Abstract
Objective
Endurance exercise training remodels skeletal muscle, leading to increased mitochondrial content and oxidative capacity. How exercise entrains skeletal muscle signaling pathways to induce adaptive responses remains unclear. In past studies, we identified Perm1 (PGC-1 and ERR induced regulator, muscle 1) as an exercise-induced gene and showed that Perm1 overexpression elicits similar muscle adaptations as endurance exercise training. The mechanism of action and the role of Perm1 in exercise-induced responses are not known. In this study, we aimed to determine the pathway by which Perm1 acts as well as the importance of Perm1 for acute and long-term responses to exercise.
Methods
We performed immunoprecipitation and mass spectrometry to identify Perm1 associated proteins, and validated Perm1 interactions with the Ca2+/calmodulin-dependent protein kinase II (CaMKII). We also knocked down Perm1 expression in gastrocnemius muscles of mice via AAV-mediated delivery of shRNA and assessed the impact of reduced Perm1 expression on both acute molecular responses to a single treadmill exercise bout and long-term adaptive responses to four weeks of voluntary wheel running training. Finally, we asked whether Perm1 levels are modulated by diet or diseases affecting skeletal muscle function.
Results
We show that Perm1 associates with skeletal muscle CaMKII and promotes CaMKII activation. In response to an acute exercise bout, muscles with a knock down of Perm1 showed defects in the activation of CaMKII and p38 MAPK and blunted induction of regulators of oxidative metabolism. Following four weeks of voluntary training, Perm1 knockdown muscles had attenuated mitochondrial biogenesis. Finally, we found that Perm1 expression is reduced in diet-induced obese mice and in muscular dystrophy patients and mouse models.
Conclusions
Our findings identify Perm1 as a muscle-specific regulator of exercise-induced signaling and Perm1 levels as tuners of the skeletal muscle response to exercise. The decreased Perm1 levels in states of obesity or muscle disease suggest that Perm1 may link pathological states to inefficient exercise responses.
Keywords: Skeletal muscle, Endurance exercise training, Mitochondrial biogenesis, CaMKII signaling, p38 MAPK regulation
Highlights
-
•
Perm1 interacts with CaMKII and activates the CaMKII-MEF2 pathway.
-
•
Perm1 is important for CaMKII activation and PGC-1α induction by an exercise bout.
-
•
In endurance training, Perm1 impacts muscle oxidative metabolism pathway responses.
-
•
Skeletal muscle levels of Perm1 are reduced in obesity and muscular dystrophy.
1. Introduction
Skeletal muscle plays key roles in metabolic homeostasis and whole body energy expenditure. Decreased physical activity is a risk factor for obesity, type 2 diabetes, and cardiovascular diseases [1]. Conversely, regular exercise, particularly when combined with dietary interventions, is effective in the prevention and treatment of type 2 diabetes and cardiovascular diseases [2], [3]. Notably, not all individuals show effective responses to a given exercise training program, indicating a high degree of variation in the pathways activated by exercise [4]. Understanding the mechanisms that determine responses to exercise is vital in finding ways to tailor exercise training and maximize exercise benefits.
Endurance exercise training remodels skeletal muscle by activating signaling pathways and regulating transcription factors that drive long-term changes in gene expression, thereby leading to increases in mitochondrial oxidative capacity and alterations in metabolic and contractile properties. Mediators of the adaptive responses to exercise include the kinases CaMKII, MAPK p38, and AMPK and the transcriptional regulators PGC-1α, PPARδ, and MEF2 [5], [6]. Notably, there is little known on muscle-specific proteins that may regulate these signaling pathways and shape responses to endurance training bouts.
The family of CaMKII kinases comprises of four distinct genes (Camk2a-d) that are widely expressed and play central roles in Ca2+ signaling pathways in many tissues, including learning and memory in the CNS, contractility in heart, and insulin secretion in pancreatic β-cells [7]. In skeletal muscle, CaMKIIβ, γ, and δ are auto-phosphorylated and activated in response to exercise [8]. CaMKII then activates MEF2-dependent transcription, inducing the expression of PGC-1α and PGC-1α driven pathways, such as mitochondrial biogenesis [9], [10], [11]. Thus, CaMKII, together with the Ca2+ -induced phosphatase calcineurin, plays a major role in transducing muscle excitation signals to changes in gene expression, a process known as “excitation-transcription coupling” [5].
Perm1, a recently identified protein, is selectively expressed in skeletal and cardiac muscles, enriched in oxidative muscle fibers, and induced by PGC-1α, Estrogen related receptors (ERRs), and endurance exercise [12], [13]. In cultured myotubes, Perm1 enables high respiratory capacity and PGC-1α-induced mitochondrial biogenesis [12]. In vivo, targeted overexpression of Perm1 in skeletal muscle leads to increases in mitochondrial content, oxidative capacity, and fatigue resistance, suggesting a role for Perm1 in exercise-induced adaptations [13]. The pathways by which Perm1 exerts these actions, and the importance of Perm1 for skeletal muscle responses to endurance exercise are not known.
To gain insights into the cellular function of Perm1, we sought Perm1 interacting proteins and used gain- and loss-of function approaches to probe the extent and pathway by which Perm1 impacts endurance exercise responses. We show that Perm1 regulates physical activity-dependent CaMKII and MAPK p38 signaling and is important for a subset of exercise-induced responses. Our findings suggest that Perm1 modulates excitation-transcription coupling and shapes the response to endurance exercise.
2. Materials and methods
2.1. Constructs and viral vectors
The MEF2C expression and TATA-MEF2-Luc reporter plasmids, the adenoviruses expressing LacZ and FLAG-tagged Perm1, and the AAV1-Perm1 vector have been described [10], [12], [13]. pAdlox-HA-CaMKIIβ expresses the HA-tagged human CaMKIIβ [cDNA sequence of pDONR-CAMK2B (Addgene #23820)]. The AAV1-shPerm1 vector was produced by the Vector Core of the University of Pennsylvania and expresses a short hairpin RNA against mouse Perm1 sequence AAGGAGGGCTTGAGGCATC [12].
2.2. Cell culture
C2C12, U2OS, and HEK293 cells were maintained in DMEM containing 10% fetal bovine serum at 37 °C and 5% CO2, and transfected using Polyethylenimine HCl MAX (Polysciences, Inc). C2C12 cells were differentiated as described [12].
2.3. Purification of Perm1 interacting proteins
C2C12 myotubes were infected with Ad-LacZ or Ad-FLAG-Perm1 at M.O.I of 50 [12]. Two days later, cell lysates were subjected to affinity purification, using beads coupled to anti-FLAG antibody (Clone M2, Sigma). Affinity-purified proteins were subjected to MudPIT for protein identification [14].
2.4. Immunoprecipitation and CaM affinity pull-down
For immunoprecipitation, HEK293 cells were transfected with expression vectors for HA-tagged CaMKIIβ and FLAG-tagged Perm1 or control vectors. Forty-eight hours later, cells were lysed; lysates were subjected to immunoprecipitation with FLAG antibody (Clone M2, Sigma) and immunoprecipitated proteins detected by western blotting. For the CaM affinity pull-down, GAS muscles were homogenized in lysis buffer [20 mM Tris-HCl pH 7.8, 150 mM NaCl, 1 mM Na3VO4, 1% Triton X-100, 5 μl/ml protease inhibitor cocktail P8340 (Sigma), 20 μg/ml phenylmethylsulfonyl fluoride], in the presence of 5 mM EGTA or 2 mM CaCl2, and mixed with CaM-agarose slurry (Upstate) for 3 h at 4 °C. CaM bound proteins were eluted with 10 mM EGTA, and subjected to western blotting.
2.5. MEF2 reporter assay
U2OS cells were transfected with MEF2-TATA-Luc reporter plasmid (50 ng/well), pCMV-beta-galactosidase (10 ng/well), and expression vectors for Perm1, CaMKIIβ, or control (5–20 ng/well). CaMKII inhibitor KN93 or its inert analogue KN92 were added to the media. Relative luciferase activities were determined as described [15].
