of cytokines in the inflammatory response induced by LPS.
3. Materials and Methods
3.1. Bacterial Strains and Mammalian Cells
The bacterial strains E. coli ATCC25922, P. aeruginosa ATCC27853, S. typhimurium ATCC14028, S. pullorum C7913, S. aureus ATCC29213, S. epidermidis ATCC12228, S. faecalis ATCC29212, and the model microbe B. subtilis CMCC63501 were obtained from the College of Veterinary Medicine, Northeast Agricultural University (Harbin, China). E. coli UB1005 was provided by the State Key Laboratory of Microbial Technology, Shandong University (Jinan, China). The hRBCs were obtained from the Northeast Agricultural University Hospital. RAW264.7 was purchased from the cell bank of the Chinese Academy of Sciences, SIBS (Shanghai, China). HEK293T was obtained from the College of Animal Science and Technology, Northeast Agricultural University (Harbin, China).
3.2. Peptides Synthesis and Sequence Analysis
All peptide-based solid phase protocols were synthesized by GL Biochem (Shanghai, China), and identified via matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS, Linear Scientific Inc., Billerica, MA, USA.) using α-cyano-4-hydroxycinnamic acid (HCCA) as the matrix. Analytical reversed-phase high-performance liquid chromatography (RP-HPLC) (LC 3000, Beijing, China) was used to confirm the purity of each peptide (>95%). The powder form peptide was dissolved in deionized water at a concentration of 2560 μM and stored at −20 °C before subsequent evaluations. The primary physicochemical parameters were calculated using the AMP database [30]. The 3D structure projection was predicted online by I-TASSER [31]. 3D hydrophobic moment (HM) vectors were calculated online by a 3D-HM calculator [32].
3.3. CD Spectroscopy
The CD spectra were performed on a J-820 spectrometer (Jasco, Tokyo, Japan). Data were recorded at 25 °C with the following parameters: 0.1 cm path length, 10 nm∙min−1 scanning rate, and 190 to 250 nm wavelength range. The final concentration of the peptide samples was 150 mM in 10 mM PBS to mimic the aqueous environment and in 30 mM SDS micelles to mimic the negatively charged prokaryotic membrane comparable environment. The acquired data were converted to mean residue ellipticity as follows: θM = (θobs × 1000)/(l × c × n), where θM is the mean residue ellipticity (deg∙cm2∙dmol−1), θobs is the measured ellipticity corrected for the buffer at a given wavelength (mdeg), l is the path length (cm), c is the peptide concentration (mM), and n is the number of amino residues.
3.4. MIC Measurements
MIC measurements were performed in 96-well plates [33]. The peptides were dissolved in 0.2% bovine serum albumin and 0.01% acetic acid, and an appropriate broth was subjected to a series of two-fold dilutions. The final concentration of microbial suspensions was from 2 × 105 to 7 × 105 colony forming units (CFU)∙mL−1. Key parameters were volume (50 μL bacterial suspension + 50 μL peptide solution), incubating time (18–24 h), and temperature (37 °C). Each test was performed in three replicates on at least two independent occasions. The Mueller Hilton broth (MHB) (AoBoX, Shanghai, China) without or with microbial cells was regarded as the negative control or positive control, respectively. MICs were recorded at the lowest peptide concentration without visible turbidity and spectrophotometrically by measuring the optical density (OD) at 492 nm (Tecan, Salzburg, Austria).
3.5. Hemolytic Activity Assay
Hemolytic activity measurements were performed to evaluate the effect of peptides on hemoglobin release levels by erythrocyte lysis [34]. The experimental protocol was reviewed and approved by the ethics committee of the Northeast Agricultural University Hospital, and the experimental method was carried out in accordance with the approved guidelines and regulations. A total of 1 mL of fresh hRBCs was collected and diluted 10-fold with PBS (pH 7.4). A quantity of 50 μL of the hRBCs solution was incubated with 50 μL of serially diluted peptides dissolved in PBS for 1 h at 37 °C. hRBC suspension treated with 0.1% Triton X-100 was employed as a positive control and an untreated hRBC suspension was used as a negative control. A total of 50 μL of the supernatant was collected from the mixture after centrifugation (1000g, 5 min, 4 °C) and transferred to a new 96-well microplate. The percent hemolysis was calculated using the following equation: Hemolysis (%) = [(OD570 of the treated sample — OD570 of the negative control)/(OD570 of the positive control — OD570 of the negative control)] × 100%.
