Abstract
Background
The prostate cancer (PCa) diagnostic pathway is undergoing a radical change with the introduction of multiparametric magnetic resonance imaging (mpMRI), genomic testing, and different prostate biopsy techniques. It has been proposed that these tests should be used in a sequential manner to optimise risk stratification.
Objective
To characterise the genomic, epigenomic, and transcriptomic features of mpMRI-visible and -nonvisible PCa in clinically localised disease.
Design, setting, and participants
Multicore analysis of fresh prostate tissue sampled immediately after radical prostatectomy was performed for intermediate- to high-risk PCa.
Intervention
Low-pass whole-genome, exome, methylation, and transcriptome profiling of patient tissue cores taken from microscopically benign and cancerous areas in the same prostate. Circulating free and germline DNA was assessed from the blood of five patients.
Outcome measurement and statistical analysis
Correlations between preoperative mpMRI and genomic characteristics of tumour and benign prostate samples were assessed. Gene profiles for individual tumour cores were correlated with existing genomic classifiers currently used for prognostication.
Results and limitations
A total of 43 prostate cores (22 tumour and 21 benign) were profiled from six whole prostate glands. Of the 22 tumour cores, 16 were tumours visible and six were tumours nonvisible on mpMRI. Intratumour genomic, epigenomic, and transcriptomic heterogeneity was found within mpMRI-visible lesions. This could potentially lead to misclassification of patients using signatures based on copy number or RNA expression. Moreover, three of the six cores obtained from mpMRI-nonvisible tumours harboured one or more genetic alterations commonly observed in metastatic castration-resistant PCa. No circulating free DNA alterations were found. Limitations include the small cohort size and lack of follow-up.
Conclusions
Our study supports the continued use of systematic prostate sampling in addition to mpMRI, as avoidance of systematic biopsies in patients with negative mpMRI may mean that clinically significant tumours harbouring genetic alterations commonly seen in metastatic PCa are missed. Furthermore, there is inconsistency in individual genomics when genomic classifiers are applied.
Patient summary
Our study shows that tumour heterogeneity within prostate tumours visible on multiparametric magnetic resonance imaging (mpMRI) can lead to misclassification of patients if only one core is used for genomic analysis. In addition, some cancers that were missed by mpMRI had genomic aberrations that are commonly seen in advanced metastatic prostate cancer. Avoiding biopsies in mpMRI-negative cases may mean that such potentially lethal cancers are missed.
Keywords: Multifocal prostate cancer, Multiparametric magnetic resonance imaging, Molecular classifiers, Genetic heterogeneity
Take Home Message
Our study supports the continued use of systematic prostate sampling in addition to multiparametric magnetic resonance imaging (mpMRI), as avoidance of systematic biopsies in patients with negative mpMRI can mean that clinically significant tumours harbouring genetic alterations commonly seen in metastatic prostate cancer can be missed. Furthermore, there is inconsistency in individual genomics when genomic classifiers are applied.
1. Introduction
Systematic transrectal ultrasound (TRUS) biopsies are associated with sampling, grading, and staging errors [1], [2]. To address this issue, two trials have now evaluated multiparametric magnetic resonance imaging (mpMRI) for target guidance to improve the diagnostic accuracy [3], [4]. The benefit suggested by these studies has increased prebiopsy mpMRI in men with suspected prostate cancer (PCa), with up to 50% of men now undergoing prebiopsy mpMRI in some countries [5]. There are further calls to use mpMRI as a triage test and dispense with systematic biopsies altogether in nearly one-quarter of the men who have negative mpMRI [6], [7]. These proposals are based on the notion that the tumours missed by mpMRI are of “low risk”, as the majority of them tend to be of low grade and <10 mm [4], [8]. However, approximately 10% of Gleason score (GS) ≥7 PCas are missed by mpMRI, and may possibly be lethal [4].
In addition, uncertainty relating to metastatic potential in GS 3 + 4 and 4 + 3 cancers, combined with the known intraprostatic tumour heterogeneity, has led to the development of genomic tests for better prognostication and prediction of clinical progression. Their combined use with mpMRI is now being proposed with mpMRI-guided biopsies to improve risk stratification and avoid overtreatment [9], [10], [11]. Therefore, a new diagnostic pathway in which mpMRI and genomic tests are used sequentially to guide PCa management is emerging [12]. Performing these tests in succession is challenging considering the multifocal nature of PCa and possible misclassifications due to intratumour heterogeneity.
This developing diagnostic sequence, which fundamentally depends on a binary decision dictated by mpMRI to trigger biopsies followed by genetic tests to augment risk stratification, raises concerns. If genomic tests are to be used widely, then the implications of intratumour heterogeneity in mpMRI-visible tumours that are biopsied need to be evaluated more comprehensively. Perhaps more important is the 10% or more potentially significant PCas that are missed because they are not detected by mpMRI; the genomic makeup of these cancers could provide insights into their metastatic capacity and help in assessing their potential lethality. To gain insights into these questions, we conducted a study correlating genomics and mpMRI findings for men undergoing radical prostatectomy to elucidate the genomic characteristics of mpMRI-visible and -nonvisible tumours and to assess the inter-relationship and reliability of currently available genomic classifiers in this setting.
2. Patients and methods
2.1. Tissue collection
Informed consent for fresh prostate tissue collection was obtained preoperatively from patients undergoing prostatectomy at The Christie NHS Foundation Trust (MCRC biobanking protocol 16_RIMA_06). The sampling protocol was adapted from Warren et al. [13]. Post-prostatectomy, the prostate was inked and then serially sectioned from apex to base. A slice ∼1 cm thick was obtained from the mid-prostate (Supplementary Fig. 1A). This slice was randomly sampled via punch biopsy to yield 8–12 cores, with core locations marked and photographed. The sampling was blinded to mpMRI. Each core was bisected and one half was flash-frozen (Supplementary Fig. 1). Frozen samples were processed according to the following procedure. Specimens were embedded in optimal cutting temperature compound and multiple sections were taken along the vertical axis (Supplementary Fig. 1B). Sections from the top, middle, and bottom of each core were stained with haemotoxylin and eosin (H&E) and submitted to pathological review. A consultant pathologist reviewed the H&E slides, marked the tumour area for the dissection process, and assigned a cellularity score (Supplementary Table 1) to evaluate tumour content. Marked tumour regions were macrodissected for simultaneous DNA/RNA extraction (AllPrep kit; Qiagen, Hilden, Germany). The remaining prostate tissue was fixed for routine histopathological reporting. PCa grading was according to the International Society of Urological Pathology 2014 recommendations [14]. Where possible (5/6 patients), whole blood was obtained before surgery and germline DNA (Qiagen DNA blood extraction kit) and circulating free DNA (cfDNA) were extracted.