2.6. Animal studies
C57BL/6J male mice were housed at 21 °C on a 12 h light-dark cycle and free access to food and water. Experimental procedures were conducted according to protocols approved by the Scripps Research Institute Institutional Animal Care and Use Committee. For intramuscular injections of AAV vectors, mice (4-week old) were anesthetized with isoflurane and injected with AAV1 vectors. For overexpression of Perm1, AAV1-LacZ or AAV1-Perm1 was injected to TA muscle, and muscles were harvested 4 weeks later, at 11 pm (active state), as described [13]. For suppression of Perm1, each mouse had the gastrocnemius (GAS) muscle of one leg injected with AAV1-shPerm1 (1.0 × 1011 viral genomes) and of the contralateral leg with AAV1-shControl (AAV1.H1.shLuc.ZsGreen). Six weeks later, mice were subjected to an acute bout of exercise on a treadmill (AEX) during the light cycle (11 am–2 pm) or given wheels for 4 weeks of voluntary wheel running training (TR). For the obese mouse model, 6-week old C57BL6/J male mice were fed for 22 weeks either control diet (10% fat, Research Diets, D12450B) or a high-fat diet (60% fat; Research Diets, D12492).
2.7. Acute exercise bout (AEX)
Mice were habituated to a motorized treadmill (Columbus Instruments, Columbus, OH) on two occasions (10 min at 10 m/min and 10° grade) prior to the experimental exercise bout. For the AEX bout, mice (fasted for 3 h) completed 55 min of running on the treadmill (10° grade) at repetitions of variable exercise intensity (10 m/min, 2.5 min; 12.5 m/min, 2.5 min; 15 m/min, 15 min; 17.5 m/min, 2.5 min; 20 m/min, 20 min; 22.5 m/min, 2.5 min; 25 m/min, 10 min). Following AEX, mice were returned to their cages without food, but with access to water, until muscle collection. Muscles were collected either immediately (0h AEX) or 3 h (3h AEX) after AEX. Sedentary (SED) littermate mice were fasted and euthanized under similar conditions.
2.8. Voluntary wheel running training (TR)
Mice were given access to running wheels for 28 days (TR, trained) with time run per day recorded via a digital recorder or assigned to a sedentary (SED) group (housed singly in similar cages as TR mice but without access to a running wheel). TR mice run on average ∼10 km per night. To minimize effects of acute exercise activity in TR mice, running wheels were removed at the end of 4 weeks and GAS muscles were harvested ∼ 48 h later, during the day (11 am–1 pm, rest state).
2.9. Histology, fiber type composition analysis, and mitochondrial enzyme activities
Samples were prepared and analyzed as described [13].
2.10. Reverse transcription and quantitative real-time PCR analysis
Total RNA was extracted from whole muscles using TRIzol reagent (Life Technologies) and cDNA was synthesized as described [16]. Relative mRNA levels were determined by quantitative PCR using cDNA, gene-specific primers (Table S1), SYBR green reagent (Affymetrix), and normalization to levels of 36B4 (Rplo) or GAPDH [16].
2.11. DNA isolation and quantification
Total DNA was prepared from whole muscles and digested with 100 μg/ml RNase A for 30 min at 37 °C. The relative copy numbers of mitochondrial and nuclear genomes were determined by quantitative PCR with primers specific to the CoxII (mitochondrial) and Dio3 (nuclear) genes [13].
2.12. Western blot and antibodies
Whole muscles were homogenized in lysis buffer containing 20 mM Tris-HCl pH 7.8, 150 mM NaCl, 1 mM Na3VO4, 5 mM EDTA, 1% Triton X-100, 5 μl/ml protease inhibitor cocktail (P8340, Sigma), and 20 μg/ml phenylmethylsulfonyl fluoride. Protein lysates were resolved by SDS-PAGE and transferred to nitrocellulose membrane (Hybond C Extra; Amersham Biosciences). Western blotting was performed using the following antibodies: anti-FLAG (Clone M2, Sigma), anti-HA antibody (sc-7392, Santa Cruz), anti-PERM1 (anti-C1orf170, #HPA031711, Sigma), anti-tubulin (#2184, Cell Signaling), anti-Rt/Ms Total OxPhos Complex Kit (Invitrogen), anti-ERRα (ab76228, Abcam), anti-PGC-1α [17], anti-Sirt3 (#5490, Cell Signaling), anti-myoglobin (sc-25607, Santa Cruz), anti-phospho-p38 MAPK (#9211, Cell Signaling), anti-p38 MAPK (#9212, Cell Signaling), anti-phospho-AMPKα (#2535, Cell Signaling), anti-AMPKα (#2532, Cell Signaling), anti-phospho-CaMKII (#12716, Cell Signaling), and anti-CaMKII (#3362, Cell Signaling). The intensity of the bands was quantified using ImageJ software. The values were then normalized by the levels of GAPDH (intensity of internal control) and expressed relative to levels in control muscle.
2.13. DMD muscles
Human muscle specimens were sourced from the Telethon Network of Genetic Biobanks (Neuromuscular Tissue Bank, Padova, Italy). Skeletal muscle biopsies were taken from the vastus lateralis muscles of patients with DMD (aged 1–9 years) or healthy control subjects (aged 18–25 years) using either a Bergstrom or open biopsy technique. For the healthy control subjects, informed consent was obtained and procedures approved by the Deakin University Human Ethics Committee. All biopsies were frozen immediately in liquid nitrogen and stored at −80 °C until analysis. Tibialis anterior muscle samples from 8 to 9 week old male C57BL/10, mdx and dystrophin/utrophin double knock-out (dko) mice were obtained as published previously [18], [19]. Procedures were approved by the Animal Ethics Committee of The University of Melbourne and conformed to the Australian code of practice for the care and use of animals for scientific purposes.
2.14. Statistics
Data shown are mean ± SEM and were analyzed with a two-tailed Student's t-test for single variables. For comparisons between multiple variables, p-values were determined using a two-way ANOVA test, followed by a Bonferroni analysis.
3. Results
3.1. Perm1 associates with CaMKII and enhances CaMKII signaling
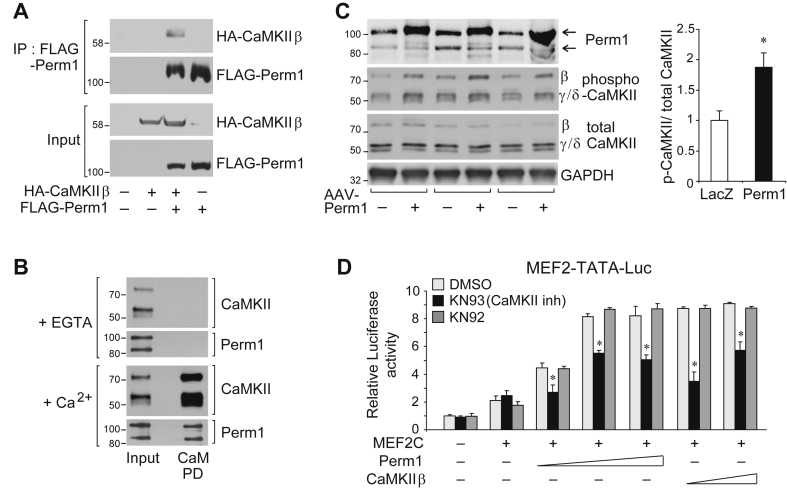
To gain insights into the pathway by which Perm1 increases oxidative capacity, we isolated Perm1-interacting proteins from C2C12 myotubes expressing FLAG-tagged Perm1 (Figure S1A). Using Mud-PIT mass spectrometric analysis [14], we identified the doublecortin-like kinase 1 (Dclk1) in immunopurified Perm1 complexes. Dclk1 is a neuronal serine/threonine protein kinase that is highly expressed in C2C12 myotubes but not present in skeletal muscle (Figure S1B) [20]. We noted that Dclk1 shares substantial homology to kinases of the CaMK family, and that the CaMKII isoforms that are highly expressed in skeletal muscle are found at comparatively lower levels in C2C12 myotubes (Figure S1B). We thus tested for interactions between Perm1 and CaMKII. First, we assessed the ability of heterologously expressed HA-tagged CaMKIIβ and FLAG-tagged Perm1 to co-immunoprecipitate. As shown in Figure 1A, CaMKII and Perm1 expressed in HEK293 cells interacted with each other. Next, we determined the ability of endogenous skeletal muscle Perm1 to co-purify with CaMKII bound at calmodulin (CaM), the activator of CaMKII. Both Perm1 and CaMKII interacted with CaM in a Ca2+ - dependent manner, suggesting that Perm1 is part of a CaM/CaMKII complex (Figure 1B). To assess the impact of Perm1 on CaMKII function, we then determined the activity state of CaMKII, using an antibody detecting CaMKII phosphorylated at Thr287 [8], in muscles injected with control or AAV1-Perm1 [13]. The increase in Perm1 levels led to increased CaMKII phosphorylation (Figure 1C), consistent with Perm1 enhancing CaMKII activity. Finally, to test the functional consequences of Perm1 overexpression on CaMKII downstream signaling, we assessed the effect of Perm1 on the activity of MEF2, a transcription factor that is activated by CaMKII [9], [21]. As shown in Figure 1D, Perm1 enhanced MEF2C transcriptional activity (∼4x), a similar effect as the one exerted by CaMKIIβ; the enhancement was suppressed by the CaMKII specific inhibitor KN93, but not the inert related compound KN92. In sum, Perm1 enhanced CaMKII activity and increased MEF2-dependent transcription in a CaMKII-dependent manner.
