Abstract
Cryptococcus neoformans is a basidiomycete fungus that is highly resistant to ionizing radiation and has been identified in highly radioactive environments. Transcription factors (TFs) are master regulators of gene expression by binding to specific DNA sequences within promoters of target genes. A library of 322 signature-tagged gene deletion strains for 155 C. neoformans TF genes has been established. Previous phenome-based functional analysis of the C. neoformans TF mutant library identified key TFs important for various phenotypes, such as growth, differentiation, virulence-factor production, and stress responses. Here, utilizing the established TF mutant library, we identified 5 TFs that are important for radiation sensitivity, including SRE1, BZP2, GAT5, GAT6, and HCM1. Interestingly, BZP2, GAT5 and GAT6 all belong to the GATA-type transcription factors. These factors regulate transcription of nitrogen catabolite repression (NCR) sensitive genes when preferred nitrogen sources are absent or limiting. In addition to radiation, we found that specific GATA factors are important for other stressors such as rapamycin, fluconazole, and hydroxyurea treatment. Using real-time PCR method, we studied the expression of GATA down-stream genes after radiation exposure and identified that AAP4, AAP5 and URO1 were differentially expressed in the GAT5 and GAT6 mutants compared to the wild type cells. In summary, our data suggest that GATA TFs are important for radiation sensitivity in C. neoformans by regulating specific downstream AAP genes.
Subject terms: Fungal biology, Transcription
Introduction
It is well established that ionizing radiation (IR) causes deleterious effects on cell survival by causing damages to cellular macromolecules including nucleic acids, proteins and lipids. However, the cellular sensitivity to IR varies greatly between different cell types. For example, mammalian cells have a D10 value (D10 is the radiation dose which decreased cell survival to 10%) about 4 Gy; most bacteria have a D10 value less than 200 Gy; interestingly some organisms in all three domains of life can withstand IR doses above several thousand Gy or even 12,000 Gy1,2. Cryptococcus neoformans is a basidiomycetous fungal pathogen that causes cryptococcal meningitis and responsible for 181,100 annual deaths globally3. Interestingly, C. neoformans was found in highly radioactive environments such as the cooling pools of nuclear reactors, the stratosphere, and the damaged nuclear reactor at Chernobyl Nuclear Power Plant4,5. The C. neoformans var. grubii (H99) strain is the most common reference strain used in the C. neoformans research community and it has a D10 value about 2000 Gy5. Many studies have been performed to understand the mechanisms of radiation resistance in C. neoformans. For example, melanin has been suggested to protect C. neoformans against IR through scavenging of free radicals generated by IR, dissipating radiation energy in the melanin polymer and converting radiation energy to metabolically useful reducing power6,7. One transcription factor, named Bdr1, has been suggested as a regulator of radiation resistance in C. neoformans by controlling the expression of DNA damage repair genes5. However, more work is still needed to understand the mechanism of radiation resistance in C. neoformans.
Transcription factors (TFs) are key regulators for gene expression coordination and their study is very important for understanding the molecular mechanisms of development and responses to environmental signals8. Many TFs, such as p53, nuclear factor κB (NF-κB), activated protein 1 (AP-1), nuclear erythroid-derived 2-related factor 2 (Nrf2), and cAMP responsive element binding protein (CREB) play important roles in cellular radiation responses9. Compared to four diverse basidiomycetes, C. neoformans are highly enriched in fungal-specific transcription factor domains10, which may help explain the radiation resistance of C.neoformans. Therefore, studying the TFs in the C. neoformans may provide important insights into their radiation resistance. Recently, Jung et al. constructed a systematic C. neoformans TF mutant library, which consists of 322 signature-tagged gene-deletion strains for 155 putative TF genes predicted by bioinformatics analysis11. Using this library, TFs important for 32 distinct growth conditions have been identified11.
In the current study, we exposed the C. neoformans TF mutant library to high dose γ-radiation, and identified several TFs that are important for radiation sensitivity. Our data showed that specific GATA family TFs are important for stress responses in C. neoformans including radiation, rapamycin, hydroxyurea and fluconazole. We also identified important amino acid permease genes that are downstream target genes of GATA TFs important for IR. In summary, our study provides better understanding of the transcriptional circuits in the C. neoformans after radiation exposure.
Materials and Methods
Strain and media
The Jung’s C. neoformans TF mutant library11 was procured from the Fungal Genetics Stock Center (FGSC, Kansas State University, Manhattan, KS). The C. neoformans H99S10 was a gift from Dr. Joe Heitman’s lab at Duke University Medical Center (Durham, North Carolina), which was the parent strain of the Jung’s TF mutant library. These strains were routinely grown at 30 °C in yeast extract peptone dextrose (YPD) medium. For other stress response tests, rapamycin, hydroxyurea or fluconazole was added in the YPD agar plates.
Irradiation with γ-radiation
For survival assays, overnight cultured fungal cells were 10-fold serially diluted in PBS and 3 µL of 102-105 dilutions were spotted onto YPD plates. The plates were irradiated in a 60Co irradiator (Model 109; J. L. Shepard and Associates, San Fernando, California) at 130 Gy/min. Cells were allowed to grow for 5 days before being photographed.
For molecular biology assays, overnight cultured fungal cells were washed in PBS and then irradiated in tubes on ice in the same irradiator. The irradiated cells were either harvested immediately or diluted in 50× volume of fresh YPD medium and harvested at selected time points after incubation with shaking.
