Abstract
The efficient use of somatic cell nuclear transfer (SCNT), in conjunction with genetic modification of donor cells provides a general means to add or inactivate genes in mammals. This strategy has substantially improved the efficacy of producing genetically identical animals carrying mutant genes corresponding to specific human disorders. Lentiviral (LV) vectors have been shown to be well suited for introducing transgenes into cells to be used as donor nuclei for SCNT. In the present study, we established an LV vector-based transgene delivery approach for producing live transgenic domestic cats by SCNT. We have demonstrated that cat fetal fibroblasts can be transduced with EGFP-encoding LV vectors bearing various promoters including the human cytomegalovirus immediate early (hCMV-IE) promoter, the human translation elongation factor 1α (hEF-1α) promoter and the human ubiquitin C (hUbC) promoter. Among the promoters tested, embryos reconstructed with donor cells transduced with a LV-vector bearing the hUbC promoter displayed sustained transgene expression at the blastocyst stage while embryos reconstructed with LV vector-transduced cells containing hCMV-IE-EGFP or hEF-1α-EGFP cassettes did not. After transfer of 291 transgenic cloned embryos into the oviducts of eight recipient domestic cats (mean = 36.5 ± 10.1), three (37.5%) were diagnosed to be pregnant, and a total of six embryos (2.1%) implanted. One live male offspring was delivered by Cesarean section on day 64 of gestation, and two kittens were born dead after premature delivery on day 55. In summary, we report the birth of transgenic cloned kittens produced by LV vector-mediated transduction of donor cells and confirm that cloned kittens express the EGFP reporter transgene in all body tissues.
Introduction
The domestic cat is a distinctive mammalian species that exhibits a close genomic relationship to the human genome. The feline genome is extensively conserved in gene content, and the level of chromosome segment conservation between the cat and human genome is among the highest observed between mammalian orders (Murphy et al., 1999, 2000; O'Brien and Nash, 1982; Rettenberger et al., 1995; Wienberg et al., 1997). Moreover, it was recently confirmed with the initial sequence of the feline genome (1.9X) that 90% of the putative genes discovered in the cat carry homologous of human genes (Pontius et al., 2007). In fact, domestic cats present a greater homology (∼90.9%) between the feline and human gene encoding cystic fibrosis transmembane conductance regulator (CFTR) than the homology between mouse and human (∼80.6%). In addition, of feline genetic diseases characterized at the molecular level, several are analogous to human diseases (O'Brien et al., 2002), including autosomal dominant polycystic kidney disease (Biller et al., 1996), retinal atrophy (Narfstrom, 1983), primary hyperoxaluria Type 2 (Danpure et al., 1989), glycogen storage disease Type IV (Fyfe et al. 1992), and hypothyroidism (Tanase et al., 1991). Domestic cats are useful models for the study of human hereditary lysosomal storage disorders, such as mucopolysaccharidosis I, VI, and VII (Fyfe et al., 1999; Haskins et al., 1983; Jackson et al., 1992), gangliosidosis (Baker et al., 1982), and spingomyelinosis C (Brown et al., 1996). Cats are also of particular interest for neurological disorders, as the cat brain has been well characterized both functionally and physiologically (Casal and Haskins, 2006), and the anatomy of the cat brain is more similar to that of a human than that of a mouse (Berman, 1968). Thus, domestic cats that carry mutant genes associated with hereditary diseases in humans provide formidable tools for the study of different human disorders and development of gene therapy strategies, with the extra benefit that the establishment of domestic cat colonies for research purposes is widespread and the maintenance of such colonies is less expensive than that of nonhuman primates.
The most widely method for producing transgenic animals has included pronuclear injection, which involves placement of DNA sequences containing the gene of interest into pronuclei of fertilized zygotes (Gordon et al., 1980; Gordon and Ruddle, 1981). However, the technique is inefficient, as no more than 1% of injected embryos integrate the transgene (Wall, 1996), and the resultant animals are often mosaic (Bosch et al., 2004). Viral vectors have also been used for the generation of transgenic domestic species. This technique is more efficient than pronuclear injection in terms of the number of transgenic animals produced per embryo manipulated. Transduction of early mouse embryos with vectors derived from gamma retroviruses, such as Moloney murine leukemia virus, resulted in the generation of transgenic founder animals (F0 generation) that transmitted the retroviral genomes to progeny (F1 generation). However, the retroviral genomes were expressed neither in the F0 nor in the F1 generation because of transcriptional silencing (Jahner et al., 1982). This block in expression following transduction using vectors based on gamma retroviruses was observed subsequently in cattle (Chan et al., 1998). Recently, a transgenic domestic cat was produced with fibroblast cells that were transduced with a DsRed2-encoding gamma retroviral vector (Yin et al., 2008). Surprisingly, transgene expression in kittens was stable, and there was no evidence for transcriptional silencing.
An alternative and more reliable approach for transgenesis involves the use of Lentiviral (LV) vectors (Park, 2007; Wiznerowicz and Trono, 2005). Like gamma retroviruses, LV vectors stably integrate their genomes into the host cells’ chromosomes to form a provirus (Reiser et al., 1996). The first LV transgenic animals were generated by transduction of murine preimplantation embryos (Lois et al., 2002; Pfeifer et al., 2002) or embryonic stem cells (ESC) (Pfeifer et al., 2002) with LV-vectors derived from human immunodeficiency virus (HIV-1). Compared to the traditional DNA microinjection technique, LV vector-mediated gene transfer resulted in a four- to eightfold higher rate of transgenic animals per embryo treated (Pfeifer, 2004), and more than 90% of F0-generation animals expressed the transgene (Lois et al., 2002; Pfeifer et al., 2002). In addition to transgenic mice, transgenic rats (Lois et al., 2002), rhesus monkeys (Wolfgang et al., 2001; Yang et al., 2008), chickens (Chapman et al., 2005; McGrew et al., 2004), birds (Scott and Lois, 2005), pigs (Whitelaw et al., 2004), cattle (Hofmann et al., 2003, 2004) and sheep (Ritchie et al., 2008) have since been produced using LV-transgenesis.
Somatic cell nuclear transfer (SCNT) offers the possibility of circumventing some limitations of other transgenic techniques. One advantage includes the use of cell culture methods for propagation of donor cells to allow stable transgene integration and expression. Large numbers of cells bearing transgene sequences can be frozen for long-term storage, and transgene structure and expression can be tested prior to SCNT (Bosch et al., 2004). SCNT facilitates not only the addition of transgene sequences at random sites but it also allows targeted insertion of DNA by homologous recombination, a procedure that forms the basis for creating gene knockouts (Bosch et al., 2004; Piedrahita, 2000). Furthermore, SCNT will ensure that 100% of the animals produced are transgenic, and will eliminate the mosaicism often observed after exposing oocytes or embryos to viral vectors (Park, 2007). In a proof of principle study, the production of a transgenic calf derived by transduction of fetal fibroblasts before nuclear transfer into enucleated oocytes has demonstrated the feasibility of combining SCNT and LV transgenesis (Hofmann et al., 2004).
