Lyme disease is caused by the spirochete Borrelia burgdorferi and is transmitted via the bite of an infected tick. B. burgdorferi enters the skin, disseminates via the bloodstream, and infects various distal tissues, leading to inflammatory sequelae, such as Lyme arthritis and Lyme carditis.
KEYWORDS: Borrelia burgdorferi, Lyme disease, dissemination, lp36, skin, vector-borne disease
ABSTRACT
Lyme disease is caused by the spirochete Borrelia burgdorferi and is transmitted via the bite of an infected tick. B. burgdorferi enters the skin, disseminates via the bloodstream, and infects various distal tissues, leading to inflammatory sequelae, such as Lyme arthritis and Lyme carditis. B. burgdorferi linear plasmid 36 (lp36) is critical for mammalian infectivity; however, the full complement of genes on lp36 that contribute to this process remains unknown. Through a targeted mutagenesis screen of the genes on lp36, we identified a novel infectivity gene of unknown function, bbk13, which encodes an immunogenic, non-surface-exposed membrane protein that is important for efficient mammalian infection. Loss of bbk13 resulted in reduced spirochete loads in distal tissues in a mouse model of infection. Through a detailed analysis of B. burgdorferi infection kinetics, we discovered that bbk13 is important for promoting spirochete proliferation in the skin inoculation site. The attenuated ability of Δbbk13 spirochetes to proliferate in the inoculation site was followed by reduced numbers of B. burgdorferi spirochetes in the bloodstream and, ultimately, consistently reduced spirochete loads in distal tissues. Together, our data indicate that bbk13 contributes to disseminated infection by promoting spirochete proliferation in the early phase of infection in the skin. This work not only increases the understanding of the contribution of the genes on lp36 to B. burgdorferi infection but also begins to define the genetic basis for B. burgdorferi expansion in the skin during localized infection and highlights the influence of the early expansion of spirochetes in the skin on the outcome of infection.
INTRODUCTION
Lyme disease is the leading arthropod-borne bacterial infection in the United States (1, 2). The causative agent of Lyme disease is the spirochete Borrelia burgdorferi (3, 4), which is transmitted to humans via the bite of an infected Ixodes scapularis tick (5). Initial symptoms of the disease include fever, nausea, and, in some cases, the appearance of the bull's-eye rash (erythema migrans) at the tick bite site. From the initial tick bite site, B. burgdorferi undergoes a brief dissemination period in the blood and colonizes various distal tissues. Lyme disease pathologies occur at sites of B. burgdorferi infection, such as the joints (Lyme arthritis), the heart (Lyme carditis), and the nervous system (neuroborreliosis) (6).
The B. burgdorferi genome is highly fragmented, with the genome being spread across a small linear chromosome and >20 linear and circular plasmids (7, 8). Linear plasmid 36 (lp36) has been shown to be dispensable for B. burgdorferi growth in culture but critical for mammalian infectivity. Spirochetes lacking lp36 demonstrate an approximately 4-log increase in the 50% infectious dose (ID50) as well as reduced bacterial loads in the tissues of infected mice (9). lp36 is approximately 37 kbp in size and carries 37 annotated genes, 28 of which are predicted protein-coding sequences (7, 8). The functions of the majority of the annotated open reading frames on lp36 remain unknown. Two genes on lp36, bbk17 and bbk32, have independently been shown to contribute to B. burgdorferi infectivity (9, 10). Gene bbk17 encodes an adenine deaminase (AdeC), which functions in the B. burgdorferi purine salvage pathway (9). Gene bbk32 encodes a surface-exposed fibronectin-binding protein (11), which facilitates spirochete adhesion to the host microenvironment, such as the blood vessel wall (12, 13) and joint tissue via a glycosaminoglycan interaction (14), as well as inhibition of the classical pathway of the host complement cascade (15). Interestingly, the ID50 shift in each single mutant background is only a fraction of what was measured upon loss of the whole plasmid (10, 16). Furthermore, deletion of both genes together does not appear to recapitulate the attenuated phenotype of the lp36-negative mutant (our unpublished results), leading to the prediction that other genes on lp36 also play a role in mammalian infection.
B. burgdorferi is introduced into the skin by the feeding activity of the Ixodes tick vector during natural infection. It has been shown that B. burgdorferi undergoes a progressive increase in spirochete number in the skin within 7 days after intradermal inoculation (17–20). Characterization of the B. burgdorferi genes important for infectivity has largely focused on measuring spirochete loads in distal tissues in the disseminated phase of infection. Also, studies that utilize the intraperitoneal route of inoculation bypass the skin altogether. Therefore, the genetic regulation of spirochete expansion in the skin is largely unexplored. Moreover, the impacts of this early event on the later phases of infection have not been studied in detail.
From a targeted mutagenesis screen of lp36 genes, we identified bbk13 to be important for efficient mammalian infection by B. burgdorferi. In the absence of bbk13, B. burgdorferi colonization of distal tissues, such as the ear, heart, and joints, was markedly reduced or absent in our mouse model. By following the progression of B. burgdorferi, we discovered that bbk13 is required early in infection for optimal spirochete expansion at the skin inoculation site. This work begins to define the genetic basis for B. burgdorferi expansion in the skin inoculation site during localized infection and demonstrates that this early event has a strong influence on spirochete loads in distal tissues during the disseminated phase of infection.
RESULTS
A targeted mutagenesis screen of lp36 identifies a possible role for bbk13 in B. burgdorferi mouse infection.
To begin to identify the genes on lp36 that contribute to B. burgdorferi infectivity, a targeted mutagenesis approach was applied. A small panel of genes, bbk09, bbk13, bbk40, and bbk41, which are annotated to encode conserved hypothetical proteins of unknown function (7, 8), was investigated. Deletion mutants were generated by replacing the target open reading frames with a streptomycin antibiotic resistance cassette using allelic exchange (21) and verified by PCR and DNA sequencing across the lesion sites. All B. burgdorferi mutants were confirmed to contain the same plasmid content as the parent clone and to have the same in vitro growth as the wild type (WT) (see Fig. S1 in the supplemental material). C3H/HeN mice were needle inoculated with 104 wild-type or mutant B. burgdorferi spirochetes. At 3 weeks postinoculation, serum and distal tissues were collected for serology and tissue reisolation analysis, respectively. All mice were positive for anti-B. burgdorferi antibodies, and all tissue reisolation cultures were positive for spirochetes by visual inspection using dark-field microscopy after 7 days of incubation (data not shown), indicating that all of the mutants were able to infect the mice. Interestingly, the reisolation cultures of tissues from Δbbk13 mutant-infected mice appeared to have lower spirochete densities than the reisolation cultures of tissues from mice infected with the wild type or the other deletion mutants, suggesting the possibility for a contribution of bbk13 to B. burgdorferi infectivity.
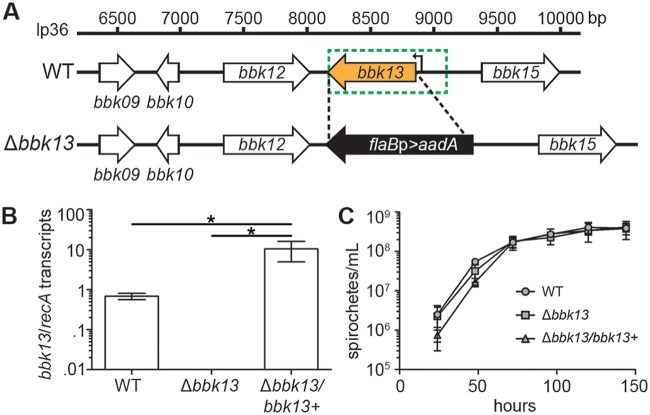
As a means to investigate the contribution of bbk13 to B. burgdorferi infectivity, the Δbbk13 mutant (Fig. 1A) was complemented in trans with a wild-type copy of bbk13, including the putative endogenous promoter region up to 221 bp upstream of the bbk13 transcription start site (22) (Fig. 1A, green dotted-line box), on a shuttle vector. Reverse transcriptase quantitative PCR (RT-qPCR) demonstrated the complete loss of bbk13 expression in Δbbk13 B. burgdorferi and restoration of expression in the Δbbk13/bbk13+ complement clone (Fig. 1B). The level of bbk13 expression in the Δbbk13/bbk13+ clone was >10-fold higher than that in the wild type (Fig. 1B), likely due to the increased copy number of the pBSV2G vector (23). In vitro growth analysis demonstrated that the in vitro growth kinetics of the Δbbk13 and Δbbk13/bbk13+ clones were similar to those of the wild type (Fig. 1C).
FIG 1.
