Abstract
Cancer remains a life-threatening disease and accounts for the major mortality rates worldwide. The practice of using biomarkers for early detection, staging, and customized therapy may increase cancer patients’ survival. Deubiquitinating enzymes (DUBs) are a family of proteases that remove ubiquitin tags from proteins of interest undergoing proteasomal degradation. DUBs play several functional roles other than deubiquitination. One of the important roles of DUBs is regulation of tumor progression. Several reports have suggested that the DUB family members were highly-elevated in various cancer cells and tissues in different stages of cancer. These findings suggest that the DUBs could be used as drug targets in cancer therapeutics. In this review, we recapitulate the role of the DUB family members, including ubiquitin-specific protease, otubain protease, and important candidates from other family members. Our aim was to better understand the connection between DUB expression profiles and cancers to allow researchers to design inhibitors or gene therapies to improve diagnosis and prognosis of cancers.
Keywords: Cancer, Deubiquitinating enzymes, Gene therapy, Inhibitors, Otubain protease, Ubiquitin-specific protease
INTRODUCTION
Cancer is one of the major diseases causing death globally, accounting for 8.2 million deaths in 2012 (1). Recent advances in molecular and cellular biology have played important roles in understanding cancer and breakthroughs that have been being translated into therapy. Because of these recent developments, genes that are involved in cancer are being unraveled (2, 3). Most of the known cancer genes were originally identified by genetic evidence. A protein that is encoded by a cancer gene typically regulates cell proliferation and differentiation and eventually leads to cell death or apoptosis. Mutations that lead to oncogenesis typically occur in genes that mediate DNA repair mechanisms (4).
Ubiquitin proteasome pathway
The post-translational attachment of ubiquitin is a modification that can determine a protein’s fate. While ubiquitin itself is a small conserved protein, its covalent conjugation to protein substrates and to other ubiquitin molecules is a tightly-controlled process involving complex cellular machinery. Perhaps the most prominent and well-known function of ubiquitin is to target a protein for degradation by the 26S proteasome. Degradation can be accomplished via an isopeptide bond formation between the carboxy-terminal glycine (Gly) site on the ubiquitin and an ɛ-amino group of the lysine (Lys) side chains of a protein substrate. The ubiquitin-substrate system is further diversified via the process of polyubiquitination, during which a ubiquitin molecule’s C-terminal Gly is conjugated with one of the seven Lys residues on another ubiquitin (Lys6, Lys11, Lys27, Lys29, Lys33, Lys48, or Lys63) or with the N-terminus to form linear chains.
The ubiquitin-proteasome protein degradation pathway is comprised of ubiquitin, a three-enzyme ubiquitination complex, the intracellular protein ubiquitination targets, and the proteasome that is the organelle of protein degradation, as well as ubiquitin-activating enzymes (E1), ubiquitin-conjugating enzymes (E2), and ubiquitin ligases (E3).
Deubiquitinating enzymes
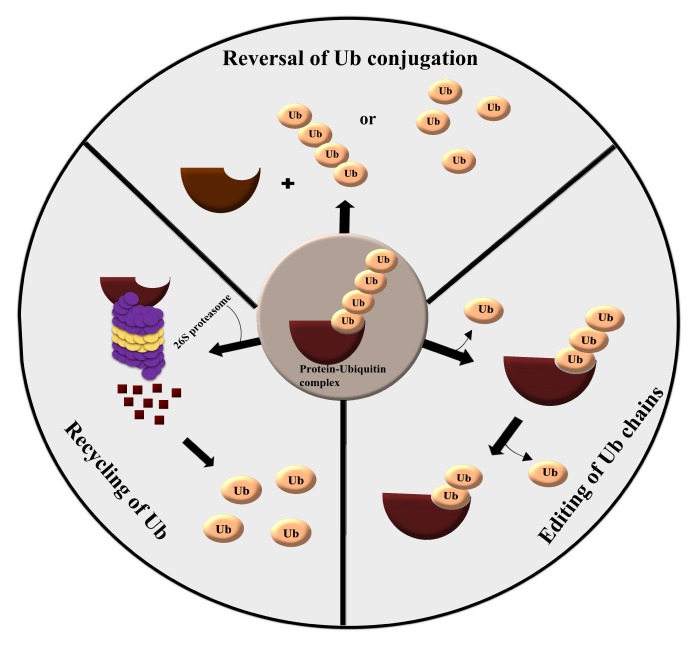
Deubiquitinating enzymes (DUBs) are proteases that reverse protein ubiquitination, a process which is significant for normal homeostasis. DUBs have four distinct mechanisms of action: 1) processing of ubiquitin precursors, 2) recycling of ubiquitin molecules during ubiquitination, 3) cleavage of polyubiquitin chains, and 4) reversal of ubiquitin conjugation (Fig. 1). DUBs regulate several cellular functions, including proteasome-dependent and lysosome-dependent proteolysis, gene expression, cell cycle progression, chromosome segregation, kinase activation, apoptosis, localization, DNA repair, maintenance of stemness, spermatogenesis, and degradation of signaling intermediates (5–10).
Fig. 1.
Various catalytic mechanisms exhibited by DUBs. DUBs can unknot ubiquitin conjugation by cleaving the bond between ubiquitin molecules and ubiquitin-target complexes, editing ubiquitin chains to remove one or more ubiquitin molecules, and finally, recycling of ubiquitin molecules in the ubiquitin-proteasome pathway.
Approximately 100 DUBs are encoded in the human genome (10). Based on the organization of the catalytic domain, DUBs are classified into distinct families, the vast majority of which are cysteine proteases. These include ubiquitin-specific proteases (USPs), ubiquitin C-terminal hydrolases (UCHs), ovarian tumor proteases (OTUs), Machado-Joseph disease proteases (MJDs), Jab1/Mov34/Mpr1 (JAMM) metalloproteases, and the recently-discovered MIU-containing novel DUB family, (MINDY) proteases (11).
UBIQUITIN-SPECIFIC PROTEASE FAMILY
USP2
USP2a, an isoform of USP2, is an androgen-regulated DUB that deubiquitinates the antiapoptotic proteins such as fatty acid synthase, Mdm2, and MdmX (12). USP2a expression is increased in glioma cells compared to normal brain tissues, which suggests that USP2a may correlate with malignant glioma progression, and therefore may be an effective marker for glioma prognosis (13). Furthermore, USP2 plays a role in tumor metastasis by modulating the activity or expression of MMP2, suggesting its use as a potential breast cancer marker (13).
