Summary
Animals rely on an internal sense of body position and movement to effectively control motor behavior. This sense of proprioception is mediated by diverse populations of mechanosensory neurons distributed throughout the body. Here, we investigate neural coding of leg proprioception in Drosophila, using in vivo two-photon calcium imaging of proprioceptive sensory neurons during controlled movements of the fly tibia. We found that the axons of leg proprioceptors are organized into distinct functional projections that contain topographic representations of specific kinematic features. Using subclass-specific genetic driver lines, we show that one group of axons encodes tibia position (flexion/extension), another encodes movement direction, and a third encodes bidirectional movement and vibration frequency. Overall, our findings reveal how proprioceptive stimuli from a single leg joint are encoded by a diverse population of sensory neurons, and provide a framework for understanding how proprioceptive feedback signals are used by motor circuits to coordinate the body.
Graphical Abstract
eTOC blurb
Proprioception, the internal sense of body position and movement, is essential for adaptive motor control. Mamiya et al. use in vivo calcium imaging to reveal the functional tuning and spatial organization of proprioceptors that monitor the femur-tibia joint of the Drosophila leg.
Introduction
Proprioception, the internal sense of body position and movement (Sherrington, 1906), is essential for the neural control of motor behavior. Sensory feedback from proprioceptive sensory neurons (i.e., proprioceptors) contributes to a wide range of behaviors, from regulation of body posture (Hasan and Stuart, 1988; Zill et al., 2004), to locomotor adaptation (Bidaye et al., 2018; Lam and Pearson, 2002) and motor learning (Isakov et al., 2016; Takeoka et al., 2014). But despite the fundamental importance of proprioception to our daily experience, it is perhaps the most poorly understood of the primary senses.
Proprioceptors are found in nearly all motile animals, from pubic lice (Graber, 1882) to sperm whales (Sierra et al., 2015). Single-unit electrophysiological recordings have revealed that proprioceptors can vary widely in their mechanical sensitivity and stimulus tuning, even within a single limb segment or muscle. For example, most vertebrate muscles contain two distinct classes of proprioceptive organs that are innervated by specialized sensory neurons: muscle spindles detect muscle length and contraction velocity, while golgi tendon organs detect mechanical load (Proske and Gandevia, 2012; Windhorst, 2007). Insects also possess proprioceptor subclasses that encode joint position, velocity, and load (Burrows, 1996; Tuthill and Wilson, 2016a) suggesting that the nervous systems of invertebrates and vertebrates may have arrived at similar evolutionary solutions to a set of common sensorimotor constraints (Tuthill and Azim, 2018).
Although individual proprioceptors may be narrowly tuned, most body movements are likely to drive activity in large numbers of proprioceptors (Proske and Gandevia, 2012; Zill et al., 2004). Characterizing the population-level structure of proprioceptive encoding has been challenging, due to the technical difficulty of recording from multiple neurons simultaneously, or identifying the same neurons across individuals. It has also been difficult to identify spatial structure or topography within proprioceptive populations from recordings of single neurons. However, a detailed understanding of proprioceptive population coding is important for understanding the function of downstream circuits and identifying the role of proprioceptive feedback in the neural control of movement.
Here, we combine genetic tools, 2-photon calcium imaging, and a magnetic leg control system to study the anatomy and function of a proprioceptor population from the leg of the fruit fly, Drosophila. Embedded within the femur of the insect leg is a cluster of proprioceptor cell bodies known collectively as the femoral chordotonal organ (FeCO; Field and Matheson, 1998). Experimental manipulation of the FeCO in locusts and stick insects has revealed a critical role for these proprioceptors during behaviors that require precise control of leg position, such as walking (Bässler, 1988) and targeted reaching (Page and Matheson, 2009). Single-unit electrophysiological recordings in these larger insect species have found that FeCO neurons monitor the position and movement of the femur-tibia joint, and that FeCO neurons may be narrowly tuned to specific kinematic features (Field and Pflüger, 1989; Hofmann et al., 1985; Kondoh et al., 1995; Matheson, 1990; Matheson, 1992; Stein and Sauer, 1999; Zill, 1985). However, it has been challenging to integrate data from single neurons to understand the population-level structure and function of the leg proprioceptive system. In Drosophila, where genetic tools enable systematic dissection of neuronal populations, the anatomy and physiology of FeCO neurons have not previously been investigated.
Using in vivo population-level calcium imaging of FeCO axons during controlled leg manipulations, we first mapped the spatial organization of proprioceptive signals in the fly ventral nerve cord (VNC). We then identified genetically and anatomically distinct FeCO subclasses that encode specific kinematic features, including position, movement direction, and vibration frequency. Overall, our results reveal the basic architecture and neural code for a key leg proprioceptive system in Drosophila. These findings provide a foundation for understanding how specialized proprioceptive feedback signals enable robust and accurate motor control.
Results
Methods to record and map proprioceptive signals in Drosophila
Positioned in the proximal femur of each Drosophila leg is a femoral chordotonal organ (FeCO) that contains ~135 cell bodies (Figure 1A). The dendrites of the FeCO neurons are connected to the cuticle and surrounding muscles by attachment cells and tendons (Shanbhag et al., 1992), and the axons of FeCO neurons project through the leg nerve into the ventral nerve cord (VNC; Phillis et al., 1996; Smith and Shepherd, 1996). Most FeCO axons arborize within the VNC neuropil, with only 3–4 cells from each leg projecting directly to the brain (Figure 1A, D) (Tsubouchi et al., 2017).
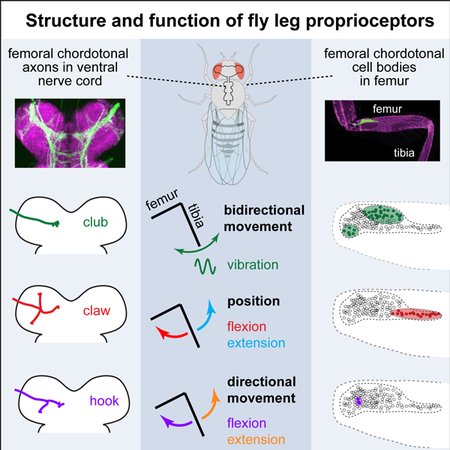
Figure 1. Investigating proprioceptive tuning of the Drosophila femoral chordotonal organ (FeCO).
A. A confocal image of the front (T1) leg of Drosophila melanogaster, showing the location of the FeCO cell bodies and dendrites (green). Magenta is auto-fluorescence from the cuticle. B. An experimental setup for 2-photon calcium imaging from the axons of FeCO neurons while controlling and tracking the femur-tibia joint. To control joint angle, we glued a pin to the tibia and positioned it using a magnet mounted on a servo motor. In vibration trials, we vibrated the tibia with a piezoelectric crystal fixed to the magnet. We backlit the tibia with an IR LED and recorded the tibia position from below using a prism and high-speed video camera. C. An example frame from a video used to track joint angle. The pin is painted black to enhance the contrast against the background. D. FeCO axon terminals (green) in the fly ventral nerve cord (VNC). Magenta is a neuropil stain (nc82). Teal box indicates region imaged by the 2-photon microscope. E. 2-photon image of FeCO axon terminals expressing GCaMP6f driven by iav-Gal4. The teal box indicates the region imaged in the example recording shown in panels G-H. F. A cross-correlation matrix showing pixel-to-pixel correlations of the changes in GCaMP6f fluorescence (∆F/F) during an example trial shown in panels G-H. Left: the cross-correlation matrix before correlation based clustering. Right: the matrix after clustering. The colors on the right of the correlation matrix correspond to the cluster colors used in G-H. G. Image of GCaMP6f fluorescence showing the recording region for the example trial shown in F and H. Each group of pixels is shaded according to its cluster identity. The colors correspond to the ∆F/F traces shown in H. H. Calcium signals from different clusters of pixels during an example trial. Each trace shows the changes in GCaMP6f fluorescence (∆F/F) for the groups of pixels shaded with the same color in the panel G. See also Figure S1.
To investigate proprioceptive signal encoding of the FeCO population, we recorded the activity of proprioceptor axons while manipulating the joint angle between the femur and tibia of the fly’s right front leg (Figure 1B–C and Movie S1). We chose to control leg kinematics, rather than force, because biomechanical studies have found that chordotonal neurons directly monitor joint displacement (Field and Matheson, 1998). For fast and accurate positioning of the leg, we designed a magnetic control system that allowed us to manipulate a pin glued to the fly’s tibia using a servo-actuated magnet (Figure 1B). This system allowed us to reproduce the range of tibia positions and speeds observed in walking flies (see Methods for details). The femur and proximal leg joints were fixed to the fly holder with UV-cured glue. We continuously recorded the position of the tibia using an IR-sensitive video camera (Figure 1C), and automatically tracked its orientation to calculate the femur-tibia joint angle. We used the Gal4-UAS system (Brand and Perrimon, 1993) to express a genetically encoded calcium indicator, GCaMP6f (Chen et al., 2013) in the majority of FeCO neurons (iav-Gal4; Kwon et al., 2010) and recorded their calcium activity in vivo with two-photon calcium imaging (Figure 1B).
To identify proprioceptor projections with shared functional tuning, we first recorded calcium activity of FeCO axons as we swung the tibia from flexion to extension at 360°/s (Figure 1F–H). To categorize calcium activity in an unbiased manner, we then calculated pairwise correlations between the calcium signal (∆F/F) in each pixel and performed k-means clustering on the resulting correlation matrix. Figure 1F shows an example of a pixel correlation matrix before and after clustering. In this trial, we identified four groups of pixels whose activity was highly correlated (for details about how the number of clusters was selected, see Figure S1A). Although this correlation-based clustering does not impose any spatial restrictions, we found that the pixels that clustered together based on their activity were also grouped together spatially (Figure 1G–H). This example illustrates that clustering of pixel correlations is sufficient to identify groups of FeCO axons that encode distinct proprioceptive stimulus features.
Functional organization of FeCO axons in the fly ventral nerve cord (VNC)
To identify functional subclasses within the FeCO population, we recorded calcium signals from four regions of interest that encompassed the different axon projections in the VNC (Figure 2 and Movie S2). For these experiments, we first clustered similarly responding pixels within each trial to identify the different response classes. Then, we combined the responses recorded from the same region in different flies and again grouped them using correlation based k-means clustering (Figure 2A–D: middle columns).
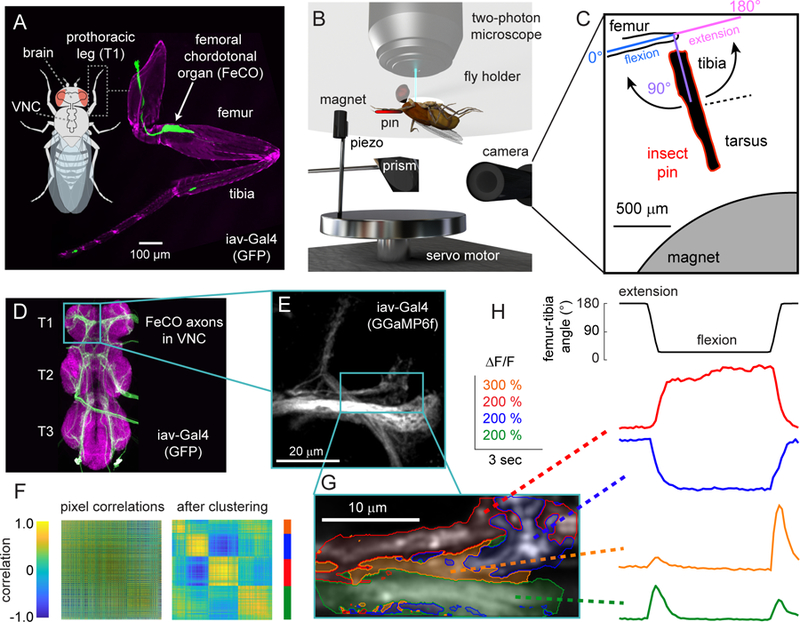
Figure 2. FeCO axons encode distinct proprioceptive features.
Each panel (A-D) shows calcium signals recorded from FeCO axons (UAS-GCamp6f; iav-Gal4) within a different region of interest. A. Imaging from an anterolateral region. Left column: example images of GCaMP6f fluorescence from three flies. In each fly, we categorized the pixels into three clusters (as in Figure 1F). The red and blue regions are tonically active when the tibia is flexed (red) or extended (blue). The green region responds phasically during tibia movement. The color scheme is maintained throughout the figure and also corresponds to the color scheme used in Figure 1G–H. A white cross represents the center of the recording region, aligned to the long axis of each axon projection. Middle column: Calcium signals for each group of pixels recorded from multiple flies. Red, blue, and green lines show GCaMP6f fluorescence (∆F/F) during the trial for each cluster of pixels (red: n= 10 flies, blue: n = 10, green: n = 9). Black lines show the response average. Right column: the location of each cluster relative to the center of the recording region across flies. The larger outlined circles represent the mean location of the center for each cluster. Each image was rotated to a common axis (see Methods for details). B. Imaging from an anteromedial region (red: n= 11 flies, blue: n = 11, green: n = 10, orange: n = 10). In addition to the three response subclasses we identified in A, this region also contained a group of pixels that responded phasically during tibia extension (orange). C. Imaging from a posterolateral region (red: n= 10 flies, blue: n = 10, purple: n = 8, orange: n = 9). Here, we identified pixels that responded phasically during joint flexion (purple). D. Imaging from a medial region (green: n = 10 flies, purple: n = 11, orange: n = 10). The scale bar in the center image is 20 µm. See also Figures S1 and S2.