2.2. mpMRI and pathology correlation
mpMRI scans were performed preoperatively as part of routine clinical care. Scans were performed using a 1.5-T Siemens Avanto or Area scanner (Siemens, Munich, Germany), with the following acquisition parameters: T2, repetition time (TR) 3600–6500 and echo time (TE) 85–107; and diffusion, TR 3500–5800 and TE 64–97. T1-weighted, T2-weighted (T2W), and diffusion-weighted images were reviewed by a specialist uroradiologist and were scored using Prostate Imaging-Reporting and Data System version 2 (PIRADS v2) [15]. Lesions with no visible areas or overall PIRADS v2 scores of 1 and 2 were assigned as nonvisible lesions, while PIRADS v2 scores 3, 4, and 5 were assigned as visible lesions (Supplementary Table 1).
mpMRI correlation to whole-mount histopathology was performed by identifying the axial plane in T2W images corresponding to the histopathology section from which the cores were obtained. The location of each core was mapped on T2W axial scans and their visibility or invisibility was determined.
2.3. Library preparation and sequencing
DNA libraries were prepared using an Accel-NGS 2S DNA Library kit (Swift Biosciences, Ann Arbor, MI, USA). In brief, 200 ng of DNA was fragmented to a target size of 200 bp using acoustic fragmentation (Covaris, Woburn, MA, USA) according to the manufacturer's protocol consisting of end-repair, adaptor ligation, and polymerase chain reaction (PCR) amplification (6 cycles). Exome capture was subsequently performed using SureSelect Exome V6 + COSMIC (Agilent Technologies, Santa Clara, CA, USA) and 750 ng of input material. In brief, libraries were hybridised to probes for 16 h at 65 °C and subjected to ten cycles of PCR amplification. Before sequencing, libraries were run on a Bioanalyser (Agilent Technologies) and quantified using the Library Quantification Kit qPCR (Kapa Biosystems, Wilmington, MA, USA), and equimolar library pools were sequenced on a MiSeq 600 cycle V3 or NextSeq Mid output (2 × 150 cycles; Illumina, San Diego, CA, USA) for low-pass whole-genome sequencing, or a HiSeq (2 × 100 cycles; Illumina) for whole-exome sequencing. RNA libraries were prepared using a Clontech SMARTer Total RNA-seq kit (Takara Bio, Kusatsu, Japan) for cases 1 and 5, and a SureSelect RNA Poly-A kit (Agilent Technologies) for the remaining cases. Libraries were prepared according to the manufacturer's protocol. All were quantified using a Library Quantification Kit (Kapa Biosystems) and sequenced using HiSeq (2 × 100 cycles; Illumina). The average coverage obtained was 0.7× for low-read whole-genome sequencing for copy number and 54× for whole-exome sequencing. Methylation analysis was performed using an Illumina Methylation EPIC BeadChip array (Eurofins Genomics, Louisville, KY, USA) with 250 ng of input DNA (data accession numbers EGAS00001002767 and GSE101908).
2.4. Percentage genome aberration and gene expression–based classifier scores
The percentage genome aberration (PGA) and a 31-locus genomic classifier were calculated using copy number aberration (CNA) data. PGA was calculated by dividing the sum of all bases altered across the whole genome by the total number of bases (3,137,144,693) in GRCh37 [16]. The 31-locus PGA was calculated by dividing the sum of all bases altered in the previously reported prognostic 31 genes by the total number of bases in these 31 genes [17].
We also generated scores based on the expression of 12 cancer genes included in the OncotypeDX test (Genomic Health, Redwood City, CA, USA) [18], 31 cell-cycle progression genes included in the Prolaris test (Myriad Genetics, Salt Lake City, UT, USA) [19], and 19 of the 22 genes included in the Decipher signature (GenomeDx, San Diego, CA, USA) [20]. Three Decipher genes are not publicly available and were not included in the analysis [20]. To expand our analysis, we also included genes from the AR signalling pathway as previously described [21], [22]. These scores were calculated as a sum of the z-scores for genes in the respective panels as previously described [23]. The z-score for genes of interest in each core was calculated by subtracting the pooled mean from the RNA sequencing (RNA-seq) expression value and dividing by the pooled standard deviation.
Additional experimental details are available in the Supplementary material.
3. Results
3.1. Correlating mpMRI to histopathology
A total of 43 cores were collected for six men with intermediate- to high-risk PCa who underwent radical prostatectomy (Table 1). Each core was microscopically evaluated for tumour content and histopathological characteristics. Of the 43 samples, 22 were classified as tumour cores, whereas the remaining samples were classified as benign; all tumour specimen characteristics are listed in Supplementary Table 1.
Table 1.
Baseline clinical and pathological characteristics for six patients who underwent radical prostatectomy for intermediate- or high-risk prostate cancer
| Case 1 | Case 2 | Case 3 | Case 4 | Case 5 | Case 6 | |
|---|---|---|---|---|---|---|
| Age at surgery (yr) | 59 | 66 | 66 | 66 | 70 | 54 |
| PSA at diagnosis (ng/ml) | 3 | 34 | 6.5 | 6.8 | 8 | 5.8 |
| Prostatectomy Gleason score | 4 + 4 | 4 + 3 | 4 + 4 | 4 + 3 | 4 + 4 | 4 + 3 |
| Pathology stage | pT3b | pT3a | pT3a | pT3a | pT3a | pT2 |
| Nodal status | pNX | pN0 | pN0 | pNX | pN0 | pNX |
| Nodes examined | 0 | 8 | 1 | 0 | 15 | 0 |
| Surgical margin | Negative | Positive (focal-apical) |
Negative | Negative | Negative | Negative |
| Perineural invasion | Yes | Yes | Yes | Yes | Yes | No |
| Lymphovascular invasion | Yes | No | Yes | No | No | No |
PSA = prostate-specific antigen.
