Abstract
The rate of decline in insulin secretion after diagnosis with type 1 diabetes (T1D) varies substantially among individuals and with age at diagnosis, but the mechanism(s) behind this heterogeneity are not well understood. We investigated the loss of pancreatic β cell function in new-onset T1D subjects using unbiased whole blood RNA-seq and verified key findings by targeted cell count measurements. We found that patients who lost insulin secretion more rapidly had immune phenotypes (“immunotypes”) characterized by higher levels of B cells and lower levels of neutrophils, especially neutrophils expressing primary granule genes. The B cell and neutrophil immunotypes showed strong age dependence, with B cell levels in particular predicting rate of progression in young subjects only. This age relationship suggested that therapy targeting B cells in T1D would be most effective in young subjects with high pretreatment B cell levels, a prediction which was supported by data from a clinical trial of rituximab in new-onset subjects. These findings demonstrate a link between age-related immunotypes and disease outcome in new-onset T1D. Furthermore, our data suggest that greater success could be achieved by targeted use of immunomodulatory therapy in specific T1D populations defined by age and immune characteristics.
Keywords: Autoimmunity
Keywords: Bioinformatics, Diabetes, Immunotherapy
Differences in neutrophils and B cells identified by gene expression profiles predict loss of insulin secretion in people with new-onset type 1 diabetes.
Introduction
Type 1 diabetes (T1D) is characterized by the autoimmune-mediated destruction of the insulin-producing β cells of the pancreatic islets (1). The resulting inability to control blood glucose levels requires lifelong intensive monitoring and treatment with exogenous insulin. Despite extensive efforts to identify therapies that can halt the loss of β cells, no immunotherapies are currently approved for clinical use to prevent or slow T1D progression.
One factor contributing to the failure to identify effective treatments is disease heterogeneity within T1D. This includes heterogeneity in loss of insulin secretion during the natural history of disease (2–7). Variation in treatment effects is also observed in clinical trials of multiple immunotherapies in new-onset T1D (8). Treated patients in these trials can often be split into a responder group, who show greater retention of endogenous insulin secretion (as measured by the insulin cleavage product C-peptide), and a nonresponder group who show loss of C-peptide similar to placebo-treated patients. The heterogeneity in treatment efficacy and in progression of untreated patients suggests that different immunological processes may have distinct contributions to disease in different patients. Investigating the underlying causes of that heterogeneity in insulin secretion could point to additional therapeutic targets, combination therapies, or personalization of treatment based on an individual’s immunological characteristics.
To address these challenges in identifying effective disease-modifying therapies for T1D, we investigated correlates of disease progression in the first 3 years after clinical diagnosis with two primary goals. First, we aimed to identify signatures of progression that could allow prediction of disease course and identification of patients most likely to benefit from therapy. Second, we aimed to characterize the immunological heterogeneity of T1D, with the goal of identifying additional therapeutic targets and/or personalizing treatment. We have previously used whole blood RNA-seq to detect correlates of C-peptide loss in clinical trials of new-onset T1D (9, 10), as applied by Linsley et al. to abatacept treatment in an accompanying article (11). This method allows unbiased examination of a large number of immune processes with a single assay. We interrogated T1D progression in control-arm subjects from 6 clinical trials in new-onset T1D using whole blood RNA-seq to determine correlates of disease progression. We identified immunological signatures in peripheral blood that may enable prediction of disease progression and/or targeted treatment of T1D patients based on immunological characteristics.
Results
Subject population and characteristics.
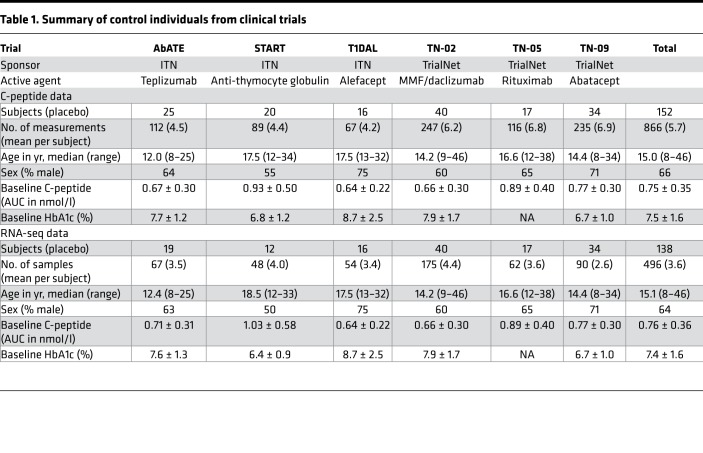
To investigate disease progression in the period immediately following diagnosis, we combined data and samples from control-arm subjects from clinical trials in new-onset T1D conducted by the Immune Tolerance Network (ITN) and Type 1 Diabetes TrialNet (TrialNet). These trials evaluated immunotherapies with a range of immunological targets but enrolled similar subject populations and collected similar clinical data. Table 1 shows the included trials, subject numbers, and clinical and demographic characteristics. We obtained C-peptide values from mixed-meal tolerance tests (MMTTs) from a total of 152 subjects (mean 5.7 visits per subject) and RNA-seq data from 138 subjects (mean 3.6 visits per subject). All clinical data were obtained from ITN and TrialNet, as previously described (5, 12–20).
Table 1. Summary of control individuals from clinical trials.
C-peptide loss in T1D patients shows exponential decay.
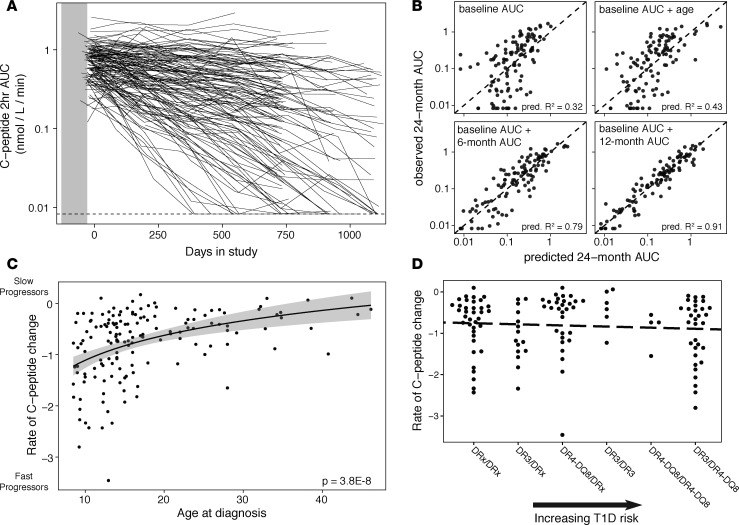
In order to better understand variation in T1D progression after diagnosis, we first assessed methods for quantifying loss of C-peptide. Retention of residual C-peptide secretion is associated with better glucose control and fewer diabetic complications (21) and is the aim of trials of disease-modifying therapy (22). C-peptide levels showed wide variation both among and within subjects over time (Figure 1A), which undermines inference from values at single time points or changes between 2 time points. To assess predictability and stability of change in C-peptide, we fit linear regression models to C-peptide AUC, with values censored at the lower limit of detection. We evaluated the ability of baseline C-peptide, age at diagnosis, and C-peptide at intermediate time points to predict 2-year C-peptide levels, comparing observed values to those predicted by leave-one-out cross-validation. C-peptide AUC at 2 years was associated with but not individually predicted by baseline C-peptide values, age at diagnosis, or these 2 variables together (Figure 1B, top). However, models using C-peptide measurements at study baseline and 6 or 12 months later enabled accurate prediction of levels at 2 years (Figure 1B, bottom). The increased accuracy of these linear models with multiple time points suggests that change in C-peptide can be approximated by a log-linear function, with the rate varying among individuals.
Figure 1. C-peptide loss follows exponential decay.
(A) C-peptide AUC from 2-hour mixed-meal tolerance test. Each line shows an individual subject’s values across multiple visits. The dashed line represents the lower limit of detection of the assay. n = 846 measurements from 152 subjects. (B) Prediction of C-peptide AUC at 2 years based on baseline C-peptide AUC, age at study entry, and AUC at 6- or 12-month visits. Predicted values are model predictions from leave-one-out cross-validation. Predictive R2 summarizes correspondence between observed and predicted values; the dashed line represents equivalence of predicted and observed values. n = 109 subjects for each model. (C) Rate of C-peptide AUC change varies with age. Model fit line is based on a logarithmic function; shading shows standard error of the model. Variance in C-peptide change is greater in younger subjects (Breusch-Pagan test, P = 0.002). n = 152 subjects. (D) Rate of C-peptide change does not vary consistently with HLA genotypes that confer T1D risk. The dashed line shows linear model fit (P = 0.4). Genotype categories are from Winkler et al. (28); DRx represents alleles that are not DR3 or DR4-DQ8. n = 124 subjects with HLA genotyping.
