Abstract
Nitrous oxide (N2O) and nitric oxide (NO) are atmospheric trace gases that contribute to climate change and affect stratospheric and ground-level ozone concentrations. Ammonia oxidizing bacteria (AOB) and archaea (AOA) are key players in the nitrogen cycle and major producers of N2O and NO globally. However, nothing is known about N2O and NO production by the recently discovered and widely distributed complete ammonia oxidizers (comammox). Here, we show that the comammox bacterium Nitrospira inopinata is sensitive to inhibition by an NO scavenger, cannot denitrify to N2O, and emits N2O at levels that are comparable to AOA but much lower than AOB. Furthermore, we demonstrate that N2O formed by N. inopinata formed under varying oxygen regimes originates from abiotic conversion of hydroxylamine. Our findings indicate that comammox microbes may produce less N2O during nitrification than AOB.
Subject terms: Microbial ecology, Bacterial physiology, Element cycles, Environmental microbiology
Ammonia-oxidizing bacteria and archaea are major producers of the gases nitrous oxide and nitric oxide. Here, Kits et al. show that a complete ammonia-oxidizing (comammox) bacterium emits nitrous oxide at levels that are comparable to those produced by ammonia-oxidizing archaea.
Introduction
Nitrous oxide (N2O) is the third most abundant greenhouse gas in the atmosphere. It contributes ~6% to the total radiative forcing and is also predicted as the dominant ozone depleting substance throughout the 21st century1. The atmospheric N2O concentration has continuously increased over the last decades at an average rate of ~0.31% per year, and this trend will continue1,2. Anthropogenic emissions of N2O make up 30–45% of the total global budget, with about two-thirds of this coming from agricultural and soil sources1, and are predicted to increase in the future as application of nitrogen fertilizers rises to feed the growing human population1,3. Microbial transformations of nitrogenous compounds, especially heterotrophic denitrification and chemolithoautotrophic aerobic nitrification, are the dominant contributor to N2O emissions from agriculture and soil management as well as wastewater treatment, the latter of which adds ~3.4% to the global N2O emission budget1,3. In addition to N2O, denitrifying and nitrifying microbes also release nitric oxide (NO) that represents an important metabolic intermediate for both guilds4. This activity is also environmentally relevant as NO contributes to the production of ground-level ozone and acid rain5. Thus, understanding the microbial players involved in NO/N2O production, the pathways that lead to the generation of these gases, and the environmental factors that control their fluxes is critical to modeling future emissions and developing appropriate mitigation strategies.
Classically, nitrification has been thought of as a two-step process. Ammonia (NH3) is oxidized via hydroxylamine (NH2OH) to nitrite (NO2−) by AOB and AOA, and subsequently nitrite oxidation to nitrate (NO3−) is catalyzed by nitrite oxidizing bacteria (NOB)4. Within the ammonia oxidizing microbes, two pathways were traditionally thought to contribute to NO and N2O emissions: (1) aerobic N2O formation from the abiotic reaction of the intermediate NH2OH with NO2− (also referred to as hybrid N2O formation), and (2) enzymatically catalyzed NO2− reduction to N2O via NO through “nitrifier-denitrification”6,7. The first pathway has been described for AOB and AOA8,9, while the latter has only been reported for AOB8,10. In AOB, hybrid N2O formation is the dominant process at atmospheric oxygen levels, while nitrifier-denitrification is more important at low O2 tension9,11–13. Very recently, two additional routes for NO/N2O production from AOB have been characterized. Firstly, the periplasmic tetraheme cyt. c P460 protein (CytL, present in most but not all AOB) oxidizes two molecules of NH2OH to N2O and water under anoxic conditions14, although the kinetics of this protein may render it inefficient at this role under physiological conditions. Additionally, this protein can bind NO and reduce it in the presence of NH2OH to N2O14. Second, NO (and not NO2− as previously considered) is formed by the activity of hydroxylamine dehydrogenase (HAO) under oxic and anoxic conditions14,15. CytL is used by AOB to detoxify NH2OH and NO, while the activity of HAO leading to NO formation, which is further oxidized by an unknown enzyme to NO2−, is essential for energy conservation in these organisms (Fig. 1). Pure culture work on marine and soil AOA, and soil microcosm studies on complex communities, strongly suggest that archaea produce lower yields of N2O than AOB (N2O/NO2− ratio %; 0.04–0.07% for AOA, 0.095–0.27% for AOB) during aerobic ammonia oxidation8,16–18.
Fig. 1.
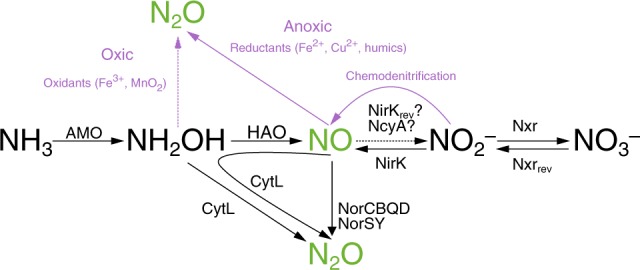
Biotic and abiotic pathways leading to NO and N2O production in bacterial nitrifiers. Black arrows depict confirmed (solid) and proposed (dashed) enzymatic reactions; confirmed or proposed enzymes are noted above or below the arrow. NcyA and NirKrev are candidates for the NO-oxidizing (NOO) enzyme bacterial ammonia-oxidizers. Violet arrows represent abiotic reactions that occur under oxic (dashed) or anoxic (solid) conditions with violet text outlining the reactants/conditions that favor those processes
Two-step nitrification requires coupling between ammonia oxidation and nitrite oxidation. Consequently, the two steps can also become uncoupled, for example, under high nitrogen load, which can inhibit NOB19,20. Additionally, mismatched nutrient affinities for NH3 and NO2− between the two guilds and varying energetic constraints often allow for the accumulation of NO2−20–22. The accumulated NO2− from uncoupled nitrification can drive production of N2O from hybrid formation, nitrifier-denitrification, and heterotrophic denitrification. In addition, it is important to keep in mind that NO and N2O can also be produced by a multitude of chemical reactions that use the key metabolites of ammonia oxidizers – NH2OH and NO2− (or its protonated form HNO2) – as the main precursors (for recent reviews see ref. 5).
Recently, the traditional perspective that the two steps of nitrification are always catalyzed by different microorganisms was refuted by the discovery of ‘comammox’ organisms that oxidize ammonia to nitrate on their own23,24. So far, comammox is restricted to the phylogenetic lineage II within the bacterial genus Nitrospira, which also contains canonical NOB25. Comammox genomes contain all genes encoding the known bacterial machineries for ammonia and nitrite oxidation – ammonia monooxygenase (AMO), HAO, the hydroxylamine ubiquinone reduction module (HURM), and nitrite oxidoreductase (NXR)23,24,26,27. Comammox Nitrospira are widely distributed and have been detected in various environments including pristine and agricultural soils, freshwater habitats, drinking water treatment systems, aquaculture biofilters, hot groundwater, and wastewater treatment plants in which they thrive, sometimes at considerable abundance23,24,27–31.
Interestingly, a potential solution to the aforementioned synchrony problem between canonical AOB/AOA and NOB is naturally provided by the comammox organisms which perform ammonia oxidation and nitrite oxidation within one cell. Consistently, recent mathematical modeling has suggested that comammox organisms should show improved efficiency in nitrogen removal and a significantly reduced production of NO and N2O32. However, it is currently not known whether comammox bacteria actually produce NO and/or N2O and if yes at which yields and via which pathways.
Recently, we obtained a pure culture of a comammox organism (Nitrospira inopinata) and characterized it kinetically33. In this study, we use this pure culture to: (1) quantify NO and N2O production by N. inopinata, (2) determine which biotic/abiotic pathways contribute to potential NO/N2O production in this organism, (3) compare the yields of NO/N2O from N. inopinata with known values for AOA and AOB, and (4) compare the genomic inventory for reactive nitrogen metabolism in various comammox organisms to gain insight into potential heterogeneity of their NO and N2O production pathways. By using a combination of micro-respirometry, gas chromatography, NO scavenging assays, N2O isotope analysis, and comparative genomics we demonstrate that Nitrospira inopinata, and possibly all other currently (meta)genomically characterized comammox organisms, produce NO as an important intermediate but cannot denitrify to N2O and thus produce low yields of N2O that are comparable to soil AOA.
Results and discussion
Comammox Nitrospira lack NO reductases
NO and N2O production by AOB, AOA, and NOB has been intensively studied and several key genes used by these nitrifiers for the production and consumption of these gases have been identified8,11,34–38 (Figs. 1 and 2). In contrast, no experimental data about the ability of comammox organisms to form NO and N2O are available and nothing is known about the potential importance of these compounds for the metabolism of complete nitrifiers.
