A hybrid technology combining endomembrane compartment isolation with large-scale glycomic analysis provides insights into structural polysaccharide and cell wall protein transport.
Abstract
The plant endomembrane system facilitates the transport of polysaccharides, associated enzymes, and glycoproteins through its dynamic pathways. Although enzymes involved in cell wall biosynthesis have been identified, little is known about the endomembrane-based transport of glycan components. This is partially attributed to technical challenges in biochemically determining polysaccharide cargo in specific vesicles. Here, we introduce a hybrid approach addressing this limitation. By combining vesicle isolation with a large-scale carbohydrate antibody arraying technique, we charted an initial large-scale map describing the glycome profile of the SYNTAXIN OF PLANTS61 (SYP61) trans-Golgi network compartment in Arabidopsis (Arabidopsis thaliana). A library of antibodies recognizing specific noncellulosic carbohydrate epitopes allowed us to identify a range of diverse glycans, including pectins, xyloglucans (XyGs), and arabinogalactan proteins in isolated vesicles. Changes in XyG- and pectin-specific epitopes in the cell wall of an Arabidopsis SYP61 mutant corroborate our findings. Our data provide evidence that SYP61 vesicles are involved in the transport and deposition of structural polysaccharides and glycoproteins. Adaptation of our methodology can enable studies characterizing the glycome profiles of various vesicle populations in plant and animal systems and their respective roles in glycan transport defined by subcellular markers, developmental stages, or environmental stimuli.
INTRODUCTION
The endomembrane system, a complex network of membrane-surrounded compartments, facilitates the transport of proteins and diverse cargo within a cell. In plants, the endomembrane system is essential for a myriad of functions including signaling, stress responses, cell wall formation, and plant growth and development (Surpin and Raikhel, 2004). While much has been accomplished in the discovery of protein cargo within endomembrane compartments (Parsons and Lilley, 2018), the elucidation of nonprotein cargo is still at its infancy. Recent insightful studies have shown that different post-Golgi transport vesicle populations contain distinct lipids (Wattelet-Boyer et al., 2016). However, beyond lipids, neither the metabolome nor the glycome profiles of specific plant endomembrane vesicles have been determined. The latter is particularly important, since glycan molecules are essential building blocks for the construction of the plant cell wall.
The cell wall, a complex macromolecular composite structure of polysaccharides, structural proteins, and other molecules, surrounds and protects plant cells and is essential for development, signal transduction, and disease resistance. This structure also plays an integral role in cell expansion, as its tensile resistance is the primary balancing mechanism against internal turgor pressure (Cosgrove, 2005, 2016). The structurally dynamic and heterogeneous primary walls of young plant cells are predominantly composed of cellulose microfibrils embedded in a matrix of pectin, hemicelluloses, and glycoproteins (McCann et al., 1992; Somerville et al., 2004; Burton et al., 2010). Although a number of cell wall biosynthetic enzymes have been identified, our understanding of how polysaccharide transport and assembly are facilitated by the endomembrane system is still elusive (Figure 1A).
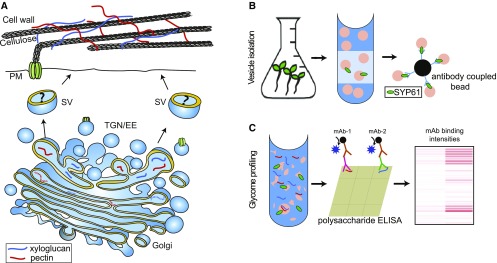
Figure 1.
Structural Polysaccharide Transport and Deposition, and Our Hybrid Methodology for Vesicle Glycomic Analysis.
(A) Schematic representation of structural polysaccharide synthesis, transport, and deposition.
The structural polysaccharides XyG and pectin are synthesized in the Golgi and transported via trans-Golgi–derived vesicles to the apoplast. The type of vesicles carrying specific polysaccharide cargo to the cell wall is unknown.
(B) Schematic representation of vesicle isolation and glycome identification. Plant extracts derived from liquid-grown plantlets are sucrose (Suc) fractionated. The enriched vesicles are isolated from the Golgi/trans-Golgi network–enriched Suc fractions with the aid of an Ab against a target protein. Green ellipsoids represent the SYP61 bait protein.
(C) Vesicle cargo release and glycome analysis. Vesicle cargo is released by sonication for glycome analysis. An ELISA-based method of glycome detection is used, and the resulting data are summarized in a heatmap for analysis. EE, early endosome; mAb, glycan-directed mAb; SV, secretory vesicle; TGN, trans-Golgi network.
Polysaccharides originate at distinct cellular locations; cellulose and callose are synthesized at the plasma membrane (PM), whereas the synthesis of hemicellulose and pectin and the glycosylation of proteins take place in the Golgi apparatus and the trans-Golgi Network (TGN; Atmodjo et al., 2013; Bashline et al., 2014; McFarlane et al., 2014; Nguema-Ona et al., 2014; Chou et al., 2015; Lund et al., 2015; Pauly and Keegstra, 2016; Lampugnani et al., 2018). Subsequently, cell wall polysaccharides, associated enzymes, and glycoproteins are carried to specific cell wall deposition sites by vesicle transport pathways, which remain poorly resolved (Driouich et al., 2012; Worden et al., 2012; Kim and Brandizzi, 2014; van de Meene et al., 2017). The highly dynamic nature of the endomembrane system makes it challenging to assign unequivocal roles to specific vesicle populations in the synthesis and assembly of the cell wall.
The TGN, the vesicle network on the trans-side of Golgi stacks, is responsible for sorting and packaging of cargo molecules for delivery to the PM and vacuoles (Roth et al., 1985; Griffiths and Simons, 1986; Kang et al., 2011; Rosquete et al., 2018), and thus represents a key intersection point of cell wall component sorting and transport (Sinclair et al., 2018). The function of the TGN is regulated by many molecular determinants including RAB GTPases, soluble N-ethylmaleimide–sensitive factor attachment protein receptors (SNAREs), tethers, and various structural and accessory proteins/regulators of pH homeostasis (Rosquete et al., 2018). SNARE proteins, including syntaxins, are required for vesicle fusion with the target membrane (Bombardier and Munson, 2015). Syntaxins are representing a subfamily of SNAREs residing in different compartments of the endomembrane pathway (Sanderfoot et al., 2000). Among these is the TGN-localized SYNTAXIN OF PLANTS61 (SYP61), which is proposed to form complexes with theVTI1-type v-SNARE VTI12 and either SYP41 or SYP42 at the TGN, and to play a key role in protein secretion to the PM (Sanderfoot et al., 2001; Li et al., 2017).
In our prior work, we established a vesicle immuno-isolation method for the characterization of vesicles displaying the TGN-localized SYP61 and determined their proteome (Drakakaki et al., 2012). Among the vesicle cargo were proteins involved in cell wall biosynthesis and metabolism, including cellulose synthases, linking SYP61 to the transport of cell wall components (Drakakaki et al., 2012). The study revealed the presence of TGN residents ECHIDNA (ECH) and the YPT/RAB GTPase-interacting protein (YIP) family members, which are implicated in the secretion of pectin and xyloglucan (XyG; Gendre et al., 2011, 2013; McFarlane et al., 2013). Taken together, these studies point to a critical role for SYP61 in post-Golgi trafficking and potentially in cell wall deposition.
Most of our current cell wall polysaccharide transport knowledge is derived from immunohistochemical electron microscopy analyses using a very limited number of antibodies (Abs) raised against polysaccharide epitopes (Moore et al., 1986, 1991; Moore and Staehelin, 1988; Lynch and Staehelin, 1992; McFarlane et al., 2008; Young et al., 2008; Kang et al., 2011). A seminal study in sycamore maple (Acer pseudoplatanus) cells points to an assembly line model, consisting of the initial biosynthesis of the XyG backbone followed by the addition of side chains in Golgi subcompartments, with the TGN largely exhibiting fully substituted XyG glycans (Zhang and Staehelin, 1992). Similarly, synthesis of the pectin backbone is thought to take place in the cis-medial Golgi, with subsequent completion in the medial cisternae (Zhang and Staehelin, 1992). However, in tobacco (Nicotiana tabacum) BY-2 cells, XyG biosynthesis was postulated to start at the cis-Golgi (Chevalier et al., 2010).
The type of vesicle used in polysaccharide transport varies with species, tissue, and cell differentiation pattern; this adds an extra layer of complexity to the dissection of polysaccharide transport (Lynch and Staehelin, 1992; Toyooka et al., 2009; Wang et al., 2017). So far, a limited number of subcellular markers have been associated with this process. In Arabidopsis (Arabidopsis thaliana), the Rab GTPase RABA4B localizes to a TGN subdomain containing fucosylated (FUC)-XyG and is overlapping with SYP61 (Kang et al., 2011). Based on immunostaining of tobacco BY-2 cells, the SECRETORY CARRIER MEMBRANE PROTEIN2 vesicles are implicated in pectin transport (Toyooka et al., 2009). Overall, following polysaccharide biosynthesis, Golgi secretory vesicles are involved in polysaccharide transport; however, their nature and cellular determinants remain largely unknown. This can be partially attributed to technical challenges in biochemically determining polysaccharide cargo in specific vesicles.
The development of methodologies that can allow large-scale analysis of glycans in isolated vesicles will clarify polysaccharide and glycoprotein transport. Numerous analytical approaches for determining carbohydrate composition at the organismal and organ scales are currently available (Nevins et al., 1967; Selvendran and O’Neill, 1987; Foster et al., 2010; Pettolino et al., 2012). These are often laborious and are unsuitable for analyzing cell wall polymers in endomembrane vesicles. An alternative technique, oligosaccharide mass profiling (OLIMP), utilizes specific glycosyl hydrolases to digest cell wall polysaccharides to soluble oligosaccharides detectable by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (Obel et al., 2009; Günl et al., 2010; Voiniciuc et al., 2018). Analyzing Arabidopsis Golgi-enriched microsomal fractions by OLIMP showed that in the Golgi apparatus, XyG oligosaccharides with a lower level of substitution are more abundant than XyG oligosaccharides with a higher degree of substitution (Obel et al., 2009; Günl et al., 2011). However, overall, this approach has limitations, since it is not possible to separate contributions of the Golgi or the endoplasmic reticulum (ER) from those of the TGN. The application of OLIMP to characterize specific vesicle populations has yet to be demonstrated. Fourier transform infrared spectroscopy can provide information about the overall composition of the cell wall, but it lacks the resolution to identify specific polymer types, particularly at the subcellular level (McCann et al., 1992; Badhan et al., 2017). Additionally, the fluorescent labeling and imaging of sugars with azido-containing fluorophores by the click reaction (Anderson et al., 2010, 2012; Wallace and Anderson, 2012; Hoogenboom et al., 2016; Wang et al., 2016) provides limited information regarding the substitutions of the glycosyl chains.
To date, no suitable glycomic approaches capture both the polysaccharide contents of specific vesicle populations and the detailed polysaccharide structures therein beyond a rough, low-resolution estimate. Cell wall glycan–directed Abs are an elegant alternative for the identification of plant cell carbohydrates (Moller et al., 2008; Pattathil et al., 2010; Pedersen et al., 2012). Monoclonal Abs (mAbs) can bind to their epitopes with high selectivity and affinity (Kd ∼10–6 M; Müller-Loennies et al., 2000; Bee et al., 2013) and are therefore highly sensitive, specific, and efficient probes. The latter aspect is of paramount importance when the quantity of the sample is severely limited, such as in the caseof endomembrane vesicles. In glycome profiling, Ab libraries are paired with an automated large-scale ELISA approach, allowing the fingerprinting of plant cell wall glycan samples with high sensitivity and a lower detection limit, falling within the range of ∼300 pg of crude carbohydrate material. The use of these Abs has yielded important insights into the biosynthesis, structure, and function of cell wall polymers and the spatiotemporal distribution of carbohydrates in species ranging from algae to Arabidopsis to biofuel feedstocks (Freshour et al., 1996; Willats et al., 1998; McCartney et al., 2005; Marcus et al., 2008; Moller et al., 2008; Pattathil et al., 2010, 2015a, 2015b; Ralet et al., 2010; Sørensen and Willats, 2011; Avci et al., 2012; Pedersen et al., 2012; Cornuault et al., 2015; Leroux et al., 2015; Raimundo et al., 2016; Peralta et al., 2017; Ruprecht et al., 2017) Thus, the exploitation of a wide collection of polysaccharide-directed mAbs has great utility in the dissection of polysaccharide transport and deposition at the subcellular level.