To capture the overall trend in C-peptide change, while limiting sensitivity to missing or anomalous data, we fit linear mixed-effects models to log-transformed C-peptide AUC over time (Supplemental Figure 1, A and B; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.125556DS1). For most subjects, C-peptide values were available up to 2 years after study entry; 29 subjects had data up to 3 years after study entry. When fitting models, we excluded any visits after a subject reached the lower limit of detection of C-peptide. The data were best fit by a model of log C-peptide AUC incorporating subject-level random effects for baseline C-peptide and rate of C-peptide change over time. This model explained 88% of the total variance in C-peptide AUC values across all visits. Adding quadratic or higher-order terms did not significantly improve the fit. Thus, C-peptide loss in newly diagnosed T1D patients follows exponential decay, which can be described using a temporal half-life.
For downstream analyses, we used the subject-specific rates of C-peptide change from the models fit to all visits. For some analyses, we classified individuals as fast or slow progressors based on a binary split of the modeled rates. Because the distribution showed a long tail of subjects with very rapid loss of C-peptide secretion, we defined fast progressors as the lowest quartile of rates and slow progressors as the upper 3 quartiles (Supplemental Figure 1, C and D). Analyses using different splits yielded qualitatively similar results.
Age of onset predicts C-peptide loss, but HLA genotype does not.
Age at T1D onset partially predicts the rate of C-peptide change (6, 7, 23–25), as previously shown in the 3 TrialNet trials included in the present study (Table 1) (26). As expected, age at onset had a strong relationship to the rate of C-peptide loss in our study, with subjects diagnosed at younger ages generally losing C-peptide secretion more rapidly (Figure 1C; P = 3.8 × 10–8 by linear regression). In addition, C-peptide change was more variable among younger subjects than among older subjects (Breusch-Pagan test, P = 0.002); while subjects diagnosed before age 18 showed a wide range in rates of decline, most subjects diagnosed after age 18 retained their C-peptide secretion over the study time frame. There were few or no older-diagnosed subjects matching the most rapid rates shown by some in the younger age groups.
Variation in the HLA class II region is strongly predictive of T1D incidence (27). To evaluate its effect on disease progression following diagnosis, we compared HLA genotype categories (28) for their ability to predict the rate of loss of C-peptide following diagnosis. We found no consistent relationship between HLA class II genotype risk categories and rates of C-peptide loss (Figure 1D). Subjects showed wide variation in rate of progression within a given risk category and no significant differences between categories; this was true even when restricted to younger ages where HLA effects on T1D risk are strongest (29) (Supplemental Figure 2). While larger sample sizes or different patient populations may reveal small differences in loss of insulin secretion among HLA genotypes, it is clear that HLA genotype does not drive variation in disease progression following diagnosis to the same extent as it drives disease risk.
The subject populations in the 6 clinical trials comprising this study showed wide variation in rates of C-peptide loss (Supplemental Figure 3). Some of this variation can be attributed to differences in age at enrollment driven by study inclusion criteria. This may have influenced the ability to detect treatment effects, as limited disease progression in control-arm participants (common in older subjects) reduces power to observe a change in the rate of decline of the treatment group.
Fast-progressing T1D subjects have high B cell gene expression and low neutrophil gene expression.
In order to interrogate immunological differences with rate of T1D progression in an unbiased manner, we used whole blood gene expression profiling by RNA-seq (see Methods). We have previously used this approach to investigate response to treatment in new-onset T1D, including variation in rates of C-peptide loss following immunotherapy (9–11). We performed RNA-seq analysis on all subjects for whom sufficient samples were available; included subjects did not differ from total study populations in age at T1D diagnosis, sex, baseline C-peptide, baseline HbA1c, or rate of C-peptide change (Table 1).
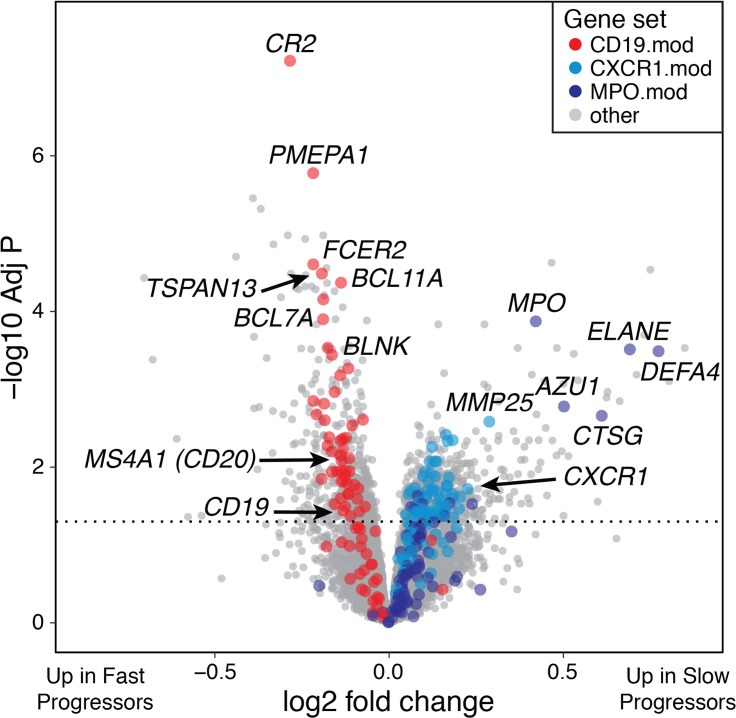
We first compared the whole blood gene expression profiles of slow-progressing and fast-progressing T1D patients using linear models (30). We treated rate of C-peptide change as a continuous variable and included subject sex, clinical trial, and sample batch as covariates. Differential expression analysis revealed a large number of genes that varied with rate of C-peptide loss (Figure 2 and Supplemental Table 1).
Figure 2. Whole blood gene expression profiles differ between T1D subjects with fast and slow loss of C-peptide.
Volcano plot showing differentially expressed genes with rate of C-peptide AUC change. Gene sets corresponding to B cells (CD19.mod) and neutrophils (CXCR1.mod and MPO.mod) are shown in red and blue, respectively, and select related genes are labeled. The dashed line represents a Benjamini-Hochberg–adjusted P = 0.05. Two genes with anomalous P values are not shown. The model used for differential expression included subject sex and RNA-seq batch as covariates. n = 471 samples from 138 subjects.
The genes significantly upregulated in fast progressors formed clusters of genes strongly enriched for the KEGG B cell receptor signaling pathway and a variety of B cell–related gene ontology processes. Genes upregulated in slow progressors formed clusters of genes strongly enriched for the gene ontology defense response and immune response terms, which contain numerous neutrophil-related and neutrophil primary/azurophilic granule genes (Figure 2). We also used predefined modules of coordinately expressed and annotated genes to capture the broad immunological signatures represented by the individual genes (31). Three modules contained many of the individual differentially expressed genes: CXCR1.mod, which comprises neutrophil genes, such as CXCR1 (32) and CEACAM3 (33); the neutrophil primary granule module MPO.mod, with numerous primary granule genes; and the B cell module CD19.mod, with canonical B cell genes, including CD19, CD79A, and CD22. Median expression of CXCR1.mod and CD19.mod were inversely correlated (r = –0.59), indicating that high levels of each represent opposite ends of a continuum. The general patterns of upregulation and downregulation were consistently detected despite extensive individual variation (Supplemental Figure 4); however, because of this variation, individual rates of progression cannot be precisely predicted from these gene expression values. The differentially expressed genes formed dense protein-protein interaction networks, more interconnected than expected by chance (Supplemental Figure 5, A and B; for genes with FDR < 0.05, P < 1 × 10–16 in genes upregulated in slow and fast progressors). These networks had highly connected nodes related to B cells in the genes upregulated in fast progressors (e.g., CD19, MS4A1/CD20, and PAX5; ref. 34) and to neutrophils in the genes upregulated in slow progressors (e.g., IL8, MPO, ELANE; ref. 35), implicating those processes as central components of the immune differences associated with variation in rates of C-peptide loss.
Neutrophil numbers and phenotype vary with rate of C-peptide loss and age at diagnosis.
