Abstract
Arsenic is a non-essential, environmentally ubiquitous toxic metalloid. In response to this pervasive environmental challenge, organisms evolved mechanisms to confer resistance to arsenicals. Inorganic pentavalent arsenate is taken into most cells adventitiously by phosphate uptake systems. Similarly, inorganic trivalent arsenite is taken into most cells adventitiously, primarily via aquaglyceroporins or sugar permeases. The most common strategy for tolerance to both inorganic and organic arsenicals is by efflux that extrude them from the cytosol. These efflux transporters span across kingdoms and belong to various families such as aquaglyceroporins, major facilitator superfamily (MFS) transporters, ATP-binding cassette (ABC) transporters and potentially novel, yet to be discovered families. This review will outline the properties and substrates of known arsenic transport systems, the current knowledge gaps in the field, and aims to provide insight into the importance of arsenic transport in the context of the global arsenic biogeocycle and human health.
Keywords: Arsenic resistance, arsenic permeases, arsenite, arsenate, methylarsenite
Introduction
Arsenic is the most prevalent environmental toxic substance. As such, the Agency for Toxic Substances and Disease Registry (ATSDR) ranks arsenic first on its priority list of Hazardous Substances (https://www.atsdr.cdc.gov/spl/index.html), above other toxic metals such as lead, cadmium and mercury. While arsenic is not more toxic than other metals on the list, it ranks above them based on a combination of frequency, toxicity, and potential for human exposure. We are exposed to arsenic daily, mostly from our food and water supplies. It enters the biosphere from geochemical sources, and to a lesser extent, from anthropogenic sources such as herbicides, growth promoters for farm animals, wood preservatives, the semiconductor industry and other industrial sources (Zhu et al., 2014). Arsenic is classified by the International Agency for Research on Cancer (IARC) as a Group 1 human carcinogen and is associated with skin, bladder and lung cancer. Other arsenic-related diseases include cardiovascular and peripheral vascular diseases, neurological disorders, diabetes mellitus and chronic kidney disease (Abernathy et al., 2003). The U.S. Food and Drug Administration recommends limiting infant consumption of baby food prepared from arsenic-contaminated rice because of the potential for serious developmental problems (https://www.fda.gov/food/foodborneillnesscontaminants/metals/ucm319870.htm).
Arsenic is not required for life; thus, no known organism has arsenic-specific uptake systems. All uptake pathways for arsenic are adventitious via several types of transporters. Their physiological functions include transport of minerals and nutrients such as phosphate, glucose and glycerol, but arsenic can mimic those natural substrates well enough to share their uptake systems. On the other hand, arsenic resistance pathways are quite specific for arsenic or other metalloids such as boron, silicon and antimony. Microbes have been exposed to arsenic since the origin of life and consequently have evolved arsenic resistance (ars) genes. Arsenic detoxification pathways are found in nearly every present-day organism, from bacteria to humans. Arsenic resistance determinants include transporters and biotransformations such as redox enzymes, methyltransferases and biosynthetic pathways for arsenosugars, arsenolipids and other nontoxic forms of arsenic (Li et al., 2016; Yang and Rosen 2016). Arsenic has two biologically-relevant oxidation states, trivalent arsenite As(III) and pentavalent arsenate As(V). Methylated arsenicals include methylarsenate (MAs(V)) and methylarsenite (MAs(III)), dimethylarsenate (DMAs(V)) and dimethylarsenite (DMAs(III)), and trimethylarsine (TMAs(III)) and trimethylarsine oxide (TMAs(V)O). In general, trivalent arsenicals are considerably more toxic than the pentavalent species, trivalent methylated arsenicals are much more toxic than inorganic As(III), and trivalent aromatic arsenicals are the most toxic (Dopp et al., 2005). MAs(III) and DMAs(III) are less stable than other arsenic species, and their stability depends on sample matrix and temperature. Low temperature conditions (4 °C) significantly improve the stability of these arsenic species over room temperature conditions. They are relatively stable in a reducing cellular environment and can be quantified in cells and tissues. It is conceivable that MAs(III) and DMAs(III) species are partly oxidized to MAs(V) and DMAs(V) during sample collection, handling, and storage. Thus, it is not surprising that these trivalent arsenic species have not been commonly detected in environmental samples. Although DMAs(III) is biosynthesized, it is very reactive and rarely detectable in biological systems. New strategies for sample handling are needed for future studies involving these organic trivalent arsenic species.
Developing analytical methods to identify more complex organoarsenical compounds has also been important for distinguishing these compounds from the more toxic inorganic arsenic species. Key organic arsenic compounds routinely found in food (depending on food type) include arsenobetaine, arsenocholine, arsenosugars, and arsenolipids. Arsenobetaine is the major form of arsenic in marine animals, and it is considered a compound that is nontoxic by itself, available information indicates it is not mutagenic, immunotoxic, or embryotoxic (Alexander et al., 2009). Arsenocholine, which is mainly found in shrimp, is chemically similar to arsenobetaine, and is also essentially nontoxic. Arsenosugars and arsenolipids have recently been identified. Exposure to these compounds and toxicological implications are currently being studied. Arsenosugars are detected mainly in seaweed but are also found to a lesser extent in marine mollusks. Concerns about the potential toxicity of arsenosugars have been raised because there is evidence that arsenosugars are metabolized to DMAs(V) (Andrewes et al., 2004). Studies addressing arsenosugar and arsenolipds toxicity, however, have largely been limited to in vitro studies, which show that they are significantly less toxic than both inorganic arsenic and trivalent methylated arsenic metabolites (Taylor et al., 2017).
The major pathway of arsenic detoxification is extrusion from cells or sequestration in intracellular compartments (Ben Fekih et al., 2018). The substrate of transporters can be either free arsenic, as is seen in most bacterial transporters and the fronds of the arsenic hyperaccumulator Pteris vittata (Poynton et al., 2004), or As(III) conjugates with GSH or other thiols, as is observed in human hepatocytes (Leslie 2011). Inorganic arsenic detoxification in prokaryotes has been well described (Ben Fekih et al., 2018); however, highly toxic organic arsenic compounds such as methylarsenite (MAs(III)) are biologically synthesized, and have the classical properties of an antibiotic (Chen et al., 2019). In response, aerobic microbes have adapted by acquiring MAs(III) resistance mechanisms, including MAs(III) transporters. While new genes found in ars operons have been suggested to be involved in organic arsenic detoxification, there is little information on the transport of these organic arsenic compounds. This review will focus on arsenic transporters for inorganic and organic arsenicals, including uptake systems that are responsible for arsenic toxicity, and efflux systems that confer arsenic tolerance.
1. Transport systems responsible for uptake and toxicity of arsenic
A. Arsenite uptake systems
Life arose when the atmosphere was anoxic, and arsenic was primarily in the form of As(III). As(III) uptake systems were responsible for arsenic toxicity long before As(V) was a problem for living cells. In 1997, the first As(III) uptake pathway was identified (Sanders et al., 1997). As(III) and Sb(III) were shown to be taken into cells of Escherichia coli by the glycerol channel GlpF. a member of major intrinsic protein (MIP) (Reizer et al., 1993) or aquaporin (AQP) (Agre et al., 2002) superfamily. GlpF was identified from a search for mutants of E. coli resistant to trivalent antimony, Sb(III), a metalloid with close chemical properties to As(III). The Sb(III)-resistant mutant had a disruption in the glpF gene. The mutant accumulated considerably less As(III) than the wild type (Meng et al., 2004).
AQPs are bidirectional channels that include classical water channels, with small pores that only allow water through, and aquaglyceroporins, with pores large enough to allow molecules such as glycerol to pass through. In neutral solution arsenite is As(OH)3, an inorganic polyol similar to glycerol. Although bidirectional, AQPs are mostly responsible for As(III) movement into cells, increasing toxicity. However, there is a single instance of an atypical aquaglyceroporin, AqpS, in the ars operon of Sinorhizobium meliloti that mediates As(III) efflux (Yang et al., 2005). In S. meliloti, As(V) is reduced to As(III), and AqpS confers resistance by downhill efflux of internally generated As(III). Sequence alignment of the putative MIPs distributed in ars operons reveals two highly conserved NPA (Asn-Pro-Ala) motifs. The x-ray crystal structure of GlpF was the first structure of an AQP, and the two NPA motifs are found in two short helices that align to span the membrane (Fu et al., 2000) (Figure 1). In AqpS orthologs the consensus NPA sequences are SGXHXNPAVT and GXXXNPAR(S/D)XG, which form constrictions in the transmembrane channel. In AQPs, a constriction is formed at the center of the pore by oppositely juxtaposing two NPA motifs. This constriction is proposed to be involved in proton exclusion. A second constriction in AQPs known as the aromatic/arginine (ar/R) selectivity filter is formed at the extracellular mouth of the pore by four residues. Variability at the ar/R selectivity filter is thought to form the basis for broad spectrum of substrates in aquaglyceroporins.
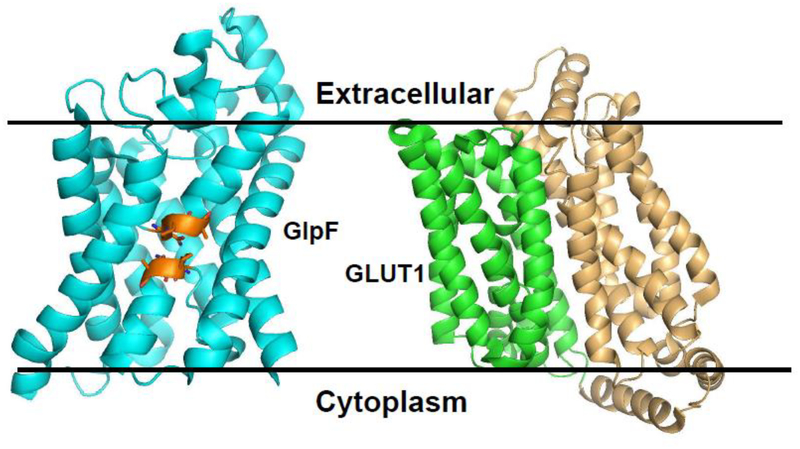
Figure 1. Structure of As(III) uptake proteins GlpF and GLUT1.
GlpF (PDB accession code 1FX8) and GLUT1 (PDB accession code 4PYP) facilitate As(III) entry into cells. Left: Aquaglyceroporins such as GlpF have six transmembrane ɑ-helices and two half-helices. The two half-helices align to span the membrane. Each half helix has an NPA motifs (orange) that form a constriction that contributes to exclusion of protons and gives selectivity for uncharged substrates. Right: MFS sugar permeases such as GLUT1 have two six-helix halves that fold together to form a central cavity into which substrate binds. In the cytosol substrate binds to the cavity in an inward-facing conformation. A conformational change reorients binding site to an outward-facing conformation, with release of substrate from the cell.
Aquaglyceroporins have also been shown to be a major route of bidirectional movement of As(III) into and out of cells of eukaryotes, including humans (Mukhopadhyay et al., 2014). The first eukaryotic AQP shown to mediate As(III) entry was the Saccharomyces cerevisiae GlpF ortholog, Fps1p (Wysocki et al., 2001). Disruption of fps1 resulted in resistance to both As(III) and antimonite. Fps1p activity is modulated by phosphorylation at position T231, which is catalyzed by the mitogen-activated protein kinase (MAPK) Hog1p that belongs to the high osmolarity glycerol (HOG) pathway of yeasts and other fungi (Tamás et al., 2000). This study suggests that the physiological function of Fps1p is to maintain turgor rather than for nutrient uptake.
