Abstract
Background: Given the considerable role played by Salmonella in the incidence of food poisoning around the world, surveillance of this infection is prioritized by both food producers and health care authorities. Data remains insufficient concerning the prevalence of Salmonella in poultry systems in Ecuador and in Latin America in general.
Methods: In this study we evaluated the prevalence and diversity of Salmonella serovars in samples taken from 21 layer farms and backyard layers in central Ecuador during August-November 2017. Salmonella was isolated following standardized methods (ISO 6579) and the serovar determination was carried out by PCR.
Results: A significant presence of Salmonella was detected, with an incidence of 76% (95% confidence interval (CI): 58–94) in farms, 33% (95%CI: 13–53) in pooled cloacal swabs from layer hens, 33% (95%CI: 12–55) on feed samples, and 10% (95%CI: 0–22) in backyard layer feces from traditional local markets. The dominant serovars detected were S. Infantis and S. Typhimurium.
Conclusions: This study forms a basis for further surveillance of Salmonella serovars in layer farms in central Ecuador.
Keywords: Salmonella, Layer Poultry, Ecuador, Serovars.
Introduction
The genus Salmonella is considered a leading cause of foodborne illnesses around the world ( WHO, 2017). These bacteria are among the most significant agents of food and water poisoning in the United States and Europe ( Bäumler et al., 2000; Callejón et al., 2015; Varma et al., 2005). Globally, it is estimated that 93.8 million cases of Salmonella-related gastroenteritis occur annually, resulting in 155,000 deaths ( Majowicz et al., 2010). In Ecuador, typhoid, together with paratyphoid fever, causes around 1,500 hospitalizations per year, while non-typhoidal salmonellosis leads to more than 2,000 hospitalizations over the same period. These infections have accounted for approximately 25% of total reported gastrointestinal illnesses in recent years ( MSP, 2018). Salmonella represents a complex and diverse genus, but only a small number of serovars are involved in human infections ( Issenhuth-Jeanjean et al., 2014; Thiennimitr et al., 2012).
Salmonella detection and the investigation of foodborne outbreaks is of maximum importance to public health, which has resulted in the establishment of epidemiological surveillance programs in many developed countries ( EFSA, 2013). These measures provide information about endemic Salmonella-serovar patterns, outbreaks, temporal trends and the monitoring of control actions ( CDC, 2018). The principal reservoir of S. enterica is the intestinal tract of livestock, representing one of the main sources of infection for humans ( Antunes et al., 2016). Despite this epidemiological and economic importance, in South America there is a paucity of information concerning Salmonella in poultry systems ( Alexandre et al., 2000; Donado-Godoy et al., 2012). Research conducted in 2016 pertaining to broiler chicken farming in Ecuador indicated the presence of Salmonella serotypes S. Infantis , S. Enteritidis and S. Corvallis ( Vinueza-Burgos et al., 2016). However, among layer hens, no information about the prevalence or diversity of Salmonella has been reported. The purpose of this study was to assess the prevalence and diversity of Salmonella bacteria present in layer farms in central Ecuador.
Methods
Samples
Samples were collected between August–November 2017 from different layer farms in central Ecuador (Tungurahua and Cotopaxi provinces), which accounts for around 60% of egg production in the country. A total of 21 farms (>1,000 birds) in Latacunga, Cevallos, Quero and Ambato (all Ecuador) were sampled, based on their willingness to provide verbal consent for this study (verbal consent was obtained over written consent owing to the farmers’ reluctance to sign their names, as they perceived this could be used to identify them). Further details for each site can be found in Dataset 1 ( Calero-Cáceres, 2019a)). One laying hen house per farm was selected. The following samples were collected in each house: 21 pooled cloacal swabs (10 cloacal swabs per pool); 21 manure drag swabs (environmental swabs, 1 per business); 21 caecum content samples (1 layer per farm). To evaluate the potential risk from contaminated feed, 18 composite samples were taken from farmyards (18 of the 21 farms consented verbally to having these samples taken). Additionally, 21 fecal samples from backyard layers were sampled in traditional local markets. All samples were transported in an icebox at 3–5°C within 2 hours of collection for bacterial isolation. The experiment was performed under supervision of the ethical committee of the Faculty of Agricultural Sciences, Universidad Técnica de Ambato.