Facile methodologies allowing oligosaccharide analysis from subcellular compartments can provide vital information for charting individual transport routes controlling polysaccharide and glycoprotein deposition into the cell wall. Yet, polysaccharide profiles of specific isolated vesicle populations are not currently available. This leaves unanswered, long-standing questions regarding the identities of distinct secretory pathways carrying polysaccharides to the apoplast (Figure 1A). In our previous studies, proteomic analysis of SYP61 vesicles revealed the presence of cargo involved in cell wall biosynthesis and metabolism, including cellulose synthases, implicating SYP61 in cell wall component transport (Drakakaki et al., 2012). The present study represents the next step in our analysis, namely, the identification of the polysaccharide cargo in Golgi/TGN-enriched fractions and isolated SYP61 TGN vesicles.
By combining our vesicle isolation methodology with a large-scale automated carbohydrate Ab arraying methodology using an ELISA amplification step, we were able to chart an initial map describing the glycome profile of the SYP61 TGN vesicles. Screening of more than 155 carbohydrate epitopes revealed trafficking and sorting of diverse glycans of pectins, XyGs, and structural cell wall glycoproteins through this compartment. Our findings were corroborated via changes in XyG- and pectin-specific epitope labeling patterns in the Arabidopsis SYP61 mutant syp61/osm1 (osmotic stress-sensitive mutant1) versus that of the wild type. Consolidation of these data allowed us to formulate the hypothesis that SYP61 resides in a TGN compartment involved in the transport of structural polysaccharides. While this approach was used to assay SYP61 vesicles in this study, extending this methodology to other vesicle types and subcellular compartments can contribute to a more comprehensive understanding of the highly dynamic, regulated transport of carbohydrates and glycoproteins through the endomembrane system and of various vesicle populations in plant as well as animal systems.
RESULTS
Development of a Hybrid Method for Large-Scale Glycome Analysis of Endomembrane Vesicles
The similar physicochemical properties and sizes of secretory vesicles do not allow for a selective separation using standard Suc gradients, a shortcoming that an immuno-isolation approach can overcome with the aid of a bait protein on the vesicle surface (Drakakaki et al., 2012). Furthermore, the currently used analytical methods for carbohydrates are unsuitable for profiling vesicle populations; thus, ELISA using carbohydrate Abs is a promising method for overcoming this limitation (Pattathil et al., 2012).
In an effort to develop a high-resolution glycome profile of a defined population of plant vesicles, we developed a hybrid method taking advantage of two complementary approaches: vesicle immuno-isolation and large-scale glycomic ELISA, using a library of Abs recognizing specific noncellulosic carbohydrate epitopes (Figures 1B and 1C). In our proof-of-concept study, we used vesicles characterized by SYP61, based on the hypothesis that the SYP61 pathway is involved in protein secretion and the trafficking of cell wall components (Drakakaki et al., 2012; Li et al., 2017). SYP61 vesicles were separated from Arabidopsis plants expressing SYP61:CFP-SYP61 (cyan fluorescent protein) using a two-step procedure comprising Suc gradient fractionation of a Golgi/TGN-enriched fraction, followed by immuno-purification with Abs against green fluorescent protein (GFP) that recognize the CFP protein, as previously described in detail (Figure 1B; Drakakaki et al., 2012; Park and Drakakaki, 2014).
We used two independent matrixes (magnetic and nonmagnetic agarose beads) for GFP-Ab coupling in our vesicle isolation step (Figures 2 and 3; Supplemental Figures 1 to 3; Supplemental Data Sets 1 and 2). In one approach, we used magnetic agarose beads coupled to anti- GFP Abs (GFP-Trap_MA, ChromoTek) and compared the three replicates of immuno-isolated SYP61 vesicles against three independent types of negative controls to demonstrate the reproducibility of the SYP61 vesicle glycome profiling. Wild type Col-0 and CFP-SYP61 seedlings were subjected to all steps of isolation, while an additional set of controls using non-Ab–coupled beads (immunoprecipitation [IP] control; Figure 2; Supplemental Data Set 1B) was used to address and rule out potential unspecific vesicle binding. Cumulatively, these controls included (1) the wild type Col-0 TGN/Golgi-enriched Suc gradient fractions immuno-purified against GFP-Trap_MA (Figure 2B, Col-0 isolates) and (controls 2 and 3) CFP-SYP61 and the wild type Col-0 TGN/Golgi-enriched fractions immuno-isolated against magnetic agarose beads not coupled to GFP Abs (IP control; Figure 2B). Each of these preparations was performed in triplicate (Figure 2; Supplemental Data Set 1B). The purity of the isolated vesicles was verified using the ER marker, the prevacuolar compartment marker SYP21, and the PM H+ATPase, detecting for possible ER, prevacuolar compartment, and PM contaminants that can potentially be present in the TGN/Golgi Suc-enriched fraction (Supplemental Figure 1). These contaminants were eliminated in the isolated vesicles (Supplemental Figure 1), as previously shown (Drakakaki et al., 2012), which is a major advantage of immuno-isolation over a simple general Suc gradient that has been commonly used for organelle purification of Golgi/TGN.
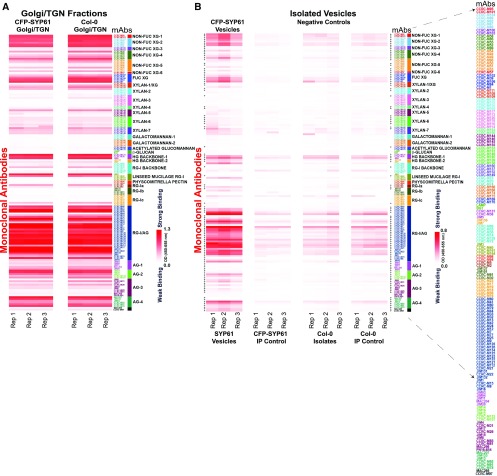
Figure 2.
Glycome Profiles of Golgi/TGN-Enriched Fraction and SYP61 Isolated Vesicles.
(A) and (B) Glycome profiling of Golgi/TGN-enriched fractions of CFP-SYP61 and wild type Col-0 (A) and the SYP61 isolated vesicles (B). Interrogation with 155 plant cell wall glycan-directed mAbs of the Golgi/TGN Suc fraction prior to vesicle immuno-isolation (A), and the isolated SYP61 vesicles and negative controls (B). White-to-red scales indicate signal intensity in the ELISAs, with white corresponding to no binding and red to strong binding. Enlarge on screen to view mAbs IDs within each Ab cluster. Heatmaps are a visual representation of the Ab binding intensities represented in Supplemental Data Set 1.
(A) Glycome profiles for the Golgi/TGN fractions of CFP-SYP61 and wild type Col-0 plants.
(B) Glycome analysis of SYP61 vesicles compared with negative controls. Three types of negative controls were included to verify the significance of the results.
SYP61 vesicles, vesicles isolated with the aid of GFP-magnetic agarose beads from CFP-SYP61 Golgi/TGN fractions. Negative controls of vesicle isolation: SYP61 IP control, vesicles isolated from CFP-SYP61 TGN/Golgi fraction with non–Ab-coupled magnetic agarose beads. Col-0 isolates, isolates with the aid of GFP-magnetic agarose beads from the wild type Col-0 Golgi/TGN fractions. Col-0 IP control, isolates from the wild type Col-0 TGN/Golgi fraction with non–Ab-coupled magnetic agarose beads. Rep, biological replicates of independently grown seedling sets.
The cross symbols in (B) adjacent to the heatmap indicate statistically significant enrichment of glycans in isolated vesicles compared with the negative controls as shown in Supplemental Data Set 3. The list of mAbs is enlarged to the right.
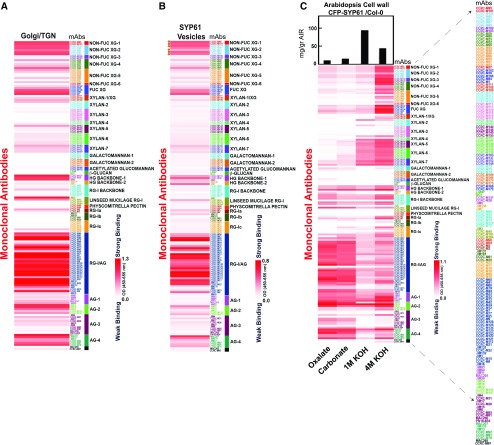
Figure 3.
Glycome Profiles of TGN, Isolated SYP61 Vesicles, and CFP-SYP61 Cell Walls.
(A) to (C) Interrogation of CFP-SYP61 Golgi/TGN (A), isolated CFP-SYP61 vesicles (B), and Arabidopsis CFP-SYP61 seedling cell walls (C) with 155 plant cell wall glycan-directed mAbs.The mean of three biological replicates is shown.
Data in (B) represent differential heatmaps, with background from all negative control vesicles subtracted to demonstrate polysaccharide enrichment in vesicles. Galactosylated XyG epitopes (XLXG, XXLG, and XLLG) recognized by the mAbs CCRC-M87, CCRC-M88, CCRC-M93, CCRC-M95, CCRC-M101, and CCRC-M104 are indicated by circles. Statistical significance of each Ab binding compared with all the negative controls is shown in Supplemental Data Set 3.
(C) Glycome profiling of CFP-SYP61 seedling cell walls. Sequentially extracted cell wall material derived from wild type Col-0 plants expressing CFP-SYP61 was analyzed using glycome profiling with the glycan-directed mAbs. Bars at the top indicate milligrams per gram of cell wall AIR. In each lane, 0.3 µg of Glc equivalent amounts of polysaccharides was applied. Labels at the bottom specify the different fractions assayed. Accompanied data are presented in Supplemental Data Set 4.
White-to-red scales indicate signal intensity in the ELISAs, with white corresponding to no binding and red to strong binding. Enlarge on screen to view mAbs IDs within each Ab cluster.The list of mAbs is enlarged to the right.
The Glycan Profile of the SYP61 Isolated Vesicles and the Robustness of the Approach
To identify the glycans that are present in SYP61 isolated vesicles, we used an ELISA-based glycome assay using a comprehensive set of 155 cell wall mAbs directed against diverse noncellulosic polysaccharide epitopes. With this approach, we charted the extensive glycome profiles of Golgi/TGN-enriched fractions and the isolated vesicles (Figures 2A and 2B; Supplemental Data Sets 1A and 1B). The heatmaps in Figure 2 are a visual representation of the ELISA intensities of polysaccharide Ab binding; the numerical values are provided in Supplemental Data Set 1. The mAbs were grouped in 32 distinct clusters, with each group representing epitopes within a particular polysaccharide class that are shared by that group of mAbs, as indicated by a color code on the right side of each heatmap (Pattathil et al., 2010, 2012). As shown in Figure 2A, a diverse array of polysaccharide glycans are present in the Suc gradient–enriched Golgi/TGN fractions, including highly substituted and less substituted XyGs, pectins, arabinogalactan proteins (AGPs), and to a lesser extent xylans and galactomannans. Comparison of the glycomes between the CFP-SYP61 and Col-0 Golgi/TGN Suc fractions using linear regression analysis illustrated the robustness of the TGN/Golgi isolation and glycan profiling procedure, and it further confirmed that the CFP-SYP61 fusion protein does not affect the glycan profiles (Figure 2A; Supplemental Figure 4A).
