Significance
STING is a key driver of monogenic interferonopathies that include STING-associated vasculopathy with onset in infancy (SAVI). Defining the molecular mechanisms and the contribution of individual cell types to disease could unveil new therapeutic targets for management of SAVI and related autoimmune diseases. Here we compare and contrast murine models representative of the two most common mutations and identify a hierarchy of lymphoid abnormalities that depend on T cell-intrinsic expression of the SAVI mutation. Although clinical manifestations result from hyperactivation of STING, they occur independently of type I interferon and necroptosis. These murine models facilitate a dissection of the role of STING in specific tissues and provide tools for evaluating STING inhibitors for the treatment of SAVI patients.
Keywords: STING, SAVI, type I interferonopathies, T cells, cell death
Abstract
Studies over the past decade have revealed a central role for innate immune sensors in autoimmune and autoinflammatory diseases. cGAS, a cytosolic DNA sensor, detects both foreign and host DNA and generates a second-messenger cGAMP, which in turn binds and activates stimulator of IFN genes (STING), leading to induction of type I interferons and inflammatory cytokines. Recently, gain-of-function mutations in STING have been identified in patients with STING-associated vasculopathy with onset in infancy (SAVI). SAVI patients present with early-onset systemic inflammation and interstitial lung disease, resulting in pulmonary fibrosis and respiratory failure. Here, we describe two independent SAVI mouse models, harboring the two most common mutations found in patients. A direct comparison of these strains reveals a hierarchy of immune abnormalities, lung inflammation and fibrosis, which do not depend on either IFN-α/β receptor signaling or mixed lineage kinase domain-like pseudokinase (MLKL)-dependent necroptotic cell death pathways. Furthermore, radiation chimera experiments reveal how bone marrow from the V154M mutant mice transfer disease to the WT host, whereas the N153S does not, indicating mutation-specific disease outcomes. Moreover, using radiation chimeras we find that T cell lymphopenia depends on T cell-intrinsic expression of the SAVI mutation. Collectively, these mutant mice recapitulate many of the disease features seen in SAVI patients and highlight mutation-specific functions of STING that shed light on the heterogeneity observed in SAVI patients.
Nucleic acids are readily detected by nucleic acid sensors that survey cells for signs of infection or tissue damage. Engagement of a diverse collection of RNA and DNA sensors trigger host-defense responses to curb pathogen replication and initiate beneficial repair responses to tissue injury. The DNA sensor cGAS (cGMP-AMP synthase) is a nucleotidyl transferase that detects double-stranded DNA and generates a novel second-messenger 2′–5′cGMP-AMP (cGAMP). cGAMP binds stimulator of IFN genes (STING), causing its dimerization leading to activation of TBK1/IRF3 and IKK/NF-κB, pathways resulting in the induction of type I IFNs and proinflammatory cytokines, respectively (1). DNases outside cells (DNase I), within the phagolysosomal compartment (DNase II) or cytosol (DNase III/Trex1), ensure that in healthy individuals self nucleic acids do not trigger DNA sensors (2). Inappropriate clearance of DNA and its subsequent detection by DNA sensors underlies the pathogenesis of debilitating human diseases, such as Aicardi-Goutiéres syndrome (3). A subset of Aicardi-Goutiéres syndrome patients have mutations in Trex1, a 3′–5′ exonuclease that degrades DNA, which accumulates from endogenous retroelements (4, 5).
Gain-of-function (GOF) mutations in components of RNA and DNA sensing pathways can also result in severe autoinflammatory and autoimmune diseases. Examples include the GOF mutations in DDX58 (RIG-I) and IFIH1 (MDA5) that lead to Singleton Merten syndrome (6–8). In addition, over 25 patients with a spectrum of GOF mutations in STING have been linked to an autoinflammatory disease called SAVI (STING-associated vasculopathy with onset in infancy) (9–13). These mutations lead to spontaneous dimerization and activation of STING in SAVI patients who exhibit early-onset systemic inflammation, severe skin vasculopathy, and interstitial lung disease, resulting in pulmonary fibrosis and respiratory failure. Despite the severe phenotypes of many SAVI patients, the same mutation in different individuals can have very different clinical manifestations (13–15). Thus, a better understanding of how, when, and where overactivation of STING causes sterile inflammation is clearly needed and animal models have been developed by investigators to explore these questions (16, 17). Unexpectedly, murine models of the two most common SAVI mutations appear to reflect different aspects of the human disease: the N153S mutant mice purportedly develop skin disease while the V154M mice were found to develop variable levels of lung inflammation. Given that these two models were generated by independent laboratories, the extent to which these differences reflect mutation-specific outcomes, off-target genetic differences, the maintenance of one line on antibiotics, or other colony specific factors is presently unclear.
To better understand these differences, we have independently generated mice expressing the two most common SAVI mutations (N153S and V154M) using CRISPR/Cas9 genome editing. Our analysis revealed that these mutations have significantly different outcomes in vivo, as the V154M mutation has more robust STING activity and leads to more severe disease phenotypes. For example, only the V154M mice developed lung fibrosis and exhibit more profound immune cell abnormalities. We have also explored pathways downstream of STING that could contribute to SAVI-associated pathology and surprisingly found minimal contribution of IFNα/βR signaling or mixed lineage kinase domain-like pseudokinase (MLKL)-dependent necroptosis. Finally, through the use of radiation chimeras, we further define inherent defects in T cell development that were much more severe in the V154M mice. Overall, our studies suggest that the variable disease outcome of the SAVI patients may reflect mutation-specific effects on STING activity and function.
Results
Reduced Survival and Spontaneous Disease in N153S and V154M SAVI Mice.
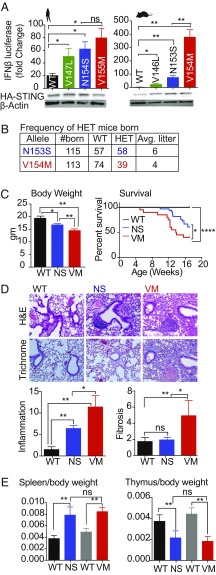
We first compared the GOF STING mutations found in patients and their murine orthologs for their ability to induce IFN promoter-driven reporter gene expression. 293T cells, which lack endogenous STING, were transfected with WT human STING (the V147L, N154S, and V155M SAVI alleles) and their murine counterparts (WT, V146L, N153S, V154M), together with an IFN-β luciferase reporter gene, and luciferase activity was measured. Compared with WT STING, the human and murine STING mutants elicited more robust reporter gene induction. Interestingly, the V154M mutant was more active than the other murine SAVI alleles and induced fourfold greater levels of reporter than the N153S mutant. STING protein levels were comparable for all of the alleles in this transient ectopic expression system (Fig. 1A). Although the human SAVI alleles induced comparable IFN-β reporter gene expression, we tested NF-kB reporter gene activation as a further readout of STING activation and observed that there was a statistically significant increase in activation of a multimerized NF-κB reporter gene with the V155M mutation compared with the N154S mutation (SI Appendix, Fig. S1A).
Fig. 1.
Reduced survival and spontaneous disease in N153S and V154M SAVI heterozygous mice. (A) 293T cells were transfected with 20 ng of plasmid encoding either human WT STING or mutant STING variants (Left) or the mouse version of WT or mutant STING variants (Right) along with an IFN-β reporter gene. Gene activation was measured by luciferase assay and fold-induction was calculated over cells transfected with IFN-β reporter only. Lysates from the same cells were probed with anti-HA for HA-tagged STING and anti–β-actin as loading control (Lower). (B) HET mutant males were bred to WT females; offspring were genotyped and litter sizes were monitored. (C) The body weight of 4- to 6-wk-old males was determined and the survival of heterozygous mice was monitored with n = 40 mice per group. (D) H&E- and trichrome-stained lungs were assessed for inflammation and fibrosis respectively, n = 5 mice per genotype were used. Representative images are shown at 20× resolution. (E) Spleen-to-body weight and thymus-to-body weight ratios were calculated for n = 8 mice per genotype at 4–6 wk of age. Black bar represents littermate control for N153S (NS) HET mice and gray bar is the littermate control for V154M (VM) HET mice. *P < 0.05; **P < 0.01; ****P < 0.0001; ns, not significant.
