Abstract
In mammals, RNA editing by site-selective adenosine deamination regulates key functional properties of neurotransmitter receptors in the central nervous system. Glutamate receptor subunit B is nearly 100% edited at one position (the Q/R-site), which is essential for normal receptor function. Its significance is apparent from mouse models in which a slightly reduced rate of Q/R-site editing is associated with early onset epilepsy and premature death. Here we report that in tissues from malignant human brain tumors, this editing position of glutamate receptor subunit B is substantially underedited compared with control tissues. We also observe alterations in editing and alternative splicing of serotonin receptor 5-HT2C transcripts. These changes correlate with a decrease in enzymatic activity of the editing enzyme adenosine deaminase acting on RNA (ADAR) 2, as deduced from analysis of ADAR2 self-editing. Our results suggest a role for RNA editing in tumor progression and may provide a molecular model explaining the occurrence of epileptic seizures in association with malignant gliomas.
The process of RNA editing is a widespread phenomenon in eukaryotes that leads to posttranscriptional base changes in mRNA and tRNA (reviewed in refs. 1 and 2). In mammals, a growing number of genes have been identified that undergo a type of RNA editing that is characterized by site-selective adenosine-to-inosine modification (3, 4). Because inosine is read as guanosine during translation, the base changes alter codons and therefore amino acid sequences, often with profound consequences for protein function. The best-studied A-to-I editing substrates are the brain-specific transcripts coding for glutamate receptor (GluR) channels (5) and G-protein-coupled serotonin receptors (6). In GluR subunit B (GluR-B), a single editing position (the Q/R-site) controls the Ca2+-permeability of the ion channel (7), whereas another position (the R/G-site) regulates the desensitization kinetics of the receptor (8). At the Q/R-site, RNA editing almost quantitatively (>99.9%) changes the glutamine (Q) codon CAG to a CIG, specifying arginine (R). This residue is the molecular determinant for low Ca-permeability of α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA)-type GluRs imparted dominantly by edited GluR-B (7). This property of AMPA receptors is critical for normal brain function, as became evident when transgenic mice that express unedited GluR-B were generated (9). Low levels of GluR-B (Q) coexpressed with GluR-B (R) lead to an epileptic phenotype and premature lethality as a consequence of the increased Ca2+-permeability of AMPA receptors (9). Excessive calcium influx is also known to be toxic and causes neuronal degeneration (10). Because of the importance of accurate RNA editing for normal brain function, the deregulation of editing activity may influence the progression of pathophysiological processes, such as neurodegenerative diseases or tumorigenesis.
Glial cells, the most numerous cells in the brain, have recently been recognized to play an active role in synaptic signaling through glial/neuronal networks (11–13). They express neurotransmitter receptors, release neurotransmitters, and propagate intracellular Ca2+-waves (reviewed in refs. 14–16). The latter, mediated by AMPA-type GluRs, have been shown to influence the activity of adjacent neurons and modulate synaptic transmission (17–19). GluR-B represents a major AMPA receptor subunit expressed in many astrocytes (14, 20).
Neoplasms of glial cells represent the most common tumors of the central nervous system, with glioblastoma multiforme (GBM), which is almost invariably fatal, being the most malignant form (reviewed in ref. 21). The molecular basis for malignant progression in gliomas is not clear but probably involves the collaborative effects of dominant and recessive lesions as well as epigenetic changes (21, 22). Epileptic seizures contribute substantially to the neurologic morbidity of patients with malignant gliomas (reviewed in ref. 23). Here we show that, in human GBM, severe alterations in RNA editing exist in Glu-B transcripts and also changes in the alternative splicing pattern of another edited gene, the 5-HT2C serotonin receptor. In all tumor-derived tissue samples, the Q/R-site of GluR-B is substantially underedited compared with control tissues from gray and white matter. In a sample of a low-grade astrocytoma [World Health Organization (WHO) grade II], similar but less severe changes are detectable. Alternative splicing of 5-HT2C-transcripts in gliomas is shifted toward a variant that results in a truncated nonfunctional receptor. The expression levels of the known editing enzymes adenosine deaminase acting on RNA (ADAR)1 and ADAR2 are not significantly altered in tumors as analyzed by quantitative real-time PCR. However, we found evidence for a specific decrease in the activity of ADAR2 through analysis of an ADAR2-specific editing site located within the ADAR2 pre-mRNA itself.
Taken together, the observed changes in RNA editing and alternative splicing predict physiological changes consistent with neurologic symptoms observed in patients with GBM.
Materials and Methods
Cell Lines, Tumor Samples, and Control Tissues.