We identified five basic subclasses of responses within the FeCO axons: two tonic (non-adapting) and three phasic (adapting). Pixels that responded tonically increased their activity when the tibia was either flexed (red) or extended (blue). Phasic pixels increased their activity transiently during the movement phase of the swing stimulus; one group responded with relatively similar amplitudes to movements in either direction (green, direction selectivity index (DSI) = 0.222±0.027; mean±sem; n = 29 clusters from 22 flies), while the two other groups responded in a more directionally selective manner to either flexion (orange, DSI = 0.494±0.039; n = 19 clusters from 17 flies) or extension (purple, DSI = −0.493±0.036; n = 29 clusters from 22 flies). We found the same basic response subclasses when we repeated this analysis on swing movements in the opposite direction (compare Figures 2 and S1B). Each response subclass was consistently located in similar positions across multiple flies and these positions corresponded well with the projection pattern of different axon bundles (Figure 2A–D, right columns; Figure S2B–C). The time courses of FeCO calcium signals were also more similar within a response subclass than across subclasses (Figure S2A). Overall, these results indicate that FeCO neurons that encode distinct kinematic features (i.e., tibia position, movement, and direction) are grouped into functional projections within the VNC.
Specific genetic driver lines delineate club, claw, and hook proprioceptors
Population-level imaging from FeCO neurons with a broad driver line (iav-Gal4) revealed that different axon projections have distinct proprioceptive tuning and are systemically organized across flies (Figure 2). We next sought to identify more specific Gal4 driver lines that would provide a genetic handle for these functional subclasses and enable more fine-grained analysis of proprioceptor anatomy and function. We screened through Gal4 lines in the Janelia FlyLight collection (Jenett et al., 2012) for lines that drove expression in each axon projection. From this screen, we identified three Gal4 lines that labeled the cardinal FeCO axon projections (Figure 3). Although we will focus on the front legs in this study, each driver line labels similar axons in all three VNC segments (Figure S3).
Figure 3. Organization of genetically-defined FeCO neuron subclasses in the VNC and leg.
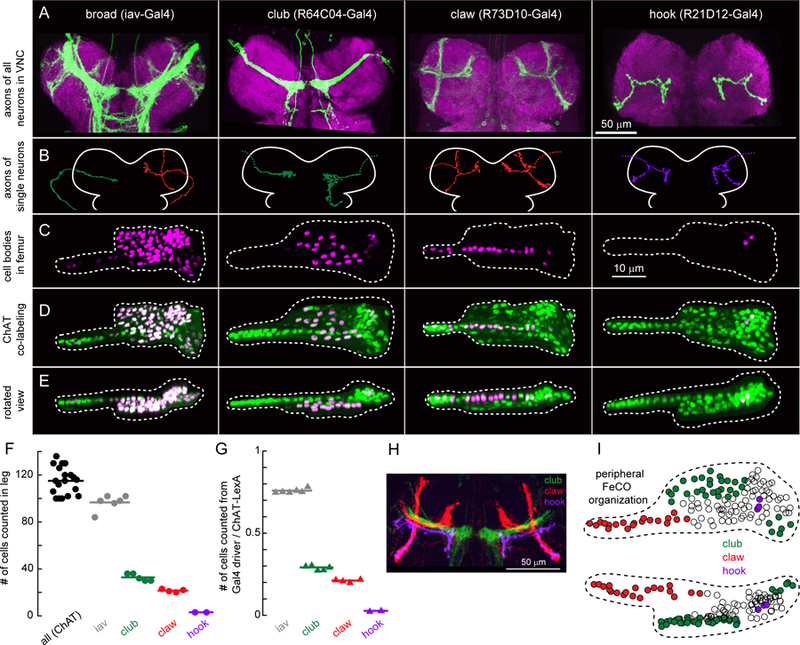
A. Four Gal4 lines label subsets of FeCO axons in the VNC. Green: GFP driven by each Gal4 line, magenta: nc82 neuropil staining. Scale bar is 50 µm. B. Example morphologies of single FeCO neurons driven by each Gal4 line, traced from images obtained by stochastic labeling with the multi-color FlpOut technique. Dotted lines indicate severed axons. C. FeCO cell bodies are clustered in characteristic locations in the fly leg. Cell bodies were labeled with UAS-Redstinger driven by each Gal4 line. Scale bars are 10 µm. D. Co-labeling of FeCO cell bodies from each Gal4 line (as in C), with a green reference marker that labels all FeCO neurons (ChAT-Lexa; LexAop - nlsGFP). E. Same as D, but viewed from the dorsal side. F. Number of neurons labeled by each Gal4 line shown in A. Circles are individual flies, lines indicate the mean (iav, n = 6 flies; club, n = 5; claw, n = 4; hook, n = 2; ChAT, n = 19). G. Ratio of neurons labeled by each Gal4 line to those labeled by ChAT-LexA in the same leg. H. in silico overlay of the axon projections of the club, claw, and hook neurons in the VNC. I. A schematic of the FeCO in the femur, showing the location of cell bodies labeled by each Gal4 line. See also Figure S3.
The first driver line (R64C04-Gal4) labels axons that run laterally through the center of the leg neuropil and form an arborization in the shape of a club (Figure 3A, 2nd column). The majority of the club axons terminate near the midline, although some also project toward the brain or other VNC segments. The second line (R73D10-Gal4) labels a group of axons shaped like a claw (club/claw nomenclature from Phillis et al., 1996). Upon entering the VNC from the leg nerve, this projection splits into three smaller branches (Figure 3A, 3rd column). We refer to these as the X, Y, and Z branches: the X branch of the claw continues to run parallel to the club, while the Y branch projects anteriorly, and the Z branch projects dorsally. The third line (R21D12-Gal4) labels axons that project along the Z branch of the claw, with only a small protuberance along the Y branch, and extend a longer arborization toward the midline, just anterior to the club (Figure 3A, 4th column). We call this projection the hook due to its resemblance to a lumberjack’s peavey hook.
To characterize the anatomy of single neurons within each driver line, we used the multicolor FlpOut technique (Nern et al., 2015) to stochastically label single cells, and manually traced their morphology (Figure 3B). Every neuron we traced from the claw Gal4 line (n = 3 cells from 3 flies) had a similar morphology, including projections to all three branches (X, Y, and Z) of the claw. Similarly, all the cells we traced from the hook Gal4 line (n = 2 cells from 2 flies) were morphologically similar, with each cell projecting an axon along all branches of the hook. In contrast, we identified three unique morphologies within the club Gal4 line (n = 14 cells from 14 flies), indicating that there may exist further functional subdivisions within the club.
For each Gal4 line, we investigated the number and distribution of proprioceptor cell bodies in the femur using confocal microscopy (Figure 3C–E). We drove expression of a nuclear-localized red fluorescent protein (UAS-RedStinger) with each Gal4 line, while expressing a nuclear-localized GFP in all FeCO neurons using a LexA line that labels all leg mechanosensory neurons (ChAT-LexA). (We were able to unambiguously identify FeCO neurons in ChAT-LexA based on their characteristic location in the leg.) In addition to the subclass-specific Gal4 lines, we performed similar co-labeling experiments using the broadly expressing line iav-gal4. Although iav-Gal4 was previously thought to drive expression in all FeCO neurons, we found that it labeled ~80% of the total population (~135 neurons; Figure 3F–G).
The claw Gal4 line labeled ~20 cell bodies (Figure 3F), which were located in a characteristic blade-shaped strip that extended along the long axis of the femur (Figure 3C–E). The club Gal4 line drove expression in ~30 neurons (Figure 3F); these cell bodies were located in the proximal part of the FeCO (Figure 3C–E). Finally, the hook Gal4 line drove expression in only 3 FeCO neurons (Figure 3F), which were located along the FeCO’s ventral edge (Figure 3C–E). The cell body location for each subclass was consistent across flies. These results suggest that the cell bodies of FeCO neurons with distinct central projections are grouped in specific locations in the FeCO (Figure 3I). Although these three Gal4 lines label less than half of the total FeCO neuron population, computational registration to a standard VNC confirmed that these three subclasses span the cardinal FeCO projections labeled by iav-Gal4 (Figure 3H).
Calcium imaging from specific Gal4 lines shows that position, movement, and direction are encoded by separate proprioceptor subclasses
We next performed calcium imaging from the claw, club, and hook neurons (Figures 4, S4 and Movie S3). Claw neurons increased their activity tonically in response to either flexion or extension of the tibia (Figure 4A). Each branch of the claw axon projection had two sub-branches that responded either to flexion or extension, which we identified with the clustering methods described above (Figure 4A). The time courses of these responses were similar across the X, Y, and Z branches of the claw (Figures 4A and S5A). Given that single claw neurons innervate all three branches (Figure 3B), these data suggest that the claw subclass can be further subdivided into separate groups of cells that encode either tibia flexion or extension. The spatial organization and functional tuning of claw neurons were also consistent with the position-tuned tonic responses recorded from population imaging (Figure 2).
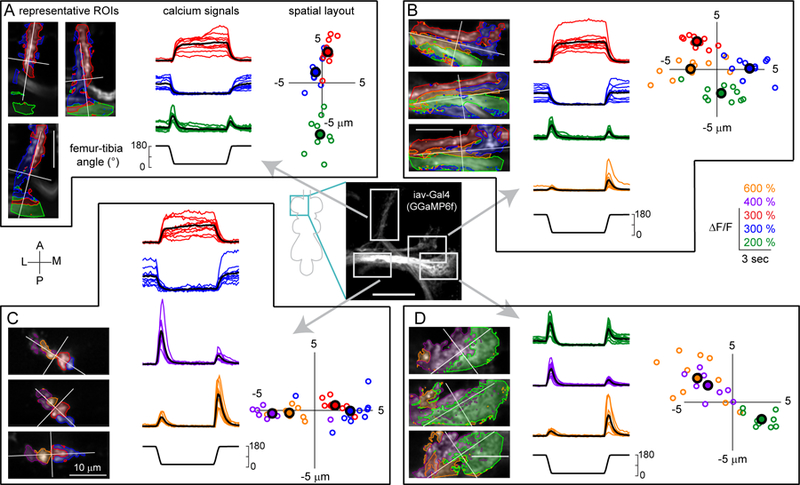
Figure 4. Three Gal4 lines delineate FeCO functional subclasses.
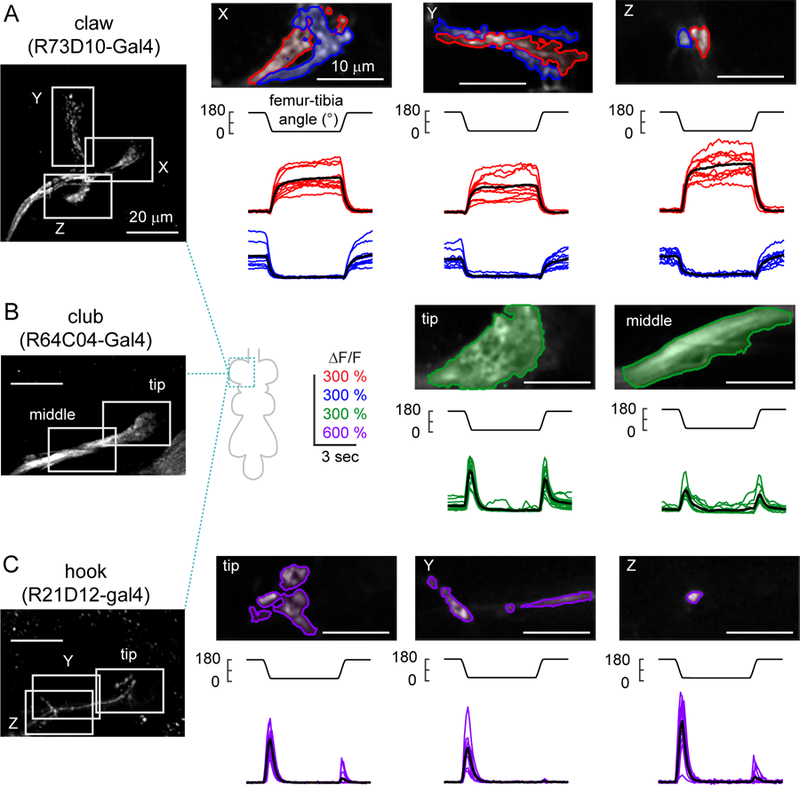
A. Claw neurons encode tibia position, with distinct pixels responding to either flexion (red) or extension (blue). Left: The claw axon projection in the VNC visualized with GCaMP6f fluorescence driven by R73D10-Gal4. White rectangles represent the imaging locations for the X, Y, and Z branches shown in the right three columns. In each column, the top image shows a representative region of interest, with flexion-encoding pixels shaded in red, extension encoding pixels in blue. The Y branch is rotated 90 degrees clockwise. The bottom rows show changes in GCaMP6f fluorescence relative to the baseline (∆F/F) recorded from each sub-region in different flies (n = 10 flies for each region). Thick black lines represent average responses. B. Same as A, but for movement-sensitive club neurons (R64C04-Gal4). The club neurons respond phasically to both the flexion and the extension of the joint (n = 14 flies for tip, 11 flies for middle). C. Same as A and B, but for directionally-tuned hook neurons (R21D12-Gal4). The hook neurons respond phasically to flexion of the joint, but not extension (n = 14 flies for tip and Z, 9 flies for Y). See also Figures S4 and S5.