The purpose of the present study was to test the usefulness of LV vector-based transgene delivery approaches for producing live transgenic domestic cats by SCNT.
Materials and Methods
Animals
Domestic short-haired cats (DSH) were used as oocyte donors and embryo recipients. The cats were group housed in environmentally controlled rooms with a 14 h:10 h light:dark cycle at 20–26°C at the Audubon Research Center (ACRES). The rooms were cleaned and cats fed once daily (Science Diet, Hill Pet Nutrition, St. Louis, MO). Fresh water was available ad libitum. All animal procedures were approved by the Institutional Animal Care and Use Committee of ACRES, as required by the Health Research Extension Act of 1985 (Public Law 99-158). DSH that received transgenic cloned embryos were housed individually until the pregnancy status was determined on day 22 by ultrasonography. DSH that were diagnosed as pregnant were maintained singly for the duration of gestation.
Chemicals
All chemicals were obtained from Sigma-Aldrich Chemical Co. (St. Louis, MO) unless otherwise stated.
Preparation of HIV-1-based LV vectors
High titer stocks of EGFP-encoding LV vectors bearing the hCMV-IE promoter, the hEF-1α promoter or the hUbC promoter (Ricks et al., 2008) were generated by calcium-phosphate transfection of human embryonic kidney 293T cells, as described previously (Reiser, 2000). A second-generation LV packaging system was used (Zhang et al., 2004). Titer of vectors stocks (transducing units, TU) were determined directly based on proviral copy numbers in transduced human osteosarcoma (HOS) target cell DNA by quantitative PCR (qPCR) using WPRE-specific primers (Ricks et al., 2008).
Test for emergence of replication-competent lentivirus (RCL)
The potential presence of RCL particles in vector stocks and in blood from cloned kittens was analyzed using a sensitive ELISA test as previously described by Mochizuki et al. (1998). A total of 105 H9 human T cells were exposed to LV–vector stocks or to serum from blood collected from cloned transgenic fetuses and a live kitten. Transduced cells were serially passaged and supernatants were collected every 7 days for a period of 4 weeks. HIV-1 p24 antigen levels were determined using an ELISA kit (ZeptoMetrix Corporation, Buffalo, NY).
Transduction of domestic cat fetal fibroblasts and cell synchronization
Domestic cat fetal fibroblasts (CFF) were generated from three fetuses collected from a single female at 30 days of gestation. Fetal tissue was finely cut, seeded in a 75-cm2 tissue culture flask and cultured in 7 mL of Glasgow's Minimal Essential Medium (GMEM) supplemented with 15% (v/v) fetal bovine serum (FBS; Hyclone, Logan, UT), gentamicin (50 μg/mL), and amphotericin B (2.5 μg/mL) at 38°C in a 5% CO2/air atmosphere. When monolayer outgrowths with fibroblastic-like morphology reached 100% confluency after 7 to 10 days of culture, cells were disaggregated with trypsin (2.5 mg/mL) and resuspended in GMEM with 10% FBS and 10% (v/v) dimethyl sulphoxide (DMSO) and cooled at 1.0°C/min to −80°C (Mr. Frosty, Nalgene, Rochester, NY) before storage in liquid nitrogen (LN2).
For transduction, frozen/thawed CFF cells (passages 1–2) were cultured in tissue culture flasks until they reached 70–80% confluency. Then, cells were transduced overnight in 3–4 mL of GMEM containing 15% FBS with LV vectors using multiplicities of infection (MOI) between 15 and 20 at 38°C in a humidified atmosphere of 5% CO2 in air. After transduction, cells were washed and cultured for 2–3 additional days and passaged at least twice to achieve stable integration of the vector genome before use for SCNT. EGFP transgene expression in vitro was determined by fluorescence imaging using a 475/490-nm filter set. Quantification of the percentage of LV transduced cells was assessed by flow cytometric analysis, as described previously (Reiser, 2000).
For contact inhibition, cells were cultured in flat-sided tissue culture tubes containing 3.0 mL of GMEM containing 15% fetal bovine serum (FBS) until cells reached 100% confluency. Then, cells were maintained in culture for an additional 5–7 days, during which time the culture medium was replaced every other day. Cells were disaggregated with 2.5 mg/mL pronase before SCNT or frozen and stored in LN2.
Collection of in vivo matured domestic cat oocytes
Domestic cats (n = 16) were treated with 3–4 IU of porcine-follicle stimulating hormone (FSH, Sioux Biochemical, Sioux Center, IA) given in decreasing daily doses (s.c.) for 4 days, followed by 3 IU of porcine-luteinizing hormone (i.m.; LH, Sioux Biochemical) on day 5. At 24–26 h after LH injection, oocytes were collected by laparoscopic aspiration of mature ovarian follicles (Gómez et al., 2000).
Production of transgenic cloned embryos
SCNT was conducted according to the method previously reported by Gómez et al. (2003). Briefly, 1–2 h after retrieval, DSH oocytes were denuded of cumulus and corona cells and stained with 25 μg/mL Hoechst 33342 to localize the second metaphase spindle (MII) and first polar using epifluorescence microscopy (1 sec of exposure). Oocytes were enucleated and single fibroblasts (cultured or frozen/thawed) from the transduced or nontransduced group (passages 4–6) were introduced into the perivitelline space. Cell fusion was induced by applying two electrical pulses (3 sec AC of 19 V, 1 MHz; followed by a 30-μsec DC pulse of 24 V), delivered with two stainless steel electrodes (LF-101; Nepa Gene, Tokyo, Japan) attached to micromanipulators. After 15–20 min, fusion was confirmed and fused couplets were cultured for 2 h before activation in 500 μL of IVC-1 medium (Gómez et al., 2000) supplemented with 7.8 mM calcium lactate and 2.5 μg Cytochalasin B (Cyt-B). Activation was performed by exposing the couplets to two 60 μsec DC pulses of 120 V in a fusion chamber (Nepa Gene) with two electrodes 1 mm apart, followed by a 4-h incubation in 30 μL of IVC-1 medium supplemented with 10 μg/mL cycloheximide and 5 μg/mL Cyt-B under mineral oil at 38°C in a 5% CO2 and air atmosphere. After activation, reconstructed couplets were cultured in 500 μL of IVC-1 medium for 12–13 h and transferred to the oviducts of recipient domestic cat females on day 1. Alternatively, couplets were cultured in IVC-2 medium until day 8, when embryo development to the blastocyst stage was evaluated.
