Abstract
Here we aim to incorporate trait-based information into the modern coexistence framework that comprises a balance between stabilizing (niche-based) and equalizing (fitness) mechanisms among interacting species. Taking the modern coexistence framework as our basis, we experimentally tested the effect of size differences among species on coexistence by using fifteen unique pairs of resident vs. invading cyanobacteria, resulting in thirty unique invasibility tests. The cyanobacteria covered two orders of magnitude differences in size. We found that both niche and fitness differences increased with size differences. Niche differences increased faster with size differences than relative fitness differences and whereas coexisting pairs showed larger size differences than non-coexisting pairs, ultimately species coexistence could not be predicted on basis of size differences only. Our findings suggest that size is more than a key trait controlling physiological and population-level aspects of phytoplankton, it is also relevant for community-level phenomena such as niche and fitness differences which influence coexistence and biodiversity.
Subject terms: Theoretical ecology, Microbial ecology, Community ecology
Introduction
The mechanisms underlying species coexistence have perplexed and fascinated ecologists for decades. Early in the 1960s, Hutchinson’s classical study ‘The paradox of the plankton’ drew attention to the surprisingly large diversity of plankton species competing for a small number of limiting resources, suggesting that non-equilibrium mechanisms are relevant for species coexistence [1]. Historically, studies on species coexistence have sought to explain biodiversity using two different perspectives. On the one hand, the classical niche-based view emphasizing that phenotypic differences among species will reduce interspecific competition and thereby allow coexistence [2–4]. On the other hand, the neutral view, suggesting that species are competitively equivalent and hence, diversity is driven by stochasticity and dispersal [5, 6]. More recently, community ecologists have looked at ways to provide a more comprehensive perspective for coexistence of species [7, 8], such as reconciling niche-based and neutral mechanisms [9–12]. Chesson [9] proposed that species coexistence could be understood as a balance between stabilizing forces (i.e., niche-based differences) and equalizing forces (i.e., those minimizing fitness differences among species; Fig. 1).
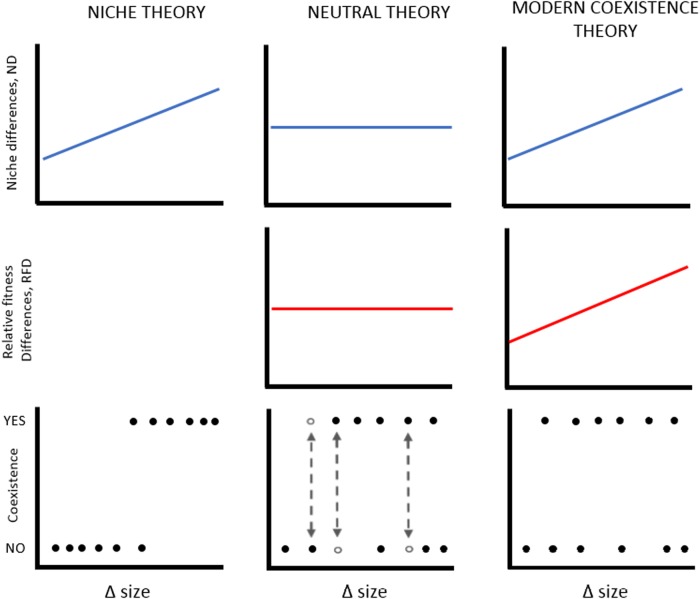
Fig. 1.
Predictions on how species coexistence is determined by differences in organisms’ size (Δ size) through the lenses of different coexistence theories. Under the niche theory perspective (left column panels), if size differences (x-axis) relate well to niche differences (ND, top panel), coexistence is possible when niche differences (i.e., size differences) among species are large (bottom panel). The niche perspective does not consider relative fitness differences. Under the neutral perspective (central column panels), all species are equivalent in terms of both niche and fitness, irrespective of their sizes. Differences in size should not affect the likelihood of coexistence. The competitive outcome in the neutral scenario is influenced by stochasticity, meaning that a pair of species may sometimes coexist and sometimes not. The empty dots and arrows illustrate such stochastic variability among replicates for a same pair of species. The modern coexistence perspective (right column panels) proposes that if size differences relate well to both niche (ND) and fitness differences (RFD), coexistence is possible when niche differences are large enough to compensate for fitness differences. Compared to the neutral scenario, the competitive outcome of the modern coexistence theory is not influenced by stochasticity and should be consistent among replicates in a same pair of species
Individual-based models accounting for effects of niche and relative fitness differences on species survival, growth, and recruitment have been developed to determine the relative importance of stabilizing and equalizing mechanisms for species coexistence [13–16]. Also, experimental studies have quantified both niche and relative fitness differences among species under controlled laboratory conditions [17]. Both theory and empirical evidence concur that neither niche, nor fitness differences alone can predict coexistence. Species coexist only if their niche differences are large enough to counteract their relative fitness differences [9, 12]. Some recent empirical studies have further explored the drivers of species coexistence by analysing the role of evolutionary relatedness on niche and relative fitness differences [17, 18]. For freshwater green microalgae, Narwani et al. [17] found that species coexistence could not be predicted based on their evolutionary relatedness. Godoy et al. [18], on the other hand, found a significant relationship between phylogenetic signal and relative fitness differences in annual plants. However, coexistence was not successfully predicted by phylogenetic relatedness due to the highly inherent variance of species relatedness traits, particularly when species were distantly related.
As an alternative to phylogenetic relatedness as a predictor for coexistence, one could directly focus on those traits that may influence niche and fitness differences [16, 19] and test their impact on coexistence. For instance, Kraft et al. [16] explored the relative importance of eleven individual plant functional traits in predicting niche and fitness differences, five of which were size-related variables. Their findings suggest that whereas phenotypic differences among plants correlated well with relative fitness differences, no correlation with niche differences was found. That is, phenotypic differences among plant species based on individual traits did not relate to niche differentiation. Only combinations of multiple traits related well to niche differentiation among species and permitted coexistence.
In phytoplankton, many individual and population level processes are size-dependent, meaning they scale with cell or colony size [20–24]. For instance, nutrient uptake, light absorption and ultimately growth rate are determined by the surface-area to-volume ratios. Smaller phytoplankton cells show a tendency to grow faster, sink slower, and be more efficient in the acquisition of limiting resources. Larger ones may dominate when nutrients are abundant or when grazing pressures are high [21, 25]. Under given nutrient and light conditions, species of different size are expected to occupy different niches, whilst the niches of species of similar size may overlap. Additional studies conclude that size differences affect fitness differences, with fitness differences that limit coexistence emerging as size differences increase [22, 24, 26, 27]. From a modern coexistence perspective, both niche and relative fitness differences will increase with size differences among species. Given this double effect, however, size differences alone may not be able to predict whether niche differences are large enough to counteract for relative fitness differences and permit coexistence. To experimentally test expectations on size as a functional trait that plays a pivotal role in species coexistence, we estimated simultaneously niche and relative fitness differences in 30 unique pairs of cyanobacteria which greatly varied in size—covering up to two orders of difference in magnitude—to amplify any potential scenario among species and guaranteeing that the effect of size is not overlooked.
