Abstract
Museum specimens play an increasingly important role in predicting the outcomes and revealing the consequences of anthropogenically driven disruption of the biosphere. As ecological communities respond to ongoing environmental change, host–parasite interactions are also altered. This shifting landscape of host–parasite associations creates opportunities for colonization of different hosts and emergence of new pathogens, with implications for wildlife conservation and management, public health, and other societal concerns. Integrated archives that document and preserve mammal specimens along with their communities of associated parasites and ancillary data provide a powerful resource for investigating, anticipating, and mitigating the epidemiological, ecological, and evolutionary impacts of environmental perturbation. Mammalogists who collect and archive mammal specimens have a unique opportunity to expand the scope and impact of their field work by collecting the parasites that are associated with their study organisms. We encourage mammalogists to embrace an integrated and holistic sampling paradigm and advocate for this to become standard practice for museum-based collecting. To this end, we provide a detailed, field-tested protocol to give mammalogists the tools to collect and preserve host and parasite materials that are of high quality and suitable for a range of potential downstream analyses (e.g., genetic, morphological). Finally, we also encourage increased global cooperation across taxonomic disciplines to build an integrated series of baselines and snapshots of the changing biosphere.
Los especímenes de museo desempeñan un papel cada vez más importante tanto en la descripción de los resultados de la alteración antropogénica de la biosfera como en la predicción de sus consecuencias. Dado que las comunidades ecológicas responden al cambio ambiental, también se alteran las interacciones hospedador-parásito. Este panorama cambiante de asociaciones hospedador-parásito crea oportunidades para la colonización de diferentes hospedadores y para la aparición de nuevos patógenos, con implicancias en la conservación y manejo de la vida silvestre, la salud pública y otras preocupaciones de importancia para la sociedad. Archivos integrados que documentan y preservan especímenes de mamíferos junto con sus comunidades de parásitos y datos asociados, proporcionan un fuerte recurso para investigar, anticipar y mitigar los impactos epidemiológicos, ecológicos y evolutivos de las perturbaciones ambientales. Los mastozoólogos que recolectan y archivan muestras de mamíferos, tienen una oportunidad única de ampliar el alcance e impacto de su trabajo de campo mediante la recolección de los parásitos que están asociados con los organismos que estudian. Alentamos a los mastozoólogos a adoptar un paradigma de muestreo integrado y holístico y abogamos para que esto se convierta en una práctica estándarizada de la obtención de muestras para museos. Con este objetivo, proporcionamos un protocolo detallado y probado en el campo para brindar a los mastozoólogos las herramientas para recolectar y preservar materiales de parásitos y hospedadores de alta calidad y adecuados para una gran variedad de análisis subsecuentes (e.g., genéticos, morfológicos, etc.). Finalmente, también abogamos por una mayor cooperación global entre las diversas disciplinas taxonómicas para construir una serie integrada de líneas de base y registros actuales de nuestra cambiante biosfera.
Keywords: emerging infectious disease, field methods, integrated collections, necropsy, parasitology, specimens
Investigations of mammals play a vital role in revealing patterns of global diversity, and in recognizing and documenting the accelerating effects of climatological and environmental perturbation across landscapes and ecosystems (e.g., Parmesan and Yohe 2003; Lawler et al. 2009). Direct effects of anthropogenically driven climate change on faunal structure and sustainability are increasingly observed, with disruptive outcomes anticipated for ecological and evolutionary processes (Harvell et al. 2002; Barnosky et al. 2012; Pecl et al. 2017). Specimen-based surveys remain necessary to define the historical foundations of diversity in terms of both pattern and process, and to set the stage for understanding the consequences of environmental change on ecological time scales. Specimen archives are and will be essential for efforts to anticipate, predict, and mitigate the impacts of biodiversity loss through the Anthropocene, now regarded as the sixth major extinction event (Wake and Vredenburg 2008; Barnosky et al. 2012; Capinha et al. 2015; Ceballos et al. 2017; Steffen et al. 2018). Pervasive landscape disturbance, ecosystem disruption, species extinctions, and emergence of infectious diseases are interrelated crises linked to expanding human population, habitat loss, and our widening footprint across the planet (Daszak et al. 2000; Brooks and Hoberg 2013; Holmes 2013; Brooks et al. 2014, 2019). However, efforts to respond to these crises are hindered by incomplete information regarding the distribution of diversity across all phylogenetic scales, as well as the scope of ecological interactions that link faunas, ecosystems, and communities through space and time.
A deep tradition of specimen-based fieldwork underlies much of what we know regarding mammalian diversity, ecology, and evolution. In mammalogy, museum collections have been critical to biodiversity discovery and documentation. Specimen preparation, once focused predominately on preservation of mammal skins and skeletons and associated data (date, locality, standard measurements), has been transformed in recent decades by the availability of new technologies (e.g., DNA sequencing, metagenomics, stable isotope analysis) that offer access to powerful approaches to address research questions that previously were intractable. Those technologies are breathing new life into old museum specimens (e.g., Bi et al. 2013), creating opportunities for discovery that are only possible because of the dedication of early scientific collectors who carefully archived specimens generations ago against the day when they might be useful to a future scientist (Grinnell 1910; Dunnum et al. 2017; Tiee et al. 2018).
We have argued elsewhere for continued development of specimen-based archives to explore mammalian diversity and ecology (McLean et al. 2016; Hope et al. 2018; Malaney and Cook 2018; Schindel and Cook 2018; Cook and Light 2019). Here, we broaden that discussion for mammalogy by emphasizing that parasites of mammals continue to be an undervalued and poorly documented component of diversity that should be directly integrated into the process of field collection and preparation of mammals worldwide. Understanding the distribution and diversity of parasites in mammals is critical, as parasites can have a profound influence on mammalian ecology (Lafferty et al. 2008), and interactions among mammals, parasites, and changing environments define the context for emerging infectious diseases that affect humans and animals around the world (Daszak et al. 2000; Cleaveland et al. 2001). Our goals are 1) to increase awareness among mammalogists of the necessity for building capacity to improve understanding of biodiversity structure and function through integrated host–parasite collections, and 2) to provide mammalogists with a practical, field-tested set of protocols for parasite sampling that can be efficiently incorporated into museum-based mammal surveys.
The case for integrated sampling
Parasites of mammals offer a rich data source that traditionally has been largely neglected by field mammalogists. Because a single mammal represents an entire ecosystem of interacting symbionts, parasites can yield insights into key processes that shape biodiversity, biogeography, and ecology, and that transcend spatial and temporal scales (Manter 1966; Hoberg 1997; Criscione et al. 2005). Parasites that rely on multiple hosts to complete their life cycle provide windows into interspecific interactions and the dynamics of complex ecosystems. They also can play important roles in regulating host populations and maintaining diverse and productive ecosystems (Dobson et al. 2008). Concurrent knowledge of mammals and their parasites facilitates understanding of the drivers of host range, host colonization, and pathogen emergence. A comprehensive understanding of parasite diversity, host associations, and geography will play an important role in addressing many current societal challenges, ranging from issues of conservation to public health (Hoberg et al. 2015; DiEuliis et al. 2016).
Over geologic time, climate change, ecological disruption, and biological invasion collectively have extensively influenced the structure of the biosphere and the distribution of parasites and diseases (e.g., Harvell et al. 2002; Hoberg and Brooks 2008; Altizer et al. 2013). Shifting ecological mosaics and faunal mixing have repeatedly instigated new parasite–host interactions, creating persistent opportunities for emergent disease (Brooks and Hoberg 2013; Araujo et al. 2015; Hoberg and Brooks 2015). New host associations for parasite lineages can also lead to novel genomic interactions (e.g., recombination between viruses) that can yield newly pathogenic strains (Parrish et al. 2008). The increasing proximity of wild mammals to agricultural, rural, and urban centers, and continued encroachment by humans on wilderness represent primary mechanisms by which zoonotic pathogens emerge in people or invade domestic animals (e.g., Daszak et al. 2000; Cleaveland et al. 2001; Woolhouse and Gowtage-Sequeria 2005; Jenkins et al. 2013; Pybus et al. 2015). Our ability to measure the scope and pace of change in host–parasite dynamics under the current regime of accelerating change requires comprehensive integrated baselines for mammal and parasite diversity (Cook et al. 2013, 2017; Hoberg et al. 2013; Brooks et al. 2014; Dunnum et al. 2017).