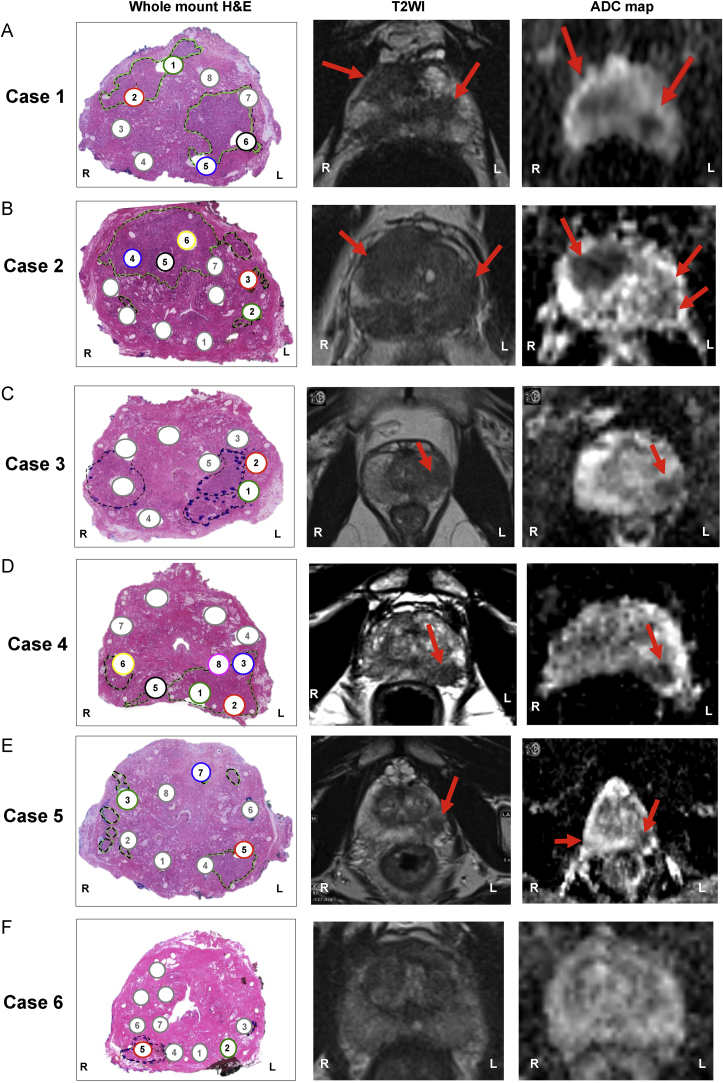
Correlation of mpMRI to whole-mount histopathology revealed that case 1 had two mpMRI-visible lesions and case 2 had three. The lesions for case 1 were sampled using two cores: both cores from one lesion were scored GS 4 + 3, and the other two had GS 4 + 4 (Fig. 1A). The three lesions for case 2 were sampled using five cores: one of the lesions was scored GS 3 + 4, while cores from the other two scored GS 3 + 3 (Fig. 1B).
Fig. 1.
Correlation between histological characteristics and visibility on multiparametric magnetic resonance imaging. Haematoxylin and eosin (H&E) whole-mount sections and corresponding axial T2-weighted image (T2WI) and apparent diffusion coefficient (ADC) maps for (A–F) cases 1–6. Tumour areas are marked with a dotted line on H&E stains and indicated by red arrows on T2WI and ADC maps. Tumour cores are numbered and labelled with colours; benign cores are labelled in grey.
Cases 3, 4, and 5 had pathologically identifiable lesions that were not visible on mpMRI, in addition to lesions that were. We obtained two cores from the mpMRI-visible lesion from case 3 (3#1 and 3#2) and these contained GS 4 + 3 disease (Fig. 1C), but the core from the nonvisible lesion contained few tumour cells and was not analysed further. Case 4 also had one visible and one nonvisible lesion on mpMRI (Fig. 1D). Two of the five cores from the visible lesion had GS 4 + 5, while three had GS 4 + 3. The core from the nonvisible lesion had GS 3 + 4. Case 5 had two visible and two nonvisible lesions. Cores were obtained from three of the lesions (one visible, two nonvisible; Fig. 1E). The core from the mpMRI-visible lesion had GS 4 + 3, whereas the two cores from the nonvisible lesions had GS 3 + 4 and 3 + 3. Finally, case 6 had no mpMRI-visible lesions, but TRUS biopsy detected GS 4 + 3 cancer. We sampled two lesions, a core with GS 4 + 3 and one with GS 3 + 3 (Fig. 1F). Thus, while most of the high-grade (GS ≥ 4 + 3) and low-grade tumours (GS 3 + 3) were visible on mpMRI, some clinically relevant lesions were not detected (case 5#3 and case 6#5; Supplementary Table 1).
3.2. Copy number landscape of mpMRI-visible and -nonvisible lesions
We performed comprehensive genomic profiling to determine genomic changes in our samples. Germline DNA from blood was used as a reference for all samples with the exception of case 3, for whom DNA was extracted from a histologically benign area of the prostate.