Mammalian AQPs were first shown to transport As(III) by expression of AQP9 of Rattus norvegicus and AQP7 of Mus musculus in a Δfps1 mutant of S. cerevisiae strain that also had deletions of As(III) resistance genes Δacr3, which encodes an As(III) efflux permease, and Δycf1, which encodes a vacuolar As(III) accumulation pump (Liu et al., 2002). When AQP9 or AQP7 was expressed in this strain, it accumulated As(III) and became As(III) hypersensitive. Similarly, Xenopus laevis oocytes expressing AQP7 showed a 10-fold increase in uptake of As(OH)3.
AQP7, the adipose tissue isoform, is responsible for glycerol release into the bloodstream, where is it taken into liver hepatocytes by AQP9, the major liver isoform, for gluconeogenesis (Lebeck 2014). Both AQP9 and AQP7 facilitate As(III) uptake and are the likely pathway for uptake of arsenic in liver and adipose. In hepatocytes As(III) is methylated to highly toxic MAs(III), which can exit into the bloodstream via AQP9 and be transported to other organs such as brain, heart and kidney (Liu et al., 2006b). It is eventually excreted in urine and oxidized in air to nontoxic MAs(V). Interestingly, AQP7 does not appear to conduct MAs(III), suggesting that AQP isoforms may differ in substrate selectivity. Aquaporins are widely expressed in tissues implicated in high rates of active fluid transport. The human AQP7 was initially described as an adipose-specific glycerol channel, but this pore is also expressed in kidneys, testes, ovaries, heart, gastrointestinal tract, and skeletal muscle. The glycerol channel AQP7 plays a pivotal role in adipose tissue enlargement and function as well as glucose homeostasis. Human AQP9 expression is most found in liver, leukocyte, lung, and spleen. In rat, AQP9 mRNA is found in liver, testis, brain and lung (Hibuse et al., 2006). The level of AQP9 expression has been shown to vary in rats due to environmental factors such as diet, suggesting that AQP9 expression in humans could also be affected by such factors, and thus affect the risk of arsenic-related diseases.
Arsenic uptake by AQPs is of considerable relevance to human health and disease, and an understanding of both metalloid chemistry and the molecular details of metalloid transport systems is essential for the rational design of new drugs and for treating drug-resistant cells and microorganisms. For example, AQP9 expression level is correlated with adjuvant chemotherapy response in colorectal cancer (CRC), particularly in stage III CRC patients (Huang et al., 2017). The role of AQP9 in regulating tumor sensitivity to adjuvant chemotherapy in CRC is of current interest. In addition, arsenic trioxide (ATO, As2O3), which forms As(III) in solution, is an important and highly effective drug in the treatment of acute promyelocytic leukemia (APL). ATO must enter cells before exerting its cytotoxic activity, thus the control of arsenic trafficking through the plasma membrane could modulate arsenic sensitivity. AQP9 has been shown to modulate transmembrane transport of ATO (Bhattacharjee et al., 2004), and an increase in AQP9 expression at the transcription and protein levels led to increased arsenic uptake and intracellular concentrations, enhancing ATO-induced cytotoxicity in human APL cells (Chau et al., 2015). Pharmacological upregulation of AQP9 may be a novel approach to increase ATO sensitivity in neoplastic cells. Additionally, AQP9 also transports MAs(III), so it is possible that MAs(III) could be a more efficient treatment for APL than inorganic arsenic.
Aquaglyceroporins have since been shown to be a universal route of As(III) uptake in plants (Bhattacharjee et al., 2008). There are many more of these channel proteins in plants than in animals, perhaps because regulation of water homeostasis is more critical to organisms that cannot move in either flood or drought. The plant MIP superfamily falls into numerous subfamilies, of which members of the nodulin 26-like intrinsic protein (NIP) subfamily are responsible for facilitating As(III) uptake into the plant root (Zhao et al., 2009). In Arabidopsis thaliana, at least five NIPs, including NIP1;1, NIP1;2, NIP5;1, NIP6;1, and NIP7;1, facilitate As(III) uptake into roots. Nip3;1 is also involved in movement of As(III) into the root and from there into the shoot (Xu et al., 2015). In addition to As(OH)3, plants aquaglyceroporin facilitate uptake of other metalloids, including B(OH)3, Si(OH)4, Sb(OH)3 and Ge(OH)3. Boron and silicon are essential minerals for plants. A. thaliana NIP5;1 facilitates boron uptake and is crucial for plant growth under boron limitation (Takano et al., 2006). In rice (Oryza sativa), OsNIP2;1 (also called Lsi1) facilitates silicon accumulation (Ma et al., 2006).
Besides Si, Lsi1 also mediates transport of a range of small neutral molecules including Sb(III), urea, B(OH)3 and As(III). In rice roots, Lsi1 which is highly expressed in the distal side of endodermal and exodermal plasma membranes where Casparian strips are formed, is a major entry route for silicic acid and As(III) (Zhao et al., 2010). Mutations in Lsi1 resulted in a 60% loss of As(III) uptake. Lsi1 also mediates the uptake of undissociated pentavalent MAs and DMAs (Alexander et al., 2009), where mutants lost 80% and 50% of the uptake capacity for MAs and DMAs respectively compared to wild-type rice. Present-day seawater contains approximately 0.4 mM borate and 0.1 mM silicate but submicromolar arsenic. We proposed that the essential metalloid oxyacids of boron and silicon, B(OH)3 and Si(OH)3, were original physiological substrates of aquaglyceroporins earlier than glycerol, which was not present in high concentrations in early oceans, whereas arsenic was taken up adventitiously.
Both As(III) and MAs(III) have also been shown to be taken up by glucose permeases, which are members of the major facilitator superfamily (MFS) (Reddy et al., 2012). Hexose permeases HXT1, HXT3, HXT4, HXT5, HXT7 and HXT9 of S. cerevisiae transport As(III) at least as well as or better than Fps1 (Liu et al., 2004). Like AQPs, these sugar transporters are bidirectional but physiologically are responsible for sugar uptake. Subsequently both human and rat glucose permease GLUT1 were shown to transport As(III) and MAs(III) (Liu et al., 2006a). The x-ray crystal structure of GLUT1 has been solved (Deng et al., 2014) (Figure 1), however the mechanism of As(III) transport cannot be deduced from the glucose translocation pathway. GLUT1 is highly expressed in heart and brain because of the high energy demand of those organs. We speculate that GLUT1 may be involved in uptake of both As(III) and MAs(III) into heart and brain and could be related to arsenic cardiotoxicity and neurotoxicity. In plants, As(III) can be transported by permeases for the sugar alcohol inositol (Duan et al., 2016). The inositol permeases AtINT2 and AtINT4 are responsible for As(III) translocation from phloem to seed. By extrapolation, we speculate that similar sugar alcohol permeases are responsible for putting arsenic into the rice grain. It is not clear how sugar permeases catalyze As(III) uptake. One possibility is that As(III) uses a different transport pathway, as has been suggested for GLUT1 (Jiang et al., 2010). It is also possible that in solution As(OH)3 polymerizes to form polyhydroxylated ring structures that are an inorganic mimetics of sugars and sugar alcohols (Meng et al., 2004), which might allow As(III) to be a substrate of sugar transporters.
B. Arsenate uptake systems
While still of worldwide concern, As(V) is much less toxic compared to trivalent arsenic species, which form strong metal-like interactions with thiol groups in protein cysteine residues and small molecule thiols such as reduced glutathione (GSH). As(III) is also more mobile in groundwater and soils than As(V) and is the substrate for several arsenic biotransformations. Knowledge of As(V) uptake and physiology is key to understanding the arsenic biogeocycle and its effects on human health worldwide.
Following the Great Oxidation Event (GOE) about 2.5 Bya, trivalent arsenicals were generally oxidized to the pentavalent species (Planavsky et al., 2014). In present day aerobic soil and water, inorganic As(V) is the prevalent species of arsenic. As(V) is chemically similar to phosphate (Pi), and entry of As(V) into most cells is primarily mediated by Pi transporters. This was originally shown in E. coli, which has two phosphate uptake systems: the high affinity Pst and low affinity Pit transporters (Rosenberg et al., 1977). Pit is the primary pathway for As(V) uptake and is mainly responsible for As(V) toxicity, although As(V) also enters cells via Pst at higher concentrations (Willsky and Malamy 1980). In rice, As(V) uptake by Pi transporters has been shown by competitive inhibition of As(V) uptake by Pi (Jiang et al., 2014). In A. thaliana, resistance to As(V) is conferred by loss-of-function mutations in Pht1;1, which encodes a high-affinity Pi transporter (Catarecha et al., 2007). The PHT1 family of plant Pi transporters are MFS transporters and are mostly, if not exclusively, expressed in roots. When exposed to arsenate, plants suppress PHT1:1 expression and remove it from the plasma membrane (Castrillo et al., 2013). Interestingly, A. thaliana Pht1 mutants also showed increased accumulation of arsenic even though the rate of uptake was lower.
This apparently paradoxical observation suggests that mutants that accumulate As(V) more slowly are able to cope with arsenic toxicity through the action of reductases such as HAC (Chao et al., 2014), which reduce As(V) to As(III), and is subsequently sequestered in vacuoles by phytochelatin-As(III) conjugate pumps such as the A. thaliana AtABCC1/2 (Song et al., 2010). This novel A. thaliana reductase, HAC1, was shown to catalyze reduction of As(V) to As(III) in the outer cell layer of the root, which allowed efflux of As(III) from the root back into the soil. Other members of this family such as OsHAC1;1/2 and OsHAC4 have also been found in rice, and also are As(V) reductases, whichare critical in restricting As accumulation in rice shoots and grain (Shi et al., 2016; Xu et al., 2017).
Interestingly, arsenic hyperaccumulators such as P. vittata are more efficient in root-to-shoot arsenic translocation, resulting in increased accumulation. Enhanced As(V) uptake of P. vittata results from both increased transporter expression (Caille et al., 2005) and increased affinity of the Pi/As(V) uptake system for As(V) (Poynton et al., 2004). The maximum influx velocity (Vmax) of As(V) into roots was much higher in P. vittata than for non-accumulator species. In P. vittata, PvPht1;3 transports As(V) with high affinity (DiTusa et al., 2016). Furthermore, Pht1:3 transporters are inhibited more strongly by addition of As(V) compared to A. thaliana transporters, suggesting that the former may have higher affinity for As(V) than the latter. Like non-accumulator species, P. vittata also relies on As(V) reduction and shuttling into cellular compartments to confer As resistance. P. vittta expresses two copies of a protein similar to the yeast As(III) efflux pump Acr3 (vide infra), which sequester As(III) in the vacuole, and when knocked-down, causes an As(III)-sensitive phenotype (Indriolo et al., 2010).. Both homologs are involved in arsenic metabolism, but have different expression patterns (Chen et al., 2017b). PvACR3 expression markedly increased As tolerance and root-to-shoot As translocation (Wang et al., 2018), and it was observed to be localized in the plasma membrane in transgenic plants, suggesting a role in mediating As(III) efflux for xylem loading (Chen et al., 2013). On the other hand, PvACR3;1 was localized to vacuolar membrane, indicating that PvACR3;1 probably mediates As sequestration into vacuoles to increase As retention in the roots, thus reducing As translocation to the shoots (Chen et al., 2017b). Mechanisms of uptake and translocation by arsenic hyperaccumulators such as P. vittata are of current interest, as these organisms offer a promising method of arsenic bioaccumulation/remediation and expand our understanding of the arsenic biogeocycle.
