Abstract
Purpose:
Telomere length-associated SNPs have been associated with incidence and survival rates for malignant brain tumors such as glioma. Here, we study the influence of genetically determined lymphocyte telomere length (LTL) by comparing telomerase associated SNPs between the most common non-malignant brain tumor, i.e. meningioma, and healthy controls.
Methods/patients:
One thousand fifty-three (1053) surgically treated meningioma patients and 4437 controls of Western European ancestry were included. Germline DNA was genotyped for 8 SNPs previously significantly associated with LTL. Genotypically-estimated LTL was then calculated by summing each SNP’s genotypically-specified telomere length increase in base pairs (bp) for each person. Odds ratios for genotypically-estimated LTL in meningioma cases and controls were evaluated using logistic regression with the first two ancestral principal components and sex as covariates.
Results:
Three out of the eight evaluated LTL SNPs were significantly associated with increased meningioma risk (rs10936599: OR: 1.14, 95%-CI: 1.01–1.28, rs2736100: OR: 1.13, 95%-CI: 1.03–1.25, rs9420907: OR: 1.22, 95%-CI: 1.07–1.39). Only rs9420907 remained significant after correction for multiple testing. Average genotypically-estimated LTL was significantly longer for those with meningioma compared to controls [mean cases: 560.2 bp (standard error (SE): 4.05bp), mean controls: 541.5 bp (SE: 2.02bp), logistic regression p-value = 2.13×10−5].
Conclusion:
Increased genotypically-estimated LTL was significantly associated with increased meningioma risk. A role for telomere length in the pathophysiology of meningioma is novel, and could lead to new insights on the etiology of meningioma.
Keywords: Leukocyte telomere length, meningioma, mendelian randomization, risk
Introduction
The telomerase complex is a group of protein subunits which build repeat sequence “caps” at the end of chromosomes.[1, 2] These caps consist of “TTAGGG” sequences to maintain telomere length, sustain renewability of cells, and prevent apoptosis.[3, 4] Telomere length varies among individuals.[5] Longer leukocyte telomere length (LTL) has been associated with longer lifespan and may also play a role in ageing,[5, 6] while shorter LTL has been associated with various age-related diseases including type 2 diabetes and cardiovascular disease.[7–11] LTL is partially genotypically determined by polymorphisms in the genes that code for protein members of the telomerase enzyme complex.[5] Longer genotypically-estimated LTL has been associated with increased risk of various cancers including B-cell lymphoma and leukemia, and also non-hematopoietic tumors such as glioma and neuroblastoma.[12–19] LTL, measured in blood, is correlated with telomere length in other body tissues.[20]
The underlying genetic basis for the genesis and recurrence of meningioma is still relatively poorly understood, although variations in the 10p12.31 and 11p15.5 regions have been associated with increased meningioma risk.[21, 22] Germline mutations in the TERT gene (located on 5p15.33) and its promotor have been associated with several malignancies and intracranial tumors.[3, 4, 23–26] Somatic TERT promotor mutations in glioma have been associated with greater telomerase activity,[27] up-regulation of TERT,[24] and even decreased survival.[28] Somatic TERT promotor mutations have also been associated with malignant progression of meningiomas.[29, 30] Furthermore, increased TERT activity in meningioma has been associated with higher WHO grade and poorer outcome.[23] Given the suggestion that TERT genes may play a role in meningioma risk and progression, we utilized a targeted panel of telomerase genes to formally assess the association between impactful telomerase gene variants in a large sample of meningioma cases and controls.
Methods
Ethics statement:
This multi-center study was approved by the Yale University, Duke University, M.D.Anderson Cancer Center, the University of California, San Francisco, and the Brigham and Woman’s Hospital institutional review boards. Written informed consent was obtained from all study participants.
Lymphocyte telomere length:
Eight SNPs previously significantly associated with LTL[13] were assessed. These SNPs were located on genes ACYP2 (rs11125529), TERC (rs10936599), NAF1 (rs7675998), TERT (rs2736100), OBFC1 (rs9420907), CTC1 (rs3027234), ZNF08 (rs8105767), and RTEL1 (rs755017), and were compared between meningioma cases and controls individually, and also combined together within a weighted genetic risk score. Individual SNP tests were performed using SNPTEST v2.5.4 under an additive frequentist model with gender and the first two ancestral principal component included as covariates.[13, 31, 32]
The genotypically-estimated LTL was estimated by summing the number of base pairs per affected allele for each sample and could, therefore, range from 0 to 1215 bp. The number of base pairs per alternate allele are described in table 1. The difference in genotypically-estimated LTL was compared between the meningioma cases and controls using a two-sample t-test (Welch’s), box and whisker plots, and logistic regression with sex, the first two ancestral principal components, and the genotyping panel (Goldengate or Axiom, see below) as covariates. Odds ratios (ORs) were calculated per 50 bp increase of genotypically-estimated LTL. A separate model that also included age was constructed without controls form the Kaiser Permanente Research Program on Genes, Environment and Health (RPGEH) study as age was not available for these samples.
Table 1:
Number of base pair effect per allele included in the analysis based on data by the ENGAGE Consortium.[13]
| SNP | Chromosome | Gene | Effect allele | Base Pairs* | EAF cases (%) | EAF controls (%) | Allelic OR | 95%-CI | P-value** |
|---|---|---|---|---|---|---|---|---|---|
| rs11125529 | 2 | ACYP2 | A | 66.9 | 12.7 | 13.3 | 0.95 | 0.82–1.10 | 0.49 |
| rs10936599 | 3 | TERC | C | 117.3 | 24.4 | 22.2 | 1.14 | 1.01–1.28 | 0.02 |
| rs7675998 | 4 | NAF1 | G | 89.7 | 21.7 | 21.7 | 1.00 | 0.89–1.13 | 0.94 |
| rs2736100 | 5 | TERT | C | 94.2 | 52.3 | 49.0 | 1.13 | 1.03–1.25 | 0.01 |
| rs9420907 | 10 | OBFC1 | C | 82.8 | 16.7 | 14.1 | 1.22 | 1.07–1.39 | 0.003*** |
| rs3027234 | 17 | CTC1 | C | 25.2 | 21.7 | 22.3 | 0.97 | 0.86–1.08 | 0.55 |
| rs8105767 | 19 | ZNF208 | G | 57.6 | 30.4 | 28.7 | 1.08 | 0.97–1.20 | 0.15 |
| rs755017 | 20 | RTEL1 | G | 74.1 | 13.5 | 12.1 | 1.15 | 1.00–1.33 | 0.05 |
Number of base pairs the affected allele increases the genotypically-estimated leukocyte telomere length (LTL)[13, 32]
p-value for risk for meningioma under an additive frequentist model
Significant after correction for multiple testing (eight degrees of freedom).