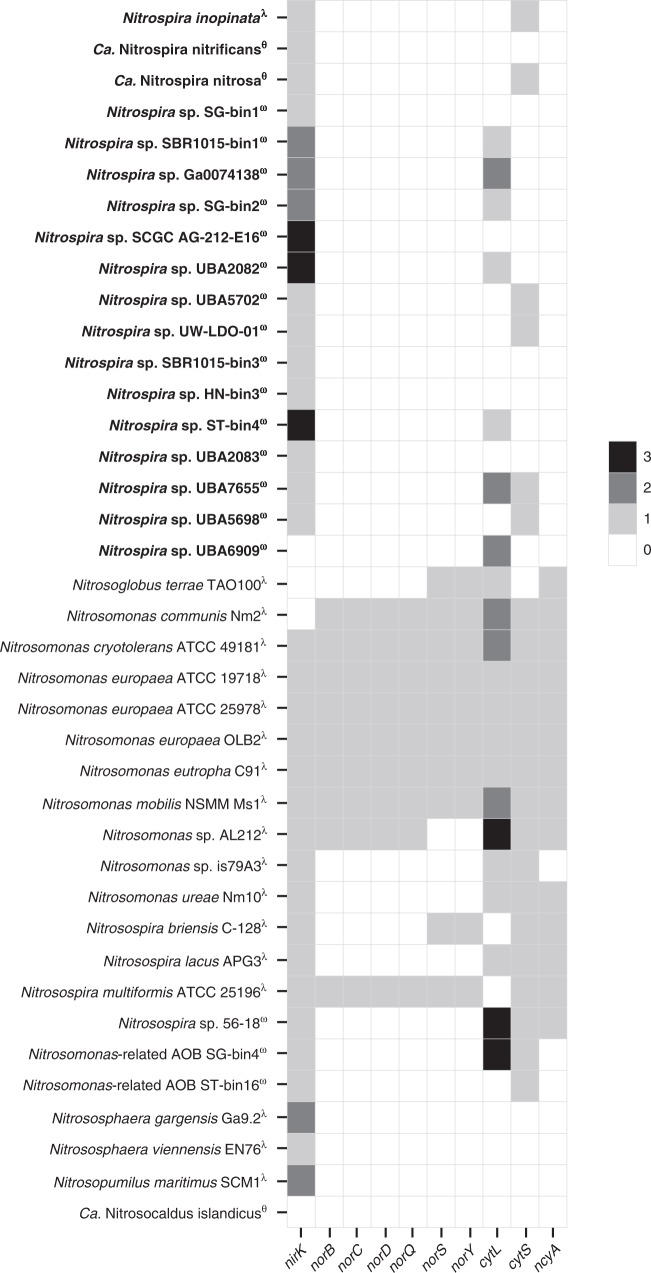
Fig. 2.
Gene inventory implicated in NOx metabolism in publicly available genomes of various nitrifiers. Comammox Nitrospira genomes are depicted in boldface. Genomes from enrichments and pure cultures are denoted by the symbols θ and ƛ, respectively. The symbol ω denotes uncultured organisms. The number of copies of each gene are denoted by the color bar. Published genomes and MAGs were compiled from the literature (see Supplementary Data 1 for sources, source data, locus tags, and bin/genome statistics) and downloaded from the Integrated Microbial Genomes website (https://img.jgi.doe.gov/cgi-bin/mer/main.cgi)
As a first step to close this knowledge gap, we updated previous comparative genomic analyses of comammox genomes26,27 and mined them for key genes that might be involved in NO and N2O metabolism (Fig. 2). These analyses included N. inopinata, the only comammox organism available as pure culture33, as well as genomes from two comammox enrichments and 15 comammox metagenome-assembled genomes (MAGs) from environmental samples retrieved via metagenomics23,26,29,30,39. All comammox organisms encode ammonia oxidation machinery that is more closely related to AOB than to AOA24. Part of this machinery is HAO that, in AOB, converts hydroxylamine (formed from NH3 by the AMO in an oxygen-dependent manner) to NO via an O2− independent, three-electron oxidation step15,24. Given its phylogenetic relationship to the HAO of AOB, and the fact that a planctomycete enzyme that shares multiple characteristics with HAO from N. europaea also produces NO40, it seems highly likely that the HAO of comammox organisms also generates NO. Under this assumption, comammox organisms, like AOB, would benefit from the enzymatic oxidation of NO to NO2− via an unknown NO oxidoreductase (NOO)41 in order to harvest a fourth electron for electron transport. For AOB the most compelling candidate for NOO is the red copper protein nitrosocyanin (NcyA), which is present in most AOB and is as highly expressed as AMO and HAO41,42. However, none of the comammox Nitrospira genomes we analyzed encoded ncyA, suggesting an alternative candidate for NOO (Fig. 2). All of the genomes, with the exception of a single MAG (with an estimated completeness of 86.84%), contained the nirK gene. NirK is encoded by many (but not all) AOB, AOA, and NOB6,27,43–46 and catalyzes the one electron reduction of NO2− to NO. NO generation by this enzyme is used by some AOB to facilitate efficient NH3 oxidation and also under hypoxic conditions to enable intracellular redox-balance via nitrifier-denitrification (Supplementary Note 1)11,13,37,47. Although NirK has been shown to operate reversibly and oxidize NO to NO2−48, the kinetics of the reaction are highly unfavorable at intracellular pH and redox potential, arguing that NirK is not an ideal candidate for the NOO.
In many AOB, the two-electron reduction of two molecules of NO to N2O is performed by two classes of cytochrome c nitric oxide reductase (NOR) – norCBQD and the alternative NO reductase norSY – while all genome-sequenced AOA lack NOR10,27. Physiological analyses of several AOB strains with or without cytochrome c-dependent NORs demonstrated that these enzymes are required for N2O formation via nitrifier-denitrification6,37. Interestingly, homologs for norCBQD and norSY are absent from all 23 currently known comammox MAGs and also absent from genomes of all cultivated strains including N. inopinata (Fig. 2)27,33. Comammox bacteria might thus not be able to reduce NO to N2O as part of the nitrifier-denitrification pathway under hypoxic conditions, similar to oligotrophic strains of AOB6.
We also queried the presence of the c cytochromes P460 (cytL) and c′-beta (cytS) in the genomes of comammox Nitrospira. The P460 enzyme from N. europaea is considered to be a detoxifying enzyme14 as it converts two equivalents of NH2OH (or one NH2OH and one NO) to N2O under anoxic conditions, though this enzyme is expressed during aerobic growth in some but not all tested AOB14,42. The function of CytS is still unknown, but for AOB it has been suggested to be involved in the oxidation/reduction of N-oxides or electron transfer for either detoxification or energy conservation49. Both the cytochrome P460 and cytochrome c′-beta are found sporadically in genomes of comammox Nitrospira (Fig. 2). All of the currently enriched or cultured comammox representatives including N. inopinata lack cytL, while the genomes of N. inopinata and Ca. N. nitrosa contain the uncharacterized cytS (Fig. 2).
Taken together, N. inopinata and all other genome-sequenced comammox microbes possess the genetic potential to produce NO via HAO or NirK activity, but lack bona fide NO reductases to form N2O. However, keeping in mind that ~46% of the N. inopinata genes have no functional annotation and several, often unrelated, enzyme classes can catalyze identical transformations of nitrogen compounds50, physiological experiments as well as protein purification and characterization are clearly required to examine formation, magnitude, and importance of NO and N2O formation and NO oxidation in comammox organisms.
N. inopinata releases and consumes NO under oxic conditions
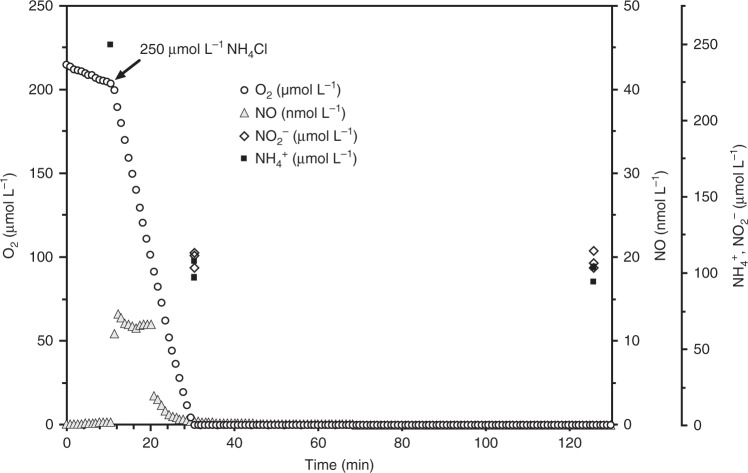
To determine whether N. inopinata produces and consumes NO, instantaneous O2 and NO kinetics were measured with microsensors during NH3 and NO2− oxidation by N. inopinata. Addition of 250 µM NH4+ into the micro-respirometry (MR) chamber led to immediate substrate-dependent O2 consumption and NO production. NO production peaked at ~13 nM after ~30% of the dissolved O2 was consumed, followed by net NO consumption (Fig. 3). Since N. inopinata is an oligotroph, we also tested how substrate concentration influences net NO flux. Lower substrate concentrations (<15 µM NH4+) resulted in significantly less NO production (<0.8 nM) (Supplementary Fig. 1). There was no measurable NO production after O2 was depleted in the presence of NH3, reflecting the dependency of NO production on O2 and strongly suggesting that N. inopinata under the conditions applied did not respire anaerobically with NO2− as an electron acceptor using storage products as electron donors. Despite comparable absolute cell numbers (~1 × 1010 cells), net production of NO (30 nM per ~1 × 1010 cells) by the N. inopinata biomass was about an order of magnitude lower during aerobic oxidation of the same amount of NH3 than by the oligotrophic AOB Nitrosomonas sp. Is79A3 or N. ureae6. Furthermore, previous work demonstrated that both of these oligotrophic AOB produce very large (>250 nM) quantities of NO after the onset of hypoxia (ascribed to the activity of their NirK enzymes). This phenomenon reflects the absence of NO reductases in these strains, which are required for NO reduction to N2O in the nitrifier-denitrification pathway6,37. In contrast, the NO reductase encoding Nitrosomonas europaea ATCC 19718 produced ~180–210 nM NO (per ~1 × 1010 cells) throughout aerobic NH3 oxidation and consumed NO via nitrifier denitrification after the onset of hypoxia (Supplementary Fig. 2). Consistently, other previously analyzed AOB also produced >50 nM NO (per 1 × 1010 total cells) during aerobic NH3 oxidation prior to hypoxia in the MR chamber6.
Fig. 3.