Our experiments showed that both niche and fitness differences increased with difference in size, which would affect the scope for coexistence. The prevalence of coexistence was not statistically determined by size differences, but coexisting species showed higher size differences than non-coexisting species. Unlike phenotypic differences based on size, phylogenetic relatedness did not relate to either niche or fitness differences.
Methods
Species selection
We selected six species of freshwater cyanobacteria: Aphanothece hegewaldii, Chroococcidiopsis cubana, Chroococcus minutus, Synechococcus leopolensis, Synechocystis pevalekii, Synechocystis PCC 6803. All taxa were obtained from the Göttingen Culture Collection (SAG, Germany). We exclusively focused on the Cyanobacteria phylum, since it is considered the most diverse and widely distributed [28] and is increasingly important in freshwater ecosystems because of the proliferation of harmful blooms [29]. Out of the main five cyanobacterial morphological groups, we excluded those with specific structures and/or cells (e.g., heterocysts, gas vacuoles) that could favor nutrient uptake and light absorption rates, which would possibly modify the effect of size in our experiment. We only selected species with similar morphology (i.e., coccoid forms) to minimize the effect of cellular shape, since it may also strongly influence coexistence [27]. All six species grew well in BG11, a common culture medium [30] and were clearly distinguishable under a microscope. Cultures were not axenic, but careful microscopic inspection revealed that the biomass of heterotrophic bacteria never exceeded 1% of total biovolume.
Experimental design
Our experimental approach consisted in allowing one species —‘invader’— to attempt to colonize an already established culture of a second species —‘resident’—. We included six resident species times five invader species = thirty unique pairwise species combinations (e.g., C. minutus invading A. hegewaldii and vice versa), each replicated three times for a total of 90 invasion cultures. We also grew the six species alone (monocultures) times six replicates = thirty-six monocultures, resulting in a total of 126 experimental flasks. We filled 50 mL-flasks with 30 mL BG11 culture medium.
Each of the resident cyanobacteria species was cultured with an initial concentration of 200 RFU (relative fluorescence units), measured with a laboratory fluorimeter (Turner BioSystems, USA), at 22 °C, with a 16:8 light:dark cycle and a light intensity of 50 μmol photons m−2 s−1 in a controlled culture chamber (INFORS Multitron, Switzerland). Because chlorophyll a levels and colony densities were highly correlated for each monoculture (R2 > 0.83), we used chlorophyll a as a proxy for density to determine when cultures reached stationary phase to start the invasibility tests (Supplementary Information, S.I. 1).
When all monocultures had reached stationary phase, 33 days after the initial inoculation, we introduced the rare invader species in a 95:5 resident-to-invader ratio, based on total biovolume. We established the resident-to-invader concentration based on the total biovolume rather than cell density because of the large differences in cell dimensions among species. Biovolume was calculated with geometric formulae based on strain morphology [31]. We measured cell/colony densities and the average cell diameter for each resident to account for the total biovolume and we calculated the cell density of each invader necessary to accomplish 5% of the resident’s biovolume. Cell densities and average cell size (diameter) were measured with a particle counter (CASYTM Roche Innovatis AG, Germany). When species could not be clearly distinguished based on their size differences, the proportional density of each species was estimated microscopically using Neubauer chambers (see Supplementary Information, S.I. 1; Intercalibration between methodologies). Growth rates of species in the invasion experiments were calculated over the exponential growth phase (between t = 41 and t = 57 days), using the following formula:
where t is the time (days) after the invasion, D0 and Dt are the cell densities at the initial and final time for each species, grown either as monoculture or as invader. The experiment lasted 60 days.
Size variability to measure trait diversity
The six cyanobacteria species used in this study covered a gradient in average size that ranged across two orders of magnitude in diameter (from 1 to 100 μm). The upper end of the size scale represents colonial growth forms, which are ultimately the ecologically relevant units for colony forming taxa [32]. We calculated differences in size as the logarithm of the differences in average cell or colony diameter between two species plus 1 (log (Δ diameter +1)). Low size differences were obtained for five combinations of species with similar average size, five combinations represent intermediate size differences (one order of magnitude difference) and five combinations large size differences (two orders of magnitude difference).
Phylogeny and calculation of phylogenetic distances
In addition to size differences, we also used phylogenetic distance as a predictor of niche and fitness differences to compare our results to a similar recent study using green algae [17]. We constructed a smoothed Maximum Likelihood phylogenetic tree (Supplementary Information, S.I. 2) using partial 16S ribosomal RNA for 23 species of freshwater cyanobacteria available on GenBank. We included other representative species from Cyanobacteria (e.g., Oscillatoriales, Nostocales) to place our pool within a broader phylogenetic framework and used Bacillus subtilis strain B6-1 as outgroup. We aligned gene sequences with ClustalX2 (Neighbor-joining distance method), excluding positions with gaps and correcting for multiple substitutions and built a tree using RAxML version 7.2.8 (Supplementary Information, S.I. 2). Phylogenetic distances between species pairs were calculated as the sum of branch lengths connecting the two species in the tree.
Measurement of coexistence mechanisms
To account for the competitive potential of each species, we calculated the “sensitivity to competition” as:
where is the growth rate of species i when growing alone (as a monoculture) and is the growth of species i as invader, that is when introduced into a steady-state population of a resident species [33]. For any species, when sensitivity both growth rates (as a monoculture and as an invader) are similar. Contrastingly, a species with a high sensitivity to competition would show a sharp drop in growth rate when invading, implying that Si approaches 1. For sensitivities Si > 1, the invasion is unsuccessful (i.e., negative growth as invader), whilst indicates facilitation (i.e., a special case in which the invader grows better in presence of the resident than in monoculture). There is mutual invasibility (i.e., both species able to invade the resident species) when the sensitivities of the two species S1,2 and S2,1 range between 0 and 1. We averaged the growth rates of the three replicates of each species. We used the approach proposed by Carroll et al. [33], in which niche differences (ND) between two species are calculated as one minus the geometric mean of the sensitivities Si of each pairwise combination, whereas their relative fitness differences (RFD) are calculated as the geometric standard deviation of the sensitivities, which can be also expressed as the square root of the ratio between sensitivities. ND and RFD formulae are noted as:
As required for geometric standard deviations, we assume that RFD ≥ 1; implying that S1 > S2 (i.e., the species with the higher sensitivity always as numerator in the ratio of RFD). High ND values (close to 1) mean that the average sensitivities to competition of both species are relatively low, meaning that, overall both species grow similarly well as monocultures and as invaders. In its original form, the coexistence threshold is curvilinear in the ND vs. RFD plane [33]. To achieve a linear threshold between the two variables and make our statistical analyses more robust, we transformed RFD into RFD* as:
If RFD is close to 1 (RFD* = 0), both species’ growth rates are equally affected by the presence of the resident, which is considered an equalizing mechanism and makes stable coexistence possible, even with a small ND [33]. High values of RFD (RFD* = 1) imply asymmetry in sensitivities to competition (e.g., only one species invades the other). Overall, competitive exclusion occurs when RFD > 1/(1−ND), while coexistence occurs when RFD < 1/(1−ND) [33]. In terms of RFD*, competitive exclusion occurs when RFD* > ND, while coexistence occurs when RFD* < ND.