Abbreviations: EAF: estimated allele frequency; bp: base pairs, OR: odds ratio, LTL: Leukocyte telomere length.
Subjects:
Subjects were non-Hispanic white ancestry to eliminate interference from population substructure. Two datasets were constructed and then merged together. The first dataset is a convenience sample of 244 persons who underwent surgery for an intracranial meningioma at Brigham and Women’s Hospital and 1141 controls from the San Francisco Adult Glioma Study which were sequenced for a panel of 953 telomere gene-related SNPs and were genotyped using Illumina GoldenGate® genotyping chemistry.[33] The second dataset consisted of 809 cases and 798 controls from the population-based Meningioma Consortium Case/Control Study[34] along with 2498 additional controls from the RPGEH study were genotyped using Affymetrix Axiom CEU World array as described elsewhere.[21, 35]
Imputation and merging datasets.
Both the Goldengate genotyped and Axiom genotyped subjects were imputed, separately, to ascertain all genotypes. The Goldengate array had rs11125529, rs10936599, rs2736100, rs9420907, rs3027234, and rs755017 on the array, and rs7675998 and rs8105767 were imputed. The Axiom array had all SNPs imputed (rs11125529, rs10936599, rs2736100, rs7675998, rs9420907, rs3027234, rs8105767, and rs755017). Imputation was performed using Minimac3 2.0.1 with the reference panel from the Haplotype Reference Consortium.[36] Imputation accuracy was determined by Pearson correlation (squared) between genotyped loci, and the same loci after being masked and imputed. A forest plot depicting the odds ratios (ORs) with 95%-CI of these 8 SNPs for meningioma case-control status was constructed in R. The data will be made available through dbGAP at a later point.
Results
The genotypes of the 8 SNPs in meningioma cases vs controls:
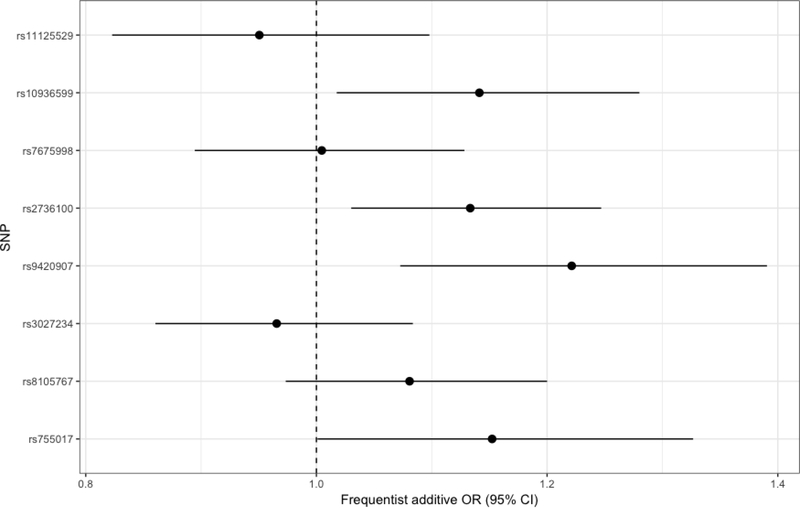
Three out of the eight evaluated SNPs were nominally significantly associated with increased meningioma risk (rs10936599: OR: 1.14, 95%-CI: 1.01–1.28, rs2736100: OR: 1.13, 95%-CI: 1.03–1.25, rs9420907: OR: 1.22, 95%-CI: 1.07–1.39, Table 1, Figure 1). Only rs9420907 remained significant with p=0.003 after application of Bonferroni correction based on 8 comparisons.
Figure 1: Forest plot depicting the association between the genotypically-estimated LTL associated SNPs and case-control status.
Odds ratios with confidence intervals for the association between the SNPs and meningioma case-control status were calculated under an additive frequentist model and were corrected for gender and the first two principal components. Abbreviations: SNP: Single nucleotide polymorphism; OR: odds ratio, CI: confidence interval.
Genotypically-estimated LTL:
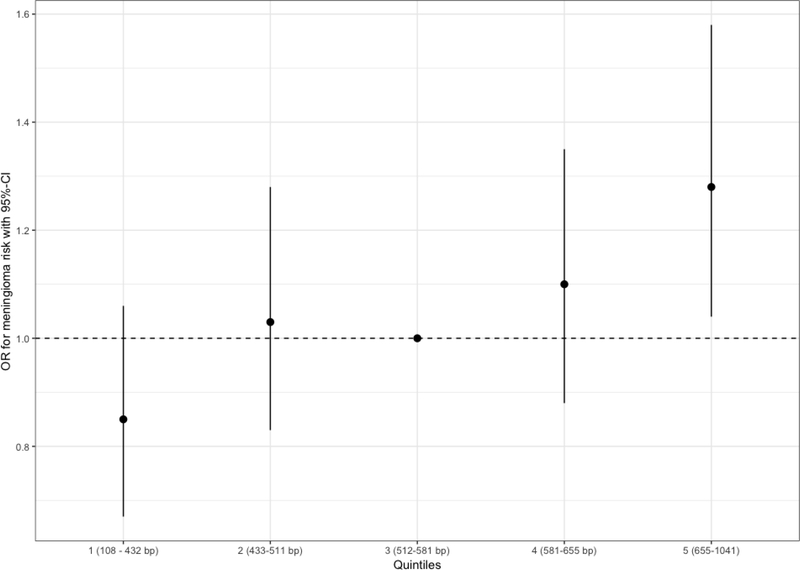
The mean genotypically-estimated LTL was 560.2 bp (standard error (SE): 4.05bp) for cases compared to 541.5 bp (SE: 2.02bp) for controls (p-value T-test: 3.62×10−5, OR per 50 bp increase in genotypically-estimated LTL: 1.06, 95%-CI: 1.03–1.09, logistic regression p-value = 2.13×10−5, Figure 2). Using quintiles of genotypically-estimated LTL based on controls, the odds ratio for meningioma increased with higher quintiles, with the highest quintile having significantly higher risk compared to the median quintile (OR: 1.28, 95%-CI: 1.04–1.58, p = 0.02, Figure 3). An additional model that also incorporated age constructed showed a very similar effect measure (OR per 50 bp increase in genotypically-estimated LTL: 1.06, 95%-CI: 1.03–1.10, logistic regression p-value = 7.31×10−5 with exclusion of RPGEH controls).
