Abstract
Here we demonstrate a mode of reciprocal regulation between GFI1 and p53 that controls the induction of apoptosis in T cells. We show that GFI1 prevents induction of p53 dependent apoptosis by recruiting LSD1 to p53, which leads to the demethylation of its C-terminal domain. This is accompanied by a decrease of the acetylation of lysine 117 within the core domain of the murine p53 protein, which is required for transcriptional induction of apoptosis. Our results support a model in which the effect of GFI1’s regulation of methylation at the c-terminus of p53 is ultimately mediated through control of acetylation at lysine 117 of p53. We propose that GFI1 acts prior to the occurrence of DNA damage to affect the post-translational modification state and limit the subsequent activation of p53. Once activated, p53 then transcriptionally activates GFI1, presumably in order to re-establish the homeostatic balance of p53 activity. These findings have implications for the activity level of p53 in various disease contexts where levels of GFI1 are either increased or decreased.
Subject terms: Tumour-suppressor proteins, Cell death, Post-translational modifications
Introduction
GFI1 is a protein mainly known as a transcription factor involved in hematopoietic cell differentiation by repressing key target genes in multi-potent precursor cells in order to direct their lineage commitment. GFI1 accomplishes this by recruiting the de-methylase LSD1 and histone de-acetylases such as HDAC1 to target promoters1–5. GFI1 also has oncogenic activity by promoting survival in leukemic T cells and is required for the development and maintenance of T cell leukemia6–8. In this role, GFI1 antagonizes the p53-dependent induction of apoptosis, in part through regulating post-translational modifications at the C-terminal domain of the p53 protein, and as a consequence through transcriptional regulation of the key apoptotic targets Bax, Bbc3 (Puma) and Pmaip1 (Noxa)8. In addition, GFI1 has been shown to promote DNA repair itself by regulating the activity of DNA repair proteins such as MRE11 and 53BP1 following damage and to contribute to T cell survival in this manner9. GFI1 has also been shown to favour the survival and accumulation of myeloid cells in myelo-proliferative disease10, potentially due to its role in repressing p53 activity.
It has long been established that the activity of p53 is regulated by the modulation of the protein’s stability by ubiquitination11–13. However, in recent years, additional post-translational modifications (PTMs) including acetylation and methylation have emerged as critical factors in the regulation of p53 activity. Transcriptional regulation by p53 of targets involved in specific biological functions has been associated with sets of corresponding PTMs. For instance, mutant mouse p53 proteins carrying lysine to arginine mutations at 3 lysine residues (117, 161, 162, corresponding to residues 120 and 164 in the human protein, which has one fewer lysine at this location) in the DNA-binding domain of p53 are deficient in inducing apoptosis, cell cycle arrest and senescence, whereas a p53 protein carrying a single lysine to arginine substitution at residue 117 (120 in the human protein) is only deficient in inducing apoptosis14.
Regulation of p53 through modification of its C-terminal Domain (CTD) has also been shown to be important for its activity, however its exact role is not clearly defined, as there is conflicting evidence in the literature. Studies have shown that the CTD is involved in the regulation of p53 DNA binding, the interaction with its negative regulator MDM2, and the regulation of its stability, among other functions15–18. Of particular interest here are several reports showing that the impact of the CTD on p53 activities is likely context- and cell type-specific. For example, through ectopic expression of mutant p53 proteins in several human cancer cell lines, it was found that p53 binding to the key apoptotic target Puma requires the CTD19, while later work using mouse models and murine primary cells showed that expression of CTD-deleted p53 actually increased Puma expression and induction of cell death in thymocytes20. Given the complexity and the number of potential residue modifications within the p53 protein, their individual roles are not yet fully understood, and neither is the interplay between modifications of different residues.
Here, we focus on detailing how GFI1 activity affects post-translational modification of p53 at both the C-terminus and at lysine 117, and how this translates into changes in induction of apoptosis in T cells. We take advantage of multiple mouse models to characterize the mechanism by which GFI1 regulates p53 activity in this way. We first use a Gfi1 KO mouse model as well as a model expressing a mutant GFI1 protein with a proline to alanine mutation at residue 2 (P2A), which affects its interaction with other proteins, notably LSD15,21. Our Gfi1 KO model features a GFP coding sequence, which is inserted in-frame with the initiation codon of Gfi1 and replaces exons 3–5 of the Gfi1 gene, resulting in the production of a GFP transcript under the control of Gfi1’s regulatory sequences and the absence of GFI1 protein22.
We also use a mouse model expressing a truncated form of the p53 protein lacking the 24 C-terminal amino acids20. This model (p53 ΔCTD) lacks several lysine residues (K370, K372, K373, K381, K382, K386 K387 in the mouse protein, corresponding to K370, K372, K373, K381, K382, K386 in the human protein) that can be modified by methylation and acetylation. Finally, we use a knock-in mouse expressing a mutant p53 in which a lysine to arginine substitution at residue 117 (p53 K117R) prevents an acetylation event necessary for the transactivation by p53 of genes critical for the induction of apoptosis14.
Here, we show that in T cells, GFI1 blocks the accumulation of methylation marks at the CTD of p53, which in turn inhibits the acetylation of K117. In addition, we demonstrate that GFI1 is activated by p53 following DNA damage. Given that GFI1 antagonizes the function of p53, our results suggest that GFI1 plays a role in post-translational negative feedback regulation of p53 induced by p53’s own transcriptional activity, as is seen with other regulators of p53, such as MDM2.