Instantaneous O2 consumption and NO production during NH3 oxidation by N. inopinata. The data shown here is a single representative of three biological replicates (n = 3). Two additional biological replicates are shown in Supplementary Fig. 6. Dissolved O2 is shown in open circles, dissolved NO in filled gray triangles, NO2− in open diamonds, and NH4+ in filled squares. The NH4+ concentration immediately after injection (~10 min) was inferred from the injected volume of a stock NH4Cl solution, otherwise NO2− and NH4+ concentrations were determined in three technical replicates (n = 3). Experiments were performed in a microrespiration (MR) chamber fitted with O2 and NO microsensors. The arrow marks the addition of 250 µM NH4Cl into the MR chamber. About 110 µM residual NO2− was present in the chamber once O2 reached a concentration below the detectable level at ~35 min. No NO formation from NH4+ was measurable in sterile media controls containing the same amount of heat-killed biomass of N. inopinata. Source data are provided as a Source Data file
The lack of measurable NO production by N. inopinata during O2 -limited conditions despite the fact that NirK was the second most abundant protein in proteomic analysis of N. inopinata even under aerobic conditions (Supplementary Fig. 3) suggests that NirK either has very weak activity under hypoxic conditions, producing NO below the detection limit of our instrument (~0.25 nM NO), or that N. inopinata does not perform NirK-based nitrifier-denitrification during hypoxia at all. The NO profile during aerobic NH3 oxidation by N. inopinata shows interesting similarities to that previously reported for the AOA strain N. viennensis, which also rapidly produces NO and then consumes it during aerobic NH3 oxidation10. However, in contrast to N. inopinata, N. viennensis additionally produces NO at the onset of hypoxia10. Although the NO production and consumption profiles of N. inopinata and the AOA strains N. viennensis and N. maritimus are different10,51,52, NirK is strongly expressed in all three organisms53,54 and the release of NO appears to be more tightly controlled by them than in AOB.
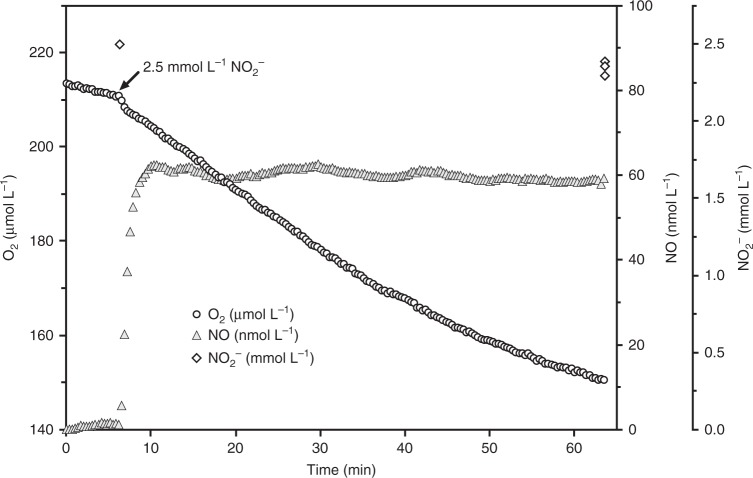
Interestingly, using NO2− instead of NH3 as the electron donor resulted in a different NO production and consumption profile by N. inopinata (Fig. 4); addition of 2.5 mM NO2− led to immediate net NO production after which NO levels reached a steady-state level of ~45 nM (Fig. 4). Net consumption of NO was not evident during further NO2− oxidation. In contrast, instantaneous NO concentrations were below our detection limit (~0.25 nM) for the closely related non-comammox Nitrospira, N. moscoviensis, during NO2− oxidation (Supplementary Fig. 4). To our knowledge, NO production in Nitrospira has not been investigated previously but previous work has shown that NO (65 nM NO in the liquid phase) inhibits NO2− oxidation to NO3− in Nitrospira dominated sludge55. The more distantly related proteobacterial nitrite-oxidizer, Nitrobacter winogradskyi produces38 and consumes NO36,38 in a NO2−-dependent manner during normal aerobic growth. However, the total NO flux is strongly influenced by its quorum sensing system and decreased when quorum sensing was quenched38.
Fig. 4.
Instantaneous O2 consumption and NO production during NO2− oxidation by N. inopinata. The data shown here represents a single replicate of three biological replicates (n = 3). Two additional biological replicates are shown in Supplementary Fig. 8. Dissolved O2 is shown in open circles, dissolved NO in filled gray triangles, and NO2− in open diamonds. The NO2− concentration immediately after injection (~6 min) were inferred from the injected volume of a stock NaNO2 solution, otherwise NO2− concentrations were determined in three technical replicates (n = 3). Experiments were performed in a 10-mL microrespiration (MR) chamber fitted with an O2 and NO microsensors. The arrow marks the addition of 2.5 mM NO2− into the MR chamber. No NO formation from NO2− was measurable in oxic sterile media controls containing heat-killed biomass. Source data are provided as a Source Data file
It is interesting to note that in N. winogradskyi the NO producing nitrite reductase NirK has been postulated to facilitate nitrite oxidation under O2-limiting conditions by maintaining redox balance via regulation of electron flow. At low oxygen tension in the presence of nitrite, NirK is strongly upregulated and more NO is produced causing reversible inhibition of the heme-copper cyt c terminal oxidase and thus intensifying reverse electron transport for NAD(P)+ reduction and storage compound formation36. In contrast, we show here that stable NO production by N. inopinata during NO2− oxidation occurs under fully oxic conditions, which is consistent with a constitutive high expression of NirK during aerobic growth of N. inopinata (Fig. 4) (Supplementary Fig. 3). Alternatively, it is conceivable that the contrasting NO profiles in N. moscoviensis and N. inopinata are caused by a reversal of the yet unknown NOO enzyme in N. inopinata at very high NO2− concentrations, a process that does not occur in N. moscoviensis because it lacks the genetic repertoire to oxidize NH3 to NO2−.
PTIO is a potent inhibitor of NH3 oxidation in N. inopinata
Both AOB and AOA produce NO as an obligate intermediate during NH3 oxidation10,41,51,52. However, AOA exhibit very tight control over the production and consumption of NO during NH3 oxidation10,51,52. This difference in the regulation of NO concentrations may explain why the AOA are selectively inhibited by low concentrations of the NO scavenger PTIO51,56. The similar aerobic NO kinetics between N. inopinata and the AOA N. viennensis raised the hypothesis that comammox Nitrospira may be sensitive to inhibition by low concentrations of PTIO as well. We tested this hypothesis by comparing instantaneous O2 consumption rates and batch culture NH3 oxidation activity of N. inopinata in the presence of various concentrations of PTIO to non PTIO-amended biomass (Supplementary Fig. 5). Indeed, PTIO was a potent inhibitor of instantaneous O2 consumption and batch culture NH3 oxidation in N. inopinata with a half-effective maximal concentrations (EC50) of 18.9 and 63.6 µM, respectively. The EC50 of PTIO for N. maritimus and N. viennensis ranges from 17.5–18.3 µM, while N. multiformis and other AOB are only inhibited by PTIO concentrations >300 µM51,56. Washing and reharvesting N. inopinata cells treated with low but inhibitory concentrations of PTIO (33–100 μM PTIO) to remove the PTIO resulted in complete recovery (>95%) of activity, while cells treated with high concentrations of PTIO only showed partial (mean ± standard deviation: 28.3 ± 11.9%, 330 μM PTIO) or no recovery of activity (1.8 ± 3.7%, 1000 μM PTIO; Supplementary Fig. 5). These results are consistent with PTIO acting as a NO-binding, reversible inhibitor at low concentrations, and a irreversible cytotoxin at high concentrations. However, it still remains a possibility that the PTIO-dependent inhibition we observed was caused by the imino nitroxides (PTIs) and NO2 formed by the reaction of PTIO and NO rather than NO chelation.
To determine whether PTIO exhibits general cytotoxicity toward Nitrospira cells, we tested the effect of PTIO on instantaneous O2 consumption and growth in the non-ammonia oxidizing N. moscoviensis. Generally, PTIO was significantly less inhibitory to N. moscoviensis, with estimated EC50 values of 385.6 μM and 100.2 μM for instantaneous activity and batch culture NO2− oxidation, respectively (Supplementary Fig. 5). However, considering that PTIO was not fully inhibitory to N. moscoviensis at even 1000 μM PTIO during batch culture growth, the calculated EC50 in this condition is not reliable. Nearly complete inhibition (<10% activity) was only evident when we measured instantaneous O2 consumption in the 1000 μM PTIO treatment and the calculated EC50 under these conditions (385.6 μM) is very similar to that of previously published values for AOB51. Finally, washing and reharvesting N. moscoviensis cells that were significantly inhibited by PTIO resulted in no measurable recovery when compared to an untreated control (6.3 ± 3.3%, 1000 μM PTIO + wash; Supplementary Fig. 5). Collectively, the inhibitory effect of PTIO on N. moscoviensis at these concentrations is permanent even after it is removed and this suggests that PTIO at high concentrations acts as a irreversible cytotoxin in N. moscoviensis.
Taken together, the NO production profile, the low EC50 of PTIO, and the reversibility of PTIO toxicity in N. inopinata suggest an essential role for NO as an intermediate in N. inopinata. Furthermore, our data reveal that PTIO can no longer be considered as a selective inhibitor of AOA in studies targeted at assessing the roles of various nitrifying microbes in environmental systems as comammox Nitrospira will also be inhibited upon addition of low concentrations of this NO scavenger. Furthermore, inhibition of other nitrifying microbes not mediated by NO scavenging but by the potential cytotoxicity of PTIO, especially at higher concentrations, has to be considered in the design of such molecular ecology experiments.