Figure 2: Boxplots for genotypically-estimated leukocyte telomere length and meningioma case-control status by sequencing panel.
The boxplots depicting the distribution of genotypically-estimated leukocyte telomere length (LTL) in meningioma patients and controls. Separate boxplots are depicted for the who different sample sources and overall. The p-values were 0.0009, 0.003, 2.13×10−5 for the telomere SNP panel, the meningioma consortium, and overall in a logistic regression model with gender and the first two ancestral principal components as covariates. The model for the overall analysis was also corrected for sequencing panel used.
Figure 3: Effect of increasing quintile of genotypically-estimated LTL on meningioma risk.
The odds ratios are relative to the median (third) quintile. Quintiles were defined based on genotypically-estimated LTL in controls. The vertical bars correspond to 95% confidence intervals. Odds ratios are corrected for the first two ancestral principal components, study, and sex.
When stratified by sex, the association was strongly significant in females and borderline significant in males (females: OR per 50 bp increase in genotypically-estimated LTL: 1.07, 95%-CI: 1.03–1.10, p-value = 9.58×10−5, males: OR per 50 bp increase in genotypically-estimated LTL: 1.04, 95%-CI: 1.00–1.09, p-value = 0.06). As rs2736100 had the lowest r2 of 0.53 (all other SNPs had an r2 greater than 0.8), the analysis was rerun using the remaining 7 SNPs. The association remained significant (OR per 50 bp increase in genotypically-estimated LTL: 1.05, 95%-CI: 1.02–1.08, p-value: 0.001). An analysis for the same 7 SNPs by sex showed that the association was significant in females but not in males (females: OR per 50 bp increase in genotypically-estimated LTL: 1.06, 95%-CI: 1.02–1.10, p-value = 0.002, males: OR per 50 bp increase in genotypically-estimated LTL: 1.03, 95%-CI: 0.98–1.09, p-value = 0.22).
Discussion
This is the first study to evaluate the relationship between genotypically-estimated LTL and meningioma risk, finding a positive association. Longer telomeres allow for more cell divisions before replicative senescence is reached and may therefore result in occurrence of mutations that allow cells to grow indefinitely and undergo malignant transformation.[19] Longer genotypically-estimated LTL based on the same SNPs used in this analysis has previously also been associated with increased risk of development of both glioma and chronic lymphocytic leukemia.[12, 37] However, longer LTL measured before ovarian cancer diagnosis has also been associated with a decreased risk of development of ovarian cancer.[38] Nevertheless, our finding that increased genotypically-estimated LTL is associated with meningioma case-control status is consistent with most other malignancies.
Reproducible genetic findings on meningioma were not reported until recent GWAS studies found associations on 10p12 and 11p15.[21, 22, 39] The alleles assessed for the current study do not have any linkage with those two known GWAS hits, nor do the known meningioma GWAS hits have any known impact on telomere length or function. The most significant gene here, OBFC1, is on chromosome 10q24, distant from the MLLT10 GWAS hit on 10p12. [21, 22] OBFC1 is part of the CST complex which helps to maintain telomere length.[40] This complex is also involved in DNA replication (DNA polymerase priming), and therefore a specific role in meningioma apart from telomere length maintenance is possible. The SNPs assessed here impart small effects which are not individually significant in any current GWAS analysis but collectively represent a phenotype contributing to genetic risk of meningioma. This analysis is a testament to the power of using combined genetic summary variables (such as polygenic risk scores and Mendelian randomization) to discover genetic based traits that impact risk of meningioma and other diseases.
This is the first study to evaluate telomere length for association with meningioma. The sample size is relatively large and the data primarily population-based, allowing for generalization of our results. The strength of using genetic variants to estimate LTL lies in the fact that genetics are determined at birth and are not influenced by external factors known to influence telomere length (e.g. age and smoking).[20, 41] Genotypically-estimated LTL does, therefore, not have to be controlled for confounding or reverse causation.
This study is also limited by several factors. Genotypically-estimated LTL is only a partial substitute for meningeal telomere length, explains a small proportion (approximately 1.23%) of variance in LTL, and is measured in leukocytes rather than meninges or their precursor cells.[13] Pleiotropy can never be truly excluded as the calculated genotypically-estimated LTL variable may reflect a different underlying disease process and act as a biomarker. It was not possible to do subgroup analyses for tumor location due to limitations of the data. The cases and controls in this study were all from Western European descent, which limits the implications of this study for other ethnicities. Therefore, further studies in other ethnicities are warranted, in particular for African Americans who suffer a 20% higher rate of meningioma compared to those of Western European descent.[42]
Acknowledgements:
The authors would like to thank all the participants in this study.
Disclosures: This work was supported by NIH R01 grants CA109468, CA109461, CA109745, CA109473, CA109475; CA52689, NIH R25 grant CA112355, NIH P50 grant CA097257, as well as by the Brain Science Foundation, the Meningioma Mommas, and the University of California, San Francisco Lewis Chair in Brain Tumor Research (MW).
Footnotes
Conflict of Interest: The authors declare that they have no conflict of interest.