Statistical analyses
We estimated the influence of size differences among pairs of species on: (1) niche differences ND, (2) transformed relative fitness differences RFD*, (3) prevalence of coexistence, and (4) sensitivities; by using linear regressions when the response variables were continuous and probit models when the response variable was binary. Additionally, we tested whether coexisting pairs differed from non-coexisting pairs regarding their size differences with a Wilcoxon signed-rank test. We also used structural equation modeling (SEM), a robust multivariate method, to test for direct and indirect links between variables. The advantages of this technique compared to conventional analyses, such as simple regressions, are given in Box 1. We used SEM to first analyse the direct effects of differences in average size between two species on: (1) niche differences ND, (2) relative fitness differences RFD*; and the direct effects of (3) ND and (4) RFD* on the prevalence of coexistence. Then we incorporated (5) indirect effects of ND (i.e., ND as covariate connecting size differences and species coexistence) and (6) indirect effects of RFD* on the likelihood of coexistence, (i.e., RFD* as a covariate). For this, a multiple mediation model was constructed using size differences between pairwise combinations (µm) as the exogenous variable, niche differences (ND) and relative fitness differences (RFD*) as mediators, and species coexistence as an endogenous binary variable (presence/absence). Data were log-transformed to fulfill normality criteria. We also ran a model including the direct effect of phylogenetic distance among species on: (1) niche differences ND, (2) relative fitness differences RFD*; the effects of (3) ND and (4) RFD* on the prevalence of coexistence; and the indirect effects of (5) ND and (6) RFD* on the likelihood of coexistence. Although two different estimators were given, we only considered the most robust estimator—WLSMV estimator, with mean and variance adjustments—, since it is even more efficient than the diagonally weighed least squares estimator (DWLS). We ran a bootstrap procedure based on 1000 replicates for each model. The SEM model was built with lavaan package, using R v. 3.4.0 (R 2017).
Box 1 Structural Equation Modeling (SEM) in ecology.
Ecologists constantly link specific system attributes to theoretical concepts using latent variables i.e. those unmeasured or unmeasurable, which is largely due to the nature of ecological data and the lack of robust generalizations in ecology. In a quest for a suitable procedure, SEM provides a comprehensive scientific framework, rather than a method, that uses a set of equations to model multivariate relationships (i.e., those involving simultaneous influences and responses, [47]). The challenge of this approach is to make strong and explicit connections between empirical data and theoretical ideas. Although SEM were developed almost three decades ago [48], their origins go back to the 1920s, when path analysis was developed to answer questions formulated in evolutionary genetics [49]. Since then, much progress has been made in more social sciences, such as sociometrics, econometrics and psychometrics [50]. In ecology, the first studies using this approach were related to natural selection (in 2000), life history strategies (2006), community ecology (2006), genomics (2006) and physiology (2007). There are several reasons why ecologists are interested in incorporating SEM in their field [51]:
SEM are not null model oriented, but theory oriented.
Hypotheses about casual networks are well characterized.
Competing models are tested.
Allows interpreting a large number of predictors and responses with complex causal connections.
The general form of expression in a SEM is a series of mathematical equations and a path diagram with all the causal connections, such as: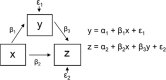 where x is an exogenous variable, y is a mediator variable and z is an endogenous variable. βi represent the regression coefficients between variables and εi are the errors.
where x is an exogenous variable, y is a mediator variable and z is an endogenous variable. βi represent the regression coefficients between variables and εi are the errors.
This generalized multi-equation framework allows ecologists to represent a broad range of multivariate hypotheses about interdependencies, in which the research questions dictate the use of structural modeling. To analyse the relations among variables, we decompose the model into several simple paths. We then estimate the parameters in the model. We assess the models fit and, if necessary, we redefine the model and start from the beginning. Lastly, results are interpreted. The model fit can be assessed by χ² (it measures the “badness” of fit), the CFI and the TLI, amongst others. The comparative fit index (CFI) is equal to the discrepancy function adjusted for sample size. CFI ranges from 0 to 1 with a larger value indicating better fit. Acceptable model fit is indicated by a CFI value similar to 0.90 or greater [52].
Differences between multiple regression and SEM are summarized as follows:
| Multiple regression | Structural Equation Modeling | |
|---|---|---|
| Number of variables | Only one dependent variable | Several variables |
| Type of variables | A variable can be either a predictor or an outcome | A variable can be either observed (being endogenous or exogenous) or latent (= unmeasurable) |
| Theoretical content | Descriptive | Confirmatory |
| Purpose/use | Exploratory studies | Evaluation of multivariate hypotheses |
| Explanatory analyses | ||
| Method specificities | Examination of 1-to-1 causal relationships | Examination of networks of causal relationships |
List of advantages and limitations of SEM:
| Advantages of SEM | Limitations of SEM |
|---|---|
| Flexibility: Permits the implications of a causally structured theory to be expressed | Does not solve all the problems associated with interpretation of multivariate relations |
| Allows the inclusion of latent (unmeasurable, ‘pure’) variables in the model | The analysis itself does not establish the causality |
| Comprehensive: Subsumes other techniques (multiple regression, confirmatory factor analysis, path analysis, ANOVA) | Statistical tests and assessment of fit more ambiguous than conventional multivariate analyses |
Results
Stable coexistence of cyanobacterial species, based on the reciprocal mutual invasion from rare criteria, occurred in ten out of the fifteen pairs tested in our experiment. Both species displayed positive growth-rates in presence of a resident species and niche differences (ND) were large enough to counteract for relative fitness differences (RFD*, Fig. 2). On one extreme of the coexistence spectrum, we found the pair S. leopolensis/A. hegewaldii with both high ND and RFD* values. On the other extreme of the gradient, we found the pair S. leopolensis/C. cubana with low ND and RFD* values (Fig. 2). The ten cases of coexistence included all five species pairs with large size differences, three out of five pairs with intermediate size differences, and only two out of five pairs with low size differences. The other three species pairs with small size differences and two pairs with intermediate size differences resulted in the incapacity of one of the two species to invade the resident species. In three out of the five non-coexisting species, the unsuccessful invader was the smaller species in the pair, whereas in the other two cases the unsuccessful invader was the larger species. Growth rates and sensitivities revealed no individual species having a general lower capacity to invade than others. Exclusion occurred when niche differences were too small to counteract for fitness differences, (e.g., in the four species pairs with the lowest ND values; Fig. 2) and in the pair S. pevalekii/C. cubana, for which both ND and RFD* values were large, but RFD* exceeded ND (Fig. 2).