Calcium signals in club axons increased phasically during both flexion and the extension of the tibia (Figure 4B). The amplitude and time course of these calcium signals were similar during extension and flexion (DSI = 0.117±0.022; mean±sem; n = 25 regions from 15 flies), and across different regions of the club (Figures 4B and S5A). These results are consistent with population imaging data (Figure 2), although the weaker direction selectivity of the club neurons labeled by a specific driver line (Figure 4B) suggests that the bidirectional clusters identified from population imaging may have been partly contaminated by axons from directionally selective (e.g., hook) neurons.
The axons of hook neurons increased their activity phasically during tibia flexion, but responded only weakly during extension (DSI = 0.811±0.028; mean±sem; n = 37 regions from 23 flies, Figure 4C). We recorded from three different locations along the hook axon projection and found that the responses were similar in all three locations (Figures 4C and S5A). The time courses of these responses were similar to those of the flexion-tuned phasic responses we observed in population-imaging experiments (Figure 2). Again, the greater direction selectivity of neurons labeled with a specific driver line (Figure 4C) suggests that the cluster identified from population imaging (Figure 2) may have been partly contaminated by nearby axons from non-directionally selective or weakly directionally selective (e.g., club) neurons.
Together, our experiments imaging from claw, club, and hook-specific Gal4 drivers reveal that these lines label distinct proprioceptor subclasses that encode different stimulus features. The response tuning of each subclass was similar for swing stimuli in the opposite direction (extension followed by flexion; Figure S4). These results match the functional and spatial organization we observed in population imaging experiments with a broad driver line (Figure 2), indicating that they represent major FeCO subclasses. The only response subclass that we failed to identify with a specific Gal4 line was the extension-tuned complement of the hook neurons (orange traces in Figure 2). Based on the location of extension-tuned pixels in population imaging experiments, we expect these neurons to have similar morphology and projections to the flexion-tuned hook neurons. Next, we will use a broader range of proprioceptive stimuli to characterize the encoding properties of each proprioceptor subclass in more detail.
Claw neurons encode tibia position, club neurons encode bidirectional tibia movement, and hook neurons encode directional tibia movement.
We used ramp-and-hold stimuli to more comprehensively explore the position-dependent tuning of each proprioceptor subclass (Figure 5). The tibia started at either a flexed (~18°) or extended (~180°) position, then moved in 18° steps to the opposite position. Claw neurons responded to these stimuli with tonic increases in calcium during either flexion (red) or extension (blue) of the tibia (Figure 5A). We identified flexion and extension-tuned pixels using the same clustering methods that we used for population imaging (Figures 1 and 2). The responses within these two regions were highly position-dependent: the activity of extension-tuned pixels increased when the tibia was between 90° and 180°, while the activity of flexion-tuned pixels increased when the tibia was between ~90° and 18°. Neither group was active in the middle of the joint range, close to 90°. Overall, the activity of claw neurons increased as a relatively linear function of the femur-tibia joint angle, although we observed some directional hysteresis (see below).
Figure 5. Claw neurons encode joint position, club neurons encode bidirectional movements, and hook neurons encode movement direction.
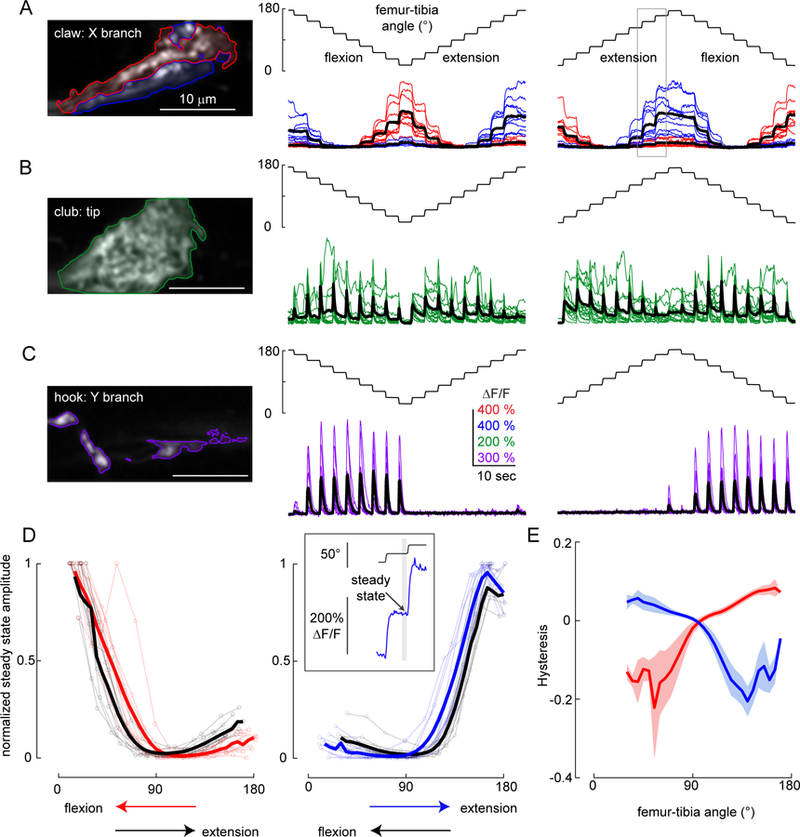
A. Responses of position-encoding claw neurons (R73D10-Gal4) to ramp-and-hold stimuli. Left column: Average GCaMP6f fluorescence from the X branch of the claw projection where the example recordings were made (red: flexion encoding, blue: extension encoding). Middle column: Responses from the two regions to a ramp-and-hold stimulus that starts with the joint extended (n = 10 flies). Right column: Same as above but starting with the joint flexed. The grey rectangle indicates the location of the trace shown in the top inset in D. B. Same as A, but for club neurons (R64C04-Gal4), which increase their activity phasically in response to each step (n = 14 flies). C. Same as A and B, but for hook neurons (driven by R21D12-Gal4), which only respond during flexion (n = 9 flies). D. Calcium signals of claw neurons depend on movement history. Left column: steady state ∆F/F at different joint angles for the flexion activated (red: during flexion, black: during extension, thick lines: average response) sub-branches of the claw, normalized by the maximum peak response recorded in each fly (n= 10 flies). Steady-state responses were measured at the end of the hold step (top inset at right). In these recordings, flexion preceded extension. Right column: Same as the left column but for the extension activated (blue: during extension, black: during flexion, thick lines: average response) sub-branches of the claw (n = 10 flies). In these recordings, extension preceded flexion. E. Hysteresis (difference between the response to the activating direction and the non-activating direction) of the steady-state response for flexion (red) and extension (blue) activated sub-branches of the claw (thick lines: average response, shading: standard error of the mean).
Club neurons responded phasically to each tibia movement, regardless of movement direction (Figure 5B). Movement responses occurred across the joint angle range but were slightly larger around 90˚ and smaller at full extension (180°). In addition to these bidirectional responses to movement steps, we occasionally observed tonic responses during the hold period, possibly due to low amplitude vibrations of the tibia (see Figure 6). These results confirm that club neurons respond to bidirectional tibia movement.
Figure 6. A map of vibration frequency in the axons of club neurons.
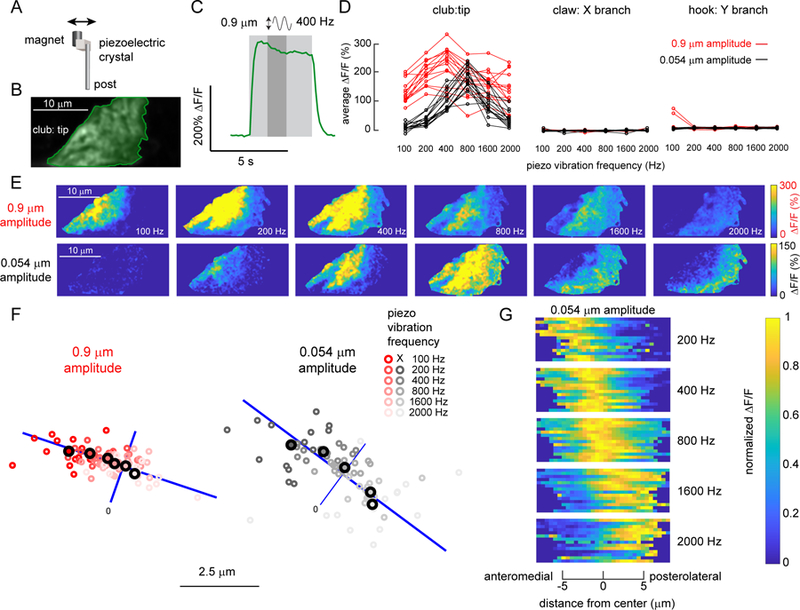
A. To vibrate the fly’s tibia, we attached one side of a piezoelectric crystal to a magnet and the other to a post fixed to a servo motor. The magnet was placed directly onto a pin glued to the tibia (see Figure S6 for details). B. Average GCaMP6f fluorescence from an example recording location, at the tip of the club axons (R64C04-Gal4). C. An example time course of the club’s response (∆F/F) to a 400 Hz, 0.9 µm vibration of the magnet. We averaged the activity level in a 1.25 second window (indicated by a darker grey shading) starting from 1.25 seconds after the stimulation onset, and used it as a measure of response amplitude in D and as activity maps in E. D. Only club neurons respond reliably to the vibration stimulus. Plots show the activity of different subsets of FeCO axons in response to tibia vibrations at different frequencies and amplitudes. Each line represents an average response from one fly. E. Example ∆F/F maps of GCaMP6f fluorescence at the tip of the club in response to tibia vibration at different frequencies and amplitudes. For both amplitudes, the responding regions shifted from the anterolateral side to the posteromedial side of the axon projection as stimulation frequency increased. F. A map of vibration frequency in club axon terminals. Smaller empty circles with different shades of red (0.9 µm amplitude) or grey (0.054 µm amplitude) represent the location of the weighted center of the responding region in different flies (n = 14 flies for both amplitudes). Larger, outlined circles represent the average location. The blue line represents the best fit line to the average locations across frequencies. We rotated the images from each fly to match the orientation of the example images shown in E. G. Distribution of activity (∆F/F) along the anterolateral to posteromedial axis (blue lines in F) of the club axons during tibia vibration. Signals were normalized by the maximum average activity during each stimulus in that fly. Responses shifted from the anterolateral to posteromedial side as vibration frequency increased. See also Figure S6
Hook neurons phasically increased their activity during tibia flexion but not extension (Figure 5C). Similar to the club neurons, hook responses were slightly weaker at fully extended positions. Phasic responses to tibia movement decayed rapidly, and we never observed tonic activity during the hold period. Overall, we found that hook neurons are directionally-selective, and encode flexion movements of the tibia.
To analyze the position-dependence of claw neurons in more detail, we measured steady state calcium activity at the end of each hold period (Figure 5D, inset) as a function of the femur-tibia angle (Figure 5D). To compare activity across flies, we normalized the response amplitudes from each fly with the largest steady state response recorded in that fly. The position tuning of claw neurons was relatively consistent across flies (Figure 5D) and branches of the claw. However, we did observe differences in claw position tuning across stimuli. For the flexion activated branch, steady state activity at each position (0□−90□) was larger when the tibia was flexed compared to when it was extended. On the other hand, extension activated branch showed larger steady state activity (at 90□−180□ range) during extension compared to flexion. (Figure 5D–E). This phenomenon, commonly referred to as hysteresis, would introduce ambiguity for downstream neurons trying to decode absolute leg angle. We did not observe directional hysteresis in the movement-tuned claw or hook neurons.
