Abstract
3- and 4- hydroxyprolines (HyP) are regioisomers that play different roles in various species and organs. Despite their distinct functions inside cells, they are generally considered indistinguishable using mass spectrometry due to their identical masses. Here, we demonstrate, for the first time, that characteristic w ions can be produced by electron-transfer/higher energy collision dissociation (EThcD) dual fragmentation technique to confidently discriminate 3-HyP/ 4-HyP isomers. An integrated and high throughput strategy was developed which combined online LC separation with EThcD for large-scale differentiation of 3-HyP/4-HyP in complex samples. An automated algorithm was developed for charge state dependent characterization of 3-HyP/ 4-HyP isomers. Using this combined discrimination approach, we identified 108 3-HyP sites and 530 4-HyP sites from decellularized pancreas, allowing more than fivefold increase of both 3-HyP and 4-HyP identifications compared to previous reports. This approach outperformed ETD and HCD in the analysis of HyP-containing peptides with unique capacity to generate w ions for HyP discrimination, improved fragmentation of precursor ions, as well as unambiguous localization of modifications. A high content of 3-HyP was observed in the C-terminal (GPP)n domain of human CO1A1, which was previously only identified in vertebrate fibrillar collagens from tendon. Unexpectedly, some unusual HyP sites at Xaa position in Gly-HyP-Ala, Gly-HyP-Val, Gly-HyP-Gln, Gly-HyP-Ser and Gly-HyP-Arg were also confirmed to be 3-hydroxylated, whose functions and enzymes are yet to be discovered. Overall, this novel discrimination strategy can be readily implemented into de novo sequencing or other proteomic search engines.
Keywords: EThcD, 3-hydroxyproline/4-hydroxyproline differentiation, decellularized pancreas, peptide sequencing, automated algorithm, PTM analysis
Graphical Abstract
Introduction
With the advancements of modern mass spectrometry (MS), researchers have gained unprecedented insights into the composition, structure, and function of proteomes using powerful MS-based technologies 1. Despite the tremendous success achieved in the proteomics field, challenges still exist for the characterization and quantification due to the presence of peptide isomers 2,3. Isomeric peptides are molecules with identical chemical formula, but different chemical structures. They play significant roles in the development of pathological conditions, such as Alzheimer’s disease 4, and affect the selectivity and affinity of receptor binding5. They can be divided into different classes, including stereoisomers (enantiomers and diastereomers), isopeptides with isomeric residue (e.g., leucine vs. isoleucine), sequence inversion (the order of amino acids is altered), and regioisomers with different localization of post-translational modifications (PTM) 6. Differentiation of sequence inversion is generally straightforward with the assistance of tandem mass spectrometry (MS/MS). The introduction of novel fragmentation techniques like electron capture dissociation (ECD) and electron transfer dissociation (ETD) have now provided the ability to distinguish the isopeptides like leucine and isoleucine 7,8. By applying ion mobility mass spectrometry (IM-MS), Jia et al. successfully mapped the localization of D-amino acid residues from the D-amino acid containing peptide (DAACP) 9. However, difficulties still persist in part for the discrimination of regioisomers where traditional MS approaches have limited capability to distinguish between 3-hydroxyproline (HyP) and 4-HyP.
HyP is a prevalent PTM catalyzed by prolyl hydroxylases. It has been found in various organisms and species, including mammals, plants, invertebrates and bacteria. Collagen makes up about 1/3 of the total protein content in human and up to 40% of them carries HyP, making prolyl hydroxylation one of the most abundant PTM. Depending on whether the hydroxylation occurs on β-C atom or γ-C atom of the cyclic ring, either 3-HyP or 4-HyP can be formed. The triple helix is the characteristic structure of collagen and collagenous-domain-containing proteins. It consists of repeating Gly-Xaa-Yaa triplets, with proline being the most frequent amino acid at both positions. Previous studies have shown that when a proline residue is present at Yaa position, it can be hydroxylated to 4-HyP, whereas a proline residue located at Xaa position can be converted to 3-HyP 10,11. However, exception was found that 4-HyP can also be identified at the Xaa position of Gly-Pro-Ala 12.
In addition to localization differences, 3-HyP and 4-HyP have distinct functional roles, which affect the stability, bioactivity and turnover of the proteins wherein they are present. It has been well established that in collagen, 4-HyP stabilizes the triple helical structure 13,14. Later studies have shown that the Gly-3-HyP-Pro destabilized the triple helix of collagen 15. Recently, differentiation of 3-HyP and 4-HyP began to attract more attention with the discovery that the loss of 3-HyP rendered catastrophic phenotypic consequences. For example, the absence of 3-HyP at P986 in collagen α1(I) and α1(II) was found in case of osteogenesis imperfecta 16,17,18. Prolyl-3-hydroxylase-2(P3H2), an enzyme responsible for catalyzing the conversion of proline to 3-HyP, was associated with severe non-syndromic high myopia19,20,21. The function of 3-HyP has also been implicated in the regulation of tendon collagen during developmental stage 22.
