SUMMARY
The diverse morphologies of primary cilia are tightly regulated as a function of cell type and cellular state. CCRK and MAK-related kinases have been implicated in ciliary length control in multiple species, although the underlying mechanisms are not fully understood. Here we show that in C. elegans, DYF-18/CCRK and DYF-5/MAK act in a cascade to generate the highly arborized cilia morphologies of the AWA olfactory neurons. Loss of kinase function results in dramatically elongated AWA cilia that lack branches. IFT motor protein localization but not velocities in AWA cilia are altered upon loss of dyf-18. We instead find that axonemal microtubules are decorated by the EBP-2 end-binding protein along their lengths, and that the tubulin load is increased, and tubulin turnover is reduced, in AWA cilia of dyf-18 mutants. Moreover, we show that predicted microtubule-destabilizing mutations in two tubulin subunits, as well as mutations in IFT proteins predicted to disrupt tubulin transport, restore cilia branching and suppress AWA cilia elongation in dyf-18 mutants. Loss of dyf-18 is also sufficient to elongate the truncated rod-like unbranched cilia of the ASH nociceptive neurons in animals carrying a microtubule-destabilizing mutation in a tubulin subunit. We suggest that CCRK/MAK activity tunes cilia length and shape in part via modulation of axonemal microtubule stability, suggesting that similar mechanisms may underlie their roles in ciliary length control in other cell types.
Keywords: CCRK, MAK, microtubules, cilia, AWA, C. elegans
eTOC Blurb
Cilia are microtubule-based organelles that exhibit cell-specific morphologies. CCRK and MAK-related kinases restrict cilia length in multiple organisms. Maurya et al. show that a CCRK and a MAK kinase act in a cascade to control cilia shape and structure by regulating axonemal microtubule dynamics in multiple sensory neuron types in C. elegans.
INTRODUCTION
Primary cilia are ubiquitous microtubule-based sensory organelles that house signaling molecules, transduce environmental stimuli, and may themselves transmit signals via the release of vesicles [1, 2]. Although the majority of cilia exhibit simple rod-like structures, cilia can also exhibit specialized morphologies [3-5]. A critical aspect of cilium morphology is its length which is regulated as a function of cell type and condition [6, 7]. Since cilium architecture dictates both the concentration and organization of signaling molecules within it, length regulation presumably contributes to precise modulation of sensory signaling, and is an active area of research.
Most cilia are formed by the conserved process of intraflagellar transport (IFT) [8]. IFT is mediated by anterograde kinesin 2 and retrograde dynein motors that traffic cargo such as axonemal tubulins via interaction with the large IFT-A and IFT-B protein complexes [9]. Tubulin is incorporated into the growing microtubule (MT) plus ends at the ciliary tip to elongate the axoneme. Experimental and theoretical work has led to multiple models for IFT-dependent mechanisms of ciliary length control. These include cilia length-dependent modulation of cargo-loading onto IFT motors as well as rates of entry (‘injection’) of motors into the cilium [10-15]. However, how these processes are regulated to ensure the generation of cilia with cell type-specific lengths is unclear.
Genetic and biochemical experiments in multiple organisms have identified several conserved kinases that regulate cilia length [16-19]. In particular, loss of function of the CCRK/LF2 and MAK/MOK/ICK/LF4 family of RCK kinases elongates cilia to varying degrees in Chlamydomonas, C. elegans, and mammalian cells [17, 19-27]. Disrupted kinase function results in increased flagella assembly rates and abolishes length-dependent modulation of tubulin transport in Chlamydomonas flagella [10, 13, 28]. How these kinases regulate IFT motors and cargoes such as tubulin to modulate cilia length and morphology remains an open question.
The cilia of C. elegans sensory neurons provide an excellent system in which to study the mechanisms that regulate the generation and maintenance of cell type-specific cilia morphologies. Eight pairs of sensory neurons in the bilateral head amphid organs of the C. elegans hermaphrodite contain rod-like cilia (‘channel cilia’) with middle and distal segments comprised of doublet (dMTs) and singlet MTs (sMTs), respectively [4, 5]. In contrast, the AWA, AWB and AWC amphid sensory neurons exhibit specialized cilia morphologies (‘wing cilia’) [4, 5, 29]. The cilia of the AWA neurons are highly arborized with a proximal axonemal stalk that splits distally into multiple short and progressively thinner ciliary branches containing few or no MTs [5]. The unique cilia of the AWB and AWC neurons are built in part via the differential deployment of IFT motors and tubulin isotypes [30-32], but additional mechanisms that generate and maintain these specialized architectures remain to be fully described.
The DYF-18/CCRK and DYF-5/MAK kinases also regulate cilia length in C. elegans. The middle segments of channel cilia in C. elegans are built by the cooperative and redundant functions of the kinesin-II and OSM-3 kinesin 2 motors, whereas the distal segment is built by OSM-3 alone [33, 34]. In dyf-18 and dyf-5 mutants, the channel cilia are slightly but significantly longer, kinesin-II inappropriately enters the distal segments, and OSM-3 speed is reduced [17, 26, 27]. DYF-5 and DYF-18 have been suggested to regulate the handover between kinesin-II and OSM-3, and regulate the docking and undocking of kinesin motors and IFT particles [17, 26]. Given the distinct morphologies and IFT mechanisms in different cilia types, it is unclear whether molecules such as CCRK and MAK also build specialized cilia, and whether they function via similar or cell-specific mechanisms.
Here we show that the DYF-18/CCRK acts via DYF-5/MAK to regulate AWA cilia morphology in part via modulation of axonemal MT stability. Loss of function of either kinase abolishes branching, and severely elongates AWA cilia. The axonemes of elongated cilia in these animals are decorated by the EBP-2 end-binding protein along their lengths instead of at their growing distal ends, and exhibit decreased tubulin turnover, suggesting that axonemal MTs are stabilized in the absence of DYF-18/DYF-5. Correspondingly, mutations in tubulin predicted to destabilize axonemal MTs, as well as mutations in IFT subunits that bind and transport tubulin, partially suppress elongation, and restore AWA ciliary branching in dyf-18 mutants. We find that the relative distribution, but not velocities, of IFT motor proteins in AWA is altered in dyf-18 mutants. Mutations in dyf-18 also partly suppress the truncated channel cilia phenotype of animals carrying MT destabilizing mutations in tubulin. Our results suggest that CCRK/MAK kinases shape specialized cilia morphology in part via regulation of axonemal MT stability, which in turn may modulate IFT properties.
RESULTS
AWA cilia are unbranched and severely elongated in dyf-18 CCRK/LF2 mutants
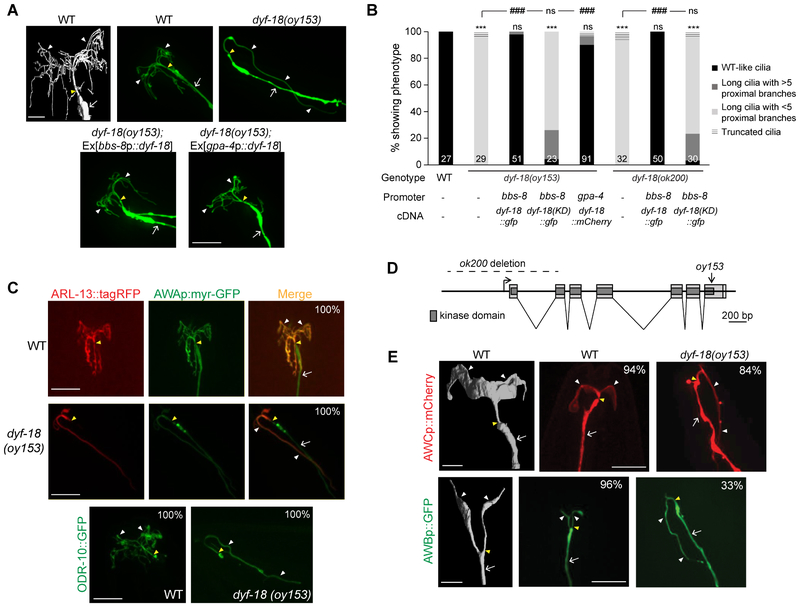
We isolated the recessive oy153 allele in a forward genetic screen for mutants exhibiting defects in the highly branched morphology of AWA cilia (Figure 1A; see STAR Methods). Few if any branched cilia-like structures were present in oy153 mutants (Figure 1A, 1B). Instead, the AWA dendrites in these animals projected a single long thin process anteriorly followed by posterior extension (Figure 1A), frequently terminating near neuronal soma, a distance of >60 μm from the worm nose. The cilia-localized GTPase ARL-13 and olfactory receptor ODR-10 proteins were specifically present in the elongated unbranched structures, suggesting that these processes are bona fide cilia (Figure 1C). Localization of the GASR-8 basal body and the MKS-5 transition zone proteins was unaltered in dyf-18 mutants (Figure S1A).
Figure 1. AWA olfactory neuron cilia are unbranched and elongated in dyf-18 CCRK mutants.
A) (Top left) 3D reconstruction model of the cilia of an AWA neuron (adapted from [5]). Scale bar: 2 μm. (Top right and bottom) Representative images of AWA cilia in the indicated genetic backgrounds. Cilia were visualized via expression of gpa-4Δ6p::myr-gfp. The cilia base and cilia are indicated by yellow and white arrowheads, respectively; the dendrite is marked by an arrow. Anterior is to the top left in all images. Scale bar: 10 μm.
B) Percentage of adult hermaphrodites of the indicated genotypes exhibiting AWA cilia phenotypes. bbs-8 and gpa-4Δ6 regulatory sequences drive expression in all ciliated neurons and in AWA alone, respectively. KD: kinase-dead. Cilia were visualized using gpa-4Δ6p::myr-gfp or gpa-4Δ6p::myr-mCherry. Numbers in each bar indicate the number of examined neurons in 2-3 independent experiments. For transgenic strains, combined data from two independent lines are shown. *** and ### indicate different from wild-type and corresponding mutant, respectively, at P<0.001; ns – not significant (Wilcoxon rank-sum test).
C) Representative images of localization of ARL-13::tagRFP (top) and ODR-10::GFP localization (bottom) in wild-type and dyf-18 animals. Cilia were visualized via expression of gpa-4Δ6p::myr-gfp in images at the top. The cilia base and cilia are indicated by yellow and white arrowheads, respectively; the dendrite is marked by an arrow (the dendrite is not visible in the bottom panels). Numbers at top right indicate the percentage of neurons exhibiting the shown phenotypes; n≥30 neurons each in 3 independent experiments. Anterior is at top or at top left in all images. Scale bars: 10 μm.
