Abstract
The role of mitochondrial DNA (mtDNA) mutations in cancer remains controversial. Ulcerative colitis is an inflammatory bowel disease that increases the risk of colorectal cancer and involves mitochondrial dysfunction, making it an ideal model to study the role of mtDNA in tumorigenesis. Our goal was to comprehensively characterize mtDNA mutations in ulcerative colitis tumorigenesis using Duplex Sequencing, an ultra-accurate next-generation sequencing method. We analyzed 46 colon biopsies from non-ulcerative colitis control patients and ulcerative colitis patients with and without cancer, including biopsies at all stages of dysplastic progression. mtDNA was sequenced at a median depth of 1,364x. Mutations were classified by mutant allele frequency: clonal > 0.95, subclonal 0.01–0.95, and very low frequency (VLF) < 0.01. We identified 208 clonal and subclonal mutations and 56,764 VLF mutations. Mutations were randomly distributed across the mitochondrial genome. Clonal and subclonal mutations increased in number and pathogenicity in early dysplasia, but decreased in number and pathogenicity in cancer. Most clonal, subclonal, and VLF mutations were C>T transitions in the heavy strand of mtDNA, which likely arise from DNA replication errors. A subset of VLF mutations were C>A transversions, which are probably due to oxidative damage. VLF transitions and indels were less abundant in the non–D-loop region and decreased with progression. Our results indicate that mtDNA mutations are frequent in ulcerative colitis preneoplasia but negatively selected in cancers.
Implications
While mtDNA mutations might contribute to early ulcerative colitis tumorigenesis, they appear to be selected against in cancer, suggesting that functional mitochondria might be required for malignant transformation in ulcerative colitis.
Introduction
While the role of nuclear DNA mutations in cancer has been extensively characterized, the contribution of mitochondrial DNA (mtDNA) mutations to carcinogenesis remains unclear. For some time, the prevailing hypothesis was that mtDNA mutations contribute to tumor progression by impairing oxidative phosphorylation and promoting aerobic glycolysis, a feature of cancer cells known as the Warburg effect (1–3). Mounting evidence, however, has challenged this idea by revealing that cancer cells rely on oxidative phosphorylation and functional mitochondria for ATP production and rapid cell growth (4, 5). Recent studies also demonstrate that mtDNA mutations accumulate randomly and clonally expand without selective pressure or, if deleterious, they are selected against (6, 7). These results call into question a driving role of mtDNA mutations in tumor progression and their contribution to the Warburg effect.
Ulcerative colitis is an inflammatory bowel disease that serves as an excellent model for studying mtDNA mutations in preneoplastic progression. Ulcerative colitis causes chronic inflammation of the colonic epithelium and affected patients have an elevated risk for colorectal cancer (8–10). Tumorigenesis in this disease follows a distinct pattern of progression from negative for dysplasia (Neg) to low-grade dysplasia (LGD), high-grade dysplasia (HGD), and finally cancer. In patients that develop colorectal cancer, molecular alterations are found not only in dysplastic tissue but in histologically normal tissue surrounding dysplasia (11–13) indicating the presence of a field effect, or field cancerization (9, 14). These premalignant fields offer a unique opportunity to study the early molecular events that contribute to tumor progression, as well as their evolution across all dysplastic stages into malignancy.
Mitochondrial dysfunction has been demonstrated in ulcerative colitis (15), but there is conflicting literature regarding its contribution to cancer progression (14). The conflict might arise from the fact that mitochondrial alterations could play different roles in early and late disease. Using cytochrome c oxidase subunit I (COXI) IHC, our group previously reported mitochondrial loss in premalignant lesions but a recovery of normal levels of mitochondria in cancer (16). On the basis of these observations, we hypothesized that while dysfunctional mitochondria might contribute to early dysplasia, functional mitochondria are essential at the cancer stage. Furthermore, this dual pattern might be mirrored in mtDNA mutations, with an increase of mutations in early dysplasia followed by negative selection of mtDNA mutations in cancer.
Previous studies of the role of mtDNA in cancer have used next-generation sequencing (NGS) technologies to analyze mutations (6, 7, 17, 18). However, conventional NGS has an error rate of 1 in 100–1,000 bp (19), which precludes the accurate detection of mutations with mutant allele frequency (MAF) < 0.01 (20). The detection of low frequency mtDNA mutations is essential to characterize the underlying mutagenic processes, as well as to detect small clones that might arise during carcinogenesis. Thus, in this study, we have utilized a double-strand molecular-tagging method called Duplex Sequencing (Fig. 1A and B), which performs error correction by scoring only mutations found on both strands of DNA independently (21). The estimated error rate is less than 1 in 107, which enables the identification of mutations at frequencies as low as 0.0001 (21, 22).
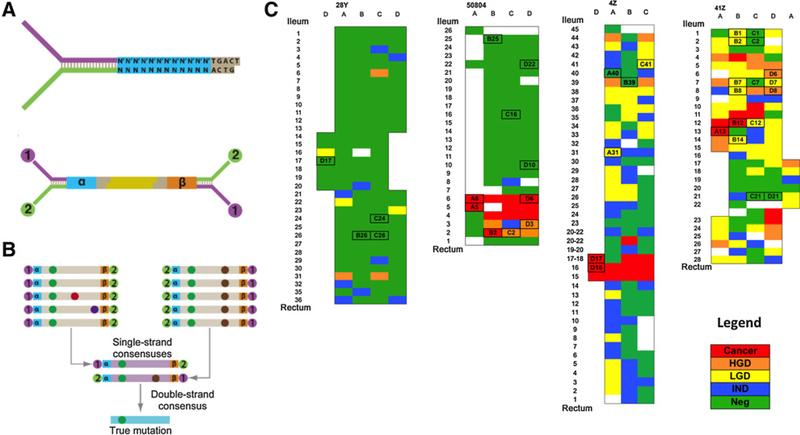
Figure 1.
Experimental design. A, Schematic of the Duplex Sequencing method. Duplex Sequencing adapters contain a molecular tag consisting of a randomized nucleotide sequence [represented as a (cyan) and b(orange)] and two universal priming sites (purple and green). DNA is fragmented (yellow) and ligated to Duplex Sequencing adapters. B, Reads from the same strand of a DNA molecule are used to produce a single-strand consensus sequence (SSCS). Then the two complementary SSCS generated from the same original DNA molecule are condensed into a double-strand consensus sequence. Only mutations found on the two complementary SSCS are considered true mutations. C, Colon maps for each of the Progressor patients as diagrammed by the pathologist after colectomy. Each box corresponds to an individual biopsy and is color-coded according to histologic findings: Neg, negative for dysplasia; IND, indefinite for dysplasia; LGD, low-grade dysplasia; HGD, high-grade dysplasia; and cancer. Biopsies are named on the basis of the coordinates defined by columns (letters) and rows (numbers). Columns correspond to the diameter of the colon divided into 3–4 sections and are approximately 2 cm apart. Rows indicate colon levels and are evenly spaced approximately 2–5cm along the length of the organ. For each colon, the biopsies analyzed are indicated with a box with the biopsy name.