Because N154S and V155M are the most common SAVI mutations identified in patients, we used CRISPR/Cas9 genome editing to introduce the corresponding N153S or V154M alleles into mice. Sanger sequencing confirmed the expected base substitutions in heterozygous mice (SI Appendix, Fig. S1 B and C). Crosses of heterozygous breeders did not yield homozygous SAVI mice as of embryonic day (E) 14 for either genotype. In general, the female heterozygous mice were poor breeders, and when pregnant, frequently had problems delivering pups. Therefore, we routinely bred SAVI heterozygous male mice to WT females to generate heterozygous experimental mice and WT littermate controls. The heterozygous mice were born at Mendelian frequencies for the N153S mutant, however, the V154M mutant offspring were born at less than the expected Mendelian frequency (Fig. 1B), indicating a significant difference between the two strains. One copy of either the N153S or V154M mutant gene resulted in reduced body weights compared with the WT littermates (SI Appendix, Fig. S1A), and the V154M mice were significantly smaller than the N153S mice (Fig. 1C). Both mutants also had shorter lifespans than their WT littermate controls, with the V154M mutants succumbing sooner over a 4-mo time course compared with the N153S strain (Fig. 1C). We did not observe any gender bias throughout our analyses; both males and females were affected similarly.
To better understand the cause of death, tissues from these mice were examined histologically. The lung was the most severly impacted organ in SAVI mice with moderate inflammation also seen in the liver (SI Appendix, Table S1). Lung disease is a common feature of SAVI in humans and both mutant strains developed lung inflammation, but only the V154M mice developed lung fibrosis as detected with a trichrome stain (Fig. 1D). As predicted by the 293T studies, STING protein levels in the WT, N153S, and V154M lungs were similar for both alleles (SI Appendix, Fig. S1D). Both the mutants had enlarged spleens (Fig. 1E and SI Appendix, Fig. S1A) and smaller thymus size by 4–6 wk of age (Fig. 1E). Taken together these findings indicate that the murine SAVI alleles spontaneously developed disease manifestations present in SAVI patients and that the V154M mutant developed more severe disease than the N153S strain.
SAVI Mutant Mice Have Elevated Type I IFN and Inflammatory Gene Signatures.
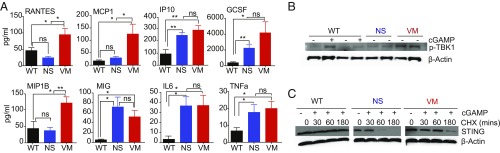
Like patients with other type I interferonopathies, SAVI patients exhibit a robust IFN gene signature and have increased levels of IFN-inducible genes in their peripheral blood (10). To determine whether the SAVI mutant mice also up-regulate genes associated with an IFN signature and inflammation, sera from 16-wk-old N153S and V154M mice were screened for cytokine levels by multiplex ELISA. We found that several chemokines and cytokines, such as IP10, MIG, and GCSF were elevated in the sera of SAVI mice compared with their WT littermate controls. Strikingly, cytokines such as RANTES, MCP1, and MIP1b were induced at higher levels in the V154M mutant compared with the N153S mutant (Fig. 2A). We also defined the gene signature of SAVI mice using a custom Nanostring probe set. Spleens from WT and SAVI mutant mice were isolated and the RNA levels of ∼100 immune response genes was measured. The SAVI gene signature was compared with the spleen of WT mice injected with the STING agonist DMXAA, an anticancer drug that induces a robust STING-dependent IFN and inflammatory gene signature in mice (18). While DMXAA elicited robust increases in IFN-stimulated genes (ISGs) and other immune genes, there was only a modest increase in expression of this gene signature in the SAVI mutants (SI Appendix, Fig. S2A). This was consistent with the findings of other investigators who also observed a minimal ISG signature as a result of SAVI mutations (16, 17). This result was surprising because murine models of two additional STING-dependent type I interferonopathies, TREX1-deficient mice (5) or DNase II-deficient mice (19), had robust ISG signatures.
Fig. 2.
SAVI mutant mice express IFN and inflammatory gene signature. (A) Sera was collected from 16-wk-old SAVI mice and serum cytokines were measured using multiplex assays with n = 6 mice per group. (B) BMDM from 4- to 6-wk-old mice were either untransfected or transfected with 2 μg/mL cGAMP for 30 min and lysates were then probed for pTBK1 and actin as loading controls. (C) BMDMs were either untreated or treated with 20 μg/mL of cycloheximide for 30 min, 60 min, or 180 min and subsequently treated with or without cGAMP for 30 min and probed for STING and actin as loading control. WT+cGAMP-treated cells were used as positive control. *P < 0.05; **P < 0.01; ns, not significant.
To evaluate the IFN signature more thoroughly, we analyzed total bone marrow (BM) from the mutant lines. We detected an increase in ISGs and other immune genes in both strains and found that the BM of V154M mutant showed higher expression of some genes compared with the N153S mutant (SI Appendix, Fig. S2B). The levels of Ifit2, Il-4, Ifi205, Icam1, nlrc4, mmp2, socs1, Irf7, Cxcl9, ccl5, IL18, and Ccl4 were all higher in the V154M BM than the N153S. To compare specific cell populations derived from the two mutants, we made BM-derived macrophages (BMDMs) and analyzed them for the activation of the STING pathway. We observed that TBK1 was phosphorylated in WT BMDMs in a cGAMP-dependent manner, whereas the mutants expressed phosphorylated TBK1 even in the absence of ligand, indicating that the STING pathway was constitutively activated in the SAVI mutant cells. Interestingly, the V154M mutant showed increased pTBK1 than the N153S mutant (Fig. 2B). Similar observations were made when the levels of IkBa phosphorylation were measured as a readout for NF-kB activity. We found more pIkBa in V154M mutant cells than in N153S cells (SI Appendix, Fig. S2C).
We also evaluated the stability of the mutant STING proteins by treating cells with cycloheximide to block new protein synthesis. We observed that the V154M mutant and N153S mutant expressed comparable levels of STING (at 0 time point); however, the V154M mutant STING protein expression was prolonged at later time points (60 and 180 min) compared with the N153S mutant in cycloheximide-treated cells (Fig. 2C). Because STING is known to be degraded through autophagy via lysosomal degradation (20), we hypothesized that enhancing the stability of STING would result in stronger STING signaling. We compared the levels of STING in cGAMP-activated WT cells coincubated or not with bafilomycin A1, an inhibitor of lysosomal acidification. STING levels decreased over time in WT cells but this effect was blocked by bafilomycin (SI Appendix, Fig. S2D). We next determined the effect of bafilomycin on the level of pIkBa following cGAMP activation. Bafilomcyin also increased phospho-IkBa levels in cGAMP-treated WT cells, as well as in SAVI mutant cells (SI Appendix, Fig. S2E). Taken together, these observations indicate that the V154M mutant is more stable than the N153S mutant, which likely accounts for the enhanced signaling and inflammation observed in the V154M mice.
V154M Mutant Mice Develop More Severe Immune Cell Alterations than N153S Mutant Mice.