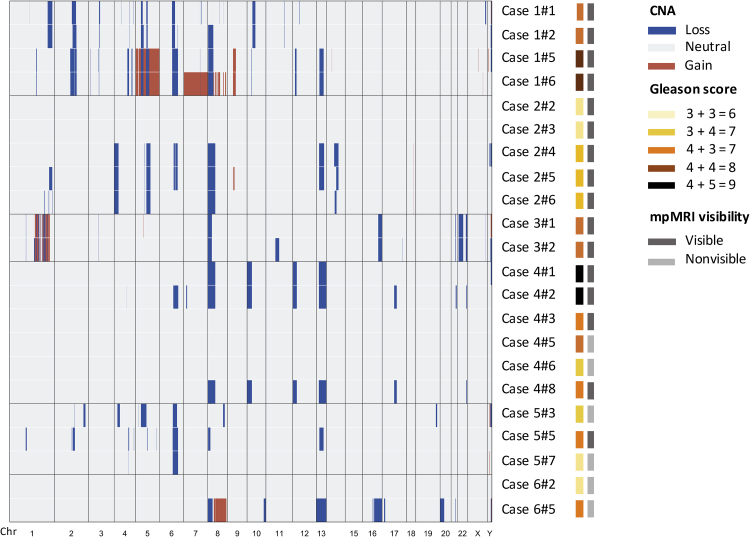
We first assessed CNA in the tumour cores. Consistent with previous studies, CNA analysis (Fig. 2) revealed high inter- and intrapatient heterogeneity associated with variable PGA (range 0–11.1%, Supplementary material). Cores obtained from mpMRI-visible lesions had a nonsignificantly higher PGA (median 3.16%) compared to nonvisible lesions (median 0.4%; p = 0.0840, Mann-Whitney test; Supplementary Fig. 2). Overall, chromosomal losses were more prevalent than gains and included deletions in chromosomes 8p (59%), 6q (45%), and 13q (41%), with the regions affected spanning genes commonly associated with PCa, such as NKX3.1, RB1, and BRCA2. Evidence of chromothripsis was observed in chromosome 1 for both tumour cores from case 3. As expected, cores with no CNA were correlated with low GS. Owing to the relatively small number of samples, we were not able to single out a specific CNA pattern for mpMRI-nonvisible tumours; however, we did find significant structural aberrations in those cores (Fig. 2, Fig. 3). For example, case 5#3 (GS 3 + 4) had deletions in known PCa-related tumour suppressors, such as MAP3K7 [24], [25] and FOXO3 [26], as well as genes involved in DNA repair pathways, such as MSH3, ERCC8, and RPC1 (Supplementary material). Case 6#5 (GS 4 + 3) had deletions in RB1, TP53, and BRCA2 loci, MYC amplification, and TMPRSS2-ERG fusion (Fig. 2, Fig. 3, Supplementary Fig. 3, Supplementary Table 2). For all the patients from whom we collected blood, we did not identify CNA in cfDNA derived from blood (Supplementary Fig. 4).
Fig. 2.
Copy number alteration profiles for cases 1–6. Copy number heatmap showing chromosomal losses (blue) and amplifications (red) for all cases. The Gleason score and multiparametric magnetic resonance imaging (mpMRI) visibility for each of the tumour cores are also indicated and described in the legends.
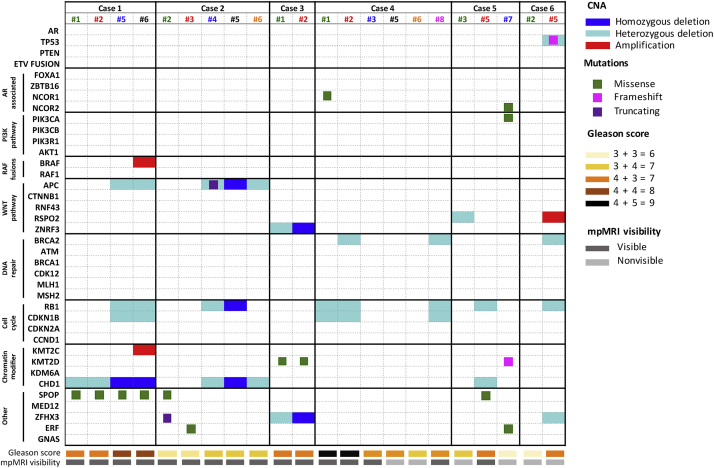
Fig. 3.
Integrative landscape analysis of somatic and copy number aberrations in tumour samples obtained from cases 1–6. Columns represent individual tumour cores from each patient included in the analysis, and rows represent specific genes grouped in pathways. Specific chromosomal aberrations and somatic alterations are described in the colour legends, together with Gleason score and magnetic resonance imaging (mpMRI) characteristics. Cases with more aberrations in a gene are represented by split colours. Cores are colour-coded according to Figure 1.
3.3. Mutational profile of mpMRI-visible and -nonvisible tumours
Marked tumour heterogeneity was also found at the level of individual somatic alterations. The number of somatic mutations varied from 104 to 182 per tumour core, with an average of 51 single-nucleotide variations per core (range 33–68) affecting exonic regions and, consequently, more likely to cause amino acid changes. SPOP mutations were found in tumour cores from four out of six patients (67% patients; 27% total tumour cores). Intriguingly, all the tumour cores from case 1 had alterations in the SPOP MATH domain [27], however, cores 1#1 and 1#2 had a missense mutation in the resulting amino acid K129E, while cores 1#5 and 1#6 in the amino acid F102 C (Fig. 3 and Supplementary material), suggesting the presence of parallel evolution. A single frameshift mutation in the TP53 gene was identified in one nonvisible core (case 6#5), while mutations in genes previously reported as commonly altered in PCa, such as IDH1, FOXA1, and MED12 [28], [29], were not detected.