RNA extraction and quantitative real-time PCR
RNA was extracted using the mirVana miRNA Isolation Kit (Thermo Fisher Scientific, Grand Island, New York) according to the manufacturer’s instructions. RNA concentration and purity were determined using a Nanodrop 1000 spectrophotometer (NanoDrop Technologies, Inc., Wilmington, DE). Reverse transcription (RT) was performed using the High-Capacity cDNA Reverse Transcription Kit. The resulting cDNA was quantitatively amplified using PowerUp SYBR® Green Master Mix with specific primers on a 7900HT Fast Real-Time PCR System (Thermo Fisher Scientific, Grand Island, New York). The primer sequences were either from literature or self-designed as shown in Table 1. The relative mRNA expression fold change was calculated using the 2−ΔΔT method with actin as the endogenous control. Each gene’s relative expression was normalized to its gene level of the H99S 1 kGy samples at 0 h.
Table 1.
Primer sequences for qRT-PCR.
| Primer | Sequence | target | References |
|---|---|---|---|
| B679 | CGCCCTTGCTCCTTCTTCTATG | actin | 5 |
| B680 | GACTCGTCGTATTCGCTCTTCG | actin | 5 |
| GAT1F1 | GTTCGCCAACGAGTGAAAAT | GAT1 | 35 |
| GAT1R1 | GGGAAGGGTTGCTTTCTTTC | GAT1 | 35 |
| GAT5F1 | GTGAGGTGGCGGAATACCAA | GAT5 | self designed |
| GAT5R1 | GAGGGTTGTGGGTTTACCAGAT | GAT5 | self designed |
| GAT6F2 | GCCTGCCACAGCCTAATACA | GAT6 | self designed |
| GAT6R2 | CCCGCAATCATCGCAAACTT | GAT6 | self designed |
| PRCP185 | GCCTTATGGTATACTCTATGATG | AAP1 | 17 |
| PRCP186 | CCGTATGCCCGAGCACCGAGG | AAP1 | 17 |
| PRCP187 | CATGGTATACGCGATGATGG | AAP2 | 17 |
| PRCP188 | TCTGGCTCCCCAGAAGTTAATG | AAP2 | 17 |
| AAP3F1 | GCTGGTGAAGCCAAGAAT | AAP3 | self designed |
| PRCP190 | ATGTACCACCGAGATAAAAG | AAP3 | 17 |
| PRCP191 | CGAGGCAAAGAACCCACG | AAP4 | 17 |
| PRCP192 | AATCAAGATGCAAGCGTTTATG | AAP4 | 17 |
| PRCP193 | ACTTACTTGGACCTCTATCCTC | AAP5 | 17 |
| PRCP194 | TTTTCGGATCAGCTTGAAACC | AAP5 | 17 |
| PRCP195 | CCTTGAAAGACCGTTTCGGC | AAP6 | 17 |
| PRCP196 | TGTCACAAGTGTTGGGTCATTG | AAP6 | 17 |
| AAP7F1 | GGATGTTTGGCCTATCTCTC | AAP7 | self designed |
| PRCP198 | CATGTATGTGAAAGCGATGG | AAP7 | 17 |
| PRCP199 | TCTCTTTCTAGGGATTCTTATC | AAP8 | 17 |
| PRCP200 | CTCCGCCATATGGGCAGAAGC | AAP8 | 17 |
| PRCP215 | AGACCGAGGAGGAGGACGCTG | TPR2 | 17 |
| PRCP216 | AAGCGTCGAATCTAGATTGGTC | TRP2 | 17 |
| PRCP217 | AGGAGATCGAGGGAATGTTCG | TRP3 | 17 |
| PRCP218 | CGTAAGCGATCTCTCCACCG | TRP3 | 17 |
| PRCP219 | GCCCAGGCCGGTGCTTTTC | TRP4 | 17 |
| PRCP221 | ATGTATCCCATCCATCGCCAC | TRP4 | 17 |
| PRCP222 | CTTGTTCATGGGCTACTACAAC | TRP5 | 17 |
| PRCP223 | ACGCCCATCTTGGAGACGAC | TRP5 | 17 |
| UQ1070 | AAAAGCTAAAGAGAACGACGTTGC | PUT1 | 20 |
| UQ1071 | TCTCACTCTTTGACTTTGGAGGTTT | PUT1 | 20 |
| UQ1072 | AGAAGGCTAAGGAGAACAACATCATT | PUT5 | 20 |
| UQ1073 | GTCCAGATCTCCTCCTTGGAAG | PUT5 | 20 |
| UQ1074 | TCATTAACGGTGAGGAGGTCAAG | PUT2 | 20 |
| UQ1075 | GCAAGAGCACCGTCAATGG | PUT2 | 20 |
| UQ613 | TGAGTACACCCTCCGAGTCCTT | URO1 | 20 |
| UQ733 | TTTACAGTGTCGGTAGCAACGAC | URO1 | 20 |
| UQ609 | CTTATCGACACCCACGTCCA | DAL1 | 20 |
| UQ736 | GCATGTCGATCAGGGTGGT | DAL1 | 20 |
| UQ739 | TAAGATCAAACTCGCTGACATGG | URE1 | 20 |
| UQ740 | CTCGAATGACCTTACCTCCACC | URE1 | 20 |
| UQ1586 | GCTTCCCAGAATGAGCTTAACG | GDH1 | 20 |
| UQ1587 | CGTCAAGTGTACAGCCCATGTT | GDH1 | 20 |
| UQ1588 | CTTATGCTTATGTCGGATGGATCTT | GDH2 | 20 |
| UQ1589 | TGGACTCATCCAGAGCATCCT | GDH2 | 20 |
| UQ1590 | TCGAATCCCCAGGCATGT | GLN1 | 20 |
| UQ1591 | CGACGAGGATGGCTGTGACT | GLN1 | 20 |
| UQ1592 | CCGTTGGATCAGAGAAATTCTACC | GLT1 | 20 |
| UQ1593 | CTGCTGAGGACCGAGGAAAC | GLT1 | 20 |
| UQ1630 | TTATGTTTTGCTGGACTACCTTGGT | AMT1 | 20 |
| UQ1631 | CGCCACCTGCATAGTCCAAT | AMT1 | 20 |
| UQ1632 | TGAGGTTAAGGGCTAGCGAAGA | AMT2 | 20 |
| UQ1633 | CGGGATCTGTGCCAACATAGT | AMT2 | 20 |
| URO2F | CTACCCAGTGGTTGAGATTACC | URO2 | self designed |
| URO2R | CCACGGTATGTGGTGTAAGAA | URO2 | self designed |
| URO3-2F | GGACATCCCATGATTGGCGA | URO3 | self designed |
| URO3-2R | TGGTTGAGATGAGCAAGTCTCT | URO3 | self designed |
| DAL233-2F | CCGTTGGCGGAGATCTCAAT | DAL233 | self designed |
| DAL233-2R | GATTGAGCGGAGGGGTATGG | DAL233 | self designed |
Phylogenetic tree analysis
Phylogenetic tree analysis was performed online using the protein sequence of each GATA factor with default settings at https://www.ebi.ac.uk/Tools/phylogeny/simple_phylogeny/12.