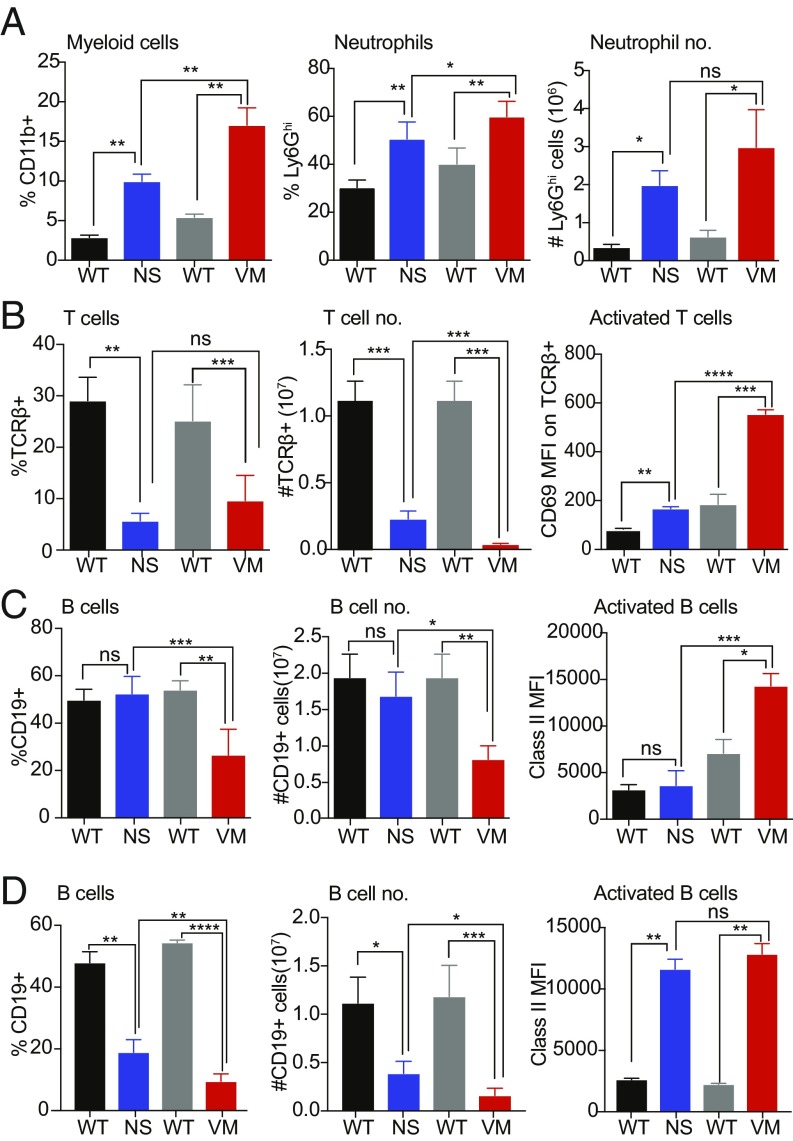
We next analyzed the immune cell composition of spleens from 6-wk-old N153S and V154M mice. Both mutants had a significantly higher percentage and total number of CD11b+ myeloid cells than their littermate controls, and the V154M mice had a higher percentage of myeloid cells than the N153S mice (Fig. 3A and SI Appendix, Fig. S3 A and B). There was also significant increase in both the percentage and number of Ly6Ghi neutrophils in the mutants, and the V154M mice had a greater percentage of neutrophils than the N153S (Fig. 3A and SI Appendix, Fig. S3C). In addition to the expansion of myeloid cell subsets, we saw a reduction in percentage and number of T cell receptor-β+ (TCR-β+) T cells. Both the mutant strains showed a significant decrease in T cells compared with WT and the V154M mutant had fewer T cells than the N153S mutant (Fig. 3B and SI Appendix, Fig. S3D). In both the mutants, the remaining splenic T cells were activated as indicated by the increased expression of CD69, and the V154M mutant T cells were significantly more activated than T cells in the N153S mice (Fig. 3B and SI Appendix, Fig. S3G). Strikingly, in these young mice, the V154M mutant—and not the N153S mutant—showed a reduced percentage and number of CD19+ B cells in the spleen compared with littermate controls. The B cells that were present in the V154M mutant were also activated, as indicated by the increased expression of MHC class II, but the N153S mutant B cells were not (Fig. 3C and SI Appendix, Fig. S3 E and G). However, when we analyzed 16-wk-old SAVI mutant mice, B cell loss was apparent in the N153S mutant as well as in V154M mutant, although the V154M mice still had significantly fewer B cells even at this time point (Fig. 3D and SI Appendix, Fig. S3F). In 16-wk-old animals, the remaining splenic B cells were activated comparably in both mutants. Taken together, these findings indicated that the V154M mutant presented with more severe immune cell abnormalities compared with the N153S mutant.
Fig. 3.
V154M mutants exhibit more severe immune cell alterations than N153S SAVI mutant. (A–C) Immune cell populations obtained from the spleens of 6-wk-old N153S and V154M HETs and their respective littermate controls were analyzed by flow cytometry, n = 8 mice per group. (A) Total myeloid cells identified as CD11b+ and neutrophils as CD11b+ Ly6Ghi. (B) T cells were identified as TCR-β+ and activation was assessed by mean fluorescence intensity of CD69. (C) B cells were identified as CD19+ and the activation status of B cells was measured using mean fluorescence intensity of MHC class II. (D) B cell number and activation status in spleens from 16- to 20-wk-old mice. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001; ns, not significant.
SAVI Mutations Cause Abnormal Lymphocyte Development.
To determine whether the reduced number of mature T and B cells in the SAVI spleens reflected defects in lymphoid cell development, we evaluated the thymus and the BM. The total number of thymocytes, including the early CD4−CD8− double-negative thymocytes, were reduced (Fig. 4A and SI Appendix, Fig. S4A). Similarly, the percentage of both immature B220+AA4.1+ and mature B220+AA4.1− B cells was reduced in the BM of both SAVI mutants compared with their littermate controls; however, the reduction in mature B cells was greater in the V154M mutant than the N153S mutant. When we compared the ratio of mature to immature B cells in the BM, V154M mutants showed a significantly greater reduction (Fig. 4B and SI Appendix, Fig. S4B). In contrast, the percentage of myeloid progenitors, identified as Lin−sca1−ckit+, was increased in the BM of both SAVI mutants and may account for the increased CD11b+ myeloid cells in the BM (Fig. 4C and SI Appendix, Fig. S4C). We conclude that the decrease in T cells and increase in neutrophils in the SAVI mice can be traced back to their progenitors, and hence the immune abnormalities occur early in development. In murine models of autoinflammatory diseases, splenomegaly is frequently associated with extramedullary erythropoiesis (21). The enlarged spleens of SAVI mutants showed an increased percentage of Ter119+ erythroid lineage cells, as well as an increased percentage of immature erythrocytes (Ter119+ CD71+) compared with their WT littermate controls, whereas the BM of the mutants exhibited reduced percentages of immature red blood cells (RBCs) (Fig. 4D and SI Appendix, Fig. S4 D and E). Therefore, we conclude that SAVI mice also exhibit extramedullary erythropoiesis as a result of systemic inflammation.
Fig. 4.
SAVI mutations cause abnormal lymphocyte development. (A) The total number of cells and percentage of double-negative (DN) thymocytes in the thymus of SAVI mutants and respective WT littermate controls was determined. of (B) The percentage of B cells, mature B cells, and frequency of immature B cells in the BM was determined. (C) The percentage myeloid progenitors in the lin− gate of the BM and the frequency of CD11b+ myeloid cells in the BM were determined. (D) The total number of Ter119+ RBCs in the spleen (Left) and the percentage of immature (CD71+) RBCs in the RBC gate in the spleen (Center) and BM (Right) was determined. n = 8 mice for each WT and mutant genotype. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001; ns, not significant.
IRF3, IFNaR, and MLKL Deficiency Fail to Rescue V154M SAVI Phenotype.
