Abstract
Osteoarthritis (OA) is an aseptic inflammatory disease which is associated with the stromal cell-derived factor 1/C-X-C chemokine receptor type 4 (SDF-1/CXCR4) axis. Accumulating studies have identified numbers of microRNAs (miRNAs) that serve important roles in the pathogenesis of OA. However, whether and how the inhibition of the SDF-1/CXCR4 axis induces alterations in miRNA expression remains largely unclear. miRNA profiling was performed in OA chondrocytes stimulated with SDF-1 alone, or SDF-1 with the CXCR4 antagonist TN14003 by miRNA microarray. Candidate miRNAs were verified by reverse transcription quantitative polymerase chain reaction. Bioinformatic analyses including target prediction, gene ontology (GO) and pathway analysis were performed to explore the potential functions of candidate miRNAs. Notably, 7 miRNAs (miR-146a-5p, miR-221-3p, miR-126-3p, miR-185-5p, miR-155-5p, miR-124-3p and miR-130a-3p) were significantly differentially expressed. GO analysis indicated that miR-146a-5p and its associated genes were enriched in receptor regulatory activity, nuclear factor-kappa-light-chain-enhancer of activated B cells (NF-κB)-inducing kinase activity, cellular response to interleukin-1, cytokine-cytokine receptor interaction, NF-κB signaling pathway and osteoclast differentiation pathways. CXCR4 was predicted to be a target of miR-146a-5p with high importance. The mRNA and protein levels of key factors involved in cartilage degeneration were measured following manipulation of the expression levels of miR-146a-5p in OA chondrocytes. CXCR4 and MMP-3 levels were negatively associated with miR-146a-5p expression, while the levels of type II collagen and aggrecan were positively associated. These data reveal that TN14003 upregulates miR-146a-5p expression, and also pinpoints a novel role of miR-146a-5p in inhibiting cartilage degeneration by directly targeting the SDF-1/CXCR4 axis.
Keywords: microRNA, osteoarthritis, chondrocyte, stromal cell-derived factor 1/C-X-C chemokine receptor type 4 axis, TN14003
Introduction
Osteoarthritis (OA) is a multifactorial articular disease characterized by cartilage degeneration, subchondral sclerosis and osteophyte formation (1–3). Normal function of articular cartilage is highly dependent on the homeostasis of the extracellular matrix (ECM), which serves as the mechanical structure and is involved in signal transduction in chondrocytes (4–7). Previous studies have demonstrated the roles of the interaction between anabolic factors, including transforming growth factor β, and catabolic factors, including matrix metalloproteinases (MMPs) and aggrecanases, in the maintenance and regeneration of ECM in chondrocytes (8–10). However, the exact molecular mechanisms involved in OA remain largely unclear. Although progress in OA therapy has been incremental, the majority of treatments only improve clinical symptoms, as opposed to restoring the damaged ECM (11). In addition, inhibition of OA by the regulation of specific genes has been an unsuccessful strategy (12,13).
Stromal cell-derived factor 1 (SDF-1) is a cytokine that is associated with inflammation, and is identified in the synovial membranes adjacent to articular cartilage (14–16). Binding of SDF-1 and its ligand C-X-C chemokine receptor type 4 (CXCR4), a G protein-coupled receptor located in the surface of chondrocytes, induces the release of MMPs from the ECM, thereby exacerbating OA (16,17). As a CXCR4 inhibitor, the compound AMD3100 blocks the SDF-1/CXCR4 axis and has been effectively utilized in the treatment of OA (18–20). However, observed side effects and the unstable nature of AMD3100 limit its clinical application (20–22). TN14003 was designed based on T140, a 14-residue peptide that possesses a high level of anti-human immunodeficiency virus (HIV) activity and antagonism of T cell line-tropic HIV-1 entry among all the antagonists of CXCR4 (23). TN14003 was generated by amidating the COOH-terminal of T140 and by substituting basic residues with non-basic polar amino acids to decrease the total-positive charges of the molecule, and is far less cytotoxic and more stable in serum compared with T140 (23,24).
MicroRNA (miRNA) belong to a class of small noncoding RNA encoded by endogenous genes, and dysregulation of miRNAs results in numerous diseases that occur in various physiological and pathological processes (25,26). Accumulating studies investigating the roles of miRNAs in bone and cartilage have identified a number of miRNAs that serve important roles in the pathogenesis of OA (27–30). Therefore, the identification of abnormally expressed miRNAs and the associated biological consequences of their targets is essential to determining the potential molecular mechanisms in the OA pathological process. Unfortunately, the changes in miRNA expression in chondrocytes as a result of inhibiting the SDF-1/CXCR4 axis by drugs including TN14003 remains largely unclear.
Using a series of bioinformatic approaches, the present study aimed to systematically evaluate the aberrant miRNA expression levels in OA chondrocytes treated with TN14003. The key miRNA miR-146a-5p was also confirmed as a differentially expressed miRNA, and the expression levels of its targets involved in the process of SDF-1/CXCR4 axis inhibition were measured, following molecular manipulation of the expression of miR-146a-5p in chondrocytes.
Materials and methods
Cartilage tissue collection and cell cultivation
OA cartilage was obtained from the weight-bearing surface of the femoral condyle and tibial plateau of 4 female and 1 male patients diagnosed with OA (using the American College of Rheumatology classification criteria), aged between 57 and 69 years old with an average age of 63.4±2.42, and undergoing total knee arthroplasty between October 2017 to March 2018 in the Department of Sports Medicine of the First Affiliated Hospital of Kunming Medical University (Kunming, China) (31,32). Written informed consent was obtained from all patients, and the present study was approved by the Ethics Committee at the First Affiliated Hospital of Kunming Medical University (Kunming, China).
Chondrocytes were digested with 0.15% collagenase and cultured in high glucose Dulbecco's Modified Eagle Medium (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal bovine serum (Gibco; Thermo Fisher Scientific, Inc.) and 100 U/ml penicillin and 100 µg/ml streptomycin. Culture medium was filtered to remove bacteria using a 0.22 µm microfilter. The first generation chondrocytes were used and divided into two groups: Treatment and control. To mimic the osteoarthritic environment of the knee joint, each group was treated with 100 ng/ml SDF-1 (PeproTech, Rocky Hill, NJ, USA). The treatment group was pretreated with 1 µM TN14003 (Scilight Biotechnology, LLC, Beijing, China) for 2 h prior to the addition of SDF-1. Each group of chondrocytes was incubated at 37°C and 5% CO2 for 2 days.
miRNA extraction and reverse transcription
For miRNA screening, total RNA was isolated from cartilage tissues with or without TN14003 treatment, purified and prepared using the Qiagen RNeasy Mini kit (Qiagen, Hilden, Germany; cat. no. 74106) according to manufacturer's protocol. The integrity and quantity of samples were determined via Nanodrop 2000 spectrophotometer (Thermo Fisher Scientific, Inc.) and Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). miRNAs were isolated from total RNA using the All-in-One™ miRNA reverse transcription quantitative polymerase chain reaction (RT-qPCR) Detection kit (GeneCopoeia, Inc., Rockville, MD, USA), according to the manufacturer's protocol.
miRNA RT-qPCR and verification
To calibrate the initial results, differentially expressed miRNAs were identified using miProfile™ human inflammatory miRNA qPCR Arrays (GC08017K18014P; GeneCopoeia, Inc.). The chondrocyte samples were isolated from 5 patients with OA and the validation was performed using samples with or without TN14003 treatment. Each well contained a forward primer for the mature miRNA sequence, and a universal adaptor reverse primer cross-linked to the 96-well plate. The primers for measuring miRNA expression were designed as summarized in Table I, and the qPCR was performed using 20 µl reaction volumes containing 1 µl reverse transcription product and SYBR Green Master Mix (Applied Biosystems; Thermo Fisher Scientific, Inc.). The amplification conditions were as follows: Pre-incubation at 50°C for 2 min, enzyme activation at 95°C for 10 min, then 40 cycles of denaturation at 95°C for 10 sec, annealing at 55°C for 30 sec and extension at 72°C for 30 sec. Detection was performed using an ABI 7500 instrument (Applied Biosystems; Thermo Fisher Scientific, Inc.), and 2−ΔΔCq was used to calculate the relative expression of miRNAs, as previously described (33). Alterations with a fold-change >2 and P<0.05 were considered to be differentially expressed.
