SUMMARY
Microcrystal electron diffraction (MicroED) allows for macromolecular structure solution from nanocrystals. To create crystals of suitable size for MicroED data collection, sample preparation typically involves sonication or pipetting a slurry of crystals from a crystallization drop. The resultant crystal fragments are fragile and the quality of the data that can be obtained from them is sensitive to subsequent sample preparation for cryoelectron microscopy as interactions in the water-air interface can damage crystals during blotting. Here, we demonstrate the use of a focused ion beam to generate lamellae of macromolecular protein crystals for continuous rotation MicroED that are of ideal thickness, easy to locate, and require no blotting optimization. In this manner, crystals of nearly any size may be scooped and milled to desired dimensions prior to data collection, thus streamlining the methodology for sample preparation for MicroED.
Graphical Abstract
In Brief
Large crystals are machined to ideal thickness for structural investigation by MicroED. The structure of a macromolecular protein crystal is solved using continuous rotation MicroED data collected from a single prepared crystalline lamella, with a total electron exposure of less than 4 e−Å−2.
INTRODUCTION
The cryoelectron microscopy (cryo-EM) method microcrystal electron diffraction (MicroED) is a relatively new method with which structures of macromolecular assemblies are solved to atomic resolution from nanocrystals about one billion times smaller than what is typically used for X-ray crystallography (Martynowycz and Gonen, 2018; Nannenga et al., 2014; Shi et al., 2013; Vergara et al., 2017). These nanocrystals either grow spontaneously in sparse matrix crystallization trays or, when large crystals are obtained, can be sonicated or broken down to small crystallite fragments that are then used for MicroED (de la Cruz et al., 2017). Once good samples are available, grid preparation for cryo-EM is arguably the most critical step before high-quality data can be collected. Typically, the crystallite solution is pipetted onto the holey carbon grid, excess solution is blotted away, and the preparation plunged into ethane to create a layer of thin, amorphous ice embedding the crystallites. Many things can go wrong during this process. For example, the blotting might mechanically destroy the crystals or could be inefficient because the crystals are embedded in a viscous solution leading to very thick preparations. Finally, the ice thickness can affect the hydration of the crystals and may deteriorate the underlying crystalline lattice (Agard et al., 2014; Taylor and Glaeser, 1974).
Electrons interact with matter more strongly than X-rays. This means that MicroED may be used to solve structures from vanishingly small crystals, but also implies that larger crystals (>1 μm thick) cannot be efficiently penetrated by the electron beam. This is unfortunate because many samples yield crystals that are too small for routine X-ray diffraction (1–20 μm) and yet are too thick for MicroED. Moreover, the vitreous ice and amorphous carbon support contribute to the diffuse background in diffraction experiments, leading to higher background noise. It would be ideal to simply shave large crystals to the desired thickness and to remove the excess ice and carbon, leaving only a thin crystal behind and this could be achieved with milling using a focused ion beam (FIB) in a scanning electron microscope (SEM). Milling away all excess material could be used to reliably make samples and would constitute a new paradigm in structure determination with MicroED.
Some MicroED data were collected recently from milled, fragmented crystals (Duyvesteyn et al., 2018). In that report, nano-crystals were prepared as typical for MicroED (de la Cruz et al., 2017; Nannenga et al., 2014; Shi et al., 2013) after which the crystals were milled to thin lamellae and used for MicroED still diffraction experiments. Since very high accumulated dose and only still diffraction was used, the data had very low completeness (Duyvesteyn et al., 2018), and the model was only refined as a rigid body against the merged intensities. Importantly, the observation was made that the milling did not appear to severely damage the underlying lattice, as some diffraction spots were observed out to 1.9Å during the first few frames of the measurement.
Here we demonstrate that large crystals, several tens of micro-meters in thickness, can be scooped directly onto an EM grid, some excess solution gently blotted manually before flash-freezing in ethane. The surrounding ice and embedded crystals are then milled using an ion beam down to any desirable thickness for analysis by continuous rotation MicroED. We show that by using this approach we could solve the structure of proteinase K from a single milled crystal with high completeness and good refinement statistics. This approach considerably widens the scope of crystals suitable for MicroED, streamlines sample preparation, and decreases the time spent screening for crystals on the grid.