Results
The intermediate domain of GFI1 interacts with the C-terminal domain of p53
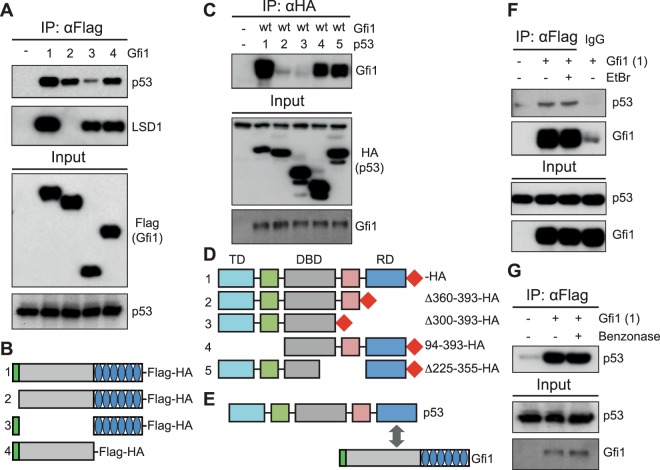
To fully characterize the interaction between GFI1 and p53, we first performed co-immunoprecipitation (Co-IP) experiments using a series of GFI1 mutants. These experiments showed that the full-length form of GFI1 is capable of interacting with p53, and that deletion of the intermediary domain of GFI1 prevents this interaction, whereas deletions of the SNAG domain or of the DNA binding domain of GFI1 had no effect on the binding to p53 (Fig. 1A,B). This is in contrast with the interaction between GFI1 and the de-methylase enzyme LSD1, which requires the SNAG domain of GFI1 for interaction. Further analysis of GFI1 variants with deletions in the intermediary domain showed that different sizeable deletions across the intermediate domain decreased the interaction with p53, without identifying a single critical region, suggesting that the domain as a whole may be involved (Suppl. Fig. 1A). Conversely, Co-IP experiments using deletion variants of the p53 protein showed that its C-terminal domain, but not the N-terminal domain of the protein, is required for its interaction with GFI1 (Fig. 1C,D), suggesting that the intermediate domain of GFI1 interacts with p53’s C-terminal end (Fig. 1E). Interestingly, further Co-IP experiments showed that the interaction between the two proteins is unaffected by the presence of benzonase or ethidium bromide (EtBr), indicating that the p53/GFI1 interaction is independent of DNA binding (Fig. 1F,G, Supplementary Fig. 1B).
Figure 1.
The intermediate domain of GFI1 interacts with the C-terminal domain of p53 (A) 293T cells were transfected with vectors expressing variants of the GFI1-Flag-HA fusion protein. Nuclear extracts from these proteins were immunoprecipitated for Flag, separated by SDS-PAGE and blotted for the indicated proteins. Blots from input controls are shown below. (B) Schematic representation of the variant GFI1 proteins used in (A). (C) 293T cells were transfected with vectors expressing variants of the p53-HA fusion protein as well as GFI1-Flag protein without an HA tag. Extracts were immunoprecipitated for HA and blotted for the indicated proteins. (D) Schematic representation of the variant p53 proteins used in (C). (E) Proposed model of the interaction between GFI1 and p53. (F) GFI1-Flag-HA fusion protein was immunoprecipitated in 293T cells in the presence or absence of ethidium bromide and blotted for p53. (G) GFI1-Flag-HA fusion protein was immunoprecipitated in 293T cells in the presence or absence of benzonase and blotted for p53.
GFI1 antagonizes p53 CTD methylation and K117 acetylation, and restrains the p53-dependent apoptotic response to DNA damage
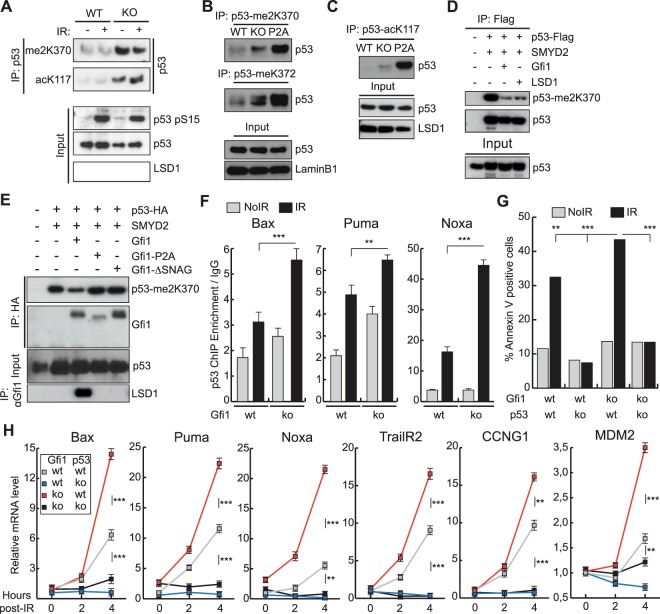
In order to measure the methylation status of the p53 CTD, we generated antibodies for p53-me2K370 and p53-me2K372 and confirmed their binding efficacy and specificity in vitro (Supplementary Fig. 2A). Using Co-IP experiments with these antibodies and others specific for post-translationally modified p53 residues, we show that, in addition to the previously reported increase in K372 mono-methylation8, K370 di-methylation as well as K117 acetylation was also increased in Gfi1 KO thymocytes. Interestingly, these increases were independent of DNA damage induction by irradiation, unlike the well-described phosphorylation of S15 (Fig. 2A). The increase in K117 acetylation is noteworthy as this PTM is well established as being required for induction of key apoptotic genes downstream of p53. Furthermore, we show that in thymocytes extracted from mice carrying the P2A mutant variant of Gfi121, which cannot interact with LSD15, both K372 mono-methylation and K370 di-methylation marks were increased (Fig. 2B), similarly to what was observed in the absence of GFI1. Furthermore, GFI1 P2A expressing cells also displayed increased K117 acetylation (Fig. 2C).
