Abstract
We demonstrate a novel strategy to engineer double-headed nanosystems by chemical modification of the carboxyl terminal polyester with a linker that offers tripodal arrangement of ligands on particle surface. The ex vivo and in vivo results suggest that the bioavailability is proportional to the ligand density rendered by double-headed nanosystems.
Graphical Abtsract
In treating any disease by oral medication a fundamental problem is the delivery of molecules across the gastrointestinal tract (GIT) so that they reach their target site in minimum therapeutic concentrations.1,2 Biodegradable nanosystems are expected to lead innovative therapeutic and diagnostic methods.3,4 In order to meet the changing paradigm in drug delivery, innovation in biomaterials is as important as discovering new drugs.5–8 Several approaches to drug delivery utilizing passive, active and physiologically responsive systems have been attempted.9–12 The best results are currently obtained when ligands that bind cell surface receptors decorate the outer shell of nanosystems.13–15 In such cases, the ligand-receptor stoichiometry plays an essential role in receptor-mediated drug delivery.16,17 Till date, in receptor-mediated drug delivery application of nanosystems, we and others have used terminal functional polyesters such as polylactide-co-glycolide (PLGA) that involves coupling with single ligand through single-headed linkers.18–20
The tripodal arrangement of ligands on a scaffold is considered ideal for molecular recognition for cell surfaces and biological applications.21 Tris(2-aminoethyl) amine (TREN), a flexible C3 symmetric linker, has been extensively utilized in co-ordination chemistry as tri- or tetradentate ligand for various metal ion interactions and sensor based applications.22–25 It has been reported that the flexibility of TREN assisted in self-assembly of the collagen mimetic scaffold due to effective adjusting of the three peptide chains to form triple helical packing.26 The TREN binding site in the artificial L-serine derivative based metallocatalyts has been reported to act as allosteric regulation sites due to conformation change upon metal binding.27 TREN-based materials also find applications in areas such as recognition and separation of tetrahedral oxo-anions,28 as a membrane transporter for chloride/bicarbonate anions,29 and as a T-junction to create self-assembled nanostructures for dual-drug delivery.30
We, herein, describe for the first time the design and development of double-headed nanosystems using TREN as a linker to carboxyl terminal end of PLGA that permits coupling with two-fold higher ligand densities as opposed to single head. In this study, we have used gambogic acid (GA) as a ligand specific for transferrin receptors (TfR) found in all regions of the intestinal tract.20,31 These functional polyesters upon emulsification lead to surface active submicron sized particles encapsulating water-insoluble bioactives e.g., curcumin. These double-headed nanosystems show increase in receptor binding ex vivo and oral bioavailability of the encapsulated curcumin in vivo that is proportional to the ligand density.
The polymer was synthesized through EDC coupling method (Scheme 1). The synthetic process is simple and scalable. The commercially available PLGA 50:50 (Resomer® 503H) was coupled with the linker (TRENdiBoc). The deprotection of boc anhydride introduced the two free –NH2 ends which became available for further coupling to ligand (GA).
Scheme 1.
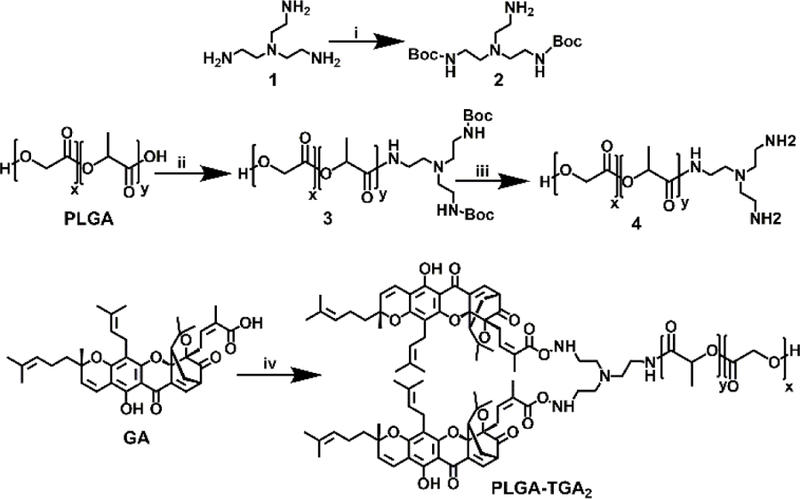
Synthesis scheme for PLGA-TGA2: (i) 20% MeOH/Et3N, 24h; (ii) EDC, 2, DIEA, 24h, N2 atm.; (iii) 30% TFA/DCM, 2 h; (iv) EDC, 4, DIEA, 24h, N2 atm.