Respirometry suggests abiotic N2O formation by N. inopinata
AOB and AOA release small amounts of NH2OH during ammonia oxidation under fully oxic conditions and the abiotic conversion of NH2OH with Fe3+, Mn4+, Cu2+, NO2−, and other components of the surrounding matrix under oxic conditions57 explains most of the N2O formation by these nitrifiers under these conditions6,7,10. Hypoxia leads to a significant increase in the N2O yield from NH3 in AOB9,12,13,58, while oxygen concentration has no major influence on the N2O yields of all tested AOA8,17,52. Main pathways that contribute to N2O formation during O2 limitation in these nitrifiers are nitrifier-denitrification –the sequential enzymatic reduction of NO2− to N2O via NIR and NOR – and chemodenitrification, whereby NO2− or NO is non-enzymatically reduced to N2O via media components or heat-killed cell moieties6,59. The NO can be derived from enzymatic NO2− reduction or from reactions of NOx with NH2OH. Studies on AOB from diverse environments and adapted to different substrate concentrations showed that all of them produce N2O within minutes of the transition from oxic to anoxic conditions6. This released N2O originates mainly from nitrifier-denitrification in AOB that encode nitric oxide reductases (norCBQD or norSY) and to a much lower extent from chemodenitrification in oligotrophic AOB that lack NO reductases and emit large quantities of NO during transition from oxic to anoxic conditions6. The soil thaumarcheaon N. viennensis, which cannot perform nitrifier-denitrification to N2O, also releases large quantities of NO during hypoxic conditions which then reacts abiotically with medium components (Cu2+ and Fe2+) to form N2O8,10.
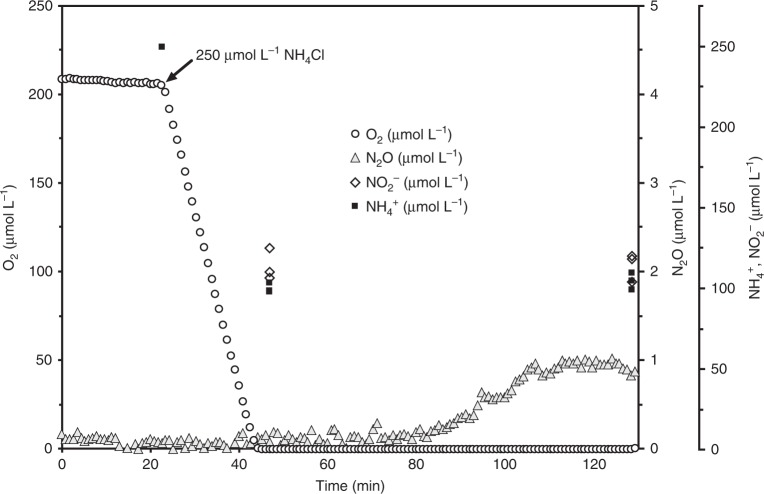
Using NH3 as the electron donor, we measured instantaneous N2O production with microsensors by N. inopinata biomass during oxic conditions and through a period of hypoxia. Interestingly, N. inopinata did not produce N2O during aerobic NH3 oxidation or during the transition from oxic to anoxic conditions (Fig. 5) that could be measured with the N2O microsensor that has a relatively low sensitivity (100 nM). Small quantities of N2O were only measurable with the sensor from N. inopinata ~40 min after O2 was depleted below the limit of detection (~300 nM dissolved O2). This delayed N2O accumulation did not coincide with NO release in replicate traces where NO was measured (Figs. 3 and S6). Unlike growth assays, these short micro-respirometry experiments with high biomass and high substrate concentrations likely result in a relatively large accumulation in intracellular reductant in the form of NH2OH52. To test whether this delayed N2O release could originate from cell lysis and abiotic formation of N2O from NO2−, NH2OH, and other media components, we incubated heat-killed N. inopinata cells under anoxic conditions in AFW media supplemented with 1.8 µM NH2OH and 100 µM NO2−. This NH2OH concentration was chosen based on determined extracellular NH2OH concentrations in enrichment cultures of N. inopinata7. These abiotic controls yielded 0.83 ± 0.1 µM N2O (mean ± standard deviation), explaining most of the observed N2O in the live cell incubations. Together, these results suggest that the delayed N2O formation under O2-limiting conditions by N. inopinata originates mainly from abiotic reactions of accumulated NH2OH and not from the enzymatic reduction of NO to N2O.
Fig. 5.
Instantaneous O2 consumption and N2O production during NH3 oxidation by N. inopinata. The data shown here is a single representative of three biological replicates (n = 3). Two additional biological replicates are shown in Supplementary Fig. 9. Dissolved O2 is shown in open circles, dissolved N2O in filled gray triangles, NO2− in open diamonds, and NH4+ in filled squares. The NH4+ concentration immediately after injection (~23 min) was inferred from the injected volume of a stock NH4Cl solution, otherwise NO2− and NH4+ concentrations were determined in three technical replicates (n = 3). Experiments were performed in a 10-mL microrespiration (MR) chamber fitted with O2 and N2O microsensors. The arrow marks the addition of 250 µM NH4Cl into the MR chamber. About 110 µM residual NO2− was present in the chamber once O2 reached below the detectable level at ~45 min. Source data are provided as a Source Data file
N. inopinata N2O originates from abiotic NH2OH conversion
The short duration of the micro-respirometry experiments and the limited sensitivity of the N2O microsensors (100 nM dissolved N2O) makes calculating the precise N2O yield during NH3 oxidation very difficult. To calculate the precise yields of N2O (as a N2O/NH3 ratio in percent) we performed batch growth experiments with N. inopinata biomass in sealed serum vials under NH3- and O2-limiting conditions and measured consumption of relevant metabolites and production of N2O using gas chromatography. N2O formation during oxic (~20.8% O2), NH3-limited growth was entirely dependent on NH3 oxidation; no N2O formation was observed during NO2− oxidation to NO3− after all of the NH3 was depleted (Fig. 6a). Similarly, NO2− oxidation by N. moscoviensis in batch growth experiments yielded only trace (mean±standard deviation: 4.3 ± 2.3 nM) amounts of N2O under fully oxic and hypoxic conditions (Supplementary Fig. 7).
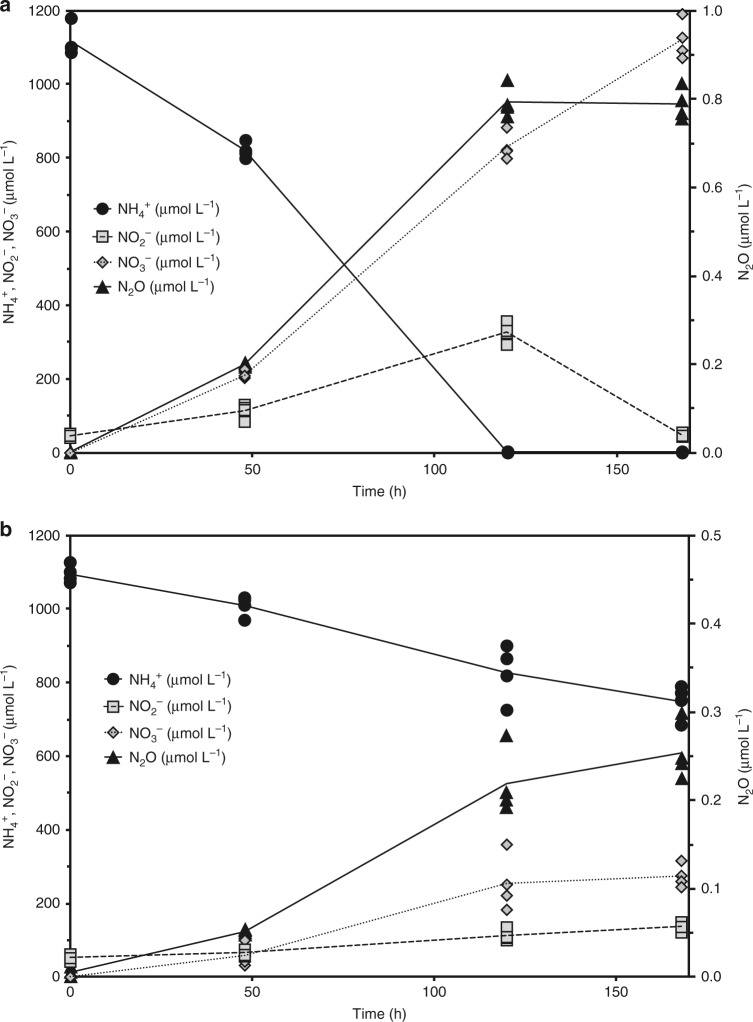
Fig. 6.
Oxidation of NH3 to NO2− and NO3− and concurrent N2O yield during growth of N. inopinata. N. inopinata was incubated in closed serum vials under fully oxic conditions (initial O2 at ~20.8%, a) or hypoxic conditions (initial O2 at ~0.89%, b). NH4+ is shown in filled circles, NO2− in filled squares, NO3− in filled diamonds, and N2O in filled triangles; for each time point, individual replicates (n = 4 biological replicates) are plotted and the means connected by a solid black line. The amount of biomass at t = 0 was the same in the oxic and hypoxic vials. NH3 oxidation to NO3− is nearly stoichiometric in panel a due to unlimiting O2; in panel b, however, limiting O2 concentrations prevented the stoichiometric oxidation of NH3 to NO3−. O2 concentrations in the hypoxic treatment at 168 h were below the limit of detection (300 nM). Source data are provided as a Source Data file
The N2O yield (as a N2O per NH3 ratio %) during aerobic NH3 oxidation for N. inopinata was 0.070 ± 0.006 (mean ± standard deviation). This measured N2O yield closely matched the predicted yield of N2O from abiotic reactions between media components and extracellular NH2OH for N. inopinata cultivated at 500 µM NH4+ (0.06%)7. This suggests that all of the N2O produced by N. inopinata during oxic growth on NH3 came from the abiotic conversion of NH2OH. Further, an N2O yield of 0.073 ± 0.033 % in O2− limited incubations that were initiated at ~1.8 µM dissolved O2 (equivalent to 0.89% O2) demonstrated that the N2O yield was not influenced by hypoxia (Fig. 6b). This matches observations in N. maritimus and N. viennensis in which the N2O yield during growth is not influenced by varying O2 concentrations and is in stark contrast to AOB like N. europaea and N. multiformis that produce ~3 times more N2O on a per mol NH3 basis during nitrifier-denitrification under low O2 conditions8,9,13,58. Furthermore, the N2O yield values for N. inopinata (0.070 ± 0.006%) are very similar to those observed for N. viennensis (0.07 to 0.09%) and other several other soil AOA (~0.080%)8.