Ethical approval: All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
References
- 1.Weinrich SL, Pruzan R, Ma L, Ouellette M, Tesmer VM, Holt SE, Bodnar AG, Lichtsteiner S, Kim NW, Trager JB, Taylor RD, Carlos R, Andrews WH, Wright WE, Shay JW, Harley CB, Morin GB (1997) Reconstitution of human telomerase with the template RNA component hTR and the catalytic protein subunit hTRT. Nat Genet 17: 498–502 doi: 10.1038/ng1297-498 [DOI] [PubMed] [Google Scholar]
- 2.Kirkpatrick KL, Mokbel K (2001) The significance of human telomerase reverse transcriptase (hTERT) in cancer. Eur J Surg Oncol 27: 754–760 doi: 10.1053/ejso.2001.1151 [DOI] [PubMed] [Google Scholar]
- 3.Griewank KG, Murali R, Puig-Butille JA, Schilling B, Livingstone E, Potrony M, Carrera C, Schimming T, Moller I, Schwamborn M, Sucker A, Hillen U, Badenas C, Malvehy J, Zimmer L, Scherag A, Puig S, Schadendorf D (2014) TERT promoter mutation status as an independent prognostic factor in cutaneous melanoma. J Natl Cancer Inst 106 doi: 10.1093/jnci/dju246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Heidenreich B, Rachakonda PS, Hemminki K, Kumar R (2014) TERT promoter mutations in cancer development. Curr Opin Genet Dev 24: 30–37 doi: 10.1016/j.gde.2013.11.005 [DOI] [PubMed] [Google Scholar]
- 5.Griffith JD, Comeau L, Rosenfield S, Stansel RM, Bianchi A, Moss H, de Lange T (1999) Mammalian telomeres end in a large duplex loop. Cell 97: 503–514 [DOI] [PubMed] [Google Scholar]
- 6.Kimura M, Hjelmborg JV, Gardner JP, Bathum L, Brimacombe M, Lu X, Christiansen L, Vaupel JW, Aviv A, Christensen K (2008) Telomere length and mortality: a study of leukocytes in elderly Danish twins. Am J Epidemiol 167: 799–806 doi: 10.1093/aje/kwm380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fitzpatrick AL, Kronmal RA, Gardner JP, Psaty BM, Jenny NS, Tracy RP, Walston J, Kimura M, Aviv A (2007) Leukocyte telomere length and cardiovascular disease in the cardiovascular health study. Am J Epidemiol 165: 14–21 doi: 10.1093/aje/kwj346 [DOI] [PubMed] [Google Scholar]
- 8.Adaikalakoteswari A, Balasubramanyam M, Mohan V (2005) Telomere shortening occurs in Asian Indian Type 2 diabetic patients. Diabet Med 22: 1151–1156 doi: 10.1111/j.1464-5491.2005.01574.x [DOI] [PubMed] [Google Scholar]
- 9.Haycock PC, Heydon EE, Kaptoge S, Butterworth AS, Thompson A, Willeit P (2014) Leucocyte telomere length and risk of cardiovascular disease: systematic review and meta-analysis. BMJ 349: g4227 doi: 10.1136/bmj.g4227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wilson WR, Herbert KE, Mistry Y, Stevens SE, Patel HR, Hastings RA, Thompson MM, Williams B (2008) Blood leucocyte telomere DNA content predicts vascular telomere DNA content in humans with and without vascular disease. Eur Heart J 29: 2689–2694 doi: 10.1093/eurheartj/ehn386 [DOI] [PubMed] [Google Scholar]
- 11.Hornsby PJ (2007) Telomerase and the aging process. Exp Gerontol 42: 575–581 doi: 10.1016/j.exger.2007.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Walsh KM, Codd V, Rice T, Nelson CP, Smirnov IV, McCoy LS, Hansen HM, Elhauge E, Ojha J, Francis SS, Madsen NR, Bracci PM, Pico AR, Molinaro AM, Tihan T, Berger MS, Chang SM, Prados MD, Jenkins RB, Wiemels JL, Group ECT, Samani NJ, Wiencke JK, Wrensch MR (2015) Longer genotypically-estimated leukocyte telomere length is associated with increased adult glioma risk. Oncotarget 6: 42468–42477 doi: 10.18632/oncotarget.6468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Codd V, Nelson CP, Albrecht E, Mangino M, Deelen J, Buxton JL, Hottenga JJ, Fischer K, Esko T, Surakka I, Broer L, Nyholt DR, Mateo Leach I, Salo P, Hagg S, Matthews MK, Palmen J, Norata GD, O’Reilly PF, Saleheen D, Amin N, Balmforth AJ, Beekman M, de Boer RA, Bohringer S, Braund PS, Burton PR, de Craen AJ, Denniff M, Dong Y, Douroudis K, Dubinina E, Eriksson JG, Garlaschelli K, Guo D, Hartikainen AL, Henders AK, Houwing-Duistermaat JJ, Kananen L, Karssen LC, Kettunen J, Klopp N, Lagou V, van Leeuwen EM, Madden PA, Magi R, Magnusson PK, Mannisto S, McCarthy MI, Medland SE, Mihailov E, Montgomery GW, Oostra BA, Palotie A, Peters A, Pollard H, Pouta A, Prokopenko I, Ripatti S, Salomaa V, Suchiman HE, Valdes AM, Verweij N, Vinuela A, Wang X, Wichmann HE, Widen E, Willemsen G, Wright MJ, Xia K, Xiao X, van Veldhuisen DJ, Catapano AL, Tobin MD, Hall AS, Blakemore AI, van Gilst WH, Zhu H, consortium CA, Erdmann J, Reilly MP, Kathiresan S, Schunkert H, Talmud PJ, Pedersen NL, Perola M, Ouwehand W, Kaprio J, Martin NG, van Duijn CM, Hovatta I, Gieger C, Metspalu A, Boomsma DI, Jarvelin MR, Slagboom PE, Thompson JR, Spector TD, van der Harst P, Samani NJ (2013) Identification of seven loci affecting mean telomere length and their association with disease. Nat Genet 45: 422–427, 427e421–422 doi: 10.1038/ng.2528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Machiela MJ, Lan Q, Slager SL, Vermeulen RC, Teras LR, Camp NJ, Cerhan JR, Spinelli JJ, Wang SS, Nieters