RESULTS
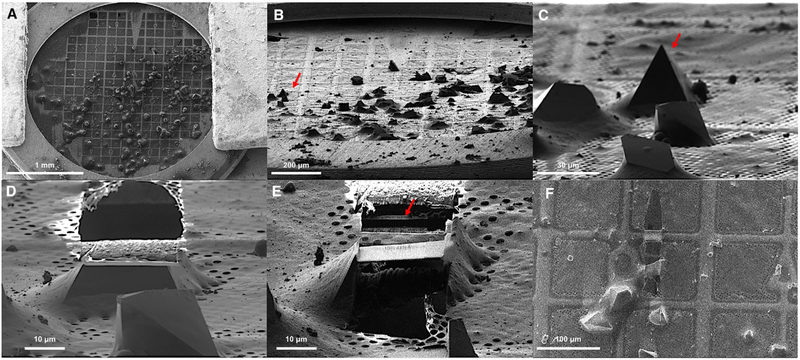
Large proteinase K crystals were grown by vapor diffusion as typically done for X-ray crystallography (Hattne et al., 2016). We took an entire 4-μL drop of crystals and transferred all of it onto a freshly glow-discharged holey carbon grid. These grids were vitrified by plunging into supercooled ethane, and transferred to an FEI Versa FIB/SEM for milling without platinum coating. An overview of the grid in the SEM and by the FIB showed the grid overlaid with many large crystals of varying size (between ~5 and 100 μm) (Figures 1A–1C), all much too large for MicroED. An ideal specimen for milling was identified in low-magnification SEM and FIB imaging by looking for a very large, sharp-edged crystals surrounded by amorphous ice that were located away from the copper grid bar (Figures 1B and 1C). Crystals like these were easily found on every TEM grid tested, allowing the milling of several lamellae on each grid. It should be noted that for this demonstration we were deliberately targeting very large crystals that typically would not be amenable for analysis by MicroED (for example, Figure 1C, arrow).
Figure 1. Preparation of Macromolecular Protein Crystals for MicroED by Focused ion Beam Milling.
(A) SEM image of a 3-mm TEM grid.
(B) FIB image of the grid at 20° tilt.
(C) FIB image of select crystals at high magnification. The arrow indicates the crystal that was milled.
(D) FIB image of select crystal from (C) after milling the top of the crystal.
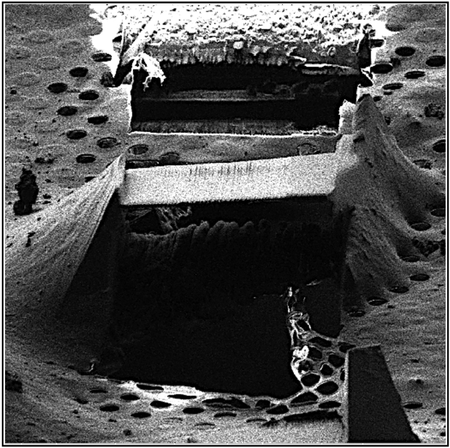
(E) FIB image of crystal after milling and cleaning both the top and bottom of the crystal leaving a 300-nm-thick lamella indicated by an arrow.
(F) SEM image of the crystal lamella at high magnification.
A focused beam of gallium ions accelerated at 30 kV was used for milling. The surrounding media and the excess crystalline area was initially gross-milled using a beam current of 300 pA to clear away the large, thick, unwanted volumes above or below the sample to a thickness of approximately 1–3 μm (Figures 1D and 1E). Subsequent reductions in the size of the crystal lamella came by milling away equal volumes above and below the initially milled volume with reduced current in a step-by-step fashion as reported previously (Mahamid et al., 2016; Zhang et al., 2016). The final lamella was approximately 300 nm thick, with the final volumes removed with a gallium beam current of 30 pA (Figure 1E). The process of milling was monitored with SEM operated at eitehr 5 kV or 10 kV and 27 pA (Figure 1F). The total time to mill a crystal of this size was approximately 10 min.