The sensitivity of the ELISA/glycome approach allowed us to chart the profile of the isolated SYP61 vesicles. Interestingly, the isolated SYP61 vesicles included a diverse array of both structural polysaccharides, namely, XyGs and pectins, as well as those of AGP glycans. Notably, FUC-XyG, as well as non-FUC, less substituted epitopes of XyG, were present in the SYP61 vesicle population (Figure 2B; Supplemental Data Set 1B) and are represented by different mAb clusters in the heatmap. In addition, pectin epitopes, including homogalacturonan (HG) backbones and a diverse array of complex glycans, represented by rhamnogalacturonan I (RG-I)/ arabinogalactan (AG) mAb clusters were detected in the isolated vesicles (Figure 2B; Supplemental Data Set 1B). To address the statistical significance of the identified glycans in the isolated SYP61 vesicles, we analyzed the intensity values of Ab binding in the isolated vesicles compared with all of the negative controls (nine repeats) using one-way analysis of variance. Statistically significant enrichment information (reported as P-values) is shown for each mAb binding in the isolated vesicles (Supplemental Data Set 3). In total, ∼70% of the Abs showed glycan enrichment in the isolated SYP61 vesicles compared with the controls. In order to contextualize the nascent glycans synthesized in the Golgi apparatus with those transported in SYP61 vesicles and the cell wall, the glycome profile of the isolated vesicles (Figure 3B) is shown in comparison with that of the Golgi/TGN (Figure 3A) and the cell wall of whole Arabidopsis CFP-SYP61 seedlings (Figure 3C). Qualitative interrogation of the carbohydrate profiles across the Golgi/TGN-enriched fractions (Supplemental Data Set 1A), the isolated SYP61 vesicles (Supplemental Data Sets 1B and 3), and the total cell wall (Figure 3; Supplemental Data Set 4) affords insights into the progression and modification of polysaccharides during biosynthesis, transport, and integration into the cell wall. Interestingly, many of the glycans present in the isolated vesicles correspond to those found in the cell wall, suggesting that they are already in their final form during transport in the SYP61 vesicles.
In an independent set of experiments, we used an agarose-based matrix coupled with GFP Abs for vesicle isolation, as previously described (Drakakaki et al., 2012; Park and Drakakaki, 2014). Polyclonal IgG-coated agarose beads were used as a control according to established methods (Kumar et al., 1997; Bonifacino et al., 2001), to allow for broad unspecific binding, which can be subtracted from the GFP-isolated fraction (Drakakaki et al., 2012; Park and Drakakaki, 2014). The overall glycome profile of the isolated SYP61 vesicles using this methodology closely mirrored that of the magnetic beads (GFP-Trap_MA) isolation (Supplemental Figures 2 and 3; Supplemental Data Set 2), demonstrating the robustness of the approach. Regression analysis between the two sets of experiments further confirmed this observation (P < 0.001; Supplemental Figure 4B), suggesting that the GFP-Ab–coupled matrix does not interfere with the vesicle isolation procedure and that the approach can be potentially adapted for other isolation matrixes.
Diverse Xyloglucan Epitopes Are Present in SYP61 Vesicles
The SYP61 vesicle glycome contained XyG epitopes with different degrees of modification (Supplemental Data Set 3). Several epitopes of FUC-XyG were significantly enriched in the isolated fraction (Figures 2 and 3; Supplemental Figures 2 and 3; see the FUC-XG cluster, detected by four different Abs), an observation supported by previous electron microscopy studies with CCRC-M1 (Zhang and Staehelin, 1992; Kang et al., 2011). This demonstrates that fully substituted XyG (constituting the major substitution of XyG in the cell wall; Figures 3B and 3C) is transported through SYP61 vesicles already in its final form before it is deposited to the apoplast and assembled into the cell wall.
Notably, our study also provided unexpected insights into XyG transit to the apoplast. Non–FUC-XyG epitopes were detected by four mAb clusters in SYP61 vesicles (Figure 3B; Supplemental Figures 2 and 3; Supplemental Data Set 3). In these clusters, some prominent Abs (CCRC-M87, CCRC-M88, CCRC-M93, CCRC-M95, CCRC-M101, and CCRC-M104; Figure 3B, indicated by circles on the left side of the heatmap) bind to galactosylated XyG epitopes (XLXG, XXLG, and XLLG; Dallabernardina et al., 2017). These galactosylated XyG epitopes were also present in the biosynthetic compartment (Golgi/TGN-enriched fraction; Figure 3A; Supplemental Data Set 1A) and cell wall extracts (Figure 3C; Supplemental Data Set 4). The consistent presence of galactosylated XyG epitopes across all compartments analyzed suggests that following biosynthesis in the Golgi apparatus, they are transported to the apoplast through SYP61 vesicles. This is reasonable, given that non–FUC-XyG constitutes a major portion of the synthesized polymer in the Golgi (Figure 3A; Obel et al., 2009; Günl et al., 2011) and is present in the cell wall (Figure 3C), and thus it is transported in the same form to the apoplast.
Altogether, our findings provide interesting insights into the diversity of XyG glycans transported to the apoplast via TGN-derived vesicles such as SYP61. This underscores the value of the large-scale aspect of our methodology in identifying diverse glycan populations transported through specific vesicle trafficking pathways. With detailed knowledge of the forms that cell wall polysaccharides are in as they leave the Golgi apparatus, and equipped with a tool to dissect the transit pathways and detect a wide variety of epitopes, we can begin to ask precise questions about where and how cell wall polysaccharide constituents are made and modified. This will help to clarify long-standing questions about what occurs between polysaccharide synthesis and their transport and deposition/modification in the cell wall.
Interestingly, the xylan clusters showed relatively low enrichment, with no statistical significance, for half of the Abs tested against the isolated vesicles (Figure 3B; Supplemental Data Set 3, see xylan clusters). Similarly, galactomannans were not significantly enriched in the isolated SYP61 vesicles. This likely reflects the low abundance of xylan and galactomannan glycans in the TGN/Golgi-enriched Suc gradient fractions compared to other structural polysaccharides (Figure 3A).
Pectin Epitopes Are Present in SYP61 Isolated Vesicles
The SYP61 vesicular glycome contained pectin epitopes with various degrees of substitution that were significantly enriched for 39 mAbs (Figure 3B; Supplemental Data Set 3, see pectin clusters). This observation complements earlier reports of RG-I components in the TGN (Zhang and Staehelin, 1992; McFarlane et al., 2008) based on a limited number of Abs (CCRC-M2, CCRC-M7, CCRC-M36, and JIM7) and identifies a wide range of epitope configurations of the pectin polymer that are transported via SYP61 vesicles. Both HG and diverse RG-I/AG (pectic-AG) epitopes, detected by a broad cluster of Abs against RG-I/AG and methyl-esterified HG backbones, were enriched in the SYP61 vesicles (Figures 2B and 3B; Supplemental Figures 2 and 3; Supplemental Data Set 3). RG-I/AG epitopes were identified in the Golgi/TGN fraction (Figure 3A), the isolated SYP61 vesicles (Figure 3B; Supplemental Figures 2 and 3), and the cell wall fraction (Figure 3C). This demonstrates that pectin backbone and pectic-AG glycans are transported from the point of synthesis (Golgi/TGN), through a SYP61 pathway to the cell wall.
Further mining of the pectin glycome of SYP61 isolated vesicles lead to interesting observations and generated hypotheses on pectin modification and transport. Both methyl-esterified and de-esterified/partially esterified pectin glycans were detected in the SYP61 isolated vesicles with the aid of CCRC-M131, CCRC-M38, JIM5, JIM7, and JIM136 Abs (Figure 3B; Supplemental Data Set 3, clusters HG-backbone 1, 2). To verify this result, we imaged pectin in double immunofluorescently labeled sections of high-pressure frozen and freeze-substituted Arabidopsis CFP-SYP61 roots. We used JIM5, one of the three mAbs (JIM5, CCRC-M38, and CCRC-M131) recognizing low-esterified HG, in SYP61 isolated vesicles (Figure 3B). Colocalization of CFP-SYP61 with JIM5 epitopes indicates transport of these glycans via SYP61 vesicles (Supplemental Figure 5). Given that JIM5 also binds strongly to the isolated cell wall extracts (Figure 3C; Supplemental Data Set 4) and that its epitope is also present in the Golgi/TGN-enriched fraction (Figure 3A; Supplemental Data Set 1A), this further demonstrates that a portion of partially esterified pectin is present in the Golgi/TGN and is transported through endomembrane vesicles to the cell wall. Interestingly, minimal or no significant enhancement of RG-I backbone epitopes was observed in the isolated vesicles (Figure 3B; Supplemental Figure 3; Supplemental Data Set 3), presumably due to their low concentration levels in the Golgi/TGN-enriched fraction prior to the vesicle isolation step (Figures 2A and 3A). Furthermore, the binding of only one Ab in the mucilage-type pectin mAb cluster was significantly enriched in the SYP61 vesicles (Figure 3B; Supplemental Data Set 3), suggesting there might be selective transport of this epitope in SYP61 vesicles.
Notably, a significant enrichment was observed for AG epitopes, which were highly abundant throughout the Golgi/TGN-enriched fraction (Figures 2A and 3A), the SYP61 vesicles (Figures 2B and 3B; Supplemental Data Set 3), and the cell wall (Figure 3C). The binding of 23 mAbs was enriched in the AG clusters. Interesting, the binding intensities of some mAbs recognizing AGPs, namely, MAC207, JIM14, and JIM133 (Pennell et al., 1989; Knox et al., 1991; Ruprecht et al., 2017), were enriched in the isolated SYP61 vesicles. This is indicative of an AGP trafficking route through the Golgi, via SYP61/TGN vesicles to the cell wall. Altogether, our data show the presence of a diverse population of pectic glycan and AGP epitopes in SYP61 TGN vesicles and indicate what may be a common polysaccharide transport pathway to the apoplast.
Pectin and XyG Deposition Is Altered in the syp61/osm1 Mutant, Validating the Glycome Profile Analysis
Analysis of the glycome profiles of the SYP61 vesicle cargo established that these vesicles carry diverse XyG and pectin glycans. To corroborate the effect of the SYP61 pathway on polysaccharide transport, we examined the pattern of polysaccharide deposition in the syp61/osm1 mutant. The osm1 mutant features a T-DNA insertion in SYP61 that results in an aberrant transcript altering SYP61 function, leading to osmotic stress hypersensitivity and trafficking defects of the PM aquaporin PIP2a;7 (Zhu et al., 2002; Hachez et al., 2014). Given that no SYP61 knockout mutant has thus far been characterized, most likely due to lethality, we reasoned that syp61/osm1 is currently the best tool to provide some insights into the impact of the SYP61 compartment on polysaccharide deposition. We hypothesized that the trafficking defects in syp61/osm1 also ultimately lead to polysaccharide changes in the cell wall.
We first examined the cell wall profile of the syp61/osm1 mutant compared with the wild type parental line C24. Cell wall analysis of the Arabidopsis syp61/osm1 mutant showed a reduction in pectin content and polymer diversity compared with the wild type C24 (Figures 4A and 4B; Supplemental Data Set 5A, cell wall content and Supplemental Data Set 5B, ratio of osm1/C24 cell wall glycomes; see oxalate and carbonate cell wall fractions). AG epitope detection was also reduced in the syp61/osm1 cell wall extracts compared with C24 (Figures 4A and 4B; Supplemental Data Sets 5A and 5B, clusters RG-I/AG through AG-4), corroborating the finding from our vesicle cargo analysis that these glycans are packaged into SYP61 vesicles en route to the cell wall.