USP4
USP4 has been strongly implicated in the regulation of tumor metastasis in breast cancer (14), liver cancer (15), and colorectal cancer (16). USP4 is significantly increased in melanoma and plays an oncogenic role by simultaneously inhibiting stress-induced cell apoptosis and promoting tumor metastasis (17). USP4 is an important protein that facilitates hepatocellular carcinoma (HCC) progression by stabilizing CYPA through deubiquitination and activating MAPK/CrkII signaling pathways, indicating that USP4 may act as a novel marker to predict prognosis and present a therapeutic opportunity for HCC (18). In breast cancer, USP4 promotes the migration and invasion of breast cancer cells via RLX-mediated TGF-β/Smad2/MMP-9 pathways, providing an attractive target for breast cancer therapy (19). The above results suggested that overexpression of USP4 in various cancers is due to the stabilization of other oncogenes in the respective cancer types. Thus, targeting USP4 as a biomarker could be useful for the early diagnosis of cancer.
USP5
USP5 acts as a exopeptidase that hydrolyzes isopeptide bonds in poly-ubiquitin from their free carboxy-terminal ends to produce monoubiquitin (20). USP5 knockdown suppressed cell proliferation, migration, and drug resistance and induced apoptosis, while USP5 overexpression promoted colony formation, migration, drug resistance, and tumorigenesis (21). USP5 plays a critical role in hepatocarcinogenesis through inactivation of the p14–p53 signaling pathway contributing to tumorigenesis and drug resistance, which provides a clue that USP5 could be a potential therapeutic target for HCC (22). In pancreatic cancer, USP5 plays a critical role in tumorigenesis and progression by stabilizing the FoxM1 protein, showing therapeutic potential against pancreatic cancer (23).
USP7
USP7, also known as herpes-associated ubiquitin-specific protease (HAUSP), was originally identified as an ubiquitin-specific protease that binds to a viral-encoded protein, called Vmw110 (24). HAUSP protein can bind to the herpes simplex virus type 1 (HSV-1) regulatory protein, which is known as infected cell polypeptide (ICPO). In epithelial ovarian cancer (EOC), USP7 and MARCH7 proteins are differentially expressed and the combination of USP7 and MARCH7 expression may function as promising biomarkers for EOC prognosis (25). HCC is one of the most dominant cancer types in the world. High expression of USP7 mRNA and protein levels in HCC tissues compared to normal liver samples has been reported (26). Cell-based assays have suggested that USP7 expression confers cell proliferation, migration, and invasion capabilities. These data suggest that USP7 could be a novel independent prognostic marker for HCC. Recently, a study suggested that USP7 deubiquitinates Ki-67, and thereby promotes cell proliferation in non-small-cell lung cancer (NSCLC) (27). Here, both Ki-67 and USP7 were expressed in NSCLC cells. Statistical data revealed a strong correlation between USP7 and Ki-67 levels. In contrast, siRNA targeting USP7 increased the ubiquitination of Ki-67 and led to delayed tumor growth. The above evidence suggests that USP7 could be an important therapeutic target in various cancer types.
USP8
USP8 belongs to the USP superfamily of DUBs targeting several substrates (28), including smoothened (29), frizzled (30), neuregulin receptor degradation protein-1, and receptor tyrosine kinase. Recently, the expression profile of USP8 in cervical squamous cell carcinoma (CSCC) has been studied (31). USP8 was upregulated in CSCC tissue samples compared to non-cancerous cervical tissues. Also, high expression of USP8 was associated with tumor stage and was recognized as an independent prognostic marker for CSCC. Elevated levels of USP8 led to cell proliferation, migration, and invasion of CSCC cell lines. Thus, USP8 could be a therapeutic and diagnostic target in CSCC patients.
USP10
USP10, also known as UBPO, is a protein consisting of 798 amino acids and was originally discovered as a DUB that interacts with the Ras-GAP SH3 domain-binding protein (32). Increased USP10 expression has been detected in some breast cancer and glioblastoma samples. Overexpression of USP10 has been associated with poor prognosis for glioblastoma multiforme patients, while decreased USP10 has been observed in gastric cancer tissues, and its downregulation has been associated with invasion, metastasis, and poor prognosis of gastric cancer. Current studies have also shown that USP10 suppressed proliferation and growth of pancreatic cancer cells. Therefore, USP10, as a novel DUB, has a crucial role in various pathological processes of tumors. In gastric cancer (GC), clinical samples and cell lines showed low-level expression of USP10, and negative USP10 expression was associated with a marked propensity toward gastric wall invasion, lymph node metastasis, highly malignant biological behavior, and poor survival. USP10 identification in GC can potentially serve as a new prognostic indicator predicting the treatment outcome for GC patients.
USP22
USP22 is a novel DUB that has been related to cell cycle progression, therapy resistance, and metastasis. The expression frequency of USP22 was extremely high in HCC compared to normal liver tissues (31). Elevated levels of USP22 represented poor HCC patient survival and have also been associated with greater mortality in patients with advanced tumor stages, shown by Kaplan-Meier analysis. As revealed by multivariate analyses, USP22 is a self-regulating prognostic marker in HCC. Several other researchers have reported that USP22 was overexpressed in salivary duct carcinoma (33) and esophageal squamous cell carcinoma (34). The above findings indicate that high USP22 expression might be an important factor in tumor progression and may serve as an independent molecular marker.
USP32
USP32 is a highly-conserved and uncharacterized gene, located on the 17q23.1–17q23.2 chromosomal band (35). USP32 was present in 22% of primary breast cancer tumors compared to non-cancerous mammary tissues, and 50% of breast cancer cell lines. Endogenously, USP32 was highly-elevated in the MCF7 cell line, and no mutation was detected in this cell line, indicating that the wild-type gene was overexpressed. Additionally, USP32 has a role in human small cell lung cancer (SCLC) (36). USP32 is highly-expressed in SCLC tissue samples compared to normal tissues. During the disease aggravation stage, USP32 has been positively correlated with SCLC expression. On the other hand, downregulation of USP32 in vitro caused reduced migration and proliferation rates of SCLC cells. Also, this downregulation arrested the cells at the G0/G1 phase by elevating p21 and decreasing CDK4-cyclin D1 complex levels. Cleaved caspase-3 and cleaved-PARP were activated when the USP32 gene was silenced, eventually leading to apoptosis by altering the epithelial-mesenchymal transition. Overall, USP32 could be a potential target for breast and lung cancers.