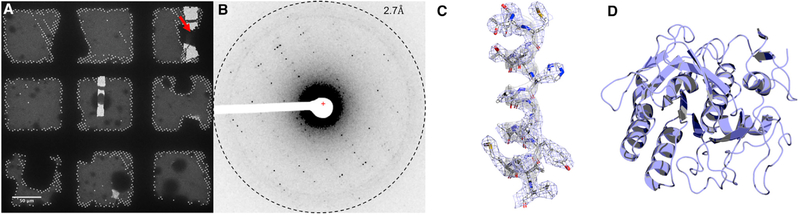
Following milling, the grids were transferred to an FEI Talos Artica 200-kV electron microscope equipped with a bottom-mount CetaD CMOS detector for MicroED data collection. Location of the crystalline lamellae after milling was apparent in low-magnification images by direct search or the inspection of a grid atlas (Figure 2A). The milled lamellae clearly stood out compared with un-milled regions and the milled crystals appeared as dark particles surrounded by the bright milled region (Figure 2A, arrow).
Figure 2. MicroED Structure of Proteinase K Determined from a Single Milled Crystal.
(A) Identification of milled crystals in low-magnification TEM. Two lamellae are easily identified, with an arrow indicating crystal from Figure 1. Scale bar is 50 μm.
(B) Single diffraction pattern from crystal in (Figures 1 and 2A).
(C) 2Fo − Fc map of the central helix (residues 223–240) contoured at 1.5 σ above the mean, carved at 2 Å from the atomic centers for clarity.
(D) Final structure of proteinase K.
Continuous rotation MicroED data were collected as described previously (Hattne et al., 2015; Nannenga et al., 2014). The milled crystals yielded diffraction to ~2.5Å resolution (Figure 2B), indicating that the milling process did not damage the underlying crystalline material. A complete dataset was collected from a single milled crystal by continuous rotation MicroED, where a single lamella was continuously rotated while the diffraction was collected as a movie in rolling shutter mode on the CetaD detector. The total exposure during the MicroED experiments was approximately 4 e− Å −2 (total dose) with a dose rate of 0.02 e− Å −2 s−1 and a rotation speed of 0.3°per second. The entire dataset was collected in less than 5 min. The structure of proteinase K was solved by molecular replacement and refined to a final resolution of 2.75Å with acceptable Rwork and Rfree of 24% and 26%, respectively. A full description of data collection and statistics of the model is given in Table 1, and in the STAR Methods sections.
Table 1.
MicroED Data Collection and Analysis of a Single, Milled Crystal
| Data | Parameter | Measure |
|---|---|---|
| Data collection | accelerating voltage (kV) | 200 |
| electron source | field emission gun | |
| total accumulated exposure (e− Å−2) | <4 | |
| no. of crystals | 1 | |
| microscope | Thermo Fischer Talos Arctica | |
| camera | CetaD | |
| rotation rate (°/s) | 0.297 | |
| wavelength (Å) | 0.0251 | |
| Data analysis |
resolution range (Å) | 43.55–2.75 (2.849–2.75) |
| space group | P 43 21 2 | |
| Unit cell | ||
| a, b, c (Å) | 67.46, 67.46, 106.67 | |
| α, β, γ (°) | 90, 90, 90 | |
| total no. of reflections | 31,013 (4,613) | |
| multiplicity | 2.9 (2.9) | |
| completeness (%) | 88.4 (88.7) | |
| mean l/σ(l) | 2.06 (0.72) | |
| Wilson B factor | 32.53 | |
| Rpim | 0.2084 | |
| CC1/2 | 0.889 | |
| Rwork | 0.2377 | |
| Rfree | 0.2626 |
DISCUSSION
We demonstrate that a complete, fully refined structure of proteinase K can be solved by continuous rotation MicroED (Nannenga et al., 2014) from a single milled crystal. The milling process did not appear to severely damage the underlying structure of the crystalline lattice. The only electron dose potentially absorbed by the lamella during FIB/SEM comes from the final image taken at low magnification used to view the lamella (Figure 1F). This dose should be surface-limited as the penetration depth of ~5-keV electrons is only a few nanometers, suggesting that any irradiation by low-energy electrons prior to milling is irrelevant (Egerton et al., 2004; Hattne et al., 2018; Matsukawa et al., 1973). Effects due to the FIB imaging or the stress from physically milling away portions of the crystal did not manifest themselves in the final density. However, these effects may have led to a reduction of the observed resolution, or global damage to the lattice, limiting the resolution to ~2.7Å, whereas previously we determined the structure of proteinase K by MicroED to 1.8Å resolution (de la Cruz et al., 2017). Gallium is expected to absorb into solid surfaces at grazing incidence angles, and might have been incorporated into the crystal similarly to gas- or liquid-phase soaking in isomorphous replacement experiments (Giannuzzi and Stevie, 1999). While gallium was not observed in the final density map, ion absorption may be a possibility for thinner lamellae. Both the effects of radiation damage and absorption of gallium into the crystal should be investigated in the future.
