The human gastrointestinal tract (GIT) is inhabited by a dense microbial community of symbionts. Enterococci are among the earliest members of this community and remain core members of the GIT microbiota throughout life.
KEYWORDS: cell surface proteins, enterococcus, intestinal colonization, sortase
ABSTRACT
The human gastrointestinal tract (GIT) is inhabited by a dense microbial community of symbionts. Enterococci are among the earliest members of this community and remain core members of the GIT microbiota throughout life. Enterococci have also recently emerged as opportunistic pathogens and major causes of nosocomial infections. Although recognized as a prerequisite for infection, colonization of the GIT by enterococci remains poorly understood. One way that bacteria adapt to dynamic ecosystems like the GIT is through the use of their surface proteins to sense and interact with components of their immediate environment. In Gram-positive bacteria, a subset of surface proteins relies on an enzyme called sortase for covalent attachment to the cell wall. Here, we show that the housekeeping sortase A (SrtA) enzyme promotes intestinal colonization by enterococci. Furthermore, we show that the enzymatic activity of SrtA is key to the ability of Enterococcus faecalis to bind mucin (a major component of the GIT mucus). We also report the GIT colonization phenotypes of E. faecalis mutants lacking selected sortase-dependent proteins (SDPs). Further examination of the mucin binding ability of these mutants suggests that adhesion to mucin contributes to intestinal colonization by E. faecalis.
INTRODUCTION
Over the past 50 years, enterococci have become a major clinical burden, causing hospital-acquired infections that are frequently complicated by their intrinsic resistance to broad-spectrum antibiotics and their ability to form biofilms at the site of infection (1, 2). In addition to being important infectious agents, these organisms are ancient members of the gastrointestinal tract (GIT) microbiota that evolved to colonize the digestive organs of diverse hosts from humans to insects (3, 4). This ability to efficiently colonize the gut is key for pathogenesis, as the GIT is the reservoir for disease-causing enterococci (5). Unfortunately, the molecular basis for GIT colonization by enterococci is poorly understood. An understanding of the mechanisms underlying GIT colonization by enterococci could therefore reveal potential therapeutic targets to interfere with enterococcal colonization and potentially provide a basis for understanding colonization by other GIT commensals.
The GIT is a highly competitive environment in which members of the microbiota compete for limited nutrients. Commensals also are also faced with a multitude of host-derived and microbe-derived antimicrobials; the ability to tolerate these antimicrobials is key to survival in the GIT (6). The continuous flushing of intestinal contents due to peristalsis poses an additional challenge to commensals. Some investigators have proposed that commensals evolved mechanisms to attach to the mucus layer overlaying the epithelium in order to overcome this challenge (7, 8). This mucus layer has a lower turnover rate than the transit time of food and therefore promotes retention of its inhabitants (9). To sense and adapt to dynamic environments like the GIT, bacteria use cell surface proteins. Cell surface proteins can also promote niche colonization by serving as adhesins for attachment to the mucosal surface and by harvesting nutrients and essential metals from the environment.
In Gram-positive bacteria, the members of one particular subclass of cell surface proteins are covalently attached to the cell wall by the activity of a membrane-bound enzyme called sortase (10). Such sortase-dependent proteins (SDPs) have a characteristic cell wall-anchoring domain near their C terminus; this domain consists of a conserved LPxTG motif followed by a stretch of hydrophobic amino acids and a positively charged tail (11). The surface-exposed N-terminal domains of SDPs exhibit functional diversity, including acting as adhesins. Deletion of sortase in most organisms does not impair cell viability but attenuates virulence, and as a result at least some SDPs are thought to perform virulence-related functions (12, 13). Enterococcus faecalis encodes two sortases: a pilus-specific class C sortase also known as Bps (biofilm- and pilus-associated sortase) and the housekeeping class A sortase SrtA (14). In E. faecalis, Bps is encoded immediately downstream of the ebp (endocarditis- and biofilm-associated pilus) locus and was previously shown to mediate polymerization of the pilus subunits whereas the base pilin (EbpB) is anchored to the cell wall by SrtA (15). This organization of the pilus locus is shared by other enterococci (16). In a previous study, Bps was shown to promote biofilm formation and pathogenesis in a urinary tract infection model in a manner similar to that exhibited by SrtA (17).
To study GIT colonization, we previously developed an experimental model in which we use E. faecalis to colonize mice harboring an unperturbed antibiotic-naive microbiota (18). Using this model, we determined that a plasmid-encoded bacteriocin can target the indigenous enterococcal population and enhance colonization by strains harboring the bacteriocin-encoding plasmid (18). We also described a transmembrane kinase, encoded by the core genome, that promotes cell envelope integrity and antimicrobial resistance and is required for GIT colonization (6). Other studies identified specific enterococcal genes that contribute to colonization (19–21) after antibiotic pretreatment to achieve long-term GIT colonization. A previous study analyzed the gene expression of E. faecalis during colonization of germfree mice by RNA sequencing and found significant changes in several genes involved in nutrient uptake and energy metabolism (22). A recent review summarized other genes and processes previously described to be important during enterococcal colonization (23).
To assess the importance of enterococcal cell surface proteins during colonization, we probed the contribution of SrtA to GIT colonization, revealing that SrtA is required for effective colonization of the GIT. We also observed that disruption of selected SDPs impairs GIT colonization and propose a mechanism whereby some of these SDPs promote colonization by enabling E. faecalis to associate with the intestinal mucus layer.
RESULTS
Sortase A promotes intestinal colonization.
To determine whether SDPs are involved in GIT colonization, we tested a mutant lacking the housekeeping sortase gene, srtA, for its ability to colonize the mouse GIT. Even though the ΔsrtA mutant was not completely eliminated from the GIT, we observed that this mutant persisted at lower levels than the parental wild-type (WT) strain over the course of a 9-week colonization experiment (Fig. 1A). To confirm that this phenotype applies to other members of the Enterococcus genus, we constructed a ΔsrtA mutant in Enterococcus faecium strain JL619 (a rifampin [Rif]-resistant derivative of E. faecium 1,141,733) and examined colonization. We found that, similarly to E. faecalis, deletion of srtA resulted in lower levels of colonization in E. faecium (see Fig. S1 in the supplemental material).
FIG 1.