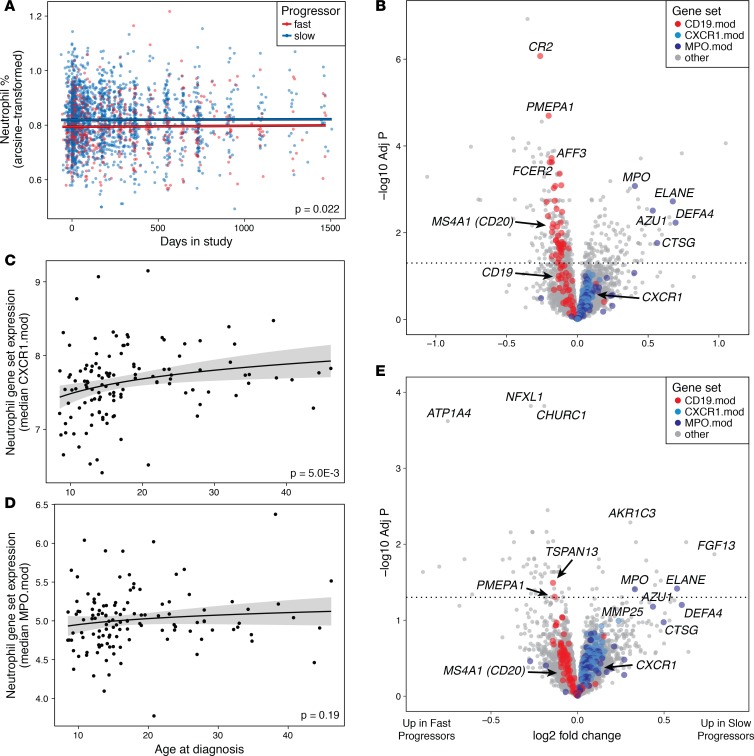
To confirm whether increased neutrophil gene counts in slow progressors corresponded to increased neutrophil proportions in peripheral blood, we used complete blood count (CBC) data to test for differences in circulating neutrophil numbers between slow and fast progressors. Though the differences were small, the neutrophil percentage throughout the course of the trials was higher in slow progressors than in fast progressors (Figure 3A, P = 0.022 by linear model). Moreover, we found no significant changes in mean neutrophil levels over time either within or between progressor groups during the time frame of this study by CBCs (Figure 3A) or RNA-seq (Supplemental Figure 6). This suggests that during the first years following diagnosis, the neutrophil levels of T1D patients as a group are relatively stable. Though changes in neutrophil levels in the peridiagnostic period have been previously reported (36), the timing of those changes generally occurs outside the window of our data, falling prior to diagnosis and not returning to normal levels until at least 5 years after diagnosis.
Figure 3. Neutrophils and primary granule genes are elevated in slow-progressing T1D subjects.
(A) Neutrophil levels from CBCs. Points show individual patient values over the study time frame; lines show model estimates for fast and slow progressors. Fit lines have identical slopes, as there was no evidence of an interaction between study day and progressor group; significance is for difference in intercept between groups from linear mixed-effects model. n = 2326 measurements from 152 subjects. (B) Volcano plot showing differential expression with rate of C-peptide change, after accounting for neutrophil and lymphocyte counts from CBCs. Multiple neutrophil granule genes (MPO.mod) are significantly upregulated after adjusting for cell counts, while general neutrophil genes (CXCR1.mod) show consistently positive but nonsignificant differences in slow progressors. In addition to CBC values, subject sex and RNA-seq batch were included as covariates. n = 420 samples from 137 subjects. (C and D) Expression of (C) neutrophil genes (CXCR1.mod) and (D) neutrophil primary granule genes (MPO.mod) at study baseline in relation to subject age. Model fit lines are based on logarithmic functions; shading shows standard error of the models. Significance values are from linear model contrasts. n = 124 subjects. (E) Volcano plot showing differential expression with rate of C-peptide change, after accounting for subject age. Multiple neutrophil primary granule genes (MPO.mod) are significantly upregulated in slow progressors after adjusting for age, while general neutrophil genes (CXCR1.mod) show consistently positive but nonsignificant differences in slow progressors. In addition to subject age, subject sex and RNA-seq batch were included as covariates. n = 471 samples from 138 subjects.
Neutrophil levels from CBCs were strongly correlated with median expression of the general neutrophil gene module CXCR1.mod (r = 0.63) and significantly but less strongly correlated with the neutrophil primary (azurophilic) granule module MPO.mod (Supplemental Figure 7; r = 0.29). In agreement with this result, multiple neutrophil primary granule genes in this module, including MPO, ELANE, AZU1, and DEFA4, were still upregulated in slow progressors after accounting for differences in neutrophil and lymphocyte counts from CBCs (Figure 3B and Supplemental Table 2).
Immunological characteristics, including relative abundance of peripheral blood leukocyte populations, change with age (37, 38). Given the differences in T1D progression with age, we hypothesized that gene expression might also vary with age, either in association with or unrelated to disease progression. As expected, comparisons with age as a continuous variable revealed numerous differentially expressed genes, including large numbers of genes related to neutrophils and B cells (Supplemental Figure 8 and Supplemental Table 3). At the gene module level, the expression of general neutrophil module genes (CXCR1.mod) was positively correlated with subject age (Figure 3C). However, expression of neutrophil primary granule genes (MPO.mod) was not associated with age (Figure 3D), and numerous genes in this set were upregulated in slow progressors after accounting for age (Figure 3E and Supplemental Table 4). Many of these differentially expressed genes code for proteins found in the primary granules of neutrophils (e.g., AZU1, MPO, DEFA4, CTSG, ELANE). These results show that both the numbers and characteristics of neutrophils varied with rate of T1D progression, with components both associated with and independent of age.
B cell gene expression predicts C-peptide loss in an age-dependent manner.
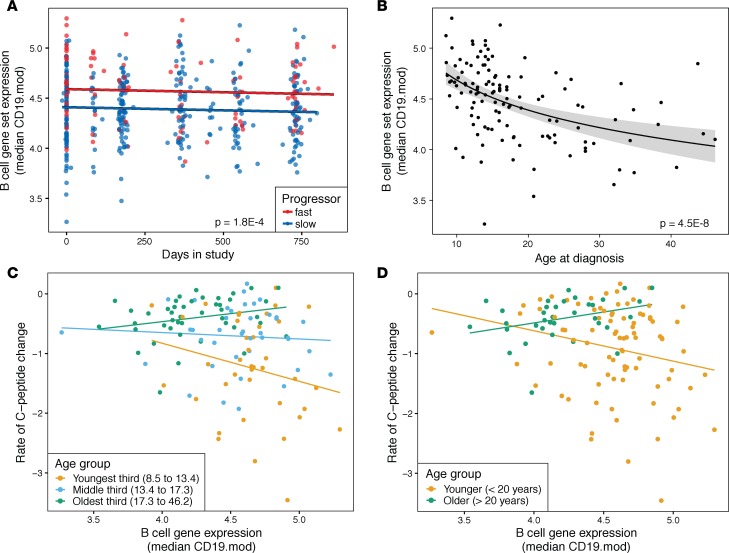
To further investigate the association of B cell gene expression with the rate of C-peptide change, we summarized gene expression using a module of B cell genes (CD19.mod). Median expression of this gene module separated fast and slow progressors (Figure 4A). Similar to the pattern seen with neutrophil levels, median expression of this module was stable both within and between groups over the time frame of the study (Figure 4A).
Figure 4. B cell gene expression varies with C-peptide loss in T1D subjects in an age-dependent manner.
(A) Median B cell gene expression levels (CD19.mod) over time. Points show individual patient values over the study time frame; lines show model estimates for fast and slow progressors. Expression values were batch-corrected to remove differences by trial and RNA-seq batch. Fit lines have identical slopes, as there was no evidence of an interaction between study day and progressor group; significance is for difference in intercept between groups from linear mixed-effects model. n = 470 measurements from 138 subjects. (B) Expression of B cell genes (CD19.mod) at study baseline in relation to subject age. Model fit line is based on a logarithmic function; shading shows standard error of the model. Significance values are from linear model contrasts. Gene expression values shown are for baseline visits only. n = 124 subjects. (C and D) Rate of change in C-peptide versus baseline B cell gene expression (CD19.mod), with subjects stratified by age at study entry. (C) Subjects were divided into 3 equal-sized groups by age. The slope of the relationship is more negative in the younger than the older patient group (P = 0.038 by linear model contrast). n = 39, 41, and 44 subjects in the youngest, middle, and oldest groups, respectively. (D) Subjects were divided into 2 groups using sliding-window analysis, which predicted a break at 20 years of age. The relationship is more negative in the younger than the older patient group (P = 0.032 by linear model contrast). n = 89 and 35 subjects in the younger and older groups, respectively.
While B cells make up only a fraction of total lymphocytes, fast and slow progressors differed in lymphocyte percentage and the ratio of lymphocyte-to-neutrophil counts from CBCs (Supplemental Figure 9). This may indicate a broader difference in lymphocytes that is dominated by, but not exclusive to, B cells; indeed, the genes upregulated in fast progressors include several specific to CD8+ or all T cells (Supplemental Figure 5A and Supplemental Table 1).
B cell gene expression showed a strong relationship with subject age, similar to but in the opposite direction of neutrophil levels (Figure 4B). To dissect the complex relationships among subject age, rate of C-peptide loss, and immunological characteristics, we tested for relationships between B cell gene expression at baseline and rate of C-peptide change by age tertile. Both B cell (Figure 4C) and neutrophil (Supplemental Figure 10) median gene set expression showed the strongest relationships in the youngest age group. These patterns were not explained by covariation of age, cell populations, and rate of progression within groups. In addition, high B cell gene expression predicted more rapid progression in younger subjects but slower progression in older subjects (P = 0.038 for difference in slope).