Genetic deletion of B. burgdorferi bbk13. (A) Schematic representation of the linear plasmid 36 (lp36) genomic locus containing bbk13 and adjacent genes. The bbk13 open reading frame (orange arrow) was replaced with the flaBp-aadA antibiotic resistance cassette (black arrow) in wild-type B. burgdorferi (WT) by allelic exchange. Deletion boundaries are indicated by the black dashed lines. Complementation of the bbk13 mutant (Δbbk13) with a wild-type copy of bbk13 under the control of its putative endogenous promoter (the DNA region indicated by the green dotted-line box) on pBSV2G was performed in trans, resulting in the Δbbk13/bbk13+ clone. The bbk13 transcription start site (22) is indicated by a bent black arrow at position 8,903 bp. (B) bbk13 expression is abolished in the Δbbk13 mutant and restored in the Δbbk13/bbk13+ complemented clone. RNA was isolated from WT, Δbbk13, and Δbbk13/bbk13+ clones, and bbk13 expression was measured by RT-qPCR. Gene expression of recA was used for normalization. The mean ± standard deviation from three biological replicates are shown. Statistical significance was tested using one-way ANOVA and Tukey’s multiple-comparison test (*, P < 0.05). (C) The loss of bbk13 does not alter B. burgdorferi growth in vitro. Triplicate cultures of the WT, Δbbk13, and Δbbk13/bbk13+ clones were inoculated at 105 spirochetes/ml. Culture densities were determined every 24 h over a 144-h period using Petroff-Hausser counting chambers. Symbols and error bars represent the mean ± standard deviation.
The gene bbk13 is constitutively expressed throughout the B. burgdorferi infectious cycle and encodes an immunogenic membrane-associated protein.
As a vector-borne pathogen, B. burgdorferi alters its gene expression profile in response to the different environments that it encounters in order to promote its survival and maintenance throughout its enzootic cycle (24, 25). To begin to understand the possible contribution of bbk13 to B. burgdorferi infectivity, we sought to determine the gene expression profile of bbk13 throughout the tick-mouse infectious cycle. RNA isolated from B. burgdorferi-infected ticks and mouse tissue was probed for expression of bbk13 as well as the constitutive flagellar gene (flaB) and the differentially expressed outer surface protein A (ospA) and outer surface protein C (ospC) genes (25). Like flaB, bbk13 was highly expressed throughout the enzootic cycle (Fig. 2A). These findings are consistent with those reported by Iyer et al. (24).
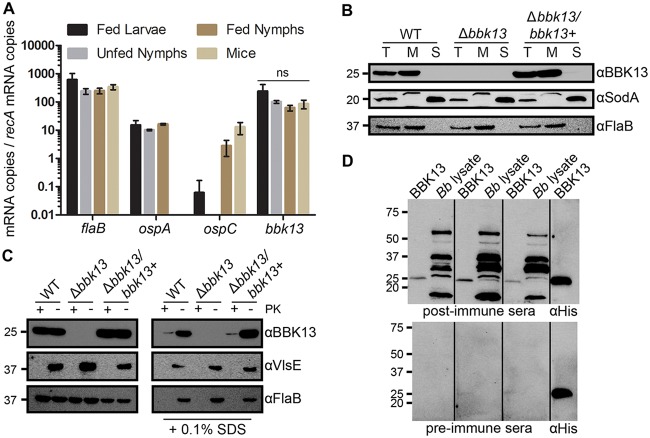
FIG 2.
Gene bbk13 is constitutively expressed throughout the spirochete’s enzootic cycle and encodes a membrane-associated, immunogenic protein. (A) RT-qPCR using gene-specific primers and a B. burgdorferi genomic standard curve was used to quantify the amount of each target mRNA transcript in infected tick and mouse tissue samples. Copy numbers for each gene target were normalized to recA mRNA copy numbers. Data are presented as the average of biological triplicate samples ± standard deviation. The genes flaB, ospA, and ospC served as controls. ns, not significant by one-way ANOVA with Tukey’s multiple-comparison test. (B) BBK13 protein localization analysis. Total protein lysates (lanes T) of the B. burgdorferi wild-type (WT), Δbbk13, and Δbbk13/bbk13+ clones were fractionated into the soluble (lanes S) and membrane (lanes M) components using ultracentrifugation. Protein fractions from equivalent numbers of spirochetes were subjected to SDS-PAGE and analyzed by immunoblotting with BBK13, periplasm-associated FlaB, or cytoplasm-associated SodA antibodies. (C) Proteinase K susceptibility of BBK13. Equal numbers of B. burgdorferi WT, Δbbk13, and Δbbk13/bbk13+ spirochetes were treated with (+) or without (−) 200 μg/ml proteinase K (PK) in the presence or absence of 0.1% SDS. Samples were subjected to SDS-PAGE and analyzed by immunoblotting with BBK13, periplasm-associated FlaB, or outer surface-associated VlsE antibodies. (D) BBK13 immunogenicity. Recombinant BBK1325–232His protein (BBK13) and B. burgdorferi protein lysate (Bb lysate) were subjected to SDS-PAGE and analyzed by immunoblotting with preimmune and postimmune serum collected from 3 different individual mice before and after infection with B. burgdorferi. Recombinant BBK1325–232His protein (BBK13) was probed with anti-His antibody as a positive control. Protein standards (in kilodaltons) are indicated to the left in panels B, C, and D.
The bbk13 gene encodes a highly conserved (97 to 100% sequence identity within B. burgdorferi isolates and 90 to 96% sequence identity across Lyme disease Borrelia species) hypothetical protein of 232 amino acids, which is predicted to contain an N-terminal transmembrane region and the conserved domain of unknown function, COG2859, also known as a SIMPL (signaling molecule that interacts with mouse pelle-like kinase) domain (26, 27). Furthermore, the BBK13 protein has recently been identified to be one of the proteins associated with inner membrane lipid rafts of B. burgdorferi (28). A protein BLAST analysis of the BBK13 amino acid sequence found that SIMPL domain-containing putative membrane-associated proteins are present across a broad number of pathogenic and nonpathogenic bacterial species. To experimentally validate BBK13 protein production and localization in the spirochete, an anti-BBK13 polyclonal antibody was generated using recombinant BBK13 lacking the first 25 amino acids containing the putative transmembrane domain along with a C-terminal His tag (rBBK1325–232His) purified from Escherichia coli. Western blot analysis of total protein lysate from wild-type and Δbbk13/bbk13+ clones using the anti-BBK13 antibody detected a protein of 24 kDa, the predicted molecular mass of BBK13 (Fig. 2B). The 24-kDa protein was not detected in the total protein lysate from the Δbbk13 mutant (Fig. 2B), indicating the specificity of the anti-BBK13 antibody for the BBK13 protein. Further, BBK13, like membrane-associated FlaB, was found to be restricted to the spirochete membrane fraction (Fig. 2B). Proteinase K treatment of intact B. burgdorferi did not affect BBK13 (Fig. 2C). Similarly, the periplasmic FlaB protein was unaffected by proteinase K treatment, whereas this treatment resulted in the complete loss of the surface-exposed outer membrane protein VlsE. In contrast, proteinase K treatment of SDS-exposed B. burgdorferi resulted in the loss of BBK13 (Fig. 2C). Together, these data support the notion that BBK13 localizes to the spirochete inner membrane. Although our data indicate that BBK13 is not surface exposed, SIMPL domain-containing proteins from Brucella spp. and Edwardsiella ictaluri have been shown to be highly immunogenic and therefore are potential diagnostic markers and/or vaccine candidates (29–32). To examine the immunogenic properties of BBK13, rBBK1325–232His and total B. burgdorferi protein lysate were separated by SDS-PAGE and Western blot analysis was performed using pre- and postimmune sera from three different wild-type B. burgdorferi-infected mice. Recombinant BBK1325–232His was also probed with an anti-His antibody as a positive control. Anti-BBK13 antibodies were found to be present in all three mouse postimmune serum samples but not the preimmune serum samples (Fig. 2D), suggesting that BBK13 is immunogenic.
The loss of bbk13 impacts B. burgdorferi loads in distal tissues as early as 2 weeks postinoculation.
