Abstract
Mechanical feedback from the tumor microenvironment regulates an array of processes underlying cancer biology. For example, increased stiffness of mammary extracellular matrix (ECM) drives malignancy and alters the phenotypes of breast cancer cells. Despite this link, the role of substrate stiffness in chemotherapeutic response in breast cancer remains unclear. This is complicated by routine culture and adaptation of cancer cell lines to unnaturally rigid plastic or glass substrates, leading to profound changes in their growth, metastatic potential and, as we show here, chemotherapeutic response. We demonstrate that primary breast cancer cells undergo dramatic phenotypic changes when removed from the host microenvironment and cultured on rigid surfaces, and that drug responses are profoundly altered by the mechanical feedback cells receive from the culture substrate. Conversely, primary breast cancer cells cultured on substrates mimicking the mechanics of their host tumor ECM have a similar genetic profile to the in situ cells with respect to drug activity and resistance pathways. These results suggest substrate stiffness plays a significant role in susceptibility of breast cancer to clinically-approved chemotherapeutics, and presents an opportunity to improve drug discovery efforts by integrating mechanical rigidity as a parameter in screening campaigns.
1. Introduction
Stiffening of tumor tissue is a hallmark of cancer, and is what allows for the identification of malignant nodules through tactile palpation. This mechanical rigidity results from aberrations in the architecture and composition of tumor extracellular matrix (ECM), leading to interstitial matrices which can be up to ten times stiffer than that of normal tissue.[1, 2] In the context of breast cancer, mechanical feedback between the mammary ECM and epithelial cells has profound and causative effects on malignancy. For example, increasing mammary stromal stiffness up-regulates integrin signaling, activates kinases involved in cell growth and has anti-apoptotic effects;[1, 3, 4] collectively promoting tumorigenesis. In fact, it has been recently demonstrated that increasing the stiffness of ECM can independently transform normal mammary epithelium into a malignant phenotype.[5] Other work has shown that changes in matrix stiffness can promote an epithelial to mesenchymal transition in pancreatic cancer,[6] and impact the chemosensitivity of leukemias.[7]
Despite the influence of substrate stiffness on cancer cell phenotype, this parameter is often overlooked in cell-based experiments employing chemical probes to elucidate biologic pathways, and in high-throughput drug discovery screening campaigns. Typical protocols employ rigid glass or polystyrene surfaces for maintaining and assaying cells that recapitulate few, if any, of the biomechanical cues present in the native tumor microenvironment.[8] Further, nearly all major screening centers utilize cell lines cultured on plastic. The continuous culture of these cells on rigid surfaces ultimately selects for sub-populations that thrive under these unnatural conditions. Consequently, the cells used in these assays can have dramatically altered phenotypes with respect to the primary tumor cells from which they were derived, and by extension may show very different chemotherapeutic responses. In support of this assertion are recent clinical studies demonstrating an early correlation between mammary tumor stiffness and patient chemotherapeutic response.[9] Ex vivo experiments have begun exploring the relationship between substrate rigidity and drug activity.[10-12] However, these studies, while provocative, have not yet identified a mechanogenetic link to define the basis of this activity.
Here, we systematically evaluate the stiffness-dependent activity of clinically-approved chemotherapeutics towards primary mammary tumor cells, as well as a panel of immortalized human breast cancer cell lines commonly used in high-throughput screens. Importantly, for our studies these lines serve as model phenotypes designed to approximate pre- and postadaptation of cells to culture on rigid synthetic substrates, respectively. Further, although the influence of substrate stiffness on drug response have been previously examined,[10-12] here we provide the first comprehensive study into the impact of substrate mechanics on chemotherapeutic response in primary tumor cells which have not undergone selection on synthetic plastic surfaces. To achieve this we directly measure the stiffness of primary tumor ECM and develop a multi-well hydrogel array designed to recapitulate the mechanical landscape of tumor tissue ex vivo. Utilizing this platform, parallel drug efficacy and gene expression profiling experiments identify a link between the mechanical feedback cells receive from the culture environment and their response to chemotherapy.
2. Materials and Methods*
2.1. Materials
Sodium dodecyl sulfate (SDS), 3-aminopropyltriethoxysilane (APTES), tetramethylethylenediamine (TEMED), ammonium persulfate (APS), 25 % glutaraldehyde solution, acrylamide, bis-acrylamide, protease inhibitor cocktail, 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide (MTT), dimethyl sulfoxide (DMSO), 50 mg/mL gentamicin solution, HEPES sodium salt, Methotrexate, 5-Fluorouracil, Tamoxifen and 200 mmol/L glutamine solution were obtained from Sigma-Aldrich. RPMI-1640 media, Alexafluor 633 goat anti-mouse antibody, and Hoechst 33342 trihydrochloride dye was purchased from Invitrogen. Heat inactivated fetal bovine serum (FBS), Hank’s Balanced Salt Solution (HBSS) and trypsin EDTA were obtained from Hyclone Laboratory Inc. EpiCult™-B Mouse Medium Kit, 0.2 % heparin sodium salt in PBS, recombinant human basic fibroblast growth factor (rh bFGF), recombinant human epidermal growth factor (rh EGF), ammonium chloride solution, 5 U/mL dispase, 1 mg/mL DNase I solution and 10X collagenase/hyaluronidase were purchased from StemCell Technologies. TRITC-labeled goat anti-rabbit IgG and biotin-labeled anti-fibronectin antibodies were purchased from Abcam. Multi-well cell culture plates, 40 μm cell strainer, falcon tubes and 96-well half area high content imaging glass bottom microplates, used to prepare the polymer gel arrays, were purchased from Fisher Scientific. Doxorubicin was purchased from AvaChem, and Paclitaxel was purchased from Alfa Aesar. Sulfosuccinimidyl-6-(4’-azido-2’-nitrophenylamino)-hexanoate (sulfo-SANPAH) was purchased from Pierce Biotechnology. Human plasma fibronectin and tetramethylrhodamine conjugated streptavidin were purchased from Life Technologies. RNeasy Micro RNA isolation kit was purchased from Qiagen.
2.2. Transgenic Animals, Tissue Processing and Primary Tumor Cell Isolation
All animal experiments and procedures were conducted following the guidelines of the National Institutes of Health Institutional Animal Care and Use Committee, under the approved Animal Study Proposal 13-314. To establish the mouse cohort, 10 breeder pairs of FVB/N-Tg(MMTV-PyVT) 634 Mul/J transgenic males (Jackson Laboratory) and FVB/J females were setup. Pregnant females were separated, and delivered pups tail-clipped and ear notched between 14 and 21 days for genotyping purposes. Female transgenic animals were genotyped, and assigned to experimental groups. Tumor growth was monitored twice per week, with multifocal mammary tumors typically developing between 5 to 8 weeks of age. Tissues were isolated when any single tumor reached >500 mm3 in volume.
For AFM studies, excised tumors were prepared into 4 mm thick sections using a microtome, and analysis was performed within 4 hours after initial isolation. In separate experiments, tumor tissue was decellularized to remove cells and isolate the ECM, before mechanical analysis. Decellularization was performed by placing sections in a 6-well dish, followed by two washes with PBS, and then submerged in 5 mL ice-cold decellularization buffer (10 mmol/L Tris, 0.5 % SDS, 0.1 % protease inhibitor, pH 8.0). Decellularization buffer was replaced every 8 hours over the course of 2 days, after which time the tissue sections were washed six times with 5 mL ice-cold PBS for 10 minutes per cycle.
Primary mouse mammary tumor cells were isolated by aseptically removing the tumors, mincing the tissue into 1 mm cubes, and suspending it in 10 volumes of dissociation solution containing 1 part 10X collagenase/hyaluronidase and 9 parts complete EpiCult-B medium (5 % FBS, 0.1 % gentamicin, 4 μg/mL heparin, 10 ng/mL rh bFGF and 10 ng/mL rh EGF). The solution was then gently shaken for 7 hours at 37 °C, with frequent vortexing. The cell suspension was centrifuged at 500 × g for 5 min., supernatant removed and tissue pellet resuspended in 5 mL of a 4:1 solution of cold ammonium chloride solution and HBSS (without phenol red) supplemented with 10 mmol/L HEPES and 2 % FBS. The mixture was gently mixed for 1 minute and centrifuged at 500 × g for 5 min. Following removal of the supernatant, 5 mL of warm trypsin-EDTA was added and solution gently mixed for 3 min. via pipette. The mixture was neutralized with 10 mL of cold HBSS (without phenol red) supplemented with 10 mmol/L HEPES and 2 % FBS, and centrifuged at 500 × g for 5 min. The supernatant was removed, 2 mL of warm dispase and 300 μL of DNase I were added, and the solution was gently mixed for 1 min. via pipette. The cell solution was then diluted with 10 mL of cold HBSS (without phenol red), supplemented with 10 mmol/L HEPES and 2 % FBS, and filtered through a 40 μm cell strainer into a sterile 50 mL falcon tube before centrifugation at 500 × g for 5 min. to pellet. Cells were then resuspended in complete EpiCult-B media, transferred to a sterile petri dish and allowed to incubate at 37 °C for 1 hour to allow fo r the adherence of stromal cells. The nonadherent fraction, containing mammary tumor cells, was collected and cell concentration determined using a hemocytometer.