Despite the distinct functions of 3-HyP and 4-HyP isomers in the cell, limited approaches have been developed to identify and discriminate these two isomeric residues. Amino acid analysis was originally employed for the discovery of 3-HyP 23. Later, Edman degradation established itself as the standard method for analysis 24. With the development of modern mass spectrometry, PTM analyses have become faster and simpler. Using a low-resolution ion trap mass spectrometer, Eyre et al. characterized several previously unknown 3-HyP along fibrillar collagen chains 25,26,27. The study revealed a high content of 3-HyP at the C-terminus of Type I collagen with repeating sequence of Gly-Pro-Pro (GPP) specifically in tendon but not in skin and bone, indicating its unique role in tissue assembly26. However, tandem MS was not performed for the 3-HyP containing peptides, which cannot confirm their sequence information. Moreover, the modifications were deduced by comparing the mass difference between experimentally measured ions and theoretically predicted ions, generating no valuable information for pinpointing the site-specific localization. To circumvent these problems, several studies have utilized multiple fragmentation techniques to interrogate collagen tryptic peptides, including collision-induced dissociation (CID), electron transfer dissociation (ETD) and higher-energy collisional dissociation (HCD) 28,29. These combined fragmentation techniques enabled identification of 98 HyP at Yaa position and 9 HyP at Xaa position from human type V collagen (CO5A1). Surprisingly, HyP at Xaa position of Gly-HyP-Ala, Gly-Hyp-Val, Gly-HyP-Gln were also reported, with no known enzyme responsible for the hydroxylation of the substrates. Therefore, these hydroxylation sites cannot be assigned to either 3-HyP or 4-HyP. Upon HCD fragmentation, the exact same mass was generated for the two isomers, making it indistinguishable between 3-HyP and 4-HyP residues. Even for the HyP located at the Xaa position of GPP motif, the assignment of 3-HyP was often inferred based on known collagen motifs. Since 4-HyP has been also identified at Xaa position 12, the assignment of hydroxyproline isomers simply depending on its localization at Xaa or Yaa position of Gly-Xaa-Yaa triplets is not accurate enough. The utilization of ETD alone, on the other hand, generates no c or z ions N-terminal to proline residue, as the ring structure of proline remains intact after the cleavage 30.
In 1990, Kassel et al. demonstrated that the w ions can be produced by a high-energy collision activated dissociation (HE CAD) method. The formation of w ions creates different masses for 3-HyP and 4-HyP, resulting in discrimination between the 3-HyP and 4-HyP isomers 31. Despite some success, only two peptides from the mussel adhesive protein were tested and the instrument used to implement the HE CAD is no longer manufactured.
In the present study, we demonstrate, for the first time, that diagnostic w ions can be produced for 3-HyP/ 4-HyP discrimination by a recently developed electron-transfer and higher-energy collision dissociation (EThcD), a dual fragmentation technique introduced by Heck and coworkers 32. The integrated strategy combines online LC separation with EThcD for large-scale differentiation of 3-HyP/4-HyP isomers from complex protein digest mixtures. This approach allows unambiguous localization of HyP residues. An automated algorithm was developed for charge dependent characterization of 3-HyP/ 4-HyP isomers. This novel discrimination strategy can be readily implemented into de novo sequencing workflows or other proteomic search engines.
Experimental Section
Pancreatic tissue protein extraction
Five discarded adult human pancreata were obtained from the University of Wisconsin Organ and Tissue Donation Services Organization within 24 hours of donation, with donor next of kin consent for research. The parenchyma was separated from vascular and adipose tissue and then cut into 1 cm3 pieces and frozen at −80 °C for storage prior to decellularization. Pancreata were treated with deoxycholate and lipase to decellularize the tissue and the resulting pancreatic extracellular matrix (ECM) was lyophilized and stored at −80 °C until analysis. A combination of physical tissue disruption (homogenization) and solubilization buffer were utilized to extract the samples. A detailed description of protein digestion and instrument operation methods can be found in supporting information and previous report 33.
Results and discussion
The discrimination of 3-HyP /4-HyP isomers has always been difficult with mass spectrometry techniques. Upon HCD fragmentation, the exactly identical mass is generated for the two isomers, making them indistinguishable between each other. ETD, on the other hand, cannot cleave the ring structure of proline to generate c or z. ions. Therefore, the assignment of hydroxyproline isomers was often inferred simply depending on its localization at Xaa or Yaa position of the Gly-Xaa-Yaa motif, which is not always true 12. To address these limitations, an EThcD approach was employed in this study. The diagnostic w ions were reliably produced to discriminate 3-HyP/ 4-HyP isomers. An improved peptide coverage and sequence-informative fragment ions were demonstrated for the characterization of ECM molecules. An automated algorithm was developed for the charge dependent characterization of 3-HyP/ 4-HyP isomers. By applying this integrated pipeline, more than fivefold of 3-HyP and 4-HyP sites were identified compared to previous studies and some unusual 3-HyP sites were also observed.