Figure 2.
GFI1 modulates the induction of apoptosis by p53 through CTD and core domain PTMs. (A) Thymocytes from GFI1 WT and KO mice were exposed to 5Gy IR or left untreated. Nuclear extracts from these cells were immunoprecipitated with p53-me2K370 or p53-acK117 specific antibodies and blotted for total p53. (B) Nuclear extracts from GFI1 WT, KO and P2A thymocytes were immunoprecipitated with p53-me2K370 or p53-meK372 specific antibodies and blotted for total p53. (C) Nuclear extracts as in B were immunoprecipitated with a p53-acK117 specific antibody and blotted for total p53. (D) 293T cells were transfected with the indicated combinations of p53-Flag, SMYD2, GFI1 and LSD1 expression vectors. Nuclear extracts were immunoprecipitated with an anti-Flag antibody and blotted for p53-me2K370 and total p53. (E) 293T cells were transfected with the indicated combinations of p53-HA, SMYD2, GFI1, GFI1-P2A and GFI1-ΔSNAG expression vectors. Nuclear extracts were immunoprecipitated with anti-HA or anti-GFI1 antibodies and blotted as indicated. (F) Thymocytes were extracted from GFI1 WT and KO mice, exposed to 5Gy IR or left untreated and fixed with formaldehyde after 2 hours. Chromatin immunoprecipitation was performed to isolate p53-bound DNA fragments. Fold enrichment of the indicated target promoters relative to a non-specific IgG control is shown. Error bars represent s.d. P values: *=<0.05, **=<0.01, ***=<0.001, calculated from a Welch corrected t-test. (G) Thymocytes were extracted from mice carrying combinations of Gfi1 KO and p53 KO. Cells were exposed to 5Gy IR or left untreated and were stained for Annexin V 4 hours later. The proportion of Annexin V positive cells as measured by FACS is shown. Statistical significance was calculated using Fisher’s exact test. (H) mRNA was extracted from thymocytes as in (G). The levels of the indicated genes were measured at the indicated time points after IR by qPCR relative to Gapdh.
We further tested the ability of GFI1 to block the accumulation of methylation marks at the C-terminal end of p53 by overexpressing p53, SMYD2, and either GFI1 or LSD1 in 293T cells. Overexpression of SMYD2 readily induced K370 di-methylation in these cells, while a catalytically inactive SMYD2-GEE mutant did not, as expected (Suppl. Fig. 2A)23. We found that concomitant overexpression of either GFI1 or LSD1 blocked the accumulation of K370 di-methylation induced by SMYD2 (Fig. 2D). This effect was dose-dependent, as overexpression of increasing levels of exogenous GFI1 showed a proportional reduction in K370 di-methylation (Suppl. Fig. 2B). Importantly, overexpression of mutant variants of GFI1 that cannot interact with LSD1, namely GFI1 P2A and GFI1 ΔSNAG, did not lead to a decrease in p53-K370 di-methylation (Fig. 2E). Together, these data demonstrate that GFI1 causes de-methylation of the C-terminal domain of p53 through recruitment of LSD1, as previously suggested8. Moreover, our results show that GFI1 antagonizes the accumulation of the acK117 mark, which is key to inducing apoptosis. As expected, the increase in C-terminal methylation and K117 acetylation in the Gfi1 KO context correlated with a greater induction of the pro-apoptotic p53 target genes Bax, Puma, Noxa and TrailR2 in response to IR exposure, as compared to WT cells (Suppl. Fig. 2C). This was likely due to increased binding of p53 to the promoter of these genes, as assessed by chromatin immunoprecipitation (Fig. 2F).
To confirm that the increased apoptotic response we observed in the Gfi1 KO cells was p53 dependent we crossed our Gfi1 KO mice with p53 KO mice. We found that the increased apoptotic response in Gfi1 KO cells was no longer observed in the context of a p53 KO, i.e. when p53 was also absent (Fig. 2G). Accordingly, the greater induction of p53 targets Bax, Puma, Noxa and TrailR2 following IR exposure in Gfi1 KO cells as compared to WT cells, was completely eliminated in Gfi1/p53 double KO cells (Fig. 2H). Interestingly, the induction of the negative feedback regulators of p53, Mdm2 and Ccng1 was also found to be increased in Gfi1 KO cells, with this effect once again being dependent on p53 itself (Fig. 2H). Taken together, these data support a model whereby GFI1 antagonizes p53 CTD methylation and K117 acetylation, which leads to a restrained p53-dependent apoptotic response to DNA damage.