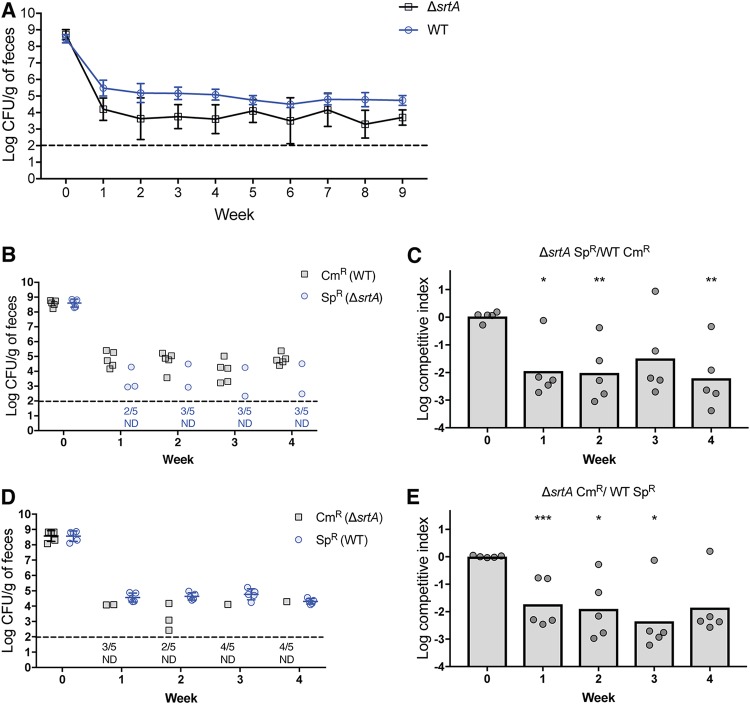
GIT colonization by sortase deletion mutants. (A) Groups of mice (n = 5) were colonized with either the E. faecalis ΔsrtA strain (IB10) or its isogenic wild-type parent (OG1RF). Colonization was assessed by enumerating viable colonies from fecal samples on rifampin-supplemented BHI agar; geometric means with standard deviations are shown for each group of mice. (B) Differentially marked strains SK11 (Cm-resistant WT strain) and IB39 (Sp-resistant ΔsrtA strain) were co-fed to a group of 5 mice for 2 weeks, after which colonization was assessed by enumerating viable colonies from fecal samples on rifampin-supplemented BHI agar containing Cm or Sp. (D) Differentially marked strains IB40 (Cm-resistant ΔsrtA) and IB38 (Sp-resistant WT) were co-fed to a group of 5 mice for 2 weeks, after which colonization was assessed as described for panel B. (C and E) Competitive indices of strains IB39 (Sp-resistant ΔsrtA strain) (C) and IB40 (Cm-resistant ΔsrtA strain) (E) were calculated relative to their WT counterparts for the competition experiments whose results are presented in panels B and D, respectively. Geometric means are shown as bar graphs; individual data points denote competitive indices for individual mice. Statistical significance was evaluated by a one-sample t test comparing means to a value of 1. *, P < 0.01; **, P < 0.001; ***, P < 0.0001. Horizontal dashed lines represent the limit of detection. Symbols for mice with undetectable colonization level were omitted; instead, the number of mice for which colonization was not detected (ND) is shown under the dashed line.
The catalytic activity of SrtA depends on the presence of a conserved cysteine residue (10, 24) corresponding to C200 in SrtA of E. faecalis. We constructed an E. faecalis mutant carrying a C200A mutation (to inactivate SrtA) in the chromosome-encoded srtA gene and observed that this mutant shared the colonization defect of the ΔsrtA mutant (Fig. S2). These results suggest that the catalytic activity of SrtA is important during colonization. We also performed competition experiments in which mice were fed equal quantities of the ΔsrtA mutant and the parental WT. In order to selectively quantify each strain by plating, we introduced chloramphenicol (Cm) or spectinomycin (Sp) resistance genes in the chromosomes of the WT strain and the ΔsrtA mutant. We performed two competition experiments, one in which the ΔsrtA mutant was marked with the Cm resistance gene (with the WT strain carrying the Sp resistance marker) and another, reciprocal experiment in which the ΔsrtA mutant was marked with the Sp resistance gene. In both experiments, the ΔsrtA mutant was quickly eliminated from the GIT of several mice (Fig. 1B and D), indicating that the competitive fitness defect of the ΔsrtA mutant is independent of the antibiotic resistance gene markers. Regardless of the mode of the colonization experiment (single strain in Fig. 1A or competition in Fig. 1B and D), WT E. faecalis exhibited greater fitness than the ΔsrtA mutant. This competitive advantage was quantified by calculating the competitive index of ΔsrtA mutants relative to WT E. faecalis (Fig. 1C and E). To determine the in vitro competitive fitness of the ΔsrtA mutant relative to the WT, we inoculated laboratory media with equal quantities of WT E. faecalis marked with a Sp resistance genes and the ΔsrtA mutant marked with a Cm resistance gene. We incubated the mixed culture at 37°C for 24 h, after which we diluted the overnight culture in fresh media and allowed continued growth for another 24 h. Two additional culture passages were performed, and, 24 h after each passage, the mixed cultures were plated on antibiotic-containing agar for selective enumeration of E. faecalis WT and ΔsrtA mutant CFUs. The competitive index of the ΔsrtA mutant was determined after each passage. As shown in Fig. S3, the ΔsrtA mutant had fitness comparable to that of WT E. faecalis in laboratory media, indicating that the colonization defect represented in Fig. 1 is GIT dependent.
We verified that the colonization defect was due to the ΔsrtA mutation, using complementation to confirm that the introduction of srtA (under the control of the constitutive P23S promoter) at an ectopic location on the chromosome of the ΔsrtA mutant restored the ability to compete for a colonization niche with WT E. faecalis (Fig. S4). Introducing the nonfunctional srtA C200A allele (also under the control of P23S) into the ΔsrtA mutant did not alter the colonization phenotype, consistent with the hypothesis that the catalytic activity of SrtA promotes intestinal colonization.
In most bacteria, deletion of the housekeeping sortase gene is well tolerated; however, in Streptococcus pyogenes and Actinomyces oris, the absence of sortase results in envelope integrity defects which lead to growth defects and susceptibility to envelope-damaging antibiotics (25, 26).To determine if the E. faecalis ΔsrtA mutant exhibits similar phenotypes, we assessed growth rates in liquid culture and fecal suspensions as well as resistance to envelope-targeting antimicrobials (cholate, lysozyme, ceftriaxone, and human defensin 5) and did not find any difference between the ΔsrtA mutant and its parental WT strain (Fig. S5 and S6). We also assessed envelope integrity by measuring permeability with respect to chlorophenol red–β-d-galactopyranoside (CPRG) as previously described (27) and did not find any difference between the ΔsrtA mutant and the WT strain (Fig. S7). Considering those findings, the colonization defect of the ΔsrtA mutant likely results from mislocalization of one or more SDPs.
E. faecalis strains encode diverse SDPs.
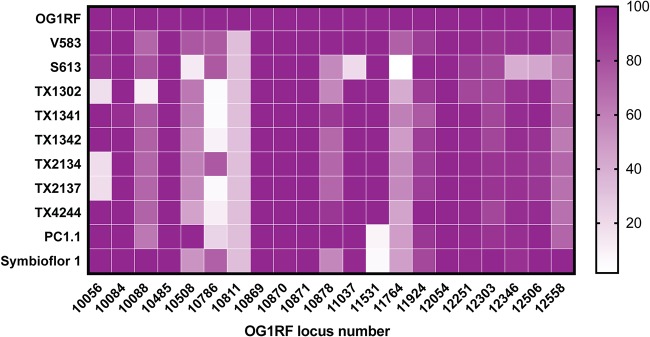
We searched the National Center for Biotechnology Information, Simple Modular Architecture Research Tool (SMART; http://smart.embl-heidelberg.de), and Interpro (https://www.ebi.ac.uk/interpro/) online databases for putative SDPs encoded in the genome of E. faecalis OG1RF and found 21 such genes (Table 1). We examined each of the 21 proteins encoded by these genes and confirmed that the C terminus of each exhibited the classic tripartite architecture of sortase substrates, which includes an LPxTG-like motif and a stretch of hydrophobic amino acids followed by basic residues at the very end of the sequence (11). We queried the genomes of GIT-isolated E. faecalis strains listed in the catalog of the Human Microbiome Project consortium (28) as well as the vancomycin-resistant strain V583 using the amino acid sequence of each SDP (Fig. 2). With the exception of OG1RF_10811 and OG1RF_10786, all the SDPs were found to be present in the majority of the queried genomes, indicating robust conservation of SDPs across E. faecalis strains. This suggests that these proteins play important roles in the biological adaptations of E. faecalis.