OTUBAIN PROTEASE FAMILY
OTUB1
The OTU domain-containing ubiquitin aldehyde-binding protein 1 (OTUB1) belongs to the OTU DUB family and is reported to be involved in various malignancies (37–40). Recently, the role of OTUB1 in human gliomas has been elucidated (41). Immunoblot and immunohistochemical experiments validated that glioma tissues overexpress OTUB1 genes and statistical studies showed that the expression pattern of OTUB1 was highly-linked to the WHO grades of the gliomas. On the other hand, downregulation of OTUB1 was linked to poor migration and also elevated EMT-related protein E-cadherin expression. Thus, OTUB1 might be involved in the regulation of ECM stability. The above results suggested that OTUB1 could be an important cancer marker in gliomas and other malignancies and could be a potential target for successful cancer therapy.
A20
A20 is a DUB that was originally found to be involved in autoimmunity and inflammation (42). However, a recent study proposed that A20 is highly involved in cancer metastasis (43). Here, A20 overexpression leads to metastasis of basal-like breast cancer by monoubiquitinating Snail1. In human basal-like breast cancers, A20 was significantly overexpressed and accounted for cancer metastasis. Additionally, A20 mediates TGF-β1-induced EMT of breast cancers by monoubiquitylating Snail1. Reports also suggested that the transient knockdown of A20 displayed decreased lung cancer metastasis in orthotopic breast cancer models and mouse xenografts.
DUB INHIBITORS
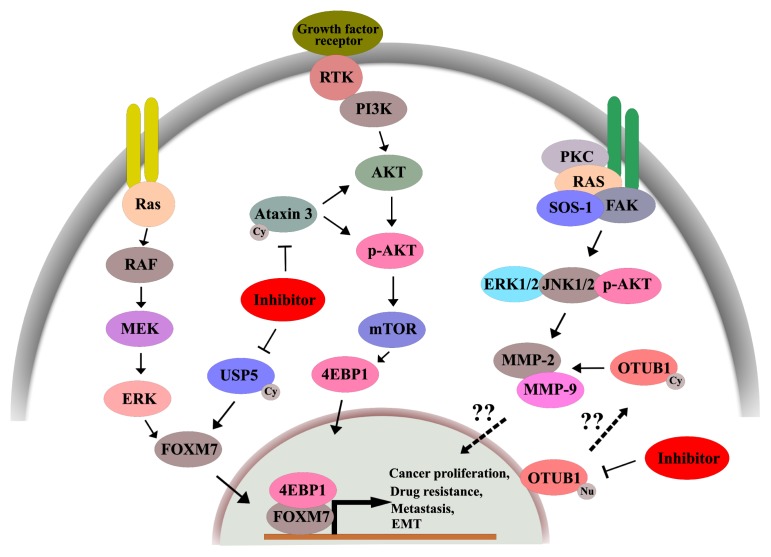
A number of reports have described the identification and utility of small molecule DUB inhibitors as anticancer agents (44). Inhibition of DUBs leads to cellular changes, such as (i) aggregation of polyubiquitinated protein molecules, (ii) reduction in the group of monomeric ubiquitin moieties, (iii) increased rates of polyubiquitin assembly, (iv) overall reduction in DUB events, and (v) altered cellular activities, such as DUB regulation of oncoproteins (45). Generally, DUB inhibition leads to impaired proteasome function and aggregation of misfolded functional proteins, resulting in cellular toxicity and death. DUBs that control oncogenic proteins can be targeted by small molecules that inhibit deubiquitinating activity via UPS degradation, while DUBs that control tumor suppressors can be targeted by increasing the deubiquitinating activity, thus inhibiting oncogenic progression. Several studies have been carried out to design small molecule DUB inhibitors because they are simpler to design than enzyme activators, using substrate modeling and competitive inhibition (46, 47). A schematic representation of DUB inhibition on relevant pathways is depicted in Fig. 2.
Fig. 2.
DUBs involved in the regulation of oncogenic pathways. Inhibition of specific DUBs leads to decreased cancer proliferation, drug resistance, and delayed metastasis.
An extensive study was carried out to discover drugs that inhibit DUBs and resulted in the discovery of ubiquitin aldehyde (Ubal) and ubiquitin vinyl sulfone (UbVS) (48). Due to their high molecular weights, peptidic nature, and lack of specificity, these compounds were not pharmacologically sustainable (48). The UCH family of proteins is involved in deubiquitination by removing ubiquitin from C-terminal adducts (48). Hence, researchers have made an effort to design their inhibitors and ended up with an isatin O-acyl oximes series (48). They are competitive and capable of directly targeting the active site with minimal IC50 values. Basically, UCH-L1 decreases cell proliferation in neuroblastoma cells; when this inhibitor is applied, cell proliferation is elevated. Thus, the data support the anti-proliferative nature of UCH-L1 proteins. A novel proteasome-inhibitory compound has been synthesized, called b-AP15 (49). The b-AP15 small molecule specifically inhibits USP14 and UCHL5 which are associated with 19S RP. Additionally, the compound b-AP15 showed effective anti-cancer responses against other refractory cancer types. Inhibitors targeting other important DUBs are described in Table 1.
Table 1.
DUBs and their inhibitors in cancer therapeutics
| DUB | Inhibitor(s) | Disease Indication | Stage of development | References |
|---|---|---|---|---|
| USP1 | ML323, Pimozide | Oncology | Preclinical | (50) |
| USP2 | ML364 | Inflammation | Preclinical | (50) |
| USP4 | Vialinin A | Inflammation and oncology | Preclinical | (51, 52) |
| USP5 | WP1130 | |||
| USP7 | ADC-01, ADC-03 | Oncology, Immuno-oncology | Preclinical | (53) |
| HBX41108 | Oncology, Immuno-oncology | Preclinical | (54, 55) | |
| P5091 | Oncology, Immuno-oncology | Preclinical | (56) | |
| P22077 | Oncology, Immuno-oncology | Preclinical | (56) | |
| USP8 | 9-(Ethoxyimino)-9H-indeno (1,2-b) pyra-zine-2,3-dicarbonitrile | Oncology | Preclinical | (56) |
| USP9X | WP1130 | Oncology | Preclinical | (57) |
| USP10 and USP13 | Spautin 1 | Inflammation | Preclinical | (57) |
| USP11 | Mitoxantrone | Oncology | Preclinical | (58) |
| USP14 | 1U1, b-AP15, VLX1570, wp1130 | Neurodegeneration | Preclinical | (59) |
| USP20 | GSK2643943A | Oncology | Preclinical | (60) |
| USP30 | 15-oxospiramilactone | Neurodegeneration | Preclinical | (61) |
| USP47 | P5091 | Cancer | Preclinical | (56) |
| UCH37 | WP1130 | Cancer | Preclinical | (57) |
| UCHL5 | b-AP15 | Neurology | Preclinical | (62) |
| UCHL1 | LDN-57444 | Cancer | Preclinical | (63) |
| UCHL3 | LDN-57444 | Cancer | Preclinical | (64) |
| UCHL5 | TCID, b-AP15 | Cancer | Preclinical | (62, 65) |
| UCH37 | WP1130 | Cancer | Preclinical | (57) |
CONCLUSION
This review delivers a comprehensive report of the DUBs for cancer diagnosis and prognosis. Recent developments of cancer therapies and the promptness of their application to clinical use for various tumors validate the prospects of exploiting DUBs as targets for drug development. The expression profiles of several DUBs in different cancer types are discussed in Table 2. Moreover, these DUBs exert their function through binding to their proteins, which can be targeted. In other cases, the DUB itself seems to be an excellent drug target. More detailed information on the roles, localization, regulation, and substrates of DUBs will help researchers understand their roles in oncogenesis and clinical applications of their inhibitors. Enhanced improvement in small molecule pharmacological development against DUBs will permit greater success in the treatment of cancer and other deadly diseases.