Human glioblastoma cell lines A172 [CRL-1620, American Type Culture Collection (ATCC)], and M095K (ATCC CRL-2365) were grown in DMEM supplemented with 10% FCS (Invitrogen). The human glioblastoma tissue samples were either obtained from the Institute of Pathology of the Friedrich Schiller University or kindly provided by David N. Louis, Molecular Neuro-Oncology Laboratory, Massachusetts General Hospital. Tumors GBM1–7 were classified as GBM (WHO grade IV), tumor AC1 as oligoastrocytoma WHO grade II. Tissue samples from postmortem human brain (coronal slab no. 9 and white matter) were obtained from the Harvard Brain Tissue Resource Center, Belmont, MA. The study was approved by the local committee on the use of humans as experimental subjects.
Stable cell lines were generated by using standard protocols. The hADAR1 cDNA (24) or the rADAR2 ORF (25) were cloned into the pEXFH plasmid vector (26), adding an in-frame FLAG tag at the N terminus and a 6-His tag at the C terminus when expressed in the transfected cell lines. Geneticin (Invitrogen)-resistant cell clones were analyzed by Western blot with monoclonal anti-FLAG antibody (Sigma) for expression of recombinant protein and nuclear extracts assayed for adenosine deaminase activity on double-stranded RNA (27).
RNA Isolation and Reverse Transcription (RT)-PCR.
Total RNA from either cultured glioblastoma cell lines or 100- to 300-mg tumor or control brain tissues was isolated with TRIzol Reagent (GIBCO), according to manufacturer's instructions, and subsequently digested with RNase-free DNase I (Roche Diagnostics) and tested for integrity. Ten micrograms of total RNA was used to synthesize cDNA by using SuperScript II reverse transcriptase (Invitrogen) and random hexamer primers. All PCRs were run including “mock”-transcribed RNA as negative control and, in addition, the PCR primers were designed to generate amplicons of the expected size only from mRNA but not from genomic DNA or pre-mRNA. For amplification of GluR-B cDNA fragments covering the Q/R and +4 editing positions, PCR with primers hRB1D 5′-TGCGGTACCTTTAGCCTATGAGATCTGGATGTGC-3′ (exon 11, sense, KpnI) and hRB2U 5′-CAGAATTCGTGTAGGAGGAGATTATGATCAGG-3′ (exon 12, antisense, EcoRI) was performed with 7 cycles of 94°C for 20 s, 57°C for 30 s, 72°C for 30 s, and 30 cycles of 94°C for 20 s, 55°C for 30 s, and 72°C for 30 s, as well as a final extension at 72°C for 10 min. For analysis of the GluR-B R/G position, the same PCR protocol was used with primers hRBRGD, 5′-TGCGGTACCACCATGAACGAGTACATTGAG-3′ (exon 13, sense, KpnI) and hRBRGU 5′-CAGAATTCAAGGATGTAGAATACTCCAGCAAC-3′ (exon 16, antisense, EcoRI). Serotonin receptor 2C subunit cDNA fragments covering the editing sites A–E were amplified with primers 5-HT1D (sense, exon 3) 5′-CAGAATTCTGGCCACTACCTAGATATTTGTGC-3′ and 5-HT2U (antisense, exon 4) 5′-CTTGCGGTACCTCAGGAAATCCAGACTTAGTCCAG-3′ by using the same PCR protocol with a 45-s extension time. For analysis of human ADAR2 alternative splicing a PCR with primers A2D 5′-GTATTTTGCCATGGATATAGAAGATG-3′ (sense, exon 1) and A2U 5′-GTACTGGGATCCAGGCTTGATCTCATTCAGCTG-3′ (antisense, exon 2) was performed which amplifies both splice variants from cDNA. Intronic editing in ADAR2 pre-mRNA was quantified from PCR products with primers A2I1D 5′-GGAATTCTATTAGTCACTAAGCAAAGTGTCAG-3′ (sense, intron 1) and A2E2U 5′-GCGGTACCCAGGTGTGCTGCCATCCTTGG-3′ (antisense, exon 2).
Analysis of RNA Editing.
RT-PCR amplicons from GluR-B were either directly sequenced after gel purification or subcloned and individual cDNA-bearing plasmid clones subjected to sequencing. Dideoxy sequencing was done by the Massachusetts Institute of Technology Cancer Center Sequencing Facility. For subcloning, PCR products were digested with restriction enzymes EcoRI and KpnI before gel purification and ligated into EcoRI/KpnI cut pBS II (Stratagene) plasmid vector. A-to-G changes in the individual clones were analyzed by single-nucleotide sequencing.
For analysis of 5-HT2C receptor editing, the 480-bp PCR amplicon was used as described for the GluR-B products. Alternative splicing was semiquantitatively determined by band densitometry of ethidium bromide stained agarose gels (Kodak software).
Alternatively, PCR amplicons of GluR-B or 5-HT2C receptor subunits were analyzed by poisoned primer extension analysis, as described (28).
ADAR Expression Analysis.
