Summary
Unc-93 homolog B1 (UNC93B1) is a key regulator of nucleic acid (NA)-sensing toll-like receptors (TLRs). Loss of NA-sensing TLR responses in UNC93B1-deficient patients facilitates HSV-1 encephalitis. UNC93B1 is thought to guide NA-sensing TLRs from the endoplasmic reticulum (ER) to their respective endosomal signaling compartments, and the flagellin receptor TLR5 to the cell surface, raising the question how UNC93B1 mediates differential TLR trafficking. Here, we report that UNC93B1 regulates a step upstream of the differential TLR trafficking process. We discovered that UNC93B1-deficiency resulted in near complete loss of TLR3 and 7 proteins in primary splenic mouse dendritic cells (DCs) and macrophages, showing that UNC93B1 is critical to maintain TLR protein expression. Notably, expression of an ER-retained UNC93B1 version was sufficient to stabilize TLRs and largely restore endosomal TLR trafficking and activity. These data are critical for an understanding of how UNC93B1 can regulate the function of a broad subset of TLRs.
Introduction
One of the strategies by which the innate immune system can detect invading microbes is by recognizing their nucleic acids (NAs). The compartmentalization of NA-sensing TLR activity to endosomes and phagosomes has been proposed to ensure efficient sensing of microbial NAs and limit autoimmunity against host NAs. In fact, endosomal acidification as well as endosomal TLR processing are required for effective NA-sensing TLR activation (Blasius and Beutler, 2010).
The polytopic membrane protein UNC93B1 is as a cofactor required for NA-sensing TLRs (Tabeta et al., 2006). According to the current model, UNC93B1 exerts its critical function by enabling trafficking of NA-sensing TLRs from the endoplasmic reticulum (ER) to endosomal compartments (Kim et al., 2008), thereby facilitating TLR cleavage and ligand recognition. This notion is mostly based on experiments using a loss-of-function mutant of mouse UNC93B1 that was identified through a forward genetic screen and termed 3d (“triple defect”) mutant (Kim et al., 2008; Tabeta et al., 2006). This mutant carries a single point mutation changing histidine 412, located in one of the predicted transmembrane domains of UNC93B1, to arginine (H412R). In the presence of UNC93B1 H412R, all UNC93B1-dependent TLR activity is abolished and neither UNC93B1 itself nor the UNC93B1-dependent TLRs are able to reach ligand-containing endosomal compartments (Kim et al., 2008). Various studies in mouse also suggest a regulatory function of UNC93B1 in the fine-tuning of NA-sensing TLR responses (Fukui et al., 2009; Lee et al., 2013) and show that dysregulation at the level of UNC93B1 leads to autoinflammatory diseases (Fukui et al., 2011). The fact that human patients lacking functional UNC93B1 develop Herpes simplex virus type 1 (HSV-1) encephalitis (HSE) emphasizes the importance of UNC93B1 in NA recognition and host defense (Casrouge et al., 2006).
Recently, it has been recognized that the requirement for UNC93B1 is not restricted to NA-sensing endosomal TLRs. Instead, UNC93B1 is also required for the cell surface expression of the flagellin sensing TLR5 (Huh et al., 2014), while other cell surface TLRs, such as TLR2 or TLR4, function independent of UNC93B1 (Huh et al., 2014; Kim et al., 2008; Tabeta et al., 2006). Considerable advancement in the development of TLR-specific antibodies provides growing evidence that mouse TLR3 (Murakami et al., 2014), TLR7 (Kanno et al., 2015) and TLR9 (Onji et al., 2013), classically defined as endosomal TLRs, also localize to the cell surface of primary splenic DCs, B cells and macrophages. Cell surface TLR3 and TLR7 are proteolytically cleaved and therefore are not immature full-length receptors that pass the cell surface on their way to endosomes (Kanno et al., 2015; Murakami et al., 2014). Whether these TLRs can be activated from the cell surface is still an open question.
The role of UNC93B1 for TLRs that are not, or not only localized to endosomal compartments, raises the question whether UNC93B1 has additional functions independent of its endosomal trafficking activity. Notably, UNC93B1 H412R is not only impaired in its ability to traffic to endosomes (Kim et al., 2008), but also unable to interact with TLRs within the ER (Brinkmann et al., 2007). Consequently, trafficking-independent functions of UNC93B1 would be missed in studies relying on this non-functional version of UNC93B1.
Here, we designed an ER-retained version of UNC93B1 that, unlike the UNC93B1 H412R mutant, still interacts with TLRs. Notably, this ER-retained version of UNC93B1 was sufficient to enable TLR trafficking and restore proinflammatory cytokine responses. Furthermore, in the absence of UNC93B1 or the presence of UNC93B1 H412R, endogenous UNC93B1-dependent TLRs were not only trafficking-defective, but absent on the protein level. Both WT UNC93B1 and ER-retained UNC93B1 rescued TLR protein expression and function. These data indicate that UNC93B1 serves a critical function independent of its endosomal trafficking activity, namely to stabilize TLR proteins and prevent their degradation.
Results
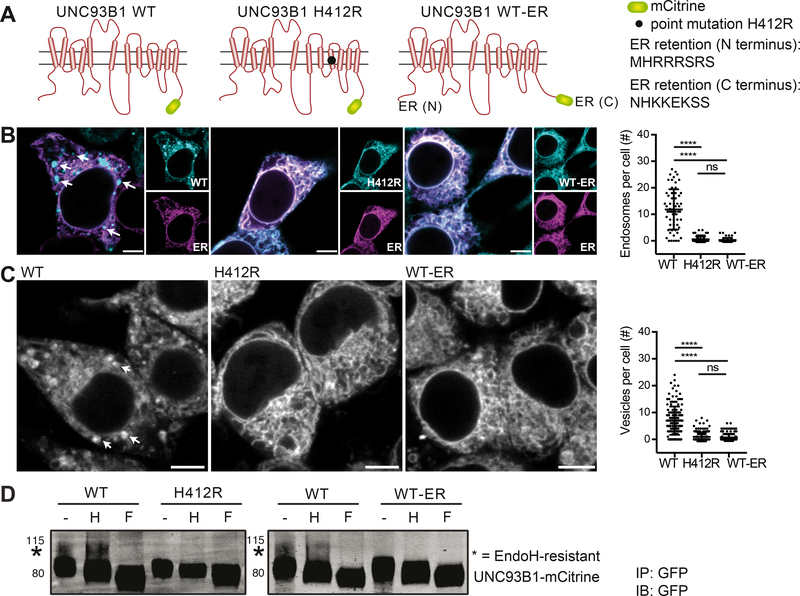
UNC93B1 WT-ER is a trafficking-defective version of UNC93B1
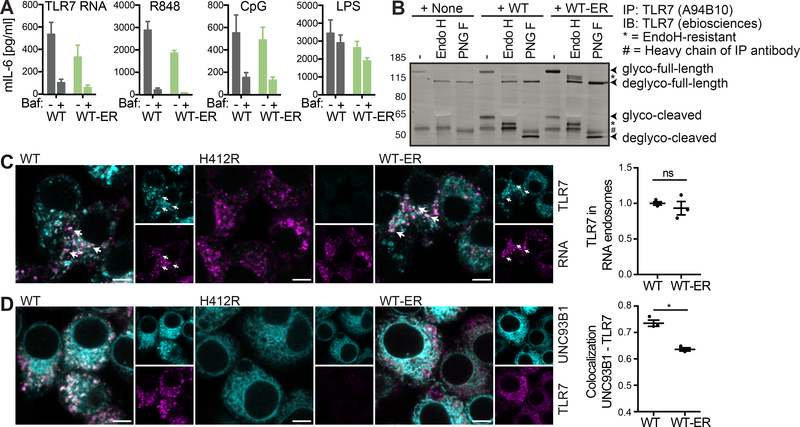
According to the current model, UNC93B1 acts as a trafficking chaperone that is crucial to guide TLRs to endosomal compartments (Kim et al., 2008; Lee et al., 2013). To discover possible trafficking-independent functions of UNC93B1, we generated a trafficking-defective version of UNC93B1 (UNC93B1 WT-ER) by adding previously described ER retention signals (Schutze et al., 1994) to both N- and C-termini of UNC93B1 (Figure 1A). These ER retention signals prevent membrane proteins from entering the secretory pathway by recruiting the COPI-dependent retrograde trafficking machinery (Lippincott-Schwartz et al., 2000). Unlike the trafficking-defective UNC93B1 H412R mutant whose internal point mutation abrogates its interaction with TLRs, the WT-ER version of UNC93B1 should be impaired in its endosomal trafficking ability, but capable of binding TLRs within the ER. Confocal microscopy of HEK 293 cells (Figure 1B) and immortalized macrophages (iMOs) derived from Unc93b1−/− mice (Figure 1C) that were reconstituted with the different human and mouse versions of UNC93B1-mCitrine indeed suggested the presence of WT UNC93B1 in endosomes, while the expression of both UNC93B1 H412R and UNC93B1 WT-ER were restricted to the ER. Cryo-electron microscopy (EM) of UNC93B1 WT, H412R and WT-ER expressing macrophages confirmed that only WT UNC93B1, but not UNC93B1 H412R or WT-ER were detectable in Golgi and endosomal compartments, while all three versions of UNC93B1 were present in the ER (Figure S1). To further analyze the trafficking defect of UNC93B1 WT-ER, the glycosylation pattern of the different UNC93B1 versions expressed in Unc93b1−/− macrophages was assessed. Proteins that traffic through the Golgi apparatus acquire oligosaccharide structures that can be experimentally removed by the enzyme Peptide-N-Glycosidase F (PNGaseF), but are resistant to cleavage by the enzyme Endoglycosidase H (EndoH). A small portion of WT UNC93B1 that was immunoprecipitated from macrophages, carried EndoH-resistant oligosaccharides, indicative of its trafficking through the Golgi apparatus (Figure 1D, star indicates EndoH-resistant fraction). In contrast, neither UNC93B1 H412R nor UNC93B1 WT-ER gained EndoH resistance, confirming that these UNC93B1 versions are retained in the ER.
Figure 1. UNC93B1 WT-ER represents a trafficking-defective version of UNC93B1.
(A) Graphical representation of the different versions of UNC93B1-mCitrine. (B) HEK 293 cell lines stably expressing human UNC93B1-mCitrine WT, H412R or WT-ER were transiently transfected with a plasmid encoding the ER marker mCherry-KDEL and analyzed by confocal microscopy. Endosomal structures in representative images are indicated by arrows and the number of endosomal structures per cell was quantified. Data are combined from three independent experiments with 20 cells per experiment; each dot represents one cell. Significance was determined by Kruskal-Wallis statistics (p<0.0001) followed by pairwise comparisons between groups (**** indicates p<0.0001, as determined by Mann-Whitney U-test statistics). Scale bars: 5 μm. (C) Unc93b1−/− iMOs stably expressing UNC93B1-mCitrine WT, H412R or WT-ER were analyzed by confocal microscopy. Endosomal structures in representative images are indicated by arrows and the number of vesicular structures per cell was quantified. Data are combined from three independent experiments with 50 cells per experiment; each dot represents one cell. Significance was determined by Kruskal-Wallis statistics (p<0.0001) followed by pairwise comparisons between groups (**** indicates p<0.0001, as determined by Mann-Whitney U-test statistics). Scale bars: 5 μm. (D) UNC93B1-mCitrine WT, H412R or WT-ER were immunoprecipitated from Unc93b1−/− iMOs stably expressing the different versions of UNC93B1. IPs were denatured and either left untreated or deglycosylated using the enzymes Endoglycosidase H (H) or Peptide-N-Glycosidase F (F), as indicated. Samples were separated on a 4–12% gradient gel and UNC93B1-mCitrine versions were detected by immunoblotting with anti-GFP antibody. EndoH-resistsant forms are indicated with a star. Data are representative of three independent experiments. Please also see Figure S1 for cryo-EM pictures of Unc93b1−/− iMOs stably expressing UNC93B1-mCitrine WT, H412R or WT-ER.