3.6. Cytotoxicity Assay
The Roswell Park Memorial Institute-1640 (RPMI-1640) (Invitrogen, Carlsbad, CA, USA) and Dulbecco’s modified Eagle’s medium-high glucose (DMEM) (Gibco, China) were supplemented with 100 μg∙mL−1 streptomycin sulfate, 100 U∙mL−1 penicillin, and 10% (v/v) fetal bovine serum (Hyclone, Logan, UT, USA). These were used to culture the RAW264.7 and HEK293T cells, respectively. The peptides were added to cell cultures (1.0–2.0 × 104 cells/well) at different final concentrations (1–64 µM) in 96-well plates for 18–24 h at 37 °C in 5% CO2, and 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) (50 µL, 0.5 mg∙mL−1) was injected and incubated with cell cultures for 4 h at 37 °C. Finally, the centrifuged cells without the supernatants were mixed with 150 µL of dimethyl sulfoxide to dissolve the formazan crystals that formed. The optical density (OD) was recorded by a microplate reader (Tecan, Salzburg, Austria) at 570 nm.
3.7. Outer Membrane (OM) Permeability Assay
An OM permeability assay was performed on an F-4500 fluorescence spectrophotometer (Hitachi, Japan). Prior to the experiment, E. coli UB1005 in mid-log phase was obtained to wash thrice with 4-(2-hydroxyethyl)piperazine-1-ethanesulfonic acid (HEPES) buffer (pH 7.4, containing 5 mM glucose) and diluted to 105 CFU∙mL−1 in the same buffer. Data were recorded (excitation λ = 350 nm, emission λ = 420 nm) in a quartz cuvette with a 2 mL bacterial suspension containing 10 μM N-phenyl-1-naphthylamine (NPN). The observed values were converted to % NPN uptake using the equation:
| % NPN uptake = (Fobs − F0) / (F100 − F0) × 100 |
where F0 is the background fluorescence of NPN in the absence of peptide; F100 is the fluorescence of NPN upon addition of 10 μg∙mL−1 polymyxin B (positive control); and Fobs is the measured fluorescence at a given peptide concentration.
3.8. Cytoplasmic Membrane Depolarization Assay
A membrane potential-sensitive fluorescent dye, diSC3-5, was used to determine the cytoplasmic membrane depolarization activity of the peptides. Coalescent preparation of E. coli UB1005 suspension was performed in different HEPES buffer (pH 7.2, 5 mM HEPES buffer, 20 mM glucose, 100 mM KCl) and reached an OD600 of 0.05. Data were recorded (excitation λ = 622 nm, emission λ = 670 nm) in a quartz cuvette with a 2 mL bacterial suspension containing different concentrations of peptides after incubation with 0.4 μM diSC3-5 for 1 h at 37 °C. Observation changes were recorded from 0 to 300 s when the cell suspension was mixed with peptides at final concentrations of 2 μM and 4 μM, respectively.
3.9. FACScan Analysis
The membrane integrity measurements were performed with a FACScan instrument (Bectone-Dickinson, San Jose, CA, USA) by using a modified method as described previously [6]. E. coli ATCC 25922 was similarly prepared to the MIC measurements, washed thrice with PBS, and diluted to 105 CFU·mL−1 in the same buffer. Two concentrations of the peptides (1/2 and 1 × MIC) were added to the bacterial suspension containing 10 μg·mL−1 propidium iodide (PI) at 4 °C for 30 min. As a control, bacteria were exposed to PBS without the peptide. The gate (P1) was defined based on the FSC and SSC properties of E. coli. The FSC threshold was appropriately raised to include the large cell events in the sample and excluded the debris population. Data acquisition was performed using FACSDIVA Software (Becton Dickinson, San Jose, CA, USA) with a stopping gate of 10,000 total events. Observations were repeated three times at a laser excitation wavelength of 488 nm. Based on the ability of flow cytometers to provide accurate quantification of cell numbers, the counts of PI-positive cells were measured for each peptide.