To obtain additional information on the pathway(s) leading to N2O production in N. inopinata, we determined the intramolecular (natural abundance) distribution of 15N within the linear N2O molecule (Nβ-Nα-O) between the central (α) and the external (β) position (also called the site preference SP; δ15N-SP, defined as δ15Nα - δ15Nβ) from headspace N2O harvested from batch yield experiments performed under NH3- and O2-limiting conditions (Fig. 6). The site preference of N2O produced during NH3-limited growth (mean ± standard deviation: 33.4 ± 0.3‰) was the same as during hypoxia (34.2 ± 1.4‰). The unvarying SP of N2O produced during the different conditions reflects that the N2O source did not change in response to hypoxia. Such a positive δ15N-SP is also consistent with a N2O source from NH2OH conversion (observed under oxic conditions to be 30.8–35.6‰60,61, for AOB and 13.1–30.8‰ for AOA16,62), whereas N2O produced via nitrifier-denitrification or heterotrophic denitrification has a SP near or below 061,63,64.
In summary, we first show that N. inopinata has very tight control over NO production during aerobic NH3 oxidation, that it also produces NO during aerobic NO2− oxidation, but does not release NO during hypoxia. These data collectively strongly suggest an important but not yet fully explored metabolic role of NO for complete nitrifiers of the genus Nitrospira. Consistently, low concentrations of the NO scavenger PTIO inhibit N. inopinata. Importantly, we then demonstrate that N. inopinata cannot denitrify to N2O due to the absence of NO reductases in the genome, that it has a low N2O yield that is comparable to cultivated soil AOA and not influenced by O2 levels, and that the N2O formed originates from abiotic NH2OH conversion. While we cannot rule out the possibility that some comammox Nitrospira encode a currently unknown NO reductase, we hypothesize that all other currently enriched and/or metagenomically characterized comammox Nitrospira also lack the ability to denitrify to N2O because they lack homologs for known NO reductases. Consequently, adoption of conditions that favor the growth of complete nitrifiers over AOB, for example, in engineered systems and in soils where comammox bacteria have been identified28,30,65, may influence nitrification-dependent N2O emissions. Quantifying the relative contribution of comammox organisms as well as canonical nitrifiers to ammonia oxidation and N2O production in natural and engineered environments will thus be an important aim for follow-up research.
Methods
Strains and cultivation
Nitrospira inopinata was routinely cultivated at 37 °C in AOM medium24 which contained (per L): 50 mg KH2PO4, 75 mg KCl, 50 mg MgSO4 7H2O, 584 mg NaCl, 1 mL specific trace element solution (TES), and 1 mL of selenium-tungstate solution (SWS). TES contained (per litre of deionized water): 34.4 mg MnSO4 × H2O, 50 mg H3BO3, 70 mg ZnCl2, 72.6 mg Na2MoO4 × 2H2O, 20 mg CuCl2 × 2H2O, 24 mg NiCl2 × 6H2O, 80 mg CoCl2 × 6H2O, and 1 g of FeSO4 × 7H2O (dissolved in 2.5 mL 37% HCl)24. SWS contained (per litre): 0.5 NaOH, 3 mg Na2SeO3 × 5H2O, and 4 mg Na2WO4 × 2H2O24. The AOM medium was supplemented with 1 mM NH4+ and 4 g L−1 of CaCO3, the latter of which acts as a solid buffering system and substrate for attachment. The final pH of the medium was ~8.0 after sterilization. AOM medium could not be used to cultivate N. inopinata for the micro-respirometry experiments because the large quantities of undissolved CaCO3 interfered mechanically with the highly sensitive NO sensor that, unlike the O2 and N2O sensors, has an extremely small signal (10−14 ampere). Consequently, Artificial Fresh Water Medium (AFW) was used to prepare N. inopinata biomass for the micro-respirometry (MR) experiments. The basal AFW (without pyruvate) containing 1 mM NH4+ was supplemented with (per L): 1 mL of 1000X non-chelated trace element solution, 1 mL of 1000X vitamin solution, and 1 mL of 7.5 mM Fe-NaEDTA solution16. The final pH of the AFW was set to ~7.2 through the addition of HEPES (prepared initially to pH 7.6) and NaHCO3, which were added at final concentrations of 4 mM and 3 mM, respectively. The pure culture of Nitrospira inopinata has been deposited in the JCM (accession no. JCM 31988). Nitrosomonas europaea ATCC 19718 was cultivated in the same basal AFW medium at pH 8.5 and supplemented with 2.5 mM total NH4Cl. Nitrospira moscoviensis was cultivated at 37 °C in mineral nitrite medium (NOM)66 amended with 5 mM NO2−. Basal NOM contained (per litre of deionized water): 10 mg CaCO3, 0.5 g NaCl, 50 mg of MgSO4 × 7H2O, 0.15 g KH2PO4, 10 mg NH4Cl, 0.034 mg MnSO4 × H2O, 0.05 mg H3BO3, 0.07 mg ZnCl2, 0.0726 mg Na2MoO4 × 2H2O, 0.02 mg CuCl2 × 2H2O, 0.024 mg NiCl2 × 6H2O, 0.08 mg CoCl2 × 6H2O, and 1 mg FeSO4 × 7H2O66.
Instantaneous NO and N2O measurement
N. inopinata, N. moscoviensis, and N. europaea cultures were monitored daily and harvested immediately once all the substrate was consumed (normally ~7–9 days) by centrifugation using 10 kDa-cutoff, Ultra-15 Centrifuge Filter units (Amicon, Darmstadt, Germany). About 500 mL of mid-exponential phase culture were harvested per replicate experiment for N. inopinata, while 300 L of culture was used per replicate experiment with N. moscoviensis and N. europaea. Harvested biomass was washed twice with substrate-free medium and then resuspended in 10 mL of the same substrate-free medium. The biomass was then transferred into a 10 mL, double-port MR chamber (allowing no headspace) that was fitted with two MR injection lids and two glass coated stir bars. All MR experiments were performed in a recirculating water-bath at 37 °C. O2 uptake was measured using a OX-MR oxygen microsensor (Unisense, Aarhus, Denmark). N2O and NO concentrations were measured using an N2O-MR sensor (Unisense) and an ami700-NO sensor (Innovative Instruments, Inc., Tampa, USA), respectively. Substrate (NH4+ or NO2−) were injected into the chamber via an injection port using either a 10-µL or 50-µL syringe (Hamilton, Reno, USA) fitted with a 26G needle. A 250-µL syringe (Hamilton, Reno, USA) fitted with a 26 G needle was also used to withdraw small aliquots (150 µL) from MR chambers during experiments to measure NH4+ and NO2− concentrations, and sterile media was always backfilled. Starting metabolite concentrations were inferred from injected amounts; otherwise, all NH4+ and NO2− concentrations were measured. NH4+, NO2−, and NO3− concentrations were quantified photometrically with the Berthelot reagent, acidic Griess reagent, and VCl2/Griess reagent, respectively24,33, using an Infinite 200 Pro spectrophotometer (Tecan Group AG, Maennedorf, Switzerland). NH4+ injections into the 10 mL MR chamber were always 250 µM for N. inopinata and 2 mM for N. europaea. NO2− injections were 1 mM for N. moscoviensis and 2.5 mM for N. inopinata. Higher concentrations of NO2− were used for N. inopinata due to the comparatively low apparent affinity of N. inopinata for NO2− (Km(app) = 449.2 ± 65.8 µM) compared to its very high affinity for ammonia (Km(app) = 63 ± 10 nM NH3)33. Further, it was not feasible to measure NO2− oxidation from fully oxic conditions to anoxic conditions due to the slow rate of NO2−-dependent O2 uptake and the poor affinity of N. inopinata for NO2−. We expected no cell doubling during the MR experiments, as the doubling time of all strains (10–40 h) is significantly greater than the duration of the longest MR trace (~2 h). The OX-MR and N2O-MR sensors were plugged directly into a microsensor multimeter while the ami700-NO sensor was polarized using a One-Channel Free Radical Analyzer (World Precision Instruments, Sarasota, USA). All electrodes were polarized for >1 day prior to use and calibrated according to the manufacturer’s instructions. All data were logged on a laptop via the microsensor multimeter using SensorTrace Logger software (Unisense). The output from One-Channel Free Radical Analyzer was run into the microsensor multimeter using a BNC/lemo adapter. Abiotic controls were performed in triplicate as described above but with heat-killed cells (autoclaved at 121 °C for 20 min) resuspended in sterile anoxic medium. Anoxically prepared aliquots of NH4+, NO2− and NH2OH were injected into the MR chamber through the injection port using a 10 µL syringe (Hamilton, Reno, USA). Anoxic AFW, NH4+, NO2−, and NH2OH were prepared by sparging the solutions with N2 gas for 1 h prior to use.