A, Vijai J, Yeager M, Wang Z, Ghesquieres H, McKay J, Conde L, de Bakker PI, Cox DG, Burdett L, Monnereau A, Flowers CR, De Roos AJ, Brooks-Wilson AR, Giles GG, Melbye M, Gu J, Jackson RD, Kane E, Purdue MP, Vajdic CM, Albanes D, Kelly RS, Zucca M, Bertrand KA, Zeleniuch-Jacquotte A, Lawrence C, Hutchinson A, Zhi D, Habermann TM, Link BK, Novak AJ, Dogan A, Asmann YW, Liebow M, Thompson CA, Ansell SM, Witzig TE, Tilly H, Haioun C, Molina TJ, Hjalgrim H, Glimelius B, Adami HO, Roos G, Bracci PM, Riby J, Smith MT, Holly EA, Cozen W, Hartge P, Morton LM, Severson RK, Tinker LF, North KE, Becker N, Benavente Y, Boffetta P, Brennan P, Foretova L, Maynadie M, Staines A, Lightfoot T, Crouch S, Smith A, Roman E, Diver WR, Offit K, Zelenetz A, Klein RJ, Villano DJ, Zheng T, Zhang Y, Holford TR, Turner J, Southey MC, Clavel J, Virtamo J, Weinstein S, Riboli E, Vineis P, Kaaks R, Boeing H, Tjonneland A, Angelucci E, Di Lollo S, Rais M, De Vivo I, Giovannucci E, Kraft P, Huang J, Ma B, Ye Y, Chiu BC, Liang L, Park JH, Chung CC, Weisenburger DD, Fraumeni JF Jr., Salles G, Glenn M, Cannon-Albright L, Curtin K, Wu X, Smedby KE, de Sanjose S, Skibola CF, Berndt SI, Birmann BM, Chanock SJ, Rothman N (2016) Genetically predicted longer telomere length is associated with increased risk of B-cell lymphoma subtypes. Hum Mol Genet 25: 1663–1676 doi: 10.1093/hmg/ddw027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bojesen SE, Pooley KA, Johnatty SE, Beesley J, Michailidou K, Tyrer JP, Edwards SL, Pickett HA, Shen HC, Smart CE, Hillman KM, Mai PL, Lawrenson K, Stutz MD, Lu Y, Karevan R, Woods N, Johnston RL, French JD, Chen X, Weischer M, Nielsen SF, Maranian MJ, Ghoussaini M, Ahmed S, Baynes C, Bolla MK, Wang Q, Dennis J, McGuffog L, Barrowdale D, Lee A, Healey S, Lush M, Tessier DC, Vincent D, Bacot F, Australian Cancer S, Australian Ovarian Cancer S, Kathleen Cuningham Foundation Consortium for Research into Familial Breast C, Gene Environment I, Breast C, Swedish Breast Cancer S, Hereditary B, Ovarian Cancer Research Group N, Epidemiological study of B, Carriers BM, Genetic Modifiers of Cancer Risk in BMC, Vergote I, Lambrechts S, Despierre E, Risch HA, Gonzalez-Neira A, Rossing MA, Pita G, Doherty JA, Alvarez N, Larson MC, Fridley BL, Schoof N, Chang-Claude J, Cicek MS, Peto J, Kalli KR, Broeks A, Armasu SM, Schmidt MK, Braaf LM, Winterhoff B, Nevanlinna H, Konecny GE, Lambrechts D, Rogmann L, Guenel P, Teoman A, Milne RL, Garcia JJ, Cox A, Shridhar V, Burwinkel B, Marme F, Hein R, Sawyer EJ, Haiman CA, Wang-Gohrke S, Andrulis IL, Moysich KB, Hopper JL, Odunsi K, Lindblom A, Giles GG, Brenner H, Simard J, Lurie G, Fasching PA, Carney ME, Radice P, Wilkens LR, Swerdlow A, Goodman MT, Brauch H, Garcia-Closas M, Hillemanns P, Winqvist R, Durst M, Devilee P, Runnebaum I, Jakubowska A, Lubinski J, Mannermaa A, Butzow R, Bogdanova NV, Dork T, Pelttari LM, Zheng W, Leminen A, Anton-Culver H, Bunker CH, Kristensen V, Ness RB, Muir K, Edwards R, Meindl A, Heitz F, Matsuo K, du Bois A, Wu AH, Harter P, Teo SH, Schwaab I, Shu XO, Blot W, Hosono S, Kang D, Nakanishi T, Hartman M, Yatabe Y, Hamann U, Karlan BY, Sangrajrang S, Kjaer SK, Gaborieau V, Jensen A, Eccles D, Hogdall E, Shen CY, Brown J, Woo YL, Shah M, Azmi MA, Luben R, Omar SZ, Czene K, Vierkant RA, Nordestgaard BG, Flyger H, Vachon C, Olson JE, Wang X, Levine DA, Rudolph A, Weber RP, Flesch-Janys D, Iversen E, Nickels S, Schildkraut JM, Silva Idos S, Cramer DW, Gibson L, Terry KL, Fletcher O, Vitonis AF, van der Schoot CE, Poole EM, Hogervorst FB, Tworoger SS, Liu J, Bandera EV, Li J, Olson SH, Humphreys K, Orlow I, Blomqvist C, Rodriguez-Rodriguez L, Aittomaki K, Salvesen HB, Muranen TA, Wik E, Brouwers B, Krakstad C, Wauters E, Halle MK, Wildiers H, Kiemeney LA, Mulot C, Aben KK, Laurent-Puig P, Altena AM, Truong T, Massuger LF, Benitez J, Pejovic T, Perez JI, Hoatlin M, Zamora MP, Cook LS, Balasubramanian SP, Kelemen LE, Schneeweiss A, Le ND, Sohn C, Brooks-Wilson A, Tomlinson I, Kerin MJ, Miller N, Cybulski C, Henderson BE, Menkiszak J, Schumacher F, Wentzensen N, Le Marchand L, Yang HP, Mulligan AM, Glendon G, Engelholm SA, Knight JA, Hogdall CK, Apicella C, Gore M, Tsimiklis H, Song H, Southey MC, Jager A, den Ouweland AM, Brown R, Martens JW, Flanagan JM, Kriege M, Paul J, Margolin S, Siddiqui N, Severi G, Whittemore AS, Baglietto L, McGuire V, Stegmaier C, Sieh W, Muller H, Arndt V, Labreche F, Gao YT, Goldberg MS, Yang G, Dumont M, McLaughlin JR, Hartmann A, Ekici AB, Beckmann MW, Phelan CM, Lux MP, Permuth-Wey J, Peissel B, Sellers TA, Ficarazzi F, Barile M, Ziogas A, Ashworth A, Gentry-Maharaj A, Jones M, Ramus SJ, Orr N, Menon U, Pearce CL, Bruning T, Pike MC, Ko YD, Lissowska J, Figueroa J, Kupryjanczyk J, Chanock SJ, Dansonka-Mieszkowska A, Jukkola-Vuorinen A, Rzepecka IK, Pylkas K, Bidzinski M, Kauppila S, Hollestelle A, Seynaeve C, Tollenaar RA, Durda K, Jaworska K, Hartikainen JM, Kosma VM, Kataja V, Antonenkova NN, Long