Real-time quantitative PCR for expression analysis of ADAR1 and ADAR2 was performed by using the SYBR Green PCR system (PE Applied Biosystems). Gene-specific PCR products were continuously measured by means of an Applied Biosystems prism 7700 sequence detection system (PE Applied Biosystems) during 40 cycles. 18S ribosomal RNA was used for normalization and relative quantification of gene expression performed according to the ΔΔCt method (PE Biosystems User Bulletin No. 2).
Results
The Q/R-Site of GluR-B Is Underedited in Malignant Gliomas.
The extent of RNA editing at the Q/R site of GluR-B is always virtually 100% throughout the mammalian brain, including white matter (7, 29). Because the status of the Q/R editing site is of major physiological importance, as evident from mouse models with decreased rates of Q/R editing (9, 30), we first analyzed the editing status of GluR-B in tissues from human gliomas, glioma-derived cell lines, and normal controls. A GluR-B cDNA fragment covering the Q/R-site as well as the +4 editing position was PCR amplified from reverse-transcribed total RNAs of the following tissues and cell lines: (i) seven GBM tissue samples (WHO grade IV); (ii) one astrocytoma tissue (WHO grade II) representing a low-grade glioma; (iii) human brain control tissues of gray matter (cortex) and white matter (WM1, WM2), the latter presumably composed mainly of glial cells; and (iv) human glioma-derived cell lines A172 and M095K.
For analysis, we directly sequenced control and tumor-derived GluR-B PCR amplicons as well as the corresponding region of the GluR-B gene from human genomic DNA. The genomically encoded glutamine (Q) codon CAG is edited to an arginine (R) codon (appears as CGG in cDNA) to ≈100% in control brain tissues of gray and white matter (Fig. 1 A and B).
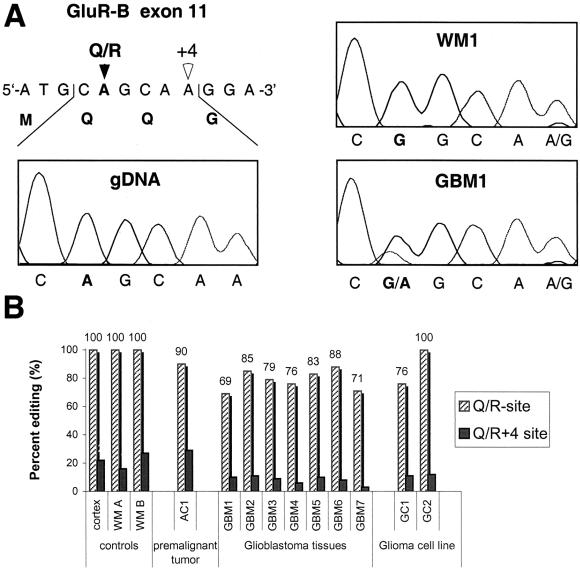
Figure 1.
GluR-B Q/R site editing in human brain and gliomas. (A) Direct sequencing of GluR-B specific PCR amplicons from human genomic DNA (gDNA), reverse-transcribed RNA from white matter of human brain (WM1), and glioblastoma tissue (GBM1). Electropherograms covering the Q/R editing position within exon 11 are shown. Mixed sequence signals (A and G) indicate RNA editing, and the ratios of the peak heights reflect the extent of Q/R site editing. (B) Quantitation of editing of the Q/R site and Q/R +4 position in human control brain from gray matter (cortex) and white matter (WM A, WM B) in comparison to values from one low-grade human astrocytoma (AC1), various high-grade glioblastomas (GBM1–7), and glioma-derived cell lines GC1 and GC2. Percent values for examined samples, each derived from direct sequencing of at least three independent amplifications, poisoned primer extension analysis as well as subsequent sequence analysis of at least 50 individually subcloned cDNAs.
In contrast, all GBM tissues show a decreased level of Q/R-site editing, ranging from 69 to 88%, indicated by the mixed signals of A and G at the Q/R position (Fig. 1 A and B). Of the two glioma-derived cell lines, GC1 also exhibits lower Q/R editing (76%), whereas the GC2 cell line edits to 100%. The +4 editing position, which does not lead to an amino acid change, displays a very similar pattern with lowered levels of editing in the GBM tissues and the glioma derived cell lines (Fig. 1B). Analysis of GluR-B transcripts from the low-grade astrocytoma (AC1) also reveals a decrease in Q/R site editing (90%), but less drastic than in GBMs.
We subsequently analyzed editing at the GluR-B R/G-site, which is known to modulate channel desensitization kinetics (8) and is subject to ontogenetic as well as cell-type-specific regulation (8). We found 0% editing in samples from white matter versus 58% in gray matter of control brain (data not shown). In the tumor specimens, R/G editing was either undetectable (0%; tumors GBMs1, 3–7) or of very low percentage (4%; tumor GBM2) (data not shown).