PTIO inhibition
Batch and micro-respirometry inhibition experiments with the NO scavenger 2–phenyl-4,4,5,5,-tetramethylimidazoline-1-oxyl 3-oxide (PTIO; purity >98%; TCI, Germany) were performed using exponential-phase cultures of N. inopinata and N. moscoviensis grown in AOM medium or mineral nitrite medium (NOM)66, respectively. For the batch experiments, cells were harvested by centrifugation (8000 × g, 20 min, 20 °C), washed once (10 mg L−1 CaCO3 AOM medium for N. inopinata and NO2−-free NOM medium for N. moscoviensis) and resuspended undiluted in the same medium containing either 1 mM NH4Cl (N. inopinata) or 1 mM NO2− (N. moscoviensis). The cultures was aliquoted (20 mL) in glass serum bottles (60 mL) sealed with crimp caps and quadruplicates were incubated in the dark at 37 °C in the presence of 0, 3.3, 10, 33, 100, and 330 µM PTIO, respectively. Only the concentrations of PTIO that inhibited N. inopinata were tested on N. moscoviensis (in addition to the unamended control) - 0, 33, 100, and 330 µM PTIO. PTIO was added in the respective amounts as a 10-mM stock solution in autoclaved MilliQ water. Abiotic controls with 0 and 330 µM PTIO and controls with heat-killed cells with 0 µM PTIO were run in triplicate. NH4+, NO2−, and NO3− concentrations were quantified with the Berthelot reagent, acidic Griess reagent, and VCl2/Griess reagent, respectively24,33, using an Infinite 200 Pro spectrophotometer (Tecan Group AG, Maennedorf, Switzerland). Activity percentage was calculated by comparing the difference in rate of NH3 (for N. inopinata) or NO2− (for N. moscoviensis) oxidation during the linear section of the substrate oxidation curve between the non-inhibited control culture and the cultures exposed to the various concentrations of PTIO.
For the micro-respirometry-based PTIO inhibition experiments, 360 mL of exponential phase culture was harvested using 10 kDa-cutoff, Ultra–15 Centrifuge Filter units as described above, washed twice with N-free medium, homogenized by vigorous vortexing, and split into 25 2.5 mL aliquots. Individual aliquots were then amended with 0, 33, 100, 330, or 1000 µM PTIO and incubated at 37 °C for at least 2 h. For each experiment, an aliquot was transferred into a 2 mL, single-port MR chamber containing a glass-coated stir bar. O2 uptake was measured using OX-MR oxygen microsensors in a recirculating water-bath at 37 °C and stirring at 400 RPM. NO2− was injected into the chamber as described above. Viability of the harvested biomass over the length of the entire experiment (~8 h) was confirmed by measuring the rate of substrate-dependent O2 uptake in biomass incubated at 37 °C without PTIO at the end of the experiment. To determine if removal of PTIO after PTIO treatment restored activity (as being expected if inhibition occurred due to NO scavenging), we harvested the biomass treated with inhibitory concentrations of PTIO using Ultra-15 Centrifuge Filter units, washed it twice with NO2−-free mineral medium using the same Ultra-15 Centrifuge filter units, resuspended it in NO2−-free mineral medium, and transferred it to a 2 mL MR chamber for re-measurement of substrate-dependent O2 uptake. To control for activity loss due to biomass loss or additional centrifugation, biomass from the 0 µM PTIO treatment group was treated identically. All experiments were run in triplicate. Activity percentage was calculated by comparing the difference in rate of NH3 (for N. inopinata) or NO2− (for N. moscoviensis) oxidation during the linear section of the substrate oxidation curve between the non-inhibited control culture and the cultures exposed to the various concentrations of PTIO.
N2O yield
Late exponential-phase cultures (containing ~0 µM NH4+ and ~350 µM remaining NO2−) of N. inopinata grown in FWM (described above) were transferred at 10% (final volume of 139 mL) to new FWM containing ~1.1 mM NH4+ and aliquoted into glass serum bottles (~255 mL) sealed with butyl rubber stoppers and crimp caps. Quadruplicates were incubated in the dark at 37 °C under a fully oxic atmosphere, with lab air as the headspace, and at low O2 conditions (~0.9% O2 with N2 as the balance, equivalent to 1.8 µM dissolved O2), respectively. A starting O2 headspace concentration of ~0.9% in the hypoxic vials was chosen so the starting mol ratio of NH4+ to O2 was 2.2:1, resulting in O2-limiting growth conditions. Hypoxic conditions were achieved by sparging the sealed serum bottles with O2-free N2 gas for 1 h and then backfilling the headspace with 4.5 mL of lab air. The final concentration of O2 in the headspace of the hypoxic vials at the beginning of the experiments was 0.89% as verified with a needle-piercing OX-NP O2 microsensor (Unisense). In addition, abiotic controls containing 1.1 mM NH4+, 250 µM NO2−, and 1 µM NH2OH with and without heat-killed N. inopinata cells were run in parallel to control for abiotic formation of N2O. Gas samples (15 mL) of the headspace were taken from the sealed serum bottles using a sterile syringe and transferred into sealed 12 mL exetainers for N2O and O2 analysis. The serum bottles were backfilled with 15 mL of O2-free N2 gas after each sampling. N2O was quantified using a TRACE GC Ultra series gas chromatograph (ThermoFisher Scientific, Waltham, USA) equipped with a pulse discharge detector (ThermoFisher Scientific, Waltham, USA), a Porapak N column, and a Al/AS1310 autosampler (S+H Analytik GmbH, Moenchengladbach, Germany). Final calculated N2O concentrations take into account N2O loss due to sampling and are N2O emissions above experimentally determined background (atmospheric) N2O levels. NH4+, NO2−, and NO3− concentrations were quantified as described previously.
For N. moscoviensis, late-exponential phase cultures were transferred at 10% to new NOM containing ~1.0 mM NaNO2 and aliquoted into glass serum bottles sealed with butyl rubber stoppers and crimp caps. Triplicates were incubated in the dark at 37 °C under a fully oxic atmosphere, with lab air as the headspace, and at hypoxic conditions (~1.0% O2). Gas samples of the headspace were taken at the beginning of the experiment and once all of the substrate was depleted, so no backfilling was necessary after sampling. N2O was quantified as described above. Final calculated N2O concentrations are N2O emissions above experimentally determined background (atmospheric) N2O levels.
Analysis of N2O isotopic signatures
For analysis of the isotopic composition of the N2O in the headspace of the N. inopinata vials, 4 mL of the headspace gas was transferred to 120 ml glass flasks filled with helium at 50 kPa overpressure. Before filling with helium, the flasks had been evacuated and flushed with helium four times. N2O was analyzed with an isotope ratio mass spectrometer (IRMS, IsoPrime 100, Elementar Analysensysteme, Hanau, Germany) coupled to an online pre-concentration unit (TraceGas, Elementar Analysensysteme) as described in detail in ref. 67. Briefly, for pre-concentration, the N2O was transferred to a cold trap in liquid nitrogen with a transfer time of 20 min. Water, CO2, and any potential CO in the sample gas were removed with magnesium perchlorate, carbosorb, and a CO oxidation catalyst (Sofnocat), respectively. Then the sample gas N2O was cryo-focused on a second cold trap, before the sample was remobilized again and separated isothermally at room temperature on a capillary column (PoraPLOT Q, 30 m length × 0.32 mm inner diameter, 10 µm coating, Varian). After GC separation, the N2O was introduced in a stream of helium to the IRMS via an open-split in continuous-flow mode. Mass-to-charge ratios (m/z) of N2O at 44 (14N14N16O), 45 (14N15N16O, 15N14N16O, and 14N15N17O), and 46 (14N14N18O), as well as NO+ fragment ions of N2O at m/z 30 (14N16O+) and 31 (15N16O+) were measured simultaneously by the IRMS. The values of δ15Nbulk, δ15Nα, and δ18O were calculated from m/z 44, 45, and 46, including a 17O correction according to68. The δ15Nβ value was calculated as δ15Nβ = 2·δ15Nbulk–δ15Nα, and 15N site preference (SP) as SP = δ15Nα–δ15Nβ. Calibration was performed as described in ref. 67. Reproducibility was 0.2‰ for δ15Nbulk, 0.3‰ for δ18O, and 0.4‰ for δ15Nα and δ15Nβ.
Comparative genomics
Genes encoding NirK, the NorCBQD complex, NorSY complex, CytL, CytS, and NcyA were initially identified by blastp and subsequently examined phylogenetically for annotation. Protein-coding genes from genomes of interest were screened using blastp against databases constructed from previously published lists6. For each gene set of interest, blastp results (default parameters) were filtered for queries that hit database entries with at least 70% of the query length. These putative genes were then aligned to the sequences that were used to construct the blastp databases using mafft69 and examined for phylogenetic placement using FastTree270. Neighborhoods of nor genes (norCBQD and norSY) were manually examined for synteny and scrutinized for genes missed by the blastp search. Alignments of CytL and CytS were further examined for the presence or absence of the diagnostic heme cross-linked lysine49 to distinguish phylogenetic clades of CytL from CytS. Putative nirK genes were aligned to a set of previously published multicopper oxidase genes53 and manually examined for phylogenetic placement into recognized clades of nirK.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Acknowledgements
We would like to thank Marton Palatinszky for providing N. inopinata biomass for the PTIO batch experiments. This work was funded by the Austrian Science Fund Grant P30570-B29 (to H.D. and M.W.), the ERC Advanced Grant NITRICARE 294343 (to M.W.), the Comammox Research Platform of the University of Vienna, and a Discovery Grant from the Natural Sciences and Engineering Research Council of Canada (to L.Y.S.).