Detection of Salmonella
Salmonella was isolated following standardized methods (ISO 6579) ( ISO, 2017). The samples were pre-enriched in buffered peptone water (Oxoid, Basingstoke, England) and then incubated at 37±1°C for 18 h ± 2 h. Next, Rappaport Vassiliadis Soy Broth (RVS Broth) (Merck Millipore, Darmstadt, Germany) was used a selective medium, being inoculated with the pre-enriched culture and incubated at 41.5±1°C for 24±3 h. One loopful of the selective enrichment medium was streaked onto xylose lysine deoxycholate agar (XLD agar) (Becton Dickinson GmbH, Heidelberg, Germany) and incubated at 37±1°C for 24±3 h. Presumptive Salmonella isolates (identified as red/yellow colonies with a black center) were purified in Mac Conkey agar (Merck, Darmstadt, Germany) and incubated at 37±1°C for 24 h. Isolates were Gram stained and the following biochemical tests were performed for confirmation the genus Salmonella: a catalase test using 30% hydrogen peroxide (Merck Millipore, Darmstadt, Germany); triple sugar iron agar test (TSI) (Becton Dickinson GmbH, Heidelberg, Germany), Simmons citrate agar test (Merck, Darmstadt, Germany), Christensen urea agar test (Britania Lab., Buenos Aires, Argentina), and indole reaction using tryptone water (Merck, Darmstadt, Germany) and Kovac’s reagent (Sigma Aldrich, St. Louis, USA). One isolate per positive sample was selected and cryopreserved using overnight growth in LB broth (Sigma Aldrich, St. Louis, USA) supplemented with 30% glycerol (Merck Millipore, Darmstadt, Germany) and maintained at -80°C until analysis.
Serovar determination by PCR
PCR assays were performed to identify the genes under specific conditions ( Table 2). DNA was extracted from overnight cultures in Casein-Peptone Soymeal-Peptone Broth (Merck, Darmstadt, Germany) as described by Muniesa et al. (2004). Approximately ≈50 ng/reaction resulting from the thermal shock of bacterial suspensions (>10 7 UFC/ml) diluted in sterile ddH 20 was used as template for the PCR reactions. The following reference Salmonella strains were used as positive controls for the six serovars under investigation: S. Enteritidis UNIETAR 1, S. Gallinarum b. Gallinarum NCTC 13346, S. Gallinarum b. Pullorum ATCC 19945®, S. Typhi ATCC® 19430, S. Typhimurium ATCC® 26930 and S. Infantis UNIETAR 3CT7.
Table 1. Salmonella-positive samples in relation to evaluated matrices.
| Location | Sample | Matrix | Samples, n | Positive samples, n (%) |
|---|---|---|---|---|
| Farms | Feed | Composite feed | 18 | 6 (33%) |
| Environment | Environmental swabs | 21 | 16 (76%) | |
| Animal | Laying hen cloacal swab (composite) | 21 | 2 (14%) | |
| Local markets | Environment | Backyard poulty feces | 21 | 7 (33%) |
Table 2. Oligonucleotides used in this study.