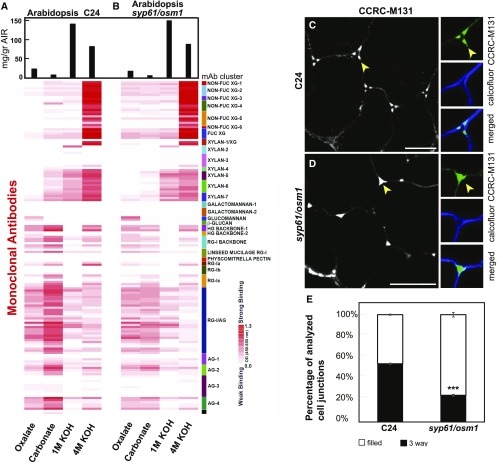
Figure 4.
Distinct Cell Wall Glycome Profiles and Patterns between Wild Type and the syp61/osm1 Mutants.
(A) and (B) Cell wall glycome profiling of wild type C24 (A) and of mutant seedlings (B). Sequentially extracted cell wall material was analyzed using glycome profiling with the glycan-directed mAbs as described in Figure 3C. A white-to-red scale indicates signal intensity in the ELISA heatmap as described before. Black bars at the top indicate milligram per gram of cell wall AIR. In each lane, 0.3 μg of Glc equivalent amounts of polysaccharides was applied. The heatmap is a visual representation of Supplemental Data Set 5.
(C) Pectin backbone labeling with CCRC-M131 in the C24 wild type background. CCRC-M131 labeling showed a distinct three-way junction pattern (arrow) in the C24 control. Insets show a close-up view of a three-way junction pattern. Green indicates staining with CCRC-M131, and blue represents cellulose staining with calcofluor white.
(D) Pectin backbone labeling with CCRC-M131 in syp61/osm1 roots. Labeling with CCRC-M131 in the syp61/osm1 mutant displays mostly a filled junction pattern (arrow) forming a different outline compared with the control. Insets show a filled junction in cell corners. Green indicates staining with CCRC-M131, and blue represents cellulose staining with calcofluor white. Images in (C) and (D) represent transverse root tip sections. Bars = 10 μm.
(E) Quantification of three-way junction labeling versus filled junction by CCRC-M131. The percentage of total analyzed cell corners displaying a three-way junction labeled by CCRC-M131 is reduced in the syp61/osm1 mutant compared with the wild type control C24 (t test, ***P < 0.001). The percentage of total cell corners displaying a filled junction pattern labeled by CCRC-M131 is significantly higher in the syp61/osm1 compared with the wild type control C24 (t test, ***P < 0.001). Thin bars represent se.
Furthermore, as proof of concept, we examined pectin deposition in Arabidopsis roots using the CCRC-M131 mAb; this mAb recognizes a de-esterified HG backbone epitope, an epitope significantly enriched in the SYP61 vesicle glycome (Supplemental Data Set 3). Our analysis of cell corners of labeled transverse root sections revealed a pattern in the syp61/osm1 mutant that differed from that of the C24 control (Figures 4C and 4D). In the C24 wild type control, CCRC-M131 discretely labeled the corners of three-way junctions (Figure 4C). By contrast, contiguous labeling of the corners was observed at the interconnections of osm1 root cells (Figure 4D). Quantitative analysis of the three-way junction labeling as a percentage of analyzed corners (C24 versus osm1) showed a significant reduction in the number of organized three-way labeling patterns in mutant cell corners and an increased number of aberrant, filled junction labeling (P < 0.001; (Figure 4E; Supplemental Table), suggesting an altered pectin deposition and assembly pattern as defined by CCRC-M131. Altogether, the data demonstrate that the SYP61 compartment plays a role in the targeted delivery of pectins and that interfering with this pathway alters the pectin deposition pattern into the cell wall.
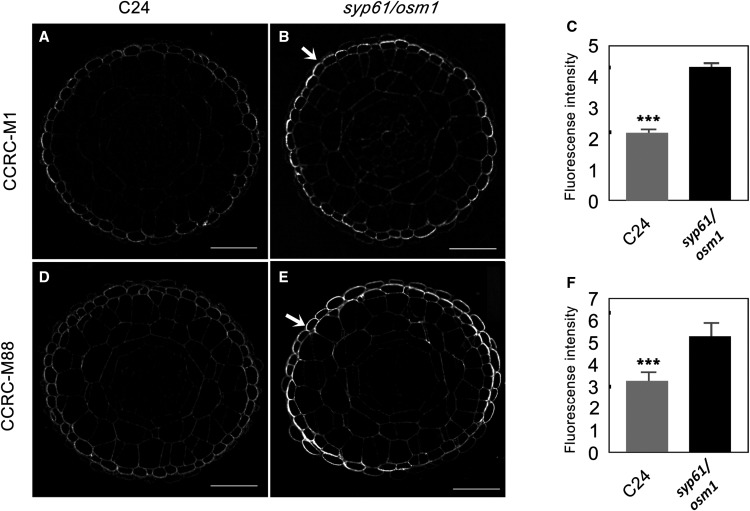
Given that SYP61 vesicles also carry XyG (Figures 2 and 3B), we expected to observe a modified glycome profile of XyG in the syp61/osm1 mutant’s cell wall fraction. We detected an increased signal for XyG mAbs in the oxalate and carbonate extracts from the syp61/osm1 mutant compared with the same extracts from the wild type C24 control (XG clusters, Figure 4B). The increase in the levels of XyG glycans compared with other polysaccharides (pectins and AGPs) might result from a compensatory mechanism that feeds back in response to cell wall modifications and trafficking defects. As a case study, we further validated cell wall changes using two selected Abs from the XyG clusters via immunocytochemistry of transverse root sections. Labeling with CCRC-M1 (Figures 5A and 5B), detecting FUC-XyG epitopes and CCRC-M88, detecting galactosylated XyG epitopes (Figures 5D and 5E; Puhlmann et al., 1994; Freshour et al., 1996; Dallabernardina et al., 2017), showed an increase in fluorescence signals in the epidermis of syp61/osm1 mutant roots compared with the C24 control. Quantitative analysis determined that the increase in CCRC-M1 and CCRC-M88 labeling in the syp61/osm1 mutant was statistically significant (P < 0.001; Figures 5C and 5F; Supplemental Table). The overall root tip morphology, visualized with propidium iodine staining (Supplemental Figure 6), did not show any discernible morphological differences for syp61/osm1 compared with the wild type C24, demonstrating that the observed differences were truly due to changes in polysaccharide deposition patterns and not an indirect effect of changes in root morphology. Nonetheless, further detailed analysis of root development in this mutant is required to examine possible phenotypic changes in syp61/osm1 that may have not been detected in the current assay. Overall, the agreement between the immunostaining analysis and the cell wall glycome profile clearly demonstrates that interfering with the SYP61 pathway affects XyG deposition into the cell wall.
Figure 5.
Xyloglucan Staining Is Enhanced in the Root Epidermis of the syp61/osm1 Mutant.
(A) and (B) Labeling with CCRC-M1 detecting FUC-XyG in transverse root sections of wild type C24 and syp61/osm1. More pronounced labeling is observed in the epidermis of the syp61/osm1 mutant (B) compared with the wild type C24 (A).
(C) Quantitative analysis of fluorescence in the epidermis shows increased labeling with CCRC-M1 in the syp61/osm1 mutant compared with the wild type C24 control (t test, ***P < 0.001). Thin bars represent se.
(D) and (E) Labeling with CCRC-M88 detecting non–FUC-XyG in root sections of wild type C24 and syp61/osm1. More pronounced labeling is observed in the syp61/osm1 mutant (E) compared with the wild type C24 (D). Images in (A), (B), (D), and (E) represent transverse root sections. Bars = 20 µm.
(F) Quantitative fluorescence analysis in the epidermis shows increased labeling with CCRC-M88 in the syp61/osm1 mutant compared with the wild type C24 control (t test, ***P < 0.001). Thin bars represent se.
DISCUSSION
Given the critical role of the endomembrane system in polysaccharide and glycoprotein biosynthesis and deposition, the identification of cargo through the various dynamic transport routes is vital for a comprehensive understanding of these processes during plant development and stress responses. Here, we show that with our hybrid approach, it is possible to identify the large-scale glycan content from a selected population of specific vesicles separated by immuno-isolation. The large-scale aspect is only limited by the number of mAbs present in the collection and can be expanded with the availability of new Abs. In mammalian systems, the use of lectin arrays has been explored to identify surface glycoproteins in extracellular vesicles (Saito et al., 2018; Williams et al., 2018), but this work did not make use of a large-scale polysaccharide Ab array. Given the high specificity of mAbs compared with lectins (Cummings and Etzler, 2009), our hybrid approach is very promising for detecting specific glycan populations in isolated vesicles. Furthermore, with the versatility and robustness of our approach, the method can be adapted for different forms of purification matrices, a field that can be further explored in the future.
The identified glycome of SYP61 vesicles revealed a diverse population of glycans including pectins, XyGs, and AGPs, suggesting that a constitutive pathway exists for polysaccharide and glycoprotein secretion in plant cells. Earlier studies of XyG and pectin epitope colocalization in transport vesicles of red clover (Trifolium pratense) root tips (Lynch and Staehelin, 1992) indicated such a scenario, corroborating our findings. This provides a fresh perspective on matrix polysaccharide transport, which now can be investigated in depth by glycome analysis of different vesicle types and immunocytochemistry using the mAbs showing modulated binding in this study.
Many important insights were gained by this large-scale analysis, although the data have not been exhaustively analyzed. The SYP61 vesicles contained several XyG epitopes, including both FUC and non-FUC types. In an earlier study, low substituted XyG oligosaccharides were identified in enriched Golgi microsomes compared with the cell wall (Obel et al., 2009; Günl et al., 2011), indicating incomplete stages of the polymer in the Golgi-enriched fraction. Since both FUC and non-FUC XyG epitopes are present in the Arabidopsis cell wall (Figure 3C; Obel et al., 2009), we propose that these diverse glycans are generated in Golgi/TGN and are transported as such to the apoplast in their final form for assembly. This is supported by the higher order complexes that reside in Golgi/TGN (Chou et al., 2012; Lund et al., 2015), producing a diverse set of XyG substitutions. The mAbs that bind to these glycans can be interrogated in depth using immunocytochemistry and genetics to pinpoint the transitions from the point of synthesis to the assembled cell wall. Given the absence of the FUCOSYLTRANSFERASE1 in the SYP61 proteome, it is unlikely that any XyG fucosylation takes place en route to the apoplast (Drakakaki et al., 2012). Presumably, these subtly different versions of XyG are also modified during their transport and incorporation into the cell wall. Glycosidases such as apoplastic fucosidases and other apoplastic enzymes contribute to the modification of polysaccharides during cell wall assembly and the final structural composition into the cell wall, creating structural cell wall diversity (Günl et al., 2011; Franková and Fry, 2013; Pauly and Keegstra, 2016).
To date, little is known about the biochemistry behind the vesicle transport pathways of pectin polysaccharides. Our study identified a wide range of pectin glycans, including the pectin backbone as well as Ara- and Gal-containing RG-I in the cargo of SYP61 vesicles. This suggests that both HG domains, along with RG-I–containing AG side chains (pectic-AG), are being sorted in the TGN and transported through a common vesicle pathway to their cell wall deposition sites. Interestingly, both methyl-esterified pectin and the partially esterified pectin HG backbone were identified in the isolated SYP61 compartment, in contrast to the current belief that pectins are secreted in a highly methylesterified form (Caffall and Mohnen, 2009). This suggests that both forms of pectin may exist throughout the endomembrane system.
