Abstract
Transmembrane channel-like protein isoform 1 (TMC1) is essential for generation of mechano-electrical transducer currents in hair cells of the inner ear. TMC1 disruption causes hair cell degeneration and deafness in mice and humans. Although thought to be expressed at the cell surface in vivo, TMC1 remains in the endoplasmic reticulum when heterologously expressed in standard cell lines, precluding determination of its roles in mechanosensing and pore formation. Here, we report that the KCNQ1 Kv channel forms complexes with TMC1 and rescues its surface expression when co-expressed in Chinese Hamster Ovary cells. TMC1 rescue is specific for KCNQ1 within the KCNQ family, is prevented by a KCNQ1 trafficking-deficient mutation, and is influenced by KCNE β subunits and inhibition of KCNQ1 endocytosis. TMC1 lowers KCNQ1 and KCNQ1-KCNE1 K+ currents, and despite surface expression does not detectably respond to mechanical stimulation or high salt. We conclude that TMC1 is not intrinsically mechano- or osmosensitive but has the capacity for cell surface expression, and requires partner protein(s) for surface expression and mechanosensitivity. We suggest that KCNQ1, expression of which is not thought to overlap with TMC1 in hair cells, is a proxy partner bearing structural elements or a sequence motif reminiscent of a true in vivo TMC1 hair cell partner. Discovery of the first reported strategy to rescue TMC1 surface expression should aid future studies of TMC1 function and native partners.
INTRODUCTION
Despite being discovered and linked to human disease more than two decades ago, the mechanisms of action of TMC gene products have remained largely enigmatic. The founder member was positionally cloned, linked to dominant and recessive forms of deafness and named transmembrane cochlear-expressed protein 1 (TMC1) because of its expression in outer hair cells of the cochlea. Since then, TMC1 and the other seven known mammalian TMC subunits have been detected in numerous other tissues, and the TMC acronym has been redesignated ‘transmembrane channel-like’ (Holt et al., 2014; Kurima et al., 2002; Pan et al., 2013; Vreugde et al., 2002). TMC1 is the most studied of the TMC genes. Despite being thought to reach the plasma membrane in vivo, TMC1 has stubbornly remained in the endoplasmic reticulum (ER) in prior over-expression studies, preventing functional analysis(Fettiplace, 2016; Labay et al., 2010). TMC proteins share no significant sequence similarity with any other known proteins, further stymying attempts to elucidate their function (Keresztes et al., 2003).
Clues to mammalian TMC1 function first came from the deafness (df) and Beethoven (Bth) mouse lines. Df and Bth mice are deaf because of abnormal maturation and reduced survival of hair cells in the cochlea, in each case arising from mutations in Tmc1. In df and Bth mice, K+ currents and mechanosensitive Ca2+ currents in the hair cells of the cochlea are disrupted(Kim and Fettiplace, 2013; Pan et al., 2013; Petersen, 2002; Vreugde et al., 2002). More recently, a distantly related C. elegans ortholog (tmc-1, a.k.a. TmcAh1) was suggested to express Na+-activated cation currents (Chatzigeorgiou et al., 2013).
Topology analysis suggests that TMC1 probably has 6 TM segments (at least in the ER membrane) (Labay et al., 2010), akin to voltage-gated potassium (Kv) channel α subunits, which form tetrameric channels, often with single TM-domain KCNE β subunits. Some members of the TMC family may, in contrast, have 8 TM segments (Figure 1A–C). The multi-TM-span topology of TMC1 and the electrical abnormalities in the cochlea of Tmc1/Tmc2-disrupted mice have fueled speculation that TMC1 forms, contributes to or regulates a mechanosensitive Ca2+ channel (Corns et al., 2016). Despite numerous recent advances in understanding of TMC1 and its requirement for mammalian hair cell function and hearing, at least two crucial questions remain unanswered: is TMC1 necessary and sufficient to form a plasma membrane ion channel, and is TMC1 intrinsically mechanosensitive (Holt et al., 2014)? To test these hypotheses directly requires heterologous expression studies, but as noted above, human TMC1 has failed to reach the plasma membrane in previous studies, so its function could not previously be evaluated(Labay et al., 2010). Here, we therefore tested the hypothesis that TMC proteins require other proteins for surface expression. By screening with co-expression of various ion channel subunits, we unexpectedly found that the KCNQ1 voltage-gated potassium (Kv) channel α subunit rescues TMC1 plasma membrane expression.
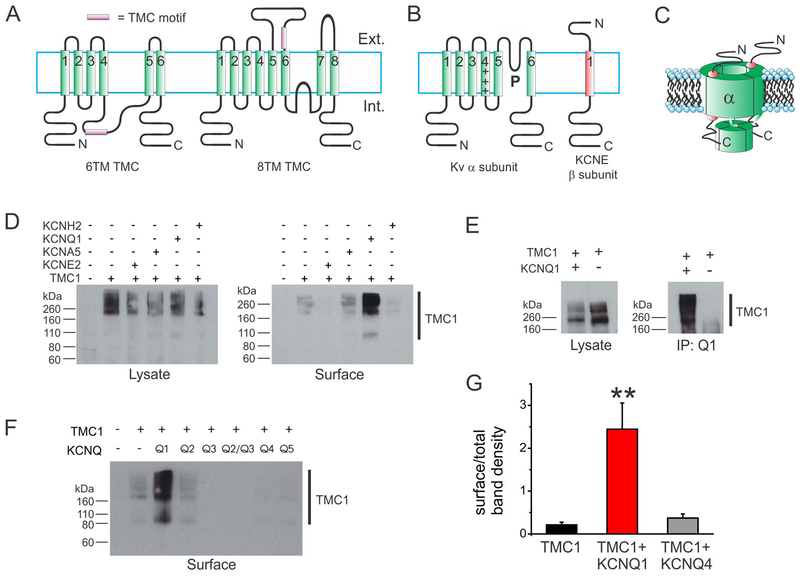
Figure 1. KCNQ1 rescues TMC1 surface expression in CHO cells.