For movement-sensitive club and hook neurons, we also examined velocity tuning using swing stimuli across a range of speeds (100–800 °/s). Because the amplitude of calcium signals reflects the integration of electrical activity over time, we compared the maximum slope of the calcium signals at different tibia movement speeds (Figure S5B). For club neurons, the slope of the calcium signal peaked around 400 °/s and decayed slightly at higher speeds (Figure S5B). In contrast, the slope of the calcium signal in hook neurons was similar across the entire speed range we tested (Figure S5B). These results suggest that in addition to their direction selectivity, club and hook neurons may also differ in their velocity sensitivity.
Club axons respond to low amplitude vibrations of the tibia and contain a spatial map of frequency.
While imaging from club neurons, we occasionally observed sustained bursts of axonal calcium following a ramp movement of the tibia. We hypothesized that these variable signals were caused by spontaneous leg movements below the spatial resolution of our leg imaging system (3.85 μm/ pixel). Indeed, previous recordings in other insects have shown that FeCO neurons can be sensitive to low amplitude vibrations (Field and Pflüger, 1989; Stein and Sauer, 1999). To test this hypothesis, we used a piezoelectric chip to vibrate the magnet in a sinusoidal pattern (peak-to-peak amplitude 0.9 μm or 0.054 μm) at different frequencies (100 to 2000 Hz) (Figure 6A) and recorded calcium signals from club, claw, and hook neurons (see Figure S6 for piezo calibration details).
Club neurons exhibited large, sustained calcium signals to tibia vibration (Figure 6). An example trace in Figure 6C shows the response of club axons to 4 seconds of a 400 Hz vibration stimulus. Claw and hook neurons did not respond to these stimuli (Figure 6D). To examine frequency tuning of club neurons, we averaged the calcium signal across the stimulus period (darkly shaded region in Figure 6C) and compared responses across different frequencies and amplitudes for multiple flies (Figure 6D). For the larger amplitude stimulus (0.9 μm), the club neurons showed significant responses at all frequencies, peaking at 400 Hz (Figure 6D, red). The responses to the smaller amplitude vibration stimulus (0.054 μm) were slightly weaker and peaked at 800 Hz (Figure 6D, black).
When we examined the distribution of calcium signals during tibia vibration, we noticed a consistent spatial shift in activity as a function of vibration frequency (Figure 6E). Specifically, the center of the calcium response moved from anterolateral to the posteromedial side of the club axon projection as frequency increased (Figure 6F). This shift was clearly visible in ∆F/F maps of the club axons for both large (0.9 μm) and small (0.054 μm) amplitude vibrations (Figure 6E). The frequency map was also consistent across flies (Figure 6F–G).
The spatial maps in Figure 6F are not merely due to additional axons being recruited at higher frequencies. When we plotted the average activity along the anterolateral to posteromedial axis of the club axon projection for each fly, we found a significant shift in the entire response distribution as vibration frequency increased (Figure 6G). This effect was bigger for smaller vibrations (0.054 μm), perhaps due to the saturation of calcium signals during larger amplitude vibrations (0.9 μm). Overall, our imaging data reveal the existence of a frequency map within the axon terminals of club proprioceptors.
Frequency and position tuning of single club and claw neurons
Using driver lines that label subsets of FeCO neurons, we identified three proprioceptor subclasses that encode distinct kinematic features: tibia position (claw), directional movement (hook), and bidirectional movement/vibration (club). However, two of these subclasses, the club and claw, are each composed of more than twenty neurons. Are cells within each subclass tuned to detect the same stimulus features, or do different cells detect different features? Imaging from 20–30 overlapping axons is likely to obscure fine-scale differences in stimulus tuning across neurons. Therefore, we sought to characterize the responses of individual club and claw neurons.
We used a FlpOut approach to stochastically express GCamp6f in single club and claw axons, and imaged their responses to swing, ramp-and-hold, and vibration stimuli (Figures 7 and S7). As we observed when imaging from the entire subclass (Figures 4 and 5), single club neurons responded to tibia movement in a bidirectional manner (Figure 7B; DSI = 0.179±0.042; n = 13 cells from 9 flies), and increased their activity in response to tibia vibration (Figure 7C–D). In addition, we found that single club neurons were tuned to different vibration frequencies — for example, one cell’s peak response (∆F/F) occurred at 200 Hz, while another cell responded maximally at 1600 Hz (Figure 7D). In cases where two club neurons were labeled from the same leg, the more posterior axon was tuned to a lower frequency than the anterior axon (Figure S7A). This suggests that the axonal map of vibration frequency we describe above (Figure 6E–G) is comprised of anatomically and functionally distinct club neurons tuned to specific frequency bands.
Figure 7. Calcium imaging from single FeCO axons reveals narrow tuning of club and claw neurons.
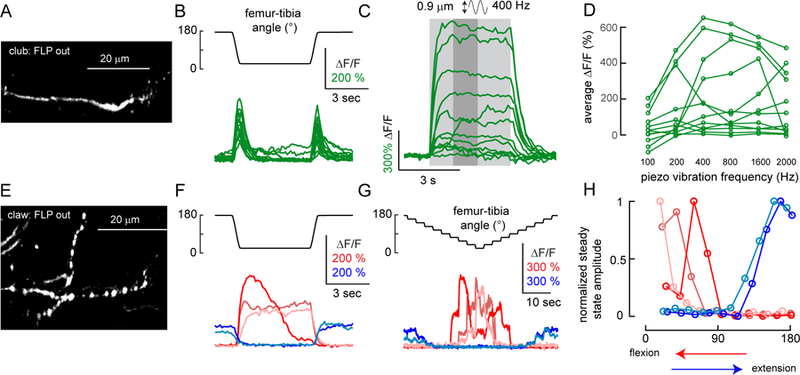
A. GCamp6f fluorescence in a single club neuron (R64C04-Gal4), imaged with a 2-photon microscope. B. Single club neurons respond to swing movements of the tibia in both directions (flexion and extension). Each trace indicates the average response of one club axon (n = 13 cells from 9 flies). C. Single club neurons respond to tibia vibration. Each trace is the average response of one axon (n = 12 cells from 7 flies). The stimulus duration is indicated by light grey shading. D. Frequency tuning of club neurons is diverse. Each line represents an average response from one neuron. We averaged the activity level in a 1.25 second window (indicated by the darker grey shading in C) starting from 1.25 seconds after the stimulation onset. E. GCamp6f fluorescence in a single claw axon (R73D10-Gal4). F. Single claw neurons encode either flexion or extension. Each trace shows the average calcium signal from a single claw axon (n = 5 cells from 5 flies, indicated by different colors). G. Position tuning of single claw neurons. Each trace shows average calcium responses to ramp-and-hold movement of the tibia. H. Single claw neurons encode different tibia angles. Each line indicates steady state activity at different joint angles for the claw neurons shown in F and G, normalized by the maximum response of each cell. Steady-state responses were measured at the end of the hold step, as in Figure 5D. To remove the effect of hysteresis, we plotted the responses during flexion for the flexion-activated neurons (red) and during extension for the extension activated neurons (blue). See also Figure S7.
We also recorded calcium activity from single claw neurons (Figure 7E–G). Each cell responded to either flexion or extension, but not both, consistent with the results we obtained by clustering across multiple claw neurons (Figures 4 and 5). However, single claw neurons were tuned to more specific femur-tibia angles — for example, one cell’s response peaked at 70°, and decreased at more flexed angles, while a second cell responded maximally at the most flexed position (20°) (Figure 7H). These results indicate that different claw neurons encode different tibia positions. Thus, calcium signals recorded from the entire claw subclass (Figure 5) likely reflect the sum of activity across FeCO neurons tuned to a narrower range of tibia positions.
Discussion
In this study, we used in vivo calcium imaging to investigate the population coding of leg proprioception in the FeCO of Drosophila. Our results reveal a basic logic for proprioceptive sensory coding: genetically distinct proprioceptor subclasses detect and encode distinct kinematic features, including tibia position, directional movement, and vibration. The cell bodies of each proprioceptor subclass reside in separate parts of the FeCO in the leg, and their axons project to distinct regions of the fly VNC. This organization suggests that different kinematic features may be processed by separate downstream circuits, and function as parallel feedback channels for the neural control of leg movement and behavior.
Neural representation of tibia position
Claw neurons encode the position of the tibia relative to the femur. Specifically, each branch consists of two sub-branches, whose calcium signals increase when the tibia is flexed or extended (Figure 5). Imaging from single claw neurons revealed that individual cells can be narrowly tuned to even more specific tibia angles (Figure 7). These data are consistent with previous reports of angular range fractionation in the locust FeCO (Matheson, 1992). Interestingly, we observed minimal activity in claw axons when the tibia was close to 90° (Figure 5), and we did not find any single claw neurons tuned to this range in our limited sample (Figure 7). Similar tuning distributions have been observed in multiunit recordings from the FeCO of locusts and stick insects (Burns, 1974; Usherwood et al., 1968). However, single unit recordings from these species also revealed the existence of a small number of position-tuned cells with peak activity in this middle range (Hofmann et al., 1985; Matheson, 1990; Matheson, 1992). It is possible that the driver lines we used did not label the FeCO neurons tuned to this range. It is also possible that this represents a real difference between Drosophila and other insects. The fly FeCO has about half as many neurons as that of the stick insect and locust, and the biomechanics of the organ may also differ between species (Field, 1991; Shanbhag et al., 1992).
How does the position tuning of claw neurons relate to natural leg kinematics? When a fly is standing still, the tibia of the front leg rests ~90° relative to the femur; during straight walking, the tibia flexes to ~40° and extends to 120° (unpublished observations). Thus, we predict that claw neurons are largely silent in a stationary fly, while extension and flexion-tuned neurons are rhythmically active during walking. Encoding deviations from the natural resting position may reflect an adaptive strategy to minimize metabolic cost.
Position-encoding claw neurons exhibit response hysteresis: both flexion and extension-tuned sub-branches of the claw showed larger steady state activity when the tibia is moved in a direction that increases their activity (Figure 5). This response asymmetry is notable because it presents a problem for downstream circuits and computations that rely on a stable readout of tibia angle. Proprioceptive hysteresis has also been described in vertebrate muscle spindles (Wei et al., 1986) and FeCO neurons of other insects (Matheson, 1992). One possible solution for solving the ambiguities created by hysteresis would be to combine the tonic activity of claw neurons with signals from directionally selective hook neurons (Figures 4 and 5). This could allow a neuron to decode tibia position based on past history of tibia movement. However, it is also possible that tibia angle hysteresis is a useful feature of the proprioceptive system, rather than a bug. For example, it has been proposed that hysteresis could compensate for the nonlinear properties of muscle activation in short sensorimotor loops (Zill and Jepson-Innes, 1988).
Neural representation of tibia movement/vibration
We identified two functional subclasses of FeCO neurons that respond phasically to tibia movement. Club neurons respond to both flexion and the extension of the tibia, while hook neurons respond only to flexion (Figures 4 and 5). In both population (Figure 2) and single neuron (Figure S7) imaging experiments, we also observed directionally selective responses to tibia extension, although we were unable to identify a specific Gal4 line for this response subclass. The movement sensitivity of the club and hook neurons resembles that of other phasic proprioceptors, including primary muscle spindle afferents (Jones et al., 2001), and movement-tuned FeCO neurons recorded in the locust and stick insect (Hofmann et al., 1985; Matheson, 1990; Matheson, 1992). Although the slow temporal dynamics of GCaMP6f did not permit a detailed analysis of velocity tuning, our results indicate that FeCO neurons respond to the natural range of leg speeds encountered during walking (Mendes et al., 2013; Pereira et al., 2018; Wosnitza et al., 2013). In the future, it will be interesting to investigate how FeCO neurons encode leg movements during walking, and how active movements may be encoded differently from passive movements, for example through presynaptic inhibition of FeCO axon terminals (Wolf and Burrows, 1995).
In addition to their directional tuning, we found that club and hook neurons differ in their sensitivity to fast (100 Hz – 2000 Hz), low amplitude (0.9 μm – 0.054 μm) tibia vibration. Club neurons are strongly activated by vibration stimuli, but hook neurons are not (Figure 6). This difference in vibration sensitivity is not likely to be caused by a difference in velocity tuning, because these differences are relatively small at the range of the speeds experienced during tibia vibration (Figure S5B). Rather, it seems that the club neurons have a lower mechanical threshold, and/or may be more sensitive to the constant acceleration produced by vibration.
The functional role of vibration-sensitive FeCO neurons is not entirely clear. Previous studies in stick insects and locusts have found that vibration-tuned FeCO neurons do not contribute to postural reflexes in the same manner as FeCO neurons tuned to joint position and directional movement (Field and Pflüger, 1989; Kittmann and Schmitz, 1992; Stein and Sauer, 1999). This raises the possibility that vibration-tuned chordotonal neurons sense external mechanosensory stimuli. For example, club neurons could monitor substrate vibrations in the environment, which serve as an important communication signal for many insect species (Hill and Wessel, 2016). Abdominal vibrations produced during courtship by male Drosophila coincide with pausing behavior in females, and hence increased receptivity to copulation (Fabre et al., 2012). These vibrations occur at frequencies that match the sensitivity of club neurons (200–2000 Hz; Caroline Fabre, personal communication). Therefore, club neurons are well-positioned to mediate intraspecific vibratory communication during courtship or other behaviors.