TABLE 1.
List of SDPs encoded in the genome of the E. faecalis strain OG1RF
| OG1RF locus no. | Annotation |
|---|---|
| 10056 | 2',3′-Cyclic-nucleotide 2'-phosphodiesterase |
| 10084 | Hypothetical protein |
| 10088 | Hypothetical protein |
| 10485 | Hypothetical protein |
| 10508 | von Willebrand factor domain protein |
| 10786 | Hypothetical protein |
| 10811 | Putative adhesin |
| 10869 | Pilus minor subunit EbpA |
| 10870 | Pilus minor subunit EbpB |
| 10871 | Pilus major subunit EbpC |
| 10878 | Collagen adhesin (Ace) |
| 11037 | Hypothetical protein |
| 11531 | Glycosyl hydrolase domain protein |
| 11764 | Hypothetical protein |
| 11924 | Hypothetical protein |
| 12054 | Hypothetical protein |
| 12251 | Hypothetical protein |
| 12303 | Family 8 polysaccharide lyase |
| 12346 | Hypothetical protein |
| 12506 | Hypothetical protein |
| 12558 | Hypothetical protein |
FIG 2.
Distribution of SDPs from E. faecalis OG1RF among E. faecalis GIT isolates. Heat map intensities represent percent similarities of best matches obtained for each SDP from OG1RF with BLASTP searches against genomes of E. faecalis GIT isolates.
E. faecalis SDPs promote mucin adhesion in vitro.
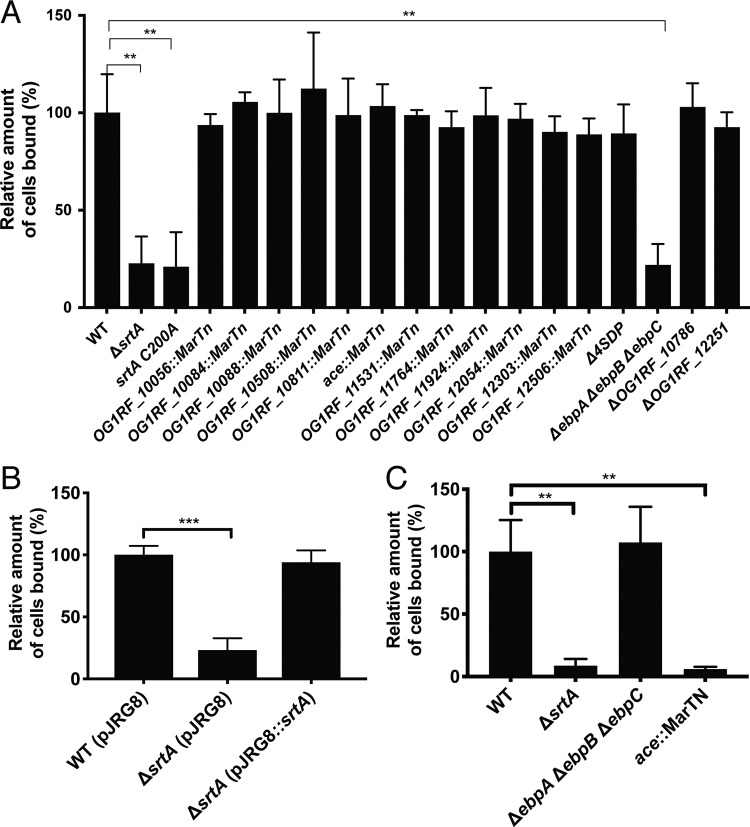
The most abundant component of intestinal mucus is mucin, a gel-forming glycoprotein secreted by goblet cells (29). We hypothesized that SDPs promote colonization by mediating adhesion to intestinal mucin. Consistent with this logic, mutants lacking srtA and those carrying the srtA C200A allele have a significantly reduced ability to bind to mucin in vitro (Fig. 3A). Expression of srtA from an ectopic locus complements the mucin binding defect, thus confirming that this binding defect was due to the sortase deletion (Fig. 3B). To determine which SDP(s) is responsible for mucin binding in WT cells, we constructed mutants harboring deletions in 1 or more of the 21 SDP-encoding genes or obtained insertional mutants from a previously described transposon library (30). We performed mucin adhesion assays using mutants lacking individual SDPs or groups of SDPs (Fig. 3A) and found that, with the exception of the mutant lacking pilin subunits [Δ(ebpA ebpB ebpC)], all mutants were able to bind mucin at levels comparable to that shown by WT E. faecalis. Pilin subunits polymerize to form the pilus, a proteinaceous surface appendage that was previously shown to mediate biofilm formation and adhesion to various extracellular matrix (ECM) proteins by E. faecalis (31, 32). That the Δ(ebpA ebpB ebpC) mutant has a reduced ability to bind mucin is consistent with the surface attachment role of pili. Ace, another SDP, is a well-studied adhesin of E. faecalis that was also previously shown to mediate binding to ECM proteins (33). The level of expression of Ace in E. faecalis is very low under laboratory conditions, but its expression can be induced by subjecting cells to a temperature of 46°C (33, 34). To determine if Ace also contributes to mucin adhesion, we assessed mucin binding using cells grown at 46°C (Fig. 3C). Under those conditions, deletion of ebpA ebpB ebpC does not affect mucin binding; disruption of ace, however, impairs mucin binding. This suggests that Ace, under inducing conditions, can also promote mucin binding.
FIG 3.
Mucin adhesion profile of E. faecalis SDP mutants. (A) Suspensions of WT E. faecalis (OG1RF) as well as of the isogenic ΔsrtA mutant and mutants lacking one or more SDPs, including the quadruple Δ4SDP mutant (IB15) grown at 37°C, were allowed to bind to mucin-coated wells; unbound cells were washed with PBS. Bound bacteria were observed by microscopy and enumerated. Numbers of counted cells were plotted relative to the WT strain. Average numbers of cells for 9 fields representing 3 biological replicates are shown. The number of bound cells for each strain is represented as a percentage relative to bound WT cells. (B) The mucin adhesion assay was performed, as described in the panel A legend, for the WT strain or the ΔsrtA mutant carrying the empty expression vector pJRG8 or expressing srtA. Values shown originated from the average counts of 9 fields representing 3 biological replicates for each strain. (C) The mucin binding assay was performed as described for panel A but using bacteria grown at 46°C. Values shown originated from the average counts of 9 fields representing 3 biological replicates for each strain. Statistical significance was evaluated by t test. **, P < 0.001; ***, P < 0.0001 (versus WT).
In vitro mucin binding activity parallels colonization phenotypes of selected SDP mutants.