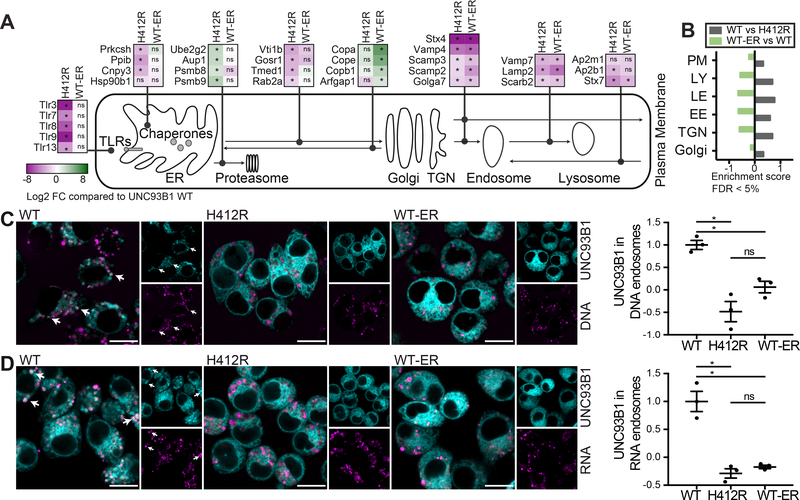
Next, we immunoprecipitated the different versions of UNC93B1 from digitonin-lysed Unc93b1−/− macrophages, a lysis procedure that keeps the intracellular membranes partly intact (Meunier and Broz, 2015), and performed quantitative proteomics in order to gain insights into the subcellular compartments that are populated by UNC93B1 WT, H412R and WT-ER. While we detected comparable amounts of endogenous TLRs 3, 7, 8, 9 and 13 in UNC93B1 WT and WT-ER immunoprecipitates (Figure 2A, shown are the log2 fold changes of UNC93B1 H412R and WT-ER compared to UNC93B1 WT), these TLRs were significantly less abundant in UNC93B1 H412R immunoprecipitates. UNC93B1 H412R immunoprecipitates also contained less ER chaperones previously identified as TLR cofactors, namely gp96 (Hsp90b1) (Yang et al., 2007) and PRAT4A (Cnpy3) (Takahashi et al., 2007) (Figure 2A). The same was true for the beta subunit of glucosidase II, also known as protein kinase C substrate 80K-H (Prkcsh), and peptidyl-prolyl cis-trans isomerase B (Ppib). Both of these ER-resident proteins are involved in the maturation of proteins and, to our knowledge, have not yet been investigated regarding their role for TLR function. Notably, proteasomal subunits, such as Psmb8 and Psmb9, and the proteins Ube2g2 and Aup1, which both are implicated in the ER degradation pathway (ERAD) (Jo et al., 2013) were significantly enriched in UNC93B1 H412R immunoprecipitates (Figure 2A). In contrast to UNC93B1 H412R, UNC93B1 WT-ER immunoprecipitates showed no significant decrease in ER chaperones and no enrichment for proteasomal components when compared to UNC93B1 WT. In accordance with the design of UNC93B1 WT-ER, we detected an enrichment of COPI coat molecules (such as Copa, Cope and Copb1), and the GTPase-activating protein Arfgap1 in UNC93B1 WT-ER immunoprecipitates (Figure 2A), indicating that UNC93B1 WT-ER is likely still exported from the ER, but then retrieved from the cis-Golgi and transported back to the ER via COPI-coated vesicles. Importantly, several proteins involved in post-Golgi trafficking pathways were significantly depleted in both UNC93B1 H412R and UNC93B1 WT-ER immunoprecipitates when compared to WT UNC93B1, among them proteins involved in the Golgi to endosome trafficking process (such as Syntaxin 4, Vamp4, Scamp2, Scamp3 and Golga7), regulators of endosome to lysosome trafficking (including Vamp7, Lamp2 and Scarb2), and trafficking molecules involved in the endocytosis pathway from the cell membrane to endosomes, namely AP2 and Syntaxin 7 (Figure 2A). The complete proteomics data set is available in Table S1. The lysosomal membrane protein Scarb2 (also known as Limp-2) and the adaptor protein 2 (AP2), which mediates clathrin-dependent endocytosis of proteins, have been previously identified in TLR and UNC93B1 trafficking pathways (Guo et al., 2015; Lee et al., 2013; Pelka et al., 2014). Furthermore, TLRs were reported to reside in and signal from Lamp2-positive compartments (Sasai et al., 2010). Hence, our proteomics approach captures known trafficking pathways of UNC93B1 and TLRs and indicates that only UNC93B1 WT, but not UNC93B1 H412R or WT-ER, is able to transit the Golgi apparatus and reach endolysosomal compartments.
Figure 2. Constitutive and stimulation-induced endosomal trafficking of UNC93B1 WT-ER is impaired.
(A and B) Quantitative proteomics of mouse UNC93B1-mCitrine WT, H412R or WT-ER immunoprecipitates from Unc93b1−/− iMOs stably expressing the respective UNC93B1 versions. (A) shows log2 fold changes of protein abundances in immunoprecipitates of mutant versus WT UNC93B1. Significant differences are indicated with * (pairwise welch t-test statistics, FDR=0.05 and S0=1). Please see Table S1 for the complete data set. (B) 1D enrichment analysis of selected Gene Ontology (GOCC) terms, namely Golgi apparatus (Golgi), trans-Golgi network (TGN), early endosomes (EE), late endosomes (LE), lysosomes (LY) and the plasma membrane (PM), for the indicated pairwise comparisons (Fisher’s exact test with Benjamini-Hochberg corrected FDR cutoff of 5%). (C) Unc93b1−/− iMOs expressing the indicated versions of UNC93B1 were fed with fluorescent CpG1826 and analyzed by confocal microscopy. Graph shows mean object-based colocalization of UNC93B1 with DNA vesicles relative to UNC93B1 WT. Each data point represents the mean of one independent experiment with thousands of DNA vesicles each. Shown are data from three independent experiments (mean + SEM). Shapiro-Wilk normality test showed normal distribution of data, Anova revealed significance (p=0.0017). * indicates p<0.05 for pairwise comparisons among groups using paired t-test statistics). Scale bars: 10 μm. (D) Unc93b1−/− iMOs expressing the indicated versions of UNC93B1 were fed with fluorescent biotinylated RNA-streptavidin complexes and analyzed by confocal microscopy. Graph shows mean object-based colocalization of UNC93B1 with RNA vesicles relative to UNC93B1 WT. Each data point represents the mean of one independent experiment with thousands of RNA vesicles each. Shown are data from three independent experiments (mean + SEM). Shapiro-Wilk normality test showed normal distribution of data, Anova revealed significance (p=0.0004). * indicates p<0.05 for pairwise comparisons among groups using paired t-test statistics). Scale bars: 10 μm.
The depletion of both UNC93B1 H412R and UNC93B1 WT-ER from Golgi apparatus, trans-Golgi network (TGN), early endosomes (EE), late endosomes (LE), lysosomes (LY) and the plasma membrane (PM) was also confirmed by Gene Ontology (GOCC) enrichment analyses. Figure 2B depicts selected significant enrichment (FDR < 5%) scores for the comparisons of WT versus H412R and WT-ER versus WT. No significant enrichments for any of these annotations were identified when UNC93B1 WT-ER was compared to UNC93B1 H412R, suggesting that indeed both of these UNC93B1 versions are equally impaired in their trans-Golgi and endosomal trafficking ability.
To further characterize the trafficking abilities of the different ER-resident UNC93B1 versions, we assessed whether they can still be recruited to endosomal TLR ligands upon stimulation. To this end, we incubated Unc93b1−/− immortalized macrophages (iMOs) expressing UNC93B1 WT, H412R or WT-ER with fluorescent TLR9-stimulatory DNA (Figure 2C) and TLR7-stimulatory RNA-streptavidin complexes (Figure 2D) and quantified the recruitment of UNC93B1 to TLR ligand containing endosomes. Unlike WT UNC93B1, both UNC93B1 H412R and UNC93B1 WT-ER were excluded from TLR-ligand containing endosomes.
Taken together, these data indicate that UNC93B1 WT-ER is defective in its constitutive and stimulation-induced post-Golgi trafficking abilities, but does not suffer from the disruptive effects of the H412R point mutation in the UNC93B1 3d mutant.
ER-retained UNC93B1 is sufficient to restore TLR-induced proinflammatory cytokine secretion
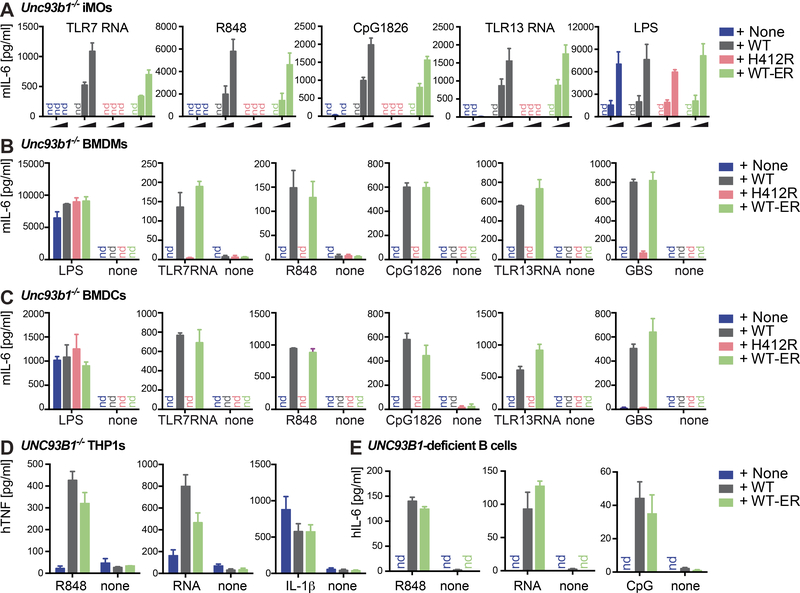
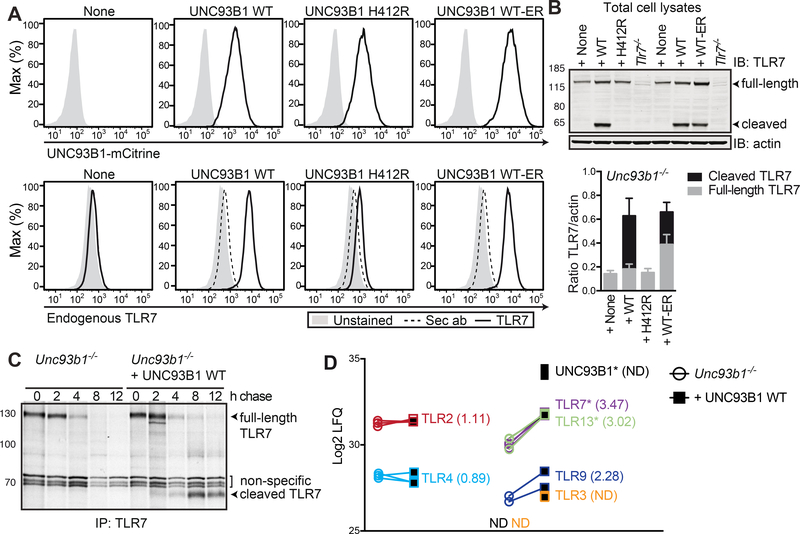
We next addressed how the expression of UNC93B1 WT-ER impacts TLR responses. We analyzed proinflammatory cytokine secretion upon stimulation with UNC93B1-dependent TLR ligands (TLR7-stimulatory RNA and R848, TLR9-stimulatory CpG1826, TLR13-stimulatory RNA derived from S. aureus ribosomal RNA) and UNC93B1–independent TLR ligands (TLR4-stimulating LPS) (Kim et al., 2008; Tabeta et al., 2006). As expected, UNC93B1 WT, but not UNC93B1 H412R, was able to restore UNC93B1-dependent TLR7, 9 and 13 activities in Unc93b1−/− iMOs (Figure 3A). Notably, UNC93B1-dependent TLR responses were largely intact in iMOs expressing UNC93B1 WT-ER. TLR4 was equally active independent of the absence or presence of UNC93B1, confirming that TLR signaling pathways and cytokine secretion in general were unimpaired in the absence of UNC93B1. To address whether overexpression of the UNC93B1 versions affected our results, we compared the protein expression of UNC93B1-mCitrine WT, H412R and WT-ER in Unc93b1−/− macrophages with that of endogenous UNC93B1 in WT macrophages by intracellular flow cytometry. We measured a moderate four-fold higher expression of UNC93B1-mCitrine compared to endogenous UNC93B1 (data not shown). Thus, we repeated our stimulation experiments in cell lines adjusted to endogenous UNC93B1 protein concentrations and confirmed that physiologic amounts of both UNC93B1 WT and UNC93B1 WT-ER were able to restore UND93B1-dependent TLR7, 9 and 13 activities in Unc93b1−/− macrophages (Figure S2).
Figure 3. ER-retained UNC93B1 is sufficient to restore TLR-induced proinflammatory cytokine secretion in mouse and human immune cells.