D) Gene structure of dyf-18. Filled light and dark gray boxes indicate exons and the kinase domain, respectively. The extent of the deletion in ok200 and the location of the oy153 mutation are indicated.
E) (Left panels) 3D reconstruction models of AWC (top) and AWB (bottom) cilia (adapted from [5]). Scale bar: 2 μm. (Right panels) Representative images of AWC and AWB cilia in the indicated genetic backgrounds. AWC and AWB cilia were visualized via expression of odr-1p::dsRed and str-1p::gfp, respectively. Numbers at top right indicate the percentage of neurons exhibiting the shown phenotypes; n≥30 neurons each in 3 independent experiments. The cilia base and cilia are indicated by yellow and white arrowheads, respectively; the dendrite is marked by an arrow. Anterior is at top in all images. Scale bars: 10 μm.
Also see Figure S1.
Genetic mapping followed by whole genome sequencing indicated the presence of a nonsense mutation in the dyf-18 CCRK/LF2 gene in oy153 animals; this mutation is predicted to delete the C-terminal 46 amino acids of DYF-18, including 12 amino acids of the predicted kinase domain (Figure 1D). Animals carrying the previously characterized dyf-18(ok200) predicted null allele [27] (Figure 1D) also exhibited AWA cilia that were unbranched and elongated (Figure 1B). The AWA cilia phenotypes of dyf-18(oy153) and/or dyf-18(ok200) were rescued upon expression of wild-type dyf-18 sequences under the ciliated neuron-specific bbs-8, as well as AWA-specific gpa-4Δ6 promoter sequences [35] (Figure 1A, 1B). No rescue was observed upon expression of the dyf-18(oy153) cDNA (Figure S1B), or a cDNA predicted to encode a kinase-dead (KD) DYF-18(K37R) protein [19] (Figure 1B). We conclude that oy153 is an allele of dyf-18, and that DYF-18/CCRK acts cell-autonomously in AWA to regulate cilia branching and length.
The proximal regions of the AWA, AWB and AWC cilia are within a channel formed proximally and distally by the processes of the amphid sheath (AMsh) and socket (AMso) glial cells, respectively, while their distal regions are fully embedded within the AMsh processes [4, 5, 29]. In contrast, both the proximal and distal segments of channel cilia are contained in the glia-formed channel [4, 5, 29]. Unlike the extreme AWA cilia elongation in dyf-18 mutants, loss of DYF-18 results in relatively modest effects on the length of channel cilia [26, 27] (see Discussion; also see Figure 7A, 7B). However, similar to the effects of dyf-18 mutations on AWA cilia, AWB and AWC neuron cilia were also markedly elongated upon loss of DYF-18 (Figure 1E). Expression of the AMsh marker F16F9.3p::mCherry [36] indicated that the distal regions of the elongated AWA cilia in dyf-18 mutants were associated with AMsh processes which also exhibited aberrant morphology (Figure S1C). The altered AMsh morphology phenotype was rescued upon expression of wild-type dyf-18 sequences in ciliated sensory neurons but not in AMsh cells (Figure S1C), indicating that neuronal cues act cell non-autonomously to modulate glial morphology in dyf-18 mutants. Together, these results indicate that DYF-18 plays a particularly critical role in regulating the morphology of specialized wing cilia in C. elegans.
Figure 7. Mutations in dyf-18 partially suppress the truncated channel cilia phenotypes of animals carrying MT destabilizing mutations in a tubulin isotype.
A, C, D) Representative images of ASH cilia in animals of the indicated genotypes expressing the shown fusion proteins. Expression was driven under the sra-6 promoter. The cilium base and cilia are indicated by yellow and white arrowheads, respectively. MS- middle segment; DS – distal segment. Anterior is at top or top left in all images. Scale bar: 5 μm. Animals were grown at 15°C.
B) Quantification of ASH cilia lengths in animals of the indicated genotypes grown at 15°C. Each dot represents a measurement from a single cilium. Horizontal line indicates the median, lower and upper bounds of box indicate the 25th and 75th percentiles, respectively, lower and upper extents of bars indicate 5th and 95th percentiles, respectively. *** indicates different between the indicated values at P<0.001 (one-way ANOVA with Bonferroni posthoc corrections for multiple comparisons).
E) Histograms of OSM-3::GFP anterograde velocities in the middle segments of ASH cilia in animals of the indicated genotypes. Fusion proteins were expressed under the sra-6 promoter. Animals were grown at 15°C.
Also see Figure S5 and Table S3.
DYF-18 potentiates the activity of the DYF-5 MAK kinase to regulate AWA cilia morphology
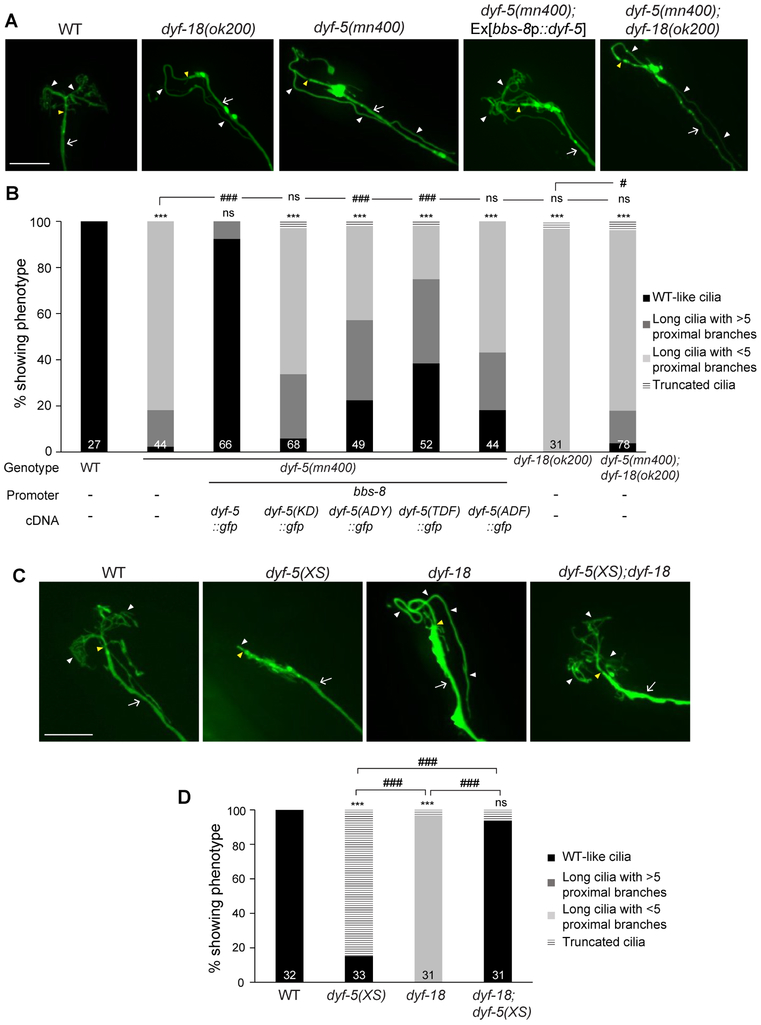
CCRK can directly phosphorylate MAK in vitro and in HEK293 cells, and these kinases act in the same pathway to control glioblastoma proliferation in a cilium-dependent manner [37, 38]. In C. elegans, DYF-18 and the DYF-5 LF4/MAK/MOK/ICK homolog have been proposed to act in the same pathway to regulate channel cilia length [26]. AWA cilia were also unbranched and severely elongated in dyf-5(mn400) putative null mutants (Figure 2A, 2B). Moreover, as in dyf-18 mutants, the morphology of AMsh processes was altered in dyf-5 mutants (Figure S1C). dyf-5; dyf-18 double mutants exhibited AWA cilia phenotypes similar to those of dyf-5 mutants alone (Figure 2A, 2B). The AWA cilia phenotype of dyf-5 mutants was rescued upon expression of sequences encoding wild-type but not kinase-dead DYF-5(K40R) under the bbs-8 promoter (Figure 2B).
Figure 2. DYF-18 acts via DYF-5 to regulate AWA cilia morphology.
A, C) Representative images of AWA cilia in the indicated genetic backgrounds. Cilia were visualized via expression of gpa-4Δ6p::myr-gfp. The cilia base and cilia are indicated by yellow and white arrowheads, respectively; the dendrite is marked by an arrow. Anterior is to the top or at top left in all images. Scale bars: 10 μm.
B, D) Percentage of adult hermaphrodites of the indicated genotypes exhibiting AWA cilia phenotypes. bbs-8 regulatory sequences drive expression in all ciliated neurons. Cilia were visualized using gpa-4Δ6p::myr-mCherry. KD: kinase-dead; dyf-5(XS): overexpressed dyf-5 [17]. For transgenic strains, combined data from two independent lines are shown. Numbers in each bar indicate the number of examined neurons in 2-3 independent experiments. *** indicates different from wild-type at P<0.001; # and ### indicate different from indicated values at P<0.05 and P<0.001, respectively; ns – not significant (Wilcoxon rank-sum test).
Also see Figure S1.
Biochemical experiments have shown that while autophosphorylation at the Y159 residue of the conserved TDY motif in the activation T-loop is required for basal activity of the ICK kinase, phosphorylation of T157 in this motif by upstream kinases is required for maximal kinase activity [39]. Consistent with this model, mutating T164 (corresponding to T157) and/or Y166 (corresponding to Y159) in DYF-5 (TDY→ADY, TDY→TDF, and TDY→ADF) in DYF-5 reduced the ability of the protein to rescue the AWA cilia phenotype of dyf-5 mutants (Figure 2B).
Overexpression of DYF-5 [dyf-5(XS)] truncates channel cilia [17]. AWA cilia were also severely truncated and unbranched in animals overexpressing DYF-5 (Figure 2C, 2D). If DYF-18-mediated phosphorylation is required for maximal activation of DYF-5, we would predict that the truncated AWA cilia phenotype observed in dyf-5(XS) animals would be partly suppressed upon loss of dyf-18. Indeed, we found that the AWA cilia morphologies of dyf-5(XS); dyf-18(ok200) double mutants resembled those of wild-type animals (Figure 2C, 2D). Defects in ciliary/dendritic processes result in a failure of a subset of amphid sensory neurons including AWB to uptake lipophilic dyes (Dyf: dyefilling defect) [4]. Although dye-filling was defective upon both loss of dyf-18 and overexpression of DYF-5, dye-filling was fully restored in dyf-5(XS); dyf-18(ok200) double mutants (Figure S1D). We infer that DYF-18-mediated phosphorylation is required for maximal activation of overexpressed DYF-5 resulting in cilia truncation. However, in the absence of DYF-18, basal activity of overexpressed DYF-5 is sufficient to promote branching and suppress elongation.