Table I.
Sequences of polymerase chain reaction primers used for the detection of miRNA expression.
| miRNA | Primer sequence, 5′-3′ | Annealing temperature, °C |
|---|---|---|
| miR-126-3p | Forward: TCGTACCGTGAGTAATAATGCG | 60 |
| Reverse: GCTGTCAACGATACGCTACGTAAC | ||
| miR-124-3p | Forward: TAAGGCACGCGGTGAATGCC | 60 |
| Reverse: GCTGTCAACGATACGCTACGTAAC | ||
| miR-130a-3p | Forward: CAGTGCAATGTTAAAAGGGCAT | 60 |
| Reverse: GCTGTCAACGATACGCTACGTAAC | ||
| miR-185-5p | Forward: TGGAGAGAAAGGCAGTTCCTGA | 60 |
| Reverse: GCTGTCAACGATACGCTACGTAAC | ||
| miR-146a-5p | Forward: TGAGAACTGAATTCCATGGGTT | 60 |
| Reverse: GCTGTCAACGATACGCTACGTAAC | ||
| miR-155-5p | Forward: TTAATGCTAATCGTGATAGGGGT | 60 |
| Reverse: GCTGTCAACGATACGCTACGTAAC | ||
| miR-221-3p | Forward: AGCTACATTGTCTGCTGGGTTTC | 60 |
| Reverse: GCTGTCAACGATACGCTACGTAAC | ||
| U6 | Forward: CTCGCTTCGGCAGCACA | 60 |
| Reverse: AACGCTTCACGAATTTGCGT | ||
| Oligo dT Adaptor | GCTGTCAACGATACGCTACGTAACGGCATG | |
| ACAGTGTTTTTTTTTTTTTTTTTV |
Bioinformatic analysis
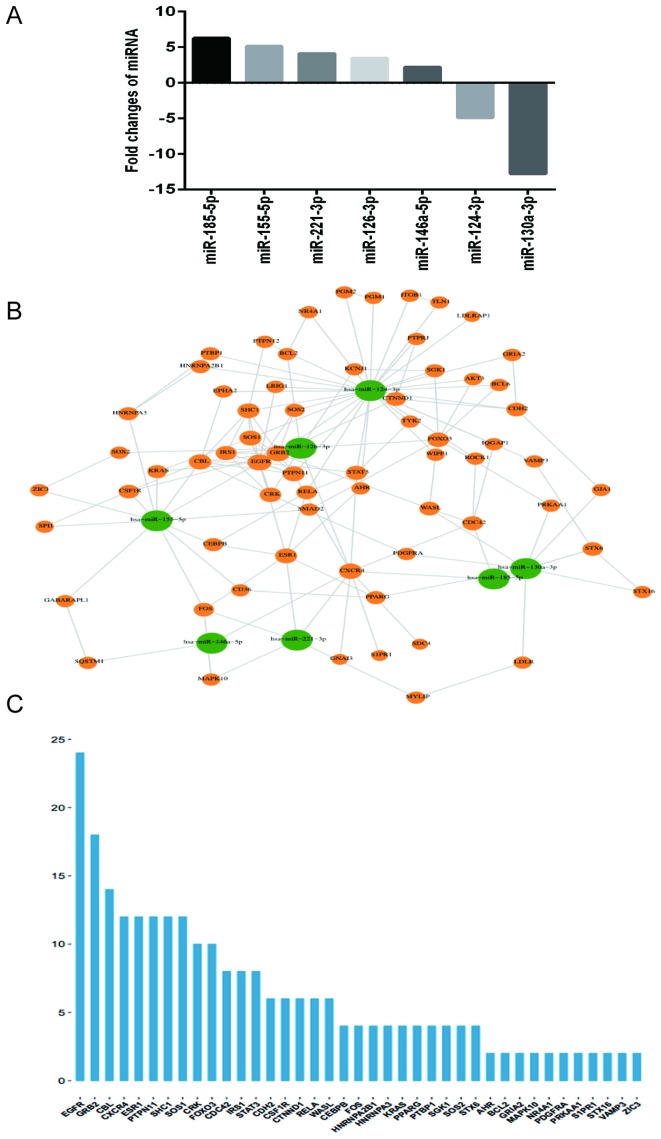
A total of 7 differentially expressed miRNAs (miR-146a-5p, miR-221-3p, miR-126-3p, miR-185-5p, miR-155-5p, miR-124-3p and miR-130a-3p) were identified (Fig. 1). The targets of these miRNAs were predicted using miRDB (34) (http://mirdb.org/miRDB/), miRTarBase (35) (http://mirtarbase.mbc.nctu.edu.tw/), miRWalk (36) (http://zmf.umm.uni-heidelberg.de/apps/zmf/mirwalk2/) and TargetScan (37) (http://www.targetscan.org/). Genes targeted by miRNAs were analyzed by Gene Ontology (GO) analysis (38) (http://geneontology.org/page/go-enrichment-analysis) and the Database for Annotation, Visualization and Integrated Discovery pathway analysis (39) (http://www.genome.jp/) to compile gene annotation terms and involved signaling pathways. Finally, miRNA-target networks were constructed to visually enrich the gene dataset. The enrichment result was visualized by Enrichment Map as a plugin of Cytoscape version 3.2.0 (40). The revised P-value was characterized by the false discovery rate (FDR) to avoid false positivity, and an FDR<0.05 was used as the cut-off value.
Figure 1.
Identification of candidate miRNAs whose alterations are in response to TN14003 treatment in SDF-1-stimulated chondrocytes. (A) Chondrocytes from patients with osteoarthritis were divided into two groups: Treatment and control. Each group was treated with 100 ng/ml SDF-1, and the treatment group was pretreated with 1 µM TN14003 for 2 h prior to the addition of SDF-1. At 2 days following SDF-1 stimulation, RNA samples isolated from these chondrocytes were subjected to miRNA microarray. The bar graph indicates the fold-changes (Treatment/Control) of miRNA levels in gene chip experiments. (B) The miRNA-target interaction network. The green nodes represent miRNAs with differential expression, while the orange nodes represent target genes of the differentially expressed miRNAs. (C) Sequence diagram of target importance. The longitudinal axis indicates the number of connected genes, while the horizontal axis indicates the individual target. miRNAs, microRNAs; SDF-1, stromal cell-derived factor 1; hsa, Homo sapiens.
Cell transfection
Chondrocytes were seeded in 6-well plates at 5×104 cells per well and incubated at 37°C for 24 h. Chondrocytes were separately transfected with miR-146a-5p mimics, miR-146a-5p inhibitors and negative controls (NC; GeneCopoeia, Inc.) at a 100 nM concentration for 48 h using Lipofectamine® 3000 (Invitrogen; Thermo Fisher Scientific, Inc, MA, USA), according to the protocol of the manufacturer. The sequences for miR-146a-5p mimics, miR-146a-5p inhibitors and the corresponding NC were as follows: miR-146a-5p mimic, 5′-UGAGAACUGAAUUCCAUGGGUU-3′ (sense) and 5′-CCCAUGGAAUUCAGUUCUCAUU-3′ (antisense); mimics NC, 5′-UUCUCCGAACGUGUCACGUTT-3′ (sense) and 5′-ACGUGACACGUUCGGGAATT-3′ (antisense); miR-146a-5p inhibitor, 5′-AACCCAUGGAAUUCAGUUCUCA-3′; inhibitor NC, 5′-CAGUACUUUUGUGUAGUACAA-3′. The chondrocytes was cultured for 48 h after transfection and then used for subsequent experiments.
RT-qPCR assay for mRNA expression
Total RNA was extracted from cartilage tissues using TRIzol® regent (Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. Total RNA (1 µg) was transcribed into cDNA using the RevertAid™ First Strand cDNA Synthesis kit (Thermo Fisher Scientific, Inc). cDNA (40 µg/µl) was used as a template for amplification of CXCR4, type II collagen (Col II), aggrecan (ACAN) and MMP-3 genes, and β-actin served as an internal reference. SYBR Green Master Mix (Applied Biosystems; Thermo Fisher Scientific, Inc.) was used for RT-qPCR analysis. The amplification conditions were as follows: Pre-incubation at 50°C for 2 min, enzyme activation at 95°C for 10 min, then 40 cycles of denaturation at 95°C for 10 sec, annealing at 55°C for 30 sec and extension at 72°C for 30 sec. All primers (Table II) were obtained from the NCBI database and designed by Premiers Express Software v1.0 (BioTools Incorporated, Edmonton, AB, Canada). Reactions for each sample were performed in at least three independent experiments. Cycle threshold values were measured and data were calculated the by 2−ΔΔCq method (33).