(A) Unc93b1−/− iMOs reconstituted with the indicated versions of UNC93B1-mCitrine were stimulated for 14 h with increasing concentrations of ligands for TLR7 (TLR7-stimulatory RNA and R848), TLR9 (CpG1826), TLR13 (TLR13-stimulatory RNA) and TLR4 (LPS) and analyzed for IL-6 secretion. Shown are combined data from three independent experiments (mean + SEM). (B) Primary Unc93b1−/− BMDMs reconstituted with the indicated versions of UNC93B1-mCitrine were stimulated for 14 h with ligands for TLR4 (1 ng/ml LPS), TLR7 (2 μM TLR7 RNA and 15 ng/ml R848), TLR9 (100 nM CpG1826), TLR13 (1 μg/ml TLR13 RNA) and heat-killed group B streptococcus (500 ng/ml GBS) and supernatants were analyzed for secreted IL-6 by ELISA. Data are representative of two independent experiments performed in triplicates (mean + SD). (C) Primary Unc93b1−/− BMDCs reconstituted with the indicated versions of UNC93B1-mCitrine were stimulated for 14 h with ligands for TLR4 (0.5 ng/ml LPS), TLR7 (1 μM TLR7 RNA and 15 ng/ml R848), TLR9 (100 nM CpG1826), TLR13 (1 μg/ml TLR13 RNA) and heat-killed group B streptococcus (125 ng/ml GBS) and analyzed for IL-6 secretion. Data are representative of two independent experiments performed in triplicates (mean + SD). (D) UNC93B1−/− THP-1 monocytes generated by CRISPR/Cas9-mediated genome editing and retrovirally transduced with human UNC93B1-mCitrine WT or WT-ER were differentiated with 10 nM PMA over night, washed and stimulated for 14 h with 2.5 μg/ml R848, 5 μM TLR7 stimulatory RNA or 2.5 ng/ml IL-1β and analyzed for TNF secretion. Data are combined from three independent experiments (mean + SEM). (E) EBV-immortalized B cells from an UNC93B1-deficient patient were retrovirally transduced with human UNC93B1-mCitrine WT or WT-ER and stimulated for 14 h with 2 μg/ml R848, 5 μM TLR7 stimulatory RNA or 2 μM CpG2006 and analyzed for IL-6 secretion. Data are combined from three independent experiments (mean + SEM). Please also see Figure S2.
In previous studies, we found that mutation of UNC93B1 can result in cell type-specific effects (Pelka et al., 2014). We therefore extended our study to primary M-CSF-derived bone marrow macrophages (BMDMs) (Figure 3B) and GM-CSF-derived bone marrow dendritic cells (BMDCs) (Figure 3C). Similar to our results in iMOs, reconstitution of primary Unc93b1−/− BMDMs and BMDCs with UNC93B1 WT-ER, but not UNC93B1 H412R, restored UNC93B1-dependent TLR responsiveness. In addition to the synthetic ligands, we also included heat-inactivated group B streptococcus (GBS) in our analysis. GBS is a gram-positive bacterium whose RNA has been previously shown to trigger TLR7- and TLR13-dependent immune responses (Mancuso et al., 2009; Signorino et al., 2014). To validate our findings in human cells, we made use of previously described UNC93B1−/− THP-1 cells (Pelka et al., 2014; Schmid-Burgk et al., 2014) (Figure 3D) and EBV-immortalized B cells (Figure 3E) derived from an UNC93B1-deficient patient with HSE (Casrouge et al., 2006) and reconstituted these cells with the respective human WT and ER-retained version of UNC93B1. Again, the ER-retained version of UNC93B1 was sufficient to largely restore immune responses.
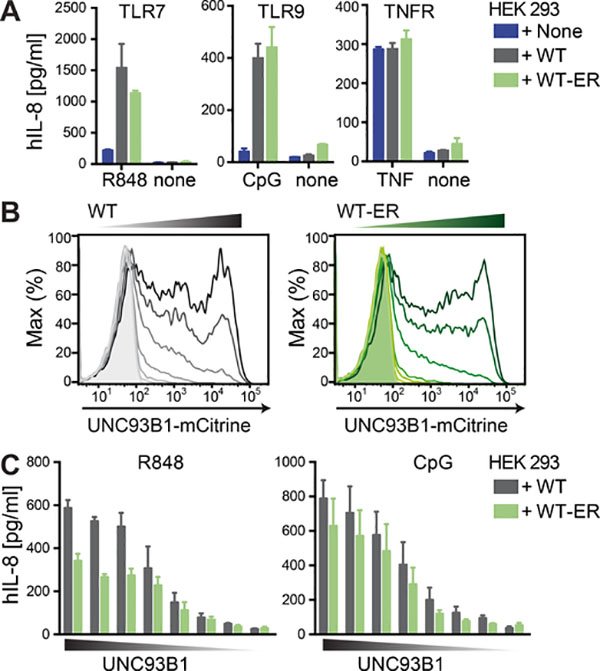
While the exclusion of both UNC93B1 H412R and WT-ER from endosomal compartments was validated using multiple different approaches including confocal microscopy, cryo-EM, glycosylation assays and proteomics, it remains possible that these ER-retained versions of UNC93B1 leak into endosomal compartments in amounts that are below the detection limit of the applied assays. Unlike UNC93B1 H412R, UNC93B1 WT-ER retains its ability to interact with TLRs. Thus, minute amounts of UNC93B1 WT-ER in endosomal compartments could potentially be functionally important. To explore the possibility that indeed small amounts of UNC93B1 could be sufficient to fully restore TLR responses, we switched to HEK 293 cells in which it is possible to tightly titrate UNC93B1 expression and analyze corresponding TLR activities. Similar to our results in mouse and human immune cells, both UNC93B1 WT and UNC93B1 WT-ER were able to confer TLR7 and TLR9 function in HEK 293 cells stably expressing the respective TLRs (Figure 4A). Tumor necrosis factor receptor (TNFR) signaling was intact independent of UNC93B1. Next, we titrated the expression of both UNC93B1-mCitrine versions (Figure 4B) and measured corresponding TLR7 and TLR9 activities (Figure 4C). Notably, TLR activities increased with increasing amounts of UNC93B1 protein suggesting that UNC93B1 boosts TLR responses in a dose-dependent manner. The maximal TLR7 response was lower in the presence of ER-retained UNC93B1 compared to WT UNC93B1. However, the minimal effective dose to increase TLR responses appeared very similar between UNC93B1 WT and UNC93B1 WT-ER. These results strongly argue against a model in which minute endosomal amounts of leaky UNC93B1 WT-ER could almost fully restore TLR responses and suggest that UNC93B1 serves critical dose-dependent functions independent of its endosomal trafficking activity.
Figure 4. UNC93B1 concentration is limiting for TLR function and similar effective doses are required for UNC93B1 WT and WT-ER.
(A) HEK cells stably expressing human TLR7 or TLR9 and human UNC93B1-mCitrine WT or WT-ER were stimulated for 14 h with 0.5 μM R848, 0.5 μM CpG2006 or 10 ng/ml TNF and analyzed for IL-8 secretion. Data are combined from three independent experiments (mean + SEM). (B and C) HEK cells stably expressing human TLR7 or TLR9 were transiently transfected with decreasing concentrations of human UNC93B1-mCitrine WT or WT-ER as shown in (B) and stimulated with 1 μM R848 or 0.5 μM CpG2006 and analyzed for IL-8 secretion. Data are combined from three independent experiments (mean + SEM).
ER-retained UNC93B1 enables TLR trafficking to endosomes
Endosomal TLR activities are known to depend on endosomal acidification (Blasius and Beutler, 2010). Experimental ER retention of UNC93B1, however, might result in an alternative signaling mode. To gain insights into the signaling requirements, we therefore treated the cells with the H+-ATPase inhibitor bafilomycin A1 and examined TLR activity upon cellular activation with TLR7-stimulatory RNA, R848, CpG1826 and LPS. In accordance with previous reports, TLR7 and 9, but not TLR4 activity were blocked by bafilomycin A1. Notably, this was true independent of UNC93B1 WT or UNC93B1 WT-ER expression (Figure 5A), suggesting that in both cases TLR activation occurred in acidified compartments.
Figure 5. ER-retained UNC93B1 is sufficient to restore TLR trafficking to TLR ligand containing compartments.
(A) Unc93b1−/− iMOs expressing UNC93B1 WT or WT-ER were pre-incubated for 30 min with 10 nM bafilomycin (where indicated), stimulated for 5 h with 12.5 ng/ml R848, 4 μM TLR7 RNA, 50 nM CpG1826 or 12.5 ng/ml LPS, and supernatants were analyzed for secreted IL-6 by ELISA. Data are combined from three independent experiments (mean + SEM). (B) TLR7 was immunoprecipitated from Unc93b1−/− iMOs expressing the indicated versions of UNC93B1-mCitrine. IPs were denatured and either left untreated or deglycosylated using the enzymes Endoglycosidase H (H) or Peptide-N-Glycosidase F (F), as indicated. Data are representative of three independent experiments. (C) Unc93b1−/− iMOs expressing the indicated versions of UNC93B1-mCitrine were incubated with biotinylated TLR7-stimulatory RNA coupled to fluorescently labeled streptavidin. 45 min upon addition of the biotinylated RNA-streptavidin complexes, cells were fixed, permeabilized and stained for TLR7 using the A94B10 anti-TLR7 antibody. The graph shows mean object-based colocalization of TLR7 with RNA vesicles relative to cells expressing UNC93B1 WT. Each data point represents the mean of one independent experiment with thousands of RNA vesicles each. Shown are data from three independent experiments (mean + SEM). Shapiro-Wilk normality test showed normal distribution of data, paired t-test showed no significance (p=0.44). Scale bars: 5 μm. (D) Unc93b1−/− iMOs expressing the indicated versions of UNC93B1-mCitrine were fixed, permeabilized and stained for TLR7 using the A94B10 anti-TLR7 antibody. The graph represents the overall correlation between UNC93B1 and TLR7 with each data point representing one independent experiment. Shown are data from three independent experiments (mean + SEM). Shapiro-Wilk normality test showed normal distribution of data, paired t-test revealed significance (p=0.019). Please also see Figure S3 for endogenous UNC93B1 and TLR7. Scale bars: 5 μm.
This led us to analyze the subcellular localization of TLR7 in the presence of the different UNC93B1 versions. In the absence of UNC93B1, TLR7 was completely EndoH-sensitive corroborating the inability of the receptor to pass through the Golgi apparatus (Figure 5B). In iMOs expressing UNC93B1 WT and UNC93B1 WT-ER, a cleaved version of TLR7 appeared and this cleaved version of TLR7 carried EndoH-resistant oligosaccharides. EndoH-resistant full-length TLR7 was hardly detectable in macrophages expressing WT UNC93B1, but a partly EndoH-resistant form accumulated in macrophages expressing the ER-retained version of UNC93B1. Of note, these TLR7 versions that carry EndoH-resistant oligosaccharides appeared in iMOs expressing the ER-retained and completely EndoH-sensitive version of UNC93B1 (Figure 1D). Hence, functional UNC93B1 prior to the Golgi is sufficient for TLR7 to readily traverse the Golgi apparatus on its own.
Given that ER-retained UNC93B1 was sufficient to support trafficking of TLR7 through the Golgi apparatus, but resulted in a stronger accumulation of partly EndoH-resistant full-length TLR7 compared to WT UNC93B1, we next analyzed the ability of TLR7 to reach RNA ligand-containing compartments. To this end, we incubated iMOs expressing UNC93B1-mCitrine WT, H412R or WT-ER with fluorescent RNA-streptavidin complexes and visualized endogenous TLR7 by immunofluorescence. TLR7 localized to RNA-containing endosomes to similar extents in both UNC93B1-mCitrine WT and WT-ER expressing iMOs (Figure 5C). The fact that TLR7, but not UNC93B1 WT-ER (Figure 2D) localized to RNA-containing compartments, suggests that the experimental retention of UNC93B1 to the ER causes the trafficking pathways of TLR and UNC93B1 to separate at some point. Indeed, overall colocalization of TLR7 and UNC93B1 WT-ER were significantly reduced compared to the colocalization of TLR7 and WT UNC93B1 (Figure 5D). We also compared WT UNC93B1-mCitrine expressing Unc93b1−/− iMOs with WT iMOs expressing endogenous UNC93B1 and found that UNC93B1 colocalized with TLR7 to similar extents in both cell lines (Figure S3). Notably, immunofluorescence staining furthermore revealed that endogenous TLR7 protein was hardly detectable in iMOs expressing the non-functional UNC93B1 H412R mutant (Figure 5C and 5D, middle panel).
Taken together, our data indicate that the presence of functional UNC93B1 prior to the Golgi apparatus is sufficient to enable TLR trafficking through the Golgi apparatus to endosomal compartments in which TLRs are proteolytically processed and TLR ligands are present.