DYF-5 localizes to AWA cilia in a DYF-18-dependent manner
Both DYF-18 and DYF-5 have previously been shown to localize diffusely throughout the cell and are weakly enriched at the cilia base in ciliated sensory neurons in C. elegans [17, 27], although a recent report indicated that both proteins may instead be enriched in distal segments of channel cilia [26]. In wild-type AWA neurons, a GFP-tagged DYF-18 fusion protein was present throughout the cell including in dendrites and in cilia proximal stalks (Figure 3A). The localization pattern of this rescuing fusion protein (Figure 1B) was similar in a dyf-18 mutant background (Figure 3A). In both cases, weak to no localization was observed in the distal ciliary branches or at the cilia tips. The localization pattern was unaltered upon loss of dyf-5 (Figure 3A). However, while expression of the kinase-dead DYF-18(K37R)::GFP protein was dramatically reduced in wild-type animals, this protein was enriched at the distal segments of the elongated AWA cilia in dyf-18 mutants (Figure 3A), suggesting that the stability of the kinase-dead protein is regulated directly or indirectly by wild-type DYF-18. These results indicate that ciliary localization of DYF-18 in AWA is regulated by its enzymatic activity such that while the wild-type protein is present in the proximal ciliary stalk, loss of enzymatic activity results in enrichment of the protein in the distal ciliary segments.
Figure 3. DYF-18 regulates ciliary localization of DYF-5.
A-B) Representative images and summary cartoons of protein localization in AWA cilia in the indicated genetic backgrounds. Cilia were visualized via expression of gpa-4Δ6p::myr-mCherry. The cilia base and cilia are indicated by yellow and white arrowheads, respectively; the dendrite is marked by an arrow. Alleles used were dyf-18(ok200) and dyf-5(mn400). Anterior is to the top or at top left in all images. KD: kinase-dead. n≥25 neurons; 2-3 independent experiments. Scale bar: 10 μm.
In contrast to the localization pattern of DYF-18::GFP, a GFP::DYF-5 fusion protein was specifically enriched in the proximal stalks of AWA cilia in wild-type animals (Figure 3B). This rescuing fusion protein exhibited a similar localization in dyf-5 mutants (Figure 3B). However, in dyf-18 mutants, GFP::DYF-5 was mislocalized throughout the elongated AWA cilia with strong enrichment in distal regions (Figure 3B). Loss of DYF-5 kinase activity resulted in weak to no expression of this protein in either a wild-type or dyf-5 mutant background (Figure 3B). We conclude that ciliary localization of DYF-5 is regulated by DYF-18, and that localization and/or stability of both kinases are dependent on their enzymatic functions. In particular, both kinases are enriched at the distal ciliary segments in the absence of DYF-18 kinase activity.
DYF-18 regulates IFT motor protein localization but not velocity in AWA cilia
DYF-5 and DYF-18 have been suggested to regulate the handover between the kinesin-II and OSM-3 motors in channel cilia in C. elegans, resulting in inappropriate entry of kinesin-II into the distal segments in animals mutant for these kinases [17, 26]. These kinases have also been proposed to regulate the activity and cargo-loading properties of OSM-3 [17, 26], and regulate IFT protein turnover/recycling [17, 23, 25, 26]. The ultrastructure of AWA cilia is distinct from those of channel cilia with no obvious middle and distal segments [4, 5], raising the question of whether DYF-18/DYF-5 regulate IFT motors and proteins similarly in AWA cilia.
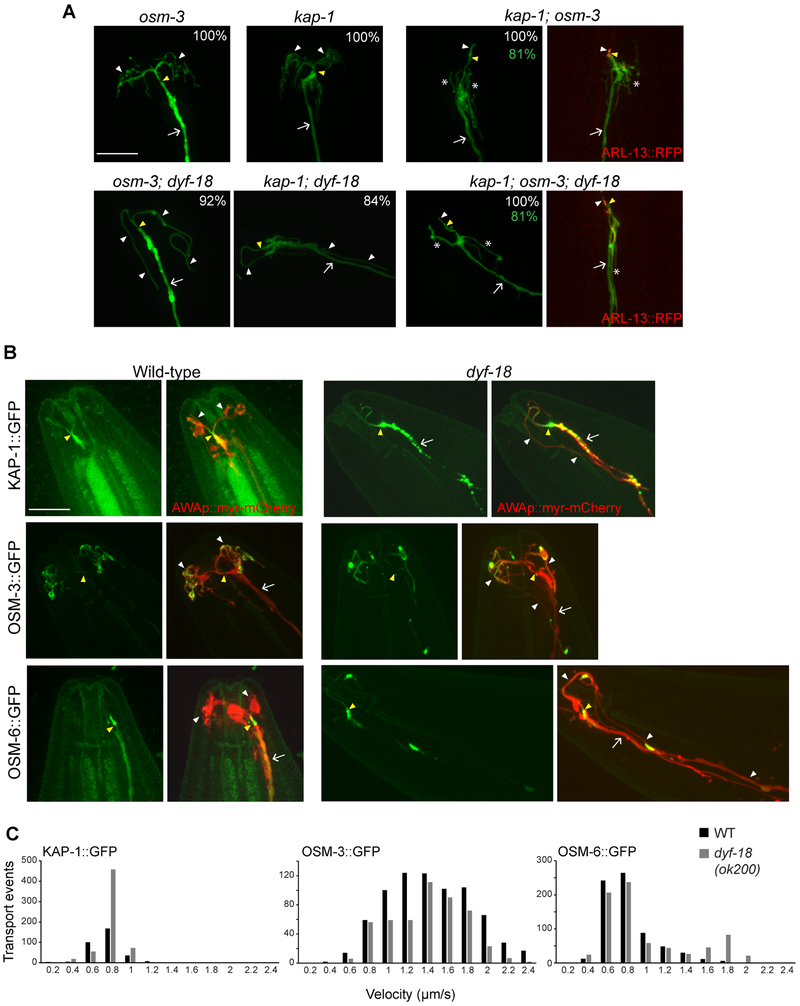
Branched or elongated unbranched AWA cilia in wild-type or dyf-18 mutants, respectively, were largely unaltered in either osm-3(p802) or kap-1(ok676) single mutants, but were severely truncated in kap-1; osm-3 double mutants (Figure 4A). In kap-1; osm-3 double mutant animals, we also observed multiple processes emanating from the distal dendritic ends of AWA (Figure 4A). These branches were qualitatively fewer and longer in kap-1; osm-3; dyf-18 triple mutant animals (Figure 4A). We conclude that similar to AWB and AWC cilia [30, 31] but in contrast to channel cilia [33, 34], kinesin-II and OSM-3 function redundantly to build the entire AWA cilium in both wild-type and dyf-18 mutants.
Figure 4. IFT motors function redundantly in the AWA cilia of wild-type and dyf-18 mutants.
A) Representative images of AWA cilia visualized via expression of gpa-4Δ6p::myr-gfp in the indicated genetic backgrounds. Two representative images each are shown for kap-1; osm-3 and kap-1; osm-3; dyf-18 mutants. Alleles used were osm-3(p802), kap-1(ok676) and dyf-18(ok200). The cilia base and cilia are indicated by yellow and white arrowheads, respectively; the dendrite is marked by an arrow; branches from the dendritic base are marked by asterisks. Numbers in white at top right indicate the percentage of neurons showing the cilia phenotype; numbers in green indicate the percentage of neurons exhibiting dendritic phenotypes; n≥ 30 each in 3 independent experiments. ARL-13::RFP (right) marks the cilium. Anterior is to the top or at top left in all images. Scale bar: 10 μm.
B) Representative images of KAP-1::GFP, OSM-3::GFP and OSM-6::GFP localization in the AWA cilia of wild-type and dyf-18(ok200) mutants. AWA cilia were visualized via expression of gpa-4Δ6p::myr-mCherry. The cilia base and cilia are indicated by yellow and white arrowheads, respectively; the dendrite is marked by an arrow. Anterior is to the top or at top left in all images. n≥30 neurons. Scale bar: 10 μm.
C) Histograms of KAP-1::GFP, OSM-3::GFP and OSM-6::GFP anterograde velocities in AWA cilia of wild-type (black bars) and dyf-18(ok200) mutants (gray bars). All fusion proteins were expressed cell-specifically in AWA under the gpa-4Δ6 promoter.
Also see Figure S2 and Table S1.
We next examined the localization of the anterograde kinesin motors in AWA cilia. Consistent with redundant functions of these motors in building the AWA cilium, KAP-1::GFP and OSM-3::GFP could be detected throughout the AWA cilia in wild-type animals (Figure 4B). However, KAP-1::GFP was preferentially enriched in the short proximal AWA ciliary stalk with low expression levels in the distal ciliary branches, whereas OSM-3::GFP accumulated strongly at the distal tips of the AWA ciliary branches (Figure 4B). A GFP-tagged OSM-6 IFT-B fusion protein was also present at low levels throughout AWA cilia but was enriched in the proximal stalk similar to KAP-1::GFP (Figure 4B). In dyf-18(ok200) mutants, both motors, as well as OSM-6::GFP, were localized similarly at low levels throughout the elongated AWA cilia (Figure 4B). We occasionally also observed enrichment of the fusion proteins at discrete membranous blebs along the elongated AWA cilia in dyf-18 mutants (Figure 4B). In contrast to observations in channel cilia [17, 26], we did not detect enrichment of IFT proteins at the distal tips of the AWA cilia in dyf-18 mutants (Figure 4B). These observations indicate that DYF-18 regulates the relative distribution of IFT motors and proteins in the AWA cilia, but in a manner distinct from that in channel cilia.