To extend these categorical age groups, we used a sliding-window analysis to more precisely determine the age dependence of the relationship between B cell gene expression and C-peptide loss (Supplemental Figure 11). A negative correlation between rate of C-peptide change and B cell gene module expression indicates that, within that age window, fast progressors generally have higher B cell gene expression. The correlation was negative in patient subsets with a median age of less than 20 years, but this relationship did not persist in patient subsets with higher median age, indicating that the connection of B cells to more rapid progression broke down at around age 20 years. Splitting the subjects into two groups at this 20-year boundary reveals a different relationship between B cell gene expression and loss of C-peptide secretion in the younger and older groups (Figure 4D; P = 0.032 for difference in slope).
To expand on these results from modular analyses, we determined the individual genes that showed the strongest age-specific associations with rate of C-peptide change (Supplemental Table 5). Genes that were upregulated in young slow progressors were strongly enriched for associations with neutrophil degranulation, secretory granule, exocytosis, and related terms. Genes upregulated in young fast progressors were enriched for associations with chromosome organization, mitotic cell cycle, and numerous related terms. Results were similar using log-transformed age or when including neutrophil and lymphocyte counts from CBCs as covariates. Taken together, these results indicate that the immunological characteristics associated with rapid loss of C-peptide vary with age, with high B cell gene expression predicting rapid disease progression in young subjects only.
B cell gene expression in younger subjects predicts response to anti–B cell therapy.
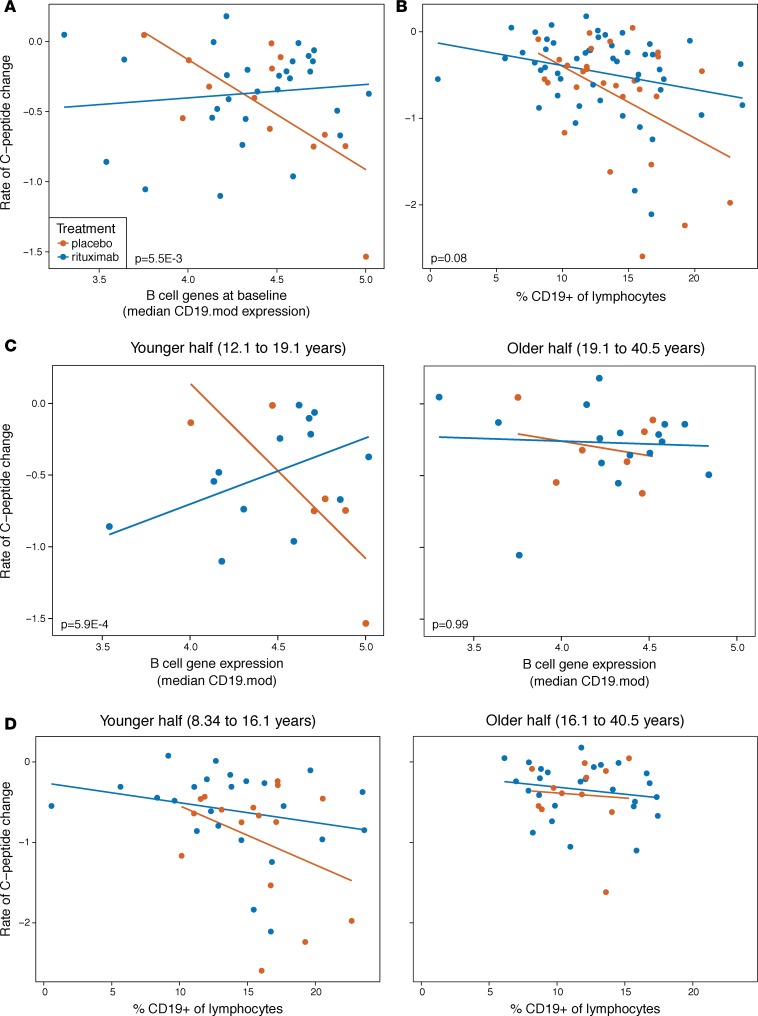
The association between B cell gene expression and rate of C-peptide decline in young subjects suggests that B cell levels may be more relevant to disease progression in younger than in older subjects. If this is the case, anti–B cell treatment should yield greater clinical benefits in younger patients; indeed, this was the trend in a trial of the anti-CD20 monoclonal antibody rituximab (12, 13). Beyond this age-dependent effect, we would expect the greatest benefits in subjects with high B cell levels prior to treatment, especially in younger patients with high pretreatment B cell levels. We tested these predictions using C-peptide and immunophenotyping data from a phase II clinical trial of the anti-CD20 monoclonal antibody rituximab (12, 13). We first reproduced the original trial finding that rituximab treatment was most pronounced in younger participants (Supplemental Figure 12). We then used additional data to evaluate the relationship between B cells and response to treatment. RNA-seq data were available for a subset of the trial participants, as described previously (9); this subset did not differ significantly from the original trial population in age, sex, or C-peptide characteristics. In the subset of participants with RNA-seq data, placebo-arm subjects with high B cell gene expression showed a more rapid loss of C-peptide (Figure 5A; P = 0.004 for slope). This relationship between B cell gene expression and loss of C-peptide secretion was absent in the rituximab-treated subjects, suggesting that rituximab was most effective in those with high B cell gene expression at baseline (P = 5.5 × 10–3 for difference in slope).
Figure 5. Effects of rituximab treatment in new-onset T1D support an increased role for B cells in some patients.
(A and B) Rituximab treatment alleviates the increased loss of C-peptide in subjects with high pretreatment B cell levels, using (A) B cell gene set expression (CD19.mod) or (B) levels of B cells from flow cytometry (n = 42 and 78 subjects, respectively). Significance values are 1-sided tests for difference in slope using linear model contrasts. (C) Treatment effect of rituximab was more strongly related to baseline B cell gene expression in younger subjects (n = 19 and 23 younger and older subjects, respectively). (D) Rituximab treatment effect shows a stronger trend in younger subjects with greater pretreatment B cell levels measured by flow cytometry (n = 40 and 38 younger and older subjects, respectively).
We next sought to expand this finding to a larger sample of study subjects with available flow cytometry data. We first verified that our B cell gene expression signature was related to B cell numbers from flow cytometry, using a subset of placebo-treated T1D patients for whom we obtained both data types. Median B cell gene set expression from RNA-seq was positively correlated with the percentage of CD19+ cells by flow cytometry (Supplemental Figure 13), supporting the use of flow cytometry data to corroborate RNA-seq results. However, the moderate correlation (r = 0.34) suggests that these measures capture overlapping but nonidentical biological characteristics. Using B cell levels quantified by flow cytometry in this larger sample of rituximab trial subjects, the rate of C-peptide change was strongly associated with baseline B cell levels in the placebo-arm subjects (Figure 5B; P = 0.003 for slope in placebo group). This relationship was partially modified by rituximab treatment (Figure 5B; P = 0.08 for difference in slope).
Finally, we compared the relationship between pretreatment B cell levels and rate of C-peptide decline in the context of age in placebo- versus rituximab-treated patients. If B cells are more important for disease progression in younger subjects, rituximab treatment should ablate the relationship between B cell levels and C-peptide loss most strongly in the younger age groups. We therefore predicted a large difference in the slope of this relationship between the placebo- and rituximab-treated groups among younger but not older individuals. This expectation was corroborated using RNA-seq data (Figure 5C), and flow cytometry data from the larger cohort also showed a trend (NS) in the expected direction in the younger patients (Figure 5D). In both cases, the older subjects showed no relationship between B cell levels and disease progression in either the placebo- or rituximab-treated groups. In summary, variation in the effects of rituximab treatment are consistent with B cells having greater relevance to disease in some individuals, particularly young patients with high pretreatment B cell levels.
Discussion
Progression of T1D varies widely among patients at all stages of disease. We used whole blood gene expression data from well-characterized control-arm subjects of new-onset T1D clinical trials to identify correlates of the loss of β cell function. We validated these findings with CBC and flow cytometry data and showed that they can be used to predict differential benefits of immunomodulatory therapy. In combination with other recent work on response to immunotherapy (9–11, 39–41), these results point toward precision targeting and modulation of therapy to achieve optimal benefits.
Using stimulated C-peptide secretion from the first 3 years after diagnosis, we found that the decline in C-peptide over time in new-onset T1D follows exponential decay (25), with the rate of decay relatively constant within individuals but variable among subjects. Because our data were limited to the immediate period after diagnosis, we cannot determine whether this pattern holds before diagnosis or into long-standing T1D. Longitudinal studies of insulin secretion are ongoing in TrialNet (42, 43); defining the temporal loss of β cell function in T1D, from events prior to seroconversion through and beyond clinical diagnosis, will help to illuminate the triggers and immunological drivers of β cell damage in the early stages of T1D.
The fact that the rate of decline in C-peptide appears to be intrinsic and constant from diagnosis to 6, 12, and 24 months suggests that earlier assessments may allow estimation of an individual’s specific rate of decline within a short time after diagnosis. Change in stimulated C-peptide in the first 6 weeks after diagnosis was previously found not to predict levels at 1 or 2 years (44). However, that study did not determine whether short-term changes in C-peptide levels predict change in C-peptide over those time frames (44). Early identification of rapid progressors would facilitate prioritization of treatment based on predicted severity of disease. Moreover, changes over time in a patient’s intrinsic rate of C-peptide loss could be used as a readout for the efficacy of a particular treatment.