By employing continuous rotation MicroED (Nannenga et al., 2014) and a sensitive CetaD camera, the total exposure of the crystalline lamella in this study was limited to <4 e− Å −2 total dose. This ultra-low dose leads to the solution of proteinase K with an overall completeness of ~90% throughout all the resolution shells. This is in contrast with recent reports demonstrating milling of crystals for MicroED. In the first, a high exposure of 25 e− Å −2 total dose was used (Duyvesteyn et al., 2018) resulting in a structure with low completeness (39%) and multiplicity (0.82). In the second, an exposure of up to 9 e− Å −2 total dose was used, and data from seven milled crystals combined resulting in a structure with very poor refinement statistics (Li et al., 2018). It is not clear why low completeness (Duyvesteyn et al., 2018) and poor refinement statistics (Li et al., 2018) were reported, but we suspect that the high exposures limited the quality and/or range of data collected (Hattne et al., 2018). In fact severe damage was observed in the reported experiment (Duyvesteyn et al., 2018), although 1.9Å resolution was recorded at the start of the exposure. Moreover, it is likely that the accuracy of the merged intensities was further limited since both studies (Duyvesteyn et al., 2018; Li et al., 2018) used still diffraction (Shi et al., 2013) rather than continuous rotation MicroED (Nannenga et al., 2014).
The results described here show that large crystals several tens of microns thick can be milled by an FIB to only 100–300 nm allowing structure determination by continuous rotation MicroED from crystals that, before milling, were otherwise not suitable for MicroED. Importantly, it appears that the underlying crystalline material was not severely affected by the FIB milling. This study broadens the capabilities of MicroED because crystals that are too fragile to survive fragmentation methods or harsh blotting conditions could become amenable for data collection after FIB milling. Likewise, milling of crystals grown in lipidic cubic phase or other viscous solvents may now be possible. Many samples that yield crystals that are too small for X-ray analysis and too big for MicroED could now be shaved down to a size allowing MicroED data collection. We believe this method greatly increases the potential scope of what can be done with MicroED, and adds a level of tunable control to all future MicroED experiments.
STAR⋆METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Tamir Gonen (tgonen@ucla.edu).
METHOD DETAILS
Protein Crystallization
Proteinase K (E. album) was purchased from Sigma and used without further purification. Crystals were grown in 24-well trays by sitting drop vapor diffusion using micro bridges (Hamilton). Drops were formed by combining 2μL of mother liqueur (1.25M ammonium sulfate 50mM Tris-HCl pH 8.5) and 2μL of protein solution at 20 mg ml−1 in 50mM Tris-HCl pH 8.5.
Protein crystals were taken directly from the drop using a 10μl pipette with the tip cut to increase its width. Approximately 4μl of protein solution was applied to the carbon side of a Quantifoil R 2/2 200 mesh finder grid (Ted Pella) in 100% humidity inside an FEI Vitrobot Mark IV after glow discharging for 60s at 15μA. Excess water on the grids was removed by gently pressing filter paper to the copper back side for 10s. Grids were then plunged directly into super cooled ethane without further blotting. Grids in ethane were transferred to liquid nitrogen and stored in a dewar until use.
Scanning Electron Microscopy (SEM) and Focused Ion Beam (FIB) Milling
Grids were clipped and loaded into a FEI Versa FIB/SEM with a cryo-transfer system (PP3010T, Quorum Technologies). During FIB milling and transfer, samples were kept at liquid nitrogen temperature. SEM and FIB images were taken at various magnifications using dwell times between 100ns and 1μs. For milling, the gallium beam was accelerated by a voltage of 30kV and the stage was tilted at angles of 17°−20°. The milling current was gradually reduced from 300pA in the first round to 30pA as lamellae were thinned to their final thicknesses. A SEM with 5 kV and 27 pA was used to monitor the milling progress.