We next quantified the movement of OSM-3, KAP-1, and OSM-6 fusion proteins. Although we could occasionally detect mobile KAP-1::GFP and OSM-6::GFP in the distal AWA cilia branches in wild-type animals, the very low expression levels of these proteins precluded analyses of their speeds in the thin distal branches. Moreover, accumulated OSM-3::GFP present at the distal tips of wild-type AWA cilia appeared to be largely immobile (Figure S2A). We thus analyzed protein movement in the proximal stalks. Both KAP-1::GFP and OSM-6::GFP moved at an average rate of ~0.7 μm/s whereas OSM-3::GFP moved at a rate of 1.3 μm/s (Figure 4C, Table S1, Figure S2B-D). Based on reported in vitro measurements and in vivo speeds of these motors in other C. elegans cilia [30, 33, 40], these results are most consistent with the hypothesis that OSM-3 moves largely independently in AWA cilia, but that a fraction of OSM-3 motors may be associated with kinesin-II to account for its higher speed. Alternatively, kinesin-II velocity may be regulated in a cell-specific manner in AWA cilia. Moreover, IFT particles appear to be transported primarily via kinesin-II.
To ask whether DYF-18 regulates IFT motor velocities, we quantified protein movement in a region of the AWA cilia approximately 7 μm from the cilia base in dyf-18 mutants. The average speeds of KAP-1::GFP and OSM-3::GFP were largely unaffected upon loss of DYF-18, whereas OSM-6::GFP moved at a slightly faster average speed of 0.9 μm/s in dyf-18 AWA cilia (Figure 4C, Table S1, Figure S2B-D). This observation implies that as in wild-type animals, IFT particles are also largely trafficked via kinesin-II in dyf-18 AWA cilia, although a fraction of these complexes may also be trafficked by OSM-3. Together, these results indicate that DYF-18 regulates the relative ciliary distributions, but not velocities, of IFT motors in AWA cilia.
AWA axonemal MT dynamics is altered in dyf-5 and dyf-18 mutants
Axonemal MT properties influence motor protein functions (eg.[41-43]). We asked whether changes in AWA axonemal MT structure or dynamics underlie the altered motor protein distribution and cilia lengthening phenotypes of dyf-18 and dyf-5 mutants. We were unable to reliably identify the MT ultrastructure of the elongated AWA cilia in dyf-5 or dyf-18 mutants using electron microscopy (Oliver E. Blacque, personal communication). We thus employed alternate approaches to examine AWA axonemal MT properties.
In channel cilia, the MT plus-end tracking protein EBP-2 is localized throughout the axoneme but is enriched at the plus-ends of the A- and B-tubules at the tips of the middle and distal segments, respectively [32]. EBP-2::GFP was present in the proximal stalk and in punctae at the distal tips of ciliary branches in wild-type AWA cilia (Figure 5A). In contrast, in dyf-5 and dyf-18 mutants, EBP-2::GFP uniformly decorated the entire elongated AWA cilium (Figure 5A). Decoration of MT bundles by EB1 proteins could be associated with increased MT stability [44, 45], or with MT fragmentation. The MT minus end-binding CAMSAP homolog PTRN-1 [46] (gift of S. Yogev) was localized similarly to bright and very faint puncta in the AWA dendrites and axonemes, respectively, of both wild-type and dyf-18 mutants (Figure 5B), suggesting that it is unlikely that the decoration by EBP-2::GFP is due to MT severing in the axoneme. Exposure to sub-lethal high temperatures has previously been shown to promote cilia destabilization and resorption in vertebrate cells, likely via promotion of tubulin deacetylation and possible Hsp90-mediated sequestration of dimeric soluble tubulin required for IFT [47, 48]. We found that although the AWA cilia of wild-type animals grown at 25°C were severely truncated, the elongated cilia of dyf-18 mutants were unaffected at this temperature (Figure S3A). Together, these observations support the notion that axonemal MT stability is increased in dyf-18 mutants.
Figure 5. AWA axonemal microtubules may be stabilized in dyf-5 and dyf-18 mutants.
A, B) Representative images of EBP-2::GFP (A) or RFP::PTRN-1 (B) localization in the AWA cilia of animals of the indicated genotypes. Insets in B show localization patterns in the axonemes; images have been overexposed to allow visualization of the faint puncta. Both fusion proteins were expressed under the gpa-4Δ6 promoter. Numbers at top right indicate the percentage of neurons exhibiting the phenotype; n≥30 each in 3 independent experiments. The cilia base and cilia are indicated by yellow and white arrowheads, respectively; the dendrite is marked by an arrow. AWA neurons were visualized via expression of gpa-4Δ6p::myr-gfp in B. Anterior is at top or at top left in all images. Scale bars: 10 μm.
C) Representative images of TBB-4::GFP localization in wild-type and dyf-18(ok200) mutants. TBB-4::GFP was expressed under the gpa-4Δ6 promoter. AWA cilia were visualized via expression of gpa-4Δ6p::myr-mCherry. Numbers at top right indicate the percentage of animals exhibiting the phenotype; n≥30 neurons each in 3 independent experiments. The cilia base and cilia are indicated by yellow and white arrowheads, respectively; the dendrite is marked by an arrow. Anterior is at left or at top left in all images. Scale bar: 10 μm.
D) (Top) Representative images of AWA cilia pre-bleach (Pre), at bleach (0h) and 12h post-bleaching (12h). The bleached area is indicated by a dotted box. Note that while the same animal was examined at all three timepoints, the animals were removed after bleaching to growth plates, and re-examined after 12h (see STAR Methods). Cilia are indicated by white arrowheads; the dendrite is marked by an arrow. Anterior is at top. Scale bar: 5 μm. (Bottom) Mean fluorescence intensity ratios (photobleached to unbleached regions within an AWA cilium; see STAR Methods) across a line segment on AWA cilia branches shown at 0 hour and 12 hour post-bleaching for individual wild-type and dyf-18(ok200) animals. Statistical significances were calculated using a paired t-test. Experiments were performed on 3 independent days.
Also see Figure S3.
To directly test this hypothesis, we examined tubulin turnover in AWA cilia. Sensory cilia in C. elegans are comprised of diverse tubulin isoforms including the TBA-5 α- and TBB-4 β-tubulins [32, 49]. In wild-type AWA cilia, GFP-tagged TBB-4 was enriched at the proximal stalks with lower levels in the distal branches consistent with their lower tubulin load [5] (Figure 5C). In contrast, TBB-4::GFP was present throughout the elongated AWA cilium in dyf-18 mutants, suggesting increased tubulin content (Figure 5C). To directly examine tubulin turnover, we examined fluorescence recovery of TBB-4::GFP after photobleaching (FRAP). We photobleached either the left or right half of the AWA cilia arbor in wild-type animals, and a region between 5-20 μm from the cilia tip in dyf-18 mutants expressing TBB-4::GFP (Figure 5D). In initial experiments, following photobleaching, we did not observe significant fluorescence recovery even after 30 mins indicating that tubulin turnover is low in steady-state cilia [13, 32, 50]. However, we found that while TBB-4::GFP fluorescence intensities recovered significantly in wild-type animals 12 hr following photobleaching (see STAR Methods), fluorescence recovery was markedly lower in the photobleached region in the cilia of dyf-18 mutants (Figure 5D). These observations suggest that tubulin turnover is reduced in the elongated AWA cilia of dyf-18 mutants.
MT destabilizing mutations in tubulin subunits and disruption of tubulin-transporting IFT-B proteins partially suppress the AWA cilia phenotype of dyf-18 mutants
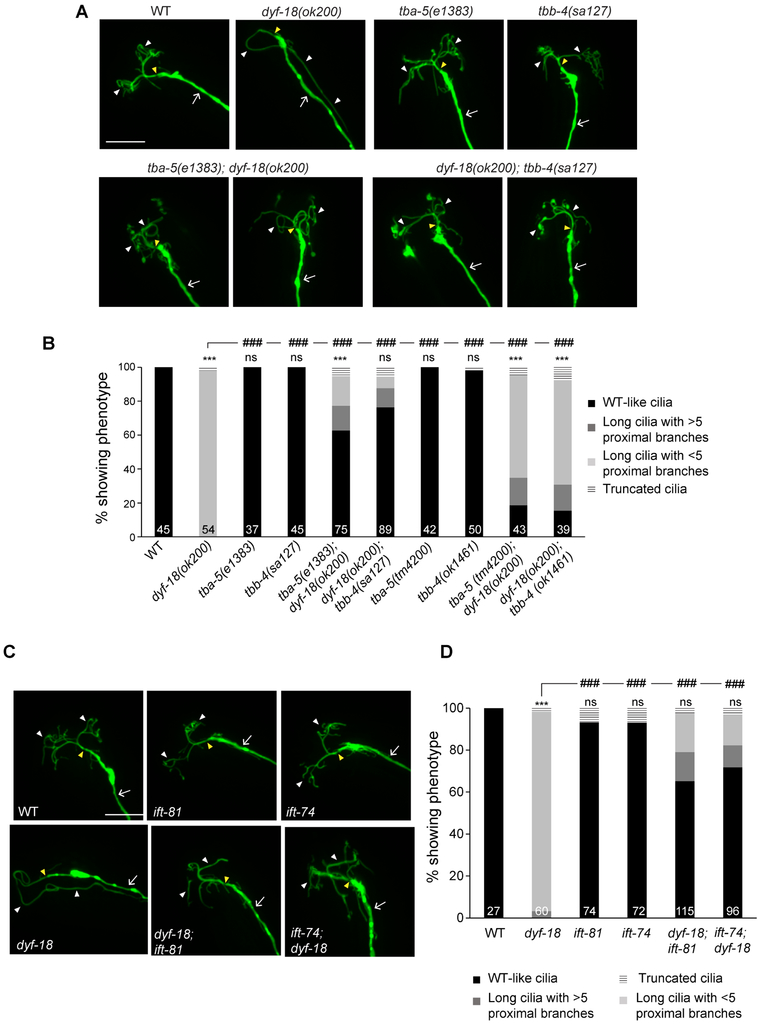
We next asked whether the observed changes in MT dynamics are causal to the elongated cilia phenotype and lack of branching in dyf-18 mutants. A previous report identified missense mutations in TBB-4 and TBA-5 tubulins that destabilize axonemal MTs; these mutations are temperature-sensitive and result in fully penetrant phenotypes at 15°C [32]. Similar destabilization was not observed in animals carrying null mutations in either gene [32, 49]. We tested whether MT destabilization is sufficient to suppress the AWA cilia phenotype of dyf-18 mutants. AWA cilia morphology was not grossly affected in animals carrying missense or null alleles of either gene alone at 15°C (Figure 6A, 6B). However, the tbb-4(sa127) or tba-5(e1383) missense mutations significantly suppressed AWA cilia elongation, and increased cilia branching in dyf-18 mutants (Figure 6A, 6B). Little to no suppression was observed in dyf-18 animals carrying the tbb-4(ok1461) or tba-5(tm4200) null mutations in either gene (Figure 6B). These observations support the hypothesis that increased MT stability underlies the AWA cilia phenotype of dyf-18 mutants.