Table 2.
DUBs expressed in various types of cancer
| Disease | DUB | References |
|---|---|---|
| Gliomas | USP2a, USP22, USP44, BAP1, OTUB1 | (41, 66, 67) |
| Breast cancer | USP2, USP22, USP37 | (68, 69) |
| Hepatocellular carcinoma | USP4, USP5, USP11, USP22, UCHL1, A20, OTUB1 | (70, 71) |
| Esophageal cancer | USP4 | (72) |
| Melanoma | USP4 | (72) |
| Pancreatic cancer | USP5 | (72) |
| Epithelial ovarian cancer | USP7 | (72) |
| Lung adenocarcinoma | USP8 | (73, 74) |
| Gastric carcinoma | USP10 | (75) |
| Endometrial cancer | USP14 | (75) |
| Non-small cell lung carcinoma | USP17, USP22, OTUD7B, OTUD6B | (76, 77) |
| Renal clear cell carcinoma | USP21 | (78) |
| Muscle invasive bladder cancer | USP18 | (79) |
| Cervical cancer | USP22 | (80) |
| Oral squamous cell carcinoma | USP22 | (81) |
| Papillary thyroid carcinoma | USP22, USP33 | (82, 83) |
| Salivary duct carcinoma | USP22 | (33) |
| Esophageal squamous cell carcinoma | USP22 | (34) |
| Salivary adenoid cystic carcinoma | USP22 | (34) |
| Bladder cancer | USP28 | (84) |
| Colorectal cancer | USP33, OTUB1, MYSM1 | (85, 86) |
| Prostate cancer | USP39 | (87) |
| Malignant peritoneal mesothelioma | BAP1 | (88) |
| Triple-negative breast cancer | OTUD7B | (89) |
| Pancreatic ductal adenocarcinoma | UCHL5 | (90) |
| Gastric cardiac adenocarcinoma | UCHL1 | (91) |
| Cholangiocarcinoma | UCHL1 | (91) |
| Aggressive multiple myeloma | UCHL1 | (92) |
| Neuronal apoptosis | USP4, UCHL1 | (93, 94) |
| Cardiac hypertrophy | USP4 | (95) |
| Aneurysmal bone cyst | USP6 | (96) |
| Pancreatic beta cells | UCHL1 | (97) |
| Aneurysmal subarachnoid hemorrhage | UCHL1 | (98) |
| Neuronal biomarker | UCHL1 | (98) |
| Traumatic brain injury | UCHL1 | (99) |
| Pancreatic neuroendocrine tumors | UCHL1 | (100) |
ACKNOWLEDGEMENTS
This research was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF), which is funded by the Ministry of Education (2017R 1A2B2008727, 2018M3A9H3022412 and 2017M3A9B3061830) and Bio and Medical Technology Development Program of the National Research Foundation (NRF) and funded by the Korean government (MSIP and MOHW) (2017M3A9E4048172).
Footnotes
CONFLICTS OF INTEREST
The authors have no conflicting interests.
REFERENCES
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics. CA Cancer J Clin. 2012;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Li J, Shen H, Himmel KL. Leukaemia disease genes: large-scale cloning and pathway predictions. Nat Genet. 1999;23:348–353. doi: 10.1038/15531. [DOI] [PubMed] [Google Scholar]
- 3.Hansen GM, Skapura D, Justice MJ. Genetic profile of insertion mutations in mouse leukemias and lymphomas. Genome Res. 2000;10:237–243. doi: 10.1101/gr.10.2.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fishel R, Lescoe MK, Rao MR, et al. The human mutator gene homolog MSH2 and its association with hereditary nonpolyposis colon cancer. Cell. 1993;75:1027–1038. doi: 10.1016/0092-8674(93)90546-3. [DOI] [PubMed] [Google Scholar]
- 5.Sharma A, Alswillah T, Singh K, et al. USP14 regulates DNA damage repair by targeting RNF168-dependent ubiquitination. Autophagy. 2018;14:1976–1990. doi: 10.1080/15548627.2018.1496877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chandrasekaran AP, Suresh B, Kim HH, Kim K-S, Ramakrishna S. Concise Review: Fate Determination of Stem Cells by Deubiquitinating Enzymes. Stem Cells. 2017;35:9–16. doi: 10.1002/stem.2446. [DOI] [PubMed] [Google Scholar]
- 7.Lork M, Verhelst K, Beyaert R. CYLD, A20 and OTULIN deubiquitinases in NF-κB signaling and cell death: so similar, yet so different. Cell Death Differ. 2017;24:1172–1183. doi: 10.1038/cdd.2017.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Darling S, Fielding AB, Sabat-Pośpiech D, Prior IA, Coulson JM. Regulation of the cell cycle and centrosome biology by deubiquitylases. Biochem Soc Trans. 2017;45:1125–1136. doi: 10.1042/BST20170087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jacq X, Kemp M, Martin NMB, Jackson SP. Deubiquitylating Enzymes and DNA Damage Response Pathways. Cell Biochem Biophys. 2013;67:25–43. doi: 10.1007/s12013-013-9635-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nijman SMB, Luna-Vargas MPA, Velds A. A genomic and functional inventory of deubiquitinating enzymes. Cell. 2005;123:773–786. doi: 10.1016/j.cell.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 11.Abdul Rehman SA, Kristariyanto YA, Choi SY, et al. MINDY-1 Is a Member of an Evolutionarily Conserved and Structurally Distinct New Family of Deubiquitinating Enzymes. Mol Cell. 2016;63:146–155. doi: 10.1016/j.molcel.2016.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Priolo C, Tang D, Brahamandan M, et al. The isopeptidase USP2a protects human prostate cancer from apoptosis. Cancer Res. 2006;66:8625–8632. doi: 10.1158/0008-5472.CAN-06-1374. [DOI] [PubMed] [Google Scholar]
- 13.Qu Q, Mao Y, Xiao G, et al. USP2 promotes cell migration and invasion in triple negative breast cancer cell lines. Tumor Biol. 2015;36:5415–5423. doi: 10.1007/s13277-015-3207-7. [DOI] [PubMed] [Google Scholar]