UNC93B1 is essential for stabilizing TLRs
The absence of endogenous TLR7 in UNC93B1 H412R expressing cells (Figure 5C and 5D) was surprising. To follow up on this finding, we analyzed protein expression of endogenous TLR7 in Unc93b1−/− iMOs and UNC93B1−/− iMOs expressing UNC93B1-mCitrine WT, H412R and WT-ER by flow cytometry. Endogenous TLR7 was undetectable in the absence of UNC93B1 (Figure 6A, lower panel). Reconstitution of Unc93b1−/− iMOs with UNC93B1 H412R resulted in only a minor increase of TLR7 protein expression, whereas reconstitution with UNC93B1 WT or WT-ER rescued the defect and normalized TLR7 expression (Figure 6A, lower panel). Furthermore, we quantified full-length versus cleaved TLR7 protein amounts in iMOs stably expressing the different versions of UNC93B1 by immunoblotting. In accordance with previous findings (Lee et al., 2013), cleaved TLR7 was not detectable in the absence of UNC93B1 or the presence of the non-functional H412R mutant UNC93B1 (Figure 6B). Reconstitution of Unc93b1−/− iMOs with UNC93B1 WT or WT-ER resulted in considerable amounts of cleaved TLR7. While cleaved TLR7 represented the major form of TLR7 in cells expressing UNC93B1 WT, the full-length TLR7 form was more abundant in cells expressing UNC93B1 WT-ER. Notably, in both UNC93B1 WT and UNC93B1 WT-ER expressing cells, total amounts of TLR7 protein were strongly increased compared to Unc93b1−/− iMOs and Unc93b1−/− iMOs reconstituted with the non-functional UNC93B1 H412R mutant. To assess whether this was due to an increased TLR half-life, we performed pulse chase experiments. Unc93b1−/− iMOs and Unc93b1−/− iMOs reconstituted with WT UNC93B1 were metabolically labeled for 30 min with [35S]methionine and [35S]cysteine and after chase periods in label-free medium of 0, 2, 4, 8 and 12 hours, endogenous TLR7 was immunoprecipitated from cell lysates (Figure 6C). The 0 h time point shows comparable amounts of the full-length (immature) TLR7 in the presence or absence of UNC93B1, suggesting that similar amounts of TLR7 were synthesized within the metabolic labeling pulse. In macrophages expressing WT UNC93B1, the signal for full-length TLR7 started to decrease after 4h of chase, while cleaved (mature) TLR7 started to appear, accumulated after 8 h and was still well detectable after 12 h of chase. In the absence of UNC93B1, full-length TLR7 disappeared with slightly faster kinetics. Furthermore and as reported previously (Lee et al., 2013), the cleaved form of TLR7 is not produced in the absence of UNC93B1, resulting in the complete lack of metabolically labeled TLR7 protein after 8 h of chase.
Figure 6. UNC93B1 WT-ER, but not UNC93B1 H412R is sufficient to stabilize TLRs.
(A) Unc93b1−/− iMOs expressing the indicated versions of UNC93B1-mCitrine were permeabilized, stained for endogenous TLR7 using the monoclonal anti-TLR7 A94B10 antibody and analyzed by flow cytometry. Dotted histograms represent negative controls stained with the secondary antibody (sec ab) alone. Data are representative of three independent experiments. (B) Total cell lysates of Unc93b1−/− iMOs expressing the indicated versions of UNC93B1-mCitrine were analyzed for the presence of full-length and cleaved TLR7 by Immunoblot. The graph shows combined data from three independent experiments (mean + SEM) quantifying the total TLR7 protein amounts (full-length plus cleaved form) normalized to the loading control β-actin. (C) Unc93b1−/− iMOs and Unc93b1−/− iMOs reconstituted with UNC93B1 WT were metabolically labeled for 30 min with [35S]methionine and [35S]cysteine and lysed upon the indicated chase periods. TLR7 was immunoprecipitated using the monoclonal anti-TLR7 A94B10 antibody. Data are representative of two independent experiments. (D) Quantitative proteomics of Unc93b1−/− iMO and Unc93b1−/− iMO reconstituted with UNC93B1 WT. Shown are log2 LFQ endogenous protein abundances for the UNC93B1-independent TLRs 2 and 4 and the UNC93B1-dependent TLRs 3, 7, 9 and 13. In Unc93b1−/− iMOs, UNC93B1 and TLR3 were not detectable. Experiment was performed in triplicates, significant differences are indicated with * (pairwise welch t-test statistics, FDR=0.05 and S0=1).
We were also interested in whether the stabilizing effect of UNC93B1 was specific to TLR7 or a general function of UNC93B1 for potentially all UNC93B1-dependent TLRs. Since specific antibodies are not available for all TLRs, we decided to analyze total cellular lysates of Unc93b1−/− iMOs and Unc93b1−/− iMOs reconstituted with WT UNC93B1 by quantitative proteomics. Protein abundances should not be compared among different TLRs, since the ionization efficiencies of peptides do not correlate directly with the amount of peptides. However, the proteomics approach enabled us to quantitatively compare the protein abundances of each TLR between the two cell lines. While TLR2 and TLR4 protein expression remained largely unchanged upon reconstitution of Unc93b1−/− iMOs with UNC93B1, the protein expression of UNC93B1-dependent TLR3, TLR7, TLR9 and TLR13 increased upon reconstitution with UNC93B1 (Figure 6D), suggesting that the stabilizing function of UNC93B1 might be common to all UNC931-dependent TLRs.
Protein expression of UNC93B1-dependent TLRs is severely reduced in Unc93b1−/− and Unc93b13d/3d mice
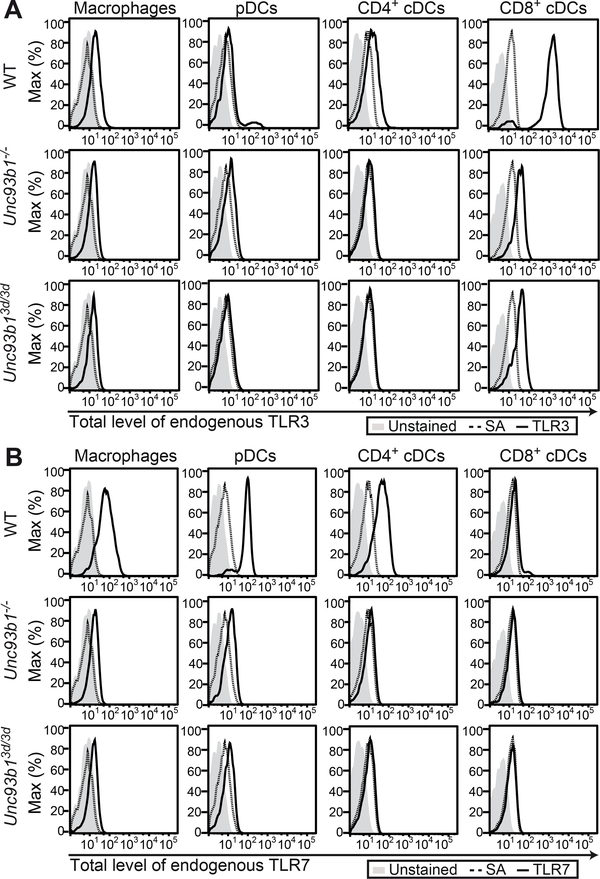
Given the profoundly decreased amounts of endogenous UNC93B1-dependent TLRs in Unc93b1−/− iMOs, we extended our study to various primary DC and macrophage cell populations in WT, Unc93b1−/− and Unc93b13d/3d mice. Flow cytometry analysis of splenocytes from WT mice stained for intracellular TLRs confirmed TLR3 expression mostly in CD8+ conventional dendritic cells (cDCs) (Figure 7A, upper panel), and TLR7 expression in macrophages, plasmacytoid DCs (pDCs) and CD4+ cDCs (Figure 7B, upper panel). The gating strategy to identify these immune cell subsets and antibody specificities are shown in Figure S4. Importantly, both TLR3 and TLR7 antibodies were shown to detect full-length (immature) and processed (mature) TLR forms (Kanno et al., 2013; Murakami et al., 2014). In accordance with our findings in iMOs, TLR3 and TLR7 proteins were hardly detectable in splenic cells from Unc93b1−/− (Figure 7A and B, middle panels) and Unc93b13d/3d mice (Figure 7A and B, lower panels). These data demonstrate that UNC93B1 is crucial to maintain expression of endogenous TLRs that functionally depend on UNC93B1 and that protein amounts of these TLRs are severely reduced in Unc93b1−/− and Unc93b13d/3d mice. In light of these data, functional studies of endosomal TLRs in cells from Unc93b13d/3d mice cannot be interpreted as TLR trafficking defects as cells are quasi-deficient in endosomal TLRs.
Figure 7. Protein expression of endogenous TLRs is severely reduced in Unc93b1−/− and Unc93b13d/3d mice.
(A and B) Splenocytes from WT, Unc93b1−/− and Unc93b13d/3d mice were isolated and stained for different immune cell subsets, namely macrophages, pDCs, CD4+ and CD8+ cDCs. Upon permeabilization, cells were stained with (A) biotinyated anti-TLR3 antibody (clone PAT3) (Murakami et al., 2014) or (B) biotinylated anti-TLR7 antibody (clone A94B10) (Kanno et al., 2013) coupled to fluorescently labeled streptavidin (SA) and analyzed by flow cytometry. Dotted histograms represent negative controls stained with fluorescently labeled SA alone. Data are representative of two independent experiments. Please see Figure S4 for gating strategies.
Discussion
The polytopic membrane protein UNC93B1 has gained researchers’ attention for two reasons. Firstly, UNC93B1 is essential for the host defense against HSV-1. UNC93B1-deficient patients are prone to develop HSV-1 encephalitis (Casrouge et al., 2006), which likely results from impaired TLR3 signaling in neurons and oligodendrocytes (Lafaille et al., 2012). Secondly, UNC93B1 has been shown to act as a key regulator of NA-sensing TLRs (Fukui et al., 2011; Fukui et al., 2009). According to the current understanding, UNC93B1 serves as a trafficking chaperone crucial to bind and guide NA-sensing TLRs to endosomal compartments where they encounter their respective ligands and initiate signaling (Brinkmann et al., 2007; Kim et al., 2008; Lee et al., 2013; Tabeta et al., 2006). Recently, however, UNC93B1 has also been proposed to be essential for TLR5 function, a receptor known to signal from the cell surface (Huh et al., 2014). Given the different subcellular signaling compartments of the various UNC93B1-dependent TLRs, the current model, in which UNC93B1 is the determining factor that dictates TLR trafficking, implies that UNC93B1 can selectively transport its client TLRs to distinct locations. Cell type- and stimulation-specific factors were speculated to influence the trafficking route of UNC93B1-TLR complexes (Huh et al., 2014). The exact molecular mechanisms, however, by which UNC93B1 enters and mediates differential trafficking pathways, remains to be determined.
The generation of an ER-retained version of UNC93B1 that is still capable of binding TLRs led us to the unexpected discovery, that NA-sensing TLR activities were largely intact in the absence of endosomal UNC93B1. Notably, this was true for all UNC93B1-dependent TLRs investigated, including TLR7, 9 and 13 and could be observed in different cell types of mouse and human origin. Hence, our findings suggest that UNC93B1 regulates TLR maturation at an early step, presumably in the ER or on the route from the ER to the Golgi apparatus, a step that most likely is shared among all UNC93B1-dependent TLRs.
In addition, UNC93B1 trafficking might be required for the access of TLRs to specific signaling compartments, such as the IRF7-signaling compartment in pDCs which has been shown to be important for the secretion of type I IFNs (Sasai et al., 2010). Indeed, UNC93B1 can itself traffic into post-Golgi compartments and partially overlaps with UNC93B1-dependent TLRs in these compartments. Furthermore, TLR cleavage was reduced and the EndoH-resistant full-length form of TLR7 accumulated upon retention of UNC93B1 to the ER, suggesting that retention of UNC93B1 did affect TLR trafficking. Nevertheless, TLR7 reached RNA-containing endosomal compartments as efficiently as in the presence of UNC93B1 WT.
One drawback of our study is that ER-retained UNC93B1 could potentially leak into endosomal compartments in amounts that are below the detection limit of the applied biochemical and imaging-based assays. Leakage of UNC93B1 H412R would be functionally irrelevant due to its inability to interact with TLRs. UNC93B1 WT-ER, however, is able to interact with TLRs and even few molecules might be functionally important. To explore this possibility, we tightly titrated the expression of UNC93B1 and assessed TLR function. Importantly, TLR activities increased with increasing protein amounts of UNC93B1 and effective doses of UNC93B1 were similar for UNC93B1 WT and WT-ER. These experiments suggest that UNC93B1 exerts a dose-dependent function that is preserved for both UNC93B1 WT and WT-ER.
Furthermore, it is worth to note that the trafficking of UNC93B1 and its client TLRs are separable events: TLR7 readily gained EndoH resistance indicative of its trafficking through the Golgi apparatus while at the same time no EndoH-resistant forms of UNC93B1 WT-ER were detectable. Colocalization studies between UNC93B1 and TLR7 further confirmed that ER retention of UNC93B1 resulted in the subcellular segregation of UNC93B1 and its client receptors. Our approach highlights the importance of TLR-specific regulatory mechanisms that operate independently of those controlling UNC93B1 trafficking. Conceptually, the individual TLR-specific regulation of post-Golgi trafficking pathways might be beneficial for fine-tuned and context-adjusted TLR responses.