Statistical analysis
Two-way ANOVA with multiple comparisons tests was performed to determine the gene expression levels compared to the wild type H99S 1 kGy group. The relative expression levels were log2 transformed. Differential gene expression was pseudocolored using the heatmap feature in GraphPad Prism 7.03 (GraphPad Software, San Diego, CA). Statistical analysis was performed using GraphPad Prism 7.03 software (GraphPad Software, San Diego, CA). All values of p < 0.05 were considered as significant differences.
Results
Important TFs for radiation sensitivity identified using the mutant TF library
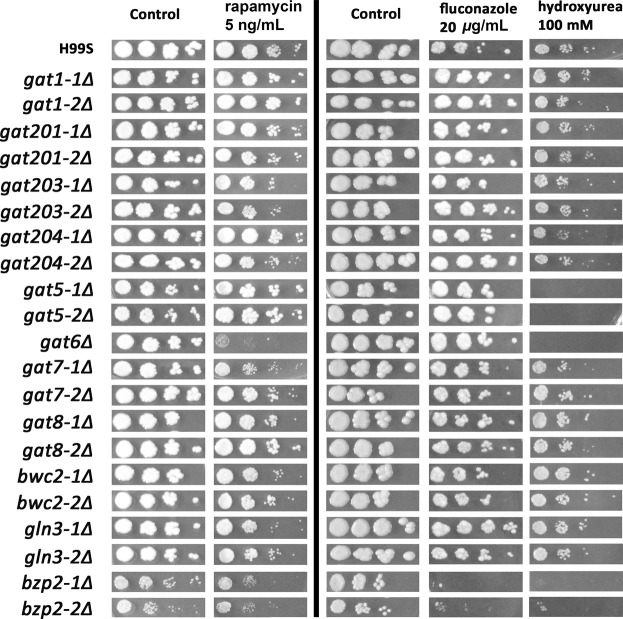
We screened the mutant TF library to identify TFs that were important for radiation sensitivity. Cells were serially diluted and plotted on YPD agar and then exposed to 1,000 and 2,000 Gy γ-radiation. Among the 155 TFs, 5 TF mutants were shown to be more sensitive to IR than the wild type control H99S cells (Fig. 1). These included sre1Δ, bzp2Δ, gat5Δ, gat6Δ and hcm1Δ. SRE1 is a TF important for sterol biosynthesis. In agreement with our findings here, it has been reported that SRE1 mutant exhibited severe growth defects when exposed to γ-radiation5. HCM1, a Forkhead TF, has been reported to regulate chromosome segregation during S-phase13, which may explain why hcm1Δ mutant was sensitive to radiation. The three remaining TFs, BZP2, GAT5 and GAT6 all belong to the GATA TFs. To further confirm and also identify other potential GATA TFs important for radiation sensitivity, all the known GATA TF mutants in the mutant TF library were exposed to γ-radiation as done previously and their survival was monitored. Note that the collection of GATA factor mutants in Jung’s mutant TF library is not complete. For example, CIR1 is a known GATA factor but it is not available in Jung’s TF library11. As shown in Fig. 2, only GAT5, GAT6 and BZP2 mutants were sensitive to radiation among all the tested GATA TFs as shown in Fig. 1.
Figure 1.
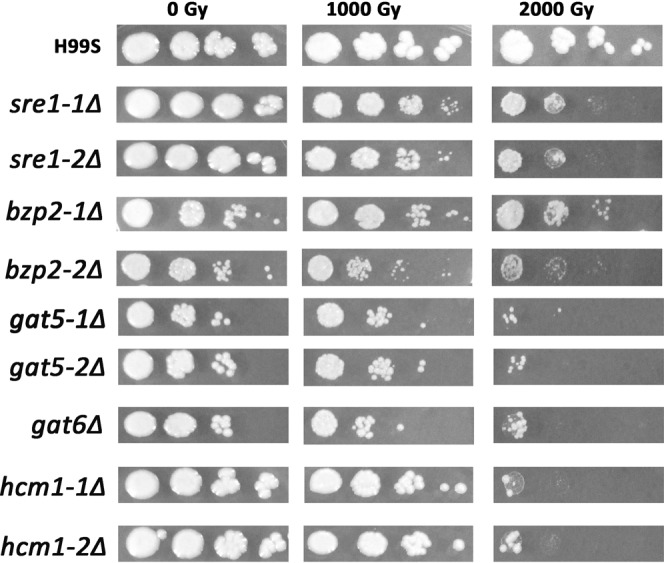
Identifying TF mutants that have different radiation sensitivity compared to the wild type C. neoformans H99S. Cells cultured overnight in liquid YPD medium were 10-fold serially diluted (102 to 105) in PBS and 3 µL of diluted solution was spotted onto the YPD plates. Cells were then exposed to the indicated doses of γ-radiation and further incubated at 30 °C for 5 days. Image was from one representative experiment of three independent experiments.