- 14.Zhang L, Zhou F, Drabsch Y, et al. USP4 is regulated by AKT phosphorylation and directly deubiquitylates TGF-β type i receptor. Nat Cell Biol. 2012;14:717–726. doi: 10.1038/ncb2522. [DOI] [PubMed] [Google Scholar]
- 15.Seibold MA, Schwartz DA. The authors reply. N Engl J Med. 2011;364:1503–1512. doi: 10.1056/NEJMoa1013660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xing C, Lu XX, Da Guo P, et al. Ubiquitin-specific protease 4-mediated deubiquitination and stabilization of PRL-3 is required for potentiating colorectal oncogenesis. Cancer Res. 2016;76:83–95. doi: 10.1158/0008-5472.CAN-14-3595. [DOI] [PubMed] [Google Scholar]
- 17.Guo W, Ma J, Pei T, et al. Up-regulated deubiquitinase USP4 plays an oncogenic role in melanoma. J Cell Mol Med. 2018;22:2944–2954. doi: 10.1111/jcmm.13603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li T, Yan B, Ma Y, et al. Ubiquitin-specific protease 4 promotes hepatocellular carcinoma progression via cyclophilin A stabilization and deubiquitination. Cell Death Dis. 2018;9:148–165. doi: 10.1038/s41419-017-0182-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cao WH, Liu XP, Meng SL, et al. USP4 promotes invasion of breast cancer cells via Relaxin/TGF-β1/Smad2/MMP-9 signal. Eur Rev Med Pharmacol Sci. 2016;20:1115–1122. [PubMed] [Google Scholar]
- 20.Hicke L. Protein regulation by monoubiquitin. Nat Rev Mol Cell Biol. 2001;2:195–201. doi: 10.1038/35056583. [DOI] [PubMed] [Google Scholar]
- 21.Tang B, Tang F, Li B, et al. High USP22 expression indicates poor prognosis in hepatocellular carcinoma. Oncotarget. 2015;6:12654–12667. doi: 10.18632/oncotarget.3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu Y, Wang W, Lu Y, et al. Usp5 functions as an oncogene for stimulating tumorigenesis in hepatocellular carcinoma. 2017;8:50655–50664. doi: 10.18632/oncotarget.16901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li XY, Wu HY, Mao XF, Jiang LX, Wang YX. USP5 promotes tumorigenesis and progression of pancreatic cancer by stabilizing FoxM1 protein. Biochem Biophys Res Commun. 2017;492:48–54. doi: 10.1016/j.bbrc.2017.08.040. [DOI] [PubMed] [Google Scholar]
- 24.Cheon KW, Baek KH. HAUSP as a therapeutic target for hematopoietic tumors (Review) Int J Oncol. 2006;28:1209–1215. [PubMed] [Google Scholar]
- 25.Zhang L, Wang H, Tian L, Li H. Expression of USP7 and MARCH7 Is Correlated with Poor Prognosis in Epithelial Ovarian Cancer. Tohoku J Exp Med. 2016;239:165–175. doi: 10.1620/tjem.239.165. [DOI] [PubMed] [Google Scholar]
- 26.Wang X, Zhang Q, Wang Y, Zhuang H, Chen B. Clinical Significance of Ubiquitin Specific Protease 7 (USP7) in Predicting Prognosis of Hepatocellular Carcinoma and its Functional Mechanisms. Med Sci Monit. 2018;24:1742–1750. doi: 10.12659/MSM.909368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang C, Lu J, Zhang Q-W, et al. USP7 promotes cell proliferation through the stabilization of Ki-67 protein in non-small cell lung cancer cells. Int J Biochem Cell Biol. 2016;79:209–221. doi: 10.1016/j.biocel.2016.08.025. [DOI] [PubMed] [Google Scholar]
- 28.Fraile JM, Quesada V, Rodríguez D, Freije JMP, López-Otín C. Deubiquitinases in cancer: New functions and therapeutic options. Oncogene. 2012;31:2373–2388. doi: 10.1038/onc.2011.443. [DOI] [PubMed] [Google Scholar]
- 29.Xia R, Jia H, Fan J, Liu Y, Jia J. USP8 promotes smoothened signaling by preventing its ubiquitination and changing its subcellular localization. PLoS Biol. 2012;10:e1001238. doi: 10.1371/journal.pbio.1001238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mukai A, Yamamoto-Hino M, Awano W, Watanabe W, Komada M, Goto S. Balanced ubiquitylation and deubiquitylation of Frizzled regulate cellular responsiveness to Wg/Wnt. EMBO J. 2010;29:2114–2225. doi: 10.1038/emboj.2010.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yan M, Zhao C, Wei N, Wu X, Cui J, Xing Y. High Expression of Ubiquitin-Specific Protease 8 (USP8) Is Associated with Poor Prognosis in Patients with Cervical Squamous Cell Carcinoma. Med Sci Monit. 2018;24:4934–4943. doi: 10.12659/MSM.909235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Soncini C, Berdo I, Draetta G. Ras-GAP SH3 domain binding protein (G3BP) is a modulator of USP10. Oncogene. 2001;20:3869–3879. doi: 10.1038/sj.onc.1204553. [DOI] [PubMed] [Google Scholar]
- 33.Piao S, Ma J, Wang W, et al. Increased expression of USP22 is associated with disease progression and patient prognosis of salivary duct carcinoma. Oral Oncol. 2013;49:796–801. doi: 10.1016/j.oraloncology.2013.03.454. [DOI] [PubMed] [Google Scholar]
- 34.Li J, Wang Z, Li Y. USP22 nuclear expression is significantly associated with progression and unfavorable clinical outcome in human esophageal squamous cell carcinoma. J Cancer Res Clin Oncol. 2012;138:1291–1297. doi: 10.1007/s00432-012-1191-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Akhavantabasi S, Akman HB, Sapmaz A, Keller J, Petty EM, Erson AE. USP32 is an active, membrane-bound ubiquitin protease overexpressed in breast cancers. Mamm Genome. 2010;21:388–397. doi: 10.1007/s00335-010-9268-4. [DOI] [PubMed] [Google Scholar]