Fig. 2.
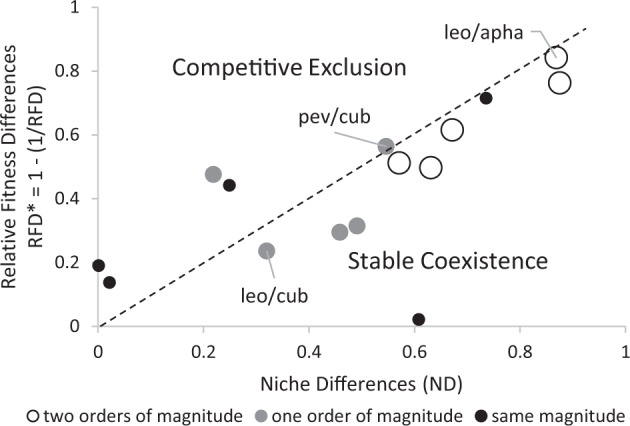
Joint effects of niche differences (ND) and relative fitness differences (RFD*) on cyanobacteria species coexistence. RFD* is calculated as 1- (1/RFD) to show niche differences and relative fitness differences at the same scale and to transform the relationship between both variables to a linear one. The experiment included fifteen pairwise combinations. Large size, open dots represent species pairs with large differences in average size (two orders of magnitude), medium size, gray dots are for species combinations with medium differences in their average size (one order of magnitude) and small size, black dots show small differences in size (same order of magnitude). As described in Adler et al. [12], coexistence occurs when RFD < 1 / (1-ND), which is equivalent to RFD* < ND (the area below the line). Low values of RFD* require low ND to allow coexistence (e.g., the pair Synechocystis leopolensis/Chroococcidiopsis cubana, leo/cub) whereas high RFD* values require high ND values (e.g., the pair Synechococcus leopolensis/Aphanothece hegewaldii, leo/apha). The pair Synechocystis pevaleckii/ Chroococcidiopsis cubana (pev/cub) did not coexist despite high ND values because RFD* was also relatively high
Size differences effects
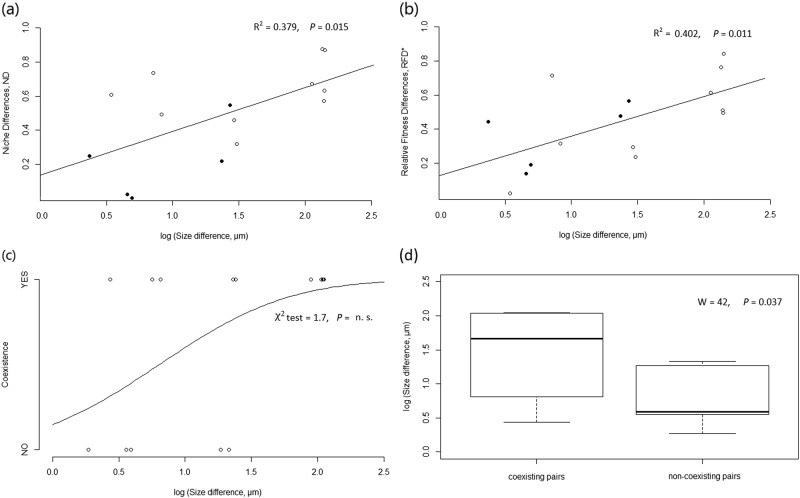
Niche differences (ND) were positively related to size differences among species (R2 = 0.379, P = 0.015, Fig. 3a) with ND values close to zero at low size differences and values close to one for large size differences. This means that species of more similar size displayed on average larger sensitivities (i.e., larger differences in growth rate when grown alone versus as invaders). In other words, the capacity of a species to invade from rare increased as size differences with the resident species increased. Relative fitness differences (RFD*) were also positively related to size differences among species (R2 = 0.402, P = 0.011, Fig. 3b) with RFD* values close to zero at low size differences and values close to one at larger size differences. Although coexistence prevalence was not determined by size differences (χ2 test = 1.7, P = 0.19; Fig. 3c), the ten pairs of species that coexisted under our experimental conditions had larger size differences than the five non-coexisting pairs (Wilcoxon signed-rank test, W = 42, P = 0.037; Fig. 3d). All species pairs with over two orders of magnitude in size differences coexisted since niche differences were large enough to counteract fitness differences, while species pairs with lower size differences (below two orders of magnitude), resulted in either coexistence or competitive exclusion (Figs. 2 and 3c).
Fig. 3.
Effects of differences in cyanobacteria interspecific size (µm, log transformed) on (a) species niche differences ND, (b) species relative fitness differences RFD*, both based on regression models, and (c) the prevalence of coexistence, based on a probit model. Filled dots represent cases where exclusion occurred and (d) size differences between coexisting and non-coexisting species pairs
In twelve out of fifteen cases, the smaller species in a pair showed lower sensitivity to competition than the larger species, revealing that in general smaller species were better invaders from rare. This happened because smaller species in a pair grew faster as invaders (11 out of 15 cases) but showed lower growth rates as monocultures (12 out of 15 cases). When competitive exclusion occurred, the smaller species excluded the larger one in three cases (out of five) and larger cells excluded smaller ones in the remaining two cases. Only sensitivities of the species with the lower sensitivities in each pair (generally the smaller species) decreased significantly as size differences decreased (Fig. 4). The sensitivities of the species with the highest sensitivities in each pair (generally the larger species) were not significantly influenced by size differences. This explains the increase in the asymmetry of sensitivities (RFD*) as size differences increased (Figs. 3b, 4). Sensitivities of the species with higher sensitivities in each pair ranged from values above one, when differences in size were low (unsuccessful invasion), to intermediate values when differences in size were large. Sensitivities of the species with lower sensitivities in each pair ranged from intermediate values, when differences in size were low, to values close to zero when differences in size were large (species grew as invaders as well as when alone, Fig. 4). Pairs of species with the highest size-differences included A. hegewaldii, the largest species in our pool, and all species invading it grew almost as well as when grown alone, resulting in sensitivities close to zero.