The PLGA-TGA2 was characterized using various spectroscopic techniques such as NMR, FT-IR, Raman, and UV-Vis. The 1H NMR of PLGA-TGA2 revealed the disappearance of –COOH peak (at 13.146 ppm) of PLGA, the disappearance of –COOH peak (12.80 ppm) of GA32 and the appearance of amide peaks (at range of 8–9 ppm) (Fig 1a). Further, the formation of amide bonds was confirmed by FT-IR spectroscopy (1670–1630 cm−1 for C=O stretching; 1650–1560 cm−1 for N-H bending in amides; 1560–1530 cm−1 for C-N stretching) (Fig 1b) and Raman spectroscopy (Fig S1).
Fig 1.
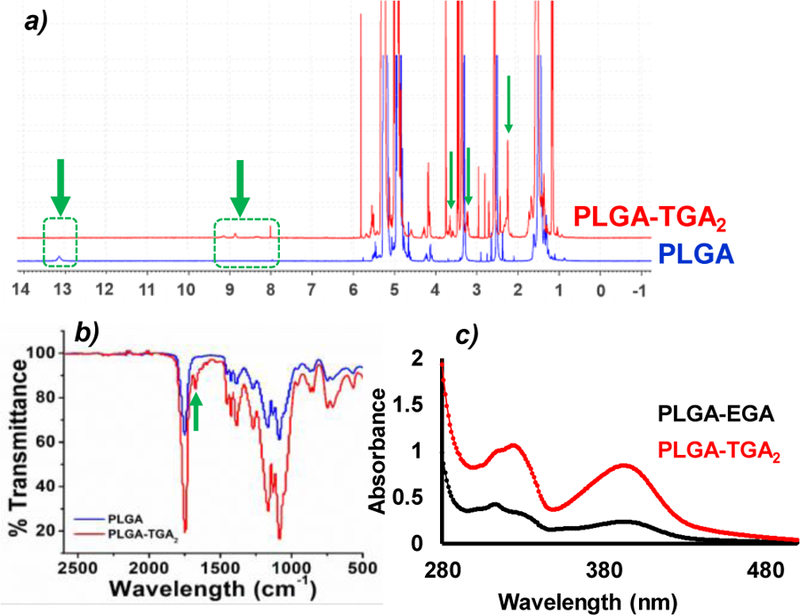
a) 1H NMR of PLGA and PLGA-TGA2; b) FT-IR of PLGA and PLGA-TGA2; c) UV-vis spectrum of PLGA-EGA and PLGA-TGA2.
The UV-vis spectrum of GA showed peak at 365 nm32, which shifted to 392 nm (bathochromic shift) upon coupling with PLGA, confirming the coupling of GA to PLGA (Fig 1c). Furthermore, the absorption intensity in UV-vis spectrum revealed the density of ligand in PLGA-TGA2 was twice as compared to PLGA-EDA (ethylenediamine)-GA (PLGA-EGA).
Further, GA concentration in PLGA-TGA2 was quantified by HPLC confirming double the amount of GA as compare to PLGA-EGA (Fig S2). The molecular mass of polymer was confirmed by gel permeation chromatography (GPC).
The two arms of tripodal linker TREN have been utilized to enhance the GA (ligand) densities on the polymer by simple EDC coupling method. It is hypothesized that the flexibility and tripodal arrangement of the linker will provide enough space and orientation to the attached GA molecules for effective interactions with TfR receptors.
PLGA-TGA2 nanosystems were prepared via oil-in-water emulsification and solvent evaporation method. The PLGA-TGA2 (50 mg) was dissolved in 2 mL ethyl acetate; curcumin, a water-insoluble drug-like compound (7.5 mg) dissolved in 0.5 mL ethyl acetate; polyvinyl alcohol (PVA, 60 mg) dissolved in 5 mL water separately. After 30 min of dissolution, PLGA-TGA2 and curcumin were mixed together and the ethyl acetate mixture was added to PVA solution dropwise forming o/w emulsion. After 7 min of emulsification at 1500 rpm, the emulsion was homogenized at 15000 rpm for 7 min. The final homogenized emulsion was poured into 20 mL of water and allowed the organic solvent to evaporate. The final o/w emulsion was centrifuged at 15000 xg for 30 min at 4 °C in order to collect the pellet that was re-suspended in 5% sucrose solution and lyophilized for further use. The curcumin-loaded nanosystems were spherical in shape as revealed by scanning electron microscopy (SEM) (Fig 2a, 2b) and the size distribution obtained by dynamic light scattering (DLS) is in the range from 210–250 nm (Fig 2c). Zeta potential of nanosystems was found to be −2.65 mV at 6.2 pH. The fresh and freeze dried nanosystems showed similar size profiles (Fig S3). PLGA-TGA2 and PLGA-EGA showed similar curcumin entrapment efficiencies of ~60% (Fig S4).