Table II.
Sequences of polymerase chain reaction primers used for detection mRNA expression.
| Target | Forward primer 5′-3′ | Reverse primer 5′-3′ |
|---|---|---|
| CXCR4 | TCAGTGGCTGACCTCCTCTT | CTTGGCCTTTGACTGTTGGT |
| Col II | ATGCACCTTGGATGCCATGA | ATGCACCTTGGATGCCATGA |
| ACAN | ACATCTCAGCAGCATCATCACC | CATCACCACGCAGTCCTCAC |
| MMP-3 | GGACAAAGGATACAACAGGGAC | TCATCTTGAGACAGGCGAAA |
| β-actin | CCACCATGTACCCAGGCATT | ACTCCTGCTTGCTGATCCAC |
CXCR4, C-X-C chemokine receptor type 4; Col II, type II collagen; ACAN, aggrecan; MMP-3, matrix metalloproteinase.
Western blot analysis
Total protein was extracted from cells following lysis with radioimmunoprecipitation assay buffer and quantified by the BCA Protein Assay kit (Thermo Fisher Scientific, Inc.). Total protein (30 µg) was separated via electrophoresis on 10% SDS-PAGE gels prior to transfer to polyvinylidene fluoride membranes. Membranes were probed with primary antibodies in TBS with 5% non-fat milk. The antibodies included were anti-CXCR4 (LifeSpan BioSciences, Inc., Seattle, WA, USA; 1:1,000 dilution; cat. no. LS-B2160-0.05), anti-Col II (Abcam, Cambridge, UK; 1:1,000 dilution; cat. no. ab188570) and anti-β-actin (Abcam, Cambridge, UK; 1:5,000 dilution; cat. no. ab8227), and horseradish peroxidase-conjugated goat anti-rabbit immunoglobulin G (Abcam, Cambridge, UK; 1:10,000 dilution; cat. no. ab97051) was used as the secondary antibody. Proteins of interest were visualized using enhanced chemiluminescent reagent (Thermo Fisher Scientific, Inc.). The band intensities were quantified by densitometry using ImageJ 1.46r software (National Institutes of Health, Bethesda, MD, USA) (41). Experiments were repeated at least 3 times.
Statistical analysis
All quantitative data were analyzed with SPSS 18.0 (SPSS, Inc. Chicago, IL, USA) and presented as mean ± standard deviation. Statistical analysis was performed using one-way analysis of variance with the Least-Significant Difference correction to determine differences between groups. P<0.05 were considered to indicate a statistically significant difference.
Results
Identification of candidate miRNAs whose alterations are in response to TN14003 treatment in SDF-1-stimulated chondrocytes
To evaluate the effects of TN14003 on chondrocytes, OA patients-derived chondrocytes were treated with SDF-1 alone or SDF-1 + TN14003 for 48 h. Cells were harvested to investigate the alteration of miRNA profile upon the inhibition of CXCR4/SDF-1 axis by TN14003. There were 7 differentially expressed miRNAs identified in cartilage samples via microarray analysis (Table III). Among these miRNAs, 5 miRNAs (miR-146a-5p, miR-221-3p, miR-126-3p, miR-185-5p and miR-155-5p) were significantly upregulated, and 2 miRNAs (miR-124-3p and miR-130a-3p) were significantly downregulated in the treatment group compared with the control group (Fig. 1A). In order to understand the mechanism of these miRNA alterations and consequently their involvement in OA treatments, the miRWalk database and Cytoscape software was used to analyze the miRNA-mRNA interactions through their visualization as a network (Fig. 1B). While the interactions between miRNAs and potential targets were built, a sequence diagram was performed to reveal the importance of targets. The importance of targets was determined according to the number of connections between each gene and miRNA and other genes in the miRNA-target network (Fig. 1C). As demonstrated in the network map, the miRNAs hsa-miR-146a-5p, hsa-miR-221-3p, hsa-miR-126-3p and hsa-miR-185-5p exhibited direct interaction with the mRNA CXCR4 (Fig. 1B). Among all the potential targets, EGFR, GRB2, CBL, CXCR4, ESR1, PTPN11, SHC1 and SOS1 were the targeted mRNAs with highest importance scores (Fig. 1C).
Table III.
Expression levels of 7 targeted miRNAs in cartilage samples with and without TN14003 treatment via microarray analysis.
| miRNA name | Fold change Treatment group/control group | P-value |
|---|---|---|
| miR-146a-5p | 2.12 | 0.00043 |
| miR-221-3p | 4.41 | 0.00024 |
| miR-126-3p | 3.49 | 0.00007 |
| miR-185-5p | 6.29 | 0.00003 |
| miR-155-5p | 5.15 | 0.00019 |
| miR-124-3p | −4.91 | 0.00017 |
| miR-130a-3p | −12.78 | 0.00022 |
miRNA, microRNA.
Verification of candidate miRNAs, GO terms assignment and pathways analysis of miR-146a-5p and its targets
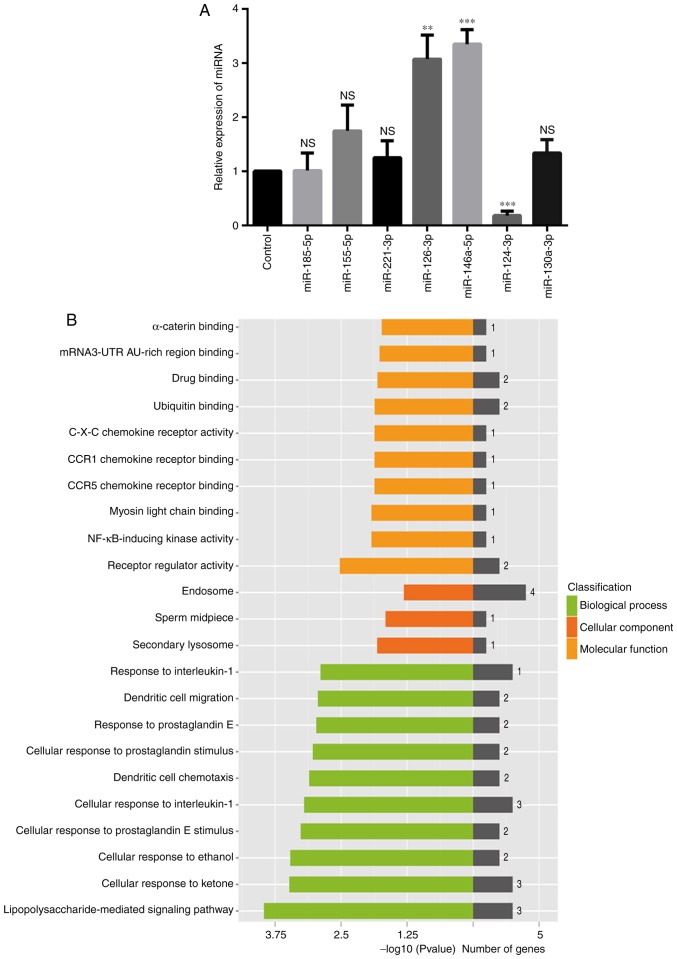
To validate the differentiated expression of the 7 miRNAs identified from the initial screening, the levels of these miRNAs in control-treated and TN14003-treated chondrocytes were measured by RT-qPCR assays. Compared with the initial microarray results, the expression levels of miR-146a-5p, miR-126-3p and miR-124-3p were validated by the RT-qPCR approach: As indicated in Fig. 1A, the expression of both miR-146a-5p and miR-126-3p was upregulated, while the expression of miR-124-3p was downregulated; the results of RT-qPCR analysis presented in Fig. 2A confirmed this observation. However, other miRNAs that did not exhibit the same changes, or exhibited no statistical differences in expression were excluded from subsequent analyses (Fig. 2A). Notably, miR-146a-5p was upregulated >3-fold. A previous study suggested an association between miR-146a-5p and OA, as evidenced by its >2-fold increased expression compared with healthy controls (42). Besides, accumulating data indicate that miR-146a-5p is a representative miRNA known to be associated with OA (43,44). Therefore, the present study focused on miR-146a-5p and its targets to delineate their associations with OA treatments by TN14003.