Figure 6. Destabilizing axonemal MTs or disrupting tubulin-IFT interactions suppresses the dyf-18 AWA cilia phenotype.
A, C) Representative images of AWA cilia morphologies in animals of the indicated genotypes. AWA neurons were visualized via expression of gpa-4Δ6p::myr-gfp. The cilium base and cilium are indicated by yellow and white arrowheads, respectively; the dendrite is marked by an arrow. Alleles used in C were dyf-18(ok200), ift-81(tm2355), ift-74(tm2393). Anterior is at top or at top left in all images. Scale bar: 10 μm. Animals were grown at 15°C in A.
B, D) Percentage of AWA neurons in adult hermaphrodites of the indicated genotypes exhibiting cilia phenotypes. Alleles used in D were dyf-18(ok200), ift-81(tm2355), ift-74(tm2393). Numbers in each bar indicate the number of examined neurons in 2-3 independent experiments. *** and ### indicate different from wild-type and indicated values, respectively, at P<0.001; ns – not significant (Wilcoxon rank-sum test). Animals were grown at 15°C in B.
Also see Figure S3, Figure S4, and Table S2.
Tubulin transport into the cilium is mediated via both IFT and diffusion [13, 14, 32]. IFT-dependent tubulin transport is in part mediated via direct interactions between tubulin and multiple core components of the IFT-B complex including IFT74, IFT81 and IFT54 [50, 51]. We asked whether disruption of tubulin transport via mutations in tubulin-binding IFT components would be sufficient to also partly bypass the dyf-18 phenotype in AWA cilia. Single mutations in either ift-74 or ift-81 [52] resulted in only minimal changes in AWA cilia morphology (Figure 6C, 6D). However, animals doubly mutant for either ift-74 or ift-81 and dyf-18 exhibited both significantly decreased AWA ciliary elongation, as well as increased ciliary branching (Figure 6C, 6D). These results imply that destabilization of axonemal MTs or inhibition of tubulin transport is able to partly suppress the effects of DYF-18/DYF-5 loss in AWA cilia.
Axonemal dynamics can be significantly influenced by post-translational tubulin modifications, as well as by depolymerizing kinesins [43, 53]. Animals mutant for the tubulin deglutamylases ccpp-1 and ccpp-6, tubulin glutamate ligase ttll-4, alpha-tubulin acetyltransferases mec-17/atat-1 and atat-2, or the depolymerizing kinesin-8 klp-13 did not exhibit altered AWA cilia morphology in the presence or absence of a dyf-18 mutation (Table S2). The CSAP protein that preferentially binds polyglutamylated MTs [54] was localized to the proximal region of AWA cilia in wild-type animals; this localization was largely unaltered in dyf-18 mutants (Figure S3B). Although we are unable to exclude redundant roles of these or other tubulin modifications or depolymerizing kinesins, these results suggest that alternative mechanisms may underlie the altered axonemal properties of dyf-18 mutants.
A MT destabilizing mutation in a tubulin subunit partly restores AWA-mediated chemosensory behaviors of dyf-18 mutants
We asked whether the altered wing cilia morphologies in dyf-18 and dyf-5 mutants affect sensory neuron responses. It has previously been shown that severe truncation of AWA cilia in IFT mutants does not affect primary responses to low concentrations of the AWA-sensed odorant diacetyl but instead decreases adaptation and desensitization to this chemical in this neuron type [55]. Consequently, IFT mutants exhibit mild to no defects in attraction to a point source of diacetyl in long-range end-point chemotaxis assays [55]. In contrast, mutations in IFT genes decrease attraction to AWC-sensed volatile odorants [31].
Consistent with observations in IFT mutants [55], dyf-5(XS) animals in which the AWA cilia are severely truncated continued to be robustly attracted to low concentrations of diacetyl in end-point chemotaxis assays but were defective in attraction to a point source of the AWC-sensed chemical isoamyl alcohol (Figure S4A). Although elongation of AWA cilia in dyf-5(mn400) mutants also did not affect attraction to diacetyl (Figure S4A), dyf-18(ok200) mutants exhibited significant defects in this behavior (Figure S4A, S4B). dyf-18(ok200) mutants also exhibited defects in chemotaxis towards isoamyl alcohol (Figure S4A, S4B). The behavioral defects in dyf-18 mutants were rescued upon expression of wild-type dyf-18 sequences under the bbs-8 promoter (Figure S4A, S4B). Interestingly, we found that restoration of wildtype-like AWA ciliary morphologies in dyf-18; tbb-4 double mutant was sufficient to partly but significantly enhance attraction to both volatile chemicals as compared to the behaviors of dyf-18 mutants alone (Figure S4B). tbb-4(sa127) single mutants retained robust attraction to both diacetyl and isoamyl alcohol (Figure S4B). These observations indicate that elongated AWA and AWC cilia in dyf-18/dyf-5 mutants are correlated with defects in sensory neuron-driven odorant attraction behaviors, and that restoration of cilia morphology in dyf-18; tbb-4 mutants is sufficient to partly restore these behaviors.
Loss of dyf-18 elongates the truncated channel cilia in tbb-4 tubulin missense mutants
While AWA cilia morphology is largely unaltered in tbb-4(sa127) missense mutants, channel cilia are truncated due to destabilization of distal segments in these animals [32]. We asked whether the proposed increased MT stability in dyf-18 mutants would suppress the length phenotype of tbb-4(sa127) mutants in channel cilia. The channel cilia of the ASH nociceptive sensory neurons were slightly elongated in dyf-18(ok200) mutants [26, 27], but were markedly shortened in tbb-4(sa127) animals grown at 15°C [32] (Figure 7A, 7B). The ASH cilia of dyf-18(ok200); tbb-4(sa127) double mutants were significantly longer than those of tbb-4(sa127) single mutants alone (Figure 7A, 7B). These observations are consistent with the hypothesis that as in AWA, DYF-18 regulates axonemal MT stability in channel cilia.
KAP-1 has been shown to inappropriately enter the distal segments of channel cilia in dyf-5 and dyf-18 mutants [17, 26]. In addition, OSM-3 speed is reduced in the middle segments of channel cilia in dyf-5 and dyf-18 mutants [17, 26]. Neither phenotype is likely to be causal to the cilia elongation phenotype of these kinase mutants, since the cilia length phenotype of dyf-5 mutants is unaltered in dyf-5; kap-1 or dyf-5; osm-3 double mutants [17]. We asked whether the observed changes in IFT could arise as a consequence of changes in MT properties in these animals.
As reported previously, while KAP-1::GFP was restricted to the middle segments of ASH cilia in wild-type and tbb-4 mutants, this fusion protein was also present in the distal segments in dyf-18 mutants (Figure 7C). In dyf-18; tbb-4 double mutants, the protein was similarly found throughout the ASH cilia (Figure 7C), suggesting that KAP-1 mislocalization may not be causal to the ciliary phenotype. However, only the middle segments may be elongated in dyf-18; tbb-4 mutants, thereby accounting for the distribution of KAP-1::GFP throughout the lengths of the cilia in these animals. Localization of OSM-3::GFP was overall similar in wild-type, dyf-18, tbb-4 and dyf-18; tbb-4 mutants, although we noted accumulation at the distal tips in tbb-4, and particularly in dyf-18; tbb-4 animals (Figure 7D).
The middle segments of channel cilia are built via the coordinated and partly redundant functions of the kinesin-II and OSM-3 motors which move together in this region [33, 34]. Consequently, OSM-3::GFP moves at a slower average speed of 0.6 μm/sec in the ASH cilia middle segments (Figure 7E, Table S3, Figure S5). Consistent with previous reports, the rate of OSM-3::GFP movement was decreased in dyf-18 mutants (average speed: 0.5 μm/sec; Figure 7E, Table S3, Figure S5) [17, 26]. Interestingly, we found that OSM-3 moved at a significantly faster rate (average speed: 0.9 μm/sec) in tbb-4(sa127) mutants grown at 15°C (Figure 7E, Table S3, Figure S5), suggesting that altering MT stability alters IFT motor properties. The speed of OSM-3 was reported to be unaltered in tbb-4(sa127) mutants in a previous report [32]; this discrepancy could arise due to differences in growth conditions and/or cell type-specific variations. The average speed of OSM-3::GFP movement in dyf-18; tbb-4 double mutants was intermediate between that in each single mutant alone (average speed: 0.7 μm/sec; Figure 7E, Table S3, Figure S5). These observations suggest that axonemal properties can influence OSM-3 motor properties, and that OSM-3 motor speed is inversely correlated with cilia length.
DISCUSSION
We show here that DYF-18/CCRK and DYF-5/MAK regulate axoneme length and cilia morphology likely via modulation of MT stability in the specialized AWA olfactory neuron cilia of C. elegans. Several lines of evidence support this hypothesis. First, the elongated cilia in dyf-18 mutants contain higher levels of the TBB-4::GFP tubulin fusion protein. Second, the localization of EBP-2::GFP along the length of the AWA cilium in dyf-18 mutants is reminiscent of the association of EB proteins with stable MT bundles in mammalian cells [45, 56]. Third, recovery of TBB-4::GFP fluorescence following photobleaching is higher in wild-type than in dyf-18 mutant AWA cilia, suggesting that axonemal TBB-4::GFP is longer-lived in dyf-18 mutants as compared to wild-type animals. Fourth, missense mutations in either TBA-5 or TBB-4 predicted to destabilize MTs reduce AWA cilia length and promote branching in dyf-18 mutants. Moreover, loss of dyf-18 significantly elongates the truncated and destabilized ASH axonemes in tbb-4 missense mutants. Interestingly, in mouse photoreceptors, MAK has also been suggested to regulate MT stability and connecting cilia length [25], suggesting CCRK/MAK-mediated regulation of MT properties may be a common feature of diverse cilia across species.