Figure 2.

Radiation sensitivity of GATA TF mutants. GATA TF mutants and H99S cells cultured overnight in liquid YPD medium were 10-fold serially diluted (102 to 105) in PBS and 3 µL of diluted solution was spotted onto the YPD plates. Cells were then exposed to the indicated doses of γ-radiation and further incubated at 30 °C for 5 days. Image was from one representative experiment of three independent experiments.
GATA TF mutants had different sensitivity to rapamycin, fluconazole or hydroxyurea treatment
To test whether the GATA TFs are important for radiation only or for other stress responses too, we exposed the GATA TF mutants to rapamycin, fluconazole or hydroxyurea treatment. Rapamycin treatment mimics nitrogen starvation through inhibiting the mammalian targets of rapamycin (mTOR). Fluconazole is the most widely used antifungal drug. And hydroxyurea is a DNA damaging agent. As shown in Fig. 3, gat6Δ mutant exhibited poorest survival after rapamycin treatment; bzp2Δ mutant exhibited poorest survival after fluconazole treatment; similar to radiation exposure, gat5Δ, gat6Δ and bzp2Δ mutants were the most sensitive mutants to hydroxyurea treatment. The data suggest that there were specific GATA TFs important for different stressors.
Figure 3.
The responses of GATA TF mutants against rapamycin, fluconazole, or hydroxyurea treatment. GATA TF mutants and H99S cells were cultured overnight in liquid YPD medium were 10-fold serially diluted (102 to 105) in PBS and 3 µL of diluted solution was spotted onto the YPD plates containing the indicated concentration of agents. Image was from one representative experiment of three independent experiments. Note that the rapamycin experiment was done at a different time with fluconazole and hydroxyurea experiment as shown divided by the black line.
Characterizing the GATA transcriptional circuits after radiation exposure
GATA TFs regulate a wide range of downstream genes involved in NCR; NCR is the physiological process that cells can preferentially utilize good nitrogen sources (such as glutamine, ammonium etc) instead of poor nitrogen sources (such as urea, proline, allantoin, glutamate, arginine, etc)14–16. In C. neoformans, GATA TF downstream targets include amino acid permeases AAP1-817; uric acid catabolic enzyme-encoding genes URO1-3, DAL1, DAL233, and URE118; proline utilization pathway genes PUT1, 2 & 519; ammonium assimilation enzyme-encoding genes GDH1-2, GLN1, GLT1, ammonium permeases AMT1-220; tryptophan biosynthetic pathway genes TRP2-517. To identify potential downstream NCR genes regulated by GAT5, GAT6 and BZP2 after radiation exposure, H99S and gat1Δ, gat5Δ, gat6Δ TF mutants were cultured and then exposed to 1 kGy γ-radiation. We included gat1Δ mutant because it was the most widely studied GATA TF in C. neoformans, even though it was not radiation sensitive. We also included the H99S un-irradiated samples to study the temporal effect on NCR gene expression. Cells were harvested at different time points after radiation. Gene expression levels were assayed using real time PCR. Both bzp2Δ mutants grew poorly in YPD broth, therefore, we didn’t study the gene expression in bzp2Δ mutants. All the gene expression levels were plotted in the heatmap shown in Fig. 4. There were no expression of GAT1, GAT5 and GAT6 in the gat1Δ, gat5Δ and gat6Δ mutant respectively, suggesting the complete deletion of GAT1, GAT5 and GAT6 genes in these mutants. Furthermore, there was no significant changes of GAT5 or GAT6 expression levels at different time points after radiation. This was in agreement with literature suggesting that GATA TFs such as Gln3 and Gat1 in Saccharomyces cerevisiae translocate to nucleus after dephosphorylation thus regulating NCR gene expression but not by changing the gene levels of GATA TF themselves14. In the gat1Δ mutant, GDH1, AMT1 and AMT2 expression was very low which is in agreement with a previous report20. Among the potential GATA downstream genes, AAP4 and AAP5 were highly elevated in the H99S and gat1Δ mutant at 22 and 28 h after radiation exposure. However, the levels of AAP4 and AAP5 were much lower in the gat5Δ and gat6Δ mutants at those time points. URO1 level in the gat6Δ mutant was very high at 22 h post radiation. To better characterize the gene expression changes after radiation in the H99S and mutants, we focused on studying the expression of these three genes as shown in Fig. 5. There were similar levels of