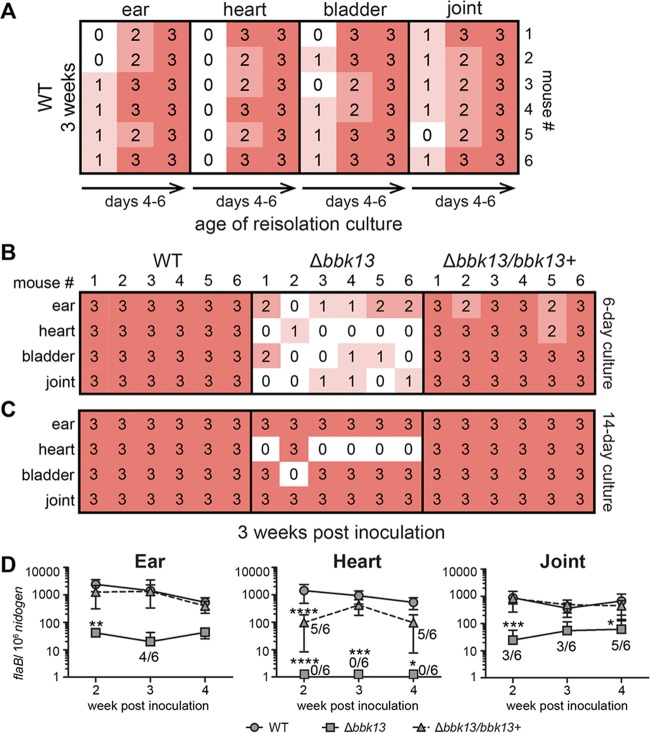
During natural infection, B. burgdorferi is introduced into the skin by tick bite, disseminates via the bloodstream, and colonizes distal tissues, such as the skin, heart, and joints (6). To confirm the role of bbk13 in mammalian infectivity, 104 spirochetes of the wild-type, Δbbk13, or Δbbk13/bbk13+ clone were delivered by intradermal injection in the dorsal thoracic skin of mice as a means to mimic the natural route of infection that initiates in the skin but control the time and dose of inoculation. At 3 weeks postinoculation, the capacity of the spirochetes to infect distal tissues was determined by tissue reisolation and quantitative PCR. To facilitate preliminary characterization and comparison of the spirochete densities in the tissue reisolation cultures, a semiquantitative numerical scoring system was developed (a score of 0 was used if spirochetes were absent, and scores of 1 to 3 were applied for increasing spirochete densities in the field of view upon inspection by dark-field microscopy; see Materials and Methods for details). Cultures of mouse tissues harvested at 3 weeks postinoculation with wild-type B. burgdorferi were monitored until the spirochete densities were high (score of 3), typically after 5 to 6 days of incubation in Barbour-Stoenner-Kelly II (BSKII) medium (Fig. 3A), at which time the reisolation cultures of tissue from the Δbbk13 and Δbbk13/bbk13+ spirochete-inoculated mice were scored (Fig. 3B). It is possible that slight variations in tissue size and/or differences in tissue type could affect the spirochete densities in the reisolation cultures. The protocol of using group sizes of six animals and monitoring the wild-type reisolation cultures to the point when all cultures of each tissue type achieved a score of 3 and then scoring the reisolation densities of the mutant clones for that tissue provided an internal control for possible tissue variations across samples. This means that different tissue types may be scored on different days of incubation. The scores of the mutant clones are therefore relative to the wild-type score of 3 for each tissue. Using this new method, the initial observation of reduced spirochete reisolation from Δbbk13 mutant-inoculated mouse tissues was recapitulated and documented in a semiquantitative manner. All of the tissues examined from the Δbbk13 mutant-inoculated mice demonstrated dramatically reduced reisolation density scores compared to those from mice inoculated with bbk13-containing spirochetes (Fig. 3B). Overall, greater than 80% of the Δbbk13 mutant reisolation cultures received a score of 0 or 1. The same reisolation cultures were also analyzed using the endpoint method of scoring for the presence or absence of spirochetes after 2 weeks of incubation. In contrast, according to this method the majority of the Δbbk13 mutant reisolation cultures were found to be positive for spirochetes (Fig. 3C), thereby strongly underestimating the infectivity defect of the mutant. These findings highlight the power of our reisolation scoring method to reveal B. burgdorferi mutants with defects in infection of distal tissues which might otherwise go undetected and be overlooked for follow-up validation analysis by qPCR. Nearly 90% of the reisolation cultures of tissues from the Δbbk13/bbk13+ spirochete-inoculated mice were given a score of 3 (Fig. 3B).
FIG 3.
Loss of bbk13 results in consistently reduced B. burgdorferi loads in distal tissue over multiple time points of infection. Groups of 6 C3H/HeN mice were inoculated intradermally in the dorsal skin with 104 spirochetes of the wild-type (WT), Δbbk13, or Δbbk13/bbk13+ clone. Infection of ear, heart, bladder, and joint tissues was assayed at 2, 3, and 4 weeks postinoculation. (A to C) Tissues collected from groups of 6 mice at 3 weeks postinoculation with B. burgdorferi clones were incubated in BSKII medium, and the spirochete density was scored by dark-field microscopy in a semiquantitative manner, where 0 indicates no spirochetes (white) and 1 to 3 indicate increasing spirochete densities (pink to red shading). (A) Reisolation cultures of tissue from WT-inoculated mice were monitored and scored for spirochete density at various time points of incubation (the age of the reisolation culture) until all cultures of each tissue type attained a score of 3. (B) Reisolation cultures of tissue from Δbbk13 and Δbbk13/bbk13+ spirochete-inoculated mice were scored for density on the time point of incubation when all WT cultures of each tissue type attained a score of 3 (6-day culture). (C) All tissue reisolation cultures were scored for endpoint density (14-day culture). (D) Spirochete loads in mouse ear, heart, and joint tissues were quantified by qPCR at 2, 3, and 4 weeks postinoculation. The data are presented as the number of flaB copies per 106 nid copies. Symbols and error bars represent the mean ± standard deviation. The Δbbk13 mutant loads were below the level of detection in heart tissue at all time points examined (symbols placed on the axis represent tissues that were tested but for which results were are not plottable on a logarithmic axis). For samples sets in which the spirochete loads in some or all of the tissues from the groups of 6 mice were below the level of detection, the number of tissues out of 6 with detectable loads are indicated. Two-way ANOVA and Tukey’s multiple-comparison test were used to test for statistical significance compared to the results for the WT (****, P < 0.0001; ***, P < 0.001; **, P < 0.01; *, P < 0.05).
The spirochete reisolation scores suggested a role for bbk13 in the ability of B. burgdorferi to efficiently colonize distal tissues. To validate these results, the levels of disseminated infection of the wild-type, Δbbk13, and Δbbk13/bbk13+ clones were assessed using qPCR to quantify the spirochete loads in the ear, heart, and joint tissues from the same set of mice at 3 weeks postinoculation. In addition, to understand the contribution of bbk13 to the kinetics of disseminated infection, additional groups of mice were inoculated with the wild type, Δbbk13, or Δbbk13/bbk13+ clone for 2 and 4 weeks and the spirochete loads in tissues were quantified. Consistent with the tissue reisolation scores, at the time point of 3 weeks postinoculation, the Δbbk13 mutant loads in the heart tissues were significantly reduced compared to those of the wild type (Fig. 3D). Although the differences in the Δbbk13 mutant and wild-type loads in the ear and joint tissues at this time point were not found to be statistically significant, only 4 out of 6 ears and 3 out 6 joints from the Δbbk13 mutant-inoculated mice had spirochete loads above the level of detection by qPCR, suggesting a trend in reduced tissue colonization by the mutant consistent with the tissue reisolation score data. At the 2- and 4-week time points postinoculation, the majority of the Δbbk13 mutant loads in the ear and joint tissues were significantly reduced compared to those of the wild type and were below the level of detection in the heart tissue (Fig. 3D). The loads of the Δbbk13/bbk13+ clone in all tissues examined were not significantly different from those of the wild type, apart from at the 2-week time point in the heart tissue (Fig. 3D, triangle). Overall, a correlation between the tissue reisolation scores and the qPCR results was observed. Together, these data demonstrate that bbk13 is important for B. burgdorferi to achieve wild-type loads in distal tissues and suggest that bbk13 contributes to events that occur during or prior to B. burgdorferi establishment in distal tissues at 2 weeks postinoculation. Furthermore, our data indicate that bbk13 is critical for B. burgdorferi infection in heart tissue.
The loss of bbk13 results in reduced spirochete numbers in the blood during dissemination.
B. burgdorferi lacking bbk13 was found at significantly reduced numbers in distal tissues as early as 2 weeks postinoculation. The bloodstream serves as a major route of dissemination of B. burgdorferi from the site of inoculation to distal tissues (33), raising the possibility that the reduced Δbbk13 mutant loads in tissues may be due to an attenuation in the numbers of disseminating spirochetes in the blood. To test this, blood was collected from cohorts of mice daily for an 11-day period following intradermal inoculation with104 wild-type, Δbbk13, or Δbbk13/bbk13+ spirochetes. The number of circulating spirochetes was determined by quantifying the CFU in BSK-agarose medium. Detection of the spirochete densities in the blood was limited to days 4 to 7 postinoculation for all three clones (Fig. 4), consistent with the transient nature of B. burgdorferi in the blood (33–35). The peak spirochete density in the blood occurred on day 6 postinoculation for all clones (Fig. 4), suggesting that blood dissemination kinetics are independent of bbk13. However, the density of circulating B. burgdorferi in the blood was bbk13 dependent. The peak density of the Δbbk13 mutant in the blood was significantly reduced compared to that of wild-type and Δbbk13/bbk13+ spirochetes (Fig. 4). This finding shows that the infection deficiency of Δbbk13 spirochetes occurs prior to distal tissue colonization and suggests that the reduced mutant loads detected in distal tissues may be due to diminished numbers of disseminating spirochetes in the blood.
FIG 4.