MicroED Data Collection and Processing
Grids were loaded from the FIB/SEM to an FEI Talos Artica operating at liquid nitrogen temperatures. Lamellae were identified in low magnification imaging using the low-dose control at a typical magnification of 155× LM mode with a spot size 11. After identification of lamellae, continuous rotation MicroED data was collected as previously described (Hattne et al., 2015; Shi et al., 2016). Briefly, a selected area aperture was used to minimize background noise, and the stage was rotated at a constant rate of 0.30 degrees per second while a parallel beam of electrons scattered from the sample.
Data was converted from FEI’s SER format to SMV using in-house developed software available from https://cryoem.ucla.edu/downloads/snapshots (Hattne et al., 2015, 2016). SMV images were indexed and integrated with XDS (Kabsch, 2010a); scaling and merging was performed using XSCALE (Kabsch, 2010b). The merged dataset was phased by molecular replacement in Phaser (McCoy et al., 2007) with 5i9s as the search model (Hattne et al., 2016), resulting in a final TFZ score >30 and LLG >1000. The structure was refined with phenix.refine (Afonine et al., 2012) using electron scattering factors to a final Rwork/Rfree of 24/26% at a resolution of 2.75Å. Protein figures were generated in PyMol (Schrödinger, 2014).
QUANTIFICATION AND STATISTICAL ANALYSIS
Number values used to calculate the statistics in Table 1 were given by the total number of reflections and the number of unique reflections. No other statistical tests were used within the manuscript.
DATA AND SOFTWARE AVAILABILITY
The accession number for the atomic coordinates reported in this paper is PDB: 6N4U. The accession number for the density maps reported in this paper is EMDB: EMD-0340.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, Peptides, and Recombinant Proteins | ||
| Proteinase K | Sigma-Aldrich | Cat#P12308; CAS: 39450-01-6 |
| Ammonium sulfate | Sigma-Aldrich | Cat#A4418; CAS: 7783-20-2 |
| Deposited Data | ||
| Atomic coordinates, proteinase K crystal structure used for molecular replacement | (Hattne et al., 2016) | PDB: 5i9s |
| Atomic coordinates and density map of proteinase K generated from a single milled lamella | this paper | PDB: 6N4U; EMDB: EMD-0343 |
| Software and Algorithms | ||
| TVIPS tools | (Hattne et al., 2015) | N/A |
| XDS | (Kabsch, 2010b) | RRID:SCR_015652 |
| XSCALE | (Kabsch, 2010a) | RRID:SCR_015652 |
| Phaser | (McCoy et al., 2007) | RRID:SCR_014219 |
| Phenix.refine | (Afonine et al., 2012) | RRID:SCR_014224 |
| PyMol | (Schrödinger, 2014) | RRID:SCR_000305 |
| Other | ||
| TEM grids | Quantifoil | N/A |
| easiGlow glow discharge cleaning system | PELCO | N/A |
| Vitrobot Mark IV plunge-freezer | Thermo Fisher | N/A |
| FEI Autoloader | Thermo Fischer | N/A |
| FEI Tabs Arctica | Thermo Fischer | N/A |
| FEI CetaD | Thermo Fischer | N/A |
| FEI Versa FIB/SEM | Thermo Fischer | N/A |
Highlights.
Large protein crystals are milled into thin lamella and imaged by FIB-SEM
Continuous rotation MicroED data are collected from FIB milled lamella
Structure of a macromolecular protein is solved from a single protein lamella
Complete dataset collected using a total exposure of less than 4 e−Å−2
ACKNOWLEDGMENTS
The Gonen lab is supported by funds from the Howard Hughes Medical Institute. Work done in the Jensen lab was supported by NIH grant R35 GM122588. We would like to thank Ikaasa Suri (UCLA) for the help with sample preparation.
Footnotes
DECLARATION OF INTERESTS
The authors declare no conflict of interests.