The strongest known predictor of loss of C-peptide is age at T1D onset (6, 7, 23–26). In agreement with this, we found C-peptide loss to be more rapid in patients diagnosed at younger ages. However, this was not a simple linear relationship: many individuals diagnosed before age 18 progressed quite slowly, with almost no measurable loss of C-peptide during the 2- to 3-year study period. The most striking pattern is the lack of adult-diagnosed subjects who rapidly lose β cell function. The causes of this age-related heterogeneity are unclear; one possibility is that T1D is in fact multiple diseases, with one form being characterized by a more rapid loss of β cell function and earlier onset. Another possibility is that the disease itself has consistent drivers, but the immunological characteristics of young people contribute to a more rapid disease course.
The HLA region, in particular the HLA class II genotype, is strongly associated with T1D (27, 45). We found that the HLA class II genotypes most associated with T1D did not predict rate of C-peptide loss after diagnosis, in alignment with previous findings that HLA class II alleles do not strongly affect progression from multiple autoantibody positivity to clinical diabetes (46–48) or C-peptide secretion in new-onset subjects (49–51). The absence of such associations indicates that the genetic factors determining risk of T1D are not the same as those determining the rate of β cell damage during T1D. As such, preventing T1D may require different interventions than halting the damage once it has begun.
While much effort has been focused on immunological differences between T1D patients and healthy controls, or with transitions between stages of T1D (52, 53), little information is available about associations between immunological features and rates of C-peptide loss following diagnosis. One notable exception is the recent discovery of a subpopulation of autoreactive CD8+ effector memory T cells in the blood whose levels parallel β cell function, suggesting that these cells are either engaged in or tracking the autoimmune response (54). In addition, using a serum-induced expression assay, Cabrera et al. (55) found that an index of immune inflammatory state predicted the rate of C-peptide loss in the first 2 years after diagnosis in a limited set of placebo-treated subjects; this relationship was ablated by CTLA4Ig therapy.
Our survey of whole blood gene expression revealed that T1D patients losing C-peptide more rapidly had elevated levels of B cell gene expression and reduced levels of general neutrophil and neutrophil primary granule gene expression. These B cell and neutrophil signatures were inversely related, both by gene module expression (r = –0.59 for median CXCR1.mod and CD19.mod), and using cell frequencies from flow cytometry and CBCs (r = –0.54). In addition, these levels did not change markedly over the first 2–3 years after diagnosis, suggesting that these are stable, individual-specific immune phenotypes (“immunotypes”) associated with the degree of damage to the β cells. In an accompanying study, Linsley et al. (11) show that alterations in B cell and neutrophil immunotypes accompany response to abatacept therapy in certain individuals with new-onset T1D. Together, these findings suggest that B cell and neutrophil immunotype alterations may contribute to both disease heterogeneity and response to therapy in T1D.
While these differences in major cell populations can been quantified by complementary assays, the unbiased approach of RNA-seq enabled their identification through a single experiment, without collection of large numbers of targeted measurements. As with our previous work in response to immunotherapy in T1D (9, 10), these findings highlight the utility of whole blood RNA-seq to broadly survey the immune system and of targeted assays to validate specific results from RNA-seq.
In addition to varying with rate of C-peptide decline, some but not all of these progression-associated signatures varied with age. The levels of B cells and neutrophils that were found in fast progressors were also found in young subjects, indicating that immunological characteristics, rate of C-peptide loss, and age are all associated. Cohorts of age-matched healthy controls will be necessary to determine to what degree these associations are specific to T1D or reflective of general immunological changes with age. However, B cell genes and especially neutrophil primary granule genes showed differences with C-peptide loss, even after accounting for the age relationship. The biological meaning of increased primary granule gene expression in slow progressors (or alternatively, lower expression in fast progressors) is unclear, but it may serve as an age-independent biomarker of disease severity.
The role of neutrophils in T1D has received renewed attention, with evidence that neutrophils preferentially infiltrate the pancreas during T1D onset in both the nonobese diabetic mouse (56, 57) and humans (36). In addition, an innate inflammatory state, potentially driven by type I interferons, is associated with risk and onset of T1D (58–61). Our finding that neutrophil gene expression is lower in fast progressors, corroborated with CBC data, supports the idea that innate immunity is dysregulated in T1D. Reduced levels of neutrophils in peripheral blood have been previously found during and immediately after onset of T1D, in association with increased infiltration of the pancreas (36), though the numbers of neutrophils in the pancreas are insufficient to explain the peripheral neutropenia. Rather, both characteristics likely reflect a more general change in immune state, which also corresponds to the peripheral deficit in neutrophils we detected via gene expression in fast-progressing new-onset subjects. While this combined evidence suggests that promoting neutrophil proliferation will not correct this dysregulation, a recent trial in new-onset T1D that included granulocyte colony-stimulating factor may provide additional information on how perturbation of neutrophils affects disease course (62). Beyond total numbers, the specific reduction in neutrophil primary granule genes in fast progressors suggests differences in the phenotype of neutrophils among T1D subjects. This downregulation of neutrophil granule genes was less age dependent than the differences in overall neutrophil and B cell gene expression, indicating that it is a more consistent characteristic of immune dysfunction in T1D.
The exact contribution of B lymphocytes to T1D remains an open question. Disease-associated autoantibodies are generally assumed to be nonpathogenic; instead, they are thought to indicate involvement of B cells in the autoimmune reaction through other mechanisms, such as antigen presentation (63, 64). Disease-related B cells in T1D subjects show loss of anergy (65) and defective central and peripheral tolerance (66), leading to increased frequencies of autoreactive cells. While we did not observe differences in genes indicative of specific B cell subsets or functions, the power of whole blood RNA-seq to detect such patterns in low-frequency cell populations is limited. As such, we cannot conclusively determine whether the differences are in B cell numbers, B cell activation, or some combination thereof. Studies using immunophenotyping, RNA-seq, or functional assays on sorted B cells from a similar cohort could clarify whether particular B cell processes are related to the variation in progression of T1D. A systemic increase in the levels of total B cells, as seen herein with rapid loss of C-peptide, may provide an elevated number of autoreactive B cells involved in disease or may reflect a more general pathogenic immune state. Investigations of insulitic infiltrate in the pancreas have shown elevated B cells in pancreata of T1D patients with more rapid loss of β cells, and this pattern is concentrated in younger individuals (67). This aligns well with our finding that high B cell gene expression in the periphery predicts more rapid progression in young individuals only. While the mechanism of this age dependency remains to be determined, it suggests a transition from a disease with greater B cell engagement in young T1D patients to a different immune dysregulation in older individuals. This points to B cell–directed therapy as a potentially more effective treatment in young people, while other therapies may be necessary for older patients.
Indeed, the anti-CD20 monoclonal antibody rituximab was most effective in younger subjects (12, 13). Using samples and data from that trial, we found that rituximab was most effective at delaying loss of β cell function in young patients with high B cell levels prior to treatment, exactly as predicted from our findings in control-arm subjects. While it is possible that these same subjects would benefit similarly from other immunotherapies that have shown mixed effects, to our knowledge this population has not been identified in responder analysis of other trials. The pattern of pretreatment B cells contrasts with elevated T cell levels during rituximab treatment that we previously found associated with more rapid loss of C-peptide and reduced pharmacodynamic activity of rituximab (9). Our age-specific B cell signature provides a predictive indicator of treatment benefit, while the T cell difference suggests a possible mechanistic explanation for lack of treatment efficacy in some patients. This combination of predictive signatures and signals of efficacy during therapy moves toward the ultimate goal of optimal treatment via precise targeting and informed modification of therapy.
Taken together, our results point to a number of potential improvements to T1D treatment. First, the disease course immediately after onset appears predictable in individual subjects, allowing for prioritization of treatment based on severity of disease. Changes to this intrinsic rate of loss of β cell function may serve as a short-term indicator of treatment efficacy, enabling modification of therapy based on its measured benefit. Second, the age-specific immunological predictors of β cell decline move beyond the simplistic understanding of old and young in T1D, toward immunotypes (68) defined by immunological characteristics that are partially explained by age. One outstanding question is whether these immunological differences are the drivers of observed variation in disease progression or simply indicators of some other underlying cause, as-yet unidentified. T1D is likely a suite of disorders, with different causes and physiological consequences that vary in frequency by genetic background, age, and environmental exposures. Better resolution of these immunotypes and determination of the optimal therapy for each will push beyond treating T1D symptoms to finding personalized cures for this challenging disease.
Methods
Subject selection.
Subjects were control-arm participants in 6 new-onset clinical T1D trials (Table 1) (5, 12–20) as well as active-treatment participants in the TrialNet phase II study of rituximab (TN-05) (9, 12, 13). The 6 new-onset T1D trials had similar inclusion criteria; subjects entered the studies within 100 days of clinical T1D diagnosis and had minimum stimulated C-peptide of 0.2 nmol/l at screening (one study had minimum 0.4 nmol/l). Age restrictions varied among trials, with minimum ages ranging from 8 to 13 years. All subjects across all studies were required to have at least one diabetes-related autoantibody.