3.10. Membrane Morphological Observations
The preparation of the bacteria suspension was similar to the FACScan analysis. The cells were incubated with the peptide (0 and 1 × MIC) at 37 °C for 30 and 120 min, respectively. After incubation, the cells were centrifuged (5000× g, 5 min) and washed three times with PBS (10 mM). Harvested cell pellets were subjected to fixation with glutaraldehyde (2.5%) for 6 to 8 h at 4 °C, followed by washing twice with PBS. Furthermore, the cells were dehydrated for 10 min in a graded ethanol series (50%, 70%, 90%, and 100%), followed by 15 min in 100% ethanol. Then, the specimens for SEM were dehydrated for 15 min in a mixture (v:v = 1:1) of 100% ethanol and tertiary butanol, and absolute tertiary butanol. Finally, the specimens were dried in a critical point dryer with liquid CO2, coated with gold-palladium, and then observed using a Hitachi S-4800 SEM (Hitachi, Japan). The TEM samples were dehydrated in a mixture (v:v = 1:1) of 100% ethanol and acetone and absolute acetone for 15 min, and then transferred to a 1:1 mixture of absolute acetone and epoxy resin for 30 min and then to pure epoxy resin and were incubated overnight at a constant temperature. Finally, the specimens were sectioned with an ultramicrotome, stained with uranyl acetate and lead citrate, and observed using a Hitachi H-7650 TEM (Hitachi, Japan).
3.11. LPS Neutralization Assay
BODIPY-TR-cadaverine fluorescent dye (BC, Sigma, USA) was used to determine the LPS neutralizing activity of the peptides. A total of 100 ng/mL LPS from E. coli O111:B4 was incubated with 5 μg/mL BC in Tris buffer (50 mM, pH 7.4) for 4 h at room temperature. The peptides were serially diluted to different concentrations (1.25–40 μg/mL) in a 96-well plate and incubated with an equal volume of the LPS-probe mixture for 1 h at 37 °C. The fluorescence was recorded (excitation λ = 580 nm, emission λ = 620 nm) with a spectrofluorophotometer (the Infinite 200 pro, Tecan, China). The values were converted to %ΔF (AU) using the following equation:
| %ΔF (AU) = (Fobs − F0) / (F100 − F0) × 100 |
where F0 is the initial fluorescence of BC with LPS; F100 is the BC fluorescence with LPS cells upon the addition of 10 μg∙mL−1 polymyxin B (the positive control); and Fobs is the observed fluorescence at a given peptide concentration.
3.12. Cytokine Measurements
The RAW264.7 cell culture was prepared using the same method as the cytotoxicity assay. The cells were seeded in 12 wells plates at a given density (5 × 105 cells per well); after overnight culture, 100 ng·mL−1 LPS from E. coli 0111:B4 was added to stimulate the generation of cytokines in the presence or absence of the peptides at concentrations of 10 μg·mL−1. The mRNA levels of IL-6 and IL-1β were determined by quantitative real-time PCR(qRT-PCR) as previously described [33]. The total RNA was isolated from the RAW264.7 cells using TRIzol according to the manufacturer’s instructions. Total RNA (1 µg) was reverse transcribed by the M-MLV Reverse Transcriptase (Thermo scientific, Waltham, MA, USA). The reactions were incubated in chronological arrangement with the following parameters: at 25 °C for 20 min, at 42 °C for 60 min, and at 70 °C for 10 min. The obtained cDNA samples were determined by qRT-PCR with SYBR green I fluorescence dye (Takara Bio Inc., Kusatsu, Japan). Relative quantifications of the mRNA expression of the target genes were calculated through the comparative threshold cycle number for each sample (2−ΔΔCT). Gene expression was normalized to the corresponding β-actin level. The primers for qRT-PCR in this study are shown below: IL-6-F: ggagaggagacttcacagagga; IL-6-R: atttccacgatttcccagaga; IL-1β-F: tgaaatgccaccttttgacag; and IL-1β-R: ccacagccacaatgagtgatac.
