Significance
Biological invasions are the second most severe threat affecting biodiversity worldwide with major economic and societal impacts. Yet, the multitude of species with the potential to be invasive makes it extremely difficult to anticipate invasions to develop efficient management plans. Predicting which species are likely to become invaders and where they are likely to invade even before their introduction outside their native range has always been a prime objective of invasion biology. Based on the example of ants, we provide a profiling method to predict future invaders and future invasions, and in this manner, we identify 18 ant species likely to become new invaders and regions at risk for these invasions.
Keywords: alien invasive species, ants, biological invasion, data imputation, Formicidae
Abstract
Invasive alien species are a great threat to biodiversity and human livelihoods worldwide. The most effective way to limit their impacts and costs is to prevent their introduction into new areas. Identifying invaders and invasions before their occurrence would arguably be the most efficient strategy. Here, we provide a profiling method to predict which species—with which particular ecological characteristics—will invade, and where they could invade. We illustrate our approach with ants, which are among the most detrimental invasive species, as they are responsible for declines of numerous taxa, are involved in local extinctions, disturb ecosystem functioning, and impact multiple human activities. Based on statistical profiling of 992 ant species from an extensive trait database, we identify 18 native ant species with an ecological profile that matches that of known invasive ants. Even though they are not currently described as such, these species are likely to become the next global invaders. We couple these predictions with species distribution models to identify the regions most at risk from the invasion of these species: Northern and Central America, Brazil, Central Africa and Madagascar, Southeast Asia, Papua New Guinea Northeast Australia, and many islands worldwide. This framework, applicable to any other taxa, represents a remarkable opportunity to implement timely and specifically shaped proactive management strategies against biological invasions.
Invasive alien species (IAS) are one of the main drivers of biodiversity loss and disruptors of ecosystem functioning and services (1–4). In addition to ecological impacts, IAS also have substantial impacts on the economy and human health (5). With increasing international trade and changing climates (6, 7), increasing numbers of invasions—and their subsequent impacts—are expected worldwide (8–12).
The most cost-effective means to reduce the impacts of IAS is to prevent their importation or implement rapid response treatment programs, which rely on early detection (13–15). Consequently, there is a pressing need to find robust ways to predict invasions before their occurrence to adjust surveillance or management policies and thereby reduce the likelihood of establishment, spread, and overall cost (16). Identifying future invaders before they become so has been a longstanding and previously unreachable objective of invasion biology (17, 18).
Until now, species distribution models have forecasted the most suitable areas, and hence the areas most at risk from known IAS (7, 19, 20). Other types of models have tried to identify what characterizes IAS based on the drivers of their invasion success, their life-history traits, or their occurrences worldwide (e.g., ref. 18). Such models have been built for various taxa, including plants, ants, fish, amphibians, reptiles, birds, and mammals (21–30). For ants, studies based on various databases characterize certain ecological traits often associated with exotic or invasive species (28, 31, 32). All these pioneer studies provide a valuable starting point, but they still have fundamental limitations. First, they remain limited in terms of coverage; notably, all studies based on interception lists rely on only a few countries. Second, the studies exclusively consider the few species that have complete data for all of the studied traits, thus removing the majority of species. Removing species with missing data drastically reduces the sample size and statistical power and also introduces biases and thus improper predictions (33–36). Finally, the existing models are still mostly limited to species of already known invasive status (i.e., invasive or exotic) or to providing traits associated with invasiveness; thus, existing models do not use these traits to predict new invaders yet to move to another area. Nevertheless, this corpus of pioneer studies shows that, by using modern statistical inference tools, improved ecological knowledge on current IAS, and large databases on species ecological traits, it is now possible to reach a higher level of prediction in invasion biology and identify in advance which species could become invasive. Establishing a list of future potential IAS also allows us to predict where they are likely to invade and where special detection efforts should be deployed.
Ants represent an excellent opportunity to take the existing predictive tools a step further by including more traits, more species, and worldwide coverage. Invasive ants are among the worst IAS (37). They have enormous ecological impacts on other ant species as well as on other taxa (38, 39), ranging from decreases in the abundance and richness of native species (37) to the disruption of species interactions (40, 41), invasion meltdown (41, 42), local species extinctions (42), and whole ecosystem functioning alteration (39). Invasive ants also have tremendous negative impacts on human activities, notably agriculture (43), human health (44), and infrastructure (39). The costs of their damage reach billions of dollars annually; for example, the economic costs of the red imported fire ant alone have been estimated to reach $211 million/y in Hawaii and exceed $1.65 billion/y in Australia (16). Finally, invasive ants are extremely difficult to control (45); more than 200 species are established outside their native range (46), of which 19 are recorded as invasive (47). There are no fewer than five ant species in the International Union for the Conservation of Nature (IUCN) list of the “100 of the world’s worst invasive alien species,” a feat equaled by no other taxonomic family (48). In this study, we will follow the definition of invasive species adopted by the Invasive Species Specialist Group of the IUCN, excluding species that are native but expanding, as well as exotic species with no impact.
Here, we have built a profiling tool that identifies which species have the ecological profile that makes them highly likely of being the next invading species. Our approach is to first define an ecological profile of the most invasive ant species and then compare it with the ecological profile of other species based on an extensive dataset of ant species traits, imputed through finely tuned and up-to-date machine-learning algorithms. Our quantitative risk assessment tool responds to two of the most crucial questions of invasion biology today: which species are likely to become invasive and where will they become invasive? This methodology is transposable to other taxa for which there is adequate ecological information on current invaders.