Analysis of Alternative Splicing and Editing of Serotonin Receptor 5HT2C Transcripts.
The pre-mRNA of the serotonin receptor 5HT2C subunit is edited at five major positions, which are located in the second intracellular loop of the G-protein-coupled transmembrane protein (6). RNA editing influences the efficiency of receptor-to-G-protein coupling on ligand binding (6). Within the normal brain, large cell-type-specific differences in editing levels have been reported (6). To determine the editing status of the 5HT2C mRNA, we amplified the corresponding cDNA region covering exons 3 and 4. With samples from control human brain, we obtained one major PCR product of the expected size (480 bp) as well as a low amount (≈5%) of an amplicon with smaller size (385 bp). Sequencing of both PCR products revealed that the short fragment originated from alternative splicing of the 5HT2C pre-mRNA, leading to the deletion of 95 bp from the 3′-region of exon 3 (Fig. 2A). Such an alternative splice form of the 5HT2C receptor had been described before (31, 32), detectable in human, rat, and mouse specimens. It was shown to produce a truncated nonfunctional receptor (31). Intriguingly, the deleted fragment of exon 3 harbors all of the known editing sites within the 5HT2C mRNA (Fig. 2A).
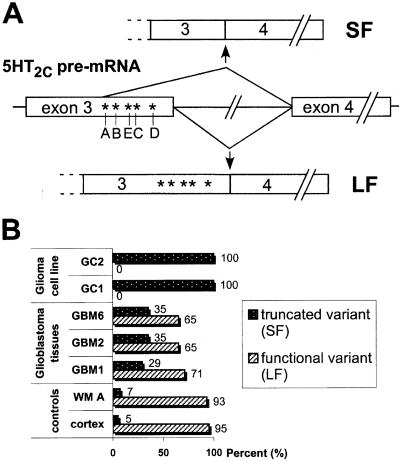
Figure 2.
(A) Editing and alternative splicing of 5HT2C receptor pre-mRNA. Schematic representation of the serotonin receptor 5HT2C pre-mRNA, with the positions of the major sites for RNA editing (A–E) depicted. The positions of alternative splicing affecting the same region are indicated. Splicing of intron 3 leads to the long splice variant (LF), resulting in a functional receptor including the edited residues. Alternative splicing with partial deletion of exon 3 gives rise to the short splice form (SF), which is translated to yield an inactive receptor molecule and also lacks the edited sites. (B) Ratios of the 5HT2C splice forms in control human brain (cortex, WM A) versus glioma tissues (GBM1, 2, 6) and glioma-derived cell lines (GC1, 2). Percent values for the truncated variant (SF) and functional variant (LF) are determined by semiquantitative RT-PCR.
Surprisingly, we found that in the tumor tissues, the ratio of the two alternative splice forms was shifted substantially toward the short variant (Fig. 2B). In samples from the glioma-derived cell lines, virtually all 5HT2C mRNA was in the short form, whereas the long splice variant (coding for the functional receptor subunit) was undetectable.
We then analyzed the editing status of positions A–E within the long splice variant from tumor samples and controls. In comparison to the control tissues, large differences in the extent of editing are evident in the tumors, however with no clear trend (data not shown). Note that several of the GBM tumors lacked detectable expression of 5HT2C mRNA.
ADAR2 Self-Editing as a Marker for ADAR2 Deaminase Activity in Vivo.
In vivo, ADAR2 is the adenosine deaminase primarily responsible for Q/R site editing (30). Underediting at this position might be due to decreased expression of ADAR2 or down-regulation of ADAR2 enzymatic activity. As judged by quantitative expression analysis 1 using real-time PCR, the expression of ADAR2 mRNA, as well as ADAR1 mRNA, is not significantly altered in the tumor tissues (data not shown).
The ADAR2 editing enzyme also edits its own transcript, leading to alternative splicing of the pre-mRNA (33) (Fig. 3A). We asked whether the extent of ADAR2 alternative splicing caused by self-editing might be an accurate and specific marker for ADAR2 activity within a cell, allowing us to monitor its enzymatic activity without the need to separate this enzyme from other adenosine deaminase activities. As a prerequisite, the ratio of edited to unedited ADAR2 RNA (and hence alternative splice form to regular splice form) should correlate with ADAR2 activity/expression, without being influenced by ADAR1 activity, which otherwise often displays overlapping substrate specificity with ADAR2 (25).
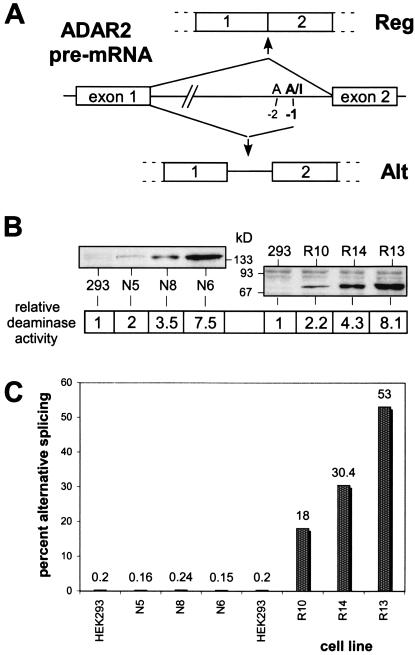
Figure 3.