Figure 2.
Verification of candidate miRNAs, GO terms assignment of miR-146a-5p and its targets. (A) The 7 differentially expressed miRNAs were validated by reverse transcription quantitative polymerase chain reaction. Chondrocytes samples were obtained from 5 patients, and the experiments were repeated 3 times with similar results. **P<0.01 and ***P<0.001 vs. the control group. (B) Histogram of enriched GO terms of miR-146a-5p and its targets. The longitudinal axis represents the GO category, while the horizontal axis represents the-log 10 (P-value) of the GO category and number of genes. miRNA, microRNA; GO, Gene Ontology; NS, not significant.
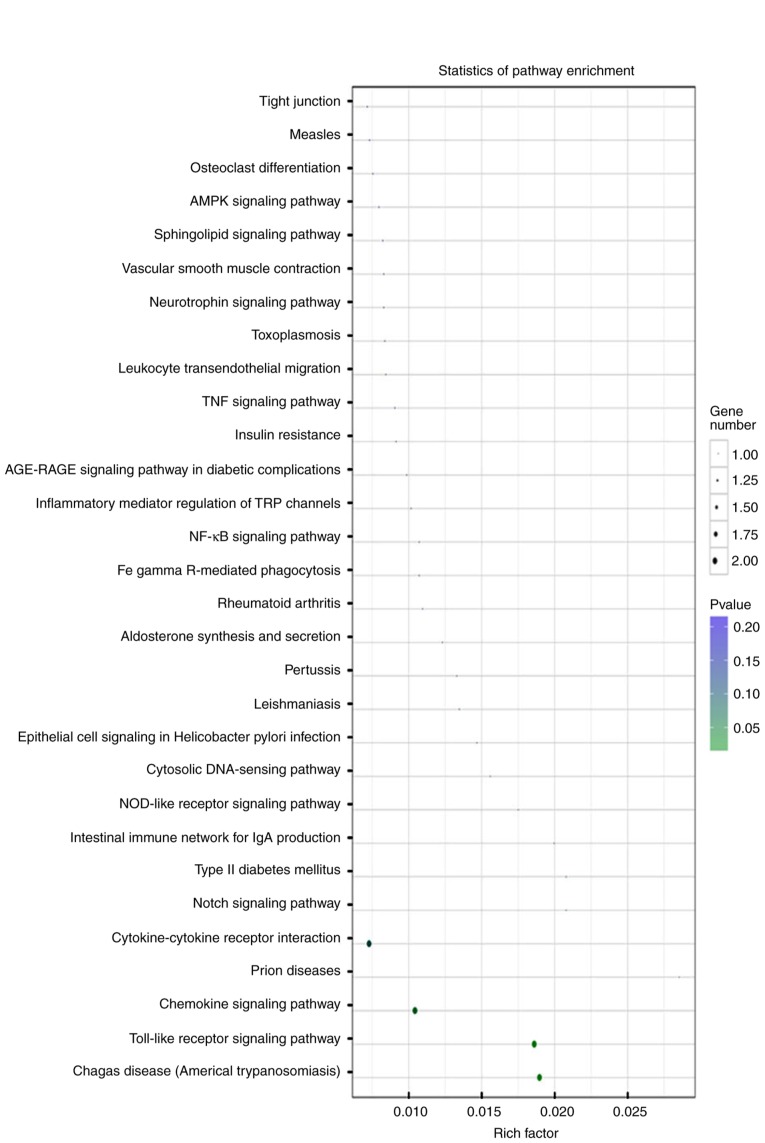
GO analysis indicated that miR-146a-5p and its targets were primarily grouped into ‘receptor regulator activity’ or ‘nuclear factor-kappa-light-chain-enhancer of activated B cells (NF-κB)-inducing kinase (NIK) activity’ in the Molecular Functions category, into ‘lipopolysaccharide-mediated signaling pathways’ or ‘cellular response to ketones’ in the Biological Processes category, and into ‘secondary lysosome’, ‘sperm midpiece’ or ‘endosomes’ in the Cellular Components category (Fig. 2B). Enriched pathways of miR-146a-5p and its targets were primarily involved in the ‘chemokine signaling pathway’ or the ‘Toll-like receptor signaling pathway’ (Fig. 3). These results indicated that these identified inflammation-associated GO terms and pathways are important to the roles of the SDF-1/CXCR4 axis in the pathogenesis of OA.
Figure 3.
Signaling pathway analysis involved in miR-146a-5p and its targets. The longitudinal axis represents the pathway category, while the horizontal axis represents the enrichment factor in each pathway. The size of a point represents the number of genes, and the color is indicative of the P-value.
Molecular manipulation of miR-146a-5p expression in chondrocytes transfected with the mimic or inhibitor of miR-146a-5p
To additionally reveal the roles of miR-146a-5p in the process of OA development, the mimic and inhibitor of miR-146a-5p were designed to examine the effects of molecular manipulation of miR-146a-5p on OA-associated molecules. First, an RT-qPCR assay was performed to verify the effect of cell transfection and successful upregulation and downregulation of miR-146a-5p expression. As demonstrated in Fig. 4, the level of miR-146a-5p was significantly increased ~12-fold compared with the mimic control in chondrocytes transfected with the miR-146a-5p mimic, and markedly downregulated to ~20% of the inhibitor control when the miR-146a-5p inhibitor was transfected. As hypothesized, there were no significant changes in miR-146a-5p expression between chondrocytes that were treated with SDF-1 only and transfected with negative controls (Fig. 4).
Figure 4.
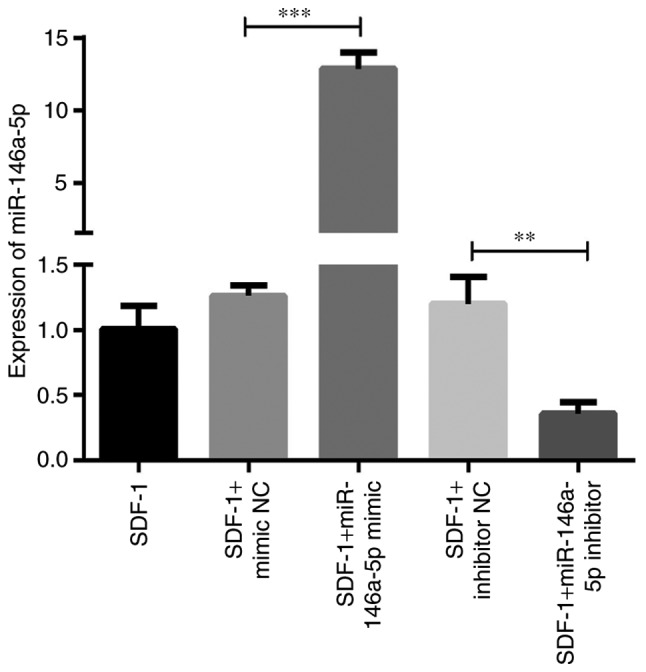
Successful manipulation of miR-146a-5p expression in chondrocytes transfected with the mimic or inhibitor of miR-146a-5p. Chondrocytes were separately transfected with miR-146a-5p mimics, miR-146a-5p inhibitors and NC for 48 h using Lipofectamine® 3000. Following stimulation with 100 ng/ml SDF-1 for 2 days, cells were harvested to evaluate the levels of miR-146a-5p by reverse transcription quantitative polymerase chain reaction. Data are summarized from 3 independent experiments with similar results. n=3 for each group. **P<0.01 and ***P<0.001, compared with the SDF-1 treated alone group. miRNA, microRNA; SDF-1, stromal cell-derived factor 1; NC, negative control.