3.4. Gene expression and DNA methylation analysis
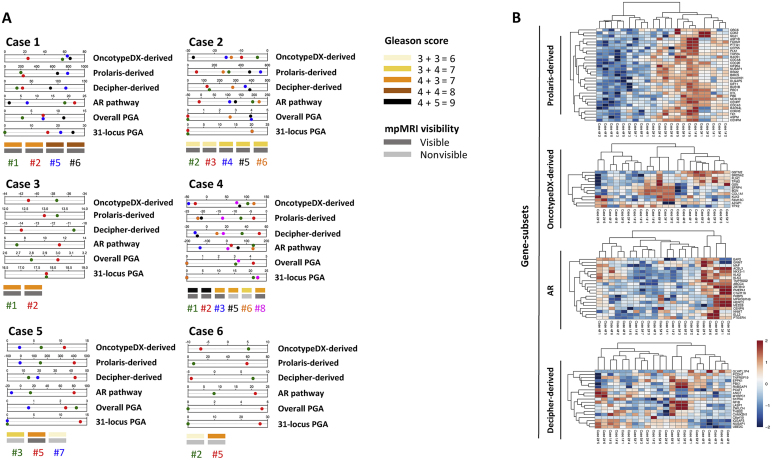
To assess the impact of tumour heterogeneity on gene expression, we performed RNA-seq analysis for tumour and benign cores from each patient in our cohort. First, principal component analysis was applied to gene expression data to identify grouping patterns for the samples. As shown in Supplementary Figure 5, benign cores, with the exception of a few cases, tended to cluster together and away from tumour specimens. Conversely, tumour samples exhibited more heterogeneous behaviour, with a trend towards clustering together, probably as a reflection of their spatial proximity in the gland. Next, we interrogated our samples for the expression of a subset of genes from commercially available genetic tests [18], [19], [20], [21], [22] that are increasingly used to stratify indolent and aggressive disease. Expression-based scores were arbitrarily generated and applied to our cohort for core stratification (Supplementary material). We found little correlation in core ranking among the different signatures, with tumours clustering together or away from each other based on the genes taken into account for the analysis, rather than mpMRI visibility (Fig. 4A). Similar results were observed when we performed unsupervised clustering of the samples (Fig. 4B).
Fig. 4.
Gene expression analysis and classification of tumour cores. (A) Tumour core ranking based on expression-derived scores arbitrarily generated using normalised RNA sequencing values for all the genes included in the OncotypeDX, Prolaris, and Decipher prognostic signatures. AR activity and percentage genomic aberration (PGA) scores were also calculated and included in the analysis. (B) Unsupervised clustering of tumour cores using gene expression data for the specified signature genes. Cores are colour-coded according to Figure 1.
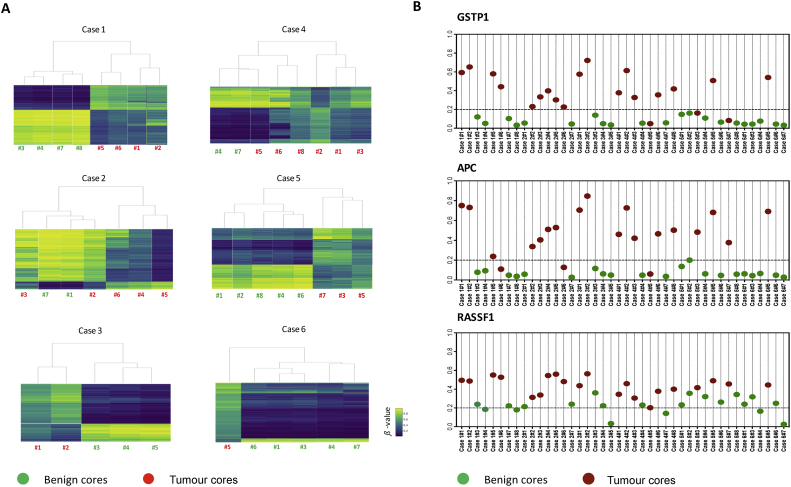
Given the high variability observed in gene expression, we performed methylation analysis to capture epigenetic changes occurring in our cohort of patients. Unsupervised clustering of tissue cores using the top 5000 variable CpG values revealed clear separation between benign and tumour samples, with the exception of case 2#3 and case 4#5 (Fig. 5A). Interestingly, the same trend was observed when multidimensional scaling analysis was used (Supplementary Fig. 6), suggesting that the molecular pressure exerted by cancerous lesions on the surrounding normal epithelium, a phenomenon described as the cancer-proximity field effect or field cancerisation [30], [31], was limited in our cohort. Finally, we looked at the specific methylation status of GSTP1, APC, and RASSF1, which have recently been proposed as a three-gene methylation signature to identify the presence of PCa lesions when obvious histopathological confirmation is not available [32]. As shown in Figure 5B, benign cores separated from tumour specimens when GSTP1 and APC were analysed, while this effect was less pronounced for RASSF1. This confirmed, at least in our cohort, that the identification of clinically relevant tumours relies on the sampling accuracy.
Fig. 5.
Methylation analysis of prostate specimens from cases 1–6. Tissue types from each individual are colour coded as green (benign) and red (tumour). (A) Unsupervised hierarchical clustering of methylation patterns for tumour and benign cores from cases 1–6. Rows of the heatmaps display the β values for the top 5000 CpG sites with the greatest intrapatient DNA methylation variability. Blue indicates low and yellow represents high methylation level (from 0 to 1). (B) Methylation array–derived β values for APC, GSTP1, and RASSF1 in benign and tumour cores.
4. Discussion
In this study we used an integrated mpMRI-histopathology-genomics approach to assess evolving methods for PCa diagnosis and risk stratification. We found that intratumour heterogeneity within mpMRI-visible lesion carries a risk of patient misclassification when using genomic biomarkers from a single biopsy. Moreover, dispensing with systematic biopsies in mpMRI-negative cases can lead to failure to detect potentially significant PCa harbouring genomic alterations that are common in metastatic disease.
Our findings are in agreement with previous studies demonstrating spatial genomic heterogeneity in localised PCa [33], [34], [35]. The implications of intratumour heterogeneity in using a single biopsy for genomic-based prognostication or prediction have also been highlighted previously [23], [36]. Our study extends the findings to mpMRI-visible tumours and evaluates the relevance of current genomic testing in combination with mpMRI-guided tissue acquisition. For instance, consider a previously validated PGA cutoff of ≥7.49%, which was associated with a multivariate adjusted hazard ratio of 3.2–4.5 for predicting relapse following radiotherapy or radical prostatectomy [37]. Then of the two cores obtained from the same visible lesion, one of the cores can classify the patient as low risk (case 1#6, PGA 7.2%) while the other core can classify the patient as high risk (case 1#5, PGA 11.1%). Similarly, intratumour transcriptomic heterogeneity within mpMRI-visible lesions can possibly lead to misclassification using RNA-based tests. As a result, patients can be misclassified using genomic analysis based on a single mpMRI-targeted biopsy. These findings have implications for management decisions made on the basis of genomic tests from a single biopsy [9], [10], [11], [38], [39], [40].