C-peptide, demographic, clinical, and other trial data.
Collection of C-peptide, CBCs, flow cytometry, and other trial data was previously described for each trial (5, 12–20). To make C-peptide levels comparable across visits and studies with different length MMTTs, we recalculated C-peptide as 2-hour AUC from MMTTs, omitting any additional time points after 2 hours. Of 866 total MMTTs, we dropped 12 due to missing data for at least one time point and 8 due to anomalous values at one or more time points, yielding a total of 846 C-peptide AUC measurements. For CBC differentials, percentages were arcsine-transformed to approximate normality.
Modeling C-peptide change over time.
We estimated individual-specific rates of change in C-peptide over time as exponential decay, using our previously described method (9). Models included subject-level random effects terms for the intercept and slopes, as log(C-peptide AUC) ~ day + (1|id) + (day-1|id). Mixed-effect models were fit using the lme4 package (69), with an unstructured random effects variance-covariance matrix. This allowed each subject to vary around a central value for both baseline C-peptide and rate of C-peptide change over time. We then extracted the subject-level coefficients from these models and used the rate of change of log C-peptide over time as a measure of rate of T1D progression. This approach provided a single continuous measure of progression per subject and allowed inclusion of subjects even if data were missing from one or more visits. A pseudo-R2 for the model was calculated as the squared correlation of the observed and fitted values.
To further validate this approach, we compared observed C-peptide AUC at 24 months to values predicted from models incorporating baseline AUC along with age and/or C-peptide measurements at 6 or 12 months. To account for the lower bound of measurements at the limit of C-peptide detection, we used tobit censored regression models, as implemented in the R package AER (70), with the response variable truncated at the lower limit of detection. Predicted values were similarly truncated to better correspond to actual measurement values. We compared performance of models by leave-one-out cross-validation; we excluded a single observation at a time and used the model fit to all other observations to generate a predicted value for the focal observation. Predicted R2 was calculated as the Pearson correlation between the actual observed values and these leave-one-out predictions.
For certain analyses, we split subjects into fast and slow progressors based on rates of C-peptide change. The bimodal distribution of rates supported treating the slowest 75% of subjects as slow progressors and the fastest 25% as fast progressors. For analyses of active- and placebo-treated subjects in the rituximab trial, rates of C-peptide change were calculated as previously described (9), using random intercepts and slopes by subject and a fixed treatment effect for slope.
Associations among age at diagnosis, HLA class II genotypes, and rates of C-peptide change.
We used linear models to test for associations among age at diagnosis, HLA class II genotypes, and rate of C-peptide change. Based on model diagnostics, we log-transformed age at diagnosis prior to fitting the final model. We evaluated nonconstant variance using the Breusch-Pagan test. HLA class II risk scores were calculated from subject genotypes based on frequency of T1D incidence for each of those genotypes. HLA genotype categories were treated as a continuous variable in linear modeling, using the coding of Winkler et al. (28).
RNA-seq data collection and processing.
Samples of RNA purified from whole blood were obtained from TrialNet and ITN. Whole blood was collected in Tempus blood RNA tubes at the clinical site. RNA was isolated, libraries were prepared and sequenced on a HiSeq 2500 or HiScanSQ sequencer (Illumina), and sequence data were processed to read counts and quality metrics, as previously described (9, 10). We ensured RNA-seq sample identity using expression of sex-specific genes and kinship comparisons using genetic variants from the RNA-seq reads (9). No samples had mismatches between predicted and annotated sex; 3 of the original 496 samples were excluded based on kinship values indicative of sample contamination or identity mismatch with annotation.
RNA-seq data analysis.
The data from the 6 trials showed slight differences in sequencing depth and quality characteristics, making it difficult to use universal quality thresholds across all samples. We therefore excluded samples from downstream analysis using trial-specific thresholds for total reads (1 × 106 to 5 × 106), the percentage of reads aligned to the human genome (80%–90%), and the median coefficient of variation of coverage (0.7–1.0). These criteria excluded 16 of the remaining 493 samples; results were insensitive to variations in these thresholds. Finally, we excluded 6 samples that duplicated subject visits, leaving 471 high-quality samples for downstream analysis. Principal component analysis showed that samples separated by trial and within several trials by RNA isolation method and/or isolation/processing site. We therefore included a batch variable in our downstream differential expression analyses for study and RNA isolation method and isolation/processing site.
We normalized RNA-seq count data using the trimmed mean of M values (TMM) (71, 72) and included genes in a given comparison if they had greater than 1 count per million in at least 15% of the libraries. Differential expression of individual genes was determined with limma-voom (30). Models included covariates for the batch variable described above, subject sex, and subject-level random (73). P values were adjusted for multiple testing using the Benjamini-Hochberg procedure (74). PPI interactions, clustering, and functional annotation enrichment were determined using STRING (75) and visualized using Cytoscape (76).
For visualizations of RNA-seq counts, we used TMM-normalized, log2-transformed counts, with batch differences due to trial and RNA-seq batch differences regressed out (30). Gene set expression was calculated as the median across genes of these batch-corrected counts.
Data and code availability.
RNA-seq data were deposited in the GEO repository (accession GSE124400). Sequencing data from the rituximab study were previously deposited in the GEO repository (accession GSE112594). Data files and R code for all analyses are available at https://github.com/mjdufort/Dufort_T1D_placebos (master branch, commit b3382eb).
Statistics.
All statistical analyses were performed using R v. 3.4.3 (77). Specific statistical tests and thresholds for significance are listed in the text and figure legends and include linear model contrasts and Breusch-Pagan tests; where not noted, tests were 2 tailed, and P < 0.05 was considered significant. Graphical visualizations were generated using R base graphics or the ggplot2 package (78).
Study approval.
All trials were conducted under protocols approved by the institutional review boards for each study site (the names and locations of these review boards are provided in the supplemental materials), as previously described (13–15, 17, 18, 20). All subjects provided informed consent to participate in the trials and subsequent ancillary studies.
Author contributions
MJD performed and interpreted analysis and prepared the manuscript. CJG and CS helped with data presentation, interpretation, and manuscript preparation. PSL conceived the study and helped with data presentation, interpretation, and manuscript preparation.
Supplementary Material
Acknowledgments
This work was supported by grants to PSL from the JDRF (1-PNF-2014-94-Q-R) and the NIH (DP3 DK104465-01) and by funding from the Immune Tolerance Network (ITN), NIH grant 5UM1AI109565, awarded to Gerald T. Nepom. We gratefully acknowledge the study subjects for their participation in the clinical trials. We thank Sarah Muller of TrialNet, and Noha Lim and Elisavet Serti of ITN, for assistance in gaining access to samples and data. We thank Vivian Gersuk, Marty Timour, Kimberly O’Brien, and Quynh-Anh Nguyen for conducting the RNA sequencing and James Eddy and Mario Rosasco for performing the RNA-seq data processing. Three anonymous reviewers provided feedback that substantially improved the manuscript. This work was supported by JDRF grants 2-PAR-2015-123-Q-R to Gerald T. Nepom and 3-SRA-2016-209-Q-R to S. Alice Long. MJD was supported by the Benaroya Research Institute Systems Immunology Discovery Fellowship. Research reported in this publication was performed as a project of the Immune Tolerance Network and supported by the National Institute of Diabetes and Digestive and Kidney Diseases and the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under award UM1AI109565. We acknowledge the support of the Type 1 Diabetes TrialNet Study Group, which identified study participants and provided samples and follow-up data for this study. The Type 1 Diabetes TrialNet Study Group is a clinical trials network funded by the NIH through the National Institute of Diabetes and Digestive and Kidney Diseases, the National Institute of Allergy and Infectious Diseases, and the Eunice Kennedy Shriver National Institute of Child Health and Human Development, through cooperative agreements U01 DK061010, U01 DK061034, U01 DK061042, U01 DK061058, U01 DK085465, U01 DK085453, U01 DK085461, U01 DK085466, U01 DK085499, U01 DK085504, U01 DK085509, U01 DK103180, U01 DK103153, U01 DK085476, U01 DK103266, U01 DK103282, U01 DK106984, U01 DK106994, U01 DK107013, U01 DK107014, UC4 DK106993, and the JDRF. The contents of this article are solely the responsibility of the authors and do not necessarily represent the official views of the NIH or the JDRF.
Version 1. 02/21/2019
Electronic publication
Footnotes
Conflict of interest: PSL received research support from Bristol-Myers Squibb and is an inventor on patent US5844095A, “CTLA4Ig fusion proteins.” CJG received research support from Janssen Inc.
License: Copyright 2019, American Society for Clinical Investigation.
Reference information: JCI Insight. 2019;4(4):e125556. https://doi.org/10.1172/jci.insight.125556.
Contributor Information
Matthew J. Dufort, Email: Mdufort@benaroyaresearch.org.
Carla J. Greenbaum, Email: cjgreen@benaroyaresearch.org.
Cate Speake, Email: CSpeake@benaroyaresearch.org.