| Organism | Name and
direction |
5’ to 3’ sequence | Gene | Annealing
temperature (°C) |
Amplicon
size (bp) |
References |
|---|---|---|---|---|---|---|
| Enteritidis | sdf-F | TGT GTT TTA TCT GAT GCA AGA G | sdf locus | 56 | 293 | ( Agron et al., 2001) |
| sdf-R | CGT TCT TCT GGT ACT TCA GAT GAC | |||||
| Enterica | bcfC-F | GGG TGG GCG GAA AAC TAT TTC | bcfC | 56 | 993 | ( Zhu et al., 2015) |
| bcfC-R | CGG CAC GGC GGA ATA GAG CAC | |||||
| Infantis | M. SinI F | CAC AAT GAA CGT GGT GAA GG | M.Sin I | 56 | 184 | ( Ranjbar et al., 2017) |
| M. SinI R | TGA ACT ACG TTC GTT CTT CTGG | |||||
| Gallinarum
biotype Gallinarum |
steB-F | TGT CGA CTG GGA CCC GCC CGC CCG C | steB | 56 | 636 | ( Pugliese et al., 2011) |
| steB-R | CCA TCT TGT AGC GCA CCA T | |||||
| Gallinarum
biotype Pullorum |
rhs-F | TCG TTT ACG GCA TTA CAC AAG TA | rhs locus | 56 | 402 | ( Zhu et al., 2015) |
| rhs-R | CAA ACC CAG AGC CAA TCT TAT CT | |||||
| Typhi | sty-1 | TGC CGG AAA CGA ATC T | 23S rRNA gene | 53 | 300 | ( Zhu et al., 1996) |
| sty-2 | GGT TGT CAT GCC AAT GCA CT | |||||
| Typhimurium | Fli15 | CGG TGT TGC CCA GGT TGG TAA T | fliC gene | 53 | 620 | ( Pui et al., 2011) |
| Typ04 | ACT GGT AAA GAT GGC T |
The reaction mixture contained: 12.5 µl of DreamTaq Green PCR Master Mix (Thermo Fisher Scientific, Massachusetts, USA), 0.5 µl of each primer (30 µM), 9 µl of nuclease-free water (Thermo Fisher Scientific, Massachusetts, USA), and 2.5 µl of crude DNA were used. PCRs were performed with an Applied Biosystems SimplyAmp Thermal Cycler (Thermo Fisher Scientific, Massachusetts, USA). A total of 10 µl of each PCR product were analyzed by agarose gel electrophoresis and stained using Sybr® Safe DNA Gel Stain (Invitrogen, Carlsbad, USA).
Results
Assessing the presence of Salmonella
Overall, 31 out of 34 isolates which showed phenotypic characteristics in accordance with Salmonella were confirmed by PCR as S. enterica ( bcfC gene amplification) ( Table 1). Of the 21 farms, 16 (76%, 95% confidence interval (CI): 58–94) showed the presence of Salmonella on environmental surfaces, and Salmonella bacteria were isolated in 7 pooled cloacal swabs (33%, 95%CI: 13–53) and in 6 of 18 composite feed samples (33%, 95%CI: 12–55). Feces from backyard layers showed the presence of Salmonella in 2 of 21 samples (10%, 95%CI: 0–22). Dataset 1 shows the location of each sample and the presence or absence of Salmonella ( Calero-Cáceres, 2019a)
Determining presence of serovars
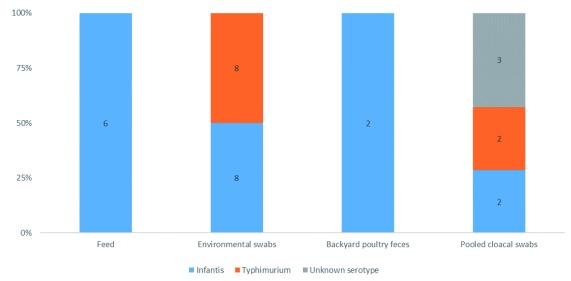
Regarding to the presence of individual serovars ( Figure 1), the highest occurrence was of S. Infantis, detected in 58% (18/31) of isolates, followed by S. Typhimurium, present in 32% of samples (10/31) ( Supplementary Figure 1 ( Calero-Cáceres, 2019c)). The serovar of 10% of Salmonella isolates (3/31) were unable to be serotyped by the panel of tests. In poultry feed samples, S. Infantis was present in 100% (6/6), while in S. enterica isolated from manure drag swabs, S. Infantis was detected in 50% (8/8) and S. Typhimurium in 50% (8/8). In backyard poultry feces from local markets, S. Infantis was detected in 100% of positive samples (2/2). The most heterogeneous diversity of Salmonella serotypes was observed in pooled cloacal swabs, with 2 S. Infantis, 2 S. Typhimurium and 3 S. enterica consisting of serovars not covered within the panel. Dataset 1 lists the identity of the serovar taken from each sampling location ( Calero-Cáceres, 2019a). Dataset 2 shows PCR gels for confirmation of Salmonella serovars ( Calero-Cáceres, 2019b).