ADAR2 self-editing as an ADAR2 activity marker. (A) Intronic editing of ADAR2 pre-mRNA and resulting splice products. A-to-I editing at the −1 position by ADAR2 leads to partial inclusion of intron1. Only the regular splice form (Reg) results in a functional enzyme. (B) Characterization of stably transformed human embryonic kidney (HEK)293 cell lines expressing increasing amounts of the editing enzyme ADAR1 (N5, N8, N6) or ADAR2 (R10, R14, R13). Immunoblot analysis of cellular extracts from stable cell lines documents different ADAR expression levels. The relative adenosine deaminase activities measured in vitro on extended double-stranded RNA of the extracts (normalized to the activity of extract from native HEK 293 cells) are indicated. (C) Analysis of ADAR2 editing in ADAR-expressing cell lines. The percent values for the alternative splice form of ADAR2 resulting from ADAR2 pre-mRNA editing are given as measured by semiquantitative RT-PCR.
To clarify this question, we generated stably transfected human cell lines that ectopically express a wide range of ADAR1 or ADAR2 protein (see Fig. 3B). We chose human embryonic kidney cells (HEK 293) as parental cell line because of their low endogenous ADAR expression (28). Analysis of endogenous ADAR2 alternative splicing in these cell lines revealed a direct correlation with ADAR2 expression levels as judged by semiquantitative RT-PCR and Western blotting (Fig. 3 B and C). Whereas we detected less than 0.5% of the alternative splice form in native HEK 293 cells, the amount increases to 18, 30, and 53% in the R10, R14, and R13 cell lines, respectively. These cell lines express increasing amounts of ADAR2. In contrast, cell lines that overexpress equivalent amounts of ADAR1 (N5, N8, N6) show no alteration in the ratio of ADAR2 alternative splice products.
Sequence analysis of PCR products of ADAR2 pre-mRNA covering the region around the ADAR2 editing site confirmed the observed alternative splicing pattern (Fig. 3A and Table 1). An elevation in the extent of modification at the −1 position from 0 to 13% (R13 cell line) occurred solely in the ADAR2-overexpressing cell lines (Table 1). The differences between −1 editing rates and percentage of alternative splicing indicate that the edited ADAR2 pre-mRNA might be spliced preferentially. We did not detect any editing at the −2 position, which would prevent alternative splicing regardless of the editing status at −1 (Fig. 3A and Table 1). Editing at another intronic position (−24) was also not affected by ADAR2 overexpression in the cell lines (Table 1).
Table 1.
ADAR2 self-editing
| Sample | Editing position
|
||
|---|---|---|---|
| −24 | −2 | −1 | |
| Cortex | 5.7 | <0.2 | 6 |
| WM | 5.2 | <0.2 | 4 |
| GBM2 | 7.3 | <0.2 | <0.2 |
| GBM3 | 2.3 | <0.2 | <0.2 |
| GBM4 | 7.1 | <0.2 | <0.2 |
| GBM5 | 8.0 | <0.2 | <0.2 |
| GBM6 | 5.6 | <0.2 | <0.2 |
| GC1 | 8.1 | <0.2 | <0.2 |
| HEK293 | 6.2 | <0.2 | <0.2 |
| N5 | 5.9 | <0.2 | <0.2 |
| N8 | 6.6 | <0.2 | <0.2 |
| N6 | 7.7 | <0.2 | <0.2 |
| R10 | 5.1 | <0.2 | 2 |
| R14 | 5.0 | <0.2 | 7 |
| R13 | 5.0 | <0.2 | 13 |
Values are in percent (%). The edited nucleotides are numbered, with position −1 representing the splice-acceptor site when edited.
We then analyzed ADAR2 self-editing and alternative splicing in control human brain and tumor tissues as well as the glioma-derived cell lines. In normal human brain, alternative splicing of ADAR2 is measured at ≈10%. In tumor specimens, we were not able to detect any alternative splicing of ADAR2 mRNA (Table 1), which argues for a substantial decrease in ADAR2 activity and corroborating the results obtained for GluR-B Q/R-site editing.