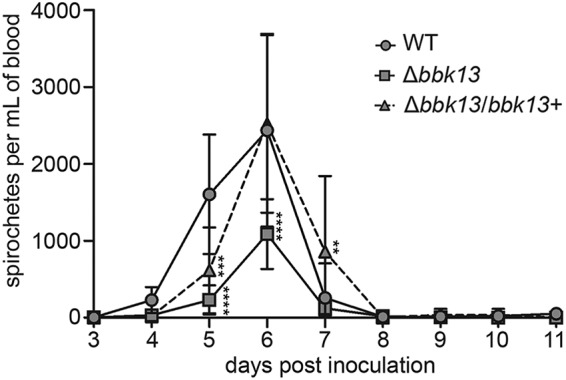
B. burgdorferi spirochetes lacking bbk13 are found at a reduced density in the blood during dissemination. Blood was collected daily from cohorts of 6 C3H/HeN mice inoculated intradermally with 104 spirochetes of the WT, Δbbk13, or Δbbk13/bbk13+ clone and plated in BSK-agarose for determination of the number of CFU. The data are reported as the number of spirochetes per milliliter of blood. Symbols and error bars represent the mean ± standard deviation. Two-way ANOVA and Tukey’s multiple-comparison test were used to test for statistical significance compared to the results for the WT (****, P < 0.0001; ***, P < 0.001; **, P < 0.01).
bbk13 is important for spirochete population expansion at the skin inoculation site.
The bbk13-dependent reduction in B. burgdorferi density in the blood may result from a defect in the ability of the Δbbk13 mutant to enter the bloodstream from the skin inoculation site and/or an attenuation that occurs at the skin inoculation site itself. To distinguish between these two possibilities, we quantified the spirochete loads in the skin inoculation site at various time points after intradermal injection of mice with 104 spirochetes of the wild-type, Δbbk13, or Δbbk13/bbk13+ clone. Starting at day 3 and up to day 7 postinoculation, the spirochete populations were detectable and continuously expanded at the skin inoculation site (Fig. 5). However, B. burgdorferi Δbbk13 failed to expand as strongly as the wild-type and Δbbk13/bbk13+ clones (Fig. 5). These findings indicate that bbk13 is important for spirochete expansion at the skin inoculation site during the earliest phase of B. burgdorferi infection.
FIG 5.
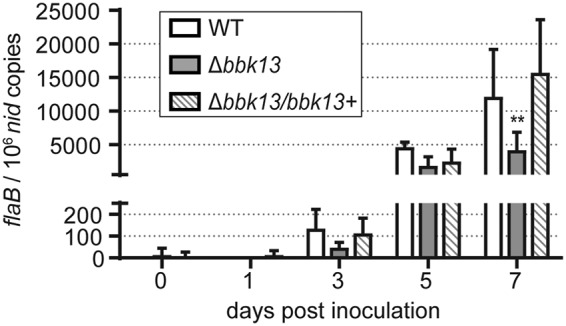
Loss of bbk13 results in deficient spirochete expansion at the skin inoculation site during early infection. Spirochete load kinetics at the skin inoculation site were determined by qPCR at different time points after intradermal inoculation of groups of 6 C3H/HeN mice with 104 spirochetes of the WT, Δbbk13, or Δbbk13/bbk13+ clone, as described in the legend to Fig. 2. The data are presented as the number of flaB copies per 106 nid copies. Early kinetics were determined by quantitating the spirochete loads in the skin inoculation sites on days 0, 1, 3, 5, and 7 postinoculation. Columns and error bars represent the mean ± standard deviation. Two-way ANOVA and Tukey’s multiple-comparison test were used to test for statistical significance compared to the results for the WT (**, P < 0.001).
DISCUSSION
B. burgdorferi lacking linear plasmid 36 (lp36) is highly attenuated for mouse infection (16). The genes bbk17 and bbk32 on lp36 each contribute to B. burgdorferi infectivity (9, 10, 13–15, 36, 37). However, the individual contribution of additional genes on lp36 to B. burgdorferi infectivity remained unknown. Through a targeted mutagenesis screen of a subset of genes on lp36, we have now established a role for bbk13 early in infection, which is critical for the generation of a robust disseminated infection. Moreover, we found that, according to the parameters measured, bbk09, bbk40, and bbk41 are dispensable for B. burgdorferi infection. For the most part, these results are consistent with those reported in a genome-scale signature tag mutagenesis (STM) study to identify B. burgdorferi genes that contribute to mouse infectivity (38). However, in the STM study, the bbk41 transposon mutant was found to be moderately attenuated for infection (38), suggesting that in the context of a mixed infection, the bbk41 gene may be important for B. burgdorferi to achieve maximal fitness.
The primary goal of this study was to identify B. burgdorferi genes on lp36 that contribute to disseminated infection, which was initially screened for by the reisolation of spirochetes from the tissues of mice at 3 weeks postinoculation. Typically, the reisolation assay is scored by microscopy analysis for the presence or absence of spirochetes following incubation of tissues in BSKII medium for up to 2 weeks (39–42). This method has been sufficient to detect strong loss of infectivity phenotypes (41) but is not likely to identify subtle phenotypes resulting from reduced numbers, rather than the complete absence, of spirochetes at distal tissues. To address this, we developed a semiquantitative method of scoring the spirochete density in the reisolation cultures. This slight modification captures more information from the reisolation assay and allows for the initial identification of attenuated B. burgdorferi mutants with reduced spirochete numbers in the tissues. Tissue reisolation scores were then subjected to follow-up verification by qPCR. Overall, our data indicated a strong correlation between the semiquantitative reisolation scores and qPCR results. Application of this scoring method was key to the discovery of bbk13 as a novel infectivity gene in our targeted screen. Moreover, broad application of this method to the analysis of the infection phenotypes of B. burgdorferi mutants is likely to lead to the discovery of additional genes that contribute to infection, which may previously have gone unrecognized.
Gene bbk13 is critical for B. burgdorferi mammalian infectivity. The Δbbk13 mutant demonstrated reduced spirochete loads in distal tissues at 3 weeks postinoculation. Furthermore, bbk13 was required for B. burgdorferi to achieve the wild-type density in the blood, though the kinetics of blood dissemination appeared to be bbk13 independent, as the timing of the peak density was the same as that of wild-type B. burgdorferi. Ultimately, our data suggest that the contribution of bbk13 to B. burgdorferi infectivity manifests at the earliest phase of infection by promoting spirochete population expansion and/or limiting spirochete clearance at the skin inoculation site.
The kinetics of spirochete loads in the skin inoculation site were dramatically different from those in the other tissues examined in this study. The spirochete loads in most distal tissues examined showed a gradual decrease over time. For example, there was a statistically significant reduction in wild-type B. burgdorferi loads over time in the ear and heart tissues (at the 2-week versus 4-week time point; P < 0.001, two-way analysis of variance [ANOVA] with Tukey’s multiple-comparison test) (Fig. 3D). In contrast, the spirochete loads in the skin inoculation site underwent a significant bbk13-dependent increase during the first week of infection. Between weeks 1 and 2, the spirochete loads in the inoculation site underwent a dramatic >100-fold reduction that was bbk13 independent. All B. burgdorferi clones remained at low but detectable levels in the skin inoculation site out to 4 weeks postinoculation (see Fig. S2 in the supplemental material). Notably, the peak wild-type spirochete loads in the skin inoculation site were higher than those in any wild-type-infected tissue examined at any time point. Conversely, the wild-type spirochete loads in the skin inoculation site at 2 to 4 weeks postinoculation were lower than those in any wild-type-infected tissue examined at any time point, including the ear, which is representative of a distal skin site. These data highlight the unique physiological context surrounding the early stage of B. burgdorferi infection in the skin.
Our data suggest that the early expansion of spirochetes in the skin inoculation site influences the spirochete loads in the blood during dissemination and, finally, in the distal tissues at later time points of infection. B. burgdorferi proliferation in the skin during primary infection has been described previously (17–20). It has also been hypothesized that B. burgdorferi replication in the skin facilitates infection and dissemination (20). Our work provides evidence to support this hypothesis and identifies a role for bbk13 in this process. Furthermore, it has been shown that infection is established at a much lower dose when B. burgdorferi is administered intradermally than when it is administered by other inoculation routes (43). This further emphasizes the significance of the early events that occur in the skin in promoting B. burgdorferi infection, as well as the relevance of using the intradermal route of inoculation in animal models of infection.
There is increasing evidence that the immunomodulatory properties of tick saliva contribute to the survival and multiplication of B. burgdorferi in the skin (19, 20). B. burgdorferi spirochetes lacking the entire lp36 plasmid are not defective for survival or replication in the tick but remain attenuated for mammalian infection by tick bite transmission (9). Future studies will focus on analysis of the skin proliferation and dissemination phenotype of the bbk13 mutant in the context of tick bite transmission.