Generation of z. ions and w ions from the HyP residues using EThcD
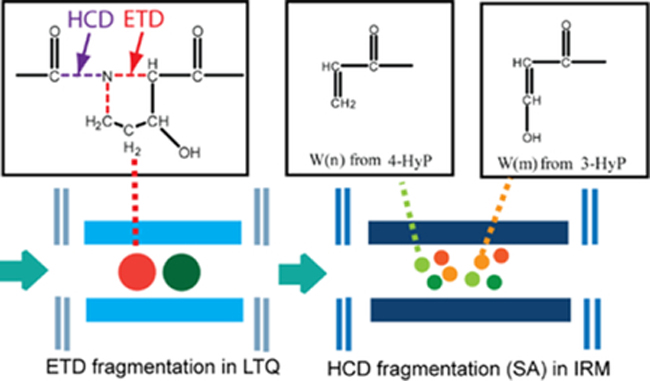
One of the caveats for ETD/ECD fragmentation is its inability to produce c and z. ions from proline/HyP residues. Due to the ring structure of proline/HyP, the peptide remains intact after the cleavage of N-Cα bond 30. Since HCD is capable of cleaving the amide bond, and the N-Cα bond can be cleaved by ETD, we reason that the generation of z. ions from HyP can be successfully achieved by applying EThcD dual fragmentation technique. Figure 1 illustrates the proposed mechanism of w ion formation from the HyP using EThcD. Multiprotonated peptide ions were first isolated and fragmented by ETD in the linear ion trap (LTQ). This created a cleavage of N-Cα bond and reduced the rigidity of cyclic structure of proline side chain. All the product ions of ETD fragmentation were further cleaved by HCD to generate the z. ions at the proline residue. The α-cleavage of the Cβ-Cγ bond of N terminal residue of z. ion is a common secondary fragmentation, leading to the formation of w ions 34. Compared to the even electron cations, the radical cations are less stable, therefore z ions with HyP at the N-terminus can be further fragmented to produce the w ions. Z. ions with 3-HyP at the N-terminus loses C2H3, while z. ions with 4-HyP is transformed to w ions with the loss of C2H3O. Due to the characteristic loss of either C2H3O (43.0184 Da) or C2H3 (27.0235Da) from the side chain of corresponding z. ions, the formation of w ions for 3-HyP/4-HyP isomers create a mass difference of 15.9949 Da, which can be used as a diagnostic ion for reliable identification of either 3-HyP or 4-HyP. To test our hypothesis, EThcD was performed on a synthetic peptide GPPGI*PGR with 4-HyP at position 6. As shown in Figure S1, comprehensive backbone fragmentation was generated in the spectrum, with the observation of both b/y and c/z ions. Both z3 and w3 ions were detected, the corresponding mass difference matched the characteristic loss of C2H3O. The result indicated the presence of 4-HyP at position 6, which correlated well with structure and localization of HyP in the synthetic peptide. Upon validating our strategy with peptide standard, we challenged ourselves to explore the application of this method in the real sample. Decellularized human pancreas was investigated in the study which contained relatively high abundance of the HyP-containing proteins. Figure 2 shows the EThcD spectrum of triply charged peptide DGLNGL*PGPIG*P*PGPR with three HyP sites at positions 7, 12 and 13 (marked with *). The presence of z4, z5, z10 ions can be revealed from the spectrum (marked with star), corresponding to the formation of z. ions from HyP residues. The fragmentation of z4 ion resulted in the generation of w4 ion with m/z of 383.2034, creating a mass difference of 43.0173. The mass difference matched to the characteristic loss of C2H3O (43.0184 Da), indicating the presence of 4-HyP at position 13. Similarly, the ejection of C2H3O from z10 ion enabled the reliable identification of 4-HyP at position 7. Due to the characteristic loss of C2H3 from z5 ion, we could confidently assign 3-HyP to position 12. Position 12 (P986) contains a well-characterized HyP residue that has been extensively examined in various studies11,16,17,25. It can be fully hydroxylated by P3H in fibrillar collagen like α1(I), α1(II), α2(V). However, the percentage of the modification is decreased or absent in recessive osteogenesis imperfecta, indicating its significant role in genetic dysfunction 17,18. In line with previous studies, our 3-HyP/ 4-HyP discrimination approach could confidently assign its identity as 3-HyP, demonstrating the feasibility of our method. It is worth noting that besides the identification of 4-HyPs at positions 7, 13 and 3-HyP at position 12, we were also able to discriminate Leu/Ile isomers in the same EThcD spectrum. The loss of isopropyl radical identified Leucine at position 6, while the ethyl radical is ejected from the z7 ion, confirming the presence of isoleucine at position 10.
Figure 1.

Formation of w ions from HyP via EThcD
Figure 2.