2.3. Polymer Gel Arrays and Cell Seeding
Polyacrylamide multi-well hydrogel arrays were prepared using a protocol adapted from Tse et al.[13] Briefly, 150 μL of a 0.1 mol/L NaOH solution was added to each well of a glass bottom 96-well microplate, with the exception of two columns that were left untreated to serve as glass control surfaces. The plates were then placed in a 100 °C drying oven for 8 hours to evaporate the solution, and repeated as necessary to develop a uniform film of NaOH on the surface of the glass. 50 μL of APTES was added to each well, and after a 5 min. reaction the solution was aspirated. Plates were rinsed with DI water, followed by two times washing of each well using 100 μL of milliQ water. 100 μL of a 0.5 wt% glutaraldehyde solution was added to each well and reacted for 30 min. at room temperature before aspiration. Plates were dried in air for 30 min., during which time the polyacrylamide solutions were prepared. To achieve gels with differing mechanical rigidity the concentration of the bis-acrylamide cross-linker was varied between 1 - 0.05 wt%, while the acrylamide concentration was held constant at 10 wt%. To initiate polymerization, a 1 % and 0.1 % solution of 10 wt% APS and TEMED, respectively, were added to the acrylamide solution and quickly inverted to mix. 50 μL of the solution was then quickly pipetted into each well of two columns of the plate, followed by slow addition of 50 μL EtOH to create a uniform and flat surface of the resulting hydrogel. Each gel was allowed to polymerize for 1 hour, followed by removal of the EtOH and two times washing with 100 μL of milliQ water to remove any unreacted acrylamide. Preparation of the gel surfaces for covalent attachment of an ECM protein was accomplished by adding 50 μL of a 0.2 mg/mL solution of sulfo-SANPAH in milliQ water to each well, and then irradiating the plate at 365 nm for 10 min. using a UV crosslinker box (Spectrolinker, Westbury, NY) at a distance of ≈ 10 cm from the source. The gels were then washed twice with 50 mmol/L HEPES (pH 8.5), followed by addition of 100 μL HEPES buffer containing either 5 μg/mL fibronectin. No additional protein was added to the untreated glass control surfaces other than that inherently contained in the culture media. The plates were then incubated overnight at 37 °C to allow for protein conjugation to the gel surface, washed twice with media and then placed in a tissue culture hood for 4 hours under UV light to sterilize before addition of cells.
MDA-MB-453, MDA-MB-231 or MCF-7 cells were plated at 25 × 103 cells/well in 96 well plates containing the polyacrylamide gel arrays and allowed to adhere overnight. Similar seeding conditions were employed for primary mammary tumor cells isolated from MMTV-PyVT transgenic females, with the exception that 50 × 103 cells/well were initially plated to enable a final seeding density of approximately 25 × 103 cells/well. This was to account for cell death that occurred when isolating MMTV-PyVT primary tumor cells from the primary tumors utilizing the stringent and lengthy isolation protocol detailed above. For proliferation studies, cells were incubated with 2 μg/mL Hoechst 33342 dye in cell culture media for 20 min. Cells were then washed with PBS and mounted onto an EVOS FL Auto fluorescent microscope (Life Technologies, Grand Island, NY) and imaged at 10x magnification using the manufacturer LED light cubes for DAPI (357/44 nm excitation, 447/60 nm emission). A series of representative images at 1, 4 and 7 days post seeding were collected, and images analyzed with a MATLAB script that utilized the Mathworks image processing toolbox and the dipimage library. Enumeration of nuclei assumed that nuclei were a similar size, and therefore a subset of nuclei were manually delineated in order to estimate the average size of nuclei. Regions of each image corresponding to nuclei were segmented by automatically thresholding the nuclear channel using the isodata method. As this method produced regions slightly larger than the nuclei, the region was reduced in area by 4 application of the erosion filter. The number of nuclei in the image was estimated as the area of the region divided by the average size of nuclei. The method was validated and its accuracy calculated by manual enumeration of nuclei in a small part of the region. Reported cell nuclei counts represent the average of three individual samples.
To assess projected cell area, phase contrast images were taken of MCF-7 or primary tumor cells 24 hours after seeding onto 5 to 60 kPa PA gels, as well as glass. Spread area was measured for 30 representative cells using ImageJ software and averaged as a function of substrate stiffness.
2.4. AFM Analysis
The stiffness profile of both the polyacrylamide gel arrays, as well as tumor tissue isolated from PyVT-MMTV transgenic females, was determined by spectroscopic force analysis via an atomic form microscope (AFM). For the polyacrylamide array, fibronectin-coated hydrogels of different stiffness were prepared on glass coverslips as described by Tse et al.,[13] and washed 3 times with PBS before mechanical analysis (n = 3 for each condition). To evaluate the mechanical properties of tumor tissue, PyVT-MMTV transgenic females were euthanized by CO2 asphyxiation, followed by dissection of the mammary carcinomas and sectioning the tissue at 2 mm intervals using a microtome. The two sections isolated closest to the midline of the tissue were retrieved and used for AFM analysis or histopathology. For mechanical analysis, the section was affixed to a glass coverslip using a silicon adhesive, followed by gluing of the opposing glass surface onto a 30 mm polystyrene petri dish (Falcon, Corning, NY).
Repeated force-displacement experiments were performed on the samples using an Asylum MFP-3D AFM instrument (Oxford Instruments, Santa Barbara, CA), and were carried out at room temperature in a fluid cell environment with samples submerged in PBS. A Bruker SNL-10-A triangular cantilever probe (Bruker, Camarillo, CA), consisting of a SiN cantilever with a sharp Si tip, was used. Prior to testing, each cantilever was calibrated using the thermal fluctuation method;[14] the cantilevers were shown to have spring constants, kc, that varied from roughly 0.25 N/m to 0.35 N/m. A minimum of 10 force-displacement curves per sample location were recorded across the sample, with a peak load of 2 nN to 10 nN and a ramp rate on the order of 1 μm/s. To determine the elastic modulus (E) for each data point the loading portion of the force-displacement (F-d) curves were converted to force-indentation (F-δ) data by subtracting the cantilever deflection (F/kc), and then fit to the Sneddon analytical model for a conical tip in contact with an elastic half-space.[15] Specifically, the expression, F = (2/π)(E/1-v2)(tan α) δ2 was fit for E using an average half-angle of the cantilever of 21.5 °for α and an assumed Poisson’s ratio of 0.33. This experimental procedure was utilized for tumor and decellularized tissue samples (n ≥ 10), as well as PA hydrogels prepared on 18mm round glass coverslips (n =3).
2.5. Visualization of ECM Proteins Conjugated to PA Hydrogel Surface
Spatial distribution of ECM proteins, following their covalent attachment to the surface of the PA hydrogels, was visualized following immunolabeling. Briefly, 100 μL of a 0.5 % Triton X-100 (v/v) solution in PBS was added to each well, followed by incubation of the plate at room temperature for 15 min. Gels were washed three times with blank PBS, followed by addition of a 100 μL of 1 % BSA, 0.1 % Tween-20 in PBS. Samples were incubated for 1 hour at room temperature, and then washed three times with PBS. A 100 μL volume of rabbit biotin-labeled anti-fibronectin antibody (1:100) was prepared in labeling buffer (0.5 % BSA, 0.1 % Tween-20 in PBS) and incubated with each well for 1 hour at room temperature. The primary antibody solution was removed and gels washed three times with PBS before addition labeling buffer containing rhodamine-conjugated streptavidin (1:100) to visualize fibronectin coating. After a 1 hour incubation at room temperature the solution was removed and gels washed three times with PBS. Plates were mounted onto an EVOS FL Auto fluorescent microscope (Life Technologies, Grand Island, NY) and imaged at 10x magnification using the manufacturer LED light cube for RFP (531/40 nm excitation, 593/40 nm emission) to visualize fibronectin. Representative images were collected from three independent samples.