In one model, altered IFT motor function in dyf-18/dyf-5 mutants may result in increased tubulin transport resulting in changes in axonemal properties and elongated cilia as has been reported in Chlamydomonas flagella [10, 12, 13]. However, it is difficult to explain whether and how altered tubulin transport is correlated with the observed changes in IFT protein recycling, OSM-3 speed, and the handover between kinesin-II and OSM-3 in channel cilia of dyf-18/dyf-5 mutants in C. elegans [17, 26]. Moreover, loss of either kap-1 or osm-3 is not sufficient to suppress the channel or AWA cilia length phenotype of dyf-18 or dyf-5 mutants ([17]; this work). We instead favor the possibility that the reported changes in IFT in dyf-18/dyf-5 mutants arise as a secondary consequence of changes in axonemal MT properties. The ‘tubulin code’ comprised of different tubulin isotypes and tubulin posttranslational modifications is interpreted by molecular motors and tubulin-associated proteins to modulate MT properties including stability, structure, and transport (eg. [41-43]). Although we did not observe an effect of mutations in individual tubulin modifying enzymes, it remains possible that tubulin is differentially modified in dyf-18/dyf-5 mutants. Indeed, MAK regulates levels of tubulin acetylation in photoreceptor cilia [25]. Since tubulin identity and posttranslational modifications modulate motor processivity and velocity [41-43], an altered tubulin code in dyf-18/dyf-5 mutants could alter MT stability but could also in parallel lead to the observed changes in IFT motor properties ([17, 26, 27]; this work). These changes in IFT properties are likely to be cell type-specific; OSM-3 speed is altered in dyf-18 mutants in ASH but not in AWA cilia. Similarly, in C. elegans males, changes in the tubulin code have been shown to alter IFT motor protein distribution and velocities in the CEM neuronal cilia [41, 42, 57]. These results suggest a fundamentally distinct mechanism by which these conserved kinases regulate cilia morphology in C. elegans.
Length-dependent feedback control of tubulin entry into flagella is disrupted in Chlamydomonas mutant for CCRK/MAK homologs [10, 13, 28]. The signal that transmits length information to modulate tubulin IFT is unknown. Although we could not detect movement of these kinases in AWA cilia, members of these kinase families have been shown to undergo IFT in channel cilia in C. elegans as well as in mammalian cells [22, 26]. It is possible that these kinases are effectors of the length control mechanism via cilia length-dependent modulation of their enzymatic activities [16, 58], or that they are components of the length sensor themselves. What are the targets of these kinases? MAK family kinases directly phosphorylate kinesin-II and the RP1 MT-binding protein [23, 25], although kinesin-II does not appear to be a target of these kinases in C. elegans [26]. Tubulin can also be phosphorylated via multiple kinases to regulate MT dynamics [53, 59], and MAK has also been suggested to target tubulin acetylases or deacetylases in photoreceptors [25]. Thus, regulation of tubulin posttranslational modifications or MT-associated proteins by these kinases together with correlated changes in IFT properties may then modulate cilia length. However, these kinases may act via alternate pathways to regulate cilia length. For instance, ICK and MOK regulate the levels of soluble tubulin via modulation of mTORC1 activity in renal epithelial cells [22, 60]. Thus, these kinases may target different proteins and mechanisms in different cell types to tune cilia length.
Wing cilia are dramatically elongated in dyf-18/dyf-5 mutants, whereas channel cilia exhibit more modest elongation. Since unlike wing cilia, channel cilia are fully contained with the glia-formed channel [4, 5, 29], elongation of these cilia might simply be limited by steric or physical constraints. It is also possible that the requirement for DYF-18/DYF-5 in regulating cilia length is distinct in different cilia types, and that this differential requirement contributes to the generation of ciliary morphological diversity.
A unique feature of AWA cilia is the presence of ciliary branches containing variable numbers of MTs [5]. In addition to regulating cilia length, loss of DYF-18/DYF-5 inhibits cilia branching. The splitting of dMTs into branches could be a secondary consequence of modulation of MT stability in AWA. Indeed, decreased MT stability has been shown to inhibit MT bundling and promote branching in the axons of rodent neurons and processes of C. elegans touch receptor neurons [61, 62]. Recently, the single transmembrane Ig-domain protein OIG-8 was shown to be expressed in and localized to the cilia of amphid wing but not channel neurons, and is both necessary and sufficient for ciliary branching [63]. Branches in amphid wing neuron cilia could be stabilized by OIG-8 via interaction with surrounding amphid sheath glia. Continued investigations into cell-specific regulation of shared pathways, as well as molecules and mechanisms unique to specific cell types, will provide a more complete description of the mechanisms that generate and maintain specialized cilia architectures and contribute to diversifying cellular functions.
STAR+METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Piali Sengupta (sengupta@brandeis.edu).
EXPERIMENTAL MODEL DETAILS
C. elegans strains
All C. elegans strains were grown on E. coli OP50 bacteria. The wild-type strain was C. elegans strain Bristol (N2). Well-fed animals were maintained for at least two generations prior to analyses. Animals were imaged on the first day of adulthood. Animals were grown at 20°C, with the exception of strains carrying the tbb-4(sa127) and tba-5(e1383) alleles which were grown at 15°C [32]. Strain genotypes were verified by PCR and sequencing. For strains generated in this work, proteins were expressed from extrachromosomal arrays (with the exception of gpa-4Δ6p::myr-gfp which was stably integrated into the genome). In all cases, expression from the same extrachromosomal arrays was compared between wild-type and mutant strains. All strains used in this work are listed in Table S4.
METHOD DETAILS
Isolation and cloning of dyf-18(oy153)
The oy153 allele was isolated in a forward genetic screen for mutants with altered AWA cilia morphology. P0 animals stably expressing gpa-4Δ6p::myr-gfp were mutagenized using EMS, and 3-5 F1 progeny placed on individual plates. 50-100 F2 animals from an individual plate were mounted on a 2% agarose pad with a coverslip to visualize AWA cilia on a compound fluorescent microscope (Zeiss Axio Imager) and examined using a 63X objective. Animals exhibiting altered cilia morphology were recovered from the slide and their progeny re-examined to confirm the phenotype.
The oy153 mutant allele was mapped to LG IV and the molecular identity of the lesion identified via paired-end Illumina whole genome sequencing (Beijing Genomics Institute). Unique variants were identified using the Galaxy server (galaxyproject.org) and the CloudMap paired-end variant identification pipeline (usegalaxy.org/cloudmap) via comparison with sequences from an independently sequenced strain. Protein coding variants were identified using snpEff v2.1 (http://snpeff.sourceforge.net/SnpEff.html) and the WS220 reference genome (wormbase.org). The mutation in dyf-18 was confirmed using Sanger sequencing.
Molecular biology
1.8 kb of gpa-4Δ6 upstream sequences driving expression specifically in AWA [35] were amplified and inserted upstream of gfp or mCherry coding sequences carrying an N-terminal myristoylation tag (5’-ATGGGATCATGTATTGGAAAA-3’) (Max Heiman, personal communication). To generate AWA-expressed transgenes, cDNAs with or without fluorescent reporter tags were subcloned from previously generated plasmids (arl-13b, odr-10, mks-5, gasr-8, osm-3::gfp, kap-1::gfp, osm-6::gfp, rfp::ptrn-1, csap::gfp) or were amplified from pooled cDNAs from mixed-stage wild-type animals (dyf-18, dyf-5, ebp-2, tbb-4) and cloned downstream of gpa-4Δ6 promoter sequences in C. elegans expression vectors containing unc-54 3’UTR sequences. To generate constructs driven under the bbs-8 promoter, cDNA sequences with or without fluorescent reporter tags were cloned into a C. elegans expression plasmid containing 0.94 kb of a bbs-8 upstream and unc-54 3’ UTR sequences. Site-directed mutagenesis was performed by reverse amplifying the relevant plasmid with the mutations incorporated into one of the oligonucleotides. The product from this amplification was self-ligated to obtain the desired mutant cDNA constructs. To visualize ASH cilia, a 3 kb promoter fragment of sra-6 was used to drive expression of myr-gfp (courtesy of Alison Philbrook). All constructs were verified by sequencing.
Microinjections
Fluorescent reporter and cDNA-expressing constructs were injected at 10-30 ng/μl and 5-30 ng/μl, respectively. Co-injection markers for transgenic strains were unc-122p::gfp or unc-122p::mCherry injected at 15-30 ng/μl. Shown data are from 1-2 independent transgenic lines each unless indicated otherwise. In all cases where expression of a transgene was compared between wild-type and mutant animals, the same transgenic array was examined in both genetic backgrounds.
Generation of the klp-13(oy154) allele
The klp-13(oy154) allele was generated via gene editing. Four sgRNAs targeting intron 1 (5’-GCAAAAGCATATATACTTAA-3’, 5’ -GGTCGGAGTTGTACTAGTAG- 3’), intron 14 (isoform a; 5’-GTTTGAAAGAACAGAGTGAA-3’) and exon 15 (isoform a; 5’-GGTTGATTCCGCAGAAGAAGT-3’) were cloned into the pU6 plasmid [64] and injected together with myo-3p::rfp and the Peft3::Cas9 plasmid [65] such that 2 sgRNAs each flanked the ~ 5kb fragment containing klp-13 coding sequences. Roller worms were selected from the progeny and animals with a ~5 kb deletion were selected by PCR. The deletion was verified by Sanger sequencing.
Dye-filling
Animals were washed off the plates with M9 into 1.5 ml or 15 ml tubes, spun at 1600g, resuspended in 100 μl or 1 ml of M9, respectively, with 10 μg/ml DiI (Thermo Fisher) and placed on a rotating rack for 0.5-2 hrs. Animals were then washed once with an excess of M9 and placed in a drop of M9 on plates with a thick bacterial lawn. Animals were examined after 20 mins using a fluorescence dissection microscope.
Microscopy
To image cilia, animals were anesthetized with 10 mM tetramisole hydrochloride (Sigma-Aldrich) and mounted on 2-10% agarose pads on microscope slides. Animals were examined on an inverted spinning disk confocal microscope using a 100X objective (Zeiss Axio Observer with a Yokogawa CSU-22 spinning disk confocal head), or on a Zeiss Axio Imager 2 epifluorescent microscope. Optical sections were acquired at 0.2 μm intervals and images were z-projected at maximum intensity (SlideBook 6.0 software; Intelligent Imaging Innovations, 3i). For optimal visualization of cilia, images were linear adjusted for brightness and contrast using ImageJ (NIH). Quantification of cilia morphology and protein localization was performed using ImageJ (NIH). All animals were imaged on at least two independent days for each data point.
IFT
IFT analyses were performed as described previously [66]. Fusion proteins were expressed from extrachromosomal arrays; expression from the same arrays were examined in wild-type and mutant strains. Examined IFT fusion proteins were expressed at low levels in AWA even from overexpressed extrachromosomal arrays, precluding examination of expression driven from single copy genomic insertions. In all cases, strains injected with the lowest DNA concentration that permitted accurate visualization and quantification of IFT without affecting AWA cilia morphology were examined.