Figure 1.
Perm1 associates with and activates CaMKII. (A) Perm1 interaction with CaMKIIβ by coIP. HEK293 cells were transfected with plasmids expressing HA-CaMKIIβ, FLAG-Perm1 or control vector (−), as indicated. Protein lysates were immunoprecipitated with anti-FLAG antibody and immunoblotted with anti-HA or anti-FLAG antibodies. IP, immunoprecipitation. (B) Skeletal muscle Perm1 co-purification with CaMKII on calmodulin (CaM)-affinity beads. Skeletal muscle lysates were subjected to CaM affinity pull-down (PD) in the presence of EGTA or CaCl2 (Ca2+). Bound proteins were eluted and analyzed by western blot, using CaMKII and Perm1 antibodies. Note the presence of two endogenous Perm1 protein isoforms, as reported earlier [12]. (C) Perm1, phospho- and total CaMKII protein levels in control (−) and AAV-Perm1 TA muscles, determined by western blot. The overexpressed Perm1 co-migrates with the high MW, full length Perm1 [13]. Quantitation of phospho-CaMKII, after normalization to total CaMKII, is shown at the right. (D) Activation of MEF2 transcription by Perm1 and CaMKII. U2OS cells were transfected with the MEF2-TATA-Luc reporter and expression vectors for MEF2C (+), Perm1 or CaMKIIβ (5–20 ng), or control vector (−), and treated with DMSO (vehicle), the CaMKII inhibitor KN93 (10 μM) or the inactive compound KN92 (10 μM). *, p < 0.05 vs DMSO.
3.2. Perm1 is required for the activation of CaMKII and p38 MAPK by exercise and shapes the transcriptional response to an acute exercise bout
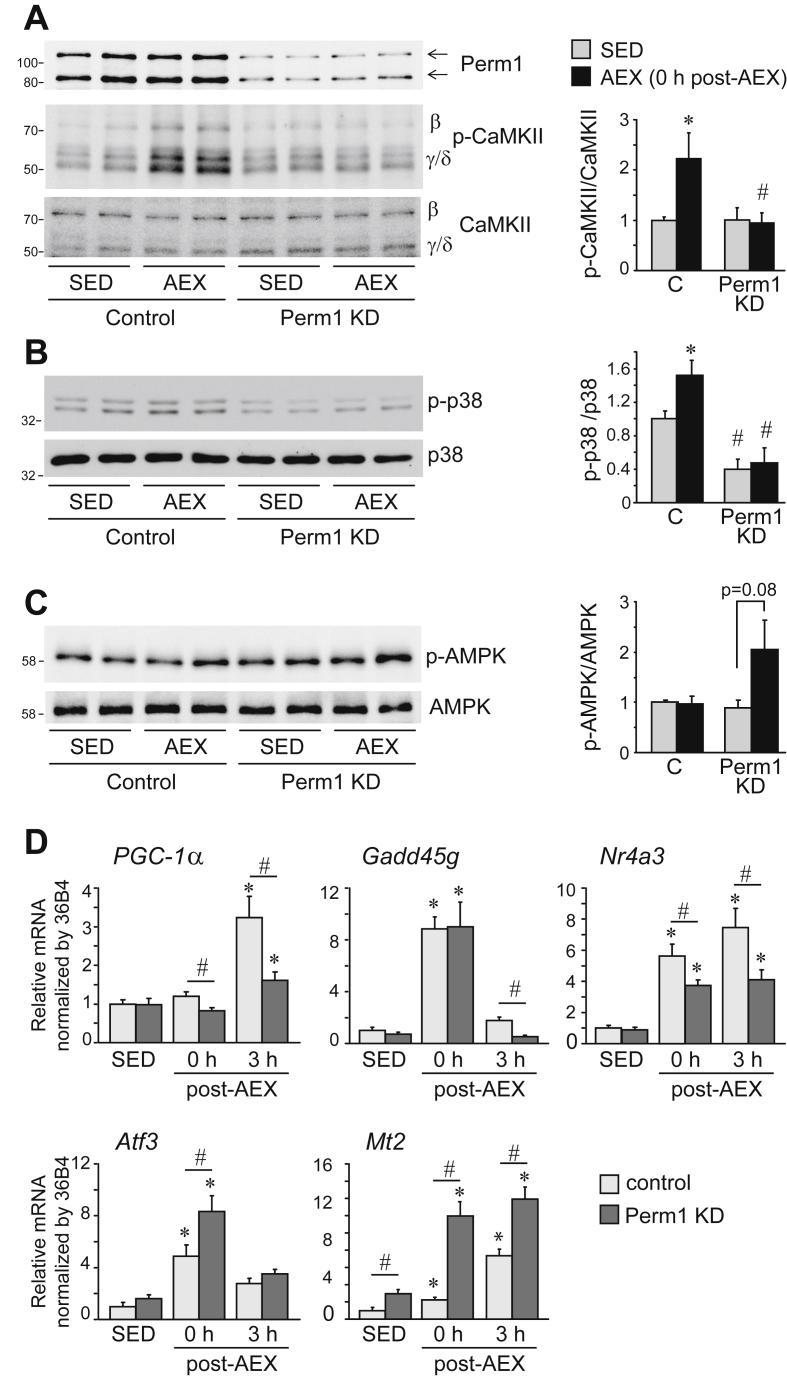
In vivo, CaMKII kinases are activated in response to an acute exercise bout [8]. To study the role of Perm1 in the exercise-induced CaMKII activation, we knocked down endogenous Perm1 expression in the gastrocnemius muscle (GAS) of one leg, by injecting AAV expressing shRNA against Perm1 (Perm1 KD). As control, the GAS muscle of the contralateral leg received an AAV expressing a control short hairpin RNA. Mice were then subjected to an acute exercise bout (AEX) or left sedentary in cages (SED). AEX increased CaMKII phosphorylation in control muscles. Perm1 KD caused a ∼4x decrease in Perm1 levels and suppressed exercise-induced phosphorylation of CaMKII (Figure 2A). It also decreased the basal activity state of MAPK p38, and suppressed exercise-induced activation of MAPK p38 (Figure 2B). We did not detect an exercise-induced activation of AMPK in control muscles, suggesting the AEX bout was not intense enough to change the myocellular energy state, and Perm1 KD did not have a significant effect on AMPK activation state [though we observed a trend for higher AMPK activity in AEX vs. SED Perm1 KD muscles (Figure 2C)]. These findings show that Perm1 is required in vivo for activation of CaMKII and p38 MAPK by AEX.