GFI1’s impact on p53 activity is ultimately mediated through changes in K117 acetylation
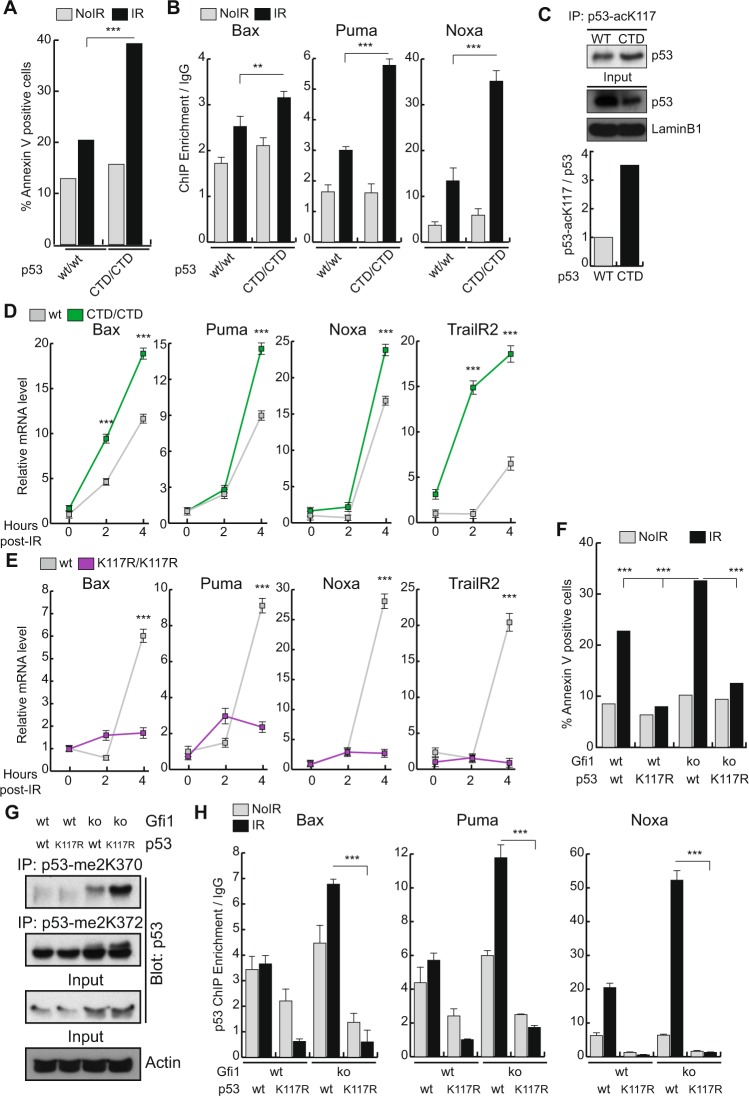
In order to better understand how the GFI1-dependent regulation of p53 PTMs affects the induction of apoptosis, we used a mouse model expressing a variant of p53 lacking its C-terminal domain20. Similarly to what we observed in the Gfi1 KO model, the absence of p53 CTD leads to increased apoptosis in thymocytes exposed to IR (Fig. 3A), as well as increased recruitment of p53 to the promoters of Bax, Puma, Noxa (Fig. 3B), an increase in the level of p53 K117 acetylation (Fig. 3C) and a greater induction of the expression of apoptotic genes following IR (Fig. 3D). These results also suggest that a p53 protein carrying an un-methylated CTD, as mediated by the activity of GFI1, is less able to induce the apoptotic response than the methylated one and that the complete removal of the CTD prevents this check on p53 activity, leading to unrestrained p53 activation, as is seen in Gfi1 KO cells.
Figure 3.
The effect of GFI1 in p53 regulation is mediated through K117 of p53. (A) Thymocytes were extracted from p53 WT and p53 ΔCTD mice. Cells were exposed to 5Gy IR or left untreated and were stained for Annexin V 4 hours later. The proportion of Annexin V positive cells as measured by FACS is shown. (B) Thymocytes were extracted from p53 WT and p53 ΔCTD mice, exposed to 5 Gy IR or left untreated and fixed with formaldehyde after 2 hours. Chromatin immunoprecipitation was performed to isolate p53-bound DNA fragments. Fold enrichment of the indicated promoter targets relative to a non-specific IgG control is shown. (C) Nuclear extracts from p53 wt and p53 CTD thymocytes were immunoprecipitated with a p53-acK117 specific antibody and blotted for total p53. (D) mRNA was extracted from thymocytes as in (A). The levels of the indicated genes were measured at the indicated time points after IR by qPCR relative to Gapdh. (E) mRNA was extracted from the thymocytes of p53 WT and p53 K117R mutant mice. The levels of the indicated genes were measured at the indicated time points after IR by qPCR relative to Gapdh. (F) Thymocytes were extracted from mice carrying combinations of GFI1 KO and p53 K117R mutation. Cells were exposed to 5Gy IR or left untreated and were stained for Annexin V 4 hours later. The proportion of Annexin V positive cells as measured by FACS is shown. (G) Thymocytes from mice carrying combinations of GFI1 KO and p53 K117R mutation were exposed to 5Gy IR or left un-irradiated. Nuclear extracts from these cells were immunoprecipitated with p53-me2K370 or p53-me2K372 specific antibodies and blotted for total p53. (H) Thymocytes were extracted from WT, p53 K117R, GFI1 KO and p53 K117R GFI1 KO mice, exposed to 5Gy IR or left untreated and fixed with formaldehyde after 2 hours. Chromatin immunoprecipitation was performed to isolate p53-bound DNA fragments. Fold enrichment of the indicated promoter targets relative to a non-specific IgG control is shown.