Fig 2.
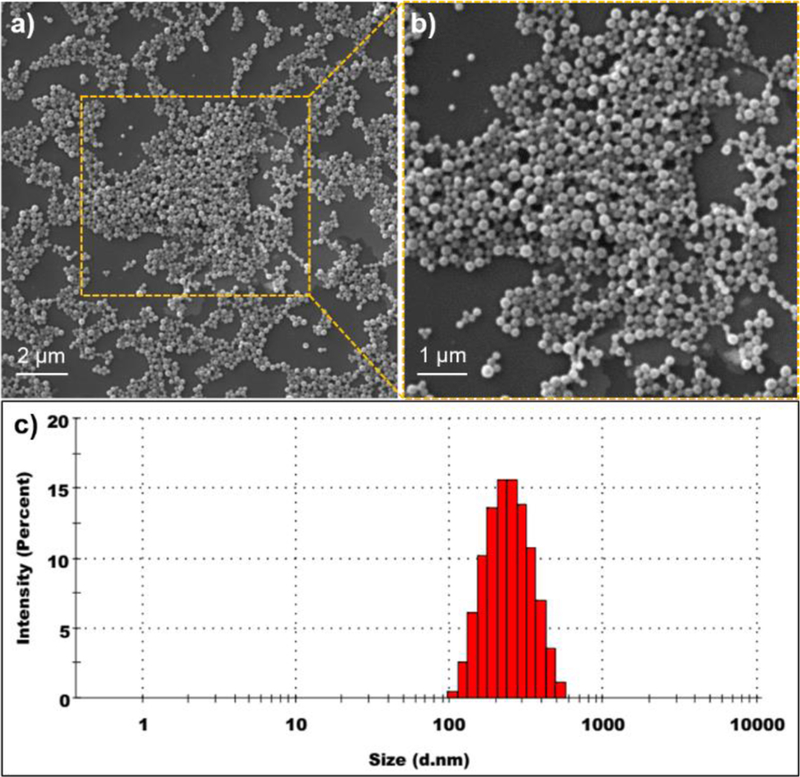
Characterization of PLGA-TGA2 nanosystems: a-b) SEM micrographs of PLGA-TGA2 nanosystems; c) Particle size distribution of PLGA-TGA2 nanosystems measured by DLS.
Curcumin (1,7-bis(4-hydroxy-3-methoxyphenyl)-1,6 heptadiene-3,5-dione) is a naturally available, water insoluble, polyphenol that is an integral part of traditional medicines, has shown promise of turning into modern medicine.33 However, extensive metabolism leading to poor systemic bioavailability is a perceived limitation of curcumin.34 The passive PLGA nanosystems have been used effectively, in the past, for enhancing the bioavailability of curcumin leading to improved outcomes in inflammatory diseases.35,36 Despite the enhanced performance of passive-nanosystems, a significant dose remained unabsorbed in the intestine, indicating potential for further improvement through active-nanosystems.37
Recently, the focus is shifting towards exploiting intestinal receptors to improve therapeutic index of drugs encapsulated into the nanosystems.38 The ligand-receptor stoichiometry plays an essential role in receptor mediated drug delivery. In order to test the hypothesis, we have conducted ex vivo ligand-receptor binding study using fluorescent nanosystems (detailed protocol in SI) and in vivo pharmacokinetics using curcumin encapsulated nanosystems. The ex vivo results demonstrate significant co-localization of PLGA-TGA2 with TfR compared to that of PLGA-EGA nanosystems (Fig 3a) suggesting an increase in ligand-receptor binding that is expected to facilitate better transport. A kinetic study was performed on healthy rodents (Sprague Dawley rats, weighing approximately 200–250 g, n=4). The rats were administered with a single dose of 20 mg/kg (curcumin equivalent) curcumin encapsulated PLGA, PLGA-EGA and PLGA-TGA2 nanosystems. The curcumin concentration (curcumin and curcumin glucuronide) in plasma was monitored from 0–48 h which revealed the maximum concentration (Cmax) was reached at 30 min (Tmax) in rats. The area under curve (AUC0–48) and Cmax for plasma concentration of curcumin were higher in case of PLGA-TGA2 as compare to PLGA-EGA and PLGA indicating the double-headed nanosystems were more actively transported across the intestinal barrier as opposed to single-headed or the respective passive nanosystems (Fig 3b). The in vivo data corroborates with the ex vivo ligand-receptor binding assay.