Figure 2.
Perm1 KD attenuates the activation of CaMKII and p38 MAPK, and induction of PGC-1α by exercise. (A–C) Levels of Perm1, phospho- and total CaMKII, phospho-p38 and total p38 MAPK, and phospho-AMPK and total AMPK in control (AAV-shCont) and Perm1 KD (AAV-shPerm1) GAS muscles of SED or AEX mice, determined by western blot analysis, and quantified. Images show 2 representative examples, and quantified data (right panels) are the mean ± SEM, expressed relative to SED control muscles, of 4–6 mice. (D) Quantitation of mRNA levels of the indicated genes in GAS muscles of SED mice or mice after AEX (0 h and 3 h post AEX). Data are the mean ± SEM, expressed relative to SED control muscles (n = 6–7). Muscles were harvested from mice immediately (0 h) and at 3 h after running for 55 min on a treadmill (AEX) or from control sedentary (SED) mice. *, p < 0.05 for the effect of acute exercise (AEX vs SED) in muscles with same Perm1 expression; #, p < 0.05 for the effect of Perm1 levels (Perm1 KD vs control) in muscles of same physical activity experience.
The exercise-induced activation of protein kinases regulates transcription factor activity and leads to gene expression changes [5], [6]. We next quantified the expression of key regulatory exercise-responsive genes, immediately and 3 h after acute exercise (0 h and 3 h post-AEX, respectively). Muscles with decreased Perm1 levels (Perm1 KD) had an attenuated induction of exercise-responsive genes that have been implicated in the control of oxidative capacity, such as PGC-1α, Nr4A3 and to a lesser degree, Gadd45g (the latter gene being at lower levels only at 3 h post-AEX). Interestingly, the decrease in Perm1 levels led to higher expression and/or induction of other exercise-responsive genes that are associated with cellular stress, such as Atf3 and Mt2, suggesting that Perm1 enables the efficient induction of select critical nodes of the exercise-induced response.
3.3. Perm1 is important for skeletal muscle adaptation to endurance training
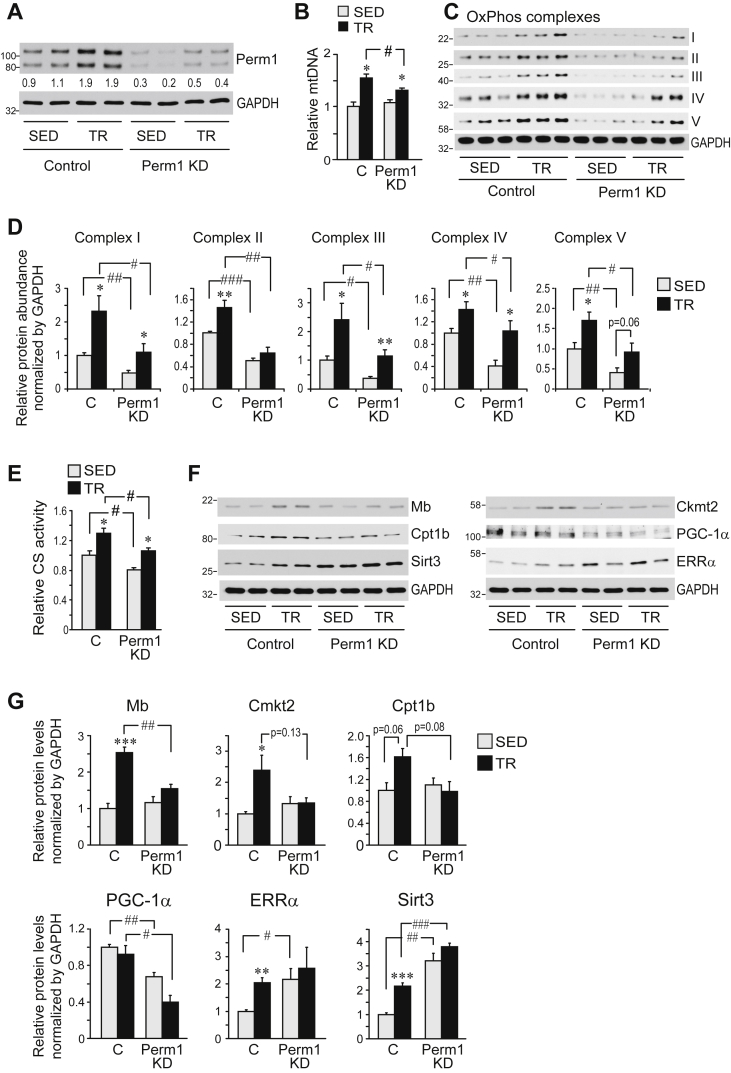
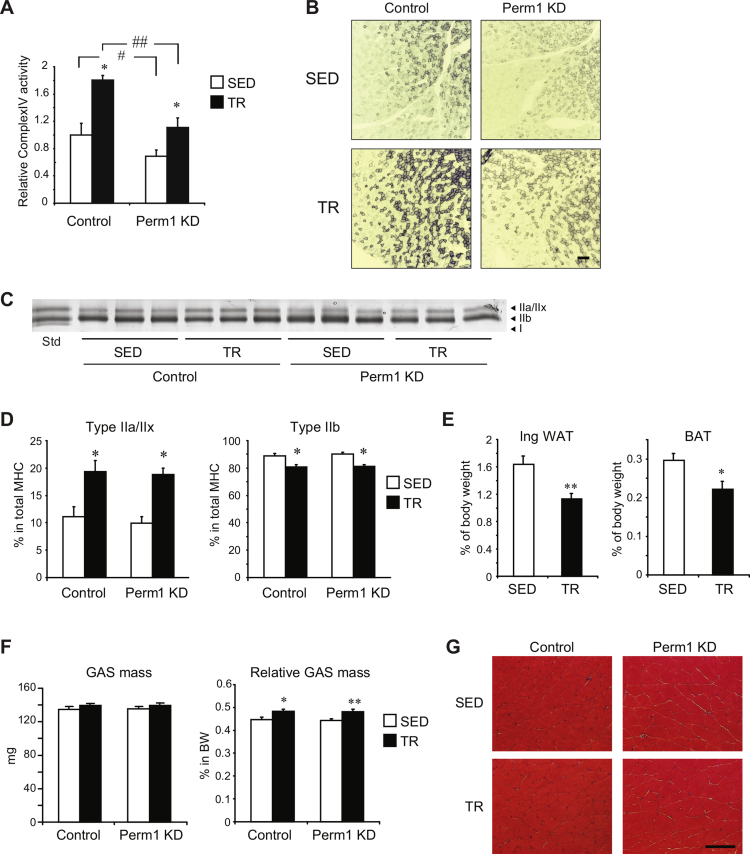
Next, we assessed the effect of Perm1 KD on long-term responses to endurance training. Mice with decreased Perm1 levels (Perm1 KD) in just one GAS muscle were given access to voluntary running wheels for 4 weeks for training (TR). Wheel use was monitored digitally and TR mice run on average ∼10 km per night. Control sedentary (SED) mice, also with Perm1 KD in just one GAS muscle, were singly housed in similar cages but without running wheels. Training led to efficient remodeling of control muscles, including increases in Perm1 protein (∼2x), mitochondrial DNA content (mtDNA, ∼50% increase), OxPhos proteins (40%–2.4x increases), activities of citrate synthase (CS, ∼30% increase), complex IV (∼80% increase) and succinate dehydrogenase (SDH), levels of other proteins important for muscle physiology (Mb, Ckmt2, Sirt3, ERRα), and a shift in contractile protein expression [Figure 3A–G, Figure S2A–D]. Perm1 KD muscles had ∼4-fold less Perm1 protein than control muscles in both SED and TR mice. Perm1 was still induced by training in Perm1 KD muscles, however, Perm1 expression was lower in TR Perm1 KD muscles compared to both SED and TR control muscles (Figure 3A). Knock down of Perm1 significantly attenuated several of the exercise training-induced adaptations. First, it led to a significant reduction in the induction of mtDNA content, without affecting mtDNA levels in SED mice (Figure 3B). Second, Perm1 KD decreased protein levels of all OxPhos complexes in both SED and TR mice; the training-induced response was lost for complex II but preserved for other complexes (Figure 3C,D). In agreement with the decreased OxPhos protein levels, we observed reduced activities of complex IV and SDH (Fig. S2A,B). Similarly, citrate synthase (CS) activity was significantly reduced in Perm1 KD muscles of both SED and TR mice (Figure 3E). Finally, knock down of Perm1 abolished the training-induced increases of myoglobin (Mb), mitochondrial creatine kinase (Ckmt2), and carnitine palmitoyl transferase Cpt1b that were seen in control muscles (Figure 3F,G). Levels of the coactivator PGC-1α were significantly decreased in both SED and TR Perm1 KD muscles (Figure 3F,G), while expression of the transcription factor ERRα and the deacetylase Sirt3 was increased.