Peter S. Linsley, Email: plinsley@benaroyaresearch.org.
References
- 1.Atkinson MA, Eisenbarth GS, Michels AW. Type 1 diabetes. Lancet. 2014;383(9911):69–82. doi: 10.1016/S0140-6736(13)60591-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Keenan HA, et al. Residual insulin production and pancreatic ß-cell turnover after 50 years of diabetes: Joslin Medalist Study. Diabetes. 2010;59(11):2846–2853. doi: 10.2337/db10-0676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Davis AK, et al. Prevalence of detectable C-Peptide according to age at diagnosis and duration of type 1 diabetes. Diabetes Care. 2015;38(3):476–481. doi: 10.2337/dc14-1952. [DOI] [PubMed] [Google Scholar]
- 4.Oram RA, et al. The majority of patients with long-duration type 1 diabetes are insulin microsecretors and have functioning beta cells. Diabetologia. 2014;57(1):187–191. doi: 10.1007/s00125-013-3067-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rigby MR, et al. Alefacept provides sustained clinical and immunological effects in new-onset type 1 diabetes patients. J Clin Invest. 2015;125(8):3285–3296. doi: 10.1172/JCI81722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Greenbaum CJ, et al. Fall in C-peptide during first 2 years from diagnosis: evidence of at least two distinct phases from composite Type 1 Diabetes TrialNet data. Diabetes. 2012;61(8):2066–2073. doi: 10.2337/db11-1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ludvigsson J, et al. Decline of C-peptide during the first year after diagnosis of Type 1 diabetes in children and adolescents. Diabetes Res Clin Pract. 2013;100(2):203–209. doi: 10.1016/j.diabres.2013.03.003. [DOI] [PubMed] [Google Scholar]
- 8.Greenbaum CJ, Schatz DA, Haller MJ, Sanda S. Through the fog: recent clinical trials to preserve β-cell function in type 1 diabetes. Diabetes. 2012;61(6):1323–1330. doi: 10.2337/db11-1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. doi: 10.1038/s41435-018-0032-1. Linsley PS, et al. Elevated T cell levels in peripheral blood predict poor clinical response following rituximab treatment in new-onset type 1 diabetes. Genes Immun . [DOI] [PMC free article] [PubMed]
- 10.Long SA, et al. Partial exhaustion of CD8 T cells and clinical response to teplizumab in new-onset type 1 diabetes. Sci Immunol. 2016;1(5):eaai7793. doi: 10.1126/sciimmunol.aai7793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Linsley PS, Greenbaum CJ, Speake C, Long SA, Dufort MJ. B lymphocyte alterations accompany abatacept resistance in new-onset type 1 diabetes. JCI Insight. 2019;4(4):e126136. doi: 10.1172/jci.insight.126136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pescovitz MD, et al. B-lymphocyte depletion with rituximab and β-cell function: two-year results. Diabetes Care. 2014;37(2):453–459. doi: 10.2337/dc13-0626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pescovitz MD, et al. Rituximab, B-lymphocyte depletion, and preservation of beta-cell function. N Engl J Med. 2009;361(22):2143–2152. doi: 10.1056/NEJMoa0904452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Herold KC, et al. Teplizumab (anti-CD3 mAb) treatment preserves C-peptide responses in patients with new-onset type 1 diabetes in a randomized controlled trial: metabolic and immunologic features at baseline identify a subgroup of responders. Diabetes. 2013;62(11):3766–3774. doi: 10.2337/db13-0345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Orban T, et al. Co-stimulation modulation with abatacept in patients with recent-onset type 1 diabetes: a randomised, double-blind, placebo-controlled trial. Lancet. 2011;378(9789):412–419. doi: 10.1016/S0140-6736(11)60886-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Orban T, et al. Costimulation modulation with abatacept in patients with recent-onset type 1 diabetes: follow-up 1 year after cessation of treatment. Diabetes Care. 2014;37(4):1069–1075. doi: 10.2337/dc13-0604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rigby MR, et al. Targeting of memory T cells with alefacept in new-onset type 1 diabetes (T1DAL study): 12 month results of a randomised, double-blind, placebo-controlled phase 2 trial. Lancet Diabetes Endocrinol. 2013;1(4):284–294. doi: 10.1016/S2213-8587(13)70111-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gitelman SE, et al. Antithymocyte globulin treatment for patients with recent-onset type 1 diabetes: 12-month results of a randomised, placebo-controlled, phase 2 trial. Lancet Diabetes Endocrinol. 2013;1(4):306–316. doi: 10.1016/S2213-8587(13)70065-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gitelman SE, et al. Antithymocyte globulin therapy for patients with recent-onset type 1 diabetes: 2 year results of a randomised trial. Diabetologia. 2016;59(6):1153–1161. doi: 10.1007/s00125-016-3917-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gottlieb PA, et al. Failure to preserve beta-cell function with mycophenolate mofetil and daclizumab combined therapy in patients with new- onset type 1 diabetes. Diabetes Care. 2010;33(4):826–832. doi: 10.2337/dc09-1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lachin JM, McGee P, Palmer JP, DCCT/EDIC Research Group Impact of C-peptide preservation on metabolic and clinical outcomes in the Diabetes Control and Complications Trial. Diabetes. 2014;63(2):739–748. doi: 10.2337/db13-0881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Palmer JP, et al. C-peptide is the appropriate outcome measure for type 1 diabetes clinical trials to preserve beta-cell function: report of an ADA workshop, 21-22 October 2001. Diabetes. 2004;53(1):250–264. doi: 10.2337/diabetes.53.1.250. [DOI] [PubMed] [Google Scholar]
- 23.Hao W, Gitelman S, DiMeglio LA, Boulware D, Greenbaum CJ, Type 1 Diabetes TrialNet Study Group Fall in C-peptide during first 4 years from diagnosis of type 1 diabetes: variable relation to age, HbA1c, and insulin dose. Diabetes Care. 2016;39(10):1664–1670. doi: 10.2337/dc16-0360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Besser REJ, Ludvigsson J, Hindmarsh PC, Cole TJ. Exploring C-peptide loss in type 1 diabetes using growth curve analysis. PLoS One. 2018;13(7):e0199635. doi: 10.1371/journal.pone.0199635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barker A, et al. Age-dependent decline of β-cell function in type 1 diabetes after diagnosis: a multi-centre longitudinal study. Diabetes Obes Metab. 2014;16(3):262–267. doi: 10.1111/dom.12216. [DOI] [PubMed] [Google Scholar]
- 26.Bundy BN, Krischer JP, Type 1 Diabetes TrialNet Study Group A model-based approach to sample size estimation in recent onset type 1 diabetes. Diabetes Metab Res Rev. 2016;32(8):827–834. doi: 10.1002/dmrr.2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Concannon P, Rich SS, Nepom GT. Genetics of type 1A diabetes. N Engl J Med. 2009;360(16):1646–1654. doi: 10.1056/NEJMra0808284. [DOI] [PubMed] [Google Scholar]
- 28.Winkler C, et al. Feature ranking of type 1 diabetes susceptibility genes improves prediction of type 1 diabetes. Diabetologia. 2014;57(12):2521–2529. doi: 10.1007/s00125-014-3362-1. [DOI] [PubMed] [Google Scholar]
- 29.Howson JM, Rosinger S, Smyth DJ, Boehm BO, ADBW-END Study Group. Todd JA. Genetic analysis of adult-onset autoimmune diabetes. Diabetes. 2011;60(10):2645–2653. doi: 10.2337/db11-0364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ritchie ME, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Linsley PS, Speake C, Whalen E, Chaussabel D. Copy number loss of the interferon gene cluster in melanomas is linked to reduced T cell infiltrate and poor patient prognosis. PLoS One. 2014;9(10):e109760. doi: 10.1371/journal.pone.0109760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cummings CJ, et al. Expression and function of the chemokine receptors CXCR1 and CXCR2 in sepsis. J Immunol. 1999;162(4):2341–2346. [PubMed] [Google Scholar]
- 33.Chen T, Gotschlich EC. CGM1a antigen of neutrophils, a receptor of gonococcal opacity proteins. Proc Natl Acad Sci USA. 1996;93(25):14851–14856. doi: 10.1073/pnas.93.25.14851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cobaleda C, Schebesta A, Delogu A, Busslinger M. Pax5: the guardian of B cell identity and function. Nat Immunol. 2007;8(5):463–470. doi: 10.1038/ni1454. [DOI] [PubMed] [Google Scholar]
- 35.Naranbhai V, et al. Genomic modulators of gene expression in human neutrophils. Nat Commun. 2015;6:7545. doi: 10.1038/ncomms8545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Valle A, et al. Reduction of circulating neutrophils precedes and accompanies type 1 diabetes. Diabetes. 2013;62(6):2072–2077. doi: 10.2337/db12-1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Castelo-Branco C, Soveral I. The immune system and aging: a review. Gynecol Endocrinol. 2014;30(1):16–22. doi: 10.3109/09513590.2013.852531. [DOI] [PubMed] [Google Scholar]