Fig. 4.
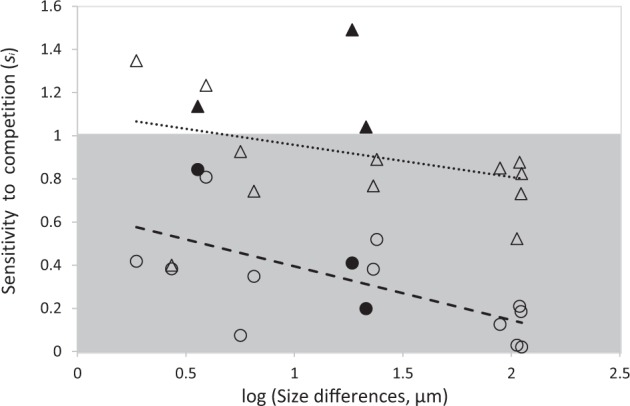
Effect of differences in cyanobacteria interspecific size on the sensitivity Si of each species in the pairwise combination, defined as Triangles represent the species with the highest sensitivity in each pair of species and circles represent the species with the lowest sensitivity in each pair of species. The gray area represents sensitivities comprised between zero and one, meaning that species in this area grew positively as invaders but less well than when grown alone. Sensitivities close to zero mean species grew as well as invaders as when alone. Sensitivities close, but below one mean species growth rates as invaders were still positive but low. Sensitivities above one mean species could not invade the resident population. The sensitivities of species with higher sensitivities were not influenced by size differences (upper line, R2 = 0.111, P = 0.225). The sensitivities of species with lower sensitivities decreased as the size differences increased (R2 = 0.416, P = 0.009). Filled symbols represent the three pairs of species for which the smallest species had a higher sensitivity than the larger species
SEM with phylogenetic distance or size differences as exogenous variables
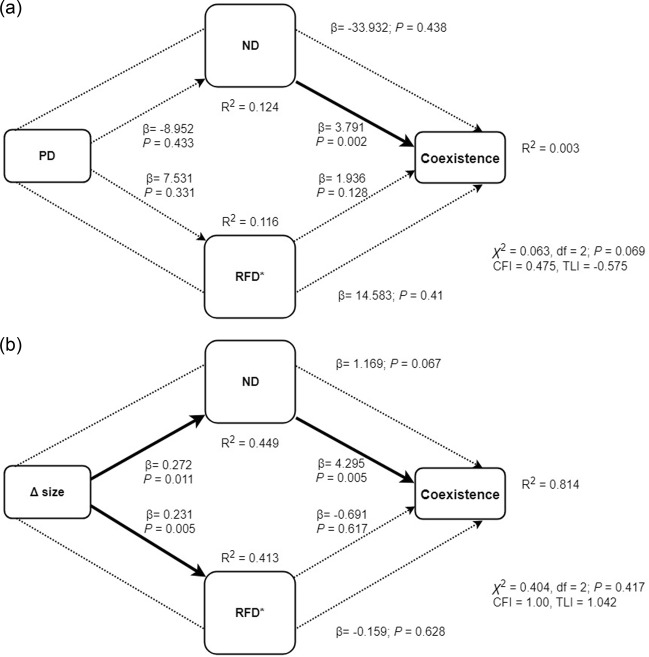
Given the lack of correlation between phylogenetic distance among species pairs (PD) and their size differences (R2 = 0.05, P = 0.4; Supplementary Information S.I. 3), we conducted two SEMs, using both PD and size differences as exogenous variables, separately.
The SEM including phylogenetic distance (PD) as the exogenous variable (χ2 = 0.063; PRobust = 0.069, CFI = 0.475, TLI = −0.575, Fig. 5a) explained the variation in the prevalence of coexistence less well compared to the model with size differences (χ2 = 0.404; PRobust = 0.417, CFI = 1.00, TLI = 1.042, Fig. 5b). Neither ND nor RFD* were determined by phylogenetic distances (β = −8.952, P > 0.433, and β = −2.936, P > 0.331 respectively, Fig. 5a). In addition, the effects of phylogenetic distances on coexistence with mediation via ND or RFD were not significant (P = 0.438 and P = 0.41 respectively). When phylogenetic distances and size differences were jointly included as exogenous variables, neither ND nor RFD* were related to phylogenetic distances (P > 0.1 in all cases; see Supplementary Information S.I. 2).
Fig. 5.
Structural equation models. In (a) the exogenous variable is the phylogenetic distance (PD), whilst in (b) the exogenous variable is differences in size (∆ size). Species coexistence is present as the endogenous variable, and niche differences (ND) and relative fitness differences (RFD) are mediator variables. β indicates unstandardized regression coefficients. Arrows indicate the direction of the interactions between variables, pointing towards endogenous variables. The weight of the arrows is proportional to the regression coefficients, with dashed lines representing no significant relationships between variables. The overall goodness-of-fit test (χ2, CFI, TLI) and the regression coefficient R2 for each variable introduced are given. CFI, comparative fit index; LTI, Tucker-Lewis index
The SEM including differences in size as an exogenous variable and ND and RFD* as mediators was significant, meaning that it explained well the variation in the prevalence of coexistence (Fig. 5b). The path analysis showed that cyanobacteria size differences (Δ size) positively influenced both niche differences (ND, β = 0.272, P = 0.011) and relative fitness differences (RFD*, β = 0.231, P = 0.005). In addition, the path analysis revealed that the prevalence of coexistence was positively related to ND (β = 4.295, P = 0.005), meaning that species coexisted more often as their niche differences increased. This can be explained by the fact that four out of five cases of exclusion were obtained at low ND values (Fig. 3a) and by the clear transition (except for only one species pair) from cases of exclusion to cases of coexistence moving along the ND gradient (Fig. 2). On the other hand, relative fitness differences (changes in the asymmetry of growth sensitivities) had no significant impact on species coexistence (β = −0.691, P = 0.617). Indeed, the five cases of exclusion were more evenly distributed along the RFD* axis (Fig. 3b) and no clear transition from exclusion to coexistence moving along the RFD* gradient could be observed (Fig. 2). The indirect effects of size differences on species’ coexistence with mediation of ND and RFD* were not significant (β = 1.169, P = 0.067 and β = −0.159, P = 0.628, respectively, Fig. 5b).