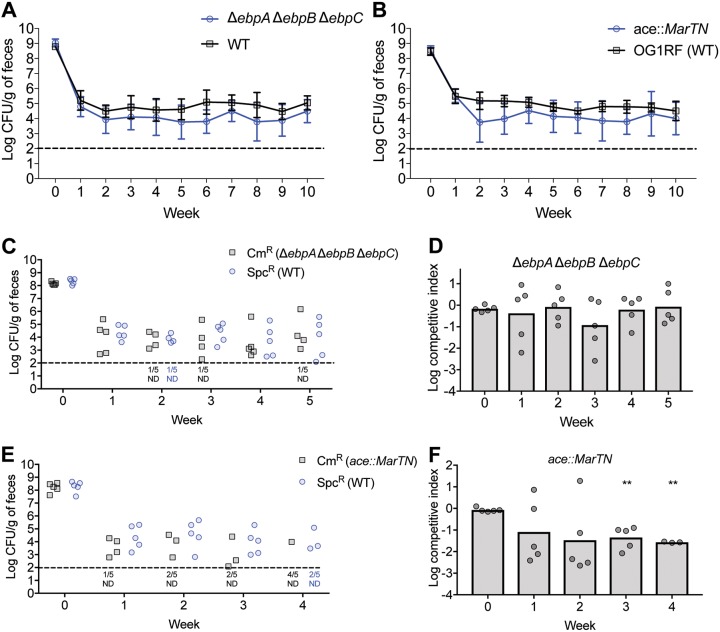
Because the Δ(ebpA ebpB ebpC) and ace::MarTn mutants exhibit mucin binding defects, we examined their ability to colonize the GIT. Single-strain colonization experiments were followed for 10 weeks, and, in that time span, the geometric means of the fecal loads for both mutants remained persistently lower than that for WT E. faecalis (Fig. 4A and B). To determine if mutation of any SDP would impair colonization, we examined the colonization of the OG1RF_10084::MarTn mutant and found that, over 10 weeks, the geometric means of fecal loads for this mutant were indistinguishable from that for WT E. faecalis (Fig. S8). In competition experiments, we found that the ace insertional mutant was outcompeted by the marked WT strain whereas the Δ(ebpA ebpB ebpC) mutant persisted at a level comparable to that of WT E. faecalis (Fig. 4C to F). As described for Fig. S3, we assessed the in vitro competitive fitness of the ace mutant and found that this mutant had fitness comparable to that of WT E. faecalis in laboratory media (Fig. S9). These data suggest that Ace enhances competitive fitness in the GIT. It is unclear why the colonization defect observed in the single-strain experiment for the ebp mutant was not detected in the context of competition with WT. One possibility might be that the presence of WT E. faecalis in this experimental setup rescues the colonization defect of the ebp mutant.
FIG 4.
GIT colonization by pilus and ace mutants. (A and B) Groups of mice (n = 5 per group) were colonized with the E. faecalis Δ(ebpA ebpB ebpC) mutant, the ace::MarTn mutant, or the isogenic parental WT E. faecalis strain (OG1RF), and colonization was assessed by enumerating viable colonies from fecal samples on rifampin-supplemented BHI agar for 10 weeks. Geometric means with standard deviations are shown for each group of mice. (C and E) Differentially marked strain IB38 (Sp-resistant WT) and either the ace::MarTn (Cm-resistant) strain or the ΔebpA-C (Cm-resistant; IB44) strain were co-fed to a group of 5 mice for 2 weeks, after which colonization was assessed by enumerating viable colonies from fecal samples on rifampin-supplemented BHI agar containing Cm or Sp. (D and F) Geometric means of competitive indices for strains IB44 and ace::MarTn relative to the Sp-resistant WT strain were calculated, and the results are shown as bar graphs in panels C and E for the competition experiments. Individual data points denote competitive indices for individual mice. Statistical significance was evaluated by one-sample t test comparing means to a value of 1. **, P < 0.01. Horizontal dashed lines represent the limit of detection. Symbols for mice with an undetectable colonization level were omitted; instead, the numbers of mice for which colonization was not detected (ND) are shown under the dotted line.
DISCUSSION
As important infectious agents, enterococci have been the subject of numerous studies investigating their pathogenesis through animal models, in vitro systems, and genetic approaches designed to identify determinants of virulence. Although it is generally accepted that enterococcal pathogenesis requires intestinal colonization, the study of intestinal colonization by these organisms has lagged behind studies investigating virulence. In E. faecalis and other Gram-positive bacteria, surface proteins, including SDPs, are often involved in virulence and host tissue colonization. In an experimental model that used antibiotic treatment to facilitate Staphylococcus aureus colonization of the GIT, deletion of srtA impaired colonization (35). Deletion of srtA was also found to compromise GIT colonization by Lactobacillus acidophilus and Lactobacillus gasseri in a colonization model of germfree mice (36). Here we showed that, through its enzymatic activity, SrtA promotes adhesion to mucin and intestinal colonization by E. faecalis in mice harboring an undisrupted antibiotic-naive microbiota. We found that deletion of srtA in E. faecium resulted in a colonization defect even more pronounced than that observed with E. faecalis. This suggests that E. faecalis is better able to compensate for the absence of SrtA than E. faecium. WxL proteins represent another class of surface proteins found in Gram-positive bacteria (37). A search for proteins containing WxL protein domains using NCBI identified 16 candidates in the genome of E. faecalis OG1RF but only 3 in the genome of E. faecium 1,141,733. It is therefore possible that the larger repertoire of WxL proteins in E. faecalis OG1RF enables enhanced compensation for the absence of SrtA.
E. faecalis OG1RF encodes 21 SDPs, several of which likely contribute to GIT colonization. We focused on SDPs with adhesin function conferring the ability to bind mucin. We found that deletion of the E. faecalis pilus locus impairs mucin binding, suggesting that E. faecalis can use its pili to attach to mucin. When the mucin adhesion experiment was performed under conditions known to induce expression of Ace (growth at 46°C), deletion of the pilus locus no longer interfered with mucin binding, suggesting that another protein mediates mucin binding under these conditions. Disruption of ace under such conditions impaired mucin binding, suggesting that Ace can also mediate attachment to mucin. It was surprising to observe that, in cells grown at 46°C, disruption of ace alone was sufficient to abolish mucin binding; the presence of the pilus locus in these cells was not sufficient to facilitate mucin adhesion, suggesting that growth at 46°C interferes with expression and/or assembly of pilin subunits. These data also raise the possibility that other SDPs for which expression (or function) requires environmental factors missing from our experimental conditions could also be involved in mucin binding. Future work will investigate this possibility. It is also unclear why, from an evolutionary perspective, growth at high temperature induces expression of Ace. It is possible that the stress of high temperature induces stress responses that are similar to the ones induced in the GIT and that colonization-promoting factors such as Ace are upregulated as a result.
As with other studies (36, 38), our adhesion assays were performed using porcine mucin. Even though galactose, N-acetylglucosamine, fucose, and sialic acid are the most common carbohydrates in both porcine and mouse mucin (39, 40), the structures of the mucin glycans differ between mouse mucin and porcine mucin. Furthermore, the mucin glycan structures differ along the intestinal tract (40). This structural variation likely affects intestinal colonization and is at least partly responsible for the differences in microbial composition along the GIT. In the case of E. faecalis, it is possible that different SDPs (or other surface proteins) mediate mucin binding in different regions along the GIT. Future studies aimed at assessing the expression of SDPs along the GIT and the association of selected SDP mutants with murine mucin from specific GIT sites will provide a more complete understanding of the role of SDPs during association with the GIT mucus.
Deletion of the pilus locus resulted in a decrease of GIT colonization in the single-strain inoculation experiment. Under conditions of competition with wild-type E. faecalis, however, we did not detect a colonization defect for the Δ(ebpA ebpB ebpC) mutant. Previous studies have shown that only a fraction of cells, in laboratory-grown WT E. faecalis populations, harbor pili (41). Thus, the presence of pili on the surface of all cells in the population is not necessary for phenotypes conferred by pili (e.g., biofilm formation) (32). It is therefore possible that the presence of WT cells in the competition experiment enhances the colonization of the Δ(ebpA ebpB ebpC) mutant, thereby masking a possible colonization defect. For example, WT cells could associate with mucins and form mixed communities within which both mutant and WT cells can persist. Alternatively, the presence of WT cells might induce the expression of additional adhesins on the surface of Δebp mutant cells, allowing them to associate with the mucus layer and effectively colonize the GIT.