The idea of host–parasite or “integrated biodiversity” informatics is not new (e.g., Rausch 1952, 1956; Gardner and Campbell 1992; Hoberg 1997; Brooks and Hoberg 2000). Among the earliest documented examples of a coordinated collection in North America is that by O. J. Murie, the renowned mammalogist and ecologist, who collected nematode parasites in a specimen of American pika (Ochotona princeps) from the Teton Range in Wyoming in 1930. The host specimen was deposited in the mammal collection of the U.S. National Museum, Smithsonian, and the parasites were forwarded to G. Dikmans at the U.S. Department of Agriculture in Washington, D.C. Dikmans went on to diagnose and describe a new genus and species (Murielus harpespiculus) from this sample of roundworms, with the specimens deposited in the United States National Parasite Collection (USNPC 30461; USNM 1332127). Many years later during extensive new biogeographic and phylogeographic studies of pikas and parasites (Galbreath et al. 2009; Galbreath and Hoberg 2012, 2015), specimens of a second undescribed species in a different genus (Ohbayashinema; the first report of this genus in the Western Hemisphere) were discovered in the original vial of archived specimens held in the USNPC for 70 years (Durette-Desset et al. 2010). Discovery of these tiny nematodes contributed directly to the larger story of episodic geographic expansion between North America and Eurasia that during the Pleistocene led to the assembly of the contemporary Holarctic fauna (Hoberg et al. 2012a). This example demonstrates three relevant points: 1) coordinated collections provide substantially greater information than the study of either mammals or parasites alone; 2) archival collections are critical for documenting mammalian and parasite diversity and for exploring the history of faunas across broad geographic scales; and 3) archival collections create unanticipated opportunities for serendipitous discoveries by future generations of scientists (Kemp 2017), including correction of past taxonomic errors (e.g., Hoberg et al. 2009).
Despite the work of individuals such as O. J. Murie, large-scale efforts to acquire collections of mammals with their associated parasites (e.g., microparasites such as protozoans, bacteria, fungi, and viruses; and macroparasites such as helminths and ectoparasites, including lice, mites, fleas, and ticks) occurred infrequently during the early 20th century. The challenge associated with collecting, curating, identifying, and archiving integrated collections while maintaining data linkages between associated specimens discouraged widespread adoption of comprehensive collection protocols. In North America, the philosophy, rationale, and outcomes for large-scale field collections of mammals and parasites were pioneered by Robert and Virginia Rausch. Robert Rausch began his studies of vertebrates and their helminth parasites in 1943, investigating pathogenicity in natural host–parasite systems in the north-central United States (Rausch 1983). From there, his work took him to Alaska, where in collaboration with Virginia Rausch, he led the development of an integrated research program for mammals and their parasites in northern and western Alaska that eventually expanded globally (e.g., Rausch 1952, 1957, 1994). As natural historians, their goal was to recognize and document diversity of mammals and parasites, directly contributing to a more complete picture of ecological community structure and the circulation of zoonoses (e.g., Rausch 1956, 1972). The “Rausch School” of coordinated field investigation was influential in the broader mammalogical and parasitological communities, establishing an infrastructure for comparative investigations based on large comprehensive collection efforts.
Building an integrated biodiversity infrastructure
In the wake of the Rausches’ work, the principle of integrated collections for mammals and parasites was expanded and codified (Gardner 1996; Gardner and Jiménez-Ruiz 2009), with a growing recognition that whenever a mammal is collected there should be coordinated and simultaneous collection of associated parasites and related data (Frey et al. 1992; DiEuliis et al. 2016). Over the past four decades, large-scale efforts to survey and archive mammal and parasite diversity have targeted remote sites across North America, South America, and parts of Asia (Table 1). The Bolivian Biodiversity Survey, which began in 1984 under the leadership of Terry Yates (Museum of Southwestern Biology, Albuquerque, New Mexico) and Sydney Anderson (American Museum of Natural History, New York, New York), demonstrated the potential for exceptional productivity in modern integrated collection programs. That project resulted in the collection of > 16,000 mammal specimens archived in the Museum of Southwestern Biology, American Museum of Natural History, and the Museo Nacional de Historia Natural, La Paz, Bolivia, along with thousands of lots of parasites deposited in the Harold W. Manter Laboratory of Parasitology at the University of Nebraska State Museum, Lincoln, Nebraska. Those collections served as the primary specimen base for hundreds of publications and continue to support ongoing research (e.g., Gardner and Campbell 1992; Anderson 1997; Salazar-Bravo et al. 2002). Other major projects that have advanced the Rausch model of integrated collections include the Mongolian Vertebrate Parasite Project (roughly 10,000 vertebrates and associated parasites archived—Tinnin et al. 2012; Gardner et al. 2013a) and the Beringian Coevolution Project (roughly 50,000 mammals and associated parasites archived—Cook et al. 2005, 2017).
Table 1.
Historical framework for development of protocols for integrated biodiversity archives. The work of diverse contributors provides context that informs recommendations for methods of collecting and archiving mammals and parasites.
| Time frame | Lead investigators | Project and geographic emphasis | Representative literature |
|---|---|---|---|
| 1947–2005 | Robert and Virginia Rausch | Mammal–parasite faunas of Alaska, northern Canada, Pacific Northwest United States, midwestern United States and Canada, and Siberia | Rausch (1952, 1957, 1994) |
| 1979–1984 | Terry Yates, Don Duszynski | Mammal–parasite faunas in Japan, American Southwest | Duszynski et al. (1982); Wash et al. (1985) |
| 1984–1993 | Terry Yates, Sydney Anderson, Joseph Cook, Scott Gardner | Mammal–parasite faunas in Bolivia | Lambert et al. (1988); Gardner and Campbell (1992); Anderson (1997); Dick et al. (2007); Gardner et al. (2013b) |
| 1991–2016 | Joseph Cook, Stephen MacDonald, Anson Koehler | Mammal–parasite faunas in Southeast Alaska—ISLES Tongass Surveys | Koehler et al. (2007, 2009); Hoberg et al. (2012b); Cook and MacDonald (2013) |
| 1993–2019 | Terry Yates, Brian Hjelle, Blas Armién, Joseph Cook, Jon Dunnum, Fernando Torres-Pérez, Eduardo Palma | Mammalian hantaviruses in the American Southwest and Central and South America | Glass et al. (2002); Yates et al. (2002); Torres-Pérez et al. (2011); Dunnum et al. (2017) |
| 1994–2017 | Susan Kutz, Emily Jenkins, Eric Hoberg | Research Group for Arctic Parasitology—lungworms of Arctic ungulates | Jenkins et al. (2005); Kutz et al. (2005, 2007, 2013); Hoberg et al. (2008); Verocai et al. (2014) |
| 1999–2017 | Joseph Cook, Eric Hoberg, Kurt Galbreath | Beringian Coevolution Project—Alaska, Siberia, northern Canada, Mongolia | Hoberg et al. (2003, 2012a, 2013); Cook et al. (2005, 2017); Makarikov et al. (2013) |
| 2003–2019 | Joseph Cook, Richard Yanagihara | Hantaviruses in non-rodent mammals | Arai et al. (2007); Kang et al. (2016) |
| 2009–2012 | Scott Gardner, Joseph Cook | Mongolian Vertebrate Parasite Project—Gobi Desert | Tinnin et al. (2012); Gardner et al. (2013a); Gardner (2014); Dursahinhan et al. (2017) |
| 2012–2018 | Andrew Hope, Vasyl Tkach | Mammal–parasite faunas in Alaska—Arctic-boreal ecotone | Hope et al. (2016); Hope (In press) |
| 2017–2018 | Stephen Greiman, Joseph Cook | Shrew–parasite faunas in the American Southwest | Greiman et al. (2018) |
As targeted sampling begins to illuminate host–parasite diversity from certain poorly documented regions, the full scope of the challenge in building the necessary specimen base for integrated investigations on a global scale comes into focus. Vast swaths of the planet remain unsampled using integrated protocols, and for those regions that were previously surveyed, resampling to evaluate faunal change through time is important. Integrated specimen archives represent snapshots in time regarding the geography of host–parasite assemblages. This record provides the basis for biodiversity discovery, assessment, and monitoring, as well as action for conservation (DAMA protocol sensu Brooks et al. 2014), which is increasingly necessary for addressing diverse challenges as environmental perturbation accelerates, including extinctions of mammals and emergence of infectious disease (Tsangaras and Greenwood 2012; Hoberg and Brooks 2015; DiEuliis et al. 2016; Brooks et al. 2019). Through integrated field approaches, we will build an increasingly nuanced picture of the structure of mammal and parasite faunas through space and time (e.g., Yates et al. 2002; Hoberg et al. 2012a; Cook et al. 2017), and the opportunities to participate in this endeavor are unlimited.