A. Two alternative topologies possible for TMC1: 6TM and 8TM. Topological position of TMC domain is speculative.
B. Topologies of a Kv channel α subunit and a KCNE subunit.
C. Cartoon of a Kv channel complex containing 4 α subunits and 2 KCNE subunits.
D. Western blots showing GFP-tagged TMC1, detected with anti-GFP antibody, in the whole cell lysate (left) and in the cell surface-expressed fraction (right). TMC1 was co-expressed with untagged Kv α or β subunits as indicated above blot.
E. Western blots showing GFP-tagged TMC1, detected with anti-GFP antibody, in the whole cell lysate (left) and in the anti-KCNQ1 antibody co-immunoprecipitated (IP:Q1) fraction as indicated. Transfections indicated above blot.
F. Western blots showing GFP-tagged TMC1, detected with anti-GFP antibody, in cell surface-expressed fraction. TMC1 was co-expressed with untagged KCNQ subunits as indicated above blot.
G. Analysis of effects of KCNQ1 versus KCNQ4 on TMC1 surface expression quantified from surface biotinylation blots as in Fig 1, n = 6 per group. **P<0.01 versus other groups. Other group comparisons P > 0.05.
MATERIALS AND METHODS
Cell culture and imaging
We seeded CHO or COS-7 cells (ATCC) onto poly-L-lysine treated glass coverslips and transfected using TransIT-LT1 (Mirus Bio LLC, Madison, WI, USA) the following day with CMV-based expression constructs containing cDNA for human TMC1 (GFP-tagged), KCNQ1–5, T587M-KCNQ1, KCNE1, KCNE2, Kv1.5, hERG, and/or K44A-dynamin. Cells were cultured in DMEM with 10% FBS and 1% penicillin/streptomycin in a 95% O2/5% CO2 humidified environment at 37°C for 48–72 hours post transfection prior to biochemical analysis, imaging or patch-clamping. Cell culture plasticware and reagents were purchased from VWR and Fisher Scientific unless otherwise stated. GFP-tagged TMC1 was visualized using epifluorescence microscopy with an Olympus BX-51 microscope fitted with a DP72 digital camera and Cell-Sens software (Olympus).
Protein biochemistry
CHO cells were transfected using Mirus LT-1 transfection reagent with a total of 15 μg cDNA per 10 cm plate and allowed 36–48 hrs expression at 37°C before analysis. For surface expression analysis, CHO cells were washed in PBS wash followed by a 30 minute incubation at 4°C in a PBS solution containing 1 mg/ml EZ-Link Sulfo-NHS-SS-Biotin (Thermo Fisher). The reaction was then quenched by subsequent wash with 50 mM Tris (pH 8.0) and PBS. Cells were then lysed in standard RIPA buffer containing a protease-inhibitor cocktail tablet (Thermo Fisher), total protein quantified, and lysates western blotted to analyze proteins.
For co-immunoprecipitation, all samples were first pre-cleared of nonspecific interaction by incubating the total lysate with protein A/G PLUS-coated agarose beads (Santa Cruz) for 1 h. Beads were then pelleted and discarded. Total protein was quantified by BCA. Immunoprecipitating antibodies were then added at a dilution of 1:100 for overnight pulldown at 4°C. The following day, antibody-antigen complexes were pulled down with fresh protein A/G PLUS agarose beads, prior to western blotting. For western blotting, proteins were resolved by SDS-PAGE and transferred onto PVDF membranes for immunoblotting with the following primary antibodies: rabbit anti-GFP (Santa Cruz Biotechnology; Rockland Immunochemicals), goat anti-KCNQ1 (Santa Cruz Biotechnology). For secondary detection, horseradish peroxidase (HRP)-conjugated antibodies (BioRad) were used in conjugation with Luminata Forte HRP substrate (Millipore). Imaging was performed using Syngene (Cambridge, UK) Gbox hardware and software. A ~30 kD nonspecific band served as a loading control (Supplementary Figure 1). All blots were performed at least twice. For quantitative comparison of effects of KCNQ1 versus KCNQ4 on TMC1 surface expression, blots were performed 6 times each, and then band densities quantified using Image J (NIH, Bethesda, MD, USA).
Whole-Cell Patch Clamp
We recorded currents in CHO and COS-7 cells in whole-cell mode at room temperature (22–25°C) with 3–6 MΩ borosilicate glass electrodes backfilled with solution containing (in mM): 90 K Acetate, 20 KCl, 40 HEPES, 3 MgCl2, 1 CaCl2, 3 EGTA-KOH, 2 MgATP; pH7.2. We perfused cells continuously at 1–2ml/min with extracellular solution (ECS) containing (in mM): 135 NaCl, 5 KCl, 5 HEPES, 1.2 MgCl2, 2.5 CaCl2, 10 glucose; pH 7.4. All chemicals were purchased from Fisher Scientific or Sigma-Millipore. We held cells at −80mV in voltage clamp before applying the voltage step protocols and recording currents in response to pulses between −80 mV and +40 or +60 mV at 20 mV intervals, followed by a single pulse to −30 mV, using a CV −7A Headstage (Axon Instruments, Foster City, CA, USA). Currents were amplified using a Multi-clamp 700B (Axon Instruments), low-pass filtered at 2–10kHz using an eight-pole Bessel filter and digitization was achieved (sampling at 10–40kHz) through a DigiData 1322A interface (Molecular Devices; Sunnyvale, CA). The pClamp8 (Molecular Devices) software package was used for data acquisition and Clampfit was used for analysis, along with Graphpad Prism 7.0 (Graphpad; La Jolla, CA, USA). Normalized tail currents were plotted versus pre-pulse voltage and fitted with a single Boltzmann function.