Here we have applied this highly accurate technology to identify the presence of mutations in mtDNA with high confidence. Our goal was to uncover the underlying mechanism of mtDNA mutagenesis in ulcerative colitis and to clarify the role of these mutations in cancer progression. We analyzed the mtDNA of 46 colon biopsies at all histologic stages of progression and detected thousands of mutations. We characterized these mutations by frequency, location, type, pathogenicity, and mutational context, thus producing a comprehensive, high-resolution analysis of mtDNA mutations in preneoplastic progression.
Materials and Methods
Patients and biopsies
The study included 10 patients: 7 with ulcerative colitis and 3 non-ulcerative colitis controls. Four of the patients with ulcerative colitis had progressed to HGD or cancer (Progressors) and the remaining 3 were cancer and dysplasia free (Nonprogressors, NP; Table 1, Supplementary Table S1; Supplementary Methods). Fresh frozen samples were collected at colectomy (patients with ulcerative colitis) or colonoscopy (controls) in accordance with Human Subjects Guidelines and the appropriate Institutional Review Board at the University of Washington (Seattle, WA). A total of 46 colon biopsies were analyzed from these patients, including 36 biopsies that represented all histologic grades in ulcerative colitis Progressors (Fig. 1C; Supplementary Table S1; Supplementary Fig. S1). Thus, we considered six biopsy types in total: normal, NP, Neg, LGD, HGD, and cancer (the last four corresponding to biopsies from Progressors). The biopsies from ulcerative colitis Progressors were selected on the basis of the colon maps generated upon colectomy (Fig. 1C) with the criteria of covering different histologic grades and different areas of progression. Formalin-fixed, paraffin-embedded biopsies adjacent to the frozen biopsies used for analysis were stained with hematoxylin and eosin and examined under a light microscope for acute inflammation (cryptitis and the presence of neutrophils in the epithelium) and chronic inflammation (lymphocytes in the lamina propria). For acute inflammation, scores were assigned the following numeric equivalents: none, 1; mild, 2; and moderate, 3. For chronic inflammation, scores were assigned the following numeric equivalents: none, 1; low, 2; and high, 3. Epithelial isolation and DNA extraction were performed as part of prior studies via EDTA shake-off, which yields approximately 90% enrichment for epithelial cells (refs. 13, 23; Supplementary Methods).
Table 1.
Study design and mutation counts
| Patient type | Number of patients | Dysplastic grade | Number of biopsies | Number of mtDNA mutations |
||
|---|---|---|---|---|---|---|
| MAF > 0.01 | MAF < 0.01 | |||||
| Normal | 3 | Negative | 3 | 5 | 6,790 | |
| Ulcerative colitis NP | 3 | Negative | 7 | 14 | 25,495 | |
| Ulcerative colitis Progressor | 4 | Negative | 15 | 77 | 7,384 | |
| Low grade | 9 | 59 | 5,026 | |||
| High grade | 4 | 22 | 5,417 | |||
| Cancer | 8 | 31 | 6,652 | |||
| Total | 10 | 46 | 208 | 56,764 | ||
Duplex sequencing
For each sample, between 50 and 150 ng of colonic epithelium DNA were processed for Duplex Sequencing of mtDNA as described previously (refs. 21, 24; Fig. 1A and B). DNA was end repaired, A-tailed, and ligated to Duplex Sequencing adapters (Integrated DNA Technologies; Supplementary Methods). To determine the optimal input of ligated DNA for amplification, samples were qPCR amplified with a Duplex Sequencing adapter specific primer (MWS13, 5′-AATGATACGGCGACCACCGAG-3′) and a primer from an internal mitochondrial sequence (MitoRev, 5′-GCGCrTACTTTGTAGCCTTCA-3′; both by Integrated DNA Technologies) and titrated against a standard DNA sample. DNA was then captured using the NimbleGen SeqCap Target Enrichment Kit (Roche) or the xGen Lockdown Target Enrichment Kit (IDT) with probes specific for the mitochondrial genome. Samples were indexed, pooled, and sequenced using 2 × 100 bp paired-end reads on the Illumina HiSeq 2500 or 2 × 0 150 bp paired-end reads on the Illumina NextSeq 550.
Data processing
Raw data files were processed as in previous studies (refs. 24, 25; https://github.com/risqueslab/DuplexSequencingScripts) with some modifications. First, consensus-making was performed prior to the alignment of reads. Second, paired read information was retained. Finally, duplex consensus sequence (DCS) reads were aligned using BWA-MEM with default parameters (bio-bwa.sourceforge.net) to a version of human reference genome v37 (GRCh37; ncbi.nlm.nih.gov/grc/human) according to the revised Cambridge reference, which corrects for an error at base 3,107 in previous versions. The Genome Analysis Tool Kit (GATK) version 3.6 (software.broadinstitute.org/gatk) Indel-Realigner was used to perform local realignment of each mapped read. GATK Clip-Reads was used to clip 10bp from both the 5′ and 3′ end of each read to remove low quality reads and artifacts created during end repair and A-tailing. DCS reads with more than 5% indeterminate bases (Ns) were removed. Indeterminate bases occur when there is no consensus. Positions with less than 100 DCS reads were not considered for analysis. The fgbio (https://github.com/fulcrumgenomics/fgbio) tool ClipOverlappingReads was then used to clip any overlapping bases from paired reads.
All samples were sequenced to an average depth of at least 600X. The frequency of Ns was calculated for each position along the genome. For each sample, positions with N ≥ 0.1 were excluded from analysis, but this never represented more than 0.5% of the mtDNA positions. The haplotype of each patient was identified with the Haplogrep Tool (http://haplogrep.uibk.ac.at). To stratify the frequency of mutational events, clonality cut-off values were established on the basis of MAF; very low frequency (VLF) mutations, MAF < 0.01; subclonal mutations, MAF ≥ 0.01 and <0.95; and clonal mutations, MAF ≥ 0.95. Clonal/subclonal mutations represent different degrees of clonal expansion within the colonic tissue. In contrast, VLF mutations could represent small clones or unique de novo events, because they were often supported by a single-mutated DCS read. Of note, mutations identified in a single-DCS read have very low probability of being artefactual (<10−7; ref. 20) because they are independently identified in the two complementary strands of DNA and are produced by the consensus of at least six raw reads (three for each DNA strand). Thus, VLF mtDNA mutations capture the ongoing mutagenic processes at the molecular level as well as small clonal expansions while clonal/subclonal mutations quantify large clonal expansions.