Figure 3.
Perm1 is important for specific skeletal muscle adaptations to endurance training. (A) Perm1 protein levels in GAS muscles of SED and TR mice, determined by western blotting and the Perm1 antibody. GAPDH is shown as loading control. Numbers below each lane show normalized Perm1 levels, expressed relative to mean Perm1 levels in SED Control muscles. (B) Relative mitochondrial DNA content, determined as the ratio of mitochondrial to genomic DNA copy numbers, and expressed relative to the ratio in SED Control muscles. Data are the mean ± SEM (n = 14). (C) Protein levels of OxPhos complexes, determined by western blot analysis and the Total OxPhos Complex antibody cocktail. (D) Quantitation of OxPhos complexes, expressed relative to the mean levels in SED Control muscles. Data are the mean ± SEM (n = 6). (E) Citrate synthase (CS) enzymatic activities in total lysates. Data are expressed relative to CS activity in SED Control muscles and are the mean ± SEM (n = 6). (F) Protein levels of Mb, Ckmt2, Cpt1b, PGC-1α, Sirt3, ERRα, and GAPDH, determined by western blot analysis. (G) Quantitation of normalized protein levels in (F), expressed relative to the mean levels of each protein in SED Control muscles. Data are the mean ± SEM (n = 4). Muscles were harvested from mice that had access to voluntary wheels for 4 weeks (TR) or sedentary (SED) control mice. *, **, ***, p < 0.05, 0.01, 0.001, respectively, for the effect of exercise (TR vs SED) in muscles with same Perm1 expression; #, ##, ###, p < 0.05, 0.01, 0.001, respectively, for the effect of Perm1 levels (Perm1 KD vs Control) in muscles of same physical activity experience.
In addition to skeletal muscle metabolic remodeling, endurance exercise training causes a shift in muscle fiber types from type IIb to type IId/x and IIa [22]. Knock down of Perm1 did not affect this switch. Voluntary wheel training led to similar decreases of IIb and increases of IIa and IIx MHC (myosin heavy chain) proteins in control and Perm1 KD muscles (Fig. S2C,D). Training also decreased the size of white and brown adipose depots and led to a relative increase in GAS muscle mass (Fig. S2E,F). Knock down of Perm1 had no effect on muscle mass or gross morphology (Fig. S2F,G).
3.4. Skeletal muscle Perm1 levels are decreased in diet-induced obese mice and in muscular dystrophy patients and mouse models
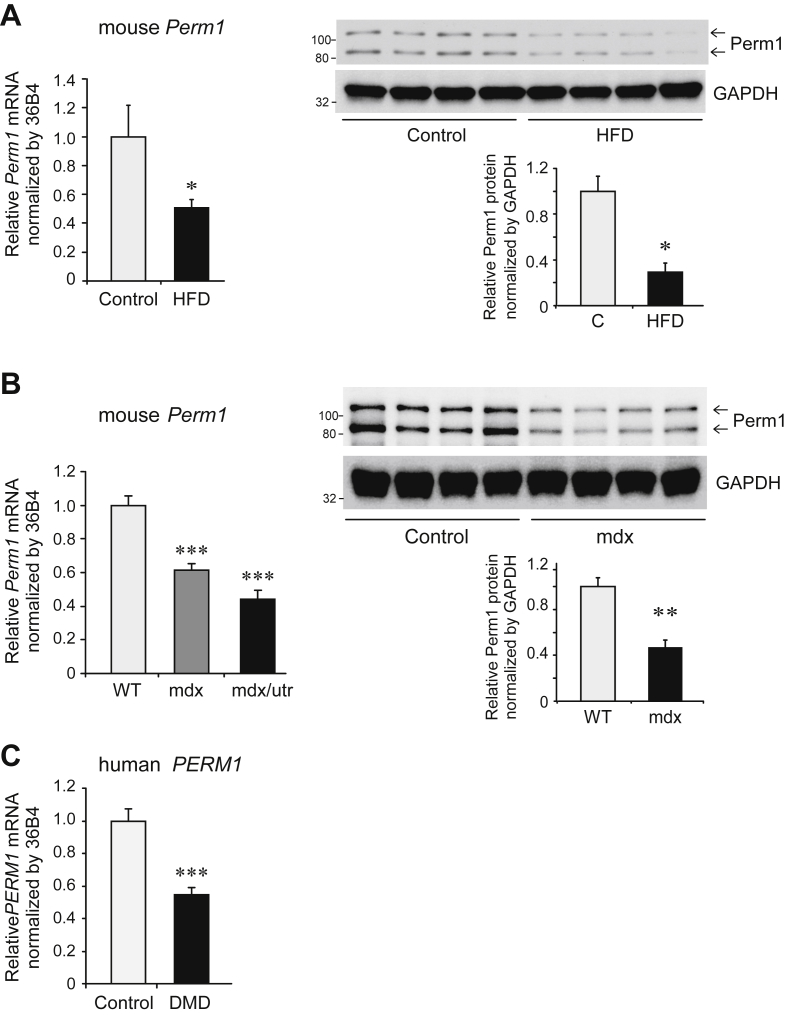
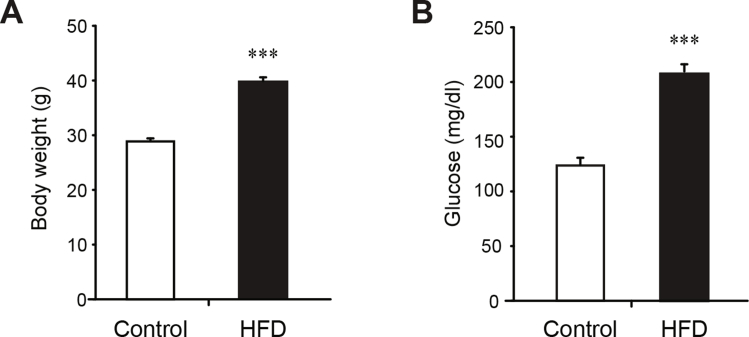
Our study shows that decreased Perm1 expression is sufficient to alter skeletal muscle responses to an acute exercise bout and to endurance training. To address whether Perm1 levels are decreased by diet or in disease states, we next quantified Perm1 in skeletal muscles of mouse models of obesity and of muscle disease. Perm1 mRNA and protein levels were down-regulated in diet-induced obese mice, as well as in mouse models of Duchenne muscular dystrophy (DMD) (Figure 4A,B; Fig. S3). Moreover, PERM1 expression was decreased in muscle of DMD human patients (Figure 4C). Our findings show that Perm1 levels are responsive to disease states, and suggest that decreased Perm1 expression may contribute to altered exercise responses in such states.
Figure 4.