The overall involvement of the SYP61 pathway in structural polysaccharide transport and deposition is supported both by our biochemical and genetic data, evidenced by changes in the cell wall profile and pectin and XyG patterns using our in situ cell wall labeling of the syp61/osm1 mutant. The differences observed using in situ labeling in the syp61/osm1 mutant may be a consequence of impaired TGN vesicle delivery due to the malfunction of the SNARE SYP61, leading to changes in the deposition and distribution of the epitope. As such, this can cause changes in CCRC-M131 labeling of de-esterified pectin, resulting in the loss of the distinct three-way pattern in cell junctions. An alternative hypothesis is that other polymers masking the CCRC-M131 epitope in the central area of the cell corners are structurally altered due to overall cell wall modifications. As a result, the full cell junctions might be exposed to CCRC-M131, leading to a loss of the organized three-way labeling. The structural modification of polysaccharides, rather than their abundance, can affect secretory trafficking, as was previously shown for the galactosyltransferase mur3-3 mutant (Kong et al., 2015). Hence, feedback mechanisms involving cell wall modifications can further affect the overall endomembrane system pathways and their polysaccharide cargo. This, in turn, can affect the polysaccharide distribution in the cell wall.
The proteome cargo of SYP61 vesicles includes PECTIN METHYLESTERASE1, which has also been identified in several TGN proteomes (Drakakaki et al., 2012; Nikolovski et al., 2012; Groen et al., 2014; Heard et al., 2015). A plausible hypothesis is that the trafficking of PECTIN METHYLESTERASE1 is altered in syp61/osm1, thus leading to localized cell wall changes in pectin modification and assembly.
While our data provide strong evidence for the role of the SYP61 pathway in the transport of structural polysaccharides, we do not yet know how many additional pathways are involved in the same process. Mutants of proteins identified in the SYP61 vesicle proteome, including ECH and the YIP family of RAB GTPase-interacting proteins, also display defects in pectin and XyG secretion (Gendre et al., 2011, 2013; McFarlane et al., 2013). Furthermore, in Arabidopsis, the Rab GTPase RABA4B localizes to a TGN subdomain containing FUC-XyG and overlaps with SYP61 (Kang et al., 2011). This leads us to put forward the hypothesis that SYP61, together with RABA4B, ECH, and YIP4s, reside in a TGN subdomain involved in structural polysaccharide transport. The SYP42/SYP43 SNARES are also part of the SYP61 proteome and regulate TGN-mediated trafficking (Uemura et al., 2012). It would be informative to investigate polysaccharide secretion in mutants or corresponding vesicles for other proteins identified in the SYP61 vesicles, such as SYP4 or RABA4B, to deepen our understanding of these trafficking pathways. Such analysis will determine whether the SYP61 pathway specifically functions in polysaccharide transport or if other pathways carry the same function. Scaling up vesicle glycomics in different vesicle types using different marker proteins of exocytic and endocytic pathways can provide insights into how different pathways are involved in polysaccharide transport and deposition and how the division of labor takes place.
The cell wall proteome includes glycoproteins such as AGPs, whose underlying secretion mechanisms into the apoplast are not well understood (Tan et al., 2012; van de Meene et al., 2017). The detection of AGP glycan structures in the SYP61 glycome and their altered profile in the syp61 mutant cell wall demonstrate the involvement of SYP61 in AGP trafficking. This is in line with the notion that AGPs are glycosylated in the Golgi apparatus and thus should follow a Golgi-mediated pathway (van de Meene et al., 2017). A Golgi-independent secretion pathway has also been implicated for AGP glycosylation (Wang et al., 2010; Poulsen et al., 2014; Davis et al., 2016). However, this requires further investigation, since both the glycosyltransferases and the AGP protein core are not leaderless proteins and are expected to follow canonical secretory pathways (van de Meene et al., 2017). Given that glycosylated proteins at the cell wall serve many functions, including signaling and responses to biotic and abiotic stress (Cantu et al., 2008; Chaliha et al., 2018; Lamport et al., 2018; Novakovic et al., 2018), understanding the mechanisms regulating their secretion under different stimuli is critical for determining their functions. The approach presented here provides supporting evidence for a TGN-mediated pathway for extracellular glycoproteins and an alternative means to conventional proteomics toward dissecting their delivery pathways via the endomembrane system.
In summary, our study delivers crucial evidence and maps out diverse glycans of XyGs, pectins, and glycoproteins in TGN vesicles isolated with the aid of the syntaxin SYP61. The large-scale data set of glycans in SYP61 isolated vesicles obtained in this study can be interrogated from different perspectives (i.e., in comparison with other glycomes of vesicles, membrane compartments, or cell walls and glycan profiles from different species or treatments) and generate new hypotheses for both polysaccharide and glycoprotein transport through the endomembrane system. Ab binding patterns in different clusters can be used to generate hypotheses for each epitope represented in a cluster. The predicted Ab binding behavior can then be investigated by biochemical and in situ analyses. This information can be used to test individual mutants and other vesicle populations for key factors involved in the biosynthesis, transport, and deposition of specific glycan cargo. Moreover, exploration of our large-scale glycan data sets in enriched Golgi/TGN and the Arabidopsis seedling cell wall can provide a holistic perspective directing research in genetic, biochemical, and cell biology studies of cell wall build up.
Future adaptation of this methodology could facilitate the characterization of additional vesicle glycome profiles using different subcellular markers, contributing to a more comprehensive understanding of how dynamic cargo shapes the endomembrane system. Identifying specific polysaccharide cargoes in different vesicle types and/or during different developmental stages, or biotic and abiotic stress responses, will aid in the construction of cell wall assembly models, incorporating information about how plants are able to adapt to specific environments and growth conditions.
Studies have underlined the role of the TGN and potentially unconventional vesicle trafficking, such as through exosomal compartments, in the plant pathogen response; however, these studies have mainly focused on the roles of proteins or RNA cargo under biotic stress (LaMontagne and Heese, 2017; Rutter and Innes, 2017; Yun and Kwon, 2017; Cai et al., 2018). Exosomal cargo has received a lot of attention with respect to the roles of protein and nucleic acid cargo in human disease (Bae et al., 2018); analysis of their glycomic cargo might provide further valuable insights (Saito et al., 2018; Williams et al., 2018). The current study lays the foundation for dissecting cellular pathways in both plants and animal systems by extending our proteomics perspective with a glycomic perspective. In toto, our methodology can lead to a more comprehensive understanding of how the endomembrane system and its polysaccharide cargo shape cell biology.
METHODS
Plant Material
The Arabidopsis (Arabidopsis thaliana) Col-0, C24 wild type plants; the transgenic line CFP-SYP61 (Robert et al., 2008); and the SYP61 mutant osm1 (Zhu et al., 2002) were used in this study. Plants were grown in temperature- and photoperiod-controlled environments, set to long-day (16-h-light/8-h-dark cycle) conditions, using fluorescent light (100 to 150 μmol quanta PAR m–2 s–1) at 22 to 24°C.
Vesicle Isolation
Vesicle isolation was performed using CFP-SYP61 seedlings as previously described (Drakakaki et al., 2012; Park and Drakakaki, 2014). Briefly, seedling cultures were grown in liquid Murashige and Skoog (MS) medium for 12 to 14 d, and Golgi/TGN vesicles were isolated by Suc gradient centrifugation. SYP61 TGN-enriched fractions were used for IP with the aid of a GFP Ab that recognizes CFP (A11122; Invitrogen) and a rabbit IgG (sc-2027; Santa Cruz Biotechnology) as control, both coupled to Protein A Agarose beads (Invitrogen).
Alternatively, CFP-SYP61 and the wild type Col-0 Golgi/TGN-enriched fractions were subjected to immuno-isolation using magnetic agarose beads coupled to anti-GFP Abs (GFP-Trap_MA; ChromoTek) and noncoupled magnetic agarose beads as controls. Immunoblot analysis was performed using standard procedures as previously described (Drakakaki et al., 2006a). The isolated SYP61 vesicles did not show contamination of ER and PM or prevacuolar compartments, as described in previous studies (Supplemental Figure 1; Drakakaki et al., 2012; Park and Drakakaki, 2014; Wattelet-Boyer et al., 2016).
Glycome Analysis
Isolated vesicles were diluted to a total of 8 mL with double distilled water and sonicated using a Branson sonicator on ice. The process was repeated three times to ensure complete disruption of vesicles. Subsequently, the samples were centrifuged for 15 min at 4000 rpm, and the resulting supernatants were immobilized onto 384-well ELISA plates and screened using cell wall glycan-directed Abs (Pattathil et al., 2010). The ELISA responses of these Abs to each vesicle isolate were compiled into a heatmap using a modified version of the R-Console software (R Development Core Team, 2006). Plant cell wall glycan-directed mAbs were obtained from laboratory stocks (CCRC, JIM, and MAC series) at the Complex Carbohydrate Research Center (available through CarboSource Services; http://www.carbosource.net) or were procured from BioSupplies (BG1, LAMP). Additional information about these Abs can be obtained from the online database WallMabDB (http://www.wallmabdb.net).
Tissue Fixation, Embedding, and Sectioning
Five-day-old Arabidopsis seedlings were grown vertically in 1/4 MS salts (4.4 g L–1; Sigma-Aldrich), 0.8% agar [w/v], pH 5.7 medium. Root tip segments of C24 and syp61/osm1 were embedded in 1% (w/v) agarose and were fixed in glutaraldehyde (0.5% [v/v]), formaldehyde (3% [v/v]) in microtubule-stabilizing buffer as previously described (Park et al., 2014). Root tip segments of CFP-SYP61 were high-pressure frozen and freeze substituted as previously described (Kang, 2010). Sections were then infiltrated with London Resin White as previously described by Drakakaki et al. (2006b). Transverse semithin root sections were used for immunofluorescence imaging with polysaccharide-specific Abs.
Immunocytochemistry
Resin-embedded sections were rinsed with 1× phosphate buffer saline solution containing 0.1% (v/v) Tween 20 (1× PBST) three times for 10 min at room temperature. Then, 3% (w/v) BSA was applied to the sections for 1 h. Following three rinses with 1× PBST, the sections were incubated with primary polysaccharide Abs diluted 1:10 in 1× PBST. After overnight incubation at 37°C, the sections were rinsed three times with 1× PBST and subsequently hybridized for 3 h with secondary Abs at 37°C. The mAbs used, were as follows: CCRC-M131 and JIM5 for de-esterified and low methyl–esterified HG epitopes, respectively (Pattathil et al., 2010); CCRC-M1 and CCRC-M88 mAbs for FUC and galactosylated XyG, respectively (Puhlmann et al., 1994; Freshour et al., 1996; Dallabernardina et al., 2017); and JL-8 for GFP (Takara). Alexa-488– and cyanine (Cy3)-conjugated anti-mouse IgG (Molecular Probes) and fluorescein isothiocyanate–conjugated anti-rat (dilution, 1:500 in 1× PBST) were used as secondary Abs. For cellulose staining, semithin sections were rinsed with 1× PBST three times and incubated with calcofluor white (1% [w/v]; Sigma-Aldrich) for 5 min in the dark. Sections were mounted in CitiFluor antifade mountant solution (Electron Microscopy Sciences) and imaged on an SP8 confocal microscope (Leica). A minimum of six sections from different biological replicates of independently grown seedlings were analyzed for each mAb and genotype.
Confocal Microscopy and Image Quantification
Images were recorded on an SP8 confocal microscope using a 40× water objective or a 100× oil objective). Fluorescent dyes were excited at 488 nm (Alexa-488) and 405 nm (calcofluor white), and emission spectra were collected over 500 to 550 nm (Alexa-488) and 420 to 460 nm (calcofluor white). Fluorescein isothiocyanate was exited at 470 nm, and emission spectra were collected over 500 to 550 nm. Cy3 was excited at 560 nm, and emission spectra were collected at 570 to 630 nm using a 0.5 Airy unit pinhole; sequential line-scanning and gating for Cy3 was used to minimize crosstalk. Three-dimensional reconstructions were obtained using IMARIS (Bitplane). Image analysis was performed using ImageJ 1.36b (http://rsbweb.nih.gov/ij/). Data were compared using a Student’s t test. Asterisks in the figures denote significant differences as follows: ***P < 0.001.