Cell-attached Patch Clamp
To record responses to mechanical stimuli, we expressed KCNQ1 and KCNQ1/KCNE1, with or without TMC1, in CHO cells as described above. Using cell-attached patch mode with glass electrodes backfilled with solution containing (in mM): 90 K Acetate, 20 KCl, 40 HEPES, 3 MgCl2, 1 CaCl2, 3 EGTA-KOH, 2 MgATP; pH7.2, we recorded electrical currents in response to membrane depolarization and/or mechanical stimulus (−40 mm Hg), the latter produced by high-speed pressure clamp system (HSPC-1, ALA Sci. Instr., Farmingdale, NY). Cells were held at either −80mV or +80 mV in voltage clamp. The extracellular solution consisted of (in mM): 5 NaCl, 140 KCl, 5 HEPES, 1.2 MgCl2, 2.5 CaCl2, 10 d-Glucose. Currents from cell-attached patches were measured using an Axopatch 200B (Molecular Devices), low-pass filtered at 1 kHz with an eight-pole Bessel filter, and digitized (sampling at 5 kHz) through a DigiData 1440A interface controlled by pClamp10 software (Molecular Devices).
Channel subunit cRNA preparation and Xenopus laevis oocyte injection
cRNA transcripts encoding human KCNQ1, TMC1 and TMC2 were generated by in vitro transcription using the T7 polymerase mMessage mMachine kit (Thermo Fisher Scientific), after vector linearization. cRNA was quantified by spectrophotometry. Defolliculated stage V and VI Xenopus laevis oocytes (Ecocyte Bioscience, Austin, TX) were injected with cRNAs (5–10 ng). The oocytes were incubated at 16 °C in Barth’s saline solution (Ecocyte) containing penicillin and streptomycin, with daily washing, for 2–12 days prior to two-electrode voltage-clamp (TEVC) recording.
Two-electrode voltage clamp (TEVC)
We performed TEVC recording at room temperature using a OC-725C amplifier (Warner Instruments, Hamden, CT) and pClamp8 software (Molecular Devices, Sunnyvale, CA) 2–5 days after cRNA injection as described in the section above. Oocytes were placed in a small-volume oocyte bath (Warner) and viewed with a dissection microscope. Unless otherwise stated, we sourced chemicals from Sigma. Standard bath solution (ND96) was (in mM): 96 NaCl, 4 KCl, 1 MgCl2, 1 CaCl2, 10 HEPES (pH 7.6). High NaCl solution was a modified ND96 solution with 250 mM NaCl. High CaCl2 solution was modified ND96 with 100 mM CaCl2 replacing the NaCl and KCl. Pipettes were of 1–2 MΩ resistance when filled with 3 M KCl. Currents were recorded in response to pulses between −80 mV and + 40 mV at 20 mV intervals, or a single pulse to +40 mV, from a holding potential of −80 mV, to yield current-voltage relationships, current magnitude, and for quantifying activation rate. Deactivation was recorded at −80 mV after a single pulse to +40 mV, from a holding potential of −80 mV. Electrophysiology data analysis was performed with Clampfit (Molecular Devices) and Graphpad Prism software (GraphPad, San Diego, CA, USA); values are stated as mean ± SEM. Raw or normalized tail currents were plotted versus prepulse voltage and fitted with a single Boltzmann function:
| Eq. 1 |
where g is the normalized tail conductance, A1 is the initial value at −∞, A2 is the final value at +∞, V1/2 is the half-maximal voltage of activation and Vs the slope factor. Activation and deactivation kinetics were fitted with single exponential functions.
Statistical analysis
All values are expressed as mean ± SEM. T-test was used for statistical comparisons. All P-values were two-sided. Statistical significance was defined as P < 0.05.
RESULTS
Co-assembly with KCNQ1 rescues TMC1 surface expression in vitro
We hypothesized that TMC1 is an ion channel subunit that requires another subunit to bring it to the cell surface and/or to function, and tested this by co-expressing Chinese Hamster Ovary (CHO) cells with cDNA encoding human TMC1 tagged with green fluorescent protein at its N-terminus (TMC1-GFP), and cDNAs for various Kv channel α and β subunits. We quantified TMC1-GFP surface expression using surface biotinylation, avidin purification, followed by western blotting using an anti-GFP antibody. This assay confirmed expression of TMC1-GFP and its inability to reach the cell surface alone. Strikingly, the assay also revealed that co-expression with the KCNQ1 α subunit resulted in the first recorded heterologous cell surface expression of TMC1 (Figure 1D). We next confirmed that TMC1-GFP co-assembles with KCNQ1 when co-expressed in CHO cells, using co-immunoprecipitation (co-IP) (Figure 1E). The surface expression rescue was specific even within the KCNQ subfamily, because other KCNQ subfamily subunits failed to rescue TMC1-GFP surface expression (Figure 1F). A quantitative comparison showed that KCNQ1 increased TMC1 surface expression >tenfold whereas KCNQ4 had no effect (Figure 1G).