Discussion
Tumorigenesis, as well as the progression from early-stage cancer to the most malignant and invasive tumor growth, involves a multitude of cellular changes with deregulation of signaling pathways that affect cell growth, survival, or proliferation. Any gene product involved in such processes represents a potential oncogene or tumor suppressor, because its hyperactivation or inactivation may contribute to the initiation or progression of cancerous growth. Aberrant posttranscriptional processing of such genes has been recognized as one possible mechanism, involved, for example, through changes in the pattern of alternative splicing (34, 35). In this study, we asked whether the cellular machinery for A-to-I RNA editing might be directly or indirectly involved in the progression of cancerous growth within the human brain, where a number of important targets for RNA editing reside. Our analysis of human glioma tissues uncovered a significant decrease in GluR-B Q/R site editing compared with control human brain, accompanied by the decrease in editing levels at the adjacent +4 adenosine. The measured percentage of GluR-B(Q) in tumors (12–31%) corresponds to an increase in the amount of unedited GluR-B of more than 300-fold. The extent of underediting at the Q/R site predicts increased Ca2+-permeability of AMPA-type GluRs with substantial cell physiologic consequences, as GluR-B constitutes a major AMPA receptor subunit expressed in many astrocytes (14, 20). It is tempting to speculate that the prevalence of epileptic seizures in patients with gliomas could be at least partly a direct result of the change in Q/R site editing. This hypothesis is supported by the fact that an epileptic phenotype was observed in transgenic mice with lowered Q/R site editing in the central nervous system (9, 30).
The editing of the R/G site of GluR-B, as well as the 5HT2C serotonin receptor sites A to E, is not as suitable for characterizing changes in editing pattern between different tissues. In both cases, there is high variability between different regions and cell types throughout the brain. Most glial cells do not seem to edit the GluR-B R/G site at all. We detected a drastic shift in the alternative splicing pattern of the 5HT2C receptor transcripts in tumor tissues versus controls. The relative increase in the splice variant, which leads to a truncated receptor, indicates a reduction in the total amount of functional serotonin receptors. It is noteworthy that complete ablation of the 5HT2C gene also leads to an epileptic phenotype in mice (36). Thus, the functional inactivation of 5HT2C receptors as a result of changes in alternative splicing could further increase the likelihood of epileptic seizures in the Q/R-editing-compromised tumor cells and surrounding tissue.
Because the Q/R site of GluR-B is targeted specifically by the ADAR2 editing enzyme, underediting at this position might be due to decreased expression of ADAR2 or down-regulation of ADAR2 enzymatic activity. A decrease in GluR-B Q/R site editing to the observed levels would require a reduction in ADAR2 expression of more than 50%, as judged from Q/R editing in gene-targeted mice with only one functional ADAR2 allele (30). In these mice, more than 99% of the GluR-B mRNA is still edited at the Q/R site. Because the expression level of ADAR2 mRNA is not significantly altered in the tumor cells, the observed changes in Q/R editing levels are not simply because of down-regulation of ADAR2 expression.
For assessment of ADAR2 deaminase activity, we analyzed the status of ADAR2 self editing, as it has been postulated to be involved in an auto-feedback loop-regulating ADAR2 activity (33). Using as a model system a set of human cell lines engineered to express different levels of the editing enzyme ADAR1 or ADAR2, we observed a direct and specific correlation between ADAR2 activity and the extent of self editing at the −1 position. Judging from the status of this ADAR2-specific target, a reduction in ADAR2 activity is evident in all tumor samples in comparison to control brain.
Taken together, our data show that, beginning in the early stages of tumorigenesis, ADAR2 activity is down-regulated, which is clearly detectable even in a low-grade astrocytoma. The cellular mechanisms that regulate ADAR2 activity are not known, but they might involve posttranslational modifications or controlled subcellular localization of the enzyme. In future work, it will be interesting to compare tumor and adjacent neuronal tissues from the same patients to see whether the underediting is confined to the tumor cells or is also induced within the neighboring tissues. It is known that neurons from cortex adjacent to tumors display abnormal electrophysiology, with an increase in spontaneous activity under resting conditions (37). This observation could explain why, in patients with gliomas, epileptic seizures often persist even after removal of the primary tumor (38, 39). It is possible that the physiological changes elicited in the tumor cells aberrantly modulate synaptic transmission of adjacent neurons through glia/neuronal networks (11, 12). In addition to its role in cell-to-cell signaling, it is known that the influx of Ca2+ on GluR activation modulates gene expression and cell proliferation in neurons and glial cells (40). The increased Ca-influx through GluRs with unedited GluR-B could therefore contribute to the aggressive growth behavior of tumor cells. Deregulation of RNA editing activity might also occur in other types of cancer, given the ubiquitous expression of ADARs. However, to date, the ADAR2 mRNA is the only well characterized editing substrate expressed in non-neuronal tissues. Only after identification of additional ADAR targets will it be possible to test if RNA editing becomes deregulated also in other cancers.
This work provides insights into the pathogenesis of human brain tumors, particularly GBMs. ADAR2 activity, as assessed by measuring the editing status of its specific targets, might represent a valid and useful molecular marker for categorization and prognosis of glioblastomas. It also constitutes a potential target for antiepileptic drugs that could be designed to enhance the editing process. Other drugs might selectively inhibit unedited GluR-B- or Ca2+-permeable AMPA receptors.