Disruption of ace resulted in a reduction of E. faecalis colonization loads, suggesting that Ace promotes intestinal colonization. Ace was previously shown to mediate adhesion to ECM proteins such as collagen and laminin (33); however, the absence of these proteins from the intestinal lumen suggests that Ace promotes intestinal colonization by mediating physical interactions with a different substrate. Our data suggest that Ace and the E. faecalis pilus contribute to both mucin adherence and intestinal colonization. However, further work demonstrating direct binding of pilus subunits and Ace to ligands present in intestinal mucin will be required to define the role of these SDPs during intestinal colonization.
Several lines of evidence support the idea that GIT commensals associate with intestinal mucus as a means to colonize the GIT (42, 43). Addition of a mucus binding coat to synthetic microspheres prolongs their transit time in the GIT (44), suggesting that mucus adhesion promotes bacterial retention in the GIT. In addition, several members of the GIT microbiota are able to use mucus carbohydrates as an energy source (45). Some investigators have even proposed that bacteria not only interact with the mucus layer but also form biofilms in this niche (43, 46, 47). Although controversial (48), these speculations were substantiated by observations of biofilm-like structures on luminal surfaces of normal intestinal tissue from mice (49) and humans (50). More recently, biofilms were observed in the GIT of germfree mice colonized with E. faecalis (51). Whether these structures are present in the GIT of conventional animals is unknown. Despite the controversy associated with the existence of biofilms in the GIT, it is widely accepted that bacteria physically associate with components of the outer mucus layer; however, fundamental questions about this arrangement remain unanswered. For example, we do not know if bacteria themselves contribute extracellular polymeric substances or whether host secretions (mucins and secretory IgA) are the only structural components of this environment. Whether bacterial species, in this environment, predominantly form pure microcolonies or freely associate with other species is also unknown. SrtA was previously shown to be necessary for biofilm formation by E. faecalis in vitro (30, 52). The finding that SrtA also promotes colonization is consistent with the idea of E. faecalis inhabiting the GIT as part of biofilm-like communities. Given the extensive research that has been done on enterococcal biofilms in vitro, E. faecalis makes an ideal model organism to investigate the possibility of microbial biofilms in the GIT and to study the nature of physical interactions between bacteria and the GIT mucosa layer as well as their role during intestinal colonization.
MATERIALS AND METHODS
Bacterial strains, growth media, and chemicals.
The bacterial strains and plasmids used in this study are listed in Table S1 in the supplemental material. The oligonucleotides used for plasmid construction were synthesized by Eurofins MWG Operon LLC. All restriction enzymes were purchased from New England BioLabs. Phusion High-Fidelity DNA polymerase (Thermo Scientific) was used for all PCRs performed for strain and plasmid construction. The Escherichia coli strains used in the study were cultured in LB medium. The E. faecalis strains were cultured in Mueller-Hinton (MH) broth or brain heart infusion (BHI) medium prepared as described by the manufacturer. When required, antibiotics were used at the following concentrations: 10 μg/ml (for E. faecalis) or 100 μg/ml (for E. coli) erythromycin (Em), 20 μg/ml (for E. coli) or 10 μg/ml (for E. faecalis) chloramphenicol (Cm), 2 mg/ml kanamycin (Kan), 200 μg/ml Rif, 80 μg/ml 5-fluorouracil (5-FU), and 50 μg/ml spectinomycin (Sp).
Animals.
The committee for animal care and use at the Medical College of Wisconsin approved all animal-related procedures and experiments. Male C57BL/6J mice (5 weeks of age) were obtained from the Jackson Laboratory (room RB08). Mice were allowed to adjust to the new environment for 1 week prior to the start of the experiments. Animals were housed under specific-pathogen-free conditions in the Medical College of Wisconsin vivarium. All mice were fed a standard chow diet (5L0D from LabDiet) ad libitum and reverse-osmosis water. Experimental sample sizes were determined by appropriate husbandry considerations as determined by the Medical College of Wisconsin vivarium. No steps were taken to randomize animals or to blind experimenters to the experimental groups.
Enterococcal mutants.
Construction of in-frame deletion mutants in enterococci was performed using the previously described markerless allelic exchange strategy (53, 54). Allelic exchange vectors were constructed by amplifying flanking regions of target genes and seamlessly inserting those amplicons into pJH082 (6) by Gibson assembly (55). Deletion alleles retained 1 to 20 codons at the 5′ and 3′ ends to minimize the risk of disrupting the expression of downstream genes. Complementation analysis performed by integration of genes into an ectopic locus on the chromosome was achieved by integrating srtA or srtAC200A preceded by the P23S promoter sequence obtained from pJRG8 (56) into the intergenic region between convergently transcribed genes OG1RF_10894 and OG1RF_10895 using pDV75-2. Stem loop terminators flanked the ectopically integrated region to prevent transcriptional read-through from (or out and into) adjacent genes (53). The chloramphenicol resistance marker consists of the cat gene from the previously described E. faecalis aMarTn (EfaMarTn) sequence (referred to as MarTn in this study), which carries a chloramphenicol acetyltransferase gene (30). The Sp resistance marker consists of the Sp adenyl transferase gene (aad9) obtained from Gram-positive shuttle vector pDL278 (57).
Mouse GIT colonization experiments.
Colonization experiments were performed as previously described (18). Stationary-phase cultures of E. faecalis were grown in Rif-supplemented MH broth, washed with sterile water, and added to sterile water to make a suspension with a final optical density at 600 nm (OD600) of 0.25. The suspension was then fed to mice as drinking water. All strains tested were confirmed to persist in drinking water at a density of between 107 and 108 CFU ml−1 over 4 days. E. faecalis-supplemented drinking water was changed every 3 to 4 days to maintain an appropriate inoculum, and mice were allowed to drink ad libitum for 14 days, after which the E. faecalis-supplemented drinking water was replaced with sterile water. The burden of the E. faecalis strain of interest was determined by plating feces specimens on BHI agar supplemented with Rif and enumerating viable colonies.
For competition experiments, mice were fed with a 1:1 mixture of the WT strain and the indicated mutant strains grown as described above; each strain was present at a final OD600 of 0.25 in drinking water. Mice were fed water supplemented with bacteria as described above. The burden of the E. faecalis strains of interest was determined by culturing feces specimens on BHI agar supplemented with Rif alone or with Rif in conjunction with Sp or Cm. Competitive indices were calculated by dividing the fecal load of mutant strains by that of WT strains recovered from each mouse. For mice in which one strain was below the limit of detection (which was 102 CFU/g), a value of 102 CFU/g was assigned to that strain for the calculation of the competitive index. Hence, for such mice, the competitive index value plotted represents an overly conservative estimate of the actual competitive index value. The competitive index calculation was omitted for mice in which both strains were below the limit of detection.
In vitro competitions.