TMC1 surface expression can be manipulated by KCNQ1 β subunits or mutagenesis
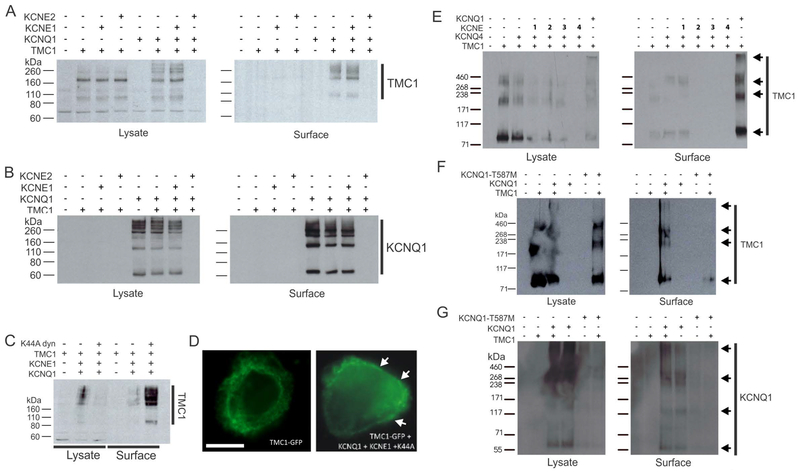
KCNQ1 is modulated by KCNE single transmembrane domain β subunits in vivo and in vitro(Abbott, 2014). Here, we found that co-expression with KCNE1 did not affects the ability of KCNQ1 to rescue TMC1 surface expression. In contrast, co-expression with KCNE2 rendered TMC1 protein undetectable, but only in the presence of KCNQ1 (Figure 2A). KCNQ1 was detectable at the cell surface as expected when expressed alone or with TMC1 with or without KCNE1, but was also undetectable in either the cell lysate or the cell surface when co-expressed with both TMC1 and KCNE2 (Figure 2B). We previously showed that KCNQ1-KCNE1 complexes are internalized from the plasma membrane by a clathrin- and dynamin-dependent endocytic process that was also KCNE1-dependent. Inhibition of this process using a dominant-negative mutant from of dynamin (K44A) greatly increased KCNQ1-KCNE1 surface expression and current (Kanda et al., 2011; Xu et al., 2009). Here, co-expression with KCNQ1, KCNE1 and K44A-dynamin increased TMC1 surface expression, compared to cells expressing TMC1, KCNQ1 and KCNE1 without K44A-dynamin (Figure 2C,D). KCNQ4 did not rescue TMC1 surface expression (Figure 1F) but given the reported expression of KCNQ4 in outer hair cells and its association with inherited deafness, we examined whether any of the KCNEs could help KCNQ4 rescue TMC1 surface trafficking. However, none of the KCNEs studied served this role, while in parallel experiments KCNQ1 again rescued TMC1 surface expression (Figure 2E). KCNQ1 was the first gene associated with the cardiac arrhythmia Long QT syndrome (LQTS), and a variety of LQTS mutations have now been characterized, including T587M, which greatly hinders KCNQ1 surface expression (Yamashita et al., 2001). Here, rescue of TMC1 surface expression by KCNQ1 was prevented by the KCNQ1-T587M mutation (Figure 2F), which we confirmed to diminish KCNQ1 surface expression as previously reported (Yamashita et al., 2001) (Figure 2G).
Figure 2. Mutation and KCNEs affect KCNQ1 rescue of TMC1 CHO cell surface expression.
A. Western blots showing GFP-tagged TMC1, detected with anti-GFP antibody, in the whole cell lysate (left) and in the cell surface-expressed fraction (right). TMC1 was co-expressed alone or with untagged KCNQ1 alone or with KCNE1 or KCNE2 as indicated above blot.
B. Western blots showing untagged KCNQ1, detected with anti-KCNQ1 antibody, corresponding to the same cell transfections as in A.
C. Western blot showing GFP-tagged TMC1, detected with anti-GFP antibody, in the whole cell lysate (left) and in the cell surface-expressed fraction (right). TMC1 was co-expressed alone or with untagged KCNQ1 alone or with KCNE1 and/or K44A dynamin as indicated above blot.
D. Representative immunofluorescence images from cells as in C, showing effects of KCNQ1, KCNE1 and K44A-dynamin co-expression on GFP-tagged TMC1 (green) surface expression (white arrows). Scale bar = 5 μm.
E. Western blots showing GFP-tagged TMC1 (arrows), detected with anti-GFP antibody, in the whole cell lysate (left) and in the cell surface-expressed fraction (right). TMC1 was co-expressed alone or with untagged KCNQ1 or KCNQ4 alone or with KCNE1–4 as indicated above blot.
F. Western blots showing GFP-tagged TMC1 (arrows), detected with anti-GFP antibody, in the whole cell lysate (left) and in the cell surface-expressed fraction (right). TMC1 was co-expressed alone or with untagged KCNQ1 or KCNQ1-T587M as indicated above blot.
G. Western blots showing untagged KCNQ1 (arrows), detected with anti-KCNQ1 antibody, corresponding to the same cell transfections as in F.
TMC1 diminishes KCNQ1 and KCNQ1-KCNE1 currents and is not responsive to high salt
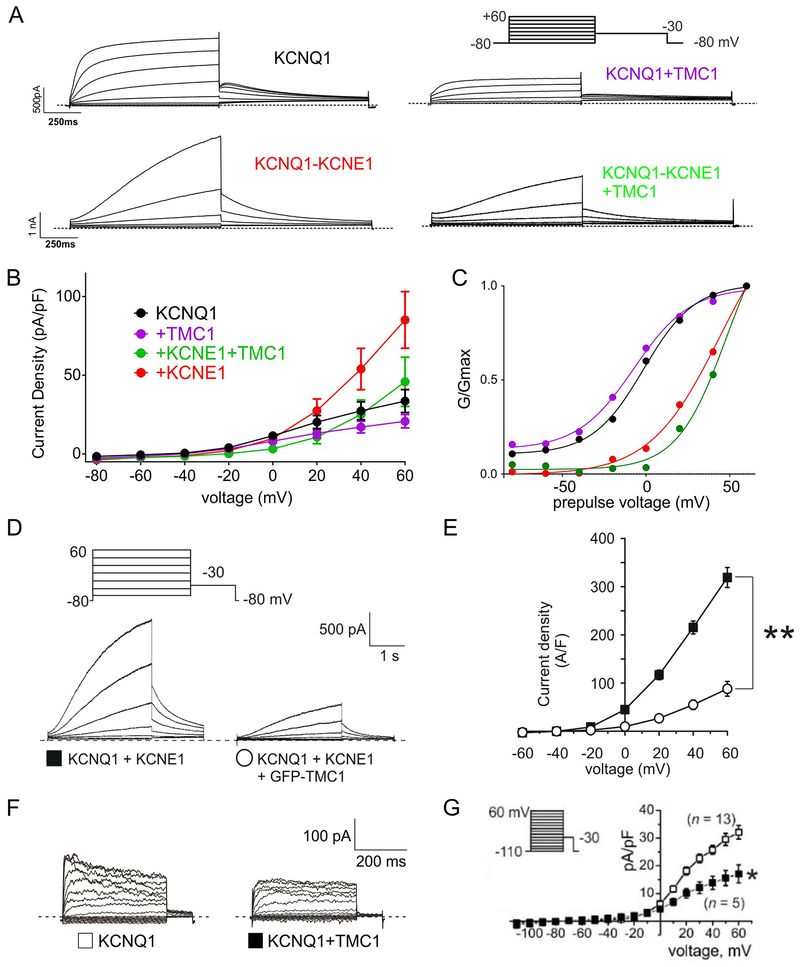
Surface-expressed TMC1-KCNQ1 complexes in CHO cells generated currents similar to those of KCNQ1 alone, but smaller. We observed a similar pattern for TMC1-KCNQ1-KCNE1 in CHO cells (Figure 3A, B). TMC1 did not alter the voltage dependence of KCNQ1 or KCNQ1-KCNE1 currents, assessed using normalized tail currents (Figure 3C). We observed qualitatively similar results using a longer pulse designed to more fully activate KCNQ1-KCNE1 channels (Figure 3 D,E). In addition, TMC1 also diminished KCNQ1 current when the two were expressed in a different mammalian cell line (COS-7) (Figure 3F,G).