Like mammalogists, parasitologists have historically maintained a tradition of collecting, but they have often not archived extensive series of specimens in accessible research collections (Hoberg et al. 2009). In the United States, this reflects in part the fact that there are relatively few parasite collections that are accessible to the scientific community. Major helminth collections of broad taxonomic scope are housed in the Smithsonian Institution’s National Museum of Natural History, Division of Invertebrate Zoology (formerly the U.S. National Parasite Collection of the Agricultural Research Service, USDA), the University of Nebraska State Museum’s Harold W. Manter Laboratory of Parasitology, and the Museum of Southwestern Biology’s Division of Parasites. Other museums house taxonomically focused parasite collections (e.g., U.S. National Tick Collection at Georgia Southern University, Statesboro, Georgia; Traub flea collection at the Carnegie Museum of Natural History, Pittsburgh, Pennsylvania). In Latin America, nine museums in seven countries curate important regional helminthological collections, with the largest collection at the Oswaldo Cruz Institute in Rio de Janeiro, Brazil (Lamothe-Argumedo et al. 2010). Globally there are fewer than 100 significant parasite collections (Lichtenfels and Pritchard 1982; Zinovieva et al. 2015; Bell et al. 2018), in contrast to at least 276 active mammal collections reported in the United States alone (Dunnum et al. 2018). The lack of a more-developed infrastructure of parasite collections among North American institutions may reflect a certain degree of historical isolation between parasitology and other organismal disciplines, which undoubtedly has resulted in missed opportunities to enhance biodiversity research infrastructure through integrated collections.
The value of museum collections increases with 1) the scope of diversity that is sampled, 2) the intensity of local sampling, 3) the geographic breadth of sampling, 4) the regularity of sampling through time, 5) the diversity and quality of ancillary data and products derived from each specimen, and 6) the accessibility of specimens for use in collaborative and integrative research. By strengthening efforts to enhance these components of existing sampling programs, we can improve the impact of our work with a proportionally modest investment, particularly when resources are already dedicated for mammal collection. At a time when resources for biodiversity research are limited (e.g., Nowogrodzki 2016) and the need for biodiversity data is increasing (e.g., Amato and DeSalle 2012), we should invest in collecting efforts that take advantage of every opportunity to maximize the taxonomic, geographic, and temporal scope of sampling (Cook et al. 2016). Given the high cost of conducting field work, especially in remote and poorly documented regions (Bradley et al. 2012), a relatively straightforward way to increase the return on our investment of time and resources is to incorporate parasite collection into standard specimen preparation protocols. This will have the added benefit of facilitating collaboration between mammalogists and parasitologists, which could help to break down the siloed nature of natural history collections. Though disciplines such as mammalogy and parasitology have historically progressed along parallel but largely isolated trajectories, interdisciplinary engagement and collaboration is increasingly important to address broad-scale questions of societal significance such as causes and consequences of extinction and factors that promote emerging infectious disease (Hoberg et al. 2015).
Scientific collectors have a responsibility to archive specimens in institutions that will maintain them for not only their own immediate use, but also for the benefit of future generations of scientists (Grinnell 1910; Morrison et al. 2017). To maximize the value of each specimen, high-quality associated data also must be preserved (Gardner and Jiménez-Ruiz 2009). As the number and diversity of research applications using museum specimens have grown, standard field protocols for mammal collecting have evolved. In addition to traditional specimens (skins, skeletons), collectors now routinely preserve mammal tissues using a variety of methods (e.g., freezing, RNAlater, ethanol, lysis buffer), each of which offers specific advantages and disadvantages for fieldwork and various downstream applications. By making an effort to maximize the utility of specimens preserved from each individual mammal, we can more efficiently leverage limited research resources and strengthen the quality, integration, and impact of our work. Thus, the role of museums continues to shift from collections of specimens to collections of information that can be applied to address issues of direct relevance to science and society through biodiversity informatics (Hoberg 2002; Hoberg et al. 2015; Dunnum et al. 2017).
Field Methods
Here, we draw from our decades of field experience spanning multiple continents and ecosystems ranging from the Neotropics to the Arctic and Patagonia to outline parasite collection protocols that would allow a mammalogist to efficiently collect parasite specimens for use in downstream applications such as morphological identification and genetic analysis. These methods complement other published protocols for preservation and documentation of host vouchers (Yates et al. 1996), preservation of symbiotypes (Frey et al. 1992), and collection of host tissues (Yates 1996). Our goal is not to offer an exhaustive description of parasitological techniques, but rather to provide a step-by-step guide to practical and productive methods that could be easily adopted by field mammalogists (Fig. 1). Other sources provide accounts of additional parasitological methods (Pritchard and Kruse 1982; Lutz et al. 2017; Tkach et al. 2019) that extend those described here. Our streamlined recommendations, summarized below and described in detail in Supplementary Data SD1, address recent developments that build upon prior descriptions of parasitological methods for use in mammals generally (Gardner 1996) and bats in particular (Gardner and Jiménez-Ruiz 2009). Although our primary emphasis is on necropsy of small mammals, we also provide in Supplementary Data SD1 a simple protocol for noninvasive collection of ungulate nematodes from fecal samples acquired in the field (Kutz et al. 2007).
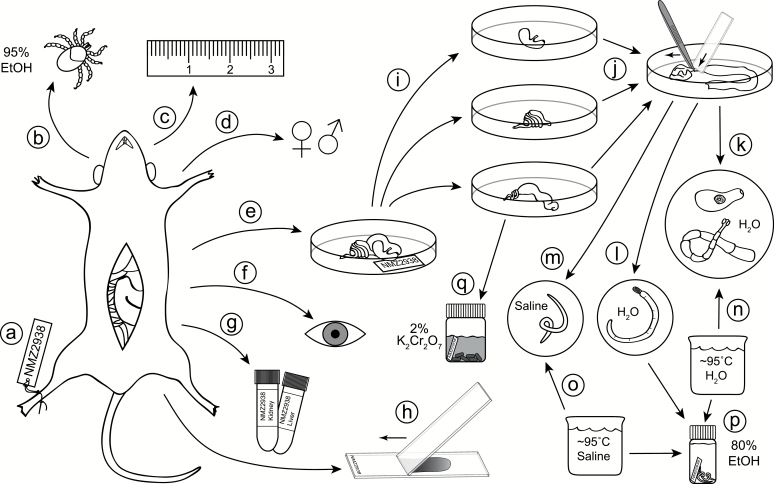
Fig. 1.
Flowchart for small mammal necropsy: a unique identifier is assigned to the mammal immediately upon capture (a); the mammal is swept for ectoparasites (b); standard mammal measurements are recorded (c); reproductive data are recorded (d); the gastrointestinal (GI) tract is transferred to a Petri dish and labeled (e); the body cavity and organs (e.g., liver) are visually inspected for parasites (e.g., encysted metacestodes, nematodes, sarcocysts) (f); host tissues are collected and preserved (g); thin blood smears can be prepared from freshly euthanized hosts to sample blood-borne pathogens (h); major sections of the GI tract are separated (stomach, small intestine, large intestine), straightened, and individually opened lengthwise (i); the lining is scraped by pulling it beneath the end of a microscope slide using forceps, and washed with saline to reveal helminths (j); after transfer of helminths to a new dish, trematodes and cestodes are washed in saline or water (k); acanthocephalans are soaked in water until the proboscis extends and the worm dies (l); and nematodes are washed in saline (m); trematodes and cestodes are simultaneously relaxed and killed by swirling in hot water (n); nematodes are killed using hot saline (o); helminths are preserved in ethanol (p); fecal pellets are collected from the colon and stored in potassium dichromate solution to sample coccidians (q).