- 36.Hu W, Wei H, Li K, Li P, Lin J, Feng R. Downregulation of USP32 inhibits cell proliferation, migration and invasion in human small cell lung cancer. Cell Prolif. 2017;50:e12343. doi: 10.1111/cpr.12343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Baietti MF, Simicek M, Abbasi Asbagh L, et al. OTUB1 triggers lung cancer development by inhibiting RAS monoubiquitination. EMBO Mol Med. 2016;8:288–303. doi: 10.15252/emmm.201505972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stanišić V, Malovannaya A, Qin J, Lonard DM, O’Malley BW. OTU Domain-containing Ubiquitin Aldehyde-binding Protein 1 (OTUB1) Deubiquitinates Estrogen Receptor (ER) α and Affects ERα Transcriptional Activity. J Biol Chem. 2009;284:16135–16145. doi: 10.1074/jbc.M109.007484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang Y, Hu R, Wu H, et al. OTUB1 Overexpression in Mesangial Cells Is a Novel Regulator in the Pathogenesis of Glomerulonephritis through the Decrease of DCN Level. PLoS One. 2012;7:e29654. doi: 10.1371/journal.pone.0029654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weng W, Zhang Q, Xu M, et al. OTUB1 promotes tumor invasion and predicts a poor prognosis in gastric adenocarcinoma. Am J Transl Res. 2016;8:2234–2244. [PMC free article] [PubMed] [Google Scholar]
- 41.Xu L, Li J, Bao Z, et al. Silencing of OTUB1 inhibits migration of human glioma cells in vitro. Neuropathology. 2017;37:217–226. doi: 10.1111/neup.12366. [DOI] [PubMed] [Google Scholar]
- 42.Catrysse L, Vereecke L, Beyaert R, van Loo G. A20 in inflammation and autoimmunity. Trends Immunol. 2014;35:22–31. doi: 10.1016/j.it.2013.10.005. [DOI] [PubMed] [Google Scholar]
- 43.Lee JH, Jung SM, Yang KM, et al. A20 promotes metastasis of aggressive basal-like breast cancers through multi-monoubiquitylation of Snail1. Nat Cell Biol. 2017;19:1260–1273. doi: 10.1038/ncb3609. [DOI] [PubMed] [Google Scholar]
- 44.Ndubaku C, Tsui V. Inhibiting the deubiquitinating enzymes (DUBs) J Med Chem. 2015;58:1581–1595. doi: 10.1021/jm501061a. [DOI] [PubMed] [Google Scholar]
- 45.D’Arcy P, Wang X, Linder S. Deubiquitinase inhibition as a cancer therapeutic strategy. Pharmacol Ther. 2015;147:32–54. doi: 10.1016/j.pharmthera.2014.11.002. [DOI] [PubMed] [Google Scholar]
- 46.Kaushal K, Antao AM, Kim KS, Ramakrishna S. Deubiquitinating enzymes in cancer stem cells: functions and targeted inhibition for cancer therapy. Drug Discov Today. 2018;23:1974–1982. doi: 10.1016/j.drudis.2018.05.035. [DOI] [PubMed] [Google Scholar]
- 47.Komander D. The emerging complexity of protein ubiquitination. Biochem Soc Trans. 2009;37:937–953. doi: 10.1042/BST0370937. [DOI] [PubMed] [Google Scholar]
- 48.Love KR, Catic A, Schlieker C, Ploegh HL. Mechanisms, biology and inhibitors of deubiquitinating enzymes. Nat Chem Biol. 2007;3:697–705. doi: 10.1038/nchembio.2007.43. [DOI] [PubMed] [Google Scholar]
- 49.Brnjic S, Mazurkiewicz M, Fryknäs M, et al. Induction of tumor cell apoptosis by a proteasome deubiquitinase inhibitor is associated with oxidative stress. Antioxid Redox Signal. 2014;21:2271–2285. doi: 10.1089/ars.2013.5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Harrigan JA, Jacq X, Martin NM, et al. Deubiquitylating enzymes and drug discovery: emerging opportunities. Nat Rev Drug Discov. 2017;17:57–78. doi: 10.1038/nrd.2017.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Okada K, Ye YQ, Taniguchi K, et al. Vialinin A is a ubiquitin-specific peptidase inhibitor. Bioorg Med Chem Lett. 2013;23:4328–4331. doi: 10.1016/j.bmcl.2013.05.093. [DOI] [PubMed] [Google Scholar]
- 52.Yang J, Xu P, Han L, et al. Cutting Edge: Ubiquitin-Specific Protease 4 Promotes Th17 Cell Function under Inflammation by Deubiquitinating and Stabilizing RORγt. J Immunol. 2015;194:4094–4097. doi: 10.4049/jimmunol.1401451. [DOI] [PubMed] [Google Scholar]
- 53.Gavory G, O’dowd C, McClelland K, et al. Abstract LB-257: Discovery and characterization of novel, highly potent and selective USP7 inhibitors. Cancer Res. 2015;75:15. doi: 10.1158/1538-7445.AM2015-LB-257. [DOI] [Google Scholar]
- 54.Reverdy C, Conrath S, Lopez R, et al. Discovery of Specific Inhibitors of Human USP7/HAUSP Deubiquitinating Enzyme. Chem Biol. 2012;19:467–477. doi: 10.1016/j.chembiol.2012.02.007. [DOI] [PubMed] [Google Scholar]
- 55.Colland F, Formstecher E, Jacq X, et al. Small-molecule inhibitor of USP7/HAUSP ubiquitin protease stabilizes and activates p53 in cells. Mol Cancer Ther. 2009;8:2286–2295. doi: 10.1158/1535-7163.MCT-09-0097. [DOI] [PubMed] [Google Scholar]
- 56.Weinstock J, Wu J, Cao P, et al. Selective Dual Inhibitors of the Cancer-Related Deubiquitylating Proteases USP7 and USP47. ACS Med Chem Lett. 2012;3:789–792. doi: 10.1021/ml200276j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kapuria V, Peterson LF, Fang D, Bornmann WG, Talpaz M, Donato NJ. Deubiquitinase Inhibition by Small-Molecule WP1130 Triggers Aggresome Formation and Tumor Cell Apoptosis. Cancer Res. 2010;70:9265–9276. doi: 10.1158/0008-5472.CAN-10-1530. [DOI] [PubMed] [Google Scholar]
- 58.Burkhart RA, Peng Y, Norris ZA, et al. Mitoxantrone Targets Human Ubiquitin-Specific Peptidase 11 (USP11) and Is a Potent Inhibitor of Pancreatic Cancer Cell Survival. Mol Cancer Res. 2013;11:901–911. doi: 10.1158/1541-7786.MCR-12-0699. [DOI] [PubMed] [Google Scholar]