2.6. Histology
Freshly isolated tumor, or decellularized tissue, was fixed in 10 % neutral buffered formalin and embedded in paraffin to facilitate sectioning of the tissue at 6 μm thickness. Sections were deparaffinized and washed with distilled water before staining. H&E staining was performed by reacting sections with hematoxylin for 2.5 min., followed by two successive washes in water and clarifier. This was followed by addition of a blueing agent, before an additional wash with water and 95 % EtOH. Sections were added to an eosin solution and reacted for 20 s, followed by four successive washes in 100 % EtOH and three washes in xylene before mounting onto a glass slide. For von Kossa calcium staining, sections were treated with a 5 % silver nitrate solution for 60 min., during which time they were exposed under a 100-watt lamp. Samples were washed with distilled water, and reacted with a 5 % sodium thiosulfate solution for 2 min. After washing with distilled water, counterstaining was performed by incubating the sections in a nuclear fast red solution for 5 min. Samples were washed a final time with distilled water, dehydrated and mounted. Representative images were collected from three independent samples.
2.7. Chemotherapeutic Activity Assay and Drug Binding
MDA-MB-453, MDA-MB-231 or MCF-7 cells were plated at 25 × 103 cells/well, while primary mammary tumor cells isolated from MMTV-PyVT transgenic females were plated at 50 × 103 cells/well, in 96 well plates containing the polyacrylamide gel arrays and allowed to adhere overnight. For cytotoxicity studies, human tumor cell lines were cultured in RPMI-1640 media containing 5 % FBS, 1 % L-glutamine and 0.1 % gentamicin, while primary mammary tumor cells were cultured in complete EpiCult-B media as described above. The culture media was then removed and cells treated with media containing Doxorubicin (0.001 μmol/L to 100 μmol/L), Paclitaxel (0.1 nmol/L to 10 μmol/L), Methotrexate (0.075 μmol/L to 750 μmol/L), 5-Fluorouracil (0.075 μmol/L to 750 μmol/L) or Tamoxifen (0.001 μmol/L to 100 μmol/L), and cultured for 4 days. Each treatment solution was prepared by dissolving an aliquot of a DMSO drug stock in the cell culture media to achieve the desired concentration, with a final DMSO concentration of 0.5 %. Blank media or 20 % DMSO containing media was used as a positive and negative control, respectively. After the treatment period, cells were washed and 100 μL of fresh serum-containing media was added to each well. 10 μL of MTT solution (5 mg/mL in PBS) was added to each well and samples incubated for 4 hours. The supernatant was then removed from each well, replaced with 100 μL of DMSO, and plate gently shaken at 37 °C for 10 min. to dissolve the formazan product. 75 μL of the colored solution was then transferred to a clean 96 well plate and absorbance read at 540 nm using a UV plate reader (Biotek, Winooski, VT). The absorbance of the negative controls was subtracted from each sample as a blank, and percent viability calculated using the equation: (Absorbancepeptide-treated cells/Absorbanceuntreated cells) × 100. GraphPad Prism 5 software was used to fit cytotoxicity curves and calculate IC50 values employing a log(inhibitor) vs. normalized response non-linear regression model. Results shown represent the average of three independent experiments, each with three replicates, ± standard deviation.
To evaluate binding of the chemotherapeutics to PA gels, a stock solution of each drug was prepared in HBSS at the maximum concentration utilized for the cellular cyototoxicity experiments (above). A 100 μL volume of each drug was added to the PA gel stiffness array, and placed in the incubator. At daily time points, a 20 μL aliquot was removed from each well and added to 80 μL of fresh HBSS in a Microtest, UV-VIS transparent 96 well plate (BD Biosciences, Franklin Lanes, NJ). Absorbance values were recorded at 230 nm, 500 nm, 370 nm, 270 nm and 280 nm for PTX, DOX, MTX, 5FU and TAM, respectively. Fraction of drug remaining in the supernatant was calculated by normalizing the results at each time point to the absorbance of the initial solution, taking into account dilution upon addition of the 100 μL drug solution with a 50 μL gel. Results shown represent the average of three independent experiments ± standard deviation.
2.8. Microarray Experiments and Biostatistics
Human MDA-MB-453 cells, or freshly isolated MMTV-PyVT mouse primary tumor cells, were seeded onto 96 well arrays containing 5kPa or 30kPa PA gels, as well as the glass control, at 25 × 103 and 50 × 103 cells/well, respectively. After 5 days of culture, the media was removed and cells collected following a 10 min. incubation with 25 μL of trypsin per well. Cells were pelleted, washed with PBS, and total RNA isolated using the RNeasy micro extraction kit, following the manufacturer’s instructions (Qiagen Sciences, Germantown, MD). RNA quality was checked on an Agilent Bioanalyzer (Agilent Technologies, Santa Clara, CA), and all samples used in microarray analysis had a high quality score (RIN >7 for mouse, RIN > 9 for human). For the human MDA-MB-453 cells, 100 ng of RNA was reverse transcribed and labeled with biotin using Affymetrix 3’ IVT Plus kit, following manufacturer’s protocol (Affymetrix, Santa Clara, CA). Four replicates from each group were prepared, labeled, and hybridized to Affymetrix GeneChip PrimeView Human Gene Expression array. For primary mouse tumor cells, 2 ng of RNA was reverse transcribed, amplified and sense strand cDNA was fragmented and labeled using WT Pico expression kit, following the manufacturer’s suggested protocol. Four replicates of each group were hybridized to Affymetrix mouse Gene ST 2.0 GeneChip in Affymetrix hybridization oven at 45 °C 60 rpm for 16 hours. Wash and staining were performed on Affymetrix Fluidics Station 450, arrays scanned using an Affymetrix GeneChip scanner 3000, and data collected using Affymetrix AGCC software. Reported data represents the average of at least three independent biologic replicates.
Affymetrix PrimeView Human Gene Expression and Mouse Gene 2.0 ST Array data were processed using the affy[16] and limma[17] bioconductor packages. For the human MDA-MB-453 cell assays, differentially expressed genes were detected in low and medium stiffness gels compared with glass. For the mouse data, gene expression changes between cells immediately after isolation from the tumor were compared those same cells plated and cultured on PA gels or glass. Differentially expressed genes were further analyzed for gene ontology enrichment using hypergeometric distribution analysis.
The datasets generated during the current study are available in the GEO repository, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE107063.
3. Results and discussion
3.1. Breast tumor ECM is mechanically heterogeneous and compliant
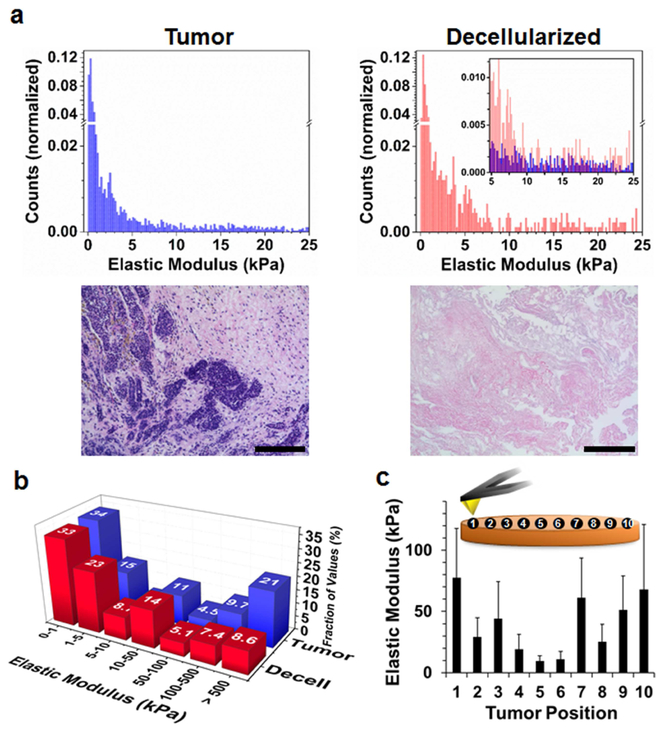
We first mapped the stiffness profile of mammary tumor ECM using atomic force microscopy (AFM) to define the rigidity landscape of primary tissue to be recapitulated in the ex vivo hydrogel array. Measurements were initially taken on intact cellularized tumor sections isolated from MMTV-PyMT transgenic mice, a standard animal model of human breast cancer.[18] Histograms shown in Fig. 1a (top left) demonstrate that freshly isolated breast tumor is highly compliant, characterized by a primary stiffness regime between 0.1 to 5 kPa that peaks at 0.11 kPa ± 0.01 kPa, and a smaller contribution of rigidities between 5 to 25 kPa. Histopathologic examination of the isolated tissues identified them primarily as late stage multifocal adenosquamos carcinoma with invasion into the surrounding fibrotic stroma (Fig. 1a, bottom left). We next decellularized the tissue to better define the ECM mechanical contribution without influence from the embedded cells (Fig. 1a, top right). Decellularized tissues displayed a trimodal rigidity landscape, with a narrow maxima at 0.52 kPa ± 0.01 kPa, and two enriched regimes between 5 to 10 kPa and 15 to 25 kPa when compared to the cellularized tissue (Fig. 1a, inset). Haemotoxylin and Eosin (H&E) staining confirmed decellularized sections were composed of only collagenous stroma (Fig. 1a, bottom right). Thus, removing the cells provides a more accurate rigidity landscape with which to develop synthetic materials that mimic the tumor ECM stiffness.