Discussion
Our results support some fundamental modern coexistence-based predictions linking size, niche and fitness differences to coexistence. The average sensitivities of species to competitors decreased with their size differences, leading to an increase in niche stabilization. Species had generally more trouble invading species of similar size, whilst invading a species of a very different size was relatively easy and some invaders grew almost as if they were growing alone. This result suggests a decrease in niche overlap between species as size differences increase, in line with classic studies proposing that differences in size promote coexistence due to niche differentiation [34–37]. The increase in niche differences with size differences was mainly due to a steep reduction in the sensitivity of the smaller species in each pair. Larger species often produce less biomass and may affect nutrient and light availability less strongly, allowing smaller species to invade relatively easily. Yet, according to modern coexistence theory, the niche-based perspective alone is insufficient to predict the outcome of competition on coexistence. Relative fitness differences in our study also showed a tendency to increase with size differences, meaning that sensitivities became more asymmetric as size differences increased. Indeed, the negative trend observed in species sensitivities with increasing size differences was particularly evident in those with low sensitivities, which mainly corresponded to the smaller species in each pair. However, our expectation that smaller species would exclude the larger ones was not always fulfilled despite most of the smaller species being less sensitive to competition —and therefore, more likely to exclude the larger ones— since niche differences must also be considered in the likelihood of coexistence.
Previous studies suggested that differences in body size might generate a competitive gradient such that certain species, due to their larger size, may exclude others [16, 38–42]. For instance, using Lotka-Volterra competition models, a positive correlation between organisms’ sizes and their competitive abilities suggests that the species with the highest carrying capacity K should exclude others [43]. To our knowledge, only one study has included size-related variables in the context of modern coexistence theory [16]. Kraft et al. considered several functional traits that account for plant size and found relatively strong correlations between single functional traits and fitness differences (that limit coexistence) but no correlations with niche differences (that ultimately promote coexistence). Probably the two orders magnitude size gradient included in our study —compared to much smaller gradients in plant studies— is responsible for the observed large niche differences among the cyanobacterial species. In phytoplankton, species’ reproductive success is tightly coupled to their nutrient uptake and resulting growth rates [25], so species with lower surface-to-volume ratios may have lower fitness, as suggested by the general decrease in abundances observed for larger species [22, 26]. In any case, in the modern coexistence perspective, the link between size-related fitness differences among species and their coexistence is not straightforward, unless one assumes that niche differences are unimportant [9–11, 14, 44]. In other words, large fitness differences by themselves lead to competitive exclusion, in the absence of sufficient stabilizing forces through niche differences. One of the novelties of our study is the inclusion of a single functional trait —size— expressed over a large gradient, covering many possible cyanobacterial species combinations that occur in nature.
Species coexistence could not be directly predicted by trait differences nor phylogenetic relatedness. However, the implications of these two results are different: Size differences had simultaneous positive effects on niche and fitness differences and were also larger when species coexisted than when they excluded each other. Contrastingly, phylogenetic relatedness was unrelated to both niche differences and relative fitness differences, implying that being closely or distantly related has no significant effect on species niche differences, nor their fitness inequalities. In a similar experiment with green algae implemented under the modern coexistence framework, Narwani et al. [17] also found non-significant relationships between phylogenetic distance and coexistence, niche- or fitness differences, concluding that there was no evolutionary signal in species coexistence, neither in the ecological mechanisms (dis)allowing coexistence (i.e., stabilizing and equalizing forces). Indeed, in our study, there was no correlation between size differences and phylogenetic distance, suggesting that cyanobacteria body size is not a phylogenetically conserved trait. These results emphasize that functional traits allow more insight into coexistence of species than phylogenetic relatedness, but in the case of phytoplankton [17] and annual plants [18], traits fail to predict coexistence.
In our study, the clearest influence of size on coexistence was observed among species pairs with large size differences, since all the combinations with size differences of two orders of magnitude could coexist thanks to their large niche differences, despite showing relatively high fitness differences. This suggests that small and large phytoplankton species are occupying different niches, which allows them to stably coexist, even when one species has a clear fitness advantage over the other. However, for species of similar size, dominance of niche differences over fitness differences was less evident. Sometimes, niche differences were not large enough to overcome even small fitness differences, leading to competitive exclusion. Strong niche-based stabilizing effects have also been demonstrated experimentally in plankton communities with high functional diversity, suggesting that differences along a functional trait axis are mainly driven by niche differences [7, 45]. In ten out of fifteen species pairs included in this experiment, the stabilizing effect of niche differences was large enough to overcome the equalizing effect of relative fitness differences among species, resulting in stable coexistence. In our experiment, coexistence rather than competitive exclusion seems to be a dominant outcome of interactions between cyanobacteria, despite the long-standing expectations resulting from the paradox of the plankton. We can only hypothesize on some underlying mechanisms that may explain the niche differences underlying coexistence in our experiment. First, larger cells are expected to have higher sedimentation rates, as described in natural ecosystems for non-motile taxa [32]. Differential sedimentation rates were observed between the smallest and the largest cyanobacteria species in our cultures, despite the continuous, but gentle shaking of the flasks. We observed that the two largest species (C. cubana and A. hegewaldii) tended to sink relatively fast, whereas the smallest species (S. pevaleckii and S. PCC 6803) showed almost no sedimentation. Such sinking differences may lead to a spatial segregation of species within the culture flask, to differences in light availability, and might be considered as a source of niche differentiation. Second, differences in surface-area to-volume ratios might also play a role, because of their strong influence on resource acquisition in phytoplankton. Small phytoplankton cells, with a large surface-area to-volume ratio, generally have superior resource uptake capacities and show higher growth rates compared to large colonial units. In this study, the smaller species in all coexisting pairs were always better invaders than larger species, as their growth rates were less affected by the presence of the resident species. This might suggest that during invasion smaller species were able to cope better with the low nutrient concentrations generated by the resident species before invasion was initiated. Third, niche differentiation causes a species to limit its own growth more than the growth of its competitors, with stronger intraspecific than interspecific competition, and this stabilizes species coexistence. In our experiment, intraspecific size differences also varied for colony forming taxa. Most of our strains were two-cell colonies, whereas the largest species formed colonies ranging from a few to hundreds of cells. We propose that possibly those species represented by larger colonies showed a higher intraspecific size variability than those species composed by two-cell colonies, affecting the scope for niche differentiation and possibly the outcome of coexistence.
Embedded within the modern coexistence framework, our study provides a more detailed understanding of the influence of species’ size differences on both stabilizing and equalizing mechanisms of coexistence. The observed positive correlations between cell size differences, niche differences and relative fitness differences corroborates the importance of size as a master functional trait that captures much of the physiology and ecology of phytoplankton [20–23, 46]. Our experiment showed that large size differences among cyanobacteria had a simultaneous strong positive influence on the niche and relative fitness differences among species.