Results
Future Invaders.
Our pool of ant species retained for the ecological profiling of invasive species was kept high on account of the setup imputation procedure, which enabled us to estimate missing values in the traits and thus have all trait values, either observed or estimated, for 992 species. This was done while keeping the imputation error very low (whole dataset imputation error: 5.14%), thus ensuring robust model estimates and predictions. We used this imputed dataset of four ecological traits to calculate the probability of species being invasive and detect ant species with a profile highly similar to known ant invaders. Our models enabled us to efficiently discriminate between likely and unlikely invasive species [Area Under the receiver operating characteristic Curve (AUC) = 0.96], and it indicated relatively strong relationships between species traits and invasion potential (pseudoR2 = 0.59). Our analyses suggest that the four most important traits for predicting ant invasiveness probability are the following: supercoloniality, generalist nesting type, a frequent association with disturbances, and to a lesser extent, independent colony foundation mode (SI Appendix, Table S3).
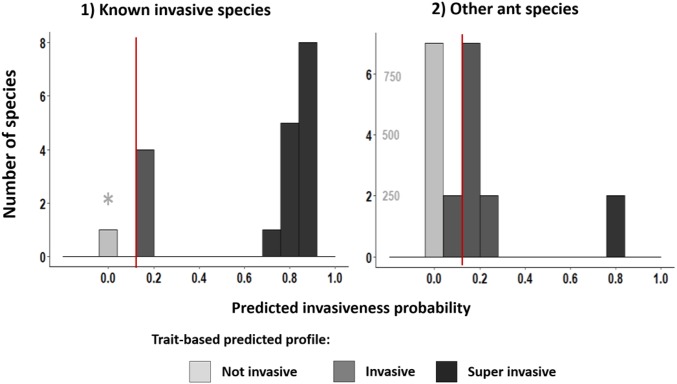
Our method also allowed us to distinguish two invasiveness profiles. Species with a probability > 0.70 (both known and predicted invasive species) are hereafter called “superinvasive.” From the 19 known invasive species, 13 have a superinvasive profile, with an invasiveness probability between 0.826 and 0.866 (mean: 0.841, Fig. 1). Five known invasive species have probabilities between 0.129 and 0.386 (mean: 0.196) and are hereafter called “invasive.” This bimodal pattern is likely linked to the colony structure of the species, as all superinvasive species are also supercolonialist while all invasive species are not. Finally, one species recorded by the IUCN as invasive, Acromyrmex octospinosus, has a significantly distinct profile, with an extremely low invasiveness probability of 0.106 × 10−4.
Fig. 1.
Predicted invasiveness probability distributions for (1) ant species recognized as invasive by IUCN and (2) ant species for which no information on invasive status exists. Dark gray bars on the right of the red lines correspond to superinvasive profiles, medium gray bars to invasive, and light gray bars to noninvasive. The asterisk (*) points to A. octospinosus, which the model shows to be wrongly classified as invasive by the IUCN. For the sake of clarity, the Right graph has two y axes: the light-gray axis corresponds to the light-gray bar of noninvasive species only.
Our model also provided us with a list of 18 species that share the trait profile of current invasive species. Among these 18 species, 2 have a superinvasive profile (invasiveness probability between 0.825 and 0.866; mean: 0.846), while 16 have an invasive profile [probabilities from 0.129 to 0.377; mean: 0.159 (Table 1)]. Note that a leave-one-out sensitivity analysis [recoding known invasive species as noninvasive (SI Appendix)] classified all known invasive species as invasive in 100% of our models, thus showing that our models were reliable for predicting invasiveness (SI Appendix, Tables S4 and S5). The remainder of the 955 species that were not identified as invasive (according to the lower fifth centile of the invasive species probability distribution) in more than 90 models of 100 were not considered to be potential future invaders. Targeted studies on some of these species may, however, be relevant to confirm that they are at risk for becoming invaders in the future.
Table 1.