Stereotypic spatial organization of leg proprioceptors
Using genetic driver lines for specific FeCO neuron subclasses, we provide the first detailed anatomical characterization of Drosophila leg proprioceptors. Our anatomy and imaging experiments revealed a systematic relationship between the functional tuning of proprioceptor subclasses and their anatomical structure. The cell bodies of the three proprioceptor subclasses are clustered in different regions of the femur, an organization that may reflect biomechanical specialization for detecting position, movement, and vibration. Proprioceptor axons then converge within the leg nerve, before branching within the VNC to form subclass-specific projections that we call the club, claw, and hook (Figures 2, 3, and 4). We found that this organization is highly stereotyped across flies.
The axons of claw neurons split into three symmetric branches, resembling a claw. This unique arborization pattern is suggestive of a Cartesian coordinate system; for example, each branch could represent a different spatial axis. However, we found that each claw neuron innervates all three branches, and that the X, Y, and Z branches all encode the same stimuli. Specifically, our calcium imaging experiments revealed that each claw branch is divided into two sub-branches that are specialized for encoding flexion or extension of the tibia (Figures 2 and 4). If each claw branch is functionally similar, what is the purpose of this tri-partite structure? Each branch may target different downstream neurons, or could be independently modulated by presynaptic inhibition. Interestingly, the axons of directionally-tuned hook neurons arborized alongside the claw, but did not innervate all three of the claw branches. Thus, the X, Y, and Z branches may facilitate integration of positional information with directionally-tuned movement signals.
We were surprised to discover a topographic map of leg vibration frequency within the axon terminals of club neurons. This structure has not previously been described in flies, but resembles the tonotopic map of sensory afferents in the cricket auditory system (Oldfield, 1983; Romer, 1983) or the cochlear nucleus in vertebrates (Cohen and Knudsen, 1999). Interestingly, the spatial layout of the frequency map in club axons was consistent across different vibration amplitudes, despite a shift in the peak frequency tuning curve (Figure 6). Recordings from single club neurons suggest that this frequency map is composed by spatially organizing individual axons that are each tuned to a narrow frequency band (Figure 7 and S7). An orderly map of vibration frequency could facilitate feature identification in downstream circuits, for example through lateral inhibition between neighboring axons with shared tuning (Suga, 1989).
Comparison with other sensory systems
Neurons in the FeCO population can be generally classified as either tonic (position-encoding) or phasic (movement-encoding). This division has been observed among proprioceptors of many animals, including other insects (Hofmann et al., 1985; Matheson, 1990; Zill, 1985), crustaceans (Burke, 1954), and mammals (Proske and Gandevia, 2012). For example, mammalian muscle spindles are innervated by both phasic (Group 1a) and tonic (Group II) afferents (Boyd, 1980). The same has been found in other primary mechanosensory neurons, including touch (Abraira and Ginty, 2013; Burrows and Newland, 1997; Juusola and French, 1998), hearing (Kamikouchi et al., 2009; Kiang, 1965; Yorozu et al., 2009), and vestibular (Cullen, 2011; Fox et al., 2010) afferents. The ubiquity of tonic and phasic neurons suggests that these two parallel information channels are essential building blocks of sensory circuits. Now that we have identified genetic tools that delineate tonic and phasic neurons in the proprioceptive system of Drosophila, these circuits have the potential to provide general insights into the utility of this sensory coding strategy.
Flies possess other chordotonal organs: the most well-studied is the Johnston’s organ (JO), which detects antennal movements produced by near-field sound, wind, gravity, and touch (Albert and Gopfert, 2015; Matsuo and Kamikouchi, 2013). Unlike the FeCO, the JO monitors rotation of a body segment that is not actively controlled by muscles or coupled to the substrate. The JO is also much larger (~500 vs. ~135 neurons). Despite these differences, the coding schemes of the two mechanosensory organs share some key similarities. JO neurons can be classified into tonic and phasic classes (Kamikouchi et al., 2009; Yorozu et al., 2009), some exhibit direction selectivity (Patella and Wilson, 2018), and their axon terminals form a rough tonotopic map of frequency (Patella and Wilson, 2018). The FeCO and JO share genetic and developmental homology (Eberl and Boekhoff-Falk, 2007), which suggests that mechanosensory specialization in these organs could arise through similar molecular or biomechanical mechanisms.
Summary
With the advent of new methods for simultaneously monitoring the activity of hundreds or thousands of neurons (Ahrens et al., 2013; Mann et al.; Sofroniew et al., 2016), a critical challenge has been to link the activity of large neuronal populations to the underlying diversity of specific cell-types (Alivisatos et al., 2013). Previous efforts have used statistical methods to compare the responses of single neurons to simultaneous optical (Tsodyks et al., 1999) or electrophysiological (Okun et al., 2015) population recordings. Here, we took a different approach, which took advantage of the fact that neurons in the fly can be reliably identified across individuals. We first used 2-photon imaging to monitor activity across a population of proprioceptive sensory neurons during controlled leg movements. From this population data, we identified spatially distinct axon branches that encode specific proprioceptive stimulus features. We then searched for genetic driver lines that specifically labeled each axon branch, and further characterized their functional tuning with targeted calcium imaging. With this approach, we were able to identify and characterize the major neuronal subclasses in a key proprioceptive organ.
With a genetic handle on position, movement, and direction pathways, it should now be possible to trace the flow of proprioceptive signals into downstream circuits, and to identify the functional role of specific proprioceptor subclasses within the broader context of motor control and behavior. We anticipate that Drosophila will provide a useful complement to other model organisms in dissecting fundamental mechanisms of proprioception and deepening our understanding of this mysterious “sixth sense”.
STAR METHODS
Contact for reagents and resource sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact John Tuthill (tuthill@uw.edu).
Experimental model and subject details
Drosophila melanogaster were raised on a standard cornmeal and molasses medium and kept at 25˚C in a 12:12 h light dark cycle. We used female flies 1 to 5 days post-eclosion for all experiments, except for FlpOut functional imaging experiments. In the FlpOut imaging experiments, we heatshocked the flies (genotype shown in the table below) 1–3 days post eclosion for 16 to 25 min at 37˚ C and imaged the flies 5 to 7 days after the heat shock. The genotypes used for each experiment are included in Table S1.
Method Details
Fly preparation for in vivo two-photon calcium imaging of FeCO axons
To gain optical access to the VNC while moving the tibia, we used a previously described fly holder (Tuthill and Wilson, 2016b; Figure 1B), but replaced the metal sheet that holds the fly’s thorax with thin, translucent plastic. The plastic sheet served as a light diffuser and provided a bright background during the automated tracking of the tibia. To place the fly into the holder, we first anesthetized the fly by cooling in a plastic tube on ice, then put the fly’s head through the hole in the holder and glued the ventral side of the thorax onto the hole using UV-cured glue (Bondic). We glued the head to the upper side of the fly holder. On the bottom side of the holder, we glued down the femur section of the right prothoracic leg so that we could control the femur-tibia joint angle by moving the tibia. When gluing the femur, we held it at a position where the movement of the tibia during the rotation of the femur-tibia joint was parallel to the plane of the fly holder. To eliminate mechanical interference, we glued down all other legs. We also pushed the abdomen to the left side and glued it at that position, so that the abdomen did not block tibia movement during flexion. To position the tibia with the magnetic control system described below, we cut a small piece of insect pin (length ~1.0 mm, 0.1 mm diameter; Living Systems Instrumentation) and glued it onto the tibia and the tarsus of the right prothoracic leg. We painted the pin with black India ink (Super Black, Speedball Art Products) to enhance contrast and improve tracking of the tibia/pin position. After immersing the preparation in Drosophila saline, we removed the cuticle above the prothoracic segment of the VNC with fine forceps and took out the digestive tract to reduce the movements of the VNC. We also removed fat bodies and larger trachea to improve the optical access to the leg neuropil. The fly remained alive throughout the experiment, as indicated by the presence of spontaneous leg movements when the magnet was removed. Fly saline contained: 103 mM NaCl, 3 mM KCl, 5 mM TES, 8 mM trehalose, 10 mM glucose, 26 mM NaHCO3, 1 mM NaH2PO4, 1.5 mM CaCl2, and 4 mM MgCl2 (pH 7.1, osmolality adjusted to 270–275 mOsm). Recordings were performed at room temperature.
Image acquisition using a two-photon excitation microscope
We used a modified version of a custom two-photon microscope previously described in detail (Euler et al., 2009). For the excitation source, we used a mode-locked Ti/sapphire laser (Mira 900-F, Coherent Inc) set at 930 nm and adjusted the laser power using a neutral density filter to keep the power at the back aperture of the objective (40x, 0.8 NA, 2.0 mm wd; Nikon Instruments Inc) below ~25 mW during the experiment. We controlled the galvo laser scanning mirrors and the image acquisition using ScanImage software (version 5.2) within Matlab (MathWorks). To detect GCaMP6f and Td-Tomato fluorescence, we used an ET510/80M (Chroma Technology Corporation) emission filter (GCaMP6f) or a 630 AF50/25R (Omega optical Inc) emission filter (Td-Tomato) and GaAsP photomultiplier tubes (H7422P-40 modified version without cooling; Hamamatsu Photonics). During the trials, we acquired images (256 × 120 pixels or 128 × 240 pixels) at 8.01 Hz. At the end of the experiment, we acquired a z-stack of the FeCO axons in the right hemisphere of the prothoracic leg neuropil to confirm the recording location. In initial pilot experiments, we sampled different imaging regions in the VNC, and selected 4 ROIs that captured all of the major response classes (shown in Figure 2). We used these 4 ROIs for all the experiments reported in this paper. These ROIs corresponded to the axon terminals of major axon bundles of the FeCO.
Moving the tibia/pin using a magnetic control system
We designed the magnetic control system to manipulate the variable most relevant for chordotonal neurons, namely joint displacement (Field and Matheson, 1998). To move the tibia/pin to different positions, we attached a rare earth magnet (1 cm height x 5 mm diameter column) to a steel post (M3×20mm flat head machine screw) and controlled its position using a programmable servo motor (SilverMax QCI-X23C-1; Max speed 24,000 ˚/s, Position resolution 0.045˚; QuickSilver Controls Inc). We placed a piezoelectric crystal (PA3JEW; ThorLabs) between the post and the magnet in order to vibrate the tibia as described below. To move the magnet in a circular trajectory centered at the femur-tibia joint, we placed the motor on a micromanipulator (MP-285, Sutter Instruments) and adjusted its position while visually inspecting the movement of the magnet and the tibia using the tibia tracking camera described below. This brought the top edge of the magnet to the same height as the tibia/pin, and the inner edge of the magnet to be 1.5 mm from the center of the femur-tibia joint (Figure 1C). Because the pin was glued slightly off the center of the joint, the distance between the pin and the magnet was approximately 300 µm. For each trial, we controlled the speed and the position of the servo motor using QuickControl software (QuickSilver Controls, Inc). During all trials, we tracked the tibia position (as described below) to confirm the tibia movement during each trial.
Because it was difficult to fully flex the femur-tibia joint without the tibia/pin and the magnet colliding with the abdomen, we only flexed the joint up to ~18˚. During swing motion trials (Figures 2 and 4), we commanded the motor to move from a fully extended position (180˚) to a fully flexed position (18˚) at 360 ˚/s and move back to the original position with the same speed. (For velocity sensitivity experiments in Figure S5, we also used 180, 720, and 1440 ˚/s movements.) We chose these range of speeds based on the available data on the kinematics of the leg movements in walking Drosophila and other insects. Although the exact kinematics of the femur-tibia joint during walking in Drosophila has not been described, quantification of gait parameters in freely walking flies show that the swing duration of the forelegs ranges from approximately 25 to 45 ms, while the stance duration ranges from 30 to 140 ms (Mendes et al., 2013; Wosnitza et al., 2013). Data from other insects suggest that the femur-tibia joint does not move through the entire range of motion (0˚ to 180˚) during walking. For example, for the mesothoracic leg of the fast walking cockroach, the maximum femur-tibia joint excursion is approximately 60˚ (Watson and Ritzmann, 1998) and our unpublished observations of tethered, walking Drosophila have found a maximum excursion of ~80⁰. These ranges would correspond to the maximum average speed of 2400–3200 ˚/s for the swing phase and approximately 2000–2670 ˚/s for the stance phase. This is comparable to the mean joint angular velocity reported for the mesothoracic leg of the cockroach during walking, which ranges from 0 to 800 ˚/s (Watson and Ritzmann, 1998).