In addition to being a major determinant of population-level processes such as resource use, growth and biomass production, body size might also be considered a key functional trait controlling community-level processes [7]. By influencing the nature and strength of species interactions and the maintenance of diversity, community ecology research may benefit from incorporating more size-related information. Further research should focus on multiple species combinations, including simulations and experiments that take more complex natural conditions into account.
Supplementary information
Acknowledgements
IG was supported by a grant from the Swiss State Secretariat for Education, Research and Innovation (SERI) to BWI, linked to the EU NETLAKE COST Action. We would like to thank Anita Narwani, Ian T. Carroll and Oscar Godoy for valuable comments on earlier versions of the manuscript.
Author contributions
All authors conceived the study. IG performed the experiment, collected data, analyzed data and wrote the first draft of the manuscript. PV contributed to data analysis, figure conception and to revisions. All authors commented on the manuscript.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Supplementary information
The online version of this article (10.1038/s41396-018-0330-7) contains supplementary material, which is available to authorized users.
References
- 1.Hutchinson GE. The paradox of the plaknton. Am Nat. 1961;95:137–45. doi: 10.1086/282171. [DOI] [Google Scholar]
- 2.Gause GF. The struggle for existence. Baltimore: The Williams & Wilkins company; 1934. [Google Scholar]
- 3.Hardin G. The competitive exclusion principle. Science. 1960;131:1292–7. doi: 10.1126/science.131.3409.1292. [DOI] [PubMed] [Google Scholar]
- 4.MacArthur R, Levins R. The limiting similarity, convergence, and divergence of coexisting species. Am Nat. 1967;101:377. doi: 10.1086/282505. [DOI] [Google Scholar]
- 5.Bell G. The distribution of abundance in neutral communities. Am Nat. 2000;155:606–17. doi: 10.1086/303345. [DOI] [PubMed] [Google Scholar]
- 6.Hubbell SP. The unified neutral theory of biodiversity and biogeography. Princeton, NJ: Princeton University Press; 2001. [Google Scholar]
- 7.Levine JM, Bascompte J, Adler PB, Allesina S. Beyond pairwise mechanisms of species coexistence in complex communities. Nature. 2017;546:56–64. doi: 10.1038/nature22898. [DOI] [PubMed] [Google Scholar]
- 8.Hart SP, Usinowicz J, Levine JM. The spatial scales of species coexistence. Nat Ecol Evol. 2017;1:1066–73. doi: 10.1038/s41559-017-0230-7. [DOI] [PubMed] [Google Scholar]
- 9.Chesson P. Mechanisms of maintenance of species diversity. Annu Rev Ecol Syst. 2000;31:343–66. doi: 10.1146/annurev.ecolsys.31.1.343. [DOI] [Google Scholar]
- 10.Holt RD. Emergent neutrality. Trends Ecol Evol. 2006;21:531–3. doi: 10.1016/j.tree.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 11.Scheffer M, van Nes EH. Self-organized similarity, the evolutionary emergence of groups of similar species. Proc Natl Acad Sci USA. 2006;103:6230–5. doi: 10.1073/pnas.0508024103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Adler PB, HilleRislambers J, Levine JM. A niche for neutrality. Ecol Lett. 2007;10:95–104. doi: 10.1111/j.1461-0248.2006.00996.x. [DOI] [PubMed] [Google Scholar]
- 13.Carroll IT, Nisbet RM. Departures from neutrality induced by niche and relative fitness differences. Theor Ecol. 2015;8:449–65. doi: 10.1007/s12080-015-0261-0. [DOI] [Google Scholar]
- 14.Li L, Chesson P. The effects of dynamical rates on species coexistence in a variable environment: the paradox of the plankton revisited. Am Nat. 2016;188:E47–58. doi: 10.1086/687111. [DOI] [PubMed] [Google Scholar]
- 15.Adler PB, Ellner SP, Levine JM. Coexistence of perennial plants: an embarrassment of niches. Ecol Lett. 2010;13:1019–29. doi: 10.1111/j.1461-0248.2010.01496.x. [DOI] [PubMed] [Google Scholar]
- 16.Kraft NJB, Godoy O, Levine JM. Plant functional traits and the multidimensional nature of species coexistence. Proc Natl Acad Sci USA. 2015;112:797–802. doi: 10.1073/pnas.1413650112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Narwani A, Alexandrou Ma, Oakley TH, Carroll IT, Cardinale BJ. Experimental evidence that evolutionary relatedness does not affect the ecological mechanisms of coexistence in freshwater green algae. Ecol Lett. 2013;16:1373–81. doi: 10.1111/ele.12182. [DOI] [PubMed] [Google Scholar]
- 18.Godoy O, Kraft NJB, Levine JM. Phylogenetic relatedness and the determinants of competitive outcomes. Ecol Lett. 2014;17:836–44. doi: 10.1111/ele.12289. [DOI] [PubMed] [Google Scholar]
- 19.Adler PB, Fajardo A, Kleinhesselink AR, Kraft NJB. Trait-based tests of coexistence mechanisms. Ecol Lett. 2013;16:1294–306. doi: 10.1111/ele.12157. [DOI] [PubMed] [Google Scholar]
- 20.Raven JA. The twelfth Tansley lecture. Small is beautiful: the picophytoplankton. Funct Ecol. 1998;12:503–13. doi: 10.1046/j.1365-2435.1998.00233.x. [DOI] [Google Scholar]
- 21.Litchman E, Klausmeier CA. Trait-based community ecology of phytoplankton. Annu Rev Ecol Evol Syst. 2008;39:615–39. doi: 10.1146/annurev.ecolsys.39.110707.173549. [DOI] [Google Scholar]
- 22.Finkel ZV, Beardall J, Flynn KJ, Quigg A, Rees TAV, Raven Ja. Phytoplankton in a changing world: cell size and elemental stoichiometry. J Plankton Res. 2010;32:119–37. doi: 10.1093/plankt/fbp098. [DOI] [Google Scholar]
- 23.Key T, McCarthy A, Campbell DA, Six C, Roy S, Finkel ZV. Cell size trade-offs govern light exploitation strategies in marine phytoplankton. Environ Microbiol. 2010;12:95–104. doi: 10.1111/j.1462-2920.2009.02046.x. [DOI] [PubMed] [Google Scholar]