Perm1 expression is decreased in diet-induced obese mice and in muscular dystrophy patients and mouse models. (A) Levels of Perm1 mRNA (left panel) and protein (right panels) in GAS muscles of mice fed control chow diet or HFD, determined by RT-qPCR and western analysis respectively, and expressed relative to Perm1 levels in mice on control chow diet (n = 7–8 for mRNA; n = 4 for protein). (B) Levels of Perm1 mRNA levels (left panel) and protein (right panels) in TA muscles of control, dystrophin (mdx), or dystrophin/utrophin double knock out mice, expressed relative to their respective corresponding control WT mice (n = 8 for RNA; n = 4 for protein). (C) PERM1 mRNA levels in vastus lateralis biopsies of male control subjects or DMD patients, expressed relative to PERM1 levels in control subjects (n = 7). Quantitative data in all panels are mean ± SEM. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
4. Discussion
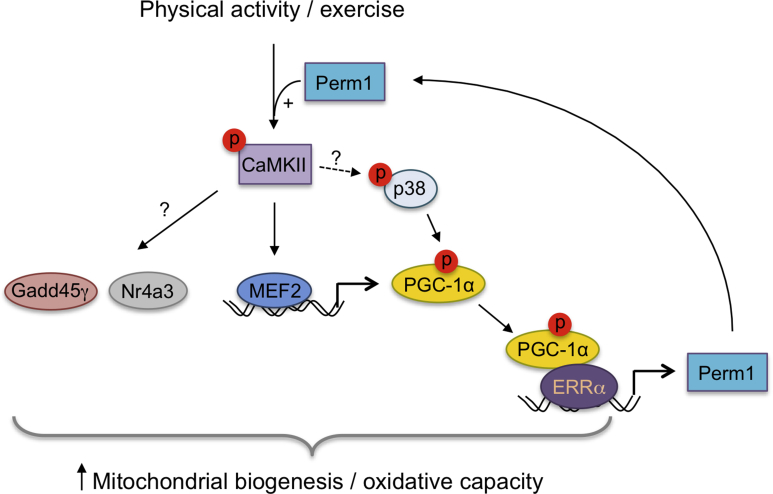
In response to endurance training, skeletal muscle undergoes structural and functional adaptations, including increased mitochondrial content, a shift from fast/glycolytic toward slower/oxidative fiber types, and changes in carbohydrate and lipid metabolism [5], [6]. Training-induced adaptations are mediated by muscle signaling pathways that modulate expression of exercise-responsive proteins. Our study shows that the muscle-specific regulator Perm1 controls activation of a subset of these signaling pathways. Perm1 is itself induced by exercise [12], suggesting a feed-forward system that entrains skeletal muscle signaling and exercise responses, and may confer memory of past exercise (Fig. S4).
We show that Perm1 levels are important determinants of the oxidative metabolism adaptations to physical activity. Knock down of Perm1 led to decreased OxPhos expression in sedentary mice, diminished CaMKII activation and PGC-1α transcription in response to acute exercise, and attenuated mitochondrial biogenesis in response to training. Conversely, overexpression of Perm1 was sufficient to enhance activation of kinases CaMKII and p38 and to drive increases in mitochondrial content and oxidative capacity (Figure 1 and [13]). Both Perm1 overexpression and knockdown selectively affect oxidative metabolism pathways, having no effect on muscle fiber type composition [13]. The selective action of Perm1 shows that mitochondrial biogenesis and fiber type switching can be dissociated from each other, a notion supported by other studies [23], [24], [25].
The sequence of Perm1 protein has revealed no similarities to other proteins and offered no hints of its molecular function [12]. Our study provides a first insight into how Perm1 acts. We find that Perm1 associates with CaMKII, regulates the activation of CaMKII, and enhances MEF2 transcription in a CaMKII-dependent manner. CaMKII - activated MEF2 can induce multiple metabolic targets, including PGC-1α, which drives mitochondrial biogenesis [5], [6]. Thus, our findings suggest that Perm1 regulates endurance training-induced increases in skeletal muscle mitochondrial content by controlling activation of CaMKII by physical activity. The exact mechanism of Perm1 action is currently unclear. Perm1 is predominantly cytoplasmic, though also present in the nucleus and possibly mitochondria [12]. Perm1 may enhance Ca2+/CaM-dependent activation of CaMKII or delay deactivation of CaMKII; it may act as a scaffold and/or direct CaMKII to specific subcellular localizations. Future experiments are required to elucidate the underlying molecular mechanism.
In addition to regulating CaMKII activation, Perm1 affects p38 MAPK activity. Muscles overexpressing Perm1 show higher p38 activity at night, when mice are physically active [13], while Perm1 KD leads to decreased p38 activity in sedentary and acutely exercised mice. The defect in p38 activity may stem from the reduction in CaMKII activity, as there is evidence for CaMKII-dependent activation of p38 in skeletal muscle and other cell types [26], [27], [28]. The decreased p38 activity is likely to contribute to the impaired response to exercise. CaMKII and p38 converge in activating PGC-1α, regulating its expression and activity [29], [30], [31], [32] (see also model in Fig. S4). Mice that lack skeletal muscle p38γ also show attenuation of the exercise-regulated increase of mitochondrial proteins [25].
An acute exercise bout elicits a characteristic profile of changes in gene expression. Central to this profile is the induction of PGC-1α, a master regulator of mitochondrial biogenesis [5], [6]. Consistent with decreased CaMKII and p38 activation, PGC-1α induction by AEX was attenuated in Perm1 KD muscle. Additional regulators of oxidative metabolism were also affected. Gadd45γ, an activator of p38 MAPK and oxidative capacity in brown fat [15], was highly induced immediately after exercise, but was significantly lower at 3 h post-exercise in Perm1 KD compared to control muscles. Induction of Nr4a3, a nuclear receptor that controls oxidative metabolism in skeletal muscle, was also blunted, possibly due to decreased CaMKII activity, as CaMKII contributes to the induction of Nr4a receptors in other cell types [33]. Mice overexpressing Nr4a3 or its close homolog Nr4a1 in skeletal muscle show similarities to Perm1 overexpressing mice, with higher mitochondrial content and increased resistance to fatigue [34], [35]. Thus, Perm1-dependent signaling may control induction of multiple regulators of oxidative capacity, including PGC-1α, Gadd45γ and Nr4a3. Notably, Perm1 is not required for induction of all exercise responsive genes. Atf3 and Mt2, genes induced by stress signals and not linked to oxidative metabolism, were more robustly induced in Perm1 KD than control muscles. The sustained inductions of Atf3 and Mt2, coupled to an intact fiber type switch response, support the notion that Perm1 controls a select subset of the transcriptional response to exercise.
Repeated bouts of endurance exercise lead to sustained increases in protein expression and long-term skeletal muscle adaptations. Consistent with the attenuated acute transcriptional responses, Perm1 KD muscles of trained mice had lower mitochondrial content, oxidative metabolism enzyme activities, and protein levels of PGC-1α, Mb and Ckmt2 (compared to contralateral control muscles of same trained mice). Interestingly, we observe three distinct patterns in the way Perm1 KD affects oxidative metabolism components and their response to training. First, some components, such as mitochondrial DNA, Mb, Ckmt2, and Cpt1b, are not affected by Perm1 KD in sedentary mice, but show an attenuated or no increase in Perm1 KD muscles following training. Second, components like OxPhos proteins, CS activity, and PGC-1α, are basally decreased in Perm1 KD muscles of sedentary mice; their response to training varies, from no induction (e.g., complex II) to strong induction (e.g., complex V), but their levels remain lower in Perm1 KD compared to control muscles in trained mice. Third, other components, such as ERRα and Sirt3, are increased in Perm1 KD compared to control muscles, in both SED and TR mice. To interpret these patterns, one must keep in mind that “sedentary” mice still have bouts of physical activity every night and that Perm1 may relay this regular physical activity to the basal expression of some components. Thus, expression of such regular physical activity-responsive genes may depend on Perm1 in “sedentary” mice, while expression of other exercise-responsive genes may become Perm1 dependent only upon physical activity of higher intensity, as seen in voluntary wheel running. Moreover, residual Perm1 expression is still induced ∼2 fold by training in Perm1 KD muscles, so different sensitivities to Perm1 levels may determine the degree by which a component responds to training. Finally, the increased ERRα and Sirt3 levels in Perm1 KD muscles are surprising, given that both ERRα and Sirt3 are PGC-1α induced genes and upregulated in muscles overexpressing Perm1 [12], [36], [37]. Their increase in Perm1 KD may reflect some compensatory response to the decreased PGC-1α and/or decreased oxidative capacity. The mechanism by which an oxidative defect may increase ERRα but not PGC-1α is currently unclear.