To confirm this hypothesis, we then used another knock-in mouse model expressing a mutant p53 with a lysine to arginine substitution at amino acid 117 (p53 K117R)14. As expected, thymocytes extracted from these mice did not show activation of the pro-apoptotic genes Bax, Puma, Noxa and TrailR2 following IR exposure (Fig. 3E). We crossed the p53 K117R line with our GFI1 KO line and found that the increase in apoptosis following IR observed in GFI1 KO cells was completely eliminated in cells that also carried the p53 K117R mutation (Fig. 3F). Most importantly, we still detected an increase in p53 C-terminal K370 di-methylation and K372 di-methylation in the GFI1 KO p53 K117R cells after IR, similar to what we observed in GFI1 KO cells (Fig. 3G). This shows that the regulation exerted at the C-Terminal domain of p53 is not sufficient to induce apoptosis on its own and strongly suggests that the effect of GFI1 on apoptosis is ultimately mediated through the regulation of p53 K117 acetylation. This is supported by p53 ChIP experiments using combinatorial GFI1 KO and p53 K117R mutant mice, which show that while GFI1 KO leads to an increase in p53 recruitment to promoters of the apoptotic genes Bax, Puma and Noxa, it does not lead to any increase in the recruitment of p53K117R to these promoters (Fig. 3H). This also suggests that the increase in C-terminal methylation of p53 in GFI1 KO cells only leads to an increase in apoptotic response after acetylation at K117.
GFI1 and p53 are part of a regulatory feedback loop
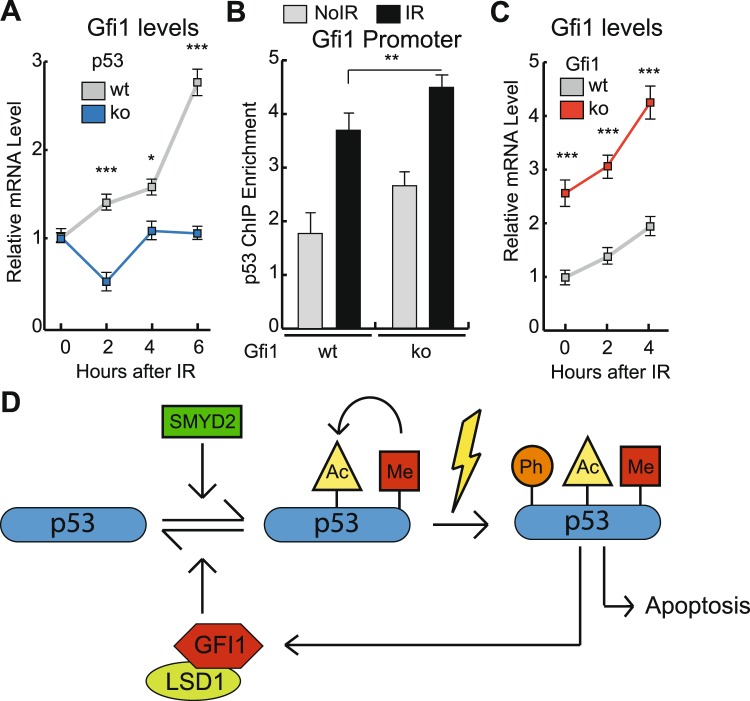
The data presented here, as well as previous reports, show that GFI1 is a critical regulator of p53 activities, at least in thymocytes8. Other studies indicated that p53 might regulate GFI1 expression, suggesting a possible regulatory feedback loop between these two proteins, in a manner similar to the well-established p53-MDM2 negative feedback loop. To determine whether p53 controls GFI1 in thymocytes, we measured the mRNA levels of Gfi1 in p53 WT and KO cells at baseline as well as following exposure to IR and observed that while Gfi1 mRNA levels were not altered by p53 KO at baseline, they increased following exposure to IR in a p53 dependent manner (Fig. 4A). Consistent with this, ChIP enrichment of p53 at the Gfi1 promoter increased following IR exposure (Fig. 4B). In order to further study the relationship between GFI1 and p53, we took advantage of the fact that the Gfi1 KO construct expresses a non-functional GFI1-GFP mutant mRNA that can still be measured using primers that recognize the 3’UTR of Gfi122. Interestingly, in cells expressing the Gfi1-GFP mRNA (and thus lacking a functional GFI1 protein), the binding of p53 to the Gfi1 promoter was increased both in the absence of DNA damage as well as following exposure to IR (Fig. 4B). Furthermore, the levels of the GFI1-GFP mRNA were higher than the baseline levels of normal Gfi1 mRNA and these levels increased even further following irradiation (Fig. 4C). These results are consistent with a model whereby GFI1 is a p53-activated negative feedback regulator of p53, since in the absence of a functional GFI1 protein p53 binds more strongly to the promoter of GFI1 and induces it without being repressed in return.
Figure 4.
GFI1 is a negative feedback regulator of p53 (A) Thymocytes were extracted from p53 WT or KO mice. Cells were exposed to 5Gy IR or left untreated. mRNA was extracted and the levels of Gfi1 were measured at the indicated time points by qPCR relative to Gapdh. (B) Thymocytes were extracted from GFI1 WT and KO mice. Cells were exposed to 5Gy IR or left untreated and fixed with formaldehyde after 2 hours. Chromatin immunoprecipitation was performed to isolate p53-bound DNA fragments. Fold enrichment amplification at the Gfi1 promoter relative to a non-specific IgG control is shown. (C) mRNA was extracted from thymocytes as in (B). The levels of KO mutant mRNA were measured at the indicated time points after IR by qPCR relative to Gapdh. (D) Diagram representing the regulatory circuit between GFI1 and p53.