Because STING activates type I interferons through IRF3, we hypothesized that blocking IRF3 to limit the production of type I interferons could rescue the lethality of SAVI mice. Therefore, we crossed the more potent SAVI V154M mutant strain to mice deficient in IRF3 or the type I IFNα/β receptor. (IFNaR). Consistent with other reports (16, 17), we found that the death observed in SAVI HET mice was not rescued by deletion of either of IRF3 or IFNaR (Fig. 5 A and B). Splenomegaly was observed in V154M HET IFNaR KO mice due to accumulation of immature RBCs, and the immune cell defects, such as an increased percentage of neutrophils, were still observed in V154M HET IFNaR KO mice (Fig. 5C, Upper). In addition, reduced thymic weight, T cell loss and activation of residual T cells were all observed at comparable levels in V154M HET IFNaR-deficient mice and IFNaR-sufficient littermate controls (Fig. 5C, Lower). To further evaluate these strains, we prepared BMDMs and evaluated the ISG and inflammation signature in SAVI V154M mice that were sufficient or deficient in IFNaR. We observed that ISGs elevated in the SAVI V154M HET were reduced in the IFNaR KO V154M HET. However, there was a subset of genes, such as Il1ra, prdm1, irf2, a20, nfkb2, and tgfb1 that still showed increased expression in SAVI mutant mice that lacked IFNaR (SI Appendix, Fig. S5). These observations indicated that pathways other than type I IFN also contribute to the clinical manifestations of the SAVI mutant strains.
Fig. 5.
IRF3, IFNaR, and MLKL deficiency fail to rescue V154M SAVI phenotype. (A–C) IRF3-deficient (IRF3−/−) V154M mutant and WT mice were compared with IRF3-sufficient Het (IRF3+/−) V154M mutant or WT mice. The same strategy was used to evaluate V154M HET IFNaR KO and littermate controls. (A) Survival curve for IRF3 mutant. n = 15 mice. (B) Survival curve for IFNaR mutant. n = 10 mice. (C) Spleen cells V154M HET IFNAR KO and littermate controls were further analyzed as described in Fig. 4. (D) Spleen cells from MLKL-deficient VM mice were compared with MLKL+/− cells using the same criteria. n = 5 mice 8–12 wk old per group. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Besides type I interferons, several studies have linked cytosolic DNA-sensing pathways to necroptosis downstream of STING activation. IFN production upon STING activation was reported to engage RIPK3 and MLKL, leading to oligomerization of MLKL and MLKL-dependent necroptotic cell death (22–24). Because we observed significant loss of lymphoid cells in these SAVI strains, we considered the possibility that necroptosis led to cell death in SAVI mice. V154M HET mice were crossed to MLKL KO mice to generate V154M HET MLKL KO mice and littermate controls. MLKL-deficiency did not change the immune abnormalities of the V154M HET mice (Fig. 5D). These data indicated that necroptotic death was not a driver of the T cell loss in these animals.
BM Chimeras Reveal Distinct Disease Outcomes Between the N153S and V154M Mutant Mice.
STING is expressed in both hematopoietic and nonhematopoetic cell types (e.g., fibroblasts, epithelial cells) (25). To determine whether the immune cell defects characteristic of SAVI were intrinsic to BM-derived hematopoietic cells or dependent on additional radio-resistant cells, we generated BM chimeras in which lethally irradiated WT mice were reconstituted with WT, N153S mutant, or V154M mutant BM cells. CD45 alleles were used to distinguish BM-derived cells (CD45.2) from radio-resistant host cells (CD45.1) (Fig. 6A). 10–12 wk postreconstitution, the chimeras were killed and evaluated for immune status and disease. Strikingly, the spleens of the V154M → WT chimeras, but not the N153S → WT or WT → WT chimeras showed an increased percentage of CD11b+ and Ly6Ghi neutrophils (Fig. 6B and SI Appendix, Fig. S6A). The CD11b+ cells in all of the chimera groups were CD45.2+, and therefore donor-derived (Fig. 6B). The V154M → WT chimeras also differed from the N153S → WT chimeras in the B cell compartment, as they were severely B cell lymphopenic, lacking mature and immature B cells in both the BM and spleen (Fig. 6C and SI Appendix, Fig. S6B). These data indicate that the expression of the V154M mutant in BM-derived myeloid cells, or a BM-derived cell that produces myeloid growth factors, was sufficient to drive the expansion of myeloid cells. Similarly, B cell development and survival was either compromised directly by B cell expression of the V154M mutant or indirectly by the inflammatory cytokines produced by non-B cells in the V154M → WT chimeras. In contrast, all of the chimeras had essentially normal percentages of TCR-β+ T cells in the spleen (Fig. 6D). As expected, in the WT → WT chimeras, the TCR-β+ T cells were donor-derived. However, quite unexpectedly, in both the N153S → WT and V154M → WT chimeras, almost all of the TCR-β+ T cells expressed CD45.1 and therefore were derived from radioresistant host cells, most likely mature T cells (Fig. 6D and SI Appendix, Fig. S6 C and D).
Fig. 6.
BM chimeras reveal distinct disease outcomes between the N153S and V154M mutant mice. (A) 107 BM cells from 8- to 10-wk-old CD45.2 WT, N153S HET, V154M HET mice were used to reconstitute lethally irradiated (850R) CD45.1 WT mice (recipient). (B) The percentage myeloid cells (CD11b+) and percentage granulocytes CD11b+ Ly6G+ in the spleens of the WT → WT (W → W), N153S → WT (N → W), and V154M → WT (V → W) chimeras, and the %CD45.1 vs. %CD45.2 of the CD11b+ population were determined by flow cytometry. (C) The %B220+ B cell in the BM %CD19+ B cells the spleen and the %CD45.1 vs. %CD45.2 of the CD19+ splenic B cell population were determined by flow cytometry. (D) The %TCR-β+ cells in the spleen and the %CD45.1 vs. %CD45.2 within the TCR-β+ population were determined by flow cytometry. Activation status of the TCR-β+ cells was assessed by CD44 mean fluorescence intensity. (E) The total cell number of Thy1.2+ cells in the thymus was measured and the %CD45.1 vs. %CD45.2 of the total thymocytes population was determined by flow cytometry. H&E-stained sections of the lung were evaluated for inflammation (Right). All mice were evaluated 10–12 wk after BM reconstitution. n = 8–10 mice per group compiled from two independent experiments. **P < 0.01; ***P < 0.001.
When we evaluated the activation status of the CD45.1 and CD45.2 TCR-β+ cells, we found that the donor-derived T cells from V154M mutant were activated, as indicated by increased expression of CD44 (Fig. 6D); however, the CD45.1+ T cells in the same chimeras were not activated. In contrast, the limited number of donor-derived T cells in the N153S → WT chimeras did not appear to be activated. The distinct impact of the V154M → WT mutation on T cell development was further reflected by the total absence of thymocytes in the V154M → WT chimeras, but not the N153S → WT chimeras (Fig. 6E). These data identify another disparity between the N153S and V154M mutants. While both mutations led to a severe defect in T cell development, in the V154M mice, the mutant STING limited early T cell development in the thymus. In contrast, in the N153S mutant mice, this defect mainly impacted mature T cells in the periphery. The presence of normal numbers of CD45.1+ nonactivated mature T cells derived from the WT host suggests that the STING mutations limited T cell development in a T cell-intrinsic manner. It further suggests that the inflammatory cytokine milieu in the STING mutant chimeras is not sufficient to drive T cell activation. Apart from differences in hematopoietic cells, when we analyzed the lungs of chimeric mice we observed inflammation in the V154M → WT but not in the other two groups (Fig. 6E). This observation indicates that the lung inflammation associated with the V154M mutation can result from immune cell dysfunction, even in mice where the radio-resistant cells express WT STING.
Mixed BM Chimeras Identify Cell-Intrinsic Defects in both V154M T Cells and Myeloid Cells.