Because Perm1 levels can qualitatively impact the response to exercise, it is important to define physiological or disease states that may lead to suppressed Perm1 expression. In addition to our previous findings of decreased Perm1 levels in muscles of ALS patients [12], we show here that Perm1 levels are reduced in DMD and mouse models of muscular dystrophy and diet-induced obesity. Consistent with these findings, human PERM1 has been found to be significantly decreased in the skeletal muscle of Type 2 diabetes subjects [38] and to be lower in muscles of untrained individuals compared to endurance trained athletes [39]. Finally, Perm1 is rapidly decreased after muscle denervation-induced atrophy in mice [40]. Though the causal relationship is unclear, (e.g., the decrease in Perm1 may be secondary to decreased physical activity in ALS, DMD and diabetic patients, and in obese mice), decreased Perm1 levels in these states may nevertheless impact responses to exercise. Interestingly, compared to healthy controls, individuals with type 2 diabetes and rats with low aerobic capacity show differential responses to exercise rather than just diminished profiles [41], [42].
5. Conclusions
Our study is the first demonstration of a muscle-specific regulator of exercise-driven activation of CaMKII and p38. The levels of Perm1 qualitatively impact skeletal muscle signaling and gene induction following an acute exercise bout, as well as oxidative metabolism pathway adaptations following endurance training. Our work provides insights into mechanisms that shape endurance exercise-activated pathways and that may contribute to the high degree of variation in exercise training responses [4].
Author contributions
Y.C. and A.K. designed experiments, analyzed data, and wrote the paper. Y.C., S.T., B.H., A.R., J. M., and B.K performed experiments and analyzed data. Y.C., B.K., R.S., A.R., E.S., and A.K. reviewed and edited the manuscript.
Acknowledgements
We thank members of the Kralli and Saez labs, A. Krstic, S. Chau, and Dr. S. Schenk for technical help and discussions; Dr. E. Olson for providing MEF2-TATA-Luc and MEF2C plasmids; Drs. L. Puri and U. E Etxaniz for mdx samples. The work was supported by NIH grant DK105126 to A.K. and American Heart Association award 14SDG17790005 to Y.C.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.molmet.2019.02.009.
Contributor Information
Yoshitake Cho, Email: cyoshitake@ucsd.edu.
Anastasia Kralli, Email: akralli1@jhmi.edu.
Conflict of interest
None declared.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
figs1.
figs2.
figs3.
figs4.
References
- 1.Booth F.W., Roberts C.K., Laye M.J. Lack of exercise is a major cause of chronic diseases. Comprehensive Physiology. 2012;2:1143–1211. doi: 10.1002/cphy.c110025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Knowler W.C., Barrett-Connor E., Fowler S.E., Hamman R.F., Lachin J.M., Walker E.A. Reduction in the incidence of type 2 diabetes with lifestyle intervention or metformin. New England Journal of Medicine. 2002;346:393–403. doi: 10.1056/NEJMoa012512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pedersen B.K., Saltin B. Exercise as medicine - evidence for prescribing exercise as therapy in 26 different chronic diseases. Scandinavian Journal of Medicine & Science in Sports. 2015;25(3):1–72. doi: 10.1111/sms.12581. [DOI] [PubMed] [Google Scholar]
- 4.Bouchard C., Rankinen T. Individual differences in response to regular physical activity. Medicine & Science in Sports & Exercise. 2001;33:S446–S451. doi: 10.1097/00005768-200106001-00013. Discussion S452-443. [DOI] [PubMed] [Google Scholar]
- 5.Egan B., Zierath J.R. Exercise metabolism and the molecular regulation of skeletal muscle adaptation. Cell Metabolism. 2013;17:162–184. doi: 10.1016/j.cmet.2012.12.012. [DOI] [PubMed] [Google Scholar]
- 6.Hawley J.A., Hargreaves M., Joyner M.J., Zierath J.R. Integrative biology of exercise. Cell. 2014;159:738–749. doi: 10.1016/j.cell.2014.10.029. [DOI] [PubMed] [Google Scholar]
- 7.Swulius M.T., Waxham M.N. Ca(2+)/calmodulin-dependent protein kinases. Cellular and Molecular Life Sciences : CMLS. 2008;65:2637–2657. doi: 10.1007/s00018-008-8086-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rose A.J., Kiens B., Richter E.A. Ca2+-calmodulin-dependent protein kinase expression and signalling in skeletal muscle during exercise. The Journal of physiology. 2006;574:889–903. doi: 10.1113/jphysiol.2006.111757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu Y., Randall W.R., Schneider M.F. Activity-dependent and -independent nuclear fluxes of HDAC4 mediated by different kinases in adult skeletal muscle. The Journal of cell biology. 2005;168:887–897. doi: 10.1083/jcb.200408128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lu J., McKinsey T.A., Nicol R.L., Olson E.N. Signal-dependent activation of the MEF2 transcription factor by dissociation from histone deacetylases. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:4070–4075. doi: 10.1073/pnas.080064097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu H., Kanatous S.B., Thurmond F.A., Gallardo T., Isotani E., Bassel-Duby R. Regulation of mitochondrial biogenesis in skeletal muscle by CaMK. Science. 2002;296:349–352. doi: 10.1126/science.1071163. [DOI] [PubMed] [Google Scholar]
- 12.Cho Y., Hazen B.C., Russell A.P., Kralli A. Peroxisome proliferator-activated receptor gamma coactivator 1 (PGC-1)- and estrogen-related receptor (ERR)-induced regulator in muscle 1 (Perm1) is a tissue-specific regulator of oxidative capacity in skeletal muscle cells. Journal of Biological Chemistry. 2013;288:25207–25218. doi: 10.1074/jbc.M113.489674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cho Y., Hazen B.C., Gandra P.G., Ward S.R., Schenk S., Russell A.P. Perm1 enhances mitochondrial biogenesis, oxidative capacity, and fatigue resistance in adult skeletal muscle. The FASEB Journal : Official Publication of the Federation of American Societies for Experimental Biology. 2016;30:674–687. doi: 10.1096/fj.15-276360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Santi L., Beys-da-Silva W.O., Berger M., Calzolari D., Guimaraes J.A., Moresco J.J. Proteomic profile of Cryptococcus neoformans biofilm reveals changes in metabolic processes. Journal of Proteome Research. 2014;13:1545–1559. doi: 10.1021/pr401075f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gantner M.L., Hazen B.C., Conkright J., Kralli A. GADD45gamma regulates the thermogenic capacity of brown adipose tissue. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:11870–11875. doi: 10.1073/pnas.1406638111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tiraby C., Hazen B.C., Gantner M.L., Kralli A. Estrogen-related receptor gamma promotes mesenchymal-to-epithelial transition and suppresses breast tumor growth. Cancer Research. 2011;71:2518–2528. doi: 10.1158/0008-5472.CAN-10-1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schreiber S.N., Knutti D., Brogli K., Uhlmann T., Kralli A. The transcriptional coactivator PGC-1 regulates the expression and activity of the orphan nuclear receptor estrogen-related receptor alpha (ERRalpha) Journal of Biological Chemistry. 2003;278:9013–9018. doi: 10.1074/jbc.M212923200. [DOI] [PubMed] [Google Scholar]
- 18.Church J.E., Trieu J., Chee A., Naim T., Gehrig S.M., Lamon S. Alterations in Notch signalling in skeletal muscles from mdx and dko dystrophic mice and patients with Duchenne muscular dystrophy. Experimental Physiology. 2014;99:675–687. doi: 10.1113/expphysiol.2013.077255. [DOI] [PubMed] [Google Scholar]