J, Shrubsole M, Deming-Halverson S, Lophatananon A, Siriwanarangsan P, Stewart-Brown S, Ditsch N, Lichtner P, Schmutzler RK, Ito H, Iwata H, Tajima K, Tseng CC, Stram DO, van den Berg D, Yip CH, Ikram MK, Teh YC, Cai H, Lu W, Signorello LB, Cai Q, Noh DY, Yoo KY, Miao H, Iau PT, Teo YY, McKay J, Shapiro C, Ademuyiwa F, Fountzilas G, Hsiung CN, Yu JC, Hou MF, Healey CS, Luccarini C, Peock S, Stoppa-Lyonnet D, Peterlongo P, Rebbeck TR, Piedmonte M, Singer CF, Friedman E, Thomassen M, Offit K, Hansen TV, Neuhausen SL, Szabo CI, Blanco I, Garber J, Narod SA, Weitzel JN, Montagna M, Olah E, Godwin AK, Yannoukakos D, Goldgar DE, Caldes T, Imyanitov EN, Tihomirova L, Arun BK, Campbell I, Mensenkamp AR, van Asperen CJ, van Roozendaal KE, Meijers-Heijboer H, Collee JM, Oosterwijk JC, Hooning MJ, Rookus MA, van der Luijt RB, Os TA, Evans DG, Frost D, Fineberg E, Barwell J, Walker L, Kennedy MJ, Platte R, Davidson R, Ellis SD, Cole T, Bressac-de Paillerets B, Buecher B, Damiola F, Faivre L, Frenay M, Sinilnikova OM, Caron O, Giraud S, Mazoyer S, Bonadona V, Caux-Moncoutier V, Toloczko-Grabarek A, Gronwald J, Byrski T, Spurdle AB, Bonanni B, Zaffaroni D, Giannini G, Bernard L, Dolcetti R, Manoukian S, Arnold N, Engel C, Deissler H, Rhiem K, Niederacher D, Plendl H, Sutter C, Wappenschmidt B, Borg A, Melin B, Rantala J, Soller M, Nathanson KL, Domchek SM, Rodriguez GC, Salani R, Kaulich DG, Tea MK, Paluch SS, Laitman Y, Skytte AB, Kruse TA, Jensen UB, Robson M, Gerdes AM, Ejlertsen B, Foretova L, Savage SA, Lester J, Soucy P, Kuchenbaecker KB, Olswold C, Cunningham JM, Slager S, Pankratz VS, Dicks E, Lakhani SR, Couch FJ, Hall P, Monteiro AN, Gayther SA, Pharoah PD, Reddel RR, Goode EL, Greene MH, Easton DF, Berchuck A, Antoniou AC, Chenevix-Trench G, Dunning AM (2013) Multiple independent variants at the TERT locus are associated with telomere length and risks of breast and ovarian cancer. Nat Genet 45: 371–384, 384e371–372 doi: 10.1038/ng.2566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Walsh KM, Whitehead TP, de Smith AJ, Smirnov IV, Park M, Endicott AA, Francis SS, Codd V, Group ECT, Samani NJ, Metayer C, Wiemels JL (2016) Common genetic variants associated with telomere length confer risk for neuroblastoma and other childhood cancers. Carcinogenesis 37: 576–582 doi: 10.1093/carcin/bgw037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang S, Chen Y, Qu F, He S, Huang X, Jiang H, Jin T, Wan S, Xing J (2014) Association between leukocyte telomere length and glioma risk: a case-control study. Neuro Oncol 16: 505–512 doi: 10.1093/neuonc/not240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Walsh KM, Wiencke JK, Lachance DH, Wiemels JL, Molinaro AM, Eckel-Passow JE, Jenkins RB, Wrensch MR (2015) Telomere maintenance and the etiology of adult glioma. Neuro Oncol 17: 1445–1452 doi: 10.1093/neuonc/nov082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Telomeres Mendelian Randomization C, Haycock PC, Burgess S, Nounu A, Zheng J, Okoli GN, Bowden J, Wade KH, Timpson NJ, Evans DM, Willeit P, Aviv A, Gaunt TR, Hemani G, Mangino M, Ellis HP, Kurian KM, Pooley KA, Eeles RA, Lee JE, Fang S, Chen WV, Law MH, Bowdler LM, Iles MM, Yang Q, Worrall BB, Markus HS, Hung RJ, Amos CI, Spurdle AB, Thompson DJ, O’Mara TA, Wolpin B, Amundadottir L, Stolzenberg-Solomon R, Trichopoulou A, Onland-Moret NC, Lund E, Duell EJ, Canzian F, Severi G, Overvad K, Gunter MJ, Tumino R, Svenson U, van Rij A, Baas AF, Bown MJ, Samani NJ, van t’Hof FNG, Tromp G, Jones GT, Kuivaniemi H, Elmore JR, Johansson M, McKay J, Scelo G, Carreras-Torres R, Gaborieau V, Brennan P, Bracci PM, Neale RE, Olson SH, Gallinger S, Li D, Petersen GM, Risch HA, Klein AP, Han J, Abnet CC, Freedman ND, Taylor PR, Maris JM, Aben KK, Kiemeney LA, Vermeulen SH, Wiencke JK, Walsh KM, Wrensch M, Rice T, Turnbull C, Litchfield K, Paternoster L, Standl M, Abecasis GR, SanGiovanni JP, Li Y, Mijatovic V, Sapkota Y, Low SK, Zondervan KT, Montgomery GW, Nyholt DR, van Heel DA, Hunt K, Arking DE, Ashar FN, Sotoodehnia N, Woo D, Rosand J, Comeau ME, Brown WM, Silverman EK, Hokanson JE, Cho MH, Hui J, Ferreira MA, Thompson PJ, Morrison AC, Felix JF, Smith NL, Christiano AM, Petukhova L, Betz RC, Fan X, Zhang X, Zhu C, Langefeld CD, Thompson SD, Wang F, Lin X, Schwartz DA, Fingerlin T, Rotter JI, Cotch MF, Jensen RA, Munz M, Dommisch H, Schaefer AS, Han F, Ollila HM, Hillary RP, Albagha O, Ralston SH, Zeng C, Zheng W, Shu XO, Reis A, Uebe S, Huffmeier U, Kawamura Y, Otowa T, Sasaki T, Hibberd ML, Davila S, Xie G, Siminovitch K, Bei JX, Zeng YX, Forsti A, Chen B, Landi S, Franke A, Fischer A, Ellinghaus D, Flores C, Noth I, Ma SF, Foo JN, Liu J, Kim JW, Cox DG, Delattre O, Mirabeau O, Skibola CF, Tang CS, Garcia-Barcelo M, Chang KP, Su WH, Chang YS, Martin NG, Gordon S, Wade TD, Lee C, Kubo M, Cha PC, Nakamura Y, Levy D, Kimura M, Hwang SJ, Hunt S, Spector T, Soranzo N, Manichaikul AW, Barr RG, Kahali B, Speliotes E, Yerges-Armstrong