Figure 1. Distribution of Salmonella serovars according to the evaluated matrices.
Discussion
The overall results showed a significant presence of Salmonella in layer farms in the central Ecuador region, one of the main sources of egg production in the country, with an incidence of more than 76% of the evaluated farms. This result is similar to that reported in Colombia (65%) ( Donado-Godoy et al., 2012), and considerably higher than the presence of Salmonella in broiler chicken farms in northern Ecuador (15.9%) ( Vinueza-Burgos et al., 2016).
The notable presence of Salmonella in poultry feed suggests that this may constitute a potential reservoir of this bacteria for poultry systems in the evaluated region, and consequently, a potential route of infection and colonization of poultry and subsequent entry into the food chain ( Fink-Gremmels, 2012; Jones, 2011). In recent research conducted in an integrated poultry farm in Ecuador, 4.1% (8/194) of samples were positive for Salmonella, particularly in animal-based feed ( Villagómez Estrada et al., 2017). Therefore, the accurate detection of this pathogen in feed is necessary in order to identify the critical points of contamination (facilities, raw materials, transport, storage, producers), that one may apply effective measures to reduce the risk of transmission.
The finding of only S. Infantis in poultry feed may be attributed to the high level of persistence of this serovar over time in poultry feed, which in turn results in its considerable presence in Ecuadorian broiler chicken ( Andino et al., 2014; Vinueza-Burgos et al., 2016). Although the origin of this serovar in feed it is not well defined, studies have pointed to cross-contamination with feces, persistent contamination of storage bins and surfaces, and poor ingredient selection as main causes of feed contamination with Salmonella ( Davies & Wray, 1997; Huss et al., 2018; Laban et al., 2014). Genomic tools, such as multilocus sequence typing, BOX-PCR, (GTG)5-PCR or whole-genome sequencing may help to identify the origin of feed contamination in a larger study.
Drag swabs revealed the presence of two different serovars in the sampled poultry farms: Infantis and Typhimurium. These non-typhoidal S. enterica serovars are commonly associated with poultry systems and are linked to outbreaks of foodborne illness ( Anderson et al., 2016; Pui et al., 2011). Serovars vary in their persistence over time and geographic distribution around the world ( Hendriksen et al., 2011), making further studies desirable in order to evaluate the variations in serovar persistence in the locations sampled in this research.
In backyard layer feces sampled at local markets, S. Infantis alone was detected (2/2), but further evaluation of Salmonella serovars in backyard flocks is recommended in order to improve surveillance of this potential source of salmonellosis ( Behravesh et al., 2014). In pooled cloacal swabs, the serovars detected were Infantis (2/7), Typhimurium (2/7) and unidentified serovars (3/7). Infantis and Typhimurium serovars are commonly detected in poultry around the world ( Foley & Lynne, 2008; Foley et al., 2011). Complementary analysis by serotyping the unclassified serovars is necessary in order to identify Salmonella bacteria not covered by the panel.