- 24.Barton AD, Pershing AJ, Litchman E, Record NR, Edwards KF, Finkel ZV, et al. The biogeography of marine plankton traits. Ecol Lett. 2013;16:522–34. doi: 10.1111/ele.12063. [DOI] [PubMed] [Google Scholar]
- 25.Litchman E, Klausmeier CA, Schofield OM, Falkowski PG. The role of functional traits and trade-offs in structuring phytoplankton communities: scaling from cellular to ecosystem level. Ecol Lett. 2007;10:1170–81. doi: 10.1111/j.1461-0248.2007.01117.x. [DOI] [PubMed] [Google Scholar]
- 26.Marañón E. Cell size as a key determinant of phytoplankton metabolism and community structure. Ann Rev Mar Sci. 2014;7:241–64. doi: 10.1146/annurev-marine-010814-015955. [DOI] [PubMed] [Google Scholar]
- 27.Roselli L, Litchman E, Stanca E, Cozzoli F, Basset A. Individual trait variation in phytoplankton communities across multiple spatial scales. J Plankton Res. 2017;39:577–88. doi: 10.1093/plankt/fbx019. [DOI] [Google Scholar]
- 28.Shih PM, Wu D, Latifi A, Axen SD, Fewer DP, Talla E, et al. Improving the coverage of the cyanobacterial phylum using diversity-driven genome sequencing. Proc Natl Acad Sci USA. 2013;110:1053–8. doi: 10.1073/pnas.1217107110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Paerl HW, Huisman J. Climate change: a catalyst for global expansion of harmful cyanobacterial blooms. Environ Microbiol Rep. 2009;1:27–37. doi: 10.1111/j.1758-2229.2008.00004.x. [DOI] [PubMed] [Google Scholar]
- 30.Stanier RY, Kunisawa R, Mandel M, Cohen-Bazire G. Purification and properties of unicellular blue-green algae (Order Chroococcales) Bacteriol Rev. 1971;35:171–205. doi: 10.1128/br.35.2.171-205.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hillebrand H, Dürselen CD, Kirschtel D, Pollingher U, Zohary T. Biovolume calculation for pelagic and benthic microalgae. J Phycol. 1999;424:403–24. doi: 10.1046/j.1529-8817.1999.3520403.x. [DOI] [Google Scholar]
- 32.Reynolds CS. Ecology of phytoplankton. Cambridge: Cambridge University Press; 2006. [Google Scholar]
- 33.Carroll IT, Cardinale BJ, Nisbet RM. Niche and fitness differences relate the maintenance of diversity to ecosystem function. Ecology. 2011;92:1157–65. doi: 10.1890/10-0302.1. [DOI] [PubMed] [Google Scholar]
- 34.Brown WL, Wilson EO. Character displacement. Syst Zool. 1956;5:49–64. doi: 10.2307/2411924. [DOI] [Google Scholar]
- 35.Hutchinson GE. Homage to Santa Rosalia or why are there so many kinds of animals? Am Nat. 1959;93:145–59. doi: 10.1086/282070. [DOI] [Google Scholar]
- 36.Schoener TW. The evolution of bill size differences among sympatric congeneric species of birds. Evolution. 1965;19:189–213. doi: 10.1111/j.1558-5646.1965.tb01707.x. [DOI] [Google Scholar]
- 37.Schoener TW. Resource partitioning in ecological communities. Science. 1974;185:27–39. doi: 10.1126/science.185.4145.27. [DOI] [PubMed] [Google Scholar]
- 38.Brooks JL, Dodson SI. Predation, body size, and composition of plankton. Science. 1965;150:28–35. doi: 10.1126/science.150.3692.28. [DOI] [PubMed] [Google Scholar]
- 39.Galbraith MG. Size‐selective predation on daphnia by rainbow trout and yellow perch. Trans Am Fish Soc. 1967;96:1–10. doi: 10.1577/1548-8659(1967)96[1:SPODBR]2.0.CO;2. [DOI] [Google Scholar]
- 40.Aarssen LW. Body size and fitness in plants: revisiting the selection consequences of competition. J Plant Ecol Evol Syst. 2015;17:236–42. doi: 10.1016/j.ppees.2015.02.004. [DOI] [Google Scholar]
- 41.Goldberg DE. Competitive ability: definitions, contingency and correlated traits. Philos Trans R Soc Lond Ser B Biol Sci. 1996;351:1377–85. doi: 10.1098/rstb.1996.0121. [DOI] [Google Scholar]
- 42.Grime JP. Plant strategies and vegetation processes. Chichester: Wiley; 1979. [Google Scholar]
- 43.MacDougall AS, Gilbert B, Levine JM. Plant invasions and the niche. J Ecol. 2009;97:609–15. doi: 10.1111/j.1365-2745.2009.01514.x. [DOI] [Google Scholar]
- 44.Vergnon R, Dulvy NK, Freckleton RP. Niches versus neutrality: uncovering the drivers of diversity in a species-rich community. Ecol Lett. 2009;12:1079–90. doi: 10.1111/j.1461-0248.2009.01364.x. [DOI] [PubMed] [Google Scholar]
- 45.Carrara F, Giometto A, Seymour M, Rinaldo A, Altermatt F. Experimental evidence for strong stabilizing forces at high functional diversity of aquatic microbial communities. Ecology. 2015;96:1340–50. doi: 10.1890/14-1324.1. [DOI] [PubMed] [Google Scholar]
- 46.Marañón E, Cermeño P, López-Sandoval DC, Rodríguez-Ramos T, Sobrino C, Huete-Ortega M, et al. Unimodal size scaling of phytoplankton growth and the size dependence of nutrient uptake and use. Ecol Lett. 2013;16:371–9. doi: 10.1111/ele.12052. [DOI] [PubMed] [Google Scholar]
- 47.Hoyle R. Structural Equation Modeling: Concepts, Issues, and Applications. Thousand Oaks: Sage Publications; 1995.
- 48.Bollen Kenneth A. Structural Equations with Latent Variables. Hoboken, NJ, USA: John Wiley & Sons, Inc.; 1989. [Google Scholar]
- 49.Wright S. Correlation and causation. J Agric Res. 1921;10:557–85.
- 50.Tomer A. A short history of structural equation models. In: Pugesek BH, Tomer A, Eye A von (eds). Structural Equation Modeling: Applications in Ecological and Evolutionary Biology. pp. 85–125. Cambridge: Cambridge University Press; 2003.
- 51.Grace James B., Anderson T. Michael, Olff Han, Scheiner Samuel M. On the specification of structural equation models for ecological systems. Ecological Monographs. 2010;80(1):67–87. doi: 10.1890/09-0464.1. [DOI] [Google Scholar]
- 52.Hu Li‐tze, Bentler Peter M. Cutoff criteria for fit indexes in covariance structure analysis: Conventional criteria versus new alternatives. Structural Equation Modeling: A Multidisciplinary Journal. 1999;6(1):1–55. doi: 10.1080/10705519909540118. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.