As(V) uptake via phosphate transporters is of relevance to human health, in particular its connection to diabetes mellitus. Epidemiological studies have previously established a correlation between arsenic exposure and prevalence of diabetes mellitus in regions of hyperendemic arsenicosis (Navas-Acien et al., 2006). Although the causality proposed by these studies has been debated, diabetes-like effects have been observed in animal models on exposure to arsenic. Human single nucleotide polymorphisms (SNPs) in the major arsenic-detoxifying enzyme As(III) S-adenosylmethionine methyltransferase (AS3MT) are linked to a higher frequency of diabetes, suggesting a correlation (Drobná et al., 2013). However, association does not imply causality; hence, there is a need to understand the mechanisms behind these observations. A recent study was designed to address this by exploring As(V)/Pi uptake by type III sodium/phosphate (Na+/Pi) cotransporters using a type I diabetes animal model (Paul et al., 2011). Transporters of this family, including PIT1 and PIT2, are of interest as they are expressed ubiquitously in various tissues and have been proposed as the predominant mechanism of phosphate transport in rats. As expected, these transporters have also been observed to transport As(V). Following administration of several doses of the beta cell-specific toxin streptozotocin to induce type I diabetes mellitus, diabetic mice accumulated significantly more As(V), with the highest concentrations being in the liver and kidney. Interestingly, transcription and expression of PIT1 and PIT2 were increased in diabetic mice in all tissues studied, suggesting that an underlying effect of diabetes may be to increase susceptibility to inorganic As(V) toxicity.
Once inside of cells, As(V) is reduced to As(III) by one of several different As(V) reductases (Mukhopadhyay and Rosen 2002). In bacteria, two unrelated ArsC enzymes reduce As(V) using either GSH and a small thiol protein glutaredoxin 2 (Grx2) or another small thiol protein thioredoxin. In yeast, an unrelated As(V) reductase Acr2 carries out the same function. Acr2 has both As(V) reductase activity and protein phosphatase activity. It is structurally similar to the catalytic domain of human Cdc25 phosphatase (Bordo and Bork 2002). Both have an active site Cys-X(5)-Arg, where the five X residues form a phosphate binding loop. Human Cdc25 is a dual specificity phosphatase involved in control of cyclin-dependent kinases and progression through the cell cycle. The catalytic domains of Cdc25b and Cdc25c isoforms have been shown to have As(V) reductase activity, although it is not clear whether this is a physiological activity of those phosphatases (Bhattacharjee et al., 2010). It may seem counter-intuitive for cells to reduce relatively nontoxic As(V) to the more toxic oxidation state As(III). Prior to the GOE, all arsenic detoxification pathways used trivalent arsenicals as substrates. Following the GOE, it was only necessary to reduce As(V) to As(III), the substrate of the efflux and biotransformation systems. Nature may not progress in the most logical path but builds on existing edifices.
C. Effect of osmolytes on arsenic uptake
Plants can also combat As-induced toxicity through the production and/or accumulation of compatible solutes or osmolytes such as glycinebetaine (Garg and Singla 2011) and proline (Mishra and Dubey 2006). Osmolyte accumulation is an important attribute for protection and survival of plants under stress (Maheshwari and Dubey 2007). Moreover, As-regulated oxidative stress could be successfully reversed in plants by exogenous supply of proline (Singh et al., 2015b). Other compounds such as sodium nitroprusside, (a source of NO) (Singh et al., 2013) and salicylic acid (Odjegba 2012; Saidi et al., 2017) have also been shown to provide tolerance against As in many plants.
Singh et al. demonstrated that exogenously applying proline to the seedlings of Solanum melongena, followed by treatment with various concentrations of As(V) under hydroponic conditions reduced toxicity of As by reducing its uptake (Singh et al., 2015b). In addition, better growth of plants under As stress was attributed to an increase in the activities of antioxidants which minimized oxidative damage to seedlings. Proline is involved in arsenic sequestering, thus conferring tolerance against As (Chandrakar et al., 2016; Chandrakar et al., 2017). Increased endogenous proline levels of have been reported under As stress, which protects plants by reducing As uptake and detoxifying OH radicals (Singh et al., 2015b; Yadav et al., 2014).
Nitric oxide has also been successfully used to reduce both growth inhibition and ROS-imposed injuries under As stress in rice (Singh et al., 2016; Singh et al., 2015a), Festuca arundinacea (Jin et al., 2010), and Vigna radiata (Ismail 2012). Acting as an antioxidant, the pretreatment of plants with NO has been reported to scavenge ROS completely and enhances tolerance against As. NO stimulates the entire biosynthetic pathway of PCs in roots. In Luffa acutangula, the application of NO boosts As tolerance by enhancing cell wall thickness in root epidermis, thereby controlling As absorption and accumulation (Singh et al., 2013). Nitric oxide provides tolerance against As by reducing As uptake through regulating various transporters, reducing chlorosis by increasing iron (Fe) concentrations in shoots, and causing vacuolar sequestration of As through enhanced synthesis of PCs.
Salicylic acid is a phenolic substance and endogenous plant growth regulator. Singh et al. found that exogenous application of SA to rice plants under As stress reduced As toxicity by increasing the endogenous concentrations of both SA and NO (Singh et al., 2017). The increased concentration of NO was attributed to the enhanced activities of nitrate reductase. Co-application of SA, compared to pre-application, was also shown to be more effective in reducing As uptake in rice due to enhanced antioxidant activity and increased concentrations of GSH and phytochelatins (PCs) (Singh et al., 2015a; Singh et al., 2015b).
2. Transport systems responsible for efflux of and tolerance to arsenic
A. Arsenite efflux systems
1). ArsB
In anaerobic environments, As(III) is the predominant form of inorganic arsenic and is significantly more toxic than its oxidized counterpart. It is also highly mobile in groundwater, making it a major source of arsenic exposure. As prokaryotes evolved in an anoxic, As(III)-rich environment throughout Earth’s early history, they acquired genes which allowed them to survive exposure to As(III) (Zhu et al., 2017). It is no surprise; therefore, that most bacterial enzymes and transporters that confer arsenic tolerance do so for the trivalent species. In bacteria and archaea, arsenic resistance genes are usually organized in ars operons that are found on both plasmids and chromosomes. Expression of the ars genes are controlled by an ArsR As(III)-responsive transcriptional repressor. Nearly every ars operon has a gene encoding one of two As(III) efflux proteins, ArsB (Meng et al., 2004) or Acr3 (Villadangos et al., 2012), which are unrelated antiporters belonging to different families (Figure 2).
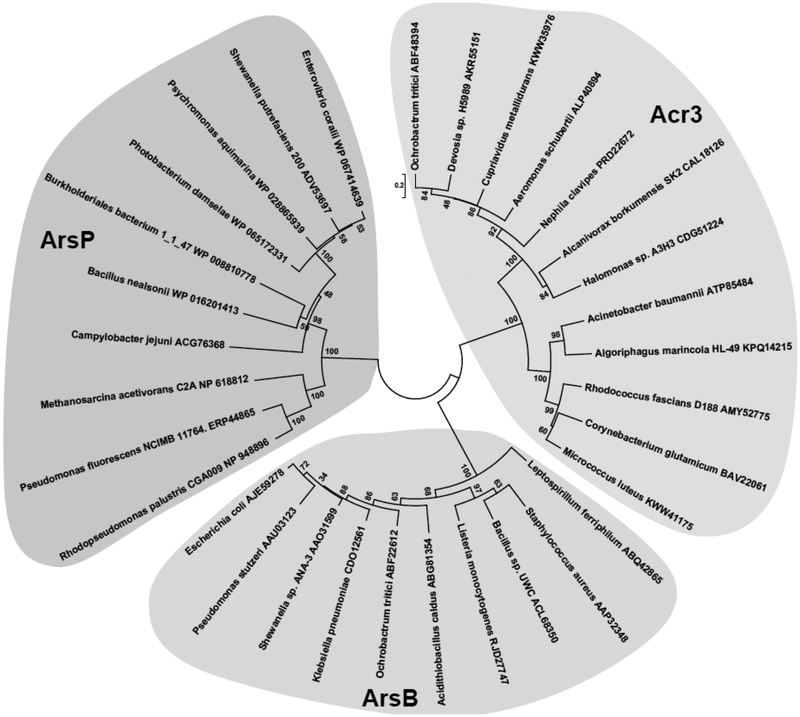
Figure 2. ArsB, Acr3, ArsP phylogenic trees.
A neighbor-joining phylogenetic tree showing the divergent from three arsenic efflux proteins: ArsB, Acr3, and ArsP. To obtain sequences of arsP genes encoding candidate orthologs, ArsP (Campylobacter jejuni RM1221), ACG76368, was used for Basic Local Alignment Search Tool (BLAST). Only sequences containing a highly conserved MAs(III) binding motif (TPFCSCSXXP) were included. To obtain sequences of arsB genes encoding candidate orthologs, ArsB (Escherichia coli), AJE59278, was used for BLAST. To obtain sequences of acr3 genes encoding candidate orthologs, Acr3 (Corynebacterium glutamicum), BAV22061, was used for blast. The tree was produced using MEGA7. Bacterial species and protein accession numbers are shown.
By extruding As(III), cells can grow in the presence of high As(III) concentrations. ArsB was the first identified As(III) efflux system and is employed by many bacteria for arsenic tolerance, followed by Ac3. Despite the similar biological roles of ArsB and Acr3 they evolved independently, with only limited sequence similarity (around 20%–40%) found between these two families. ArsB is classified in the ion transporter superfamily. A preliminary study of genome databases revealed a wide distribution of Acr3 homologues in bacteria, archaea and eukaryotes (mainly fungi and some lower plants) (Achour et al., 2007), while fewer ArsB homologues are found outside the bacteria kingdom. Thus, it appears that ArsB and Acr3 are the products of convergent evolution. Although it has not been identified in animals, an ArsB homologue exists in plants that is responsible for transcellular translocation of As(III) (Ma et al., 2007b; Ma et al., 2008a). Nearly every prokaryote has either an arsB or acr3 gene, which are found in roughly equal frequencies. Often there are multiple copies of either, and sometimes both within a single organism. For example, E. coli has a chromosomal arsB gene as well as plasmids that have additional copy or copies.
The best characterized member of the family is the 429-residue ArsB permease from plasmid R773 (Yang et al., 2012). From membrane topology studies, ArsB has been proposed to have 12 transmembrane (TM) segments in two groups of six with a central cytosolic loop (Figure 3A). While ArsB has little sequence similarity with MFS transporters, its transmembrane topology may place it in that superfamily. ArsB is an antiporter that extrudes As(III) or Sb(III) from cells by H+/As(OH)3 exchange coupled to the electrochemical proton gradient (Meng et al., 2004). Many proteins with As(III) binding sites have groups or pairs of cysteine residues in the binding site (Chen et al., 2017a; Huang et al., 2018; Yoshinaga and Rosen 2014). In contrast, ArsB has only a single cysteine residue that could be changed to either a serine or alanine residue without effect and so is not an essential residue (Rosen et al., 1995). Thus, unlike most other As(III) detoxification proteins, ArsB does not use soft metal chemistry in As(III) transport but has been proposed to transport a polymeric ring composed of three As(OH)3 molecules (Meng et al., 2004). ArsB is unique in having a dual mode of energy coupling depending on its association with ArsA, a 583-residue soluble enzyme with As(III)-stimulated ATPase activity (Rosen et al., 1995). From the crystal structure of ArsA, the As(III) binding site is composed of three cysteine residues (Zhou et al., 2000). Although ArsB can catalyze As(III) efflux coupled to the electrochemical proton gradient, when it binds ArsA, it is converted into a primary ATP-coupled As(III) efflux pump. In vivo studies on the energetics of As(III) extrusion showed that expression of both the arsA and arsB genes resulted in ATP-coupled As(III) extrusion independent of the electrochemical proton gradient. To confirm this, everted membrane vesicles from E. coli cells expressing the ArsA–ArsB complex were prepared and shown to catalyze ATP-coupled uptake of As(III). These results demonstrate that ArsB has the potential to be either a secondary antiporter coupled to the proton motive force or an obligatory ATP-coupled pump, depending on whether ArsA is part of the transport complex. As an antiporter, the maximum concentration gradient of As(III) outside the cell to inside the cytosol can be calculated to be 103. In contrast, depending on the number of ATP molecules hydrolyzed per As(III) transported, an ATP coupled pump can be calculated to create a concentration ratio of 106, a thousand times more efficient, providing a driving force for the evolution of the ArsAB complex.