Fig. 1.
Mechanical profiling of breast tumor ECM. (a) Top Left: Stiffness profile of primary mammary tumor tissue, as measured by AFM analysis and reported as the elastic modulus (units of kPa). Top right: Mechanical profile of decellularized tumor sections. Inset: Cellularized (blue) stiffness data overlaid onto decellularized (red) results between 5 to 25 kPa (n ≥ 10). Bottom: H&E staining reveal tumor sections are composed of multi-focal regions of cancer cells (purple; cell nuclei) encapsulated by a dense fibrotic stroma (pink; collagen), while decellularized tissues are primarily composed of tumor ECM (scale bar = 400 μm; n = 3). (b) Fraction of stiffness measurements collected from primary tumor tissue (blue) or decellularized sections (red), binned as distinct regimes of elastic moduli. (c) Average stiffness values recorded at ten positions along the central axis of the cellularized tumor sections; error bars represent one standard deviation from the mean (n ≥ 10).
Figure 1b groups the AFM data to include higher order stiffness regimes, showing that 30 % of the stiffness values measured in the intact tumor were >100 kPa, while this range accounted for only 16 % of the decellularized samples. These highly stiff regions likely result from contact of the AFM tip with calcium microdeposits observed in the intact tumor sections (Fig. S1), which are disrupted during the decellularization process. We next measured stiffness values at spatially resolved locations across the central axis of intact tumor sections, showing that the periphery of the tissue is ≈7 times more stiff than the interior (Fig. 1c). These results support previous studies in which human breast tumor biopsies possessed a collagen-rich invasive front that is characterized by increased mechanical stiffness over that of both adjacent normal tissue,[19] and the highly cellularized and ECM-deficient necrotic core.[20] Importantly, together this data shows that the ECM of decellularized tissue is much more compliant than commonly employed cell culture substrates, including plastic (103 MPa) and glass (105 MPa), and that the heterogeneous non-mineralized ECM within the tumor microenvironment can be adequately mimicked by ex vivo substrates with stiffness of 1 to 50 kPa.
3.2. Chemotherapeutic response of breast cancer mediated by substrate mechanics
We next prepared 96-well polyacrylamide gel arrays (Fig. 2a) designed to recapitulate the breast tumor ECM stiffness profile identified by the AFM measurements. Protein-laminated polyacrylamide (PA), which is a widely utilized material in mechanobiology,[3, 21-23] was chosen as substrate rigidity can be readily modulated by controlling the amount of the bis-acrylamide cross-linker included during polymerization.[13] Although simple, these gels serve as an ideal material to isolate the influence of substrate stiffness, using a simple 2D format, on cancer cell response to various stimuli without other confounding variables. There are a litany of factors, including biochemical cues and 3D microenvironment architecture, that impact cancer cell biology. For example, it is becoming clear that the genetic phenotype of cells differs when grown as a 2D monolayer versus 3D cultures.[24, 25] In our study, it is important that we separate the effects of mechanics before further exploring synergistic influences of multiple variables of the tumor microenvironment, including 3D architecture. Further, the chemical simplicity, facile production and low cost of PA gels make them tractable materials to be scaled for future high throughput applications in drug screening.
Fig. 2.
Breast tumor ECM mimetic stiffness array. (a) 96 well PA hydrogel stiffness arrays encumbering the mechanical profile of primary tumor ECM. Here, stiffness is systematically increased across the plate, with untreated glass included as a rigid control (moduli ≈105 MPa). To prepare the multi-well array, glass bottom plates are chemically treated (red) to covalently attach PA gels formed in each well (purple). Functionalization of the gel surface with fibronectin (orange) allows for attachment of seeded cells. (b) Stiffness array gels were prepared with elastic moduli values that matched the rigidity profile of primary mammary tumor ECM (5 to 60 kPa) by controlling the weight percent of bis-acrylamide (bis) crosslinker added to the polymer solution (n = 3). (c) Representative immnofluorescent image demonstrating homogenous display of covalently linked fibronectin (red) on the gel surface (scale bar = 400 m). Associated z-stacks confirm protein coating is limited to the top of the gels. (d) Proliferation of primary mammary tumor cells isolated from MMTV-PYMT mice on 5 to 30 kPa PA gels, as measured by cell nuclei counting (n = 3). (e) Cell spreading as a function of substrate stiffness for primary tumor cells (1° tumor) or the human breast cancer cell li ne MCF-7 (n = 30). Error bars represent one standard deviation
We prepared PA gels with elastic moduli between 5 to 60 kPa (Fig. 2b), representing a stiffness manifold that closely matches that of the tumor ECM (Fig. 1b). Attempts to prepare PA substrates with elastic moduli ≈1 kPa resulted in highly viscous gels that were too compliant to be used in our multi-well screening platform. The surface of each gel was then functionalized with fibronectin to promote cell attachment, resulting in uniform display of the ECM protein (Fig. 2c), and a constant protein loading density (Fig. S2), across the stiffness array. Primary cancer cells isolated from mouse MMTV-PyMT mammary tumors, as well as a panel of immortalized human breast cancer cell lines, seeded onto the fibronectin-coated gels were able to adhere and proliferate (Fig. 2d and Fig. S3). Interestingly, we observed no change in the spreading behavior of primary tumor cells as a function of the substrate’s mechanical stiffness (Fig. 2e). Conversely, the projected cell area of the model human breast cancer cell line MCF-7 linearly increased with substrate rigidity. This suggests that the prior maintenance of MCF-7 cells on rigid plastic may have conditioned these cells to more readily adopt a spread morphology when seeded onto stiff surfaces. On the contrary, maintenance of primary cells in the compliant mammary tumor microenvironment may not induce the phenotypic changes that are required to cause anchorage-dependent spread phenotypes. Taken together, this may indicate that substrate stiffness alters the genetic profile of cultured cells, an assertion we explore later. Although it has been shown that not all cell types display stiffness dependent spreading,[23] the fact that these cells share similar tissues of origin supports the assertion that spreading behavior is likely derived from phenotypic alterations induced during long-term culture on rigid non-natural substrates.