Culture tubes containing 2 ml of fresh MH broth were inoculated with stationary-phase-grown marked strains to reach a final density of ∼105 CFU/ml per strain. Mixed cultures were incubated at 37°C for 24 h, after which they were diluted 1:1,000,000 in fresh media to allow continued growth; two additional culture passages were performed in a similar way. After each 24-h passage, the mixed cultures were plated on BHI agar supplemented with Rif alone or in conjunction with Sp or Cm for viable colony enumeration. Competitive indices were calculated by dividing the colony counts of mutant strains by those of WT strains from each mixed culture.
Growth curves.
(i) Growth in laboratory medium. Microtiter plates were inoculated from stationary-phase cultures to reach a final density of ∼105 CFU/ml in fresh MH broth and incubated at 37°C, and growth was monitored by measuring OD600 values using a Bioscreen C plate reader.
(ii) Growth in fecal suspensions. Freshly collected mouse feces specimens were subjected to homogenization in phosphate-buffered saline (PBS) to reach a final concentration of 100 mg/ml. Homogenates were gently centrifuged at a relative centrifugal force (RCF) level of 29 to pellet debris but not bacteria. Stationary-phase cultures were used to inoculate fecal bacterium-containing homogenates to reach a final concentration of ∼105 CFU/ml and incubated at 37°C. Growth was monitored by plating on Rif-containing agar and enumerating viable colonies.
Antibiotic susceptibility determinations. MICs were determined after 24 h at 37°C with 2-fold serial dilutions of antimicrobials in MH broth. Microtiter plates were inoculated from stationary-phase cultures to reach a final density of ∼105 CFU/ml, and growth was monitored using a Bioscreen C plate reader. The lowest antibiotic concentration that prevented growth was recorded as the MIC.
CPRG hydrolysis to assess cell envelope integrity.
Hydrolysis of CPRG was monitored to assess cell envelope integrity, as described previously (27). Bacterial cultures were grown at 37°C to the stationary phase in MH broth supplemented with 40 μg/ml CPRG and erythromycin (for the maintenance of pCJK205). CPRG hydrolysis was quantified by measuring the absorbance of cell-free supernatants at 570 nm and was normalized to bacterial density (optical density at 630 nm).
Mucin adhesion assays.
The mucin adhesion assay was adapted from a previous study (36). A type III porcine mucin (Sigma) suspension in 1× phosphate buffer saline (PBS) (10 mg/ml) was immobilized on flat-bottom 96-well plates by incubation at 4°C overnight. Excess mucin was washed three times with PBS, and coated wells were blocked with PBS containing 2% bovine serum albumin (BSA) and 1% Tween 20 for 4 h at 4°C; excess blocking solution was washed once with PBS. Strains of interest were cultured in Rif-supplemented BHI medium overnight at 37°C or 46°C (Em was supplemented for plasmid-carrying strains). Cells were then washed and adjusted to reach a final OD600 value of 1.0. A 100-μl volume of bacterial suspension was dispensed into each well and incubated at 4°C for 1 h. Wells were washed 7 times with PBS to remove unbound cells, after which adhered cells were fixed by drying the plate at 65°C for 1 h. Fixed cells were incubated with 1 mg/ml crystal violet (100 μl) for 30 min, washed thrice with PBS, and visualized by light microscopy. Numbers of bound cells were determined using the ImageJ built-in particle counter.
Searches for SDPs and distribution of SDPs among E. faecalis strains.
SDPs encoded in the genome of E. faecalis OG1RF were obtained by searching for the term “cell wall surface anchor” in the list of OG1RF proteins in NCBI. In addition, we searched for OG1RF proteins containing the “Pfam:Gram_pos_anchor” domain in the SMART database. Finally, we searched for OG1RF proteins containing the IPR019948 (Gram-positive anchor) domain in the Interpro database. A list of GIT-collected E. faecalis isolates was obtained from the reference genome catalog of the Human Microbiome Project. Amino acid sequences for SDPs were obtained from NCBI and used as queries in blastP searches against indicated reference genomes. The percentage of positive amino acid matches relative to the length of given SDPs for the top blast result was determined and illustrated as a heat map using PRISM.
Supplementary Material
ACKNOWLEDGMENTS
We thank Barbara Murray and Jaime Little for the generous provision of mutant strains for this study.
This study was supported by grants R01 AI121552 and AI132927 from the National Institutes of Health (NIH) to C.J.K. as well as by grants R01 GM099526 and R35 GM122503 to N.H.S. This work was also supported by the Children’s Research Institute of the Children’s Hospital of Wisconsin and by the Advancing a Healthier Wisconsin Endowment Research and Education Program (N.H.S.). This work was supported in part by the American Heart Association Midwest Affiliate Predoctoral Fellowship 16PRE29700011 (L.I.B.).
The content of this work is solely our responsibility and does not necessarily represent the official views of the NIH. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/IAI.00853-18.
REFERENCES
- 1.Yuen GJ, Ausubel FM. 2014. Enterococcus infection biology: lessons from invertebrate host models. J Microbiol 52:200–210. doi: 10.1007/s12275-014-4011-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gilmore MS, Lebreton F, van Schaik W. 2013. Genomic transition of enterococci from gut commensals to leading causes of multidrug-resistant hospital infection in the antibiotic era. Curr Opin Microbiol 16:10–16. doi: 10.1016/j.mib.2013.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mundt JO. 1963. Occurrence of enterococci in animals in a wild environment. Appl Microbiol 11:136–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Martin JD, Mundt JO. 1972. Enterococci in insects. Appl Microbiol 24:575–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Taur Y, Xavier JB, Lipuma L, Ubeda C, Goldberg J, Gobourne A, Lee YJ, Dubin KA, Socci ND, Viale A, Perales MA, Jenq RR, van den Brink MR, Pamer EG. 2012. Intestinal domination and the risk of bacteremia in patients undergoing allogeneic hematopoietic stem cell transplantation. Clin Infect Dis 55:905–914. doi: 10.1093/cid/cis580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Banla IL, Kommineni S, Hayward M, Rodrigues M, Palmer KL, Salzman NH, Kristich CJ. 19 December 2018. Modulators of Enterococcus faecalis cell envelope integrity and antimicrobial resistance influence stable colonization of the mammalian gastrointestinal tract. Infect Immun doi: 10.1128/IAI.00381-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smith HF, Fisher RE, Everett ML, Thomas AD, Randal Bollinger R, Parker W. 2009. Comparative anatomy and phylogenetic distribution of the mammalian cecal appendix. J Evol Biol 22:1984–1999. doi: 10.1111/j.1420-9101.2009.01809.x. [DOI] [PubMed] [Google Scholar]
- 8.Randal Bollinger R, Barbas AS, Bush EL, Lin SS, Parker W. 2007. Biofilms in the large bowel suggest an apparent function of the human vermiform appendix. J Theor Biol 249:826–831. doi: 10.1016/j.jtbi.2007.08.032. [DOI] [PubMed] [Google Scholar]