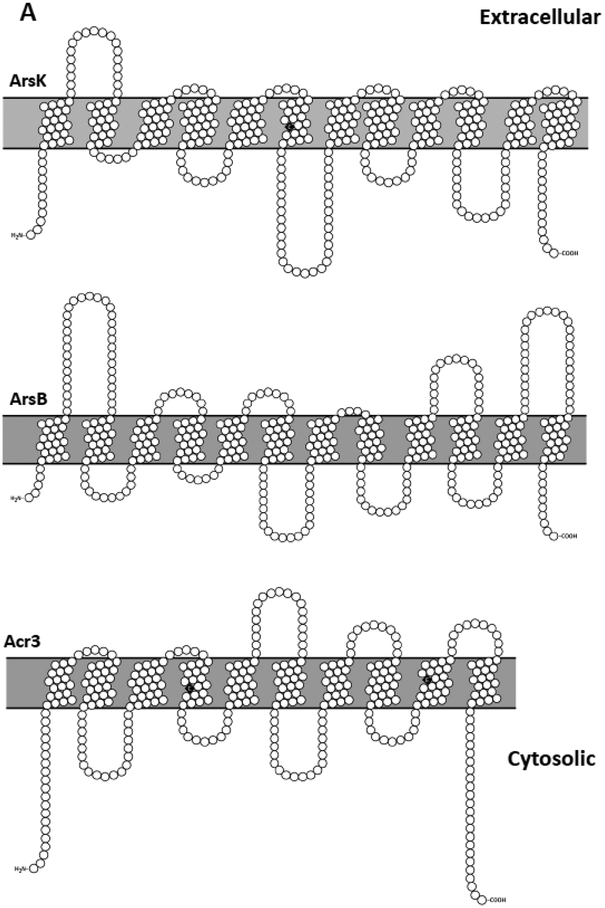
Figure 3. Transmembrane topology of arsenic permeases.
While the number of predicted TMs for ArsB and ArsK (A) supports their classification as MFS transporters, other pumps such as Acr3 (A), ArsJ, and ArsP (B) are predicted to have fewer than 12 TMs. The ArsB and Acr3 topologies are based on experimental data. ArsJ, ArsP and ArsK topologies are predicted from in silico analysis. Transmembrane topology models were calculated with TMHMM and illustrated with Protter (Omasits et al., 2014). Residues in black are predicted to be involved in the catalytic mechanism.
In addition to As(III), Sb(III) has also been shown to be a substrate for the ArsB transporter from E. coli and plasmid R773. ArsB from Campylobacter jejuni appears to be different. First, its predicted transmembrane topology has only 11 transmembrane domains, instead of 12 for the R773 ArsB. Second, CjArsB does not appear to catalyze Sb(III) extrusion (Shen et al., 2013). It is not clear what produces the difference in substrate specificity; perhaps the difference in the number of transmembrane helices contributes.
2). Acr3
Even though ArsB and Acr3 are both efflux systems for As(III), they have key functional and structural differences. With 357 residues, Acr3 is considerably smaller than ArsB. The Acr3 permease is a member of the bile/arsenite/riboflavin transporter (BART) superfamily (Mansour et al., 2007). The transmembrane topology of Acr3–1 from the iron-reducing alkaliphile Alkaliphilus metalliredigens has been determined by a combination of cysteine scanning mutagenesis and accessibility to thiol-reactive probes (Fu et al., 2009). The results clearly demonstrate that AmAcr3–1 has 10 transmembrane spanning segments, with the N- and C-termini localized in the cytosol (Figure 3A), like other BART superfamily proteins. This is consistent with the smaller size of Acr3 compared with ArsB, which has two additional transmembrane spanning segments. Two residues may be involved in metalloid translocation, a highly conserved cysteine residue in the P/R-C-T/I-AMV motif in TM3 of CgAcr3–1, and a conserved glutamate residue in TM9. Replacing either Cys129 or Glu305 led to loss of transport activity in CgAcr3–1. The possible requirement for a thiol suggests that the transport mechanism of Acr3 may be different from ArsB, which has no required cysteine residues. The role of these residues in mediating transport is still unknown, but it is reasonable to speculate that both residues may serve as a selectivity filter for As(III). There is, however, debate over the substrate specificity for Acr3. Fungal members of this family include the ScAcr3p metalloid efflux protein from S. cerevisiae, which was proposed to be selective for As(III) over Sb(III). ScAcr3p has been suggested to transport Sb(III) in vivo as well, and yeast plasma membrane vesicles accumulate both As(III) and Sb(III) by exchange with protons (Maciaszczyk-Dziubinska et al., 2012). However, a separate study found no Sb(III) transport in yeast plasma membrane. Bacterial Acr3 orthologues appear to be specific for As(III) and do not transport Sb(III). Cells of E. coli expressing AmAcr3–1 or CgAcr3–1 confer As(III) resistance (Fu et al., 2009). In addition, everted membrane vesicles prepared from C. glutamicum expressing Acr3–1 were shown to transport As(III) but not Sb(III) using NADH and an electron donor for the respiratory chain.
Some plants also have proteins related to Acr3 that confer arsenic tolerance. For example, an Acr3 ortholog was identified in the vacuole of P. vittata and is responsible for arsenic tolerance in this plant (Indriolo et al., 2010). In contrast, rice, in which arsenic in the rice grain is a serious health hazard, does not have Acr3 (Zhao et al., 2010). In an attempt to mitigate this problem, transgenic rice plants expressing the S. cerevisiae Acr3 gene increased As(III) efflux and also lowered arsenic accumulation in rice grain (Duan et al., 2012). Expression of PvAcr3;1 from P. vittata in A. thaliana and tobacco (Nicotiana tabacum) was shown to increase arsenic retention in the roots, thereby decreasing arsenic accumulation in the shoots (Chen et al., 2017b).
B. Arsenate efflux systems
1). ArsJ
As described above, As(V) is taken up adventitiously by phosphate transporters and is reduced to As(III) to become the substrate of the ArsB, Acr3 or ArsK efflux permeases. If first methylated to MAs(III) it becomes the substrate of ArsP or ArsK permeases. Until recently there were no known efflux permeases that confer resistance to pentavalent As(V).
A new bacterial mechanism for As(V) tolerance was identified that in effect transports As(V) out of cells even though As(V) is not the actual substrate (Chen et al., 2016). Two genes, gapdh and arsJ, encoding the glycolytic enzyme glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and a predicted MFS protein (Figure 4), respectively were found to link together in ars operons in several bacterial species. The two genes were cloned from the chromosome of Pseudomonas aeruginosa DK2 and expressed in E. coli. Together, but not individually, they conferred resistance to As(V) but not to As(III) or any trivalent organoarsenical. These results indicated that resistance is specific to As(V) and that the products of the two genes work synergistically to enhance As(V) resistance. Everted membrane vesicles prepared from cells expressing arsJ accumulated As(V) when purified rabbit GAPDH and its substrates were present. These results indicate that ArsJ transports an As(V)-containing product of the GAPDH reaction. ArsJ is a 410-residue membrane protein with 10 predicted TMs (Figure 3B). It has a large putative extracellular loop between TMs 3 and 4, and a smaller intracellular loop between TMs 4 and 5. It is possible that the some of these form two additional TMs, which would give ArsJ 12 TMs, similar to other MFS transporters. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) plays a major role in glycolysis. It fulfills a catabolic function, allowing oxidative phosphorylation of glyceraldehyde-3-phosphate (G3P) into1,3-bisphosphoglycerate (1,3-BPG) and reduction of NAD+ to NADH (Iddar et al., 2002). GAPDH can use As(V) instead of inorganic phosphate (Pi), converting G3P into 1-arseno-3-phosphoglycerate (1As3PGA) rather than 1,3-BPG (Byers et al., 1979). This arsenylated metabolite is extremely unstable and spontaneously hydrolyzes with a half-life of less than 2.5s. In everted membrane vesicles, As(V) uptake was observed only when purified GAPDH, G3P, and NADH were present. Little uptake occurred in the absence of GAPDH, G3P or NAD+ (Chen et al., 2016). Thus the GAPDH reaction is required for As(V) accumulation in vesicles. Energy-dependent transport by ArsJ was confirmed by addition of an uncoupling agent of the proton motive force. Since the genes for ArsJ and GAPDH are linked, it seemed reasonable to consider that ArsJ could be an efflux system for 1As3PGA. After uptake by Pi transporters, intracellular As(V) would be converted to 1As3PGA by GAPDH, followed by extrusion from cells by ArsJ, conferring As(V) resistance. Once outside the cell, 1As3PGA immediate hydrolyzes to generate external As(V), making ArsJ effectively an As(V) efflux system. Coupling of a glycolytic enzyme and an organoarsenical efflux permease is a novel transport pathway for As(V) resistance. To date ArsJ represents the sole efflux permease for a pentavalent arsenical.
Figure 4.
A neighbor-joining phylogenetic tree showing the evolutionary relationship of MFS proteins encoded in ars operons. To obtain sequences of mfs1 genes encoding candidate orthologs, ArsJ (Shewanella putrefaciens 200), WP_014610147, was used for BLAST. To obtain sequences of mfs2 genes encoding candidate orthologs, ArsK (Agrobacterium tumefaciens GW4), KDR86814, was used for BLAST. To obtain sequences of mfs3 genes encoding candidate orthologs, MFS3 from Cupriavidus metallidurans CH34, WP_011515208, was used for BLAST. Only mfs genes in arsenic operons were included for phylogenetic analysis. ArsJ, predicted to transport the organorarsenical 1-arseno-3-phosphoglycerate (1As, 3PGA), is a found in the MFS1 branch. ArsK, which has broad substrate specificity for both inorganic and organic trivalent arsenicals, is found in the MFS2 branch. Substrates of MFS3 and MFS4 transporters have not yet been determined. The tree was produced by MEGA7. The bacterial species and protein accession numbers are shown.
C. Methylarsenite efflux systems
1). ArsP
While ArsB and Acr3 have been recognized as the dominant efflux permeases for inorganic arsenic in most bacteria, they do not transport organoarsenicals. ArsP was identified as an MAs(III)-selective permease. ArsP was first identified as encoded by an ars operon of Campylobacter jejuni (Shen et al., 2014; Wang et al., 2009). ArsP was originally reported to be an organic arsenic transporter for the pentavalent forms of roxarsone and nitarsone (Shen et al., 2014). The substrate specificity was subsequently demonstrated to be for trivalent organoarsenicals (Chen et al., 2015). E. coli expressing CjarsP conferred high resistance to MAs(III) and Rox(III) but not to inorganic As(III) or pentavalent organoarsenicals. Everted membrane vesicles from those cells accumulated MAs(III) energized by the proton motive force, consistent with ArsP catalyzing efflux from cells. ArsP was selective for the natural product MAs(III) over synthetic trivalent aromatic arsenicals such as Rox(III). ArsP is unrelated to other ars operon permeases (Figure 2) and is the smallest, with only 315 residues. It is smaller than MFS transporters and is predicted to have only 8 TMs (Figure 3B), which is atypical for bacterial permeases. There is a vicinal cysteine pair, Cys65 and Cys67, present in a highly conserved motif (TPFCSCSXXP) in the middle of predicted TM2 that appears to be involved in the transport reaction. Vicinal cysteine pairs form strong binding sites for arsenicals. Given the substrate specificity, it would have to be a binding site for MAs(III) and not As(III). If one assumes that TM2 is a straight α-helix, the distance between the two thiols would be too long to form a binding site for MAs(III), where the As-S distance would be predicted to be on the order of 2.2 – 2.4 Å. However, there are proline residues on each side of the two cysteine residues that could produce a bend in the helix and bring the thiols close enough to each other to bind MAs(III) on the cytosolic side of the membrane. Reorientation of the binding site to the medium side of the membrane could be predicted to allow the helix to straighten, releasing MAs(III) into the external medium (Chen et al., 2015).