LM, Cheng CY, Jonas JB, Wong TY, Fogh I, Lin K, Powell JF, Rice K, Relton CL, Martin RM, Davey Smith G (2017) Association Between Telomere Length and Risk of Cancer and Non-Neoplastic Diseases: A Mendelian Randomization Study. JAMA Oncol 3: 636–651 doi: 10.1001/jamaoncol.2016.5945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Daniali L, Benetos A, Susser E, Kark JD, Labat C, Kimura M, Desai K, Granick M, Aviv A (2013) Telomeres shorten at equivalent rates in somatic tissues of adults. Nat Commun 4: 1597 doi: 10.1038/ncomms2602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Claus EB, Cornish AJ, Broderick P, Schildkraut JM, Dobbins SE, Holroyd A, Calvocoressi L, Lu L, Hansen HM, Smirnov I, Walsh KM, Schramm J, Hoffmann P, Nothen MM, Jockel KH, Swerdlow A, Larsen SB, Johansen C, Simon M, Bondy M, Wrensch M, Houlston R, Wiemels JL (2018) Genome-wide association analysis identifies a meningioma risk locus at 11p15.5. Neuro Oncol doi: 10.1093/neuonc/noy077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dobbins SE, Broderick P, Melin B, Feychting M, Johansen C, Andersson U, Brannstrom T, Schramm J, Olver B, Lloyd A, Ma YP, Hosking FJ, Lonn S, Ahlbom A, Henriksson R, Schoemaker MJ, Hepworth SJ, Hoffmann P, Muhleisen TW, Nothen MM, Moebus S, Eisele L, Kosteljanetz M, Muir K, Swerdlow A, Simon M, Houlston RS (2011) Common variation at 10p12.31 near MLLT10 influences meningioma risk. Nat Genet 43: 825–827 doi: 10.1038/ng.879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Langford LA, Piatyszek MA, Xu R, Schold SC Jr., Wright WE, Shay JW (1997) Telomerase activity in ordinary meningiomas predicts poor outcome. Hum Pathol 28: 416–420 [DOI] [PubMed] [Google Scholar]
- 24.Heidenreich B, Rachakonda PS, Hosen I, Volz F, Hemminki K, Weyerbrock A, Kumar R (2015) TERT promoter mutations and telomere length in adult malignant gliomas and recurrences. Oncotarget 6: 10617–10633 doi: 10.18632/oncotarget.3329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eckel-Passow JE, Lachance DH, Molinaro AM, Walsh KM, Decker PA, Sicotte H, Pekmezci M, Rice T, Kosel ML, Smirnov IV, Sarkar G, Caron AA, Kollmeyer TM, Praska CE, Chada AR, Halder C, Hansen HM, McCoy LS, Bracci PM, Marshall R, Zheng S, Reis GF, Pico AR, O’Neill BP, Buckner JC, Giannini C, Huse JT, Perry A, Tihan T, Berger MS, Chang SM, Prados MD, Wiemels J, Wiencke JK, Wrensch MR, Jenkins RB (2015) Glioma Groups Based on 1p/19q, IDH, and TERT Promoter Mutations in Tumors. N Engl J Med 372: 2499–2508 doi: 10.1056/NEJMoa1407279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Furtjes G, Kochling M, Peetz-Dienhart S, Wagner A, Hess K, Hasselblatt M, Senner V, Stummer W, Paulus W, Brokinkel B (2016) hTERT promoter methylation in meningiomas and central nervous hemangiopericytomas. J Neurooncol 130: 79–87 doi: 10.1007/s11060-016-2226-6 [DOI] [PubMed] [Google Scholar]
- 27.Arita H, Narita Y, Fukushima S, Tateishi K, Matsushita Y, Yoshida A, Miyakita Y, Ohno M, Collins VP, Kawahara N, Shibui S, Ichimura K (2013) Upregulating mutations in the TERT promoter commonly occur in adult malignant gliomas and are strongly associated with total 1p19q loss. Acta Neuropathol 126: 267–276 doi: 10.1007/s00401-013-1141-6 [DOI] [PubMed] [Google Scholar]
- 28.Arita H, Yamasaki K, Matsushita Y, Nakamura T, Shimokawa A, Takami H, Tanaka S, Mukasa A, Shirahata M, Shimizu S, Suzuki K, Saito K, Kobayashi K, Higuchi F, Uzuka T, Otani R, Tamura K, Sumita K, Ohno M, Miyakita Y, Kagawa N, Hashimoto N, Hatae R, Yoshimoto K, Shinojima N, Nakamura H, Kanemura Y, Okita Y, Kinoshita M, Ishibashi K, Shofuda T, Kodama Y, Mori K, Tomogane Y, Fukai J, Fujita K, Terakawa Y, Tsuyuguchi N, Moriuchi S, Nonaka M, Suzuki H, Shibuya M, Maehara T, Saito N, Nagane M, Kawahara N, Ueki K, Yoshimine T, Miyaoka E, Nishikawa R, Komori T, Narita Y, Ichimura K (2016) A combination of TERT promoter mutation and MGMT methylation status predicts clinically relevant subgroups of newly diagnosed glioblastomas. Acta Neuropathol Commun 4: 79 doi: 10.1186/s40478-016-0351-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goutagny S, Nault JC, Mallet M, Henin D, Rossi JZ, Kalamarides M (2014) High incidence of activating TERT promoter mutations in meningiomas undergoing malignant progression. Brain Pathol 24: 184–189 doi: 10.1111/bpa.12110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen HJ, Liang CL, Lu K, Lin JW, Cho CL (2000) Implication of telomerase activity and alternations of telomere length in the histologic characteristics of intracranial meningiomas. Cancer 89: 2092–2098 [DOI] [PubMed] [Google Scholar]
- 31.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D (2006) Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet 38: 904–909 doi: 10.1038/ng1847 [DOI] [PubMed] [Google Scholar]