Determination of EGFP transgene expression in cloned embryos and confirmation of vector copy numbers in transgenic cloned blastocysts
Transgene expression in DSH cloned embryos was evaluated in blastomeres on days 1, 2, 5, and 8 by epifluorescence microscopy using a 475/490 nm filter. Embryos were classified as EGFP positive if blastomeres displayed green fluorescence.
To analyze LV vector copies, qPCR quantification was performed on genomic DNA derived from single blastocysts. Genomic DNA was extracted from frozen blastocysts using a ChargeSwitch Forensic DNA Purification kit (Invitrogen, Carlsbad, CA). A primer/probe set specific for the WPRE element present in the LV vector was used as described previously (Ricks et al., 2008). Standards were generated from vector plasmids. Individual PCR reactions consisted of 25 μL of 2 × TaqMan Universal PCR Master Mix (Applied Biosystems, Foster City, CA), 10 pmol of forward primer, 10 pmol of reverse primer, and 10 pmol of the probe each targeting the WPRE sequence. PCR reactions were carried out using a Stratagene Mx3000p PCR machine (Stratagene, La Jolla, CA) with cycle conditions of 10 min at 95°C and 40 cycles of 95°C for 30 sec, 60°C for 1 min, and 72°C for 1 min.
Embryo transfer, pregnancy detection, and parturition
Cloned embryos (days 1–2) were transferred into the oviducts of domestic cat recipients, some of which had served as oocyte donors on day 0 (n = 4) and others that served as recipients after induction of ovulation, but without oocyte retrieval (n = 4) as previously described (Gómez et al., 2004). Briefly, sterile polyethylene tubing (PE-10, #427400, Becton Dickinson, Sparks, MD) containing one-cell or two-cell reconstructed cybrid/embryos near the lower tip were threaded through a 14-cm tomcat catheter that had been inserted into the upper ampullary portion of the oviduct by laparoscopy. Then, using a 1-mL threaded plunger syringe, cybrids/embryos were expelled into the oviduct with in 10–15 μL of IVC-1 medium.
Pregnancy status of recipient cats was determined by abdominal ultrasonography on day 22 after ovulation or oocyte aspiration. Pregnant recipients were monitored weekly by ultrasonography until delivery of term kittens by Caesarean section. In an effort to reduce the onset of premature parturition, pregnant cats received an orally active progestin (0.08 mg/kg, ReguMate, DPT Laboratories, San Antonio, TX) daily starting on day 55 of gestation. In addition, to reduce perinatal kitten mortality, bethametasone (0.1 mg/kg, i.m.; CelestoneSoluspan, Schering Corporation, Kenilworth, NJ) was administered at various times after day 60 of gestation, as described previously (Gómez et al., 2004).
Clonal status and presence of LV vector in the genome of cloned kittens
To determine the clonal status of kittens, DNA was extracted from donor fetal fibroblast cells, umbilical cord blood of the live kitten, and spleens of dead kittens by using the DNeasy Blood & Tissue Kit (Qiagen, Valencia, CA). Sterile cytological brushes were used to obtain buccal (cheek) cells noninvasively from the live kitten. DNA was isolated using the QIAamp DNA mini kit (Qiagen). Feline microsatellite loci were amplified as reported previously by Gómez et al. (2004) using standard ABI fluorescent chemistries on an ABI 3730 DNA Analyzer (Applied Biosystems, Foster City, CA). The results were analyzed using the STRand software (Hughes, Germantown, MD).
EGFP expression was detected by fluorescence imaging of live and dead kittens and their placentae. Direct fluorescence was applied to the whole animal body and excitation of green fluorescence was achieved by using a black and blue flash light adapted with a 485-nm filter. The emitted fluorescence was visualized using a long pass filter of 500-nm.
The presence of the LV vector genomes in dead cloned kittens was confirmed by Southern blotting as described by Bao et al. (2004). Genomic DNA was isolated from tissue using Genomic tip 100 columns (Qiagen). To excise vector sequences, genomic DNA was initially digested with the restriction enzyme BglII (New England Biolabs, Ipswich, MA). Vector sequences were cut internally using BamHI, NsiI, or SpeI. Standars for vector copy numbers were produced by digesting the pNL-EGFP/UbC/WPREΔU3 plasmid (Ricks et al., 2008) with BglII. The EGFP probe was produced by digesting the pNL-EGFP/UbC/WPREΔU3 plasmid with BamHI and NotI and labeling the purified EGFP fragment with α32P-dCTP using the Rediprime II random labeling system (Amersham, Piscataway, NJ).
Statistical analyses
One-way ANOVA was used to analyze the data related to transgene expression in CFF. The data for cleavage rates, blastocyst development, and transgene expression in cloned embryos was analyzed using chi-square.
Results
Cell transduction, embryo production, and transgene expression
To study transgene expression in CFF and DSH cloned embryos, three EGFP-encoding LV vectors bearing hCMV-IE, hEF1-α, or hUbC promoters were designed (Fig. 1). After transduction of CFF, we observed that cells transduced with each of the LV vectors started to express the transgene as early as 48 h after exposure to the LV vectors. Using MOIs around 15 to 20, up to 99.5% of the cell transduced with LV vectors bearing the hUbC promoter expressed the EGFP-transgene as judged by fluorescence microscopy (Fig. 2A) and FACS analysis (data not shown). To determine whether LV-mediated expression of the transgene was sustained in cloned embryos and whether embryo development was affected by the presence of the LV vector genome, we reconstructed embryos with CFF that were previously transduced with the three LV vectors carrying different promoters. As shown in Table 1, embryo cleavage (day 2) and development to the blastocyst stage (day 8) of reconstructed embryos were not affected by either the LV vector genome or the type of promoter used. Also, there was no significant difference in development between SCNT embryos reconstructed with transduced or nontransduced cells (Table 1).
FIG. 1.
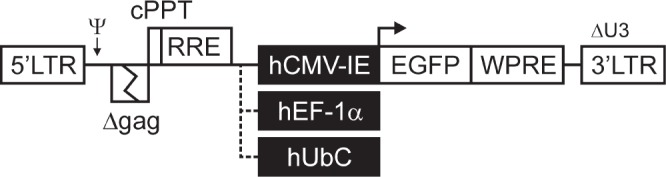
LV vectors used to transduce cat fetal fibroblasts. LTR: viral long terminal repeat. Ψ: Packaging signal. cPPT: central polypurine tract. RRE: Rev response element. hCMV-IE: human cytomegalovirus immediate early promoter; hEF-1α: human translation elongation factor 1 alpha promoter; hUbC: human ubiquitin C promoter. EGFP: enhanced green fluorescent protein. WPRE: woodchuck hepatitis virus post-transcriptional regulatory element.