2). ArsK:
Quite recently a novel MFS transporter (Figure 4), ArsK, was identified in an arsenic gene island in the chromosome of Agrobacterium tumefaciens GW4 (Shi et al., 2018) and a Bacillus sp. strain (Jia et al., 2019). ArsK is a 411 amino acid residue transmembrane protein that can be predicted to have 12 TMs (Figure 3A). Genome analysis showed ArsK orthologs are widely distributed in arsenic-resistant bacteria, but not in archaea, fungi, plants or animals. ArsK is in a distinct group that is highly phylogenetically divergent from other arsenic transporters. ArsK confers resistance to As(III), Sb(III), Rox(III) and MAs(III) by reducing cellular accumulation of these compounds. It does not confer resistance to As(V), DMAs(III) or Cd(II). There are three cysteine residues in ArsK from A. tumefaciens GW4, but only one cysteine is conserved in other putative orthologs. Whether this cysteine residue is involved in ArsK function was not investigated. The ars operon containing arsK is negatively regulated by an ArsR repressor. ArsK expression is induced by As(III), Sb(III), Rox(III) and MAs(III) and As(V) (perhaps after reduction to As(III)). ArsK has the broadest substrate specificity of any known trivalent arsenic efflux permease, including both inorganic and organic species.
D. ABC transporters
The ATP-binding cassette (ABC) superfamily is one of the oldest group of transporters found in species of every kingdom. Some of these ATP-coupled pumps confer resistance to multiple drugs and metals, including As(III). ABC transporters normally contain 12–14 transmembrane helices and are among the biggest membrane super families across all domains of life (Rees et al., 2009). ABC transporters facilitating arsenic extrusion have been found in several eukaryotes. One example is the S. cerevisiae ABC pump Ycf1p that was originally isolated as a cadmium resistance transporter. Ycf1p pumps the Cd-glutathione complex (Cd(GS)2) into the yeast vacuole, and this sequestration confers a resistant phenotype. Ycf1p also transports As(III) and Sb(III) from the cytoplasm into the vacuole, as the glutathione adducts As(GS)3 and Sb(GS)3. In addition to Ycf1p, S. cerevisiae has a cluster of three ACR genes that also confer arsenic resistance (Bobrowicz et al., 1997). Acr1p is a transcription factor, and Acr2p is the yeast As(V) reductase. Acr3p is a plasma membrane carrier protein that catalyzes extrusion of As(III) from cytosol. Thus, these two systems are independent of each other and provide parallel pathways for arsenic detoxification in yeast. Acr3p homologs are present in bacteria and archaea but have not yet been found in mammals.
The related mammalian liver ABC pump is MRP2, which belongs to the multidrug resistance-associated protein (MRP) group of the ABC superfamily. Like the yeast pumps, it extrudes arsenic–glutathione complexes into bile and may be a major route of arsenic detoxification in human liver, and it is responsible for biliary excretion of not only As(GS)3 but also the methylated species, MAs(GS)2 (Leslie 2011). These findings suggest that arsenic excretion by MRP transporter is in part dependent on GSH. Previous studies show that the liver plays a primary role in inorganic As metabolism (Drobná et al., 2010). Liver cells methylate arsenic to MAs(III), DMA(III), and their pentavalent forms. In addition, the capacity of hepatocytes to metabolize inorganic As is closely linked to the expression of specific membrane transporters. To date, several MRP orthologs have been shown to be involved in arsenic transport. In vivo studies show that MRP1 is also responsible for biliary excretion of As(GS)3 and MA(GS)2. However, despite the roles MRP1 and MRP2 play in protecting cells and tissues from arsenic, neither protein is localized to the basolateral surface of human hepatocytes (Banerjee et al., 2014). Thus, MRP1 and MRP2 are not responsible for the transport of hepatic arsenic metabolites into sinusoidal blood, leading to urinary clearance. Instead, other transporters of this family such as MRP3, MRP4, MRP5 are expressed at the basolateral surface of hepatocytes. Interestingly, MRP3–5 are not capable of reducing the cytotoxicity of inorganic arsenic. HEK293 cells overexpressing MRP3 or MRP5 did not exhibit resistance to As(III), As(V), MAs(III), MAs(V), DMAs(III), or DMAs(V), suggesting that MRP3 and MRP5 do not provide cellular protection against arsenicals. MRP4 reduced the toxicity and cellular accumulation of As(V), MAs(V), and DMAs(III), but were not directly transported by MRP4. Instead, MRP4 resistance to As(V) was completely GSH-dependent, suggesting that MRP4 reduced the toxicity and accumulation of As(V) by extruding As-GSH complexes. MRP4 was also found to transport DMAs(V), elucidating a novel detoxification pathway for this As metabolite. MRP expression levels in tissues of humans and rats has been characterized, and differences in expression of the individual MRP family members exist in various tissues, with age, and with gender (Maher et al., 2005). Humans have high expression of MRP1 in lung, bladder, spleen, testes, thyroid, and adrenal glands, and little variation among these tissues. Interestingly, MRP1 was not expressed in the liver (Sugawara et al., 1997), whereas MRP2 is highly expressed in the liver and intestine. MRP3–5 are expressed in the liver; however, tissues where the most expression is observed is different between each transporter, with such as the large intestine (MRP3), kidney and lung (MRP4), and skeletal muscle (MRP5).
In plants, phytochelatins (PCs) are glutathione-derived peptides that chelate heavy metals and metalloids such as arsenic, thereby functioning as the first step in their detoxification (Dhankher et al., 2002). Most of the As(III) in the root and shoot tissues of plants is coordinated with the sulfhydryl groups of thiol-rich peptides such as glutathione (GSH) and phytochelatins (PCs), as has been observed in A. thaliana and Brassica juncea (Schmöger et al., 2000).
Like GSH complexes, metal(loid)–PC complexes are sequestered into vacuoles, protecting cellular components against exposure. The most prominent complexes are (GS)As(III)-PC2, As(III)-PC3, As(III)-(PC2)2, and As(GS)3. In A. thaliana, two ABC transporters were identified that are required for arsenic detoxification (Song et al., 2010). In the absence of the ABCC-type transporters AtABCC1 and AtABCC2, A. thaliana is extremely sensitive to arsenic and arsenic-based herbicides. Heterologous expression of these ABCC transporters in phytochelatin-producing S. cerevisiae also enhanced arsenic tolerance and accumulation. Thus, AtABCC1 and AtABCC2 are major vacuolar PC transporters.
In addition to the previously discussed Lsi1 transporter, which catalyzes uptake of As(III) and Sb(III), a second Si(OH)4 transporter, Lsi2, was reported in rice (Ma et al., 2007a). Lsi2 shows homology to plant citrate-efflux transporters and to the bacterial As(III) efflux transporter ArsB. E. coli ArsB and Lsi2 are both efflux transporters of arsenic and antimony. Lsi2 is localized to the proximal side of the plasma membranes of the exodermis and endodermis cells, allowing solute efflux toward the stele for xylem loading. Mutations in Lsi2 had a dramatic effect on As(III) transport to the xylem and As accumulation in the shoots. However, rice Lsi2 is not involved in the efflux of MAs or DMAs, possibly because most MAs and DMAs is dissociated at cytoplasmic pH (Ma et al., 2008b).
Another C-type ABC transporter (OsABCC), OsABCC1 was found to be involved in the detoxification and reduction of As in rice grains (Song et al., 2014). OsABCC1 is localized to the phloem region of vascular bundles. Furthermore, OsABCC1 is localized to the tonoplast and confers phytochelatin-dependent As resistance in yeast. In addition to As detoxification, OsABCC1 is also involved in reducing the allocation of As to the rice grain, particularly during the reproductive growth stage. Knockout of OsABCC1 in rice resulted in decreased tolerance to As and significantly lower As concentrations in the grain. OsABCC1 in the upper nodes of rice plants restricts distribution of As to the grain by sequestering it in the vacuoles of the phloem companion cells of diffuse vascular bundles directly connected to the grain. Overexpression of AtPCS1 and AtABCC1 resulted in plants exhibiting increased arsenic tolerance. Modification of ABCC transporter and PCS expression in plants may therefore enable future engineering of reduction in the accumulation of arsenic in edible organs of plants, a strategy for limiting As accumulation in rice grains and thereby reducing human As exposure.
Summary and future prospects
Arsenic efflux systems are found in nearly every organism and evolved to rid cells of this toxic metalloid. As described in this review, the most frequently employed stratagem for resistance is efflux of inorganic and organic arsenicals from cells (Figure 5). The majority of these transporters are related to ArsB, Acr3, ArsP, ArsK, ArsJ or AqpS that confer resistance to environmental As(III), As(V) and MAs(III) and to synthetic trivalent aromatic arsenicals. The existence of multiple permeases demonstrates that life evolves multiple solutions to similar environmental challenges.
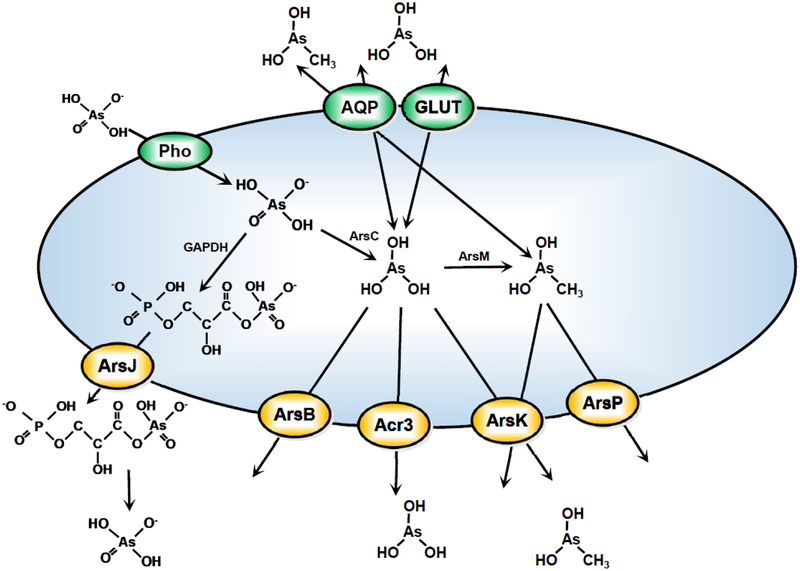
Figure 5. Model of transporters.
As arsenic compounds adventitiously enter the cell through nutrient transporters such as aquaglyceroporins (AQP), glucose permeases (GLUT) or phosphate transporters (Pho), they are either directly extruded by specialized arsenic efflux pumps such as ArsB, Acr3, ArsK or ArsP, or are transformed by enzymes such as ArsC, ArsM, or GAPDH prior to efflux.
Even as we learn more about these known efflux systems, it is obvious that there are probably more. As whole genome sequencing has become increasingly available, a number of additional ars genes that encode membrane proteins of as-yet unknown function have been observed. Approximately 2,500 genomes have been analyzed to date, with more added daily, and over 700 ars genes encoding membrane proteins have been identified. Not all of these fall into known classes. We predict that some, if not all, of these are transporters for inorganic or organic arsenicals. The recent identification of ArsK as a broad-substrate efflux permease is an example. However, the identification of ArsJ as an organoarsenical efflux permease for 1As3PGA shows that the substrates or physiological function of putative permeases cannot be deduced solely from its presence in ars operons. Originally annotated simply as a membrane protein in the major facilitator family, without additional information – in this case, association of the arsJ and gapdh genes in the ars operons – it is doubtful that the substrate of the ArsJ permease would have been identified.