We emphasize examination and preservation of organs and tissues that have traditionally received little attention from mammalogists. Our methods are intended to complement standard mammal collecting protocols, and therefore do not address aspects of parasitological examination that require destruction of anatomical features that are commonly preserved by mammalogists. We especially focus on sampling of metazoan ecto- and endoparasites of small terrestrial mammals, as these represent obvious targets for mammalogists who are interested in maximizing the diversity of parasites collected from a host organism, while minimizing additional investment in time, training, and equipment. We advocate conducting field necropsies using an assembly line model, in which each specimen moves through a succession of stations at which individual workers are responsible for completing specific necropsy tasks (e.g., ectoparasite sweeps, measurements, tissue pulling, gut examination). This approach maximizes the efficiency with which specimen preparation is completed to minimize limitations on the number of specimens that can be processed in a given field day. Our parasitological recommendations fit well into the assembly line model of specimen preparation.
Procedural overview
To ensure the highest quality of preserved material, mammal specimens should ideally be processed as soon as possible after they are euthanized. Useful, but lower-quality material can be acquired from frozen specimens assuming that they have passed through no more than a single freeze-thaw cycle. From the parasitological perspective, necropsy of whole fluid-preserved hosts is a suboptimal alternative given that helminth morphology is almost always compromised and ectoparasites can be lost or accidentally transferred among such specimens, but useful data can potentially still be acquired (e.g., genomic data, estimates of parasite intensity and prevalence—Greiman et al. 2018).
Given that a major goal of our protocol is to create a pipeline from the field to the museum that maximizes the preservation of data for biodiversity informatics as well as taxonomic applications, the first step in performing a mammal necropsy must be to assign a unique identifier to the specimen (e.g., museum tissue or catalog number). The identifier must be permanently affixed to all datasheets and physical products (e.g., skeleton, skin, tissues, parasites) derived from the specimen, which should be traceable through an accessible museum database and archive. To facilitate tracking of these parts derived from individual specimens, application of preprinted barcode labels with unique codes is strongly recommended (see Supplementary Data SD1). Such labels help to reduce or correct errors that are common through hand-written transcription. As new data are generated from the specimen over the course of future investigations, the unique identifier will unite the expanding network of data points that results (e.g., DNA sequences, morphometric data, stable isotope profiles). Data that become decoupled from the network by losing this critical linkage lose all the advantages that the integrated specimen-based data set confers. All published products derived from specimen-based research, such as data sets in public data repositories (e.g., GenBank), should report data linked to specimen identifiers using data structures that are designed to be both human- and machine-readable to simplify subsequent analyses (Verde Arregoitia et al. 2018). Each new link between a specimen and its products progressively increases the value of the specimen itself.
Once a unique identifier has been assigned, the specimen necropsy begins with screening for ectoparasites. Fleas, ticks, mites, and lice represent the most common and abundant ectoparasites that many mammalogists are likely to encounter. Bats may also harbor parasitic flies of the families Streblidae and Nycteribiidae. Botfly larvae (family Oestridae) are also commonly found in subcutaneous tissues of mammals, but must be either carefully excised through their air hole or removed when the host is skinned. Freshly captured mammal specimens should be fumigated using a chemical inhalant (e.g., chloroform) prior to sweeping, which facilitates ectoparasite collection and reduces risk of exposure to arthropod-vectored pathogens. Many arthropods will be shed quickly after fumigation, though embedded mites and ticks will require care to remove without damaging delicate mouthparts that are diagnostic for species identification. To maintain quality of these arthropod specimens for molecular and morphological analysis, preserve them in ≥ 95% ethanol. Record the type and ideally the number of collected ectoparasites on the host’s datasheet.
With the ectoparasite sweep complete, standard measurements for the host can be recorded, the abdomen opened via a midventral incision, and reproductive condition assessed. At this point the gastrointestinal (GI) tract between the esophagus and rectum should be carefully transferred to a Petri dish labeled with the specimen identifier, and host tissues harvested. As organs are removed, they and the rest of the body cavity should be inspected for parasites. The liver and gall bladder, in particular, have potential to harbor diverse parasites, including larval cestodes and adult trematodes. Hard nodules in the lungs may indicate encysted helminths. Larval cestodes and nematodes can be free in the body cavity or embedded within the mesenteries. Nematodes can occupy the urinary bladder. White sarcocysts of tissue-dwelling coccidians (Sarcocystidae) can be present in muscle tissue. Any obvious helminths or sarcocysts should be immediately preserved using methods appropriate to the taxon (see Supplementary Data SD1). Suspected helminths or organs requiring additional examination (e.g., urinary bladder) should be transferred to the Petri dish with the GI tract for further inspection.
Dissection of the GI tract begins with separating the major sections of the gut (stomach, small intestine, large intestine), followed by careful straightening of each organ using forceps to strip off the mesenteries that bind them. Care must be taken not to pinch the GI tract with forceps, as it may damage larger worms (e.g., cestodes and nematodes). Each organ should then be dissected separately to ensure that the anatomical origin of any collected parasites can be traced correctly. Each section of the GI tract should be opened lengthwise beginning at the posterior end and working anteriorly, occasionally washing the gut contents with 0.9% saline while exercising care to avoid damaging emerging helminths (nematodes, cestodes, trematodes, acanthocephalans). Once opened, the gut lining and any adhering parasites should be scraped off the muscle wall of the gut by pulling the gut wall beneath the end of a microscope slide that is pressed against the floor of the Petri dish (Tkach et al. 2019). Parasites should be transferred to a clean Petri dish using either a transfer pipette or by scooping from beneath (never pinch helminths in forceps) and rinsed clean with ample saline.
The most critical element of the field necropsy protocol for ensuring the collection of good quality parasite material is handling of helminth specimens prior to preservation. Although in most cases helminths will ultimately be preserved in ethanol, they must first be relaxed and euthanized using appropriate methods. Depositing a live helminth directly into ethanol induces muscle contractions that distort the specimen, often rendering it useless for morphological examination. To relax and euthanize helminths prior to preservation, we recommend the following methods (described in full detail in Supplementary Data SD1; also see Lutz et al. 2017; Tkach et al. 2019). Douse trematodes and cestodes in hot (steaming, but not boiling) freshwater. Especially large and robust cestodes can alternatively be held in freshwater until death occurs by osmotic shock, but this method will lead to degradation of small cestodes, for which heat-killing is preferred. Douse nematodes in hot saline. Hold acanthocephalans in freshwater until death occurs by osmotic shock. Once euthanized, helminths should be placed in labeled vials with 80% ethanol.
In addition to this streamlined protocol for routine collection of ectoparasites and endoparasites, other methods can be incorporated into field collection protocols to target specific parasites and pathogens (see Supplementary Data SD1). For example, adult tissue-dwelling nematodes of large mammals (e.g., artiodactyls) are difficult to sample directly, but larvae are easily collected from feces using a modified beaker-Baermann method (Forrester and Lankester 1997). To build an archive of samples for screening blood-borne pathogens, thin blood smears are not difficult to prepare in the field from freshly euthanized mammals. Whole blood samples for a variety of downstream applications can be easily collected and archived using FTA paper and blood serum can be collected using Nobuto strips, which allow tests of the presence of antibodies associated with various microbial pathogens (Nobuto 1963; Dusek et al. 2011). Flash-freezing tissues in liquid nitrogen ensure the highest quality preservation of host, parasite, and pathogen DNA and RNA. For RNA viruses (e.g., hantaviruses in lung tissue), RNAlater solution offers an alternative tool for preservation when cryogenic storage is unavailable, though care should be taken to follow the manufacturer’s recommendations for proper use to ensure long-term RNA preservation. Preservation of whole or partial guts, or fecal material, in ≥ 95% ethanol or frozen can provide options for using metagenomic methods to detect both macro- and microparasites and other symbionts (Greiman et al. 2018).