Data remains insufficient concerning the prevalence of Salmonella in poultry systems in Ecuador and in Latin America in general. The coordination of similar future studies may provide a starting point for surveillance of zoonotic bacteria within a defined public health area, leading to an improvement in policies and safe practices in the food industry. Such studies may reduce the risk of infection and establish protocols for corrective measures to be implemented in key upstream points of the chain as indicated by the data.
At the same time, the intervention of public health authorities may be required in order to ensure the participation of a fully representative range of poultry businesses in future research. This study depended on the voluntary consent of farm owners to providing samples, which may have led to selection bias. In order to establish a thorough program of surveillance, all potential sources of Salmonella infection in the region need to be made accessible to researchers.
These findings show a significant presence of Salmonella in layer farms in the central zone of Ecuador. The predominant serovars are S. Infantis and S. Typhimurium, typified by PCR. The principal source of infection could be related with poultry feed. From a public health perspective, it is necessary to establish adequate surveillance of Salmonella, including protocols covering biosecurity practices, antibiotic usage and random sampling programs.
Data availability
Underlying data
Figshare: Dataset 1. Origin, biochemical tests, PCR results and serovar of Salmonella isolates. https://doi.org/10.6084/m9.figshare.7726163.v2 ( Calero-Cáceres, 2019a)
Figshare: Dataset 2. PCR results of bcfC, fliC and M.SinI genes. https://doi.org/10.6084/m9.figshare.7726217.v2 ( Calero-Cáceres, 2019b)
Extended data
Figshare: Supplementary figure 1. Multiplex PCR amplification of serovar-specific genes. a) bcfC and mSinI fragments ( S. Infantis), b) bcfC and fliC fragments ( S. Typhimurium).
https://doi.org/10.6084/m9.figshare.7732934.v1 ( Calero-Cáceres, 2019c.)
Data are available under the terms of the Creative Commons Zero "No rights reserved" data waiver (CC0 1.0 Public domain dedication).
Funding Statement
This study was supported by the Dirección de Investigación y Desarrollo DIDE-Universidad Técnica de Ambato (Project 1568-CU-P-2017).
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
[version 1; peer review: 2 approved with reservations]
References
- Agron PG, Walker RL, Kinde H, et al. : Identification by subtractive hybridization of sequences specific for Salmonella enterica serovar enteritidis. Appl Environ Microbiol. 2001;67(11):4984–4991. 10.1128/AEM.67.11.4984-4991.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandre SM, Pozo MC, González GV, et al. : Detección de Salmonella enteritidis en muestras de productos avícolas de consumo humano en la Región Metropolitana. Revista Médica de Chile. 2000;128(10):1075–1083. 10.4067/S0034-98872000001000001 [DOI] [PubMed] [Google Scholar]
- Anderson TC, Nguyen TA, Adams JK, et al. : Multistate outbreak of human Salmonella Typhimurium infections linked to live poultry from agricultural feed stores and mail-order hatcheries, United States 2013. One Health. 2016;2:144–149. 10.1016/j.onehlt.2016.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andino A, Pendleton S, Zhang N, et al. : Survival of Salmonella enterica in poultry feed is strain dependent. Poult Sci. 2014;93(2):441–447. 10.3382/ps.2013-03401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antunes P, Mourão J, Campos J, et al. : Salmonellosis: the role of poultry meat. Clin Microbiol Infect. 2016;22(2):110–121. 10.1016/j.cmi.2015.12.004 [DOI] [PubMed] [Google Scholar]