The molecular function of bbk13 remains unknown. Our data and the data of others (24) indicate that bbk13 is constitutively expressed at high levels throughout the enzootic cycle. Consistent with the identification of BBK13 as one of the proteins associated with inner membrane lipid rafts of B. burgdorferi (28), we have shown that BBK13 is a non-surface-exposed membrane-associated protein. Overall, the inner membrane lipid rafts were found to be associated with proteins with transport and signaling functions (28), perhaps suggesting that the function of BBK13 lies within one or both of these categories. Despite not being surface exposed, BBK13, like other bacterial SIMPL domain-containing proteins (30–32), was found to be immunogenic. SIMPL domain-containing protein homologues are found broadly across pathogenic and nonpathogenic bacterial species. This may suggest that the members of this protein family are not direct virulence determinants. B. burgdorferi, however, is a unique bacterial pathogen in that it lacks classical virulence factors and physiological functions that contribute to survival mechanisms that have been shown to be significant components of B. burgdorferi infectivity (39, 40, 42, 44, 45).
In sum, we have identified bbk13 as a novel gene on lp36 that contributes to B. burgdorferi infectivity. This work represents a detailed analysis of the kinetics of B. burgdorferi mouse infection and the contribution of bbk13 to B. burgdorferi replication in the skin during early infection. Moreover, this work demonstrates that the early expansion of spirochetes in the skin inoculation site has a strong influence on dissemination and the distal tissue load. Further studies focused on the function of bbk13 as well as the pathogen, vector, and host factors important for B. burgdorferi proliferation in the skin will improve our understanding of early B. burgdorferi infection. Future approaches aimed at blocking this initial, localized expansion may serve as novel therapeutics to reduce or prevent pathogen dissemination and, therefore, the debilitating clinical manifestations of Lyme disease.
MATERIALS AND METHODS
Bacterial clones and growth conditions.
The B. burgdorferi clones used in this study were derived from the low-passage infectious clone B31 A3. Low-passage infectious clone B31 A3-68 Δbbe02, which lacks plasmids cp9 and lp56, as well as the bbe02 gene on lp25, was used for all genetic manipulations (46). B. burgdorferi cultures were grown at 35°C in liquid Barbour-Stoenner-Kelly II (BSKII) medium containing gelatin and 6% rabbit serum in screw-cap tubes. Individual clones were isolated by plating in solid BSK-agarose medium, as described previously (47, 48). The plates were incubated at 35°C under 2.5% CO2. The following antibiotics were used for selection, as needed: kanamycin (200 μg/ml), streptomycin (50 μg/ml), and gentamicin (40 μg/ml). Cloning steps were performed using Escherichia coli DH5α, and recombinant protein was produced in Escherichia coli NiCo21(DE3) (New England Biolabs), grown in LB broth liquid cultures or on LB agar plates. Kanamycin was used in E. coli cultures at 50 μg/ml, spectinomycin was used at 300 μg/ml, and/or gentamicin was used at 10 μg/ml, as appropriate.
Borrelia burgdorferi genetic manipulations.
Target gene deletion constructs were engineered to completely replace each open reading frame with the constitutively expressed spectinomycin/streptomycin resistance cassette flaBp-aadA using a PCR-based overlap extension strategy described previously (21) and the gene-specific primers listed in Table S1 in the supplemental material. The allelic exchange constructs were verified by PCR, restriction digestion, and DNA sequencing. B. burgdorferi A3-68 Δbbe02 was transformed with 20 μg of a linearized allelic exchange plasmid. B. burgdorferi transformants were plated in BSK-agarose medium with streptomycin. Streptomycin-resistant B. burgdorferi colonies were isolated and screened by PCR and DNA sequencing to verify the gene deletions. The plasmid profiles of all B. burgdorferi deletion mutants were determined by PCR (49) to maintain consistency with the parent clones.
Complementation of the Δbbk13 mutant was done in trans using the B. burgdorferi shuttle vector pBSV2G (50). A DNA fragment containing the gene bbk13 with an additional 221 bp upstream of the transcription start site (22) was amplified from B31 A3 genomic DNA using Phusion enzyme (Thermo Scientific) and primers 2156 and 2157. The bbk13 DNA fragment was cloned into pBSV2G using a SalI restriction site. The pBSV2G bbk13 plasmid structure was verified by restriction enzyme digestion and DNA sequencing. The Δbbk13 mutant clone was transformed with 20 μg pBSV2G bbk13 or pBSV2G alone, and positive transformants were selected. The clones that retained the B. burgdorferi plasmid content of the parent clone were selected for use in further experiments and are designated the Δbbk13 (Δbbk13/pBSV2G) and Δbbk13/bbk13+ (Δbbk13/pBSV2G bbk13) clones herein.
In vitro gene expression analysis.
RNA was isolated from B. burgdorferi log-phase cultures of the wild-type, Δbbk13, and Δbbk13/bbk13+ clones in biological triplicate, as described previously (42). Total cDNA was generated from 1 μg of RNA using an iScript Select cDNA synthesis kit (Bio-Rad) and random primers. Parallel cDNA reactions were carried out in the absence of reverse transcriptase. One hundred nanograms of cDNA was used as the template in subsequent real-time quantitative PCRs. qPCR was performed on a Bio-Rad CFX Connect real-time PCR system using iQ SYBR Green Supermix (Bio-Rad) and the following primer pairs: primers 2416 and 2417 (bbk13) and primers 1123 and 1124 (recA) (Table S1). The amounts of bbk13 and recA transcripts were determined using a genomic DNA standard curve for each gene target with 100-ng, 10-ng, 1-ng, and 0.1-ng DNA concentrations. Both standard curve reactions and reactions using cDNA from each biological triplicate were performed in technical triplicate. mRNA transcript copy numbers for bbk13 were normalized to recA copy numbers. The data sets were analyzed using the Kruskal-Wallis test with Dunn’s multiple-comparison test (GraphPad Prism, version 6.0, software).
Ethics statement.
The University of Central Florida is accredited by the International Association for Assessment and Accreditation of Laboratory Animal Care. Protocols for all animal experiments were prepared according to the guidelines of the National Institutes of Health and were reviewed and approved by the University of Central Florida Institutional Animal Care and Use Committee.
Gene expression analysis in ticks and mouse tissue.
Six- to 8-week-old female C3H/HeN mice were needle inoculated with B. burgdorferi B31 A3 at a dose of 105 spirochetes. At 3 weeks postinoculation, Ixodes scapularis ticks (Centers for Disease Control and Prevention, BEI Resources) were allowed to feed on infected mice to repletion. At approximately 1 week after feeding, triplicate groups of 25 to 50 fed larvae or 13 to 15 fed nymphs were flash frozen in liquid nitrogen. Triplicate groups of 25 unfed nymphs were flash frozen at 5 weeks postfeeding of the larvae. Flash-frozen ticks were homogenized in TE buffer (10 mM Tris, 1 mM EDTA, pH 8.0) containing 0.5 mg/ml lysozyme using a gentleMACS dissociator at the RNA 2 setting (Miltenyi Biotech). Triplicate groups of 3 bladders from infected mice were flash frozen in liquid nitrogen and homogenized as described above for infected ticks. Mouse and tick samples were incubated in 1% SDS at 64°C for 2 min, which was followed by the addition of 0.1 M sodium acetate, pH 5.2. Total RNA was isolated using hot phenol extraction. Fifty micrograms of RNA was treated twice with 10 U DNase I (Roche) and 80 U rRNAsin (Promega) at 47°C for 15 min to remove contaminating DNA. cDNA was synthesized from the purified RNA using the iScript Select cDNA synthesis kit and random primers (Bio-Rad). Parallel reactions without reverse transcriptase were also performed. Quantitative PCR was used to measure the expression of bbk13, recA, flaB, ospA, and ospC, as described above, using primers 2416 and 2417 (bbk13), 1123 and 1124 (recA), 1875 and 1776 (flaB), 2138 and 2139 (ospA), and 2203 and 2204 (ospC) (Table S1). All minus reverse transcriptase samples were verified to contain no significant amplification and were represented in technical duplicates for each sample. mRNA copy numbers for bbk13, flaB, ospA, and ospC were normalized to the recA copy numbers.
In vitro growth analysis.
Spirochetes were inoculated in triplicate cultures at a starting density of 1 × 105 spirochetes/ml in 5 ml of BSKII medium containing the appropriate antibiotics. Spirochete densities were determined every 24 h over a 144-h time period by cell enumeration under a dark-field microscope using a Petroff-Hausser chamber.
Recombinant BBK13 protein purification and anti-BBK13 antibody production.