Conclusion
For integrated collections to meet their potential as resources for investigating complex ecological interactions between mammals and parasites, information on linkages between specimens and all associated data must be accessible to the scientific community. Archiving these specimens and data in natural history museum collections is a necessary first step, but functionality as a resource for research requires that the interconnecting relationships be traceable via globally accessible databases. The large collection of mammal and parasite specimens amassed by Robert and Virginia Rausch over six decades of fieldwork in Alaska, Siberia, and elsewhere (Hoberg 2014), exemplifies both the opportunities and challenges of curating and disseminating data associated with integrated mammal–parasite collections. This collection, now archived at the Museum of Southwestern Biology, includes approximately 6,000 mammal specimens and tens of thousands of associated parasite specimens.
Relationships between host and parasite data in the Rausch Helminthological Collection are tracked via the Arctos database (arctos.database.museum), which allows specimen records linked across separate collections and different museums to be discoverable through a single data portal. Though data entry continues for this large collection, to date > 13,000 parasite records and associated host data are electronically linked. Roughly 1,000 Rausch parasite voucher specimens are now linked to their mammal voucher specimens distributed across four different institutions, including the Museum of Southwestern Biology, University of Alaska Museum of the North (Fairbanks, Alaska), Museum of Vertebrate Zoology (Berkeley, California), and University of Colorado Museum of Natural History (Boulder, Colorado). Thus, modern museum database systems now permit investigators to extract information on relationships among specimens, specimen parts, geographic localities, dates of collection, and other data associated with each collecting event (Dunnum et al. 2017). More broadly within the Arctos database network, > 34,000 parasite records (both voucher and documented observations) are linked to physical host specimens and nearly 23,000 host specimens are linked to one or more parasite records distributed across multiple institutions. As more mammalogists collect and archive parasites along with specimens of their primary study organisms, this resource will grow at an unprecedented rate. Future investment in museum database infrastructure should emphasize enhancements to tools for analyzing relational data to yield new insights into geographic and temporal patterns of interaction among hosts and parasites. Such a resource for biodiversity informatics will open doors to new computational approaches for investigating patterns and processes that have shaped interspecific interactions over space and time.
As naturalists who are routinely engaged in scientific collecting of wild mammals, mammalogists have a unique opportunity to build both the mammalogical and parasitological records, which will create new opportunities for collaboration and discovery. By embracing a holistic sampling paradigm, we can develop integrated data sets of diverse host materials, their symbionts, and associated data. To address broad-scale questions regarding ecological dynamics and evolutionary processes that have structured faunas across the globe, the integrated sampling model must be extended to the remote regions of the planet through ambitious and intensive field collection programs (e.g., Cook et al. 2017). Documenting the identities, distributions, and interactions of mammals, parasites, and pathogens in poorly studied regions will further lay the foundation for understanding the effects of climate change and other anthropogenically driven disruptions that will have important consequences for local populations of people, domestic animals, and wildlife (Hoberg et al. 2015; DiEuliis et al. 2016). To build this global specimen base, it is imperative that we reach across taxonomic, disciplinary, and international boundaries to establish strong cooperative networks that unite diverse expertise and experience. In particular, international partnerships that leverage strengths and contributions of collaborators to yield mutually advantageous outcomes will be critical for success (Grieneisen et al. 2014; Dangles et al. 2016). If implemented widely, the protocols described here (and in Supplementary Data SD1) will yield a rich resource for parasitological investigations that will launch a new phase of discovery regarding the intersection of ecology, evolution, geography, hosts, parasites, pathology, and public health.
Supplementary Data
Supplementary data are available at Journal of Mammalogy online.
Supplementary Data SD1.—Stand-alone field manual describing protocols for mammal necropsy and parasite collection and preservation.
Supplementary Material
Acknowledgments
This paper has its origins in the workshop in field parasitology that was held during the 2018 Annual Meeting of the American Society of Mammalogists at Kansas State University. We extend our sincere appreciation to Dr. M. Dryden, Dr. B. Herrin, and the Kansas State University College of Veterinary Medicine for hosting and providing facilities for the workshop. We also thank all the workshop participants, whose enthusiasm for learning parasitological methods bodes well for a future in which integrated collections of mammal–parasite assemblages become standard practice. The ideas and methods that we describe here have been influenced by numerous colleagues, including many whose participation in past NSF-funded field expeditions (to JAC and EPH: DEB 9972154, 0196095, 0415668, 1258010; to JAC and SLG: DEB 0717214; to SLG: BSR 8612329, 9024816; to KEG: DEB 1256943; to SEG: DBI 1523410) helped us to identify strategies for enhancing efficiency and productivity on the processing line. We appreciate their many contributions over the years. We additionally acknowledge support from FONDECYT to FT-P (1171280), from Sistema Nacional de Investigación (SNI) SENACYT to BA, and from NSF-DUE 1564969 to FAJ. Additional support for this project was provided by the Kansas State Division of Biology to AGH and the Department of Biology and College of Arts and Sciences at Northern Michigan University to KEG. Finally, our work owes a great debt to Robert and Virginia Rausch, whose vision and leadership in advancing studies of mammals and their parasites provided us with an extraordinary legacy and continuing model for integrated research.
Literature Cited
- Altizer S., R. S. Ostfeld P. T. Johnson S. Kutz, and Harvell C. D.. 2013. Climate change and infectious diseases: from evidence to a predictive framework. Science 341:514–519. [DOI] [PubMed] [Google Scholar]
- Amato G., and DeSalle R.. 2012. Assessing biodiversity funding during the sixth extinction. BioEssays 34:658–660. [DOI] [PubMed] [Google Scholar]
- Anderson S. 1997. Mammals of Bolivia: taxonomy and distribution. Bulletin of the American Museum of Natural History 231:1–652. [Google Scholar]
- Arai S.,, et al. 2007. Hantavirus in northern short-tailed shrew, United States. Emerging Infectious Diseases 13:1420–1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araujo S. B.,, et al. 2015. Understanding host-switching by ecological fitting. PLoS One 10:e0139225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnosky A. D.,, et al. 2012. Approaching a state shift in Earth’s biosphere. Nature 486:52–58. [DOI] [PubMed] [Google Scholar]
- Bell K. C., C. J. Carlson, and Phillips A. J.. 2018. Parasite collections: overlooked resources for integrative research and conservation. Trends in Parasitology 34:637–639. [DOI] [PubMed] [Google Scholar]
- Bi K., T. Linderoth D. Vanderpool J. M. Good R. Nielsen, and Moritz C.. 2013. Unlocking the vault: next-generation museum population genomics. Molecular Ecology 22:6018–6032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley R. D., Bradley L. C., Garner H. J., and Baker R. J.. 2012. Cost of collection and preparing mammal voucher specimens for natural history collections. Occasional Papers Museum of Texas Tech University 313:1–14. [Google Scholar]