Predicted invasiveness probabilities, or “invasion profiles,” of 19 invasive species from the IUCN red list (in boldface) and 18 potential future invaders identified with our model
| Species | P | ± | % |
| Superinvasive profiles | |||
| Technomyrmex difficilis | 0.87 | 0.02 | 100 |
| Lasius neglectus | 0.87 | 0.02 | 100 |
| Solenopsis geminata | 0.87 | 0.02 | 100 |
| Solenopsis invicta | 0.87 | 0.02 | 100 |
| Technomyrmex albipes | 0.87 | 0.02 | 100 |
| Trichomyrmex destructor | 0.87 | 0.02 | 100 |
| Lepisiota canescens | 0.83 | 0.01 | 100 |
| Anoplolepis gracilipes | 0.83 | 0.01 | 100 |
| Linepithema humile | 0.83 | 0.01 | 100 |
| Monomorium pharaonis | 0.83 | 0.01 | 100 |
| Myrmica rubra | 0.83 | 0.01 | 100 |
| Nylanderia pubens | 0.83 | 0.01 | 100 |
| Paratrechina longicornis | 0.83 | 0.01 | 100 |
| Tapinoma melanocephalum | 0.83 | 0.01 | 100 |
| Wasmannia auropunctata | 0.83 | 0.01 | 100 |
| Invasive profiles | |||
| Pheidole megacephala | 0.39 | 0.04 | 100 |
| Anoplolepis custodiens | 0.38 | 0.04 | 98 |
| Formica yessensis | 0.23 | 0.01 | 100 |
| Tapinoma litorale | 0.17 | 0.01 | 100 |
| Tapinoma sessile | 0.17 | 0.01 | 100 |
| Brachyponera chinensis | 0.17 | 0.01 | 100 |
| Solenopsis richteri | 0.17 | 0.01 | 100 |
| Ochetellus glaber | 0.17 | 0.16 | 100 |
| Lasius fuliginosus | 0.14 | 0.02 | 100 |
| Aphaenogaster spinosa | 0.13 | 0.01 | 100 |
| Cardiocondyla emeryi | 0.13 | 0.01 | 100 |
| Cardiocondyla minutior | 0.13 | 0.01 | 100 |
| Dolichoderus bispinosus | 0.13 | 0.01 | 100 |
| Lasius sabularum | 0.13 | 0.01 | 100 |
| Monomorium minimum | 0.13 | 0.01 | 100 |
| Neivamyrmexnigrescens | 0.13 | 0.01 | 100 |
| Neivamyrmex pilosus | 0.13 | 0.01 | 100 |
| Tetramorium bicarinatum | 0.13 | 0.01 | 100 |
| Tetramorium simillimum | 0.13 | 0.01 | 100 |
| Monomorium floricola | 0.13 | 0.01 | 100 |
| Solenopsis papuana | 0.13 | 0.01 | 100 |
| Acromyrmex octospinosus | 0.00 | 0.00 | 100 |
“P”: invasiveness probability (mean: 100 models); “±”: invasiveness variability (SD: 100 models); “%”: percentage of models. Note that the values of A. octospinosus are P = 0.0001 ± 9.07E-05.
Areas at Risk.
Species distribution models allowed us to identify regions of climatic suitability for each of the 18 (16 invasives and 2 superinvasives) likely future invaders for which we had sufficient data (SI Appendix, Fig. S11). These predictions were of high quality: True Skill Statistics (TSS) values were between 0.88 and 0.98 and AUC values were between 0.96 and 0.99, indicating good model performances.
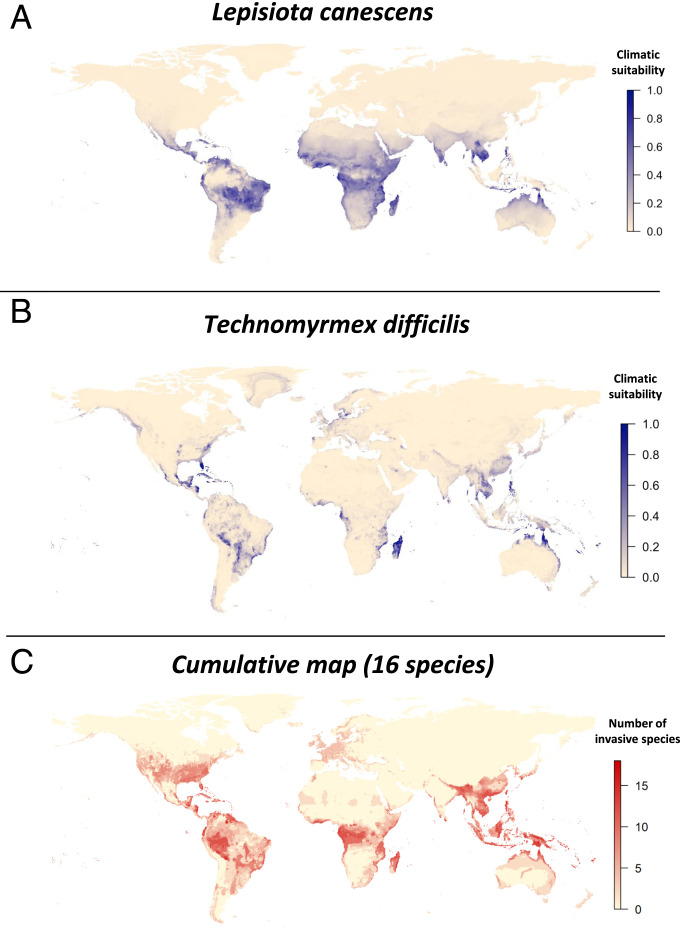
The potential invasions are more likely in tropical regions, as illustrated in the cumulated map (Fig. 2). When overlapping all potential distributions, invasion hotspots (i.e., areas with more than 15 invasive species) were predicted to occur in Northern and Central America, Brazil, Central Africa and Madagascar, Southeast Asia, Papua New Guinea, Northeast Australia, and many islands worldwide. Areas characterized by temperate and extreme climatic conditions (i.e., ice, hot desert, tundra), especially in the Palearctic and Nearctic (SI Appendix, Fig. S1) seemed less at risk from these novel ant invasive species. This was also the pattern observed for the two identified potential superinvasive species (Fig. 2), although Lepisiota canescens could also invade almost all of Africa and South East Asia, and Technomyrmex difficilis had a much more restricted potential distribution.
Fig. 2.