A DNA fragment containing the BBK13 open reading frame, excluding the N-terminal transmembrane domain, was amplified from B31 A3 genomic DNA using Phusion enzyme (Thermo Scientific) and primers 2335 and 2336 (Table S1). The DNA fragment was cloned into the C-terminal His tag protein expression vector pET28A (EMD Millipore) using the restriction enzymes BamHI and SacI. The BBK1325–232His expression clone was verified by restriction digestion and sequence analysis. An overnight culture of NiC021(DE3)/pET28A-BBK1325–232His in LB was diluted 1:500 in two flasks each containing 150 ml of LB and grown at 37°C with shaking for 4 h or until the optical density at 600 nm was ∼1.3. Protein expression was induced with the addition of 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) per culture, and the cultures were incubated at 18°C with shaking for 16 h. The bacteria were pelleted by centrifugation at 3,210 × g for 10 min at 4°C. The pellets were each resuspended in 30 ml of His binding buffer (GE Healthcare) and sonicated twice at 20 A for 2 min. The sonicated cells were spun at 10,000 × g for 20 min at 4°C, and the soluble lysate fractions were each combined with 990 μl of Ni Sepharose high-performance histidine-tagged protein purification resin (GE Healthcare), which had been washed twice with 500 μl of His binding buffer (GE Healthcare) according to the manufacturer’s instructions. The soluble lysate-Ni resin mixtures were incubated at 4°C overnight rotating. Each soluble lysate-Ni resin mixture was divided between 3 5-ml disposable plastic columns with filters (GE Healthcare), and the lysate was allowed to flow through the columns. The columns were washed with 5 ml of His binding buffer (GE Healthcare). One milliliter of His elution buffer was added to each column and allowed to incubate for 10 min at room temperature, inverting every 2 min. The eluted protein was collected in a 1.5-ml tube. Immediately after elution the 1-ml protein samples were each added to an Ultracel 10K centrifugal filter unit (EMD Millipore) and spun at 3,210 × g for 12 min. Two milliliters of phosphate-buffered saline (PBS; 137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4, pH 7.4) was added to the filter units, and the samples were spun at 3,210 × g for 12 min. This step was repeated twice. Finally, 1.5 ml of PBS was added to each concentrated protein sample. The recombinant BBK1325–232His protein was further purified via cation exchange using strong cation-exchange minispin columns (Pierce). The columns were washed and protein sample was added according to the manufacturer’s instructions. The columns were washed one time each with purification buffer, 0.5 M NaCl elution buffer, and 0.8 M NaCl elution buffer (Pierce). Protein was eluted with 6 additions of 0.9 M NaCl elution buffer (Pierce). Elution fractions were pooled and run through Zeba Spin 7,000-molecular-weight-cutoff desalting columns (Thermo Scientific), according to the manufacturer’s instructions. The rBBK1325–232His protein was confirmed by Coomassie-stained SDS-PAGE and Western blot analysis using anti-His antibody (1:5,000; GenScript). Polyclonal rabbit anti-BBK13 antibody against purified rBBK1325–232His protein was generated by Cocalico Biologicals, Inc.
BBK13 protein localization.
Fractionation of B. burgdorferi lysates was performed as described previously (16). Briefly, spirochetes were grown to log phase (3 × 107 to 7 × 107/ml) in BSKII medium, spun at 3,210 × g for 10 min, washed twice with cold HN buffer (50 mM HEPES, 50 mM NaCl, pH 7.5), and resuspended in 1 ml HN buffer and 100 μl Halt protease inhibitor cocktail (Thermo Scientific). Spirochetes were lysed via sonication and spun at 125,000 × g to collect the soluble and membrane components of the lysate.
Proteinase K digestions were performed as described previously (51, 52). Briefly, 109 log-phase-grown (3 × 107/ml) B. burgdorferi spirochetes were resuspended in PBS-Mg2+ (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4, 5 mM MgCl2, pH 7.4) with or without 200 μg/ml proteinase K and incubated at 20°C for 1 h. Simultaneously, incubations were performed under the same conditions with the addition of 0.1% SDS. Reactions were stopped with the addition of 1 mg/ml phenylmethylsulfonyl fluoride.
Protein samples were combined with equal parts HN buffer and 2× Laemmli sample buffer (Bio-Rad) with 5% β-mercaptoethanol, incubated for 5 min at 100°C, and separated on a 10% SDS-PAGE gel. The gels were subsequently transferred to nitrocellulose membranes (Bio-Rad) and blocked with 5% skim milk in Tris-buffered saline–Tween (TBS-T; 25 mM Tris, 150 mM NaCl, 0.5% Tween). The antibodies polyclonal rabbit anti-BBK13 UCF34 (1:10,000), monoclonal mouse anti-FlaB H9724 (53) (1:200), polyclonal mouse anti-SodA NIH754 (16) (1:1,000), and polyclonal rabbit anti-VlsE (1:1,000) (Rockland Immunochemicals) detected BBK13, FlaB, SodA, and VlsE, respectively. Horseradish peroxidase-conjugated goat anti-mouse or anti-rabbit immunoglobulin (1:10,000; EMD Millipore) was used as the secondary antibody. The membranes were washed between incubations with TBS-T, developed using Supersignal West Pico chemiluminescent substrate (Thermo Scientific), and visualized using a Bio-Rad ChemiDoc imager.
BBK13 immunoreactivity.
Six hundred nanograms of rBBK1325–232His protein and total protein lysate generated from 107 wild-type B. burgdorferi spirochetes was separated by SDS-PAGE. Western blotting was performed using preimmune and postimmune sera collected from B. burgdorferi-infected mice at 3 weeks postinoculation (1:500) (see the methods in “Mouse infection experiments” below) or anti-His antibody (1:5,000) (GenScript).
Mouse infection experiments.
B. burgdorferi cultures were grown from frozen glycerol stocks to stationary phase (1 × 108/ml) in BSKII medium with the appropriate antibiotics. The B. burgdorferi culture density was determined using a Petroff-Hausser chamber under a dark-field microscope. B. burgdorferi cultures were diluted in BSKII to achieve the desired number of spirochetes in the inoculum. Groups of six 6- to 8-week-old female C3H/HeN mice (Envigo) were inoculated (i) intraperitoneally (200 μl) and subcutaneously under the upper dorsal skin (50 μl) (lp36 mutant screen) or (ii) intradermally into the shaved dorsal skin (100 μl) (all other infection studies) with 104 B. burgdorferi spirochetes. All inoculum cultures were analyzed for plasmid content by PCR. Furthermore, the inoculum cultures were plated in solid BSK-agarose medium, and individual colonies were verified for the presence of virulence plasmids lp25, lp28-1, and lp36 (9). All inoculum cultures carried the expected endogenous plasmid content, and 80 to 100% of the individual colonies from each clone were confirmed to contain all three virulence plasmids. At various time points postinoculation, the mice were assessed for infection by serology (40) and/or reisolation of spirochetes from tissues (see below).
Semiquantitative tissue reisolation scoring method.
The mouse inoculation site, ear, heart, bladder, and joint tissues were dissected at various time points postinoculation and placed in BSKII containing an antibiotic cocktail of rifampin (50 μg/ml), amphotericin B (2.5 μg/ml), and phosphomycin (20 μg/ml). B. burgdorferi is naturally resistant to these antibiotics. Reisolation cultures were incubated at 35°C. Cultures were visually inspected for the presence of spirochetes using dark-field microscopy. A numerical scoring system was developed for preliminary screening assessment of the spirochete loads in tissue while viewing at a ×200 magnification, as follows: 0, absence of spirochetes; 1, low spirochete density (<5 spirochetes in the field of view); 2, medium spirochete density (5 to 15 spirochetes in the field of view); and 3, high spirochete density (>15 spirochetes in the field of view). Cultures of tissues from the wild-type B. burgdorferi-inoculated mice were monitored daily for the presence of spirochetes using dark-field microscopy until the spirochete densities were high (score of 3), typically after 5 to 6 days of incubation in BSKII medium (Fig. 3A), at which time the reisolation cultures of the tissues from the Δbbk13 mutant- and Δbbk13/bbk13+ clone-inoculated mice were scored (Fig. 3B). It is important to note that this method is a tool to screen B. burgdorferi mutants for possible tissue colonization defects, which then requires follow-up verification by qPCR analysis of spirochete loads in tissues.
Quantification of B. burgdorferi loads in mouse tissues.
Skin inoculation site, ear, heart, and joint tissues paired to those used for reisolation culture scoring were collected from the mice at various time points postinoculation, and total DNA was extracted, as previously described (9). TaqMan probe and primers (IDT DNA) specific to the B. burgdorferi flaB (primers 1137, 1138, 1139) and mouse nid (primers 1140, 1141, 1142) (Table S1) genes were used to measure their respective copy numbers by qPCR and a standard curve approach as described above, using 100 ng of DNA extracted from mouse tissues as the template (9). The qPCR was performed using iQ Supermix (Bio-Rad) on a Bio-Rad CFX Connect real-time PCR system. flaB copy numbers were normalized against 106 mouse nid copy numbers to allow for comparison across mice and B. burgdorferi clones. Technical triplicates were performed for each tissue sample. Statistical significance compared to the results for the wild type was determined by two-way ANOVA with Tukey’s multiple-comparison test (GraphPad Prism, version 6.0, software).
Quantification of B. burgdorferi density in the blood.