Clonal and subclonal mutation analysis
Clonal and subclonal mutations were analyzed jointly and compared across the spectrum of biopsy types in the study, that is, normal, NP, Neg, LGD, HGD, and cancer. Mutations found in all samples from a given colon and with >75% of samples having a mutation frequency ≥ 0.80 were considered constitutional to the patient and removed from consideration. For colons where only one sample was analyzed, mutations with a frequency ≥ 0.99 that were commonly identified polymorphisms in the human population were also considered constitutional and thus removed. Clonal and subclonal mutation location was visualized using the Circlize package in R (https://CRAN.R-project.org/package=circlize). Clonal and subclonal mutations were compared across samples based on D-loop mutation frequency, clonality, number of mutations per biopsy, pathogenicity based on MitImpact (26), and mutational signature (Supplementary Methods). The mutational signature analysis was based on the substitution rate, which calculated the number of observed mutations of each type (e.g., C>A, C>G, C>T, T>A, T>C, and T>G) in each mtDNA strand and divided it by the number of expected mutations assuming equal probability for all substitutions.
VLF mutation analysis
Mutations with a MAF < 0.01 were considered VLF. Different mutations identified at the same nucleotide position were independently counted. Similar to clonal/subclonal mutations, VLF mutations were compared across the six biopsy types in the study. However, there were three major differences in the analysis. First, to calculate the frequency of MAF < 0.01 mutations in each biopsy, the number of mutations was divided by the total amount of mtDNA nucleotides sequenced in each biopsy. This was critical to correct for sequencing depth because higher depth results in finding more VLF mutations. Second, to calculate the frequency of each mutation type, the number of mutations for each possible nucleotide substitution was divided by the number of times that nucleotide was sequenced in each given sample. This takes into consideration the depth of sequencing of each sample, as well as the nucleotide composition of the mtDNA. This calculation was done separately for mutations in the D-loop and non–D-loop. Third, due to the much larger number of VLF mutations than clonal/subclonal mutations, the mutational signature analysis could be performed taking into consideration not only the six possible nucleotide substitutions in the heavy and light strand of DNA, but also the trinucleotide context of each substitution, for a total of 96 substitution types in each strand.
Statistical analysis
To account for the possibility of correlation between observations from the same individual (or biopsy), we applied the method of generalized estimating equations (GEE). However, GEE relies on large sample theory for the validity of the estimates, particularly the SE estimates. Because the sample size here is modest, we also applied resampling with GEE (see Supplementary Methods).
Data access
Sequencing data that supports the findings of this study have been deposited in the Sequence Read Archive (SRA: SRP139857, BioProject ID: PRJNA449763).
Results
Duplex sequencing identifies abundant mtDNA mutations in ulcerative colitis biopsies
Mutations in mtDNA were identified by performing Duplex Sequencing on DNA extracted from colonic epithelium from 46 biopsies covering different stages of preneoplastic and neoplastic progression (Table 1). Samples were sequenced at a median depth of 1,364x with a minimum depth of 600x (Supplementary Table S2). Because Duplex Sequencing enables ultra-accurate deep sequencing (21), we were able to detect and classify mtDNA mutations in three groups according to their MAF: clonal ≥ 0.95; subclonal ≥ 0.01 and <0.95; and VLF mutations <0.01. We used Haplogrep2 (haplogrep.uibk.ac.at) to identify each patient’s haplotype (Supplementary Table S1), which allowed us to discount haplotype-specific polymorphisms and constitutional polymorphisms. In total, we identified 208 clonal/subclonal mutations and 56,764 VLF mutations (Table 1).
Clonality increases with progression
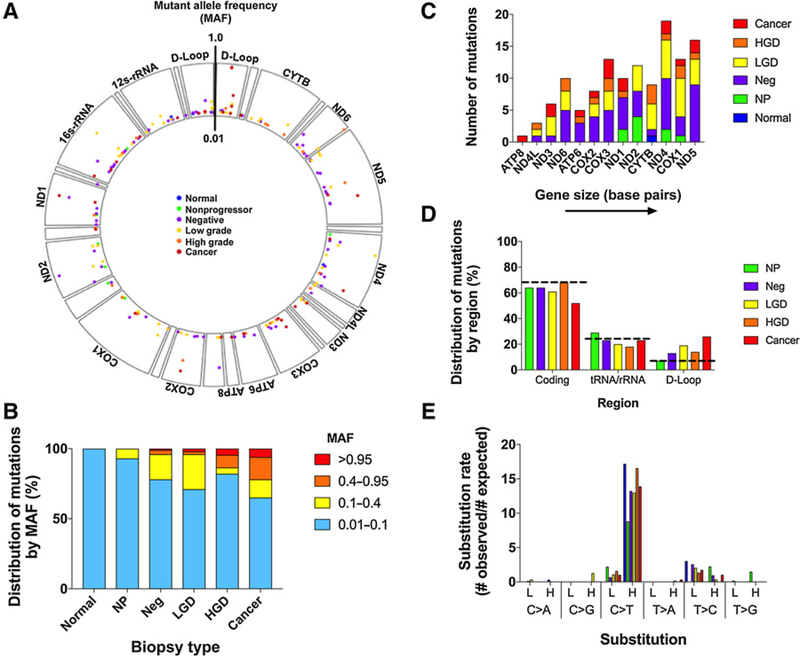
The overall distribution of clonal and subclonal mutations across the mitochondrial genome as well as their MAF is shown in Fig. 2A. While most mutations were low frequency (0.01 < MAF < 0.1), a subset of mutations appeared at larger frequencies (MAF > 0.1). The proportion of these large frequency mutations as well as their MAF increased with progression (Fig. 2B), consistent with larger clones progressively expanding during tumorigenesis. A detailed analysis of these mutations revealed that in the colons from ulcerative colitis Progressors, some mutations were shared at different frequencies in adjacent and relatively distant biopsies (~25 cm), often spanning colonic epithelium of different histologic grades (Supplementary Fig. S2). These findings confirm the clonal nature of the expansions and the presence of large fields of cancerization in ulcerative colitis (14). The analysis of individual biopsies (Supplementary Fig. S3A) indicated that the majority of ulcerative colitis Progressor biopsies (29/36 = 80.5%) harbored a clonal expansion in which a mtDNA mutation was present at MAF > 0.1, whereas these expansions were less frequent in colon from ulcerative colitis NPs or non-ulcerative colitis colon (1/10 = 10%; P = 9 × 10−5 by Fisher exact test). Importantly, the number of mutations within both the MAF > 0.1 and MAF > 0.01 categories did not correlate with the total amount of DCS nucleotides sequenced (Supplementary Fig. S4A), indicating that differences in sequencing depth did not explain the variation in number of subclonal mutations observed across biopsies. Clonal/subclonal mutations were slightly higher in older patients with ulcerative colitis (Supplementary Fig. S5A) however, at all ages, they were more frequent in ulcerative colitis Progressors than in NPs. There were no associations between clonal and subclonal mutations and sex, disease duration, active inflammation, and chronic inflammation (Supplementary Fig. S5B–S5E). Within histologic grades, the number of mutations was not associated to inflammation scores (Supplementary Fig. S5F).