expression of AAP4, AAP5 and URO1 in H99S cells with or without radiation and gat1Δ mutant at each time point. For AAP4 gene (Fig. 5A), there were no significant differences between each group at 0, 2, and 4 h after radiation. However, at 22 h and 28 h after radiation the expression levels of AAP4 were significantly lower in the gat5Δ and gat6Δ mutants compared to the H99S irradiated group (at 22 h, the relative expression for AAP4 was 15.51 ± 1.44, 1.46 ± 0.34, and 1.09 ± 0.10 respectively in H99S, gat5Δ and gat6Δ mutants. At 28 h, the relative expression for AAP4 was 66.09 ± 5.40, 2.31 ± 0.53, and 4.63 ± 0.22 respectively in H99S, gat5Δ and gat6Δ mutants). For AAP5 gene (Fig. 5B), there were small but significantly increased expressions at early time points in gat5Δ mutant at 0 h, 2 h and 4 h (relative expressions were 8.16 ± 1.21, 5.08 ± 0.92, and 8.88 ± 1.50 in the gat5Δ mutant vs 1.12 ± 0.12, 1.97 ± 0.26, and 1.21 ± 0.23 in the H99S 1 kGy at 0 h, 2 h, and 4 h respectively), and in gat6Δ mutant at 0 h compared to the H99S 1 kGy samples (relative expressions were 4.26 ± 0.71 vs 1.12 ± 0.12); at later time points, there were significantly decreased levels of AAP5 in the gat5Δ and gat6Δ mutants (The relative expression at 22 h was 52.43 ± 7.78, 18.70 ± 7.20, and 15.80 ± 2.69 respectively in H99S, gat5Δ and gat6Δ mutants; the relative expression at 28 h was148.28 ± 18.92, 22.62 ± 2.60, and 17.75 ± 2.14 respectively in H99S, gat5Δ and gat6Δ mutants). The URO1 gene had significantly higher level at 0 h post radiation in the gat5Δ mutant compared to the H99S 1kGy (36.12 ± 12.29 in gat5Δ vs 1.96 ± 0.96 in H99S); and it also had significantly higher level at 4 h post radiation in the gat5Δ and gat6Δ mutants (3.93 ± 0.88, 33.62 ± 8.66, and 19.34 ± 8.12 in H99S, gat5Δ and gat6Δ mutants respectively). In Fig. 5C, URO1 gene was significantly higher in the gat6Δ mutant at 22 h post radiation compared to the H99S 1kGy group (7.14 ± 3.14 vs 124.30 ± 33.32 in H99S and gat6Δ mutant). Radiation alone did not change the levels of AAP4, AAP5 and URO1 at multiple time points as shown by the insignificant difference between H99S 1 kGy and 0 Gy samples, implying that radiation had no direct effect on the levels of these genes. The data suggest that GAT5 and GAT6 specifically regulate AAP4, AAP5 and URO1 genes with different dynamics. The GAT5 and GAT6 up-regulate AAP4 gene expression at later time points; while GAT5 and GAT6 down-regulate AAP5 gene expression at early time points, and then up-regulate AAP5 expression at later time points. GAT5 inhibits URO1 expression at early time points and GAT6 inhibits URO1 expression at later time points.
Figure 4.
Heat map diagram of differential gene expression in wild type H99S and gat1Δ, gat5Δ, and gat6Δ mutants after radiation exposure and also H99S without radiation exposure. The relative gene expression level was log2 transformed. The key for the heatmap coloring is included on the right. Each square represents the average of at least two biological repeats.
Figure 5.
Differential gene expression of AAP4, AAP5 and URO1 in H99S and GATA mutants after radiation exposure and also H99S without radiation exposure. Gene expression was determined using qRT-PCR and normalized to actin. Gene expression levels were log2 transformed. Each group was compared to the H99S 1 kGy group at each time point. Each bar represents mean ± SEM of 4 biological replicates. (*P < 0.05, **P < 0.01, ***P < 0.001).
Fungal GATA TFs recognize an HGATAR (5′-(T/A/C) GATA(A/G)-3′) motif upstream of their target genes21. We analyzed 2 kb DNA sequences upstream of AAP4 and AAP5 genes, and identified 4 GATA binding sites for AAP4 gene and 8 GATA binding sites for AAP5 gene as shown in the Supplementary file.
Phylogenetic analysis of the GATA factors in C. neoformans
To obtain information about the evolutionary relationships of the GATA factors in C. neoformans, phylogenetic analysis were performed based on the protein sequence alignments of all transcribed proteins of the GATA family members (Fig. 6). Compared to other GATA factors, GAT5 and GAT6 were mostly closely related to each other. BZP2 and GAT7 were the next closely related GATA factors to GAT5/6.
Figure 6.
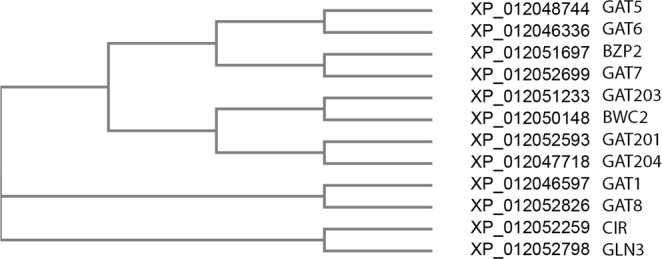
Phylogenetic tree of GATA TFs in C. neoformans. The protein sequences of GATA factors were used for phylogenetic tree analysis. Each GATA factor’s NCBI reference number is listed before the GATA TF names.