- Brooks D. R., and Hoberg E. P.. 2000. Triage for the biosphere: the need and rationale for taxonomic inventories and phylogenetic studies of parasites. Comparative Parasitology 67:1–25. [Google Scholar]
- Brooks D. R., and Hoberg E. P.. 2013. The emerging infectious diseases crisis and pathogen pollution: a question of ecology and evolution. Pp. 215–229 in The balance of nature and human impact (Rohde K., ed.). Cambridge University Press, Cambridge, United Kingdom. [Google Scholar]
- Brooks D. R., Hoberg E. P., and Boeger W. A.. 2019. The Stockholm paradigm, climate change and emerging disease. University of Chicago Press, Chicago, Illinois. [Google Scholar]
- Brooks D. R., et al. 2014. Finding them before they find us: informatics, parasites, and environments in accelerating climate change. Comparative Parasitology 81:155–164. [Google Scholar]
- Capinha C., F. Essl H. Seebens D. Moser, and Pereira H. M.. 2015. BIOGEOGRAPHY. The dispersal of alien species redefines biogeography in the Anthropocene. Science 348:1248–1251. [DOI] [PubMed] [Google Scholar]
- Ceballos G., Ehrlich P. R., and Dirzo R.. 2017. Biological annihilation via the ongoing sixth mass extinction signaled by vertebrate population losses and declines. Proceedings of the National Academy of Sciences 114:E6089–E6096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleaveland S., M. K. Laurenson, and Taylor L. H.. 2001. Diseases of humans and their domestic mammals: pathogen characteristics, host range and the risk of emergence. Philosophical Transactions of the Royal Society of London, B. Biological Sciences 356:991–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook J. A., et al. 2013. Genetics. Pp. 514–539 in Arctic biodiversity assessment: status and trends in Arctic biodiversity (Meltofte H., ed.). Conservation of Arctic Floral and Fauna, Arctic Council, Akureyi, Iceland. [Google Scholar]
- Cook J. A., et al. 2017. The Beringian Coevolution Project: holistic collections of mammals and associated parasites reveal novel perspectives on evolutionary and environmental change in the North. Arctic Science 3:585–617. [Google Scholar]
- Cook J. A., et al. 2016. Transformational principles for NEON sampling of mammalian parasites and pathogens: a response to Springer and colleagues. BioScience 66:917–919. [Google Scholar]
- Cook J. A., et al. 2005. Beringia: intercontinental exchange and diversification of high latitude mammals and their parasites during the Pliocene and Quaternary. Mammal Study 30:S33–S44. [Google Scholar]
- Cook J. A., and Light J. E.. 2019. The emerging role of mammal collections in 21st century mammalogy. Journal of Mammalogy. [Google Scholar]
- Cook J. A., and MacDonald S. O.. 2013. Island life: coming to grips with the insular nature of North Pacific coastal forests. Pp. 19–42 in Conservation of North Pacific coastal forests (Orians G. H. and Schoen J. W., eds.). University of Washington Press, Seattle, Washington. [Google Scholar]
- Criscione C. D., R. Poulin, and Blouin M. S.. 2005. Molecular ecology of parasites: elucidating ecological and microevolutionary processes. Molecular Ecology 14:2247–2257. [DOI] [PubMed] [Google Scholar]
- Dangles O., J. Loirat C. Freour S. Serre J. Vacher, and Le Roux X.. 2016. Research on biodiversity and climate change at a distance: collaboration networks between Europe and Latin America and the Caribbean. PLoS One 11:e0157441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daszak P., A. A. Cunningham, and Hyatt A. D.. 2000. Emerging infectious diseases of wildlife–threats to biodiversity and human health. Science 287:443–449. [DOI] [PubMed] [Google Scholar]
- Dick C. W., Gettinger D., and Gardner S. L.. 2007. Bolivian ectoparasites: a survey of bats (Mammalia: Chiroptera). Comparative Parasitology 74:372–378. [Google Scholar]
- DiEuliis D., Johnson K. R., Morse S. S., and Schindel D. E.. 2016. Opinion: specimen collections should have a much bigger role in infectious disease research and response. Proceedings of the National Academy of Sciences 113:4–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobson A., Lafferty K. D., Kuris A. M., Hechinger R. F., and Jetz W.. 2008. Homage to Linnaeus: how many parasites? How many hosts? Proceedings of the National Academy of Sciences 105:11482–11489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunnum J. L., B. S., McLean R. C., Dowler, and Systematic Collections Committee of the American Society of Mammalogists 2018. Mammal collections of the Western Hemisphere: a survey and directory of collections. Journal of Mammalogy 99:1307–1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunnum J. L.,, et al. 2017. Biospecimen repositories and integrated databases as critical infrastructure for pathogen discovery and pathobiology research. PLoS Neglected Tropical Diseases 11:e0005133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durette-Desset M. C., K. E. Galbreath, and Hoberg E. P.. 2010. Discovery of new Ohbayashinema spp. (Nematoda: Heligmosomoidea) in Ochotona princeps and Ochotona cansus (Lagomorpha: Ochotonidae) from western North America and Central Asia, with considerations of historical biogeography. The Journal of Parasitology 96:569–579. [DOI] [PubMed] [Google Scholar]
- Dursahinhan A. T., Nyamsuren B., Tufts D. M., and Gardner S. L.. 2017. A new species of Catenotaenia (Cestoda: Catenotaeniidae) from Pygeretmus pumilio Kerr, 1792 from the Gobi of Mongolia. Comparative Parasitology 84:124–135. [Google Scholar]
- Dusek R. J., J. S. Hall S. W. Nashold J. L. TeSlaa, and Ip H. S.. 2011. Evaluation of Nobuto filter paper strips for the detection of avian influenza virus antibody in waterfowl. Avian Diseases 55:674–676. [DOI] [PubMed] [Google Scholar]
- Duszynski D. W., Eastham G., and Yates T. L.. 1982. Eimeria from jumping mice (Zapus spp.): a new species and genetic and geographic features of Z. hudsonius luteus. Journal of Parasitology 68:1146–1148. [Google Scholar]
- Forrester S. G., and Lankester M. W.. 1997. Extracting protostrongylid nematode larvae from ungulate feces. Journal of Wildlife Diseases 33:511–516. [DOI] [PubMed] [Google Scholar]
- Frey J. K., Yates T. L., Duszynski D. W., Gannon W. L., and Gardner S. L.. 1992. Designation and curatorial management of type host specimens (symbiotypes) for new parasite species. Journal of Parasitology 78:930–932. [Google Scholar]
- Galbreath K. E., D. J. Hafner, and Zamudio K. R.. 2009. When cold is better: climate-driven elevation shifts yield complex patterns of diversification and demography in an alpine specialist (American pika, Ochotona princeps). Evolution 63:2848–2863. [DOI] [PubMed] [Google Scholar]
- Galbreath K. E., and Hoberg E. P.. 2012. Return to Beringia: parasites reveal cryptic biogeographic history of North American pikas. Proceedings of the Royal Society of London, B. Biological Sciences 279:371–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galbreath K. E., and Hoberg E. P.. 2015. Host responses to cycles of climate change shape parasite diversity across North America’s Intermountain West. Folia Zoologica 64:218–232. [Google Scholar]
- Gardner S. L. 1996. Field parasitology techniques for use with mammals. Pp. 291–298 in Measuring and monitoring biological diversity: standard methods for mammals (Wilson D. E., Cole F. R., Nichols J. D., Rudran R., and Foster M. S., eds.). Smithsonian Institution Press, Washington, D.C. [Google Scholar]
- Gardner S. L. 2014. New species of Ctenomys Blainville 1826: (Rodentia: Ctenomyidae) from the lowlands and central valleys of Bolivia. Special Publication, Museum of Texas Tech University 62:1–34. [Google Scholar]
- Gardner S., et al. 2013a. Sylvatic species of Echinococcus from rodent intermediate hosts in Asia and South America. Occasional Papers, Museum of Texas Tech University 318:1–13. [Google Scholar]
- Gardner S. L., and Campbell M. L.. 1992. Parasites as probes for biodiversity. The Journal of Parasitology 78:596–600. [PubMed] [Google Scholar]