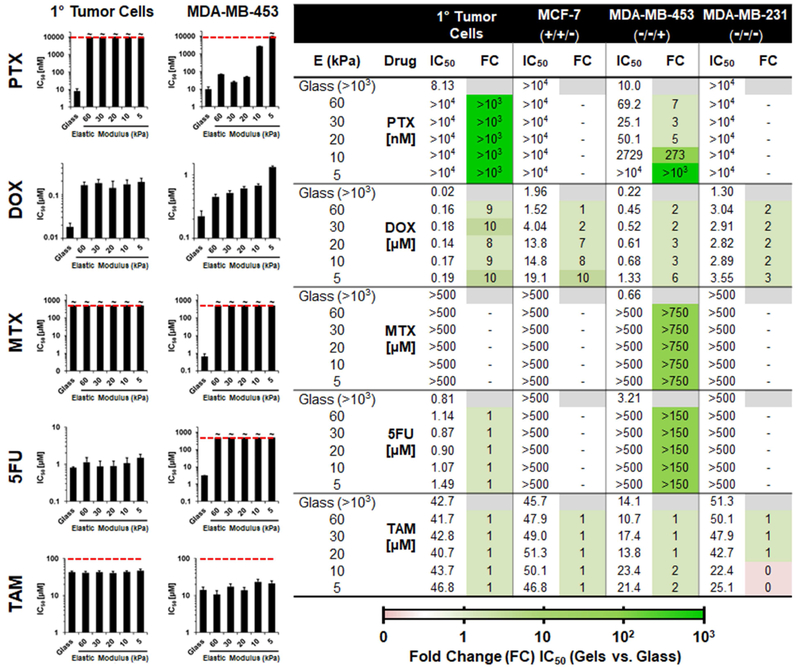
Using this stiffness array, we evaluated the influence of substrate mechanical rigidity on the activity of five clinically approved chemotherapeutic drugs towards a panel of breast cancer cell lines seeded onto the array (Fig. 3), which include primary MMTV-PyMT mouse tumor cells and the human breast cancer cell lines MCF-7, MDA-MB-453 and MDA-MB-231. Importantly, the human cell lines selected represent three different molecular subtypes of breast cancer, defined by their status of the estrogen receptor (ER), progesterone receptor (PR) and human epithelial receptor 2 (HER2). Based on this conventional classification, the subtype of each cell line, in order of increasing aggressiveness, is reported as luminal (MCF-7; ER+PR+HER2−), HER2 positive (MDA-MB-453; ER−PR−HER2+) and triple negative (MDA-MB-231; ER−PR−HER2−).[26] In general, breast cancer cells cultured on ‘soft’ substrates that mimic the rigidity of tumor tissue displayed a dramatic reduction in chemotherapeutic response when compared to rigid control surfaces, many by multiple orders of magnitude. For example, Paclitaxel (PTX) has a half-maximal inhibitor concentration (IC50) of 8 to 10 nM towards primary tumor and MDA-MB-453 cells cultured on glass, but is essentially inactive when these cells are cultured on the softest (5 kPa) hydrogel substrate. Further, as the culture surface becomes more stiff we observed a restoration of PTX activity for the immortalized cell line. Similarly, Doxorubicin (DOX) cytotoxicity is reduced by 3 to 10 fold change towards all four of the tested cell lines when cultured on the softest substrate versus glass (see table in Fig. 3). Methotrexate (MTX) and 5-Fluorouracil (5FU) also show a stiffness-dependent reduction in therapeutic efficacy, but only towards MDA-MB-453 cells. Although, MTX was active against the immortalized cells on glass, previous animal studies show the drug to be inactive in the MMTV-PyMT mouse breast cancer model.[27] Interestingly, in our studies primary MMTV-PyMT tumor cells are recalcitrant towards MTX when grown on all tested substratum. Thus, in this case employing immortalized cells cultured on rigid substrates is unable to accurately predict in vivo drug activity. Adsorption experiments confirmed that the observed variations in drug susceptibility were not due to sequestration of the chemotherapeutics within the PA gels (Fig. S4), but rather a cellular response to available drug. Collectively this data suggests that the therapeutic activity of antiproliferative (e.g. DOX and PTX) and antimetabolite (e.g. MTX and 5FU) drugs are vastly different between cells grown on unnaturally rigid surfaces and those cultured on substrates of similar stiffness to the native tumor microenvironment. More importantly, these studies support the assertion that drug screening protocols utilizing unnaturally rigid culture substrates can lead to in vitro drug responses that are either grossly overestimated, or all together inaccurate, when compared to their in vivo activity.
Fig. 3.
Stiffness-dependent chemotherapeutic response of breast cancer cells. Left: Plots demonstrating the change in activity of selected small molecule chemotherapeutics after a 72 hour incubation with primary tumor cells, or the human breast cancer cell line MDA-MB-453, as a function of the culture substrate’s elastic modulus. PTX = Paclitaxel; DOX = Doxorubicin; MTX = Methotrexate; 5FU = 5-Fluorouracil; TAM = Tamoxifen. Dashed red line represents highest concentration tested, with the symbol ‘~’ indicating that an IC50 could not be reached at this concentration for the specified condition. Error bars represent one standard deviation from the mean (n = 9). Right: Table of IC50 values for each drug towards the panel of breast cancer cell lines tested following their culture on substrates of varying elastic modulus (E). Molecular sub-type status (ER/PR/HER2) is shown below the name of the tested cell line. Fold change (FC) in IC50 of cells cultured on each of the gels versus the glass control surface is tabulated to the right of the IC50 values, and color coded to visually symbolize order of magnitude.
Conversely, the activity of Tamoxifen (TAM) was found to be invariant across the stiffness array for the entire panel of cell lines. TAM, and its active metabolite 4- hydroxytamoxifen, are estrogen receptor antagonists which bind to cell-surface growth factor receptors and ultimately inhibit the expression of estrogen-responsive genes. With this background in mind, the toxicity of TAM observed in our studies towards the ER-deficient MDA-MB-453 and MDA-MB-231 cell lines is unexpected. Yet, recent work provides some precedence, demonstrating that TAM treatment of even ER-negative tumors can generate anticancer responses.[28] This is attributed to the drug’s ability to disrupt lysosome maturation and activate autophagic cell death pathways.[29, 30] In our studies, genetic analyses, presented later, indicate that cellular response to drugs which act by inducing apoptosis are highly influenced by substrate stiffness. Taken together, our data suggests that, while apoptosis may be impacted by microenvironment rigidity, lysosomal functions and autophagy in cancer cells is likely regulated in a stiffness-independent manner.
3.3. Expression of drug resistance genes is substrate stiffness-dependent
Through parallel microarray experiments we next explored stiffness-dependent changes in cellular gene expression to identify the role of substrate rigidity in the activation of pathways important to drug action. Two independent experiments were performed. In the first, we introduce the MDA-MB-453 human breast cancer cell line, which has been conditioned to grow on plastic, onto compliant surfaces that recapitulate the rigidity of the tumor microenvironment. In the second experiment, we transplant isolated MMTV-PyMT primary tumor cells to both a rigid culture surface (glass) and compliant substrates that better mimic the stiffness of their native tissue. For both studies, change in gene expression after five days of growth on 5kPa or 30kPa gels is measured via microarray analysis relative to control cells cultured on glass.
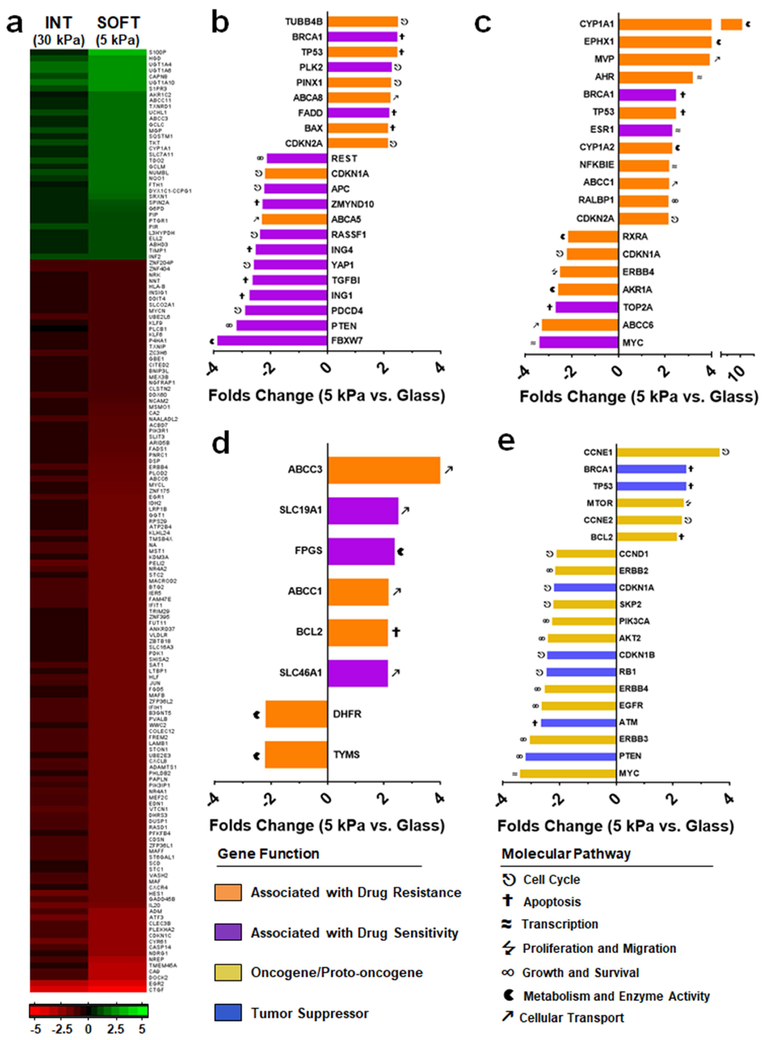
When immortalized MDA-MB-453 cells are introduced onto 5 kPa gels we observed large changes in global gene expression compared to those cells cultured on glass, with >16,000 genes (33 % of total, cutoff p<0.01) differentially expressed. We also found that the general direction of gene expression change (up or down regulated) was similar between cells cultured on soft (5 kPa) and intermediate (30 kPa) stiffness substrates compared to glass (Fig. 4a). Interestingly, the overall magnitude of these changes increased as material rigidity is reduced. Hence, stiffness-dependent changes in gene expression appear to scale proportionally with substrate rigidity.