For the data in Figure 2, there was a 5-second interval between the flexion and the extension movement. We repeated this swing motion 3 times with a 5-second inter-trial interval. Responses to each repetition of the swing motion were similar, and thus we averaged the responses within each fly prior to averaging across flies. In the experiments using iav-Gal4, the mean ratio of the amplitude of the response to the 2nd stimulus compared to the 1st, and 3rd stimulus compared to the 2nd, was 0.953±0.035 (mean±sem) and 0.9748±0.034, respectively, for bidirectional phasic neurons responding to flexion, 0.907±0.033 and 0.998±0.023, respectively, for bidirectional phasic neurons responding to extension, 0.987±0.023 and 1.001±0.025, respectively, for flexion selective phasic neurons, 0.984±0.025 and 1.024±0.039, respectively, for extension selective phasic neurons, 0.927±0.043 and 1.016±0.118, respectively, for flexion selective tonic neurons, and 1.099±0.036 and 1.176±0.054, respectively, for extension selective tonic neurons. For the experiments using R73D10-Gal4, they were 0.894±0.029 and 0.969±0.026, respectively, for flexion selective tonic neurons, and 1.179±0.036 and 1.061±0.031, respectively, for extension selective tonic neurons. For the club neurons (expressed using R64C04-Gal4), they were 1.058±0.038 and 0.997±0.021, respectively, for responses to flexion, and 1.015±0.036 and 0.975±0.046, respectively, for responses to extension. For the hook neurons (expressed using R21D12-Gal4), they were 0.971±0.017 and 0.984±0.019, respectively, for responses to flexion.
For ramp-and-hold motion trials (Figures 5 and 7), we programmed the motor to move in 18˚ steps between full extension and flexion at 240 ˚/s. There was a 3-second hold step between each ramp movement. We used two types of movements for a ramp-and-hold motion: one that started with a flexion of the joint and extended back to the original position and another that started with an extension of the joint and flexed back to the original position. We repeated each type of trial twice and averaged the responses within each fly before averaging across flies. We set the acceleration of the motor to 72000 ˚/s2 for all movements. Movements of the tibia during each trial varied slightly due to several factors, including a small offset between the center of the motor rotation and the femur-tibia joint, and the acceleration and deceleration of the tibia movement in response to the magnet motion. Because these variations were relatively small (judging from how the responses changed with speed in Figure S5B), we did not consider these differences in the initial summary of the responses to swing motion and ramp-and-hold motion (Figures 2, 4, and 5A-C). However, for quantifying the positional dependence of the responses, we plotted the response against the actual position of the tibia during each trial (Figure 5D). Because the actual position of the tibia differed slightly between preparations, we interpolated the data at 5˚ intervals when averaging position dependent responses across preparations.
Tracking the femur-tibia joint angle
To track the position of the tibia, we backlit the tibia/pin with an 850 nm IR LED (M850F2, ThorLabs) and recorded video using an IR sensitive high speed video camera (Basler Ace A800–510um, Basler AG) with a 1.0x InfiniStix lens (94 mm wd, Infinity) equipped with 900 nm short pass filter (Edmund optics) to filter out the two-photon laser light (Figure 1B). Because the servo motor was directly underneath the fly, we placed the camera to the side and used a prism to capture the view from below. We recorded the images at 180 Hz for the ramp-and-hold motion, and at 200 Hz for the swing motion (exposure time 2.5 ms for both types of motion). To synchronize the images taken by the camera with those taken by the two-photon microscope, we acquired both the camera exposure signal and the position of the galvo scanning mirrors at 20 kHz. After acquiring the images (Figure 1C), we identified the position of the dark tibia/pin against the bright background by thresholding the image. We then approximated the orientation of the leg as the long axis of an ellipse with the same normalized second central moments as the thresholded image (Haralick and Shapiro, 1992). The spatial resolution of the image was 3.85 µm per pixel and assuming circular movement of the tibia/pin, 1-pixel movement at the edge of the tibia/pin (~1.2 mm from the center of the rotation) corresponded to 0.18˚.
Vibrating the tibia using a piezoelectric crystal
To vibrate the tibia at high frequencies, we moved the magnet using a piezoelectric crystal (PA3JEW, Max displacement 1.8 µm; ThorLabs) (Figure 6A). For controlling the movement of the piezo, we generated sine waves of different frequencies in Matlab (sampling frequency 10 kHz) and sent them to the piezo through a single channel open-loop piezo controller (Thorlabs). Because tibia movements induced by the piezo electric crystal were below the resolution of our tibia tracking system (3.85 µm/pixel), we first calibrated the piezo induced tibia movements using a separate tracking system equipped with a long working distance high magnification objective (50x, 0.45 NA, Nikon) connected to a high speed video camera (A800–510um, Basler) via InfiniTube FM-100 (Infinity) (Figure S6). In this setup, we were able to record images at 4000 Hz (exposure 190 µs) with a spatial resolution of 0.106 µm/pixel. For better control of the tibia position during high-frequency vibration, we attached the magnet to the pin for all vibration experiments (magnet overlapped with the pin for ~200 µm). For each vibration frequency, we measured the amplitude of the tibia oscillation envelope (Figure S6B, D) and power spectrum of the tibia movement (Figure S6C, E; Thomson’s multitaper power spectral density estimate with time-half bandwidth product = 4). The power spectrum of tibia movement showed a large peak at the command frequency of the piezo electric crystal, suggesting that most of the vibrations were indeed at the target frequency (Figure S6C). Both the amplitude of the oscillation envelope and the power of the oscillation at the target frequency decreased greatly at around 2000 Hz (Figure S6D, E). Thus, we decided to use vibration stimuli up to 2000 Hz.
For each stimulus, we presented 4 seconds of vibration twice with an inter-stimulus interval of 8 seconds. We averaged the responses within each fly before averaging across flies. For calculating ∆F/F maps and average response amplitudes, we used the activity level in a 1.25-second window starting 1.25 seconds after the vibration onset.
Immunohistochemistry and anatomy
For confocal imaging of the FeCO neuron axons driven by each Gal4 line in the VNC (Figure 3A), we crossed flies carrying the Gal4 driver to flies carrying pJFRC7–20XUAS-IVS-mCD8::GFP and dissected the VNC out of the thorax in Drosophila saline. We first fixed the VNC in a 4% formaldehyde PBS solution for 15 minutes. After rinsing the VNC in PBS three times, we put it in blocking solution (5% normal goat serum in PBS with 0.2% Triton-X) for 20 minutes, then incubated it with a solution of primary antibody (anti-CD8 rat antibody 1:50 concentration; anti-brp mouse for neuropil staining; 1:50 concentration) in blocking solution for 24 hours at room temperature. At the end of the first incubation, we washed the VNC in PBS with 0.2% Triton-X (PBST) three times, then incubated the VNC in a solution of secondary antibody (anti-rat-Alexa 488 1:250 concentration; anti-mouse-Alexa 633 1:250 concentration) dissolved in blocking solution for 24 hours at room temperature. Finally, we washed the VNC in PBST three times and then mounted it on a slide with Vectashield (Vector Laboratories). We acquired a z-stack image of the slides on a confocal microscope (FV1000; Olympus) to capture the axonal projection pattern relative to the neuropil.
For multicolor FlpOut experiments (Figure 3B), we crossed flies carrying the multicolor FlpOut cassettes and Flp recombinase drivers (Nern et al., 2015) to flies carrying different Gal4 drivers, and dissected out the VNCs of resulting progeny. For temperature induced expression of Flp, we placed adult flies in a plastic tube and incubated them in a 37˚C water bath for 13 to 15 minutes (up to 1 hour for the R21D12-Gal4 flies). We dissected the VNC three days after the Flp induction and followed the procedure described in Nern et al. (2015) to detect HA (using anti-HA-rabbit antibody and anti-Rabbit-Alexa 594 secondary antibody), V5 (using DyLight 549-conjugated anti-V5), and FLAG (using anti-FLAG-rat antibody and anti-Rat-Alexa 647 secondary antibody) labels expressed due to Flp induction in individual neurons. VNCs were mounted in Vectashield and imaged on a confocal microscope (Leica SP8). Single neurons were manually traced using the Simple Neurite Tracer in Fiji (Longair et al., 2011).
For co-labeling of FeCO cell bodies (Figure 3C–F), we crossed flies carrying UAS-RedStinger, LexAop-nlsGFP, and ChAT-LexA to each of the three FeCO Gal4 lines. Legs from the resulting flies were removed with forceps, mounted in Vectashield, and imaged on a confocal microscope (FluoView 1000; Olympus). To improve image quality, we imaged each leg from both the dorsal and ventral sides and stitched the resulting images together using the pairwise stitching function in Fiji (Preibisch et al., 2009). Cell bodies were then counted manually with Fiji.
For in silico overlay of the expression patterns of specific Gal4 lines (Figure 3G), we used confocal stacks of each Gal4 line with neuropil counterstaining (from the Janelia FlyLight database; Jenett et al., 2012) and used the neuropil staining to align the expression pattern in the VNC using the Computational Morphometry Toolkit (CMTK; Jefferis et al., 2007).
Quantification and statistical analyses
Image processing, K-means clustering of the responses, and analyses of clustered responses
We performed all image processing and analyses using scripts written in Matlab (MathWorks). After acquiring the images for a trial, we first applied a Gaussian filter (size 5×5 pixel, σ = 3) and aligned each frame to a mean image of the trial using a sub-pixel registration algorithm (registered to ¼ pixel; Guizar-Sicairos et al., 2008). For alignment of images, we used the red channel tdTomato fluorescence, which should not change as a function of calcium. TdTomato fluorescence remained stable over the course of each experiment (data not shown), indicating that movement artifacts were absent or small. For detecting calcium signals, we chose pixels whose mean GCaMP6f fluorescence was above a set threshold. Based on the level of GCaMP expression in each Gal4 line, as well as the background fluorescence level in our recordings, we used a uniform threshold of 150 (fluorescence intensity, arbitrary unit) for all recordings except for the hook neurons (21D12 Gal4 x GCaMP6f; threshold set to 100) and FlpOut experiments (threshold set to 60), which had a lower expression level. For all experiments except for FlpOut experiments, we used correlation based k-means clustering to group pixels based on the similarity of their intensity change during the trial (Figure 1F). We initially chose the number of clusters based on visual inspection of the response patterns during individual trials. After this initial clustering, we manually adjusted the number of clusters to verify response separability. When the cluster number was too small, clustering failed to capture some of the response features (Figure S1A, 1–3 clusters). On the other hand, when we increased the number of clusters above a certain number, we started to see clusters with similar response features (Figure S1A, 5–6 clusters). We chose the minimal number of clusters that captured all the main features of the response. For FlpOut experiments, we manually selected pixels that belong to an axon branch of a single cell.
For calculating the GCaMP6f fluorescence change relative to the baseline (∆F/F), we used the lowest average fluorescence level in a 10-frame window as the baseline fluorescence during that trial. To group similarly-responding clusters across flies in an unbiased fashion, we collected the ∆F/F traces recorded from each region in all flies and performed another correlation based k-means clustering. As in the clustering for each trial, we initially chose the number of groups based on visual inspection of the responses recorded from that region. We varied the group numbers for each region and selected the number of groups that best categorized the response patterns we observed. The results of this second clustering exercise are indicated by the red, blue, green, purple, and orange response classes in Figure 2. In the anterolateral region, we found red, blue, and green response classes in n = 10, 10, and 9 flies, respectively. In the anteromedial region, we found red, blue, green, and orange response classes in n = 11, 11, 10, and 10 flies, respectively. In the posterolateral region, we found red, blue, purple, and orange response classes in n = 10, 10, 8, and 9 flies, respectively. In the medial region, we found green, purple, and orange response classes in n = 10, 11, and 10 flies, respectively.
When comparing the time course of the same class of responses recorded from different axonal regions during swing motion, we first normalized each response time course (74 frames starting from 10 frames before the onset of the flexion) with the maximum amplitude for that response. After the normalization, we calculated the root mean square error for each response time course against the average response time course for that region, or for all the recording regions (Figures S2A and S5A). If the root mean square errors in both cases were not statistically different (t-test), we considered that the time course of the same response class recorded from different regions were not statistically different. As a reference, we also calculated the root mean square error against the overall average response time course of different response classes (Figures S2A and S5A). For Figure S2A, extension selective tonic responses in anteromedial, anterolateral, and posterolateral regions were recorded from 11, 10, and 10 flies, respectively. Flexion selective tonic responses in anteromedial, anterolateral, and posterolateral regions were recorded from 11, 10, and 10 flies, respectively. Bi-directional phasic responses in medial, anteromedial, and anterolateral regions were recorded from 10, 10, and 9 flies, respectively. Extension selective phasic responses in medial, anteromedial, and posterolateral regions were recorded from 10, 10, and 9 flies, respectively. Flexion selective phasic responses in medial and posterolateral regions were recorded from 11 and 8 flies, respectively. For Figure S5A, flexion and extension selective claw responses in X, Y, and Z branches were all recorded from 10 flies. In hook neurons, n= 14 flies for tip and Z, and 9 flies for Y. In club neurons, n = 14 flies for tip and 11 flies for middle.