- 19.Malecova B., Gatto S., Etxaniz U., Passafaro M., Cortez A., Nicoletti C. Dynamics of cellular states of fibro-adipogenic progenitors during myogenesis and muscular dystrophy. Nature Communications. 2018;9:3670. doi: 10.1038/s41467-018-06068-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Burgess H.A., Martinez S., Reiner O. KIAA0369, doublecortin-like kinase, is expressed during brain development. Journal of Neuroscience Research. 1999;58:567–575. [PubMed] [Google Scholar]
- 21.Smith J.A., Kohn T.A., Chetty A.K., Ojuka E.O. CaMK activation during exercise is required for histone hyperacetylation and MEF2A binding at the MEF2 site on the Glut4 gene. American Journal of Physiology. Endocrinology and Metabolism. 2008;295:E698–E704. doi: 10.1152/ajpendo.00747.2007. [DOI] [PubMed] [Google Scholar]
- 22.Booth F.W., Thomason D.B. Molecular and cellular adaptation of muscle in response to exercise: perspectives of various models. Physiological Reviews. 1991;71:541–585. doi: 10.1152/physrev.1991.71.2.541. [DOI] [PubMed] [Google Scholar]
- 23.Geng T., Li P., Okutsu M., Yin X., Kwek J., Zhang M. PGC-1alpha plays a functional role in exercise-induced mitochondrial biogenesis and angiogenesis but not fiber-type transformation in mouse skeletal muscle. American Journal of Physiology. Cell Physiology. 2010;298:C572–C579. doi: 10.1152/ajpcell.00481.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.O'Neill H.M., Maarbjerg S.J., Crane J.D., Jeppesen J., Jorgensen S.B., Schertzer J.D. AMP-activated protein kinase (AMPK) beta1beta2 muscle null mice reveal an essential role for AMPK in maintaining mitochondrial content and glucose uptake during exercise. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:16092–16097. doi: 10.1073/pnas.1105062108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pogozelski A.R., Geng T., Li P., Yin X., Lira V.A., Zhang M. p38gamma mitogen-activated protein kinase is a key regulator in skeletal muscle metabolic adaptation in mice. PLoS One. 2009;4:e7934. doi: 10.1371/journal.pone.0007934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lim N.R., Thomas C.J., Silva L.S., Yeap Y.Y., Yap S., Bell J.R. Cardioprotective 3',4'-dihydroxyflavonol attenuation of JNK and p38(MAPK) signalling involves CaMKII inhibition. Biochemical Journal. 2013;456:149–161. doi: 10.1042/BJ20121538. [DOI] [PubMed] [Google Scholar]
- 27.Takeda K., Matsuzawa A., Nishitoh H., Tobiume K., Kishida S., Ninomiya-Tsuji J. Involvement of ASK1 in Ca2+-induced p38 MAP kinase activation. EMBO Reports. 2004;5:161–166. doi: 10.1038/sj.embor.7400072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wright D.C., Geiger P.C., Han D.H., Jones T.E., Holloszy J.O. Calcium induces increases in peroxisome proliferator-activated receptor gamma coactivator-1alpha and mitochondrial biogenesis by a pathway leading to p38 mitogen-activated protein kinase activation. Journal of Biological Chemistry. 2007;282:18793–18799. doi: 10.1074/jbc.M611252200. [DOI] [PubMed] [Google Scholar]
- 29.Akimoto T., Li P., Yan Z. Functional interaction of regulatory factors with the Pgc-1alpha promoter in response to exercise by in vivo imaging. American Journal of Physiology Cell Physiology. 2008;295:C288–C292. doi: 10.1152/ajpcell.00104.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Handschin C., Rhee J., Lin J., Tarr P.T., Spiegelman B.M. An autoregulatory loop controls peroxisome proliferator-activated receptor gamma coactivator 1alpha expression in muscle. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:7111–7116. doi: 10.1073/pnas.1232352100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Knutti D., Kressler D., Kralli A. Regulation of the transcriptional coactivator PGC-1 via MAPK-sensitive interaction with a repressor. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:9713–9718. doi: 10.1073/pnas.171184698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Puigserver P., Rhee J., Lin J., Wu Z., Yoon J.C., Zhang C.Y. Cytokine stimulation of energy expenditure through p38 MAP kinase activation of PPARgamma coactivator-1. Molecular Cell. 2001;8:971–982. doi: 10.1016/s1097-2765(01)00390-2. [DOI] [PubMed] [Google Scholar]
- 33.Kovalovsky D., Refojo D., Liberman A.C., Hochbaum D., Pereda M.P., Coso O.A. Activation and induction of NUR77/NURR1 in corticotrophs by CRH/cAMP: involvement of calcium, protein kinase A, and MAPK pathways. Molecular Endocrinology. 2002;16:1638–1651. doi: 10.1210/mend.16.7.0863. [DOI] [PubMed] [Google Scholar]
- 34.Pearen M.A., Eriksson N.A., Fitzsimmons R.L., Goode J.M., Martel N., Andrikopoulos S. The nuclear receptor, Nor-1, markedly increases type II oxidative muscle fibers and resistance to fatigue. Molecular Endocrinology. 2012;26:372–384. doi: 10.1210/me.2011-1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chao L.C., Wroblewski K., Ilkayeva O.R., Stevens R.D., Bain J., Meyer G.A. Skeletal muscle Nur77 expression enhances oxidative metabolism and substrate utilization. The Journal of Lipid Research. 2012;53:2610–2619. doi: 10.1194/jlr.M029355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Giralt A., Hondares E., Villena J.A., Ribas F., Diaz-Delfin J., Giralt M. Peroxisome proliferator-activated receptor-gamma coactivator-1alpha controls transcription of the Sirt3 gene, an essential component of the thermogenic brown adipocyte phenotype. Journal of Biological Chemistry. 2011;286:16958–16966. doi: 10.1074/jbc.M110.202390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schreiber S.N., Emter R., Hock M.B., Knutti D., Cardenas J., Podvinec M. The estrogen-related receptor alpha (ERRalpha) functions in PPARgamma coactivator 1alpha (PGC-1alpha)-induced mitochondrial biogenesis. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:6472–6477. doi: 10.1073/pnas.0308686101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu C., Xu G., Tsai S.A., Freed W.J., Lee C.T. Transcriptional profiles of type 2 diabetes in human skeletal muscle reveal insulin resistance, metabolic defects, apoptosis, and molecular signatures of immune activation in response to infections. Biochemical and Biophysical Research Communications. 2017;482:282–288. doi: 10.1016/j.bbrc.2016.11.055. [DOI] [PubMed] [Google Scholar]
- 39.Schild M., Ruhs A., Beiter T., Zugel M., Hudemann J., Reimer A. Basal and exercise induced label-free quantitative protein profiling of m. vastus lateralis in trained and untrained individuals. Journal of Proteomics. 2015;122:119–132. doi: 10.1016/j.jprot.2015.03.028. [DOI] [PubMed] [Google Scholar]
- 40.Lang F., Aravamudhan S., Nolte H., Turk C., Holper S., Muller S. Dynamic changes in the mouse skeletal muscle proteome during denervation-induced atrophy. Disease Models & Mechanisms. 2017;10:881–896. doi: 10.1242/dmm.028910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hansen J.S., Zhao X., Irmler M., Liu X., Hoene M., Scheler M. Type 2 diabetes alters metabolic and transcriptional signatures of glucose and amino acid metabolism during exercise and recovery. Diabetologia. 2015;58:1845–1854. doi: 10.1007/s00125-015-3584-x. [DOI] [PubMed] [Google Scholar]
- 42.Lessard S.J., Rivas D.A., Alves-Wagner A.B., Hirshman M.F., Gallagher I.J., Constantin-Teodosiu D. Resistance to aerobic exercise training causes metabolic dysfunction and reveals novel exercise-regulated signaling networks. Diabetes. 2013;62:2717–2727. doi: 10.2337/db13-0062. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.