Fig. 4.
Substrate stiffness alters the expression of drug resistance genes. (a) Gross changes in MDA-MB-453 gene expression when cultured on 5 kPa or 30 kPa PA gels compared to cells grown on glass, in units of log2 (p<1×10−6). Cutoffs were set at ≥2 folds up-regulated (green) or ≤−2 folds down-regulated (red). (b-d) Fold change in the expression of genes associated with resistance (orange) or sensitivity (purple) of cells towards (b) PTX, (c) DOX or (d) MTX/5FU, following culture of MDA-MB-453 on 5 kPa gels versus glass. (e) Fold change in selected oncogene/proto-oncogenes (yellow) and tumor suppressor genes (blue). P value cutoff for panels b–e set at p<0.05. Data is averaged from n = 4 biologic replicates.
Next, we examined the influence of substrate rigidity on the expression of selected genes implicated in drug resistance.[31-35] Genes whose molecular action imparts PTX and DOX resistance (Fig. 4b and 4c, respectively; orange color) were found to be largely overexpressed when MDA-MB-453 cells are cultured on the 5 kPa substrate versus glass, while those genes that impart sensitivity (purple) were down regulated under these conditions. Notably, we observed a ≈4 fold decrease in FBXW7 expression between cells seeded onto soft substrates versus glass (Fig. 4b). Previous studies have demonstrated that loss of FBXW7 expression, a regulator of cellular apoptosis, directly transforms PTX sensitive breast cancer cells into a resistant phenotype, and correlates with adverse prognoses in PTX-treated breast cancer patients.[36] Similarly, in our experiments we observed a 10 fold over expression of CYP1A1 (drug metabolism) in MDA-MB-453 cells on soft gels as compared to glass, while TOP2A (apoptosis) was down-regulated by over 3 fold (Fig. 4c). Both of these genes have been shown to play a role in the transformation of DOX-sensitive breast cancer cells into a resistant phenotype when over and under-expressed, respectively.[32]
In the context of MTX and 5FU resistance, nearly all of the selected genes that regulate drug sensitivity were found to be overexpressed (Fig. 4d), particularly ABCC3 which encodes for an ABC efflux transporter involved in multi-drug resistance.[37, 38] Finally, no discernable pattern in the change of TAM resistance genes was observed between cells cultured on soft substrates and glass (Fig. S5); not surprising as TAM activity was found to be stiffness-independent (Fig. 3).
Gene ontology enrichment analysis reveals that pathways involved in cellular response to stress, toxins and drugs, multi-drug efflux (ABC) transporters, and a number of metabolic processes and negative regulators of apoptosis, were all upregulated following culture of these immortalized cells on the soft substrates versus glass (Fig. S6). While under these conditions, down regulated genes fell into categories that include cell-cell interactions and development of focal adhesions, as well as multiple growth signaling pathways (Fig. S7). When taken together with the resistance gene expression data (Fig. 4b and 4c), these results suggest that immortalized human breast cancer cells initiate a broad range of stress responses and survival mechanisms when transplanted from the rigid substratum they have been conditioned to grow on, and seeded onto soft materials that mimic the mechanical properties of native tumor tissue. Changes in cell phenotype resulting from this stress response ultimately modulate the expression of key genes involved in chemotherapeutic resistance, despite no previous drug exposure. Thus, substrate stiffness alters the genetic profile of immortalized breast cancer cells resulting in a variation of their drug susceptibility. These changes most likely account for the weakened, and often negligible, drug responses we observed when these immortalized cells are cultured on compliant versus rigid substrates (Fig. 3). Importantly, within the tumor microenvironment poor oxygenation and nutrient deprivation leads to stressed conditions,[39] suggesting that removal of this pressure by culturing immortalized cells on rigid substrates may lead to drug activities that are grossly overestimated in screening campaigns.
Interestingly, we also found that substrate stiffness modulates the expression of oncogenes and proto-oncogenes, as well as a number of well-known breast cancer tumor suppressors (Fig. 4e). Notably, members that encode for the epidermal growth factor receptor subfamily were found to be widely down-regulated in cells grown on soft substrates as compared to glass, including EGFR (HER1) and ERBB2 (HER2). This suggests that the potency of monoclonal antibodies that inhibit these growth factor signaling pathways may also be influenced by substrate stiffness.
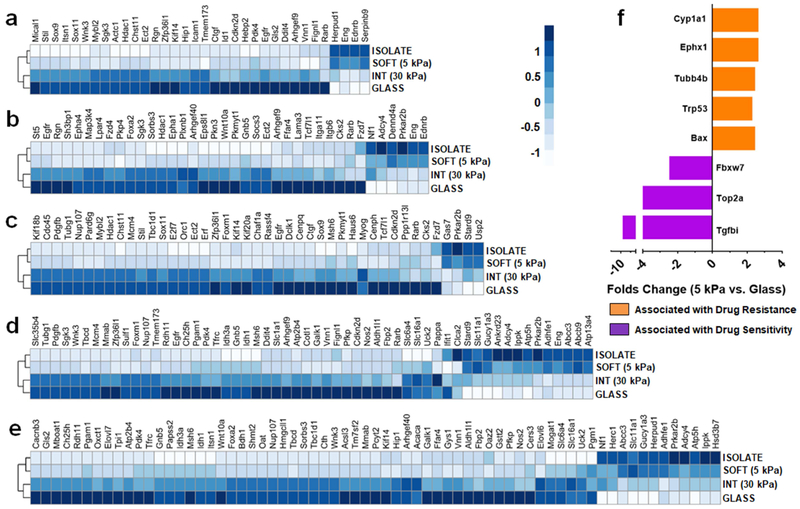
We next investigated the effect of substrate stiffness on the gene expression of primary mammary tumor cells from MMTV-PyMT transgenic mice. The data in Figure 5 is derived from an experiment where the genetic profile of freshly isolated cells is compared to those same cells cultured on soft, intermediate and glass substrates. Panels a-e explore the expression of gene orthologs broadly involved in apoptosis, cell signaling, cell cycle and growth, response to drug, and small molecule metabolism and transport, respectively. Irrespective of category, we observe that gene expression levels dramatically change when isolated primary tumor cells are introduced to glass. Remarkably, as the stiffness of the substrate decreases there is a gradual shift in gene expression that culminates in restoration of expression levels closely resembling that of the primary isolated cells. For example, in panel d the blue coloration (indicative of expression levels) of drug response genes in glass cultured cells is gradually restored to the color profile of isolated cells as substrate stiffness decreases. The remaining panels show a similar trend. This strongly suggests that breast cancer cells seeded onto soft substrates that approximate the tumor microenvironment stiffness are able to better maintain the in situ cancer phenotype.
Fig. 5.
Cancer cells seeded onto gels that mimic tumor ECM stiffness recapitulate in situ gene expression. (a-e) Z-normalized expression levels of selected mouse genes whose human orthologs are involved in (a) apoptosis, (b) cell signaling, (c) cell cycle and growth, (d) response to drug and (e) small molecule metabolism and transport pathways. Gene expression of primary mammary tumor cells is shown for the in situ cells (isolate) immediately after removal, or following their culture for 5 days on soft gels (5 kPa), intermediate stiffness gels (30 kPa) or the highly rigid glass control surface. (f) Folds change (units of log2; p<1×10−6) in the expression of genes associated with resistance (orange) or sensitivity (purple) of cells towards PTX and DOX between cells seeded onto soft (5 kPa) gels versus glass. Data is averaged from n ≥ 3 biologic replicates.