Avoiding systematic biopsies in patients with negative mpMRI is a strategy with significant risk. While there are a range of options for consideration, the emerging enthusiasm for proceeding without biopsy on the basis of an argument that tumours missed by mpMRI can be regarded as “low risk” [8] may be misguided. This approach will expose patients to the risk of misclassification of clinically significant nonvisible tumours. Our study shows that PCa tumours undetected by mpMRI can harbour genomic alterations that are commonly seen in metastatic castration-resistant PCa (mCRPC) [41], [42]. Using our mpMRI-blind sampling approach, we collected six mpMRI-nonvisible cores from three patients, three of which had one or more genomic aberrations commonly associated with aggressive biology and mCRPC. While two of these patients had other mpMRI-visible lesions, case 6 had a negative mpMRI. This patient had GS 4 + 3 (case 6#5) cancer with copy number changes including RB1 and TP53 loss, as well as MYC amplification, which are commonly seen in mCRPC [42]. This shows that mpMRI-nonvisible tumours could be regarded as genomically aggressive and could potentially give rise to lethal clones. Moreover, previous studies have reported the presence of genomic alterations in histomorphologically benign prostate that could provide a backdrop against which PCa develops from benign prostate glands [34]. These findings based on “cancer field effect” are relevant in cases with false-negative mpMRI, for whom histologically benign biopsies could be used to predict occult PCa [30], [43]. Two studies, MATLOC and DOCUMENT, have evaluated this approach in benign biopsies using a methylation-based test to predict adverse pathology [32], [44]. However, we did not observe a consistent cancer-field effect at the epigenetic level, again suggesting that molecular evaluation of a single benign biopsy may be insufficient to predict occult clinically significant PCa.
Our study has inherent limitations. The co-registration of histopathology and mpMRI was performed cognitively, so some tumours might have been missed. However, using our single-slice protocol we captured the index tumour in all six patients. Tighter spatial correlation can be attempted when designing future studies using three-dimensional prostate moulds with guides corresponding to mpMRI slice thickness and scanning orientation to improve precision [45], [46]. Another shortcoming is the relatively small number of patients and short-term follow-up. However, detailed studies such as this are both time-consuming and expensive, and it would be difficult to validate our findings on a large scale and include long-term natural history. We also used derived RNA-based expression scores rather than commercial tests. While the proprietary algorithms used by these tests are not publicly available, results supporting our findings have been published previously [23]. Another study comparing the results of the commercial Decipher test suggested that approximately one in five patients could be potentially misclassified using mpMRI-targeted biopsies [9].
5. Conclusions
Our study emphasises the diagnostic complexities for clinically localised PCa. Importantly, it highlights the shortcomings of the new diagnostic method that uses mpMRI on its own as a triage test and avoids systematic sampling when visible lesions are present and avoids biopsy when they are not. This clearly carries a risk of missing potentially aggressive lesions that is not mitigated by the use of current genomic classifiers. A single targeted biopsy does not eliminate the problem of intraprostatic heterogeneity and the results obtained using this approach should be interpreted with care to minimise the possibility of disease misclassification.
Author contributions: Esther Baena had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Study concept and design: Parry, Ali, Srivastava, Clarke, Marais, Baena.
Acquisition of data: Parry, Ali, Antonello, Cannistraci, Barros-Silva, Ubertini, Ramani, Lau, Shanks, Nonaka, Oliveira.
Analysis and interpretation of data: Srivastava, Parry, Ali, Cannistraci, Antonello, Ubertini, Hambrock, Brady, Clarke, Marais, Baena.
Drafting of the manuscript: Cannistraci, Ali, Parry, Srivastava, Dhomen, Clarke, Marais Baena.
Critical revision of the manuscript for important intellectual content: All authors.
Statistical analysis: Srivastava, Ali, Leong.
Obtaining funding: Miller, Brady, Dive, Clarke, Marais, Baena.
Administrative, technical, or material support: Ramani, Lau, Shanks, Nonaka, Oliveira.
Supervision: Clarke, Marais, Baena.
Other: None.
Financial disclosures: Esther Baena certifies that all conflicts of interest, including specific financial interests and relationships and affiliations relevant to the subject matter or materials discussed in the manuscript (eg, employment/affiliation, grants or funding, consultancies, honoraria, stock ownership or options, expert testimony, royalties, or patents filed, received, or pending), are the following: None.
Funding/Support and role of the sponsor: This research was supported by funds from CRUK Manchester Institute to R.M and E.B. (C5759/A20971) and funds from Prostate Cancer UK (CE013_2-004) to N.W.C and R.M. The sponsors played no direct role in the study.
Acknowledgments: We thank the Manchester Cancer Research Centre Biobank and The Christie Foundation Trust Pathology Department, in particular Paul Montgomery, Sarah Brown, Nickolaos Katsfados, and Susan Williams for coordinating the sample collection. We thank the CRUK Manchester Institute Histology, Molecular Biology, and Imaging units for assistance, especially Garry Ashton and Wolfgang Breitwieser for assistance with genomic DNA/RNA extraction and sequencing. We would like to express our deepest gratitude to patients for contributing to our study.
Associate Editor: Gianluca Giannarini
Footnotes
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.euo.2018.08.005.
Contributor Information
Noel W. Clarke, Email: noel.clarke@christie.nhs.uk.
Richard Marais, Email: richard.marais@cruk.manchester.ac.uk.
Esther Baena, Email: esther.baena@cruk.manchester.ac.uk.