Fig 3.
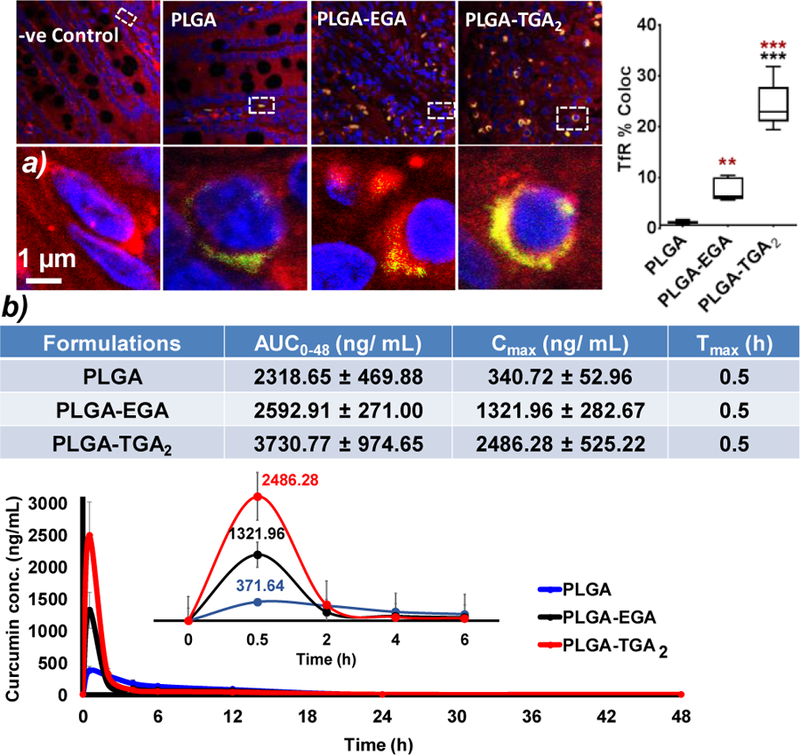
a) Ex vivo ligand-receptor binding study using intestine tissue section. Representative confocal images of small intestine sections incubated with fluorescent nanosystems (Red: TfR; nucleus: blue and nanosystems: green), images acquired at 60X magnification. The co-localization analysis was performed with default M1 and M2 coefficients in ImageJ software. Five to six images were used for fluorescent quantification and all data are presented as mean ± SEM of three independent experiments. Statistical comparisons were made with ordinary one-way ANOVA, Tukey’s multiple comparisons test p value (p < 0.05). The brown stars are comparison with PLGA nanosystems and black PLGA-EGA vs PLGA-TGA2 nanosystems; b) Pharmacokinetic parameters and plasma curcumin concentration profile over 48 h (n=4), (inset: plasma concentration for 6 h)
PLGA-TGA2 improved the Cmax by almost 7-fold and 2-fold compared to PLGA and PLGA-EGA nanosystems respectively. On the other hand, the AUC between PLGA and PLGA-EGA are comparable, while PLGA-TGA2 showed about 1.5 times higher than PLGA as well as PLGA-EGA, probably because curcumin is a very low half-life compound. In order to better understand the curcumin bio-distribution profile, we have conducted a second study, in which the rodents were sacrificed at the time point (Tmax 30 min) showing maximum plasma levels (Cmax). The tissue concentrations of curcumin (curcumin and curcumin glucuronide) was found 2-times higher for PLGA-TGA2 in plasma and various tissues such as intestine, liver, kidney, brain and eye (Fig S5). The plasma levels in terminal sacrifice study seems to be higher compared to those observed in multiple blood withdrawal study, because the former does not account for elimination, while in both the cases we estimate free and encapsulated curcumin (Fig S5). It is interesting to note that the eye and brain levels in PLGA-TGA2 group is about 1.5 times higher compared to PLGA and PLGA-EGA proportional to ligand density. However, the difference between PLGA and PLGA-EGA curcumin levels in multiple withdrawal study did not translate statistically, but qualitative to terminal sacrifice study for the above reasons as well as possible inter-individual variability. The intestine levels indicate that the PLGA-TGA2 are still making their way to systemic circulation and a 30 min sacrifice doesn’t seem to be addressing the question we pose, and may need a few time points to better understand the curcumin disposition. While this data does not indicate if PLGA-TGA2 nanosystems circumvent blood-retinal or blood-brain barriers, our prior studies did show that these nanosystems have the ability to permeate across these barriers.31
In conclusion, we demonstrate a method by which higher ligand densities can be achieved for a terminal functional polyesters such as PLGA, which otherwise was not possible. Our findings suggest that the use of TREN as a linker allowed coupling of two GA molecules as opposed to one with the routinely used EDA linker. The increase in ligand density led to proportional increase in oral bioavailability of encapsulated curcumin, which otherwise is poorly bioavailable. The PLGA-TGA2 offers renewed hope for curcumin, alone or in combination with standard therapeutics, can be effectively used in the treatment of autoimmune, infectious, vascular, and inflammatory disease conditions, with an exception to cancer. Further, this method of double-headed nanosystems not only holds great promise for introducing high ligand densities but also allows dual-ligand coupling for synergistic transport efficiencies. Thus, the methods described here represent a unique platform for effective delivery of drugs with enhanced capacity for targeted drug delivery.