Discussion
In the current study, we identified TFs and their downstream genes that play important roles in radiation resistance in C. neoformans, which is a highly radiation-resistant fungus found in highly radioactive environment. Radiation-resistant microbes utilize many different strategies to counteract the damaging effects of ionizing radiation, such as high-levels of H-Mn2+ complexes to protect proteasome in D. radiodurans1,22 and efficient repair of DNA damages in cyanobacterium Chroococcidiopsis23 and D. radiodurans24. The molecular mechanisms of C. neoformans radiation-resistance are still not well understood. Transcription factors regulate coordinated gene expressions in response to various physiological or environmental signals. In our current study, we found that specific GATA TFs play important roles in C. neoformans after radiation exposure. GATA TFs are a class of highly conversed TFs present in fungi, metazoans and plants16. The fungal GATA TFs regulate many functions such as nitrogen metabolism, light induction, siderophore biosynthesis and mating-type switching16. There were few studies of GATA TFs in C. neoformans. Our data showed that even though they belong to the same TF family, each GATA TF may have specific functions. GAT1 is the most widely studied GATA TF in C. neoformans, which regulates nitrogen uptake and controls the transcription of genes involved in NCR, ergosterol biosynthesis, iron uptake, cell wall organization and capsule biosynthesis25. However, GAT1 mutant had the same sensitivity to rapamycin treatment as the wild type25, which is further confirmed in our current study. We showed that GAT6 mutant was most sensitive to rapamycin treatment in the GATA family. Therefore, the TOR signaling pathway, which regulates cell proliferation and is the target of inhibition by rapamycin, may act through GAT6 in C. neoformans. GAT5, GAT6, and BZP2 mutants were all sensitive to IR and hydroxyurea, suggesting they are important for DNA damage responses. BZP2 is the only GATA TF that is sensitive to fluconazole treatment. Our data suggest that specific GATA TF or their combination is important for different stressors. Furthermore, we showed that among the 28 downstream genes of GATA TFs we studied, only AAP4, AAP5 and URO1 were significantly changed in the gat5Δ and gat6Δ mutants after radiation (Fig. 5).
Studies in D. radiodurans radiation has suggested that IR resistance is predominantly a metabolic phenomenon, because D. radiodurans can grow on rich medium in the presence of continuous radiation (60 Gy/h) without lethality while D. radiodurans loses viability coupled with severe DNA degradation if grown in minimal medium and exposed to 60 Gy/h chronic IR, unless high concentrations of amino acids are provided22,26,27. Interestingly, it was shown in the 1960s that radiation inhibits amino acid uptakes in Escherichia coli28. In agreement with literature, our findings suggest that certain micronutrients are important for radiation resistance. We hypothesis that in response to IR, GAT5 and GAT6 in C. neoformans are required for the expression of amino acid permeases such as AAP4 and AAP5, thus transporting more specific micronutrients to the cells, and increasing cell survival following radiation. The data suggest that supplementing specific micronutrients may increase cell survival following radiation as shown in D. radiodurans. Not only in microbes, it has been shown that micronutrients such as amino acid mixture mitigates radiation-induced gastrointestinal toxicity in mice29,30 and oral supplementation with amino acid preparation helps hepatocellular carcinoma patients undergoing radiotherapy31. It would be interesting to check whether GATA TFs and AAPs in other organisms are also important for radiation sensitivity. If that is the case, then identifying the specific substrates for the AAPs may provide better radio-protection reagents for cells or even animals.
In addition to their importance in radiation, GAT5, GAT6 and BZP2 were shown to be important for C. neoformans virulence11. Interestingly, GAT5, GAT6 and BZP2 had no effect on virulence-factor (capsule, melanin, and urease) production as shown in the phenotype database at tf.cryptococcus.org11. C. neoformans has to handle the various stresses found at infection sites such as nutrient limitation, high temperature, and other innate factors for successful proliferation, dissemination, and colonization32,33. Therefore, even though these three GATA factors are not required for virulence factor production, they may up-regulate downstream NCR genes, such as AAP4 and AAP5, thus enabling cells to utilize poor nitrogen source for better survival. In other words, GAT5, GAT6 and BZP2 are important for virulence not by regulating the virulence factor production, but by regulating downstream NCR genes for better survival. In agreement with this hypothesis, it has been shown that AAP4 and AAP5 were essential for virulence in C. neoformans against high temperature and oxidative stress by mediating amino acid uptake during these stress conditions34.
The AAP4 and AAP5 gene expression showed a similar level of time-dependent increase in both the un-irradiated and irradiated H99S samples as shown in Fig. 5, suggesting that nutrition is a major regulator of AAP4 and AAP5 expression. We hypothesize that as the nutrients are being depleted in culture with time, AAP4 and AAP5 are being transcribed so that unfavorable amino acids can be transported into cells for better survival. Radiation will not directly regulate the expression of AAP4 and AAP5 genes. However, in irradiated cells, amino acids transported by permeases AAP4 and AAP5 are not only needed for starvation-related survival, but also needed for the repair of radiation-induced macromolecular damages. Therefore, cell survival after radiation is being determined by the availability of specific amino acids transported by AAP4 and AAP5.
The phylogenetic analysis showed the close relationship between GAT5 and GAT6 (Fig. 6). Interestingly, previous work also showed that AAP4 and AAP5 are mostly closely related within the 8 amino acid permeases17. So potentially a subfamily of GATA factors exists to respond to different stressors. For example, the GAT5/6-AAP4/5 subfamily is most important in response to ionizing radiation; while the GAT1-GDH1/AMT1/2 subfamily may be more important in response to nitrogen starvation.
We summarized the findings of GATA TF circuits in response to different stressors in Fig. 7. Specific GATA TFs are important for C. neoformans in response to different stressors. GAT1 is responsible for nitrogen uptake and metabolism but not important for rapamycin treatment as reported earlier25, mixed results for virulence11,20, also not important for radiation resistance. GDH1, AMT1 and AMT2 are important down-stream genes of GAT1. GAT5, GAT6 and BZP2 are important GATA TFs for radiation resistance and virulence. GAT5 and GAT6 regulate AAP4, AAP5 and URO1 genes in response to radiation. GAT5 and GAT6 regulate AAP4 and AAP5 in virulence11,34. GAT5, GAT6 and BZP2 are also important GATA TFs for another DNA damaging agent hydroxyurea treatment. GAT6 is important for rapamycin treatment while BZP2 is important for fluconazole treatment.