Author contributions
H.D. and M.W. conceived the study. H.D., M.W., and K.D.K. wrote the manuscript with input from all authors. K.D.K., M.Y.J., and C.J.S. performed the micro-respirometry and batch incubations. J.V. and P.P. performed the PTIO batch experiments. S.L. performed the batch N. moscoviensis experiments. C.H. made the comparative genome analysis. L.Y.S. assisted with data analysis. A.R., H.W., and N.B. performed the GC and N2O site preference measurements.
Data availability
The proteomics data used for Supplementary Fig. 3 is publicly available at the PRIDE/ProteomeXchange database under the accession code PXD013103 (ref. 24). The source data underlying Figs. 3–6 and Supplementary Figs. 1, 2, 4–9 are provided as a Source Data file.
Competing interests
The authors declare no competing interests.
Footnotes
Journal peer review information: Nature Communications would like to thank Boran Kartal and other anonymous reviewer(s) for their contribution to the peer review of this work. Peer review reports are available.
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information accompanies this paper at 10.1038/s41467-019-09790-x.
References
- 1.Myhre, G. et al. In Climate Change 2013: The Physical Science Basis. Contribution of Working Group I to the Fifth Assessment Report of the Intergovernmental Panel on Climate Change (eds. Stocker, T. F., et al.) 659–740 (Cambridge University Press, Cambridge, United Kingdom and New York, NY, USA 2013).
- 2.Wuebbles DJ. Nitrous oxide: no laughing matter. Science. 2009;326:56–57. doi: 10.1126/science.1179571. [DOI] [PubMed] [Google Scholar]
- 3.Duan H, Ye L, Erler D, Ni BJ, Yuan Z. Quantifying nitrous oxide production pathways in wastewater treatment systems using isotope technology – a critical review. Water Res. 2017;122:96–113. doi: 10.1016/j.watres.2017.05.054. [DOI] [PubMed] [Google Scholar]
- 4.Kuypers MMM, Marchant HK, Kartal B. The microbial nitrogen-cycling network. Nat. Rev. Microbiol. 2018;16:263–276. doi: 10.1038/nrmicro.2018.9. [DOI] [PubMed] [Google Scholar]
- 5.Heil J, Vereecken H, Brüggemann N. A review of chemical reactions of nitrification intermediates and their role in nitrogen cycling and nitrogen trace gas formation in soil. Eur. J. Soil Sci. 2015;67:23–39. doi: 10.1111/ejss.12306. [DOI] [Google Scholar]
- 6.Kozlowski, J. A., Dimitri Kits, K. & Stein, L. Y. Comparison of nitrogen oxide metabolism among diverse ammonia-oxidizing bacteria. Front. Microbiol. 7, 1090 (2016). [DOI] [PMC free article] [PubMed]
- 7.Liu S, et al. Abiotic conversion of extracellular NH2OH contributes to N2O Emission during ammonia oxidation. Environ. Sci. Technol. 2017;51:13122–13132. doi: 10.1021/acs.est.7b02360. [DOI] [PubMed] [Google Scholar]
- 8.Stieglmeier M, et al. Aerobic nitrous oxide production through N-nitrosating hybrid formation in ammonia-oxidizing archaea. ISME J. 2014;8:1135–1146. doi: 10.1038/ismej.2013.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Poth M, Focht DD. 15N kinetic analysis of N2O production by Nitrosomonas europaea: an examination of nitrifier denitrification. Appl. Environ. Microbiol. 1985;49:1134–1141. doi: 10.1128/aem.49.5.1134-1141.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kozlowski JA, Stieglmeier M, Schleper C, Klotz MG, Stein LY. Pathways and key intermediates required for obligate aerobic ammonia-dependent chemolithotrophy in bacteria and Thaumarchaeota. ISME J. 2016;10:1836–1845. doi: 10.1038/ismej.2016.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schmidt I, van Spanning RJM, Jetten MSM. Denitrification and ammonia oxidation by Nitrosomonas europaea wild-type, and NirK- and NorB-deficient mutants. Microbiology. 2004;150:4107–4114. doi: 10.1099/mic.0.27382-0. [DOI] [PubMed] [Google Scholar]
- 12.Goreau TJ, et al. Production of NO2− and N2O by nitrifying bacteria at reduced concentrations of oxygen. Appl. Environ. Microbiol. 1980;40:526–532. doi: 10.1128/aem.40.3.526-532.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wrage-Mönnig N, et al. The role of nitrifier denitrification in the production of nitrous oxide revisited. Soil Biol. Biochem. 2018;123:A3–A16. doi: 10.1016/j.soilbio.2018.03.020. [DOI] [Google Scholar]
- 14.Caranto JD, Vilbert AC, Lancaster KM. Nitrosomonas europaea cytochrome P460 is a direct link between nitrification and nitrous oxide emission. Proc. Natl Acad. Sci. USA. 2016;113:14704–14709. doi: 10.1073/pnas.1611051113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Caranto JD, Lancaster KM. Nitric oxide is an obligate bacterial nitrification intermediate produced by hydroxylamine oxidoreductase. Proc. Natl Acad. Sci. USA. 2017;114:8217–8222. doi: 10.1073/pnas.1704504114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jung MY, et al. Isotopic signatures of N2O produced by ammonia-oxidizing archaea from soils. ISME J. 2014;8:1115–1125. doi: 10.1038/ismej.2013.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hink L, Nicol GW, Prosser JI. Archaea produce lower yields of N2O than bacteria during aerobic ammonia oxidation in soil. Environ. Microbiol. 2016;19:4829–4837. doi: 10.1111/1462-2920.13282. [DOI] [PubMed] [Google Scholar]
- 18.Hink L, Gubry-Rangin C, Nicol GW, Prosser JI. The consequences of niche and physiological differentiation of archaeal and bacterial ammonia oxidisers for nitrous oxide emissions. ISME J. 2018;12:1084–1093. doi: 10.1038/s41396-017-0025-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vadivelu VM, Keller J, Yuan Z. Effect of free ammonia on the respiration and growth processes of an enriched Nitrobacter culture. Water Res. 2007;41:826–834. doi: 10.1016/j.watres.2006.11.030. [DOI] [PubMed] [Google Scholar]
- 20.Ushiki N, et al. Nitrite oxidation kinetics of two Nitrospira strains: the quest for competition and ecological niche differentiation. J. Biosci. Bioeng. 2017;123:581–589. doi: 10.1016/j.jbiosc.2016.12.016. [DOI] [PubMed] [Google Scholar]
- 21.Bristow LA, et al. Ammonium and nitrite oxidation at nanomolar oxygen concentrations in oxygen minimum zone waters. Proc. Natl Acad. Sci. USA. 2016;113:10601–10606. doi: 10.1073/pnas.1600359113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zakem EJ, et al. Ecological control of nitrite in the upper ocean. Nat. Commun. 2018;9:1206. doi: 10.1038/s41467-018-03553-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Van Kessel MAHJ, et al. Complete nitrification by a single microorganism. Nature. 2015;528:555–559. doi: 10.1038/nature16459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Daims H, et al. Complete nitrification by Nitrospira bacteria. Nature. 2015;528:504–509. doi: 10.1038/nature16461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Daims H, Lücker S, Wagner M. A new perspective on microbes formerly known as nitrite-oxidizing bacteria. Trends Microbiol. 2016;24:699–712. doi: 10.1016/j.tim.2016.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Camejo PY, Domingo JS, McMahon KD, Noguera DR. Genome-enabled insights into the ecophysiology of the comammox bacterium “Candidatus Nitrospira nitrosa”. mSystems. 2017;2:e00059–17. doi: 10.1128/mSystems.00059-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Palomo A, et al. Comparative genomics sheds light on niche differentiation and the evolutionary history of comammox Nitrospira. ISME J. 2018;12:1779–1793. doi: 10.1038/s41396-018-0083-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pjevac P, et al. AmoA-targeted polymerase chain reaction primers for the specific detection and quantification of comammox Nitrospira in the environment. Front. Microbiol. 2017;8:1508. doi: 10.3389/fmicb.2017.01508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pinto AJ, et al. Metagenomic evidence for the presence of comammox Nitrospira-like bacteria in a drinking water system. mSphere. 2015;1:e00054–15. doi: 10.1128/mSphere.00054-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Y, et al. Comammox in drinking water systems. Water Res. 2017;116:332–341. doi: 10.1016/j.watres.2017.03.042. [DOI] [PubMed] [Google Scholar]
- 31.Bartelme RP, McLellan SL, Newton RJ. Freshwater recirculating aquaculture system operations drive biofilter bacterial community shifts around a stable nitrifying consortium of ammonia-oxidizing archaea and comammox Nitrospira. Front. Microbiol. 2017;8:101. doi: 10.3389/fmicb.2017.00101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alexandra Pokhilko OE. A mathematical model of bacteria capable of complete oxidation of ammonium predicts improved nitrogen removal and reduced production of nitrous oxide. JMEST. 2017;4:7099–7108. [Google Scholar]