Appendix A. Supplementary data
The following are the supplementary data to this article:
References
- 1.Bjurlin M.A., Taneja S.S. Standards for prostate biopsy. Curr Opin Urol. 2014;24:155–161. doi: 10.1097/MOU.0000000000000031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Serefoglu E.C., Altinova S., Ugras N.S., Akincioglu E., Asil E., Balbay M.D. How reliable is 12-core prostate biopsy procedure in the detection of prostate cancer? In: Can Urol Assoc J. 2013;7:E293–E298. doi: 10.5489/cuaj.11224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kasivisvanathan V., Rannikko A.S., Borghi M. MRI-targeted or standard biopsy for prostate-cancer diagnosis. N Engl J Med. 2018;378:1767–1777. doi: 10.1056/NEJMoa1801993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ahmed H.U., El-Shater Bosaily A., Brown L.C. Diagnostic accuracy of multi-parametric MRI and TRUS biopsy in prostate cancer (PROMIS): a paired validating confirmatory study. Lancet. 2017;389:815–822. doi: 10.1016/S0140-6736(16)32401-1. [DOI] [PubMed] [Google Scholar]
- 5.National Prostate Cancer Audit. NPCA annual report 2017 www.npca.org.uk/content/uploads/2018/02/NPCA-2017-Annual-Report_final_211117.pdf.
- 6.Ahmed H.U., Kirkham A., Arya M. Is it time to consider a role for MRI before prostate biopsy? Nat Rev Clin Oncol. 2009;6:197–206. doi: 10.1038/nrclinonc.2009.18. [DOI] [PubMed] [Google Scholar]
- 7.Brizmohun Appayya M., Adshead J., Ahmed H.U. National implementation of multi-parametric MRI for prostate cancer detection—recommendations from a UK consensus meeting. BJU Int. 2018;122:13–25. doi: 10.1111/bju.14361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Le J.D., Tan N., Shkolyar E. Multifocality and prostate cancer detection by multiparametric magnetic resonance imaging: correlation with whole-mount histopathology. Eur Urol. 2015;67:569–576. doi: 10.1016/j.eururo.2014.08.079. [DOI] [PubMed] [Google Scholar]
- 9.Radtke J.P., Takhar M., Bonekamp D. Transcriptome wide analysis of magnetic resonance imaging-targeted biopsy and matching surgical specimens from high-risk prostate cancer patients treated with radical prostatectomy: the target must be hit. Eur Urol Focus. 2018;4:540–546. doi: 10.1016/j.euf.2017.01.005. [DOI] [PubMed] [Google Scholar]
- 10.Renard-Penna R., Cancel-Tassin G., Comperat E. Multiparametric magnetic resonance imaging predicts postoperative pathology but misses aggressive prostate cancers as assessed by cell cycle progression score. J Urol. 2015;194:1617–1623. doi: 10.1016/j.juro.2015.06.107. [DOI] [PubMed] [Google Scholar]
- 11.Salmasi A., Said J., Shindel A.W. A 17-gene Genomic Prostate Score assay provides independent information on adverse pathology in the setting of combined mpMRI fusion-targeted and systematic prostate biopsy. J Urol. 2018;3:564–572. doi: 10.1016/j.juro.2018.03.004. [DOI] [PubMed] [Google Scholar]
- 12.Ali A., Hoyle A., Baena E., Clarke N.W. Identification and evaluation of clinically significant prostate cancer: a step towards personalized diagnosis. Curr Opin Urol. 2017;27:217–224. doi: 10.1097/MOU.0000000000000385. [DOI] [PubMed] [Google Scholar]
- 13.Warren A.Y., Whitaker H.C., Haynes B. Method for sampling tissue for research which preserves pathological data in radical prostatectomy. Prostate. 2013;73:194–202. doi: 10.1002/pros.22556. [DOI] [PubMed] [Google Scholar]
- 14.Epstein J.I., Egevad L., Amin M.B., Delahunt B., Srigley J.R., Humphrey P.A. The 2014 International Society of Urological Pathology (ISUP) consensus conference on Gleason grading of prostatic carcinoma: definition of grading patterns and proposal for a new grading system. Am J Surg Pathol. 2016;40:244–252. doi: 10.1097/PAS.0000000000000530. [DOI] [PubMed] [Google Scholar]
- 15.Weinreb J.C., Barentsz J.O., Choyke P.L. PI-RADS Prostate Imaging-Reporting and Data System: 2015, version 2. Eur Urol. 2017;69:16–40. doi: 10.1016/j.eururo.2015.08.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hieronymus H., Schultz N., Gopalan A. Copy number alteration burden predicts prostate cancer relapse. Proc Natl Acad Sci U S A. 2014;111:11139–11144. doi: 10.1073/pnas.1411446111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lalonde E., Alkallas R., Chua M.L.K. Translating a prognostic DNA genomic classifier into the clinic: retrospective validation in 563 localized prostate tumors. Eur Urol. 2017;72:22–31. doi: 10.1016/j.eururo.2016.10.013. [DOI] [PubMed] [Google Scholar]
- 18.Knezevic D., Goddard A.D., Natraj N. Analytical validation of the Oncotype DX prostate cancer assay—a clinical RT-PCR assay optimized for prostate needle biopsies. BMC Genomics. 2013;14:690. doi: 10.1186/1471-2164-14-690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cuzick J., Swanson G.P., Fisher G. Prognostic value of an RNA expression signature derived from cell cycle proliferation genes in patients with prostate cancer: a retrospective study. Lancet Oncol. 2011;12:245–255. doi: 10.1016/S1470-2045(10)70295-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Erho N., Crisan A., Vergara I.A. Discovery and validation of a prostate cancer genomic classifier that predicts early metastasis following radical prostatectomy. PLoS One. 2013;8:e66855. doi: 10.1371/journal.pone.0066855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hieronymus H., Lamb J., Ross K.N. Gene expression signature-based chemical genomic prediction identifies a novel class of HSP90 pathway modulators. Cancer Cell. 2006;10:321–330. doi: 10.1016/j.ccr.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 22.The Cancer Genome Atlas Research Network The molecular taxonomy of primary prostate cancer. Cell. 2015;163:1011–1025. doi: 10.1016/j.cell.2015.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wei L., Wang J., Lampert E. Intratumoral and intertumoral genomic heterogeneity of multifocal localized prostate cancer impacts molecular classifications and genomic prognosticators. Eur Urol. 2016;71:183–192. doi: 10.1016/j.eururo.2016.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kluth M., Hesse J., Heinl A. Genomic deletion of MAP3K7 at 6q12-22 is associated with early PSA recurrence in prostate cancer and absence of TMPRSS2:ERG fusions. Mod Pathol. 2013;26:975–983. doi: 10.1038/modpathol.2012.236. [DOI] [PubMed] [Google Scholar]