3.13. Statistical Analysis
Data were analyzed by the unpaired Student’s t-test or one-way ANOVA using SPSS 22.0 software (IBM, Chicago, IL, USA). Quantitative data were presented as the mean ± standard deviation of the mean. Differences were defined as significant when the P-value was less than 0.05.
4. Conclusions
In this study, the minimized dermaseptin peptide was optimized by symmetry modification in light of the physicochemical properties. The derivatives were folded into α-helical structures in membrane-mimetic environments, but were unstructured in water. Optimized peptides demonstrated an enhanced and extended spectrum of antimicrobial activities against both Gram-positive and Gram-negative bacteria. The peptide WW was found to possess a high cell selectivity with TI = 64.0. WW exhibited the ability to permeabilize the outer membrane of bacterial cells and to depolarize the cytoplasmic membrane, which damaged the membrane integrity and caused intracellular content leakage. Additionally, it displayed the capacity to modulate LPS-induced inflammatory response in RAW264.7 cells by avoiding the overexpression of cytokines. In short, all of the data we presented are helpful in the design of AMPs and the centrosymmetric modification of minimized peptide WW has potential as a broad-spectrum antimicrobial, antisepsis, or topical endotoxin neutralization agent.
Author Contributions
Conceptualization, C.S. and W.L.; Methodology, W.L.; Software, C.S.; Validation, A.S. and D.M.; Formal Analysis, P.T.; Investigation, D.M.; Resources, A.S.; Data Curation, X.D.; Writing—Original Draft Preparation, C.S.; Writing—Review and Editing, A.S.; Visualization, P.T.; Supervision, D.M.; Project Administration, C.L.; Funding Acquisition, A.S. and W.L.
Funding
This study was supported by the National Natural Science Foundation of China (31672434, 31472104, 31872368); Agriculture Research System of China (CARS-35); Weifang Science and Technology Development Program (2016GX022), and Weifang University Doctoral Foundation Program (2017BS10).
Conflicts of Interest
The authors declare no conflicts of interest.
References
- 1.Sommer M.O.A., Munck C., Toft-Kehler R.V., Andersson D.I. Prediction of antibiotic resistance: Time for a new preclinical paradigm? Nat. Rev. Microbiol. 2017;15:689–696. doi: 10.1038/nrmicro.2017.75. [DOI] [PubMed] [Google Scholar]
- 2.Zhang L.J., Gallo R.L. Antimicrobial peptides. Curr. Biol. 2016;26:R14–R19. doi: 10.1016/j.cub.2015.11.017. [DOI] [PubMed] [Google Scholar]
- 3.Omardien S., Brul S., Zaat S.A. Antimicrobial activity of cationic antimicrobial peptides against gram-positives: Current progress made in understanding the mode of action and the response of bacteria. Front. Cell Dev. Biol. 2016;4:111. doi: 10.3389/fcell.2016.00111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ong Z.Y., Gao S.J., Yang Y.Y. Short syntheticβ-sheet forming peptide amphiphiles as broad spectrum antimicrobials with antibiofilm and endotoxin neutralizing capabilities. Adv. Funct. Mater. 2013;23:3682–3692. doi: 10.1002/adfm.201202850. [DOI] [Google Scholar]
- 5.Ong Z.Y., Wiradharma N., Yang Y.Y. Strategies employed in the design and optimization of synthetic antimicrobial peptide amphiphiles with enhanced therapeutic potentials. Adv. Drug Deliv. Rev. 2014;78:28–45. doi: 10.1016/j.addr.2014.10.013. [DOI] [PubMed] [Google Scholar]