Global distribution of predicted invasive species for (A) the predicted superinvasive L. canescens, (B) the predicted superinvasive T. difficilis, and (C) the cumulation of the individual climatic suitability of the 11 species with sufficient data (see main text). Since the exact native range is often unknown, the projected climatic suitability includes the native range of species.
Discussion
Using a large database on ant species traits and an up-to-date phylogeny of ant genera, we developed a robust methodology to quantify the relative importance of ecological traits in shaping their invasiveness probability and used it to both characterize the ecological profile of current invasive ants and identify 18 noninvasive species that share this profile. We distinguished between invasive and highly invasive species based on their probability of invasiveness. We complemented this prediction by identifying regions most at risk for invasion from these species: tropical regions, both mainland and islands. Our predictive approach directly points to species that have not yet invaded and areas that are not yet invaded, but are likely to do so in the future.
Pioneer risk-assessment models are all based on datasets that only comprise information on exotic ant species, i.e., species that have already been moved and detected outside of their native zone (24, 29), or pest species, i.e., species that are already known to have concerning impacts (49, 50). As a result, these models cannot point to novel species of potential risk. In contrast, our screening tool is based on a large and collaborative dataset that contains information for 25 ecological traits for the 19 ant species recognized as invasive by the IUCN (47) as well as over 2,000 native species, of which 992 had all information (observed or inferred) for the traits selected in our model. The fact that this dataset comprises species from 47% of the ant family originating from all of the world’s ecozones (SI Appendix, Fig. S1) makes our predictive model genuinely compatible with the anticipation of future invasions worldwide.
Unlike most current trait-based predictive models, our methodology includes a robust and accurate way of dealing with missing values, which are commonplace in life-history trait datasets in general. Indeed, the removal of cases with missing values is known to result in reduced statistical power and biased parameter estimates (31, 33, 51). We overcame this issue by bringing together the most recent and accurate imputation algorithms (52, 53) and finely calibrating them to fit our data. We comprehensively examined the performance of tuning parameters for multiple imputation and included the most relevant number of phylogenetic eigenvectors to impute each trait. The crucial issue of how many phylogenetic eigenvectors should be included for trait imputation, which we carefully consider in the present paper, has been rarely discussed in the literature (54). Our analysis, in accordance with previous work, also revealed the existence of some patterns of phylogenetic signal above the species level (SI Appendix, Table S4), supporting the fact that our genus phylogeny was a useful predictor to include when imputing the ant species dataset (20). After imputing the dataset multiple times, the multimodel inference enabled us to establish strong relationships between species traits and invasion potential and to efficiently discriminate between likely and unlikely invasive species.
Our results highlighted several traits of importance that were previously highlighted as commonly found in invasive ant species: supercoloniality, generalist nesting type, affinity for disturbed environments, and expansion through independent colony founding (37, 55). Moreover, our study goes beyond trait analysis by using it to calculate species invasiveness probability and applying it to a large set of species not yet known to be invasive. This led us to distinguish two invasive profiles—superinvasive and invasive—and detect two potential superinvasive species (L. canescens and T. difficilis) and 16 potential invasive species (Table 1). This result seems in accordance with information available from experts who describe these two superinvasive species as having the characteristics to become future invaders and impact native ecosystems (56–58). However, the 16 other species have, to our knowledge, never been described as problematic, which highlights the proactive potential of our method. There are in fact relatively few studies available on most of them, emphasizing the need to further study and monitor them. Although they have not yet been categorized as invasive, all 18 predicted invaders should be carefully surveyed at points of entry in at-risk regions and become the target of serious monitoring programs. In this regard, our cumulated map highlights the mismatch between the large regions most at risk and those most likely to put proactive measures into place (59).
The fact that one of the known invasive species, A. octospinosus, showed a radically distinct profile from the other invasive species illustrates an additional strength of our methodology; in addition to detecting new invaders, it can also question the established invasive status of certain species. Despite being on the IUCN list of invasive ants (47), A. octospinosus is characterized as invasive by only one scientific paper (60), yet without providing evidence. This leaf-cutting ant species has the potential to destroy agricultural crops and has been observed to damage the fern Cyathea arborea on one island (Guadeloupe) in its native ecozone. However, its traits (strict monogyny, highly aggressive attitude, independent colony foundation, strict mutualist relationship with fungus species, and long generation time), its slow spread in Guadeloupe (0.51 km/y) (60), its limited ability to be transported inadvertently by humans (55), and the climatic and geographic proximity between its native zone and the only island where it has been introduced seem to demonstrate that this species is not invasive, but rather an agricultural pest species that is still currently filling its suitable niche. This species is, however, known to have major impacts on agriculture (32), which may explain why it was proposed and accepted for the IUCN list of invasive species. We suggest that, in the absence of further evidence, its presence on the IUCN list should be questioned, as it does not correspond to the ecological definition of an invasive alien species but rather to that of an agricultural pest. We nevertheless recognize that ant species with a high potential to become invasive may exist with a profile very different from known invasive species. These potential invaders would be detected neither by our model nor by any trait-based model calibrated with invasive species characteristics, which illustrates the complex and idiosyncratic nature of biological invasions.