- Bäumler AJ, Hargis BM, Tsolis RM: Tracing the origins of Salmonella outbreaks. Science. 2000;287(5450):50–52. 10.1126/science.287.5450.50 [DOI] [PubMed] [Google Scholar]
- Behravesh CB, Brinson D, Hopkins BA, et al. : Backyard poultry flocks and salmonellosis: a recurring, yet preventable public health challenge. Clin Infect Dis. 2014;58(10):1432–1438. 10.1093/cid/ciu067 [DOI] [PubMed] [Google Scholar]
- Callejón RM, Rodríguez-Naranjo MI, Ubeda C, et al. : Reported foodborne outbreaks due to fresh produce in the United States and European Union: trends and causes. Foodborne Pathog Dis. 2015;12(1):32–38. 10.1089/fpd.2014.1821 [DOI] [PubMed] [Google Scholar]
- Calero-Cáceres W: Dataset 1. Origin, biochemical tests, PCR results and serovar of Salmonella isolates. figshare.Dataset.2019a. 10.6084/m9.figshare.7726163.v2 [DOI] [Google Scholar]
- Calero-Cáceres W: Dataset 2. PCR results of bcfC, fliC and M.SinI genes. figshare.Figure.2019b. 10.6084/m9.figshare.7726217.v2 [DOI] [Google Scholar]
- Calero-Cáceres W: Supplementary figure 1. figshare.Figure.2019c. 10.6084/m9.figshare.7732934.v1 [DOI] [Google Scholar]
- CDC: Salmonella Atlas. Reports and Publications. Salmonella CDC [Online]. [Accessed: 7 February 2019].2018. Reference Source [Google Scholar]
- Davies RH, Wray C: Distribution of Salmonella contamination in ten animal feedmills. Vet Microbiol. 1997;57(2–3):159–169. 10.1016/S0378-1135(97)00114-4 [DOI] [PubMed] [Google Scholar]
- Donado-Godoy P, Gardner I, Byrne BA, et al. : Prevalence, risk factors, and antimicrobial resistance profiles of Salmonella from commercial broiler farms in two important poultry-producing regions of Colombia. J Food Prot. 2012;75(5):874–883. 10.4315/0362-028X.JFP-11-458 [DOI] [PubMed] [Google Scholar]
- EFSA: EU Summary Report on antimicrobial resistance in zoonotic and indicator bacteria from humans, animals and food in 2013. [Online]. [Accessed: 7 February 2019].2013. Reference Source [Google Scholar]
- Fink-Gremmels J: Animal feed contamination: effects on livestock and food safety.2012. Reference Source [Google Scholar]
- Foley SL, Lynne AM: Food animal-associated Salmonella challenges: pathogenicity and antimicrobial resistance. J Anim Sci. 2008;86(14 Suppl):E173–87. 10.2527/jas.2007-0447 [DOI] [PubMed] [Google Scholar]
- Foley SL, Nayak R, Hanning IB, et al. : Population dynamics of Salmonella enterica serotypes in commercial egg and poultry production. Appl Environ Microbiol. 2011;77(13):4273–4279. 10.1128/AEM.00598-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendriksen RS, Vieira AR, Karlsmose S, et al. : Global monitoring of Salmonella serovar distribution from the World Health Organization Global Foodborne Infections Network Country Data Bank: results of quality assured laboratories from 2001 to 2007. Foodborne Pathog Dis. 2011;8(8):887–900. 10.1089/fpd.2010.0787 [DOI] [PubMed] [Google Scholar]
- Huss A, Cochrane R, Muckey M, Jones C, et al. : Animal Feed Mill Biosecurity: Prevention of Biological Hazards. In Food and feed safety systems and analysis2018; Elsevier,63–81. 10.1016/B978-0-12-811835-1.00004-X [DOI] [Google Scholar]
- ISO: ISO 6579-1:2017 - Microbiology of the food chain -- Horizontal method for the detection, enumeration and serotyping of Salmonella -- Part 1: Detection of Salmonella spp. International Organization for Standardization.2017. Reference Source [DOI] [PubMed] [Google Scholar]