Three cohorts of six mice each per B. burgdorferi clone (the wild-type, Δbbk13, and Δbbk13/bbk13+ clones) were intradermally inoculated with 104 spirochetes, as described above. To prevent complications due to oversampling, approximately 50 μl of blood/mouse was obtained every day from one of the three cohorts via submandibular bleed and collected in EDTA collection tubes (RAM Scientific). In this way, each cohort was bled every 3 days over a time period of 11 days. Whole blood was serially diluted in BSKII medium and plated in solid BSK-agarose medium for determination of the number of CFU, and the B. burgdorferi density in the blood was calculated. Statistical significance compared to the results for the wild type was determined by two-way ANOVA with Tukey’s multiple-comparison test (GraphPad Prism, version 6.0, software).
Supplementary Material
ACKNOWLEDGMENTS
We appreciate all of the technical and intellectual contributions of Travis Jewett. Thank you to Katelan Yap for technical assistance. Thank you to the NAF animal care staff. The following reagent was provided by the Centers for Disease Control and Prevention for distribution by BEI Resources, NIAID, NIH: Ixodes scapularis Larvae (Live), NR-44115.
This work was supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health (R01AI099094 to M.W.J.) and a Deborah and Mark Blackman-Global Lyme Alliance postdoctoral fellowship (to G.F.A.).
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/IAI.00887-18.
REFERENCES
- 1.Bacon RM, Kugeler KJ, Mead PS, Centers for Disease Control and Prevention . 2008. Surveillance for Lyme disease—United States, 1992–2006. MMWR Surveill Summ 57(SS-10):1–9. [PubMed] [Google Scholar]
- 2.Hubalek Z. 2009. Epidemiology of Lyme borreliosis. Curr Probl Dermatol 37:31–50. doi: 10.1159/000213069. [DOI] [PubMed] [Google Scholar]
- 3.Burgdorfer W, Barbour AG, Hayes SF, Benach JL, Grunwaldt E, Davis JP. 1982. Lyme disease—a tick-borne spirochetosis? Science 216:1317–1319. doi: 10.1126/science.7043737. [DOI] [PubMed] [Google Scholar]
- 4.Steere AC, Grodzicki RL, Kornblatt AN, Craft JE, Barbour AG, Burgdorfer W, Schmid GP, Johnson E, Malawista SE. 1983. The spirochetal etiology of Lyme disease. N Engl J Med 308:733–740. doi: 10.1056/NEJM198303313081301. [DOI] [PubMed] [Google Scholar]
- 5.Burgdorfer W, Keirans JE. 1983. Ticks and Lyme disease in the United States. Ann Intern Med 99:121. doi: 10.7326/0003-4819-99-1-121. [DOI] [PubMed] [Google Scholar]
- 6.Hu LT. 2016. Lyme disease. Ann Intern Med 165:677. doi: 10.7326/L16-0409. [DOI] [PubMed] [Google Scholar]
- 7.Casjens S, Palmer N, van Vugt R, Huang WM, Stevenson B, Rosa P, Lathigra R, Sutton G, Peterson J, Dodson RJ, Haft D, Hickey E, Gwinn M, White O, Fraser CM. 2000. A bacterial genome in flux: the twelve linear and nine circular extrachromosomal DNAs in an infectious isolate of the Lyme disease spirochete Borrelia burgdorferi. Mol Microbiol 35:490–516. [DOI] [PubMed] [Google Scholar]
- 8.Fraser CM, Casjens S, Huang WM, Sutton GG, Clayton R, Lathigra R, White O, Ketchum KA, Dodson R, Hickey EK, Gwinn M, Dougherty B, Tomb JF, Fleischmann RD, Richardson D, Peterson J, Kerlavage AR, Quackenbush J, Salzberg S, Hanson M, van Vugt R, Palmer N, Adams MD, Gocayne J, Weidman J, Utterback T, Watthey L, McDonald L, Artiach P, Bowman C, Garland S, Fuji C, Cotton MD, Horst K, Roberts K, Hatch B, Smith HO, Venter JC. 1997. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature 390:580–586. doi: 10.1038/37551. [DOI] [PubMed] [Google Scholar]
- 9.Jewett MW, Lawrence K, Bestor AC, Tilly K, Grimm D, Shaw P, VanRaden M, Gherardini F, Rosa PA. 2007. The critical role of the linear plasmid lp36 in the infectious cycle of Borrelia burgdorferi. Mol Microbiol 64:1358–1374. doi: 10.1111/j.1365-2958.2007.05746.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Seshu J, Esteve-Gassent MD, Labandeira-Rey M, Kim JH, Trzeciakowski JP, Höök M, Skare JT. 2006. Inactivation of the fibronectin-binding adhesin gene bbk32 significantly attenuates the infectivity potential of Borrelia burgdorferi. Mol Microbiol 59:1591–1601. doi: 10.1111/j.1365-2958.2005.05042.x. [DOI] [PubMed] [Google Scholar]
- 11.Probert WS, Johnson BJ. 1998. Identification of a 47 kDa fibronectin-binding protein expressed by Borrelia burgdorferi isolate B31. Mol Microbiol 30:1003–1015. doi: 10.1046/j.1365-2958.1998.01127.x. [DOI] [PubMed] [Google Scholar]
- 12.Moriarty TJ, Shi M, Lin YP, Ebady R, Zhou H, Odisho T, Hardy PO, Salman-Dilgimen A, Wu J, Weening EH, Skare JT, Kubes P, Leong J, Chaconas G. 2012. Vascular binding of a pathogen under shear force through mechanistically distinct sequential interactions with host macromolecules. Mol Microbiol 86:1116–1131. doi: 10.1111/mmi.12045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Niddam AF, Ebady R, Bansal A, Koehler A, Hinz B, Moriarty TJ. 2017. Plasma fibronectin stabilizes Borrelia burgdorferi-endothelial interactions under vascular shear stress by a catch-bond mechanism. Proc Natl Acad Sci U S A 114:E3490–E3498. doi: 10.1073/pnas.1615007114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin YP, Chen Q, Ritchie JA, Dufour NP, Fischer JR, Coburn J, Leong JM. 2015. Glycosaminoglycan binding by Borrelia burgdorferi adhesin BBK32 specifically and uniquely promotes joint colonization. Cell Microbiol 17:860–875. doi: 10.1111/cmi.12407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garcia BL, Zhi H, Wager B, Hook M, Skare JT. 2016. Borrelia burgdorferi BBK32 inhibits the classical pathway by blocking activation of the C1 complement complex. PLoS Pathog 12:e1005404. doi: 10.1371/journal.ppat.1005404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jewett MW, Byram R, Bestor A, Tilly K, Lawrence K, Burtnick MN, Gherardini F, Rosa PA. 2007. Genetic basis for retention of a critical virulence plasmid of Borrelia burgdorferi. Mol Microbiol 66:975–990. doi: 10.1111/j.1365-2958.2007.05969.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bernard Q, Wang Z, Di Nardo A, Boulanger N. 2017. Interaction of primary mast cells with Borrelia burgdorferi (sensu stricto): role in transmission and dissemination in C57BL/6 mice. Parasit Vectors 10:313. doi: 10.1186/s13071-017-2243-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hodzic E, Feng S, Freet KJ, Borjesson DL, Barthold SW. 2002. Borrelia burgdorferi population kinetics and selected gene expression at the host-vector interface. Infect Immun 70:3382–3388. doi: 10.1128/IAI.70.7.3382-3388.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Horka H, Cerna-Kyckova K, Skallova A, Kopecky J. 2009. Tick saliva affects both proliferation and distribution of Borrelia burgdorferi spirochetes in mouse organs and increases transmission of spirochetes to ticks. Int J Med Microbiol 299:373–380. doi: 10.1016/j.ijmm.2008.10.009. [DOI] [PubMed] [Google Scholar]
- 20.Kern A, Collin E, Barthel C, Michel C, Jaulhac B, Boulanger N. 2011. Tick saliva represses innate immunity and cutaneous inflammation in a murine model of Lyme disease. Vector Borne Zoonotic Dis 11:1343–1350. doi: 10.1089/vbz.2010.0197. [DOI] [PubMed] [Google Scholar]
- 21.Ellis TC, Jain S, Linowski AK, Rike K, Bestor A, Rosa PA, Halpern M, Kurhanewicz S, Jewett MW. 2014. In vivo expression technology identifies a novel virulence factor critical for Borrelia burgdorferi persistence in mice. PLoS Pathog 10:e1004260. doi: 10.1371/journal.ppat.1003567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Adams PP, Flores Avile C, Popitsch N, Bilusic I, Schroeder R, Lybecker M, Jewett MW. 2017. In vivo expression technology and 5′ end mapping of the Borrelia burgdorferi transcriptome identify novel RNAs expressed during mammalian infection. Nucleic Acids Res 45:775–792. doi: 10.1093/nar/gkw1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kasumba IN, Bestor A, Tilly K, Rosa PA. 2015. Use of an endogenous plasmid locus for stable in trans complementation in Borrelia burgdorferi. Appl Environ Microbiol 81:1038–1046. doi: 10.1128/AEM.03657-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Iyer R, Caimano MJ, Luthra A, Axline D Jr, Corona A, Iacobas DA, Radolf JD, Schwartz I. 2015. Stage-specific global alterations in the transcriptomes of Lyme disease spirochetes during tick feeding and following mammalian host adaptation. Mol Microbiol 95:509–538. doi: 10.1111/mmi.12882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Samuels DS. 2011. Gene regulation in Borrelia burgdorferi. Annu Rev Microbiol 65:479–499. doi: 10.1146/annurev.micro.112408.134040. [DOI] [PubMed] [Google Scholar]