Our data support a model describing a novel regulatory loop between GFI1 and p53, which controls p53 activity in T cells. While methyltransferases- and acetylases-dependent PTMs prime p53 for potent pro-apoptotic responses after DNA damage, GFI1 favours the removal of these marks, through the recruitment of the lysine de-methylase LSD1 at K370 and K372, and through an unknown mechanism at K117, which leads to restrained p53 activities. Upon exposure to DNA damage, the modified form of p53 is more strongly activated than its unmodified counterpart and it promotes both a potent apoptotic response and the transactivation of GFI1, which in turn leads to a de-methylation of p53 and its inactivation hence forming a novel negative feedback loop (Fig. 4D).
Discussion
Our results indicate that a regulatory circuit exists between GFI1 and p53 that controls the induction of apoptosis in T cells. We speculate that this link may exist in other cell types that express GFI1, such as myeloid cells, where a link between the two proteins has also been suggested10.
Our study builds on a previously suggested mechanism of regulation of p53 activity by GFI1, which proposed that the two proteins form a complex with the lysine de-methylase LSD1. Using deletion mutants and biochemical approaches we were able to characterize the domains critical for the interaction between GFI1 and p53 proteins, i.e. the intermediate domain of GFI1 and the C-Terminal domain of p53. Interestingly, the intermediate domain of GFI1 has also recently been shown to mediate its interaction with the DNA repair protein MRE11 and the arginine methyltransferase PRMT1 that targets it9. This suggests that this domain of GFI1 is involved in the regulation of functions related to p53 activity and DNA repair, whereas the SNAG domain at the very N-terminal end of GFI1 is more closely, but not exclusively, associated with its transcriptional activity. This also indicates that GFI1 has two very different mechanisms of action, one as a DNA binding transcription factor and another one as a bridging factor facilitating contact between enzymes and their protein substrates and enabling specific PTMs to take place.
We provide new evidence that the recruitment of LSD1 to p53 is necessary for the effect of GFI1 on p53 CTD methylation and consequently on apoptosis. Furthermore, our work with the p53ΔCTD mouse model shows that this form of p53 is overactive and induces apoptosis more readily than wild-type p53. This is in agreement with the original report describing this mouse model and defines the CTD as a negative regulator of p53 activity in thymocytes20. These data also suggest that a non-methylated CTD, as seen in GFI1 expressing cells, acts in a way that represses p53 activation.
Importantly, we show that C-terminal methylation on its own is insufficient to control p53-induced apoptosis but also requires acetylation at lysine 117 to have an effect of p53 activity. Our experiment with a K117R mutant variant of p53 demonstrates this, since cells from this mutant do not undergo apoptosis regardless whether Gfi1 is present or not. This mutation effectively renders the increase in C-terminal domain methylation seen in Gfi1 KO cells inoperative or incapable of increasing p53’s activity. These results support the notion that GFI1’s effect on programmed cell death is ultimately dependent on its ability to control acetylation of the K117 residue of p53 via the regulation of CTD methylation. It also suggests that other regulatory pathways that involve methylation events at p53’s CTD may ultimately be mediated through PTM of other p53 domains. Notably, this idea is consistent with findings indicating that the CTD of p53 may be involved in conformational changes of the DNA binding domain, which contains the K117 residue24.
All effects of GFI1 on post-translational modifications of p53 that we analyzed here are independent of both exposure to irradiation and interaction with DNA, suggesting that GFI1’s role is to control the extent to which p53 is “primed” to induce apoptosis once DNA damage occurs. In the context of GFI1 KO, there appears to be excessive priming of p53, leading to greater induction of apoptosis, although a death-inducing stimulus, such as exposure to IR, is still required to fully activate p53. Although this function of GFI1 is distinct from its canonical role as a DNA binding transcription factor, it remains to be determined whether GFI1 additionally plays a direct transcriptional role in regulating the induction of apoptosis and to which extent both functions overlap.
Although data exist in the literature showing that p53 can regulate GFI1, other conflicting evidence suggests that this regulation is context specific. For instance, p53 KO BM Lineage-Sca+c-Kit+ progenitor cells have been shown to have lower baseline levels of GFI125, while another group has shown this effect occurs in the spleen and thymus, but not in the bone marrow20. However, others have shown that p53 represses GFI1 expression specifically following DNA damage26. This is in contrast with the data presented here showing that GFI1 is transcriptionally activated following IR exposure in a p53-dependent manner. This discrepancy may be explained by the different cell systems used, i.e. murine primary T cells for the present study versus a murine pro B cell line and several human cancer cell lines in the previous study26. Moreover, our own experiments were carried out within a much shorter time frame after exposure to DNA damage (2–6 h vs. >24 h). Nevertheless, our data otherwise agree with findings from the literature that GFI1 antagonizes the p53-dependent apoptotic response. Given the data present here and the fact that the activation of key pro-apoptotic targets of p53 is dampened by the action of GFI1, it is likely that GFI1 is a negative feedback regulator of p53 in T cells. Indeed, the increased binding of p53 at the Gfi1 promoter, and the increased Gfi1 transcript levels in the absence of a functional GFI1 protein strongly support this notion and a model in which p53 and GFI1 are members of a regulatory circuit controlling apoptosis in T-cells.
Methods
Mouse Strains
Gfi1 KO and Gfi1 P2A mice used in this study have been previously described5,10,27,28. p53CTD Mutant Mice20 and p53 K117R14 mutant mice were graciously provided by the laboratories of James Manfredi and Wei Gu, respectively. Mice have been bred on to C57BL/6 genetic background and were maintained in a Specific-Pathogen-Free Plus environment at the Institut de recherches cliniques de Montreal (IRCM). The Institutional Review Board of the IRCM approved all animal protocols and experimental procedures were performed in compliance with IRCM and CCAC (Canadian Council of Animal Care) guidelines.