Figure 3. TMC1 diminishes KCNQ1 and KCNQ1-KCNE1 currents in CHO and COS-7 cells.
A. Currents generated in CHO cells by transfection with cDNAs encoding subunits as indicated, recorded by whole-cell patch-clamp. Averaged traces are shown (n = 11–18). Dotted lines indicate zero current level.
B. Mean peak currents from recordings as in A, n = 11–18. Inhibitory effects of TMC1: versus KCNQ1 alone, P = 0.12; versus KCNQ1-KCNE1: P = 0.1.
C. Mean normalized peak tail currents (G/Gmax) versus prepulse voltage from recordings as in A, n = 11–18. KCNQ1: V0.5, −1.9 ± 1.0 mV; slope, 14.6 ± 1.0 mV. KCNQ1-TMC1: V0.5, −8.2 ± 0.81 mV; slope, 16.5 ± 0.8 mV; P > 0.05. GV relationships for cells expressing KCNE1: fit values not reported as saturation does not occur.
D. Currents generated in CHO cells by transfection with cDNAs encoding subunits as indicated, recorded by whole-cell patch-clamp. Exemplar traces are shown (n = 9). Voltage protocol upper inset. Dotted lines indicate zero current level.
E. Mean peak currents from recordings as in D, n = 9. **P<0.01 at +60 mV.
F. Currents generated in COS-7 cells by transfection with cDNAs encoding subunits as indicated, recorded by whole-cell patch-clamp. Exemplar traces are shown (n = 5–13). Dotted lines indicate zero current level.
G. Mean peak currents from recordings as in F, n = 5–13. *P<0.05 at +60 mV. Voltage protocol upper inset.
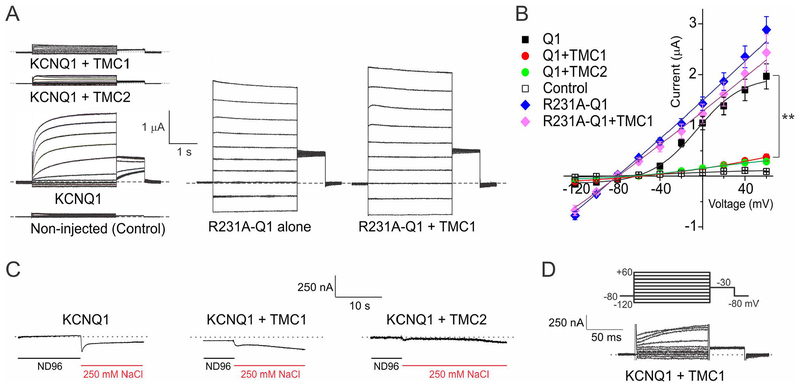
We also generated cRNAs for TMC1 and TMC2, and examined their effects when expressed in Xenopus laevis oocytes, using two-electrode voltage clamp (TEVC). Expression of TMC1 or TMC2 alone or together, in the absence of KCNQ1, made the oocytes unhealthy and precluded interpretable recordings. Co-expression of either TMC1 or TMC2 with KCNQ1 resulted in current inhibition as we observed in mammalian cell lines. Interestingly, the constitutively active R231A-KCNQ1 mutant channel (Panaghie and Abbott, 2007) was not inhibited by TMC1 co-expression (Figure 4A, B). Elevation of extracellular Na+ to 250 mM did not activate TMC1 or TMC2, in contrast to reports for C. elegans tmc-1 (Chatzigeorgiou et al., 2013) (Figure 4C). Similarly, oocytes expressing KCNQ1 + TMC1 did not exhibit inward Ca2+ currents when bathed with a solution containing 100 mM CaCl2+ (Figure 4 D). We conclude that TMC1 or TMC2 co-expressed with KCNQ1 in oocytes are not activated by membrane depolarization or by high extracellular salt.
Figure 4. TMC1 and TMC2 diminish KCNQ1 currents in Xenopus oocytes.
A. Currents generated in Xenopus oocytes by transfection with cRNAs encoding subunits as indicated, recorded by two-electrode voltage-clamp. Exemplar traces are shown, n = 6–9. Dotted lines indicate zero current level. Voltage protocol, upper inset.
B. Mean peak currents from recordings as in A, n = 6–9. **P<0.01 KCNQ1 versus KCNQ1 + TMC1 or TMC2. R231A-KCNQ1 versus R231A-KCNQ1 + TMC1, P > 0.05.