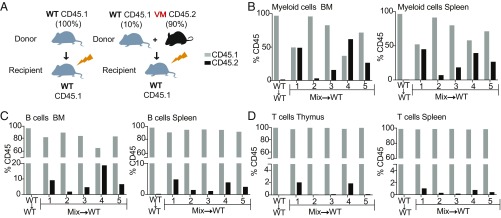
Our ability to directly compare mutant and WT T cells in the single-donor chimeras was highly informative, but the absence of WT myeloid cells in the two mutant chimeras and WT B cells in the V154M chimeras did not allow us to distinguish the direct role of the mutant STING in myeloid cells from indirect effects of the immune environment on myeloid cell activation. To further evaluate the role of STING mutants in the development and activation of BM-derived cells, we used a mixed BM chimera strategy that involved reconstituting irradiated mice with a combination of WT and mutant stem cells. We initially injected the irradiated mice with a mixture of 50% CD45.1 WT + 50% CD45.2 N153S stem cells, but 2 mo later we found that the recipients had been mostly engrafted by the WT stem cells, potentially due to the reduced frequency of pluripotent stem cells in the mutant donor mice as a result of ongoing BM inflammation. To generate mixed chimeras for the more severe V154M strain, we used a 10% CD45.1 WT + 90% CD45.2 V154M BM cell mixture (Fig. 7A) and were able to obtain a variable but significant level of V154M stem cell engraftment as detected by the presence of CD45.2+ V154M cells in the BM 10–11 wk postreconstitution. As a staining control, no CD45.2+ cells were present in any of the five WT → WT chimeras. Importantly, in both mixed chimera #1 and mixed chimera #4, 50% or more of the CD11b+ myeloid lineage cells in the BM were derived from V154M stem cells (Fig. 7B). The same chimeras also had a high frequency of CD11b+ cells in the spleen. V154M stem cells also contributed to B lineage cells in the BM and spleen, but to a much lesser extent than the myeloid compartment (Fig. 7C). There were even fewer V154M-derived T cells in the thymus and only chimeras #1 and #4 had detectable numbers of CD45.2+ thymocytes and minimal numbers of CD45.2+ mature T cells in the spleen (Fig. 7D). Overall these data indicate that V154M stem cells were able to reconstitute the myeloid compartment much more effectively than the B cell compartment, and made an even smaller contribution to the thymic and peripheral T cell compartments.
Fig. 7.
Mixed BM chimeras identify cell intrinsic defects in both T cells and myeloid cells. (A) 107 BM cells were isolated from 8- to 10-wk-old CD45.1 WT and CD45.2 V154M HET mice. A total of 107 cells were injected that were either WT only (CD45.1+) or a combination of 90% V154M HET and 10% WT were used to reconstitute lethally irradiated (850R) CD45.1+ WT mice. (B) One representative WT → WT chimera is compared with the five mice that received the 90:10 mix. The %CD45.1 and %CD45.2 of the CD11b+ cells in the BM and spleen was determined by flow cytometry. (C) The %CD45.1 and %CD45.2 of the CD19+ cells in the BM and spleen. (D) The %CD45.1 and %CD45.2 of the Thy1.2 cells in the thymus and TCR-β+ cells in the spleen. n = 5 mice per group analyzed 8–10 wk after BM reconstitution.
Discussion
A variety of mutations in genes associated with the metabolism or sensing of endogenous nucleic acids lead to an assortment of clinical syndromes that are collectively referred to as “type I interferonopathies,” because these patients share an elevated IFN gene signature (3, 26). To what extent type I IFNs and IFN-induced genes promote disease manifestations or simply provide a surrogate signature of nucleic acid sensor activation remains controversial. One of the more recent additions to the interferonopathy roster is SAVI, a disease impacting young children who either inherit or acquire one of the several GOF mutations in STING. To better understand the STING-regulated pathways that contribute to immune activation and the clinical features of SAVI patients, two investigators have recently developed murine models. Remarkably N153S and V154M SAVI mice have been reported to have very distinct phenotypes but because they were studied in different laboratories, it was not clear if these phenotypes reflected molecular or environmental differences. We now have independently generated our own mutant lines and made direct comparisons of both the N153S and V154M alleles side-by-side in the same colony. Our studies have identified clear differences in STING activation, clinical parameters, and dependence on radio-resistant cells in these two lines that may account for some of the heterogeneity seen in SAVI patient populations.
The N153S and V154M mutations are in adjacent amino acids that are absolutely conserved across STING orthologs across a broad range of species. These mutations likely impact STING dimerization. Indeed, the corresponding recombinant human N154S and V155M proteins each form stable dimers (10). In ectopic expression systems described here, both the N153S and V154M mutants are expressed at similar levels and are both GOF in terms of their ability to turn on IFN-β reporter genes. The V154M allele, however, was considerably more active than the N153S allele. In cells from these animals, both SAVI alleles are associated with constitutive TBK1 activation, IkBa phosphorylation, and ISG expression, although the V154M cells were more strongly activated. While both proteins are expressed at similar levels by immunoblotting in either 293T transfectants or in BM-derived macrophages or lung homogenates from animals, studies using cycloheximide to evaluate protein stability indicated that the stability of the V154M protein was enhanced compared with the N153S protein upon activation, which may account for the more potent phenotypes observed in all of our analyses. Our studies with bafilomycin A1 treatment to prevent autophagy-mediated degradation of active WT or mutant STING alleles revealed heightened signaling capability and are consistent with this possibility.
The more robust STING activity of the V154M mice corresponds with a more severe clinical phenotype by a variety of criteria. (i) There was a significant difference in birthrate. In crosses between SAVI heterozygous males and WT females, N153S mice were born close to the expected 50% Mendelian ratio, while the frequency of V154M offspring was closer to 35%. (ii) While both strains failed to thrive, the V154M offspring weighed less and had a reduced lifespan compared with the N153S mice. (iii) The immune status of the V154M mice was more compromised. By 6 wk of age, the V154M mice already had a significantly higher percentage of CD11b+ myeloid cells and lower percentage of B cells in the spleen than the N153S mice; the N153S mice only developed lymphopenia later in life. (iv) Finally, there was a difference in lung disease. While both strains developed pulmonary inflammation, only the V154M mice developed pulmonary fibrosis, as demonstrated by the trichrome stain of lung sections from mice. Because lung fibrosis is a major complication of SAVI patients, the V154M mice should provide a useful experimental system for better understanding the factors that contribute to the transition from inflammation to fibrosis. Lung fibrosis was not detected in the previous description of the V154M mice where the mice used for long-term studies were continuously dosed with antibiotics (16). It is possible that the antibiotics may have modified the microbiota of the lung (or other tissues) in ways that constrained the fibrotic response. Alternatively, other environmental factors may be involved.
Intriguingly the difference between the two strains was dramatically illustrated by their capacity to engraft lethally irradiated mice, as shown in the single-donor chimera studies. The V154M → WT chimeras, and not the N153S → WT chimeras, had a significantly expanded CD11b+ compartment, while at the same time only the V154M → WT chimeras were severely B cell lymphopenic. As for T cells, neither the N153S nor the V154M stem cells were able to repopulate the splenic T cell compartment; although both chimera groups had normal numbers of T cells, the peripheral T cells were almost entirely derived from an expanded pool of radio-resistant host cells. In contrast, N153S-derived stem cells but not V154M stem cells were able to successfully engraft the thymus. Overall, these data point to a hierarchy of outcomes that correlate with the greater STING activation of the V154M mutant. Apparently the N153S mutant signaling is sufficient to promote the death of mature T cells in the spleen, but the greater signaling activity of the V154M mutant-enhanced myeloid expansion blocked B cell development in the BM and thymocyte development in the thymus. The single-donor chimeras strongly support the notion that inherent signals emanating from the GOF STING mutations are responsible for the death of mature T cells because radio-resistant WT T cells exposed to the same immune/cytokine milieu persisted and repopulated the spleen. It is more difficult to eliminate the role of the immune environment in regulating the myeloid and B cell lineages, because these compartments were entirely repopulated by the donor stem cells. The single-donor chimeras further demonstrate that V154M hematopoietic cells are sufficient to drive the entire SAVI phenotype without any factors produced by radio-resistant cells; the same is not true for the N153S strain. STING expression in different cell types may contribute to these phenotypes. For example, in T cells it was shown that high expression of STING resulted in stronger T lymphocyte defects than in macrophages where STING is expressed at lower levels (27). However, whether STING is truly higher in T cells is unclear, as other groups found comparable STING levels in T cells and macrophages (28). Hence, the assessment of STING protein levels and degree of STING activation in different human/murine cells is needed to further understand cell- and tissue-type–specific phenotypes in SAVI disease.