Figure 2.
Clonal and subclonal mtDNA mutations A, Genomic positions of clonal and subclonal mtDNA mutations. Biopsy types are color-coded. MAF from 0.01–1.0 is indicated by position along the vertical axis. B, Distribution of clonal and subclonal mutations by MAF in each biopsy type. C, Association between number of mutations and gene size (bp). Biopsy types are color-coded. D, Distribution of clonal and subclonal mutations by region compared with the expected distribution based on composition of the mitochondrial genome. E, Substitution rate for clonal and subclonal mutations shown by biopsy type.
Clonal and subclonal mutations are randomly distributed in the coding region but tend to cluster in the D-loop with advanced disease
Clonal and subclonal mutations appeared randomly distributed across the mtDNA coding region (Fig. 2A), an observation that was confirmed by plotting the number of mutations in each mtDNA encoded gene sorted by ascending size (Fig. 2C). Larger genes had more mutations and no significant clustering by gene was observed (P = 0.36 by χ2 test of homogeneity). The proportion of D-loop mutations, however, increased with progression (Fig. 2D). The D-loop is a noncoding region that represents 6.7% of the mitochondrial genome, but as much as 19%, 14%, and 26% of clonal/subclonal mutations in LGD, HGD and cancer, respectively, were found in the D-loop. Mutations in tRNA and rRNA did not significantly change with progression, but the percentage of mutations in the coding region decreased in cancers. Individual analysis of all the biopsies in the study confirmed that these results were not driven by a single biopsy or by biopsies from a single colon (Supplementary Fig. S3B). These results suggest that mtDNA mutations in the coding region are selected against in ulcerative colitis cancer progression.
Clonal and subclonal mutations display a mutational signature indicative of mtDNA replication errors
Previous studies have demonstrated that most mtDNA mutations that accumulate with aging and cancer correspond to C>T transitions that occur almost exclusively in the heavy strand of the mtDNA (6, 7, 27). These mutations are attributed to mtDNA replication errors. To determine whether the same mutational mechanisms are operative in the inflammatory setting of ulcerative colitis, we quantified the mutation substitution rate for each of the six possible mutation types in each of the two strands of mtDNA. The mutation substitution rate was calculated as the ratio of the number of observed mutations divided by the number of expected mutations. Indeed, C>T transitions in the heavy strand were the most predominant type of mutation across all six biopsy types, observed between 8- to 16-fold times more than what would be expected by chance (Fig. 2E). Of note, clonal and subclonal mutations did not show a significant contribution from C>A transversions, the signature of oxidative damage.
The number of clonal/subclonal mutations spikes in early stages of progression but decreases in later stages
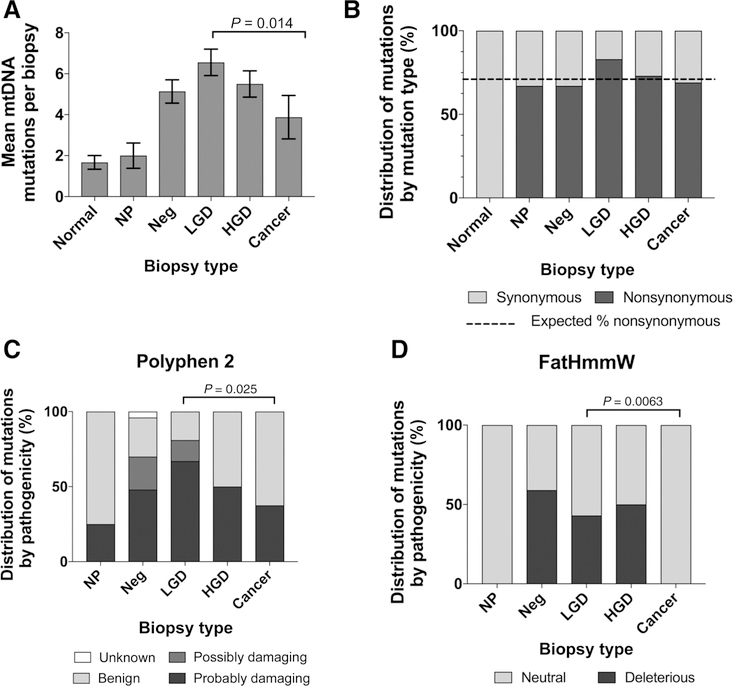
To better characterize the role of mtDNA mutations in ulcerative colitis clonal expansions, we performed a detailed analysis of the number, MAF, and mutational consequence of mtDNA mutations by biopsy type (Supplementary Fig. S6). We observed that normal colon biopsies had low frequency subclonal mutations that were either noncoding or synonymous. Ulcerative colitis NPs also featured low frequency subclonal mutations, but they were often nonsynonymous. The number and the frequency of mutations dramatically increased in negative for dysplasia biopsies from ulcerative colitis Progressors compared with normal and NPs. However, with advanced progression, the proportion of mutations with high MAF increased (Fig. 2B) but the overall number of mutations appeared to decrease. To better quantify this finding, we compared the mean number of mtDNA mutations for each biopsy type (Fig. 3A). While biopsies from normal and ulcerative colitis NP colons only harbored, on average, about two mtDNA mutations (MAF > 0.01), this number increased to five and six in biopsies from ulcerative colitis Progressors negative for dysplasia and LGD, respectively. However, the number of mutations decreased in HGD and even more in cancers. This decrease was statistically significant [P = 0.014 for linear effect over LGD, HGD, and CA; P = 3.6 × 10−5 for a quadratic effect (inverse V-shape) over all biopsy types, by GEE permutation tests]. Overall these results indicate that (i) clonal expansions that carry mtDNA mutations are a feature of ulcerative colitis preneoplastic progression, (ii) the maximum number of mutations is achieved in LGD and decreases in HGD and cancer, showing an inverse V-shape that is in agreement with prior findings of mitochondrial alterations in ulcerative colitis (16).
Figure 3.
Comparison of clonal and subclonal mutations by biopsy type. A, Mean number of clonal and subclonal mtDNA mutations for each biopsy type. Error bars indicate SE of the mean. P value corresponds to the linear effect by GEE permutation tests. B, The proportion of synonymous and nonsynonymous clonal and subclonal mutations is shown by biopsy type. Dashed line indicates expected percentage of nonsynonymous mutations in the mtDNA in the absence of selection. Proportion of clonal and subclonal mutations of each biopsy type predicted to have pathogenic impact by Polyphen 2 (C) and FatHmmW (D) algorithms. P values were calculated using GEE permutation tests.
Clonal and subclonal mutations are enriched for nonsynonymous and pathogenic mutations in LGD but not in cancer
We next quantified the frequency of nonsynonymous mutations for each biopsy type (Fig. 3B). Interestingly, for all ulcerative colitis biopsy types except LGD the frequency of nonsynonymous mutations was less than 71%, which is the expected frequency given the composition of the mitochondrial genome. For LGD, however, the frequency was 81%. While the test for a decreasing linear trend from LGD to HGD to cancer was not significant (P = 0.11), there was a nominally significant difference when comparing the frequency for LGD, 81%, to nonsynonymous frequency over all other grades, 62% (P = 0.026, n = 109 total mutations). These results suggest that damaging mtDNA mutations might be positively selected in LGD, but they appear to be selected against at other stages.