REFERENCES
- Afonine PV, Grosse-Kunstleve RW, Echols N, Headd JJ, Moriarty NW, Mustyakimov M, Terwilliger TC, Urzhumtsev A, Zwart PH, and Adams PD (2012). Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D Biol. Crystallogr 68, 352–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agard D, Cheng Y, Glaeser RM, and Subramaniam S (2014). Single-particle Cryo-electron Microscopy (Cryo-EM): Progress, Challenges, and Perspectives for Further Improvement (Elsevier). [Google Scholar]
- de la Cruz MJ, Hattne J, Shi D, Seidler P, Rodriguez J, Reyes FE, Sawaya MR, Cascio D, Weiss SC, Kim SK, et al. (2017). Atomic-resolution structures from fragmented protein crystals with the cryoEM method MicroED. Nat. Methods 14, 399–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duyvesteyn HME, Kotecha A, Ginn HM, Hecksel CW, Beale EV, de Haas F, Evans G, Zhang P, Chiu W, and Stuart DI (2018). Machining protein microcrystals for structure determination by electron diffraction. Proc. Natl. Acad. Sci. U S A 115, 9569–9573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egerton RF, Li P, and Malac M (2004). Radiation damage in the TEM and SEM. Micron 35, 399–409. [DOI] [PubMed] [Google Scholar]
- Giannuzzi LA, and Stevie FA (1999). A review of focused ion beam milling techniques for TEM specimen preparation. Micron 30, 197–204. [Google Scholar]
- Hattne J, Reyes FE, Nannenga BL, Shi D, De La Cruz MJ, Leslie AGW, and Gonen T (2015). MicroED data collection and processing. Acta Crystallogr. A Found. Adv 71, 353–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattne J, Shi D, De La Cruz MJ, Reyes FE, and Gonen T (2016). Modeling truncated pixel values of faint reflections in MicroED images. J. Appl. Crystallogr 49, 1029–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattne J, Shi D, Glynn C, Zee C, Gallagher-Jones M, Martynowycz MW, Rodriguez JA, and Gonen T (2018). Analysis of global and site-specific radiation damage in cryo-EM. Structure 26, 759–766.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W (2010a). XDS. Acta Crystallogr. D Biol. Crystallogr 66, 125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W (2010b). Integration, scaling, space-group assignment and post-refinement. Acta Crystallogr. D Biol. Crystallogr 66, 133–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Zhang S, and Sun F (2018). In situ protein micro-crystal fabrication by cryo-FIB for electron diffraction. Biophys. Reports 4, 339–347. https://link.springer.com/article/10.1007/s41048-018-0075-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahamid J, Pfeffer S, Schaffer M, Villa E, Danev R, Cuellar LK, Förster F, Hyman AA, Plitzko JM, and Baumeister W (2016). Visualizing the molecular sociology at the HeLa cell nuclear periphery. Science 351, 969–972. [DOI] [PubMed] [Google Scholar]
- Martynowycz MWMW, and Gonen T (2018). From electron crystallography of 2D crystals to MicroED of 3D crystals. Curr. Opin. Colloid Interface Sci 34, 9–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsukawa T, Murata K, and Shimizu R (1973). Investigation of electron penetration and X-ray production in solid targets. Phys. Status Solidi B Basic Solid State Phys 55, 371–383. [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, and Read RJ (2007). Phaser crystallographic software. J. Appl. Crystallogr 40, 658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nannenga BL, Shi D, Leslie AGW, and Gonen T (2014). High-resolution structure determination by continuous-rotation data collection in MicroED. Nat. Methods 11, 927–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrödinger LLC (2014). The PyMOL molecular graphics system. https://www.schrodinger.com/pymol/
- Shi D, Nannenga BL, Iadanza MG, and Gonen T (2013). Three-dimensional electron crystallography of protein microcrystals. Elife 2013, e01345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi D, Nannenga BL, de la Cruz MJ, Liu J, Sawtelle S, Calero G, Reyes FE, Hattne J, and Gonen T (2016). The collection of MicroED data for macromolecular crystallography. Nat. Protoc 11, 895–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor KA, and Glaeser RM (1974). Electron diffraction of frozen, hydrated protein crystals. Science 186, 1036–1037. [DOI] [PubMed] [Google Scholar]
- Vergara S, Lukes DA, Martynowycz MW, Santiago U, Plascencia-Villa G, Weiss SC, De La Cruz MJ, Black DM, Alvarez MM, López-Lozano X, et al. (2017). MicroED structure of Au146(p-MBA)57at subatomic resolution reveals a twinned FCC cluster. J. Phys. Chem. Lett 8, 5523–5530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Ji G, Huang X, Xu W, and Sun F (2016). An improved cryo-FIB method for fabrication of frozen hydrated lamella. J. Struct. Biol 194, 218–223. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The accession number for the atomic coordinates reported in this paper is PDB: 6N4U. The accession number for the density maps reported in this paper is EMDB: EMD-0340.