FIG. 2.
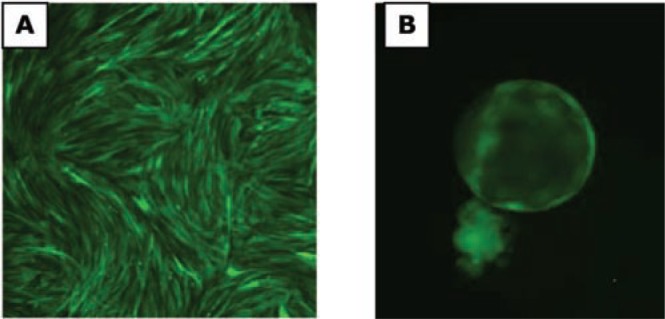
(A) LV vector-transduced cat fetal fibroblasts expressing EGFP from the hUbC promoter. (B) Cloned blastocyst (day 8) reconstructed with cat fetal fibroblasts transduced with EGFP-expressing LV vectors bearing the hUbC promoter.
Table 1.
In Vitro Development of Cat Cloned Embryos Reconstructed with Cat Fetal Fibroblasts Transduced with LV Vectors
| Promoter | Cleavage frequency/total fused couplets day 2 n (%) | Blastocysts/total cleaved embryos day 8 n (%) |
|---|---|---|
| hCMV-IE | 34/43 (79)a | 4/34 (12)a,b |
| hEF1-α | 34/44 (77)a | 10/34 (29)a |
| hUbC | 44/52 (85)a | 5/44 (11)b |
| None | 21/26 (81)a | 4/21 (19)a,b |
Different superscripts within the same column indicate significant differences (p < 0.05).
Green fluorescence in DSH cloned embryos reconstructed with cells transduced with either of the three LV vectors was undetectable for at least 24 h after reconstruction. By day 2 of culture, a higher percentage of embryos reconstructed with CFF transduced with LV vectors bearing the hUbC promoter started to express EGFP (32%) compared to embryos reconstructed with LV vectors bearing the hCMV-IE (12%) or hEF1-α (9%) promoters. On day 5 of culture, a substantial number of embryos derived from cells transduced with LV vectors bearing the hUbC (41%) and hEF1-α (18%) promoters displayed green fluorescence. One embryo derived from cells transduced with LV vectors bearing the hCMV-IE promoter displayed faint fluorescence on day 5 (3%), but it disappeared by day 8. In contrast, all day-8 embryos derived with LV vectors bearing the hUbC promoter displayed sustained transgene expression and blastocysts (n = 5) derived from this vector exhibited green fluorescence (Fig. 2B), while none of the blastocysts obtained using LV vectors bearing the hCMV-IE and hEF1-α promoters (n = 14) exhibited green fluorescence (Table 2).
Table 2.
EGFP Protein Expression of Cat Cloned Embryos at Days 2, 5, and 8 after Reconstruction with Cat Fetal Fibroblasts Transduced with LV Vectors
| Promoter | Total cleaved embryos n | 2–4 Cells day 2 n (%) | Premorula/morula day 5 n (%) | Blastocysts day 8 n (%) |
|---|---|---|---|---|
| hCMV | 34 | 4 (12)a | 1 (3)a | 0/4 (0)a |
| hEF1-α | 34 | 3 (9)a | 6 (18)b | 0/6 (0)a |
| hUbC | 44 | 14 (32)b | 18 (41)c | 5/5 (100)b |
Different superscripts within the same column indicate significant differences (p < 0.05).
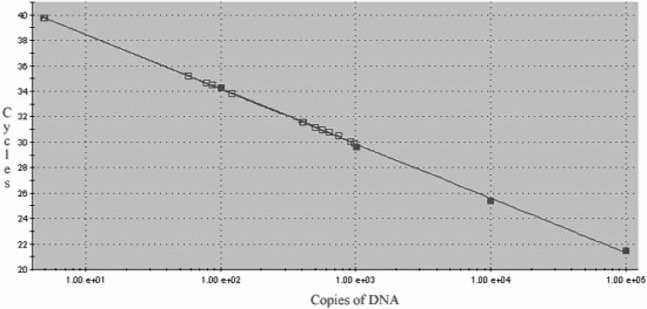
To determine LV vector copy numbers in blastocysts, a qPCR analysis of genomic DNA extracted from single blastocysts was carried out with a primer/probe set specific for the WPRE sequence present in the LV vector (Fig. 3). The results showed that cloned blastocysts reconstructed with cells transduced with LV vectors bearing hCMV-IE, hEF1-α and hUbC promoters that did not express the EGFP transgene had between three and nine vector copies per cell. Similar vector copy numbers ranging from 0.7 to 4 copies per cell were obtained with blastocysts reconstructed with LV vectors bearing hUbC promoter, while cloned blastocysts reconstructed with nontransduced cells did not display any vector DNA (Fig. 3). Thus, the lack of transgene expression observed with blastocysts obtained using LV vectors containing the hCMV-IE or hEF1-α promoters was not caused by the absence of vector sequences.
FIG. 3.
Quantitative PCR analysis of LV vector genomes in cloned blastocysts. Solid squares represent DNA standard and open squares represent genomic DNA samples extracted from blastocysts.
Embryo transfer, pregnancies, and birth of transgenic cloned kittens
Cloned embryos (days 1–2) reconstructed using CFF transduced with LV vectors bearing the hUbC promoter were next transferred into domestic cat recipients. CFF transduced with LV vectors bearing the hUbC promoter were used because expression of the EGFP transgene was stable during all stages of development. A total of 291 transgenic cloned embryos were transferred into the oviducts of eight recipient cats (mean = 36.5 ± 10.1), of which three (37.5%) were diagnosed to be pregnant when examined by ultrasonography at day 22.
On day 22, a total of six embryos (2.1%) had implanted in three of the domestic cat recipients. Four embryos implanted in one recipient and one embryo in each of the other two recipients. One of the four embryos that implanted in one recipient was reabsorbed by day 30, while a second embryo was reabsorbed by day 39, but its placenta was not expelled until day 55. The two remaining kittens died in utero at 55 and 56 days of gestation, possibly as a result of uterine contractions from expulsion of the remaining placenta. An embryo that implanted in a second recipient was reabsorbed by day 30 of gestation. The third pregnant recipient began exhibiting early signs of labor (milk letdown, vaginal discharge) and one live male kitten was delivered by elective Cesarean section on day 64 (Fig. 4A). The birth weight of the live kitten (106.7 g) was higher than the birth weight of the two dead kittens (54.7 and 59.5 g, respectively). In addition, the placental weight of the live kitten (32.7 g) was higher than the weight of placentae from the dead kittens (18.3 g and 20.8 g). All kittens had a normal physical appearance. Microsatellite analysis confirmed that the clones were identical to the donor CFF cells (Table 3).