While nonsynonymous mutations are a first indication of potential for pathogenicity, they often lead to amino acid changes that are inconsequential. Thus, a better estimate of pathogenicity can be achieved by utilizing computational algorithms to predict the functional impact of a specific missense variant. To comprehensively address this issue, we used MitImpact 2.9 (mitimpact.css-mendel.it; ref. 26), which is a collection of precomputed pathogenicity predictions for all possible nucleotide changes that cause nonsynonymous substitutions in human mitochondrial protein-coding genes. We interrogated six different algorithms (Polyphen2, Fathmmw, CADD, Mutation Assessor, SIFT, and Provean) that categorized missense clonal and subclonal mutations into different pathogenicity groups. Two of the algorithms, Polyphen2 (28) and FatHmmW (29), identified significant differences with progression (Fig. 3C and D; P = 0.025 and P = 0.006 for decrease in pathogenicity from LGD to CA by GEE permutation tests, respectively). The six algorithms measure different aspects of pathogenicity using different mathematical approaches and, thus, they vary in their predictions. Polyphen2 predicts structural and functional impact of missense mutations using a probabilistic classifier whereas FatHmmW predicts functional impact by combining sequence conservation with hidden Markov models. Interestingly, both algorithms showed increased pathogenicity in early stages of progression and decreased pathogenicity in cancer (Fig. 3C and D). These findings complement our previous observations based on number of mutations (Fig. 3A) and frequency of nonsynonymous mutations (Fig. 3B). Overall, these data indicate that the clones in early progression tend to carry more mtDNA mutations and these are more pathogenic. However, the clones that eventually evolve to cancer tend to carry mutations that are not coding or nonpathogenic, suggesting selection against deleterious mtDNA mutations.
VLF mutations display mutational signatures corresponding to mtDNA replication errors and oxidative damage
In contrast to clonal and subclonal mutations, VLF mutations were very abundant in all biopsies (Table 1) and their number was highly associated with the total amount of sequenced nucleotides (Supplementary Fig. S4B). Thus, to compare between biopsies we calculated the VLF mutation frequency as the number of VLF mutations divided by the total DCS nucleotides sequenced. A subset of biopsies showed a disproportionately large number of VLF mutations, which corresponded mostly to C>A transversions, the signature caused by oxidative damage (Supplementary Fig. S7). All the biopsies from ulcerative colitis NPs, the normal biopsy with Hirschprung disease, and 7 of 10 biopsies from one of the ulcerative colitis Progressors harbored a high frequency of C>A mutations in both the heavy and the light strand of mtDNA (Supplementary Fig. S7). VLF mutations were not associated with age, sex, disease duration, acute inflammation, or chronic inflammation (Supplementary Fig. S8A–S8E). Importantly, within NPs, the high level of C>A mutations was not associated with higher levels of inflammation in those biopsies (Supplementary Fig. S8F).
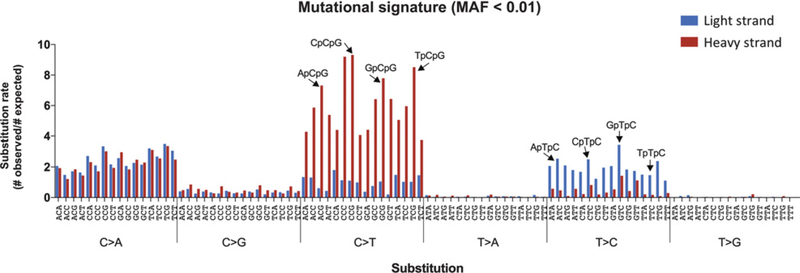
To further investigate the mutational signatures operative in VLF mutations, we analyzed the trinucleotide context in which each of the six possible substitutions occurred in the heavy or light strand of the mtDNA. This analysis generates 96 possible mutational events (six substitutions × 16 flanking nucleotide combinations) and has been extensively used to elucidate mutagenic processes in both nuclear and mitochondrial tumor DNA (6, 7, 30). The combined analysis of all samples revealed two overlapping mutational signatures (Fig. 4): (i) C>A trans-versions in both strands of DNA and independent of nucleotide context; and (ii) C>T transitions in the heavy strand of DNA and T>C transitions in the light strand of DNA, both with markedly increased frequency in certain trinucleotide contexts. Specifically, C>T in the heavy strand were enriched in NpCpG contexts and T>C in the light strand were enriched in NpTpC contexts. These mutational events correspond to the ones previously identified in mtDNA from cancer samples (6, 7) and have been attributed to DNA replication errors. Mutational signature analysis by biopsy type (Supplementary Fig. S9) demonstrated that the mtDNA “replication error” signature is not exclusive to cancers but is also found in preneoplastic biopsies and normal colon. In biopsies from ulcerative colitis NPs, the signature was also present but overshadowed by an excess of C>A transversions (Supplementary Fig. S9).
Figure 4.
VLF (MAF < 0.01) mtDNA mutational signature. The substitution rate of each of the 96 possible substitution classes is shown for mtDNA mutations with MAF < 0.01. Substitution rate is calculated as the ratio of the number of observed mutations to the number of expected mutations. Black arrows indicate CpG and TpC trinucleotides, which are among the most frequently mutated in the heavy strand and light strand of mtDNA, respectively.
VLF transitions and indels are more common in the D-loop than non–D-loop and decrease with progression
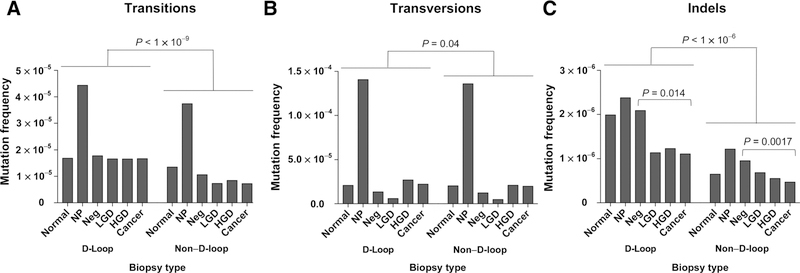
Because of the prominent role of C>A mutations in some biopsies, the comparison of mutation frequency between biopsies and within D-loop and non–D-loop regions was best performed by separating transitions and transversions (Fig. 5). We observed that in NPs, not only transversions were disproportionally high, so were transitions, pointing to an excessive mutational load beyond oxidative damage. For all biopsy types, the frequency of transitions was lower in the non–D-loop region than in the D-loop (Fig. 5A; mean difference = 7.8 × 10−6; P < 1 × 10−9). Remarkably, in the non–D-loop, C>T transitions in the heavy strand and T>C transitions in the light strand, which correspond to the predominant mtDNA mutational signature, significantly decreased with progression (Supplementary Fig. S10C; P = 6.6 × 10−4). Transversions (Fig. 5B) displayed a much smaller difference in frequencies in the D-loop and non–D-loop over all biopsy types (mean difference = 2.3 × 10−6; P = 0.04) and did not show any changes with progression. Indels (Fig. 5C) showed a similar pattern to transitions, presenting at higher frequency in D-loop than in non–D-loop (P < 1 × 10−6).They were highest in NPs and decreased with progression both in the non–D-loop (P = 0.0017) as well as in the D-loop (P = 0.014).