C. Exemplar traces showing lack of TMC-dependent effects of 250 mM bath NaCl on currents generated by membrane depolarization in Xenopus oocytes expressing KCNQ1 and/or TMC1 or TMC2, n = 4–5. Dotted lines indicate zero current level.
D. Exemplar trace showing lack of inward current with extracellular 100 mM Ca2+ in Xenopus oocytes expressing KCNQ1 and TMC1. Representative of 3 cells each for KCNQ1-TMC1 and KCNQ1-TMC2.
Surface-expressed TMC1 is not intrinsically mechanosensitive
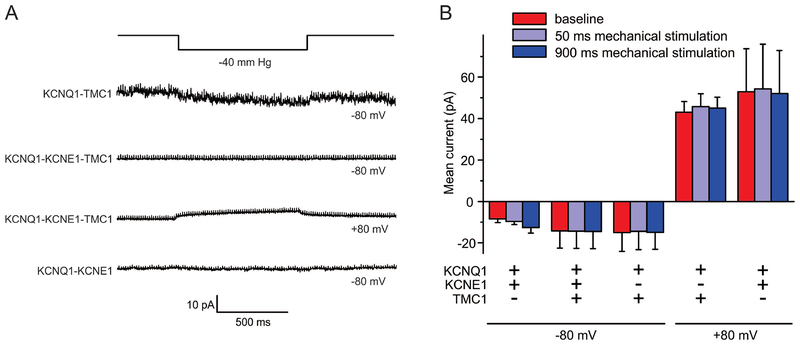
We next utilized quantitative, piezo-driven membrane displacement in conjunction with patch-clamping, as previously described for other channels (Pathak et al., 2014), in cell-attached patch mode in CHO cells expressing TMC1, co-expressed with KCNQ1, with or without KCNE1. We did not detect responses to mechanical stimulus (Figure 5A,B), either at voltages at which KCNQ1 is closed (−80 mV) or at which it is activated (+80 mV). We conclude that surface-expressed TMC1 is not intrinsically mechanosensitive.
Figure 5. CHO cell surface-expressed TMC1 is not intrinsically mechanosensitive.
A. Currents generated at various membrane potentials and in response to increased membrane tension generated by a high-speed pressure clamp system in CHO cells transfected with cDNAs encoding subunits as indicated, recorded by cell-attached patch-clamp. Exemplar traces are shown, n = 3–9.
B. Mean peak currents from recordings as in A, n = 3–9.
DISCUSSION
The TMCs are transmembrane proteins of uncertain function and no sequence similarity with other genes, yet with several links to inherited human diseases. The lack of knowledge of how TMCs function has severely hampered study of this gene family. Most have not been studied at all aside from genetic studies of disease linkage, and PCR analysis of transcript expression. The exceptions are that TMC1 and TMC2 expression in the ear has been studied in detail, TMC1 topology has been determined, and the cellular and auditory defects of df and Bth mice have been thoroughly dissected(Holt et al., 2014; Kawashima et al., 2011; Kurima et al., 2002; Lin, 2011; Marcotti et al., 2006; Pan et al., 2013; Vreugde et al., 2002). Given their expression in tissues including the auditory system, heart, brain, kidney and colon, and association of several of the TMC genes with inherited human diseases, the eight TMC genes are likely to be of considerable importance in human physiology and disease.
A critical barrier to progress in human TMC research has been the inability to study their function by heterologous expression (Fettiplace, 2016). The native current characteristics of ion channels, for example the voltage-gated K+ (Kv), Ca2+ (Cav) and Na+ (Nav) channels, are diversified by, and in many cases absolutely require, association with β subunits. For Kv channels, these are primarily the cytosolic Kvβ and KChIP subunits (An et al., 2000; Dolly and Parcej, 1996) and the single TM-segment DPPX (Nadal et al., 2003) and KCNE subunits (Abbott and Goldstein, 1998). KCNEs form stable complexes with Kv α subunits, generating functionally distinct K+ currents (Abbott et al., 2001; Abbott et al., 1999; Barhanin et al., 1996; Sanguinetti et al., 1996; Sesti et al., 2000a; Sesti and Goldstein, 1998; Sesti et al., 2000b; Takumi et al., 1988), and KCNE gene mutations cause human disease (Abbott et al., 1999; Delpon et al., 2008; Lundby et al., 2008; Splawski et al., 1997). Here, we found that TMC1, which is thought to have similar topology to that of Kv α subunits, does not appear to be directly regulated by KCNEs, but is brought to the cell surface by KCNQ1 co-expression. We show that this is via direct interaction, and that TMC1 surface expression can be altered by factors that similarly alter co-expressed KCNQ1 surface expression.
KCNQ1 is an osmosensitive channel that is activated by changes in cell volume, but not by stretch activation using negative hydrostatic pressure applied via the pipette (unlike, e.g., BK channels)(Hammami et al., 2009). Here, we show that although KCNQ1 rescues TMC1 surface expression, KCNQ1 does not rescue TMC1 activity. Thus, KCNQ1-TMC1 complexes (with or without KCNE1) did not respond to mechanical stimulation or high extracellular sodium. Currents generated by co-expression of TMC1 with KCNQ1 were scaled down versions of KCNQ1 currents, suggesting that the two do not form heteromeric pores together, but that TMC1, by co-assembling with KCNQ1, partially hampers KCNQ1 forward trafficking.
As KCNQ1 is not thought to co-localize with TMC1 in outer hair cells, we suggest that KCNQ1 is a proxy for another protein that is required by TMC1 for surface expression in vivo. This may be because KCNQ1 carries a sequence motif reminiscent of a true native partner of TMC1, permitting interaction and co-migration to the cell surface; KCNQ1 might harbor a motif that obscures a possible ER retention motif on TMC1. Further analysis may identify this motif, possibly leading to identification of the requisite native partner for TMC1 and/or TMC2 in outer hair cells. However, it is interesting to note that TMC1 is expressed in many other tissues outside the ear, including for example the colon, where KCNQ1 is also expressed. Thus, it is possible that KCNQ1 and TMC1 interact in vivo in locations other than the ear.