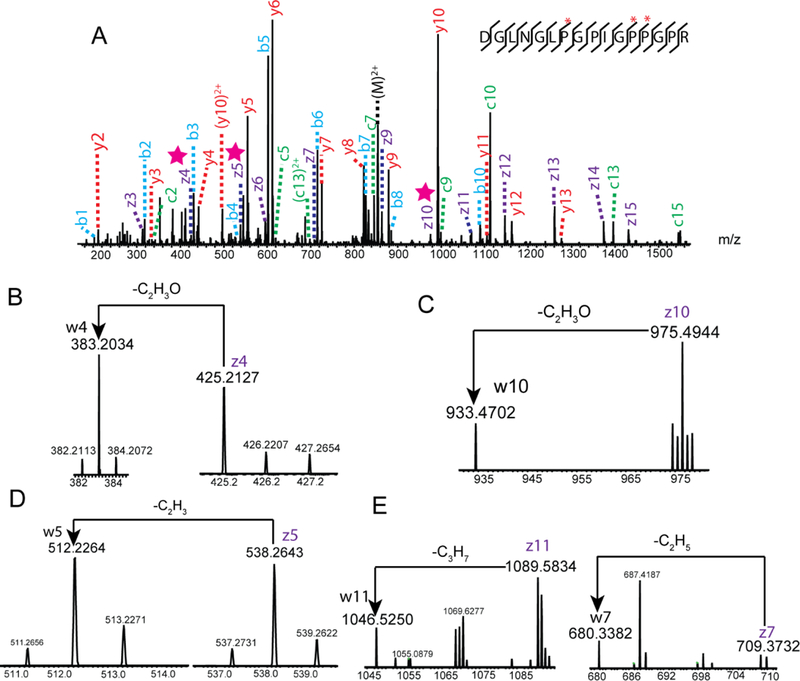
(A) An EThcD fragmentation spectrum of the triply charged peptide DGLNGLPGPIGPPGPR, (B) Formation of w4 ion from z4 ion at position 13, (C) Formation of w10 ion from z10 ion at position 7, (D) Formation of w5 ion from z5 ion at position 12, and (E) Differentiation of Ile/Leu at position 6, 10 (HyP sites are marked with *, z. ions are marked with star).
Optimization of NCE (Normalized Collision Energy)
To obtain the w ions for the differentiation of hydroxyproline isomers, it is very important to choose the right NCE. Lebedev and coworkers were able to use NCE ranging from 0~25 to generate the w ions, allowing confident discrimination of Leu/Ile isomers 8. Compared with the side chain of Leu/Ile, the cyclic structure of proline is more rigid. Therefore, we reasoned that a higher NCE was needed for the formation of w ions from HyP. Figure 3 shows the fragmentation patterns of resulting EThcD spectra of a triply charged ion of GIPGPVGAAGATGAR peptide at various levels of collisional energy. At NCE 10, neither z13 ion nor w13 ion was observed for position 3 in the EThcD spectrum, with limited sequence information due to the absence of fragment ion at positions 5 (Data not shown). At NCE 25, both z13 ion and w13 ion were present, with isotopic cluster for z13. The mass of w13 ion was reduced by 43.0171 Da due to the C2H3O loss, confirming the presence of 4-HyP. With confident identification of a series of fragment ions at positions 5, the complete sequence assignment could be achieved. A further increase of NCE to 33 kept the presence of both z13 and w13 ions, with the addition of isotopic cluster of w13. The relative intensity of the zoomed-in spectrum also reached the maximum value (1.08 E5). By applying higher NCE of 40, w13 and z13 ions can still be observed, with slight decrease of their intensities (5.39 E4). The intensities of both w13 and z13 ions were further diminished with the increase of NCE to 45 and 50. As expected, the formation of w ions and z. ions for proline and HyP requires higher NCE compared to the formation of w ions for Leu/Ile. On the other hand, multiple fragmentation pathways could be activated at higher NCE like 45 and 50, resulting in more complicated mass spectra. Similar phenomenon was observed for other peptides as well. Since the best performance was achieved at NCE 33 and 40, these two NCE were chosen for further analysis.
Figure 3.
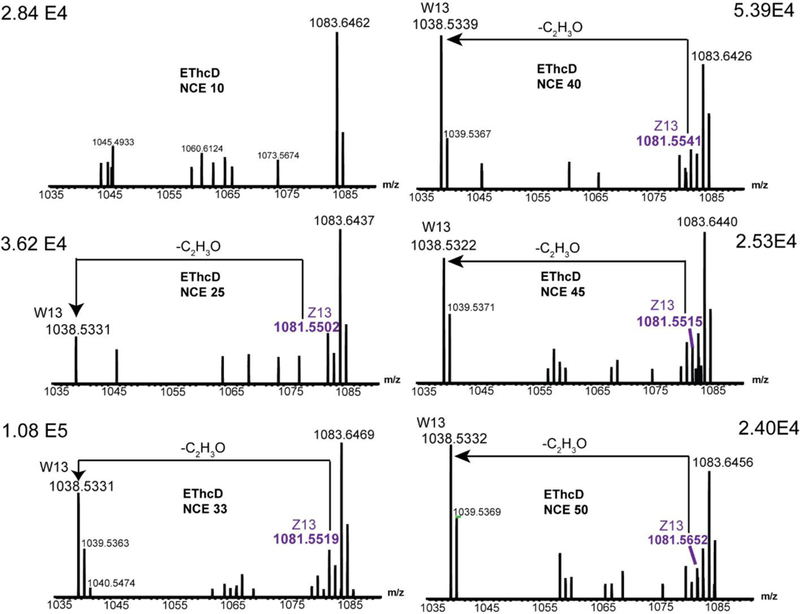
EThcD fragmentation spectra of a triply charged ion of GIPGPVGAAGATGAR peptide. Different NCE values were applied (Left panels, NCE 0, 25, 33. Right panels, NCE, 40, 45, 50).