FIG. 4.
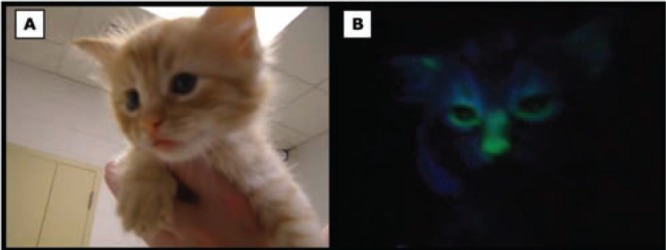
(A) Four-week-old live cloned kitten under normal day light, and (B) Cloned kitten expressing EGFP.
Table 3.
Analysis of Feline Genetic Markers (FCA) of Transgenic Domestic Cloned Kittens
| Feline markers | Donor cat fetal fibroblasts | Live clone kitten | Surrogate cat live clone kitten | Dead kitten #1 | Dead kitten #2 | Surrogate cat dead clone kittens #1, 2 |
|---|---|---|---|---|---|---|
| FCA078 | 188/202 | 188/202 | 194/200 | 188/202 | 202/202 | 198/200 |
| FCA097 | 148/150 | 148/150 | 138/146 | 148/150 | 148/150 | 146/146 |
| FCA105 | 193/199 | 199/201 | 146/— | 201/203 | 199/201 | 146/191 |
| 201/203 | ||||||
| FCA123 | 133/135 | 135/139 | 135/135 | 133/135 | 133/139 | 133/137 |
| FCA201 | 141/155 | 141/155 | 153/155 | 141/155 | 141/155 | 153/153 |
| FCA210 | 158/166 | 158/166 | 170/170 | 158/166 | 158/166 | 166/170 |
| FCA220 | 216/216 | 216/216 | 214/216 | 216/216 | 216/216 | 214/214 |
| FCA240 | 172/158 | 172/172 | 170/— | 172/158 | 172/172 | 170/156 |
| FCA310 | 126/— | — | — | 126/128 | 126/136 | 124/136 |
| 128/136 | ||||||
| FCA649 | 126/— | 126/144 | — | 126/140 | 126/144 | 132/132 |
| 140/144 |
Donor cat fetal fibroblasts were generated from three fetuses collected from a single female at 30 days of gestation.
The presence and expression of the EGFP transgene in cloned kittens was confirmed by fluorescence imaging of live (Fig. 4B) and dead kittens and placentae. Southern blot analysis of DNA extracted from placentae of the two dead kittens confirmed that between five and six vector copies were integrated into the genomic DNA (Fig. 5). The absence of RCL in the original LV vector stock and in serum derived from the cloned kitten and fetus was confirmed using a sensitive p24 ELISA assay (Fig. 6).
FIG. 5.
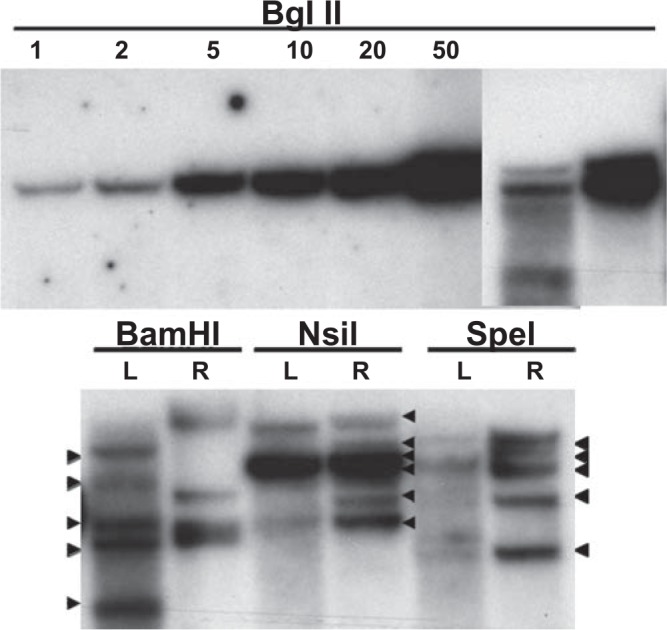
Southern blot analysis of genomic DNA extracted from two placentae (L) and (R) of a domestic cat uterus. Top: DNAs were digested using Bgl II to excise vector sequences. Top left panel: plasmid copy number standards corresponding to 1, 2, 5, 10, 20, and 50 copies. Top right panel: genomic from L and R placentas. Bottom panel: genomic DNA from L and R placentas digested with BamHI, NsiI, and SpeI, which cut once within the vector sequence. DNA fragments were blotted onto a nitrocellulose membrane and hybridized to a 32P-labeled probe corresponding to the EGFP coding region region. This Southern blot analysis indicates that between five and six LV vector copies were present in the genome of both placentas (black arrowheads).
FIG. 6.
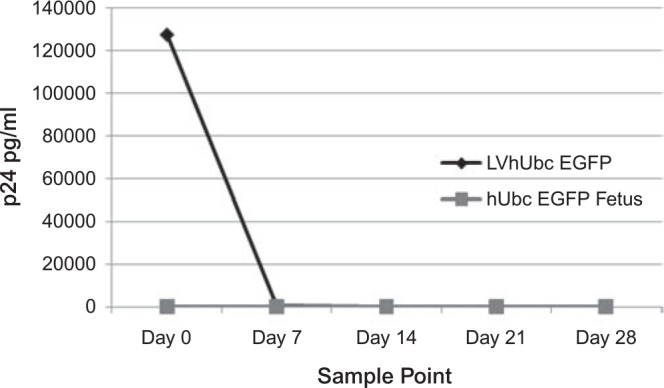
RCL analysis of LV vector samples and serum from a cloned transgenic fetus. Cells of the human H9 T cell line were exposed either to an aliquot of the original LV vector stock (black line) or to serum obtained from a cloned fetus following LV transgenesis (gray line). Cells were serially passaged and monitored for the production of p24 antigen.