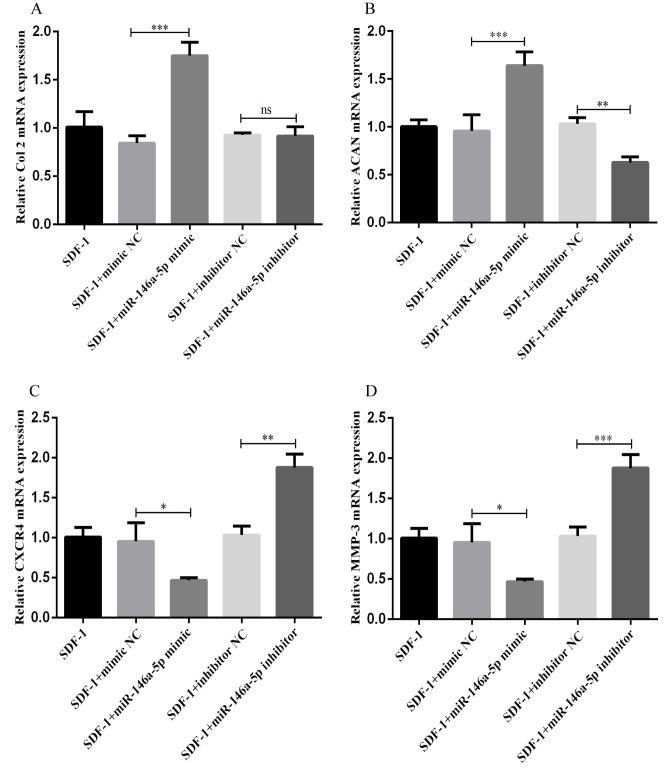
Association between miR-146a-5p expression and the mRNA levels of Col II, ACAN, CXCR4 and MMP-3
Next, RT-qPCR was performed to measure the expression of cartilage degeneration-associated factors (including Col II, ACAN, CXCR4 and MMP-3) following transfection of the chondrocytes with the mimic and inhibitor of miR-146a-5p, and corresponding NCs. As indicated in Fig. 5A and B, in the chondrocytes transfected with the miR-146a-5p mimic, the expression levels of Col II and ACAN were significantly increased. However, there was no significant difference in Col II expression between chondrocytes transfected with the inhibitor NC and the miR-146a-5p inhibitor. The expression levels of ACAN decreased markedly in the chondrocytes transfected with the miR-146a-5p inhibitor. In contrast, as indicated in Fig. 5C and D, the mRNA expression levels of CXCR4 and MMP-3 decreased in the chondrocytes following transfection with the miR-146a-5p mimic, and increased following transfection with the miR-146a-5p inhibitor. These results demonstrate a positive association between miR-146a-5p expression and the mRNA expression levels of Col II and ACAN, and a negative association between miR-146a-5p expression and the mRNA expression of CXCR4 and MMP-3.
Figure 5.
Association between miR-146a-5p expression and the mRNA levels of Col II, ACAN, CXCR4 and MMP-3. (A-D) Chondrocytes were separately transfected with miR-146a-5p mimics, miR-146a-5p inhibitors and NC for 48 h using Lipofectamine® 3000. Following stimulation with 100 ng/ml SDF-1 for 2 days, cells were harvested to evaluate the levels of (A) Col II, (B) ACAN, (C) CXCR4 and (D) MMP-3 by reverse transcription quantitative polymerase chain reaction. Data are summarized from 3 independent experiments with similar results. n=3 for each group. *P<0.05, **P<0.01 and ***P<0.001 vs. the corresponding control group. miRNA, miRNA; Col II, type II collagen; ACAN, aggrecan; CXCR4, C-X-C chemokine receptor type 4; MMP-3, matrix metalloproteinase-3; SDF-1, stromal cell-derived factor 1; NC, negative control; NS, not significant.
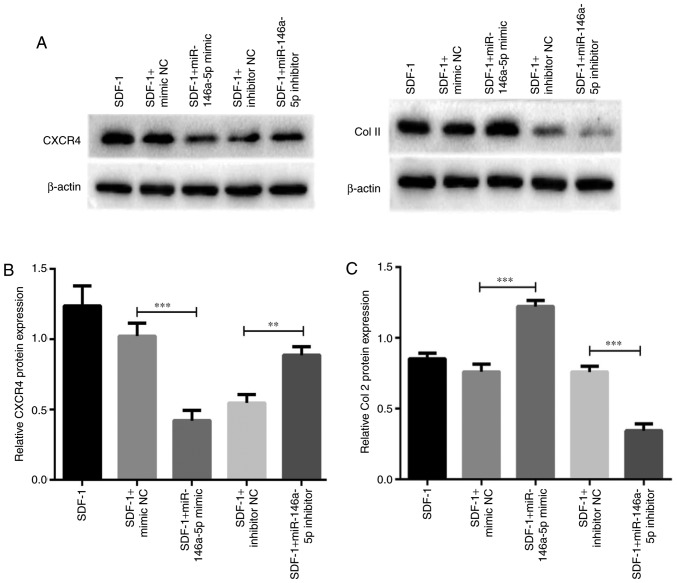
Association between miR-146a-5p expression and the protein levels of CXCR4 and Col II
As CXCR4 was confirmed to be a target of miR-146a-5p, and Col II is the principal component of cartilage ECM, effects of miR-146a-5p expression on the protein levels of these 2 factors were additionally assessed. Consistent with the alteration of mRNA expression, expression of the CXCR4 protein was downregulated as a result of miR-146a-5p mimic transfection (Fig. 6A and B). Concomitantly, the protein level of CXCR4 was significantly upregulated following miR-146a-5p inhibitor transfection. As hypothesized, the opposite pattern of expression was observed for Col II protein in chondrocytes transfected with the mimic or inhibitor of miR-146a-5p (Fig. 6A and C). Taken together, these results highlight the critical roles of miR-146a-5p on regulating the expression of cartilage degeneration-associated factors.
Figure 6.
Association between miR-146a-5p expression and the protein levels of CXCR4 and Col II. Chondrocytes were separately transfected with miR-146a-5p mimics, miR-146a-5p inhibitors and NC for 48 h using Lipofectamine® 3000. Following stimulation with 100 ng/ml SDF-1 for 2 days, cells were harvested to evaluate the protein levels of CXCR4 and Col II. (A) Represent western blot showing the protein levels of CXCR4 and Col II. (B and C) Summary data from 3 independent western blot analysis experiments on the protein levels of (B) CXCR4 and (C) Col II. **P<0.01 and ***P<0.001 vs. the corresponding control group. miRNA, miRNA; Col II, type II collagen; CXCR4, C-X-C chemokine receptor type 4; SDF-1, stromal cell-derived factor 1; NC, negative control.
Discussion
Antagonists of CXCR4 (AMD3100, T140 and TN14003) have been demonstrated to successfully inhibit the SDF-1/CXCR4 axis by competing with CXCR4 for its ligand SDF-1 (45). These antagonists have been utilized in the treatment of HIV infection and various types of cancer (46,47). AMD3100 and T140 were indicated to be efficient in the management of OA in previous studies (48,49). However, TN14003 is recommended above AMD3100 and T140, as AMD3100 is a weak partial antagonist, and T140 possesses unstable properties, limiting their clinical applications (50,51). In addition, previous studies have demonstrated that TN14003 is a more effective in inhibiting MMP-3, MMP-9 and MMP-13 release, and in Col-II and ACAN degradation, when compared to AMD3100 and T140 (52,53). The present study investigated whether TN14003 therapy may elicit an alteration in miRNA profile in chondrocytes derived from patients with OA, and also identified that miR-146a-5p was a CXCR4/SDF-1 axis inhibitor that induced differentially expressed miRNA, which regulated the expression of cartilage degeneration-associated factors, including CXCR4, Col II, ACAN and MMP-3.
It has been established that miRNAs serve key roles in the pathological processes of OA (54,55). Kopańska et al (42) identified 4 miRNAs (miR-138-5p, miR-146a-5p, miR-335-5p and miR-9-5p) in OA cartilage that were upregulated >2-fold compared with healthy controls, indicating an association between miRNA and OA. Zheng et al (56) demonstrated that miR-221-3p was significantly downregulated in OA compared with normal controls, and that upregulating miR-221-3p may inhibit interleukin 1β (IL-1β)-induced cartilage degradation via targeting of the SDF-1/CXCR4 axis. The present study indicated that 84 miRNAs were differentially expressed in OA chondrocytes, and miR-146a-5p, miR-126-3p and miR-124-3p were validated, suggesting that these miRNAs may exert their effects via inhibition of SDF-1/CXCR4 with TN14003 treatment.
miR-146a-5p is a representative miRNA known to be associated with OA (43,44). In addition to the data from Kopańska et al (42), Genemaras et al (57) suggested that following stimulation with IL-1β and tumor necrosis factor-α (TNF-α), miR-146a was significantly upregulated in pig chondrocytes, indicating an interaction between miR-146a and inflammatory cytokines in the promotion of OA. In addition, Spinello et al (58) detected a parallel effect between miR-146a and the CXCR4 antagonist. The present study determined that CXCR4 protein expression was decreased following AMD3100 treatment. The sensitivity of leukemic blast cells to cytotoxic drugs was demonstrated to be increased, and this effect was augmented with the overexpression of miR-146a. However, unlike miR-146-5p, which has been extensively studied, few studies have explored the role of miR-126-3p and miR-124-3p in the process of OA.