One major drawback in the analysis of NA-sensing TLR function in recent years has been the lack of sensitive antibodies against these TLRs. Consequently, previous studies have been performed with overexpressed tagged versions of these TLRs, or relied on metabolic labeling of endogenous TLRs which enables their detection by immunoprecipitation and radiography. Heterologous expression of UNC93B1 and TLRs in HEK 293 cells suggested that the presence of UNC93B1 prolongs the protein half-lives of TLR3 and 9 (Pohar et al., 2013; Qi et al., 2012). On the other hand, WT and Unc93b13d/3d mutant mice were found to express equal amounts of metabolically labeled endogenous TLR7 (Brinkmann et al., 2007). Notably, only the newly synthesized immature (full-length), but not the mature (cleaved) pool of TLR7 was detected at the time. Thus, these results are consistent with our present finding that the initial biosynthesis of TLR7 is not impaired in the absence of UNC93B1. The question, however, whether the total protein amounts of endogenous UNC93B1-dependent TLRs are affected by the absence of UNC93B1, remained open.
The generation of antibodies suitable for the detection of endogenously expressed TLR3 (Murakami et al., 2014) and TLR7 (Kanno et al., 2013) enabled us to analyze the role of UNC93B1 for the expression and stability of these TLRs in their endogenous context and to answer this essential question. We discovered that UNC93B1 was crucial to maintain protein expression of endogenous TLR3 and 7 in various primary immune cell types. Total expression proteomes revealed that the same was true for endogenous TLR13 and to slightly less extent TLR9, but not TLR2 and 4, which are known to function independent of UNC93B1.
The severely reduced protein expression of endogenous UNC93B1-dependent TLRs in the presence of the H412R mutant UNC93B1 compared to WT UNC93B1 has implications for previous (Brinkmann et al., 2007) as well as current interaction proteomics analyses. The fact, that UNC93B1-dependent TLR proteins were less abundant in UNC93B1 H412R immunoprecipitates, for example, could simply reflect the reduced total protein amounts of UNC93B1-dependent TLRs within these cells rather than the TLR binding defect of UNC93B1 H412R. Importantly however, the defect of UNC93B1 H412R to interact with NA-sensing TLRs has been confirmed in HEK 293 cells heterologously expressing both UNC93B1 and TLRs (Brinkmann et al., 2007).
Previous studies have proposed that competition between TLR7 and TLR9 for UNC93B1-mediated trafficking is vital to keep NA-sensing TLRs in check and prevent uncontrolled inflammation (Fukui et al., 2011; Fukui et al., 2009). Our data propose that regulating the protein expression of UNC93B1 might be a key mechanism to modulate UNC93B1-dependent TLR function. In a clinical context, elevated UNC93B1 expression might stabilize NA-sensing TLRs and render them hyperactive. Notably, both type I IFN (Panchanathan et al., 2013; Pohar et al., 2013) and estrogen (Panchanathan et al., 2013) regulate UNC93B1 expression and UNC93B1 is upregulated in female lupus-prone B6.Nba2 mice (Panchanathan et al., 2013) as well as in B cells of patients with active systemic lupus erythematosus (Nakano et al., 2010). Hence, dysregulation on the level of UNC93B1 might contribute to autoimmunity by enhancing the protein expression and overall reactivity of UNC93B1-dependent TLRs.
Given the fundamental function of UNC93B1 in maintaining TLR protein expression, it furthermore appears likely that the major defect of UNC93B1-deficient patients is the instability of TLR proteins in the absence of UNC93B1. Whether UNC93B1 is involved in the folding process of TLRs or important for the ER export event itself is currently unknown. Immature glycoproteins undergo constant glycosylation and deglycosylation cycles, allowing the binding of chaperones such as calnexin and calreticulin (Helenius and Aebi, 2004). Glucosidase II which was enriched in immunoprecipitates of UNC93B1 WT and WT-ER, but not of UNC93B1 H412R, is involved in the deglycosylation process necessary to release glycoproteins from ER chaperones and enables the ER exit of mature proteins (Helenius and Aebi, 2004). These quality control mechanisms ensure that only correctly folded proteins leave the ER while misfolded proteins will be targeted to the ER degradation pathway. Recently, so-called corrector agents (e.g. lumacaftor) greatly improved the treatment of cystic fibrosis patients carrying the F508del mutation in the cystic fibrosis transmembrane conductance regulator (CFTR) protein (Davies, 2015). These small molecule compounds stabilize misfolded CFTRs and thereby enable the trafficking of this ion channel to its site of action (the cell membrane) and restore function (Davies, 2015). It is tempting to speculate that similar approaches might be successful in stabilizing TLRs in the absence of UNC93B1 and rescue NA-sensing TLR activity in UNC93B1-deficient patients.
STAR Methods
Contact for Reagent and Resource Sharing
Further information and requests for reagents may be directed to, and will be fulfilled by the Lead Contact, Eicke Latz (eicke.latz@uni-bonn.de).
Experimental Model and Subject Details
Cells
Unc93b1−/− M-CSF-derived bone marrow macrophages BMDMs were immortalized as described in the detailed methods section. UNC93B1−/− THP-1 monocytes (Pelka et al., 2014; Schmid-Burgk et al., 2014) and UNC93B1-deficient EBV-immortalized B cells (clone 185) (Casrouge et al., 2006) were reported previously, hTLR7HA (RRID:CVCL_Y410) and hTLR9HA HEK 293 cell lines (RRID:CVCL_Y412) were purchased from Invivogen. Stable cell lines were generated by retroviral transduction and sorted for similar protein expression of UNC93B1 versions by flow cytometry as described in the detailed methods. M-CSF-derived bone marrow macrophages (BMDMs) and GM-CSF-derived bone marrow dendritic cells (BMDCs) were differentiated for 7 days in complete RPMI 1640 medium supplemented with 40 ng/ml M-CSF (R&D Systems) or 20 ng/ml GM-CSF (Immunotools), respectively, and retrovirally transduced with the different UNC93B1 versions on day 2 of the differentiation procedure. Immortalized mouse macrophages, BMDMs, BMDCs, hTLR7HA and hTLR9HA HEK 293 cell lines were cultured in complete DMEM containing 10 % (v/v) heat-inactivated and sterile filtered FBS and 1% (v/v) penicillin-streptomycin. THP-1 monocytes and EBV-immortalized B cells were cultured in complete RPMI containing 10 % (v/v) heat-inactivated and sterile filtered FBS and 1 % (v/v) penicillin-streptomycin. All cells were cultured at 37 °C, 5 % CO2. Cell lines were regularly tested for mycoplasma contamination and were found to be negative.
Mice
WT C57BL/6J mice were purchased from Jackson Laboratory. Unc93b1−/− mice were generated by Regeneron Pharmaceuticals, Inc. as part of the Knockout Mouse Project (KOMP ID VG10049) as described previously (Valenzuela et al., 2003). Unc93b1−/−, Tlr3−/− and Tlr7−/− mice were bred under specific pathogen-free conditions at the University of Massachusetts Medical School in accordance with the regulations of the American Association for the Accreditation of Laboratory Animal Care. Experiments were performed with both male and female mice and results were not affected by the sex of the mice.
Method Details
Immortalization of Unc93b1−/− BMDMs
Femurs and tibiae of one Unc93b1−/− mouse were removed, briefly sterilized using 70 % ethanol and washed in 1x PBS. Bone marrow was harvested and passed through a 70 μm cell strainer, red blood cells were lysed. Cells were resuspended in complete DMEM medium supplemented with 40 ng/ml M-CSF, and divided into two T75 flasks. Cells were incubated for 3 days at 37 °C, 5 % CO2 until macrophages started to adhere to the flask. In order to immortalize the BMDMs, cell culture medium from one T75 flask was removed and replaced with 12 ml complete DMEM medium containing 50 % (v/v) J2 recombinant retrovirus (Roberson and Walker, 1988) and 40 ng/ml M-CSF. Cells in the other T75 were left untouched as a control. After approximately 24 h, the virus-containing medium was removed and replaced with virus-free complete DMEM medium supplemented with 40 ng/ml M-CSF. Cells were allowed to recover for 24 h before the viral transduction procedure was repeated. After removal of the virus-containing medium, cells were cultured in complete DMEM supplemented with 40 ng/ml M-CSF for another week. During this time, the non-immortalized control cells slowly started to die, while the immortalized cells started to form small colonies. Cells were passaged by using trypsin-EDTA with the control and the immortalized cells being treated exactly the same way. Approximately two weeks after the bone marrow harvest, the M-CSF concentration was started to be reduced in 1:2 dilution steps to 1 ng/ml M-CSF approximately three months after the immortalization. Up from then, the cells were grown in M-CSF-free complete DMEM. Frozen stocks of the Unc93b1−/− immortalized macrophages were prepared from early passages by resuspending approximately 3 × 106 cells in 1 ml heat-inactivated FBS containing 10 % (v/v) DMSO. Immediately upon resuspension, cells were cooled down to −80 °C at standardized cooling rates of −1 °C /min in cell freezing containers. For long-term storage, frozen vials were transferred to −150 °C. Immortalized Unc93b1−/− macrophages were reconstituted with mouse UNC93B1-mCitrine versions using retroviral transduction as described in the next section.
Retroviral transduction and fluorescence activated cell sorting
For each viral supernatant to be produced, 3 × 106 HEK293T cells were plated in 10 ml complete DMEM in one 10 cm tissue culture dish. After 16 – 24 h, HEK293T cells were transfected with retroviral constructs encoding the gene of interest (10 μg per dish) as well as the retroviral packaging plasmids gag-pol (10 μg per dish) and VSV-G (1 μg per dish) using GeneJuice transfection reagent (Novagen). Cells were incubated at 37 °C, 5 % CO2 for 5 – 8 h, then 650 μl heat-inactivated FBS was carefully added to the cells and cells were incubated for another 24 h. After 24 h, the medium was exchanged with 10 ml of fresh complete DMEM. 48 h upon transfection of the HEK293T cells, the viral supernatant was harvested and target cells (seeded to be 80 % confluent on the day of transduction) were transduced. To this end, viral supernatant was collected using a 10 ml luer-lock syringe attached to a blunt 18G needle and filtered through a 0.45 μm filter unit into a 50 ml falcon. Medium on target cells was removed and undiluted viral supernatant was added immediately. Target cells were incubated with the viral supernatant for approximately 24 h at 37 °C, 5 % CO2. The medium was changed to complete DMEM and transduced cells were passaged three times before frozen stocks were prepared. UNC93B1-mCitrine-positive cells were selected and normalized for comparable protein expression among the different verisons of UNC93B1 using fluorescence assisted cell sorting on a FACS Aria cell sorter.
Deglycosylation assay
Cells were lysed for 1 h on ice in a buffer containing 50 mM Tris-HCl pH 7.4, 150 mM NaCl, 5 mM EDTA, 5% glycerol, 1% digitonin and the complete EDTA-free protease inhibitor cocktail from Roche. Immunoprecipitations were performed from equal amounts of cleared protein lysate per sample (as measured by BCA) for at least 2 h at 4 °C using TrapGFP plates (ChromoTek) to precipitate UNC93B1-mCitrine and anti-mTLR7 A94B10 antibody coupled to protein G dynabeads to precipitate endogenous TLR7. Denaturation and deglycosylation was performed using EndoHf and PNGaseF enzymes from NEB following the manufacturer’s instructions. Proteins were separated on a Nupage Novex 4–12% Bis-Tris Protein Gel (Invitrogen) in MOPS buffer and transferred onto methanol-activated PVDF membranes (pore size 0.45 μm) using the Mini-Blot module from Thermo Fisher Scientific. The transfer was performed at 30 V for 1 h in transfer buffer containing 20 % (v/v) methanol. Membranes were incubated in TBS-T containing 3 % (w/v) BSA for 1 h at room temperature before adding the primary antibody diluted in TBS-T containing 1 % BSA, followed by three washes with TBS-T and incubation with fluorescently labeled secondary antibodies from Licor for 1 h in the dark at room temperature. Membranes were washed twice with TBS-T, once with TBS (10 min each wash), and scanned on the Odyssey Imaging System from Licor.