- 25.Rodrigues L.U., Rider L., Nieto C. Coordinate loss of MAP3K7 and CHD1 promotes aggressive prostate cancer. Cancer Res. 2015;75:1021–1034. doi: 10.1158/0008-5472.CAN-14-1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Das T.P., Suman S., Alatassi H., Ankem M.K., Damodaran C. Inhibition of AKT promotes FOXO3a-dependent apoptosis in prostate cancer. Cell Death Dis. 2016;7:e2111. doi: 10.1038/cddis.2015.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhuang M., Calabrese M.F., Liu J. Structures of SPOP-substrate complexes: insights into molecular architectures of BTB-Cul3 ubiquitin ligases. Mol Cell. 2009;36:39–50. doi: 10.1016/j.molcel.2009.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barbieri C.E., Baca S.C., Lawrence M.S. Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nat Genet. 2012;44:685–689. doi: 10.1038/ng.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.The Molecular Taxonomy of Primary Prostate Cancer Cell. 2015;163(4):1011–1025. doi: 10.1016/j.cell.2015.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Slaughter D.P., Southwick H.W., Smejkal W. Field cancerization in oral stratified squamous epithelium; clinical implications of multicentric origin. Cancer. 1953;6:963–968. doi: 10.1002/1097-0142(195309)6:5<963::aid-cncr2820060515>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 31.Dakubo G.D., Jakupciak J.P., Birch-Machin M.A., Parr R.L. Clinical implications and utility of field cancerization. Cancer Cell Int. 2007;7:2. doi: 10.1186/1475-2867-7-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stewart G.D., Van Neste L., Delvenne P. Clinical utility of an epigenetic assay to detect occult prostate cancer in histopathologically negative biopsies: results of the MATLOC study. J Urol. 2013;189:1110–1116. doi: 10.1016/j.juro.2012.08.219. [DOI] [PubMed] [Google Scholar]
- 33.Boutros P.C., Fraser M., Harding N.J. Spatial genomic heterogeneity within localized, multifocal prostate cancer. Nat Genet. 2015;47:736–745. doi: 10.1038/ng.3315. [DOI] [PubMed] [Google Scholar]
- 34.Cooper C.S., Eeles R., Wedge D.C. Analysis of the genetic phylogeny of multifocal prostate cancer identifies multiple independent clonal expansions in neoplastic and morphologically normal prostate tissue. Nat Genet. 2015;47:367–372. doi: 10.1038/ng.3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brocks D., Assenov Y., Minner S. Intratumor DNA methylation heterogeneity reflects clonal evolution in aggressive prostate cancer. Cell Rep. 2014;8:798–806. doi: 10.1016/j.celrep.2014.06.053. [DOI] [PubMed] [Google Scholar]
- 36.VanderWeele DJ, Finney R, Katayama K, et al. Genomic heterogeneity within individual prostate cancer foci impacts predictive biomarkers of targeted therapy. Eur Urol Focus. In press. 10.1016/j.euf.2018.01.006. [DOI] [PMC free article] [PubMed]
- 37.Lalonde E., Ishkanian A.S., Sykes J. Tumour genomic and microenvironmental heterogeneity for integrated prediction of 5-year biochemical recurrence of prostate cancer: a retrospective cohort study. Lancet Oncol. 2014;15:1521–1532. doi: 10.1016/S1470-2045(14)71021-6. [DOI] [PubMed] [Google Scholar]
- 38.Hiew K, Hart CA, Ali A, et al. Primary mutational landscape linked with pre-docetaxel lactate dehydrogenase levels predicts docetaxel response in metastatic castrate-resistant prostate cancer. Eur Urol Focus. In press. 10.1016/j.euf.2018.04.006. [DOI] [PubMed]
- 39.Gilson C., Chowdhury S., Parmar M., Sydes M. Incorporating biomarker stratification into STAMPEDE: an adaptive multi-arm, multi-stage trial platform. Clin Oncol. 2017;28:778–786. doi: 10.1016/j.clon.2017.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mateo J., Carreira S., Sandhu S. DNA-repair defects and olaparib in metastatic prostate cancer. N Engl J Med. 2015;373:1697–1708. doi: 10.1056/NEJMoa1506859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Abida W, Armenia J, Gopalan A, et al. Prospective genomic profiling of prostate cancer across disease states reveals germline and somatic alterations that may affect clinical decision making. JCO Precis Oncol. In press. 10.1200/PO.17.00029. [DOI] [PMC free article] [PubMed]
- 42.Robinson D., Van Allen E.M., Wu Y.M. Integrative clinical genomics of advanced prostate cancer. Cell. 2015;162:454. doi: 10.1016/j.cell.2015.06.053. [DOI] [PubMed] [Google Scholar]
- 43.Trujillo K.A., Jones A.C., Griffith J.K., Bisoffi M. Markers of field cancerization: proposed clinical applications in prostate biopsies. Prostate Cancer. 2012;2012:302894. doi: 10.1155/2012/302894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Partin A.W., Van Neste L., Klein E.A. Clinical validation of an epigenetic assay to predict negative histopathological results in repeat prostate biopsies. J Urol. 2014;192:1081–1087. doi: 10.1016/j.juro.2014.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Costa D.N., Chatzinoff Y., Passoni N.M. Improved magnetic resonance imaging-pathology correlation with imaging-derived, 3D-printed, patient-specific whole-mount molds of the prostate. Invest Radiol. 2017;52:507–513. doi: 10.1097/RLI.0000000000000372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Turkbey B., Mani H., Shah V. Multiparametric 3T prostate magnetic resonance imaging to detect cancer: histopathological correlation using prostatectomy specimens processed in customized magnetic resonance imaging based molds. J Urol. 2011;186:1818–1824. doi: 10.1016/j.juro.2011.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.