To facilitate a direct side-by-side comparison of various cell types, where both mutant and WT cells were exposed to the same immune environment, we used mixed BM chimeras. When we used a 50:50 mix of CD45.1 WT and CD45.2 N153S mutant BM cells, the T cell compartment of the resulting chimeras was almost entirely derived from the WT stem cells. Similar findings were reported in published studies that used Thy1.1 vs. Thy1.2 to track the relative engraftment of WT and N153S cells (17). However, here in our chimeras the B cells and myeloid cells were also mostly derived from the WT mice, consistent with the notion that the mutant stem cells had not engrafted to a measurable extent. Therefore, to study the more severe V154M mutant, we used 90% mutant to 10% WT to reconstitute the lethally irradiated hosts. In these 90:10 chimeras, up to 50% of the CD11b+ cells in the BM and spleen came from the V154M mutant donor, while a much smaller percentage of the B and T cell compartment came from the V154M donor. This suggests that the V154M CD11b+ lineage cells survive or expand to a greater extent than the V154M lymphocytes and that the expansion of CD11b+ cells in the mutant mice does not simply reflect increased production of myeloid growth factors. Whether intrinsic signals in the CD11b mutant cells enhance the survival/expansion of myeloid cells remains to be determined and will require a better understanding of the relative frequency of pluripotent hematopoietic stem cells in the two donor BM compartments. However, our data indicate that the outcome of constitutive STING activation is highly cell-type specific. The extent to which CD11b+ WT and V154M cells have distinct functions will, however, warrant further investigation. Moreover, stem cells from the two strains have dramatically different capacities to transfer these clinical manifestations to the lethally irradiated WT recipients. STING signaling has previously been shown to modulate hematopoietic stem cells and their niches during bacterial infection (29), consistent with our observations that STING activation affects hematopoesis. While both mutants fail to engraft the mature T cell compartment, only the V154M → WT chimeras show CD11b+ cell expansion, B cell lymphopenia, and loss of thymocytes. Explanations for this hierarchy of outcomes may include cell-type–specific differences in the level of STING activation or cell-specific differences in downstream signaling components.
In this study we also confirmed the minimal role played by IRF3 and type I IFNs in disease outcomes. There are currently no effective therapies for patients with SAVI, although JAK inhibitors have been effective in suppressing STAT1 phosphorylation in lymphocytes from children with SAVI (10, 30, 31). Consistent with these observations, we also observe that SAVI IFNaR-deficient BMDM temper the ISG signature. However, the expression of other inflammatory mediators was not modulated by IFNaR, indicating that additional downstream signaling events are still unleashed in SAVI mutant cells. Ectopic expression of the human SAVI alleles in 293T cells revealed robust NF-κB promoter activation and the V155M mutant had stronger activity. It is likely that the NF-κB pathway, which drives a broad array of inflammatory cytokines, may contribute to SAVI disease. Spontaneous cell death has been observed in T cells, monocytes, and endothelial cells from SAVI patients, suggesting that cell death pathways may also be at play in vivo (10, 32). The STING signaling pathway has been linked to necroptosis, apoptosis, and pyroptosis, the latter being particularly relevant in human cells (22–24, 27, 28, 32, 33). By crossing SAVI V154M to MLKL KO mice, the executioner of necroptosis, we further demonstrate that STING-dependent necroptosis does not contribute to SAVI phenotypes either. It remains to be determined whether apoptotic or pyroptotic death pathways contribute. In addition, STING has also been linked to autophagy, endoplasmic reticulum stress, and inflammasome activation; therefore it remains possible that multiple pathways could be activated and contribute to SAVI pathogenesis. Additionally, interferons other than type I IFN could also play a role in STING dependent disease manifestations (34).
In summary, the N153S and V154M mutations result in distinct clinical phenotypes that reflect, to varying extents, the clinical manifestations of SAVI patients. To understand if this hierarchy of severity is relevant for humans that have N154S versus V155M mutations, we reviewed all published studies on different SAVI mutations (SI Appendix, Table S2). Altogether, 10 of 11 V155M patients developed interstitial lung disease (fibrosis was confirmed in 9 of these) and 3 of 5 N153S patients developed interstitial lung disease with only two cases of fibrosis. The study by Liu et al. (10) included both N154S and V155 patients and reported that only two of four N154S patients had respiratory symptoms, whereas one of one V155M SAVI patient examined developed interstitial lung disease associated with fibrosis. In the same report, the authors performed an IFN-reporter assay and found that the V155M mutant was more active than the N154S allele. Another study demonstrated that G166E SAVI mutant patients develop skin disease but show no evidence of lung disease; in vitro assays demonstrated that the G166E patients had even lower IFN-inducing activity than the N154S patients (35). The authors conclude that the differences in the SAVI alleles correlate with increased IFN levels and greater disease severity. In summary, these clinical studies are consistent with our murine models, where the V154M mouse mutant has more robust STING activation than the N153S mutant resulting in the development of lung fibrosis. In light of these data, it may be reasonable to treat SAVI patients in an allele-specific manner; patients with a specific SAVI allele may have less severe clinical manifestations than those with other mutations and may need a different dose, duration, or kind of treatment. Because SAVI patient populations are rare and have diverse genetic backgrounds, a comparison of murine models with different mutations on genetically identical backgrounds may provide a useful platform for testing tissue specific therapies.
Materials and Methods
Reagents.
The –110 IFNb-reporter gene was from T. Maniatis, Columbia University, New York (36). Full-length human and murine STING with HA tag were cloned int pcDNA3.1 vector, respectively. The human and murine SAVI alleles were generated by site directed mutagenesis using New England Biolabs Q5 Mutagenesis Kit (cat #E0554S).
Mice.
To generate SAVI mutant mice, a single sgRNA 5′-GTTAAATGTTGCCCACGGGC TGG-3′ was used based on specificity and proximity to the targeted mutation site. A single-stranded oligonucleotide donor (ssODN) encoding the N153S mutation with the homology arm was synthesized as follows: tcaggcccttctgctgtcttcagAGCTTGACTCCAGCGGAAGTCTCTGCAGTCTGTGAAGAAAAGAAGTTAAGTGTTGCCCATGGGCTGGCCTGGTCATACTACATTGGGTACTTGCGGTTGATCTTACCAGgtagggcacctctggatg and one encoding the V154M mutant was as follows: tcaggcccttctgctgtcttcagAGCTTGACTCCAGCGGAAGTCTCTGCAGTCTGTGAAGAAAAGAAGTTAAATATGGCCCATGGGCTTGCCTGGTCATACTACATTGGGTACTTGCGGTTGATCTTACCAGgtagggcacctctggatg. The underlined regions show the mutations introduced to generate the SAVI allele; uppercase represents exonic region and lowercase the intronic region. Female C57BL/6J mice, 4 wk of age, were superovulated and mated with C57BL/6J males. Single-cell embryos were isolated and injected with a combination of Cas9, gRNA, and ssODN in DNase/RNase-free microinjection buffer. Modified embryos were transferred into pseudopregnant females. For detection of heterozygous mice for both the SAVI strains, a 554 base pair product was amplified using 5′GGTCCTCTATAAGTCCCTAAG 3′ forward primer and 5′GGTCAC CCTCAAATAAATAGG 3′ reverse primer; the PCR product was purified and used for Sanger sequencing using 5′TCCTGGGCCTTCAGGTATGA3′ as the sequencing primer. Independent founder lines were back-crossed to WT animals. SAVI N153S or V154M mice and WT littermate controls for each strain were generated by crossing heterozygous male mice with WT females. WT littermate controls were used for all experiments. Irf3−/− mice (37) were kindly provided by T. Taniguchi, University of Tokyo, Tokyo, IFNAR−/− (38) were provided by J. Sprent, Scripps, La Jolla, CA. Mlkl−/− mice (39) were provided by M. Kelliher, University of Massachusetts Medical School, Worcester, MA. All animal experiments were conducted in accordance with the Institutional Animal Care and Use Committees at the University of Massachusetts Medical School.