Sample preparation for LC-MS
Unc93b1−/− iMOs expressing UNC93B1-mCitrine WT, H412R or WT-ER were lysed for 1 h on ice in a buffer containing 50 mM Tris-HCl pH 7.4, 150 mM NaCl, 5 mM EDTA, 5% glycerol, 1% digitonin and complete EDTA-free protease inhibitor cocktail (Roche). Lysates were cleared by centrifugation (4,000xg, 5 min) and UNC93B1-mCitrine versions were immunoprecipitated on TrapGFP plates (ChromoTek) for 2 h at 4°C. Immunoprecipitates were washed twice with lysis buffer containing 0.1% digitonin and four times with detergent-free buffer containing 50 mM Tris-HCl pH 7.4 and 150 mM NaCl and resuspended in 2 M Urea, 20 mM Tris-HCl pH 8.0. Cysteines were reduced with DTT and alkylated with iodoacetamide. Proteins were digested with trypsin for 16 h. Resulting peptide mixtures were desalted on C18 Stage Tips (Rappsilber et al., 2007) and analyzed in single shots.
LC-MS
Peptides were separated on a Thermo Scientific EASY-nLC 1000 HPLC system (Thermo Fisher Scientific, Odense, Denmark) via in-house packed columns (75 μm inner diameter, 20 cm length, 1.9 μm C18 particles, Dr. Maisch GmbH, Germany) in a 100 min gradient from 2 % acetonitrile, 0.5 % formic acid to 80 % acetonitrile, 0.5 % formic acid at 250 nl/min. The column temperature was set to 45°C. An Orbitrap mass spectrometer (Q Exactive Plus, Thermo Fisher Scientific (Scheltema et al., 2014)) was directly coupled to the LC via nano electrospray source and operated in a data-dependent mode using the Xcalibur software (Thermo Scientific). The survey scan range was set to 300 – 1,650 m/z, with a resolution of 70,000. Up to 10 most abundant isotope patterns with a charge ≥ 2 were subjected to HCD fragmentation at a normalized collision energy of 25, an isolation window of 1.4 Th and a resolution of 17,500. Thresholds for ion injection time and ion target values were set to 20 ms and 3*106 for the survey scans and 120 ms and 1*105 for the MS/MS scans, respectively. To limit repeated sequencing, dynamic exclusion of sequenced peptides was set to 20 s.
MS data analysis
MS raw files were processed with MaxQuant (v1.4.3.10) (Cox and Mann, 2008) and Andromeda search engine (Cox et al., 2011), against the UniProtKB mouse FASTA database (06/2012) using default settings. Enzyme specificity was set to trypsin allowing cleavage N-terminal to proline and up to 2 miss-cleavages. Carbamidomethylation was set as fixed modification, acetylation (N-terminus) and methionine oxidation were set as variable modifications. A false discovery rate (FDR) cut-off of 1 % was applied at the peptide and protein level. Initial precursor mass deviation of up to 4.5 ppm and fragment mass deviation up to 20 ppm were allowed. ‘Match between runs’, which allows transfer of peptide identifications in the absence of sequencing was enabled, and a sample-matched peptide library was used. Proteins were quantified using MaxLFQ (Cox et al., 2014). Protein identification required at least 1 razor peptide. Data were filtered for common contaminants (n=247). Peptides only identified by site-modification were excluded from further analysis. Data were filtered for at least two valid values in at least one experimental group and missing values were imputed with a normal distribution (width=0.3; shift=1.5) where indicated, as described (Hubner et al., 2010). For the determination of significant differences in protein abundances pairwise welch t-test statistics assuming unequal variances were applied with a permutation-based FDR of 5 % and S0 of 1 (Tusher et al., 2001). For bioinformatic analysis as well as visualization we used the open PERSEUS environment, which is part of MaxQuant and the R framework (Team, R Development Core, 2008).
Total proteome and SCX methods
Cells were lysed in a buffer containing 50mM Tris-HCl, pH 8.0, 0.1 M DTT, and 2% SDS (wt/vol) at 95 °C for 5 min. The lysates were sonicated using a Branson type sonicator following which cysteines were alkylated with 55mM mM iodoacetamide for 20min at RT in the dark. 100 μg of proteins were precipitated using acetone to facilitate removal of detergent and then re-solubilized in 6M urea. Proteins were then digested with LysC for 2hr at RT, diluted to 2M urea with a 50mM ammonium bicarbonate solution and then digested with Trypsin overnight in a two-step digestion protocol. For single shot analysis, 10ug of peptides were desalted on C18 StageTips (Rappsilber et al., 2007).
To gain deeper proteome coverage, peptides were also subjected to strong cation exchange (SCX) fractionation. 60ug peptides were loaded onto 6 disks of SCX material and eluted in three steps with buffers of increasing pH values of 4.3 (SCX buffer 1: 125 mM Ammonium Acetate, 0.5% Formic Acid, 20% Acetonitrile), 5.2 (SCX buffer 2: 300 mM Ammonium Acetate, 0.5% Formic Acid, 20% Acetonitrile) and 12 (SCX buffer 3: 5% Ammonia, 80% Acetonitrile). Fractionated peptides were further desalted with C18 StageTips. Peptides were analyzed as described in LC-MS with the following modifications: Peptides were separated on an EASY-nLC 1000 HPLC system (Thermo Fisher Scientific) coupled online to the Q Exactive HF mass spectrometer via a nanoelectrospray source (Thermo Fisher Scientific) (Scheltema et al., 2014). A nonlinear 180 min gradient of 5–60% buffer B (80% ACN, 0.5% formic acid) at a flow rate of 250 nl/min and a column temperature of 55 °C was used to elute peptides. The Q Exactive HF was operated in a data dependent mode with a survey scan range of 300–1650 m/z and a resolution of 60,000 at m/z 200. 15 of the most abundant isotope patterns with a charge > 1 were isolated with a 1.4 Thomson (Th) isolation window and subjected to higher-energy collisional dissociation (HCD) fragmentation at a normalized collision energy of 27. Fragmentation spectra were acquired with a resolution of 15,000 at m/z 200. Dynamic exclusion of sequenced peptides was set to 30 s to reduce repeated peptide sequencing.
Confocal imaging experiments and analysis
Unc93b1−/− macrophages expressing either mouse UNC93B1-mCitrine WT, H412R or WT-ER were incubated with complexes consisting of biotinylated TLR7 RNA and streptavidin-AlexaFluor647 (RNA-SA complexes, 0.5 μM final concentration) or fluorescent CpG 1826 (0.5 μM final concentration) as indicated for 45 min at 37 °C. RNA-SA complexes were formed by incubating a 15-fold molar excess of RNA with tetrameric streptavidin-FluorAlexa647 for 15 min at room temperature. Upon incubation, cells were fixed with 2 % PFA in PBS over night at 4 °C. PFA was quenched by applying 50 mM NH4Cl in PBS for 10 min at room temperature. Specimens were washed twice with PBS and permeabilized with a solution containing 0.1 % Saponin (w/v) and 0.5 % BSA (w/v) for 5 minutes at room temperature. Endogenous mouse TLR7 was stained with anti-mTLR7 A94B10 antibody at a final concentration of 1 μg/ml diluted in PBS containing 0.1 % Saponin (w/v) and 1.5 % (w/v) BSA for 2 h at room temperature. Specimens were washed three times with PBS and incubated with anti-mouse AlexaFluor647 antibody diluted 1:2000 in PBS containing 0.1 % (w/v) Saponin and 1.5 % (w/v) BSA for 30 min at room temperature. Cells were washed three times with PBS and imaged on a Leica SP5 SMD confocal microscope with a 63x/1.2 water immersion objective. For the imaging analysis in Figure 2C, 2D and 5C, Cell profiler (version 6c2d896) (Carpenter et al., 2006) was used to identify all RNA or DNA vesicles (as indicated) in 8 different fields of view for each experiment. RNA or DNA vesicles were defined as objects and colocalization of UNC93B1 or TLR7 with RNA or DNA objects was determined for each field of view. Each data point represents the mean colocalization for each experiment relative to UNC93B1 WT. For Figure 5D and Figure S3, overall colocalization between UNC93B1 and TLR7 was determined by Cell profiler (version 6c2d896). Each data point represents the mean colocalization for each experiment.
Cell stimulation and cytokine measurements
Cells were stimulated as indicated with TLR7-stimulatory RNA (5’-ACUG1CG1AG1CUU-X-UUCG1AG1CG1UCA-5’, G1 is 7-deazaguanosine, X is 1,2,3-propanetriol) from Idera Pharmaceuticals, R848 (InvivoGen), TLR13-stimulatory RNA (ORN Sa19, InvivoGen), LPS (InvivoGen), CpG1826 PTO (Metabion), CpG2006 (Metabion), heat-killed GBS (kind gift from Dr. G. Teti), human TNF (R&D Systems) or human IL-1β (R&D Systems). Transient transfections were performed using Lipofectamine 2000 transfection reagent (Thermo Fisher Scientific). Where indicated, cells were pre-incubated with Bafilomycin A1 (biomol). Cytokines were measured using commercially available ELISA kits (R&D Systems and BD Biosciences).
Pulse chase experiment
Unc93b1−/− iMOs and Unc93b1−/− iMOs stably expressing WT UNC93B1 were starved in DMEM devoid of cysteine and methionine (cys−met−) for 40 min, pulsed for 30 min in DMEM (cys−met−) containing 10% dialysed FCS (Sigma) and 100μCi/ml [35S]methionine and [35S]cysteine (Hartmann Analytic). Cells were washed once in DMEM and chased in DMEM supplemented with 10% FCS, 50 μM β-ME, glutamine and Pen-Strep for 2, 4, 8 and 12 h. Cells were washed once and lysed in buffer containing 50 mM Tris-HCl pH 7.4, 150 mM NaCl, 0.5 mM EDTA, 1% NP40 and protease inhibitors (Roche). TLR7 was immunoprecipitated from cleared lysates using the anti-mTLR7 A94B10 antibody and protein A agarose beads (RepliGen). Immunoprecipitates were subjected to SDS-PAGE without heating the samples and polypeptides were visualized by radiography.
Flow cytometry
Spleens of WT, Unc93b1−/−, Tlr3−/− or Tlr7−/− mice were isolated and injected with 1 ml non-supplemented RPMI 1640 without phenol red containing 80 μl Liberase TL (Roche, stock concentration 2.5 mg/ml) and 20 μl DNase (Roche, stock concentration 10 mg/ml) and digested for 20 min at 37 °C. Digested spleens were squashed through sterile 70 μm strainers and centrifuged (350 g, 5 min, 4 °C). Erythrocytes were lysed for 2 min on ice. Cells were washed with PBS containing 0.5% BSA, 2 mM EDTA and depleted from B cells using CD19 MicroBeads (Miltenyi) following the manufacturer’s instructions. Immunostainings were performed in V-bottom 96 well plates with approximately 1 × 106 cells/well. Upon centrifugation (300 x g, 5 min, 4 °C) cells were resuspended in 30 μl/well FcR block diluted 1:100 in PBS containing 0.5% BSA, 2 mM EDTA and incubated for 15 min at 4 °C. Next, 10 μl/well of antibody master mixes were added. To identify macrophages, cells were incubated with anti-CD11b PerCP-Cy5.5 (diluted 1:1000), anti-F4/80 PE (diluted 1:50) and anti-CD3 Fitc (diluted 1:100). To identify cDCs, cells were incubated with anti-CD11c PECy7 (diluted 1:1000), anti-CD8 eFluor405 (diluted 1:100), anti-CD4 PE (diluted 1:100) anti-CD3 Fitc (diluted 1:100). pDCs were identified upon staining with anti-CD11c PECy7 (diluted 1:1000), anti-PDCA1 PE (diluted 1:100), anti-B220 eFluor450 (diluted 1:100 and anti-CD3 Fitc (diluted 1:100). Gating strategies are shown in Figure S4. Immunostaining of cell surface molecules was performed for 25 min in the dark at 4 °C. Cells were washed twice with PBS containing 0.5 % (w/v) BSA, 2 mM EDTA, resuspended in 150 μl/well 2 % (v/v) PFA in PBS and incubated for 15 min at room temperature. Cells were washed once with PBS containing 0.5 % (w/v) BSA, 2 mM EDTA and once with PBS containing 0.5 % (w/v) BSA, 0.1 % (w/v) Saponin. For intracellular stainings, cells were resuspended in PBS containing 0.5 % (w/v) BSA, 0.1 % (w/v) Saponin and FcR Block (1:100) and incubated for 15 min at room temperature prior to addition of biotinylated anti-TLR7 or biotinylated anti-TLR3 antibody at a final concentration of 1 μg/ml. Cells were incubated for 2 h at room temperature in the dark, washed twice with PBS containing 0.5 % (w/v) BSA, 0.1 % (w/v) Saponin and incubated with streptavidin-APC diluted 1:10 000 in PBS containing 0.5 % (w/v) BSA, 0.1 % (w/v) Saponin for 30 min at room temperature. Cells were washed three times, measured on a LSRII flow cytometer, and analyzed using the FlowJo software. The monoclonal anti-mTLR7 A94B10 (Kanno et al., 2013) and anti-mTLR3 PAT3 (Murakami et al., 2014) antibodies used for flow cytometry, IP and IF experiments were described previously and detect both the cleaved and the full-length receptor (N-terminal epitopes) (Kanno et al., 2013; Murakami et al., 2014).