To investigate the velocity sensitivity of the club and hook neurons, we first calculated the maximum slope of the ∆F/F curves for these neurons during swing motion (both flexion and extension for club neurons, flexion only for hook neurons) based on the frame-by-frame change in the ∆F/F value. We reasoned that the maximum slope of the ∆F/F curves more accurately represents the maximum activity level of these neurons than the maximum amplitude of the ∆F/F curves, because the calcium signal integrates the activity of the neuron over time. For each response, we calculated the average tibia speed during the inter-frame interval that showed the maximum slope, and plotted the maximum slope against this speed (Figure S5D, E). In a minority of cases, the maximum slope of the ∆F/F curve occurred very early in the stimulus, when the angular velocity of the tibia had not yet reached the target velocity. This resulted in multiple angular velocities for each stimulus speed. We recorded flexion and extension responses of the club neurons from 14 flies for all speeds. For hook neurons, we recorded from 9 flies for all speeds.
To more quantitatively evaluate directional tuning of phasic responses to swing motion of the tibia, we calculated a direction selectivity index for each response cluster (Figure 2), or for club and hook neurons (Figure 4). DSI ranges from 1 for a purely flexion selective response to −1 for a purely extension selective response. We used the maximum response amplitude during a 6-frame period after the onset of the tibia movement as a measure of the response amplitude to the flexion and the extension of the tibia. In Figure 2, for green clusters, we calculated DSI using 29 clusters recorded from 22 flies. For orange clusters, we used 19 clusters recorded from 19 flies. For purple clusters, we used 29 clusters recorded from 22 flies. In Figure 4, for club neurons, we calculated DSI using recordings from 25 regions in 15 flies. For hook neurons, we used recordings from 37 regions in 23 flies.
Analysis of the spatial organization of each response class
After identifying different response classes for each trial, we inspected the spatial organization of pixels that belonged to each response class. Because these pixels were grouped together in space, we used the center of mass of these pixels as a measure of their location and calculated their position relative to the center of mass of all the GCaMP6f positive pixels in the image. We used the characteristic shapes of the axon projections as landmarks and recorded from similar locations in different flies, but the anterior-posterior axis of the VNC in the image slightly differed across flies (Figure 2A–D, left columns). Before analyzing the spatial organization of each response classes, we compensated for these differences by rotating the images. For each recording region, we rotated the images from different flies to align the orientation of the claw axon projection (for recordings near the X and Y branches), the anterior edge of the club axon projection (for recordings near the tip of the club), or the axis connecting the Z branch and the directionally selective branch (for recordings near the Z branch) in the images (Figure 2A–D, right columns). Figure S2A shows outlines of the GCaMP6f positive pixels from different iav-Gal4 x UAS-GCaMP6f flies for each recording location after these compensations (anterolateral, n = 10 flies; anteromedial, n = 11 flies; posterolateral, n = 10 flies; medial, n = 11 flies). As can be seen from the Figure, these rotations were able to account for most of the differences in the recording region orientations. For a statistical comparison of the locations of the different response classes in iav-Gal4 x UAS-GCamp6f flies, we calculated the relative orientation and distance between each cluster’s center of mass and constructed a vector plot (Figure S2C). For each cluster pair, we first calculated a mean vector and took each vector’s component along the mean vector orientation. Then, we ran one-tailed Wilcoxon signed rank test on those components. If they were significantly positive, we considered the centers of mass for these two clusters to be statistically different. For Figure S2C anteromedial region, we recorded red, blue, green, and orange response clusters from n = 11, 11, 10, and 10 flies, respectively. For medial region, we recorded green, purple, and orange response clusters from n = 10, 11, and 10 flies, respectively. For anterolateral region, we recorded red, blue, and green response clusters from n = 10, 10, and 9 flies, respectively. For posterolateral region, we recorded red, blue, purple, and orange response clusters from n = 10, 10, 8, and 9 flies, respectively.
Analyzing frequency tuning map within the club axon projection
Because the orientation of the club axon projection in the recorded image differed slightly between preparations, we first rotated the images so that the orientation of the anterior edge of the club axon projection matched the example images in Figure 6E. In each fly, we defined the center of the response for each vibration frequency as the weighted center of mass of the average ∆F/F map for each stimulus. To see how the location of the response centers changed with the vibration frequency, we plotted the response centers relative to the center of mass for all the GCaMP6f positive pixels in the image (Figure 6F). After calculating the location of the average response centers for each vibration frequency, we fit a line through them by taking the first principal component of the x, y coordinates of the average response centers as the slope of the line. This line minimizes the total distance between the line and average response centers. To calculate the response distribution along this line (Figure 6G), we binned (5 pixels/bin) the pixels based on the distance along this line and averaged their ∆F/F values. For each vibration frequency in each fly, we normalized the response distribution by the maximum response. For both the 0.9 µm and 0.054 µm amplitude vibrations, we recorded responses from 14 flies for each frequency.
Supplementary Material
Top: A view of a fly from below during ramp-and-hold movement of the femur-tibia joint, recorded with an IR-sensitive high-speed video camera. To control joint angle, we glued an insect pin (painted black) to the tibia (indicated by a red line) and positioned it using a magnet mounted on a servo motor. The blue line indicates the femur. Bottom: Femur-tibia angle during the ramp-and-hold trial, automatically tracked from the video shown above.
Example recordings of GCaMP6f signals from each region shown in Figure 2. For each region, we first display the representative ROIs, calcium signals, and spatial layout as shown in Figure 2, followed by an example recording during a swing stimulus. Red lines in the example recording represent the femur-tibia joint angle, with the red dot indicating the distal tip of the tibia.
Left column: Example recordings of GCaMP6f signals from each region show in the left column of Figure 5A–C during a ramp-and-hold stimulus. Red lines in the example recording represent the femur-tibia joint angle during the trial. Right Column: same as the middle column of the Figure 5A–C.
Figure S1, related to Figures 1 and 2. Example of cluster analysis and population responses for opposite swing stimuli.
Figure S2, related to Figure 2. Additional quantification of population responses.
Figure S3, related to Figure 3. Anatomy of Gal4 driver lines used to label FeCO neurons.
Figure S4, related to Figure 4. Responses of neurons labeled by specific Gal4 lines to swing stimuli.
Figure S5, related to Figure 4. Additional quantification of subclass-specific responses.
Figure S6, related to Figure 6. Calibration of vibration stimuli.
Figure S7, related to Figure 7. Examples responses of single club, claw, and extension selective phasic neurons.
Table S1, Table of genotypes, related to STAR Methods experimental model and subject details.
Highlights.
Proprioceptors in the fly leg encode tibia position, movement, and vibration
Proprioceptor axons are organized topographically in the fly ventral nerve cord
Genetic tools subdivide proprioceptors that encode distinct kinematic features
Single proprioceptors are tuned to specific joint angles and vibration frequencies
Acknowledgements
We thank Eiman Azim, Richard Mann, Jim Truman, Julijana Gjorgjieva, Caroline Fabre, and members of the Tuthill laboratory for helpful discussions and comments on the manuscript. We thank Sophia Tintori for making the schematic in Figure 1B, Peter Detwiler, Fred Rieke, and Rachel Wong for generous sharing of equipment, Shellee Cunnington for preparation of solutions, Aljoscha Nern for advice on multicolor FlpOut, Greg Jefferis for assistance with VNC anatomy registration, and Barrett Pfeiffer, Michael Reiser, Peter Weir, and Michael Dickinson for sharing fly stocks. We acknowledge support from the NIH (S10 OD016240) to the Keck Imaging Center at UW, and the assistance of its manager, Nathaniel Peters. This work was funded by a Searle Scholar Award, a UW Innovation Award, a Klingenstein-Simons Fellowship, and NIH grant R01NS102333 to J.C.T.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interests
The authors declare no competing interests.
Data and software availability
All calcium imaging data is available for download from our lab website.
References
- Abraira VE, and Ginty DD (2013). The sensory neurons of touch. Neuron 79, 618–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahrens MB, Orger MB, Robson DN, Li JM, and Keller PJ (2013). Whole-brain functional imaging at cellular resolution using light-sheet microscopy. Nat Methods 10, 413–420. [DOI] [PubMed] [Google Scholar]
- Albert JT, and Gopfert MC (2015). Hearing in Drosophila. Current opinion in neurobiology 34, 79–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alivisatos AP, Chun M, Church GM, Deisseroth K, Donoghue JP, Greenspan RJ, McEuen PL, Roukes ML, Sejnowski TJ, Weiss PS, et al. (2013). Neuroscience. The brain activity map. Science 339, 1284–1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bässler U (1988). Functional principles of pattern generation for walking movements of stick insect forelegs: the role of the femoral chordotonal organ afferences. Journal of Experimental Biology 136, 125–147. [Google Scholar]
- Bidaye SS, Bockemuhl T, and Buschges A (2018). Six-legged walking in insects: how CPGs, peripheral feedback, and descending signals generate coordinated and adaptive motor rhythms. J Neurophysiol 119, 459–475. [DOI] [PubMed] [Google Scholar]
- Boyd IA (1980). The isolated mammalian muscle spindle. Trends in Neurosciences 3, 258–265. [Google Scholar]
- Brand AH, and Perrimon N (1993). Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118, 401–415. [DOI] [PubMed] [Google Scholar]
- Burke W (1954). An Organ for Proprioception and Vibration Sense in Carcinus Maenas. Journal of Experimental Biology 31, 127–138. [Google Scholar]
- Burns MD (1974). Structure and physiology of the locust femoral chordotonal organ. J Insect Physiol 20, 1319–1339. [DOI] [PubMed] [Google Scholar]
- Burrows M (1996). Neurobiology of an Insect Brain (Oxford: Oxford University Press; ). [Google Scholar]
- Burrows M, and Newland PL (1997). Processing of tactile information in neuronal networks controlling leg movements of the Locust. J Insect Physiol 43, 107–123. [DOI] [PubMed] [Google Scholar]
- Chen TW, Wardill TJ, Sun Y, Pulver SR, Renninger SL, Baohan A, Schreiter ER, Kerr RA, Orger MB, Jayaraman V, et al. (2013). Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature 499, 295–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen YE, and Knudsen EI (1999). Maps versus clusters: different representations of auditory space in the midbrain and forebrain. Trends Neurosci 22, 128–135. [DOI] [PubMed] [Google Scholar]
- Cullen KE (2011). The neural encoding of self-motion. Current opinion in neurobiology 21, 587–595. [DOI] [PubMed] [Google Scholar]
- Eberl DF, and Boekhoff-Falk G (2007). Development of Johnston’s organ in Drosophila. The International journal of developmental biology 51, 679–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Euler T, Hausselt SE, Margolis DJ, Breuninger T, Castell X, Detwiler PB, and Denk W (2009). Eyecup scope--optical recordings of light stimulus-evoked fluorescence signals in the retina. Pflugers Arch 457, 1393–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabre, Caroline C, Hedwig B, Conduit G, Lawrence, Peter A, Goodwin, Stephen F, and Casal J (2012). Substrate-Borne Vibratory Communication during Courtship in Drosophila melanogaster. Current Biology 22, 2180–2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field LH (1991). Mechanism for range fractionation in chordotonal organs of Locusta migratoria (L) and Valanga sp. (Orthoptera : Acrididae). International Journal of Insect Morphology & Embryology 20, 25–39. [Google Scholar]
- Field LH, and Matheson T (1998). Chordotonal Organs of Insects. In Advances in Insect Physiology, Evans PD, ed. (San Diego: Academic Press Inc; ), pp. 1–228. [Google Scholar]
- Field LH, and Pflüger HJ (1989). The femoral chordotonal organ: A bifunctional orthopteran (Locusta migratoria) sense organ? Comp Biochem Physiol A Comp Physiol 93, 729–743. [Google Scholar]