OA is an aseptic inflammatory disease (59,60). Several miRNAs, including miR-146a-5p, have been demonstrated to be genetic markers of inflammation, and to function as promoters of OA (61,62). Notably, miR-146a-5p was upregulated in the treatment group in the present study, indicating that it may serve a parallel role with TN14003. Although a number of studies have investigated the role of miR-146a-5p by comparing miRNA profiles between OA and normal chondrocytes, few studies have focused on miRNA expression profile following therapy with specific inhibitors, including CXCR4 antagonists. Through a computational approach to mine miR-146a-5p associated genes and pathways, the present study revealed that the ‘receptor regulatory activity’ or ‘NIF activity’ (Molecular Functions), ‘cellular response to interleukin-1’ (Biological Processes), ‘cytokine-cytokine receptor interaction’, ‘NF-κB signaling pathway’ and ‘osteoclast differentiation pathways’ were involved. Activation of the SDF-1/CXCR4 signaling axis has been verified to be a process of cytokine-to-receptor transmembrane transport, and this activity may regulate disease progress via the NF-κB pathway (63). This indicated that miR-146a-5p may be associated with the SDF-1/CXCR4 axis through the regulation of the NF-κB pathway.
Numerous genes are negatively regulated by complementary pairing with miRNAs, and dysregulation of genes may affect OA (64). Additionally, OA therapy based on miRNAs has been developed in previous years, and may result in high-efficiency treatment with less biological toxicity (65). Yang et al (61) predicted that CXCR4 may function as a direct target of miR-146a-5p, as verified by the fact that CXCR4 expression was decreased and miR-146a-5p was upregulated in endometrial tissue samples. In addition, Labbaye et al (51) determined that two ‘seed’ regions of the 3′-untranslated region in CXCR4 mRNA directly interacted with miR-146a, thereby demonstrating that CXCR4 mRNA translation was inhibited by miR-146a. In the present study, CXCR4 was predicted to be a target of miR-146a-5p with high importance. Then, RT-qPCR and western blot analysis were used to determine whether several key factors in chondrocytes associated with the SDF-1/CXCR4 axis were regulated by miR-146a-5p. It was identified that the expression levels of Col II and ACAN were positively associated with miR-146a-5p expression, and levels of CXCR4 and MMP-3 were negatively associated with miR-146a-5p expression. The results suggest that miR-146a-5p may serve a parallel and additive role with TN14003 in blocking the SDF-1/CXCR4 axis and inhibiting cartilage degeneration.
There are certain recognized limitations of the present study that must be considered. The effect of miR-146a-5p on cartilage degeneration was determined in the present study, but a Cell Counting Kit-8 assay should be performed to evaluate the effect of miR-146a-5p on chondrocyte survival. In addition, the primary aim of the present study to investigate chondrocytes, and may not capture the role of miR-146a-5p on the neighboring tissues containing synovium and subchondral bone. Finally, in order to fully demonstrate the function of miR-146a-5p in SDF-1-induced cartilage degeneration by targeting CXCR4, an in vivo investigation should be included in future studies.
In conclusion, the present study provided compelling evidence for the critical roles of miRNAs in SDF-1-induced cartilage degradation by miRNA microarray profiling in OA chondrocytes following TN14003 treatment. miR-146a-5p was detected to be differentially expressed and it most likely represents the key miRNA that participates in the regulation of the SDF-1/CXCR4 axis through the inhibition of CXCR4. Although additional work involving the biocompatibility of miR-146a-5p mimics in vitro and in vivo may be required to fully delineate its roles in OA pathogenesis, the present study offers a promising framework through which diagnostic and therapeutic biomarkers of OA may be determined. The combined use of TN14003 and miR-146a-5p mimics may represent an approach for developing effective OA-targeted therapies with decreased side effects.
Acknowledgements
Not applicable.
Glossary
Abbreviations
- OA
osteoarthritis
- SDF-1
stromal cell-derived factor 1
- CXCR4
C-X-C chemokine receptor type 4
- RT-qPCR
reverse transcription quantitative polymerase chain reaction
- GO
gene ontology
- NF-κB
nuclear factor-κ-light-chain-enhancer of activated B cells
- ECM
extracellular matrix
- MMPs
matrix metalloproteinase
- mRNA
messenger RNA
- IL-1β
interleukin 1β
- TNF-α
tumor necrosis factor α
- Col II
collagen type II
- ACAN
aggrecan
- MMP-3
matrix metalloproteinases 3
Funding
The present study was supported by the National Natural Science Foundation of China (grant nos., 81460340 and 81760403).
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
YLL designed the study. DJ performed the research and wrote the paper. RH and KW analyzed data. GFC and CH collated the data and checked the results. LJY helped perform the research. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Written informed consent was obtained from all patients and the present study was approved by the Ethics Committee at the First Affiliated Hospital of Kunming Medical University.
Patient consent for publication
Written informed consent was obtained from all patients.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Loeser RF, Goldring SR, Scanzello CR, Goldring MB. Osteoarthritis: A disease of the joint as an organ. Arthritis Rheum. 2012;64:1697–1707. doi: 10.1002/art.34453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li G, Yin J, Gao J, Cheng TS, Pavlos NJ, Zhang C, Zheng MH. Subchondral bone in osteoarthritis: insight into risk factors and microstructural changes. Arthritis Res Ther. 2013;15:223. doi: 10.1186/ar4405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Man GS, Mologhianu G. Osteoarthritis pathogenesis-a complex process that involves the entire joint. J Med Life. 2014;7:37–41. [PMC free article] [PubMed] [Google Scholar]
- 4.Gao Y, Liu S, Huang J, Guo W, Chen J, Zhang L, Zhao B, Peng J, Wang A, Wang Y, et al. The ECM-cell interaction of cartilage extracellular matrix on chondrocytes. Biomed Res Int. 2014;2014:648459. doi: 10.1155/2014/648459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maldonado M, Nam J. The role of changes in extracellular matrix of cartilage in the presence of inflammation on the pathology of osteoarthritis. Biomed Res Int. 2013;2013:284873. doi: 10.1155/2013/284873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haslauer CM, Elsaid KA, Fleming BC, Proffen BL, Johnson VM, Murray MM. Loss of extracellular matrix from articular cartilage is mediated by the synovium and ligament after anterior cruciate ligament injury. Osteoarthritis Cartilage. 2013;21:1950–1957. doi: 10.1016/j.joca.2013.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sophia Fox AJ, Bedi A, Rodeo SA. The basic science of articular cartilage: Structure, composition, and function. Sports Health. 2009;1:461–468. doi: 10.1177/1941738109350438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aigner T, Soeder S, Haag J. IL-1beta and BMPs-interactive players of cartilage matrix degradation and regeneration. Eur Cell Mater. 2006;12:49–56. doi: 10.22203/eCM.v012a06. [DOI] [PubMed] [Google Scholar]