Of particular significance, Abcb9 and Abcc3 were both highly expressed when primary tumor cells are cultured on soft gels, while these genes are down-regulated in cells seeded onto glass (see Fig. 5d). These genes encode for multi-drug resistance proteins of the ATP-binding cassette (ABC) transporter superfamily, and have been implicated in the efflux-mediated detoxification of PTX[40, 41] and DOX,[42, 43] respectively. Further, in panel f we show changes in the expression of genes relevant to PTX and DOX resistance between cells seeded onto soft substrates (5 kPa) versus glass. On the soft substrate, genes that impart resistance are overexpressed (orange), and those that are associated with drug sensitivity are decreased (purple). These changes likely account for the high activity of PTX and DOX towards primary tumor cells on glass, compared to the low activity of these drugs towards cells seeded onto the soft substrates (Fig. 3). More importantly, this signifies that breast cancer cells lose their original drug-resistant phenotype when removed from the compliant tumor microenvironment and plated onto rigid surfaces such as plastic or glass. The ramification of these changes will not be equal across all classes of drugs, and is dependent on the relative change of specific genes and pathways. For example, our data indicates that stiffness-dependent changes in cellular phenotype significantly affects the activity of PTX and DOX. Conversely, for MTX, 5FU and TAM this change in phenotype shows little effect in altering the relative sensitivity of primary tumor cells to these drugs.
4. Conclusions
In summary, this work demonstrates that the mechanical stiffness of the tumor microenvironment has profound and causative effects on the response of breast cancer cells to clinically-approved chemotherapeutics. Adding to the current paradigm that genetic mutations cultivate heterogeneous tumor cell populations, our work suggests that the mechano-feedback cells receive from the mechanically diverse tumor microenvironment may also give rise to subclonal populations with altered drug sensitivity. This assertion is supported by the fact that primary breast cancer cells undergo dramatic changes in phenotype when removed from the soft host environment and cultured on rigid substrates. This leads to significant changes in pathways important to drug transport, metabolism and activity, making these cells more susceptible to some chemotherapies. Conversely, these same cells cultured on soft substrates have similar gene expression profiles to the in situ tumor cells, and demonstrate low sensitivity towards PTX and DOX.
More broadly, our work helps to further substantiate the assertion that screening cancer cell lines on un-naturally rigid plastic substrates does not accurately approximate the in situ cellular response to drug. Further, in addition to substrate mechanics, there are a litany of other factors that impact chemotherapeutic potency, including media selection,[44] method of measuring drug response (IC50 vs. GI50),[45] and the 3D nature of the tumor microenvironment.[46-48] In regards to the latter, 3D cell cultures (e.g. spheroids, organoids, etc.) have been shown to more closely recapitulate the tumor microenvironment and better model the in situ cellular phenotype.[24, 25, 47, 48] Yet, despite the design of sophisticated 3D tissue culture systems, the vast majority of high throughput drug screening centers still employ 2D cell monolayers grown on plastic or glass. Thus, new screening methods that employ 2D cultures, but better recapitulate key genetic traits, are valuable. For example, a 2D surface that mimics the stiffness of the tumor, along with defined culture media, could be used relatively inexpensively at scale after assurance that the cells maintain the desired genetic profile. Routine sequencing could be performed to periodically confirm that assayed cells match the genetic profile of the original tumor. Nevertheless, it is impractical to expect ex vivo culture platforms will maintain cells lines that comprehensively phenocopy the in situ tumor. Instead, a more tractable solution may be to implement platforms that can maintain the expression of key genes involved in cellular response to drug. Here, we help to advance this goal by providing an example methodology by which 2D arrays of variable mechanical rigidity can be integrated into high-throughput strategies to better recapitulate drug responses that may occur in native tumor environments.
Supplementary Material
Acknowledgements
This work was supported by funds provided by Penn State Biomedical Engineering laboratory startup to S.H.M., as well as Intramural Research Program of the National Cancer Institute, National Institutes of Health (1ZIABC01131308).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Data Availability
The raw data required to reproduce these findings are available to download from https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE107063.
Appendix A. Supplementary data
Supplementary data to this article can be found online.
Specific equipment, instruments, and materials identified in the paper are listed in order to adequately describe the experimental procedure. Such identification does not imply recommendation by NIST, nor does it imply the equipment and materials are the best available for the purpose.
References
- [1].Levental KR, Yu H, Kass L, Lakins JN, Egeblad M, Erler JT, Fong SFT, Csiszar K, Giaccia A, Weninger W, Yamauchi M, Gasser DL, Weaver VM, Matrix Crosslinking Forces Tumor Progression by Enhancing Integrin Signaling, Cell 139(5) (2009) 891–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Lu P, Weaver VM, Werb Z, The extracellular matrix: A dynamic niche in cancer progression, Int. J. Biochem. Cell Biol. 196(4) (2012) 395–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Paszek MJ, Zahir N, Johnson KR, Lakins JN, Rozenberg GI, Gefen A, Reinhart-King CA, Margulies SS, Dembo M, Boettiger D, Hammer DA, Weaver VM, Tensional homeostasis and the malignant phenotype, Cancer Cell 8(3) (2005) 241–254. [DOI] [PubMed] [Google Scholar]
- [4].Mouw JK, Yui Y, Damiano L, Bainer RO, Lakins JN, Acerbi I, Ou G, Wijekoon AC, Levental KR, Gilbert PM, Hwang ES, Chen Y-Y, Weaver VM, Tissue mechanics modulate microRNA-dependent PTEN expression to regulate malignant progression, Nat. Med. 20(4) (2014) 360–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Chaudhuri O, Koshy ST, Branco da Cunha C, Shin J-W, Verbeke CS, Allison KH, Mooney DJ, Extracellular matrix stiffness and composition jointly regulate the induction of malignant phenotypes in mammary epithelium, Nat. Mater. 13(10) (2014) 970–978. [DOI] [PubMed] [Google Scholar]
- [6].Rice AJ, Cortes E, Lachowski D, Cheung BCH, Karim SA, Morton JP, del Rio Hernandez A, Matrix stiffness induces epithelial-mesenchymal transition and promotes chemoresistance in pancreatic cancer cells, Oncogenesis 6 (2017) e352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Shin J-W, Mooney DJ, Extracellular matrix stiffness causes systematic variations in proliferation and chemosensitivity in myeloid leukemias, Proc. Natl. Acad. Sci. 113(43) (2016) 12126–12131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Infanger DW, Lynch ME, Fischbach C, Engineered Culture Models for Studies of Tumor-Microenvironment Interactions, Annu. Rev. Biomed. Eng. 15(1) (2013) 29–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Hayashi M, Yamamoto Y, Ibusuki M, Fujiwara S, Yamamoto S, Tomita S, Nakano M, Murakami K, Iyama K.-i., Iwase H, Evaluation of tumor stiffness by elastography is predictive for pathologic complete response to neoadjuvant chemotherapy in patients with breast cancer, Ann. Surg. Oncol. 19(9) (2012) 3042–3049. [DOI] [PubMed] [Google Scholar]
- [10].Schrader J, Gordon-Walker TT, Aucott RL, van Deemter M, Quaas A, Walsh S, Benten D, Forbes SJ, Wells RG, Iredale JP, Matrix stiffness modulates proliferation, chemotherapeutic response, and dormancy in hepatocellular carcinoma cells, Hepatology 53(4) (2011) 1192–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Rehfeldt F, Engler AJ, Eckhardt A, Ahmed F, Discher DE, Cell responses to the mechanochemical microenvironment—Implications for regenerative medicine and drug delivery, Adv. Drug Delivery Rev. 59(13) (2007) 1329–1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Tokuda EY, Leight JL, Anseth KS, Modulation of matrix elasticity with PEG hydrogels to study melanoma drug responsiveness, Biomaterials 35(14) (2014) 4310–4318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Tse JR, Engler AJ, Preparation of Hydrogel Substrates with Tunable Mechanical Properties, Curr. Protoc. Cell Biol, John Wiley & Sons, Inc; 2001. [DOI] [PubMed] [Google Scholar]