The major facilitator superfamily is an ancient and ubiquitous transporter group, consisting of more than 15,000 sequenced members, and this number is growing rapidly with the continuing emergence of genome sequences (Reddy et al., 2012). Members of the MFS have an extraordinarily broad spectrum of substrates, including inorganic and organic ions, nucleosides, amino acids, short peptides, and lipids. MFS members comprise facilitators, symporters, and antiporters that move substrates across membranes via facilitated diffusion, cotransport, or exchange. Genomics analysis reveals at least four distinct MFS clusters (Figure 4). ArsJ and ArsK are members two of the clusters, and other putative arsenic transporters belong to two other clusters. However, there are no experimental data that demonstrate involvement of those uncharacterized permeases in arsenic resistance. For example, an MFS3 transporter has been designated as arsQ in Rhodobacter sphaeroides ATCC17025, but an arsenic-related function is not known (Chauhan et al., 2009). In our MFS phylogenetic tree analysis, it appears to be more closely related to the MFS4 subgroup. Another MFS gene is found downstream of the gapdharsJ genes in Halomonas sp. strain GFAJ-1 (Wu et al., 2018). This MFS transporter likely belongs to the MFS3 group. Another representative of MFS3 is a transporter in an ars operon found in Cupriavidus metallidurans CH34. MFS3 transporters have several highly conserved sequences, but there is only one conserved cysteine residue, suggesting that it is not an ArsP-like MAs(III) transporter, which has two conserved cysteines in its MAs(III) binding site (Chen et al., 2015). MFS4 transporter genes are not as widely distributed as other ars operon MFS genes. MFS transporters in ars operon are quite different in terms of their diversity and occurrence, suggesting that they may have diversity in arsenical substrate specificity.
In summary, a general observation is that arsenic uptake systems are adventitious. For the most part, organisms would prefer not to accumulate arsenic, but both As(V) and As(III) As(V) enter cells via uptake systems for other nutrients and minerals such as glucose permeases and aquaglyceroporins. On the other hand, arsenic efflux systems show considerable specificity and appear to have evolved to confer resistance to inorganic and organic arsenicals. However, members of the MRP subfamily of the ABC superfamily are nonspecific arsenic conjugate efflux pumps. With only one exception, the substrates of these efflux systems are trivalent arsenicals, reflecting their early evolution in an anoxic world. The only efflux system identified for a pentavalent arsenical is ArsJ, which extrudes the very unstable pentavalent organoarsenical 1-arseno-3-phosphoglycerate, which rapidly hydrolyzes to release free As(V) outside of the cell, conferring As(V) resistance.
There is still much to be learned about arsenic transport. First, the identification of ArsJ and genes for other MFS transporters illustrates that there may be other novel and highly unusual permeases to be discovered. Second, the molecular mechanism of arsenic transport has yet to be determined. Efflux permeases like ArsB and Acr3 do not appear to use cysteine motifs for binding of As(III). Cysteine pairs or triads form binding sites for trivalent arsenicals in ArsR repressors, the ArsA ATPase subunit of the ArsAB pump, and the soluble enzymes of arsenic biotransformation ArsM and ArsI. It was thought that if membrane proteins such as ArsB or Acr3 used vicinal cysteine pairs for substrate binding, they might bind trivalent arsenical with such high affinity that they could not be released, yet ArsP uses a vicinal cysteine pair to bind MAs(III). The binding helix is predicted to have a bend that brings the two cysteines close enough to each other to bind MAs(III) on the cytosolic side of the membrane. In a conformational change, the helix straightens out on the periplasmic side, releasing the highly toxic organoarsenical into the medium. Thus, the transport mechanism cannot be predicted from primary sequence. A third remaining obstacle is the lack of structural information about arsenic transporters. The two-dimensional transmembrane structures have been estimated from a combination of molecular and chemical methods for ArsB and Acr3 (Figure 3A), and can be predicted from various algorithms for ArsK, ArsJ and ArsP (Figure 3B). The number of TMs appears to be variable, from 8–12. However, the predictions for ArsK, ArsJ and ArsP have not been experimentally verified, and their actual transmembrane topology could be different. Except for ArsP, the topology provides little predictive value about the substrate binding site. The arrangement of the helices in the membrane and the localization of specific residues is absolutely required to deduce a catalytic mechanism. This has been accomplished for membrane proteins such as the lactose permease, potassium channel, bacteriorhodopsin, to name only a few. The number of crystal structures of membrane proteins is increasing rapidly, and it is only a matter of time before this is expanded to elucidate the structure and function of arsenic permeases.
Highlights:
Arsenic is a toxic metalloid that enters cells adventitiously through uptake systems for nutrients such phopshate permeases (arsenate), and or aquaglyceroporins or glucose permeases (arsenite).
Cells detoxify arsenic primarily by extrusion from cells.
Efflux permeases for arsenite include ArsB and Acr3.
Efflux permeases for methylarsenite include ArsP and ArsK.
Novel efflux proteins ArsJ and AqpS confer resistance to arsenate even though it is not the substrate of those transporters.
Funding:
This work was supported by grants GM55425 and ES023779 from the National Institutes of Health.
Abbreviations:
- ABC
ATP-binding cassette
- As(III)
arsenite
- As(V)
arsenate
- AQP
aquaporin
- DMAs(III)
dimethylarsenite
- GSH
glutathione
- GAPDH
glyceraldehyde 3-phosphate dehydrogenase, methylarsenite
- MAs(III)
methylarsenite
- MAs(V)
methylarsenate
- MFS
major facilitator superfamily
- PC
phytochelatin
- Sb(III)
antimonite
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declarations of interest: none.
References:
- Abernathy CO; Thomas DJ; Calderon RL Health Effects and Risk Assessment of Arsenic. J Nutr 2003;133:1536S–1538S [DOI] [PubMed] [Google Scholar]
- Achour AR; Bauda P; Billard P Diversity of arsenite transporter genes from arsenic-resistant soil bacteria. Res Microbiol 2007;158:128–137 [DOI] [PubMed] [Google Scholar]
- Agre P; King LS; Yasui M; Guggino WB; Ottersen OP; Fujiyoshi Y; Engel A; Nielsen S Aquaporin water channels – from atomic structure to clinical medicine. J Physiol 2002;542:3–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander J; Benford D; Boobis A; Ceccatelli S; Cravedi J-P; Di Domenico A; Doerge D; Dogliotti E; Edler L; Farmer P; Filipič M; Fink-Gremmels J; Fürst P; Guerin T; Knutsen HK; Machala M; Mutti A; Schlatter J; van Leeuwen R; Verger P Scientific Opinion on Arsenic in Food. EFSA Journal 2009;7:1351 [Google Scholar]
- Andrewes P; DeMarini DM; Funasaka K; Wallace K; Lai VWM; Sun H; Cullen WR; Kitchin KT Do Arsenosugars Pose a Risk to Human Health? The Comparative Toxicities of a Trivalent and Pentavalent Arsenosugar. Environ Sci Technol 2004;38:4140–4148 [DOI] [PubMed] [Google Scholar]
- Banerjee M; Carew MW; Roggenbeck BA; Whitlock BD; Naranmandura H; Le XC; Leslie EM A Novel Pathway for Arsenic Elimination: Human Multidrug Resistance Protein 4 (MRP4/ABCC4) Mediates Cellular Export of Dimethylarsinic Acid (DMAV) and the Diglutathione Conjugate of Monomethylarsonous acid (MMAIII). Mol Pharmacol 2014:mol.113.091314 [DOI] [PubMed] [Google Scholar]
- Ben Fekih I; Zhang C; Li YP; Zhao Y; Alwathnani HA; Saquib Q; Rensing C; Cervantes C Distribution of Arsenic Resistance Genes in Prokaryotes. Front Microbiol 2018;9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharjee H; Carbrey J; Rosen BP; Mukhopadhyay R Drug uptake and pharmacological modulation of drug sensitivity in leukemia by AQP9. Biochem Biophys Res Commun 2004;322:836–841 [DOI] [PubMed] [Google Scholar]
- Bhattacharjee H; Mukhopadhyay R; Thiyagarajan S; Rosen BP Aquaglyceroporins: ancient channels for metalloids. J Biol 2008;7:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharjee H; Sheng J; Ajees AA; Mukhopadhyay R; Rosen BP Adventitious Arsenate Reductase Activity of the Catalytic Domain of the Human Cdc25B and Cdc25C Phosphatases. Biochemistry 2010;49:802–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobrowicz P; Wysocki R; Owsianik G; Goffeau A; Ułaszewski S Isolation of Three Contiguous Genes, ACR1, ACR2 and ACR3, Involved in Resistance to Arsenic Compounds in the Yeast Saccharomyces cerevisiae. Yeast 1997;13:819–828 [DOI] [PubMed] [Google Scholar]
- Bordo D; Bork P The rhodanese/Cdc25 phosphatase superfamily. EMBO Reports 2002;3:741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byers LD; She H,S; Alayoff A. Interaction of Phosphate Analogues with Glyceraldehyde-3-phosphate Dehydrogenase. Biochemistry 1979;18:2471–2480 [DOI] [PubMed] [Google Scholar]
- Caille N; Zhao FJ; McGrath SP Comparison of root absorption, translocation and tolerance of arsenic in the hyperaccumulator Pteris vittata and the nonhyperaccumulator Pteris tremula. New Phytol 2005;165:755–761 [DOI] [PubMed] [Google Scholar]
- Castrillo G; Sánchez-Bermejo E; de Lorenzo L; Crevillén P; Fraile-Escanciano A; TC M; Mouriz A; Catarecha P; Sobrino-Plata J; Olsson S; Leo del Puerto Y; Mateos I; Rojo E; Hernández LE; Jarillo JA; Piñeiro M; Paz-Ares J; Leyva A WRKY6 Transcription Factor Restricts Arsenate Uptake and Transposon Activation in Arabidopsis. The Plant Cell 2013;25:2944–2957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catarecha P; Segura MD; Franco-Zorrilla JM; García-Ponce B; Lanza M; Solano R; Paz-Ares J; Leyva A A Mutant of the Arabidopsis Phosphate Transporter PHT1;1 Displays Enhanced Arsenic Accumulation. Plant Cell 2007;19:1123–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrakar V; Naithani Subhas C; Keshavkant S Arsenic-induced metabolic disturbances and their mitigation mechanisms in crop plants: A review. Biologia; 2016 [Google Scholar]
- Chandrakar V; Yadu B; Meena RK; Dubey A; Keshavkant S Arsenic-induced genotoxic responses and their amelioration by diphenylene iodonium, 24-epibrassinolide and proline in Glycine max L. Plant Physiol Biochem 2017;112:74–86 [DOI] [PubMed] [Google Scholar]