Supplementary Material
Acknowledgements
This work was supported by National Institutes of Health (Grant No. R01EY028169). Use of the TAMU Integrated Metabolomics Analysis Core is acknowledged.
Footnotes
Conflicts of interest
There are no conflicts to declare.
Please see the ESI for details on synthesis methods, experimental protocols and characterization (Figs S6–11).
Electronic Supplementary Information (ESI) available: See DOI: 10.1039/x0xx00000x
Notes and references
- 1.Traverso G and Langer R, Nature, 2015, 519, S19. [DOI] [PubMed] [Google Scholar]
- 2.Tibbitt MW, Dahlman JE and Langer R, J. Am. Chem. Soc, 2016, 138, 704–717. [DOI] [PubMed] [Google Scholar]
- 3.Kirtane AR, Abouzid O, Minahan D, Bensel T, Hill AL, Selinger C, Bershteyn A, Craig M, Mo SS, Mazdiyasni H, Cleveland C, Rogner J, Lee Y-AL, Booth L, Javid F, Wu SJ, Grant T, Bellinger AM, Nikolic B, Hayward A, Wood L, Eckhoff PA, Nowak MA, Langer R and Traverso G, Nat. Commun, 2018, 9, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bellinger AM, Jafari M, Grant TM, Zhang S, Slater HC, Wenger EA, Mo S, Lee Y-AL, Mazdiyasni H, Kogan L, Barman R, Cleveland C, Booth L, Bensel T, Minahan D, Hurowitz HM, Tai T, Daily J, Nikolic B, Wood L, Eckhoff PA, Langer R and Traverso G, Sci. Transl. Med, 2016, 8, 365ra157 LP–365ra157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Darnell M and Mooney DJ, Nat. Mater, 2017, 16, 1178–1184. [DOI] [PubMed] [Google Scholar]
- 6.Nicolas J, Mura S, Brambilla D, MacKiewicz N and Couvreur P, Chem. Soc. Rev, 2013, 42, 1147–1235. [DOI] [PubMed] [Google Scholar]
- 7.Fenton OS, Olafson KN, Pillai PS, Mitchell MJ and Langer R, Adv. Mater, 2018, 30, 1705328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tibbitt MW and Langer R, Acc. Chem. Res, 2017, 50, 508–513. [DOI] [PubMed] [Google Scholar]
- 9.de Greef TFA and Meijer EW, Nature, 2008, 453, 171. [DOI] [PubMed] [Google Scholar]
- 10.Aida T, Meijer EW and Stupp SI, Science (80-. ), 2012, 335, 813 LP–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dong R, Zhou Y, Huang X, Zhu X, Lu Y and Shen J, Adv. Mater, 2015, 27, 498–526. [DOI] [PubMed] [Google Scholar]
- 12.Webber MJ and Langer R, Chem. Soc. Rev, 2017, 46, 6600–6620. [DOI] [PubMed] [Google Scholar]
- 13.Russell-Jones GJ, Adv. Drug Deliv. Rev, 2001, 46, 59–73. [DOI] [PubMed] [Google Scholar]
- 14.Pridgen EM, Alexis F, Kuo TT, Levy-Nissenbaum E, Karnik R, Blumberg RS, Langer R and Farokhzad OC, Sci. Transl. Med, DOI: 10.1126/scitranslmed.3007049. [DOI] [PMC free article] [PubMed]
- 15.Pridgen OC, Alexis EM, Farokhzad F and, Expert Opin. Drug Deliv, 2015, 12, 1459–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mura S, Nicolas J and Couvreur P, Nat. Mater, 2013, 12, 991–1003. [DOI] [PubMed] [Google Scholar]