Our species distribution models allowed us to map the world regions with the most suitable climatic conditions for the two predicted superinvasive species and 16 predicted invasive species. These maps showed that future invasions by these predicted invaders are more likely in tropical regions, most of which are at risk for a large number of these species. Tropical islands, already invaded or at risk for invasion by many of the 19 current invasive species, are also generally at high risk of invasion by these potential invaders, emphasizing the importance of biosecurity in these biodiversity-rich ecosystems. It is worth mentioning here that, although occurrence data used for Species Distribution Models (SDMs) were thoroughly checked for issues typical of such data (SI Appendix, Supplementary Material and Methods), some spatiotemporal bias due to the lack of knowledge of the sampling effort remains unavoidable (61). Therefore, the resulting distribution maps should, as usual, be considered with caution.
Our list of predicted invaders crossed with climatic niche modeling offers a worldwide risk assessment of the areas that are the most at risk and informs the relevant countries about the necessity to pay attention to the introduction of ants to prevent invasions. This should be viewed together with the corresponding distribution studies of the already established invasive ant species (e.g., ref. 7). Although climatic suitability is a prerequisite for a successful invasion, the biotic and abiotic characteristics of the receiving community also condition the species establishment and spread and may modulate the local environmental suitability of these species (23). Climate and land use changes may also slightly alter the predictions and create new opportunities for these species to spread in temperate regions and at higher latitudes, as observed for many invasive species (19, 62). A promising future step would be to predict the potential impacts of species predicted to be likely future invaders (63, 64).
Our results are based on the definition of ecological traits. For example, one could argue that using the trait “supercolonial” in our model may hinder its ability to detect new potential invaders, since this behavior has mostly been observed in ant colonies after they have become invasive. However, supercoloniality has already been outlined as an important characteristic of invasive species (37, 65). It might be worth, in this context, completing the Antprofiler database (that currently contains about one-sixth of the known ant species), as having more data is the first step to obtaining more (and more accurate) identifications. A complementary and relevant study would be to analyze which traits condition each step of the invasion process (introduction, establishment, spread, and impact), thus providing some information on which invasion stage is the most critical in order to design a targeted control strategy for each species according to its specific combination of traits (30).
Invasive ant populations, once established in a climatically suitable area, are extremely difficult to control or eradicate (45). A range of control measures exists (e.g., use of chemical insecticides incorporated into baits, biocontrol), but the majority have low efficiency, high negative impacts on native taxa and ecosystems (39), or high costs (13). Overall, our tool can greatly reduce border detection efforts and costs by precisely informing management strategies and preventing some of the potential impacts of invasive ants through the implementation of proactive ant detection programs. Preventing biological invasions is generally believed to be more cost-effective and efficient than the eradication of an established population, long-term control, and repairing the damage caused by invasions (3). This study provides a methodology to predict potential invaders and likely regions of invasions, and we recommend extending and transposing it to assess the invasion potential and areas at risk for other taxa with existing trait databases such as plants, fish, and terrestrial vertebrates.
Materials and Methods
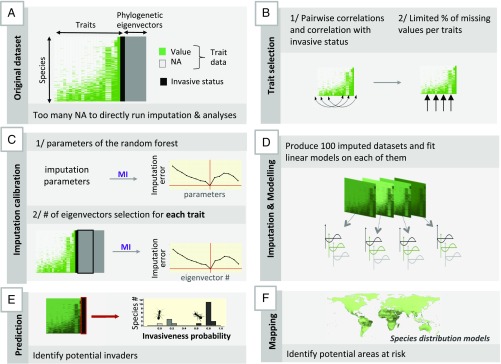
To simultaneously identify the most relevant traits and keep the highest number of species, we proceeded in two steps. First, we identified and selected traits that performed well at predicting invasiveness. Second, we optimized our usable dataset by removing species with too many missing values and by estimating the values of the selected traits for the species with few missing values (using all available traits). We then used the dataset with the selected traits, either observed or estimated, to predict invasiveness probability with Generalized Linear Models. Finally, we predicted areas at risk for invasion of species identified as potential invaders, using Species Distribution Models. Our step-by-step process is illustrated Fig. 3 and is presented below, with further details in SI Appendix.
Fig. 3.
Diagram summarizing our approach with (A) the initial trait dataset (invasive status is only known for species on IUCN list); (B) trait selection according to different criteria; (C) imputation calibration to maintain the error below a given threshold; (D) multiple imputation (using the best parameters identified in B) and modeling of the imputed datasets; (E) use of the model to predict any species invasiveness probability based on its trait data; and (F) mapping of potential areas at risk using species distribution models.
Trait Data Selection and Imputation.
We searched for trait combinations that are associated with invasive species. We extracted ant species traits from our AntProfiler online database (https://antprofiler.ese.u-psud.fr). This database includes information on 2,176 ant species (134 genera) and ecological traits related to their occurrence, morphology, behavior, and invasive status (66). From this database, we selected traits noncorrelated to each other but correlated to the invasive status. The full description of the trait selection process is given in SI Appendix, Supplementary Material and Methods and illustrated in SI Appendix, Fig. S3.
Despite our data collection efforts, the database contained many missing values. We therefore used an imputation technique (missForest) to estimate the missing values (53). Before imputation, we removed the species with too many missing values to keep a dataset with a maximum of ∼60% of missing information (following ref. 52). We then explored how missing data could be imputed into our database to minimize bias and maximize prediction accuracy. To improve the estimation of missing values, we used phylogenetic information and all traits initially downloaded from Antprofiler. We used out-of-bag error estimations to calibrate the models as these were shown to accurately estimate imputation error, and we repeated the imputation 100 times (Fig. 3 and SI Appendix, Supplementary Material and Methods).