- 9.Mariani E, Pulsatelli L, Facchini A. Signaling pathways in cartilage repair. Int J Mol Sci. 2014;15:8667–8698. doi: 10.3390/ijms15058667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fosang AJ, Beier F. Emerging frontiers in cartilage and chondrocyte biology. Best Pract Res Clin Rheumatol. 2011;25:751–766. doi: 10.1016/j.berh.2011.11.010. [DOI] [PubMed] [Google Scholar]
- 11.Goldring MB, Marcu KB. Epigenomic and microRNA-mediated regulation in cartilage development, homeostasis, and osteoarthritis. Trends Mol Med. 2012;18:109–118. doi: 10.1016/j.molmed.2011.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hart LE. In knee OA, intraarticular triamcinolone increased cartilage loss and did not differ from saline for knee pain. Ann Intern Med. 2017;167:JC27. doi: 10.7326/ACPJC-2017-167-6-027. [DOI] [PubMed] [Google Scholar]
- 13.McAlindon TE, LaValley MP, Harvey WF, Price LL, Driban JB, Zhang M, Ward RJ. Effect of intra-articular triamcinolone vs saline on knee cartilage volume and pain in patients with knee osteoarthritis: A randomized clinical trial. JAMA. 2017;317:1967–1975. doi: 10.1001/jama.2017.5283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thomas NP, Wu WJ, Fleming BC, Wei F, Chen Q, Wei L. Synovial inflammation plays a greater role in post-traumatic osteoarthritis compared to idiopathic osteoarthritis in the Hartley guinea pig knee. BMC Musculoskelet Disord. 2017;18:556. doi: 10.1186/s12891-017-1913-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Scanzello CR. Chemokines and inflammation in osteoarthritis: Insights from patients and animal models. J Orthop Res. 2017;35:735–739. doi: 10.1002/jor.23471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yellowley C. CXCL12/CXCR4 signaling and other recruitment and homing pathways in fracture repair. Bonekey Rep. 2013;2:300. doi: 10.1038/bonekey.2013.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kanbe K, Takagishi K, Chen Q. Stimulation of matrix metalloprotease 3 release from human chondrocytes by the interaction of stromal cell-derived factor 1 and CXC chemokine receptor 4. Arthritis Rheum. 2002;46:130–137. doi: 10.1002/1529-0131(200201)46:1<130::AID-ART10020>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 18.Thomas NP, Li P, Fleming BC, Chen Q, Wei X, Xiao-Hua P, Li G, Wei L. Attenuation of cartilage pathogenesis in post-traumatic osteoarthritis (PTOA) in mice by blocking the stromal derived factor 1 receptor (CXCR4) with the specific inhibitor, AMD3100. J Orthop Res. 2015;33:1071–1078. doi: 10.1002/jor.22862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang XY, Chen Y, Tang XJ, Jiang LH, Ji P. AMD3100 attenuates matrix metalloprotease-3 and −9 expressions and prevents cartilage degradation in a monosodium iodo-acetate-induced rat model of temporomandibular osteoarthritis. J Oral Maxillofac Surg. 2016;74:927.e1–927.e13. doi: 10.1016/j.joms.2015.12.018. [DOI] [PubMed] [Google Scholar]
- 20.Hendrix CW, Flexner C, MacFarland RT, Giandomenico C, Fuchs EJ, Redpath E, Bridger G, Henson GW. Pharmacokinetics and safety of AMD-3100, a novel antagonist of the CXCR-4 chemokine receptor, in human volunteers. Antimicrob Agents Chemother. 2000;44:1667–1673. doi: 10.1128/AAC.44.6.1667-1673.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stamatopoulos B, Meuleman N, De Bruyn C, Pieters K, Mineur P, Le Roy C, Saint-Georges S, Varin-Blank N, Cymbalista F, Bron D, Lagneaux L. AMD3100 disrupts the cross-talk between chronic lymphocytic leukemia cells and a mesenchymal stromal or nurse-like cell-based microenvironment: Pre-clinical evidence for its association with chronic lymphocytic leukemia treatments. Haematologica. 2012;97:608–615. doi: 10.3324/haematol.2011.052779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liles WC, Broxmeyer HE, Rodger E, Wood B, Hübel K, Cooper S, Hangoc G, Bridger GJ, Henson GW, Calandra G, Dale DC. Mobilization of hematopoietic progenitor cells in healthy volunteers by AMD3100, a CXCR4 antagonist. Blood. 2003;102:2728–2730. doi: 10.1182/blood-2003-02-0663. [DOI] [PubMed] [Google Scholar]
- 23.Tamamura H, Omagari A, Hiramatsu K, Gotoh K, Kanamoto T, Xu Y, Kodama E, Matsuoka M, Hattori T, Yamamoto N, et al. Development of specific CXCR4 inhibitors possessing high selectivity indexes as well as complete stability in serum based on an anti-HIV peptide T140. Bioorg Med Chem Lett. 2001;11:1897–1902. doi: 10.1016/S0960-894X(01)00323-7. [DOI] [PubMed] [Google Scholar]
- 24.Tamamura H, Fujisawa M, Hiramatsu K, Mizumoto M, Nakashima H, Yamamoto N, Otaka A, Fujii N. Identification of a CXCR4 antagonist, a T140 analog, as an anti-rheumatoid arthritis agent. FEBS Lett. 2004;569:99–104. doi: 10.1016/j.febslet.2004.05.056. [DOI] [PubMed] [Google Scholar]
- 25.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 26.Hernando E. microRNAs and cancer: Role in tumorigenesis, patient classification and therapy. Clin Transl Oncol. 2007;9:155–160. doi: 10.1007/s12094-007-0029-0. [DOI] [PubMed] [Google Scholar]
- 27.Sondag GR, Haqqi TM. The role of microRNAs and their targets in osteoarthritis. Curr Rheumatol Rep. 2016;18:56. doi: 10.1007/s11926-016-0604-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu XM, Meng HY, Yuan XL, Wang Y, Guo QY, Peng J, Wang AY, Lu SB. MicroRNAs' involvement in osteoarthritis and the prospects for treatments. Evid Based Complement Alternat Med. 2015;2015:236179. doi: 10.1155/2015/236179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jingsheng S, Yibing W, Jun X, Siqun W, Jianguo W, Feiyan C, Gangyong H, Jie C. MicroRNAs are potential prognostic and therapeutic targets in diabetic osteoarthritis. J Bone Miner Metab. 2015;33:1–8. doi: 10.1007/s00774-014-0628-0. [DOI] [PubMed] [Google Scholar]
- 30.Wu C, Tian B, Qu X, Liu F, Tang T, Qin A, Zhu Z, Dai K. MicroRNAs play a role in chondrogenesis and osteoarthritis (review) Int J Mol Med. 2014;34:13–23. doi: 10.3892/ijmm.2014.1743. [DOI] [PubMed] [Google Scholar]
- 31.Castell MV, van der Pas S, Otero A, Siviero P, Dennison E, Denkinger M, Pedersen N, Sanchez-Martinez M, Queipo R, van Schoor N, et al. Osteoarthritis and frailty in elderly individuals across six European countries: Results from the European Project on OSteoArthritis (EPOSA) BMC Musculoskelet Disord. 2015;16:359. doi: 10.1186/s12891-015-0807-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Damen J, van Rijn RM, Emans PJ, Hilberdink WKHA, Wesseling J, Oei EHG, Bierma-Zeinstra SMA. Prevalence and development of hip and knee osteoarthritis according to American College of Rheumatology criteria in the CHECK cohort. Arthritis Res Ther. 2019;1:4. doi: 10.1186/s13075-018-1785-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 34.Wong N, Wang X. miRDB: An online resource for microRNA target prediction and functional annotations. Nucleic Acids Res. 2015;43:D146–D152. doi: 10.1093/nar/gku1104. Database Issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chou CH, Shrestha S, Yang CD, Chang NW, Lin YL, Liao KW, Huang WC, Sun TH, Tu SJ, Lee WH, et al. miRTarBase update 2018: A resource for experimentally validated microRNA-target interactions. Nucleic Acids Res. 2018;46(D1):D296–D302. doi: 10.1093/nar/gkx1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dweep H, Sticht C, Pandey P, Gretz N. miRWalk-database: Prediction of possible miRNA binding sites by ‘walking’ the genes of three genomes. J Biomed Inform. 2011;44:839–847. doi: 10.1016/j.jbi.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 37.