- 59.Lee B-H, Lee MJ, Park S, et al. Enhancement of proteasome activity by a small-molecule inhibitor of USP14. Nature. 2010;467:179–184. doi: 10.1038/nature09299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Deng H, O’Keefe H, Davie CP, et al. Discovery of Highly Potent and Selective Small Molecule ADAMTS-5 Inhibitors That Inhibit Human Cartilage Degradation via Encoded Library Technology (ELT) J Med Chem. 2012;55:7061–7079. doi: 10.1021/jm300449x. [DOI] [PubMed] [Google Scholar]
- 61.Yue W, Chen Z, Liu H, et al. A small natural molecule promotes mitochondrial fusion through inhibition of the deubiquitinase USP30. Cell Res. 2014;24:482–496. doi: 10.1038/cr.2014.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tian Z, D’Arcy P, Wang X, et al. A novel small molecule inhibitor of deubiquitylating enzyme USP14 and UCHL5 induces apoptosis in multiple myeloma and overcomes bortezomib resistance. Blood. 2014;123:706–716. doi: 10.1182/blood-2013-05-500033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gu Y, Ding X, Huang J, et al. The deubiquitinating enzyme UCHL1 negatively regulates the immunosuppressive capacity and survival of multipotent mesenchymal stromal cells. Cell Death Dis. 2018;9:459. doi: 10.1038/s41419-018-0532-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.D’Arcy P, Wang X, Linder S. Deubiquitinase inhibition as a cancer therapeutic strategy. Pharmacol Ther. 2015;147:32–54. doi: 10.1016/j.pharmthera.2014.11.002. [DOI] [PubMed] [Google Scholar]
- 65.Lei H, Shan H, Wu Y. Targeting deubiquitinating enzymes in cancer stem cells. Cancer Cell Int. 2017;17:101. doi: 10.1186/s12935-017-0472-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zou Y, Qiu G, Jiang L, et al. Overexpression of ubiquitin specific proteases 44 promotes the malignancy of glioma by stabilizing tumor-promoter securin. Oncotarget. 2017;8:58231–58246. doi: 10.18632/oncotarget.16447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Boustani MR, Khoshnood RJ, Nikpasand F, et al. Overexpression of ubiquitin-specific protease 2a (USP2a) and nuclear factor erythroid 2-related factor 2 (Nrf2) in human gliomas. J Neurol Sci. 2016;363:249–252. doi: 10.1016/j.jns.2016.03.003. [DOI] [PubMed] [Google Scholar]
- 68.Zhang QX, Wang XC, Chen SP, Qin XT. Predictive value of deubiquitination enzymes USP37 in the prognosis of breast cancer. Zhonghua Yi Xue Za Zhi. 2016;96:944–948. doi: 10.3760/cma.j.issn.0376-2491.2016.12.008. [DOI] [PubMed] [Google Scholar]
- 69.Qu Q, Mao Y, Xiao G, et al. USP2 promotes cell migration and invasion in triple negative breast cancer cell lines. Tumor Biol. 2015;36:5415–5423. doi: 10.1007/s13277-015-3207-7. [DOI] [PubMed] [Google Scholar]
- 70.Ni Q, Chen J, Li X, et al. Expression of OTUB1 in hepatocellular carcinoma and its effects on HCC cell migration and invasion. Acta Biochim Biophys Sin (Shanghai) 2017;49:680–688. doi: 10.1093/abbs/gmx056. [DOI] [PubMed] [Google Scholar]
- 71.Liu R, Zhao D, Zhang X, et al. A20 enhances the radiosensitivity of hepatocellular carcinoma cells to 60Co-γ ionizing radiation. Oncotarget. 2017;8:93103–93116. doi: 10.18632/oncotarget.21860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yao R, Pu J, Fan R, et al. Ubiquitin-specific protease 4 improves the prognosis of the patients in esophageal cancer. Cancer Biomarkers. 2017;20:317–323. doi: 10.3233/CBM-170308. [DOI] [PubMed] [Google Scholar]
- 73.Baykara M, Yaman M, Buyukberber S, et al. Clinical and prognostic importance of XIAP and USP8 in advanced stages of non-small cell lung cancer. J BUON. 2009;18:921–927. [PubMed] [Google Scholar]
- 74.Kim Y, Shiba-Ishii A, Nakagawa T, et al. Ubiquitin-specific protease 8 is a novel prognostic marker in early-stage lung adenocarcinoma. Pathol Int. 2017;67:292–301. doi: 10.1111/pin.12546. [DOI] [PubMed] [Google Scholar]
- 75.Zeng Z, Wu HX, Zhan N, et al. Prognostic significance of USP10 as a tumor-associated marker in gastric carcinoma. Tumor Biol. 2014;35:3845–3853. doi: 10.1007/s13277-013-1509-1. [DOI] [PubMed] [Google Scholar]
- 76.Zhang B, Wang H, Yang L, et al. OTUD7B and NIK expression in non-small cell lung cancer: Association with clinicopathological features and prognostic implications. Pathol Res Pract. 2016;212:893–898. doi: 10.1016/j.prp.2016.07.011. [DOI] [PubMed] [Google Scholar]
- 77.McFarlane C, McFarlane S, Paul I, et al. The deubiquitinating enzyme USP17 is associated with non-small cell lung cancer (NSCLC) recurrence and metastasis. Oncotarget. 2013;4:1836–1843. doi: 10.18632/oncotarget.1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Peng L, Hu Y, Chen D, Jiao S, Sun S. Ubiquitin specific peptidase 21 regulates interleukin-8 expression, stem-cell like property of human renal cell carcinoma. Oncotarget. 2016;7:42007–42016. doi: 10.18632/oncotarget.9751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kim Y-H, Kim WT, Jeong P, et al. Novel Combination Markers for Predicting Survival in Patients with Muscle Invasive Bladder Cancer: USP18 and DGCR2. J Korean Med Sci. 2014;29:351. doi: 10.3346/jkms.2014.29.3.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yang M, Liu YD, Wang YY, Liu TB, Ge TT, Lou G. Ubiquitin-specific protease 22: a novel molecular biomarker in cervical cancer prognosis and therapeutics. Tumor Biol. 2014;35:929–934. doi: 10.1007/s13277-013-1121-4. [DOI] [PubMed] [Google Scholar]