Acknowledgments
We thank Dr. D. Louis for GBM tissue specimens, Dr. K. Nishikura for hADAR1 cDNA, and Dr. P. H. Seeburg for rADAR2 cDNA. Postmortem human brain control tissues were provided by the Harvard Brain Tissue Resource Center, which is supported in part by Public Health Service Grant no. MH/NS 31862. This work was supported by an Anna Fuller fellowship in Molecular Oncology (to S.M.) and by grants from The National Institutes of Health, the National Science Foundation, and the National Foundation for Cancer Research.
Abbreviations
- GluR
glutamate receptor
- GluR-B
GluR subunit B
- AMPA
α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid
- ADAR
adenosine deaminase acting on RNA
- WHO
World Health Organization
- GBM
glioblastoma multiforme
- RT
reverse transcription
References
- 1.Maas S, Rich A. BioEssays. 2000;22:790–802. doi: 10.1002/1521-1878(200009)22:9<790::AID-BIES4>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 2.Gerber A P, Keller W. Trends Biochem Sci. 2001;26:376–384. doi: 10.1016/s0968-0004(01)01827-8. [DOI] [PubMed] [Google Scholar]
- 3.Bass B L. Trends Biochem Sci. 1997;22:157–162. doi: 10.1016/s0968-0004(97)01035-9. [DOI] [PubMed] [Google Scholar]
- 4.Rueter S M, Emeson R B. In: Modification and Editing of RNA. Grosjean H, Benne R, editors. Washington, DC: Am. Soc. Microbiol.; 1998. pp. 343–361. [Google Scholar]
- 5.Seeburg P H, Higuchi M, Sprengel R. Brain Res Brain Res Rev. 1998;26:217–229. doi: 10.1016/s0165-0173(97)00062-3. [DOI] [PubMed] [Google Scholar]
- 6.Burns C M, Chu H, Rueter S M, Hutchinson L K, Canton H, Sanders-Bush E, Emeson R B. Nature (London) 1997;387:303–308. doi: 10.1038/387303a0. [DOI] [PubMed] [Google Scholar]
- 7.Sommer B, Kohler M, Sprengel R, Seeburg P H. Cell. 1991;67:11–19. doi: 10.1016/0092-8674(91)90568-j. [DOI] [PubMed] [Google Scholar]
- 8.Lomeli H, Mosbacher J, Melcher T, Hoger T, Geiger J R, Kuner T, Monyer H, Higuchi M, Bach A, Seeburg P H. Science. 1994;266:1709–1713. doi: 10.1126/science.7992055. [DOI] [PubMed] [Google Scholar]
- 9.Brusa R, Zimmermann F, Koh D S, Feldmeyer D, Gass P, Seeburg P H, Sprengel R. Science. 1995;270:1677–1680. doi: 10.1126/science.270.5242.1677. [DOI] [PubMed] [Google Scholar]
- 10.Choi D W. Trends Neurosci. 1995;18:58–60. [PubMed] [Google Scholar]
- 11.Parpura V, Haydon P G. Proc Natl Acad Sci USA. 2000;97:8629–8634. doi: 10.1073/pnas.97.15.8629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.LoTurco J J. Proc Natl Acad Sci USA. 2000;97:8196–8197. doi: 10.1073/pnas.97.15.8196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bergles D E, Roberts J D, Somogyi P, Jahr C E. Nature (London) 2000;405:187–191. doi: 10.1038/35012083. [DOI] [PubMed] [Google Scholar]
- 14.Steinhauser C, Gallo V. Trends Neurosci. 1996;19:339–345. doi: 10.1016/0166-2236(96)10043-6. [DOI] [PubMed] [Google Scholar]
- 15.Cotrina M L, Lin J H, Lopez-Garcia J C, Naus C C, Nedergaard M. J Neurosci. 2000;20:2835–2844. doi: 10.1523/JNEUROSCI.20-08-02835.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Araque A, Parpura V, Sanzgiri R P, Haydon P G. Trends Neurosci. 1999;22:208–215. doi: 10.1016/s0166-2236(98)01349-6. [DOI] [PubMed] [Google Scholar]
- 17.Smit A B, Syed N I, Schaap D, van Minnen J, Klumperman J, Kits K S, Lodder H, van Der Schors R C, van Elk R, Sorgedrager B, et al. Nature (London) 2001;411:261–268. doi: 10.1038/35077000. [DOI] [PubMed] [Google Scholar]
- 18.Parpura V, Basarsky T A, Liu F, Jeftinija K, Jeftinija S, Haydon P G. Nature (London) 1994;369:744–747. doi: 10.1038/369744a0. [DOI] [PubMed] [Google Scholar]