Several proteins may interact directly or indirectly with TMCs in outer hair cells and other cells, and could contribute to mechanosensitivity or transduce mechanical signals to facilitate TMC gating (Maeda et al., 2014; Xiong et al., 2012; Zhao et al., 2014). Future studies could incorporate co-expression of TMC1 and TMC1-KCNQ1 with, for example, outer hair cell proteins TMIE, TMHS and PCDH15 to determine if this induces mechanosensitivity. However, we consider it most likely that TMC1 (and TMC2) require the hair cell tip link apparatus in its entirety for mechanosensitivity.
Supplementary Material
ACKNOWLEDGEMENTS
This work was funded by the National Institutes of Health, National Institute of Deafness and Communication Disorders (DC015982 to GWA). We are grateful to Dr. Francesco Tombola (University of California, Irvine) for his invaluable advice and help with the mechanosensitivity experiments.
REFERENCES
- Abbott GW. 2014. Biology of the KCNQ1 potassium channel. New Journal of Science 2014(Article ID 237431):26. [Google Scholar]
- Abbott GW, Butler MH, Bendahhou S, Dalakas MC, Ptacek LJ, Goldstein SA. 2001. MiRP2 forms potassium channels in skeletal muscle with Kv3.4 and is associated with periodic paralysis. Cell 104(2):217–231. [DOI] [PubMed] [Google Scholar]
- Abbott GW, Goldstein SA. 1998. A superfamily of small potassium channel subunits: form and function of the MinK-related peptides (MiRPs). Quarterly reviews of biophysics 31(4):357–398. [DOI] [PubMed] [Google Scholar]
- Abbott GW, Sesti F, Splawski I, Buck ME, Lehmann MH, Timothy KW, Keating MT, Goldstein SA. 1999. MiRP1 forms IKr potassium channels with HERG and is associated with cardiac arrhythmia. Cell 97(2):175–187. [DOI] [PubMed] [Google Scholar]
- An WF, Bowlby MR, Betty M, Cao J, Ling HP, Mendoza G, Hinson JW, Mattsson KI, Strassle BW, Trimmer JS, Rhodes KJ. 2000. Modulation of A-type potassium channels by a family of calcium sensors. Nature 403(6769):553–556. [DOI] [PubMed] [Google Scholar]
- Barhanin J, Lesage F, Guillemare E, Fink M, Lazdunski M, Romey G. 1996. K(V)LQT1 and lsK (minK) proteins associate to form the I(Ks) cardiac potassium current. Nature 384(6604):78–80. [DOI] [PubMed] [Google Scholar]
- Chatzigeorgiou M, Bang S, Hwang SW, Schafer WR. 2013. tmc-1 encodes a sodium-sensitive channel required for salt chemosensation in C. elegans. Nature 494(7435):95–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corns LF, Johnson SL, Kros CJ, Marcotti W. 2016. Tmc1 Point Mutation Affects Ca2+ Sensitivity and Block by Dihydrostreptomycin of the Mechanoelectrical Transducer Current of Mouse Outer Hair Cells. The Journal of neuroscience : the official journal of the Society for Neuroscience 36(2):336–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delpon E, Cordeiro JM, Nunez L, Thomsen PE, Guerchicoff A, Pollevick GD, Wu Y, Kanters JK, Larsen CT, Hofman-Bang J, Burashnikov E, Christiansen M, Antzelevitch C. 2008. Functional effects of KCNE3 mutation and its role in the development of Brugada syndrome. Circulation Arrhythmia and electrophysiology 1(3):209–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolly JO, Parcej DN. 1996. Molecular properties of voltage-gated K+ channels. Journal of bioenergetics and biomembranes 28(3):231–253. [DOI] [PubMed] [Google Scholar]
- Fettiplace R. 2016. Is TMC1 the Hair Cell Mechanotransducer Channel? Biophysical journal 111(1):3–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammami S, Willumsen NJ, Olsen HL, Morera FJ, Latorre R, Klaerke DA. 2009. Cell volume and membrane stretch independently control K+ channel activity. The Journal of physiology 587(Pt 10):2225–2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt JR, Pan B, Koussa MA, Asai Y. 2014. TMC function in hair cell transduction. Hearing research. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanda VA, Purtell K, Abbott GW. 2011. Protein kinase C downregulates I(Ks) by stimulating KCNQ1-KCNE1 potassium channel endocytosis. Heart rhythm : the official journal of the Heart Rhythm Society 8(10):1641–1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawashima Y, Geleoc GS, Kurima K, Labay V, Lelli A, Asai Y, Makishima T, Wu DK, Della Santina CC, Holt JR, Griffith AJ. 2011. Mechanotransduction in mouse inner ear hair cells requires transmembrane channel-like genes. The Journal of clinical investigation 121(12):4796–4809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keresztes G, Mutai H, Heller S. 2003. TMC and EVER genes belong to a larger novel family, the TMC gene family encoding transmembrane proteins. BMC genomics 4(1):24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KX, Fettiplace R. 2013. Developmental changes in the cochlear hair cell mechanotransducer channel and their regulation by transmembrane channel-like proteins. The Journal of general physiology 141(1):141–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurima K, Peters LM, Yang Y, Riazuddin S, Ahmed ZM, Naz S, Arnaud D, Drury S, Mo J, Makishima T, Ghosh M, Menon PS, Deshmukh D, Oddoux C, Ostrer H, Khan S, Deininger PL, Hampton LL, Sullivan SL, Battey JF Jr., Keats BJ, Wilcox ER, Friedman TB, Griffith AJ. 2002. Dominant and recessive deafness caused by mutations of a novel gene, TMC1, required for cochlear hair-cell function. Nature genetics 30(3):277–284. [DOI] [PubMed] [Google Scholar]