Electron microscopy - morphology
Unc93b1−/− iMOs reconstituted with UNC93B1-mCitrine WT, H412R and WT-ER were fixed in 2.5% gluteraldehyde, 3% paraformaldehyde with 5% sucrose in 0.1M sodium cacodylate buffer (pH 7.4) for 1 hour, pelleted in a small eppendorf tube, washed three times with 0.1M sodium cacodylate buffer, and post fixed in 1% OsO4 in veronal-acetate buffer for 1 hour. The pellet was stained overnight with 0.5% uranyl acetate in veronal-acetate buffer, dehydrated and embedded in Embed 812 resin. Sections were cut on a Leica Ultracut UCT microtome with a Diatome diamond knife at a thickness setting of 50 nm, stained with uranyl acetate, and lead citrate. The sections were examined using a FEI Tecnai Spirit at 80KV.
Electron microscopy – cryo-EM
Unc93b1−/− iMOs reconstituted with UNC93B1-mCitrine WT, H412R and WT-ER were fixed mildly using PLP (paraformaldehyde/lysine/sodium periodate) fixative for 4 hours. Cells were pelleted and trimmed into small (<1mmsq) blocks. Blocks were infused with a cryo-protectant for at least one hour (PVP/sucrose), mounted onto cryo-pins, and snap frozen in liquid nitrogen. Ultrathin sections were cut at −110 ° C with a Leica UC7 equipped with a FC7 cryo-stage using a glass knife, and immunolabeled for GFP (Invitrogen Catalognr. A11122), stained and embedded using the Tokuyasu technique. The material was examined using a FEI Tecnai Spirit bioTWIN.
Quantification and Statistical Analysis
Statistical parameters including the exact value of n, the definition of center, dispersion and precision measures (mean ± SEM) and statistical significance are reported in the Figures and Figure Legends. Data is judged to be statistically significant when p < 0.05 by two-tailed Student’s t test (if normally distributed); nonparametric data are analyzed using a Mann-Whitney U-test. To compare several groups, a one-way ANOVA is used. In figures, asterisks denote statistical significance (*, p < 0.05; **, p < 0.01; ***, p < 0.001, ****, p < 0.0001). Statistical analysis was performed in GraphPad PRISM 7 (Graph Pad Software Inc.) for imaging quantifications. For proteomics data, statistical methods are described in detail in the MS data analysis section and were performed with R (Team, R Development Core, 2008).
Data And Software Availability
The quantitative proteomics data set of UNC93B1 WT, H412R and WT-ER immunoprecipitates (related to Figure 2A and 2B) is available in Table S1.
Supplementary Material
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal anti-mouse actin | Licor Biosciences | Cat#926–42212 |
| Mouse monoclonal anti-GFP antibody (clone JL-8) | Clontech | Cat#632381 |
| Rabbit polyclonal anti-GFP antibody | Thermo Fisher Scientific | Cat#A-11122 |
| Rabbit polyclonal anti-mouse TLR7 antibody (epitope: 14 amino acid residues near the top of mouse TLR7) | eBioscience | Cat#14–9079–81 |
| IRDye 680RD Donkey anti-mouse IgG (H + L) | Odyssey Li-cor | Cat#926–68072 |
| IRDye 680RD Donkey anti-rabbit IgG (H+L) | Odyssey Li-cor | Cat#926–68073 |
| IRDye 800CW Donkey Anti-mouse | Odyssey Li-cor | Cat#926–32212 |
| IRDye 800CW Donkey Anti-rabbit IgG (H+L) | Odyssey Li-cor | Cat#926–32213 |
| FcR Block | BD Pharmingen | Cat#553142 |
| Rat anti-mouse CD11b PerCP-Cy5.5 (clone M1/70) | eBioscience | Cat#45–0112–82 |
| Rat anti-mouse F4/80 PE (clone BM8) | Biolegend | Cat#123109 |
| Syrian hamster anti-mouse CD3e Fitc (clone eBio500A2) | eBioscience | Cat#11–0033–82 |
| Armenian hamster anti-mouse CD11c PECy7 (clone N418) | eBioscience | Cat#25–0114–81 |
| Rat anti-mouse CD8a eFluor405 (clone 53–6.7) | eBioscience | Cat#48–0081–80 |
| Rat anti-mouse CD4 PE (clone GK1.5) | eBioscience | Cat#12–0041–82 |
| Rat anti-mouse PDCA1 PE (clone eBio129c) | eBioscience | Cat#12–3171–81 |
| Rat anti-mouse B220 eFluor450 (clone RA3–6B2) | eBioscience | Cat#48–0452–82 |
| Mouse monoclonal anti-mouse TLR7 antibody (clone A94B10) | (Kanno et al., 2013) | N/A |
| Biotinylated mouse monoclonal anti-mouse TLR7 antibody (clone A94B10) | (Kanno et al., 2013) | N/A |
| Biotinylated mouse monoclonal anti-mouse TLR3 antibody (clone PAT3) | (Murakami et al., 2014) | N/A |
| Affinity purified anti-mouse UNC93B1 antibody (UNC-C) | (Brinkmann et al., 2007) | N/A |
| F(ab)-Goat anti-mouse antibody Alexa568 | LifeTechnologies | Cat#A-11019 |
| F(ab)-Goat anti-mouse antibody Alexa647 | LifeTechnologies | Cat#A-21237 |
| F(ab)-Goat anti-rabbit antibody Alexa647 | LifeTechnologies | Cat#A-21246 |
| CD19 MicroBeads mouse | Miltenyi | Cat#130–052–201 |
| Biological Samples | ||
| J2 recombinant retrovirus | (Roberson and Walker, 1988) | N/A |
| Heat-inactivated group B streptococcus (GBS) | Dr. G. Teti, (Mancuso et al., 2009; Signorino et al., 2014) | N/A |
| LPS-EB Ultrapure (E.coli 0111:B4) | InvivoGen | Cat#tlrl-3pelps |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Recombinant human M-CSF | R&D Systems | Cat#216-MC-025 |
| Recombinant mouse GM-CSF | Immunotools | Cat#12343123 |
| Recombinant human TNF | R&D Systems | Cat#210-TA-010 |
| Recombinant human IL-1β | R&D Systems | Cat#201-LB-005 |
| Bafilomycin A1 | biomol | Cat#BVT-0252-C100 |
| R848 | InvivoGen | Cat#tlrl-r848 |
| GeneJuice Transfection Reagent | Novagen | Cat#70967–3 |
| Lipofectamine 2000 Transfection Reagent | Thermo Fisher Scientific | Cat#11668–019 |
| Dynabeads Protein G | Thermo Fisher Scientific | Cat#100–04D |
| Protein A Agarose Beads | RepliGen | Cat#CA-PRI-0005 |
| Complete EDTA-free Protease Inhibitor Cocktail | Roche | Cat#4693132001 |
| Liberase TL | Roche | Cat#5401020001 |
| DNase | Roche | Cat#11284932001 |
| Streptavidin Alexa Fluor 647 Conjugate | Thermo Fisher Scientific | Cat#S32357 |
| 100μCi/ml [35S]methionine-[35S]cysteine | Hartmann Analytic | Cat#ARS0110A |
| Critical Commercial Assays | ||
| Human IL-8 ELISA set | BD Biosciences | Cat#555244 |
| Human IL-6 DuoSet | R&D Systems | Cat#DY206 |
| Human TNF DuoSet | R&D Systems | Cat#DY210 |
| Mouse IL-6 DuoSet | R&D Systems | Cat#DY406 |
| EndoHf | NEB | Cat#P0703S |
| PNGaseF | NEB | Cat#P0704S |
| TrapGFP plates | ChromoTek | Cat# GFP-Trap-multiTrap |
| Pierce BCA Protein Assay Kit | Thermo Fisher Scientific | Cat#23225 |
| Deposited Data | ||
| Quantitative proteomics data set of UNC93B1 WT, H412R, and WT-ER immunoprecipitates (related to Figure 2A and 2B) | This paper | Table S1 |
| Experimental Models: Cell Lines | ||
| Immortalized Unc93b1−/− M-CSF-derived BMDMs (Unc93b1−/− iMO) | This paper | N/A |
| Immortalized Unc93b1−/− M-CSF-derived BMDMs (Unc93b1−/− iMO) with mouse UNC93B1-mCitrine WT | This paper | N/A |
| Immortalized Unc93b1−/− M-CSF-derived BMDMs (Unc93b1−/− iMO) with mouse UNC93B1-mCitrine H412R | This paper | N/A |
| Immortalized Unc93b1−/− M-CSF-derived BMDMs (Unc93b1−/− iMO) with mouse UNC93B1-mCitrine WT-ER | This paper | N/A |
| UNC93B1−/− THP-1 monocytes | (Pelka et al., 2014; Schmid-Burgk et al., 2014) | N/A |
| UNC93B1−/− THP-1 monocytes with human UNC93B1-mCitrine WT | This paper | N/A |
| UNC93B1−/− THP-1 monocytes with human UNC93B1-mCitrine WT-ER | This paper | N/A |
| UNC93B1-deficient EBV-immortalized B cells (clone 185) | (Casrouge et al., 2006) | N/A |
| UNC93B1-deficient EBV-immortalized B cells (clone 185) with human UNC93B1-mCitrine WT | This paper | N/A |
| UNC93B1-deficient EBV-immortalized B cells (clone 185) with human UNC93B1-mCitrine WT-ER | This paper | N/A |
| 293XL/hTLR7-HA cell line | Invivogen | RRID:CVCL_Y410 |
| 293XL/hTLR7-HA cell line (RRID:CVCL_Y410) with human UNC93B1-mCitrine WT | This paper | N/A |
| 293XL/hTLR7-HA cell line (RRID:CVCL_Y410) with human UNC93B1-mCitrine WT-ER | This paper | N/A |
| 293XL/hTLR9-HA cell line | Invivogen | RRID:CVCL_Y412 |
| 293XL/hTLR9-HA cell line (RRID:CVCL_Y412) with human UNC93B1-mCitrine WT | This paper | N/A |
| 293XL/hTLR9-HA cell line (RRID:CVCL_Y412) with human UNC93B1-mCitrine WT-ER | This paper | N/A |
| Experimental Models: Organisms/Strains | ||
| Mouse: C57BL/6J | Jackson Laboratory | Stock No.: 00664 |
| Mouse: Unc93b1−/− (KOMP ID VG10049) | Regeneron Pharmaceuticals, Inc. (Valenzuela et al., 2003) | KOMP ID VG10049 |
| Mouse: Tlr3−/− | In house breeding | N/A |
| Mouse: Tlr7−/− | In house breeding | N/A |
| Oligonucleotides | ||
| TLR7-stimulatory RNA (5'-ACUG1CG1AG1CUU-X-UUCG1AG1CG1UCA-5', G1 is 7-deazaguanosine, X is 1,2,3-propanetriol) | Idera Pharmaceuticals | N/A |
| Biotinylated TLR7-stimulatory RNA (5'-ACUG1CG1AG1CUU-X-UUCG1AG1CG1UCA-5', G1 is 7-deazaguanosine, X is 1,2,3-propanetriol) | Idera Pharmaceuticals | N/A |
| TLR13-stimulatory RNA (ORN Sa19) | InvivoGen | Cat#tlrl-orn19 |
| CpG1826 PTO (TCCATGACGTTCCTGACGTT) | Metabion | N/A |
| CpG1826 PTO (TCCATGACGTTCCTGACGTT) AlexaFlour 647 | Metabion | N/A |
| CpG2006 PTO (TCGTCGTTTTGTCGTTTTGTCGTT) | Metabion | N/A |
| Recombinant DNA | ||
| Human UNC93B1-mCitrine WT in pR backbone | This paper | N/A |
| Human UNC93B1-mCitrine WT-ER in pR backbone | This paper | N/A |
| Mouse UNC93B1-mCitrine WT in pR backbone | This paper | N/A |
| Mouse UNC93B1-mCitrine H412R in pR backbone | This paper | N/A |
| Mouse UNC93B1-mCitrine WT-ER in pR backbone | This paper | N/A |
| mCherry-KDEL in pLenti III CMV backbone | This paper | N/A |
| Gag-pol packaging plasmids for retroviral transduction | This paper | N/A |
| VSV-G packaging plasmids for retroviral transduction | This paper | N/A |
| Software and Algorithms | ||
| Cell profiler (version 6c2d896) | (Carpenter et al., 2006) | http://cellprofiler.org/releases/ |
| ImageJ (v1.44o) | Rasband, W.S., ImageJ, U. S. NIH | https://imagej.net/Downloads |
| Image Studio 3.1.4 | LI-COR | In house license |
| Xcalibur software | Thermo Scientific | In house license |
| MaxQuant (v1.4.3.10) | (Cox and Mann, 2008) | http://www.coxdocs.org/doku.php?id=maxquant:common:download_and_installation |
| Andromeda search engine | (Cox et al., 2011) | http://www.coxdocs.org/doku.php?id=maxquant:andromeda |
| PERSEUS (part of MaxQuant and the R framework) | (Team, R Development Core, 2008) | http://www.coxdocs.org/doku.php?id=perseus:common:download_and_installation |
| FACS Diva | BD Biosciences | In house license |
| Flow Jo (v10.0.8) | TreeStar | In house license |
| GraphPad Prism7 | GraphPad Software | In house license |
Acknowledgements
Heat-killed GBS were kindly provided by Dr. G. Teti, the TLR7-stimulatory RNA ligand by Idera Pharmaceuticals. EBV-immortalized UNC93B1-deficient B cells were kindly provided by Dr. Jean-Laurent Casanova. The Unc93b1−/− mouse strain used for this research project was created from ES cell clone 10049A-G9, obtained from the NCRR-NIH supported KOMP Repository (www.komp.org) and generated by Regeneron Pharmaceuticals, Inc. This work was supported by NIH grant 1R01AR066808–01A1 (to A.M.R. and E.L.), the Excellence Cluster ImmunoSensation (to K.Pelka and E.L.), German Research Foundation Grants SFB670 (to A.H. and E.L.), SFB900 and BR 3432/3–1 (to M.M.B.), and the Initiative and Networking Fund of the Helmholtz Association (to M.M.B.). We thank G. Engels for technical support and Dr. F. Adolf, Dr. F. Wieland, Dr. Terje Espevik and the whole Latz Lab for helpful discussions.