- 33.Kits KD, et al. Kinetic analysis of a complete nitrifier reveals an oligotrophic lifestyle. Nature. 2017;549:269–272. doi: 10.1038/nature23679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Beaumont HJE, et al. Nitrite reductase of Nitrosomonas europaea is not essential for production of gaseous nitrogen oxides and confers tolerance to nitrite. J. Bacteriol. 2002;184:2557–2560. doi: 10.1128/JB.184.9.2557-2560.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Beaumont HJE, van Schooten B, Lens SI, Westerhoff HV, van Spanning RJM. Nitrosomonas europaea expresses a nitric oxide reductase during nitrification. J. Bacteriol. 2004;186:4417–4421. doi: 10.1128/JB.186.13.4417-4421.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Starkenburg SR, Arp DJ, Bottomley PJ. Expression of a putative nitrite reductase and the reversible inhibition of nitrite-dependent respiration by nitric oxide in Nitrobacter winogradskyi Nb-255. Environ. Microbiol. 2008;10:3036–3042. doi: 10.1111/j.1462-2920.2008.01763.x. [DOI] [PubMed] [Google Scholar]
- 37.Kozlowski JA, Price J, Stein LY. Revision of N2O-producing pathways in the ammonia-oxidizing bacterium Nitrosomonas europaea ATCC 19718. Appl. Environ. Microbiol. 2014;80:4930–4935. doi: 10.1128/AEM.01061-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mellbye, B. L., Giguere, A. T., Bottomley, P. J. & Sayavedra-Soto, L. A. Quorum quenching of Nitrobacter winogradskyi suggests that quorum sensing regulates fluxes of nitrogen oxide(s) during nitrification. MBio7, (2016). 10.1128/mBio.01753-16. [DOI] [PMC free article] [PubMed]
- 39.Parks DH, et al. Recovery of nearly 8,000 metagenome-assembled genomes substantially expands the tree of life. Nat. Microbiol. 2017;2:1533–1542. doi: 10.1038/s41564-017-0012-7. [DOI] [PubMed] [Google Scholar]
- 40.Maalcke WJ, et al. Structural basis of biological NO generation by octaheme oxidoreductases. J. Biol. Chem. 2014;289:1228–1242. doi: 10.1074/jbc.M113.525147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lancaster KM, Caranto JD, Majer SH, Smith MA. Alternative bioenergy: updates to and challenges in nitrification metalloenzymology. Joule. 2018;2:421–441. doi: 10.1016/j.joule.2018.01.018. [DOI] [Google Scholar]
- 42.Zorz JK, Kozlowski JA, Stein LY, Strous M, Kleiner M. Comparative proteomics of three species of ammonia-oxidizing bacteria. Front. Microbiol. 2018;9:938. doi: 10.3389/fmicb.2018.00938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Abby SS, et al. Candidatus Nitrosocaldus cavascurensis, an ammonia oxidizing, extremely thermophilic archaeon with a highly mobile genome. Front. Microbiol. 2018;9:28. doi: 10.3389/fmicb.2018.00028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Daebeler A, et al. Cultivation and genomic analysis of “Candidatus Nitrosocaldus islandicus”, an obligately thermophilic, ammonia-oxidizing Thaumarchaeon from a hot spring biofilm in Graendalur Valley, Iceland. Front. Microbiol. 2018;9:193. doi: 10.3389/fmicb.2018.00193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kozlowski JA, Kits KD, Stein LY. Genome sequence of Nitrosomonas communis strain Nm2, a mesophilic ammonia-oxidizing bacterium isolated from mediterranean soil. Genome Announc. 2016;4:e01541–15. doi: 10.1128/genomeA.01541-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hayatsu M, et al. An acid-tolerant ammonia-oxidizing γ-proteobacterium from soil. ISME J. 2017;11:1130–1141. doi: 10.1038/ismej.2016.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cantera JJL, Stein LY. Role of nitrite reductase in the ammonia-oxidizing pathway of Nitrosomonas europaea. Arch. Microbiol. 2007;188:349–354. doi: 10.1007/s00203-007-0255-4. [DOI] [PubMed] [Google Scholar]
- 48.Wijma HJ, Canters GW, de Vries S, Verbeet MP. Bidirectional catalysis by copper-containing nitrite reductase. Biochemistry. 2004;43:10467–10474. doi: 10.1021/bi0496687. [DOI] [PubMed] [Google Scholar]
- 49.Elmore BO, Bergmann DJ, Klotz MG, Hooper AB. Cytochromes P460 and c’-beta; a new family of high-spin cytochromes c. FEBS Lett. 2007;581:911–916. doi: 10.1016/j.febslet.2007.01.068. [DOI] [PubMed] [Google Scholar]
- 50.McEwan AG, Ridge JP, McDevitt CA, Hugenholtz P. The DMSO reductase family of microbial molybdenum enzymes; molecular properties and role in the dissimilatory reduction of toxic elements. Geomicrobiol. J. 2002;19:3–21. doi: 10.1080/014904502317246138. [DOI] [Google Scholar]
- 51.Martens-Habbena W, et al. The production of nitric oxide by marine ammonia-oxidizing archaea and inhibition of archaeal ammonia oxidation by a nitric oxide scavenger. Environ. Microbiol. 2015;17:2261–2274. doi: 10.1111/1462-2920.12677. [DOI] [PubMed] [Google Scholar]
- 52.Hink L, et al. Kinetics of NH3-oxidation, NO-turnover, N2O-production and electron flow during oxygen depletion in model bacterial and archaeal ammonia oxidisers. Environ. Microbiol. 2017;19:4882–4896. doi: 10.1111/1462-2920.13914. [DOI] [PubMed] [Google Scholar]
- 53.Kerou M, et al. Proteomics and comparative genomics of Nitrososphaera viennensis reveal the core genome and adaptations of archaeal ammonia oxidizers. Proc. Natl Acad. Sci. USA. 2016;113:E7937–E7946. doi: 10.1073/pnas.1601212113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Qin W, et al. Stress response of a marine ammonia-oxidizing archaeon informs physiological status of environmental populations. ISME J. 2018;12:508–519. doi: 10.1038/ismej.2017.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Courtens ENP, et al. Nitric oxide preferentially inhibits nitrite oxidizing communities with high affinity for nitrite. J. Biotechnol. 2015;193:120–122. doi: 10.1016/j.jbiotec.2014.11.021. [DOI] [PubMed] [Google Scholar]
- 56.Shen T, Stieglmeier M, Dai J, Urich T, Schleper C. Responses of the terrestrial ammonia-oxidizing archaeon Ca. Nitrososphaera viennensis and the ammonia-oxidizing bacterium Nitrosospira multiformis to nitrification inhibitors. FEMS Microbiol. Lett. 2013;344:121–129. doi: 10.1111/1574-6968.12164. [DOI] [PubMed] [Google Scholar]
- 57.Heil J, Liu S, Vereecken H, Brüggemann N. Abiotic nitrous oxide production from hydroxylamine in soils and their dependence on soil properties. Soil Biol. Biochem. 2015;84:107–115. doi: 10.1016/j.soilbio.2015.02.022. [DOI] [Google Scholar]
- 58.Zhu X, Burger M, Doane TA, Horwath WR. Ammonia oxidation pathways and nitrifier denitrification are significant sources of N2O and NO under low oxygen availability. Proc. Natl Acad. Sci. USA. 2013;110:6328–6333. doi: 10.1073/pnas.1219993110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhu-Barker X, Cavazos AR, Ostrom NE, Horwath WR, Glass JB. The importance of abiotic reactions for nitrous oxide production. Biogeochemistry. 2015;126:251–267. doi: 10.1007/s10533-015-0166-4. [DOI] [Google Scholar]
- 60.Sutka RL, Ostrom NE, Ostrom PH, Gandhi H, Breznak JA. Nitrogen isotopomer site preference of N2O produced by Nitrosomonas europaea and Methylococcus capsulatus Bath. Rapid Commun. Mass Spectrom. 2003;17:738–745. doi: 10.1002/rcm.968. [DOI] [PubMed] [Google Scholar]
- 61.Sutka RL, et al. Distinguishing nitrous oxide production from nitrification and denitrification on the basis of isotopomer abundances. Appl. Environ. Microbiol. 2006;72:638–644. doi: 10.1128/AEM.72.1.638-644.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Santoro AE, Buchwald C, McIlvin MR, Casciotti KL. Isotopic signature of N2O produced by marine ammonia-oxidizing archaea. Science. 2011;333:1282–1285. doi: 10.1126/science.1208239. [DOI] [PubMed] [Google Scholar]
- 63.Frame CH, Casciotti KL. Biogeochemical controls and isotopic signatures of nitrous oxide production by a marine ammonia-oxidizing bacterium. Biogeosci. Discuss. 2010;7:3019–3059. doi: 10.5194/bgd-7-3019-2010. [DOI] [Google Scholar]
- 64.Yamazaki T, et al. Isotopomeric characterization of nitrous oxide produced by reaction of enzymes extracted from nitrifying and denitrifying bacteria. Biogeosciences. 2014;11:2679–2689. doi: 10.5194/bg-11-2679-2014. [DOI] [Google Scholar]
- 65.Annavajhala MK, Kapoor V, Santo-Domingo J, Chandran K. Comammox functionality identified in diverse engineered biological wastewater treatment systems. Environ. Sci. Technol. Lett. 2018;5:110–116. doi: 10.1021/acs.estlett.7b00577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Koch H, et al. Growth of nitrite-oxidizing bacteria by aerobic hydrogen oxidation. Science. 2014;345:1052–1054. doi: 10.1126/science.1256985. [DOI] [PubMed] [Google Scholar]
- 67.Wei J, Zhou M, Vereecken H, Brüggemann N. Large variability in CO2 and N2O emissions and in 15N site preference of N2O from reactions of nitrite with lignin and its derivatives at different pH. Rapid Commun. Mass Spectrom. 2017;31:1333–1343. doi: 10.1002/rcm.7912. [DOI] [PubMed] [Google Scholar]
- 68.Kaiser, J., Röckmann, T. & Brenninkmeijer, C. A. M. Complete and accurate mass spectrometric isotope analysis of tropospheric nitrous oxide. J. Geophys. Res. 108, 1–17 (2003).
- 69.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Price MN, Dehal PS, Arkin AP. FastTree 2 – Approximately maximum-likelihood trees for large alignments. PLoS One. 2010;5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
The proteomics data used for Supplementary Fig. 3 is publicly available at the PRIDE/ProteomeXchange database under the accession code PXD013103 (ref. 24). The source data underlying Figs. 3–6 and Supplementary Figs. 1, 2, 4–9 are provided as a Source Data file.