Discussion
We report the birth of transgenic cloned kittens produced by LV vector-mediated transduction of donor cells. We also provide evidence that cloned kittens express the EGFP reporter transgene in all body tissues. Although LV vectors carrying the EGFP transgene under the control of the hCMV-IE, hEF1-α, and hUbC promoters resulted in efficient transduction of domestic cat fetal fibroblasts, only the hUbC promoter was able to sustain transgene expression in reconstructed embryos.
At present, it is not clear why sustained transgene expression was not observed in blastocysts reconstructed with donor cells transduced with LV vectors carrying the hCMV-IE and hEF1-α promoters, because vector copy numbers in blastocysts reconstructed with such donor cells were similar. Thus, transgene copy number effects, as seen in transgenic pigs, are unlikely to be the cause for the lack of transgene expression (Hofmann et al., 2006).
It is well established that epigenetic factors, such as DNA hypermethylation, CpG levels and histone acetylation, can affect transgene expression levels. For example, a repressive chromatin environment (Mohamedali et al., 2004), a high percentage of CpG dinucleotides (Hofmann et al., 2006; Yao et al., 2004) and hypoacetylated histone H3 or the presence of histone H1 in the LV vector genome may lead to silencing of LV vector integrants (He et al., 2005). Cloned embryos frequently fail to follow the demethylation patterns observed during normal embryogenesis. For instance, bovine cloned embryos are hypermethylated in various genomic regions throughout preimplantation development (Dean et al., 2001; Kang et al., 2001), as compared to embryos derived by IVF. A similar abnormal methylation pattern has been associated with abnormal expression of imprinted and pluripotency related genes (Bortvin et al., 2003; Mann et al., 2003). It has also been demonstrated that DNA hypermethylation plays an important role in embryonic transgene silencing (Kasamatsu et al., 2006) and in the silencing of LV vector genomes in transgenic animals (Hofmann et al., 2003). Overall, these studies suggest that gene silencing can be observed when LV vectors are used for the production of transgenic animals, and that DNA hypermethylation plays an important role in LV silencing. Although, DNA methylation patterns of transgenic cat cloned blastocysts that did not express the transgene were not investigated, we surmise that abnormal methylation patterns in cloned embryos may have affected transgene expression of embryos reconstructed with somatic cells transduced with LV vectors carrying the EGFP transgene. DNA methylation levels in cloned embryos, the number of LV vector copies and transgene expression levels will need to be evaluated to clarify this point.
Although EGFP-transgenic animals from different species have been successfully produced, EGFP transgene sequences may impact the viability of transfected/transduced cells (Arat et al., 2001). In the present study, development to the blastocyst stage of transgenic (11–29%) and nontransgenic (19%) cloned cat embryos was more efficient compared to results reported recently using transfected domestic cat cells (3–6.5%; Yin et al., 2008) or nontransduced domestic cat cells as donor nuclei (3–8%; Gómez et al., 2003; Kitiyanant et al., 2003; Skrzyszowska, 2002; Thongphakdee et al., 2006; Yin et al., 2006, 2005).
In addition, in the present study, the pregnancy rate (37.5%) after the transfer of DSH transgenic cloned embryos into domestic cat recipients was higher than that reported involving the transfer of transgenic domestic cat embryos carrying the DsRed2 transgene into domestic cat recipients (27.3%; Yin et al., 2008). However, embryo implantation rates (2.1%) of transferred DSH transgenic cloned embryos were comparable to the rates observed in previous studies involving the transfer of transgenic cloned embryos (1.7%; Yin et al., 2008) or nontransgenic domestic cat cloned embryos (1–2.3%; Shin et al., 2002; Yin et al., 2005, 2007) into domestic cat recipients. Also, we observed robust EGFP transgene expression in one live and two dead cloned kittens. All together, these results clearly indicate that the EGFP transgene or the LV vector genome does not reduce embryo development or survival rates of transgenic cloned cats.
In summary, we demonstrated for the first time the feasibility of combining SCNT and LV transgenesis to produce a healthy transgenic cloned cat that expresses the EGFP transgene. LV transgenesis in the context of SCNT is a potent technique for producing transgenic animals. Future studies will be needed to determine whether the cloned kitten has stably incorporated the transgene sequences into the germ line and whether cats of the F1 generation produced by mating nontransgenic cats with transgenic cats exhibit sustained expression of the EGFP transgene.
Acknowledgments
We are grateful to Dr. Bob MacLean and Dr. John Edwards for providing excellent surgical expertise and for veterinary assistance with the transgenic cloned kitten, and; to all graduate students, veterinary technicians, and animal care personnel at ACRES for their technical assistance and round-the-clock care of the cloned kitten.
Author Disclosure Statement
The authors declare that no competiting financial interests exist.
References
- Arat S., Rzucidlo S.J., Gibbons J., et al. (2001). Production of transgenic bovine embryos by transfer of transfected granulosa cells into enucleated oocytes. Mol. Reprod. Dev. 60, 20–26 [DOI] [PubMed] [Google Scholar]
- Baker H.J., Walkley S.U., Rattazzi M. et al. (1982). Feline gangliosidoses as models of human lysosomal storage diseases. Prog. Clin. Biol. Res. 94, 203–212 [PubMed] [Google Scholar]
- Bao L., Jaligam V., Zhang X.Y., et al. (2004). Stable transgene expression in tumors and metastases after transduction with lentiviral vectors based on human immunodeficiency virus type 1. Hum. Gene. Ther. 15, 445–456 [DOI] [PubMed] [Google Scholar]
- Berman A. (1968). The Brain Stem of the Cat, a Cytoarchitectonic Atlas with Stereotaxic Coordinates. (University of Wisconsin Press: Madison, WI) [Google Scholar]
- Biller D.S., DiBartola S.P., Eaton K.A. et al. (1996). Inheritance of polycystic kidney disease in Persian cats. J. Hered. 87, 1–5 [DOI] [PubMed] [Google Scholar]