- 9.Lebeer S, Vanderleyden J, De Keersmaecker SCJ. 2008. Genes and molecules of Lactobacilli supporting probiotic action. Microbiol Mol Biol Rev 72:728–764. doi: 10.1128/MMBR.00017-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mazmanian SK, Liu G, Ton-That H, Schneewind O. 1999. Staphylococcus aureus sortase, an enzyme that anchors surface proteins to the cell wall. Science 285:760–763. doi: 10.1126/science.285.5428.760. [DOI] [PubMed] [Google Scholar]
- 11.Navarre WW, Schneewind O. 1999. Surface proteins of Gram-positive bacteria and mechanisms of their targeting to the cell wall envelope. Microbiol Mol Biol Rev 63:174–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mazmanian SK, Liu G, Jensen ER, Lenoy E, Schneewind O. 2000. Staphylococcus aureus sortase mutants defective in the display of surface proteins and in the pathogenesis of animal infections. Proc Natl Acad Sci U S A 97:5510–5515. doi: 10.1073/pnas.080520697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bierne H, Mazmanian SK, Trost M, Pucciarelli MG, Liu G, Dehoux P, Jänsch L, Portillo FG-d, Schneewind O, Cossart P; European Listeria Genome Consortium. 2002. Inactivation of the srtA gene in Listeria monocytogenes inhibits anchoring of surface proteins and affects virulence. Mol Microbiol 43:869–881. doi: 10.1046/j.1365-2958.2002.02798.x. [DOI] [PubMed] [Google Scholar]
- 14.Mandlik A, Swierczynski A, Das A, Ton-That H. 2008. Pili in Gram-positive bacteria: assembly, involvement in colonization and biofilm development. Trends Microbiol 16:33–40. doi: 10.1016/j.tim.2007.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sillanpaa J, Chang C, Singh KV, Montealegre MC, Nallapareddy SR, Harvey BR, Ton-That H, Murray BE. 2013. Contribution of individual Ebp pilus subunits of Enterococcus faecalis OG1RF to pilus biogenesis, biofilm formation and urinary tract infection. PLoS One 8:e68813. doi: 10.1371/journal.pone.0068813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nallapareddy SR, Sillanpaa J, Mitchell J, Singh KV, Chowdhury SA, Weinstock GM, Sullam PM, Murray BE. 2011. Conservation of Ebp-type pilus genes among enterococci and demonstration of their role in adherence of Enterococcus faecalis to human platelets. Infect Immun 79:2911–2920. doi: 10.1128/IAI.00039-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kemp KD, Singh KV, Nallapareddy SR, Murray BE. 2007. Relative contributions of Enterococcus faecalis OG1RF sortase-encoding genes, srtA and bps (srtC), to biofilm formation and a murine model of urinary tract infection. Infect Immun 75:5399–5404. doi: 10.1128/IAI.00663-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kommineni S, Bretl DJ, Lam V, Chakraborty R, Hayward M, Simpson P, Cao Y, Bousounis P, Kristich CJ, Salzman NH. 2015. Bacteriocin production augments niche competition by enterococci in the mammalian gastrointestinal tract. Nature 526:719–722. doi: 10.1038/nature15524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Top J, Paganelli FL, Zhang X, van Schaik W, Leavis HL, van Luit-Asbroek M, van der Poll T, Leendertse M, Bonten MJ, Willems RJ. 2013. The Enterococcus faecium enterococcal biofilm regulator, EbrB, regulates the esp operon and is implicated in biofilm formation and intestinal colonization. PLoS One 8:e65224. doi: 10.1371/journal.pone.0065224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang X, Top J, de Been M, Bierschenk D, Rogers M, Leendertse M, Bonten MJ, van der Poll T, Willems RJ, van Schaik W. 2013. Identification of a genetic determinant in clinical Enterococcus faecium strains that contributes to intestinal colonization during antibiotic treatment. J Infect Dis 207:1780–1786. doi: 10.1093/infdis/jit076. [DOI] [PubMed] [Google Scholar]
- 21.Rigottier-Gois L, Madec C, Navickas A, Matos RC, Akary-Lepage E, Mistou MY, Serror P. 2015. The surface rhamnopolysaccharide epa of Enterococcus faecalis is a key determinant of intestinal colonization. J Infect Dis 211:62–71. doi: 10.1093/infdis/jiu402. [DOI] [PubMed] [Google Scholar]
- 22.Lindenstrauss AG, Ehrmann MA, Behr J, Landstorfer R, Haller D, Sartor RB, Vogel RF. 2014. Transcriptome analysis of Enterococcus faecalis toward its adaption to surviving in the mouse intestinal tract. Arch Microbiol 196:423–433. doi: 10.1007/s00203-014-0982-2. [DOI] [PubMed] [Google Scholar]
- 23.Banla LI, Salzman NH, Kristich CJ. 2018. Colonization of the mammalian intestinal tract by enterococci. Curr Opin Microbiol 47:26–31. doi: 10.1016/j.mib.2018.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ton-That H, Liu G, Mazmanian SK, Faull KF, Schneewind O. 1999. Purification and characterization of sortase, the transpeptidase that cleaves surface proteins of Staphylococcus aureus at the LPXTG motif. Proc Natl Acad Sci U S A 96:12424–12429. doi: 10.1073/pnas.96.22.12424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Raz A, Tanasescu AM, Zhao AM, Serrano A, Alston T, Sol A, Bachrach G, Fischetti VA. 2015. Streptococcus pyogenes sortase mutants are highly susceptible to killing by host factors due to aberrant envelope physiology. PLoS One 10:e0140784. doi: 10.1371/journal.pone.0140784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu C, Huang IH, Chang C, Reardon-Robinson ME, Das A, Ton-That H. 2014. Lethality of sortase depletion in Actinomyces oris caused by excessive membrane accumulation of a surface glycoprotein. Mol Microbiol 94:1227–1241. doi: 10.1111/mmi.12780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Djoric D, Kristich CJ. 2015. Oxidative stress enhances cephalosporin resistance of Enterococcus faecalis through activation of a two-component signaling system. Antimicrob Agents Chemother 59:159–169. doi: 10.1128/AAC.03984-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.NIH HMP Working Group, Peterson J, Garges S, Giovanni M, McInnes P, Wang L, Schloss JA, Bonazzi V, McEwen JE, Wetterstrand KA, Deal C, Baker CC, Di Francesco V, Howcroft TK, Karp RW, Lunsford RD, Wellington CR, Belachew T, Wright M, Giblin C, David H, Mills M, Salomon R, Mullins C, Akolkar B, Begg L, Davis C, Grandison L, Humble M, Khalsa J, Little AR, Peavy H, Pontzer C, Portnoy M, Sayre MH, Starke-Reed P, Zakhari S, Read J, Watson B, Guyer M. 2009. The NIH Human Microbiome Project. Genome Res 19:2317–2323. doi: 10.1101/gr.096651.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Johansson ME, Larsson JM, Hansson GC. 2011. The two mucus layers of colon are organized by the MUC2 mucin, whereas the outer layer is a legislator of host-microbial interactions. Proc Natl Acad Sci U S A 108:4659–4665. doi: 10.1073/pnas.1006451107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kristich CJ, Nguyen VT, Le T, Barnes AMT, Grindle S, Dunny GM. 2008. Development and use of an efficient system for random mariner transposon mutagenesis to identify novel genetic determinants of biofilm formation in the core Enterococcus faecalis genome. Appl Environ Microbiol 74:3377–3386. doi: 10.1128/AEM.02665-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nallapareddy SR, Singh KV, Sillanpaa J, Zhao M, Murray BE. 2011. Relative contributions of Ebp pili and the collagen adhesin Ace to host extracellular matrix protein adherence and experimental urinary tract infection by Enterococcus faecalis OG1RF. Infect Immun 79:2901–2910. doi: 10.1128/IAI.00038-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nallapareddy SR, Singh KV, Sillanpaa J, Garsin DA, Hook M, Erlandsen SL, Murray BE. 2006. Endocarditis and biofilm-associated pili of Enterococcus faecalis. J Clin Invest 116:2799–2807. doi: 10.1172/JCI29021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nallapareddy SR, Qin X, Weinstock GM, Höök M, Murray BE. 2000. Enterococcus faecalis adhesin, Ace, mediates attachment to extracellular matrix proteins collagen type IV and laminin as well as collagen type I. Infect Immun 68:5218–5224. doi: 10.1128/IAI.68.9.5218-5224.