- Issenhuth-Jeanjean S, Roggentin P, Mikoleit M, et al. : Supplement 2008-2010 (no. 48) to the White-Kauffmann-Le Minor scheme. Res Microbiol. 2014;165(7):526–530. 10.1016/j.resmic.2014.07.004 [DOI] [PubMed] [Google Scholar]
- Jones FT: A review of practical Salmonella control measures in animal feed. J Appl Poult Res. 2011;20(1):102–113. 10.3382/japr.2010-00281 [DOI] [Google Scholar]
- Laban SE, Moustafa GZ, Anwer W, et al. : Microbial Load of Poultry By-Products Following Rendering Process. Global Veterinaria. 2014;12:756–759. Reference Source [Google Scholar]
- Majowicz SE, Musto J, Scallan E, et al. : The global burden of nontyphoidal Salmonella gastroenteritis. Clin Infect Dis. 2010;50(6):882–889. 10.1086/650733 [DOI] [PubMed] [Google Scholar]
- MSP: Gaceta Epidemiológica Ecuador SIVE-ALERTA – Ministerio de Salud Pública. [Online]. [Accessed: 6 February 2019].2018. Reference Source [Google Scholar]
- Muniesa M, Blanco JE, De Simón M, et al. : Diversity of stx 2 converting bacteriophages induced from Shiga-toxin-producing Escherichia coli strains isolated from cattle. Microbiology. 2004;150(Pt 9):2959–2971. 10.1099/mic.0.27188-0 [DOI] [PubMed] [Google Scholar]
- Pugliese N, Circella E, Pazzani C, et al. : Validation of a seminested PCR approach for rapid detection of Salmonella enterica subsp. enterica serovar Gallinarum. J Microbiol Methods. 2011;85(1):22–27. 10.1016/j.mimet.2011.01.003 [DOI] [PubMed] [Google Scholar]
- Pui CF, Wong WC, Chai LC, et al. : Simultaneous detection of Salmonella spp., Salmonella Typhi and Salmonella Typhimurium in sliced fruits using multiplex PCR. Food Control. 2011;22(2):337–342. 10.1016/j.foodcont.2010.05.021 [DOI] [Google Scholar]
- Ranjbar R, Mortazavi SM, Mehrabi Tavana A, et al. : Simultaneous Molecular Detection of Salmonella enterica Serovars Typhi, Enteritidis, Infantis, and Typhimurium. Iran J Public Health. 2017;46(1):103–111. [PMC free article] [PubMed] [Google Scholar]
- Thiennimitr P, Winter SE, Bäumler AJ: Salmonella, the host and its microbiota. Curr Opin Microbiol. 2012;15(1):108–114. 10.1016/j.mib.2011.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varma JK, Greene KD, Ovitt J, et al. : Hospitalization and antimicrobial resistance in Salmonella outbreaks, 1984-2002. Emerg Infect Dis. 2005;11(6):943–946. 10.3201/eid1106.041231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villagómez Estrada S, Logacho Pilataxi M, Vinueza Burgos C: Presencia y Resistencia a los Antimicrobianos de serovariedades de Salmonella enterica aisladas en una empresa avícola integrada del Ecuador. Rev Ecuat Med Cienc Biol. 2017;38(1). 10.26807/remcb.v38i1.17 [DOI] [Google Scholar]
- Vinueza-Burgos C, Cevallos M, Ron-Garrido L, et al. : Prevalence and Diversity of Salmonella Serotypes in Ecuadorian Broilers at Slaughter Age. PLoS One. 2016;11(7):e0159567. 10.1371/journal.pone.0159567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO: Salmonella. WHO. [Online]. [Accessed: 7 February 2019].2017. Reference Source [Google Scholar]
- Zhu C, Yue M, Rankin S, et al. : One-Step Identification of Five Prominent Chicken Salmonella Serovars and Biotypes. J Clin Microbiol. 2015;53(12):3881–3883. 10.1128/JCM.01976-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Q, Lim CK, Chan YN: Detection of Salmonella Typhi by polymerase chain reaction. J Appl Bacteriol. 1996;80(3):244–251. 10.1111/j.1365-2672.1996.tb03216.x [DOI] [PubMed] [Google Scholar]