Figure 5.
Quantification of very low frequency mutations (MAF < 0.01) in D-loop versus non–D-loop and by progression. The total frequency of mutations for variants with MAF < 0.01 is shown for transitions (A), transversions (B), and indels (C) for each biopsy type and by D-Loop or non–D-Loop regions. P values were calculated using GEE permutation tests.
To further investigate the mutational pattern by biopsy type within the D-loop and non–D-loop region, we separated transitions and transversions into the corresponding nucleotide substitutions in each strand of DNA (Supplementary Fig. S10). This analysis allowed us to determine that the C>T and T>C strand biases were exclusive of the non–D-loop region, in agreement with prior findings in aging and cancer (ref. 6, 27; Supplementary Fig. S10A–S10C). Transitions in the D-loop had no strand bias and did not change in frequency with progression. However, transitions in the non–D-loop were strongly biased according to the “replication error” signature described previously (Fig. 4) and sharply declined with progression. In contrast, transversions, which were almost exclusively C>A, were found at similar frequencies in the heavy and light strand and in the D-loop and non–D-loop region (Supplementary Fig. S10D–S10F), consistent with the widespread effect of oxidative damage.
VLF mutations are randomly distributed in the coding region and tend to be enriched for synonymous mutations during progression
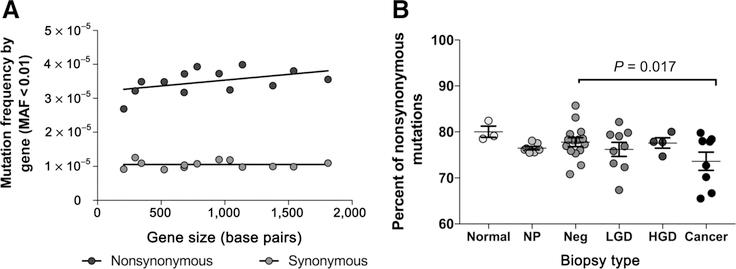
The mean frequency of nonsynonymous and synonymous mutations was constant across all genes indicating that mutations accumulated randomly, without any detectable clustering by gene (Fig. 6A). The same was true when tested for each biopsy type (Supplementary Fig. S11) indicating no preferential incidence of VLF mutations in any given gene during progression. Regarding the percentage of nonsynonymous mutations, we observed a decreasing trend during progression (P = 0.017; Fig. 6B), although there was substantial variability within biopsy type.
Figure 6.
VLF (MAF < 0.01) mutation selection A, Nonsynonymous and synonymous mutation frequency for each mtDNA encoded gene plotted by gene size. Mutation frequency was calculated as the number of mutations with MAF < 0.01 (nonsynonymous or synonymous) within each given gene divided by the total number of DCS nucleotides sequenced. B, Proportion of nonsynonymous mutations (MAF < 0.01) for each biopsy by histologic grade. P values were calculated using a GEE permutation test.
Discussion
The contribution of mtDNA mutations to tumorigenesis has been an area of controversy for some time. The work presented here helps to explain this contribution in the context of ulcerative colitis-associated colorectal tumorigenesis: mtDNA mutations increase in early ulcerative colitis carcinogenesis, but appear to be selected against in cancer. Previous studies of mtDNA mutations have been limited by the sensitivity issues inherent to standard NGS (20) and few have been able to detect mutations with MAF < 0.1 (6). The accuracy of Duplex Sequencing allowed us to obtain reliable estimates of MAF ranging from 1 down to 0.0005. This provided a comprehensive analysis of mtDNA mutations with progression because we could accurately quantify not only the number of mutations but their clonality. In addition, because VLF mutations are extremely frequent in the mitochondrial genome, in spite of the relatively low number of biopsies in the study, we identified thousands of mutations that enabled us to perform detailed mutational signature analyses.
The main finding of our study is the selection against mtDNA mutations in cancer compared with early stages of progression, which was supported by multiple lines of evidence. In cancers, we observed: (i) fewer mtDNA mutations, both subclonal and VLF, (ii) decreased proportion of distinct subclonal and VLF mutations (transitions and indels) in the coding region, (iii) fewer nonsynonymous mutations, and (iv) fewer subclonal pathogenic mutations. Although our findings are derived from ulcerative colitis–associated cancer, they are in agreement with a prior report of decreased mtDNA mutagenesis in sporadic colorectal cancer (18) and with the detailed mutational analysis of mtDNA from TCGA data, which demonstrated negative selection of deleterious mitochondrial mutations in cancers (6, 7). Collectively, these results support the notion that cancer cells require functional mitochondria. This notion is consistent with a novel view of mitochondria as essential organelles in cancer (4, 5), which not only supply energy and intermediate metabolites, but are critical to enable the metabolic reprogramming characteristic of cancer cells (31).
A limitation of our study is the small number of patients with ulcerative colitis. However, multiple biopsies were included from each patient and a large number of mutations were analyzed, enabling a detailed characterization of the mutational profile of mtDNA in ulcerative colitis tumorigenesis. Importantly, our results are in agreement with our previous work in ulcerative colitis. We previously demonstrated the widespread loss of mitochondrial function in ulcerative colitis via IHC staining for mitochondrial proteins (16). In ulcerative colitis Progressors, we identified a V-shaped pattern with maximum mitochondrial loss in LGD and a recovery of normal levels in cancer. This pattern was confirmed by mtDNA copy number quantification (16). On the basis of these findings, we hypothesized an initial increase and a later decrease in the burden of mtDNA mutations over the course of dysplastic progression. Our results have now confirmed this hypothesis, strongly suggesting that, while mitochondrial dysfunction might be associated with the earlier stages of the disease, cancer cells tend to feature functional mitochondria. Several nonexclusive mechanisms are possible: (i) mitochondria with damaged DNA might be removed by autophagy and mitochondrial biogenesis activated via PGC1α (16); (ii) a genetic bottleneck might be bypassed only by premalignant cells with intact mitochondria; or (iii) cells with damaged mtDNA might acquire whole functional mitochondria by horizontal transfer from neighboring tissue (32).