- Fox JL, Fairhall AL, and Daniel TL (2010). Encoding properties of haltere neurons enable motion feature detection in a biological gyroscope. Proc Natl Acad Sci U S A 107, 3840–3845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graber V (1882). Die chordotonalen Sinnesorgane und das Gehör der Insecten. Archiv für mikroskopische Anatomie 20, 506–640. [Google Scholar]
- Guizar-Sicairos M, Thurman ST, and Fienup JR (2008). Efficient subpixel image registration algorithms. Opt Lett 33, 156–158. [DOI] [PubMed] [Google Scholar]
- Haralick RM, and Shapiro LG (1992). Computer and robot vision (Reading, Mass.: Addison-Wesley Pub. Co.). [Google Scholar]
- Hasan Z, and Stuart DG (1988). Animal Solutions to Problems of Movement Control - the Role of Proprioceptors. Annual Review of Neuroscience 11, 199–223. [DOI] [PubMed] [Google Scholar]
- Hill PSM, and Wessel A (2016). Biotremology. Current Biology 26, R187–R191. [DOI] [PubMed] [Google Scholar]
- Hofmann T, Koch UT, and Bässler U (1985). Physiology of the femoral chordotonal organ in the stick insect, Cuniculina impigra. Journal of Experimental Biology 114, 207–223. [Google Scholar]
- Isakov A, Buchanan SM, Sullivan B, Ramachandran A, Chapman JK, Lu ES, Mahadevan L, and de Bivort B (2016). Recovery of locomotion after injury in Drosophila depends on proprioception. J Exp Biol 219, 1760–1771. [DOI] [PubMed] [Google Scholar]
- Jefferis GS, Potter CJ, Chan AM, Marin EC, Rohlfing T, Maurer CR Jr., and Luo L (2007).Comprehensive maps of Drosophila higher olfactory centers: spatially segregated fruit and pheromone representation. Cell 128, 1187–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenett A, Rubin GM, Ngo TT, Shepherd D, Murphy C, Dionne H, Pfeiffer BD, Cavallaro A, Hall D, Jeter J, et al. (2012). A GAL4-driver line resource for Drosophila neurobiology. Cell Rep 2, 991–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones KE, Wessberg J, and Vallbo ÅB (2001). Directional tuning of human forearm muscle afferents during voluntary wrist movements. The Journal of Physiology 536, 635–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juusola M, and French AS (1998). Adaptation properties of two types of sensory neurons in a spider mechanoreceptor organ. J Neurophysiol 80, 2781–2784. [DOI] [PubMed] [Google Scholar]
- Kamikouchi A, Inagaki HK, Effertz T, Hendrich O, Fiala A, Göpfert MC, and Ito K (2009). The neural basis of Drosophila gravity-sensing and hearing. Nature 458, 165–171. [DOI] [PubMed] [Google Scholar]
- Kiang NY-S (1965). Stimulus Coding in the Auditory Nerve and Cochlear Nucleus. Acta Oto-Laryngologica 59, 186–200. [Google Scholar]
- Kittmann R, and Schmitz J (1992). Functional specialization of the scoloparia of the femoral chordotonal organ in stick insects. Journal of Experimental Biology 173, 91–108. [Google Scholar]
- Kondoh Y, Okuma J, and Newland PL (1995). Dynamics of neurons controlling movements of a locust hind leg: Wiener kernel analysis of the responses of proprioceptive afferents. J Neurophysiol 73, 1829–1842. [DOI] [PubMed] [Google Scholar]
- Kwon Y, Shen WL, Shim HS, and Montell C (2010). Fine Thermotactic Discrimination between the Optimal and Slightly Cooler Temperatures via a TRPV Channel in Chordotonal Neurons. Journal of Neuroscience 30, 10465–10471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam T, and Pearson KG (2002). The role of proprioceptive feedback in the regulation and adaptation of locomotor activity. Adv Exp Med Biol 508, 343–355. [DOI] [PubMed] [Google Scholar]
- Longair MH, Baker DA, and Armstrong JD (2011). Simple Neurite Tracer: Open Source software for reconstruction, visualization and analysis of neuronal processes. Bioinformatics 27(17), 2453–2454. [DOI] [PubMed] [Google Scholar]
- Mann K, Gallen CL, and Clandinin TR (2017). Whole-Brain Calcium Imaging Reveals an Intrinsic Functional Network in Drosophila. Current Biology 27, 2389–2396.e2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matheson T (1990). Responses and locations of neurones in the locust metathoracic femoral chordotonal organ. Journal of Comparative Physiology A 166, 915–927. [Google Scholar]
- Matheson T (1992). Range fractionation in the locust metathoracic femoral chordotonal organ. Journal of Comparative Physiology a-Sensory Neural and Behavioral Physiology 170, 509–520. [Google Scholar]
- Matsuo E, and Kamikouchi A (2013). Neuronal encoding of sound, gravity, and wind in the fruit fly. Journal of comparative physiology A, Neuroethology, sensory, neural, and behavioral physiology 199, 253–262. [DOI] [PubMed] [Google Scholar]
- Mendes CS, Bartos I, Akay T, Marka S, and Mann RS (2013). Quantification of gait parameters in freely walking wild type and sensory deprived Drosophila melanogaster. Elife 2, e00231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nern A, Pfeiffer BD, and Rubin GM (2015). Optimized tools for multicolor stochastic labeling reveal diverse stereotyped cell arrangements in the fly visual system. Proc Natl Acad Sci U S A 112, E2967–2976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okun M, Steinmetz N, Cossell L, Iacaruso MF, Ko H, Bartho P, Moore T, Hofer SB, Mrsic-Flogel TD, Carandini M, et al. (2015). Diverse coupling of neurons to populations in sensory cortex. Nature 521, 511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldfield BP (1983). Central projections of primary auditory fibres in Tettigoniidae (Orthoptera: Ensifera). J Comp Physiol A 151, 389–395. [Google Scholar]
- Page KL, and Matheson T (2009). Functional Recovery of Aimed Scratching Movements after a Graded Proprioceptive Manipulation. Journal of Neuroscience 29, 3897–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patella P, and Wilson RI (2018). Functional Maps of Mechanosensory Features in the Drosophila Brain. Current Biology 28, 1189–+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira TD, Aldarondo DE, Willmore L, Kislin M, Wang SS-H, Murthy M, and Shaevitz JW (2018). Fast animal pose estimation using deep neural networks. bioRxiv [DOI] [PMC free article] [PubMed]
- Phillis R, Statton D, Caruccio P, and Murphey RK (1996). Mutations in the 8 kDa dynein light chain gene disrupt sensory axon projections in the Drosophila imaginal CNS. Development 122, 2955–2963. [DOI] [PubMed] [Google Scholar]
- Preibisch S Saalfeld S, and Tomancak P (2009) Globally optimal stitching of tiled 3D microscopic image acquisitions, Bioinformatics, 25(11):1463–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proske U, and Gandevia SC (2012). The proprioceptive senses: their roles in signaling body shape, body position and movement, and muscle force. Physiol Rev 92, 1651–1697. [DOI] [PubMed] [Google Scholar]
- Romer H (1983). Tonotopic organization of the auditory neuropile in the bushcricket Tettigonia viridissima. Nature 306, 60–62. [Google Scholar]
- Schindelin J, Arganda-Carreras I, and Frise E et al. (2012). Fiji: an open-source platform for biological-image analysis, Nature methods 9(7): 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanbhag SR, Singh K, and Naresh Singh R (1992). Ultrastructure of the femoral chordotonal organs and their novel synaptic organization in the legs of Drosophila melanogaster Meigen (Diptera : Drosophilidae). International Journal of Insect Morphology and Embryology 21, 311–322. [Google Scholar]
- Sherrington CS (1906). The integrative action of the nervous system (New Haven, CT: Yale University Press; ). [Google Scholar]
- Sierra E, Fernández A, Espinosa de los Monteros A, Díaz-Delgado J, Bernaldo de Quirós Y, García-Álvarez N, Arbelo M, and Herráez P (2015). Comparative histology of muscle in free ranging cetaceans: shallow versus deep diving species. Scientific Reports 5, 15909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SA, and Shepherd D (1996). Central afferent projections of proprioceptive sensory neurons in Drosophila revealed with the enhancer-trap technique. J Comp Neurol 364, 311–323. [DOI] [PubMed] [Google Scholar]
- Sofroniew NJ, Flickinger D, King J, and Svoboda K (2016). A large field of view two-photon mesoscope with subcellular resolution for in vivo imaging. Elife 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein W, and Sauer AE (1999). Physiology of vibration-sensitive afferents in the femoral chordotonal organ of the stick insect. J Comp Physiol A 184, 253–263. [Google Scholar]
- Suga N (1989). Principles of auditory information-processing derived from neuroethology. J Exp Biol 146, 277–286. [DOI] [PubMed] [Google Scholar]
- Takeoka A, Vollenweider I, Courtine G, and Arber S (2014). Muscle spindle feedback directs locomotor recovery and circuit reorganization after spinal cord injury. Cell 159, 1626–1639. [DOI] [PubMed] [Google Scholar]
- Tsodyks M, Kenet T, Grinvald A, and Arieli A (1999). Linking spontaneous activity of single cortical neurons and the underlying functional architecture. Science 286, 1943–1946. [DOI] [PubMed] [Google Scholar]
- Tsubouchi A, Yano T, Yokoyama TK, Murtin C, Otsuna H, and Ito K (2017). Topological and modality-specific representation of somatosensory information in the fly brain. Science 358, 615–623. [DOI] [PubMed] [Google Scholar]
- Tuthill JC, and Azim E (2018). Proprioception. Curr Biol, R8–r12. [DOI] [PubMed]
- Tuthill JC, and Wilson RI (2016a). Mechanosensation and adaptive motor control in insects. Current Biology [DOI] [PMC free article] [PubMed]
- Tuthill JC, and Wilson RI (2016b). Parallel transformation of tactile signals in central circuits of Drosophila. Cell 164, 1046–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usherwood PN, Runion HI, and Campbell JI (1968). Structure and Physiology of a Chordotonal Organ in Locust Leg. Journal of Experimental Biology 48, 305–323. [Google Scholar]
- Watson JT, and Ritzmann RE (1998). Leg kinematics and muscle activity during treadmill running in the cockroach, Blaberus discoidalis: II. Fast running. J Comp Physiol A 182, 23–33. [DOI] [PubMed] [Google Scholar]
- Wei JY, Simon J, Randić M, and Burgess PR (1986). Joint angle signaling by muscle spindle receptors. Brain Research 370, 108–118. [DOI] [PubMed] [Google Scholar]
- Windhorst U (2007). Muscle proprioceptive feedback and spinal networks. Brain Res Bull 73, 155–202. [DOI] [PubMed] [Google Scholar]
- Wolf H, and Burrows M (1995). Proprioceptive sensory neurons of a locust leg receive rhythmic presynaptic inhibition during walking. J Neurosci 15, 5623–5636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wosnitza A, Bockemuhl T, Dubbert M, Scholz H, and Büschges A (2013). Inter-leg coordination in the control of walking speed in Drosophila. J Exp Biol 216, 480–491. [DOI] [PubMed] [Google Scholar]
- Yorozu S, Wong A, Fischer BJ, Dankert H, Kernan MJ, Kamikouchi A, Ito K, and Anderson DJ (2009). Distinct sensory representations of wind and near-field sound in the Drosophila brain. Nature 458, 201–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zill S, Schmitz J, and Büschges A (2004). Load sensing and control of posture and locomotion. Arthropod structure & development 33, 273–286. [DOI] [PubMed] [Google Scholar]
- Zill SN (1985). Plasticity and proprioception in insects. I. Responses and cellular properties of individual receptors of the locust metathoracic femoral chordotonal organ. Journal of Experimental Biology 116, 435–461. [DOI] [PubMed] [Google Scholar]
- Zill SN, and Jepson-Innes K (1988). Evolutionary adaptation of a reflex system: sensory hysteresis counters muscle ‘catch’ tension. J Comp Physiol A 164, 43–48. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Top: A view of a fly from below during ramp-and-hold movement of the femur-tibia joint, recorded with an IR-sensitive high-speed video camera. To control joint angle, we glued an insect pin (painted black) to the tibia (indicated by a red line) and positioned it using a magnet mounted on a servo motor. The blue line indicates the femur. Bottom: Femur-tibia angle during the ramp-and-hold trial, automatically tracked from the video shown above.
Example recordings of GCaMP6f signals from each region shown in Figure 2. For each region, we first display the representative ROIs, calcium signals, and spatial layout as shown in Figure 2, followed by an example recording during a swing stimulus. Red lines in the example recording represent the femur-tibia joint angle, with the red dot indicating the distal tip of the tibia.
Left column: Example recordings of GCaMP6f signals from each region show in the left column of Figure 5A–C during a ramp-and-hold stimulus. Red lines in the example recording represent the femur-tibia joint angle during the trial. Right Column: same as the middle column of the Figure 5A–C.
Figure S1, related to Figures 1 and 2. Example of cluster analysis and population responses for opposite swing stimuli.
Figure S2, related to Figure 2. Additional quantification of population responses.
Figure S3, related to Figure 3. Anatomy of Gal4 driver lines used to label FeCO neurons.
Figure S4, related to Figure 4. Responses of neurons labeled by specific Gal4 lines to swing stimuli.
Figure S5, related to Figure 4. Additional quantification of subclass-specific responses.
Figure S6, related to Figure 6. Calibration of vibration stimuli.
Figure S7, related to Figure 7. Examples responses of single club, claw, and extension selective phasic neurons.
Table S1, Table of genotypes, related to STAR Methods experimental model and subject details.