- 6.Ma Z., Wei D., Yan P., Zhu X., Shan A., Bi Z. Characterization of cell selectivity, physiological stability and endotoxin neutralization capabilities of alpha-helix-based peptide amphiphiles. Biomaterials. 2015;52:517–530. doi: 10.1016/j.biomaterials.2015.02.063. [DOI] [PubMed] [Google Scholar]
- 7.Lyu Y., Yang Y., Lyu X., Na D., Shan A. Antimicrobial activity, improved cell selectivity and mode of action of short PMAP-36-derived peptides against bacteria and Candida. Sci. Rep. 2016;6:27258. doi: 10.1038/srep27258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dong N., Zhu X., Chou S., Shan A., Li W., Jiang J. Antimicrobial potency and selectivity of simplified symmetric-end peptides. Biomaterials. 2014;35:8028–8039. doi: 10.1016/j.biomaterials.2014.06.005. [DOI] [PubMed] [Google Scholar]
- 9.Li W., Tan T., Xu W., Xu L., Dong N., Ma D., Shan A. Rational design of mirror-like peptides with alanine regulation. Amino Acids. 2016;48:403–417. doi: 10.1007/s00726-015-2094-y. [DOI] [PubMed] [Google Scholar]
- 10.Fimland G., Eijsink V.G.H., Nissen-Meyer J. Mutational analysis of the role of tryptophan residues in an antimicrobial peptide. Biochemistry. 2002;41:9508–9515. doi: 10.1021/bi025856q. [DOI] [PubMed] [Google Scholar]
- 11.Chan D.I., Prenner E.J., Vogel H.J. Tryptophan-and arginine-rich antimicrobial peptides: Structures and mechanisms of action. Biochim. Et Biophys. Acta (Bba) Biomembr. 2006;1758:1184–1202. doi: 10.1016/j.bbamem.2006.04.006. [DOI] [PubMed] [Google Scholar]
- 12.Yang S.T., Shin S.Y., Hahm K.S., Kim J.I. Design of perfectly symmetric Trp-rich peptides with potent and broad-spectrum antimicrobial activities. Int. J. Antimicrob. Agents. 2006;27:325–330. doi: 10.1016/j.ijantimicag.2005.11.014. [DOI] [PubMed] [Google Scholar]
- 13.Sung-Tae Y., Yub S.S., Jae Il K. Interaction mode of a symmetric Trp-rich undeca peptide PST11-RK with lipid bilayers. Febs Lett. 2007;581:157–163. doi: 10.1016/j.febslet.2006.12.003. [DOI] [PubMed] [Google Scholar]
- 14.Podorieszach A.P., Huttunen-Hennelly H.E. The effects of tryptophan and hydrophobicity on the structure and bioactivity of novel indolicidin derivatives with promising pharmaceutical potential. Org. Biomol. Chem. 2010;8:1679–1687. doi: 10.1039/b921248e. [DOI] [PubMed] [Google Scholar]
- 15.Papanastasiou E.A., Hua Q., Sandouk A., Son U.H., Christenson A.J., Van Hoek M.L., Bishop B.M. Role of acetylation and charge in antimicrobial peptides based on human β-defensin-3. Apmis. 2009;117:492–499. doi: 10.1111/j.1600-0463.2009.02460.x. [DOI] [PubMed] [Google Scholar]
- 16.Cunningham B.C., Wells J.A. Minimized proteins. Curr. Opin. Struct. Biol. 1997;7:457–462. doi: 10.1016/S0959-440X(97)80107-8. [DOI] [PubMed] [Google Scholar]
- 17.Takahashi D., Shukla S.K., Prakash O., Zhang G. Structural determinants of host defense peptides for antimicrobial activity and target cell selectivity. Biochimie. 2010;92:1236–1241. doi: 10.1016/j.biochi.2010.02.023. [DOI] [PubMed] [Google Scholar]
- 18.Reisser S., Strandberg E., Steinbrecher T., Ulrich A.S. 3D hydrophobic moment vectors as a tool to characterize the surface polarity of amphiphilic peptides. Biophys. J. 2014;106:2385–2394. doi: 10.1016/j.bpj.2014.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wiradharma N., Khoe U., Hauser C.A., Seow S.V., Zhang S., Yang Y.-Y. Synthetic cationic amphiphilic α-helical peptides as antimicrobial agents. Biomaterials. 2011;32:2204–2212. doi: 10.1016/j.biomaterials.2010.11.054. [DOI] [PubMed] [Google Scholar]