We selected a final subset of four traits: colony foundation, colony structure, nesting type, and association with disturbed areas and 992 species (of 2,176).
Predictive Model Building.
Potential future invasive species.
We modeled the invasive profile using generalized linear models with our four selected traits as predictors and a binomial distribution. We ran 100 models for each of the previously imputed datasets and used model predictions to determine future invasive species. For each model, we identified potentially new invasive ant species as those with a predicted invasiveness probability above the lower fifth centile of the 19 known invasive species probability distribution. Each identified species was considered to be potentially invasive if it was selected in at least 90 of the 100 models.
Areas at risk.
SDMs are based on correlations between environmental variables and geolocalized species records and can be used to delineate potential species distributions (67). We built SDMs for the ant species found to have similar profiles to those of known invasive species to identify the areas that present suitable environmental conditions for these species and thus areas at risk for invasion. A full description of the SDM process is given in SI Appendix, Supplementary Material and Methods. Two metrics were used to evaluate SDM accuracy: the TSS and the AUC.
Ensemble models—i.e., averages of all models used, weighted by the TSS—were run for each of the predicted invaders with sufficient occurrence points to produce individual climatic suitability distribution maps (SI Appendix, Fig. S11). It is noteworthy that two species (Formica yessensis and Aphaenogaster spinosa) had very few occurrence points (24 and 33, respectively), meaning that the resulting distribution maps should be taken with additional caution. Finally, we also combined these individual predictions by summing each species potential distribution as binary maps to obtain a cumulative invasion risk map from these future invaders. Binary transformation was based on the threshold that maximized the TSS for each species.
Supplementary Material
Acknowledgments
We thank Cleo Bertelsmeier and Sebastien Ollier with whom we worked on different statistical approaches for a preliminary study and who provided useful comments on an earlier version of the manuscript; five anonymous reviewers who provided useful comments that led to improved analyses; Guillaume Guénard and Daniel J. Stekhoven for their advice, which helped improve our imputation methodology; Benoît Guénard and James Wetterer for sharing their ant occurrence data; and Corrie Moreau and Philip Ward who provided information and data on ant phylogeny.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1803456116/-/DCSupplemental.
Change History
July 26, 2021: The text of this article, Figs. 1 and 2, and the SI file have been updated; please see accompanying Correction for details.
References
- 1.Vilà M, et al. How well do we understand the impacts of alien species on ecosystem services? A pan-European, cross-taxa assessment. Front Ecol Environ. 2010;8:135–144. [Google Scholar]
- 2.Bellard C, Cassey P, Blackburn TM. Alien species as a driver of recent extinctions. Biol Lett. 2016;12:20150623. doi: 10.1098/rsbl.2015.0623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Simberloff D, et al. Impacts of biological invasions: What’s what and the way forward. Trends Ecol Evol. 2013;28:58–66. doi: 10.1016/j.tree.2012.07.013. [DOI] [PubMed] [Google Scholar]
- 4.Clavero M, García-Berthou E. Invasive species are a leading cause of animal extinctions. Trends Ecol Evol. 2005;20:110. doi: 10.1016/j.tree.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 5.Bradshaw CJA, et al. Massive yet grossly underestimated global costs of invasive insects. Nat Commun. 2016;7:12986. doi: 10.1038/ncomms12986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bertelsmeier C, Ollier S, Liebhold A, Keller L. Recent human history governs global ant invasion dynamics. Nat Ecol Evol. 2017;1:0184. doi: 10.1038/s41559-017-0184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bertelsmeier C, Luque GM, Hoffmann BD, Courchamp F. Worldwide ant invasions under climate change. Biodivers Conserv. 2015;24:117–128. [Google Scholar]
- 8.Seebens H, et al. Global trade will accelerate plant invasions in emerging economies under climate change. Glob Change Biol. 2015;21:4128–4140. doi: 10.1111/gcb.13021. [DOI] [PubMed] [Google Scholar]
- 9.Seebens H, et al. No saturation in the accumulation of alien species worldwide. Nat Commun. 2017;8:14435. doi: 10.1038/ncomms14435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Capinha C, Essl F, Seebens H, Moser D, Pereira HM. The dispersal of alien species redefines biogeography in the Anthropocene. Science. 2015;348:1248–1251. doi: 10.1126/science.aaa8913. [DOI] [PubMed] [Google Scholar]
- 11.Butchart SH, et al. Global biodiversity: Indicators of recent declines. Science. 2010;328:1164–1168. doi: 10.1126/science.1187512. [DOI] [PubMed] [Google Scholar]
- 12.Walther GR, et al. Alien species in a warmer world: Risks and opportunities. Trends Ecol Evol. 2009;24:686–693. doi: 10.1016/j.tree.2009.06.008. [DOI] [PubMed] [Google Scholar]
- 13.Kaiser BA, Burnett KM. Spatial economic analysis of early detection and rapid response strategies for an invasive species. Resour Energy Econ. 2010;32:566–585. [Google Scholar]
- 14.Keller RP, Lodge DM, Finnoff DC. Risk assessment for invasive species produces net bioeconomic benefits. Proc Natl Acad Sci USA. 2007;104:203–207. doi: 10.1073/pnas.0605787104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Finnoff D, Shogren JF, Leung B, Lodge D. Take a risk: Preferring prevention over control of biological invaders. Ecol Econ. 2007;62:216–222. [Google Scholar]