Cell Culture
293T cells were maintained in RPMI media (Multicell) supplemented with 10% Bovine Growth Serum (RMBIO Fetalgro) and 100 IU Penicillin and 100 μg/ml Streptomycin (Multicell). We verified that none of the cell lines used in this study were found in the Register of Misidentified Cell Lines maintained by the International Cell Line Authentication Committee (http://iclac.org/databases/cross-contaminations/).
Annexin V Staining
Cells were extracted from mouse thymi and treated with red blood cell lysis buffer for 5 min at RT. Cells were then resuspended in RPMI media as for cell culture. Cells were exposed to 5 Gy IR and incubated for 4 hours prior to staining using the FITC Annexin V Apoptosis Kit I (BD Pharmingen) according to the manufacturer’s instruction. Cell staining was measured on a FACSCalibur.
Antibodies
For the detection of p53-me2K370 and p53-me2K372, polyconal antibodies were generated in rabbits against the peptide sequences CSSHLK(me2)SKKGQS and CSHLKSK(me2)KGQST. The following commercial antibodies were used: LaminB1 ab16048 (Abcam), p53 1C12 (Cell Signaling Technology), p53-meK372 ab16033 (Abcam), GFI1 AF3540 (R&D Systems), GFI1 H-200 (Santa-Cruz), LSD1 EPR6826 (Epitomics 5839-1), HA 12CA5 (Sigma), Flag M2 (Sigma).
Co-Immunoprecipitation
For each Immunoprecipitation, 10 million cells were lysed in buffer I (0.5% NP-40, 10 mM Hepes, 10 mM KCl, 2 mM EDTA, 10% Glycerol, Complete protease inhibitor (Roche), pH7.5), incubated on ice for 10 min and centrifuged for 10 min at 13,000 rpm. Pellets were lysed in 500 μl buffer II (50 mM Sodium Phosphate, 300 mM NaCl, 1 mM β-mercaptoethanol, 10% Glycerol, 0.5% NP-40, 0.5% Triton X-100, Complete protease inhibitor (Roche), pH 7.5), mixed by vortexing and sonicated twice on a Brason digital sonifier for 10 sec at 50% output followed by 10 min incubation on ice and centrifuged for 10 min at 13,000 rpm. Supernatant was incubated for 2 h using the antibody of interest followed by 1 h incubation with Protein-A or -G agarose beads (Roche). Beads were washed 4 times with buffer II and proteins were extracted by boiling the beads for 5 min in SDS-PAGE sample loading buffer prior to separation by SDS-PAGE electrophoresis.
Expression level measurement
Total mRNA was extracted from cells using the RNeasy Micro Kit (Qiagen) according to the manufacturer’s instructions. mRNA was reverse transcribed to cDNA using superscript II (Thermo Fisher Scientific).
Supplementary information
Acknowledgements
We would like to thank the staff of the IRCM Animal Facility. We thank professor Wei Gu from the Institute of Cancer Genetics of Columbia University for kindly providing us with the p53 K117R mouse line. This work was supported by a CIHR (Canadian Institutes of Health Research) Foundation grant (FDN-148372) and a Canada Research Chair Tier 1 to T.M. C.V. was funded by a post-doctoral fellowship from FRQS (Fonds de Recherche Santé Québec) and by a post-doctoral fellowship from CIHR. J. M. was supported by grant R01 CA200256 from the National Cancer Institute.
Author Contributions
Conceptualization, C.V. and T.M.; Investigation, C.V., R.C. and J.F.; Resources, P.-J.H., J.M. and T.M. Writing - Review & Editing, C.V., P.-J.H., J.M. and T.M.
Data Availability
No datasets were generated or analyzed as part of this study.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-41684-2.
References
- 1.Moroy T, Vassen L, Wilkes B, Khandanpour C. From cytopenia to leukemia: the role of Gfi1 and Gfi1b in blood formation. Blood. 2015;126:2561–2569. doi: 10.1182/blood-2015-06-655043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Phelan JD, Shroyer NF, Cook T, Gebelein B, Grimes HL. Gfi1-cells and circuits: unraveling transcriptional networks of development and disease. Curr Opin Hematol. 2010;17:300–307. doi: 10.1097/MOH.0b013e32833a06f8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van der Meer LT, Jansen JH, van der Reijden BA. Gfi1 and Gfi1b: key regulators of hematopoiesis. Leukemia. 2010;24:1834–1843. doi: 10.1038/leu.2010.195. [DOI] [PubMed] [Google Scholar]
- 4.Duan Z, Zarebski A, Montoya-Durango D, Grimes HL, Horwitz M. Gfi1 coordinates epigenetic repression of p21Cip/WAF1 by recruitment of histone lysine methyltransferase G9a and histone deacetylase 1. Mol Cell Biol. 2005;25:10338–10351. doi: 10.1128/MCB.25.23.10338-10351.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Saleque S, Kim J, Rooke HM, Orkin SH. Epigenetic regulation of hematopoietic differentiation by Gfi-1 and Gfi-1b is mediated by the cofactors CoREST and LSD1. Mol Cell. 2007;27:562–572. doi: 10.1016/j.molcel.2007.06.039. [DOI] [PubMed] [Google Scholar]
- 6.Grimes HL, Gilks CB, Chan TO, Porter S, Tsichlis PN. The Gfi-1 protooncoprotein represses Bax expression and inhibits T-cell death. Proc Natl Acad Sci USA. 1996;93:14569–14573. doi: 10.1073/pnas.93.25.14569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Karsunky H, Mende I, Schmidt T, Moroy T. High levels of the onco-protein Gfi-1 accelerate T-cell proliferation and inhibit activation induced T-cell death in Jurkat T-cells. Oncogene. 2002;21:1571–1579. doi: 10.1038/sj.onc.1205216. [DOI] [PubMed] [Google Scholar]