- 81.Piao S, Liu Y, Hu J, et al. USP22 Is Useful as a Novel Molecular Marker for Predicting Disease Progression and Patient Prognosis of Oral Squamous Cell Carcinoma. PLoS One. 2012;7:e42540. doi: 10.1371/journal.pone.0042540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jia M, Guo Y, Lu X. USP33 is a Biomarker of Disease Recurrence in Papillary Thyroid Carcinoma. Cell Physiol Biochem. 2018;45:2044–2053. doi: 10.1159/000488041. [DOI] [PubMed] [Google Scholar]
- 83.Wang H, Li YP, Chen JH, et al. Prognostic significance of USP22 as an oncogene in papillary thyroid carcinoma. Tumor Biol. 2013;34:1635–1639. doi: 10.1007/s13277-013-0696-0. [DOI] [PubMed] [Google Scholar]
- 84.Guo G, Xu Y, Gong M, Cao Y, An R. USP28 is a potential prognostic marker for bladder cancer. Tumor Biol. 2014;35:4017–4022. doi: 10.1007/s13277-013-1525-1. [DOI] [PubMed] [Google Scholar]
- 85.Li Y, Li J, Liu H, Liu Y, Cui B. Expression of MYSM1 is associated with tumor progression in colorectal cancer. PLoS One. 2017;12:e0177235. doi: 10.1371/journal.pone.0177235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Liu H, Zhang Q, Li K, et al. Prognostic significance of USP33 in advanced colorectal cancer patients: new insights into arrestin-dependent ERK signaling. Oncotarget. 2016;7:81223–81240. doi: 10.18632/oncotarget.13219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Huang Y, Pan XW, Li L, et al. Overexpression of USP39 predicts poor prognosis and promotes tumorigenesis of prostate cancer via promoting EGFR mRNA maturation and transcription elongation. Oncotarget. 2016;7:22016–22030. doi: 10.18632/oncotarget.7882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Testa JR, Cheung M, Pei J, et al. Germline BAP1 mutations predispose to malignant mesothelioma. Nat Genet. 2011;43:1022–1025. doi: 10.1038/ng.912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chiu HW, Lin HY, Tseng IJ, Hsiao M, Lin YF. OTUD7B upregulation predicts a poor response to paclitaxel in patients with triple-negative breast cancer. Oncotarget. 2018;9:553–565. doi: 10.18632/oncotarget.23074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Arpalahti L, Saukkonen K, Hagström J, et al. Nuclear ubiquitin C-terminal hydrolase L5 expression associates with increased patient survival in pancreatic ductal adenocarcinoma. Tumor Biol. 2017;39 doi: 10.1177/1010428317710411. 101042831771041. [DOI] [PubMed] [Google Scholar]
- 91.Yang H, Zhang C, Fang S, Ou R, Li W, Xu Y. UCH-LI acts as a novel prognostic biomarker in gastric cardiac adenocarcinoma. Int J Clin Exp Pathol. 2015;8:13957–13967. [PMC free article] [PubMed] [Google Scholar]
- 92.Hussain S, Bedekovics T, Chesi M, Bergsagel PL, Galardy PJ. UCHL1 is a biomarker of aggressive multiple myeloma required for disease progression. Oncotarget. 2015;6:40704–40718. doi: 10.18632/oncotarget.5727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kwon J, Mochida K, Wang YL, et al. Ubiquitin C-Terminal Hydrolase L-1 Is Essential for the Early Apoptotic Wave of Germinal Cells and for Sperm Quality Control During Spermatogenesis. Biol Reprod. 2005;73:29–35. doi: 10.1095/biolreprod.104.037077. [DOI] [PubMed] [Google Scholar]
- 94.Liu C, Liu C, Liu H, et al. Increased Expression of Ubiquitin-Specific Protease 4 Participates in Neuronal Apoptosis After Intracerebral Hemorrhage in Adult Rats. Cell Mol Neurobiol. 2017;37:427–435. doi: 10.1007/s10571-016-0375-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.He B, Zhao YC, Gao LC, et al. Ubiquitin-Specific Protease 4 Is an Endogenous Negative Regulator of Pathological Cardiac Hypertrophy. Hypertension. 2016;67:1237–1248. doi: 10.1161/HYPERTENSIONAHA.116.07392. [DOI] [PubMed] [Google Scholar]
- 96.Oliveira AM, Chou MM. USP6-induced neoplasms: the biologic spectrum of aneurysmal bone cyst and nodular fasciitis. Hum Pathol. 2014;45:1–11. doi: 10.1016/j.humpath.2013.03.005. [DOI] [PubMed] [Google Scholar]
- 97.Brackeva B, De Punt V, Kramer G, et al. Potential of UCHL1 as biomarker for destruction of pancreatic beta cells. J Proteomics. 2015;117:156–167. doi: 10.1016/j.jprot.2015.01.009. [DOI] [PubMed] [Google Scholar]
- 98.Lewis SB, Wolper R, Chi YY, et al. Identification and preliminary characterization of ubiquitin C terminal hydrolase 1 (UCHL1) as a biomarker of neuronal loss in aneurysmal subarachnoid hemorrhage. J Neurosci Res. 2010;88:1475–1484. doi: 10.1002/jnr.22323. [DOI] [PubMed] [Google Scholar]
- 99.Papa L, Akinyi L, Liu MC, et al. Ubiquitin C-terminal hydrolase is a novel biomarker in humans for severe traumatic brain injury. Crit Care Med. 2010;38:138–144. doi: 10.1097/CCM.0b013e3181b788ab. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Song YL, Yu R, Qiao X-W, et al. Prognostic relevance of UCH-L1 and α-internexin in pancreatic neuroendocrine tumors. Sci Rep. 2017;7:2205. doi: 10.1038/s41598-017-02051-1. [DOI] [PMC free article] [PubMed] [Google Scholar]