- Bortvin A., Eggan K., Skaletsky H., et al. (2003). Incomplete reactivation of Oct4-related genes in mouse embryos cloned from somatic nuclei. Development 13, 1673–1680 [DOI] [PubMed] [Google Scholar]
- Bosch P., Hodges C.A., and Stice S.L. (2004). Generation of transgenic livestock by somatic cell nuclear transfer. Biotecnol. Aplicada 21,128–136 [Google Scholar]
- Brown D.E., Thrall M.A., Walkley S.U., et al. (1996). Metabolic abnormalities in feline Niemann-Pick type C heterozygotes. J. Inherit. Metab. Dis. 19, 319–330 [DOI] [PubMed] [Google Scholar]
- Casal M., and Haskins M. (2006). Large animal models and gene therapy. Eur. J. Hum. Genet. 14, 266–272 [DOI] [PubMed] [Google Scholar]
- Chan A.W., Homan E.J., Ballou L.U., et al. (1998). Transgenic cattle produced by reverse transcribed gene transfer in oocytes. Proc. Natl. Acad. Sci. USA 95, 14028–14033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman S.C., Lawson A., Macarthur W.C., et al. (2005). Ubiquitous GFP expression in transgenic chickens using a lentiviral vector. Development 132, 935–940 [DOI] [PubMed] [Google Scholar]
- Danpure C.J., Jennings P.R., Mistry J., et al. (1989). Enzymological characterization of a feline analogue of primary hyperoxaluria type 2: a model for the human disease. J. Inherit. Metab. Dis. 12, 403–414 [DOI] [PubMed] [Google Scholar]
- Dean W., Santos F., Stojkovic M., et al. (2001). Conservation of methylation reprogramming in mammalian development: aberrant reprogramming in cloned embryos. Proc. Natl. Acad. Sci. USA 98, 13734–13738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fyfe J.C., Giger U., Van Winkle T.J., et al. (1992). Glycogen storage disease type IV: inherited deficiency of branching enzyme activity in cats. Pediatr. Res. 32, 719–725 [DOI] [PubMed] [Google Scholar]
- Fyfe J.C., Kurzhals R.L., Lassaline M.E., et al. (1999). Molecular basis of feline beta-glucuronidase deficiency: an animal model of mucopolysaccharidosis VII. Genomics 58, 121–128 [DOI] [PubMed] [Google Scholar]
- Gómez M.C., Pope C.E., Harris R., et al. (2000). Births of kittens produced by intracytoplasmic sperm injection of domestic cat oocytes matured in vitro. Reprod. Fertil. Dev. 12, 423–433 [DOI] [PubMed] [Google Scholar]
- Gómez M.C., Jenkins J.A., Giraldo A., et al. (2003). Nuclear transfer of synchronized African wild cat somatic cells into enucleated domestic cat oocytes. Biol. Reprod. 69, 1032–1041 [DOI] [PubMed] [Google Scholar]
- Gómez M.C., Pope C.E., Giraldo A., et al. (2004). Birth of African Wildcat cloned kittens born from domestic cats. Cloning Stem Cells 6, 247–258 [DOI] [PubMed] [Google Scholar]
- Gordon J.W., and Ruddle F.H. (1981). Integration and stable germ line transmission of genes injected into mouse pronuclei. Science 214, 1244–1246 [DOI] [PubMed] [Google Scholar]
- Gordon J.W., Scangos G.A., Plotkin D.J., et al. (1980). Genetic transformation of mouse embryos by microinjection of purified DNA. Proc. Natl Acad Sci USA 77, 7380–7384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haskins M.E., Aguirre G.D., Jezyk P.F., et al. (1983). The pathology of the feline model of mucopolysaccharidosis I. Am. J. Pathol. 112, 27–36 [PMC free article] [PubMed] [Google Scholar]
- He J., Yang Q., and Chang L.J. (2005). Dynamic DNA methylation and histone modifications contribute to lentiviral transgene silencing in murine embryonic carcinoma cells. J. Virol. 79, 13497–13508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann A., Kessler B., Ewerling S., et al. (2003). Efficient transgenesis in farm animals by lentiviral vectors. EMBO Rep. 4, 1054–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann A., Zakhartchenko V., Weppert M., et al. (2004). Generation of transgenic cattle by lentiviral gene transfer into oocytes. Biol. Reprod. 71, 405–409 [DOI] [PubMed] [Google Scholar]
- Hofmann A., Kessler B., Ewerling S., et al. (2006). Epigenetic regulation of lentiviral transgene vectors in a large animal model. Mol. Ther. 13, 59–66 [DOI] [PubMed] [Google Scholar]
- Jackson C.E., Yuhki N., Desnick R.J., et al. (1992). Feline aryl-sulfatase B (ARSB): isolation and expression of the cDNA, comparison with human ARSB, and gene localization to feline chromosome A1. Genomics 14, 403–411 [DOI] [PubMed] [Google Scholar]
- Jahner D., Stuhlmann H., Stewart C.L., et al. (1982). De novo methylation and expression of retroviral genomes during mouse embryogenesis. Nature 298, 623–628 [DOI] [PubMed] [Google Scholar]
- Kang Y.K., Koo B.D., Park J.S., et al. (2001). Aberrant DNA methylation of donor genome in cloned embryos. Nat. Genet. 28, 173–177 [DOI] [PubMed] [Google Scholar]
- Kasamatsu A., Saeki K., Iwamoto D., et al. (2006). Bovine somatic cell nuclear transfer embryos: effect of the gene expression ability and methylation on their development to the blastocyst stage. Proc. Soc. Stud. Reprod. 74(special Issue)111 [abst. 176]
- Kitiyanant Y., Saikhun J., and Pavasuthipaisit K. (2003). Somatic cell nuclear transfer in domestic cat oocytes treated with IGF-I for in vitro maturation. Theriogenology 59, 1775–1786 [DOI] [PubMed] [Google Scholar]
- Lois C., Hong E.J., Pease S., et al. (2002). Germline transmission and tissue-specific expression of transgenes delivered by lentiviral vectors. Science 295, 868–872 [DOI] [PubMed] [Google Scholar]
- Mann M.R., Chung Y.G., Nolen L.D., et al. (2003). Disruption of imprinted gene methylation and expression in cloned preimplantation stage mouse embryos. Biol. Reprod. 69, 902–914 [DOI] [PubMed] [Google Scholar]
- McGrew M.J., Sherman A., Ellard F.M., et al. (2004). Efficient production of germline transgenic chickens using lentiviral vectors. EMBO Rep. 5, 728–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki H., Schwartz J.P., Tanaka K., et al. (1998). High-titer human immunodeficiency virus type 1-based vector systems for gene delivery into nondividing cells. J. Virol. 72:8873–8883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohamedali A., Moreau-Gaudry F., Richard E., et al. (2004). Self-inactivating lentiviral vectors resist proviral methylation but do not confer position-independent expression in hematopoietic stem cells. Mol. Ther. 10, 249–259 [DOI] [PubMed] [Google Scholar]