- Chao D-Y; Chen Y; Chen J; Shi S; Chen Z; Wang C; Danku JM; Zhao F-J; Salt DE Genome-wide Association Mapping Identifies a New Arsenate Reductase Enzyme Critical for Limiting Arsenic Accumulation in Plants. PLoS Biol 2014;12:e1002009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chau D; Ng K; Chan TSY; Cheng YY; Fong B; Tam S; Kwong YL; Tse E Azacytidine sensitizes acute myeloid leukemia cells to arsenic trioxide by up-regulating the arsenic transporter aquaglyceroporin 9. J Hematol Oncol 2015;8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauhan NS; Ranjan R; Purohit HJ; Kalia VC; Sharma R Identification of genes conferring arsenic resistance to Escherichia coli from an effluent treatment plant sludge metagenomic library. FEMS Microbiol Ecol 2009;67:130–139 [DOI] [PubMed] [Google Scholar]
- Chen J; Madegowda M; Bhattacharjee H; Rosen BP ArsP: a methylarsenite efflux permease. Mol Microbiol 2015;98:625–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J; Nadar VS; Rosen BP A novel MAs(III)-selective ArsR transcriptional repressor. Mol Microbiol 2017a;106:469–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J; Yoshinaga M; Garbinski LD; Rosen BP Synergistic interaction of glyceraldehydes-3-phosphate dehydrogenase and ArsJ, a novel organoarsenical efflux permease, confers arsenate resistance. Mol Microbiol 2016;100:945–953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J; Yoshinaga M; Rosen BP The antibiotic action of methylarsenite is an emergent property of microbial communities. Mol Microbiol 2019;111:487–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y; Hua C-Y; Jia M-R; Fu J-W; Liu X; Han Y-H; Liu Y; Rathinasabapathi B; Cao Y; Ma LQ Heterologous Expression of Pteris vittata Arsenite Antiporter PvACR3;1 Reduces Arsenic Accumulation in Plant Shoots. Environ Sci Technol 2017b;51:10387–10395 [DOI] [PubMed] [Google Scholar]
- Chen Y; Xu W; Shen H; Yan H; Xu W; He Z; Ma M Engineering Arsenic Tolerance and Hyperaccumulation in Plants for Phytoremediation by a PvACR3 Transgenic Approach. Environ Sci Technol 2013;47:9355–9362 [DOI] [PubMed] [Google Scholar]
- Deng D; Xu C; Sun P; Wu J; Yan C; Hu M; Yan N Crystal structure of the human glucose transporter GLUT1. Nature 2014;510:121. [DOI] [PubMed] [Google Scholar]
- Dhankher OP; Li Y; Rosen BP; Shi J; Salt D; Senecoff JF; Sashti NA; Meagher RB Engineering tolerance and hyperaccumulation of arsenic in plants by combining arsenate reductase and γ-glutamylcysteine synthetase expression. Nat Biotechnol 2002;20:1140. [DOI] [PubMed] [Google Scholar]
- DiTusa SF; Fontenot EB; Wallace RW; Silvers MA; Steele TN; Elnagar AH; Dearman KM; Smith AP A member of the Phosphate transporter 1 (Pht1) family from the arsenic-hyperaccumulating fern Pteris vittata is a high-affinity arsenate transporter. New Phytol 2016;209:762–772 [DOI] [PubMed] [Google Scholar]
- Dopp E; Hartmann LM; von Recklinghausen U; Florea AM; Rabieh S; Zimmermann U; Shokouhi B; Yadav S; Hirner AV; Rettenmeier AW Forced Uptake of Trivalent and Pentavalent Methylated and Inorganic Arsenic and Its Cyto-/genotoxicity in Fibroblasts and Hepatoma Cells. Toxicol Sci 2005;87:46–56 [DOI] [PubMed] [Google Scholar]
- Drobná Z; Del Razo LM; García-Vargas GG; Sánchez-Peña LC; Barrera-Hernández A; Stýblo M; Loomis D Environmental exposure to arsenic, AS3MT polymorphism and prevalence of diabetes in Mexico. Journal Of Exposure Science And Environmental Epidemiology 2013;23:151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drobná Z; Walton FS; Paul DS; Xing W; Thomas DJ; Stýblo M Metabolism of arsenic in human liver: the role of membrane transporters. Arch Toxicol 2010;84:3–16 [DOI] [PubMed] [Google Scholar]
- Duan G-L; Hu Y; Schneider S; McDermott J; Chen J; Sauer N; Rosen BP; Daus B; Liu Z; Zhu Y-G Inositol transporters AtINT2 and AtINT4 regulate arsenic accumulation in Arabidopsis seeds. Nat Plants 2016;2:15202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan G; Kamiya T; Ishikawa S; Arao T; Fujiwara T Expressing ScACR3 in Rice Enhanced Arsenite Efflux and Reduced Arsenic Accumulation in Rice Grains. Plant Cell Physiol 2012;53:154–163 [DOI] [PubMed] [Google Scholar]
- Fu D; Libson A; Miercke LJW; Weitzman C; Nollert P; Krucinski J; Stroud RM Structure of a Glycerol-Conducting Channel and the Basis for Its Selectivity. Science 2000;290:481–486 [DOI] [PubMed] [Google Scholar]
- Fu H-L; Meng Y; Ordóñez E; Villadangos AF; Bhattacharjee H; Gill JA; Mateos LM; Rosen BP Properties of arsenite efflux permeases (Acr3) from Alkaliphilus metalliredigens and Corynebacterium glutamicum. J Biol Chem 2009;284:19887–19895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg N; Singla P Arsenic toxicity in crop plants: physiological effects and tolerance mechanisms. Environ Chem Lett 2011;9:303–321 [Google Scholar]
- Hibuse T; Maeda N; Nagasawa A; Funahashi T Aquaporins and glycerol metabolism. Bba-Biomembranes 2006;1758:1004–1011 [DOI] [PubMed] [Google Scholar]
- Huang DD; Feng XZ; Liu YT; Deng YH; Chen H; Chen DC; Fang LK; Cai Y; Liu HL; Wang L; Wang JP; Yang ZH AQP9-induced cell cycle arrest is associated with RAS activation and improves chemotherapy treatment efficacy in colorectal cancer. Cell Death Dis 2017;8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang K; Xu Y; Packianathan C; Gao F; Chen C; Zhang J; Shen Q; Rosen BP; Zhao F-J Arsenic methylation by a novel ArsM As(III) S-adenosylmethionine methyltransferase that requires only two conserved cysteine residues. Mol Microbiol 2018;107:265–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iddar A; Serrano A; Soukri A A phosphate-stimulated NAD(P)+-dependent glyceraldehyde-3-phosphate dehydrogenase in Bacillus cereus. FEMS Microbiol Lett 2002;211:29–35 [DOI] [PubMed] [Google Scholar]
- Indriolo E; Na G; Ellis D; Salt DE; Banks JA A Vacuolar Arsenite Transporter Necessary for Arsenic Tolerance in the Arsenic Hyperaccumulating Fern Pteris vittata Is Missing in Flowering Plants. Plant Cell 2010;22:2045–2057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ismail GSM Protective role of nitric oxide against arsenic-induced damages in germinating mung bean seeds. Acta Physiol Plant 2012;34:1303–1311 [Google Scholar]
- Jia M-R; Tang N; Cao Y; Chen Y; Han Y-H; Ma LQ Efficient arsenate reduction by As-resistant bacterium Bacillus sp. strain PVR-YHB1–1: Characterization and genome analysis. Chemosphere 2019;218:1061–1070 [DOI] [PubMed] [Google Scholar]
- Jiang W; Hou Q; Yang Z; Zhong C; Zheng G; Yang Z; Li J Evaluation of potential effects of soil available phosphorus on soil arsenic availability and paddy rice inorganic arsenic content. Environ Pollut 2014;188:159–165 [DOI] [PubMed] [Google Scholar]
- Jiang X; McDermott JR; Ajees AA; Rosen BP; Liu Z Trivalent arsenicals and glucose use different translocation pathways in mammalian GLUT1. Metallomics 2010;2:211–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J-W; Xu Y-F; Huang Y-F Protective effect of nitric oxide against arsenic-induced oxidative damage in tall fescue leaves. Afr J Biotechnol 2010;9:1619–1627 [Google Scholar]
- Lebeck J Metabolic impact of the glycerol channels AQP7 and AQP9 in adipose tissue and liver. J Mol Endocrinol 2014;52:R165–178 [DOI] [PubMed] [Google Scholar]
- Leslie EM Arsenic–glutathione conjugate transport by the human multidrug resistance proteins (MRPs/ABCCs). J Inorg Biochem 2011;108:141–149 [DOI] [PubMed] [Google Scholar]
- Li J; Pawitwar SS; Rosen BP The organoarsenical biocycle and the primordial antibiotic methylarsenite. Metallomics 2016;8:1047–1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z; Boles E; Rosen BP Arsenic Trioxide Uptake by Hexose Permeases in Saccharomyces cerevisiae. J Biol Chem 2004;279:17312–17318 [DOI] [PubMed] [Google Scholar]
- Liu Z; Sanchez MA; Jiang X; Boles E; Landfear SM; Rosen BP Mammalian glucose permease GLUT1 facilitates transport of arsenic trioxide and methylarsonous acid. Biochem Biophys Res Commun 2006a;351:424–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z; Shen J; Carbrey JM; Mukhopadhyay R; Agre P; Rosen BP Arsenite transport by mammalian aquaglyceroporins AQP7 and AQP9. Proc Natl Acad Sci USA 2002;99:6053–6058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z; Styblo M; Rosen BP Methylarsonous Acid Transport by Aquaglyceroporins. Environ Health Perspect 2006b;114:527–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma JF; Tamai K; Yamaji N; Mitani N; Konishi S; Katsuhara M; Ishiguro M; Murata Y; Yano M A silicon transporter in rice. Nature 2006;440:688. [DOI] [PubMed] [Google Scholar]
- Ma JF; Yamaji N; Mitani N; Tamai K; Konishi S; Fujiwara T; Katsuhara M; Yano M An efflux transporter of silicon in rice. Nature 2007a;448:209–U212 [DOI] [PubMed] [Google Scholar]
- Ma JF; Yamaji N; Mitani N; Tamai K; Konishi S; Fujiwara T; Katsuhara M; Yano M An efflux transporter of silicon in rice. Nature 2007b;448:209. [DOI] [PubMed] [Google Scholar]
- Ma JF; Yamaji N; Mitani N; Xu X-Y; Su Y-H; McGrath SP; Zhao F-J Transporters of arsenite in rice and their role in arsenic accumulation in rice grain. Proc Natl Acad Sci USA 2008a;105:9931–9935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma JF; Yamaji N; Mitani N; Xu XY; Su YH; McGrath SP; Zhao FJ Transporters of arsenite in rice and their role in arsenic accumulation in rice grain. P Natl Acad Sci USA 2008b;105:9931–9935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maciaszczyk-Dziubinska E; Wawrzycka D; Sloma E; Migocka M; Wysocki R The yeast permease Acr3p is a dual arsenite and antimonite plasma membrane transporter. Biochim Biophys Acta 2012;1798:2170–2175 [DOI] [PubMed] [Google Scholar]
- Maher JM; Slitt AL; Cherrington NJ; Cheng X; Klaassen CD Tissue distribution and hepatic and renal ontogeny of the multidrug resistance-associated protein (Mrp) family in mice. Drug Metab Disposition 2005; [DOI] [PubMed] [Google Scholar]
- Maheshwari R; Dubey RS Nickel toxicity inhibits ribonuclease and protease activities in rice seedlings: protective effects of proline. Plant Growth Regul 2007;51:231–243 [Google Scholar]
- Mansour NM; Sawhney M; Tamang DG; Vogl C; Saier MH Jr The bile/arsenite/riboflavin transporter (BART) superfamily. FEBS J 2007;274:612–629 [DOI] [PubMed] [Google Scholar]
- Meng Y-L; Liu Z; Rosen BP As(III) and Sb(III) Uptake by GlpF and Efflux by ArsB in Escherichia coli. J Biol Chem 2004;279:18334–18341 [DOI] [PubMed] [Google Scholar]