- 8.Khandanpour C, et al. Growth factor independence 1 antagonizes a p53-induced DNA damage response pathway in lymphoblastic leukemia. Cancer Cell. 2013;23:200–214. doi: 10.1016/j.ccr.2013.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vadnais C, et al. GFI1 facilitates efficient DNA repair by regulating PRMT1 dependent methylation of MRE11 and 53BP1. Nat Commun. 2018;9:1418. doi: 10.1038/s41467-018-03817-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fraszczak Jennifer, Vadnais Charles, Rashkovan Marissa, Ross Julie, Beauchemin Hugues, Chen Riyan, Grapton Damien, Khandanpour Cyrus, Möröy Tarik. Reduced expression but not deficiency of GFI1 causes a fatal myeloproliferative disease in mice. Leukemia. 2018;33(1):110–121. doi: 10.1038/s41375-018-0166-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Toledo F, Wahl GM. Regulating the p53 pathway: in vitro hypotheses, in vivo veritas. Nat Rev Cancer. 2006;6:909–923. doi: 10.1038/nrc2012. [DOI] [PubMed] [Google Scholar]
- 12.Kruse JP, Gu W. Modes of p53 regulation. Cell. 2009;137:609–622. doi: 10.1016/j.cell.2009.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pant V, Lozano G. Dissecting the p53-Mdm2 feedback loop in vivo: uncoupling the role in p53 stability and activity. Oncotarget. 2014;5:1149–1156. doi: 10.18632/oncotarget.1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li T, et al. Tumor suppression in the absence of p53-mediated cell-cycle arrest, apoptosis, and senescence. Cell. 2012;149:1269–1283. doi: 10.1016/j.cell.2012.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Laptenko O, Tong DR, Manfredi J, Prives C. The Tail That Wags the Dog: How the Disordered C-Terminal Domain Controls the Transcriptional Activities of the p53 Tumor-Suppressor Protein. Trends Biochem Sci. 2016;41:1022–1034. doi: 10.1016/j.tibs.2016.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Poyurovsky MV, et al. The C terminus of p53 binds the N-terminal domain of MDM2. Nat Struct Mol Biol. 2010;17:982–989. doi: 10.1038/nsmb.1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim H, et al. p53 requires an intact C-terminal domain for DNA binding and transactivation. J Mol Biol. 2012;415:843–854. doi: 10.1016/j.jmb.2011.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Laptenko O, et al. The p53 C terminus controls site-specific DNA binding and promotes structural changes within the central DNA binding domain. Mol Cell. 2015;57:1034–1046. doi: 10.1016/j.molcel.2015.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hamard PJ, Lukin DJ, Manfredi JJ. p53 basic C terminus regulates p53 functions through DNA binding modulation of subset of target genes. J Biol Chem. 2012;287:22397–22407. doi: 10.1074/jbc.M111.331298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hamard PJ, et al. The C terminus of p53 regulates gene expression by multiple mechanisms in a target- and tissue-specific manner in vivo. Genes Dev. 2013;27:1868–1885. doi: 10.1101/gad.224386.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fiolka K, et al. Gfi1 and Gfi1b act equivalently in haematopoiesis, but have distinct, non-overlapping functions in inner ear development. EMBO Rep. 2006;7:326–333. doi: 10.1038/sj.embor.7400618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yucel R, Kosan C, Heyd F, Moroy T. Gfi1:green fluorescent protein knock-in mutant reveals differential expression and autoregulation of the growth factor independence 1 (Gfi1) gene during lymphocyte development. J Biol Chem. 2004;279:40906–40917. doi: 10.1074/jbc.M400808200. [DOI] [PubMed] [Google Scholar]
- 23.Abu-Farha M, et al. The tale of two domains: proteomics and genomics analysis of SMYD2, a new histone methyltransferase. Mol Cell Proteomics. 2008;7:560–572. doi: 10.1074/mcp.M700271-MCP200. [DOI] [PubMed] [Google Scholar]
- 24.D’Abramo M, et al. The p53 tetramer shows an induced-fit interaction of the C-terminal domain with the DNA-binding domain. Oncogene. 2016;35:3272–3281. doi: 10.1038/onc.2015.388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu Y, et al. p53 regulates hematopoietic stem cell quiescence. Cell Stem Cell. 2009;4:37–48. doi: 10.1016/j.stem.2008.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Du P, Tang F, Qiu Y, Dong F. GFI1 is repressed by p53 and inhibits DNA damage-induced apoptosis. PLoS One. 2013;8:e73542. doi: 10.1371/journal.pone.0073542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fraszczak J, et al. Threshold Levels of Gfi1 Maintain E2A Activity for B Cell Commitment via Repression of Id1. PLoS One. 2016;11:e0160344. doi: 10.1371/journal.pone.0160344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Khandanpour C, et al. The human GFI136N variant induces epigenetic changes at the Hoxa9 locus and accelerates K-RAS driven myeloproliferative disorder in mice. Blood. 2012;120:4006–4017. doi: 10.1182/blood-2011-02-334722. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
No datasets were generated or analyzed as part of this study.