- 38.Peters MJ, et al. The transcriptional landscape of age in human peripheral blood. Nat Commun. 2015;6:8570. doi: 10.1038/ncomms9570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Long SA, et al. Remodeling T cell compartments during anti-CD3 immunotherapy of type 1 diabetes. Cell Immunol. 2017;319:3–9. doi: 10.1016/j.cellimm.2017.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Herold KC, et al. Increased T cell proliferative responses to islet antigens identify clinical responders to anti-CD20 monoclonal antibody (rituximab) therapy in type 1 diabetes. J Immunol. 2011;187(4):1998–2005. doi: 10.4049/jimmunol.1100539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Orban T, et al. Reduction in CD4 central memory T-cell subset in costimulation modulator abatacept-treated patients with recent-onset type 1 diabetes is associated with slower C-peptide decline. Diabetes. 2014;63(10):3449–3457. doi: 10.2337/db14-0047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mahon JL, et al. The TrialNet Natural History Study of the Development of Type 1 Diabetes: objectives, design, and initial results. Pediatr Diabetes. 2009;10(2):97–104. doi: 10.1111/j.1399-5448.2008.00464.x. [DOI] [PubMed] [Google Scholar]
- 43.Battaglia M, et al. Understanding and preventing type 1 diabetes through the unique working model of TrialNet. Diabetologia. 2017;60(11):2139–2147. doi: 10.1007/s00125-017-4384-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.DiMeglio LA, et al. Changes in beta cell function during the proximate post-diagnosis period in persons with type 1 diabetes. Pediatr Diabetes. 2016;17(4):237–243. doi: 10.1111/pedi.12271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Erlich H, et al. HLA DR-DQ haplotypes and genotypes and type 1 diabetes risk: analysis of the type 1 diabetes genetics consortium families. Diabetes. 2008;57(4):1084–1092. doi: 10.2337/db07-1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lipponen K, et al. Effect of HLA class I and class II alleles on progression from autoantibody positivity to overt type 1 diabetes in children with risk-associated class II genotypes. Diabetes. 2010;59(12):3253–3256. doi: 10.2337/db10-0167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Achenbach P, Hummel M, Thümer L, Boerschmann H, Höfelmann D, Ziegler AG. Characteristics of rapid vs slow progression to type 1 diabetes in multiple islet autoantibody-positive children. Diabetologia. 2013;56(7):1615–1622. doi: 10.1007/s00125-013-2896-y. [DOI] [PubMed] [Google Scholar]
- 48.Ilonen J, et al. Genetic susceptibility to type 1 diabetes in childhood - estimation of HLA class II associated disease risk and class II effect in various phases of islet autoimmunity. Pediatr Diabetes. 2016;17 Suppl 22:8–16. doi: 10.1111/pedi.12327. [DOI] [PubMed] [Google Scholar]
- 49.Schiffrin A, Suissa S, Weitzner G, Poussier P, Lalla D. Factors predicting course of beta-cell function in IDDM. Diabetes Care. 1992;15(8):997–1001. doi: 10.2337/diacare.15.8.997. [DOI] [PubMed] [Google Scholar]
- 50.Schiffrin A, Suissa S, Poussier P, Guttmann R, Weitzner G. Prospective study of predictors of beta-cell survival in type I diabetes. Diabetes. 1988;37(7):920–925. doi: 10.2337/diab.37.7.920. [DOI] [PubMed] [Google Scholar]
- 51.Mortensen HB, et al. Multinational study in children and adolescents with newly diagnosed type 1 diabetes: association of age, ketoacidosis, HLA status, and autoantibodies on residual beta-cell function and glycemic control 12 months after diagnosis. Pediatr Diabetes. 2010;11(4):218–226. doi: 10.1111/j.1399-5448.2009.00566.x. [DOI] [PubMed] [Google Scholar]
- 52.van Belle TL, Coppieters KT, von Herrath MG. Type 1 diabetes: etiology, immunology, and therapeutic strategies. Physiol Rev. 2011;91(1):79–118. doi: 10.1152/physrev.00003.2010. [DOI] [PubMed] [Google Scholar]
- 53.Wållberg M, Cooke A. Immune mechanisms in type 1 diabetes. Trends Immunol. 2013;34(12):583–591. doi: 10.1016/j.it.2013.08.005. [DOI] [PubMed] [Google Scholar]
- 54.Yeo L, et al. Autoreactive T effector memory differentiation mirrors β cell function in type 1 diabetes. J Clin Invest. 2018;128(8):3460–3474. doi: 10.1172/JCI120555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cabrera SM, et al. Innate immune activity as a predictor of persistent insulin secretion and association with responsiveness to CTLA4-Ig treatment in recent-onset type 1 diabetes. Diabetologia. 2018;61(11):2356–2370. doi: 10.1007/s00125-018-4708-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Diana J, et al. Crosstalk between neutrophils, B-1a cells and plasmacytoid dendritic cells initiates autoimmune diabetes. Nat Med. 2013;19(1):65–73. doi: 10.1038/nm.3042. [DOI] [PubMed] [Google Scholar]
- 57.Diana J, Lehuen A. Macrophages and β-cells are responsible for CXCR2-mediated neutrophil infiltration of the pancreas during autoimmune diabetes. EMBO Mol Med. 2014;6(8):1090–1104. doi: 10.15252/emmm.201404144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kallionpää H, et al. Innate immune activity is detected prior to seroconversion in children with HLA-conferred type 1 diabetes susceptibility. Diabetes. 2014;63(7):2402–2414. doi: 10.2337/db13-1775. [DOI] [PubMed] [Google Scholar]
- 59.Ferreira RC, et al. A type I interferon transcriptional signature precedes autoimmunity in children genetically at risk for type 1 diabetes. Diabetes. 2014;63(7):2538–2550. doi: 10.2337/db13-1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen YG, et al. Molecular signatures differentiate immune states in type 1 diabetic families. Diabetes. 2014;63(11):3960–3973. doi: 10.2337/db14-0214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cabrera SM, Henschel AM, Hessner MJ. Innate inflammation in type 1 diabetes. Transl Res. 2016;167(1):214–227. doi: 10.1016/j.trsl.2015.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Haller MJ, et al. Low-dose anti-thymocyte globulin (ATG) preserves β-cell function and improves HbA1c in new-onset type 1 diabetes. Diabetes Care. 2018;41(9):1917–1925. doi: 10.2337/dc18-0494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hinman RM, Cambier JC. Role of B lymphocytes in the pathogenesis of type 1 diabetes. Curr Diab Rep. 2014;14(11):543. doi: 10.1007/s11892-014-0543-8. [DOI] [PubMed] [Google Scholar]
- 64.Wong FS, et al. Investigation of the role of B-cells in type 1 diabetes in the NOD mouse. Diabetes. 2004;53(10):2581–2587. doi: 10.2337/diabetes.53.10.2581. [DOI] [PubMed] [Google Scholar]
- 65.Smith MJ, et al. Loss of B-cell anergy in type 1 diabetes is associated with high-risk HLA and non-HLA disease susceptibility alleles. Diabetes. 2018;67(4):697–703. doi: 10.2337/db17-0937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Menard L, et al. The PTPN22 allele encoding an R620W variant interferes with the removal of developing autoreactive B cells in humans. J Clin Invest. 2011;121(9):3635–3644. doi: 10.1172/JCI45790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Leete P, et al. Differential insulitic profiles determine the extent of β-cell destruction and the age at onset of type 1 diabetes. Diabetes. 2016;65(5):1362–1369. doi: 10.2337/db15-1615. [DOI] [PubMed] [Google Scholar]
- 68.Kaczorowski KJ, et al. Continuous immunotypes describe human immune variation and predict diverse responses. Proc Natl Acad Sci USA. 2017;114(30):E6097–E6106. doi: 10.1073/pnas.1705065114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bates D, Mächler M, Bolker B, Walker S. Fitting linear mixed-effects models using lme4. J Stat Softw. 2015;67(1):1–48. [Google Scholar]
- 70. Kleiber C, Zeileis A. Applied econometrics with R. New York, NY: Springer New York; 2008. [Google Scholar]
- 71.Robinson MD, Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010;11(3):R25. doi: 10.1186/gb-2010-11-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Smyth GK, Michaud J, Scott HS. Use of within-array replicate spots for assessing differential expression in microarray experiments. Bioinformatics. 2005;21(9):2067–2075. doi: 10.1093/bioinformatics/bti270. [DOI] [PubMed] [Google Scholar]
- 74.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol. 1995;57(1):289–300. [Google Scholar]
- 75.Szklarczyk D, et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43(Database issue):D447–D452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Shannon P, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. http://www.R-project.org/ Accessed January 28, 2019.
- 78. Wickham H. ggplot2: elegant graphics for data analysis. New York, NY: Springer-Verlag; 2016. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq data were deposited in the GEO repository (accession GSE124400). Sequencing data from the rituximab study were previously deposited in the GEO repository (accession GSE112594). Data files and R code for all analyses are available at https://github.com/mjdufort/Dufort_T1D_placebos (master branch, commit b3382eb).