- Gardner S. L., and Jiménez-Ruiz F. A.. 2009. Methods of endoparasite analysis. Pp. 795–805 in Ecological and behavioral methods for the study of bats (Kunz T. and Parsons S., eds.). Johns Hopkins University Press, Baltimore, Maryland. [Google Scholar]
- Gardner S. L., Jiménez-Ruiz F. A., and Campbell M. L.. 2013b. Pritchardia boliviensis n. gen., n. sp. (Anoplocephalidae: Linstowinae), a tapeworm from opossums (Didelphidae) in the Yungas and lowlands of Bolivia and Atlantic Forest of Paraguay. Occasional Papers, Museum of Texas Tech University 319:1–8. [Google Scholar]
- Glass G. E., et al. 2002. Satellite imagery characterizes local animal reservoir populations of Sin Nombre virus in the southwestern United States. Proceedings of the National Academy of Sciences 99:16817–16822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greiman S. E.,, et al. 2018. Museum metabarcoding: a novel method revealing gut helminth communities of small mammals across space and time. International Journal for Parasitology 48:1061–1070. [DOI] [PubMed] [Google Scholar]
- Grieneisen M. L., Zhan Y. U., Potter D., and Zhang M.. 2014. Biodiversity, taxonomic infrastructure, international collaboration, and new species discovery. BioScience 64:322–332. [Google Scholar]
- Grinnell J. 1910. The methods and uses of a research museum. Popular Science Monthly 77:163–169. [Google Scholar]
- Harvell C. D.,, et al. 2002. Climate warming and disease risks for terrestrial and marine biota. Science 296:2158–2162. [DOI] [PubMed] [Google Scholar]
- Hoberg E. 1997. Phylogeny and historical reconstruction: host-parasite systems as keystones in biogeography and ecology. Pp. 243–261 in Biodiversity II (Reaka-Kudla M. L., Wilson D. E., and Wilson E. O., eds.). Joseph Henry Press, Washington, D.C. [Google Scholar]
- Hoberg E. P. 2002. Foundations for an integrative parasitology: collections, archives, and biodiversity informatics. Comparative Parasitology 69:124–131. [Google Scholar]
- Hoberg E. P. 2014. Robert Lloyd Rausch–a life in nature and field biology: 1921-2012. The Journal of Parasitology 100:547–552. [DOI] [PubMed] [Google Scholar]
- Hoberg E. P., S. J. Agosta W. A. Boeger, and Brooks D. R.. 2015. An integrated parasitology: revealing the elephant through tradition and invention. Trends in Parasitology 31:128–133. [DOI] [PubMed] [Google Scholar]
- Hoberg E. P., and Brooks D. R.. 2008. A macroevolutionary mosaic: episodic host-switching, geographical colonization and diversification in complex host-parasite systems. Journal of Biogeography 35:1533–1550. [Google Scholar]
- Hoberg E. P., and Brooks D. R.. 2015. Evolution in action: climate change, biodiversity dynamics and emerging infectious disease. Philosophical Transactions of the Royal Society of London, B. Biological Sciences 370:20130553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoberg E. P., Galbreath K. E., Cook J. A., Kutz S. J., and Polley L.. 2012a. Northern host–parasite assemblages: history and biogeography on the borderlands of episodic climate and environmental transition. Advances in Parasitology 79:1–97. [DOI] [PubMed] [Google Scholar]
- Hoberg E. P., Koehler A. V. A., And Cook J.. 2012. Complex host-parasite systems in Martes: implications for conservation biology of endemic faunas. Pp. 39–57 in Biology and conservation of Martens, Sables and Fishers: a new synthesis (Aubry K. B., Zielinski W. J., Raphael M. G., Proulx G., and Buskirk S. W., eds.). Cornell University Press, Ithaca, New York. [Google Scholar]
- Hoberg E. P., et al. 2013. Parasites in terrestrial, freshwater and marine systems. Pp. 476–505 in Arctic biodiversity assessment: status and trends in Arctic biodiversity (Meltofte H., ed.). Conservation of Arctic Floral and Fauna, Arctic Council, Akureyi, Iceland. [Google Scholar]
- Hoberg E. P., Kutz S. J., Galbreath K. E., and Cook J. A.. 2003. Arctic biodiversity: from discovery to faunal baselines - revealing the history of a dynamic ecosystem. Journal of Parasitology 89(Suppl.):S84–S95. [Google Scholar]
- Hoberg E. P., P. A. Pilitt, and Galbreath K. E.. 2009. Why museums matter: a tale of pinworms (Oxyuroidea: Heteroxynematidae) among pikas (Ochotona princeps and O. collaris) in the American West. The Journal of Parasitology 95:490–501. [DOI] [PubMed] [Google Scholar]
- Hoberg E. P., L. Polley E. J. Jenkins S. J. Kutz A. M. Veitch, and Elkin B. T.. 2008. Integrated approaches and empirical models for investigation of parasitic diseases in northern wildlife. Emerging Infectious Diseases 14:10–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes E. C. 2013. What can we predict about viral evolution and emergence? Current Opinion in Virology 3:180–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hope A. G. In press. Arctic tundra mammals. In Encyclopedia of the world’s biomes (Elias S., ed.). [Google Scholar]
- Hope A., Greiman S., Tkach V., Hoberg E. P., and Cook J.. 2016. Shrews and their parasites: small species indicate big changes. Arctic Report Card. www.arctic.noaa.gov/Report-Card. [Google Scholar]
- Hope A. G., Sandercock B. K., and Malaney J. L.. 2018. Collection of scientific specimens: benefits for biodiversity sciences and limited impacts on communities of small mammals. BioScience 68:35–42. [Google Scholar]
- Jenkins E. J.,, et al. 2005. Geographic distribution of the muscle-dwelling nematode Parelaphostrongylus odocoilei in North America, using molecular identification of first-stage larvae. The Journal of Parasitology 91:574–584. [DOI] [PubMed] [Google Scholar]
- Jenkins E. J.,, et al. 2013. Tradition and transition: parasitic zoonoses of people and animals in Alaska, northern Canada, and Greenland. Advances in Parasitology 82:33–204. [DOI] [PubMed] [Google Scholar]
- Kang H. J., S. H. Gu J. A. Cook, and Yanagihara R.. 2016. Dahonggou Creek virus, a divergent lineage of hantavirus harbored by the long-tailed mole (Scaptonyx fusicaudus). Tropical Medicine and Health 44:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemp C. 2017. The lost species: great expeditions in the collections of natural history museums. University of Chicago Press, Chicago, Illinois. [Google Scholar]
- Koehler A. V., E. P. Hoberg N. E. Dokuchaev, and Cook J. A.. 2007. Geographic and host range of the nematode Soboliphyme baturini across Beringia. The Journal of Parasitology 93:1070–1083. [DOI] [PubMed] [Google Scholar]
- Koehler A. V., et al. 2009. Phylogeography of a Holarctic nematode, Soboliphyme baturini, among mustelids: climate change, episodic colonization, and diversification in a complex host-parasite system. Biological Journal of the Linnean Society 96:651–663. [Google Scholar]
- Kutz S. J., et al. 2007. Serendipitous discovery of a novel protostrongylid (Nematoda: Metastrongyloidea) in caribou, muskoxen, and moose from high latitudes of North America based on DNA sequence comparisons. Canadian Journal of Zoology 85:1143–1156. [Google Scholar]
- Kutz S. J.,, et al. 2013. Invasion, establishment, and range expansion of two parasitic nematodes in the Canadian Arctic. Global Change Biology 19:3254–3262. [DOI] [PubMed] [Google Scholar]
- Kutz S. J., Hoberg E. P., Polley L., and Jenkins E. J.. 2005. Global warming is changing the dynamics of Arctic host-parasite systems. Proceedings of the Royal Society of London, B. Biological Sciences 272:2571–2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafferty K. D.,, et al. 2008. Parasites in food webs: the ultimate missing links. Ecology Letters 11:533–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert C. R., S. L. Gardner, and Duszynski D. W.. 1988. Coccidia (Apicomplexa: Eimeriidae) from the subterranean rodent Ctenomys opimus Wagner (Ctenomyidae) from Bolivia, South America. The Journal of Parasitology 74:1018–1022. [PubMed] [Google Scholar]
- Lamothe-Argumedo R., Damborenea C., García-Prieto L., Lunaschi L. I., and Osorio-Sarabia D.. 2010. Guide to helminthological collections of Latin America. Instituto De Biologia, Universidad Nacional Autónoma De México, and Museo De La Plata, Universidad De La Plata, Argentina. [Google Scholar]