- 16.Wylie FR, Janssen-May S. Red imported fire ant in Australia: What if we lose the war? Ecol Manage Restor. 2017;18:32–44. [Google Scholar]
- 17.Ward DF. Modelling the potential geographic distribution of invasive ant species in New Zealand. Biol Invasions. 2007;9:723–735. [Google Scholar]
- 18.Kolar CS, Lodge DM. Progress in invasion biology: Predicting invaders. Trends Ecol Evol. 2001;16:199–204. doi: 10.1016/s0169-5347(01)02101-2. [DOI] [PubMed] [Google Scholar]
- 19.Bellard C, et al. Will climate change promote future invasions? Glob Change Biol. 2013;19:3740–3748. doi: 10.1111/gcb.12344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bomford M, Kraus F, Barry SC, Lawrence E. Predicting establishment success for alien reptiles and amphibians: A role for climate matching. Biol Invasions. 2009;11:713–724. [Google Scholar]
- 21.Sol D, et al. Unraveling the life history of successful invaders. Science. 2012;337:580–583. doi: 10.1126/science.1221523. [DOI] [PubMed] [Google Scholar]
- 22.Allen WL, Street SE, Capellini I, Letters E. Fast life history traits promote invasion success in amphibians and reptiles. Ecol Lett. 2017;20:222–230. doi: 10.1111/ele.12728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Capellini I, Baker J, Allen WL, Street SE, Venditti C. The role of life history traits in mammalian invasion success. Ecol Lett. 2015;18:1099–1107. doi: 10.1111/ele.12493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Suarez AV, Holway DA, Ward PS. The role of opportunity in the unintentional introduction of nonnative ants. Proc Natl Acad Sci USA. 2005;102:17032–17035. doi: 10.1073/pnas.0506119102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jeschke JM, Strayer DL. Determinants of vertebrate invasion success in Europe and North America. Glob Change Biol. 2006;12:1608–1619. [Google Scholar]
- 26.Fujisaki I, et al. Risk assessment of potential invasiveness of exotic reptiles imported to South Florida. Biol Invasions. 2010;12:2585–2596. [Google Scholar]
- 27.Goodwin BJ, Allister AJMC, Fahrig L. Predicting invasiveness of plant species based on biological information. Conserv Biol. 1999;13:422–426. [Google Scholar]
- 28.Lloret F, et al. Species attributes and invasion success by alien plants on Mediterranean islands. J Ecol. 2014;93:512–520. [Google Scholar]
- 29.Lester PJ. Determinants for the successful establishment of exotic ants in New Zealand. Divers Distrib. 2005;11:279–288. [Google Scholar]
- 30.Marchetti MP, Moyle PB, Levine R. Invasive species profiling? Exploring the characteristics of non-native fishes across invasion stages in California. Freshw Biol. 2004;49:646–661. [Google Scholar]
- 31.Nakagawa S, Freckleton RP. Model averaging, missing data and multiple imputation: A case study for behavioural ecology. Behav Ecol Sociobiol. 2011;65:103–116. [Google Scholar]
- 32.Wittenborn D, Jeschke JM. Characteristics of exotic ants in North America. NeoBiota. 2011;10:47–64. [Google Scholar]
- 33.Nakagawa S, Freckleton RP. Missing inaction: The dangers of ignoring missing data. Trends Ecol Evol. 2008;23:592–596. doi: 10.1016/j.tree.2008.06.014. [DOI] [PubMed] [Google Scholar]
- 34.Veron S, et al. Integrating data-deficient species in analyses of evolutionary history loss. Ecol Evol. 2016;6:8502–8514. doi: 10.1002/ece3.2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hadfield JD, Nakagawa S. General quantitative genetic methods for comparative biology: Phylogenies, taxonomies and multi-trait models for continuous and categorical characters. J Evol Biol. 2010;23:494–508. doi: 10.1111/j.1420-9101.2009.01915.x. [DOI] [PubMed] [Google Scholar]
- 36.Lach L, Hooper-Bui LM. Consequences of ant invasions. In: Lach L, Parr CL, Abbott KL, editors. Ant Ecology. Oxford Univ Press; Oxford: 2010. pp. 261–286. [Google Scholar]
- 37.Holway DA, Lach L, Suarez AV, Tsutsui ND, Case TJ. The causes and consequences of ant invasions. Annu Rev Ecol Syst. 2002;33:181–233. [Google Scholar]
- 38.Lach L, Thomas ML. Invasive ants in Australia: Documented and potential ecological consequences. Aust J Entomol. 2008;47:275–288. [Google Scholar]
- 39.Rabitsch W. The hitchhiker’s guide to alien ant invasions. BioControl. 2011;56:551–572. [Google Scholar]
- 40.Lach L. Floral visitation patterns of two invasive ant species and their effects on other hymenopteran visitors. Ecol Entomol. 2008;33:155–160. [Google Scholar]
- 41.Green PT, et al. Invasional meltdown: Invader-invader mutualism facilitates a secondary invasion. Ecology. 2011;92:1758–1768. doi: 10.1890/11-0050.1. [DOI] [PubMed] [Google Scholar]
- 42.O’Dowd DJ, Green PT, Lake PS. Invasional “meltdown” on an oceanic island. Ecol Lett. 2003;6:812–817. [Google Scholar]
- 43.Addison P, Samways MJ. Surrogate habitats demonstrate the invasion potential of the African pugnacious ant. Biodivers Conserv. 2006;15:411–428. [Google Scholar]
- 44.Nelder MP, Paysen ES, Zungoli PA, Benson EP. Emergence of the introduced ant Pachycondyla chinensis (Formicidae: Ponerinae) as a public health threat in the southeastern United States. J Med Entomol. 2006;43:1094–1098. doi: 10.1603/0022-2585(2006)43[1094:eotiap]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 45.Hoffmann BD, Luque GM, Bellard CC, Holmes ND, Donlan CJ. Improving invasive ant eradication as a conservation tool: A review. Biol Conserv. 2016;198:37–49. [Google Scholar]
- 46.Suarez AV, McGlynn TP, Tsutsui ND. Biogeographic and taxonomic patterns of introduced ants. In: Lach L, Parr CL, Abbott KL, editors. Ant Ecology. Oxford Univ Press; Oxford: 2010. pp. 233–244. [Google Scholar]
- 47.ISSG 2017. Global Invasive Species Database. Available at http://www.iucngisd.org/gisd/. Accessed March 15, 2019.