Cell Wall Extraction and Glycome Analysis
Arabidopsis seedlings were grown for 14 d either in liquid 1/2 MS medium in seedling cultures (for CFP-SYP61) or for 10 d vertically in 1/2 MS salt (0.8% agar, pH 5.7) medium. Cell wall extraction and preparation for glycomic analysis were performed as previously described (Pattathil et al., 2012). Briefly, alcohol-insoluble residues (AIRs) from Arabidopsis were extracted in 80% (v/v) ethanol and washed twice in absolute ethanol and acetone and then air-dried. AIR fractionation was performed via sequential extraction, using 50 mM ammonium oxalate, 50 mM sodium carbonate (containing 0.5% [w/v] sodium borohydride), and 1 and 4 M KOH (both containing 1% [w/v] sodium borohydride). Cell wall extracts were dialyzed with deionized water (sample:water ratio, ∼1:60) at room temperature for a total of 48 h and then lyophilized and weighed. Cell wall extracts were dissolved in deionized water, and total sugar levels were determined via a phenol-sulfuric acid microplate assay (Dubois et al., 1956; Masuko et al., 2005). Finally, the plates were coated with equal Glc amounts of polysaccharides (Pattathil et al., 2012). An ELISA was used to probe the cell wall extracts against a set of ∼155 mAbs directed against noncellulosic cell wall glycans (Pattathil et al., 2010). The ELISA responses of these Abs to each extract were summarized into a heatmap using a modified version of R-Console software (R Development Core Team, 2006). Background noise data generated using no antigen (water) controls were subtracted from the mean values obtained, making the data further error resilient and statistically significant. ELISAs were performed with an automated platform (Thermo Fisher Scientific), making the data generated free of any human errors, particularly for the color development step. Glycomic analyses using the comprehensive suite of cell wall glycan-directed mAbs were previously demonstrated to generate data that reveal significant differences in the abundance of cell wall glycan epitopes (Liang et al., 2013; Pattathil et al., 2015a). These studies involved in-depth statistical analyses using advanced statistical analytical tools such as JMP Genomics 6.1 (SAS Institute) and revealed that any variation in the OD of raw data values (ranging from 0.05 to 0.1 OD) are indeed statistically significant and thus demonstrated a significant variation in the relative abundance of epitopes detected (Liang et al., 2013).
Statistical Analysis
One-way analysis of variance was used using SAS 9.1 statistical software (SAS Institute) to evaluate the difference between SYP61 vesicle isolates compared with the controls. Values of SYP61 vesicle isolates were compared with values of all the negative controls. Linear regression trends were fitted to compare values of Golgi/TGN fractions and the reproducibility of different types of isolated vesicle glycomes from two different experimental sets using either agarose- or magnetic-based beads.
Accession Numbers
Sequence data from this article can be found in The Arabidopsis Information Resource database under the following accession numbers: AT1G28490 (AtSYP61), AT1G53840 (PECTIN METHYLESTERASE1).
Supplemental Data
Supplemental Figure 1. Purity of SYP61 isolated vesicles.
Supplemental Figure 2. Glycome profiles of Golgi/TGN-enriched fraction and SYP61 vesicles.
Supplemental Figure 3. Glycan enrichment in SYP61 vesicles isolated in experimental set 2.
Supplemental Figure 4. Regression plots between the glycome profiles of Golgi/TGN and between various SYP61 vesicle isolations.
Supplemental Figure 5. SYP61 partially colocalizes with JIM5 labelling of pectin glycans.
Supplemental Figure 6. Root structure morphology of C24 and syp61/osm1.
Supplemental Table. t Test analyses in Figures 4 and 5.
Supplemental Data Set 1. Glycome profiles of isolated compartments: Experimental set 1 using magnetic beads for vesicle isolation.
Supplemental Data Set 2. Glycome profiles of isolated compartments: Experimental set 2 using non-magnetic beads for vesicle isolation.
Supplemental Data Set 3. Statistical analysis SYP61 vesicle glycome profiles.
Supplemental Data Set 4. Glycome analysis of CFP-SYP61 seedling cell walls.
Supplemental Data Set 5. Cell wall glycomes of syp61/osm1 and WT C24 seedlings.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank Shahab Madahhosseini for statistical analysis and Tereza Ticha for her assistance to G.R. We thank Dan Kliebenstein for his critical comments and suggestions. We thank John Labavitch and Victoria G. Pook for critically reading this article. This work was supported by the National Science Foundation awards IOS 1258135 and MCB 1818219 to G.D. and U.S. Department of Agriculture award CA-D-PLS-2132-H to G.D. The CCRC series of plant cell wall glycan-directed Abs were generated with the support of the National Science Foundation Plant Genome Program (awards DBI-0421683 and IOS-0923992 to M.G.H.). Immunological screening of cell wall and Golgi samples was also supported in part by the United States Department of Energy-funded Center for Plant and Microbial Complex Carbohydrates (award DE-SC0015662). W.B. was partially supported by the China Scholarship Council.
AUTHOR CONTRIBUTIONS
G.D., S.P., T.W. designed the research. A.G.P., G.R., W.B., D.J.D., D.S.D., D.D., G.D., S.P., and T.W. performed research. A.G.P., W.B., G.D., D.J.D., D.S.D., M.G.H., S.P., and T.W. analyzed data. T.W. and G.D. wrote the manuscript. All authors read revised and approved the article.
Footnotes
Articles can be viewed without a subscription.
References
- Anderson C.T., Carroll A., Akhmetova L., Somerville C. (2010). Real-time imaging of cellulose reorientation during cell wall expansion in Arabidopsis roots. Plant Physiol. 152: 787–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson C.T., Wallace I.S., Somerville C.R. (2012). Metabolic click-labeling with a fucose analog reveals pectin delivery, architecture, and dynamics in Arabidopsis cell walls. Proc. Natl. Acad. Sci. USA 109: 1329–1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atmodjo M.A., Hao Z., Mohnen D. (2013). Evolving views of pectin biosynthesis. Annu. Rev. Plant Biol. 64: 747–779. [DOI] [PubMed] [Google Scholar]
- Avci U., Pattathil S., Hahn M.G. (2012). Immunological approaches to plant cell wall and biomass characterization: Immunolocalization of glycan epitopes. Methods Mol. Biol. 908: 73–82. [DOI] [PubMed] [Google Scholar]
- Badhan A., Wang Y., McAllister T.A. (2017). Analysis of complex carbohydrate composition in plant cell wall using Fourier transformed mid-infrared spectroscopy (FT-IR). Methods Mol. Biol. 1588: 209–214. [DOI] [PubMed] [Google Scholar]
- Bae S., Brumbaugh J., Bonavida B. (2018). Exosomes derived from cancerous and non-cancerous cells regulate the anti-tumor response in the tumor microenvironment. Genes Cancer 9: 87–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashline L., Lei L., Li S., Gu Y. (2014). Cell wall, cytoskeleton, and cell expansion in higher plants. Mol. Plant 7: 586–600. [DOI] [PubMed] [Google Scholar]
- Bee C., Abdiche Y.N., Pons J., Rajpal A. (2013). Determining the binding affinity of therapeutic monoclonal antibodies towards their native unpurified antigens in human serum. PLoS One 8: e80501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bombardier J.P., Munson M. (2015). Three steps forward, two steps back: Mechanistic insights into the assembly and disassembly of the SNARE complex. Curr. Opin. Chem. Biol. 29: 66–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonifacino J.S., Dell’Angelica E.C., Springer T.A. (2001). Immunoprecipitation. Curr. Protoc. Immunol. 41: 8.3.1–8.3.28. [DOI] [PubMed] [Google Scholar]
- Burton R.A., Gidley M.J., Fincher G.B. (2010). Heterogeneity in the chemistry, structure and function of plant cell walls. Nat. Chem. Biol. 6: 724–732. [DOI] [PubMed] [Google Scholar]
- Caffall K.H., Mohnen D. (2009). The structure, function, and biosynthesis of plant cell wall pectic polysaccharides. Carbohydr. Res. 344: 1879–1900. [DOI] [PubMed] [Google Scholar]
- Cai Q., Qiao L., Wang M., He B., Lin F.M., Palmquist J., Huang S.D., Jin H. (2018). Plants send small RNAs in extracellular vesicles to fungal pathogen to silence virulence genes. Science 360: 1126–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantu D., Vicente A.R., Greve L.C., Dewey F.M., Bennett A.B., Labavitch J.M., Powell A.L. (2008). The intersection between cell wall disassembly, ripening, and fruit susceptibility to Botrytis cinerea. Proc. Natl. Acad. Sci. USA 105: 859–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaliha C., Rugen M.D., Field R.A., Kalita E. (2018). Glycans as modulators of plant defense against filamentous pathogens. Front. Plant Sci. 9: 928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevalier L., Bernard S., Ramdani Y., Lamour R., Bardor M., Lerouge P., Follet-Gueye M.L., Driouich A. (2010). Subcompartment localization of the side chain xyloglucan-synthesizing enzymes within Golgi stacks of tobacco suspension-cultured cells. Plant J. 64: 977–989. [DOI] [PubMed] [Google Scholar]
- Chou Y.H., Pogorelko G., Zabotina O.A. (2012). Xyloglucan xylosyltransferases XXT1, XXT2, and XXT5 and the glucan synthase CSLC4 form Golgi-localized multiprotein complexes. Plant Physiol. 159: 1355–1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou Y.H., Pogorelko G., Young Z.T., Zabotina O.A. (2015). Protein-protein interactions among xyloglucan-synthesizing enzymes and formation of Golgi-localized multiprotein complexes. Plant Cell Physiol. 56: 255–267. [DOI] [PubMed] [Google Scholar]
- Cornuault V., Buffetto F., Rydahl M.G., Marcus S.E., Torode T.A., Xue J., Crépeau M.J., Faria-Blanc N., Willats W.G., Dupree P., Ralet M.C., Knox J.P. (2015). Monoclonal antibodies indicate low-abundance links between heteroxylan and other glycans of plant cell walls. Planta 242: 1321–1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove D.J. (2005). Growth of the plant cell wall. Nat. Rev. Mol. Cell Biol. 6: 850–861. [DOI] [PubMed] [Google Scholar]
- Cosgrove D.J. (2016). Plant cell wall extensibility: Connecting plant cell growth with cell wall structure, mechanics, and the action of wall-modifying enzymes. J. Exp. Bot. 67: 463–476. [DOI] [PubMed] [Google Scholar]
- Cummings R.D., Etzler M.E. (2009). Antibodies and lectins in glycan analysis. In Essentials of Glycobiology, Varki A., Cummings R.D., Esko J.D., Freeze H.H., Stanley P., Bertozzi C.R., Hart G.W., and Etzler M.E., eds, 2nd ed (New York: Cold Spring Harbor Laboratory Press; ). [PubMed] [Google Scholar]
- Dallabernardina P., Ruprecht C., Smith P.J., Hahn M.G., Urbanowicz B.R., Pfrengle F. (2017). Automated glycan assembly of galactosylated xyloglucan oligosaccharides and their recognition by plant cell wall glycan-directed antibodies. Org. Biomol. Chem. 15: 9996–10000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis D.J., Kang B.H., Heringer A.S., Wilkop T.E., Drakakaki G. (2016). Unconventional protein secretion in plants. Methods Mol. Biol. 1459: 47–63. [DOI] [PubMed] [Google Scholar]