EThcD enhances the characterization of ECM peptides
Trypsin has become one of the most commonly used proteases in MS-based bottom up proteomics. However, this enzyme often suffers from insufficient cleavage of peptide chain at carboxyl side of arginine and lysine when either is followed by proline 35. A known limitation of ETD is its inability to cleave the N-Cα bond of proline, whereas HCD often suffers from insufficient dissociation of peptide backbone when an internal residue like arginine or lysine is present. Collagens are major components of ECM, and contain many different isoforms, therefore developing an effective method for characterizing collagen would help unravel the complexity of ECM composition. Given the high content of proline and HyP in the collagen, many tryptic digested peptides may contain internal residues whose sequence information could not be comprehensively characterized by using either ETD or HCD fragmentation method alone. However, the utilization of EThcD dual fragmentation technique could overcome these limitations due to the generation of information-rich tandem mass spectrum and the more comprehensive backbone fragmentation. As exemplified in Figure 4, the 16-residue tryptic peptide AGEDGH*PGKPGR*PGER has two HyP sites at positions 7 and 13. The peptide contains two missed cleavages at positions 9 and 12. After ETD fragmentation, 23 of the sequence-informative fragment ions were detected in the spectrum, corresponding to cleavage of twelve of the fifteen total inter-residue positions (80% sequence coverage). The c and z. ions not observed belonged to the bond adjacent to the proline residue. Similarly, HCD produces 17 b- and y- type product ions, providing incomplete sequence information (86% sequence coverage) due to the absence of b and y ions derived from the arginine fragmentation. The dual fragmentation technique EThcD generates complementary c/z. and b/y ions, resulting in highly confident peptide sequence assignment (100% sequence coverage). More than two-fold of the sequence-informative fragment ions (48 of c/z. and b/y ions) can be observed compared to ETD or HCD alone. The improvement of spectral quality is also reflected by the establishment of two HyP sites at positions 7 and 13 (marked in stars), through the generation of doubly charged z10 ion with the m/z of 606.3396 and the singly charged z4 ion with the m/z of 458.2018. When the 4-HyP is located at position 13, C2H3O should be ejected from z4 ion and the generated w4 ion should have a mass difference of 43.0184 Da compared to z4. An intensive peak with m/z of 415.1936 was observed in the EThcD spectrum, corresponding to the w4 ion. Therefore, we were able to confirm that the HyP at position 13 belongs to 4-HyP. Similarly, the doubly charged w10 ion with the isotopic cluster was identified in the EThcD spectrum. The w10 ion peak of m/z of 584.8326 corresponded to the loss of C2H3O, allowing the confident assignment of residue 7 as 4-HyP.
Figure 4.

Fragmentation of a quadruply charged ion of AGEDGHpGKPGRpGER by (A) ETD, (B) HCD and (C) EThcD (HyP sites are marked with *, z. ions are marked with star).
An automated algorithm for retrieval of HyP- containing peptides
An automated algorithm to retrieve HyP-containing peptides and their spectral information was developed. A detailed description of database search can be found in the online method. Briefly, we first searched the Uniprot Human protein database using pFind 2.8 36. Then those peptides with hydroxylated proline were filtered out under the 1% false discovery rate (FDR) at the spectral level for later analysis. The discriminative marker ions before 3-HyP and 4-HyP were then searched, matched, and retrieved automatically using an in-house program. Development of the HyP algorithm as a plug-in for the pFind software is under way.
Hydrogen rearrangement (HR) is a common phenomenon in ETD and ECD based fragmentation 37,38. It was proposed that upon generation of c and z. ions through electron transfer, they were held together in close proximity via non-covalent interaction. This can result in the hydrogen abstraction and rearrangement, which generally leads to a mass addition of one hydrogen to the regular z. ions (z.+H) whereas the hydrogen rearranged c ions lose one hydrogen compared to the regular c ions (c-H). It was proven that consideration of hydrogen rearranged ions can significantly improve the number of peptides identified when searching the protein database 36. Given the large-scale high-quality peptide EThcD spectra identified, we can retrieve specific fragment ions related to the two types of hydroxyproline, one is 3-HyP and the other is 4-HyP. Specifically, we attempted to find the fragmentation differences between these two types of hydroxylated proline. Provided that the peptide with n amino acids is AnAn-1…Aj+1PjAj-1…A2A1, and P (Proline) is hydroxylated, we can compute the w ions with mass as zj-1+C3H4NO (70.0293 Da), zj-1 +C3H4NO2 (86.0242 Da), which corresponds to 4-HyP and 3-HyP cleavages, respectively (Figure S2). We then searched each EThcD spectrum from all those hydroxylated peptides to find which peak can be matched to those w ions under the mass tolerance of ±0.01 Da. If a peptide precursor is highly charged, such as +3 or +4, the fragment ions may also carry multiple charges. Therefore, we also calculated its +2 fragment ions in this study. Due to the presence of HR in the EThcD spectra, the hydrogen rearranged ions zj-1+C3H5NO (71.0371 Da), zj-1 +C3H5NO2 (87.0320 Da) were taken into considerations as well when searching the w ions (Figure S3).