Cell Culture.
BM-derived macrophages were generated from BM in the presence of L929 supernatants for 7 d. Cells were transfected with 2 μg/mL cGAMP using Lipofectamine 2000 (Invitrogen) and cells were treated with 5 μM bafilomycin A1 (Invivogen) and 100 ng LPS (Sigma).
Reporter Assays.
The 293T cells (2 × 105 cells per well) were seeded into 96-well plates and transfected on the following day with 40 ng of luciferase reporter genes (IFNb luciferase or NF-κB luciferase) using Genejuice transfection reagent (Millipore). The Renilla-luciferase reporter gene (Promega; 40 ng) was cotransfected for normalization. In all cases, cell lysates were prepared 24 h later and reporter gene activity measured using a Dual Luciferase Assay System (Promega). Data are expressed as mean relative stimulation ± SEM for a representative experiment from a minimum of three separate experiments, each performed in triplicate.
Nanostring.
Total RNA was isolated (RNeasy kit; Qiagen) and quantitated via a Nanodrop ND-1000 spectrophotometer (Thermo Scientific). Next, 100 ng of RNA was hybridized and quantified with a custom probe set using the NanoString nCounter analysis system (NanoString Technologies). Gene-expression data were normalized to internal positive and negative controls. All values were scaled by a log2(x−min(x)+1) function and a heatmap generated using R-based software.
Cytokine Analysis.
Cytokines levels in serum were measured by multiplex protein analysis using Mouse Cytokine/Chemokine Array 21-Plex (EVE Technologies) and LUNARIS Multiplex Protein Analysis by (AYOXXA Biosystems).
Immunoblotting.
Cells were lysed in 50 µL of ice-cold Pierce RIPA lysis buffer (ThermoFisher Scientific) supplemented with 1× complete protease and phosphatase mixture inhibitor (Sigma). Protein concentration was measured using a protein DC assay kit. Whole-cell lysates were denatured for 5 min at 85 °C in presence of 1× Sample Buffer and reducing agent (Invitrogen). Fifty micrograms of samples were separated by SDS/PAGE on 10% gels. Each gel was run initially for 15 min at 80 V and then at 120 V. Transfer onto nitrocellulose membranes (Bio-Rad) was done using a Trans-Blot Turbo Transfer system for 10 min. Membranes were blocked for 1 h with 5% skim-milk (Sigma Aldrich) at room temperature in PBS supplemented with 0.05% Tween-20 (PBST). Membranes were probed overnight at 4 °C with the following primary antibodies in PBST: anti-HA (# 3724S Cell Signaling 1:1,000) antiphospho-TBK1/NAK (5483; Cell Signaling 1:1,000), anti-STING (13647; Cell Signaling 1:1,000), anti-pIkBa (9246S; Cell Signaling 1:1,000), anti–b-actin Peroxidase (A3854; Sigma-Aldrich). All membranes were washed with PBST and exposed using the SuperSignal West Pico PLUS chemiluminescent substrate (ThermoScientific) on ImageQuant LAS4000 mini Imager (GE Healthcare).
H&E and Trichrome Staining of Tissue.
All tissues were fixed in 10% neutral buffered formalin for 24–48 h before being processed and paraffin-embedded. Five-micrometer-thin sections were stained by H&E in an automated stainer (Leica Autostainer XL), or stained with trichrome staining kit manually following the manufacturer’s instructions (IW-3006; IHC World). Histomorphology of each H&E slides was evaluated by Applied Pathology Systems at low and high-power field on Olympus BX40 microscope, and the images were captured with Olympus cellSens Entry software at indicated magnifications. Inflammation was semiquantified at high-power field on the H&E slides using multitier system, as previously described with some modifications (40, 41).
Flow Cytometry.
The following conjugated anti-mouse Abs were used: CD11b PB, Ly6C APC, Ly6G FITC, CD45.1 APCCy7, CD45.2 PerCP, TCRb PerCPCy5.5, CD4 PECy7, CD8 PE CD45.1 PB, CD45.2 FITC, CD19 Pacific Blue, CD44 FITC, CD25 PerCPCy5.5, CD69 APC, Class II APCCY7, CD45.1 PE CD45.2 PerCPCy5.5, B220 Pacific Blue, AA4.1 (CD93) PE, Ter119 APC, CD71 PECy7, c-KIT APC-Cy7, and SCA1 Pacific Blue. Cells were incubated in CD16/32 (Fc Block; BD Biosciences) and stained with antibodies. Cells were acquired on a LSR II (BD Biosciences) and analyzed with FlowJo software (Tree Star).
Generation of BM Chimeras.
Lethally irradiated (850R) 8- to 12-wk-old female recipients were reconstitued by intravenous injection of 107 total BM cells from 8- to 10-wk-old female mice. The extent of reconstitution was determined by flow cytometry for CD45.1/CD45.2+ cells in total peripheral blood and confirmed by FACS analysis of the spleen at the time of killing, which was 8–12 wk after the intravenous injections of the BM.
Statistical Analysis.
Data are reported as mean ± SEM. Statistical significance was analyzed with the unpaired Mann–Whitney test using Prism software (GraphPad). Statistical significance is represented by the following notation in the figures: *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Supplementary Material
Acknowledgments
We thank Zhaozhao Jiang for help with genotyping of the mice and Richard Kumaran Kandasamy for nanostring data analysis. This study was supported by a Target Identification in Lupus Grant from the Lupus Research Alliance and the NIH (R01AI128358). The K.A.F. laboratory is part of the Li Weibo Institute For Rare Diseases Research, a consortium of researchers at UMass Medical School studying Rare Diseases.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1818281116/-/DCSupplemental.