- Drakakaki G., Marcel S., Arcalis E., Altmann F., Gonzalez-Melendi P., Fischer R., Christou P., Stoger E. (2006b). The intracellular fate of a recombinant protein is tissue dependent. Plant Physiol. 141: 578–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drakakaki G., Zabotina O., Delgado I., Robert S., Keegstra K., Raikhel N. (2006a). Arabidopsis reversibly glycosylated polypeptides 1 and 2 are essential for pollen development. Plant Physiol. 142: 1480–1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drakakaki G., van de Ven W., Pan S., Miao Y., Wang J., Keinath N.F., Weatherly B., Jiang L., Schumacher K., Hicks G., Raikhel N. (2012). Isolation and proteomic analysis of the SYP61 compartment reveal its role in exocytic trafficking in Arabidopsis. Cell Res. 22: 413–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driouich A., Follet-Gueye M.L., Bernard S., Kousar S., Chevalier L., Vicré-Gibouin M., Lerouxel O. (2012). Golgi-mediated synthesis and secretion of matrix polysaccharides of the primary cell wall of higher plants. Front. Plant Sci. 3: 79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois M., Gilles K.A., Hamilton J.K., Rebers P.A., Smith F. (1956). Colorimetric method for determination of sugars and related substances. Anal. Chem. 28: 350–356. [Google Scholar]
- Foster C.E., Martin T.M., Pauly M. (2010). Comprehensive compositional analysis of plant cell walls (lignocellulosic biomass) part II: Carbohydrates. J. Vis. Exp. 37: 1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franková L., Fry S.C. (2013). Biochemistry and physiological roles of enzymes that ‘cut and paste’ plant cell-wall polysaccharides. J. Exp. Bot. 64: 3519–3550. [DOI] [PubMed] [Google Scholar]
- Freshour G., Clay R.P., Fuller M.S., Albersheim P., Darvill A.G., Hahn M.G. (1996). Developmental and tissue-specific structural alterations of the cell-wall polysaccharides of Arabidopsis thaliana roots. Plant Physiol. 110: 1413–1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendre D., Oh J., Boutté Y., Best J.G., Samuels L., Nilsson R., Uemura T., Marchant A., Bennett M.J., Grebe M., Bhalerao R.P. (2011). Conserved Arabidopsis ECHIDNA protein mediates trans-Golgi-network trafficking and cell elongation. Proc. Natl. Acad. Sci. USA 108: 8048–8053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendre D., McFarlane H.E., Johnson E., Mouille G., Sjödin A., Oh J., Levesque-Tremblay G., Watanabe Y., Samuels L., Bhalerao R.P. (2013). Trans-Golgi network localized ECHIDNA/Ypt interacting protein complex is required for the secretion of cell wall polysaccharides in Arabidopsis. Plant Cell 25: 2633–2646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths G., Simons K. (1986). The trans Golgi network: Sorting at the exit site of the Golgi complex. Science 234: 438–443. [DOI] [PubMed] [Google Scholar]
- Groen A.J., Sancho-Andrés G., Breckels L.M., Gatto L., Aniento F., Lilley K.S. (2014). Identification of trans-golgi network proteins in Arabidopsis thaliana root tissue. J. Proteome Res. 13: 763–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Günl M., Gille S., Pauly M. (2010). OLIgo mass profiling (OLIMP) of extracellular polysaccharides. J. Vis. Exp. 40: 2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Günl M., Neumetzler L., Kraemer F., de Souza A., Schultink A., Pena M., York W.S., Pauly M. (2011). AXY8 encodes an α-fucosidase, underscoring the importance of apoplastic metabolism on the fine structure of Arabidopsis cell wall polysaccharides. Plant Cell 23: 4025–4040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hachez C., Laloux T., Reinhardt H., Cavez D., Degand H., Grefen C., De Rycke R., Inzé D., Blatt M.R., Russinova E., Chaumont F. (2014). Arabidopsis SNAREs SYP61 and SYP121 coordinate the trafficking of plasma membrane aquaporin PIP2;7 to modulate the cell membrane water permeability. Plant Cell 26: 3132–3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heard W., Sklenář J., Tomé D.F., Robatzek S., Jones A.M. (2015). Identification of regulatory and cargo proteins of endosomal and secretory pathways in Arabidopsis thaliana by proteomic dissection. Mol. Cell. Proteomics 14: 1796–1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoogenboom J., Berghuis N., Cramer D., Geurts R., Zuilhof H., Wennekes T. (2016). Direct imaging of glycans in Arabidopsis roots via click labeling of metabolically incorporated azido-monosaccharides. BMC Plant Biol. 16: 220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang B.H. (2010). Electron microscopy and high-pressure freezing of Arabidopsis. Methods Cell Biol. 96: 259–283. [DOI] [PubMed] [Google Scholar]
- Kang B.H., Nielsen E., Preuss M.L., Mastronarde D., Staehelin L.A. (2011). Electron tomography of RabA4b- and PI-4Kβ1-labeled trans Golgi network compartments in Arabidopsis. Traffic 12: 313–329. [DOI] [PubMed] [Google Scholar]
- Kim S.J., Brandizzi F. (2014). The plant secretory pathway: an essential factory for building the plant cell wall. Plant Cell Physiol. 55: 687–693. [DOI] [PubMed] [Google Scholar]
- Knox J.P., Linstead P.J., Cooper J.P.C., Roberts K. (1991). Developmentally regulated epitopes of cell surface arabinogalactan proteins and their relation to root tissue pattern formation. Plant J. 1: 317–326. [DOI] [PubMed] [Google Scholar]
- Kong Y., et al. (2015). Galactose-depleted xyloglucan is dysfunctional and leads to dwarfism in Arabidopsis. Plant Physiol. 167: 1296–1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A., Wetzler E., Berger M. (1997). Isolation and characterization of complement receptor type 1 (CR1) storage vesicles from human neutrophils using antibodies to the cytoplasmic tail of CR1. Blood 89: 4555–4565. [PubMed] [Google Scholar]
- LaMontagne E.D., Heese A. (2017). Trans-Golgi network/early endosome: A central sorting station for cargo proteins in plant immunity. Curr. Opin. Plant Biol. 40: 114–121. [DOI] [PubMed] [Google Scholar]
- Lamport D.T.A., Tan L., Held M., Kieliszewski M.J. (2018). The role of the primary cell wall in plant morphogenesis. Int. J. Mol. Sci. 19: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampugnani E.R., Khan G.A., Somssich M., Persson S. (2018). Building a plant cell wall at a glance. J. Cell Sci. 131: 131. [DOI] [PubMed] [Google Scholar]
- Leroux O., Sørensen I., Marcus S.E., Viane R.L., Willats W.G., Knox J.P. (2015). Antibody-based screening of cell wall matrix glycans in ferns reveals taxon, tissue and cell-type specific distribution patterns. BMC Plant Biol. 15: 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R., Rodriguez-Furlan C., Wang J., van de Ven W., Gao T., Raikhel N.V., Hicks G.R. (2017). Different endomembrane trafficking pathways establish apical and basal polarities. Plant Cell 29: 90–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y., Basu D., Pattathil S., Xu W.L., Venetos A., Martin S.L., Faik A., Hahn M.G., Showalter A.M. (2013). Biochemical and physiological characterization of fut4 and fut6 mutants defective in arabinogalactan-protein fucosylation in Arabidopsis. J. Exp. Bot. 64: 5537–5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund C.H., Bromley J.R., Stenbæk A., Rasmussen R.E., Scheller H.V., Sakuragi Y. (2015). A reversible Renilla luciferase protein complementation assay for rapid identification of protein-protein interactions reveals the existence of an interaction network involved in xyloglucan biosynthesis in the plant Golgi apparatus. J. Exp. Bot. 66: 85–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M.A., Staehelin L.A. (1992). Domain-specific and cell type-specific localization of two types of cell wall matrix polysaccharides in the clover root tip. J. Cell Biol. 118: 467–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcus S.E., Verhertbruggen Y., Hervé C., Ordaz-Ortiz J.J., Farkas V., Pedersen H.L., Willats W.G., Knox J.P. (2008). Pectic homogalacturonan masks abundant sets of xyloglucan epitopes in plant cell walls. BMC Plant Biol. 8: 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuko T., Minami A., Iwasaki N., Majima T., Nishimura S., Lee Y.C. (2005). Carbohydrate analysis by a phenol-sulfuric acid method in microplate format. Anal. Biochem. 339: 69–72. [DOI] [PubMed] [Google Scholar]
- McCann M.C., Wells B., Roberts K. (1992). Complexity in the spatial localization and length distribution of plant cell-wall matrix polysaccharides. J. Microsc. 166: 123–136. [Google Scholar]
- McCartney L., Marcus S.E., Knox J.P. (2005). Monoclonal antibodies to plant cell wall xylans and arabinoxylans. J. Histochem. Cytochem. 53: 543–546. [DOI] [PubMed] [Google Scholar]
- McFarlane H.E., Young R.E., Wasteneys G.O., Samuels A.L. (2008). Cortical microtubules mark the mucilage secretion domain of the plasma membrane in Arabidopsis seed coat cells. Planta 227: 1363–1375. [DOI] [PubMed] [Google Scholar]
- McFarlane H.E., Watanabe Y., Gendre D., Carruthers K., Levesque-Tremblay G., Haughn G.W., Bhalerao R.P., Samuels L. (2013). Cell wall polysaccharides are mislocalized to the vacuole in echidna mutants. Plant Cell Physiol. 54: 1867–1880. [DOI] [PubMed] [Google Scholar]
- McFarlane H.E., Döring A., Persson S. (2014). The cell biology of cellulose synthesis. Annu. Rev. Plant Biol. 65: 69–94. [DOI] [PubMed] [Google Scholar]
- Moller I., Marcus S.E., Haeger A., Verhertbruggen Y., Verhoef R., Schols H., Ulvskov P., Mikkelsen J.D., Knox J.P., Willats W. (2008). High-throughput screening of monoclonal antibodies against plant cell wall glycans by hierarchical clustering of their carbohydrate microarray binding profiles. GlycoconJ. J. 25: 37–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore P.J., Staehelin L.A. (1988). Immunogold localization of the cell-wall-matrix polysaccharides rhamnogalacturonan I and xyloglucan during cell expansion and cytokinesis in Trifolium pratense L.; implication for secretory pathways. Planta 174: 433–445. [DOI] [PubMed] [Google Scholar]
- Moore P.J., Darvill A.G., Albersheim P., Staehelin L.A. (1986). Immunogold localization of xyloglucan and rhamnogalacturonan I in the cell walls of suspension-cultured sycamore cells. Plant Physiol. 82: 787–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore P.J., Swords K.M., Lynch M.A., Staehelin L.A. (1991). Spatial organization of the assembly pathways of glycoproteins and complex polysaccharides in the Golgi apparatus of plants. J. Cell Biol. 112: 589–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller-Loennies S., MacKenzie C.R., Patenaude S.I., Evans S.V., Kosma P., Brade H., Brade L., Narang S. (2000). Characterization of high affinity monoclonal antibodies specific for chlamydial lipopolysaccharide. Glycobiology 10: 121–130. [DOI] [PubMed] [Google Scholar]
- Nevins D.J., English P.D., Albersheim P. (1967). The specific nature of plant cell wall polysaccharides. Plant Physiol. 42: 900–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguema-Ona E., Vicré-Gibouin M., Gotté M., Plancot B., Lerouge P., Bardor M., Driouich A. (2014). Cell wall O-glycoproteins and N-glycoproteins: Aspects of biosynthesis and function. Front. Plant Sci. 5: 499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolovski N., Rubtsov D., Segura M.P., Miles G.P., Stevens T.J., Dunkley T.P., Munro S., Lilley K.S., Dupree P. (2012). Putative glycosyltransferases and other plant Golgi apparatus proteins are revealed by LOPIT proteomics. Plant Physiol. 160: 1037–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novakovic L., Guo T., Bacic A., Sampathkumar A., Johnson K.L. (2018). Hitting the wall-sensing and signaling pathways involved in plant cell wall remodeling in response to abiotic stress. Plants 7: 89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obel N., Erben V., Schwarz T., Kühnel S., Fodor A., Pauly M. (2009). Microanalysis of plant cell wall polysaccharides. Mol. Plant 2: 922–932. [DOI] [PubMed] [Google Scholar]