- 17.Cheng CJ, Tietjen GT, Saucier-Sawyer JK and Saltzman WM, Nat. Rev. Drug Discov, 2015, 14, 239–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gu F, Zhang L, Teply BA, Mann N, Wang A, Radovic-Moreno AF, Langer R and Farokhzad OC, Proc. Natl. Acad. Sci, 2008, 105, 2586 LP–2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bertrand N, Grenier P, Mahmoudi M, Lima EM, Appel EA, Dormont F, Lim J-M, Karnik R, Langer R and Farokhzad OC, Nat. Commun, 2017, 8, 777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ganugula R, Arora M, Guada M, Saini P and Kumar MNVR, ACS Macro Lett, 2017, 6, 161–164. [DOI] [PubMed] [Google Scholar]
- 21.Humblet V, Misra P, Bhushan KR, Nasr K, Sen Ko Y, Tsukamoto T, Pannier N, Frangioni JV and Maison W, J. Med. Chem, 2009, 52, 544–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sietzen M, Batke S, Merz L, Wadepohl H and Ballmann J, Organometallics, 2015, 34, 1118–1128. [Google Scholar]
- 23.Brown JL, Gaunt AJ, King DM, Liddle ST, Reilly SD, Scott BL and Wooles AJ, Chem. Commun, 2016, 52, 5428–5431. [DOI] [PubMed] [Google Scholar]
- 24.Robinson SG, Burns PT, Miceli AM, Grice KA, Karver CE and Jin L, Dalt. Trans, 2016, 45, 11817–11829. [DOI] [PubMed] [Google Scholar]
- 25.Lee MH, Kang G, Kim JW, Ham S and Kim JS, Supramol. Chem, 2009, 21, 135–141. [Google Scholar]
- 26.Kwak J, De Capua A, Locardi E and Goodman M, J. Am. Chem. Soc, 2002, 124, 14085–14091. [DOI] [PubMed] [Google Scholar]
- 27.Scarso A, Zaupa G, Houillon FB, Prins LJ and Scrimin P, J. Org. Chem, 2007, 72, 376–385. [DOI] [PubMed] [Google Scholar]
- 28.Custelcean R, Chem. Commun, 2013, 49, 2173–2182. [DOI] [PubMed] [Google Scholar]
- 29.Busschaert N, Gale PA, Haynes CJE, Light ME, Moore SJ, Tong CC, Davis JT and Harrell WA, Chem. Commun, 2010, 46, 6252–6254. [DOI] [PubMed] [Google Scholar]
- 30.Tai W, Mo R, Lu Y, Jiang T and Gu Z, Biomaterials, 2014, 35, 7194–7203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saini P, Ganugula R, Arora M and Kumar MNVR, Sci. Rep, 2016, 6, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ren Y, Yuan C, Chai H, Ding Y, Li X-C, Ferreira D and Kinghorn AD, J. Nat. Prod, 2011, 74, 460–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Corson TW and Crews CM, Cell, 2007, 130, 769–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Esatbeyoglu T, Huebbe P, Ernst IMA, Chin D, Wagner AE and Rimbach G, Angew. Chemie - Int. Ed, 2012, 51, 5308–5332. [DOI] [PubMed] [Google Scholar]
- 35.Ganugula R, Arora M, Jaisamut P, Wiwattanapatapee R, Jørgensen HG, Venkatpurwar VP, Zhou B, Rodrigues Hoffmann A., Basu R, Guo S and Majeti NVRK, Br. J. Pharmacol, 2017, 174, 2074–2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Grama CN, Suryanarayana P, Patil MA, Raghu G, Balakrishna N, Ravi Kumar MNV and Reddy GB, PLoS One, 2013, 8, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lamprou DA, Venkatpurwar V and Kumar MNVR, PLoS One, 2013, 8, 8–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roger E, Kalscheuer S, Kirtane A, Guru BR, Grill AE, Whittum-Hudson J and Panyam J, Mol. Pharm, 2012, 9, 2103–2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