Large-scale identification of hydroxyproline sites
Upon demonstrating the capability of our strategy, we applied the integrated 3-HyP/4-HyP discrimination approach to determine the 3-HyP and 4-HyP sites in the decellularized pancreas, the tissue scaffold which has an enrichment of ECM proteins. The samples were digested by trypsin and analyzed by online LC-MS method. In total, we identified 108 3-HyP sites in the X position of Gly-Xaa-Yaa triplets and 530 4-HyP sites in the Y position of Gly-Xaa-Yaa triplets. This is, by far, the largest dataset of 3-HyP and 4-HyP identifications in a single experiment (Figure S4). Unlike previous MS analysis that deduced the 3-HyP/4-HyP isomers based on their sequence information, our study utilized the w ions as a signature to confidently assign these isomers to their respective positions. By comparing the hydroxyproline sites identified in our study with those reported by Weis et al. 25, we were able to detect all hydroxyproline sites from CO1A1, CO1A2, CO3A1, CO5A2 except P470 from CO5A2. Since the samples we used in this work underwent extensive decellularization with detergent, we believe that peptide sequence not observed was not well retained during the decellularization process. As exemplified in Figure S5, trypsin digestion allows the identification of 111 4-HyP sites and 28 3-HyP sites in CO1A1, with the sequence coverage of 91.3% for the triple helical domain. Of the 28 3-HyP sites, 19 of them were found at Xaa position of Gly-HyP-HyP motif, including the well characterized 3-HyP at P986 25,26,27. Here we observed a high content of 3-HyP at the C-terminal (GPP)n domain of human CO1A1, whose presence was originally found only in vertebrates, in a tissue specific manner by Eyre and the coworkers 26,39. Unexpectedly, 3-HyP was also identified in several unusual triple helical motifs, including Gly-HyP-Ala, Gly-HyP-Val and Gly-HyP-Gln. As prolyl-4-hydroxylase is responsible for proline hydroxylation at Yaa position of Gly-Xaa-Yaa triplets and prolyl-3-hydroxylase is thought to hydroxylate only proline in Gly-Pro-HyP triplets, it is unclear which enzyme is responsible for these unique HyP sites. Similar observations were also reported by previous studies 28,29. Due to the lack of diagnostic ion, these HyP sites were not confidently assigned to either 3-HyP or 4-HyP. By using our 3-HyP/4-HyP discrimination strategy, we were able to not only confirm the identity of these HyP sites as 3-HyP, but also to identify two novel 3-HyP sites in Gly-HyP-Ser at P608, P710, and one in Gly-HyP-Arg in P989 of Col1A1 protein.
Conclusions
We demonstrate, for the first time, that diagnostic w ions can be produced by hybrid fragmentation technique EThcD to confidently discriminate 3-HyP/ 4-HyP isomers in collagen proteins. We developed an integrated strategy which combined online LC separation with EThcD for the large-scale differentiation of 3-HyP/4-HyP in complex protein digest mixtures. This approach outperformed ETD and HCD alone in the analysis of HyP-containing peptides with discrimination of HyP isomers, improved fragmentation of precursor ions, allowing unambiguous localization. An automated algorithm was developed for charge state dependent characterization of 3-HyP/ 4-HyP isomers. Using this integrated 3-HyP/4-HyP discrimination approach to analyze the decellularized pancreas, we identified 108 3-HyP sites in the X position of Gly-Xaa-Yaa triplets and 530 4-HyP sites in the Y position of Gly-Xaa-Yaa triplets, which significantly boosted the identifications of both 3-HyP and 4-HyP sites reported previously by more than fivefold. A high content of 3-HyP was observed from the C-terminal (GPP)n domain of human CO1A1, which was previously only identified in vertebrate fibrillar collagens from certain tissues. Unexpectedly, some unusual HyP sites at Xaa position in Gly-HyP-Ala, Gly-HyP-Val, Gly-HyP-Gln, Gly-HyP-Ser and Gly-HyP-Arg were also confirmed to be 3-hydroxylated, whose functions and enzymes are yet to be discovered. Taken together, this novel discrimination strategy can be readily implemented into de novo sequencing or other proteomic search engines. It is also applicable for reliable production of z. ions for proline residues and other cyclic amino acids.
Additionally, HyP has been found in a multiplicity of non-collagenous proteins (e.g. HIF-1α, prion protein, conotoxins, etc.). Unlike the GPP triplets of collagen, the corresponding substrate recognition sites and the type of hydroxylases for non-collagenous proteins are mostly unknown. Therefore, our 3-HyP/4-HyP discrimination approach opens up a new avenue for the characterization of HyP isomers for these proteins. As an ancient and conserved modification, various unsolved mysteries and correlations have been found between HyP and other PTMs. In plants, the cell wall consists of hydroxyproline-rich glycoproteins, and the type and extent of O-glycosylation is related to the location and context of HyP residues 10,40. Phosphorylation of hydroxyproline (O-phosphohydroxyproline) was recently found in the protein α-crystallin A of rat heart tissue, with undetermined position of phosphate group and HyP isomer 41,42. Since EThcD has been successfully implemented for characterizing phosphoproteome and glycoproteome 43,44, 45,46, we reason that our method is applicable to simultaneously capture various types of PTMs in a single experiment. With the capacity to uncover the interplay among different PTMs, our approach possesses the potential to enable a better understanding of the molecular and cellular processes inside the cell and translate various PTM cross-talks into valuable biological and mechanistic insights.