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hall AE, Gorovits EL, Syribeys PJ, Domanski PJ, Ames BR, Chang CY, Vernachio JH, Patti JM, Hutchins JT. 2007. Monoclonal antibodies recognizing the Enterococcus faecalis collagen-binding MSCRAMM Ace: conditional expression and binding analysis. Microb Pathog 43:55–66. doi: 10.1016/j.micpath.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 35.Misawa Y, Kelley KA, Wang X, Wang L, Park WB, Birtel J, Saslowsky D, Lee JC. 2015. Staphylococcus aureus colonization of the mouse gastrointestinal tract is modulated by wall teichoic acid, capsule, and surface proteins. PLoS Pathog 11:e1005061. doi: 10.1371/journal.ppat.1005061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Call EK, Goh YJ, Selle K, Klaenhammer TR, O’Flaherty S. 2015. Sortase-deficient lactobacilli: effect on immunomodulation and gut retention. Microbiology 161:311–321. doi: 10.1099/mic.0.000007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brinster S, Furlan S, Serror P. 2007. C-terminal WxL domain mediates cell wall binding in Enterococcus faecalis and other gram-positive bacteria. J Bacteriol 189:1244–1253. doi: 10.1128/JB.00773-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Carasi P, Racedo SM, Jacquot C, Elie AM. Serradell MdlÁ, Urdaci MC. 2017. Enterococcus durans EP1 a promising anti-inflammatory probiotic able to stimulate sIgA and to increase Faecalibacterium prausnitzii abundance. Front Immunol 8:88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Glenister DA, Salamon KE, Smith K, Beighton D, Keevil CW. 1988. Enhanced growth of complex communities of dental plaque bacteria in mucin-limited continuous culture. Microb Ecol Health Dis 1:31–38. doi: 10.3109/08910608809140176. [DOI] [Google Scholar]
- 40.Holmén Larsson JM, Thomsson KA, Rodríguez-Piñeiro AM, Karlsson H, Hansson GC. 2013. Studies of mucus in mouse stomach, small intestine, and colon. III. Gastrointestinal Muc5ac and Muc2 mucin O-glycan patterns reveal a regiospecific distribution. Am J Physiol Gastrointest Liver Physiol 305:G357–G363. doi: 10.1152/ajpgi.00048.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kline KA, Kau AL, Chen SL, Lim A, Pinkner JS, Rosch J, Nallapareddy SR, Murray BE, Henriques-Normark B, Beatty W, Caparon MG, Hultgren SJ. 2009. Mechanism for sortase localization and the role of sortase localization in efficient pilus assembly in Enterococcus faecalis. J Bacteriol 191:3237–3247. doi: 10.1128/JB.01837-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sonnenburg JL, Angenent LT, Gordon JI. 2004. Getting a grip on things: how do communities of bacterial symbionts become established in our intestine? Nat Immunol 5:569–573. doi: 10.1038/ni1079. [DOI] [PubMed] [Google Scholar]
- 43.Sicard JF, Le Bihan G, Vogeleer P, Jacques M, Harel J. 2017. Interactions of intestinal bacteria with components of the intestinal mucus. Front Cell Infect Microbiol 7:387. doi: 10.3389/fcimb.2017.00387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Akiyama Y, Nagahara N, Kashihara T, Hirai S, Toguchi H. 1995. In vitro and in vivo evaluation of mucoadhesive microspheres prepared for the gastrointestinal tract using polyglycerol esters of fatty acids and a poly(acrylic acid) derivative. Pharm Res 12:397–405. doi: 10.1023/A:1016208703380. [DOI] [PubMed] [Google Scholar]
- 45.Ouwerkerk JP, de Vos WM, Belzer C. 2013. Glycobiome: bacteria and mucus at the epithelial interface. Best Pract Res Clin Gastroenterol 27:25–38. doi: 10.1016/j.bpg.2013.03.001. [DOI] [PubMed] [Google Scholar]
- 46.de Vos WM. 2015. Microbial biofilms and the human intestinal microbiome. NPJ Biofilms Microbiomes 1:15005. doi: 10.1038/npjbiofilms.2015.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Donaldson GP, Lee SM, Mazmanian SK. 2016. Gut biogeography of the bacterial microbiota. Nat Rev Microbiol 14:20–32. doi: 10.1038/nrmicro3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Swidsinski A, Loening-Baucke V, Theissig F, Engelhardt H, Bengmark S, Koch S, Lochs H, Dörffel Y. 2007. Comparative study of the intestinal mucus barrier in normal and inflamed colon. Gut 56:343–350. doi: 10.1136/gut.2006.098160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Swidsinski A, Loening-Baucke V, Lochs H, Hale LP. 2005. Spatial organization of bacterial flora in normal and inflamed intestine: a fluorescence in situ hybridization study in mice. World J Gastroenterol 11:1131–1140. doi: 10.3748/wjg.v11.i8.1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Palestrant D, Holzknecht ZE, Collins BH, Parker W, Miller SE, Bollinger RR. 2004. Microbial biofilms in the gut: visualization by electron microscopy and by acridine orange staining. Ultrastruct Pathol 28:23–27. doi: 10.1080/01913120490275196. [DOI] [PubMed] [Google Scholar]
- 51.Barnes AM, Dale JL, Chen Y, Manias DA, Greenwood Quaintance KE, Karau MK, Kashyap PC, Patel R, Wells CL, Dunny GM. 2016. Enterococcus faecalis readily colonizes the entire gastrointestinal tract and forms biofilms in a germ-free mouse model. Virulence doi: 10.1080/21505594.2016.1208890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Guiton PS, Hung CS, Kline KA, Roth R, Kau AL, Hayes E, Heuser J, Dodson KW, Caparon MG, Hultgren SJ. 2009. Contribution of autolysin and sortase a during Enterococcus faecalis DNA-dependent biofilm development. Infect Immun 77:3626–3638. doi: 10.1128/IAI.00219-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vesić D, Kristich CJ. 2013. A Rex family transcriptional repressor influences H2O2 accumulation by Enterococcus faecalis. J Bacteriol 195:1815–1824. doi: 10.1128/JB.02135-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kristich CJ, Djorić D, Little JL. 2014. Genetic basis for vancomycin-enhanced cephalosporin susceptibility in vancomycin-resistant enterococci revealed using counterselection with dominant-negative thymidylate synthase. Antimicrob Agents Chemother 58:1556–1564. doi: 10.1128/AAC.02001-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gibson DG, Young L, Chuang R-Y, Venter JC, Hutchison CA, Smith HO. 2009. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 56.Kristich CJ, Little JL, Hall CL, Hoff JS. 2011. Reciprocal regulation of cephalosporin resistance in Enterococcus faecalis. mBio 2:e00199-11. doi: 10.1128/mBio.00199-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chandler JR, Dunny GM. 2008. Characterization of the sequence specificity determinants required for processing and control of sex pheromone by the intramembrane protease Eep and the plasmid-encoded protein PrgY. J Bacteriol 190:1172–1183. doi: 10.1128/JB.01327-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.