In brief, movies of mobile GFP particles within the proximal stalk of wild-type AWA cilia or in a region approximately ~7 μm from the cilia base in dyf-18 mutants were acquired on a spinning disk confocal microscope for 0.5–2 mins with 300 ms exposure. IFT was analyzed only in animals in which the proximal AWA ciliary stalk or the distal tips (for examining movement of accumulated OSM-3::GFP) were in the focal plane of the spinning disk confocal microscope. Animals whose cilia were oriented at an angle to the z-plane were excluded from acquisition and analysis. IFT particles were identified manually and line segments over the tracks were drawn to derive velocity measurements. Kymograph analyses were performed using the Multi Kymograph plugin in ImageJ (NIH).
Fluorescence recovery after photobleaching (FRAP)
Animals were mounted on 3% agar pads containing 10 mM tetramisole and imaged at 100X with 0.5 μm z-steps on a Nikon Ni-E upright microscope with a Yokogawa CSU-W1 spinning disk head, and an Andor iXon 897U EMCCD camera with Nikon Elements AR software. Cilia were photobleached using a 405-nm laser (50% power, 2 s), directed by an Andor Mosaic 3 digital micromirror device. The left or right half of the AWA cilia arbor, and an area 5-20 μm from the AWA cilia tip in wild-type and dyf-18 mutants, respectively, were photobleached. Post-bleach images were acquired within 2-5 mins of photobleaching. Animals were recovered, and placed on NGM plates containing OP50 bacteria at 20°C. Cilia were re-imaged after 12 hours. We note that while only ~20% of animals exhibited locomotion at 6 hours following recovery on growth plates, locomotion was restored to all animals after 12 hours.
To quantify fluorescence, line segments were drawn on photobleached and unbleached segments of cilia using the AWA-expressed myr-tagRFP fusion protein as a guide. In wild-type animals, the left or right half of the cilia was photobleached; in dyf-18 animals, a 20 μm region from the distal tip was photobleached. On average, the fluorescence values decreased to 33% in wild-type animals, and to 23% in dyf-18 mutants, from their pre-bleach levels immediately following photobleaching. Average fluorescence intensities were measured along line segments, and the ratio of bleached vs unbleached fluorescence intensities were compared and plotted. Areas to be measured were kept constant to the best of our abilities, and identified following recovery using morphological landmarks of AWA dendrites and cilia via expression of gpa-4Δ6p::myr-tag-rfp.
Chemotaxis assays
Well-fed young adult animals were washed twice with S-basal and once with water and placed on 10 cm square plates containing agar. 1 μl of odor diluted in 100% ethanol or ethanol were placed on either ends of assay plates together with 1 μl 1M sodium azide. Diacetyl (DIA) was diluted to 10−3 and isoamyl alcohol (IAA) was diluted to 10−2. Chemotaxis index = (number of animals in plate segments containing the odor) – (number of animals in plate segments containing ethanol)/total number of animals. The number of animals in each region and the total number of animals on the plate were counted after one hour.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical analyses were performed using the SPSS 25.0 statistical analyses package (IBM) or GraphPad PRISM version 7.0 software (GraphPad Software). Cilia phenotypic categories were designated as ordinal variables. Statistical tests used depended on whether the data were normally or non-normally distributed. The Mann-Whitney/Wilcoxon rank-sum or Kruskal-Wallis nonparametric tests were used for data with non-normal distributions. Posthoc corrections for multiple comparisons were performed when appropriate. For each experiment, data are presented from 2-3 independent trials with total sample sizes of ≥25 cilia. Data acquisition and analyses were not blinded. Further details of statistical tests, numbers of cilia/animals analyzed, and significance values are provided in each figure legend.
Supplementary Material
Table S4. List of strains used in this work. Related to the Key Resources Table
Highlights.
DYF-18 CCRK and DYF-5 MAK kinases regulate cilia length and branching
Axonemal microtubules (MTs) are stabilized upon loss of kinase function
Destabilizing MTs can partly restore cilia length and branching in kinase mutants
These kinases modulate MT dynamics in multiple cilia types in C. elegans
Acknowledgements
We thank the Caenorhabditis Genetics Center and Shohei Mitani (National BioResource Project, Japan) for strains, and Shai Shaham, Gert Jansen, Cori Bargmann, Alison Philbrook, and Shaul Yogev for reagents. We are grateful to Mike O’Donnell for assistance with mapping dyf-18(oy153), Kassandra Ori-McKenney for experimental assistance, and Avi Rodal for advice on FRAP analyses. We thank the Sengupta lab for general advice and the Sengupta lab ‘cilia squad’ and Oliver Blacque for comments on the manuscript. We apologize for the omission of citations due to constraints on the number of permitted references. This work was funded in part by the NIH [R35 GM122463 (P.S.) and T32 GM007122 (T.R.)].
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Hilgendorf KI, Johnson CT, and Jackson PK (2016). The primary cilium as a cellular receiver: organizing ciliary GPCR signaling. Curr Opin Cell Biol 39, 84–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang J, and Barr MM (2016). Ciliary extracellular vesicles: Txt msg organelles. Cell Mol Neurobiol 36, 449–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Silverman MA, and Leroux MR (2009). Intraflagellar transport and the generation of dynamic, structurally and functionally diverse cilia. Trends Cell Biol 19, 306–316. [DOI] [PubMed] [Google Scholar]
- 4.Perkins LA, Hedgecock EM, Thomson JN, and Culotti JG (1986). Mutant sensory cilia in the nematode Caenorhabditis elegans. Dev Biol 117, 456–487. [DOI] [PubMed] [Google Scholar]
- 5.Doroquez DB, Berciu C, Anderson JR, Sengupta P, and Nicastro D (2014). A high-resolution morphological and ultrastructural map of anterior sensory cilia and glia in C. elegans. eLife 3, e01948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Besschetnova TY, Kolpakova-Hart E, Guan Y, Zhou J, Olsen BR, and Shah JV (2010). Identification of signaling pathways regulating primary cilium length and flow-mediated adaptation. Curr Biol 20, 182–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rosenbaum JL, Moulder JE, and Ringo DL (1969). Flagellar elongation and shortening in Chlamydomonas. The use of cycloheximide and colchicine to study the synthesis and assembly of flagellar proteins. J Cell Biol 41, 600–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rosenbaum JL, and Witman GB (2002). Intraflagellar transport. Nat Rev Mol Cell Biol 3, 813–825. [DOI] [PubMed] [Google Scholar]
- 9.Taschner M, and Lorentzen E (2016). The intraflagellar transport machinery. Cold Spring Harb Perspect Biol 8, pii:a028092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ludington WB, Wemmer KA, Lechtreck KF, Witman GB, and Marshall WF (2013). Avalanche-like behavior in ciliary import. Proc Natl Acad Sci USA 110, 3925–3930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Engel BD, Ludington WB, and Marshall WF (2009). Intraflagellar transport particle size scales inversely with flagellar length: revisiting the balance-point length control model. J Cell Biol 187, 81–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wren KN, Craft JM, Tritschler D, Schauer A, Patel DK, Smith EF, Porter ME, Kner P, and Lechtreck KF (2013). A differential cargo-loading model of ciliary length regulation by IFT. Curr Biol 23, 2463–2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Craft JM, Harris JA, Hyman S, Kner P, and Lechtreck KF (2015). Tubulin transport by IFT is upregulated during ciliary growth by a cilium-autonomous mechanism. J Cell Biol 208, 223–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harris JA, Van De Weghe JC, Kubo T, Witman GB, and Lechtrek KF (2018). Diffusion rather than IFT provides most of the tubulin required for axonemal assembly. bioRxiv, doi: 10.1101/268573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marshall WF, and Rosenbaum JL (2001). Intraflagellar transport balances continuous turnover of outer doublet microtubules: implications for flagellar length control. J Cell Biol 155, 405–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Luo M, Cao M, Kan Y, Li G, Snell W, and Pan J (2011). The phosphorylation state of an aurora-like kinase marks the length of growing flagella in Chlamydomonas. Curr Biol 21, 586–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Burghoorn J, Dekkers MP, Rademakers S, de Jong T, Willemsen R, and Jansen G (2007). Mutation of the MAP kinase DYF-5 affects docking and undocking of kinesin-2 motors and reduces their speed in the cilia of Caenorhabditis elegans. Proc Natl Acad Sci USA 104, 7157–7162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Muthaiyan Shanmugam M, Bhan P, Huang HY, Hsieh J, Hua TE, Wu GH, Punjabi H, Lee Aplicano VD, Chen CW, and Wagner OI (2018). Cilia length and intraflagellar transport regulation by kinases PKG-1 and GCK-2 in C. elegans sensory neurons. Mol Cell Biol 38, pii:e00612–00617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tam LW, Wilson NF, and Lefebvre PA (2007). A CDK-related kinase regulates the length and assembly of flagella in Chlamydomonas. J Cell Biol 176, 819–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Asleson CM, and Lefebvre PA (1998). Genetic analysis of flagellar length control in Chlamydomonas reinhardtii: a new long-flagella locus and extragenic suppressor mutations. Genetics 148, 693–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Berman SA, Wilson NF, Haas NA, and Lefebvre PA (2003). A novel MAP kinase regulates flagellar length in Chlamydomonas. Curr Biol 13, 1145–1149. [DOI] [PubMed] [Google Scholar]
- 22.Broekhuis JR, Verhey KJ, and Jansen G (2014). Regulation of cilium length and intraflagellar transport by the RCK-kinases ICK and MOK in renal epithelial cells. PLoS One 9, e108470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chaya T, Omori Y, Kuwahara R, and Furukawa T (2014). ICK is essential for cell type-specific ciliogenesis and the regulation of ciliary transport. EMBO J 33, 1227–1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moon H, Song J, Shin JO, Lee H, Kim HK, Eggenschwiller JT, Bok J, and Ko HW (2014). Intestinal cell kinase, a protein associated with endocrine-cerebro-osteodysplasia syndrome, is a key regulator of cilia length and Hedgehog signaling. Proc Natl Acad Sci USA 111, 8541–8546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Omori Y, Chaya T, Katoh K, Kajimura N, Sato S, Muraoka K, Ueno S, Koyasu T, Kondo M, and Furukawa T (2010). Negative regulation of ciliary length by ciliary male germ cell-associated kinase (Mak) is required for retinal photoreceptor survival. Proc Natl Acad Sci USA 107, 22671–22676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yi P, Xie C, and Ou G (2018). The kinases male germ cell-associated kinase and cell cycle-related kinase regulate kinesin-2 motility in Caenorhabditis elegans neuronal cilia. Traffic 19, 522–535. [DOI] [PubMed] [Google Scholar]