- 20.Mecke A., Lee D.-K., Ramamoorthy A., Orr B.G., Holl M.M.B. Membrane thinning due to antimicrobial peptide binding: An atomic force microscopy study of MSI-78 in lipid bilayers. Biophys. J. 2005;89:4043–4050. doi: 10.1529/biophysj.105.062596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao J., Zhao C., Liang G., Zhang M., Zheng J. Engineering antimicrobial peptides with improved antimicrobial and hemolytic activities. J. Chem. Inf. Modeling. 2013;53:3280–3296. doi: 10.1021/ci400477e. [DOI] [PubMed] [Google Scholar]
- 22.Zasloff M. Antimicrobial peptides of multicellular organisms. Nature. 2002;415:389–395. doi: 10.1038/415389a. [DOI] [PubMed] [Google Scholar]
- 23.Jenssen H., Hamill P., Hancock R.E. Peptide antimicrobial agents. Clin. Microbiol. Rev. 2006;19:491–511. doi: 10.1128/CMR.00056-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Epand R.M., Walker C., Epand R.F., Magarvey N.A. Molecular mechanisms of membrane targeting antibiotics. Biochim. Et Biophys. Acta (Bba) Biomembr. 2016;1858:980–987. doi: 10.1016/j.bbamem.2015.10.018. [DOI] [PubMed] [Google Scholar]
- 25.Teixeira V., Feio M.J., Bastos M. Role of lipids in the interaction of antimicrobial peptides with membranes. Prog. Lipid Res. 2012;51:149–177. doi: 10.1016/j.plipres.2011.12.005. [DOI] [PubMed] [Google Scholar]
- 26.Lv Y., Wang J., Gao H., Wang Z., Dong N., Ma Q., Shan A. Antimicrobial properties and membrane-active mechanism of a potential alpha-helical antimicrobial derived from cathelicidin PMAP-36. PLoS ONE. 2014;9:e86364. doi: 10.1371/journal.pone.0086364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fjell C.D., Hiss J.A., Hancock R.E., Schneider G. Designing antimicrobial peptides: Form follows function. Nat. Rev. Drug Discov. 2012;11:37–51. doi: 10.1038/nrd3591. [DOI] [PubMed] [Google Scholar]
- 28.Zong X., Song D., Wang T., Xia X., Hu W., Han F., Wang Y. LFP-20, a porcine lactoferrin peptide, ameliorates LPS-induced inflammation via the MyD88/NF-kappaB and MyD88/MAPK signaling pathways. Dev. Comp. Immunol. 2015;52:123–131. doi: 10.1016/j.dci.2015.05.006. [DOI] [PubMed] [Google Scholar]
- 29.Lee W., Yoo H., Kim J.A., Lee S., Jee J.G., Lee M.Y., Lee Y.M., Bae J.S. Barrier protective effects of piperlonguminine in LPS-induced inflammation in vitro and in vivo. Food Chem. Toxicol. Int. J. Publ. Br. Ind. Biol. Res. Assoc. 2013;58:149–157. doi: 10.1016/j.fct.2013.04.027. [DOI] [PubMed] [Google Scholar]
- 30.AMP Database. [(accessed on 25 January 2019)]; Available online: http://aps.unmc.edu/AP/main.php.
- 31.I-TASSER. [(accessed on 12 December 2018)]; Available online: http://zhanglab.ccmb.med.umich.edu/I-TASSER/
- 32.3D-HM Calculator. [(accessed on 25 January 2019)]; Available online: http://www.ibg.kit.edu/HM/
- 33.Shao C., Tian H., Wang T., Wang Z., Chou S., Shan A., Cheng B. Central beta-turn increases the cell selectivity of imperfectly amphipathic alpha-helical peptides. Acta Biomater. 2018;69:243–255. doi: 10.1016/j.actbio.2018.01.009. [DOI] [PubMed] [Google Scholar]
- 34.Tan T., Wu D., Li W., Zheng X., Li W., Shan A. High Specific selectivity and membrane-active mechanism of synthetic cationic hybrid antimicrobial peptides based on the peptide FV7. Int. J. Mol. Sci. 2017;18:339. doi: 10.3390/ijms18020339. [DOI] [PMC free article] [PubMed] [Google Scholar]