- 32.Mangino M, Hwang SJ, Spector TD, Hunt SC, Kimura M, Fitzpatrick AL, Christiansen L, Petersen I, Elbers CC, Harris T, Chen W, Srinivasan SR, Kark JD, Benetos A, El Shamieh S, Visvikis-Siest S, Christensen K, Berenson GS, Valdes AM, Vinuela A, Garcia M, Arnett DK, Broeckel U, Province MA, Pankow JS, Kammerer C, Liu Y, Nalls M, Tishkoff S, Thomas F, Ziv E, Psaty BM, Bis JC, Rotter JI, Taylor KD, Smith E, Schork NJ, Levy D, Aviv A (2012) Genome-wide meta-analysis points to CTC1 and ZNF676 as genes regulating telomere homeostasis in humans. Hum Mol Genet 21: 5385–5394 doi: 10.1093/hmg/dds382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wrensch M, Jenkins RB, Chang JS, Yeh RF, Xiao Y, Decker PA, Ballman KV, Berger M, Buckner JC, Chang S, Giannini C, Halder C, Kollmeyer TM, Kosel ML, LaChance DH, McCoy L, O’Neill BP, Patoka J, Pico AR, Prados M, Quesenberry C, Rice T, Rynearson AL, Smirnov I, Tihan T, Wiemels J, Yang P, Wiencke JK (2009) Variants in the CDKN2B and RTEL1 regions are associated with high-grade glioma susceptibility. Nat Genet 41: 905–908 doi: 10.1038/ng.408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Claus E, Calvocoressi L, Schildkraut J, Walsh K, Hansen H, Smirnov I, McCoy L, Lu L, Ma X, Bondy M, Wrensch M, Wiemels J (2016) MNGO-11. REPORT FROM THE MENINGIOMA CONSORTIUM: CONFIRMATION OF A MENINGIOMA RISK LOCUS AT 10p12. Neuro-Oncology 18: vi103–vi103 doi: 10.1093/neuonc/now212.432 [DOI] [Google Scholar]
- 35.Wiemels JL, Wrensch M, Sison JD, Zhou M, Bondy M, Calvocoressi L, Black PM, Yu H, Schildkraut JM, Claus EB (2011) Reduced allergy and immunoglobulin E among adults with intracranial meningioma compared to controls. Int J Cancer 129: 1932–1939 doi: 10.1002/ijc.25858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McCarthy S, Das S, Kretzschmar W, Delaneau O, Wood AR, Teumer A, Kang HM, Fuchsberger C, Danecek P, Sharp K, Luo Y, Sidore C, Kwong A, Timpson N, Koskinen S, Vrieze S, Scott LJ, Zhang H, Mahajan A, Veldink J, Peters U, Pato C, van Duijn CM, Gillies CE, Gandin I, Mezzavilla M, Gilly A, Cocca M, Traglia M, Angius A, Barrett JC, Boomsma D, Branham K, Breen G, Brummett CM, Busonero F, Campbell H, Chan A, Chen S, Chew E, Collins FS, Corbin LJ, Smith GD, Dedoussis G, Dorr M, Farmaki AE, Ferrucci L, Forer L, Fraser RM, Gabriel S, Levy S, Groop L, Harrison T, Hattersley A, Holmen OL, Hveem K, Kretzler M, Lee JC, McGue M, Meitinger T, Melzer D, Min JL, Mohlke KL, Vincent JB, Nauck M, Nickerson D, Palotie A, Pato M, Pirastu N, McInnis M, Richards JB, Sala C, Salomaa V, Schlessinger D, Schoenherr S, Slagboom PE, Small K, Spector T, Stambolian D, Tuke M, Tuomilehto J, Van den Berg LH, Van Rheenen W, Volker U, Wijmenga C, Toniolo D, Zeggini E, Gasparini P, Sampson MG, Wilson JF, Frayling T, de Bakker PI, Swertz MA, McCarroll S, Kooperberg C, Dekker A, Altshuler D, Willer C, Iacono W, Ripatti S, Soranzo N, Walter K, Swaroop A, Cucca F, Anderson CA, Myers RM, Boehnke M, McCarthy MI, Durbin R, Haplotype Reference C (2016) A reference panel of 64,976 haplotypes for genotype imputation. Nat Genet 48: 1279–1283 doi: 10.1038/ng.3643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ojha J, Codd V, Nelson CP, Samani NJ, Smirnov IV, Madsen NR, Hansen HM, de Smith AJ, Bracci PM, Wiencke JK, Wrensch MR, Wiemels JL, Walsh KM, Group ECT (2016) Genetic Variation Associated with Longer Telomere Length Increases Risk of Chronic Lymphocytic Leukemia. Cancer Epidemiol Biomarkers Prev 25: 1043–1049 doi: 10.1158/1055-9965.EPI-15-1329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang M, Prescott J, Poole EM, Rice MS, Kubzansky LD, Idahl A, Lundin E, De Vivo I, Tworoger SS (2017) Prediagnosis Leukocyte Telomere Length and Risk of Ovarian Cancer. Cancer Epidemiol Biomarkers Prev 26: 339–345 doi: 10.1158/1055-9965.EPI-16-0466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Egan KM, Baskin R, Nabors LB, Thompson RC, Olson JJ, Browning JE, Madden MH, Monteiro AN (2015) Brain tumor risk according to germ-line variation in the MLLT10 locus. Eur J Hum Genet 23: 132–134 doi: 10.1038/ejhg.2014.70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Casteel DE, Zhuang S, Zeng Y, Perrino FW, Boss GR, Goulian M, Pilz RB (2009) A DNA polymerase-{alpha}{middle dot}primase cofactor with homology to replication protein A-32 regulates DNA replication in mammalian cells. J Biol Chem 284: 5807–5818 doi: 10.1074/jbc.M807593200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huzen J, Wong LS, van Veldhuisen DJ, Samani NJ, Zwinderman AH, Codd V, Cawthon RM, Benus GF, van der Horst IC, Navis G, Bakker SJ, Gansevoort RT, de Jong PE, Hillege HL, van Gilst WH, de Boer RA, van der Harst P (2014) Telomere length loss due to smoking and metabolic traits. J Intern Med 275: 155–163 doi: 10.1111/joim.12149 [DOI] [PubMed] [Google Scholar]
- 42.Ostrom QT, Gittleman H, Liao P, Vecchione-Koval T, Wolinsky Y, Kruchko C, Barnholtz-Sloan JS (2017) CBTRUS Statistical Report: Primary brain and other central nervous system tumors diagnosed in the United States in 2010–2014. Neuro Oncol 19: v1–v88 doi: 10.1093/neuonc/nox158 [DOI] [PMC free article] [PubMed] [Google Scholar]