We detected signs of positive selection for mtDNA mutations in LGD, including enrichment for nonsynonymous and pathogenic mutations. Others have reported mtDNA mutations in precancerous lesions, suggesting a potential contribution to early transformation (1). It is well known that the carcinogenic process in ulcerative colitis is histologically and genetically different from sporadic colorectal cancer (14) and is possible for mitochondria to play differential roles in these processes. However, it is remarkable that in sporadic colorectal carcinogenesis, mtDNA mutations have also been observed to increase in adenomas and decrease in colon cancer (18), in agreement with our data. Thus, it appears that the opposite role of mtDNA mutations in early and late cancer might occur in sporadic carcinogenesis as well and might explain some of the contradictions in the field.
Regarding the causes of mtDNA mutations, our results support mtDNA replication as the major mechanism of mutation, in agreement with previous results from cancer (6, 7, 33) and aging (27). The resemblance of the mutational signature reported here to those reported by Ju and colleagues is striking (6) and indicates that the same mutational processes operative in the mitochondria of tumors take place in the mitochondria of normal, inflamed, and preneoplastic colon. Although the exact mechanism of mutagenesis is unknown, based on the exclusive non–D-loop location of the strand bias (also observed here), Ju and colleagues proposed three explanations: (i) the parent heavy strand might be more prone to cytosine and adenine deamination while being single-stranded during replication, (ii) endogenous POLG errors might occur on the leading strand preferentially, and (iii) different repair mechanisms might be at play in the leading versus lagging strand (6). A major difference with Ju and colleagues is that in a subset of samples in our study we did observe an important contribution from oxidative damage, although only at the level of VLF mutations. In the presence of reactive oxygen species (ROS), guanine oxidizes to 8-oxo-guanine, which results in C>A transversions (34). Oxidative damage is an important pathogenic factor in ulcerative colitis (35, 36), and, thus, we expected to observe this mutational signature. Surprisingly, C>A mutations were not widespread among all biopsies from ulcerative colitis patients, but were restricted to biopsies from ulcerative colitis NPs, most biopsies (negative, LGD, and cancer) from a single Progressor patient, and the non-ulcerative colitis colon that had Hirschprung disease. These results were not explained by variability in inflammation levels because C>A mutations, as well as mtDNA mutations in general, were not associated with acute or chronic inflammation. However, these results might be explained by inter-individual or interregional variations in the generation of ROS or in the production of antioxidant defenses, an interesting hypothesis that deserves further investigation with a larger number of patients.
In the context of ulcerative colitis Progressors, a critical finding is the presence of large clonal expansions in nondys-plastic epithelium. mtDNA mutations with MAF > 0.1 were abundant in nondysplastic biopsies from ulcerative colitis Progressors, but were rare in NPs. While these clonal expansions might be driven by a pathogenic mtDNA mutation that confers a selective advantage, in many cases mtDNA mutations might be carried as passengers and arrive to homoplasmy by genetic drift (6, 37). In any case, these mutations could be used as markers of clonal expansions, which are an essential component of preneoplastic fields in ulcerative colitis (9, 14). This study was not designed to assess differences between Progressors and NPs and the number of cases in each group is insufficient to make these group comparisons. However, we have previously demonstrated the potential value of clonal expansions to detect ulcerative colitis cancer progression (23, 38) and the utility of mtDNA for this purpose warrants further investigation.
Our findings support a model in which mtDNA mutations accumulate and clonally expand in early tumorigenesis but are subject to purifying selection in cancer (Supplementary Fig. S12). During normal aging, mtDNA mutations accumulate and clonally expand in the colon epithelium (39), but this process might be accelerated in ulcerative colitis due to the increased cellular proliferation necessary to regenerate the ulcerated epithelium. This increased cellular proliferation would lead to extensive replication of the mtDNA, which appears to be the main cause of mutation not only in ulcerative colitis tumorigenesis, but also in most cancers (6). Cells with pathogenic mtDNA mutations might clonally expand in early progression, leading to multiple small clones. However, progression to malignancy appears to be characterized by a decrease in the number and pathogenicity of mtDNA mutations, possibly due to the outgrowth of one or few clones carrying nonpathogenic mtDNA mutations that drift to homoplasmy. Further research is necessary to elucidate the role of mitochondrial epigenetic regulation and metabolic reprogramming during this process (16) and to determine to what extent this model is applicable to other cancer types.
Supplementary Material
Acknowledgments
The authors thank Jesse J. Salk and Jeffrey D. Krimmel for their preliminary contributions to this work, Jake G. Hoekstra and Monica Sanchez-Contreras for their advice and expertise analyzing mitochondrial DNA, Kelly Jin for her assistance with data visualization, and Rebecca Ortega for her helpful comments and suggestions. This work was supported by NIH grants R01CA181308 (to R.A. Risques) and R01CA160674 (to T.A. Brentnall). K.T. Baker was a recipient of the predoctoral fellowship from the Molecular Medicine Predoctoral Training Program, NIH T32GM95421.
Disclosure of Potential Conflicts of Interest
S.R. Kennedy has ownership interest (including stock, patents, etc.) and is a consultant/advisory board member for TwinStrand Biosciences. R.A. Risques reports receiving a commercial research grant (SBIR grant) from TwinStrand Biosciences and has ownership interest (including stock, patents, etc.) in NanoString Technologies Inc. No potential conflicts of interest were disclosed by the other authors.
Footnotes
Note: Supplementary data for this article are available at Molecular Cancer Research Online (http://mcr.aacrjournals.org/).