- 26.Marchler-Bauer A, Bo Y, Han L, He J, Lanczycki CJ, Lu S, Chitsaz F, Derbyshire MK, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Lu F, Marchler GH, Song JS, Thanki N, Wang Z, Yamashita RA, Zhang D, Zheng C, Geer LY, Bryant SH. 2017. CDD/SPARCLE: functional classification of proteins via subfamily domain architectures. Nucleic Acids Res 45:D200–D203. doi: 10.1093/nar/gkw1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marchler-Bauer A, Derbyshire MK, Gonzales NR, Lu S, Chitsaz F, Geer LY, Geer RC, He J, Gwadz M, Hurwitz DI, Lanczycki CJ, Lu F, Marchler GH, Song JS, Thanki N, Wang Z, Yamashita RA, Zhang D, Zheng C, Bryant SH. 2015. CDD: NCBI's conserved domain database. Nucleic Acids Res 43:D222–D226. doi: 10.1093/nar/gku1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Toledo A, Huang Z, Coleman JL, London E, Benach JL. 2018. Lipid rafts can form in the inner and outer membranes of Borrelia burgdorferi and have different properties and associated proteins. Mol Microbiol 108:63–76. doi: 10.1111/mmi.13914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gupta VK, Radhakrishnan G, Harms J, Splitter G. 2012. Invasive Escherichia coli vaccines expressing Brucella melitensis outer membrane proteins 31 or 16 or periplasmic protein BP26 confer protection in mice challenged with B. melitensis. Vaccine 30:4017–4022. doi: 10.1016/j.vaccine.2012.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moore MM, Fernandez DL, Thune RL. 2002. Cloning and characterization of Edwardsiella ictaluri proteins expressed and recognized by the channel catfish Ictalurus punctatus immune response during infection. Dis Aquat Org 52:93–107. doi: 10.3354/dao052093. [DOI] [PubMed] [Google Scholar]
- 31.Qiu J, Wang W, Wu J, Zhang H, Wang Y, Qiao J, Chen C, Gao GF, Allain JP, Li C. 2012. Characterization of periplasmic protein BP26 epitopes of Brucella melitensis reacting with murine monoclonal and sheep antibodies. PLoS One 7:e34246. doi: 10.1371/journal.pone.0034246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rossetti OL, Arese AI, Boschiroli ML, Cravero SL. 1996. Cloning of Brucella abortus gene and characterization of expressed 26-kilodalton periplasmic protein: potential use for diagnosis. J Clin Microbiol 34:165–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wormser GP. 2006. Hematogenous dissemination in early Lyme disease. Wien Klin Wochenschr 118:634–637. doi: 10.1007/s00508-006-0688-9. [DOI] [PubMed] [Google Scholar]
- 34.Halpern MD, Jain S, Jewett MW. 2013. Enhanced detection of host response antibodies to Borrelia burgdorferi using immuno-PCR. Clin Vaccine Immunol 20:350–357. doi: 10.1128/CVI.00630-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang G, Ojaimi C, Iyer R, Saksenberg V, McClain SA, Wormser GP, Schwartz I. 2001. Impact of genotypic variation of Borrelia burgdorferi sensu stricto on kinetics of dissemination and severity of disease in C3H/HeJ mice. Infect Immun 69:4303–4312. doi: 10.1128/IAI.69.7.4303-4312.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Caine JA, Coburn J. 2015. A short-term Borrelia burgdorferi infection model identifies tissue tropisms and bloodstream survival conferred by adhesion proteins. Infect Immun 83:3184–3194. doi: 10.1128/IAI.00349-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hyde JA, Weening EH, Chang M, Trzeciakowski JP, Hook M, Cirillo JD, Skare JT. 2011. Bioluminescent imaging of Borrelia burgdorferi in vivo demonstrates that the fibronectin-binding protein BBK32 is required for optimal infectivity. Mol Microbiol 82:99–113. doi: 10.1111/j.1365-2958.2011.07801.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lin T, Gao L, Zhang C, Odeh E, Jacobs MB, Coutte L, Chaconas G, Philipp MT, Norris SJ. 2012. Analysis of an ordered, comprehensive STM mutant library in infectious Borrelia burgdorferi: insights into the genes required for mouse infectivity. PLoS One 7:e47532. doi: 10.1371/journal.pone.0047532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jain S, Showman AC, Jewett MW. 2015. Molecular dissection of a Borrelia burgdorferi in vivo essential purine transport system. Infect Immun 83:2224–2233. doi: 10.1128/IAI.02859-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jain S, Sutchu S, Rosa PA, Byram R, Jewett MW. 2012. Borrelia burgdorferi harbors a transport system essential for purine salvage and mammalian infection. Infect Immun 80:3086–3093. doi: 10.1128/IAI.00514-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jewett MW, Lawrence KA, Bestor A, Byram R, Gherardini F, Rosa PA. 2009. GuaA and GuaB are essential for Borrelia burgdorferi survival in the tick-mouse infection cycle. J Bacteriol 191:6231–6241. doi: 10.1128/JB.00450-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Showman AC, Aranjuez G, Adams PP, Jewett MW. 2016. Gene bb0318 is critical for the oxidative stress response and infectivity of Borrelia burgdorferi. Infect Immun 84:3141–3151. doi: 10.1128/IAI.00430-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Barthold SW. 1991. Infectivity of Borrelia burgdorferi relative to route of inoculation and genotype in laboratory mice. J Infect Dis 163:419–420. doi: 10.1093/infdis/163.2.419. [DOI] [PubMed] [Google Scholar]
- 44.Khajanchi BK, Odeh E, Gao L, Jacobs MB, Philipp MT, Lin T, Norris SJ. 2016. Phosphoenolpyruvate phosphotransferase system components modulate gene transcription and virulence of Borrelia burgdorferi. Infect Immun 84:754–764. doi: 10.1128/IAI.00917-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ouyang Z, He M, Oman T, Yang XF, Norgard MV. 2009. A manganese transporter, BB0219 (BmtA), is required for virulence by the Lyme disease spirochete, Borrelia burgdorferi. Proc Natl Acad Sci U S A 106:3449–3454. doi: 10.1073/pnas.0812999106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rego RO, Bestor A, Rosa PA. 2011. Defining the plasmid-borne restriction-modification systems of the Lyme disease spirochete Borrelia burgdorferi. J Bacteriol 193:1161–1171. doi: 10.1128/JB.01176-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rosa PA, Hogan D. 1992. Colony formation by Borrelia burgdorferi in solid medium: clonal analysis of osp locus variants, p 95–103. In Munderloh UG, Kurtti TJ (ed.), Proceedings of the First International Conference on Tick Borne Pathogens at the Host-Vector Interface. University of Minnesota, St. Paul, MN. [Google Scholar]
- 48.Samuels DS. 1995. Electrotransformation of the spirochete Borrelia burgdorferi. Methods Mol Biol 47:253–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Elias AF, Stewart PE, Grimm D, Caimano MJ, Eggers CH, Tilly K, Bono JL, Akins DR, Radolf JD, Schwan TG, Rosa P. 2002. Clonal polymorphism of Borrelia burgdorferi strain B31 MI: implications for mutagenesis in an infectious strain background. Infect Immun 70:2139–2150. doi: 10.1128/IAI.70.4.2139-2150.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Elias AF, Bono JL, Kupko JJ III, Stewart PE, Krum JG, Rosa PA. 2003. New antibiotic resistance cassettes suitable for genetic studies in Borrelia burgdorferi. J Mol Microbiol Biotechnol 6:29–40. doi: 10.1159/000073406. [DOI] [PubMed] [Google Scholar]
- 51.Bono JL, Tilly K, Stevenson B, Hogan D, Rosa P. 1998. Oligopeptide permease in Borrelia burgdorferi: putative peptide-binding components encoded by both chromosomal and plasmid loci. Microbiology 144:1033–1044. doi: 10.1099/00221287-144-4-1033. [DOI] [PubMed] [Google Scholar]
- 52.Bunikis J, Barbour AG. 1999. Access of antibody or trypsin to an integral outer membrane protein (P66) of Borrelia burgdorferi is hindered by Osp lipoproteins. Infect Immun 67:2874–2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Barbour AG, Hayes SF, Heiland RA, Schrumpf ME, Tessier SL. 1986. A Borrelia-specific monoclonal antibody binds to a flagellar epitope. Infect Immun 52:549–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.