Supplementary Material
ACKNOWLEDGMENTS
This research was supported in part by the National Institutes of Health grants R21AI126419, R01DK071801, R01AG052324, and P41GM108538. The Orbitrap instruments were purchased through the support of an NIH shared instrument grant (NIH-NCRR S10RR029531) and Office of the Vice Chancellor for Research and Graduate Education at the University of Wisconsin-Madison. LL acknowledges a Vilas Distinguished Achievement Professorship and Janis Apinis Professorship with funding provided by the Wisconsin Alumni Research Foundation and University of Wisconsin-Madison School of Pharmacy.
Footnotes
COMPETING FINANCIAL INTEREST
The authors declare no competing financial interests.
Supporting Information
The supporting information is available and noted in the text. The material is available free of charge via the Internet at http://pubs.acs.org.
Reference
- (1).Aebersold R; Mann M Nature 2016, 537 (7620), 347–355. [DOI] [PubMed] [Google Scholar]
- (2).Tao WA; Cooks RG Anal. Chem 2003, 75 (1), 25A–31A. [DOI] [PubMed] [Google Scholar]
- (3).Shvartsburg AA; Creese AJ; Smith RD; Cooper HJ Anal. Chem 2010, 82 (19), 8327–8334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Roher AE; Lowenson JD; Clarke S; Wolcow C; Wang R; Cotter RJ; Reardon IM; Zurcher-Neely HA; Heinrikson RL; Ball MJ; Greenberg BD J. Biol. Chem 1993, 268 (5), 3072–3083. [PubMed] [Google Scholar]
- (5).Fomby P; Cherlin AJ Bioanal. Rev 2009, 72 (2), 181–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Creese AJ; Smart J; Cooper HJ Anal. Chem 2013, 85 (10), 4836–4843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Kjeldsen F; Haselmann KF; Sørensen ES; Zubarev RA Anal. Chem 2003, 75 (6), 1267–1274. [DOI] [PubMed] [Google Scholar]
- (8).Lebedev AT; Damoc E; Makarov AA; Samgina TY Anal. Chem 2014, 86 (14), 7017–7022. [DOI] [PubMed] [Google Scholar]
- (9).Jia C; Lietz CB; Yu Q; Li L Anal. Chem 2013, 86, 2972–2981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Gorres KL; Raines RT Prolyl 4-Hydroxylase; 2010; Vol. 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Eyre DMH and D. R. October 2013, 141 (4), 520–529. [Google Scholar]
- (12).Kimuraeii T; Cheahllll KSE; Chanll SDH; Luili VCH; Mattei M; Rest M Van Der; Ono K; Solomonllll E; Olsenll R J. Biol. Chem 1989, 264 (23), 13910–13916. [PubMed] [Google Scholar]
- (13).Berg RA; Prockop DJ Biochem. Biophys. Res. Commun 1973, 52 (1), 115–120. [DOI] [PubMed] [Google Scholar]
- (14).Kotch Frank W., Guzei Ilia A., R. T. R. J. Am. Chem. Soc 2008, 130, 2952–2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Inouye Katsuhiko, Kobayashi Y; Prockop J Arch. Biochem. Biophys 1982, 219 (1), 198–203. [DOI] [PubMed] [Google Scholar]
- (16).Morello R; Bertin TK; Chen Y; Hicks J; Tonachini L; Monticone M; Castagnola P; Rauch F; Glorieux FH; Vranka J; Bachinger HP; Pace JM; Schwarze U; Byers PH; Weis MA; Fernandes RJ; Eyre DR; Yao Z; Boyce BF; Lee B Cell 2006, 127 (2), 291–304. [DOI] [PubMed] [Google Scholar]
- (17).Cabral W. a; Chang W; Barnes AM; Weis M; Scott M. a; Leikin S; Makareeva E; Kuznetsova NV; Rosenbaum KN; Tifft CJ; Bulas DI; Kozma C; Smith P. a; Eyre DR; Marini JC Nat. Genet 2007, 39 (3), 359–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Dijk F. S. Van; Nesbitt IM; Zwikstra EH; Nikkels PGJ; Piersma SR; Fratantoni SA; Jimenez CR; Huizer M; Morsman AC; Cobben JM; Roij M. H. H. Van; Elting MW; Verbeke JIML; Wijnaendts LCD; Mckeown C; Sistermans EA; Dalton A; Shaw NJ; Ho W Am. J. Hum. Genet 2009, 85 (4), 521–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Hudson DM; Joeng KS; Werther R; Rajagopal A; Weis M; Lee BH; Eyre DR J. Biol. Chem 2015, 290 (130), 8613–8622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Mordechai S; Gradstein L; Pasanen A; Ofir R; Amour K. El; Levy J; Belfair N; Lifshitz T; Joshua S; Narkis G; Elbedour K; Myllyharju J; Birk OS Am. J. Hum. Genet 2011, 89 (3), 438–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Guo H; Tong P; Peng Y; Wang T; Liu Y; Chen J; Li Y; Tian Q; Zheng Y; Xiao L; Xiong W; Pan Q; Hu Z; Homozygous XK Clin. Genet 2014, 86, 575–579. [DOI] [PubMed] [Google Scholar]