- Park E., Drakakaki G. (2014). Proteomics of endosomal compartments from plants case study: Isolation of trans-Golgi network vesicles. Methods Mol. Biol. 1209: 179–187. [DOI] [PubMed] [Google Scholar]
- Park E., Díaz-Moreno S.M., Davis D.J., Wilkop T.E., Bulone V., Drakakaki G. (2014). Endosidin 7 specifically arrests late cytokinesis and inhibits callose biosynthesis, revealing distinct trafficking events during cell plate maturation. Plant Physiol. 165: 1019–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons H.T., Lilley K.S. (2018). Mass spectrometry approaches to study plant endomembrane trafficking. Semin. Cell Dev. Biol. 80: 123–132. [DOI] [PubMed] [Google Scholar]
- Pattathil S., et al. (2010). A comprehensive toolkit of plant cell wall glycan-directed monoclonal antibodies. Plant Physiol. 153: 514–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattathil S., Avci U., Miller J.S., Hahn M.G. (2012). Immunological approaches to plant cell wall and biomass characterization: Glycome profiling. Methods Mol. Biol. 908: 61–72. [DOI] [PubMed] [Google Scholar]
- Pattathil S., Avci U., Zhang T., Cardenas C.L., Hahn M.G. (2015b). Immunological approaches to biomass characterization and utilization. Front. Bioeng. Biotechnol. 3: 173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattathil S., Hahn M.G., Dale B.E., Chundawat S.P. (2015a). Insights into plant cell wall structure, architecture, and integrity using glycome profiling of native and AFEXTM-pre-treated biomass. J. Exp. Bot. 66: 4279–4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauly M., Keegstra K. (2016). Biosynthesis of the plant cell wall matrix polysaccharide xyloglucan. Annu. Rev. Plant Biol. 67: 235–259. [DOI] [PubMed] [Google Scholar]
- Pedersen H.L., et al. (2012). Versatile high resolution oligosaccharide microarrays for plant glycobiology and cell wall research. J. Biol. Chem. 287: 39429–39438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennell R.I., Knox J.P., Scofield G.N., Selvendran R.R., Roberts K. (1989). A family of abundant plasma membrane-associated glycoproteins related to the arabinogalactan proteins is unique to flowering plants. J. Cell Biol. 108: 1967–1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peralta A.G., Venkatachalam S., Stone S.C., Pattathil S. (2017). Xylan epitope profiling: An enhanced approach to study organ development-dependent changes in xylan structure, biosynthesis, and deposition in plant cell walls. Biotechnol. Biofuels 10: 245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettolino F.A., Walsh C., Fincher G.B., Bacic A. (2012). Determining the polysaccharide composition of plant cell walls. Nat. Protoc. 7: 1590–1607. [DOI] [PubMed] [Google Scholar]
- Poulsen C.P., Dilokpimol A., Mouille G., Burow M., Geshi N. (2014). Arabinogalactan glycosyltransferases target to a unique subcellular compartment that may function in unconventional secretion in plants. Traffic 15: 1219–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puhlmann J., Bucheli E., Swain M.J., Dunning N., Albersheim P., Darvill A.G., Hahn M.G. (1994). Generation of monoclonal antibodies against plant cell-wall polysaccharides. I. Characterization of a monoclonal antibody to a terminal alpha-(1-->2)-linked fucosyl-containing epitope. Plant Physiol. 104: 699–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raimundo S.C., Avci U., Hopper C., Pattathil S., Hahn M.G., Popper Z.A. (2016). Immunolocalization of cell wall carbohydrate epitopes in seaweeds: Presence of land plant epitopes in Fucus vesiculosus L. (Phaeophyceae). Planta 243: 337–354. [DOI] [PubMed] [Google Scholar]
- Ralet M.C., Tranquet O., Poulain D., Moïse A., Guillon F. (2010). Monoclonal antibodies to rhamnogalacturonan I backbone. Planta 231: 1373–1383. [DOI] [PubMed] [Google Scholar]
- R Development Core Team (2006). R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria.http://www.R-project.org/
- Robert S., Chary S.N., Drakakaki G., Li S., Yang Z., Raikhel N.V., Hicks G.R. (2008). Endosidin1 defines a compartment involved in endocytosis of the brassinosteroid receptor BRI1 and the auxin transporters PIN2 and AUX1. Proc. Natl. Acad. Sci. USA 105: 8464–8469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosquete M.R., Davis D.J., Drakakaki G. (2018). The plant trans-Golgi network: Not just a matter of distinction. Plant Physiol. 176: 187–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth J., Taatjes D.J., Lucocq J.M., Weinstein J., Paulson J.C. (1985). Demonstration of an extensive trans-tubular network continuous with the Golgi apparatus stack that may function in glycosylation. Cell 43: 287–295. [DOI] [PubMed] [Google Scholar]
- Ruprecht C., Bartetzko M.P., Senf D., Dallabernadina P., Boos I., Andersen M.C.F., Kotake T., Knox J.P., Hahn M.G., Clausen M.H., Pfrengle F. (2017). A synthetic glycan microarray enables epitope mapping of plant cell wall glycan-directed antibodies. Plant Physiol. 175: 1094–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutter B.D., Innes R.W. (2017). Extracellular vesicles isolated from the leaf apoplast carry stress-response proteins. Plant Physiol. 173: 728–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito S., Hiemori K., Kiyoi K., Tateno H. (2018). Glycome analysis of extracellular vesicles derived from human induced pluripotent stem cells using lectin microarray. Sci. Rep. 8: 3997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderfoot A.A., Assaad F.F., Raikhel N.V. (2000). The Arabidopsis genome. An abundance of soluble N-ethylmaleimide-sensitive factor adaptor protein receptors. Plant Physiol. 124: 1558–1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderfoot A.A., Kovaleva V., Bassham D.C., Raikhel N.V. (2001). Interactions between syntaxins identify at least five SNARE complexes within the Golgi/prevacuolar system of the Arabidopsis cell. Mol. Biol. Cell 12: 3733–3743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selvendran R.R., O’Neill M.A. (1987). Isolation and analysis of cell walls from plant material. Methods Biochem. Anal. 32: 25–153. [DOI] [PubMed] [Google Scholar]
- Sinclair R., Rosquete M.R., Drakakaki G. (2018). Post-Golgi trafficking and transport of cell wall components. Front. Plant Sci. 9: 1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somerville C., Bauer S., Brininstool G., Facette M., Hamann T., Milne J., Osborne E., Paredez A., Persson S., Raab T., Vorwerk S., Youngs H. (2004). Toward a systems approach to understanding plant cell walls. Science 306: 2206–2211. [DOI] [PubMed] [Google Scholar]
- Sørensen I., Willats W.G. (2011). Screening and characterization of plant cell walls using carbohydrate microarrays. Methods Mol. Biol. 715: 115–121. [DOI] [PubMed] [Google Scholar]
- Surpin M., Raikhel N. (2004). Traffic jams affect plant development and signal transduction. Nat. Rev. Mol. Cell Biol. 5: 100–109. [DOI] [PubMed] [Google Scholar]
- Tan L., Showalter A.M., Egelund J., Hernandez-Sanchez A., Doblin M.S., Bacic A. (2012). Arabinogalactan-proteins and the research challenges for these enigmatic plant cell surface proteoglycans. Front. Plant Sci. 3: 140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyooka K., Goto Y., Asatsuma S., Koizumi M., Mitsui T., Matsuoka K. (2009). A mobile secretory vesicle cluster involved in mass transport from the Golgi to the plant cell exterior. Plant Cell 21: 1212–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uemura T., Kim H., Saito C., Ebine K., Ueda T., Schulze-Lefert P., Nakano A. (2012). Qa-SNAREs localized to the trans-Golgi network regulate multiple transport pathways and extracellular disease resistance in plants. Proc. Natl. Acad. Sci. USA 109: 1784–1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Meene A.M., Doblin M.S., Bacic A. (2017). The plant secretory pathway seen through the lens of the cell wall. Protoplasma 254: 75–94. [DOI] [PubMed] [Google Scholar]
- Voiniciuc C., Pauly M., Usadel B. (2018). Monitoring polysaccharide dynamics in the plant cell wall. Plant Physiol. 176: 2590–2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace I.S, Anderson C.T. (2012). Small molecule probes for plant cell wall polysaccharide imaging. Front. Plant Sci. 10.3389/fpls.2012.00089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B., McClosky D.D., Anderson C.T., Chen G. (2016). Synthesis of a suite of click-compatible sugar analogs for probing carbohydrate metabolism. Carbohydr. Res. 433: 54–62. [DOI] [PubMed] [Google Scholar]
- Wang J., Ding Y., Wang J., Hillmer S., Miao Y., Lo S.W., Wang X., Robinson D.G., Jiang L. (2010). EXPO, an exocyst-positive organelle distinct from multivesicular endosomes and autophagosomes, mediates cytosol to cell wall exocytosis in Arabidopsis and tobacco cells. Plant Cell 22: 4009–4030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P., Chen X., Goldbeck C., Chung E., Kang B.H. (2017). A distinct class of vesicles derived from the trans-Golgi mediates secretion of xylogalacturonan in the root border cell. Plant J. 92: 596–610. [DOI] [PubMed] [Google Scholar]
- Wattelet-Boyer V., Brocard L., Jonsson K., Esnay N., Joubès J., Domergue F., Mongrand S., Raikhel N., Bhalerao R.P., Moreau P., Boutté Y. (2016). Enrichment of hydroxylated C24- and C26-acyl-chain sphingolipids mediates PIN2 apical sorting at trans-Golgi network subdomains. Nat. Commun. 7: 12788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willats W.G., Marcus S.E., Knox J.P. (1998). Generation of monoclonal antibody specific to (1-->5)-alpha-L-arabinan. Carbohydr. Res. 308: 149–152. [DOI] [PubMed] [Google Scholar]
- Williams C., Royo F., Aizpurua-Olaizola O., Pazos R., Boons G.J., Reichardt N.C., Falcon-Perez J.M. (2018). Glycosylation of extracellular vesicles: Current knowledge, tools and clinical perspectives. J. Extracell. Vesicles 7: 1442985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worden N., Park E., Drakakaki G. (2012). Trans-Golgi network: An intersection of trafficking cell wall components. J. Integr. Plant Biol. 54: 875–886. [DOI] [PubMed] [Google Scholar]
- Young R.E., McFarlane H.E., Hahn M.G., Western T.L., Haughn G.W., Samuels A.L. (2008). Analysis of the Golgi apparatus in Arabidopsis seed coat cells during polarized secretion of pectin-rich mucilage. Plant Cell 20: 1623–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun H.S., Kwon C. (2017). Vesicle trafficking in plant immunity. Curr. Opin. Plant Biol. 40: 34–42. [DOI] [PubMed] [Google Scholar]
- Zhang G.F., Staehelin L.A. (1992). Functional compartmentation of the Golgi apparatus of plant cells: Immunocytochemical analysis of high-pressure frozen- and freeze-substituted sycamore maple suspension culture cells. Plant Physiol. 99: 1070–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Gong Z., Zhang C., Song C.P., Damsz B., Inan G., Koiwa H., Zhu J.K., Hasegawa P.M., Bressan R.A. (2002). OSM1/SYP61: A syntaxin protein in Arabidopsis controls abscisic acid-mediated and non-abscisic acid-mediated responses to abiotic stress. Plant Cell 14: 3009–3028. [DOI] [PMC free article] [PubMed] [Google Scholar]