- Murphy W.J., Sun S., Chen Z.Q., et al. (1999). Extensive conservation of sex chromosome organization between cat and human revealed by parallel radiation hybrid mapping. Genome Res 9:1223–1230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy W.J., Sun S., Chen Z-Q., et al. (2000). Radiation hybrid map of the cat genome: implications for comparative mapping. Genome Res. 10, 691–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narfstrom K. (1983). Hereditary progressive retinal atrophy in the Abyssinian cat. J. Hered. 74, 273–276 [DOI] [PubMed] [Google Scholar]
- O'Brien S.J., and Nash W.G. (1982). Genetic mapping in mammals: chromosome map in domestic cat. Science 216, 257–265 [DOI] [PubMed] [Google Scholar]
- O'Brien S.J., Menotti-Raymond M., Murphy W.J., et al. (2002). The Feline Genome Project. Annu. Rev. Genet. 36, 657–686 [DOI] [PubMed] [Google Scholar]
- Park F. (2007). Lentiviral vectors: are they the future of animal transgenesis? Physiol Genomics 31, 159–73 [DOI] [PubMed] [Google Scholar]
- Pfeifer A. (2004). Lentiviral transgenesis. Transgenic Res. 13, 513–522 [DOI] [PubMed] [Google Scholar]
- Pfeifer A., Ikawa M., Dayn Y., et al. (2002). Transgenesis by lentiviral vectors: lack of gene silencing in mammalian embryonic stem cells and preimplantation embryos. Proc. Natl. Acad. Sci. USA 99, 2140–2145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piedrahita J.A. (2000). Gene targeting in domestic species: a new beginning. Transgenic Res. 9, 261–262 [DOI] [PubMed] [Google Scholar]
- Pontius J.U., Mullikin J.C., Smith D.R., et al. (2007). Initial sequence and comparative analysis of the cat genome. Genome Res. 17:1675–1689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiser J. (2000). Production and concentration of pseudotyped HIV-1-based gene transfer vectors. Gene Ther. 7, 910–913 [DOI] [PubMed] [Google Scholar]
- Reiser J., Harmison G., Kluepfel-Stahl S., et al. (1996). Transduction of nondividing cells using pseudotyped defective high-titer HIV type 1 particles. Proc. Natl. Acad. Sci. USA 93, 15266–15271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rettenberger G., Klett Ch., Zechner U., et al. (1995). ZOO-FISH analysis: cat and human karyotypes closely resemble the putative ancestral mammalian karyotype. Chromosome Res, 3, 479–486 [DOI] [PubMed] [Google Scholar]
- Ricks D., Kutner R., Zhang XY., et al. (2008). Optimized lentiviral transduction of mouse bone marrow-mesenchymal stem cells. Stem Cells Dev. 17, 441–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie W.A, King T., Neil C., et al. (2008). Transgenic sheep designed for transplantation studies. Mol. Reprod. Dev. (DOI 10.1002/mrd.20930) [DOI] [PubMed]
- Scott B.B., and Lois C. (2005). Generation of tissue-specific transgenic birds with lentiviral vectors. Proc. Natl. Acad. Sci. USA 102, 16443–16447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin T., Kraemer D., Pryor J., et al. (2002). A cat cloned by nuclear transplantation. Nature 415, 859. [DOI] [PubMed] [Google Scholar]
- Skrzyszowska M., Katska L., Ry§ska B., et al. (2002). In vitro developmental competence of domestic cat embryos after somatic cloning: a preliminary report. Theriogenology 58, 1615–1621 [DOI] [PubMed] [Google Scholar]
- Tanase H., Kudo K., Horikoshi H., et al. (1991). Inherited primary hypothyroidism with thyrotrophin resistance in Japanese cats. J. Endocrinol. 129, 245–251 [DOI] [PubMed] [Google Scholar]
- Thongphakdee A., Numchaisrika P., Omsongkram S., et al. (2006). In vitro development of marbled cat embryos derived from interspecies somatic cell nuclear transfer. Reprod. Dom. Anim. 41, 219–226 [DOI] [PubMed] [Google Scholar]
- Wall R.J. (1996). Transgenic livestock: progress and prospects for the future. Theriogenology 45, 57–68 [Google Scholar]
- Whitelaw C.B., Radcliffe P.A., Ritchie W.A., et al. (2004). Efficient generation of transgenic pigs using equine infectious anemia virus (EIAV) derived vector. FEBS Lett. 571, 233–236 [DOI] [PubMed] [Google Scholar]
- Wienberg J., Stanyon R., Nash W.G., et al. (1997). Conservation of human vs. feline genome organization revealed by reciprocal chromosome painting. Cytogenet. Cell Genet. 77, 211–217 [DOI] [PubMed] [Google Scholar]
- Wiznerowicz M., and Trono D. (2005). Harnessing HIV for therapy, basic research and biotechnology. Trends Biotechnol. 23, 42–47 [DOI] [PubMed] [Google Scholar]
- Wolfgang M.J., Eisele S.G., Browne M.A., et al. (2001). Rhesus monkey placental transgene expression after lentiviral gene transfer into preimplantation embryos. Proc. Natl. Acad. Sci. USA 98, 10728–10732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S-H., Cheng P-H., Banta H., et al. (2008). Towards a transgenic model of Huntington's disease in a non-human primate. Nature doi: 10.1038/nature06975 [DOI] [PMC free article] [PubMed]
- Yao S., Sukonnik T., Kean T., et al. 2004. Retrovirus silencing, variegation, extinction, and memory are controlled by a dynamic interplay of multiple epigenetic modifications. Mol. Ther. 10, 27–36 [DOI] [PubMed] [Google Scholar]
- Yin X.J., Lee H.S., Lee Y.H., et al. (2005). Cats cloned from fetal and adult somatic cells by nuclear transfer. Reproduction 129, 245–249 [DOI] [PubMed] [Google Scholar]
- Yin X.J., Lee Y.H., Jin J.Y., et al. (2006). Nuclear and microtubule remodeling and in vitro development of nuclear transferred cat oocytes with skin fibroblasts of the domestic cat (Felis silvestris catus) and leopard cat (Prionailurus bengalensis). Anim. Reprod. Sci. 95, 307–315 [DOI] [PubMed] [Google Scholar]
- Yin J.Y., Lee H.S., Kim L.H., et al. (2007). Effect of serum starvation on the efficiency of nuclear transfer using odd-eyed white cat fibroblasts. Theriogenology 67, 816–823 [DOI] [PubMed] [Google Scholar]
- Yin X.J., Lee H.S., Yu X.F., et al. (2008). Generation of cloned transgenic cats expressing red fluorescence protein. Biol. Reprod. 78, 425–431 [DOI] [PubMed] [Google Scholar]
- Zhang X.Y., La Russa V.F., and Reiser J. (2004). Transduction of bone-narrow-derived mesenchymal stem cells by using lentivirus vectors pseudotyped with modified RD114 envelope glycoproteins. J. Virol. 78, 1219–1229 [DOI] [PMC free article] [PubMed] [Google Scholar]