- 48.Lowe S, Browne M, Boudjelas S, De Poorter M. 2000. 100 of the World’s Worst Invasive Alien Species: A selection from the Global Invasive Species Database. Available at https://www.iucn.org/content/100-worlds-worst-invasive-alien-species-a-selection-global-invasive-species-database. Accessed March 15, 2019.
- 49.Worner SP, Gevrey M. Modelling global insect pest species assemblages to determine risk of invasion. J Appl Ecol. 2006;43:858–867. [Google Scholar]
- 50.Paini DR, Worner SP, Cook DC, De Barro PJ, Thomas MB. Threat of invasive pests from within national borders. Nat Commun. 2010;1:115–116. doi: 10.1038/ncomms1118. [DOI] [PubMed] [Google Scholar]
- 51.Ellington EH, et al. Using multiple imputation to estimate missing data in meta-regression. Methods Ecol Evol. 2015;6:153–163. [Google Scholar]
- 52.Penone C, et al. Imputation of missing data in life-history trait datasets: Which approach performs the best? Methods Ecol Evol. 2014;5:1–10. [Google Scholar]
- 53.Stekhoven DJ, Bühlmann P. MissForest: Non-parametric missing value imputation for mixed-type data. Bioinformatics. 2012;28:112–118. doi: 10.1093/bioinformatics/btr597. [DOI] [PubMed] [Google Scholar]
- 54.Diniz-Filho JAF, et al. On the selection of phylogenetic eigenvectors for ecological analyses. Ecography. 2012;35:239–249. [Google Scholar]
- 55.McGlynn TP. The worldwide transfer of ants: Geographical distribution and ecological invasions. J Biogeogr. 1999;26:535–548. [Google Scholar]
- 56.Wetterer JK. Worldwide spread of the difficult white-footed ant, Technomyrmex difficilis (Hymenoptera : Formicidae) Myrmecol News. 2013;18:93–97. [Google Scholar]
- 57.Wetterer JK. Technomyrmex difficilis (Hymenoptera: Formicidae) in the West Indies. Fla Entomol. 2008;91:428–430. [Google Scholar]
- 58.Sorger DM, Booth W, Wassie Eshete A, Lowman M, Moffett MW. Outnumbered : A new dominant ant species with genetically diverse supercolonies in Ethiopia. Insectes Soc. 2016;64:141–147. [Google Scholar]
- 59.Early R, et al. Global threats from invasive alien species in the twenty-first century and national response capacities. Nat Commun. 2016;7:12485. doi: 10.1038/ncomms12485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mikheyev AS. History, genetics and pathology of a leaf-cutting ant introduction: A case study of the Guadeloupe invasion. Biol Invasions. 2008;10:467–473. [Google Scholar]
- 61.Syfert MM, Smith MJ, Coomes DA. The effects of sampling bias and model complexity on the predictive performance of MaxEnt species distribution models. PLoS One. 2013;8:e55158. doi: 10.1371/journal.pone.0055158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bellard C, Leroy B, Thuiller W, Rysman J-F, Courchamp F. Major drivers of invasion risks throughout the world. Ecosphere. 2016;7:1–14. [Google Scholar]
- 63.Bacher S, et al. Socio-economic impact classification of alien taxa (SEICAT) Methods Ecol Evol. 2018;9:159–168. [Google Scholar]
- 64.Blackburn TM, et al. A proposed unified framework for biological invasions. Trends Ecol Evol. 2011;26:333–339. doi: 10.1016/j.tree.2011.03.023. [DOI] [PubMed] [Google Scholar]
- 65.Vogel V, et al. The worldwide expansion of the Argentine ant. Divers Distrib. 2010;16:170–186. [Google Scholar]
- 66.Bertelsmeier C, Luque GM, Confais A, Courchamp F. Ant Profiler: A database of ecological characteristics of ants (Hymenoptera: Formicidae) Myrmecol News. 2013;18:73–76. [Google Scholar]
- 67.Araújo MB, Peterson AT. Uses and misuses of bioclimatic envelope modeling. Ecology. 2012;93:1527–1539. doi: 10.1890/11-1930.1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.