- [14].Butt HJ, Jaschke M, Calculation of thermal noise in atomic force microscopy, Nanotechnology 6(1) (1995) 1. [Google Scholar]
- [15].Sneddon IN, The relation between load and penetration in the axisymmetric boussinesq problem for a punch of arbitrary profile, Int. J. Eng. Sci. 3(1) (1965) 47–57. [Google Scholar]
- [16].Gautier L, Cope L, Bolstad BM, Irizarry RA, affy–analysis of Affymetrix GeneChip data at the probe level, Bioinformatics 20(3) (2004) 307–315. [DOI] [PubMed] [Google Scholar]
- [17].Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK, limma powers differential expression analyses for RNA-sequencing and microarray studies, Nucleic Acids Res. 43(7) (2015) e47–e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Lin EY, Jones JG, Li P, Zhu L, Whitney KD, Muller WJ, Pollard JW, Progression to Malignancy in the Polyoma Middle T Oncoprotein Mouse Breast Cancer Model Provides a Reliable Model for Human Diseases, Am. J. Pathol. 163(5) (2003) 2113–2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Acerbi I, Cassereau L, Dean I, Shi Q, Au A, Park C, Chen YY, Liphardt J, Hwang ES, Weaver VM, Human breast cancer invasion and aggression correlates with ECM stiffening and immune cell infiltration, Integr. Biol. 7(10) (2015) 1120–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Plodinec M, Loparic M, Monnier CA, Obermann EC, Zanetti-Dallenbach R, Oertle P, Hyotyla JT, Aebi U, Bentires-Alj M, LimRoderick YH, Schoenenberger C-A, The nanomechanical signature of breast cancer, Nat. Nano. 7(11) (2012) 757–765. [DOI] [PubMed] [Google Scholar]
- [21].Engler AJ, Sen S, Sweeney HL, Discher DE, Matrix Elasticity Directs Stem Cell Lineage Specification, Cell 126(4) (2006) 677–689. [DOI] [PubMed] [Google Scholar]
- [22].Discher DE, Janmey P, Wang Y.-l., Tissue Cells Feel and Respond to the Stiffness of Their Substrate, Science 310(5751) (2005) 1139–1143. [DOI] [PubMed] [Google Scholar]
- [23].Yeung T, Georges PC, Flanagan LA, Marg B, Ortiz M, Funaki M, Zahir N, Ming W, Weaver V, Janmey PA, Effects of substrate stiffness on cell morphology, cytoskeletal structure, and adhesion, Cell Motil. Cytoskeleton 60(1) (2005) 24–34. [DOI] [PubMed] [Google Scholar]
- [24].Sachs N, de Ligt J, Kopper O, Gogola E, Bounova G, Weeber F, Balgobind AV, Wind K, Gracanin A, Begthel H, A living biobank of breast cancer organoids captures disease heterogeneity, Cell 172(1-2) (2018) 373–386. e10. [DOI] [PubMed] [Google Scholar]
- [25].Clevers H, Modeling development and disease with organoids, Cell 165(7) (2016) 1586–1597. [DOI] [PubMed] [Google Scholar]
- [26].Dai X, Cheng H, Bai Z, Li J, Breast Cancer Cell Line Classification and Its Relevance with Breast Tumor Subtyping, J. Cancer 8(16) (2017) 3131–3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Hansen MF, Jensen SO, Fuchtbauer E-M, Martensen PM, High folic acid diet enhances tumour growth in PyMT-induced breast cancer, Br. J Cancer 116(6) (2017) 752–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Wu S-T, Sun G-H, Cha T-L, Kao C-C, Chang S-Y, Kuo S-C, Way T-D, CSC-3436 switched tamoxifen-induced autophagy to apoptosis through the inhibition of AMPK/mTOR pathway, J. Biomed. Sci. 23(1) (2016) 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Kim LA, Amarnani D, Gnanaguru G, Tseng WA, Vavvas DG, D'Amore PA, Tamoxifen Toxicity in Cultured Retinal Pigment Epithelial Cells Is Mediated by Concurrent Regulated Cell Death Mechanisms, Invest. Ophthalmol. Visual Sci. 55(8) (2014) 4747–4758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Altan N, Chen Y, Schindler M, Simon SM, Tamoxifen inhibits acidification in cells independent of the estrogen receptor, Proc. Natl. Acad. Sci. 96(8) (1999) 4432–4437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Xu J-H, Hu S-L, Shen G-D, Shen G, Tumor suppressor genes and their underlying interactions in paclitaxel resistance in cancer therapy, Cancer Cell Int. 16 (2016) 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].AbuHammad S, Zihlif M, Gene expression alterations in doxorubicin resistant MCF7 breast cancer cell line, Genomics 101(4) (2013) 213–20. [DOI] [PubMed] [Google Scholar]
- [33].Bertino JR, Göker E, Gorlick R, Li WW, Banerjee D, Resistance Mechanisms to Methotrexate in Tumors, The Oncologist 1(4) (1996) 223–226. [PubMed] [Google Scholar]
- [34].Wang W, Cassidy J, O’Brien V, Ryan KM, Collie-Duguid E, Mechanistic and Predictive Profiling of 5-Fluorouracil Resistance in Human Cancer Cells, Cancer Res. 64(22) (2004) 8167–8176. [DOI] [PubMed] [Google Scholar]
- [35].Huber-Keener KJ, Liu X, Wang Z, Wang Y, Freeman W, Wu S, Planas-Silva MD, Ren X, Cheng Y, Zhang Y, Vrana K, Liu C-G, Yang J-M, Wu R, Differential Gene Expression in Tamoxifen-Resistant Breast Cancer Cells Revealed by a New Analytical Model of RNA-Seq Data, PLoS One 7(7) (2012) e41333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Gasca J, Flores ML, Giráldez S, Ruiz-Borrego M, Tortolero M, Romero F, Japón MA, Sáez C, Loss of FBXW7 and accumulation of MCL1 and PLK1 promote paclitaxel resistance in breast cancer, Oncotarget 7(33) (2016) 52751–52765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Balaji SA, Udupa N, Chamallamudi MR, Gupta V, Rangarajan A, Role of the Drug Transporter ABCC3 in Breast Cancer Chemoresistance, PLoS One 11(5) (2016) e0155013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Zelcer N, Saeki T, Reid G, Beijnen JH, Borst P, Characterization of Drug Transport by the Human Multidrug Resistance Protein 3 (ABCC3), J. Biol. Chem. 276(49) (2001) 46400–46407. [DOI] [PubMed] [Google Scholar]
- [39].Fulda S, Gorman AM, Hori O, Samali A, Cellular stress responses: cell survival and cell death, Int. J. Cell Biol. 2010 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Ohashi-Kobayashi A, Ohashi K, Du W.-b., Omote H, Nakamoto R, Al-shawi M, Maeda M, Examination of drug resistance activity of human TAP-like (ABCB9) expressed in yeast, Biochem. Biophys. Res. Commun. 343(2) (2006) 597–601. [DOI] [PubMed] [Google Scholar]
- [41].Gong J-P, Yang L, Tang J-W, Sun P, Hu Q, Qin J-W, Xu X-M, Sun B-C, Tang J-H, Overexpression of microRNA-24 increases the sensitivity to paclitaxel in drug-resistant breast carcinoma cell lines via targeting ABCB9, Oncology Lett. 12(5) (2016) 3905–3911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Gottesman MM, Fojo T, Bates SE, Multidrug resistance in cancer: role of ATP-dependent transporters, Nat. Rev. Cancer 2(1) (2002) 48–58. [DOI] [PubMed] [Google Scholar]
- [43].Kool M, van der Linden M, de Haas M, Scheffer GL, de Vree JML, Smith AJ, Jansen G, Peters GJ, Ponne N, Scheper RJ, Elferink RPJO, Baas F, Borst P, MRP3, an organic anion transporter able to transport anti-cancer drugs, Proc. Natl. Acad. Sci. 96(12) (1999) 6914–6919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Muir A, Danai LV, Vander Heiden MG, Microenvironmental regulation of cancer cell metabolism: implications for experimental design and translational studies, Dis. Models Mech. 11(8) (2018) dmm035758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Hafner M, Niepel M, Chung M, Sorger PK, Growth rate inhibition metrics correct for confounders in measuring sensitivity to cancer drugs, Nat. Methods 13(6) (2016) 521–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Wilson KM, Mathews-Griner LA, Williamson T, Guha R, Chen L, Shinn P, McKnight C, Michael S, Klumpp-Thomas C, Binder ZA, Mutation Profiles in Glioblastoma 3D Oncospheres Modulate Drug Efficacy, SLAS Technol. 24(1) (2019) 28–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Debnath J, Brugge JS, Modelling glandular epithelial cancers in three-dimensional cultures, Nat. Rev. Cancer 5(9) (2005) 675. [DOI] [PubMed] [Google Scholar]
- [48].Lee S-Y, Bissell MJ, A Functionally Robust Phenotypic Screen that Identifies Drug Resistance-associated Genes Using 3D Cell Culture, Bio-Protoc. 8(22) (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.