- Lawler J. J.,, et al. 2009. Projected climate-induced faunal change in the Western Hemisphere. Ecology 90:588–597. [DOI] [PubMed] [Google Scholar]
- Lichtenfels J. R., and Pritchard M. H.. 1982. A guide to the parasite collections of the world. Allen Press, Lawrence, Kansas. [Google Scholar]
- Lutz H. L., Tkach V. V., and Weckstein J. D.. 2017. Methods for specimen-based studies of avian symbionts. Pp. 157–183 in The extended specimen: emerging frontiers in collections-based ornithological research (Webster M. S., ed.). CRC Press, Boca Raton, Florida. [Google Scholar]
- Makarikov A. A., K. E. Galbreath, and Hoberg E. P.. 2013. Parasite diversity at the Holarctic nexus: species of Arostrilepis (Eucestoda: Hymenolepididae) in voles and lemmings (Cricetidae: Arvicolinae) from greater Beringia. Zootaxa 3608:401–439. [DOI] [PubMed] [Google Scholar]
- Malaney J. L., and Cook J. A.. 2018. A perfect storm for mammalogy: declining sample availability in a period of rapid environmental degradation. Journal of Mammalogy 99:773–788. [Google Scholar]
- Manter H. W. 1966. Parasites of fishes as biological indicators of recent and ancient conditions. Pp. 59–71 in Host-parasite relationships (McCauley J. E., ed.). Oregon State University Press, Corvallis, Oregon. [Google Scholar]
- McLean B. S.,, et al. 2016. Natural history collections-based research: progress, promise, and best practices. Journal of Mammalogy 97:287–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison S. A., T. S. Sillett W. C. Funk C. K. Ghalambor, and Rick T. C.. 2017. Equipping the 22nd-century historical ecologist. Trends in Ecology & Evolution 32:578–588. [DOI] [PubMed] [Google Scholar]
- Nobuto K. 1963. Toxoplasmosis in animal and laboratory diagnosis. Proceedings of the Society of Plant Protection of North Japan 14:45–46. [Google Scholar]
- Nowogrodzki A. 2016. Biological specimen troves threatened by funding pause. Nature 531:561. [DOI] [PubMed] [Google Scholar]
- Parmesan C., and Yohe G.. 2003. A globally coherent fingerprint of climate change impacts across natural systems. Nature 421:37–42. [DOI] [PubMed] [Google Scholar]
- Parrish C. R.,, et al. 2008. Cross-species virus transmission and the emergence of new epidemic diseases. Microbiology and Molecular Biology Reviews 72:457–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pecl G. T., et al. 2017. Biodiversity redistribution under climate change: impacts on ecosystems and human well-being. Science 355:eaai9214. [DOI] [PubMed] [Google Scholar]
- Pritchard M. H., and Kruse G. O.. 1982. The collection and preservation of animal parasites. University of Nebraska Press, Lincoln, Nebraska. [Google Scholar]
- Pybus O. G., Tatem A. J., and Lemey P.. 2015. Virus evolution and transmission in an ever more connected world. Proceedings of the Royal Society of London, B. Biological Sciences 282:20142878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rausch R. 1952. Studies on the helminth fauna of Alaska. XI. Helminth parasites of microtine rodents; taxonomic considerations. The Journal of Parasitology 38:415–444. [PubMed] [Google Scholar]
- Rausch R. 1956. Studies on the helminth fauna of Alaska. XXX. The occurrence of Echinococcus multilocularis Leuckart, 1863, on the mainland of Alaska. The American Journal of Tropical Medicine and Hygiene 5:1086–1092. [DOI] [PubMed] [Google Scholar]
- Rausch R. L. 1957. Distribution and specificity of helminths in microtine rodents: evolutionary implications. Evolution 11:361–368. [Google Scholar]
- Rausch R. L. 1972. Observations on some natural-focal zoonoses in Alaska. Archives of Environmental Health 25:246–252. [DOI] [PubMed] [Google Scholar]
- Rausch R. L. 1983. The biology of avian parasites: helminths. Pp. 367–442 in Avian biology (Farner D. S., King J. R., and Parkes K. C., eds.). Academic Press, New York, New York. [Google Scholar]
- Rausch R. L. 1994. Transberingian dispersal of cestodes in mammals. International Journal for Parasitology 24:1203–1212. [DOI] [PubMed] [Google Scholar]
- Salazar-Bravo J., J. W. Dragoo M. D. Bowen C. J. Peters T. G. Ksiazek, and Yates T. L.. 2002. Natural nidality in Bolivian hemorrhagic fever and the systematics of the reservoir species. Infection, Genetics and Evolution 1:191–199. [DOI] [PubMed] [Google Scholar]
- Schindel D. E., and Cook J. A.. 2018. The next generation of natural history collections. PLoS Biology 16:e2006125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffen W., et al. 2018. Trajectories of the Earth system in the Anthropocene. Proceedings of the National Academy of Sciences 115:8252–8259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiee M. S., R. J. Harrigan H. A. Thomassen, and Smith T. B.. 2018. Ghosts of infections past: using archival samples to understand a century of monkeypox virus prevalence among host communities across space and time. Royal Society Open Science 5:171089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinnin D. S., Jensen E., and Gardner S. L.. 2012. Coccidia (Apicomplexa: Eimeriidae) from Vespertilio murinus and Eptesicus gobiensis (Chiroptera: Vespertilionidae) in Mongolia and how many species of Coccidia occur in bats? Erforschung Biologischer Ressourcen Der Mongolei (Halle/Saale) 12:117–124. [Google Scholar]
- Tkach V. V., Hope A. G., and Greiman S. E.. 2019. Method for the rapid fixation of gastrointestinal helminths in small mammals. Acta Parasitologica. [DOI] [PubMed] [Google Scholar]
- Torres-Pérez F., R. E. Palma B. Hjelle E. C. Holmes, and Cook J. A.. 2011. Spatial but not temporal co-divergence of a virus and its mammalian host. Molecular Ecology 20:4109–4122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsangaras K., and Greenwood A. D.. 2012. Museums and disease: using tissue archive and museum samples to study pathogens. Annals of Anatomy - Anatomischer Anzeiger 194:58–73. [DOI] [PubMed] [Google Scholar]
- Verde Arregoitia L. D., Cooper N., and D’Elía G.. 2018. Good practices for sharing analysis-ready data in mammalogy and biodiversity research. Hystrix, the Italian Journal of Mammalogy 29:155–161. [Google Scholar]
- Verocai G. G., S. J. Kutz M. Simard, and Hoberg E. P.. 2014. Varestrongylus eleguneniensis sp. n. (Nematoda: Protostrongylidae): a widespread, multi-host lungworm of wild North American ungulates, with an emended diagnosis for the genus and explorations of biogeography. Parasites & Vectors 7:556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wake D. B., and Vredenburg V. T.. 2008. Are we in the midst of the sixth mass extinction? A view from the world of amphibians. Proceedings of the National Academy of Sciences 105:11466–11473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wash C. D., D. W. Duszynski, and Yates T. L.. 1985. Eimerians from different karyotypes of the Japanese wood mouse (Apodemus spp.), with descriptions of two new species and a redescription of Eimeria montgomeryae Lewis and Ball, 1983. The Journal of Parasitology 71:808–814. [PubMed] [Google Scholar]
- Woolhouse M. E., and Gowtage-Sequeria S.. 2005. Host range and emerging and reemerging pathogens. Emerging Infectious Diseases 11:1842–1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates T. L. 1996. Tissues, cell suspensions and chromosomes. Pp. 275–278 in Measuring and monitoring biological diversity - standard methods for mammals (Wilson D. E., Cole F. R., Nichols J. D., Rudran R., and Foster M. S., eds.). Smithsonian Institution Press, Washington, D.C. [Google Scholar]
- Yates T. L., Jones C., and Cook J. A.. 1996. Preservation of voucher specimens. Pp. 265–273 in Measuring and monitoring biological diversity - standard methods for mammals (Wilson D. E., Cole F. R., Nichols J. D., Rudran R., and Foster M. S., eds.). Smithsonian Institution Press, Washington, D.C. [Google Scholar]
- Yates T. L., et al. 2002. The ecology and evolutionary history of an emergent disease: hantavirus pulmonary syndrome. BioScience 52:989–998. [Google Scholar]
- Zinovieva S. V., et al. 2015. World collections of parasitic worms. Biology Bulletin 42:540–545. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.