- (22).Taga Y; Kusubata M; Ogawa-Goto K; Hattori SJ Biol. Chem 2016, 291 (2), 837–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).James Ogle, Arlinghaus.Ralph LM J. Biol. Chem 1962, 237 (12), 3667–3673.13939597 [Google Scholar]
- (24).Fietzek PP; Rexrodt FW; Hopper KE; Kuhn K Eur. J. Biochem 1973, 400 (38), 396–400. [DOI] [PubMed] [Google Scholar]
- (25).Weis MA; Hudson DM; Kim L; Scott M; Wu JJ; Eyre DR J. Biol. Chem 2010, 285 (4), 2580–2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Eyre DR; Weis M; Hudson DM; Wu J; Kim LJ Biol. Chem 2011, 286 (10), 7732–7736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Fernandes RJ; Farnand AW; Traeger GR; Weis MA; Eyre DR J. Biol. Chem 2011, 286 (35), 30662–30669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Yang C; Park AC; Davis NA; Russell JD; Kim B; Brand DD; Lawrence MJ; Ge Y; Westphall MS; Coon JJ; Greenspan DS J. Biol. Chem 2012, 287 (48), 40598–40610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Song E; Mechref Y J. pr 2013, 12, 3599–3609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Mikesh LM; Ueberheide B; Chi A; Coon JJ; Syka JEP; Shabanowitz J; Hunt DF Biochimica et Biophysica Acta - Proteins and Proteomics 2006, pp 1811–1822. [DOI] [PMC free article] [PubMed]
- (31).Kassel DB; Biemann K Anal. Chem 1990, 62 (15), 1691–1695. [DOI] [PubMed] [Google Scholar]
- (32).Frese CK; Altelaar AFM; Van Den Toorn H; Nolting D; Griep-Raming J; Heck AJR; Mohammed S Anal. Chem 2012, 84 (22), 9668–9673. [DOI] [PubMed] [Google Scholar]
- (33).Liu F; Ma F; Wang Y; Hao L; Zeng H; Jia C; Wang Y; Liu P; Ong IM; Li B; Chen G; Jiang J; Gong S; Li L; Xu W Nat. Cell Biol 2017, 19 (11), ncb3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Li X; Lin C; Han L; Costello CE J. Am. Soc. mass Spectrom 2010, 21 (4), 646–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Olsen JV; Ong S; Mann M Mol. Cell. Proteomics 2004, 608–614. [DOI] [PubMed]
- (36).Wang Le-heng, Li De-Quan, Fu Yan, Wang Hai-Peng, J.-F. Z.; Yuan Zuo-Fei, Sun Rui-Xiang, Zeng Rong, He Si-Min, and W. G. Rapid Commun. Mass Spectrom 2010, 24 (24), 3567–3577.21108305 [Google Scholar]
- (37).Sun R; Dong M; Song C; Chi H; Yang B; Xiu L; Tao L; Jing Z; Liu C; Wang L; Fu Y; He S J. Proteome Res 2010, 6354–6367. [DOI] [PubMed]
- (38).Savitski MM; Kjeldsen F; Nielsen ML; Zubarev RA J. Am. Soc. mass Spectrom 2007. [DOI] [PubMed]
- (39).Hudson DM; Werther R; Weis M; Wu JJ; Eyre DR PLoS One 2014, 9 (4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Shpak E; Leykam JF; Kieliszewski MJ Proc. Natl. Acad. Sci. U. S. A 1999, 96 (26), 14736–14741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Kühlberg A; Haid M; Metzger SJ Biol. Chem 2010, 285 (41), 31484–31490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Piggott MJ; Attwood PV Amino Acids 2017, 49 (8), 1309–1323. [DOI] [PubMed] [Google Scholar]
- (43).Frese CK; Zhou H; Taus T; Altelaar AFM; Mechtler K; Heck AJR; Mohammed SJ Proteome Res 2013, 12 (3), 1520–1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Yu Q; Wang B; Chen Z; Urabe G; Glover MS; Shi X; Guo LW; Kent KC; Li LJ Am. Soc. Mass Spectrom 2017, 28 (9), 1751–1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Liu MQ; Zeng WF; Fang P; Cao WQ; Liu C; Yan GQ; Zhang Y; Peng C; Wu JQ; Zhang XJ; Tu HJ; Chi H; Sun RX; Cao Y; Dong MQ; Jiang BY; Huang JM; Shen HL; Wong CCL; He SM; Yang PY Nat. Commun 2017, 8 (1), 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Tian Yuan; Kelly-Spratt KS; Kemp CJ; Zhang H J. Proteome Res 2011, 9 (11), 5837–5847. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