References
- 1.Chatterjee A, Dasgupta S, Sidransky D. Mitochondrial subversion in cancer. Cancer Prev Res 2011;4:638–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Larman TC, DePalma SR, Hadjipanayis AG, Cancer Genome Atlas Research Network, Protopopov A, Zhang J, et al. Spectrum of somatic mitochondrial mutations in five cancers. Proc Natl Acad Sci U S A 2012;109:14087–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yu M Somatic mitochondrial DNA mutations in human cancers. Adv Clin Chem 2012;57:99–138. [DOI] [PubMed] [Google Scholar]
- 4.Zong WX, Rabinowitz JD, White E. Mitochondria and cancer. Mol Cell 2016;61:667–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wallace DC. Mitochondria and cancer. Nat Rev Cancer 2012;12:685–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ju YS, Alexandrov LB, Gerstung M, Martincorena I, Nik-Zainal S, Ramakrishna M, et al. Origins and functional consequences of somatic mitochondrial DNA mutations in human cancer. eLife 2014;3:e02935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stewart JB, Alaei-Mahabadi B, Sabarinathan R, Samuelsson T, Gorodkin J, Gustafsson CM, et al. Simultaneous DNA and RNA mapping of somatic mitochondrial mutations across diverse human cancers. PLoS Genet 2015;11:e1005333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hanauer SB. Inflammatory bowel disease: epidemiology, pathogenesis, and therapeutic opportunities. Inflamm Bowel Dis 2006;12:S3–9. [DOI] [PubMed] [Google Scholar]
- 9.Choi CR, Bakir IA, Hart AL, Graham TA. Clonal evolution of colorectal cancer in IBD. Nat Rev Gastroenterol Hepatol 2017;14:218–29. [DOI] [PubMed] [Google Scholar]
- 10.Dyson JK, Rutter MD. Colorectal cancer in inflammatory bowel disease: what is the real magnitude of the risk? World J Gastroenterol 2012;18: 3839–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brentnall TA, Crispin DA, Rabinovitch PS, Haggitt RC, Rubin CE, Stevens AC, et al. Mutations in the p53 gene: an early marker of neoplastic progression in ulcerative colitis. Gastroenterology 1994;107: 369–78. [DOI] [PubMed] [Google Scholar]
- 12.Rabinovitch PS, Dziadon S, Brentnall TA, Emond MJ, Crispin DA, Haggitt RC, et al. Pancolonic chromosomal instability precedes dysplasia and cancer in ulcerative colitis. Cancer Res 1999;59:5148–53. [PubMed] [Google Scholar]
- 13.Risques RA, Lai LA, Himmetoglu C, Ebaee A, Li L, Feng Z, et al. Ulcerative colitis-associated colorectal cancer arises in a field of short telomeres, senescence, and inflammation. Cancer Res 2011;71:1669–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baker KT, Salk JJ, Brentnall TA, Risques RA. Precancer in ulcerative colitis: the role of the field effect and its clinical implications. Carcinogenesis 2018;39:11–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Novak EA, Mollen KP. Mitochondrial dysfunction in inflammatory bowel disease. Front Cell Develop Biol 2015;3:62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ussakli CH, Ebaee A, Binkley J, Brentnall TA, Emond MJ, Rabinovitch PS, et al. Mitochondria and tumor progression in ulcerative colitis. J Natl Cancer Inst 2013;105:1239–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.He Y, Wu J, Dressman DC, Iacobuzio-Donahue C, Markowitz SD, Velculescu VE, et al. Heteroplasmic mitochondrial DNA mutations in normal and tumour cells. Nature 2010;464:610–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ericson NG, Kulawiec M, Vermulst M, Sheahan K, O’Sullivan J, Salk JJ, et al. Decreased mitochondrial DNA mutagenesis in human colorectal cancer. PLoS Genet 2012;8:e1002689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goodwin S, McPherson JD, McCombie WR. Coming of age: ten years of next-generation sequencing technologies. Nat Rev Genet 2016;17:333–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Salk JJ, Schmitt MW, Loeb LA. Enhancing the accuracy of next-generation sequencing for detecting rare and subclonal mutations. Nat Rev Genet 2018;19:269–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schmitt MW, Kennedy SR, Salk JJ, Fox EJ, Hiatt JB, Loeb LA. Detection of ultra-rare mutations by next-generation sequencing. Proc Natl Acad Sci U S A 2012;109:14508–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krimmel JD, Schmitt MW, Harrell MI, Agnew KJ, Kennedy SR, Emond MJ, et al. Ultra-deep sequencing detects ovarian cancer cells in peritoneal fluid and reveals somatic TP53 mutations in noncancerous tissues. Proc Natl Acad Sci U S A 2016;113:6005–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Salk JJ, Salipante SJ, Risques RA, Crispin DA, Li L, Bronner MP, et al. Clonal expansions in ulcerative colitis identify patients with neoplasia. Proc Natl Acad Sci U S A 2009;106:20871–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kennedy SR, Schmitt MW, Fox EJ, Kohrn BF, Salk JJ, Ahn EH, et al. Detecting ultralow-frequency mutations by Duplex Sequencing. Nat Protoc 2014; 9:2586–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nachmanson D, Shenyi L, Schmidt EK, Hipp MJ, Baker KT, Zhang Y, et al. Targeted genome fragmentation with CRISPR/Cas9 enables fast and efficient enrichment of small genomic regions and ultra-accurate sequencing with low DNA input (CRISPR-DS). Genome Res 2018;10:1589–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Castellana S, Ronai J, Mazza T. Mitlmpact: an exhaustive collection of precomputed pathogenicity predictions of human mitochondrial nonsynonymous variants. Hum Mutat 2015;36:E2413–22. [DOI] [PubMed] [Google Scholar]
- 27.Kennedy SR, Salk JJ, Schmitt MW, Loeb LA. Ultra-sensitive sequencing reveals an age-related increase in somatic mitochondrial mutations that are inconsistent with oxidative damage. PLoS Genet 2013;9:e1003794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, et al. A method and server for predicting damaging missense mutations. Nat Methods 2010;7:248–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shihab HA, Gough J, Cooper DN, Stenson PD, Barker GL, Edwards KJ, et al. Predicting the functional, molecular, and phenotypic consequences of amino acid substitutions using hidden Markov models. Hum Mutat 2013;34:57–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alexandrov LB, Stratton MR. Mutational signatures: the patterns of somatic mutations hidden in cancer genomes. Curr Opin Genet Dev 2014;24:52–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011;144:646–74. [DOI] [PubMed] [Google Scholar]
- 32.Dong LF, Kovarova J, Bajzikova M, Bezawork-Geleta A, Svec D, Endaya B, et al. Horizontal transfer of whole mitochondria restores tumorigenic potential in mitochondrial DNA-deficient cancer cells. eLife 2017;6: e22187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ahn EH, Lee SH, Kim JY, Chang CC, Loeb LA. Decreased mitochondrial mutagenesis during transformation of human breast stem cells into tumorigenic cells. Cancer Res 2016;76:4569–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Delaney S, Jarem DA, Volle CB, Yennie CJ. Chemical and biological consequences of oxidatively damaged guanine in DNA. Free Radical Res 2012;46:420–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roessner A, Kuester D, Malfertheiner P, Schneider-Stock R. Oxidative stress in ulcerative colitis-associated carcinogenesis. Pathol Res Pract 2008;204:511–24. [DOI] [PubMed] [Google Scholar]
- 36.Jena G, Trivedi PP, Sandala B. Oxidative stress in ulcerative colitis: an old concept but a new concern. Free Radical Res 2012;46:1339–45. [DOI] [PubMed] [Google Scholar]
- 37.Coller HA, Bodyak ND, Khrapko K. Frequent intracellular clonal expansions of somatic mtDNA mutations: significance and mechanisms. Ann N Y Acad Sci 2002;959:434–47. [DOI] [PubMed] [Google Scholar]
- 38.Salk JJ, Bansal A, Lai LA, Crispin DA, Ussakli CH, Horwitz MS, et al. Clonal expansions and short telomeres are associated with neoplasia in early-onset, but not late-onset, ulcerative colitis. Inflamm Bowel Dis 2013;19:2593–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Greaves LC, Preston SL, Tadrous PJ, Taylor RW, Barron MJ, Oukrif D, et al. Mitochondrial DNA mutations are established in human colonic stem cells, and mutated clones expand by crypt fission. Proc Natl Acad Sci U S A 2006;103:714–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.