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 38.Gene Ontology Consortium. Gene ontology consortium: Going forward. Nucleic Acids Res. 2015;43:D1049–D1056. doi: 10.1093/nar/gku1179. Database Issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 40.Isserlin R, Merico D, Voisin V, Bader GD. Enrichment Map-a Cytoscape app to visualize and explore OMICs pathway enrichment results. F1000Res. 2014;3:141. doi: 10.12688/f1000research.4536.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kopańska M, Szala D, Czech J, Gabło N, Gargasz K, Trzeciak M, Zawlik I, Snela S. MiRNA expression in the cartilage of patients with osteoarthritis. J Orthop Surg Res. 2017;12:51. doi: 10.1186/s13018-017-0577-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yamasaki K, Nakasa T, Miyaki S, Ishikawa M, Deie M, Adachi N, Yasunaga Y, Asahara H, Ochi M. Expression of MicroRNA-146a in osteoarthritis cartilage. Arthritis Rheum. 2009;60:1035–1041. doi: 10.1002/art.24404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhong JH, Li J, Liu CF, Liu N, Bian RX, Zhao SM, Yan SY, Zhang YB. Effects of microRNA-146a on the proliferation and apoptosis of human osteoarthritis chondrocytes by targeting TRAF6 through the NF-κB signalling pathway. Biosci Rep. 2017;37(pii):BSR20160578. doi: 10.1042/BSR20160578. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 45.Burger JA, Stewart DJ, Wald O, Peled A. Potential of CXCR4 antagonists for the treatment of metastatic lung cancer. Expert Rev Anticancer Ther. 2011;4:621–30. doi: 10.1586/era.11.11. [DOI] [PubMed] [Google Scholar]
- 46.Tamamura H, Omagari A, Hiramatsu K, Kanamoto T, Gotoh K, Kanbara K, Yamamoto N, Nakashima H, Otaka A, Fujii N. Synthesis and evaluation of bifunctional anti-HIV agents based on specific CXCR4 antagonists-AZT conjugation. Bioorg Med Chem. 2001;9:2179–2187. doi: 10.1016/S0968-0896(01)00128-6. [DOI] [PubMed] [Google Scholar]
- 47.Green MM, Chao N, Chhabra S, Corbet K, Gasparetto C, Horwitz A, Li Z, Venkata JK, Long G, Mims A, et al. Plerixafor (a CXCR4 antagonist) following myeloablative allogeneic hematopoietic stem cell transplantation enhances hematopoietic recovery. J Hematol Oncol. 2016;9:71. doi: 10.1186/s13045-016-0301-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li P, Deng J, Wei X, Jayasuriya CT, Zhou J, Chen Q, Zhang J, Wei L, Wei F. Blockade of hypoxia-induced CXCR4 with AMD3100 inhibits production of OA-associated catabolic mediators IL-1β and MMP-13. Mol Med Rep. 2016;2:1475–1482. doi: 10.3892/mmr.2016.5419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang K, Li Y, Han R, Cai G, He C, Wang G, Jia D. T140 blocks the SDF-1/CXCR4 signaling pathway and prevents cartilage degeneration in an osteoarthritis disease model. PLoS One. 2017;12:e0176048. doi: 10.1371/journal.pone.0176048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang WB, Navenot JM, Haribabu B, Tamamura H, Hiramatu K, Omagari A, Pei G, Manfredi JP, Fujii N, Broach JR, Peiper SC. A point mutation that confers constitutive activity to CXCR4 reveals that T140 is an inverse agonist and that AMD3100 and ALX40-4C are weak partial agonists. J Biol Chem. 2002;277:24515–24521. doi: 10.1074/jbc.M200889200. [DOI] [PubMed] [Google Scholar]
- 51.Labbaye C, Spinello I, Quaranta MT, Pelosi E, Pasquini L, Petrucci E, Biffoni M, Nuzzolo ER, Billi M, Foà R, et al. A three-step pathway comprising PLZF/miR-146a/CXCR4 controls megakaryopoiesis. Nat Cell Biol. 2008;10:788–801. doi: 10.1038/ncb1741. [DOI] [PubMed] [Google Scholar]
- 52.Yu T, Wu Y, Helman JI, Wen Y, Wang C, Li L. CXCR4 promotes oral squamous cell carcinoma migration and invasion through inducing expression of MMP-9 and MMP-13 via the ERK signaling pathway. Mol Cancer Res. 2011;9:161–172. doi: 10.1158/1541-7786.MCR-10-0386. [DOI] [PubMed] [Google Scholar]
- 53.Villalvilla A, Gomez R, Roman-Blas JA, Largo R, Herrero-Beaumont G. SDF-1 signaling: A promising target in rheumatic diseases. Expert Opin Ther Targets. 2014;18:1077–1087. doi: 10.1517/14728222.2014.930440. [DOI] [PubMed] [Google Scholar]
- 54.Vicente R, Noël D, Pers YM, Apparailly F, Jorgensen C. Deregulation and therapeutic potential of microRNAs in arthritic diseases. Nat Rev Rheumatol. 2016;12:211–220. doi: 10.1038/nrrheum.2015.162. [DOI] [PubMed] [Google Scholar]
- 55.Wang S, Zhou C, Zheng H, Zhang Z, Mei Y, Martin JA. Chondrogenic progenitor cells promote vascular endothelial growth factor expression through stromal-derived factor-1. Osteoarthritis Cartilage. 2017;25:742–749. doi: 10.1016/j.joca.2016.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zheng X, Zhao FC, Pang Y, Li DY, Yao SC, Sun SS, Guo KJ. Downregulation of miR-221-3p contributes to IL-1β-induced cartilage degradation by directly targeting the SDF1/CXCR4 signaling pathway. J Mol Med (Berl) 2017;6:615–627. doi: 10.1007/s00109-017-1516-6. [DOI] [PubMed] [Google Scholar]
- 57.Genemaras AA, Ennis H, Kaplan L, Huang CY. Inflammatory cytokines induce specific time- and concentration-dependent MicroRNA release by chondrocytes, synoviocytes, and meniscus cells. J Orthop Res. 2016;34:779–790. doi: 10.1002/jor.23086. [DOI] [PubMed] [Google Scholar]
- 58.Spinello I, Quaranta MT, Riccioni R, Riti V, Pasquini L, Boe A, Pelosi E, Vitale A, Foà R, Testa U, Labbaye C. MicroRNA-146a and AMD3100, two ways to control CXCR4 expression in acute myeloid leukemias. Blood Cancer J. 2011;1:e26. doi: 10.1038/bcj.2011.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Huang ZY, Stabler T, Pei FX, Kraus VB. Both systemic and local lipopolysaccharide (LPS) burden are associated with knee OA severity and inflammation. Osteoarthritis Cartilage. 2016;24:1769–1775. doi: 10.1016/j.joca.2016.01.590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Choi YS, Park JK, Kang EH, Lee YK, Kim TK, Chung JH, Zimmerer JM, Carson WE, Song YW, Lee YJ. Cytokine signaling-1 suppressor is inducible by IL-1beta and inhibits the catabolic effects of IL-1beta in chondrocytes: Its implication in the paradoxical joint-protective role of IL-1beta. Arthritis Res Ther. 2013;15:R191. doi: 10.1186/ar4381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yang RQ, Teng H, Xu XH, Liu SY, Wang YH, Guo FJ, Liu XJ. Microarray analysis of microRNA deregulation and angiogenesis-related proteins in endometriosis. Genet Mol Res. 2016;15 doi: 10.4238/gmr.15027826. [DOI] [PubMed] [Google Scholar]
- 62.Lu Y, Cao DL, Jiang BC, Yang T, Gao YJ. MicroRNA-146a-5p attenuates neuropathic pain via suppressing TRAF6 signaling in the spinal cord. Brain Behav Immun. 2015;49:119–129. doi: 10.1016/j.bbi.2015.04.018. [DOI] [PubMed] [Google Scholar]
- 63.Liu Z, Ma C, Shen J, Wang D, Hao J, Hu Z. SDF-1/CXCR4 axis induces apoptosis of human degenerative nucleus pulposus cells via the NF-κB pathway. Mol Med Rep. 2016;14:783–789. doi: 10.3892/mmr.2016.5341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang H, Song B, Pan Z. Downregulation of microRNA-9 increases matrix metalloproteinase-13 expression levels and facilitates osteoarthritis onset. Mol Med Rep. 2018;17:3708–3714. doi: 10.3892/mmr.2017.8340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Si HB, Zeng Y, Liu SY, Zhou ZK, Chen YN, Cheng JQ, Lu YR, Shen B. Intra-articular injection of microRNA-140 (miRNA-140) alleviates osteoarthritis (OA) progression by modulating extracellular matrix (ECM) homeostasis in rats. Osteoarthritis Cartilage. 2017;25:1698–1707. doi: 10.1016/j.joca.2017.06.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.