- 27.Phirke P, Efimenko E, Mohan S, Burghoorn J, Crona F, Bakhoum MW, Trieb M, Schuske K, Jorgensen EM, Piasecki BP, et al. (2011). Transcriptional profiling of C. elegans DAF-19 uncovers a ciliary base-associated protein and a CDK/CCRK/LF2p-related kinase required for intraflagellar transport. Dev Biol 357, 235–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hilton LK, Gunawardane K, Kim JW, Schwarz MC, and Quarmby LM (2013). The kinases LF4 and CNK2 control ciliary length by feedback regulation of assembly and disassembly rates. Curr Biol 23, 2208–2214. [DOI] [PubMed] [Google Scholar]
- 29.Ward S, Thomson N, White JG, and Brenner S (1975). Electron microscopical reconstruction of the anterior sensory anatomy of the nematode Caenorhabditis elegans. J Comp Neurol 160, 313–337. [DOI] [PubMed] [Google Scholar]
- 30.Mukhopadhyay S, Lu Y, Qin H, Lanjuin A, Shaham S, and Sengupta P (2007). Distinct IFT mechanisms contribute to the generation of ciliary structural diversity in C. elegans EMBO J. 26, 2966–2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Evans JE, Snow JJ, Gunnarson AL, Ou G, Stahlberg H, McDonald KL, and Scholey JM (2006). Functional modulation of IFT kinesins extends the sensory repertoire of ciliated neurons in Caenorhabditis elegans. J Cell Biol 172, 663–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hao L, Thein M, Brust-Mascher I, Civelekoglu-Scholey G, Lu Y, Acar S, Prevo B, Shaham S, and Scholey JM (2011). Intraflagellar transport delivers tubulin isotypes to sensory cilium middle and distal segments. Nat Cell Biol 13, 790–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Signor D, Wedaman KP, Rose LS, and Scholey JM (1999). Two heteromeric kinesin complexes in chemosensory neurons and sensory cilia of Caenorhabditis elegans. Mol Biol Cell 10, 345–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Snow JJ, Ou G, Gunnarson AL, Walker MR, Zhou HM, Brust-Mascher I, and Scholey JM (2004). Two anterograde intraflagellar transport motors cooperate to build sensory cilia on C. elegans neurons. Nat Cell Biol 6, 1109–1113. [DOI] [PubMed] [Google Scholar]
- 35.Ryan DA, Miller RM, Lee K, Neal SJ, Fagan KA, Sengupta P, and Portman DS (2014). Sex, age, and hunger regulate behavioral prioritization through dynamic modulation of chemoreceptor expression. Curr Biol 24, 2509–2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bacaj T, Tevlin M, Lu Y, and Shaham S (2008). Glia are essential for sensory organ function in C. elegans. Science 322, 744–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fu Z, Larson KA, Chitta RK, Parker SA, Turk BE, Lawrence MW, Kaldis P, Galaktionov K, Cohn SM, Shabanowitz J, et al. (2006). Identification of yin-yang regulators and a phosphorylation consensus for male germ cell-associated kinase (MAK)-related kinase. Mol Cell Biol 26, 8639–8654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang Y, Roine N, and Makela TP (2013). CCRK depletion inhibits glioblastoma cell proliferation in a cilium-dependent manner. EMBO Rep 14, 741–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fu Z, Schroeder MJ, Shabanowitz J, Kaldis P, Togawa K, Rustgi AK, Hunt DF, and Sturgill TW (2005). Activation of a nuclear Cdc2-related kinase within a mitogen-activated protein kinase-like TDY motif by autophosphorylation and cyclin-dependent protein kinase-activating kinase. Mol Cell Biol 25, 6047–6064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pan X, Ou G, Civelekoglu-Scholey G, Blacque OE, Endres NF, Tao L, Mogilner A, Leroux MR, Vale RD, and Scholey JM (2006). Mechanism of transport of IFT particles in C. elegans cilia by the concerted action of kinesin-II and OSM-3 motors. J Cell Biol 174, 1035–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Silva M, Morsci N, Nguyen KC, Rizvi A, Rongo C, Hall DH, and Barr MM (2017). Cell-specific alpha-tubulin isotype regulates ciliary microtubule ultrastructure, intraflagellar transport, and extracellular vesicle biology. Curr Biol 27, 968–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.O'Hagan R, Silva M, Nguyen KCQ, Zhang W, Bellotti S, Ramadan YH, Hall DH, and Barr MM (2017). Glutamylation regulates transport, specializes function, and sculpts the structure of cilia. Curr Biol 27, 3430–3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sirajuddin M, Rice LM, and Vale RD (2014). Regulation of microtubule motors by tubulin isotypes and post-translational modifications. Nat Cell Biol 16, 335–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sayas CL, Tortosa E, Bollati F, Ramirez-Rios S, Arnal I, and Avila J (2015). Tau regulates the localization and function of End-binding proteins 1 and 3 in developing neuronal cells. J Neurochem 133, 653–667. [DOI] [PubMed] [Google Scholar]
- 45.Nakata T, and Hirokawa N (2003). Microtubules provide directional cues for polarized axonal transport through interaction with kinesin motor head. J Cell Biol 162, 1045–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Richardson CE, Spilker KA, Cueva JG, Perrino J, Goodman MB, and Shen K (2014). PTRN-1, a microtubule minus end-binding CAMSAP homolog, promotes microtubule function in Caenorhabditis elegans neurons. Elife 3, e01498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Prodromou NV, Thompson CL, Osborn DP, Cogger KF, Ashworth R, Knight MM, Beales PL, and Chapple JP (2012). Heat shock induces rapid resorption of primary cilia. J Cell Sci 125, 4297–4305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weis F, Moullintraffort L, Heichette C, Chretien D, and Garnier C (2010). The 90-kDa heat shock protein Hsp90 protects tubulin against thermal denaturation. J Biol Chem 285, 9525–9534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hurd DD, Miller RM, Nunez L, and Portman DS (2010). Specific alpha- and beta-tubulin isotypes optimize the functions of sensory cilia in Caenorhabditis elegans. Genetics 185, 883–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bhogaraju S, Cajanek L, Fort C, Blisnick T, Weber K, Taschner M, Mizuno N, Lamla S, Bastin P, Nigg EA, et al. (2013). Molecular basis of tubulin transport within the cilium by IFT74 and IFT81. Science 341, 1009–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Taschner M, Weber K, Mourao A, Vetter M, Awasthi M, Stiegler M, Bhogaraju S, and Lorentzen E (2016). Intraflagellar transport proteins 172, 80, 57, 54, 38, and 20 form a stable tubulin-binding IFT-B2 complex. EMBO J 35, 773–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kobayashi T, Gengyo-Ando K, Ishihara T, Katsura I, and Mitani S (2007). IFT-81 and IFT-74 are required for intraflagellar transport in C. elegans. Genes Cells 12, 593–602. [DOI] [PubMed] [Google Scholar]
- 53.Wloga D, Joachimiak E, Louka P, and Gaertig J (2017). Posttranslational modifications of tubulin and cilia. Cold Spring Harb Perspect Biol 9, pii:a028159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Backer CB, Gutzman JH, Pearson CG, and Cheeseman IM (2012). CSAP localizes to polyglutamylated microtubules and promotes proper cilia function and zebrafish development. Mol Biol Cell 23, 2122–2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Larsch J, Flavell SW, Liu Q, Gordus A, Albrecht DR, and Bargmann CI (2015). A circuit for gradient climbing in C. elegans chemotaxis. Cell Rep 12, 1748–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Leterrier C, Vacher H, Fache MP, d'Ortoli SA, Castets F, Autillo-Touati A, and Dargent B (2011). End-binding proteins EB3 and EB1 link microtubules to ankyrin G in the axon initial segment. Proc Natl Acad Sci USA 108, 8826–8831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.O'Hagan R, Piasecki BP, Silva M, Phirke P, Nguyen KC, Hall DH, Swoboda P, and Barr MM (2011). The tubulin deglutamylase CCPP-1 regulates the function and stability of sensory cilia in C. elegans. Curr Biol 21, 1685–1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cao M, Meng D, Wang L, Bei S, Snell WJ, and Pan J (2013). Activation loop phosphorylation of a protein kinase is a molecular marker of organelle size that dynamically reports flagellar length. Proc Natl Acad Sci USA 110, 12337–12342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Song Y, and Brady ST (2015). Post-translational modifications of tubulin: pathways to functional diversity of microtubules. Trends Cell Biol 25, 125–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wu D, Chapman JR, Wang L, Harris TE, Shabanowitz J, Hunt DF, and Fu Z (2012). Intestinal cell kinase (ICK) promotes activation of mTOR complex 1 (mTORC1) through phosphorylation of Raptor Thr-908. J Biol Chem 287, 12510–12519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dan W, Gao N, Li L, Zhu JX, Diao L, Huang J, Han QJ, Wang S, Xue H, Wang Q, et al. (2018). alpha-tubulin acetylation restricts axon overbranching by dampening microtubule plus-dnd dynamics in neurons. Cerebr Cortex 28, 3332–3346. [DOI] [PubMed] [Google Scholar]
- 62.Topalidou I, Keller C, Kalebic N, Nguyen KC, Somhegyi H, Politi KA, Heppenstall P, Hall DH, and Chalfie M (2012). Genetically separable functions of the MEC-17 tubulin acetyltransferase affect microtubule organization. Curr Biol 22, 1057–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Howell K, and Hobert O (2017). Morphological diversity of C. elegans sensory cilia instructed by the differential expression of an immunoglobulin domain protein. Curr Biol 27, 1782–1790 [DOI] [PubMed] [Google Scholar]
- 64.Kim H, Ishidate T, Ghanta KS, Seth M, Conte D Jr., Shirayama M, and Mello CC (2014). A co-CRISPR strategy for efficient genome editing in Caenorhabditis elegans. Genetics 197, 1069–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Friedland AE, Tzur YB, Esvelt KM, Colaiacovo MP, Church GM, and Calarco JA (2013). Heritable genome editing in C. elegans via a CRISPR-Cas9 system. Nat Methods 10, 741–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cornils A, Maurya AK, Tereshko L, Kennedy J, Brear AG, Blacque OE, and Sengupta P (2016). Structural and functional recovery of sensory cilia in C. elegans IFT mutants upon aging. PLoS Genet 12, e1006325. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S4. List of strains used in this work. Related to the Key Resources Table