- 19.Cornell-Bell A H, Finkbeiner S M, Cooper M S, Smith S J. Science. 1990;247:470–473. doi: 10.1126/science.1967852. [DOI] [PubMed] [Google Scholar]
- 20.Patneau D K, Wright P W, Winters C, Mayer M L, Gallo V. Neuron. 1994;12:357–371. doi: 10.1016/0896-6273(94)90277-1. [DOI] [PubMed] [Google Scholar]
- 21.Louis D N, Gusella J F. Trends Genet. 1995;11:412–415. doi: 10.1016/s0168-9525(00)89125-8. [DOI] [PubMed] [Google Scholar]
- 22.Fearon E R. Science. 1997;278:1043–1050. doi: 10.1126/science.278.5340.1043. [DOI] [PubMed] [Google Scholar]
- 23.Villemure J G, de Tribolet N. Curr Opin Neurol. 1996;9:424–428. doi: 10.1097/00019052-199612000-00005. [DOI] [PubMed] [Google Scholar]
- 24.Kim U, Wang Y, Sanford T, Zeng Y, Nishikura K. Proc Natl Acad Sci USA. 1994;91:11457–11461. doi: 10.1073/pnas.91.24.11457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Melcher T, Maas S, Herb A, Sprengel R, Seeburg P H, Higuchi M. Nature (London) 1996;379:460–464. doi: 10.1038/379460a0. [DOI] [PubMed] [Google Scholar]
- 26.Maas S, Gerber A P, Rich A. Proc Natl Acad Sci USA. 1999;96:8895–8900. doi: 10.1073/pnas.96.16.8895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.O'Connell M A, Keller W. Proc Natl Acad Sci USA. 1994;91:10596–10600. doi: 10.1073/pnas.91.22.10596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maas S, Melcher T, Herb A, Seeburg P H, Keller W, Krause S, Higuchi M, O'Connell M A. J Biol Chem. 1996;271:12221–12226. doi: 10.1074/jbc.271.21.12221. [DOI] [PubMed] [Google Scholar]
- 29.Paschen W, Hedreen J C, Ross C A. J Neurochem. 1994;63:1596–1602. doi: 10.1046/j.1471-4159.1994.63051596.x. [DOI] [PubMed] [Google Scholar]
- 30.Higuchi M, Maas S, Single F N, Hartner J, Rozov A, Burnashev N, Feldmeyer D, Sprengel R, Seeburg P H. Nature (London) 2000;406:78–81. doi: 10.1038/35017558. [DOI] [PubMed] [Google Scholar]
- 31.Canton H, Emeson R B, Barker E L, Backstrom J R, Lu J T, Chang M S, Sanders-Bush E. Mol Pharmacol. 1996;50:799–807. [PubMed] [Google Scholar]
- 32.Xie E, Zhu L, Zhao L, Chang L S. Genomics. 1996;35:551–561. doi: 10.1006/geno.1996.0397. [DOI] [PubMed] [Google Scholar]
- 33.Rueter S M, Dawson T R, Emeson R B. Nature (London) 1999;399:75–80. doi: 10.1038/19992. [DOI] [PubMed] [Google Scholar]
- 34.Ekstrand B C, Mansfield T A, Bigner S H, Fearon E R. Oncogene. 1995;11:2393–2402. [PubMed] [Google Scholar]
- 35.Ge K, DuHadaway J, Du W, Herlyn M, Rodeck U, Prendergast G C. Proc Natl Acad Sci USA. 1999;96:9689–9694. doi: 10.1073/pnas.96.17.9689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tecott L H, Sun L M, Akana S F, Strack A M, Lowenstein D H, Dallman M F, Julius D. Nature (London) 1995;374:542–546. doi: 10.1038/374542a0. [DOI] [PubMed] [Google Scholar]
- 37.Strowbridge B W, Masukawa L M, Spencer D D, Shepherd G M. Brain Res. 1992;587:158–163. doi: 10.1016/0006-8993(92)91440-p. [DOI] [PubMed] [Google Scholar]
- 38.Patt S, Steenbeck J, Hochstetter A, Kraft R, Huonker R, Haueisen J, Haberland N, Ebmeier K, Hliscs R, Fiehler J, et al. Neurobiol Dis. 2000;7:260–269. doi: 10.1006/nbdi.2000.0288. [DOI] [PubMed] [Google Scholar]
- 39.Beaumont A, Whittle I R. Acta Neurochir. 2000;142:1–15. doi: 10.1007/s007010050001. [DOI] [PubMed] [Google Scholar]
- 40.Pende M, Holtzclaw L A, Curtis J L, Russell J T, Gallo V. Proc Natl Acad Sci USA. 1994;91:3215–3219. doi: 10.1073/pnas.91.8.3215. [DOI] [PMC free article] [PubMed] [Google Scholar]