Figure 7.
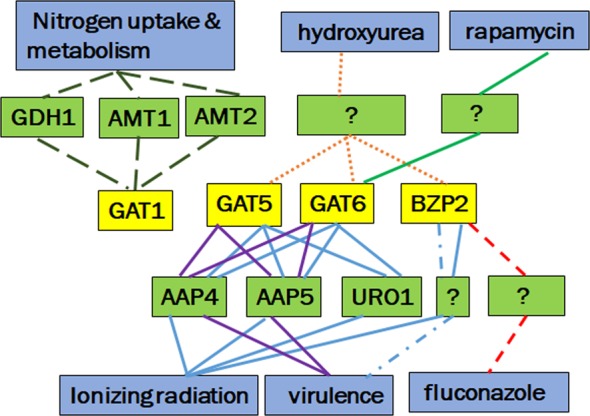
Summary of the GATA TF circuits important for different stressors in C. neoformans. The yellow boxes are GATA factors, green boxes are downstream genes of GATA factors, and the blue boxes are related biological processes. The subfamily of GATA factors important for one biological process is linked by the lines with same color/pattern.
In conclusion, we identified important TFs for radiation sensitivity in C. neoformans. Not only important for radiation response, specific GATA TFs are also important in response to other stressors. Particularly, GAT5 and GAT6 may control radiation sensitivity by regulating the expression of downstream genes such as AAP4, AAP5 and URO1.
Supplementary information
Acknowledgements
The views expressed here do not necessarily represent those of the Armed Forces Radiobiology Research Institute (AFRRI), the Uniformed Services University of the Health Sciences (USUHS) or the Department of Defense (DoD). We would like to acknowledge the help and support from Dr. William F. Blakely at AFRRI, and Dr. Michael Daly’s lab at USUHS, particularly Dr. Rok Tkavc and Robert Volpe. The work was supported by DEFENSE THREAT REDUCTION AGENCY Grant HDTRA-1-15-1-7335 to MX. The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author Contributions
Conceptualization: W.C. and M.X. Data curation: W.C. Formal analysis: W.C. Funding acquisition: M.X. Investigation: W.C., X.L., L.H. and M.X.
Data Availability
All data generated or analyzed during this study are included in this published article.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-42778-7.
References
- 1.Daly MJ. Death by protein damage in irradiated cells. DNA repair. 2012;11:12–21. doi: 10.1016/j.dnarep.2011.10.024. [DOI] [PubMed] [Google Scholar]
- 2.Jung KW, Lim S, Bahn YS. Microbial radiation-resistance mechanisms. J Microbiol. 2017;55:499–507. doi: 10.1007/s12275-017-7242-5. [DOI] [PubMed] [Google Scholar]
- 3.Rajasingham R, et al. Global burden of disease of HIV-associated cryptococcal meningitis: an updated analysis. Lancet Infect Dis. 2017;17:873–881. doi: 10.1016/S1473-3099(17)30243-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dadachova E, Casadevall A. Ionizing radiation: how fungi cope, adapt, and exploit with the help of melanin. Current opinion in microbiology. 2008;11:525–531. doi: 10.1016/j.mib.2008.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jung, K. W. et al. Unraveling Fungal Radiation Resistance Regulatory Networks through the Genome-Wide Transcriptome and Genetic Analyses of Cryptococcus neoformans. mBio7, 10.1128/mBio.01483-16 (2016). [DOI] [PMC free article] [PubMed]
- 6.Dadachova E, et al. The radioprotective properties of fungal melanin are a function of its chemical composition, stable radical presence and spatial arrangement. Pigment cell & melanoma research. 2008;21:192–199. doi: 10.1111/j.1755-148X.2007.00430.x. [DOI] [PubMed] [Google Scholar]
- 7.Eisenman HC, Casadevall A. Synthesis and assembly of fungal melanin. Applied microbiology and biotechnology. 2012;93:931–940. doi: 10.1007/s00253-011-3777-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Todeschini AL, Georges A, Veitia RA. Transcription factors: specific DNA binding and specific gene regulation. Trends Genet. 2014;30:211–219. doi: 10.1016/j.tig.2014.04.002. [DOI] [PubMed] [Google Scholar]
- 9.Hellweg CE, Spitta LF, Henschenmacher B, Diegeler S, Baumstark-Khan C. Transcription Factors in the Cellular Response to Charged Particle Exposure. Frontiers in oncology. 2016;6:61. doi: 10.3389/fonc.2016.00061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Janbon G, et al. Analysis of the genome and transcriptome of Cryptococcus neoformans var. grubii reveals complex RNA expression and microevolution leading to virulence attenuation. PLoS genetics. 2014;10:e1004261. doi: 10.1371/journal.pgen.1004261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jung KW, et al. Systematic functional profiling of transcription factor networks in Cryptococcus neoformans. Nature communications. 2015;6:6757. doi: 10.1038/ncomms7757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chojnacki S, Cowley A, Lee J, Foix A, Lopez R. Programmatic access to bioinformatics tools from EMBL-EBI update: 2017. Nucleic acids research. 2017;45:W550–W553. doi: 10.1093/nar/gkx273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pramila T, Wu W, Miles S, Noble WS, Breeden LL. The Forkhead transcription factor Hcm1 regulates chromosome segregation genes and fills the S-phase gap in the transcriptional circuitry of the cell cycle. Genes & development. 2006;20:2266–2278. doi: 10.1101/gad.1450606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cooper TG. Transmitting the signal of excess nitrogen in Saccharomyces cerevisiae from the Tor proteins to the GATA factors: connecting the dots. FEMS microbiology reviews. 2002;26:223–238. doi: 10.1111/j.1574-6976.2002.tb00612.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Broach JR. Nutritional control of growth and development in yeast. Genetics. 2012;192:73–105. doi: 10.1534/genetics.111.135731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Scazzocchio C. The fungal GATA factors. Current opinion in microbiology. 2000;3:126–131. doi: 10.1016/S1369-5274(00)00063-1. [DOI] [PubMed] [Google Scholar]