- Labay V, Weichert RM, Makishima T, Griffith AJ. 2010. Topology of transmembrane channel-like gene 1 protein. Biochemistry 49(39):8592–8598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X 2011. Perception of sound and gravity by TMC1 and TMC2. The Journal of clinical investigation 121(12):4633–4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundby A, Ravn LS, Svendsen JH, Hauns S, Olesen SP, Schmitt N. 2008. KCNE3 mutation V17M identified in a patient with lone atrial fibrillation. Cellular physiology and biochemistry : international journal of experimental cellular physiology, biochemistry, and pharmacology 21(1–3):47–54. [DOI] [PubMed] [Google Scholar]
- Maeda R, Kindt KS, Mo W, Morgan CP, Erickson T, Zhao H, Clemens-Grisham R, Barr-Gillespie PG, Nicolson T. 2014. Tip-link protein protocadherin 15 interacts with transmembrane channel-like proteins TMC1 and TMC2. Proceedings of the National Academy of Sciences of the United States of America 111(35):12907–12912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcotti W, Erven A, Johnson SL, Steel KP, Kros CJ. 2006. Tmc1 is necessary for normal functional maturation and survival of inner and outer hair cells in the mouse cochlea. The Journal of physiology 574(Pt 3):677–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadal MS, Ozaita A, Amarillo Y, Vega-Saenz de Miera E, Ma Y, Mo W, Goldberg EM, Misumi Y, Ikehara Y, Neubert TA, Rudy B. 2003. The CD26-related dipeptidyl aminopeptidase-like protein DPPX is a critical component of neuronal A-type K+ channels. Neuron 37(3):449–461. [DOI] [PubMed] [Google Scholar]
- Pan B, Geleoc GS, Asai Y, Horwitz GC, Kurima K, Ishikawa K, Kawashima Y, Griffith AJ, Holt JR. 2013. TMC1 and TMC2 are components of the mechanotransduction channel in hair cells of the mammalian inner ear. Neuron 79(3):504–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panaghie G, Abbott GW. 2007. The role of S4 charges in voltage-dependent and voltage-independent KCNQ1 potassium channel complexes. The Journal of general physiology 129(2):121–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathak MM, Nourse JL, Tran T, Hwe J, Arulmoli J, Le DT, Bernardis E, Flanagan LA, Tombola F. 2014. Stretch-activated ion channel Piezo1 directs lineage choice in human neural stem cells. Proceedings of the National Academy of Sciences of the United States of America 111(45):16148–16153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen MB. 2002. Non-syndromic autosomal-dominant deafness. Clinical genetics 62(1):1–13. [DOI] [PubMed] [Google Scholar]
- Sanguinetti MC, Curran ME, Zou A, Shen J, Spector PS, Atkinson DL, Keating MT. 1996. Coassembly of K(V)LQT1 and minK (IsK) proteins to form cardiac I(Ks) potassium channel. Nature 384(6604):80–83. [DOI] [PubMed] [Google Scholar]
- Sesti F, Abbott GW, Wei J, Murray KT, Saksena S, Schwartz PJ, Priori SG, Roden DM, George AL Jr., Goldstein SA. 2000a. A common polymorphism associated with antibiotic-induced cardiac arrhythmia. Proceedings of the National Academy of Sciences of the United States of America 97(19):10613–10618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sesti F, Goldstein SA. 1998. Single-channel characteristics of wild-type IKs channels and channels formed with two minK mutants that cause long QT syndrome. The Journal of general physiology 112(6):651–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sesti F, Tai KK, Goldstein SA. 2000b. MinK endows the I(Ks) potassium channel pore with sensitivity to internal tetraethylammonium. Biophysical journal 79(3):1369–1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Splawski I, Tristani-Firouzi M, Lehmann MH, Sanguinetti MC, Keating MT. 1997. Mutations in the hminK gene cause long QT syndrome and suppress IKs function. Nature genetics 17(3):338–340. [DOI] [PubMed] [Google Scholar]
- Takumi T, Ohkubo H, Nakanishi S. 1988. Cloning of a membrane protein that induces a slow voltage-gated potassium current. Science 242(4881):1042–1045. [DOI] [PubMed] [Google Scholar]
- Vreugde S, Erven A, Kros CJ, Marcotti W, Fuchs H, Kurima K, Wilcox ER, Friedman TB, Griffith AJ, Balling R, Hrabe De Angelis M, Avraham KB, Steel KP. 2002. Beethoven, a mouse model for dominant, progressive hearing loss DFNA36. Nature genetics 30(3):257–258. [DOI] [PubMed] [Google Scholar]
- Xiong W, Grillet N, Elledge HM, Wagner TF, Zhao B, Johnson KR, Kazmierczak P, Muller U. 2012. TMHS is an integral component of the mechanotransduction machinery of cochlear hair cells. Cell 151(6):1283–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Kanda VA, Choi E, Panaghie G, Roepke TK, Gaeta SA, Christini DJ, Lerner DJ, Abbott GW. 2009. MinK-dependent internalization of the IKs potassium channel. Cardiovascular research 82(3):430–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita F, Horie M, Kubota T, Yoshida H, Yumoto Y, Kobori A, Ninomiya T, Kono Y, Haruna T, Tsuji K, Washizuka T, Takano M, Otani H, Sasayama S, Aizawa Y. 2001. Characterization and subcellular localization of KCNQ1 with a heterozygous mutation in the C terminus. Journal of molecular and cellular cardiology 33(2):197–207. [DOI] [PubMed] [Google Scholar]
- Zhao B, Wu Z, Grillet N, Yan L, Xiong W, Harkins-Perry S, Muller U. 2014. TMIE Is an Essential Component of the Mechanotransduction Machinery of Cochlear Hair Cells. Neuron 84(5):954–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.