- Mishra S; Dubey RS Inhibition of ribonuclease and protease activities in arsenic exposed rice seedlings: Role of proline as enzyme protectant. J Plant Physiol 2006;163:927–936 [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay R; Bhattacharjee H; Rosen BP Aquaglyceroporins: Generalized metalloid channels. Biochimica et Biophysica Acta (BBA) - General Subjects 2014;1840:1583–1591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay R; Rosen BP Arsenate reductases in prokaryotes and eukaryotes. Environ Health Perspect 2002;110:745–748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navas-Acien A; Silbergeld EK; Streeter RA; Clark JM; Burke TA; Guallar E Arsenic Exposure and Type 2 Diabetes: A Systematic Review of the Experimental and Epidemiologic Evidence. Environ Health Perspect 2006;114:641–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odjegba VJ Exogenous salicylic acid alleviates arsenic toxicity in Arabidopsis thaliana. Indian J Innov Dev 2012;1:515–522 [Google Scholar]
- Omasits U; Ahrens CH; Muller S; Wollscheid B Protter: interactive protein feature visualization and integration with experimental proteomic data. Bioinformatics 2014;30:884–886 [DOI] [PubMed] [Google Scholar]
- Paul DS; Walton FS; Saunders RJ; Stýblo M Characterization of the Impaired Glucose Homeostasis Produced in C57BL/6 Mice by Chronic Exposure to Arsenic and High-Fat Diet. Environ Health Perspect 2011;119:1104–1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planavsky NJ; Reinhard CT; Wang X; Thomson D; McGoldrick P; Rainbird RH; Johnson T; Fischer WW; Lyons TW Low Mid-Proterozoic atmospheric oxygen levels and the delayed rise of animals. Science 2014;346:635–638 [DOI] [PubMed] [Google Scholar]
- Poynton CY; Huang JW; Blaylock MJ; Kochian LV; Elless MP Mechanisms of arsenic hyperaccumulation in Pteris species: root As influx and translocation. Planta 2004;219:1080–1088 [DOI] [PubMed] [Google Scholar]
- Reddy VS; Shlykov MA; Castillo R; Sun EI; Saier MH Jr The major facilitator superfamily (MFS) revisited. FEBS J 2012;279:2022–2035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rees DC; Johnson E; Lewinson O ABC transporters: the power to change. Nature Reviews Molecular Cell Biology 2009;10:218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reizer J; Reizer A; Saier MH The MIP Family of Integral Membrane Channel Proteins: Sequence Comparisons, Evolutionary Relationships, Reconstructed Pathway of Evolution, and Proposed Functional Differentiation of the Two Repeated Halves of the Proteins. Crit Rev Biochem Mol Biol 1993;28:235–257 [DOI] [PubMed] [Google Scholar]
- Rosen BP; Bhattacharjee H; Shi W Mechanisms of metalloregulation of an anion-translocating ATPase. J Bioenerg Biomembr 1995;27:85–91 [DOI] [PubMed] [Google Scholar]
- Rosenberg H; Gerdes RG; Chegwidden K Two systems for the uptake of phosphate in Escherichia coli. J Bacteriol 1977;131:505–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saidi I; Yousfi N; Borgi MA Salicylic acid improves the antioxidant ability against arsenic-induced oxidative stress in sunflower (Helianthus annuus) seedling. J Plant Nutr 2017;40:2326–2335 [Google Scholar]
- Sanders OI; Rensing C; Kuroda M; Mitra B; Rosen BP Antimonite is accumulated by the glycerol facilitator GlpF in Escherichia coli. J Bacteriol 1997;179:3365–3367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmöger MEV; Oven M; Grill E Detoxification of Arsenic by Phytochelatins in Plants. Plant Physiol 2000;122:793–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Z; Han J; Wang Y; Sahin O; Zhang Q The Contribution of ArsB to Arsenic Resistance in Campylobacter jejuni. PLoS One 2013;8:e58894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Z; Luangtongkum T; Qiang Z; Jeon B; Wang L; Zhang Q Identification of a Novel Membrane Transporter Mediating Resistance to Organic Arsenic in Campylobacter jejuni. Antimicrob Agents Chemother 2014;58:2021–2029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi K; Li C; Rensing C; Dai X; Fan X; Wang G Efflux Transporter ArsK Is Responsible for Bacterial Resistance to Arsenite, Antimonite, Trivalent Roxarsone, and Methylarsenite. Appl Environ Microbiol 2018;84:e01842–01818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi S; Wang T; Chen Z; Tang Z; Wu Z; Salt DE; Chao D-Y; Zhao F-J OsHAC1;1 and OsHAC1;2 Function as Arsenate Reductases and Regulate Arsenic Accumulation. Plant Physiol 2016;172:1708–1719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh AP; Dixit G; Kumar A; Mishra S; Kumar N; Dixit S; Singh PK; Dwivedi S; Trivedi PK; Pandey V; Dhankher OP; Norton GJ; Chakrabarty D; Tripathi RD A protective role for nitric oxide and salicylic acid for arsenite phytotoxicity in rice (Oryza sativa L.). Plant Physiol Biochem 2017;115:163–173 [DOI] [PubMed] [Google Scholar]
- Singh AP; Dixit G; Kumar A; Mishra S; Singh PK; Dwivedi S; Trivedi PK; Chakrabarty D; Mallick S; Pandey V; Dhankher OP; Tripathi RD Nitric Oxide Alleviated Arsenic Toxicity by Modulation of Antioxidants and Thiol Metabolism in Rice (Oryza sativa L.). Front Plant Sci 2016;6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh AP; Dixit G; Mishra S; Dwivedi S; Tiwari M; Mallick S; Pandey V; Trivedi PK; Chakrabarty D; Tripathi RD Salicylic acid modulates arsenic toxicity by reducing its root to shoot translocation in rice (Oryza sativa L.). Front Plant Sci 2015a;6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh M; Pratap Singh V; Dubey G; Mohan Prasad S Exogenous proline application ameliorates toxic effects of arsenate in Solanum melongena L. seedlings. Ecotoxicol Environ Saf 2015b;117:164–173 [DOI] [PubMed] [Google Scholar]
- Singh VP; Srivastava PK; Prasad SM Nitric oxide alleviates arsenic-induced toxic effects in ridged Luffa seedlings. Plant Physiol Biochem 2013;71:155–163 [DOI] [PubMed] [Google Scholar]
- Song W-Y; Park J; Mendoza-Cózatl DG; Suter-Grotemeyer M; Shim D; Hörtensteiner S; Geisler M; Weder B; Rea PA; Rentsch D; Schroeder JI; Lee Y; Martinoia E Arsenic tolerance in Arabidopsis is mediated by two ABCC-type phytochelatin transporters. Proc Natl Acad Sci USA 2010;107:21187–21192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song W-Y; Yamaki T; Yamaji N; Ko D; Jung K-H; Fujii-Kashino M; An G; Martinoia E; Lee Y; Ma JF A rice ABC transporter, OsABCC1, reduces arsenic accumulation in the grain. Proc Natl Acad Sci USA 2014;111:15699–15704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugawara I; Akiyama S; Scheper RJ; Itoyama S Lung resistance protein (LRP) expression in human normal tissues in comparison with that of MDR1 and MRP. Cancer Lett 1997;112:23–31 [DOI] [PubMed] [Google Scholar]
- Takano J; Wada M; Ludewig U; Schaaf G; von Wirén N; Fujiwara T The Arabidopsis Major Intrinsic Protein NIP5;1 Is Essential for Efficient Boron Uptake and Plant Development under Boron Limitation. Plant Cell 2006;18:1498–1509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamás MJ; Rep M; Thevelein JM; Hohmann S Stimulation of the yeast high osmolarity glycerol (HOG) pathway: evidence for a signal generated by a change in turgor rather than by water stress. FEBS J 2000;472:159–165 [DOI] [PubMed] [Google Scholar]
- Taylor V; Goodale B; Raab A; Schwerdtle T; Reimer K; Conklin S; Karagas MR; Francesconi KA Human exposure to organic arsenic species from seafood. Sci Total Environ 2017;580:266–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villadangos AF; Fu H-L; Gil JA; Messens J; Rosen BP; Mateos LM Efflux Permease CgAcr3–1 of Corynebacterium glutamicum Is an Arsenite-specific Antiporter. J Biol Chem 2012;287:723–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C; Na G; Bermejo ES; Chen Y; Banks JA; Salt DE; Zhao F-J Dissecting the components controlling root-to-shoot arsenic translocation in Arabidopsis thaliana. New Phytol 2018;217:206–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L; Jeon B; Sahin O; Zhang Q Identification of an Arsenic Resistance and Arsenic-Sensing System in Campylobacter jejuni. Appl Environ Microbiol 2009;75:5064–5073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willsky GR; Malamy MH Effect of arsenate on inorganic phosphate transport in Escherichia coli. J Bacteriol 1980;144:366–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S; Wang L; Gan R; Tong T; Bian H; Li Z; Du S; Deng Z; Chen S Signature Arsenic Detoxification Pathways in Halomonas sp. Strain GFAJ-1. mBio 2018;9:e00515–00518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wysocki R; Chéry CC; Wawrzycka D; Van Hulle M; Cornelis R; Thevelein JM; Tamás MJ The glycerol channel Fps1p mediates the uptake of arsenite and antimonite in Saccharomyces cerevisiae. Mol Microbiol 2001;40:1391–1401 [DOI] [PubMed] [Google Scholar]
- Xu J; Shi S; Wang L; Tang Z; Lv T; Zhu X; Ding X; Wang Y; Zhao F-J; Wu Z OsHAC4 is critical for arsenate tolerance and regulates arsenic accumulation in rice. New Phytol 2017;215:1090–1101 [DOI] [PubMed] [Google Scholar]
- Xu W; Dai W; Yan H; Li S; Shen H; Chen Y; Xu H; Sun Y; He Z; Ma M Arabidopsis NIP3;1 Plays an Important Role in Arsenic Uptake and Root-to-Shoot Translocation under Arsenite Stress Conditions. Molecular Plant 2015;8:722–733 [DOI] [PubMed] [Google Scholar]
- Yadav G; Srivastava PK; Singh VP; Prasad SM Light Intensity Alters the Extent of Arsenic Toxicity in Helianthus annuus L. Seedlings. Biol Trace Elem Res 2014;158:410–421 [DOI] [PubMed] [Google Scholar]
- Yang H-C; Cheng J; Finan TM; Rosen BP; Bhattacharjee H Novel Pathway for Arsenic Detoxification in the Legume Symbiont Sinorhizobium meliloti. J Bacteriol 2005;187:6991–6997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H-C; Fu H-L; Lin Y-F; Rosen BP Chapter Twelve - Pathways of Arsenic Uptake and Efflux in: Argüello JM, Lutsenko S, eds. Curr Top Membr: Academic Press; 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H-C; Rosen BP New mechanisms of bacterial arsenic resistance. Biomedical Journal 2016;39:5–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshinaga M; Rosen BP A C. As lyase for degradation of environmental organoarsenical herbicides and animal husbandry growth promoters. Proc Natl Acad Sci USA 2014;111:7701–7706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao F-J; McGrath SP; Meharg AA Arsenic as a Food Chain Contaminant: Mechanisms of Plant Uptake and Metabolism and Mitigation Strategies. Annu Rev Plant Biol 2010;61:535–559 [DOI] [PubMed] [Google Scholar]
- Zhao FJ; Ma JF; Meharg AA; McGrath SP Arsenic uptake and metabolism in plants. New Phytol 2009;181:777–794 [DOI] [PubMed] [Google Scholar]
- Zhou T; Radaev S; Rosen BP; Gatti DL Structure of the ArsA ATPase: the catalytic subunit of a heavy metal resistance pump. EMBO J 2000;19:4838–4845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y-G; Xue X-M; Kappler A; Rosen BP; Meharg AA Linking Genes to Microbial Biogeochemical Cycling: Lessons from Arsenic. Environ Sci Technol 2017;51:7326–7339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y-G; Yoshinaga M; Zhao F-J; Rosen BP Earth Abides Arsenic Biotransformations. Annu Rev Earth and Planet Sci 2014;42:443–467 [DOI] [PMC free article] [PubMed] [Google Scholar]