Footnotes
Declaration of interests
E.L. is a co-founder and consultant and D.B. and R.S. are employees of IFM Therapeutics.
References
- Blasius AL, and Beutler B (2010). Intracellular toll-like receptors. Immunity 32, 305–315. [DOI] [PubMed] [Google Scholar]
- Brinkmann MM, Spooner E, Hoebe K, Beutler B, Ploegh HL, and Kim YM (2007). The interaction between the ER membrane protein UNC93B and TLR3, 7, and 9 is crucial for TLR signaling. J Cell Biol 177, 265–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter AE, Jones TR, Lamprecht MR, Clarke C, Kang IH, Friman O, Guertin DA, Chang JH, Lindquist RA, Moffat J, et al. (2006). CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol 7, R100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casrouge A, Zhang SY, Eidenschenk C, Jouanguy E, Puel A, Yang K, Alcais A, Picard C, Mahfoufi N, Nicolas N, et al. (2006). Herpes simplex virus encephalitis in human UNC-93B deficiency. Science 314, 308–312. [DOI] [PubMed] [Google Scholar]
- Cox J, Hein MY, Luber CA, Paron I, Nagaraj N, and Mann M (2014). Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol Cell Proteomics 13, 2513–2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J, and Mann M (2008). MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol 26, 1367–1372. [DOI] [PubMed] [Google Scholar]
- Cox J, Neuhauser N, Michalski A, Scheltema RA, Olsen JV, and Mann M (2011). Andromeda: a peptide search engine integrated into the MaxQuant environment. J Proteome Res 10, 1794–1805. [DOI] [PubMed] [Google Scholar]
- Davies JC (2015). The future of CFTR modulating therapies for cystic fibrosis. Curr Opin Pulm Med 21, 579–584. [DOI] [PubMed] [Google Scholar]
- Fukui R, Saitoh S, Kanno A, Onji M, Shibata T, Ito A, Onji M, Matsumoto M, Akira S, Yoshida N, and Miyake K (2011). Unc93B1 restricts systemic lethal inflammation by orchestrating Toll-like receptor 7 and 9 trafficking. Immunity 35, 69–81. [DOI] [PubMed] [Google Scholar]
- Fukui R, Saitoh S, Matsumoto F, Kozuka-Hata H, Oyama M, Tabeta K, Beutler B, and Miyake K (2009). Unc93B1 biases Toll-like receptor responses to nucleic acid in dendritic cells toward DNA- but against RNA-sensing. J Exp Med 206, 1339–1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Zhang J, Zhang X, Wang Y, Yu H, Yin X, Li J, Du P, Plumas J, Chaperot L, et al. (2015). SCARB2/LIMP-2 Regulates IFN Production of Plasmacytoid Dendritic Cells by Mediating Endosomal Translocation of TLR9 and Nuclear Translocation of IRF7. J Immunol 194, 4737–4749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helenius A, and Aebi M (2004). Roles of N-linked glycans in the endoplasmic reticulum. Annu Rev Biochem 73, 1019–1049. [DOI] [PubMed] [Google Scholar]
- Hubner NC, Bird AW, Cox J, Splettstoesser B, Bandilla P, Poser I, Hyman A, and Mann M (2010). Quantitative proteomics combined with BAC TransgeneOmics reveals in vivo protein interactions. J Cell Biol 189, 739–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh JW, Shibata T, Hwang M, Kwon EH, Jang MS, Fukui R, Kanno A, Jung DJ, Jang MH, Miyake K, and Kim YM (2014). UNC93B1 is essential for the plasma membrane localization and signaling of Toll-like receptor 5. Proc Natl Acad Sci U S A 111, 7072–7077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo Y, Hartman IZ, and DeBose-Boyd RA (2013). Ancient ubiquitous protein-1 mediates sterol-induced ubiquitination of 3-hydroxy-3-methylglutaryl CoA reductase in lipid droplet-associated endoplasmic reticulum membranes. Mol Biol Cell 24, 169–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanno A, Tanimura N, Ishizaki M, Ohko K, Motoi Y, Onji M, Fukui R, Shimozato T, Yamamoto K, Shibata T, et al. (2015). Targeting cell surface TLR7 for therapeutic intervention in autoimmune diseases. Nat Commun 6, 6119. [DOI] [PubMed] [Google Scholar]
- Kanno A, Yamamoto C, Onji M, Fukui R, Saitoh S, Motoi Y, Shibata T, Matsumoto F, Muta T, and Miyake K (2013). Essential role for Toll-like receptor 7 (TLR7)-unique cysteines in an intramolecular disulfide bond, proteolytic cleavage and RNA sensing. Int Immunol 25, 413–422. [DOI] [PubMed] [Google Scholar]
- Kim YM, Brinkmann MM, Paquet ME, and Ploegh HL (2008). UNC93B1 delivers nucleotide-sensing toll-like receptors to endolysosomes. Nature 452, 234–238. [DOI] [PubMed] [Google Scholar]
- Lafaille FG, Pessach IM, Zhang SY, Ciancanelli MJ, Herman M, Abhyankar A, Ying SW, Keros S, Goldstein PA, Mostoslavsky G, et al. (2012). Impaired intrinsic immunity to HSV-1 in human iPSC-derived TLR3-deficient CNS cells. Nature 491, 769–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee BL, Moon JE, Shu JH, Yuan L, Newman ZR, Schekman R, and Barton GM (2013). UNC93B1 mediates differential trafficking of endosomal TLRs. Elife 2, e00291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippincott-Schwartz J, Roberts TH, and Hirschberg K (2000). Secretory protein trafficking and organelle dynamics in living cells. Annu Rev Cell Dev Biol 16, 557–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancuso G, Gambuzza M, Midiri A, Biondo C, Papasergi S, Akira S, Teti G, and Beninati C (2009). Bacterial recognition by TLR7 in the lysosomes of conventional dendritic cells. Nat Immunol 10, 587–594. [DOI] [PubMed] [Google Scholar]
- Meunier E, and Broz P (2015). Quantification of Cytosolic vs. Vacuolar Salmonella in Primary Macrophages by Differential Permeabilization. J Vis Exp, e52960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami Y, Fukui R, Motoi Y, Kanno A, Shibata T, Tanimura N, Saitoh S, and Miyake K (2014). Roles of the cleaved N-terminal TLR3 fragment and cell surface TLR3 in double-stranded RNA sensing. J Immunol 193, 5208–5217. [DOI] [PubMed] [Google Scholar]
- Nakano S, Morimoto S, Suzuki S, Watanabe T, Amano H, and Takasaki Y (2010). Up-regulation of the endoplasmic reticulum transmembrane protein UNC93B in the B cells of patients with active systemic lupus erythematosus. Rheumatology (Oxford) 49, 876–881. [DOI] [PubMed] [Google Scholar]
- Onji M, Kanno A, Saitoh S, Fukui R, Motoi Y, Shibata T, Matsumoto F, Lamichhane A, Sato S, Kiyono H, et al. (2013). An essential role for the N-terminal fragment of Toll-like receptor 9 in DNA sensing. Nat Commun 4, 1949. [DOI] [PubMed] [Google Scholar]
- Panchanathan R, Liu H, and Choubey D (2013). Expression of murine Unc93b1 is up-regulated by interferon and estrogen signaling: implications for sex bias in the development of autoimmunity. Int Immunol 25, 521–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelka K, Phulphagar K, Zimmermann J, Stahl R, Schmid-Burgk JL, Schmidt T, Spille JH, Labzin LI, Agrawal S, Kandimalla ER, et al. (2014). Cutting edge: the UNC93B1 tyrosine-based motif regulates trafficking and TLR responses via separate mechanisms. J Immunol 193, 3257–3261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohar J, Pirher N, Bencina M, Mancek-Keber M, and Jerala R (2013). The role of UNC93B1 protein in surface localization of TLR3 receptor and in cell priming to nucleic acid agonists. J Biol Chem 288, 442–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi R, Singh D, and Kao CC (2012). Proteolytic processing regulates Toll-like receptor 3 stability and endosomal localization. J Biol Chem 287, 32617–32629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappsilber J, Mann M, and Ishihama Y (2007). Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat Protoc 2, 1896–1906. [DOI] [PubMed] [Google Scholar]
- Roberson SM, and Walker WS (1988). Immortalization of cloned mouse splenic macrophages with a retrovirus containing the v-raf/mil and v-myc oncogenes. Cell Immunol 116, 341–351. [DOI] [PubMed] [Google Scholar]
- Sasai M, Linehan MM, and Iwasaki A (2010). Bifurcation of Toll-like receptor 9 signaling by adaptor protein 3. Science 329, 1530–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheltema RA, Hauschild JP, Lange O, Hornburg D, Denisov E, Damoc E, Kuehn A, Makarov A, and Mann M (2014). The Q Exactive HF, a Benchtop mass spectrometer with a pre-filter, high-performance quadrupole and an ultra-high-field Orbitrap analyzer. Mol Cell Proteomics 13, 3698–3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid-Burgk JL, Schmidt T, Gaidt MM, Pelka K, Latz E, Ebert TS, and Hornung V (2014). OutKnocker: a web tool for rapid and simple genotyping of designer nuclease edited cell lines. Genome Res 24, 1719–1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schutze MP, Peterson PA, and Jackson MR (1994). An N-terminal double-arginine motif maintains type II membrane proteins in the endoplasmic reticulum. EMBO J 13, 1696–1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Signorino G, Mohammadi N, Patane F, Buscetta M, Venza M, Venza I, Mancuso G, Midiri A, Alexopoulou L, Teti G, et al. (2014). Role of Toll-like receptor 13 in innate immune recognition of group B streptococci. Infect Immun 82, 5013–5022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabeta K, Hoebe K, Janssen EM, Du X, Georgel P, Crozat K, Mudd S, Mann N, Sovath S, Goode J, et al. (2006). The Unc93b1 mutation 3d disrupts exogenous antigen presentation and signaling via Toll-like receptors 3, 7 and 9. Nat Immunol 7, 156–164. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Shibata T, Akashi-Takamura S, Kiyokawa T, Wakabayashi Y, Tanimura N, Kobayashi T, Matsumoto F, Fukui R, Kouro T, et al. (2007). A protein associated with Toll-like receptor (TLR) 4 (PRAT4A) is required for TLR-dependent immune responses. J Exp Med 204, 2963–2976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, and Chu G (2001). Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A 98, 5116–5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenzuela DM, Murphy AJ, Frendewey D, Gale NW, Economides AN, Auerbach W, Poueymirou WT, Adams NC, Rojas J, Yasenchak J, et al. (2003). High-throughput engineering of the mouse genome coupled with high-resolution expression analysis. Nat Biotechnol 21, 652–659. [DOI] [PubMed] [Google Scholar]
- Yang Y, Liu B, Dai J, Srivastava PK, Zammit DJ, Lefrancois L, and Li Z (2007). Heat shock protein gp96 is a master chaperone for toll-like receptors and is important in the innate function of macrophages. Immunity 26, 215–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The quantitative proteomics data set of UNC93B1 WT, H412R and WT-ER immunoprecipitates (related to Figure 2A and 2B) is available in Table S1.