References
- 1.Chen Q, Sun L, Chen ZJ. Regulation and function of the cGAS-STING pathway of cytosolic DNA sensing. Nat Immunol. 2016;17:1142–1149. doi: 10.1038/ni.3558. [DOI] [PubMed] [Google Scholar]
- 2.Ahn J, Barber GN. Self-DNA, STING-dependent signaling and the origins of autoinflammatory disease. Curr Opin Immunol. 2014;31:121–126. doi: 10.1016/j.coi.2014.10.009. [DOI] [PubMed] [Google Scholar]
- 3.Crow YJ, Manel N. Aicardi-Goutières syndrome and the type I interferonopathies. Nat Rev Immunol. 2015;15:429–440. doi: 10.1038/nri3850. [DOI] [PubMed] [Google Scholar]
- 4.Gall A, et al. Autoimmunity initiates in nonhematopoietic cells and progresses via lymphocytes in an interferon-dependent autoimmune disease. Immunity. 2012;36:120–131. doi: 10.1016/j.immuni.2011.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stetson DB, Ko JS, Heidmann T, Medzhitov R. Trex1 prevents cell-intrinsic initiation of autoimmunity. Cell. 2008;134:587–598. doi: 10.1016/j.cell.2008.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bursztejn AC, et al. Unusual cutaneous features associated with a heterozygous gain-of-function mutation in IFIH1: Overlap between Aicardi-Goutières and Singleton-Merten syndromes. Br J Dermatol. 2015;173:1505–1513. doi: 10.1111/bjd.14073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rutsch F, et al. A specific IFIH1 gain-of-function mutation causes Singleton-Merten syndrome. Am J Hum Genet. 2015;96:275–282. doi: 10.1016/j.ajhg.2014.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jang MA, et al. Mutations in DDX58, which encodes RIG-I, cause atypical Singleton-Merten syndrome. Am J Hum Genet. 2015;96:266–274. doi: 10.1016/j.ajhg.2014.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jeremiah N, et al. Inherited STING-activating mutation underlies a familial inflammatory syndrome with lupus-like manifestations. J Clin Invest. 2014;124:5516–5520. doi: 10.1172/JCI79100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu Y, et al. Activated STING in a vascular and pulmonary syndrome. N Engl J Med. 2014;371:507–518. doi: 10.1056/NEJMoa1312625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Munoz J, et al. Stimulator of interferon genes-associated vasculopathy with onset in infancy: A mimic of childhood granulomatosis with polyangiitis. JAMA Dermatol. 2015;151:872–877. doi: 10.1001/jamadermatol.2015.0251. [DOI] [PubMed] [Google Scholar]
- 12.Chia J, et al. Failure to thrive, interstitial lung disease, and progressive digital necrosis with onset in infancy. J Am Acad Dermatol. 2016;74:186–189. doi: 10.1016/j.jaad.2015.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Melki I, et al. Disease-associated mutations identify a novel region in human STING necessary for the control of type I interferon signaling. J Allergy Clin Immunol. 2017;140:543–552.e5, and erratum (2017) 140:1757. doi: 10.1016/j.jaci.2016.10.031. [DOI] [PubMed] [Google Scholar]
- 14.Picard C, et al. Severe pulmonary fibrosis as the first manifestation of interferonopathy (TMEM173 mutation) Chest. 2016;150:e65–e71. doi: 10.1016/j.chest.2016.02.682. [DOI] [PubMed] [Google Scholar]
- 15.Saldanha RG, et al. CIRCA; AADRY A mutation outside the dimerization domain causing atypical STING-associated vasculopathy with onset in infancy. Front Immunol. 2018;9:1535. doi: 10.3389/fimmu.2018.01535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bouis D, et al. Severe combined immunodeficiency in stimulator of interferon genes (STING) V154M/wild-type mice. J Allergy Clin Immunol. 2018;143:712–725.e5. doi: 10.1016/j.jaci.2018.04.034. [DOI] [PubMed] [Google Scholar]
- 17.Warner JD, et al. STING-associated vasculopathy develops independently of IRF3 in mice. J Exp Med. 2017;214:3279–3292. doi: 10.1084/jem.20171351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Conlon J, et al. Mouse, but not human STING, binds and signals in response to the vascular disrupting agent 5,6-dimethylxanthenone-4-acetic acid. J Immunol. 2013;190:5216–5225. doi: 10.4049/jimmunol.1300097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yoshida H, Okabe Y, Kawane K, Fukuyama H, Nagata S. Lethal anemia caused by interferon-beta produced in mouse embryos carrying undigested DNA. Nat Immunol. 2005;6:49–56. doi: 10.1038/ni1146. [DOI] [PubMed] [Google Scholar]
- 20.Gonugunta VK, et al. Trafficking-mediated STING degradation requires sorting to acidified endolysosomes and can be targeted to enhance anti-tumor response. Cell Rep. 2017;21:3234–3242. doi: 10.1016/j.celrep.2017.11.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baum R, et al. Synergy between hematopoietic and radioresistant stromal cells is required for autoimmune manifestations of DNase II-/-IFNaR-/- mice. J Immunol. 2016;196:1348–1354. doi: 10.4049/jimmunol.1502130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brault M, Olsen TM, Martinez J, Stetson DB, Oberst A. Intracellular nucleic acid sensing triggers necroptosis through synergistic type I IFN and TNF signaling. J Immunol. 2018;200:2748–2756. doi: 10.4049/jimmunol.1701492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen D, et al. PUMA amplifies necroptosis signaling by activating cytosolic DNA sensors. Proc Natl Acad Sci USA. 2018;115:3930–3935. doi: 10.1073/pnas.1717190115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sarhan J, et al. Constitutive interferon signaling maintains critical threshold of MLKL expression to license necroptosis. Cell Death Differ. 2018;26:332–347. doi: 10.1038/s41418-018-0122-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhu Q, et al. Cutting edge: STING mediates protection against colorectal tumorigenesis by governing the magnitude of intestinal inflammation. J Immunol. 2014;193:4779–4782. doi: 10.4049/jimmunol.1402051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Crow YJ. Type I interferonopathies: Mendelian type I interferon up-regulation. Curr Opin Immunol. 2015;32:7–12. doi: 10.1016/j.coi.2014.10.005. [DOI] [PubMed] [Google Scholar]
- 27.Gulen MF, et al. Signalling strength determines proapoptotic functions of STING. Nat Commun. 2017;8:427. doi: 10.1038/s41467-017-00573-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Larkin B, et al. Cutting edge: Activation of STING in T cells induces type I IFN responses and cell death. J Immunol. 2017;199:397–402. doi: 10.4049/jimmunol.1601999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kobayashi H, et al. Bacterial c-di-GMP affects hematopoietic stem/progenitors and their niches through STING. Cell Rep. 2015;11:71–84. doi: 10.1016/j.celrep.2015.02.066. [DOI] [PubMed] [Google Scholar]
- 30.Sanchez GAM, et al. JAK1/2 inhibition with baricitinib in the treatment of autoinflammatory interferonopathies. J Clin Invest. 2018;128:3041–3052. doi: 10.1172/JCI98814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Frémond ML, et al. Efficacy of the Janus kinase 1/2 inhibitor ruxolitinib in the treatment of vasculopathy associated with TMEM173-activating mutations in 3 children. J Allergy Clin Immunol. 2016;138:1752–1755. doi: 10.1016/j.jaci.2016.07.015. [DOI] [PubMed] [Google Scholar]
- 32.Cerboni S, et al. Intrinsic antiproliferative activity of the innate sensor STING in T lymphocytes. J Exp Med. 2017;214:1769–1785. doi: 10.1084/jem.20161674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gaidt MM, et al. The DNA inflammasome in human myeloid cells is initiated by a STING-cell death program upstream of NLRP3. Cell. 2017;171:1110–1124.e18. doi: 10.1016/j.cell.2017.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hamming OJ, Gad HH, Paludan S, Hartmann R. Lambda interferons: New cytokines with old functions. Pharmaceuticals (Basel) 2010;3:795–809. doi: 10.3390/ph3040795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.König N, et al. Familial chilblain lupus due to a gain-of-function mutation in STING. Ann Rheum Dis. 2017;76:468–472. doi: 10.1136/annrheumdis-2016-209841. [DOI] [PubMed] [Google Scholar]
- 36.Fitzgerald KA, et al. IKKepsilon and TBK1 are essential components of the IRF3 signaling pathway. Nat Immunol. 2003;4:491–496. doi: 10.1038/ni921. [DOI] [PubMed] [Google Scholar]
- 37.Sato M, et al. Distinct and essential roles of transcription factors IRF-3 and IRF-7 in response to viruses for IFN-alpha/beta gene induction. Immunity. 2000;13:539–548. doi: 10.1016/s1074-7613(00)00053-4. [DOI] [PubMed] [Google Scholar]
- 38.Kolumam GA, Thomas S, Thompson LJ, Sprent J, Murali-Krishna K. Type I interferons act directly on CD8 T cells to allow clonal expansion and memory formation in response to viral infection. J Exp Med. 2005;202:637–650. doi: 10.1084/jem.20050821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Murphy JM, et al. The pseudokinase MLKL mediates necroptosis via a molecular switch mechanism. Immunity. 2013;39:443–453. doi: 10.1016/j.immuni.2013.06.018. [DOI] [PubMed] [Google Scholar]
- 40.Curtis JL, Byrd PK, Warnock ML, Kaltreider HB. Requirement of CD4-positive T cells for cellular recruitment to the lungs of mice in response to a particulate intratracheal antigen. J Clin Invest. 1991;88:1244–1254. doi: 10.1172/JCI115428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ashcroft T, Simpson JM, Timbrell V. Simple method of estimating severity of pulmonary fibrosis on a numerical scale. J Clin Pathol. 1988;41:467–470. doi: 10.1136/jcp.41.4.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.