- 17.Fernandes JD, Martho K, Tofik V, Vallim MA, Pascon RC. The Role of Amino Acid Permeases and Tryptophan Biosynthesis in Cryptococcus neoformans Survival. PloS one. 2015;10:e0132369. doi: 10.1371/journal.pone.0132369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee IR, et al. Characterization of the complete uric acid degradation pathway in the fungal pathogen Cryptococcus neoformans. PloS one. 2013;8:e64292. doi: 10.1371/journal.pone.0064292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee IR, et al. Reactive oxygen species homeostasis and virulence of the fungal pathogen Cryptococcus neoformans requires an intact proline catabolism pathway. Genetics. 2013;194:421–433. doi: 10.1534/genetics.113.150326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee IR, Chow EW, Morrow CA, Djordjevic JT, Fraser JA. Nitrogen metabolite repression of metabolism and virulence in the human fungal pathogen Cryptococcus neoformans. Genetics. 2011;188:309–323. doi: 10.1534/genetics.111.128538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hwang LH, Seth E, Gilmore SA, Sil A. SRE1 regulates iron-dependent and -independent pathways in the fungal pathogen Histoplasma capsulatum. Eukaryotic cell. 2012;11:16–25. doi: 10.1128/EC.05274-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sharma A, et al. Across the tree of life, radiation resistance is governed by antioxidant Mn(2+), gauged by paramagnetic resonance. Proceedings of the National Academy of Sciences of the United States of America. 2017;114:E9253–E9260. doi: 10.1073/pnas.1713608114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Billi D, Friedmann EI, Hofer KG, Caiola MG, Ocampo-Friedmann R. Ionizing-radiation resistance in the desiccation-tolerant cyanobacterium Chroococcidiopsis. Applied and environmental microbiology. 2000;66:1489–1492. doi: 10.1128/AEM.66.4.1489-1492.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krisko, A. & Radman, M. Biology of extreme radiation resistance: the way of Deinococcus radiodurans. Cold Spring Harb Perspect Biol5, 10.1101/cshperspect.a012765 (2013). [DOI] [PMC free article] [PubMed]
- 25.Kmetzsch L, et al. The GATA-type transcriptional activator Gat1 regulates nitrogen uptake and metabolism in the human pathogen Cryptococcus neoformans. Fungal genetics and biology: FG & B. 2011;48:192–199. doi: 10.1016/j.fgb.2010.07.011. [DOI] [PubMed] [Google Scholar]
- 26.Venkateswaran A, et al. Physiologic determinants of radiation resistance in Deinococcus radiodurans. Applied and environmental microbiology. 2000;66:2620–2626. doi: 10.1128/AEM.66.6.2620-2626.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Slade D, Radman M. Oxidative stress resistance in Deinococcus radiodurans. Microbiol Mol Biol Rev. 2011;75:133–191. doi: 10.1128/MMBR.00015-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kempner ES, Pollard EC. Radiation inhibition of amino acid uptake by Escherichia coli. Biophysical journal. 1961;1:265–277. doi: 10.1016/S0006-3495(61)86888-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yin L, et al. An amino acid-based oral rehydration solution (AA-ORS) enhanced intestinal epithelial proliferation in mice exposed to radiation. Scientific reports. 2016;6:37220. doi: 10.1038/srep37220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yin L, et al. An amino acid mixture mitigates radiation-induced gastrointestinal toxicity. Health physics. 2014;106:734–744. doi: 10.1097/HP.0000000000000117. [DOI] [PubMed] [Google Scholar]
- 31.Lee IJ, et al. Effect of Oral Supplementation with Branched-chain Amino Acid (BCAA) during Radiotherapy in Patients with Hepatocellular Carcinoma: A Double-Blind Randomized Study. Cancer research and treatment: official journal of Korean Cancer Association. 2011;43:24–31. doi: 10.4143/crt.2011.43.1.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Perfect JR. Cryptococcus neoformans: the yeast that likes it hot. FEMS yeast research. 2006;6:463–468. doi: 10.1111/j.1567-1364.2006.00051.x. [DOI] [PubMed] [Google Scholar]
- 33.Perfect, J. R. The triple threat of cryptococcosis: it’s the body site, the strain, and/or the host. mBio3, 10.1128/mBio.00165-12 (2012). [DOI] [PMC free article] [PubMed]
- 34.Martho KF, et al. Amino Acid Permeases and Virulence in Cryptococcus neoformans. PloS one. 2016;11:e0163919. doi: 10.1371/journal.pone.0163919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ngamskulrungroj P, Chang Y, Roh J, Kwon-Chung KJ. Differences in nitrogen metabolism between Cryptococcus neoformans and C. gattii, the two etiologic agents of cryptococcosis. PloS one. 2012;7:e34258. doi: 10.1371/journal.pone.0034258. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article.